Significance
Single-cell transcriptomics has revealed specific glial activation states associated with the pathogenesis of neurodegenerative diseases, such as Alzheimer’s and Parkinson’s disease (AD and PD). What is still needed are clinically relevant biomarkers for deciphering such glial states in AD and PD patients. To this end, we applied proteome analysis in cerebrospinal fluid (CSF) of mouse models of AD and PD pathology. This allowed us to identify a panel of glial CSF proteins that largely match the transcriptomic changes. The identified proteins can also be quantified in human CSF and show changes in AD patients, supporting their relevance as biomarker candidates to stage glial activation in patients with neurodegenerative diseases.
Keywords: CSF biomarkers, Alzheimer’s disease, Parkinson’s disease, glial activation, mass spectrometry
Abstract
Single-cell transcriptomics has revealed specific glial activation states associated with the pathogenesis of neurodegenerative diseases, such as Alzheimer’s and Parkinson’s disease. While these findings may eventually lead to new therapeutic opportunities, little is known about how these glial responses are reflected by biomarker changes in bodily fluids. Such knowledge, however, appears crucial for patient stratification, as well as monitoring disease progression and treatment responses in clinical trials. Here, we took advantage of well-described mouse models of β-amyloidosis and α-synucleinopathy to explore cerebrospinal fluid (CSF) proteome changes related to their respective proteopathic lesions. Nontargeted liquid chromatography-mass spectrometry revealed that the majority of proteins that undergo age-related changes in CSF of either mouse model were linked to microglia and astrocytes. Specifically, we identified a panel of more than 20 glial-derived proteins that were increased in CSF of aged β-amyloid precursor protein- and α-synuclein-transgenic mice and largely overlap with previously described disease-associated glial genes identified by single-cell transcriptomics. Our results also show that enhanced shedding is responsible for the increase of several of the identified glial CSF proteins as exemplified for TREM2. Notably, the vast majority of these proteins can also be quantified in human CSF and reveal changes in Alzheimer’s disease cohorts. The finding that cellular transcriptome changes translate into corresponding changes of CSF proteins is of clinical relevance, supporting efforts to identify fluid biomarkers that reflect the various functional states of glial responses in cerebral proteopathies, such as Alzheimer’s and Parkinson’s disease.
Fluid biomarkers have become instrumental for the clinical diagnosis and assessment of the progression of neurodegenerative diseases (1, 2). For Alzheimer’s disease (AD), most useful biomarkers in cerebrospinal fluid (CSF) and blood are related to the pathological hallmark lesions β-amyloid (Aβ) plaques and tau inclusions (2). Fluid biomarkers are more elusive for Parkinson’s disease (PD) (3, 4). From single-cell transcriptomics, there is now growing evidence for distinct molecular changes related to various disease stages (5, 6). In particular, microglia and astrocytes are at the forefront of attention since they show characteristic transcriptomic alterations with aging and disease progression (6–8). In addition, the majority of AD risk genes relate to microglia (7, 8). Whether such glial transcriptomic changes translate to corresponding protein changes that can be gathered in bodily fluids and be used as molecular biomarkers is of high clinical relevance.
CSF proteome analysis using nontargeted liquid chromatography-mass spectrometry (LC-MS) is a powerful technique and allows the unbiased quantification of hundreds of proteins (9). However, large interindividual variations due to genetic and disease heterogeneity and comorbidities complicate such analyses in human cohorts. In contrast, genetically defined mouse models show minimal interindividual variability and rarely exhibit other confounding factors (10, 11). Indeed, transgenic (tg) mice have already proven instrumental for the molecular dissection of CSF biomarker changes in different neurodegenerative diseases (12–18).
To search for fluid biomarker candidates, we studied age-related CSF proteome changes in established mouse models of β-amyloidosis (APPPS1 mice) (19) and of α-synucleinopathy (A30P-αS mice) (20) in comparison with non-tg mice using unbiased LC-MS/MS and label-free quantification (LFQ). Besides proteome changes within each mouse model, we also aimed to characterize changes that are common or different between the two models to highlight the specificity of the biomarker signatures. Finally, to translate the findings to a clinical setting, we studied whether the CSF proteins identified in the mouse models can also be quantified in human CSF and show changes in AD.
Results
Age-Dependent Changes in the CSF Proteome of APPPS1 Mice.
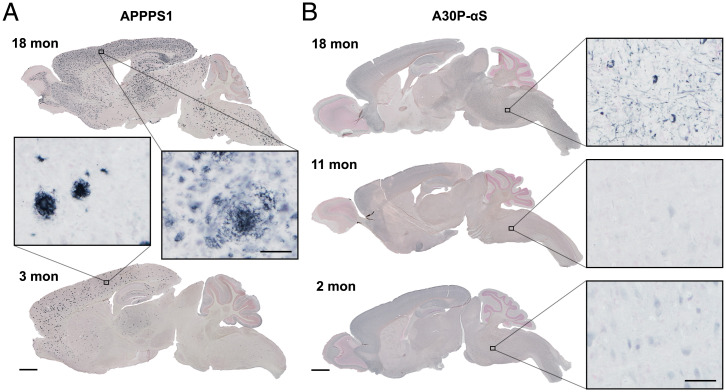
Male 3- and 18-mo-old APPPS1 mice (19) and age-matched non-tg controls were used to study CSF proteome changes related to β-amyloidosis (Fig. 1). While 3-mo-old APPPS1 mice have a low amyloid load, 18-mo-old mice APPPS1 mice have a high load and are already in the plateau phase of amyloid deposition (21).
Fig. 1.
APPPS1 and A30P-αS mice show progressing proteopathic brain lesions. (A) Representative images of immunostained Aβ deposits in young (3-mo-old) and aged (18-mo-old) APPPS1 mice. Note the distinct Aβ plaques at 3 mo mainly in the neocortex and the appearance of Aβ deposition all over the brain at 18 mo of age. (B) Immunostained phosphorylated (pS129) αS in young (2-mo-old), adult (11-mo-old), and aged (end-stage disease; 18-mo-old) A30P-αS mice. (Scale bars, 1 mm for brain sections and 50 µm for Insets.) Counterstaining with nuclear fast red.
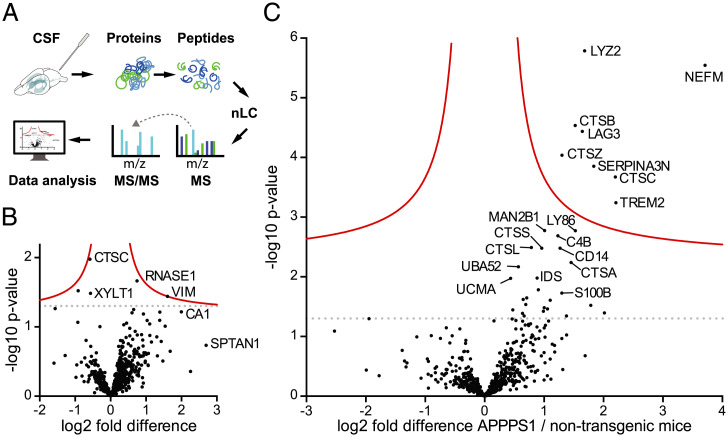
The CSF proteome was analyzed using LFQ to identify protein abundance differences (Fig. 2A and Dataset S1A). Only proteins that could be relatively quantified in at least three animals per group were used for pairwise comparisons. No significant differences in the CSF proteome of APPPS1 vs. non-tg control mice at 3 mo of age were found (Fig. 2B). In contrast, of the 665 relatively quantified proteins at 18 mo of age, 8 proteins—namely lysozyme C-2 (LYZ2), neurofilament medium polypeptide (NEFM), cathepsin B (CTSB), lymphocyte activation gene 3 protein (LAG3), cathepsin Z (CTSZ), serine protease inhibitor A3N (SERPINA3N), dipeptidyl peptidase 1 (CTSC), and the triggering receptor expressed on myeloid cells 2 (TREM2)—showed a significant increase ranging from 2.5- (CTSZ) to 13-fold (NEFM) (Fig. 2C).
Fig. 2.
CSF proteome changes in APPPS1 mice at 3 and 18 mo of age compared to age-matched controls. (A) Workflow for mouse CSF proteome analysis: CSF was collected from the cisterna magna of anesthetized mice. After denaturation and alkylation, proteins were digested with Lys-C and trypsin. Resulting peptides were separated and eluted on a nano high-performance LC system. Subsequently, proteins underwent electrospray-ionization followed by MS analysis on a Q-Exactive Orbitrap Mass Spectrometer. The MS measurements were analyzed using MaxQuant software and UniProt database. (B) Protein abundances between 3-mo-old APPPS1 and non-tg control mice were compared (seven and eight male mice per group, respectively; note, one APPPS1 animal could not be measured due to an instrument failure). Each dot represents a protein that could be quantified in at least three CSF samples per group (SI Appendix, SI Materials and Methods). The –log10 of the P value of each protein is plotted against its log2 fold difference between APPPS1 and non-tg mice. No significant differences were found at 3 mo. (C) The same analysis was performed for 18-mo-old male APPPS1 mice and age-matched non-tg mice (eight animals per group). Dots above the solid red line indicate proteins that showed a significantly higher abundance in APPPS1 compared to age-matched non-tg mice. Significance cutoff (red line) is based on permutation-based FDR-corrected t tests (P < 0.05 and S0 = 0.1). The dotted lines indicate the threshold P = 0.05 (i.e., without correction for multiple hypothesis testing) (compare with Fig. 4A).
Soluble human amyloid precursor protein (APP) was exclusively detected in the APPPS1 mice, whereas soluble murine APP was detected in all samples but did not show differences between APPPS1 and non-tg mice (SI Appendix, Fig. S1). Consistent with the previously reported decrease of CSF Aβ42 with increasing brain amyloid (16), a decrease of a tryptic peptide corresponding to amino acids 17–28 of Aβ was found in the aged APPPS1 mice (SI Appendix, Fig. S1). Endogenous mouse tau (mTau) was below the limit of quantification although two tryptic peptides from the proline-rich mid-domain (mTau184–198 and 201–213, corresponding to human Tau195–209 and 212–224 of the 2N4R isoform, respectively) were detected mainly or exclusively in aged APPPS1 tg mice (SI Appendix, Fig. S1), which is in line with the age-dependent increase of CSF tau observed in APPPS1 tg mice (14, 16, 18).
Age-Dependent Changes in the CSF Proteome of A30P-αS Mice.
Male and female 3-, 11-, and 18-mo-old A30P-αS mice were used to study CSF protein changes related to α-synucleinopathy (20) (Fig. 1). While 3- and 11-mo-old A30P-αS mice do not reveal α-synuclein (αS) inclusions, 18-mo-old mice A30P-αS mice have a very high lesion load in the brainstem and some other subcortical areas (22, 23). The mean life span of A30P-αS mice is around 19 mo and no sex-related difference has been reported (23).
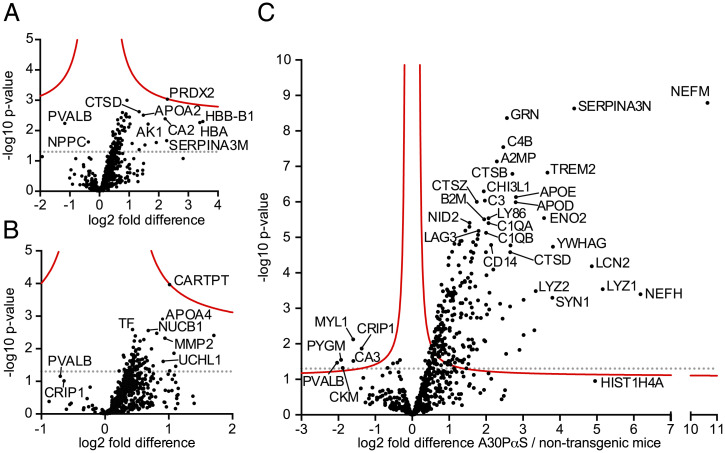
Overall, 545, 526, and 636 proteins were relatively quantified between A30P-αS and non-tg mice at 3, 11, and 18 mo, respectively (again, only proteins that could be relatively quantified in at least three animals per group were used for pairwise comparisons) (Dataset S2A). There were no significant differences between A30P-αS and non-tg mice at 3 and 11 mo of age (Fig. 3 A and B). At the late stage of α-synucleinopathy, 220 proteins showed significant abundance changes, with the vast majority being elevated in A30P-αS compared to non-tg mice (Fig. 3C and Dataset S2A). Among them, several proteins that were also significantly increased in the aged APPPS1 mice—namely NEFM, SERPINA3N, TREM2, LYZ2, LAG3, CTSB, and CTSZ—indicating commonalities between both proteopathies. The increase in the A30P-αS mouse model was between 3-fold (CTSZ) and more than 1,000-fold (NEFM) (Fig. 3C).
Fig. 3.
CSF Proteome changes in A30P-αS mice at 3, 11, and 18 mo of age compared to age-matched controls. (A) Protein abundances between 3-mo-old A30P-αS and non-tg control mice (six and nine mice, respectively; for unequal mouse number justification, see SI Appendix, SI Materials and Methods) were compared. Each dot represents a protein that could be quantified in at least three CSF samples per group (SI Appendix, SI Materials and Methods). The –log10 of the P value of each protein is plotted against its log2 fold difference between A30P-αS and non-tg mice. No significant differences were found. (B) Protein abundances between 11-mo-old A30P-αS and non-tg control mice (six and five mice, respectively) were compared in the same way. Again, no significant differences were observed. (C) CSF analysis of 18-mo-old A30P-αS and age-matched non-tg mice (six vs. ten mice; for unequal mouse number justification, see SI Appendix, SI Materials and Methods) revealed a transgene-related abundance change for 220 proteins (dots above the red threshold). The vast majority was increased in A30P-αS mice. Significance cutoff (red line) is based on permutation-based FDR-corrected t tests (P < 0.05 and S0 = 0.1). The dotted lines indicate the threshold P = 0.05 (i.e., not corrected for multiple hypothesis testing) (compare with Fig. 4A).
Other prominent CSF proteins with an increase in 18-mo-old A30P-αS mice compared to aged-matched non-tg mice were granulins (GRN), complement C4-B (C4B), α-2-macroglobulin-P (A2M or A2MP), chitinase-3-like protein 1 (CHI3L1, also known as YKL-40), apolipoprotein D and E (APOD, APOE), complement C3 (C3), neuron-specific enolase (ENO2), lymphocyte antigen 86 (LY86), β-2-microglobulin (B2M), nidogen-2 (NID2), complement C1q subcomponent subunit A and B (C1QA, C1QB), vitamin K-dependent protein S (PROS1), and EGF-containing fibulin-like extracellular matrix protein 2 (EFEMP2). The highest fold-difference was found for NEFM (see above), neurofilament heavy polypeptide (NEFH), lysozyme C-1 (LYZ1), and neutrophil gelatinase-associated lipocalin (LCN2) (Fig. 3C and Dataset S2A). Consistent with the overexpression of αS (SNCA) in the A30P-αS mice, peptides derived from SNCA were much more abundant or exclusively quantified in A30P-αS mice (SI Appendix, Fig. S1). Although most of the 220 identified CSF proteins showed an increase, six proteins were identified with significantly lower concentrations in the 18-mo-old A30P-αS mice compared to the age-matched controls: cysteine-rich protein 1 (CRIP1), glycogen phosphorylase, muscle form (PYGM), parvalbumin-α (PVALB), carbonic anhydrase 3 (CA3), creatine kinase M-type (CKM), and myosin light chain 1/3 (MYL1) (Fig. 3C). Interestingly, most of these proteins are involved in muscle or energy metabolism (24). The sex of the animals had no influence on the abundance of the top 20 proteins that showed the most significant changes (SI Appendix, Fig. S2).
Enrichment analysis of the 220 significantly changed proteins in comparison to all quantified CSF proteins revealed “innate immune response” and “proteolysis” as primary Gene Ontology (GO) biological processes. The Kyoto Encyclopedia of Genes and Genomes (KEGG) suggests that lysosome-related processes are the most affected cellular pathway (Dataset S2C).
Several proteins were exclusively detected in aged A30P-αS mice, and thus presumably were below detection in young and non-tg mice. Among them were several microtubule-associated proteins, MAP1B, MAP1LC3A, MAP2, MAP6, and MAPT (i.e., tau), and intermediate filament proteins, peripherin (PRPH) and NEFL (Table 1). Thus, all neurofilament proteins (NEFH, NEFM, NEFL), including α-internexin (INA, also known as neurofilament-66) and PRPH, were increased in CSF of aged A30P-αS mice (SI Appendix, Fig. S3). Also detected in aged but not young tg and not in non-tg mice was cystatin F (CST7; see also “stage 2 DAM" proteins (Fig. 5B)), synaptosomal-associated protein 25 (SNAP25), a previously described candidate biomarker for synaptic degeneration (25, 26), and reticulon-4 (RTN4; also known as Nogo) an inhibitor of neurite outgrowth and axonal regeneration (27) (Table 1).
Table 1.
Proteins exclusively detected in aged (18-mo-old) A30P-αS
| Uniprot accession | Protein name | Gene name | LFQ values in A30P-αS (18 mo) | Unique peptides (n) |
|---|---|---|---|---|
| P08551 | Neurofilament light polypeptide | Nefl | 6 | 27 |
| P14873 | Microtubule-associated protein 1B; MAP1B heavy chain; MAP1 light chain LC1 | Map1b | 6 | 25 |
| Q99P91 | Transmembrane glycoprotein NMB | Gpnmb | 6 | 10 |
| P10637 | Microtubule-associated protein tau | Mapt | 3 | 5 |
| Q7TSJ2 | Microtubule-associated protein 6 | Map6 | 4 | 6 |
| P60879 | Synaptosomal-associated protein 25 | Snap25 | 5 | 6 |
| P24288 | Branched-chain-amino-acid aminotransferase, cytosolic | Bcat1 | 4 | 3 |
| Q91VR7 | Microtubule-associated proteins 1A/1B light chain 3A | Map1lc3a | 4 | 2 |
| Q08331 | Calretinin | Calb2 | 5 | 7 |
| P15331 | Peripherin | Prph | 6 | 9 |
|
P63328/ P48453 |
Serine/threonine-protein phosphatase 2B catalytic subunit-α isoform/ Serine/threonine-protein phosphatase 2B catalytic subunit-β isoform |
Ppp3ca/
Ppp3cb |
3 | 6 |
| P20357 | Microtubule-associated protein 2 | Map2 | 6 | 6 |
| Q9Z121 | C-C motif chemokine 8 | Ccl8 | 5 | 3 |
| Q99P72 | Reticulon-4 | Rtn4 | 4 | 2 |
| P25119 | Tumor necrosis factor receptor superfamily member 1B | Tnfrsf1b | 5 | 2 |
| O89098 | Cystatin-F | Cst7 | 6 | 1 |
| Q6S5C2 | N-acetylglucosamine-1-phosphotransferase subunit gamma | Gnptg | 4 | 4 |
| P30275 | Creatine kinase U-type, mitochondrial | Ckmt1 | 3 | 6 |
| Q9R1Q8 | Transgelin-3 | Tagln3 | 4 | 2 |
The analysis of the A30P-αS dataset revealed a group of 19 proteins that were exclusively quantified in the oldest age group (n ≥ 3 mice; see also SI Appendix, SI Materials and Methods). The absence of LFQ signals in any of the other experimental groups, including age-matched controls, implies that these proteins were below detection limit. Hence, an increase could not be computed for these proteins. Proteins are ranked according to the average LFQ intensity in A30P-αS at 18 mo (Dataset S2A).
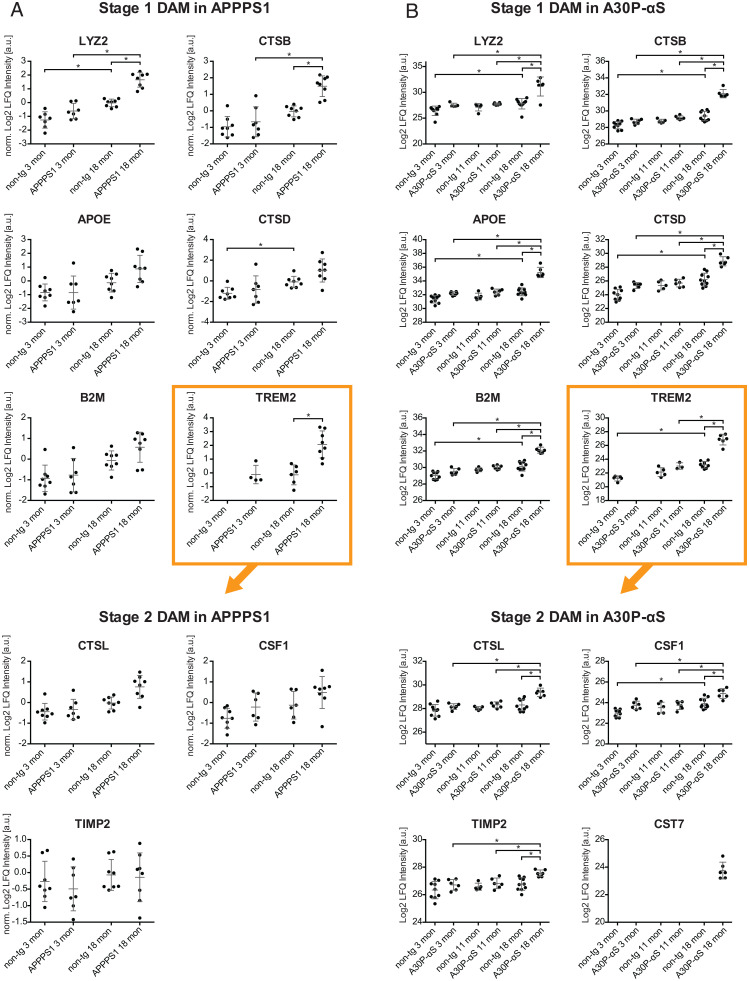
Fig. 5.
Stage 1 and stage 2 DAM related proteins. (A) LFQ intensities of various stage 1 and stage 2 DAM proteins in the CSF of 3- and 18-mo-old APPPS1 mice and age-matched controls (the same mice as presented in Figs. 2 and 4) based on MS data. LFQ intensities were normalized to the average LFQ value for 18-mo-old non-tg mice (log2 scale) since APPPS1 mice were analyzed in two batches. (B) LFQ intensities of various stage 1 and 2 DAM proteins in the CSF of 3-, 11-, and 18-mo-old A30P-αS mice and age-matched non-tg mice (the same mice as presented in Figs. 3 and 4) based on MS data. Significant differences based on t tests with permutation-based FDR correction (*P < 0.05 and S0 = 0.1) are indicated for tg vs. age-matched non-tg mice and for comparisons between different age-groups of the same genotype (see also Datasets S1B and S2B).
The CSF Proteome of APPPS1 and A30P-αS Mice Indicate Glial Activation.
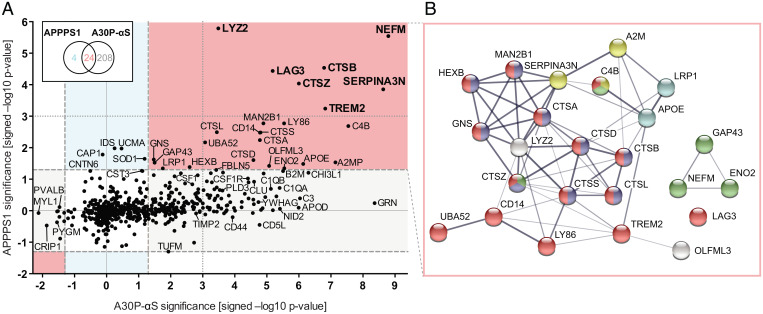
To evaluate potential CSF biomarkers associated with common downstream pathomechanisms of β-amyloidosis and α-synucleinopathy, we compared results from the aged (18-mo-old) APPPS1 and A30P-αS datasets with each other. For this purpose, we plotted the probability value (P value) for the abundance change of each CSF protein in aged APPPS1 mice against the corresponding P value in aged A30P-αS mice (Fig. 4A; note that for this comparison, significance levels were set at 5%; i.e., without false-discovery rate [FDR] correction as done in Figs. 2 and 3). Only four proteins showed a tendency to be specifically increased in APPPS1 but not in A30P-αS mice, namely iduronate 2-sulfatase (IDS), unique cartilage matrix-associated protein (UCMA), adenylyl cyclase-associated protein 1 (CAP1), and superoxide dismutase [Cu-Zn] (SOD1). In contrast, 208 proteins were mainly or exclusively altered in the A30P-αS mice, including GRN, APOD, C3, NID2, C1QA, and C1QB.
Fig. 4.
Meta-analysis of CSF proteome changes. (A) Probability values (displayed as –log10 P values) of individual CSF protein changes in the aged (18-mo-old) A30P-αS versus age-matched non-tg mice are plotted against those in aged APPPS1 versus age-matched non-tg mice for each analyte. These are the same mice as presented in Figs. 2 and 3. Note: positive values indicate an increase while negative values signify a decrease. Proteins with P < 0.05 (i.e., >1.3 on the –log10 scale) in both datasets reflect a general increase: that is, both mouse models showed an abundance difference between tg and non-tg animals at 18 mo of age (red sector), while P < 0.05 in either the A30P-αS (gray) or the APPPS1 (blue) dataset rather point at pathology-specific modulations. Note: Although LCN2 also appeared to be increased in both aged APPPS1 and A30P-αS mice, it escaped the APPPS1 analysis since it was quantified in less than three 18-mo-old wild-type mice; dashed lines indicate the threshold of P = 0.05 (i.e., not corrected for multiple hypothesis testing) (compare with Figs. 2 and 3). Bold letters indicate the proteins that were significantly up-regulated in aged tg mice of both disease models after FDR correction. (B) Protein–protein interaction of CSF analytes changing in response to pathology in both mouse models (as displayed in A, red sector) revealed 7.5 times more interactions than would be predicted for a random set of proteins of comparable size (i.e., 60 vs. 8 edges, respectively; protein–protein interaction enrichment P < 1.0e-16). Line thickness corresponds to the strength of data support. Color code based on functional enrichment analysis: “immune system” (red; reactome pathway), “lysosome” and “axon” (blue and green, respectively; GO Cellular Components), “serine-type endopeptidase inhibitor activity” (yellow; GO Molecular Function), and “cholesterol metabolism” (cyan; KEGG pathway).
Common changes in both mouse models were found for 24 proteins (Fig. 4A). Predicted protein interactions using Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) revealed a larger complex of immune-associated analytes (including TREM2, LY86, C4B, monocyte differentiation antigen CD14 [CD14], ubiquitin-60S ribosomal protein L40 [UBA52], and lysosomal proteins, i.e., β-hexosaminidase subunit B [HEXB], lysosomal α-mannosidase [MAN2B1], N-acetylglucosamine-6-sulfatase [GNS], and six cathepsins) together with APOE, LYZ2, olfactomedin-like protein 3 (OLFML3), and astrocyte- or oligodendrocyte-related proteins SERPINA3N, A2M, and its receptor, Prolow-density lipoprotein receptor-related protein 1 (LRP1) (Fig. 4B). A separate network of axonal proteins was found, including ENO2, neuromodulin (GAP43), and NEFM (Fig. 4B). Noteworthy, LCN2 also appeared to be increased in both, aged APPPS1 and A30P-αS mice, but escaped the APPPS1 analysis since it was quantified in less than three 18-mo-old wild-type mice (SI Appendix, SI Materials and Methods).
More than half of these commonly elevated proteins have been associated with microglia (28–31). Two activation stages of disease-associated microglia (DAM) have been postulated (8). While the molecular factors for the transition from homeostatic to DAM stage 1 are poorly understood, the initiation of stage 2 is dependent on TREM2 signaling (32). The present results revealed that stage 1 DAM proteins (LYZ2, CTSB, APOE, CTSD, and B2M) were mostly up-regulated in both mouse models (Fig. 5), whereas stage 2 proteins (macrophage colony-stimulating factor 1 [CSF1], metalloproteinase inhibitor 2 [TIMP2], and CTSL) were predominantly increased in aged A30P-αS mice (Fig. 5B). Consistently, CST7, a stage 2 DAM gene product, was exclusively detected in 18-mo-old A30P-αS mice.
Validation of Selected Glia-Associated Hits.
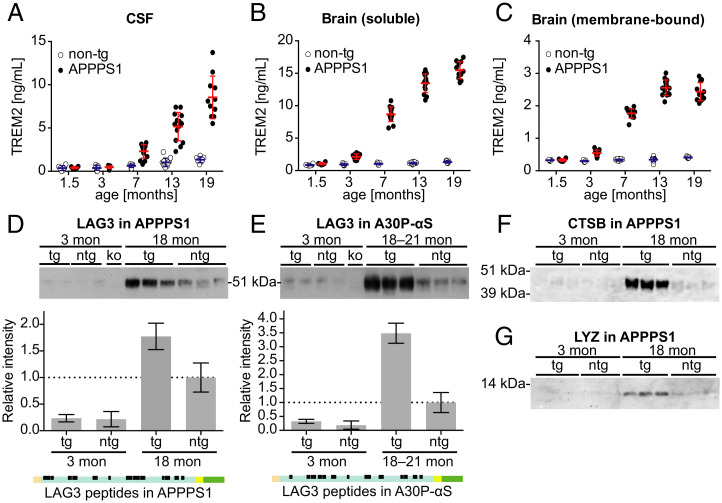
The proteomic analysis indicated that TREM2 was 4.6-fold (= 22.21) higher in 18-mo-old APPPS1 compared to the age-matched non-tg mice (Fig. 2C and Dataset S1A). Using a sandwich-immunoassay and separate batches of APPPS1 and non-tg mice, a 6.5-fold increase of TREM2 was found in the CSF of 19-mo-old APPPS1 compared to the non-tg littermates (Fig. 6A). Quantification of the microglia-specific protein TREM2 in the brains of the same mice revealed an age-related increase for both soluble TREM2 released by proteolytic shedding, as well as membrane-bound forms of TREM2 (Fig. 6 B and C).
Fig. 6.
Biochemical validation of age- and transgene-related changes of selected glia-associated CSF proteins in APPPS1 and A30P-αS mice. (A–C) Using an ECL-based immunoassay, TREM2 was measured in the CSF (A), and soluble (B) and membrane-bound brain fraction (C) from a new cohort of APPPS1 and non-tg mice ranging from 1.5 to 19 mo of age (males, n = 6–15 per group) (see also SI Appendix, SI Materials and Methods). Bars indicate mean ± SD. (A) Two-way ANOVA revealed a significant age × genotype interaction for CSF TREM2; F(4, 89) = 21.4, P < 0.0001. Tukey's post hoc test showed a significant increase in 7-mo-old APPPS1 compared to age-matched non-tg and 1.5-mo-old APPPS1 mice (P < 0.0001; note, values of one 1.5- and two 3-mo-old non-tg mice were below limit of detection). (B) Quantification of TBS-soluble brain TREM2 levels was done in the same mice. Two-way ANOVA revealed a significant age × genotype interaction; F(4, 92) = 241.8, P < 0.0001. Tukey's post hoc test showed that soluble brain TREM2 in APPPS1 mice was significantly increased starting at 3 mo of age compared to age-matched non-tg and 1.5-mo-old APPPS1 mice (P < 0.0001). (C) Quantification of membrane-associated brain TREM2 levels (TBS/Triton-soluble fraction) in the same mouse cohort. Two-way ANOVA revealed a significant age × genotype interaction F(4, 89) = 210.4, P < 0.0001. Tukey's post hoc test showed that membrane-bound TREM2 was significantly increased in 3-mo-old APPPS1 mice compared to age-matched non-tg and to1.5-mo-old tg mice (P < 0.0001) but reached a plateau at 13 mo. Note: Two samples (1.5-mo-old non-tg and 7-mo-old APPPS1 mouse) could not be measured and one value (7-mo-old non-tg mouse) was excluded because it was a significant outlier (i.e., 6.4 SD greater than the mean of all non-tg mice). All P values of post hoc comparisons (for A–C) are reported in SI Appendix, Table S1. (D) Immunoblotting of CSF samples from APPPS1 tg mice using a LAG3 antibody revealed the expected 54-kDa band corresponding to soluble LAG3 (88). Semiquantitative analysis revealed that 18-mo-old APPPS1 mice had about 77% more CSF LAG3 than age-matched non-tg controls (ntg), compared to a threefold difference based on MS (compare with Fig. 2C). LAG3 was barely detectable in the CSF of non-tg and 3-mo-old APPPS1 tg mice and no signal appeared in the Lag3 knockout (ko) mouse (n = 2 and 3 mice per group in young and aged mice, respectively). (E) The same LAG3-related protein band was detected in CSF samples of A30P-αS tg mice. Semiquantitative analysis revealed that A30P-αS mice had about 3.5 times more LAG3 in the CSF than non-tg mice at 18 to 21 mo of age (the same difference that was quantified based on MS data; compare with Fig. 3C). Again, LAG3 was barely detectable in the CSF of non-tg and 3-mo-old A30P-αS tg mice, and no signal was seen in the Lag3 knockout (ko) control. Mean ± SD of signal intensities (n = 2 and 3 mice per group in young and aged mice, respectively). Topological representation of peptides mapped on LAG3 as determined by QARIP analysis (compare with Dataset S3). All detected peptides originate from the extracellular domain of LAG3 (D and E: orange, signal peptide; blue, extracellular domain; yellow, transmembrane domain; green, cytoplasmic domain). (F and G) Western blots for CTSB and LYZ in APPPS1 tg and non-tg mice at 3 and 18 mo of age (n = 3 per group). While in young animals no signal was detected, aged APPPS1 mice showed the expected bands for the two proteins (note: CSF samples from the same mice were used in F and G).
Additional up-regulated glia-related CSF markers were validated in independent mouse CSF samples using immunoblotting. LAG3 in the CSF of aged (18-mo-old) APPPS1 and A30P-αS tg mice was elevated 1.8-fold and 3.5-fold, respectively, both compared to age-matched non-tg mice (Fig. 6 D and E). As for LAG3, the vast majority of identified peptides from single-pass transmembrane proteins were assigned to the extracellular domain (Dataset S3). Similarly, immunoblotting confirmed the prominent increase of CTSB and LYZ in aged APPPS1 mice found in the proteome analysis (Fig. 6 F and G).
Glia-Associated CSF Markers Are Also Changed in Human CSF.
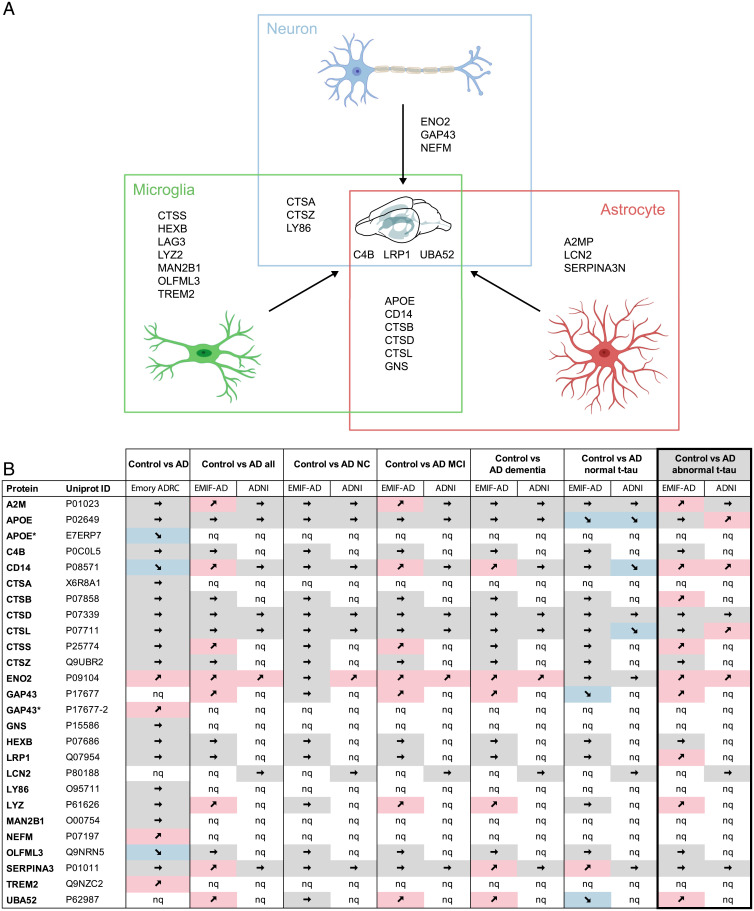
To assess whether the 25 (24 plus LCN2, see above) commonly elevated CSF proteins in the mouse models (Fig. 7A) can also be identified in human and specifically AD CSF, we explored datasets from two recently published studies comparing the CSF proteome of AD cohorts and healthy controls from the Emory Goizueta Alzheimer’s Disease Research Center (ADRC) (33), and from the European Medical Information Framework for Alzheimer’s Disease Multimodal Biomarker Discovery (EMIF-AD) and Alzheimer’s Disease Neuroimaging Initiative (ADNI) (34). In fact, homologs of all these mouse CSF proteins, except LAG3, were also detected in human CSF, and many of them also revealed changes in AD (Fig. 7B).
Fig. 7.
Common CSF protein changes in aged APPPS1 and A30P-αS mice are linked to glial cells and translate to human AD. (A) The majority of CSF proteins found to be increased in both mouse models (APPPS1 and A30P-αS) are linked to microglia (green) and/or astrocytes (red) and only a few are of neuronal origin (blue). See Results for details. Homologs of 24 of the 25 mouse proteins (all except LAG3) could also be measured in human CSF samples using MS-based technologies (compare with B). Partially created with bioRender. (B) Direct comparison of the murine CSF proteins with recently published human CSF proteome datasets including three different cohorts—Emory-ADRC (33) or EMIF-AD and ADNI (34)—revealed that several proteins of the mouse panel also show disease-related changes in AD patients. The best match between human and mouse data were seen for AD patients with abnormal CSF t-tau levels (based on study-specific cutoff values; bold frame). A total of 11 proteins showed a significant increase (9 in the EMIF-AD and additional 2 in the ADNI cohort) compared to healthy controls (P < 0.05; note that CD14 and ENO2 were increased in both cohorts). MCI, mild cognitive impairment; NC, normal cognition; nq, not quantified; ➚ increase, ➘ decrease, ➙ no significant change (*proteins highlighted with a star are [potential] isoforms of proteins listed in the table, according to UniProt). Note that due to stringent data filtering or targeted approach, only 707 and 204 proteins were relatively quantified in the EMIF-AD and ADNI cohort, respectively. Definition of AD in the EMIF-AD and ADNI cohort was based on abnormal CSF Aβ1–42 concentrations and cognitive function was assessed according to international consensus criteria (for details see ref. 34).
The best match between glial proteins elevated in the mouse models and AD was observed in AD subcohorts with abnormal CSF total tau (t-tau) levels. In these AD subcohorts, 11 of the 25 proteins were also increased, with 8 related to glial cells (Fig. 7B). Of note, also aged APPPS1 mice reveal an increase in CSF t-tau in magnitude similar to AD (14, 16, 18). The observation that the match between mouse and human CSF protein changes was dependent on the AD subcohort corroborates that different disease stages reveal different glial CSF protein signatures and that AD is more than a combination of Aβ and αS lesions. In addition, methodological variations may also contribute to the observed differences: for example, the ADRC and ADNI CSF samples were depleted of the most abundant proteins or the use of tandem mass tag technique in the ADRC and EMIF-AD cohorts vs. multiple reaction monitoring targeted MS or rules-based medicine multiplex in the ADNI cohort (33, 34).
Discussion
An unbiased proteomic approach was used to compare CSF protein changes in genetically defined mouse models with distinct proteopathic lesions (β-amyloidosis and α-synucleinopathy). Similar approaches in humans are more difficult because disease heterogeneity due to comorbidities and biological variability require large numbers of patient samples (35, 36). Although the volume of murine CSF is very small, technical advances in proteomic analysis allowed us to identify changes in more than 600 proteins using only 2 μL of CSF per technical replicate.
The most remarkable finding of the present study was the predominance of microglia-associated proteins among the CSF proteins identified to change in aged APPPS1 and A30P-αS mice. The importance of microglial activation in the pathogenesis of neurological disorders including AD and PD has been extensively documented (8, 37–39). There is evidence for both destructive and beneficial effects of microglial activation, which are likely dependent on both disease stage and type of proteopathic lesions (40, 41). Glial cells in brain are not directly accessible for clinical diagnostics. However, a first indication that glial transcriptomic changes translate into corresponding CSF protein changes came from observations that key transcriptomic hits often represent secreted or shed glial proteins. The prime example of a shed protein is soluble TREM2, which is released from the microglia surface into the interstitial fluid/CSF by proteolytic cleavage through ADAM10 and ADAM17 (42). The observed plateau in concentration of membrane-bound TREM2 in the aged APPPS1 mice suggests that the persistent increase of TREM2 in CSF is attributed to enhanced shedding activity. Soluble TREM2 is a CSF biomarker for microglia activation and antibody-mediated attenuation of soluble TREM2 shedding appears a promising therapeutical approach (43). Other glial-derived CSF protein fragments found in the present study and known to be shed from the cell surface by ADAM10 and ADAM17 are soluble forms of CSF1, LAG3, and LRP1 (44–46).
Microglia activation states have been tried to be classified into DAM or activated response microglia phenotypes based on cellular transcriptomic analysis (8, 39). Moreover, microglial Aβ response proteins have been identified (47). However, such microglia subpopulations were not only reported in mouse models of cerebral β-amyloidosis but also in tauopathies, amyotrophic lateral sclerosis, multiple sclerosis, or healthy aging (8, 39, 47–56). The increased abundance of many DAM markers in the CSF of A30P-αS mice now suggests a DAM-like phenotype also in α-synucleinopathies. The same is true for the recently described amyloid plaque-induced gene signature (57) that also shows great overlap. Notably, microglia have been reported to modulate the cell-to-cell propagation of αS lesions and seeding-prone αS species have been isolated from microglia-derived exosomes in CSF of patients with α-synucleinopathies (23, 58–60).
While our proteomic analysis reveals a CSF signature of microglial activation associated with both Aβ and αS lesions, it is important to note that the microglial secretome shows distinct overlap with the secretome of astrocytes (61–63) (Datasets S1B and S2B). APOE, CSF1, CTSD, CTSB, CTSL, and TIMP2 all found to be up-regulated at least in aged A30P-αS mice are also part of the astrocytic secretome (Fig. 7). In analogy to DAM, a concept of disease-associated astrocytes (DAA) has emerged with an overlap of several genes between DAM and DAA (64). In contrast, C4B, SERPINA3N A2M(P), CHI3L1, and LCN2 appear to be more specific for the activation of astrocytes (64, 65). Astrocytosis in response to activated microglia is a common feature of multiple neurodegenerative diseases (66), and vice versa, secreted astrocyte proteins, such as LCN2, have been reported to impact microglial gene expression (67). Both, microgliosis and astrocytosis are distinct and prominent features in aged APPPS1 and A30P-αS mice (19, 22). The finding that both microglia- and astrocyte-associated proteins are increased in CSF makes such markers promising candidates for monitoring onset and progression of gliosis in β-amyloidoses and α-synucleinopathies. Indeed, human α1-antichymotrypsin (SERPIN A3), YKL-40 (CHI3L1), and LCN2 are already considered potential biomarkers for AD and (atypical) PD progression (68–71). Such observations in humans not only corroborate our data but also demonstrate that biomarker candidates identified in the present study have great relevance for human diseases.
Beyond the glial markers described above, several neuronal proteins associated with neurodegeneration were found to increase in the CSF of the two mouse models. Common changes were found for NEFM, ENO2, or GAP43, while intermediate filament proteins (NEFH, NEFL, INA, PRPH), microtubule-associated proteins (MAP1B, MAP1LC3A, MAP2, MAP6, and MAPT), and SNAP25 and RTN4 were predominantly or exclusively detected in aged A30P-αS mice. An increase in SOD1 was only found in the APPPS1 mouse model and SOD1 was recently reported to be elevated also in AD CSF (36). However, before suggesting distinct differences in the molecular cascade of neurodegeneration between the two mouse models, it has to be ruled out that the difference is caused by the more severe neurodegeneration in the A30P-αS mice compared to the less severely affected APPPS1 mice. Such a view is consistent with CSF NfL levels quantified by immunoassays to be more than 10-fold higher in aged A30P-αS compared to APPPS1 mice (12).
Despite the vast majority of proteins being increased in the CSF of aged transgenic compared to non-tg mice, some proteins—CA3, CKM, CRIP1, MYL1, PVALB, and PYGM—were significantly lower in 18-mo-old A30P-αS than in age-matched control mice. All of these proteins, except CRIP1, were shown to be involved in the molecular response to prolonged hypoxia in skeletal muscles (24) and might be related to the locomotor phenotype in aged A30P-αS tg mice (22). CA3 is also expressed and secreted by microglia (63, 72) (Dataset S2B) and gene expressions of CKM and MYL1 were decreased in PD patient-derived dopamine neurons with αS gene triplication (73). Moreover, loss of PVALB+ GABAergic interneurons along with microgliosis was observed in mice overexpressing human wild-type αS (74).
Current LC-MS/MS technologies are limited by the dynamic range of the investigated proteome and often do not reach the sensitivity of state-of-the-art immunoassays for low abundant proteins but provide a more comprehensive picture by relatively quantifying hundreds of proteins. Thus, in the present study only the most abundant CSF proteins could be quantified. Nevertheless, our study revealed an increase of proteins associated with either Aβ or αS lesions and a panel of more than 20 biomarker candidates related to glial activation in models of β-amyloidosis and α-synucleinopathy with the vast majority also being detected in human CSF. The finding that many of these glial proteins are also changed in AD and that changes are dependent on the AD subcohorts, and thus presumably on the disease stages, implies that CSF biomarker changes are bimodal rather than unidirectional (e.g., ref. 75). This is in line with mouse models of β-amyloidosis to represent early AD disease stages, while most AD CSF comes from symptomatic and thus late disease stages including tau pathology and other comorbid lesions.
In conclusion, neuroinflammation is a consistent pathological hallmark of AD and PD and glial cells adopt various functional states. The identified panel of CSF proteins has clinical relevance and the potential to differentiate and monitor neuroinflammatory disease stages in humans.
Materials and Methods
Further details for materials and methods described here are provided in SI Appendix.
Mice.
Hemizygous APPPS1 (Tg[Thy1-APPSw,Thy1-PSEN1*L166P]) (19) and homozygous A30P-αS (Tg[Thy1-SNCA*A30P]) (20) and age-matched non-tg mice were used in this study. Lymphocyte activation gene 3 protein knockout (Lag3 ko) mice (B6.129S2-Lag3tm1Doi/J) (76) were used to validate the specificity of anti-LAG3 antibody (4-10-C9). The experimental procedures were carried out in accordance with the veterinary office regulations of Baden-Wuerttemberg (Germany) and approved by the local Animal Care and Use Committee.
Mouse CSF Sampling.
A standardized protocol for CSF collection was used as described previously (18). The CSF samples were centrifuged at 2,000 × g for 10 min, controlled for blood contamination, aliquoted (5 µL), and finally stored at –80 °C.
Collection and Processing of Mouse Brain Tissue.
After PBS perfusion, brains were removed. One hemisphere was snap-frozen and the other was fixed-frozen and stored at –80 °C. Fresh frozen mouse brain hemispheres were subsequently homogenized in TBS with protease and phosphatase inhibitors. After ultracentrifugation, supernatants were collected as TBS-soluble fractions and pellets were further extracted using TBS supplemented with 1% Triton X-100.
Mouse CSF Proteomics.
Mouse CSF samples were analyzed by non-targeted LC-MS/MS. LFQ of the mouse CSF proteome data was performed using the MaxQuant software package (v1.5.3.12) (77) and Perseus (v1.5.2.6) was used for additional data analysis (78, 79).
Ontological Enrichment Analysis.
DAVID bioinformatics resources 6.8 (david.ncifcrf.gov) (80, 81) were used to functionally annotate the mouse CSF proteome data (Datasets S1C and S2C).
Protein–Protein Interaction Analysis.
Protein–protein interaction analysis was performed using STRING v11.0 (82), including functional enrichment analysis based on the reactome pathway, GO terms, and KEGG annotations.
Quantitative Analysis of Regulated Intramembrane Proteolysis.
The identified peptides of single-pass transmembrane proteins were matched to the respective protein sequences with indication of topological domains using the web-based tool Quantitative Analysis of Regulated Intramembrane Proteolysis (QARIP) (83).
Immunoblotting.
Mouse CSF samples were run on 4–12% Bis-Tris NuPAGE gels and electroblotted onto nitrocellulose membranes. Subsequently, target proteins were marked with 4-10-C9 mouse monoclonal antibody against murine LAG3 (MABF954; Merck Millipore), ST50-02 rabbit monoclonal antibody against lysozyme C (NBP2-67507; Novus Biologicals), or D1C7Y rabbit monoclonal antibody against cathepsin B (XP Rabbit mAb #31718, Cell Signaling Technology) followed by horseradish peroxidase-conjugated secondary antibodies.
Immunohistochemistry.
For immunohistochemistry, 25-µm-thick mouse brain sections were stained with rabbit polyclonal antibody NT12 against Aβ (84) or rabbit monoclonal antibody against pS129-αS (EP1536Y, Abcam).
CSF αS Measurement.
Human CSF αS was measured on a Sector Imager 6000 (Meso Scale Discovery, MSD) using a commercial electrochemiluminescence-linked immunoassay (Cat# K151TGD) according to the manufacturer’s instructions. MSD Discovery Workbench software v2.0 was used for data analysis.
TREM2 Measurement.
Quantification of murine TREM2 was performed on the MesoScale platform using a customized assay, as described previously (15). Samples were measured on an Imager 2400 reader using the Discovery Workbench software v4 (Meso Scale Discovery) according to the manufacturer’s instructions.
Statistical Analysis of Immunoassay Results.
Statistical analyses were done using JMP software (SAS Institute Inc., v15.2). Non-normally distributed variables were logarithmically transformed prior to statistical calculations. Graphpad Prism (v6) was used to generate the graphs. Statistical significance was set at P < 0.05. All values are expressed as means ± SD.
Supplementary Material
Acknowledgments
We thank Anika Bühler, Carina Leibssle, Jörg Odenthal, and Jessica Wagner for experimental assistance, and Henrik Zetterberg (Univ Gothenburg, Sweden) and Ida Pesämaa (German Center for Neurodegenerative Diseases, Munich) for advice with the human proteome studies. This work was made possible by generous funding from the Cure Alzheimer’s Fund (to S.K. and M.J.), from the network Neuroinflammation and Neurodegeneration, State of Baden-Württemberg, Germany (to M.J.), from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy (EXC 2145 SyNergy – ID 390857198, to S.F.L. and C.H.) and through consortium FOR2290 (263531414, to S.F.L.), and from the Bundesministerium für Bildung und Forschung (JPND PMG AD, to S.F.L. and C.H., and CLINSPECT-M, FKZ161L0214C, to S.F.L.).
Footnotes
Competing interest statement: C.H. collaborates with Denali Therapeutics Inc., participated on one advisory board meeting of Biogen, received a speaker honorarium from Novartis and Roche, and is chief advisor of ISAR Bioscience. G.K. is a former employee of ISAR Bioscience. M. Staufenbiel is a former employee of Novartis.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2119804119/-/DCSupplemental.
Data Availability
The mouse mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (85) partner repository with the dataset identifiers PXD021322 (86) and PXD021356 (87). Previously published data were used for this work (33, 34).
References
- 1.Ashton N. J., et al. , An update on blood-based biomarkers for non-Alzheimer neurodegenerative disorders. Nat. Rev. Neurol. 16, 265–284 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Zetterberg H., Bendlin B. B., Biomarkers for Alzheimer’s disease-preparing for a new era of disease-modifying therapies. Mol. Psychiatry 26, 296–308 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blennow K., Biscetti L., Eusebi P., Parnetti L., Cerebrospinal fluid biomarkers in Alzheimer’s and Parkinson’s diseases-From pathophysiology to clinical practice. Mov. Disord. 31, 836–847 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Llorens F., et al. , Cerebrospinal α-synuclein in α-synuclein aggregation disorders: Tau/α-synuclein ratio as potential biomarker for dementia with Lewy bodies. J. Neurol. 263, 2271–2277 (2016). [DOI] [PubMed] [Google Scholar]
- 5.De Strooper B., Karran E., The cellular phase of Alzheimer’s disease. Cell 164, 603–615 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Smajić S.et al., . Single-cell sequencing of human midbrain reveals glial activation and a Parkinson-specific neuronal state. Brain 145, 964–978 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardy J., Escott-Price V., Genes, pathways and risk prediction in Alzheimer’s disease. Hum. Mol. Genet. 28 (R2), R235–R240 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Keren-Shaul H., et al. , A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290.e17 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Galasko D., Expanding the repertoire of biomarkers for Alzheimer’s disease: Targeted and non-targeted approaches. Front. Neurol. 6, 256 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chesselet M. F., Carmichael S. T., Animal models of neurological disorders. Neurotherapeutics 9, 241–244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jucker M., The benefits and limitations of animal models for translational research in neurodegenerative diseases. Nat. Med. 16, 1210–1214 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Bacioglu M., et al. , Neurofilament light chain in blood and CSF as marker of disease progression in mouse models and in neurodegenerative diseases. Neuron 91, 56–66 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Barten D. M., et al. , Tau transgenic mice as models for cerebrospinal fluid tau biomarkers. J. Alzheimers Dis. 24 (suppl. 2), 127–141 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Kaeser S. A., et al. , CSF p-tau increase in response to Aβ-type and Danish-type cerebral amyloidosis and in the absence of neurofibrillary tangles. Acta Neuropathol. 143, 287–290 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleinberger G., et al. , The FTD-like syndrome causing TREM2 T66M mutation impairs microglia function, brain perfusion, and glucose metabolism. EMBO J. 36, 1837–1853 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maia L. F., et al. , Changes in amyloid-β and Tau in the cerebrospinal fluid of transgenic mice overexpressing amyloid precursor protein. Sci. Transl. Med. 5, 194re2 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Maia L. F., et al. , Increased CSF Aβ during the very early phase of cerebral Aβ deposition in mouse models. EMBO Mol. Med. 7, 895–903 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schelle J., et al. , Prevention of tau increase in cerebrospinal fluid of APP transgenic mice suggests downstream effect of BACE1 inhibition. Alzheimers Dement. 13, 701–709 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Radde R., et al. , Abeta42-driven cerebral amyloidosis in transgenic mice reveals early and robust pathology. EMBO Rep. 7, 940–946 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kahle P. J., et al. , Subcellular localization of wild-type and Parkinson’s disease-associated mutant alpha-synuclein in human and transgenic mouse brain. J. Neurosci. 20, 6365–6373 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye L., et al. , Aβ seeding potency peaks in the early stages of cerebral β-amyloidosis. EMBO Rep. 18, 1536–1544 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neumann M., et al. , Misfolded proteinase K-resistant hyperphosphorylated alpha-synuclein in aged transgenic mice with locomotor deterioration and in human alpha-synucleinopathies. J. Clin. Invest. 110, 1429–1439 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanriöver G., et al. , Prominent microglial inclusions in transgenic mouse models of α-synucleinopathy that are distinct from neuronal lesions. Acta Neuropathol. Commun. 8, 133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capitanio D., et al. , TCA cycle rewiring fosters metabolic adaptation to oxygen restriction in skeletal muscle from rodents and humans. Sci. Rep. 7, 9723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bereczki E., et al. , Synaptic proteins in CSF relate to Parkinson’s disease stage markers. NPJ Parkinsons Dis. 3, 7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brinkmalm A., et al. , SNAP-25 is a promising novel cerebrospinal fluid biomarker for synapse degeneration in Alzheimer’s disease. Mol. Neurodegener. 9, 53 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jurewicz A., Matysiak M., Raine C. S., Selmaj K., Soluble Nogo-A, an inhibitor of axonal regeneration, as a biomarker for multiple sclerosis. Neurology 68, 283–287 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Hickman S. E., et al. , The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 16, 1896–1905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tay T. L., et al. , A new fate mapping system reveals context-dependent random or clonal expansion of microglia. Nat. Neurosci. 20, 793–803 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Thomas D. M., Francescutti-Verbeem D. M., Kuhn D. M., Gene expression profile of activated microglia under conditions associated with dopamine neuronal damage. FASEB J. 20, 515–517 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y., et al. , An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deczkowska A., et al. , Disease-associated microglia: A universal immune sensor of neurodegeneration. Cell 173, 1073–1081 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Higginbotham L., et al. , Integrated proteomics reveals brain-based cerebrospinal fluid biomarkers in asymptomatic and symptomatic Alzheimer’s disease. Sci. Adv. 6, eaaz9360 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tijms B. M., et al. ; Alzheimer’s Disease Neuroimaging Initiative (ADNI), Pathophysiological subtypes of Alzheimer’s disease based on cerebrospinal fluid proteomics. Brain 143, 3776–3792 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guldbrandsen A., et al. , CSF-PR 2.0: An interactive literature guide to quantitative cerebrospinal fluid mass spectrometry data from neurodegenerative disorders. Mol. Cell. Proteomics 16, 300–309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bader J. M., et al. , Proteome profiling in cerebrospinal fluid reveals novel biomarkers of Alzheimer’s disease. Mol. Syst. Biol. 16, e9356 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hickman S., Izzy S., Sen P., Morsett L., El Khoury J., Microglia in neurodegeneration. Nat. Neurosci. 21, 1359–1369 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kannarkat G. T., Boss J. M., Tansey M. G., The role of innate and adaptive immunity in Parkinson’s disease. J. Parkinsons Dis. 3, 493–514 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sala Frigerio C., et al. , The major risk factors for Alzheimer’s disease: Age, sex, and genes modulate the microglia response to Aβ plaques. Cell Rep. 27, 1293–1306.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerrits E., et al. , Distinct amyloid-β and tau-associated microglia profiles in Alzheimer’s disease. Acta Neuropathol. 141, 681–696 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewcock J. W., et al. , Emerging microglia biology defines novel therapeutic approaches for Alzheimer’s disease. Neuron 108, 801–821 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Schlepckow K., et al. , An Alzheimer-associated TREM2 variant occurs at the ADAM cleavage site and affects shedding and phagocytic function. EMBO Mol. Med. 9, 1356–1365 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlepckow K., et al. , Enhancing protective microglial activities with a dual function TREM2 antibody to the stalk region. EMBO Mol. Med. 12, e11227 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li N., et al. , Metalloproteases regulate T-cell proliferation and effector function via LAG-3. EMBO J. 26, 494–504 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Q., et al. , LRP1 shedding in human brain: Roles of ADAM10 and ADAM17. Mol. Neurodegener. 4, 17 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang J., et al. , Neutrophil and macrophage cell surface colony-stimulating factor 1 shed by ADAM17 drives mouse macrophage proliferation in acute and chronic inflammation. Mol. Cell. Biol. 38, e00103-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sebastian Monasor L., et al. , Fibrillar Aβ triggers microglial proteome alterations and dysfunction in Alzheimer mouse models. eLife 9, e54083 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ajami B., et al. , Single-cell mass cytometry reveals distinct populations of brain myeloid cells in mouse neuroinflammation and neurodegeneration models. Nat. Neurosci. 21, 541–551 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chiu I. M., et al. , A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep. 4, 385–401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Friedman B. A., et al. , Diverse brain myeloid expression profiles reveal distinct microglial activation states and aspects of Alzheimer’s disease not evident in mouse models. Cell Rep. 22, 832–847 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Holtman I. R., et al. , Induction of a common microglia gene expression signature by aging and neurodegenerative conditions: A co-expression meta-analysis. Acta Neuropathol. Commun. 3, 31 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krasemann S., et al. , The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 47, 566–581.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leyns C. E. G., et al. , TREM2 deficiency attenuates neuroinflammation and protects against neurodegeneration in a mouse model of tauopathy. Proc. Natl. Acad. Sci. U.S.A. 114, 11524–11529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mrdjen D., et al. , High-dimensional single-cell mapping of central nervous system immune cells reveals distinct myeloid subsets in health, aging, and disease. Immunity 48, 380–395.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Olah M., et al. , A transcriptomic atlas of aged human microglia. Nat. Commun. 9, 539 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Safaiyan S., et al. , White matter aging drives microglial diversity. Neuron 109, 1100–1117.e10 (2021). [DOI] [PubMed] [Google Scholar]
- 57.Chen W. T., et al. , Spatial transcriptomics and in situ sequencing to study Alzheimer’s disease. Cell 182, 976–991.e19 (2020). [DOI] [PubMed] [Google Scholar]
- 58.George S., et al. , Microglia affect α-synuclein cell-to-cell transfer in a mouse model of Parkinson’s disease. Mol. Neurodegener. 14, 34 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olanow C. W., Savolainen M., Chu Y., Halliday G. M., Kordower J. H., Temporal evolution of microglia and α-synuclein accumulation following foetal grafting in Parkinson’s disease. Brain 142, 1690–1700 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Guo M., et al. , Microglial exosomes facilitate α-synuclein transmission in Parkinson’s disease. Brain 143, 1476–1497 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jeon H., Lee S., Lee W. H., Suk K., Analysis of glial secretome: The long pentraxin PTX3 modulates phagocytic activity of microglia. J. Neuroimmunol. 229, 63–72 (2010). [DOI] [PubMed] [Google Scholar]
- 62.Suk K., Combined analysis of the glia secretome and the CSF proteome: Neuroinflammation and novel biomarkers. Expert Rev. Proteomics 7, 263–274 (2010). [DOI] [PubMed] [Google Scholar]
- 63.Tüshaus J., et al. , An optimized quantitative proteomics method establishes the cell type-resolved mouse brain secretome. EMBO J. 39, e105693 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Habib N., et al. , Disease-associated astrocytes in Alzheimer’s disease and aging. Nat. Neurosci. 23, 701–706 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zamanian J. L., et al. , Genomic analysis of reactive astrogliosis. J. Neurosci. 32, 6391–6410 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liddelow S. A., et al. , Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jang E., et al. , Secreted protein lipocalin-2 promotes microglial M1 polarization. FASEB J. 27, 1176–1190 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Craig-Schapiro R., Fagan A. M., Holtzman D. M., Biomarkers of Alzheimer’s disease. Neurobiol. Dis. 35, 128–140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Craig-Schapiro R., et al. , YKL-40: A novel prognostic fluid biomarker for preclinical Alzheimer’s disease. Biol. Psychiatry 68, 903–912 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Magdalinou N. K., et al. , Identification of candidate cerebrospinal fluid biomarkers in parkinsonism using quantitative proteomics. Parkinsonism Relat. Disord. 37, 65–71 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Nielsen H. M., et al. , Plasma and CSF serpins in Alzheimer disease and dementia with Lewy bodies. Neurology 69, 1569–1579 (2007). [DOI] [PubMed] [Google Scholar]
- 72.Nógrádi A., Differential expression of carbonic anhydrase isozymes in microglial cell types. Glia 8, 133–142 (1993). [DOI] [PubMed] [Google Scholar]
- 73.Lin M., et al. , In Parkinson’s patient-derived dopamine neurons, the triplication of α-synuclein locus induces distinctive firing pattern by impeding D2 receptor autoinhibition. Acta Neuropathol. Commun. 9, 107 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Torres E. R. S., et al. , Alpha-synuclein pathology, microgliosis, and parvalbumin neuron loss in the amygdala associated with enhanced fear in the Thy1-aSyn model of Parkinson’s disease. Neurobiol. Dis. 158, 105478 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suárez-Calvet M., et al. , sTREM2 cerebrospinal fluid levels are a potential biomarker for microglia activity in early-stage Alzheimer’s disease and associate with neuronal injury markers. EMBO Mol. Med. 8, 466–476 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miyazaki T., Dierich A., Benoist C., Mathis D., Independent modes of natural killing distinguished in mice lacking Lag3. Science 272, 405–408 (1996). [DOI] [PubMed] [Google Scholar]
- 77.Cox J., et al. , Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cox J., Mann M., 1D and 2D annotation enrichment: A statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics 13 (suppl. 16), S12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tyanova S., et al. , The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 80.Huang W., Sherman B. T., Lempicki R. A., Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 82.Szklarczyk D., et al. , STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47 (D1), D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ivankov D. N., et al. , QARIP: A web server for quantitative proteomic analysis of regulated intramembrane proteolysis. Nucleic Acids Res. 41, W459–W464 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eisele Y. S., et al. , Peripherally applied Abeta-containing inoculates induce cerebral beta-amyloidosis. Science 330, 980–982 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Perez-Riverol Y., et al. , The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47 (D1), D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.S. A. Müller, S. F. Lichtenthaler, Cerebrospinal Fluid (CSF) Proteomics of APPPS1 mice. PRIDE (Proteomics IDEntifications Database). https://www.ebi.ac.uk/pride/archive/projects/PXD021322/. Deposited 4 September 2020. [Google Scholar]
- 87.S. A. Müller, S. F. Lichtenthaler, Cerebrospinal Fluid (CSF) Proteomics of A30P-αS mice. PRIDE (Proteomics IDEntifications Database). https://www.ebi.ac.uk/pride/archive/projects/PXD021356/. Deposited 4 September 2020. [Google Scholar]
- 88.Li N., Workman C. J., Martin S. M., Vignali D. A., Biochemical analysis of the regulatory T cell protein lymphocyte activation gene-3 (LAG-3; CD223). J. Immunol. 173, 6806–6812 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mouse mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (85) partner repository with the dataset identifiers PXD021322 (86) and PXD021356 (87). Previously published data were used for this work (33, 34).