Significance
Higher plants produce specialized metabolites to cope with biotic and abiotic challenges faced in natural environments. The diversity and complexity of specialized metabolism is often beneficial, providing functional synergisms and evolutionary stability when metabolic solutions to ecological problems are deployed as mixtures. These benefits of diversity have recently been realized in the deployment of antibiotics, insecticides, fungicides, and herbicides. However, the functional downside of metabolite diversity is rarely studied, in part because the mechanisms of post-ingestive metabolite interactions are largely unknown. Here we showed that larvae of the tobacco hornworm, Manduca sexta, rearrange key constituents of two distinct defense pathways in its wild tobacco host plant, Nicotiana attenuata, to thwart the defensive properties of both pathways.
Keywords: plant-specialized metabolites, frass metabolites, synergy effects, chlorogenic acid, diterpenoids
Abstract
Deploying toxins in complex mixtures is thought to be advantageous and is observed during antagonistic interactions in nature. Toxin mixtures are widely utilized in medicine and pest control, as they are thought to slow the evolution of detoxification counterresponses in the targeted organisms. Here we show that caterpillars rearrange key constituents of two distinct plant defense pathways to postingestively disable the defensive properties of both pathways. Specifically, phenolic esters of quinic acid, chlorogenic acids (CAs), potent herbivore and ultraviolet (UV) defenses, are reesterified to decorate particular sugars of 17-hydroxygeranyllinalool diterpene glycosides (HGL-DTGs) and prevent their respective anti–herbivore defense functions. This was discovered through the employment of comparative metabolomics of the leaves of Nicotiana attenuata and the frass of this native tobacco’s specialist herbivore, Manduca sexta larvae. Feeding caterpillars on leaves of transgenic plants abrogated in each of the two pathways, separately and together, revealed that one of the fully characterized frass conjugates, caffeoylated HGL-DTG, originated from ingested CA and HGL-DTGs and that both had negative effects on the defensive function of the other compound class, as revealed by rates of larval mass gain. This negative defensive synergy was further explored in 183 N. attenuata natural accessions, which revealed a strong negative covariance between the two defense pathways. Further mapping analyses in a biparental recombinant inbred line (RIL) population imputed quantitative trait loci (QTLs) for the two pathways at distinct genomic locations. The postingestive repurposing of defense metabolism constituents reveals a downside of deploying toxins in mixtures, a downside which plants in nature have evolved to counter.
Being at the foundation of most food webs on the planet, autotrophic plants require sophisticated defenses to survive in a world full of heterotrophs. Chemicals allow plants to cope not only with attack from heterotrophic organisms (1) but also with abiotic challenges (drought, extreme temperature, and irradiation) (2) and even to communicate with neighbors (3). Plants have evolved diverse specialized metabolomes to solve these ecological challenges. Several influential hypotheses invoke functional interpretations for this metabolite diversity, such as increasing the likelihood of producing a bioactive compound against a natural enemy (4) and providing the functional redundancy that facilitates the evolution of compounds that target specific natural enemies (5). In short, producing multiple specialized metabolites often increase a plant’s Darwinian fitness in environments with diverse natural enemies (5, 6). The responsible mechanisms for the adaptive value of metabolic diversity are also diverse, including defensive synergisms (7) and the evolutionary stability that mixtures provide in resistance management of insecticides, herbicides, and antibiotics in the face of coevolving antagonists. However, the possible functional downsides of diversity have received less attention.
Specialist insect herbivores that have evolved the ability to feed on only a few closely related host plant taxa provide ideal natural systems with which to study functional interactions among diverse plant-specialized metabolites. These specialist herbivores have evolved multiple adaptive mechanisms that allow them to degrade and detoxify ingested defenses, avoid contact and ingestion, evolve target-site insensitivity, rapidly excrete ingested defenses, and even sequester plant defenses to co-opt them for their own defense (8, 9). For example, although glucosinolates, the main chemical defense of Brassicaceae, are hydrolyzed to release toxic isothiocyanates in response to attack, many insects readily feed and perform well on glucosinolate-accumulating plants by thwarting glucosinolate breakdown or deactivating toxic products (10, 11). Whether these diverse plant defenses interact after ingestion remains poorly studied.
The wild tobacco, Nicotiana attenuata, an ecological model plant with a rich portfolio of specialized metabolites, has long interacted with the voracious larvae of the lepidopteran Manduca sexta, a specialist that feeds largely on Solanaceae plants. N. attenuata plants accumulate large quantities of defensive pyridine alkaloids, proteinase inhibitors, 17-hydroxygeranyllinalool diterpene glycosides (HGL-DTGs), phenolamides, and phenolics (12–16) and hence constitute an excellent system in which to explore functional interactions among diverse specialized metabolites. Some of these interactions can result in defensive synergies, as has been shown with the neurotoxin nicotine and the antidigestive defense of proteinase inhibitors, where nicotine prevents herbivores from consuming more plant material to compensate for the effects of the proteinase inhibitors (17).
Here, a metabolomic comparison of N. attenuata leaves and M. sexta larval frass revealed a class of compounds derived from quinic acid conjugates and HGL-DTGs, plant defenses which are synthesized through two distinct pathways. Performance assays revealed that this specialist herbivore produced the compounds in the process of counteracting the defensive function of the two distinct defense pathways. A strong negative genetic covariance in the regulation of these two plant defense pathways was uncovered in natural accessions and a recombinant inbred population, suggesting that this herbivore’s detoxification strategy has influenced selection for metabolic diversity in its hostplant.
Results and Discussion
Compounds Appear in the Frass of Insect Herbivores.
The coyote tobacco, N. attenuata, is native to the Great Basin Desert in the southwestern United States and is an ecological model used to study the function of different sectors of specialized metabolism. HGL-DTGs are a class of abundant defensive specialized metabolites in Nicotiana plants (13), and a recent study revealed that metabolomic analyses of herbivore frass provided a robust means of understanding the defensive mechanisms and cellular targets of ingested HGL-DTGs (18). To better understand the metabolism of HGL-DTGs in herbivore guts, we employed a tandem mass spectrometry (MS/MS) analytical dereplication workflow (19) to compare the metabolomes of N. attenuata leaves and the frass of a specialist herbivore, M. sexta larvae, feeding on the leaves of this native tobacco.
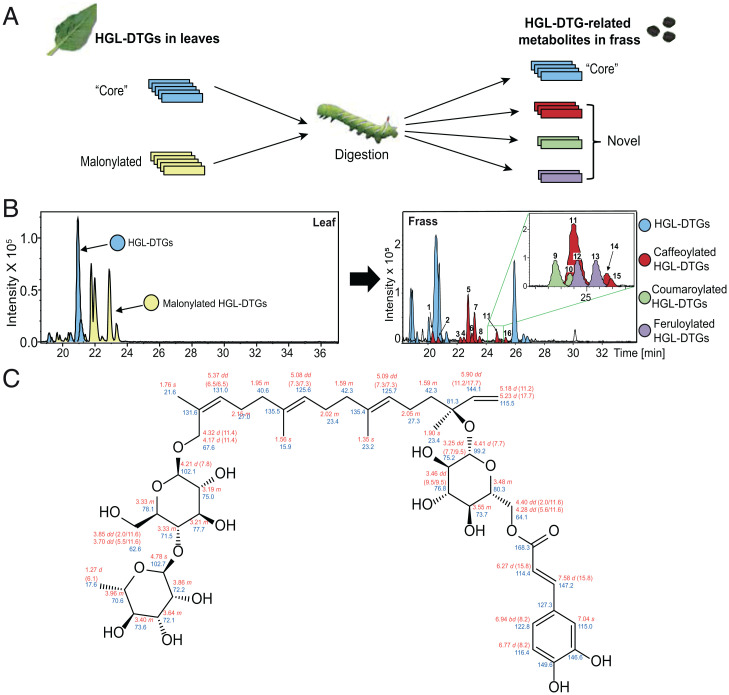
Consistent with previous research, this comparative analysis revealed that the malonyl moieties of HGL-DTGs were lost in the alkaline condition of the larval midgut (20) and all malonylated HGL-DTGs were converted into the core glycosides (Fig. 1 A and B). In addition to the previously identified 3-O-α-rhamnopyranosyl-(1→4)-β-glucopyranosyl-17-hydroxygeranyllinalool (RGHGL), a deglycosylation and detoxification product of HGL-DTGs (20), we detected a group of unknown metabolites with the diagnostic fragment corresponding to the aglycone of HGL-DTGs (SI Appendix, Fig. S1A). Our rapid dereplication and identification workflow detected three known fragments diagnostic of hydroxycinnamoyl (HC) derivatives: mass-to-charge ratio (m/z) 163.0389 ([M+H]+) corresponding to caffeoyl, m/z 147.0440 ([M+H]+) corresponding to coumaroyl, and m/z 177.0546 ([M+H]+) corresponding to feruloyl, as previously verified (21). A detailed mass spectrometry (MS) analysis and putative structure descriptions suggested that the HC moieties were linked to the sugar moieties of the HGL-DTGs to form hydroxycinnamoylated HGL-DTGs (HC-HGL-DTGs) (SI Appendix, Fig. S1B). To test this inference, we purified one of the most abundant peaks (peak 5 in Fig. 1B) from the frass of M. sexta larvae that fed on N. attenuata plants and conducted NMR analysis, which confirmed the structure as a caffeoylated HGL-DTG (Fig. 1C).
Fig. 1.
HC-HGL-DTGs appear in the frass of M. sexta larvae that fed on N. attenuata plants. (A) The scheme of the comparative metabolomics analysis of the leaves of N. attenuata and frass of M. sexta larvae fed on N. attenuata leaves. (B) Comparative chromatographic analysis between leaves and M. sexta frass confirmed the loss of malonyl moieties of HGL-DTGs and revealed a class of HC-HGL-DTGs in M. sexta frass that are not detected in the leaves ingested by the larvae. All the detected HC-HGL-DTGs were numbered following their retention times, and their corresponding annotated mass spectra are shown in SI Appendix, Fig. S1. (C) The NMR-verified chemical structure of the peak 5 HC-HGL-DTG, which is a caffeoylated HGL-DTG. Red: 1H chemical shifts δ (ppm, multiplet, 3JHH in hertz). Blue: 13C chemical shifts (δ ppm).
Moieties of the Compounds Are from Two Distinct Plant Defense Pathways.
The HGL-DTG part of HC-HGL-DTGs likely originates from plant-produced HGL-DTGs, a diverse metabolic sector of N. attenuata. To test this inference, we fed M. sexta larvae several lines of transformed plants silenced in different HGL-DTG biosynthetic genes by RNA inference, including geranylgeranyl pyrophosphate synthase (irGGPPS) (22) and uridine diphosphate glycosyltransferases (irUGT74P5, irUGT74P3&5, and irUGT91T1) (23), and compared their frass with that from larvae fed wild-type (WT) N. attenuata plants. In all cases, the abundances of HC-HGL-DTGs in the frass were dramatically decreased when larvae fed on HGL-DTG–depleted plants (SI Appendix, Fig. S2A), demonstrating the plant-based origin of HGL-DTG moieties and leading to the question of the origin of the HC part of the HC-HGL-DTGs in frass.
N. attenuata plants produce two major classes of compounds that contain HC moieties: caffeoylquinic acids and phenolamides. To parse the potential origins of the HC moieties, we fed M. sexta larvae leaves of transgenic plants silenced in the expression of HC–coenzyme A (CoA) quinate transferase (irHQT), which controls the last step of the conjugation between HC moieties and quinic acid (15), or the phenolamide master regulatory transcription factor NaMYB8 (irMYB8) (14) and quantified metabolites in larval frass (Fig. 2A). While both lines of transgenic plants have WT levels of HGL-DTGs in their leaves, the contents of quinic acid conjugates and phenolamides were specifically regulated in irHQT and irMYB8 leaves, respectively (Fig. 2B and SI Appendix, Fig. S2 B and C). HC-HGL-DTG levels were dramatically decreased only in the frass of larvae that were fed irHQT plants, not of those fed irMYB8 plants, although the levels of core HGL-DTGs in the frass were not significantly affected (Fig. 2C and SI Appendix, Fig. S2 B and C). From these results, we inferred that the origin of the HC moieties is from ingested HC quinate conjugates.
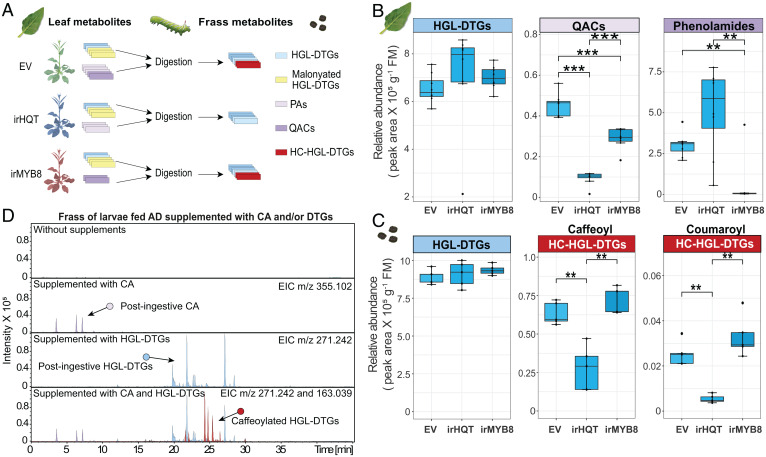
Fig. 2.
The caffeoyl moieties of caffeoylated HGL-DTGs arise from ingested CAs. (A) Specialized metabolomes of leaves of three different transgenic wild tobacco plants were compared with the frass metabolomes of caterpillars fed these leaves; different colors reflect different classes of metabolites; PA, phenolamides. (B) The concentrations ( n = 6, in fresh mass (FM)) of HGL-DTGs (blue), quinic acid conjugates (QACs; pink), and phenolamides (purple) in homozygous transgenic plants that were silenced in CA accumulations [irHQT (15)] or in the expression of the phenolamide master regulator [irMYB8 (14)], with plants transformed with an EV as controls. (C) The concentrations (n = 6) of HGL-DTGs and two major forms of HC-HGL-DTGs (red), caffeoylated and coumaroylated, in the frass of M. sexta larvae fed different transgenic plants. (D) Supplementing AD with CA and HGL-DTGs results in the accumulation of caffeoylated HGL-DTGs in the frass of M. sexta larvae feeding on these diets. Asterisks indicate significant differences compared with data from EV plants (**P < 0.01; ***P < 0.001; Student t test with Bonferroni correction). (B and C) The upper whisker extends from the hinge to the largest value no further than 1.5 * interquartile range (IQR) from the hinge and the lower whisker extends from the hinge to the smallest value at most 1.5 * IQR of the hinge.
To further evaluate the origins of HC-HGL-DTGs, we fed M. sexta larvae artificial diet (AD) supplemented with a caffeoyl quinate ester, chlorogenic acid (CA); HGL-DTGs; or both. The HC-HGL-DTGs were only detected in frass of larvae fed AD supplemented with both CA and HGL-DTGs (Fig. 2D). Notably, CA can isomerize to cryptochlorogenic acid and neochlorogenic acid when ingested by M. sexta larvae, likely due to the alkaline environment of the larval midgut (SI Appendix, Fig. S3A). Alkaline buffer alone can isomerize CA but does not lead to the production of HC-HGL-DTGs (SI Appendix, Fig. S3B). Additionally, HC-HGL-DTGs accumulate mainly in the gut content, followed by hindgut and midgut body tissues (SI Appendix, Fig. S3 C–E). From these results, we inferred that HC-HGL-DTGs are postingestively produced from ingested CA and HGL-DTGs, and their conjugation is likely an active process controlled by the larvae, considering that M. sexta lacks a resident gut microbiome (24).
Postingestive Conjugation Thwarts Defense Functions of Two Distinct Pathways.
Semiquantitative comparisons of HC-HGL-DTGs and HGL-DTGs in the frass of M. sexta larvae that fed on N. attenuata plants revealed that about 10% of ingested HGL-DTGs are conjugated to produce HC-HGL-DTGs (Fig. 2C). Considering the large amounts of HGL-DTGs in N. attenuata aboveground tissues [∼6 mg g−1 fresh mass (20)], HC-HGL-DTGs are quantitatively abundant in frass. To investigate the defensive function of this conjugation, we performed a larval performance bioassay with AD supplemented with CA, HGL-DTG, or both compounds. CA supplementation significantly decreased larval growth; this effect was marginally, but not significantly, attenuated when both HGL-DTGs and CA were added into AD (Fig. 3A). Consistent with the qualitative analysis, the caffeoylated HGL-DTGs only accumulated in the frass of M. sexta larvae that fed on AD supplemented with both CA and HGL-DTGs (Fig. 3B). Frass CA levels were slightly decreased when larvae were fed AD supplemented with both CA and HGL-DTGs, compared with that of larvae fed AD supplemented only with CA (Fig. 3B). From these data, we inferred that ingested HGL-DTGs can detoxify ingested CA. Consistent with previous research that the defensive function of HGL-DTGs requires their metabolism into hydroxylated derivatives, a process not occurring when larvae feed on AD (18), HGL-DTG supplementation of AD did not affect M. sexta larval growth (Fig. 3A). Although AD have long been used to evaluate defensive functions of purified metabolites or plant extracts, their drawbacks are well described. Insect AD not only contain large quantities of antibiotics and antioxidants to enhance their shelf life, but they are also commonly based on unnatural components (wheat germ and milk protein) that provide larvae with unrealistically high protein/carbohydrate ratios, which are known to influence a larva’s ability to detoxify defense metabolites (25). Thus, performance bioassays with plants genetically modified in their metabolite accumulations provide a superior means of evaluating the defensive function of specialized metabolites.
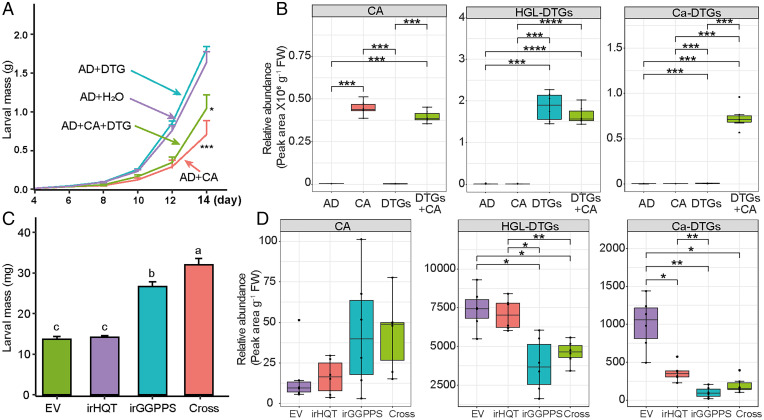
Fig. 3.
M. sexta larvae use CA and HGL-DTGs to detoxify each other. (A) Mass (mean + standard error (SE), n = 13–15) of M. sexta larvae fed AD supplemented with 1.2 mg g−1 CA, 2 mg g−1 HGL-DTGs (also abbreviated DTG or DTGs in the figure image), or both CA and HGL-DTGs, compared with AD supplied with only water (H2O) as control (Con). (B) Relative abundances (n = 5) of CA, HGL-DTGs, and caffeoylated DTGs (Ca-DTGs) in the frass of M. sexta larvae that fed on different AD. (C) Mass (mean + SE, n = 36–40) of M. sexta larvae fed hemizygous transgenic irHQT, irGGPPS plants, or an irHQT x irGGPPS Cross for 4 d. (D) The relative abundances (n = 5) of CA, HGL-DTGs, and Ca-DTGs in the frass of M. sexta larvae that fed on different transgenic plants. Asterisks indicate significant differences among different groups (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; Student t test with Bonferroni correction). (B and D) The upper whisker extends from the hinge to the largest value no further than 1.5 * interquartile range (IQR) from the hinge and the lower whisker extends from the hinge to the smallest value at most 1.5 * IQR of the hinge.
To parse functional consequences of the interaction between HGL-DTGs and CA in the natural dietary context of a plant, we conducted larval performance assays on irGGPPS plants which are silenced in HGL-DTG production, irHQT plants which are silenced in CA production, the hemizygous cross of both defense pathway-silenced lines (Cross), and empty vector (EV) plants as control so as to provide plant diets in which the HGL-DTGs and CA were manipulated independently or simultaneously (SI Appendix, Fig. S4). Consistent with previous research, HGL-DTGs proved to be one of the most robust defensive compounds in N. attenuata (26): M. sexta larvae fed irGGPPS plants grew much better than larvae fed EV plants (Fig. 3C). There are three copies of NaGGPPS in the N. attenuata genome, with the copy that dominates HGL-DTG synthesis specifically silenced in irGGPPS plants (22). To further test the inference about the defensive function of HGL-DTGs, we performed M. sexta larval bioassays on virus-induced gene-silenced (VIGS) plants, in which the expression of the cytochrome P450 gene (NaCYP736A) that controls a key branch point in HGL-DTG biosynthesis was silenced (18). Similar to their performance on irGGPPS plants, larvae grew much better on NaCYP736A-silenced plants than on EV plants. The HC-HGL-DTG contents of the frass of M. sexta larvae that fed on different transgenic plants followed the same pattern as another detoxification product, RGHGL (SI Appendix, Fig. S5). These results demonstrate the defensive function of HGL-DTGs in planta.
In contrast, the defensive function of CA, while apparent in the experiments conducted with supplemented AD (Fig. 3A), was not apparent when comparing larvae fed EV and irHQT plants. This result is consistent with the results of a series of earlier experiments with CA biosynthesis-modified transgenic Nicotiana tabacum plants, which revealed no negative effects of CA on larval growth (27–29). However, M. sexta larvae grew significantly better on the Cross plants than on the irGGPPS plants, indicating a context-dependent defensive function of CA in planta. The different performances of larvae fed EV vs. irHQT plants and larvae fed irGGPPS vs. Cross plants suggest an interaction between CA and HGL-DTGs after ingestion. In contrast with the HGL-DTG–producing Nicotiana plants, CA’s defensive function has been demonstrated in chrysanthemum plants which do not produce HGL-DTGs (30). Taken together, these in planta bioassays reveal that the defensive function of CA is thwarted when HGL-DTGs are present, likely by their use in the detoxification of HGL-DTGs to form HC-HGL-DTGs. The trend of increased CA in the frass of larvae feeding on irGGPPS and Cross plants is consistent with this inference (Fig. 3D). In addition, the hydroxylated products of HC-HGL-DTGs, which are the defensive form of HGL-DTGs (18), were not detected in the frass (SI Appendix, Fig. S3F). Hence, the apparent lack of CA defensive function is the integrated outcome of CA’s defensive and detoxification functions. Previously, the quinic acid moieties of CA were reported as conjugated to saligenin, a defensive phenolic glucoside in salicaceous plants, in the frass of another lepidopteran specialist herbivore (31), suggesting that several lepidopteran herbivores can use CA as a versatile detoxification agent. In short, the specialist herbivore M. sexta uses two distinct defensive compounds to detoxify each other.
Natural Populations Revealed the Signatures of Counterresponses.
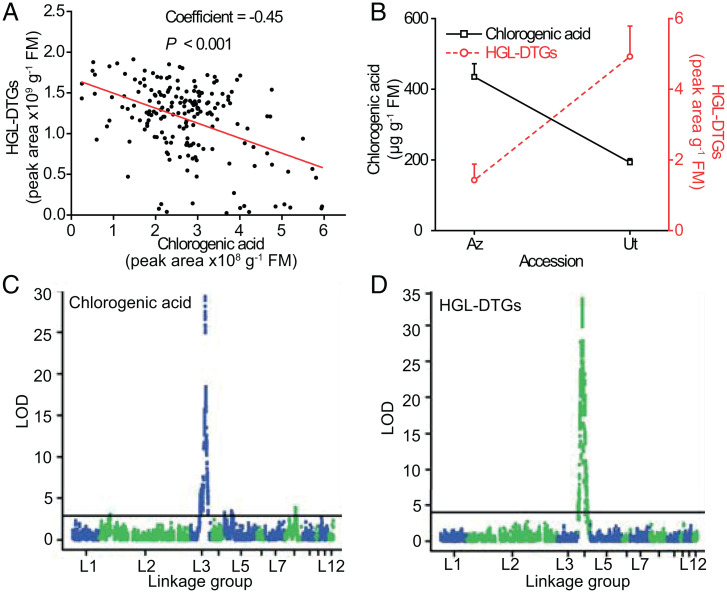
N. attenuata plants have likely interacted with M. sexta larvae for the majority of their 24 million years of existence (32). To evaluate if natural populations harbor the expected signatures of a strong negative defensive synergy driven by a specialist herbivore’s detoxification strategy, we quantified CA and HGL-DTGs in 183 natural accessions of N. attenuata derived from seeds collected in the Great Basin Desert (33). Interestingly, there was a significant negative relationship between the abundances of CA and HGL-DTGs in leaves (Fig. 4A). Among these accessions, two well-studied accessions originate from Utah (Ut) and Arizona (Az), respectively. The absolute concentration of CA and HGL-DTGs in Ut and Az plants were quantified: Ut plants accumulated higher levels of HGL-DTGs and lower levels of CA, while Az plants showed the opposite pattern (Fig. 4B). At higher altitudes, with higher ultraviolet B (UV-B) fluence, plants from the Az accession would be selected for higher CA levels due to well-established sunscreen functions of CA (34). However, due to the detoxification process described here, Az plants would also be selected for lower HGL-DTG levels (Fig. 4B). Considering the more robust defense function of HGL-DTGs than CA (Fig. 3C), Az plants would be expected to be more susceptible to herbivore attack than UT plants. Az plants are also known to have higher nicotine levels and a disabled proteinase inhibitor defense system (35, 36), which may help to compensate for their lower HGL-DTG levels.
Fig. 4.
The abundances of CA and HGL-DTGs are genetically negatively correlated in natural accessions of N. attenuata and impute different loci in a RIL population. (A) Relative abundances of CA and HGL-DTGs in leaves of 183 natural accessions of N. attenuata are negatively correlated. (B) Abundances (mean + SE, n = 5) of CA and HGL-DTGs in two well-studied accessions of N. attenuata, Ut and Az. The QTL mapping using an AI-RIL population generated from intercrosses of the Ut and Az accessions and subsequent inbreeding revealed separate loci regulating the abundances of CA (C) and HGL-DTGs (D). Three genes imputed in the CA QTL—namely, a MYBb308 transcription factor (TF), a PAL biosynthetic gene, and a bHLH TF—underwent VIGS, but only the MYB308 TF significantly altered CA contents (SI Appendix, Fig. S6).
To elucidate the possible genetic basis underlying this negative correlation, we performed a quantitative trait locus (QTL) analysis using an advanced intercross recombinant inbred line (AI-RIL) population developed by crossing Ut and Az plants (37). The QTL mapping analysis revealed that the loci controlling CA and HGL-DTGs are on linkage groups 3 and 4, respectively (Fig. 4 C and D), of the published assembly of the N. attenuata genome (38). Genetic analysis of the genomic loci imputed by CA levels uncovered three candidate genes: a MYB (myeloblastosis virus) transcription factor, a basic helix-loop-helix (bHLH) transcription factor, and an upstream biosynthetic gene of CA, phenylalanine ammonia-lyase (PAL). Sequence analysis revealed that the MYB transcription factor, namely, NaMYB308, contained a premature stop codon in Ut plants but that Az plants had an intact coding region (SI Appendix, Fig. S6B). VIGS was performed in Ut and Az plants to specifically silence the candidate genes, and only silencing NaMYB308 expression abolished the differences in CA contents between Ut and Az plants (SI Appendix, Fig. S6 C and D). Thus, NaMYB308 likely controls CA accumulation differences between Ut and Az plants. Although a single sharp LOD (logarithm of odds) peak was imputed by the HGL-DTG contents, we were unsuccessful in identifying a protein-coding locus responsible for the differences in HGL-DTGs between Ut and Az plants. Previous research revealed that HGL-DTGs are positively regulated by jasmonate (JA) signaling (22), and here, we show that JA negatively regulates CA in Az plants through its influence on NaMYB308 expression (SI Appendix, Fig. S6), suggesting that the covariance of CA and HGL-DTGs is regulated by JA signaling. While substantially more research is required to flesh out the mechanisms and evolutionary history of this negative genetic covariance, these results are consistent with its association with the detoxification strategy of Manduca larvae described here.
Conclusion.
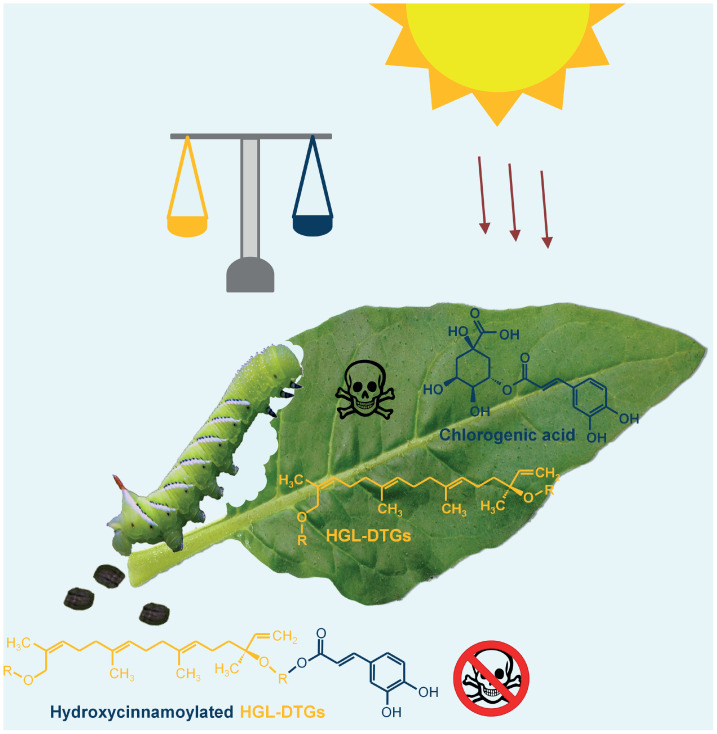
The benefits of diversity in plant-specialized metabolites have long been recognized in early theoretical deliberations (6, 7, 39), and the synergistic effects of deploying mixtures are thought to accelerate the evolution of diversification (40). Through the study of frass metabolites, we found that the caffeoyl moieties of CA are reesterified to glucose of HGL-DTGs and reduce the growth-retardant effects of the two distinct metabolisms. This study presents a downside to metabolic diversity, which through the relentless evolutionary arms races among plants and their attackers could counter selection for enhanced metabolic diversity (Fig. 5).
Fig. 5.
Postingestive interactions between diterpenoids and phenolics allow caterpillars to detoxify both defense pathways, while plants balance opposing selection pressures to minimize this detoxification process. Plants produce diverse specialized metabolites to deal with biotic and abiotic stresses from environments, such as diterpenoids (yellow) and phenolic CA (blue), both of which have deterrent effects on caterpillar feeding. Phenolics also function as sunscreens that protect plants from UV-B radiation. When fed these two toxic compounds, larvae of specialized insects can reesterify the phenolic moiety of CA onto the diterpenoid defenses and prevent the antiherbivore function of both defense compound classes. In nature, plants can balance the abundances of diterpenoids and phenolics to circumvent the detoxification of specialized insects and optimize UV protection.
Method Details
Plants and Growth Conditions.
The 31st inbred generation of N. attenuata was used as Ut WT genotype plants. Seeds were germinated on a mixture of plant agar with Gamborg’s B5 medium in sterile Petri dishes, and seedlings were transferred to pots and grown under 19 to 35 °C with 16 h of light (supplemental lighting by Philips Sun-T Agro 400- and 600-W sodium lights) and 60 to 65% relative humidity as previously described (41). Stably transformed and fully characterized RNA interference plants, including irUGT91T1 [A-11-538-05 (23)], irUGT74P5 [A-11-544-11 (23)], irUGT74_3&5 [A-12-076-07 (23)], irGGPPS [A-08-230-5 (22)], irHQT [A-14-153-6 (15)], and irMYB8 [A-07-810 (14)], were germinated and grown under glasshouse conditions. Cross plants were produced by crossing the homozygous irGGPPS and irHQT in both parental directions. Hemizygous irGGPPS and irHQT were produced by crossing the homozygous irGGPPS and irHQT with EV (A-03-009-1). The VIGS procedure was used with Ut WT plants and Az plants of the 22nd inbred generation (42), following the procedures described by Galis et al. (43). Briefly, leaves of young rosette-stage plants were pressure infiltrated with a mixture of Agrobacterium tumefaciens containing pBINTRA and either pTV-MYB308 or pTV00 (EV control). The VIGS experiments for NaCYP736A on EV or irGGPPS plants are described in Li et al. (18). VIGS experiments were repeated at least three times.
M. sexta Growth Conditions and Larval Performance Assays.
M. sexta eggs were obtained from an in-house colony in which insects are reared in a growth chamber (Snijders Scientific, Tilburg, the Netherlands) at 26 °C:16-h light and 24 °C:8-h dark, 65% relative humidity, until hatching.
To measure the performance of M. sexta larvae on plants (EV, irHQT, irGGPPS, and Cross), a single newly hatched neonate was introduced onto the first stem leaf of each early elongated N. attenuata plant and allowed to feed freely. The larvae were weighed 4 d after release.
AD assays were performed in plastic polyethylen (PE)-packing cups containing AD based on the recipes of Machado et al. (44). Additionally, neonates were fed AD containing 2 mg g−1 fresh weight (FW)-purified HGL-DTGs (aqueous in 5 mL), 3.4 µmol g−1 FW CA (aqueous in 5 mL), and a combination of 2 mg g−1 FW HGL-DTGs and 3.4 µmol g−1 FW CA. The AD was exchanged with fresh diet after each weighing.
Frass Collection.
Two freshly hatched neonates of M. sexta were reared on the first fully elongated stem leaf (S1; younger leaves are subsequently labeled S2, S3, and S4 proceeding up the main stalk) of transgenic 43-d-old flowering N. attenuata plants and retained on the S1 leaf with clip cages for 3 d. Approximately, 10 to 20 mg of frass were collected either from the top of the leaves with forceps or from the bottom of the clip cage. Frass from AD was collected using forceps from the PE-packing cups. All materials were flash frozen in liquid nitrogen and stored at −80 °C until use. In general, frass was collected from caterpillars while they were in the first to third instars.
Extraction and Analysis of CA, HGL-DTGs, and HC-HGL-DTGs.
Plant samples were ground in liquid nitrogen to a fine powder and ca. 100 mg of leaf tissue were aliquoted for extraction. CA and HGL-DTGs were extracted using 1 mL 80% methanol aqueous buffer in a 2-mL Eppendorf tube containing two steel balls and were agitated twice at 1,200 strokes min-1 for 60 s using a GenoGrinder 2000. Homogenized samples were centrifuged twice at 16,000 × g for 20 min at 4 °C, and the supernatants were analyzed on a micrOTOF-Q II or Impact II system (Bruker) as previously described (19).
Collected frass was weighed and extracted using 80% aqueous methanol with the ratio of 1 mL per 100 mg or 1 mL per 10 mg of frass (experiment using the cross) and analyzed using the same method used for plant samples.
Rapid Screening of HC-HGL-DTGs.
Chromatographic separation was performed using a Dionex UltiMate 3000 rapid separation LC system (Thermo Fisher), combined with a Thermo Acclaim RSLC 120 C18 column (particle size, 2.2 µm; average pore diameter, 120 Å; column dimension, 2.1 × 150 mm). Solvent A: water with 0.1% acetonitrile and 0.05% formic acid, and solvent B: acetonitrile with 0.05% formic acid, was used. Sample elution steps were as follows: 0 to 3 min at 10% solvent B, 3 to 12 min at 20% solvent B, 12 to 17 min at 35% solvent B, 17 to 23 min at 40% solvent B, 23 to 25 min at 45% solvent B, 25 to 30 min at 50% solvent B, 30 to 40 min at 90% solvent B, and 40 to 45 min at 90% solvent B, followed by column equilibration steps and a return to the starting conditions. A second gradient was also used with the same solvents for more rapid screening (experiment using the cross). Sample elution steps were as follows: 0 to 0.5 min at 10% solvent B, 0.5 to 23.5 min at 90% solvent B, and 23.5 to 25 min at 90% solvent B, followed by column equilibration steps and a return to the starting conditions. The injection volume was 2 µL, and the flow rate was 0.4 mL min-1.
MS detection was performed using a micrOTOF-Q II and an Impact II ultra high resolution quadrupole time-of-flight mass spectrometer (UHR-Q-TOF-MS) system, all equipped with an electrospray ionization (ESI) source and operated in positive ion mode. ESI conditions for the micrOTOF-Q II system were endplate offset of 500 V, capillary voltage of 4,500 V, capillary exit of 130 V, dry temperature of 180 °C, and dry gas flow of 10 L min−1. ESI conditions for the Impact II UHR-Q-TOF-MS system were endplate offset of 500 V, capillary voltage of 4,500 V, nebulizer of 2 bar, dry temperature of 200 °C, and dry gas flow of 8 L min−1. MS data were collected over a range of m/z 100 to 1,600. Mass calibration was performed using sodium formate (50 mL of isopropanol, 200 µL of formic acid, and 1 mL of 1 M NaOH in water). Data files were calibrated using the Bruker high-precision calibration algorithm.
MS/MS experiments were performed using AutoMS/MS runs at various collision-induced dissociation (CID) voltages from 12 to 25 eV for HGL-DTGs and HC-HGL-DTGs. Instrument control, data acquisition, and reprocessing were performed using HyStar 3.1 (Bruker Daltonics). Peak areas were integrated with QuantAnalysis (Bruker Daltonics).
Dereplication and Annotation of HGL-DTGs.
The dereplication workflow relies on a well-established and extensive MS and MS/MS database constructed from previously identified HGL-DTGs from 35 solanaceous species (19). It includes a detailed rule set for the annotation of fragmentation patterns of the different moieties decorating the 17-hydroxygeranyllinalool (17-HGL) aglycone. To visualize HGL-DTG profiles, we computed the extracted ion chromatogram (EIC) of m/z 271.2420, which corresponds to the 17-HGL aglycone fragment lacking both hydroxyl groups at the C-3 and C-17 positions. The definition of malonylated HGL-DTGs or core HGL-DTGs is based on whether the compounds contain malonyl moieties (20). The identification levels are based on community standards reported in Sumner et al. (45).
Isolation of HGL-DTGs from N. attenuata Plants.
N. attenuata leaf material (500 g) was ground in liquid nitrogen and extracted in 2.5 L of 80% methanol overnight. Further extraction steps were performed as previously described (18). The supernatants were fractionated by reversed-phase fast protein liquid chromatography (Pharmacia Biotech LCC 501 Plus Core) at a flow rate of 5 mL min-1. Mixtures of solvent A (Millipore water) and solvent B (methanol, gradient grade; Merck) were used to elute analytes from the column as follows: 0 to 10 min at 20% of solvent B, 10 to 15 min at 40% of solvent B, 15 to 25 min at 40% of solvent B, 25 to 30 min at 60% of solvent B, 30 to 40 min at 60% of solvent B, 40 to 45 min at 80% of solvent B, 45 to 55 min at 80% of solvent B, 55 to 60 min at 100% of solvent B, 60 to 85 min at 100% of solvent B, and 85 to 95 min at 100 to 0% of solvent B. Highly enriched fractions of HGL-DTGs were collected from fractions 15 to 17 at ∼50 to 60% methanol; these were subsequently dried to produce about 220 mg of HGL-DTG crude mixture. The mixture was used for M. sexta bioassays on irGGPPS plants and AD.
Purification of Caffeoylated HGL-DTG from M. sexta Frass.
Frass of M. sexta larvae reared on N. attenuata WT plants was collected and purified using high performance liquid chromatography (HPLC) as previously described for the hydroxylated HGL-DTGs (18). The HPLC fractions containing caffeoylated HGL-DTGs were further separated using the same HPLC systems with HPLC-grade acetonitrile (VWR International) as solvent B instead of methanol and the following chromatographic solvent gradient: 0 to 3 min at 42% of solvent B, 3 to 11 min at 50% of solvent B, 11 to 15 min at 98% of solvent B, 15 to 17 min at 98% of solvent B, and 17 to 20 min at 42% of solvent B. The obtained pure metabolites were used for NMR analysis.
NMR Analysis.
1H NMR, 13C NMR, 1H-1H correlation spectroscopy (COSY), 1H-13C heteronuclear single quantum correlation (HSQC), 1H-13C heteronuclear single quantum correlation - total correlation spectroscopy (HSQC-TOCSY), and 1H-13C heteronuclear multiple bond correlation (HMBC) experiments were measured on Bruker 500-MHz Avance III HD and 700-MHz Avance III HD NMR spectrometers (Bruker Biospin, Germany). A 5-mm proton-optimized triple resonance NMR ‘inverse’ (TCI) cryoprobe (500 MHz) or a 1.7-mm TCI cryoprobe (700 MHz) at a probe temperature of 298 K was used. Samples were prepared in methanol-d3, and 1H and 13C chemical shifts (δ) were referenced to the residual solvent signals of methanol-d3 at δH 3.31 and δC 49.15, respectively.
Correlation of CA and HGL-DTGs in N. attenuata Natural Population.
For the analysis of HGL-DTGs and CA in 183 accessions, we mined the published data by Li et al. (33). The correlation between HGL-DTGs and CA was performed using Pearson correlation across all 183 accessions. Significance levels for correlation values (r) were determined following the number of metabolite pairs (n) using the equation t = r × (n − 2)0.5/(1 − r2)0.5, with t indicating t-value required for the test of significance.
QTL Mapping.
We used a biparental (Ut and Az parental genotypes) AI-RIL population previously described by Zhou et al. (37) to map the genetic loci underlying the variation in CA and HGL-DTG levels. CA and HGL-DTG tissue concentrations were analyzed in fully developed stem leaves of elongated nonflowering plants in the F11 generation (five generations of inbreeding after the F2 to F6 intercrossing procedure) (37). Three 6-mm leaf discs (average fresh mass of 26 ± 3 mg per leaf disc) were harvested from each of the 261 recombinant inbred line (RIL) plants, flash frozen in liquid nitrogen, extracted, and analyzed on micrOTOF-Q II as described above.
QTL mapping was performed on the fresh mass-normalized peak area data for total HGL-DTGs and CA after log transformation using the QTLRel package (46) in R version 3.3.1 as described by Zhou et al. (37).
Quantitative Real-Time PCR Analysis of Transcript Levels.
Total RNA was extracted from leaves of VIGS N. attenuata plants using the Plant RNA purification kit (Macherey-Nagel) according to the manufacturer’s instructions. Complementary DNA (cDNA) libraries were prepared using the PrimeScript RT Reagent kit (Takara Bio Inc., Japan). Quantitative real-time PCR (qRT-PCR) were performed on a Stratagene 500 MX3005P (Agilent) using a SYBR green reaction mix (Euro-gentec, Belgium) in accordance with the manufacturer’s instructions. The mRNA of the N. attenuata eukaryotic translation initiation factor 5α-2 gene (IF5α-2,NIATv7_g37283) was used as internal control. The delta-delta Ct method was used for data analysis (47). The amplification specificity of primers was confirmed by single peaks in a dissociation curve following qRT-PCR. Primers used in this project are listed in SI Appendix, Table S1.
Statistical Analysis.
Data were analyzed using SPSS 20.0 (SPSS Inc.). Unless otherwise stated, parametric data were compared using ANOVA followed by Honestly Significant Difference (HSD) or Least Significant Difference (LSD) tests as described in figure legends of corresponding data.
Supplementary Material
Acknowledgments
We thank Drs. Meredith Schuman and Heidi Dalton for experimental help, Rishav Ray for the QTL mapping analysis, and Youngsung Joo for fruitful discussion. This work was supported by the Max Planck Society, the Shanghai Institute of Plant Physiology and Ecology/Center for Excellence in Molecular Plant Sciences, a Deutsche Forschungsgemeinschaft grant (SFB 1127/2 ChemBioSys, 239748522) and an ERC advanced grant (293926) to I.T.B., and Pujiang Talent Program (21PJ1414200) to J.L.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2122808119/-/DCSupplemental.
Data Availability
MS metabolomics data were submitted and have been deposited in the MetaboLights public database (accession no. MTBLS3676) (48). All other study data are included in the article and/or SI Appendix.
References
- 1.Kessler A., Baldwin I. T., Defensive function of herbivore-induced plant volatile emissions in nature. Science 291, 2141–2144 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Isah T., Stress and defense responses in plant secondary metabolites production. Biol. Res. 52, 39 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meents A. K., Mithöfer A., Plant-plant communication: Is there a role for volatile damage-associated molecular patterns? Front. Plant Sci. 11, 583275 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Firn R. D., Jones C. G., Natural products—A simple model to explain chemical diversity. Nat. Prod. Rep. 20, 382–391 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Berenbaum M. R., Zangerl A. R., “Phytochemical diversity: Adaptation or random variation” in Phytochemical Diversity and Redundancy in Ecological Interactions, Romeo J. T., Saunders J. A., Barbosa P., Eds. (Springer, Boston, MA, 1996), vol. 30, pp. 319. [Google Scholar]
- 6.Firn R. D., Jones C. G., A Darwinian view of metabolism: Molecular properties determine fitness. J. Exp. Bot. 60, 719–726 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Berenbaum M., Brementown revisited: Interactions among allelochemicals in plants. Chemically Mediated Interactions Between Plants and Other Organisms, Cooper-Driver G. A., Swain T., Eds. (Springer Science, New York, 1985), vol. 19. [Google Scholar]
- 8.Kumar P., Pandit S. S., Steppuhn A., Baldwin I. T., Natural history-driven, plant-mediated RNAi-based study reveals CYP6B46's role in a nicotine-mediated antipredator herbivore defense. Proc. Natl. Acad. Sci. U.S.A. 111, 1245–1252 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heckel D. G., “Insect detoxification and sequestration strategies” in Annual Plant Reviews, Voelckel C., Jander G., Eds. (Wiley Blackwell, 2014), vol. 47, chap. 3, pp. 77–114. [Google Scholar]
- 10.Jeschke V., Gershenzon J., Vassão D. G., “Insect detoxification of glucosinolates and their hydrolysis products” in Glucosinolates, Kopriva S., Ed. (Elsevier, 2016), chap. 8, pp. 199–245. [Google Scholar]
- 11.Malka O., et al. , Glucosylation prevents plant defense activation in phloem-feeding insects. Nat. Chem. Biol. 16, 1420–1426 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Steppuhn A., Gase K., Krock B., Halitschke R., Baldwin I. T., Nicotine’s defensive function in nature. PLoS Biol. 2, E217 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jassbi A. R., Gase K., Hettenhausen C., Schmidt A., Baldwin I. T., Silencing geranylgeranyl diphosphate synthase in Nicotiana attenuata dramatically impairs resistance to tobacco hornworm. Plant Physiol. 146, 974–986 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaur H., Heinzel N., Schöttner M., Baldwin I. T., Gális I., R2R3-NaMYB8 regulates the accumulation of phenylpropanoid-polyamine conjugates, which are essential for local and systemic defense against insect herbivores in Nicotiana attenuata. Plant Physiol. 152, 1731–1747 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee G., Joo Y., Kim S. G., Baldwin I. T., What happens in the pith stays in the pith: Tissue-localized defense responses facilitate chemical niche differentiation between two spatially separated herbivores. Plant J. 92, 414–425 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Zavala J. A., Patankar A. G., Gase K., Hui D., Baldwin I. T., Manipulation of endogenous trypsin proteinase inhibitor production in Nicotiana attenuata demonstrates their function as antiherbivore defenses. Plant Physiol. 134, 1181–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steppuhn A., Baldwin I. T., Resistance management in a native plant: Nicotine prevents herbivores from compensating for plant protease inhibitors. Ecol. Lett. 10, 499–511 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Li J., et al. , Controlled hydroxylations of diterpenoids allow for plant chemical defense without autotoxicity. Science 371, 255–260 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Heiling S., et al. , Using the knowns to discover the unknowns: MS-based dereplication uncovers structural diversity in 17-hydroxygeranyllinalool diterpene glycoside production in the Solanaceae. Plant J. 85, 561–577 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Poreddy S., et al. , Detoxification of hostplant’s chemical defence rather than its anti-predator co-option drives β-glucosidase-mediated lepidopteran counteradaptation. Nat. Commun. 6, 8525 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onkokesung N., et al. , MYB8 controls inducible phenolamide levels by activating three novel hydroxycinnamoyl-coenzyme A:polyamine transferases in Nicotiana attenuata. Plant Physiol. 158, 389–407 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heiling S., et al. , Jasmonate and ppHsystemin regulate key Malonylation steps in the biosynthesis of 17-Hydroxygeranyllinalool Diterpene Glycosides, an abundant and effective direct defense against herbivores in Nicotiana attenuata. Plant Cell 22, 273–292 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heiling S., et al. , Specific decorations of 17-hydroxygeranyllinalool diterpene glycosides solve the autotoxicity problem of chemical defense in Nicotiana attenuata. Plant Cell 33, 1748–1770 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammer T. J., Janzen D. H., Hallwachs W., Jaffe S. P., Fierer N., Caterpillars lack a resident gut microbiome. Proc. Natl. Acad. Sci. U.S.A. 114, 9641–9646 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berenbaum M. R., Zangerl A. R., Costs of inducible defense: Protein limitation, growth, and detoxification in parsnip webworm. Ecology 75, 2311–2317 (1994). [Google Scholar]
- 26.Lou Y., Baldwin I. T., Manduca sexta recognition and resistance among allopolyploid Nicotiana host plants. Proc. Natl. Acad. Sci. U.S.A. 100 (suppl. 2), 14581–14586 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bi J. L., et al. , Do plant phenolics confer resistance to specialist and generalist insect herbivores? J. Agric. Food Chem. 45, 4500–4504 (1997). [Google Scholar]
- 28.Eichenseer H., Bi J. L., Felton G. W., Indiscrimination of Manduca sexta larvae to overexpressed and underexpressed levels of phenylalanine ammonia-lyase in tobacco leaves. Entomol. Exp. Appl. 87, 73–78 (1998). [Google Scholar]
- 29.Ali M. I., Bi J. L., Young S. Y., Felton G. W., Do foliar phenolics provide protection to Heliothis virescens from a baculovirus? J. Chem. Ecol. 25, 2193–2204 (1999). [Google Scholar]
- 30.Leiss K. A., Maltese F., Choi Y. H., Verpoorte R., Klinkhamer P. G., Identification of chlorogenic acid as a resistance factor for thrips in chrysanthemum. Plant Physiol. 150, 1567–1575 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feistel F., Paetz C., Menezes R. C., Veit D., Schneider B., Acylated quinic acids are the main salicortin metabolites in the Lepidopteran specialist herbivore Cerura vinula. J. Chem. Ecol. 44, 497–509 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Särkinen T., Bohs L., Olmstead R. G., Knapp S., A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): A dated 1000-tip tree. BMC Evol. Biol. 13, 214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li D., Baldwin I. T., Gaquerel E., Navigating natural variation in herbivory-induced secondary metabolism in coyote tobacco populations using MS/MS structural analysis. Proc. Natl. Acad. Sci. U.S.A. 112, E4147–E4155 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reine Judesse Soviguidi D., et al. , Chlorogenic acid metabolism: The evolution and roles in plant response to abiotic stress. Phyton 91, 239–255 (2022). [Google Scholar]
- 35.Wu J., Hettenhausen C., Schuman M. C., Baldwin I. T., A comparison of two Nicotiana attenuata accessions reveals large differences in signaling induced by oral secretions of the specialist herbivore Manduca sexta. Plant Physiol. 146, 927–939 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu J., Kang J. H., Hettenhausen C., Baldwin I. T., Nonsense-mediated mRNA decay (NMD) silences the accumulation of aberrant trypsin proteinase inhibitor mRNA in Nicotiana attenuata. Plant J. 51, 693–706 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Zhou W., et al. , Tissue-specific emission of (E)-alpha-bergamotene helps resolve the dilemma when pollinators are also herbivores. Curr. Biol. 27, 1336–1341 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Xu S., et al. , Wild tobacco genomes reveal the evolution of nicotine biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 114, 6133–6138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones C. G., Firn R. D., On the evolution of plant secondary chemical diversity. Philos T R Soc B 333, 273–280 (1991). [Google Scholar]
- 40.Richards L. A., et al. , Phytochemical diversity and synergistic effects on herbivores. Phytochem. Rev. 15, 1153–1166 (2016). [Google Scholar]
- 41.Krugel T., Lim M., Gase K., Halitschke R., Baldwin I. T., Agrobacterium-mediated transformation of Nicotiana attenuata, a model ecological expression system. Chemoecology 12, 177–183 (2002). [Google Scholar]
- 42.Guo H., Halitschke R., Wielsch N., Gase K., Baldwin I. T., Mate selection in self-compatible wild tobacco results from coordinated variation in homologous self-incompatibility genes. Curr. Biol. 29, 2020–2030.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Galis I., et al. , The use of VIGS technology to study plant-herbivore interactions. Methods Mol. Biol. 975, 109–137 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Machado R. A. R., Arce C. C. M., Ferrieri A. P., Baldwin I. T., Erb M., Jasmonate-dependent depletion of soluble sugars compromises plant resistance to Manduca sexta. New Phytol. 207, 91–105 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Sumner L. W., et al. , Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3, 211–221 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng R., Abney M., Palmer A. A., Skol A. D., QTLRel: An R package for genome-wide association studies in which relatedness is a concern. BMC Genet. 12, 66 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Heiling S., Li J., Halitschke R., Paetz C., Baldwin I. T., MTBLS3676: The downside of metabolic diversity: post-ingestive rearrangements by specialized insects. MetaboLights. https://www.ebi.ac.uk/metabolights/MTBLS3676. Deposited 12 May 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MS metabolomics data were submitted and have been deposited in the MetaboLights public database (accession no. MTBLS3676) (48). All other study data are included in the article and/or SI Appendix.