Significance
Maternal asthma is one of the most replicated risk factors for childhood-onset asthma. However, the underlying mechanisms are unknown. We identified DNA methylation signatures in bronchial epithelial cells from adults with asthma that were specific to those with a mother with asthma. These maternal asthma-associated methylation signatures were correlated with distinct gene regulatory pathways and clinical features. Genes in 16 pathways discriminated cases with and without maternal asthma and suggested impaired T cell signaling and responses to viral and bacterial pathogens in asthmatic children of an asthmatic mother. Our findings suggest that the prenatal environment in pregnancies of mothers with asthma alters epigenetically mediated developmental programs that may lead to severe asthma in their children through diverse gene regulatory pathways.
Keywords: maternal asthma, epigenetics, transcriptomics
Abstract
Maternal asthma (MA) is among the most consistent risk factors for asthma in children. Possible mechanisms for this observation are epigenetic modifications in utero that have lasting effects on developmental programs in children of mothers with asthma. To test this hypothesis, we performed differential DNA methylation analyses of 398,186 individual CpG sites in primary bronchial epithelial cells (BECs) from 42 nonasthma controls and 88 asthma cases, including 56 without MA (NMA) and 32 with MA. We used weighted gene coexpression network analysis (WGCNA) of 69 and 554 differentially methylated CpGs (DMCs) that were specific to NMA and MA cases, respectively, compared with controls. WGCNA grouped 66 NMA-DMCs and 203 MA-DMCs into two and five comethylation modules, respectively. The eigenvector of one MA-associated module (turquoise) was uniquely correlated with 85 genes expressed in BECs and enriched for 36 pathways, 16 of which discriminated between NMA and MA using machine learning. Genes in all 16 pathways were decreased in MA compared with NMA cases (P = 7.1 × 10−3), a finding that replicated in nasal epithelial cells from an independent cohort (P = 0.02). Functional interpretation of these pathways suggested impaired T cell signaling and responses to viral and bacterial pathogens. The MA-associated turquoise module eigenvector was additionally correlated with clinical features of severe asthma and reflective of type 2 (T2)-low asthma (i.e., low total serum immunoglobulin E, fractional exhaled nitric oxide, and eosinophilia). Overall, these data suggest that MA alters diverse epigenetically mediated pathways that lead to distinct subtypes of severe asthma in adults, including hard-to-treat T2-low asthma.
Asthma is among the most common chronic diseases, affecting 5.5 million children and 19.2 million adults in the United States and with annual health care costs exceeding $80 billion (1). Overall, there is significant interindividual variation in the clinical presentations of asthma, with nearly 15% of asthmatics having severe manifestations of the disease, including persistent exacerbations and difficult-to-control symptoms (2). Approximately half of adult severe asthma patients do not have features of type 2 (T2) inflammation and do not respond to T2 cytokine-targeting therapies (3). Among the many epidemiologic risk factors for asthma, having a mother with asthma is one of the most reproducible (4), although the mechanisms underlying this risk are unknown and the relationship to severe asthma has not been investigated.
It has been suggested that the in utero environment modifies the epigenetic landscape of the fetus and impacts downstream developmental programs (5–7). This idea has been supported by previous studies in asthma (8–12). For example, a longitudinal birth cohort study revealed 589 differentially methylated regions between cord blood immune cells from 18 children with maternal asthma (MA) who developed asthma by age 9 compared with 18 children with MA who did not have a diagnosis of asthma by age 9 (10). The genes near the differentially methylated regions clustered in immunoregulatory and proinflammatory pathways that were most connected by the transcription factor SMAD3, whose gene resides within a highly replicated asthma genome-wide association study locus (13). The promoter of SMAD3 was hypermethylated in cord blood cells from asthmatic children with MA compared with asthmatic children with a nonasthmatic mother (NMA), a finding that was replicated in two additional birth cohorts (10). This study supports the idea that MA modifies childhood asthma risk through epigenetic mechanisms that are detectable as early as birth. Yet, no previous studies have examined the effects of MA on DNA methylation patterns in airway cells or in adult children of mothers with asthma, or included paired gene-expression data to characterize the transcriptional pathways associated with epigenetic programs in the child.
Here, we fill these gaps by performing an epigenetic study of MA and DNA methylation at individual CpG sites in primary bronchial epithelial cells (BECs) sampled from 96 adult asthma cases and 46 adult nonasthma controls. We integrated those data with transcriptional profiling from the same cells and clinical characteristics in the same subjects. Using systems biology and machine-learning approaches, our results support the hypothesis that DNA methylation signatures in BECs reflect different transcriptional programs and endotypes of severe asthma that may have their origins in prenatal life.
Results
Differential DNA Methylation Analyses of MA Risk Groups.
The overarching hypothesis of our study is that MA affects the development of asthma in the mother’s children through epigenetic (DNA methylation) changes that occur during pregnancy and persist throughout life. In this study, we specifically asked whether DNA methylation signatures in airway cells differ between adult asthmatics with and without a mother with asthma and whether these signatures reflect different transcriptional networks and endotypes of asthma (see SI Appendix, Fig. S1 for our study overview). To this end, we obtained array-based DNA methylation measures of individual CpG sites in freshly isolated BECs from 150 adults who participated in genetic and epigenetic studies of asthma at the University of Chicago (11, 14–16). After processing and quality control of DNA methylation (Materials and Methods), we retained 398,186 CpGs from 130 subjects (42 nonasthma controls without MA, 56 asthma cases without MA, and 32 asthma cases with MA) (SI Appendix, Fig. S2). Both groups of cases (with and without MA) differed from the controls with respect to most clinical measures except for body mass index (BMI) and blood eosinophils (Table 1 and SI Appendix, Table S1). Overall, BMI was higher in NMA cases compared with nonasthma controls (34.64 vs. 28.52; P = 4.5 × 10−4) and blood eosinophil levels, a marker of T2 inflammation (17), were higher in MA cases compared with nonasthma controls (225 vs. 70 cells per microliter; P = 8.4 × 10−4), but neither differed between NMA and MA cases after multiple test corrections.
Table 1.
Characteristics of 130 subjects at the time of bronchoscopy by risk group
| MA risk groups | |||
|---|---|---|---|
| Characteristic | Control | Case NMA | Case MA |
| (n = 42) | (n = 56) | (n = 32) | |
| Age (mean y ± SD) | 37.45 ± 11.75 | 41.41 ± 12.24 | 37.56 ± 13.39 |
| Gender (% female) | 62 | 79 | 66 |
| Ethnicity (%) (Af Am/Eur Am/Other) | 64/29/7 | 55/45/0** | 66/34/0* |
| Percent smoker at bronchoscopy (%) | 17 | 4* | 6 |
| Clinical measures | |||
| ICS use (%) | — | 75 | 75 |
| OCS use (%) | — | 45 | 25 |
| STEP category (% mild/% moderate/% severe) |
— | 22/23/55 | 28/25/47 |
| Mean FEV1% predicted (± SD) | 95.36 ± 11.63 | 75.45*** ± 19.73 | 70.41*** ± 18.15 |
| Mean FEV1/FVC (± SD) | 0.82 ± 0.05 | 0.73*** ± 0.10 | 0.83*** ± 0.48 |
| Median total serum IgE (IU/mL) (lower, upper quartile) |
56.50 (22.00, 169.00) |
117.50 (22.00, 305.25) |
156.50** (70.25, 626.75) |
| Median FeNO (ppb) (lower, upper quartile)† |
14.00 (10.50, 17.75) |
23.50** (13.00, 45.75) |
27.00** (13.25, 54.75) |
| Median BAL eosinophilia (%) (lower, upper quartile)‡ |
0.0 (0.0, 0.4) |
2.9*** (1.5, 5.6) |
3.6*** (1.5, 7.1) |
| Median BAL neutrophilia (%) (lower, upper quartile)§ |
4.9 (3.0, 5.9) |
5.2 (4.2, 6.8) |
3.5 (2.2, 6.4) |
| Median blood eosinophilia (cells/µL) (lower, upper quartile)¶ |
70 (100, 170) |
170 (70, 283) |
225*** (108, 323) |
| Mean BMI (± SD) | 28.52 ± 5.71 | 34.64*** ± 8.83 | 33.68* ± 10.48 |
Severity was defined by STEP classification (18) of mild, moderate and severe asthma. Af Am, African American; Eur Am, European American; ICS, inhaled corticosteroid; OCS, oral corticosteroid. Asthma cases without an asthmatic mother (Case NMA) and cases with an asthmatic mother (Case MA) were each compared with controls (*P < 0.05, **P < 0.01, ***P < 0.001). Continuous variables were tested with a Wilcoxon rank-sum test and categorical variables were tested using a Fisher’s exact test. See SI Appendix, Table S1 for all pairwise comparisons.
†Fifty-two Case NMA subjects and 30 Case MA subjects had FeNO measurements.
‡Forty-one controls and 55 Case NMA had BAL eosinophil measurements.
§Forty-one controls and 55 Case NMA subjects had BAL neutrophil measurements.
¶Forty-one controls had blood eosinophil measurements.
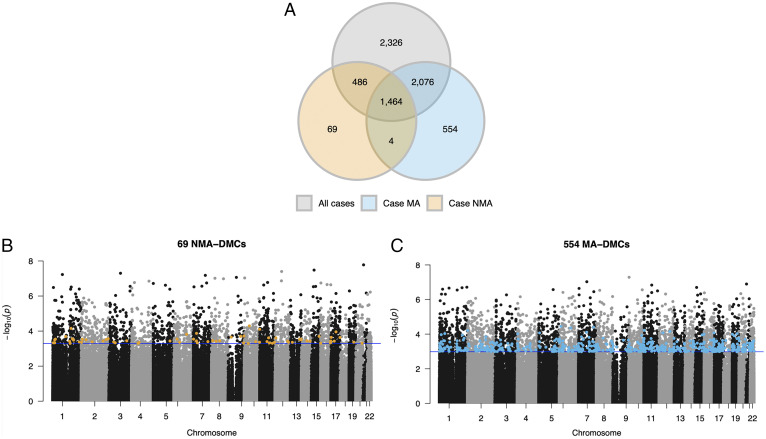
To identify differentially methylated CpGs (DMCs), we compared DNA methylation levels (covariate-adjusted M-values for each CpG) in nonasthma controls (n = 42) with all asthma cases (n = 88), with NMA cases (n = 56), and with MA cases (n = 32). All analyses included age, gender, current smoking status, and the first three ancestry principal components (PCs) to account for population structure as covariates. The three comparisons revealed 6,352, 2,023, and 4,098 DMCs, respectively (false-discovery rate [FDR] < 0.10) (Fig. 1A). Among the 6,352 DMCs in the analysis including all cases, 2,326 were not DMCs at FDR < 0.10 in the other two analysis. However, of those 2,326 DMCs, 92.30% (n = 2,147) had larger effect sizes in the MA cases compared with NMA cases, despite the smaller sample size of MA cases (paired signed-rank test; P < 2.2 × 10−16) (SI Appendix, Fig. S3). Among the 2,023 DMCs between controls and NMA cases, only 69 CpGs (3.41%) (orange points in Fig. 1B) were specific to NMA (i.e., were not DMCs in the other two analyses). In contrast, among the 4,098 DMCs between controls and MA cases, 554 CpGs (13.52%) (blue points in Fig. 1C) were not DMCs in the other two analyses. We refer to these as “NMA-associated DMCs” (NMA-DMCs; n = 69) and “MA-associated DMCs” (MA-DMCs; n = 554), respectively, and consider the remaining 6,356 DMCs to be “shared asthma DMCs.” These analyses revealed widespread DNA methylation differences in BECs from adult asthma cases with and without MA, supporting the hypothesis that MA is associated with an altered epigenetic landscape of cells from exposed children.
Fig. 1.
Comparisons of DMCs in MA risk groups. (A) Venn diagrams show the number of DMCs (FDR < 0.10) in each of three analyses: controls (n = 42) vs. all asthma cases (n = 88; all cases; gray), controls vs. asthma cases without an asthmatic mother (n = 56; Case NMA; orange), and controls vs. asthma cases with an asthmatic mother (n = 32; Case MA; blue). These comparisons revealed 6,352, 2,023, and 4,098 DMCs, respectively. Manhattan plots show the distribution and P values of 398,186 CpGs compared between nonasthma controls to Case NMA (B) and nonasthma controls to Case MA (C). P values (y axis) correspond to the differences in methylation between asthma cases and nonasthma controls. Blue horizontal lines correspond to the q value threshold of 0.10. Orange points in B indicate the 69 CpGs that were differentially methylated in Case NMA at FDR < 0.10 and blue points in (C) indicate the 554 CpGs that were differentially methylated in Case MA at FDR < 0.10.
MA-Associated Comethylation Signatures Highlight Diverse Transcriptional Pathways.
To characterize the correlation structure of the 69 NMA-DMCs and 554 MA-DMCs and further examine their relationship to gene regulatory networks in BECs, we used a systems biology approach, as implemented in Weighted Gene Co-Expression Network Analysis (WGCNA) (19). WGCNA grouped 66 (95.65%) NMA-DMCs and 203 (36.64%) MA-DMCs into two and five comethylation modules, respectively, with 15 to 74 CpGs per module (Table 2 and SI Appendix, Figs. S4 and S5). The DMCs in each comethylation module were distributed across chromosomes and functional categories, reflecting the genome-wide methylation responses to MA (SI Appendix, Figs. S6 and S7). The remaining 3 NMA-DMCs and 351 MA-DMCs were uncorrelated and therefore assigned to a gray module (SI Appendix, Figs. S4 and S5). These DMCs were not considered in downstream analyses.
Table 2.
Associations among comethylation modules, transcriptional pathways, and clinical measures
| Variables | WGCNA comethylation modules (No. of DMCs) | ||||||
|---|---|---|---|---|---|---|---|
| NMA associated | MA associated | ||||||
| Orange | Green | Black | Blue | Red | Turquoise | Yellow | |
| (31) | (35) | (15) | (74) | (17) | (73) | (24) | |
| Correlated genes and pathways | |||||||
| No. of correlated genes | 2,217 | 2,511 | 3,752 | 2,825 | 1,721 | 1,133 | 2,351 |
| No. of unique genes | 115 | 110 | 794 | 234 | 46 | 85 | 391 |
| No. of pathways | 0 | 0 | 11 | 0 | 0 | 36 | 33 |
| No. of hub DMCs | 0 | 4 | 9 | 8 | 7 | 19 | 9 |
| First pathway | — | — | Gene expression | — | — | S. aureus infection | Eukaryotic translation elongation |
| Second pathway | — | — | T cell receptor signaling pathway | — | — | Rheumatoid arthritis | Cytoplasmic ribosomal proteins |
| Third pathway | — | — | Cancer immunotherapy by PD-1 blockade | — | — | Tuberculosis | Peptide chain elongation |
| Clinical measures | |||||||
| Asthma severity (n = 142) |
5.81
4.1 × 10−8 |
−6.02
1.5 × 10−8 |
−5.20
6.9 × 10−7 |
−5.69
7.4 × 10−8 |
4.72
5.6 × 10−6 |
−5.24
6.0 × 10−7 |
4.60
9.6 × 10−6 |
| FEV1% predicted (n = 142) |
−0.36
1.2 × 10−5 |
0.38
3.4 × 10−6 |
0.40
7.6 × 10−7 |
0.40
6.7 × 10−7 |
−0.37
7.8 × 10−6 |
0.38
3.4 × 10−6 |
−0.33
5.1 × 10−5 |
| FEV1/FVC (n = 142) |
−0.29
5.2 × 10−4 |
0.29
4.0 × 10−4 |
0.26
2.2 × 10−3 |
0.20 0.018 |
−0.21 0.013 |
0.25
2.6 × 10−3 |
ns |
| Total serum IgE (n = 141) |
0.21 0.013 |
−0.23 6.9 × 10−3 |
−0.30
2.8 × 10−4 |
−0.31
1.7 × 10−4 |
0.26
1.8 × 10−3 |
−0.21 0.011 |
0.29
4.53 × 10−4 |
| FeNO (n = 135) |
0.18 0.037 |
ns |
−0.27
2.9 × 10−3 |
−0.22
9.7 × 10−3 |
0.26
2.0 × 10−3 |
ns |
0.28
9.4 × 10−4 |
| BAL eosinophilia (n = 140) |
0.22 8.7x10−3 |
ns |
−0.24
4.5 × 10−3 |
−0.26
2.1 × 10−3 |
0.24
4.6 × 10−3 |
−0.17 0.039 |
0.30
3.5 × 10−4 |
| BAL neutrophilia (n = 140) |
ns | ns | ns | ns | ns | ns | ns |
| Blood eosinophilia (n = 141) |
ns | ns | ns | ns | ns | ns | ns |
| BMI (n = 142) |
0.26
1.9 × 10−3 |
ns | ns | ns | ns | ns |
0.24
3.5 × 10−3 |
Of the 69 NMA-DMCs and the 554 MA-DMCs, 66 (95.65%) and 203 (36.64%) formed two and five comethylation modules, respectively (15 to 74 CpGs in each module). See SI Appendix, Figs. S4 and S5 for cluster dendrograms of the seven modules. Correlated genes and pathways: For each module, the number of correlated genes (FDR < 0.10), the number of uniquely correlated genes (FDR < 0.10 only in that module), the number of hub DMCs, and the top three enriched pathways for the uniquely correlated genes are shown. SI Appendix, Table S3 lists all the pathway enrichments in each module. Clinical measures: Correlation coefficients and P values are shown between clinical measures and module eigenvectors. Significant P values after Bonferroni-correcting for nine clinical phenotypes (P < 5.56 × 10−3) are shown in bold. The numbers of subjects with measurements for each variable are shown. Asthma severity is determined by STEP classification (18). ns, not significant (P > 0.05).
To characterize transcriptional networks of genes that are potentially regulated by the CpGs assigned to each module, and to assign biological function to each module, we next considered the sets of genes whose expression was correlated with the eigenvector of each module. Among the 13,757 genes that were detected as expressed in the same BECs, 5,458 genes were significantly correlated (FDR < 0.10) with the eigenvector of one or more modules. We defined a set of uniquely correlated genes for each module as those that were correlated with only that module at FDR < 0.10, and then subjected those module-specific genes to pathway analyses using TopFunn (20) (Table 2, Correlated genes and pathways, and SI Appendix, Table S2). The genes specifically correlated with three MA-associated modules (black, turquoise, and yellow) were enriched for pathways with known roles in asthma pathobiology and epithelial cell integrity and proliferation (SI Appendix, Table S3).
The 794 unique genes correlated with the black module eigenvector were significantly enriched for 11 pathways, with the 3 most significant pathways being “Gene expression” (FDR-corrected P = 1.91 × 10−3; 111 genes), “T cell receptor signaling” (FDR-corrected P = 0.015; 15 genes), and “Cancer immunotherapy by PD-1 blockade” (FDR-corrected P = 0.015; 7 genes). The latter two, together with five additional pathways, were all involved in T cell signaling and activation or other adaptive immune processes. The 85 unique genes correlated with the turquoise module eigenvector were significantly enriched in 36 pathways, with the top being “Staphylococcus aureus infection” (FDR-corrected P = 3.24 × 10−6; 7 genes). Most of the 36 pathways were largely comprised of the same five HLA class II genes (HLA-DMA, HLA-DOA, HLA-DPB1, HLA-DRA, and HLA-DRB5). Other enriched pathways of the turquoise module genes included asthma, autoimmune and infectious diseases, and immune-mediated pathways of viral response, such as “type II interferon signaling.” The 391 unique genes correlated with the yellow module eigenvector were significantly enriched in 33 pathways, with the three most significant being “Eukaryotic translation elongation” (FDR-correct P = 3.95 × 10−16; 23 genes), “Cytoplasmic ribosomal proteins” (FDR-corrected P = 4.74 × 10−16; 22 genes), and “Peptide chain elongation” (FDR-corrected P = 5.12 × 10−16; 22 genes). Most pathways included the same 22 genes that primarily encode ribosomal proteins with significant roles in protein synthesis, cell growth, apoptosis, and immunosurveillance (21). Overall, these results suggest that the CpGs in each module regulate diverse gene-expression networks, potentially reflecting different mechanistic pathways (or endotypes) of asthma.
A Gene Signature Distinguishes MA and NMA Cases.
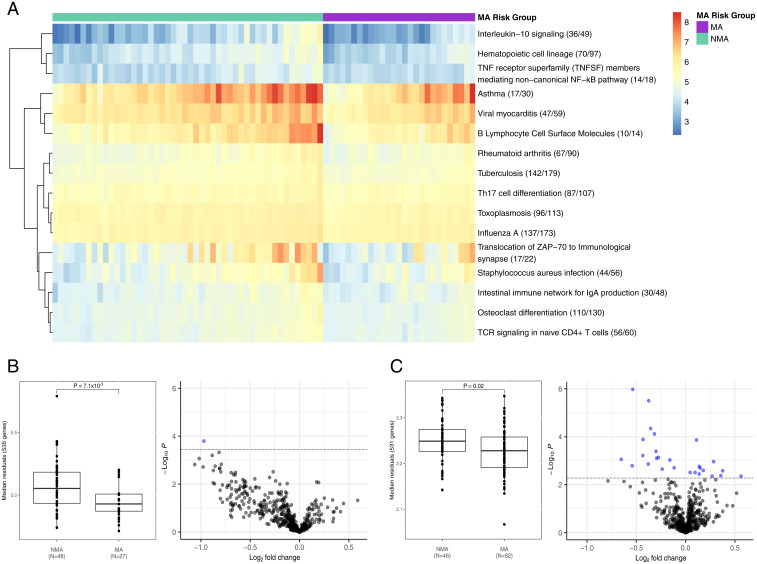
To gain greater insight into each module with pathway enrichments, we used machine-learning algorithms to identify the pathways that discriminated MA and NMA cases using all the genes in each of the 11, 36, and 33 pathways from the black, turquoise, and yellow modules, separately, that were expressed in BECs (n = 3 to 1,844 genes per pathway). This identified 3, 16, and 18 discriminatory pathways for the black, turquoise, and yellow modules, respectively (Fig. 2A and SI Appendix, Table S4). Only the median gene expression of the 535 genes in the 16 pathways of the turquoise module was significantly different in MA cases compared with NMA cases (Fig. 2B) (P = 7.1 × 10−3). The median gene expression of the 231 genes of the black module and the 675 genes of the yellow module did not differ between MA and NMA cases (SI Appendix, Fig. S8) (P ≥ 0.10). The median expression of the 231 genes of the turquoise module were on average decreased in MA cases compared with NMA cases and implicated “TCR signaling in naïve CD4+ T cells,” Th17 cell differentiation, and additional chemokine and cytokine signaling pathways, all of which are known to be impaired in asthma (22). The 16 pathways also included “Staphylococcus aureus infection” and “Influenza A,” supporting a role for impaired responses to bacterial and viral pathogens in asthma (23). Consistent with these findings, comparison of the expression levels of each of the 535 genes in NMA and MA cases revealed one differentially expressed gene (ICOS; inducible T cell costimulator) (Fig. 2B and SI Appendix, Table S5), which enhances T cell signaling responses to foreign antigens (24). Overall, these results link the DMCs in the MA-associated turquoise module to a 535-gene signature that is down-regulated in MA compared with NMA cases and reflects both impaired T cell signaling and response to bacterial and viral infection.
Fig. 2.
MA-discriminatory pathways implicate a T2-low gene signature. (A) RandomForests selected 16 of 36 pathways of the turquoise module that discriminated between asthma cases with an asthmatic mother (MA; n = 27) and without an asthmatic mother (NMA; n = 48). The heatmap represents the median normalized and covariate-adjusted expression of annotated genes (expressed/total annotated genes) for each pathway enrichment. The median normalized and covariate-adjusted expression (residuals) for all 535 unique genes across the 16 pathways was compared between NMA and MA cases in (B) bronchial epithelial cells (discovery) and in (C) nasal epithelial cells (replication) using a Wilcoxon rank-sum test. Four genes (CCL3L1, CALML6, EPX, RAC3) were not expressed in nasal epithelial cells. See SI Appendix, Table S4 for median normalized and covariate-adjusted expression by MA risk group and list of all annotated genes for the 16 pathways. Of the 535 genes, 1 and 27 were differentially expressed between MA and NMA cases in bronchial and nasal epithelial cells, respectively (FDR < 0.10; blue points) (SI Appendix, Table S5). Log2 fold-change is the log-ratio of log2(expression in MA cases/expression in NMA cases). Higher and lower gene expression in MA cases compared with NMA cases are reflected by log2 fold-changes > 0 and < 0, respectively.
To replicate this finding, we used RNA-sequencing (RNA-seq) from nasal epithelial cells collected at age 11 y from children in the Urban Environment and Childhood Asthma (URECA) birth cohort (25) and compared median expression of the 535 genes between MA and NMA cases. Four of 535 genes were not expressed in nasal epithelial cells. Indeed, the 531-gene signature in nasal epithelial cells was significantly decreased in MA cases compared with NMA cases (Fig. 2C) (P2-tailed = 0.02). Differential expression of each of the 531 genes revealed 15 (decreased) and 12 (increased) in MA cases compared with NMA cases (Fig. 2C and SI Appendix, Table S5). The pathways that were enriched among the differential expressed genes included “Hematopoietic cell lineage,” “Influenza A,” “TCR signaling in naïve CD4+ T cells,” “Th17 cell differentiation,” “Staphylococcus aureus infection,” and “Toxoplasmosis,” supporting dysregulation of pathways relating to T cell signaling and response to bacterial and viral infection (22, 23, 26). Taken together, these findings reveal a gene signature in both the lower and upper airways in adults and children, respectively, that is down-regulated in MA compared with NMA cases.
Comethylation Signatures Reflect Clinical Heterogeneity and Endotypes of Asthma.
To test directly the hypothesis that the MA-DMCs in different modules underlie distinct clinical features or endotypes of asthma, we examined correlations between the eigenvectors of each module and nine clinical measures that were available in most subjects (Table 2, Clinical measures). This revealed distinct patterns of correlated phenotypes. Notably, the eigenvectors of all seven modules of NMA-DMCs and MA-DMCs were correlated with asthma severity, defined by STEP classification of mild, moderate, and severe asthma (18), and forced expiratory volume at 1 s (FEV1) percent predicted, a measure of lung function that is used in STEP classification. The ratio of FEV1 to forced vital capacity (FEV1/FVC), a measure of lung obstruction and marker of more refractory asthma, was significantly correlated with two NMA-associated (orange and green) and two MA-associated (black and turquoise) module eigenvectors, and nominally correlated with the MA-associated blue and red module eigenvectors but not with the yellow module eigenvector. Total serum immunoglobulin (Ig) E was significantly correlated with four MA-associated module eigenvectors (black, blue, red, and yellow) and nominally correlated with the other three module eigenvectors. Fractional exhaled nitric oxide (FeNO) and bronchoalveolar lavage (BAL) eosinophilia were also significantly correlated with the MA-associated black, blue, red, and yellow module eigenvectors and nominally correlated with the NMA-associated orange module; BAL eosinophilia was also nominally correlated with the MA-associated turquoise module eigenvector. IgE, FeNO, and eosinophilia are features of T2 asthma (27). Thus, whereas all modules were associated with asthma severity and FEV1, only four MA-associated modules (black, blue, red, and yellow) were also associated with features of T2-high asthma. BMI was correlated with the NMA-associated orange module eigenvector and one MA-associated yellow (T2-high) module eigenvector. The module with the greatest number of pathway enrichments, the MA-associated turquoise module, reflected features of severe asthma in the absence of T2-high markers. Taken together with the gene-expression data, we suggest that the DMCs in the turquoise module reflect a T2-low endotype of severe asthma with impaired T cell signaling and responses to bacterial and viral pathogens. Overall, these studies support mechanistic links between NMA- and MA-associated DNA methylation patterns in BECs with diverse asthma endotypes.
Hub DMCs Drive Associations with Gene Expression and Clinical Measures.
Each module is characterized by key driver (hub) DMCs or the DMCs that are most connected to the other DMCs in the module, and potentially contribute most to the correlations with gene expression and clinical phenotypes. We therefore assessed whether the correlations between each module’s eigenvector with gene expression and clinical measures of asthma were driven by hub DMCs. There were 0 to 19 hub DMCs per module (module membership > 0.80) that were distributed across chromosomes and functional categories (Table 2 and SI Appendix, Fig. S9 and Table S6). To assess the relative importance of these hub DMCs to the observed correlations with gene expression or clinical measures in each module, we repeated all association tests conditioned on DNA methylation levels at the hub DMCs. The eigenvectors of these modules were no longer correlated with the expression of their assigned genes after including each module’s hub DMCs as covariates in the model (FDR-corrected P > 0.999) (SI Appendix, Table S7), suggesting that the hub DMCs are the primary drivers of correlations with gene expression. In addition, no significant correlations remained for FEV1% predicted, FeNO, BAL eosinophilia, or BMI in any of the modules. Some correlations remained between the MA-associated blue module and the FEV1/FVC ratio and between the MA-associated black and yellow modules with total serum IgE, although none were significant after multiple testing. In contrast, asthma severity remained significantly associated with the eigenvectors of the NMA-associated green module and the MA-associated blue, turquoise, and yellow modules after correcting for hub DMCs. These results indicate that the hub DMCs as well as the nonhub DMCs in these modules contribute to variation in asthma severity, FEV1/FVC, and total serum IgE. However, overall the hub DMCs are the primary drivers of association with gene expression and many of the associations with clinical measures.
Discussion
In this study, we explored the relationship between exposure to an MA environment in utero and endotypes of asthma in adults by testing the hypothesis that this relationship is facilitated through epigenetic changes in airway cells that modulate downstream transcriptional programs. In an earlier study (11), in a subset of the subjects included in this study, we reported differential expression of a microRNA that down-regulated the expression of an asthma-associated gene, HLA-G, in BECs from adults with MA but not from adults without MA. We suggested then that prenatal exposures in fetuses of asthmatic mothers had effects on the regulatory landscape of BECs. In this study, we extended this mechanism to include a widespread epigenetic mark, DNA methylation. Our study revealed DNA methylation patterns that differed between BECs from adult asthmatics with and without an asthmatic mother. Moreover, we show that the comethylated module eigenvectors of NMA-DMCs and MA-DMCs were correlated with the expression of different genes that highlighted distinct biological pathways and harbored a signature that distinguished cases with and without a mother with asthma. The associated pathways, gene signature, and clinical features of one module, the MA-associated turquoise module, reflected features of a T2-low endotype of asthma and impaired T cell signaling and responses to viral and bacterial pathogens.
The four additional MA-associated module eigenvectors (black, blue, red, and yellow) were correlated with clinical features reflective of a T2-high severe asthma endotype (3, 28); one (yellow) was additionally associated with BMI. This distinction, along with the different sets of uniquely correlated genes, suggests that DMCs in these modules may represent different forms of T2-high asthma. The genes uniquely correlated with the black module eigenvector were enriched in pathways related to T cell signaling and activation, while those uniquely correlated with the yellow module were enriched in pathways that suggested potential mechanisms related to apoptosis and immunosurveillance processes. The two NMA-associated (orange, green) module eigenvectors may also be associated with a T2-low severe asthma endotype but neither had any pathway enrichments. Overall, our results suggest that epigenetic programs in BECs may underlie a spectrum of features from T2-high to T2-low severe asthma, both with and without obesity, and reveal transcriptional pathways that may drive the features defining these endotypes.
Whereas symptoms among severe asthmatics with a T2-high endotype can be managed with corticosteroids and T2 cytokine-targeting therapies (3, 28), T2-low asthmatics are generally nonresponsive to such treatments and experience more exacerbations and hospitalizations. Elucidating the pathways that are perturbed in T2-low severe asthma could lead to novel therapeutic targets or interventions. To explore this further, we examined the genes in the enriched pathways for each module eigenvector of MA-associated DMCs. The 85 genes correlated with the turquoise module were uniquely enriched for diverse immune-related pathways, reflecting responses to viral and bacterial pathogens, inflammatory conditions, and antigen presentation (SI Appendix, Table S3). Using machine-learning algorithms, we further showed that the expression levels of the genes in 16 of these pathways discriminated asthma cases with MA from asthma cases without MA (Fig. 2). The median expression of a 535-gene score was significantly reduced in BECs from adult asthmatics with an asthmatic mother compared with asthmatics without an asthmatic mother in this study, a finding that was replicated in nasal epithelial cells from asthmatic children with and without an asthmatic mother from an independent cohort (Fig. 2C). These findings suggested that innate and adaptive immune responses to viruses and bacteria may be impaired in both adults and children with a history of MA. Impaired responses to respiratory pathogens can lead to damaged epithelium, asthma exacerbations, and more severe asthma (29–31). These data support the idea that the CpGs in the turquoise module represent a DNA methylation signature in bronchial and nasal epithelial cells that modulates the suppression of immunomodulatory gene networks in offspring of mothers with asthma and promotes a severe, hard-to-treat T2-low subtype of asthma. This supposition raises the possibility that immune boosting therapies may be more effective than immunosuppressive therapies in these individuals.
Overall, the results of our study support the hypothesis that the epigenetic landscape in airway epithelial cells differs between adult asthmatics with and without an asthmatic mother, and further suggest that DNA methylation signatures modulate genes underlying different endotypes of asthma. However, there are some limitations. First, MA was determined from participants’ report and is subject to recall bias. However, a recent study of 6,752 parent (ages 39 to 66 y)–child (ages 18 to 51 y) pairs from northern Europe, Spain, and Australia demonstrated that the agreement between adult offspring’s recollection of whether the mother ever had asthma was 92% (32). Thus, it is likely that the subjects reporting MA in our study did indeed have a mother with asthma. Second, we did not have a sufficient number of controls with MA to assess whether the observed MA effects on DNA methylation patterns would be similar or not in nonasthma controls. We also could not disentangle the effects of MA from other variables that may be correlated with MA. Third, although cells from bronchial brushings are composed largely of epithelial cells, we did not have actual counts of different epithelial cell types. However, our successful replication of the 531-gene signature in nasal epithelial cells that included proportions of measured epithelial cells as a covariate suggests that our results were not driven by differences in cell proportions between MA and NMA cases. In particular, the DMCs from the MA-associated turquoise module was associated with an MA-discriminatory transcriptional signature that was not explained by different nasal epithelial cell types. Fourth, because this was a cross-sectional study with all assessments performed in adulthood, we were not able to determine the longitudinal development of MA-associated DNA methylation patterns and their effects on gene expression prior to the inception of asthma. Thus, we could not resolve DNA methylation signatures that are the cause or effect of severe asthma in the cases. However, the fact that these signatures differed between asthma cases with MA compared with controls, but not between asthma cases without MA compared with controls, or vice versa, suggests that they may reflect in utero exposures that are distinct to pregnancies of mothers with or without asthma. Finally, our studies surveyed only a fraction of the epigenome. Therefore, we may have missed important CpG sites that are not included on the arrays, as well as their correlated genes, that differ between offspring of mothers with and without asthma. Future studies using more advanced technologies or bisulfite sequencing to assess DNA methylation or including other epigenetic marks, such as ATAC-seq, may better capture the MA-associated epigenetic signatures and their gene regulation and clinical correlates. Addressing these important questions in both longitudinal and functional studies will provide additional insights into MA effects on linked epigenetic and transcriptional programs and their causal relationships with clinical features of severe asthma.
In summary, our study of DNA methylation profiles in lower airway epithelial cells revealed distinct epigenetic patterns in adult children of mothers with asthma that may mediate the effects of MA on immunomodulatory gene-expression pathways and subsequent risk of severe asthma in her children. We suggest that the MA environment alters epigenetically mediated developmental pathways that lead to different subtypes of severe asthma that represent a spectrum of T2-high to T2-low endotypes.
Materials and Methods
Study Design.
Our study design has been previously described (11, 14, 33–36). Briefly, adults (>18 y of age) were recruited from asthma clinics and through recruitment postings around the University of Chicago Medical Center between January 2010 and March 2014. All subjects were clinically evaluated at a first visit, at which time their case or control status was confirmed by pulmonary function and methacholine challenge testing. Demographic variables and measures of FeNO were ascertained, and blood samples were collected for measurements of total serum IgE and complete blood counts with differentials. Eligible subjects underwent bronchoscopy at a second visit. Endobronchial brushings and BAL fluid were obtained during bronchoscopy, as previously described (11, 14, 16, 33–36). See SI Appendix, Supplementary Methods for a list of all inclusion and exclusion criteria.
A total of 169 subjects had either available genotypes for estimating ancestry PCs (n = 162), RNA sequences (n = 128), or array-based DNA methylation measurements (n = 150). See SI Appendix, Supplementary Methods for a description of our pipeline for processing and quality control of the genotyping and RNA-seq, and for the generation of ancestry PCs from genotype data. A flowchart of the sample selection for this study is shown in SI Appendix, Fig. S1. These studies were approved by the University of Chicago’s Institutional Review Board. Written informed consent was obtained from all research participants.
Processing and Quality Control of DNA Methylation Arrays.
DNA was isolated and purified from epithelial cells using the QIAzol lysis reagent or the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen). DNA methylation profiles were measured on the Infinium Human Methylation 450K Bead Chip (n = 482,421) or the Illumina Infinium MethylationEPIC BeadChip (n = 853,307) at the University of Chicago Functional Genomics Core. Only probes that were present on both arrays were retained using the combineArrays function in the minfi package (37) (n = 398,186; 105,882 were type I [two probes each] and 292,304 were type II [one probe each]). Probes located on the sex chromosomes and those with detection P values greater than 0.01 in more than 10% of samples were removed from the analysis. Cross-reactive probes and probes within 2 nt of an SNP with a minor allele frequency greater than 0.05 were removed using the function rmSNPandCH() from the R package DMRcate (38), retaining 398,186 probes for preprocessing. A preprocessing control normalization function was applied to correct for raw probe values or background and single-sample Noob (ssNoob) was used to correct for technical differences between the Infinium type I and type II probes and across the two generations of Infinium methylation arrays (39). Finally, eight outlier samples (>2.5 absolute deviations from the median of methylation) were removed, retaining DNA methylation profiles for 142 subjects and 398,186 CpGs (105,882 type I [two probes each] and 292,304 type II [one probe each]) for downstream analyses. PC analysis was used to determine the effects of 11 known confounding variables on global DNA methylation profiles. See SI Appendix, Table S8 for tests of association between the first 10 DNA methylation PCs and 11 potential confounding variables. After regressing out the effects of chip (ComBat) (40), age, gender, current smoking status, and the first three ancestry PCs, none of the remaining variables were significantly associated with any of the 10 PCs.
Statistical Analyses.
Our initial analyses focused on differential methylation at single CpG sites between the NMA- and MA-asthma groups compared with the nonasthma controls. To both accommodate our small sample size and generate more inclusive lists of DMCs and their correlated genes, we used liberal significance thresholds of FDR < 0.10, using the method of Benjamini and Hochberg (41) for analyses of DNA methylation and gene expression. P values for the correlations between DNA methylation module eigenvectors and clinical phenotypes were Bonferroni-corrected for the number of tests (phenotypes) performed.
Differential methylation analysis was performed using a linear model in limma (42), corrected for age, gender, current smoking status, and the first three ancestry PCs. Our study was not large enough to perform analyses separately in males and females, so we included gender as a covariate in all analyses. To assure that this appropriately corrected for any gender differences in methylation levels at individual autosomal CpGs, we compared DNA methylation levels between males and females, with and without including gender as a covariate. Of the 398,186 CpGs that passed quality control, 3,036 CpGs (0.7%) were differentially methylated between males and females (FDR < 0.10) when gender was not included as a covariate. After adjusting for gender, no CpGs were differentially methylated between males and females (all P values prior to FDR correction were >0.99), indicating that gender was properly adjusted for in our analyses.
Three differential methylation analyses were performed to identify DMCs: nonasthma controls without MA were compared with all asthma cases, to cases without MA, and to cases with MA. We observed inflation of DMC P values in all three analyses (λ = 1.68, 1.26, and 1.59, respectively) and adjusted for the genomic control inflation factor accordingly (43) (λ = 1 for all analyses after correction). SI Appendix, Fig. S10 shows the Q-Q plots and λs before and after correcting for inflation. We required DMCs to be significant in each group at FDR < 0.10. Eight subjects without MA information and four control subjects with MA were excluded from these analyses, retaining 130 subjects (SI Appendix, Fig. S2).
The CpGs that were differentially methylated only between controls and cases without MA (NMA) or only between controls and cases with MA (FDR < 0.10) were referred to as NMA-DMCs and MA-DMCs, respectively, and were separately clustered into comethylation modules using WGCNA (19). NMA-DMCs and MA-DMCs that did not cluster into a comethylation module with at least 15 DMCs were assigned to gray modules and were not considered in downstream analyses (SI Appendix, Figs. S4 and S5). The soft thresholding power was 10 and 20 for the NMA- and MA-focused analyses, respectively (SI Appendix, Fig. S11). Modules that had highly correlated eigenvectors were merged using the mergeCloseModules function. Spearman’s rank correlation coefficient was used to assess correlations between the eigenvectors for each comethylation module with all genes that were expressed in BECs (CPM > 1 in at least 25% of 118 subjects with both DNA methylation and gene expression data). Sequencing batch, age, sex, current smoking status, and the first three ancestry PCs were regressed out from the gene-expression data and the residuals were used to test for Spearman correlations with comethylation module eigenvectors. Only significantly correlated genes (FDR < 0.10) that were not correlated with the eigenvectors of the other six modules (FDR > 0.10) were retained for pathway analysis using the TopFunn function within the ToppGene Suite (20). The correlated genes were required to have the same direction of effect in all subjects and in nonasthma controls. Significant pathways with FDR < 0.10 were retained.
Comethylation module eigenvectors were also tested for associations with nine clinical phenotypes. Age, sex, current smoking status, and the first three ancestry PCs were regressed out from each of eight clinical phenotypes (excluding asthma severity [STEP classification]) and the residuals were used to test for Spearman correlations with module eigenvectors. For STEP severity categories (control, STEP = 0; mild, STEP = 1, 2; moderate, STEP = 3, 4; severe, STEP = 5, 6) (18), the eigenvectors were tested for association using an ordinal logistic regression adjusting for the same covariates. Significant correlations were determined after Bonferroni-corrections of the P values, considering nine tests.
For the modules that were correlated with genes enriched for pathways, RandomForests from the RandomForests package in R was used to identify pathways that discriminated asthma cases with (n = 27) and without (n = 48) MA using genes that were expressed and included in the annotation for all pathways as input. For each subject, each pathway was defined by the median normalized and covariate-adjusted expression of all annotated genes (n = 3 to 1,844 genes per pathway). Reactions with a mean decrease in accuracy greater than 1 were extracted and plotted in a heatmap using pheatmap from the pheatmap package in R. Using the 535 unique genes across the 13 MA-discriminatory pathways, an overall median score was calculated and compared between cases with and without MA using a Wilcoxon rank-sum test. Each of the 535 genes were also tested for differential expression between cases with and without MA; differentially expressed genes were detected at FDR < 0.10. As replication, processed RNA-seq was available from nasal epithelial cells sampled from a birth cohort of children at high risk for asthma (46 NMA and 82 MA), as described previously (15, 25). A 531-gene score (four genes were not expressed in nasal epithelial cells) was compared between asthma cases with and without MA and analysis of differential expression was performed, as detailed above, including sex, study site, batch ID, epithelial cell proportion, and 12 latent factors (44) as covariates.
Supplementary Material
Acknowledgments
We thank Dan Nicolae for statistical advice and comments on the manuscript and the coordinators and subjects who participated in these studies. This study was funded by NIH grants U19 AI095230 (to C.O and S.R.W. at University of Chicago) and UM1 AI160040 (to D.J.J. at University of Wisconsin, Madison). K.M.M. was supported by an NIH predoctoral training grant F31 HL143891 (University of Chicago).
Footnotes
Competing interest statement: D.K.H. reports personal fees from Olympus/Spiration and personal fees from PulmonX during the conduct of the study. Outside of the submitted work, D.K.H. reports: personal fees and other from Auris; personal fees from Ambu; personal fees, nonfinancial support, and other from Body Vision; personal fees and other from Eolo; other from Eon; other from Gravitas; personal fees and other from Noah Medical; personal fees and other from LX-Medical; other from Med-Opsys; other from Monogram Orthopedics; personal fees and other from Preora; other from VIDA; other from Viomics; grants and personal fees from Boston Scientific; personal fees from Johnson and Johnson; personal fees from Oncocyte; personal fees from Veracyte; personal fees and other from Broncus; grants and personal fees from Gala; personal fees from Heritage Biologics; personal fees from IDbyDNA; personal fees from Level-Ex; personal fees from Medtronic; personal fees from Neurotronic; personal fees from Olympus; personal fees from PulmonX; personal fees from Astra-Zeneca; personal fees from Biodesix; personal fees from Genetech; personal fees from Grifols; personal fees from Takeda; personal fees from CSL; personal fees from InhibRX; and personal fees and other from Prothea-X. M.C.A. reports consulting fees from Regeneron outside the submitted work.
This article is a PNAS Direct Submission. E.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2116467119/-/DCSupplemental.
Data Availability
DNA methylation and RNA-seq datasets supporting the findings in this article are available in the Gene Expression Omnibus (GEO) repositories under the accession numbers GSE201872 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE201872) and GSE201955 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE201955), respectively.
References
- 1.Nurmagambetov T., Kuwahara R., Garbe P., The economic burden of asthma in the United States, 2008–2013. Ann. Am. Thorac. Soc. 15, 348–356 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Chung K. F., et al. , International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur. Respir. J. 43, 343–373 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Lambrecht B. N., Hammad H., Fahy J. V., The cytokines of asthma. Immunity 50, 975–991 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Lim R. H., Kobzik L., Dahl M., Risk for asthma in offspring of asthmatic mothers versus fathers: A meta-analysis. PLoS One 5, e10134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perera F., et al. , Relation of DNA methylation of 5′-CpG island of ACSL3 to transplacental exposure to airborne polycyclic aromatic hydrocarbons and childhood asthma. PLoS One 4, e4488 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tobi E. W., et al. , DNA methylation of IGF2, GNASAS, INSIGF and LEP and being born small for gestational age. Epigenetics 6, 171–176 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGowan P. O., et al. , Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat. Neurosci. 12, 342–348 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gunawardhana L. P., et al. , Differential DNA methylation profiles of infants exposed to maternal asthma during pregnancy. Pediatr. Pulmonol. 49, 852–862 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Rothers J., et al. , Maternal cytokine profiles during pregnancy predict asthma in children of mothers without asthma. Am. J. Respir. Cell Mol. Biol. 59, 592–600 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeVries A., et al. , Epigenome-wide analysis links SMAD3 methylation at birth to asthma in children of asthmatic mothers. J. Allergy Clin. Immunol. 140, 534–542 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicodemus-Johnson J., et al. , Maternal asthma and microRNA regulation of soluble HLA-G in the airway. J. Allergy Clin. Immunol. 131, 1496–1503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeVries A., Vercelli D., The neonatal methylome as a gatekeeper in the trajectory to childhood asthma. Epigenomics 9, 585–593 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoettler N., Rodríguez E., Weidinger S., Ober C., Advances in asthma and allergic disease genetics: Is bigger always better? J. Allergy Clin. Immunol. 144, 1495–1506 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicodemus-Johnson J., et al. , DNA methylation in lung cells is associated with asthma endotypes and genetic risk. JCI Insight 1, e90151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ober C., et al. ; Environmental Influences on Child Health Outcomes-Children’s Respiratory Research Workgroup, Expression quantitative trait locus fine mapping of the 17q12-21 asthma locus in African American children: A genetic association and gene expression study. Lancet Respir. Med. 8, 482–492 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magnaye K. M., et al. , A-to-I editing of miR-200b-3p in airway cells is associated with moderate-to-severe asthma. Eur. Respir. J. 58, 2003862 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Gould H. J., Sutton B. J., IgE in allergy and asthma today. Nat. Rev. Immunol. 8, 205–217 (2008). [DOI] [PubMed] [Google Scholar]
- 18.National Asthma Education and Prevention Program, Expert panel report 3 (EPR-3): Guidelines for the diagnosis and management of asthma-summary report 2007. J. Allergy Clin. Immunol. 120(5, suppl.)S94–S138 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Langfelder P., Horvath S., WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J., Bardes E. E., Aronow B. J., Jegga A. G., ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37, W305-11 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei J., et al. , Ribosomal proteins regulate MHC class I peptide generation for immunosurveillance. Mol. Cell 73, 1162–1173.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson D. S., The role of the T cell in asthma. J. Allergy Clin. Immunol. 126, 1081–1091, quiz 1092–1093 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Busse W. W., Lemanske R. F. Jr., Gern J. E., Role of viral respiratory infections in asthma and asthma exacerbations. Lancet 376, 826–834 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coyle A. J., Gutierrez-Ramos J. C., The role of ICOS and other costimulatory molecules in allergy and asthma. Springer Semin. Immunopathol. 25, 349–359 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Gern J. E., The Urban Environment and Childhood Asthma Study. J. Allergy Clin. Immunol. 125, 545–549 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCauley K., et al. ; National Institute of Allergy and Infectious Diseases–sponsored Inner-City Asthma Consortium, Distinct nasal airway bacterial microbiotas differentially relate to exacerbation in pediatric patients with asthma. J. Allergy Clin. Immunol. 144, 1187–1197 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fahy J. V., Type 2 inflammation in asthma—Present in most, absent in many. Nat. Rev. Immunol. 15, 57–65 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pavord I., et al. , Severe T2-high asthma in the biologics era: European experts’ opinion. Eur. Respir. Rev. 28, 190054 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwards M. R., et al. , Viral infections in allergy and immunology: How allergic inflammation influences viral infections and illness. J. Allergy Clin. Immunol. 140, 909–920 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Message S. D., et al. , Rhinovirus-induced lower respiratory illness is increased in asthma and related to virus load and Th1/2 cytokine and IL-10 production. Proc. Natl. Acad. Sci. U.S.A. 105, 13562–13567 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu J., et al. , Bronchial mucosal IFN-α/β and pattern recognition receptor expression in patients with experimental rhinovirus-induced asthma exacerbations. J. Allergy Clin. Immunol. 143, 114–125.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuiper I. N., et al. , Agreement in reporting of asthma by parents or offspring—The RHINESSA generation study. BMC Pulm. Med. 18, 122 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharma A., et al. , Associations between fungal and bacterial microbiota of airways and asthma endotypes. J. Allergy Clin. Immunol. 144, 1214–1227.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White S. R., et al. , Evidence for an IL-6-high asthma phenotype in asthmatic patients of African ancestry. J. Allergy Clin. Immunol. 144, 304–306.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White S. R., et al. , Levels of soluble human leukocyte antigen-G are increased in asthmatic airways. Eur. Respir. J. 35, 925–927 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White S. R., et al. , Elevated levels of soluble humanleukocyte antigen-G in the airways are a marker for a low-inflammatory endotype of asthma. J. Allergy Clin. Immunol. 140, 857–860 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aryee M. J., et al. , Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30, 1363–1369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peters T. J., et al. , De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin 8, 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fortin J. P., Triche T. J. Jr., Hansen K. D., Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics 33, 558–560 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leek J. T., Johnson W. E., Parker H. S., Jaffe A. E., Storey J. D., The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hochberg Y., Benjamini Y., More powerful procedures for multiple significance testing. Stat. Med. 9, 811–818 (1990). [DOI] [PubMed] [Google Scholar]
- 42.Ritchie M. E., et al. , limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Devlin B., Roeder K., Genomic control for association studies. Biometrics 55, 997–1004 (1999). [DOI] [PubMed] [Google Scholar]
- 44.McKennan C., Nicolae D., Accounting for unobserved covariates with varying degrees of estimability in high-dimensional biological data. Biometrika 106, 823–840 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA methylation and RNA-seq datasets supporting the findings in this article are available in the Gene Expression Omnibus (GEO) repositories under the accession numbers GSE201872 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE201872) and GSE201955 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE201955), respectively.