Significance
The sudden explosion of seed pods in popping cress (Cardamine hirsuta) takes less than 3 ms to accelerate seeds away from the plant. This explosive mechanism relies on polar deposition of the cell-wall polymer lignin. To investigate the genetic basis for polar lignin deposition, we conducted a mutant screen and identified SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE 7 (SPL7)—a transcriptional regulator of copper homeostasis. We discovered three multicopper laccases, LAC4, 11, and 17, that precisely colocalize with, and are required for, the polar deposition of lignin in explosive seed pods. Activity of these three laccases depends on SPL7 to acclimate to copper deficiency. Our findings demonstrate how mineral nutrition is integrated with polar lignin deposition to facilitate dispersal.
Keywords: Cardamine hirsuta, lignin, laccases, squamosa promoter-binding protein-like 7, seed dispersal
Abstract
Exploding seed pods evolved in the Arabidopsis relative Cardamine hirsuta via morphomechanical innovations that allow the storage and rapid release of elastic energy. Asymmetric lignin deposition within endocarpb cell walls is one such innovation that is required for explosive seed dispersal and evolved in association with the trait. However, the genetic control of this novel lignin pattern is unknown. Here, we identify three lignin-polymerizing laccases, LAC4, 11, and 17, that precisely colocalize with, and are redundantly required for, asymmetric lignification of endocarpb cells. By screening for C. hirsuta mutants with less lignified fruit valves, we found that loss of function of the transcription factor gene SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE 7 (SPL7) caused a reduction in endocarpb cell-wall lignification and a consequent reduction in seed dispersal range. SPL7 is a conserved regulator of copper homeostasis and is both necessary and sufficient for copper to accumulate in the fruit. Laccases are copper-requiring enzymes. We discovered that laccase activity in endocarpb cell walls depends on the SPL7 pathway to acclimate to copper deficiency and provide sufficient copper for lignin polymerization. Hence, SPL7 links mineral nutrition to efficient dispersal of the next generation.
Exploding seed pods are one of many different adaptations that plants have evolved to disperse their seeds. This huge diversity reflects the important ecological and evolutionary consequences of dispersal, including the ability to change or expand a species’ range (1, 2). Cardamine hirsuta is a small, ruderal weed, related to the plant model Arabidopsis thaliana (3). Unlike A. thaliana, it uses an explosive mechanism to disperse its seeds. In the seed pods of C. hirsuta, two exploding valves coil back rapidly, firing the seeds at speeds greater than 10 m⋅s−1 to disperse over a large area (4). This mechanism requires localized lignin deposition in a single-cell layer of the fruit valve called the endocarpb (endb), as mutant fruit that lack endb cells fail to explode (4). A striking feature of endb cells in C. hirsuta is the asymmetric deposition of lignin in three stiff rods connected by thin hinges (4). Endb cell walls are uniformly lignified in A. thaliana and other species with nonexplosive fruit in the Brassicaceae family (5). Explosive seed dispersal evolved once in this family, in the Cardamine genus, and asymmetric lignin deposition is strictly associated with this trait in Cardamine species (4). Currently, the genetic basis of this localized lignin pattern is unknown.
Asymmetric lignin deposition is known to play a key role in the mechanics of exploding seed pods. Simulations of a mathematical model that described the elastic energy in the fruit valve were compared using an asymmetrically hinged- versus a uniformly lignified-wall geometry. These results showed that the hinged-wall geometry is critical for the explosive release of stored elastic potential energy, allowing the fruit valve to employ a rapid release mechanism like a toy slap bracelet (4). Predictions from these model simulations were tested genetically, by creating transgenic plants with uniformly lignified endb cells. These seed pods failed to explode, showing that the asymmetric pattern of lignin deposition in endb cells is required for explosive seed dispersal (4).
Lignin is a polymer that imparts stiffness and hydrophobicity to the plant cell wall. It is composed of monolignols, which are synthesized from phenylalanine in the cytosol and exported to the cell wall where they are activated by oxidation (6). Laccases (LACs) and type III peroxidases (PERs) are two different types of secreted enzymes that catalyze this oxidation. The lignin polymer is then formed by nonenzymatic random coupling of activated monolignols. While the biosynthesis of monolignols is well-understood, it is less clear how localized patterns of lignin deposition are produced. Targeted export of monolignols to specific cell-wall domains is unlikely to contribute to lignin patterning, because monolignols are highly mobile in the apoplast and do not perturb lignin patterning when applied exogenously (7–9). In contrast to this, LAC and PER enzymes often colocalize precisely with subcellular patterns of lignin (10–12). Type III PER and LAC enzymes are encoded by large gene families with 73 and 17 members, respectively, in A. thaliana (13–15). Genetic and biocatalytic redundancy within and between these large enzyme families has made it difficult to ascribe biological functions to individual genes. Although a mixed set of oxidative enzymes is likely to contribute to lignification in different cells and tissues (10), the relative requirement for LACs versus PERs tends to differ between cell types. For example, lignification of Casparian strips in the root endodermis is PER- rather than LAC-dependent, as a quintuple PER mutant (per3 9 39 72 64) abolished Casparian strips, while a nonuple LAC mutant (lac1 3 5 7 8 9 12 13 16) had no effect (16). By contrast, the lignification of xylem tissues depends on laccases. Lignin content was reduced in the stems of lac4 17 double mutants and further reduced in lac4 11 17 triple mutants, which failed to grow past the seedling stage (17, 18).
LACs and PERs are secreted glycoproteins that require metal cofactors for enzymatic activity. Type III PERs are heme-containing proteins that reduce H2O2 to oxidize monolignols. For this reason, PER-dependent lignification requires reactive oxygen species (ROS) produced by NADPH oxidases (respiratory burst oxidase homologs) (12, 19). On the other hand, LACs are multicopper proteins that, unlike PERs, reduce O2 to H2O in order to oxidize monolignols, and do not require ROS. Laccases bind four Cu ions: One Cu ion participates in the oxidation of the substrate and a cluster of three participates in the reduction of O2 to H2O (20). The cycling of Cu between an oxidized (Cu2+) and a reduced state (Cu1+) is used in many different redox reactions and electron transport. Thus, Cu is an essential micronutrient in nearly all eukaryotic organisms. However, when Cu ions are present in excess, redox cycling can catalyze the production of highly toxic ROS and cause cellular damage. Therefore, Cu homeostasis is tightly controlled in plants (21).
Plants take up Cu from the soil, and the bioavailability of Cu varies depending on soil type, though natural soils with high Cu levels are rare (22). To acclimate to Cu deficiency, A. thaliana regulates Cu homeostasis via the SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE 7 (SPL7) transcription factor. SPL7 is one of 16 SPL proteins in A. thaliana, which all bind DNA sequences with a core GTAC motif (23). The majority of this gene family are targeted for posttranscriptional degradation by microRNAs (miRNAs) 156/157 but SPL7 is not (24). SPL7 is homologous to COPPER RESPONSE REGULATOR 1 (CRR1) in the green alga Chlamydomonas reinhardtii (25, 26). Under copper-limiting conditions, this evolutionarily conserved switch activates the transcription of genes that increase Cu uptake and economize on the use of available Cu (27–29). In this way, the SPL7 pathway ensures that when Cu is limiting, sufficient Cu is available for the function of essential cuproproteins such as plastocyanin, which is required for photosynthetic electron transfer and cannot be replaced (30).
Here, we describe the genetic control of localized lignin deposition by three multi-Cu laccases in endb cell walls of explosive fruit. LAC4, 11, and 17 precisely colocalize with, and are required for, the unique pattern of lignin in these cells. The nonlignified endb cell walls found in lac4 11 17 triple mutants were phenocopied by loss of SPL7 function. Our findings show that laccase activity in endb cell walls requires SPL7 in order to acclimate to Cu deficiency and provide sufficient Cu to polymerize lignin.
Results
SPL7 Is Required for endb Lignification.
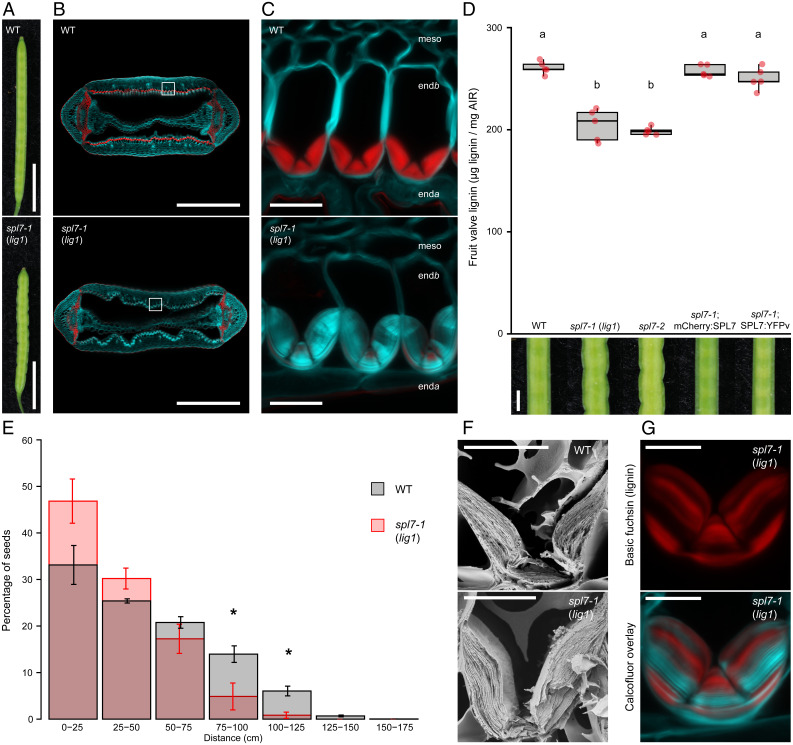
To identify genes required for localized lignin deposition in endb cells, we performed a forward genetics screen in C. hirsuta for mutants with less lignified fruit valves. The less lignin 1 (lig1) mutant showed reduced lignification of endb secondary cell walls (SCWs) (Fig. 1 A–C). The phenotype was particularly pronounced in endb cells compared with other lignified cell types in the fruit (Fig. 1 B and C). By quantifying acetylbromide-soluble lignin, we found that lignin content was significantly reduced in the fruit valves of lig1 compared with wild type (Fig. 1D). In addition, mature lig1 fruit buckled along their edge, compared with the straight edge of wild-type fruit (Fig. 1 A and D). The timing of this buckling coincided with the lignification of endb cells, suggesting it may be a consequence of reduced lignin. We also found a significant reduction in the seed dispersal range of lig1 plants (Fig. 1E). Maximum dispersal distance was reduced by 0.5 m in lig1 and significantly fewer lig1 than wild-type seeds were dispersed farther than 0.75 m (Fig. 1E). Therefore, lignification of valve endb SCWs is critical for explosive seed dispersal in C. hirsuta.
Fig. 1.
SPL7 is required for endb lignification in C. hirsuta fruit. (A) Mature wild-type and spl7-1 (lig1) fruit. (B and C) Lignin, stained red with basic fuchsin, in cross-sections of wild-type and spl7-1 (lig1) fruit (B) and endb cells (C), cell walls stained cyan with calcofluor white. Cells in C correspond to regions of the valve indicated by white boxes in B. (D) Boxplot of lignin concentration, shown as microgram of acetylbromide-soluble lignin per milligram of alcohol-insoluble residue (AIR), in mature fruit valves, and close-up views of fruit margin, in wild type, spl7-1 (lig1), spl7-2, spl7-1; mCherry:SPL7, and spl7-1; SPL7:YFPv. Plot shows median (thick black lines); n = 5 biological replicates per genotype (red dots) where each replicate contains 16 valves from two plants; different letters denote statistical significance at P < 0.05 using Kruskal–Wallis and Fisher’s least significant difference as post hoc analysis. (E) Barplot of seed dispersal in wild type (gray) and spl7-1 (red), showing the percentage of seeds in each distance bin. Error bars indicate SEM; n = 10,039 total seeds dispersed by four plants per genotype; * denotes statistical significance at P < 0.05 using Student’s t test. (F) Scanning electron micrographs of wild-type and spl7-1 cryofractured fruit showing endb SCW layers. (G) Variable lignin deposition (stained red with basic fuchsin) in spl7-1 endb cells, overlaid with calcofluor white cell-wall stain (cyan). Confocal micrographs show z-axis sum projections of transverse fruit sections (B, C, and G). All plants were supplemented with 0.5 mM CuSO4 to ensure plant growth and fruit development of spl7 mutants. enda, endocarpa; meso, mesocarp. (Scale bars, 5 mm [A], 500 µm [B], 20 µm [C], 1 mm [D], and 10 µm [F and G].).
We used mapping by sequencing to identify the causal mutation of lig1 as a C>T mutation that introduced a P443S substitution in the C. hirsuta ortholog of the transcription factor SPL7 (CARHR194170) (SI Appendix, Fig. S1). To verify that loss of SPL7 function caused a similar phenotype to the recessive lig1 allele, we used CRISPR-Cas9 to introduce a single-nucleotide deletion that resulted in a truncated 66–amino acid (aa) protein lacking the conserved SQUAMOSA PROMOTER-BINDING PROTEIN (SBP) DNA-binding domain (SI Appendix, Fig. S1). These spl7 mutant fruit valves had reduced lignin content and less lignified endb SCWs, indistinguishable from lig1 (Fig. 1D and SI Appendix, Fig. S1). Moreover, an allelism test showed that lig1 is a spl7 allele and likely to represent complete loss of SPL7 function in C. hirsuta (SI Appendix, Fig. S1). We showed that all lig1 phenotypes were fully complemented by expressing the wild-type SPL7 genomic locus, tagged at either the N terminus with mCherry (pSPL7::mCherry:SPL7) or the C terminus with Venus yellow fluorescent protein (YFP) (pSPL7::SPL7:YFPv) (Fig. 1D and SI Appendix, Fig. S1). Moreover, a 253-aa truncated SPL7 protein, including the SBP DNA-binding domain, nuclear localization signal (NLS), and transcriptional activation domain, tagged with green fluorescent protein (GFP) (pSPL7::SBP:GFP), was sufficient to fully rescue the fruit defects of lig1 to wild type (SI Appendix, Fig. S1). A similar finding was reported for A. thaliana SPL7 (31) and suggests that lignification of endb SCWs requires transcriptional responses that are regulated by SPL7. Based on these findings, we renamed lig1 as C. hirsuta spl7-1 and the CRISPR-Cas9 allele as spl7-2.
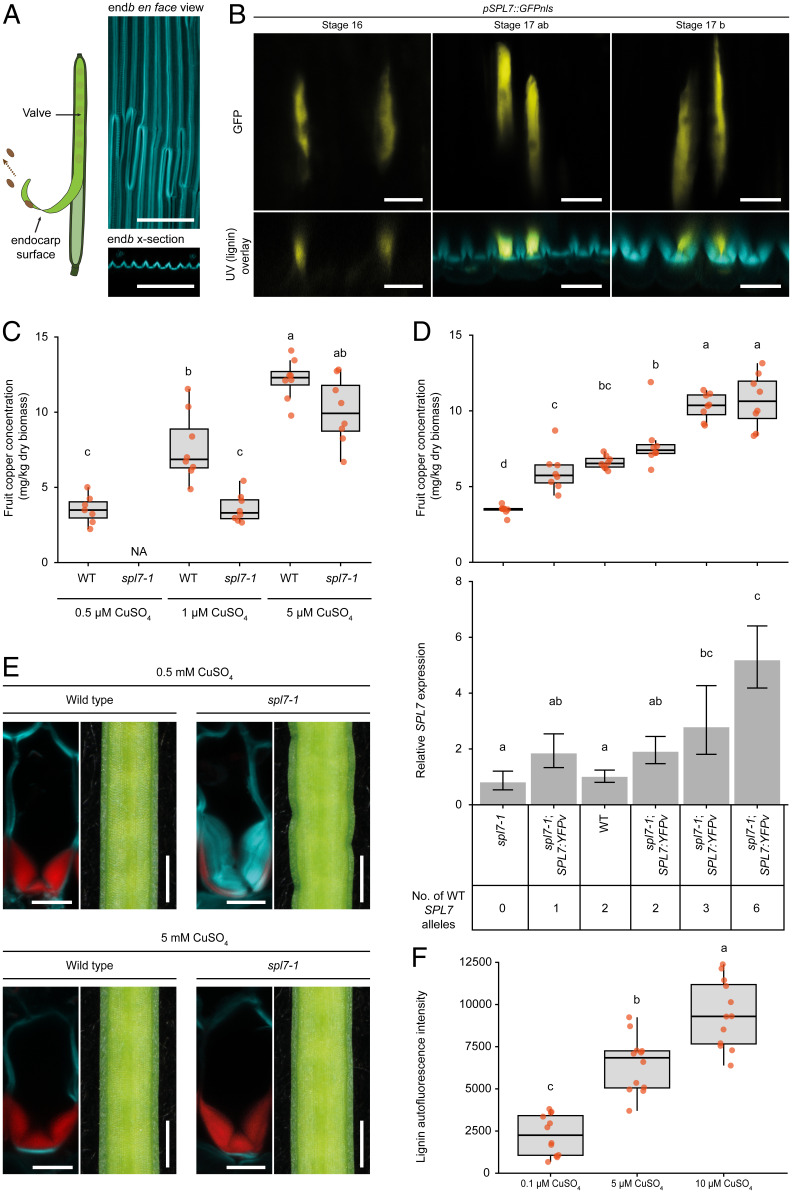
To investigate the localization of SPL7 in C. hirsuta fruit during endb SCW formation, we analyzed transcriptional SPL7 reporters and functional SPL7 protein fusions. pSPL7::GFPnls localized to endb cells before and during the deposition of lignified SCWs in C. hirsuta fruit valves (Fig. 2 A and B). We observed an identical pattern in complementing pSPL7::SBP:GFP lines (SI Appendix, Fig. S2). We could detect SPL7 transcripts in most C. hirsuta plant tissues (SI Appendix, Fig. S2). However, we could not reliably detect the tagged full-length SPL7 proteins that complemented C. hirsuta spl7-1, similar to previous reports for A. thaliana SPL7 (31). In summary, SPL7 accumulates in endb cells where the lignification defect is observed in spl7 mutants.
Fig. 2.
SPL7 is necessary for Cu accumulation in C. hirsuta fruit. (A) Schematic indicating the endocarp surface of the fruit valve (Left) and endb SCWs imaged en face (Top Right) and shown as transverse optical sections (Bottom Right) to indicate the orientation in B. Lignin autofluorescence is shown in cyan. (B) pSPL7::GFPnls expression (yellow) in endb cell nuclei of stage 16, 17ab, and 17b fruit valves, imaged en face (Top) and shown together with lignin autofluorescence (cyan) in transverse optical sections (Bottom). UV, ultraviolet. (C) Boxplot of Cu concentration (mg/kg dry biomass) in mature fruits of wild-type and spl7-1 plants grown in an aeroponics system and irrigated with solutions containing either 0.5, 1, or 5 µM CuSO4; n ≥ 7 biological replicates per condition (red dots) where each replicate contains five to seven pooled fruits from multiple plants; NA (not applicable) indicates the absence of spl7-1 fruit in this condition. (D) Dose–response between SPL7 gene expression and Cu concentration in mature fruit of spl7-1, wild-type, and spl7-1; SPL7:YFPv complementation lines with an increasing number of wild-type SPL7 alleles. Boxplot of Cu concentration (mg/kg dry biomass) in mature fruit; n = 8 biological replicates per genotype (red dots) where each replicate contains five pooled fruits from multiple plants (Top). SPL7 gene expression in mature fruit of the same genotypes measured by qRT-PCR, normalized against the housekeeping gene TIP41 (CARHR242510), and expressed relative to wild type; error bars indicate SD; n = 3 biological replicates per genotype where each replicate contains two pooled fruit (Bottom). The number of wild-type SPL7 alleles present in each genotype is indicated. (E) Endb cells and fruit margins of wild type and spl7-1 mutant. All plants were supplemented with 0.5 mM CuSO4 for 4 wk; 5 mM CuSO4 was provided as additional supplementation during fruit development. Lignin is stained with basic fuchsin (red); cell walls are stained with calcofluor white (cyan). (F) Boxplot of lignin autofluorescence in endb cells of spl7-1 fruit excised and grown on Murashige and Skoog (MS) media containing either 0.1, 5, or 10 µM CuSO4; n = 12 endb cell regions per treatment (red dots) from three fruits from multiple plants. Plants were grown on soil and supplemented with 0.2 to 0.5 mM CuSO4 to ensure growth and development before fruit were excised. Boxplots show medians (thick black lines); different letters denote statistical significance at P < 0.05 using one-way ANOVA and Tukey’s test as post hoc analysis (D and F) or Kruskal–Wallis and Fisher’s least significant difference as post hoc analysis (C). Confocal micrographs show z-axis sum projections of fruit valves en face (A and B) or transverse sections (E). (Scale bars, 100 µm [A], 20 µm [B], 10 µm [E, Left], and 1 mm [E, Right].)
SPL7 Regulates Cu Concentration in Fruit.
In A. thaliana, SPL7 is required for plants to acclimate to Cu deprivation (29). To test this in C. hirsuta, we grew wild-type and spl7 plants on soil in low-Cu conditions and found that plant size was severely reduced in spl7 but unaffected in wild type (SI Appendix, Fig. S2). To test whether SPL7 regulates Cu homeostasis in C. hirsuta fruit, we grew plants in aeroponic chambers to directly supply bioavailable Cu, and measured Cu concentration in the fruit by inductively coupled plasma mass spectrometry (ICP-MS). We found that spl7 fruit accumulated significantly less Cu than wild type under low-Cu conditions (1 µM CuSO4; Fig. 2C). In contrast to wild type, no fruit were produced by spl7 plants in 0.5 µM CuSO4, indicating that minimal Cu supplementation is required to restore spl7 fertility (Fig. 2C) (32, 33). The concentration of Cu in both wild-type and spl7 fruit increased in response to the concentration of Cu supplied to the roots, over a range of 0.5 to 5 µM CuSO4 (Fig. 2C). At high concentrations of Cu, the difference in fruit Cu concentration was no longer significant between genotypes, indicating that sufficient Cu supplementation can bypass the need for SPL7 (Fig. 2C). Transgenic expression of SPL7 fully restored the Cu concentration in spl7 fruit to wild-type levels (Fig. 2D and SI Appendix, Fig. S2). Using six different SPL7 genotypes grown in Cu-limiting conditions, we found a dose–response between increasing SPL7 activity and increasing Cu levels in the fruit (Fig. 2D). Therefore, SPL7 is necessary for Cu to accumulate in fruit tissues in low-Cu conditions, and is also sufficient to increase Cu above wild-type levels in the fruit.
The thick, asymmetric, endb SCW in C. hirsuta is built up by sequential deposition of lignified SCW layers (Fig. 1F) (4). In spl7 fruit, we observed variable amounts of lignin in individual SCW layers, suggesting that SPL7 likely buffers lignification against fluctuations in Cu availability (Fig. 1G). To understand whether different phenotypes of the spl7 mutant are all attributable to defective Cu homeostasis, we grew plants on soil in high-Cu conditions. This rescued the buckling of the fruit margin and the reduced lignification of endb cell walls and caused a significant increase in the height and biomass of spl7 mutants (Fig. 2E and SI Appendix, Fig. S2). To directly quantify the effect of Cu concentration in the fruit on lignin deposition in endb cells, we grew spl7 fruit in Cu-containing media. We observed a dose–response between increasing Cu concentration and increasing amounts of lignin autofluorescence in endb SCWs over a range of 0.1 to 10 µM CuSO4 (Fig. 2F). Therefore, the mechanism of localized lignin deposition in endb cells is conditional on SPL7 activity and shows a dose dependence on Cu concentration in the fruit.
Polar Localization of LAC4, 11, and 17 Precisely Predicts endb Lignification.
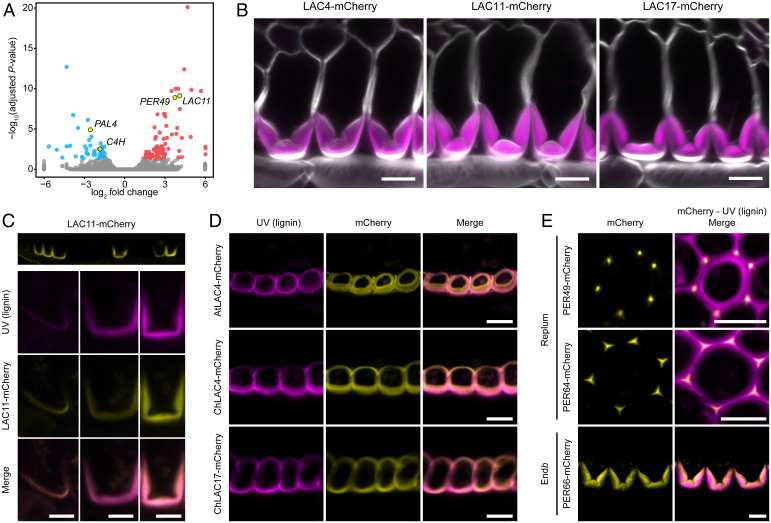
To identify genes required for endb lignification, we took advantage of the less lignified fruit valves of spl7 mutants for RNA sequencing. Among the 10 most significant differentially expressed genes between spl7 and wild-type fruit valves are 2 genes encoding lignin-polymerizing enzymes (Fig. 3A and Dataset S1). LACCASE 11 (LAC11, CARHR210090) and PEROXIDASE 49 (PER49, CARHR244760) are both significantly up-regulated in spl7 (Fig. 3A and SI Appendix, Fig. S3). On the other hand, phenylalanine ammonia-lyase (PAL, CARHR084820) and cinnamate-4-hydroxylase (C4H, CARHR123940)—genes encoding enzymes that catalyze the first steps of monolignol biosynthesis—are significantly down-regulated in spl7 (Fig. 3A and SI Appendix, Fig. S3). To investigate whether reduced monolignol biosynthesis might contribute to the reduced lignification of spl7 endb cells, we grew spl7 fruit in low-Cu media containing monolignols (coniferyl and sinapyl alcohols). The addition of monolignols had no effect on endb SCW lignification (SI Appendix, Fig. S3), suggesting that the down-regulation of PAL and C4H gene expression in spl7 fruit valves is not the cause of less lignin in endb cells but rather a consequence of feedback mechanisms. The up-regulation of LAC11 and PER49 gene expression in spl7 fruit valves may reflect similar feedbacks.
Fig. 3.
LACCASE4, 11, and 17 colocalize with lignin in endb cell walls. (A) Volcano plot of differential gene expression between wild-type and spl7-1 fruit valves with up-regulated genes (log2 fold change > 1.5) shown in red and down-regulated genes (log2 fold change < −1.5) shown in blue; adjusted P < 0.05. Genes related to lignin biosynthesis are indicated in yellow. For visualization purposes, values of log2 fold change <−6 or >6 were set to −6 and 6, respectively. (B) C. hirsuta LAC4, LAC11, and LAC17 protein fusions (magenta; pLAC::LAC:mCherry) localize to C. hirsuta endb SCWs; cell walls are stained with calcofluor white. (C) pLAC11::LAC11:mCherry expression (yellow) in C. hirsuta endb SCWs as they start to lignify (Top) and close-up views of individual endb SCWs (Bottom) shown together with lignin autofluorescence (magenta). (D) A. thaliana LAC4 and C. hirsuta LAC4 and LAC17 protein fusions (yellow; pLAC::LAC:mCherry) colocalize with nonpolar lignin autofluorescence (magenta) in A. thaliana endb SCWs. (E) C. hirsuta PER49, PER64, and PER66 protein fusions (yellow; pPER::PER:mCherry) colocalize with lignin autofluorescence (magenta) in replum cells (PER49, PER64) or endb cells (PER66) in C. hirsuta fruit. Confocal micrographs show z-axis sum projections of transverse fruit sections (B–E). (Scale bars, 10 µm [B, D, and E] and 5 µm [C].)
Interestingly, laccases are multi-Cu glycoproteins and thus good candidates to control Cu-dependent lignin deposition in endb cells. We detected transcripts for 6 out of 15 LAC gene family members in C. hirsuta fruit valves (mean of wild-type and spl7-1 normalized read counts >5), with LAC4 and LAC17 showing the highest expression (SI Appendix, Fig. S3). Type III peroxidases are heme-containing glycoproteins, encoded by a large gene family with 62 members annotated in the current C. hirsuta genome assembly (34). We detected transcripts for 9 out of 62 PER gene family members in C. hirsuta fruit valves (mean of wild-type and spl7-1 normalized read counts >5), including PER64 which is required for Casparian strip lignification in A. thaliana (12, 16), with PER42 and PER66 showing the highest expression in wild type (SI Appendix, Fig. S3). We selected LAC4, LAC11, LAC17, PER49, PER64, and PER66 for further study and generated reporters to visualize where and when these genes and their protein products are expressed in C. hirsuta fruit.
Similar to SPL7, we found expression of LAC4, LAC11, and LAC17 (pLAC::GFPnls) in endb cells in the fruit valve (SI Appendix, Fig. S3). Given the asymmetric deposition of lignin within C. hirsuta endb SCWs, we predicted that enzymes required for lignin polymerization should prepattern this asymmetry. Fluorescent protein fusions of LAC4, LAC11, and LAC17 (pLAC::LAC:mCherry) precisely colocalized with asymmetric lignin deposits in endb SCWs (Fig. 3B). LAC11 localized preferentially to lignified cell walls in the valve of the fruit, while LAC4 and LAC17 also localized to lignified cell walls in the replum (SI Appendix, Fig. S3). LAC11 first accumulated asymmetrically in a thin layer of the endb SCW, forming a U shape in cross-section (Fig. 3C). This asymmetric pattern colocalized perfectly in time and space with lignin (Fig. 3C), indicating that the required monolignols are present as substrates in the apoplast for lignin polymerization. LAC11 continued to accumulate throughout the U-shaped SCW as it rapidly thickened and started to form characteristic hinges at the base of the U (Fig. 3C). We next investigated whether these C. hirsuta laccases maintained their polar localization when transferred into A. thaliana. In the nonexplosive fruit of A. thaliana, pAtLAC4::AtLAC4:mCherry colocalized with the symmetrically lignified endb SCWs (Fig. 3D). Similarly, we observed a symmetric localization of C. hirsuta LAC4 and 17 fusion proteins in A. thaliana endb cell walls, but we could not detect C. hirsuta LAC11 (Fig. 3D and SI Appendix, Fig. S3). These results suggest that the asymmetric localization of C. hirsuta LAC4 and 17 is not determined in cis to these genes but depends on additional polarity determinants that are present in C. hirsuta, but not A. thaliana, endb cells.
To understand whether peroxidases show a similar pattern to laccases, we localized mCherry C-terminal fusions of PER49, PER66, and PER64 (pPER::PER:mCherry) in C. hirsuta fruit (Fig. 3E). We could not detect PER49 or PER64 in endb SCWs in the valve but rather in lignified cell walls in the replum. PER49 and PER64 localized to the middle lamella at cell corners and did not accumulate throughout the lignified SCWs (Fig. 3E). However, PER66 localized precisely to endb SCWs in the valve of the fruit, similar to LAC11 (Fig. 3E and SI Appendix, Fig. S3). Therefore, all three laccases, but only one of three peroxidases that we examined, localized to endb SCWs.
LACCASE4, 11, and 17 Are Required for endb Lignification.
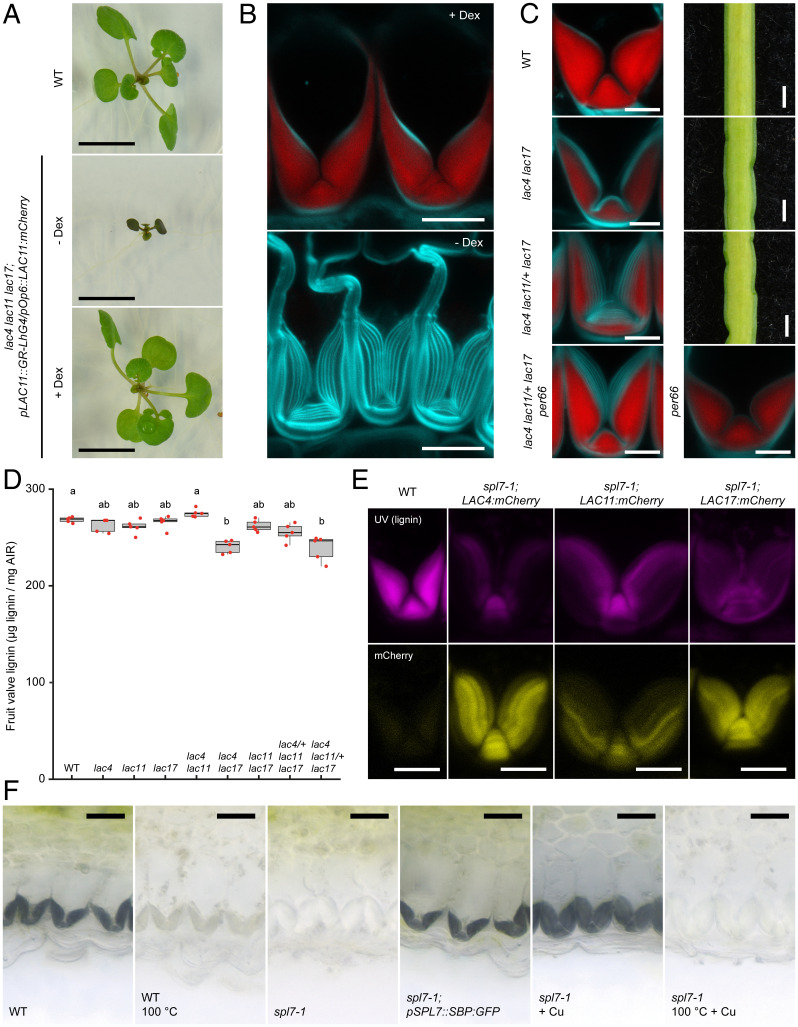
To directly assess whether LAC4, 11, and 17 are needed for endb lignification, we generated loss-of-function alleles for all three genes using a CRISPR-Cas9 multiplex guide RNA strategy. Premature stop codons in these alleles result in truncated proteins that lack aa residues required to coordinate the Cu ions that are essential for laccase activity (SI Appendix, Fig. S4). These triple mutants, therefore, knock out LAC4, 11, and 17 function and show severe growth arrest, which can be partially rescued in culture to produce stems with reduced lignification and collapsed xylem vessels (Fig. 4A and SI Appendix, Fig. S5). Dexamethasone induction of a pLAC11::LhGR≫LAC11:mCherry construct (pLAC11::GR-LhG4/pOp6::LAC11:mCherry) was sufficient to rescue the growth arrest of triple lac4 11 17 mutants, demonstrating that this phenotype is LAC-dependent (Fig. 4A).
Fig. 4.
Cu-dependent laccase activity is necessary for endb lignification in C. hirsuta fruit. (A and B) Triple lac4 11 17 mutants expressing pLAC11::LhGR≫LAC11:mCherry. Two-week-old seedlings grown on 1/2 MS plates without (− Dex) or with 10 µM dexamethasone (+ Dex), compared with wild type (A). Endb cells of plants treated with 100 µM dexamethasone daily until fruit matured (+ Dex) or only until plants started to bolt, such that fruit developed in the absence of dexamethasone (− Dex) (B). Lignin is stained red with basic fuchsin; cell walls are stained cyan with calcofluor white. (C) Endb cells and close-up views of fruit margin in wild type, lac4 17, and lac4 11/+ 17. Endb cells in lac4 11/+ 17 per66 and per66 fruit. Lignin is stained red with basic fuchsin; cell walls are stained cyan with calcofluor white. (D) Boxplot of lignin concentration shown as microgram of acetylbromide-soluble lignin per milligram of AIR in mature fruit valves of wild type and allelic combinations of lac4, lac11, and lac17 mutants. Plot shows median (thick black lines); n = 5 biological replicates per genotype (red dots) where each replicate contains 16 valves from two plants; different letters denote statistical significance at P < 0.05 using Kruskal–Wallis and Dunn’s test as post hoc analysis. (E) LAC4, LAC11, and LAC17 protein fusions (yellow; pLAC::LAC:mCherry) accumulate in spl7-1 endb SCWs, which have less lignin (magenta; autofluorescence) than wild type. (F) Laccase activity (indicated by black precipitate produced by oxidation of 4-hydroxyindole substrate) in fruit valves of wild type (± enzyme inactivation by heat), spl7-1, spl7-1; pSPL7::SBP:GFP, and spl7-1 incubated with 5 mM CuSO4 (± enzyme inactivation by heat). Confocal micrographs show z-axis sum projections of transverse fruit sections (B, C, and E). (Scale bars, 10 mm [A], 10 µm [B and E], 5 µm [C], 1 mm [fruit; C], and 20 µm [F].).
To observe endb SCW lignification in lac4 11 17 triple mutants, we took advantage of the rescue conferred by pLAC11::LhGR≫LAC11:mCherry to grow triple mutants through to flowering. By removing dexamethasone induction at this point, we recovered lac4 11 17 fruit that no longer expressed LAC11:mCherry. Endb SCWs in these fruits failed to lignify, whereas continued dexamethasone induction produced fully lignified endb SCWs (Fig. 4B). Therefore, LAC4, 11, and 17 are redundantly required for localized lignin deposition in endb cells of C. hirsuta. We used various allelic combinations of lac4, 11, and 17 to assess their relative contribution to endb lignification. We found significantly less lignin in the fruit valves of lac4 17 double mutants and lac4 11/+ 17 plants that contained five mutant alleles (Fig. 4D). We observed lignin in only a portion of the endb SCW in these genotypes (Fig. 4C). Newly deposited SCW layers lacked lignin in a similar way to Cu-deprived fruit of spl7 mutants (Fig. 1 C and G). Therefore, LAC4 and 17 contribute redundantly to endb lignification and LAC11 is required with both genes to fully lignify the endb SCW. We found additional similarities between the fruit phenotypes of spl7 mutants and lac4 17 and lac4 11/+ 17 mutants. For example, buckling occurred along the edges of mature fruit in both lac genotypes (Fig. 4C) and these fruit valves had significantly elevated ratios of syringyl-to-guaiacyl (S/G) lignin monomers, which was also found in spl7 alleles and is indicative of laccase deficiency (SI Appendix, Fig. S5) (17). Therefore, the Cu dependence of endb lignification in spl7 fruit is likely to reflect the contribution of multicopper laccases to polymerizing lignin in this cell type.
PER66 showed a similar, polar localization to LAC4, 11, and 17 in the SCW of C. hirsuta endb cells (Fig. 3E). To investigate any additional contribution of this peroxidase to endb SCW lignification, we used CRISPR-Cas9 to generate a per66 loss-of-function allele in a segregating lac4 lac11/+ lac17 background. A premature stop codon in per66-1 resulted in a truncated 121-aa protein (SI Appendix, Fig. S4). The defects in endb SCW lignification observed in lac4 lac11/+ lac17 were not enhanced by per66, and per66 mutants did not differ from wild type (Fig. 4C). Moreover, endb lignification was unaffected by addition of the peroxidase inhibitor salicylhydroxamic acid (SHAM) (12, 35) when we grew spl7 fruit in Cu-containing media (SI Appendix, Fig. S3). Therefore, the highly localized patterns of LAC4, 11, and 17, but not PER66, are required for endb SCW lignification. This is reminiscent of Casparian strip formation, where it is laccases that are replaceable for lignification, while peroxidases are absolutely required (16). In both cases, genetic evidence indicates that despite colocalization, only one class of oxidative enzymes is required for local lignin deposition.
SPL7-Dependent Cu Homeostasis Is Required for Laccase Activity in endb SCWs.
Our findings suggest that SPL7 is needed to ensure sufficient Cu in the fruit for the activity of LAC4, 11, and 17 in endb cell walls to form lignin. To test this hypothesis, we first verified that LAC4, 11, and 17 protein fusions accumulate in the endb cell walls of spl7 fruit (Fig. 4E). We also verified that the laccase-specific oxidation of 4-hydroxyindole was suitable to detect laccase activity in glycoprotein extracts of wild-type fruit (SI Appendix, Fig. S5). Using this assay in situ, we detected high levels of laccase activity localized to endb SCWs in wild-type fruit (Fig. 4F). Heat inactivation showed that this activity is enzyme-dependent (Fig. 4F). We found no laccase activity in the endb SCWs of spl7 fruit grown in low-Cu conditions (Fig. 4F). The absence of laccase activity matched the reduced lignin in these SCWs (Fig. 1C). Laccase activity was restored in spl7 endb SCWs by complementation with a pSPL7::SBP:GFP transgene (Fig. 4F). Strikingly, enzyme-dependent laccase activity was restored in spl7 endb SCWs by direct application of CuSO4 to spl7 fruit tissue (Fig. 4F). This result indicates that although laccases are present in spl7 endb SCWs, they require Cu supplementation for enzymatic activity. Therefore, localized lignin deposition in endb SCWs requires three laccases (LAC4, 11, and 17), which depend on SPL7 to provide sufficient Cu for their activity.
Discussion
Explosive seed dispersal in C. hirsuta depends on the precise subcellular deposition of lignin in endb cells of the fruit valves. Here, we identified four lignin-polymerizing enzymes—PER66 and LAC4, 11, and 17—that colocalize with the asymmetric pattern of lignin in endb SCWs. We used conditional gene expression to bypass the growth arrest of lac4 11 17 triple mutants and show that LAC4, 11, and 17 are required to lignify the distinctive endb SCW. The requirement for multi-Cu laccases rather than peroxidases to polymerize lignin in this cell type explains why SPL7, a key regulator of Cu homeostasis, is essential for robust endb SCW lignification.
The polar deposition and hinged pattern of lignin in endb cells of explosive fruit are an evolutionary novelty of Cardamine, associated with the appearance of explosive seed dispersal in this genus of the Brassicaceae family (4). We found that C. hirsuta LAC4 and 17 adopted a nonpolar localization when expressed under their native promoters in A. thaliana, matching the nonpolar lignification of A. thaliana endb SCWs. These findings show conservation between species in LAC gene expression but not protein localization. Therefore, the polarity and pattern of C. hirsuta LAC4, 11, and 17 endb localization are likely to be determined by trans factors that are specific to C. hirsuta. Identifying such factors will be an important follow-up to this study.
Apoplastic laccases are synthesized in the endomembrane system and are secreted by exocytosis (36). Hence, targeting laccase-loaded vesicles to specific regions of the plasma membrane may be important to pattern their localization in the cell wall. Laccases are also immobile in the dense SCW matrix and their localization may be fixed by anchoring to specific SCW components (11). LAC11 localization in C. hirsuta endb SCWs is coincident with the initial, asymmetric deposition of lignin throughout the SCW. In contrast to this, C. hirsuta LAC4, 17, and PER66 accumulate to higher levels in distinct layers of the lignified endb SCW. This may reflect spatial regulation within the SCW or temporal regulation during the sequential deposition of SCW layers in C. hirsuta endb cells (4). Interestingly, dexamethasone induction of pLAC11::LhGR≫LAC11:mCherry can overcome the restriction of LAC11 to the polar SCW domain and cause ectopic lignification throughout the endb cell wall (SI Appendix, Fig. S5). This suggests that levels of LAC11 expression in endb cells can influence protein localization, and further suggests that monolignols are available for lignin polymerization throughout the endb cell wall. It is an open question whether the hinged pattern of lignin deposition in C. hirsuta endb cells is also regulated by the same factors that determine polarity, or by an independent mechanism.
The reduced range of seed dispersal in spl7 mutants suggests that the material properties of lignin in the endb cell wall influence explosive dispersal. The tension that generates elastic energy for explosion is produced by differential contraction of valve tissues in C. hirsuta fruit (4). This puts the endb layer under compression and the exocarp layer under tension, and results in the storage of potential elastic energy. Material properties of the endb SCW will determine its compressive strength. We observed that less lignified endb SCWs in spl7, lac4 17, and lac4 11/+ 17 genotypes resulted in buckling of the fruit valve along its margin under load. Consequently, the amount of stored potential elastic energy released in this buckling would no longer be available for explosive valve coiling. This offers one explanation for the reduction in seed dispersal range in spl7 mutants. Future experiments will be useful to distinguish between alternative hypotheses.
Homeostatic regulation of Cu is a conserved function of SPL7 between C. hirsuta and A. thaliana. In A. thaliana, LAC4 and 17, but not LAC11, are targeted by SPL7-activated Cu miRNAs for posttranscriptional degradation in response to Cu deprivation (28). Although this regulation may also be conserved in C. hirsuta, our findings show that it is laccase activity, rather than the abundance of LAC4 and 17, that determines Cu-dependent endb lignification in spl7. Future work to dissect the SPL7 transcriptional response to Cu deprivation in C. hirsuta fruit will help to identify the precise mechanisms through which Cu is made available for LAC4, 11, and 17 activity in endb SCWs via the SPL7 pathway. The Cu dependence of laccase activity means that lignification of individual layers of endb SCWs in C. hirsuta spl7 fruit provides a cell-level readout of Cu availability (Fig. 1F). This readout gives some insight into the large variation in Cu availability that cells experience during growth and development, in the absence of SPL7. This variation in Cu availability in different plant tissues, throughout development and between different growing conditions, is buffered by SPL7 activity. The functional conservation of SPL7 homologs from green algae to flowering plants is indicative of how essential this regulation of Cu homeostasis is for green plant life (22, 26, 29).
In summary, we have identified a module that links mineral nutrition with seed dispersal. Localized lignin deposition is critical for the mechanism of explosive seed dispersal in C. hirsuta. Cu-requiring laccases regulate this lignification, making explosive dispersal dependent on the homeostatic control of Cu by SPL7. In this way, a SPL7/LAC4/11/17 module integrates mineral nutrition with polar lignin deposition to facilitate dispersal.
Materials and Methods
Plant Material.
C. hirsuta (Ox), herbarium specimen voucher Hay 1 (OXF) (3), and A. thaliana (Col-0) were used as wild type. The lig1 (spl7-1) allele was identified in a previous mutant screen in C. hirsuta (4), and the causal mutation was identified by mapping-by-sequencing. The following mutant alleles were generated in C. hirsuta using CRISPR-Cas9: spl7-2, per66-1, lac4-1, lac4-2, lac11-1, and lac17-1 (see SI Appendix for more details). pAtLAC4::AtLAC4:mCherry; lac4 lac17 was previously described (8). The following transgenic lines were generated in C. hirsuta for this study (constructs are described in SI Appendix): pSPL7::GFP-NLS, pSPL7::mCherry:SPL7, pSPL7::SPL7:YFPv, pSPL7::ΔSPL7(SBP):GFP, pLAC11::LhGR::pOp6::LAC11:mCherry, pLAC4::GFP-NLS, pLAC11::GFP-NLS, pLAC17::GFP-NLS, pLAC4::LAC4:mCherry, pLAC11::LAC11:mCherry, pLAC17::LAC17:mCherry, pPER49::PER49:mCherry, pPER64::PER64:mCherry, and pPER66::PER66:mCherry. Plants were grown on soil, in vitro and using aeroponics, both with and without CuSO4 supplementation. Dexamethasone was supplied in solid media and spray solution. Fruits were grown in vitro and treated with CuSO4, the peroxidase inhibitor SHAM, and the monolignols coniferyl alcohol and sinapyl alcohol. More details are described in SI Appendix.
Seed Dispersal.
Seed dispersal distance was measured in concentric rings around wild-type and spl7-1 plants as previously described (4) with slight modifications (SI Appendix).
Microscopy.
A Leica TCS SP8 was used for confocal laser scanning microscopy (CLSM) with the following excitation (ex) and emission (em) parameters (nm): lignin autofluorescence ex: 405, em: 440 to 510; calcofluor ex: 405, em: 425 to 475; GFP ex: 488, em: 500 to 550 or 492 to 540; basic fuchsin ex: 561, em: 600 to 650; mCherry ex: 594, em: 600 to 640; chlorophyll ex: 488, em: 650 to 730. Epifluorescence and bright-field microscopy were performed using a Zeiss Axio Imager M2 microscope. Cryofracture scanning electron microscopy was performed using an Emitech K1250X cryounit and a Zeiss Supra 40VP microscope. CLSM images were processed, and lignin autofluorescence intensity was quantified, using the Fiji package of ImageJ (https://fiji.sc). More details are described in SI Appendix.
Histochemistry.
The ClearSee protocol (37) was used to visualize fluorescent proteins and cell-wall stains (basic fuchsin and calcofluor white) by CLSM. Phloroglucinol staining of lignin was performed as described (4). Fresh fruit sections were treated with 4-hydroxyindole to visualize laccase activity; pretreatments included 100 °C or CuSO4.
Cu Quantification by ICP-MS.
Cu content of fruits was measured using an Agilent 7700 ICP mass spectrometer and expressed as mg/kg dry biomass. More details are described in SI Appendix.
Lignin Quantification and Monomer Analysis.
Acetylbromide-soluble lignin and lignin composition via thioacidolysis was determined as previously described (38). More details are described in SI Appendix.
RNA Sequencing.
RNA was extracted from three biological replicates of wild-type and spl7-1 stage 17 fruit valves using the Spectrum Total RNA Kit, and complementary DNA (cDNA) was synthesized using SuperScript III reverse transcriptase. Libraries (100-bp) were prepared and sequenced using the HiSeq 2500 Illumina platform at the Max Planck Institute for Plant Breeding Research Genome Centre (39). Paired-end reads were quality-checked, aligned to the C. hirsuta reference genome, and quantified (34). Differential gene expression was analyzed using DESeq from Bioconductor (40). More details are described in SI Appendix.
qRT-PCR.
RNA was extracted from three biological replicates of stage 17 fruit per genotype and used for cDNA synthesis as described above. qPCR was performed on a QuantStudio5 thermocycler using SYBR Green Supermix. The housekeeping gene TIP41 (CARHR242510) was used as reference and relative expression of SPL7 was calculated using the 2−ΔΔCt method. Primers used are listed in SI Appendix, Table S1.
Statistical Analyses.
Statistical analyses were done with R statistical software (41). More details are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank P. Huijser, M. Tsiantis, and A. Emonet for comments; K. Lufen for lignin analyses; P. Sarchet for conducting the mutant screen; L. Samuels and C. Kamei for sharing materials; X. Gan for bioinformatic services; A. Stamatakis for greenhouse support; R. Franzen for scanning electron microscopy; and W. Faigl for laccase purification. This work was supported by an International Max Planck Research School studentship (to M.P.-A.), the Deutsche Forschungsgemeinschaft (DFG) under Germany’s Excellence Strategy—EXC 2048/1—Project ID No. 390686111 (to M.P.), and a DFG FOR2581 Plant Morphodynamics grant (to A.H.). Portions of the paper were developed from the thesis of M.P.-A.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2202287119/-/DCSupplemental.
Data Availability
Short-sequence read data for this study has been deposited in the European Nucleotide Archive (ENA) at the European Molecular Biology Laboratory's European Bioinformatics Institute (EMBL-EBI) under accession number PRJEB50935 (39).
All other study data are included in the article and/or supporting information.
References
- 1.Hamilton W. D., May R. M., Dispersal in stable habitats. Nature 269, 578–581 (1977). [Google Scholar]
- 2.Kokko H., López-Sepulcre A., From individual dispersal to species ranges: Perspectives for a changing world. Science 313, 789–791 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Hay A., Tsiantis M., The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 38, 942–947 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Hofhuis H., et al. , Morphomechanical innovation drives explosive seed dispersal. Cell 166, 222–233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spence J., Vercher Y., Gates P., Harris N., ‘Pod shatter’ in Arabidopsis thaliana, Brassica napus and B. juncea. J. Microsc. 181, 195–203 (1996). [Google Scholar]
- 6.Dixon R. A., Barros J., Lignin biosynthesis: Old roads revisited and new roads explored. Open Biol. 9, 190215 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tobimatsu Y., Schuetz M., Lignin polymerization: How do plants manage the chemistry so well? Curr. Opin. Biotechnol. 56, 75–81 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Schuetz M., et al. , Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiol. 166, 798–807 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naseer S., et al. , Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc. Natl. Acad. Sci. U.S.A. 109, 10101–10106 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann N., Benske A., Betz H., Schuetz M., Samuels A. L., Laccases and peroxidases co-localize in lignified secondary cell walls throughout stem development. Plant Physiol. 184, 806–822 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yi Chou E., et al. , Distribution, mobility, and anchoring of lignin-related oxidative enzymes in Arabidopsis secondary cell walls. J. Exp. Bot. 69, 1849–1859 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee Y., Rubio M. C., Alassimone J., Geldner N., A mechanism for localized lignin deposition in the endodermis. Cell 153, 402–412 (2013). [DOI] [PubMed] [Google Scholar]
- 13.McCaig B. C., Meagher R. B., Dean J. F., Gene structure and molecular analysis of the laccase-like multicopper oxidase (LMCO) gene family in Arabidopsis thaliana. Planta 221, 619–636 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Tognolli M., Penel C., Greppin H., Simon P., Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 288, 129–138 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Welinder K. G., et al. , Structural diversity and transcription of class III peroxidases from Arabidopsis thaliana. Eur. J. Biochem. 269, 6063–6081 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Rojas-Murcia N., et al. , High-order mutants reveal an essential requirement for peroxidases but not laccases in Casparian strip lignification. Proc. Natl. Acad. Sci. U.S.A. 117, 29166–29177 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berthet S., et al. , Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23, 1124–1137 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Q., et al. , Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25, 3976–3987 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barros J., Serk H., Granlund I., Pesquet E., The cell biology of lignification in higher plants. Ann. Bot. 115, 1053–1074 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J., et al. , Lignin engineering through laccase modification: A promising field for energy plant improvement. Biotechnol. Biofuels 8, 145 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Printz B., Lutts S., Hausman J. F., Sergeant K., Copper trafficking in plants and its implication on cell wall dynamics. Front. Plant Sci. 7, 601 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burkhead J. L., Gogolin Reynolds K. A., Abdel-Ghany S. E., Cohu C. M., Pilon M., Copper homeostasis. New Phytol. 182, 799–816 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Birkenbihl R. P., Jach G., Saedler H., Huijser P., Functional dissection of the plant-specific SBP-domain: Overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 352, 585–596 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Huijser P., Schmid M., The control of developmental phase transitions in plants. Development 138, 4117–4129 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Quinn J. M., Merchant S., Two copper-responsive elements associated with the Chlamydomonas Cyc6 gene function as targets for transcriptional activators. Plant Cell 7, 623–628 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kropat J., et al. , A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. U.S.A. 102, 18730–18735 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernal M., et al. , Transcriptome sequencing identifies SPL7-regulated copper acquisition genes FRO4/FRO5 and the copper dependence of iron homeostasis in Arabidopsis. Plant Cell 24, 738–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abdel-Ghany S. E., Pilon M., MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 283, 15932–15945 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamasaki H., Hayashi M., Fukazawa M., Kobayashi Y., Shikanai T., SQUAMOSA promoter binding protein-like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21, 347–361 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weigel M., et al. , Plastocyanin is indispensable for photosynthetic electron flow in Arabidopsis thaliana. J. Biol. Chem. 278, 31286–31289 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Molina A., Xing S., Huijser P., Functional characterisation of Arabidopsis SPL7 conserved protein domains suggests novel regulatory mechanisms in the Cu deficiency response. BMC Plant Biol. 14, 231 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahmati Ishka M., Vatamaniuk O. K., Copper deficiency alters shoot architecture and reduces fertility of both gynoecium and androecium in Arabidopsis thaliana. Plant Direct 4, e00288 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan J., et al. , Arabidopsis pollen fertility requires the transcription factors CITF1 and SPL7 that regulate copper delivery to anthers and jasmonic acid synthesis. Plant Cell 29, 3012–3029 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gan X., et al. , The Cardamine hirsuta genome offers insight into the evolution of morphological diversity. Nat. Plants 2, 16167 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee Y., et al. , A lignin molecular brace controls precision processing of cell walls critical for surface integrity in Arabidopsis. Cell 173, 1468–1480.e9 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Hoffmann N., King S., Samuels A. L., McFarlane H. E., Subcellular coordination of plant cell wall synthesis. Dev. Cell 56, 933–948 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Ursache R., Andersen T. G., Marhavý P., Geldner N., A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. Plant J. 93, 399–412 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Foster C. E., Martin T. M., Pauly M., Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part I: Lignin. J. Vis. Exp. (37), 1745 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.M. Pérez-Antón, A. Hay, Project: PRJEB50935. European Nucleotide Archive. https://www.ebi.ac.uk/ena/browser/view/PRJEB50935. Deposited 15 April 2022. [Google Scholar]
- 40.Anders S., Huber W., Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.R Core Team, R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2019). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Short-sequence read data for this study has been deposited in the European Nucleotide Archive (ENA) at the European Molecular Biology Laboratory's European Bioinformatics Institute (EMBL-EBI) under accession number PRJEB50935 (39).
All other study data are included in the article and/or supporting information.