Significance
The intricate membranous network inside cells, with the endoplasmic reticulum (ER) at its center, allows coordinated cellular function by transmitting information across membrane contact sites (MCSs) that form between diverse kinds of cellular organelles and the ER. Unfortunately, these submicroscopic MCSs remain poorly understood and difficult to visualize and manipulate inside cells. We developed an in vitro assay to form MCSs between lipid droplets and an ER-mimicking microsomal membrane purified from rat liver. An optical trap is used to demonstrate physical tethering at the MCS, which changes dramatically in response to metabolic state and immune activation under control of Rab18 GTPase and phosphatidic acid. This assay can potentially be adapted to understand abnormal MCS formation in various disorders.
Keywords: lipid droplets, membrane contact sites, optical trap, supported lipid bilayer, Rab18
Abstract
Coordinated cell function requires a variety of subcellular organelles to exchange proteins and lipids across physical contacts that are also referred to as membrane contact sites. Such organelle-to-organelle contacts also evoke interest because they can appear in response to metabolic changes, immune activation, and possibly other stimuli. The microscopic size and complex, crowded geometry of these contacts, however, makes them difficult to visualize, manipulate, and understand inside cells. To address this shortcoming, we deposited endoplasmic reticulum (ER)-enriched microsomes purified from rat liver or from cultured cells on a coverslip in the form of a proteinaceous planar membrane. We visualized real-time lipid and protein exchange across contacts that form between this ER-mimicking membrane and lipid droplets (LDs) purified from the liver of rat. The high-throughput imaging possible in this geometry reveals that in vitro LD–ER contacts increase dramatically when the metabolic state is changed by feeding the animal and also when the immune system is activated. Contact formation in both cases requires Rab18 GTPase and phosphatidic acid, thus revealing common molecular targets operative in two very different biological pathways. An optical trap is used to demonstrate physical tethering of individual LDs to the ER-mimicking membrane and to estimate the strength of this tether. These methodologies can potentially be adapted to understand and target abnormal contact formation between different cellular organelles in the context of neurological and metabolic disorders or pathogen infection.
Lipid droplets (LDs) are triglyceride-rich cellular organelles that act as an energy depot, a reservoir for membrane biosynthesis, and a location where proteins are sequestered in response to specific stimuli (1–3). These functions require LDs to be dynamic, wherein the LDs exchange proteins and lipids with the endoplasmic reticulum (ER), mitochondria, peroxisomes, endosomes, and other organelles via physical contacts (4, 5). Abnormal LD–organelle interactions are implicated in neurological disease, metabolic disorder, fatty liver, and pathogen infection (5–7). Multispectral imaging showed that LD–ER contacts are the most promiscuous, with the ER contacting a majority of LDs (∼85%) irrespective of their location inside the cell (8). Accordingly, many researchers have used cell culture models to extract a wealth of information on the factors that sustain ER–LD contacts (1, 9–11). Most of these assays aim to understand the biogenesis of LDs, which is now well accepted to proceed through a triglyceride-rich, lens-like structure that expands in the ER lumen, eventually forming mature LDs on the cytosolic side of the ER lumen. Proteins such as seipin, sorting nexins (Snx14), and the NRZ-SNARE complex are required to sustain ER–LD contacts during these steps of LD-biogenesis (12–14).
As an alternate line of investigation to these previous studies, we have attempted to address the physiologically relevant mechanisms that catabolize LDs inside mammalian liver to produce very-low-density lipoproteins (VLDL). We proposed that LD catabolism and ER–LD interactions are enhanced several-fold in the liver after feeding the animal (15, 16). Feeding stimulates insulin release, whereafter insulin activates the ARF1-GTPase and phospholipase-D to generate phosphatidic acid (PA) on the LD membrane. PA directly binds to kinesin-1 and recruits this motor to LDs. Kinesin induces rapid transport of LDs along microtubules to peripheral regions of the ER in hepatocytes, where ER–LD contacts likely form for catabolizing LDs and for delivering the resultant triglyceride to the ER lumen for assembling VLDL (15, 16). This entire pathway is toned down when insulin levels drop after fasting, causing ER–LD contacts to diminish and the liver to sequester LDs in a fasted state to protect other organs from lipotoxicity. Indeed, disrupting this homeostatic pathway caused massive accumulation of triglycerides in cardiac and skeletal tissue with potentially harmful consequences (15). In another exciting development, LDs were found to be a hub for immune activation in the liver, wherein the LDs could harbor and deliver antibacterial proteins for killing bacteria (3). It appeared to us that immune activation also promotes intimate ER–LD contacts in the liver (see supplementary figure 6 in ref. 3). We therefore wondered if LDs in liver cells first acquire antibacterial proteins from the ER via ER–LD contacts before these proteins can be used by the LDs to kill bacteria. Notably, this possibility was not explored by Bosch et al. (3). Two very different kinds of stimuli, namely feeding and immune activation, may therefore induce ER–LD contacts in the liver. Because the liver is a key player in lipid metabolism and systemic immune response, we saw an opportunity to understand how ER–LD contacts are modulated in the animal to serve physiologically important functions outside the well-understood and widely studied pathway of LD biogenesis.
To address these questions, we developed an assay to deposit ER-enriched microsomes purified from rat liver, and also from cultured cells, on a coverslip in the form of an ER-mimicking planar membrane hereafter called the microsomal supported lipid bilayer (mSLB). We demonstrate that LDs purified from rat liver adhere to the mSLB via physical contacts, across which lipids and proteins are exchanged. Our results suggest formation of LD–mSLB fusion intermediates that mediate this exchange. In line with the above-discussed findings (3, 15), LD–mSLB contacts are dramatically enhanced after feeding or immune activation, thus bringing out their native-like nature and physiological relevance. Taking advantage of this in vitro assay, we show that the Rab18 GTPase and PA are common molecular players that engineer LD–mSLB contacts after feeding as well as after immune activation. This assay opens up possibilities to understand, in a controlled, in vitro environment, the molecular pathways that sustain LD–ER contacts in processes other than LD biogenesis. If adapted to other organelles, it may provide a tool to interrogate metabolic disorder, fatty liver, pathogen infection, and other human diseases arising out of abnormal organelle-to-organelle contact formation of diverse kinds.
Results
In Vitro Formation of LD–ER Contacts.
The conventional technique of preparing planar supported lipid bilayers (SLBs) employs synthetic liposomes that are prepared using purified lipids in the absence of proteins (17). In order to replicate a more native-like membrane that contains proteins and lipids, we replaced synthetic liposomes with ER-enriched microsomal vesicles isolated from rat liver or also from cultured cells; these are mSLBs. The purity of microsomal preparations was confirmed (SI Appendix, Fig. S1A) as described previously (15). BODIPY-C12 was chosen as a probe to visualize the mSLBs because this fluorescent fatty acid analog incorporates spontaneously into membranes and also into the hydrophobic core of LDs (18). Microsomes were first doped with trace amounts of BODIPY-C12 and then introduced into a flow cell made by sticking a hydrophilic plasma-cleaned coverslip to a slide with double-stick tape (Fig. 1A). Excess microsomes were washed off, and the surface of the flow cell was imaged on a confocal microscope. A uniform layer of microsomal membrane labeled with BODIPY-C12 was observed, suggesting the formation of a membranous structure on the coverslip (Fig. 1A; red pseudocolor). The ER-marker protein calnexin (SI Appendix, Fig. S1B) was detected on the mSLB, suggesting that membrane-bound proteins of the ER may be retained on the mSLB. Fluorescence recovery after photobleaching (FRAP) of BODIPY-C12 confirmed that the mSLB is a continuous membrane with a high mobility of lipids (SI Appendix, Fig. S1 C and D), as also reported for artificial membrane tubes (19). The FRAP experiment was also done for protein-free conventional SLBs prepared using phosphatidylcholine (PC; SI Appendix, Fig. S1 E and F). The diffusion coefficient of BODIPY-C12 in mSLBs (2.3 ± 0.17 μm2 sec−1) was lower than in PC-SLBs (3.0 ± 0.32 μm2 sec−1), as shown in SI Appendix, Fig. S1G. Slower diffusion in the mSLB may be attributed to the presence of ER-associated proteins that act as a diffusion barrier.
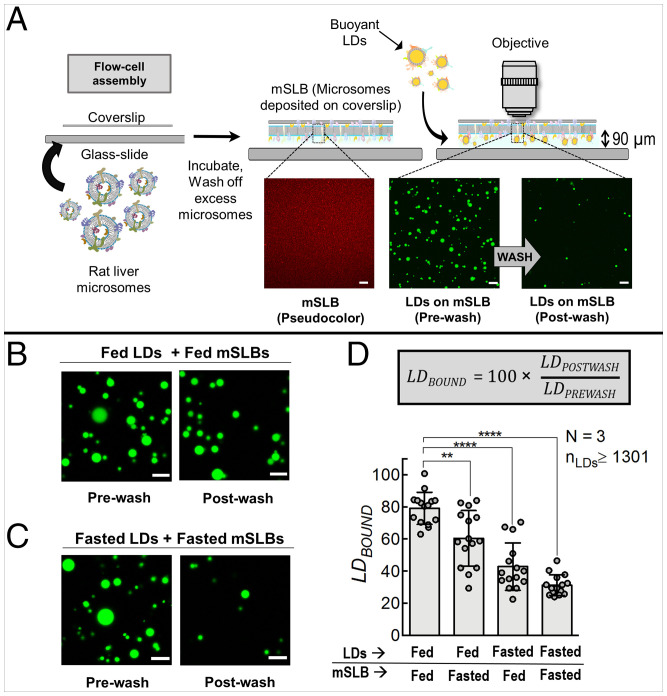
Fig. 1.
In vitro reconstitution of LD–mSLB contacts that are sensitive to the fed or fasted state of the animal. A) Schematic depicting formation of ER-mimicking mSLB. Microsomes are prepared from rat liver, sonicated, and introduced inside a chamber containing a plasma-cleaned coverslip. Spontaneous fusion of microsomes to the coverslip leads to formation of a continuous mSLB. The Bodipy-C12–labeled mSLB is shown in red pseudocolor to distinguish it from green LDs. Excess microsomes are washed off before introducing LDs into the chamber. Due to their buoyancy, LDs float up and come in contact with mSLBs. A fraction of the LDs bind stably to the mSLB. Unbound or transiently bound LDs are washed off after incubation (see prewash and postwash LD images). B) Representative images of fed-LDs (green) incubated with fed-mSLB. Prewash and postwash images are shown. C) Representative images of fasted-LDs (green) incubated with fasted-mSLB. Prewash and postwash images are shown. Scale bar, 10 μm. D) Quantitation of LDBOUND under different metabolic states. Each point on the graph represents the LDBOUND calculated from a single field of view (area, 18,090 µm2) containing multiple LDs. Data represent mean ± SD. ****P < 0.0001, **P = 0.003 using Mann–Whitney test. All scale bars, 10 μm. N, the number of independent pair of animals used in different experiments; nLDs, total number of LDs observed.
We next purified LDs from the liver of normally fed rats (fed-LDs), as we described in previous work (15, 16, 20). Purity of LDs was verified using an antibody against perilipin-2, a well-known marker for hepatic LDs (SI Appendix, Fig. S1A; last lane). Fed-LDs were added to mSLBs prepared from liver of fed rat (fed-mSLBs) in a flow cell, followed by incubation for 15 min in the orientation shown in Fig. 1A (coverslip on top). This geometry allowed buoyant LDs to float up and make contact with the mSLB on the coverslip. We then imaged the flow cell looking down through an upright confocal microscope, which allowed us to detect the transfer of lipids after contact formation and onward. An upright confocal is needed only when the initial LD–mSLB contact and earliest events following contact are to be imaged. When this is not necessary, an inverted confocal microscope will suffice.
To estimate the probability of LD–mSLB contact formation, LDs were first imaged after the incubation (Fig. 1A; prewash). The flow cell was then washed to remove the unbound LDs. LDs still retained on the mSLB were again imaged (Fig. 1A; postwash) with the assumption that these LDs have made stable LD–mSLB contacts. LD numbers were counted in prewash (LDPREWASH) and postwash (LDPOSTWASH) images. We then calculated LDBOUND (= 100 × LDPOSTWASH/LDPREWASH) to measure the efficiency of forming stable LD–mSLB contacts. Upon flowing in a dilution series of LDs onto mSLBs, LDPREWASH was expectedly lower for dilute samples (SI Appendix, Fig. S2A); however, LDBOUND was unchanged (SI Appendix, Fig. S2B). The use of this ratio (i.e., LDBOUND) therefore makes our estimate of LD–mSLB contacts largely independent of variation in LD numbers across samples. It may therefore be expected that changes in LDBOUND, if any, could reflect modulations in ER–LD binding affinity arising from cellular pathways that are modulated in response to specific stimuli.
Smaller LDs that experience a lower buoyant force would float up more slowly across the flow-cell chamber (vertical distance = thickness of double-stick tape [∼90 µm]; Fig. 1A). The diameter of fed-LDs is 1.96 ± 0.76 µm (21). Calculation of terminal velocity shows that an average-sized LD at the bottom of the chamber will reach the mSLB in approximately 7 min, well within the 15-min incubation time. The smallest LD (within 1 SD) at the bottom of the chamber would complete this journey in approximately 17 min. To avoid possible degradation of proteins or lipids over longer incubation, a 15-min incubation period was chosen. Longer incubations did not increase LDPREWASH significantly. The buoyant density of the organelle and dimensions of the flow cell should be kept in mind and the incubation period adjusted accordingly for other organelles and/or flow-cell types.
LD–mSLB Contact Formation Is Sensitive to Fed and Fasted States and Immune Activation.
We next used the liver of rats that had been fasted for 16 h to prepare LDs (fasted-LDs) and mSLBs (fasted-mSLBs). Note that fasting according to this procedure causes massive accumulation of LDs (i.e., LD biogenesis) in the liver, with the hepatic LDs containing adipose tissue–derived triglycerides (16, 22). Fed-LD and fasted-LD samples were adjusted to equal optical density by dilution to ensure that they had an approximately equal number of LDs per unit volume (15, 16). As expected because of this normalization, LDPREWASH was comparable between fed-LD and fasted-LD samples (SI Appendix, Fig. S2C). The normalization was further confirmed by measuring equal triglyceride content in fed-LD and fasted-LD samples (SI Appendix, Fig. S2D). Note that purified fed-LDs and fasted-LDs have similar size (SI Appendix, Fig. S2E), as also reported previously (21, 23). Fed-LDs or fasted-LDs were next added to the flow cell containing fed-mSLBs or fasted-mSLBs in four possible combinations and allowed to incubate for 15 min in a humid chamber. This duration was chosen because longer incubations did not change LDBOUND any further and also in order to minimize degradation of proteins and lipids during incubation. The samples were then imaged (Fig. 1 B and C; prewash images [only two combinations shown]). Each flow cell was then washed in an identical manner with equal volume of buffer, and LDs were imaged again (Fig. 1 B and C; postwash images). Significantly more fed-LDs remained bound to fed-mSLBs after the wash (Fig. 1D; LDBOUND, ∼80%), supporting the proposal that ER–LD contacts are enhanced in the fed state to supply triglycerides for VLDL assembly inside hepatocytes (15, 16). We observed a graded reduction of LDBOUND when moving from fed-fed to fasted-fasted combination (Fig. 1D). This observation can be explained if specific factors play a role on both membranes (LD and ER) for ER–LD contact formation, and these factors are reduced or inactivated after fasting.
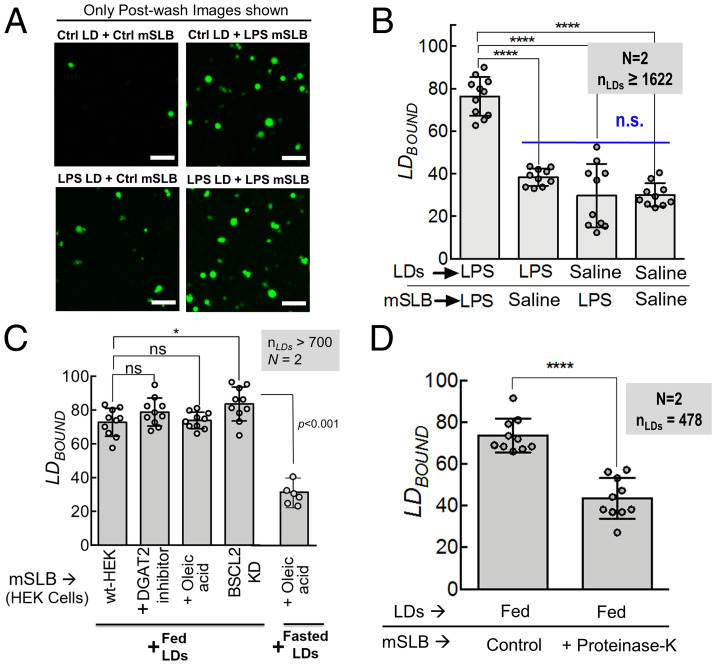
We next activated innate immunity in fasted rats by lipopolysaccharide (LPS) injection (3), followed by LD and mSLB preparation from LPS-injected rats. LDs and mSLB were also prepared from fasted rats injected with saline as a control. These LDs and mSLBs were again assayed in four possible combinations to measure LDBOUND in each case (Fig. 2A; only postwash images shown). Very few LD–mSLB contacts were seen when either LD or mSLB was prepared from control animals (Fig. 2B). This is expected because fasted-LDs show minimal contacts with fasted-mSLB (Fig. 1 C and D). Contact formation was, however, significantly enhanced when both LDs and mSLB were prepared from fasted rats injected with LPS (Fig. 2 A and B). This finding supports the earlier mentioned possibility (based on supplementary figure 6 in ref. 3) that ER–LD contacts are enhanced following immune activation in the liver, perhaps permitting LDs to acquire antibacterial proteins that the LDs then use for killing bacteria.
Fig. 2.
LD–mSLB contacts are sensitive to activation of immune response, but not to LD biogenesis. A) Representative fluorescence micrographs of LDs isolated from control (saline-injected, fasted) and LPS (LPS-injected, fasted) rat liver. LDs are labeled with BODIPY (green). LDs were incubated with mSLBs prepared from control (saline-injected, fasted) or LPS (LPS-injected, fasted) rats and imaged in four possible combinations. The flow cell was washed and LDs were imaged again. Only postwash images are shown. Scale bar, 10 μm. B) LDBOUND calculated as described from experiments in A. Each point on the graph represents the LDBOUND calculated from a single field of view (area, 18,090 μm2) containing multiple LDs. Data represent mean ± SD. ****P < 0.001, Mann–Whitney test. C) Quantitation of LDBOUND for fed-LDs or fasted-LDs binding to mSLBs isolated from HEK293T cells under different treatments. Each point on the graph represents the LDBOUND calculated from a single field of view (area, 18,090 µm2) containing multiple LDs. N represents number of independent experiments. Data represent mean ± SD. Mann–Whitney test. D) Quantitation of LDBOUND for fed-LDs binding to fed-mSLBs treated with proteinase K to digest exposed proteins on the membrane. Each point on the graph represents the LDBOUND calculated from a single field of view containing multiple LDs across independent experiments. Data represent mean ± SD. KD, knockdown; N, the number of independent pair of animals used in different experiments; nLDs, total number of LDs observed; ns, not significant; wt, wild type.
LD-mSLB Contacts Do Not Pertain to LD Biogenesis and Require Protein Factors.
Many researchers have focused on LD biogenesis at ER–LD contacts (1, 11). Although fasting induces massive LD biogenesis in the liver (16, 22), LDBOUND was lowest when fasted-LDs were added to fasted-mSLBs (Fig. 1D). This finding suggests the interesting possibility that LD–mSLB contacts do not pertain to LD biogenesis. To test this further, we prepared mSLBs from 1) untreated HEK-293T cells; 2) cells previously loaded with oleic acid to induce LD biogenesis; 3) cells treated with an inhibitor of diacylglycerol acyltransferase-2, an ER-associated enzyme required for triglyceride synthesis in the ER lumen; and 4) cells treated with short hairpin RNA (shRNA) against Seipin/Bscl2, the canonical tether that sustains ER–LD contacts during LD biogenesis (SI Appendix, Fig. S3A for seipin knockdown). LDs were introduced over flow cells containing different kinds of mSLBs, and LDBOUND was determined for each case (Fig. 2C). Fed-LDs formed contacts efficiently with the mSLB even in the presence of diacylglycerol acyltransferase (DGAT) inhibitor or after Seipin knockdown. Therefore, LD-mSLB contacts likely do not pertain to LD biogenesis because they are insensitive to generation of nascent LDs or to presence of seipin in the ER membrane. Accordingly, fasted-LDs also did not form contacts even after cells were loaded with oleic acid (Fig. 2C).
From these experiments, it appears that the mSLB assay may provide an opportunity to identify novel molecular players that engineer ER–LD contacts outside of the widely studied pathway of LD biogenesis. Before investigating further, we tested if protein factors are, indeed, involved in LD–mSLB contact formation. To do this, we digested proteins that are exposed to the solution on the mSLB, using proteinase K. Proteinase K–containing solution was introduced in the chamber containing fed-mSLBs and incubated at 37 °C for 90 min before being washed off with 1× phosphate-buffered saline (PBS). For control mSLBs, 1× PBS was used during incubation. Fed-LDs were then added to treated or control mSLBs. Proteinase K treatment reduced LDBOUND, suggesting that protein factors on the mSLB play a role in forming LD–mSLB contacts (Fig. 2D). Next, we attempted to identify such a protein factor.
Rab18 and PA Promote LD–mSLB Contacts after Feeding.
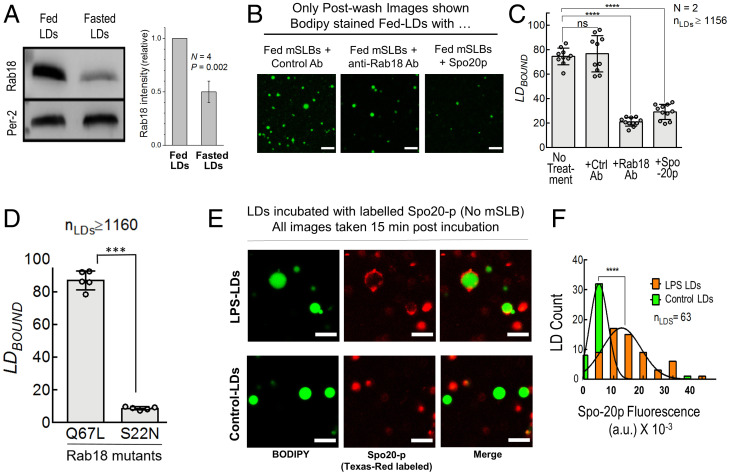
The Rab18 GTPase is present on the ER and LDs, associating with the NRZ-SNARE complex to form ER–LD contacts (24). The function of Rab18 appears controversial because some authors suggest that Rab18 aids LD growth (24), whereas others suggest that Rab18 functions in lipolysis and LD catabolism (25, 26) and has no role in LD biogenesis (27). Because LDs are catabolized with high efficiency at the ER membrane in hepatocytes in the fed state (15, 16), it is possible that Rab18 promotes ER–LD contacts toward catabolism of LDs for VLDL production. Indeed, Rab18 levels were dramatically enhanced on fed-LDs (Fig. 3A), suggesting that Rab18 appears on hepatic LDs when they are being catabolized (i.e., in the fed state), and not when they are being generated (i.e., in the fasted state). Fed-LD to fed-mSLB contacts were inhibited by an Rab18 antibody (Fig. 3 B and C), supporting that Rab18 facilitates ER–LD contacts in hepatocytes in the fed state. Incubating the Rab18 antibody only with fed-LDs, or only with fed-mSLB, or with both caused a graded reduction in LDBOUND (SI Appendix, Fig. S3B). This observation suggests a function of Rab18 on both LD and ER membranes during LD–ER interactions. To investigate the GTP-dependence of Rab18 in this context, mSLBs were prepared from microsomes of cells overexpressing constitutively active (Q67L; a GTP mimic) or inactive (S22N; a GDP mimic) mutants of Rab18 (28). Both forms of overexpressed Rab18 were equally abundant on microsomes (SI Appendix, Fig. S3C). When fed-LDs were added to these mSLBs, LDBOUND was significantly higher on mSLBs containing Rab18-Q67L (Fig. 3D). Therefore, ER–LD contacts are likely promoted in the fed state by GTP-bound Rab18 to induce LD catabolism in the liver, as also evidenced by increase of Rab18 on fed-LDs (Fig. 3A).
Fig. 3.
Rab18 and PA regulate metabolic and immune-dependent LD–mSLB contacts. A) Immunoblot for Rab18 and perilipin-2 (LD marker; loading control) on fed and fasted LDs isolated from rat liver. Graph at right quantifies Rab18 levels on LDs. Rab18 intensity on fed-LDs was always taken equal to 1. Result of a one-sample t test is shown across four independent experiments. B) Representative images of postwash fed-LDs added to fed-mSLB treated with anti-FLAG–IgG antibody (control), anti-Rab18 antibody, or Spo20p protein. LDs are labeled in green. Scale bar, 10 μm. C) LDBOUND for fed-LDs bound to fed-mSLB under different conditions. Each point on the graph represents the LDBOUND calculated from a single field of view (area, 18,090 µm2) containing multiple LDs across independent experiments. Data represent mean ± SD. ****P < 0.001, Mann–Whitney test. D) LDBOUND for fed-LDs on mSLBs prepared from COS7 cells overexpressing active (Q67L; a GTP mimic) or inactive (S22N; a GDP mimic) mutants of Rab18. Each point on the graph represents the LDBOUND calculated from a single field of view (area, 18,090 μm2) containing multiple LDs across independent experiments. Data represent mean ± SD. ***P < 0.001, Mann–Whitney test. E) Representative fluorescence micrographs of Texas-Red maleimide–labeled Spo20p binding to LDs isolated from the liver of control or LPS-treated rats. Images are taken 15-min postincubation. LDs were incubated with labeled Spo20p prepared from fasted control or LPS-injected rats. LDs labeled with BODIPY are in green. Spo20p is fluorescently tagged with Texas-Red C-2 maleimide (red). Scale bar, 5 μm. F) Frequency distribution of Spo20p fluorescence intensity on LPS-treated (orange bars; mean ± SD, 12,700 ± 6,521) and control (green bars; mean ± SD, 5,014 ± 3,022) LDs. Black trace represents Gaussian fit. ****P < 0.0001, Student’s t test. Ab, antibody; a.u., arbitrary unit; N, the number of independent pair of animals used in different experiments; nLDs, total number of LDs observed; ns, not significant.
We showed that PA on the LD directly binds to kinesin-1 to recruit this motor to LDs (15, 16). We also found that Rab18-containing microsomal vesicles associate strongly with PA-containing artificial LDs (ALDs), causing Rab18 to be transferred from microsomes to the ALD membrane (15). It is therefore possible that Rab18 and PA function together for ER–LD contact formation. The yeast SNARE protein Spo20p binds to PA with very high affinity (29, 30), and by doing so can displace other proteins (e.g., kinesin) that are bound to PA on LDs with relatively lower affinity (15). Treatment of fed-LDs with Spo20p-GST significantly reduced LD–mSLB contacts (Fig. 3 B and C). This observation may suggest that Spo20p binds to PA on the LD membrane, thereby removing or blocking certain LD-bound proteins that are needed for ER–LD contact formation.
Rab18 and PA Also Promote LD–mSLB Contacts after Immune Activation.
As noted, immune activation appears to promote ER–LD contacts in the liver (3). We speculate that these contacts could result in transfer of antibacterial proteins from ER to LDs, enabling LDs to kill bacteria. Very interestingly, immune activation was also found to enhance Rab18 on LDs (3), which is reminiscent of increased Rab18 on LDs after feeding (Fig. 3A). Because Rab18 is needed for ER–mSLB contacts (Fig. 3 C and D), Rab18 may also have a role in promoting ER–LD contacts after immune activation. The details of this process and identity of other proteins trafficking to LDs from the ER in response to immune activation remain open to investigation. We have reported comprehensive lipidomic profiling of hepatic LDs (15), wherein we found that the PA sensor Spo20p (29, 30) can be used to reliably measure the abundance of PA on purified LDs. We therefore labeled Spo20p with Texas-Red maleimide and incubated the PA sensor with LDs purified from liver of fasted rat (control) and LDs purified from liver of fasted rat injected with LPS. Spo20p labeling was significantly greater on LDs from LPS-treated animals, suggesting enrichment of PA on LDs after immune activation (Fig. 3 E and F). This finding is reminiscent of the enhancement of PA on LDs in the fed state (15). Taken together with the increase of LDBOUND after LPS treatment (Fig. 2 A and B), it appears that two very different biological responses, namely feeding and immune activation, require a molecular pathway involving Rab18 and PA to induce ER–LD contacts in the liver.
Optical Tweezers to Verify Physical Tethering and Measure Force of Single LD–mSLB Contacts.
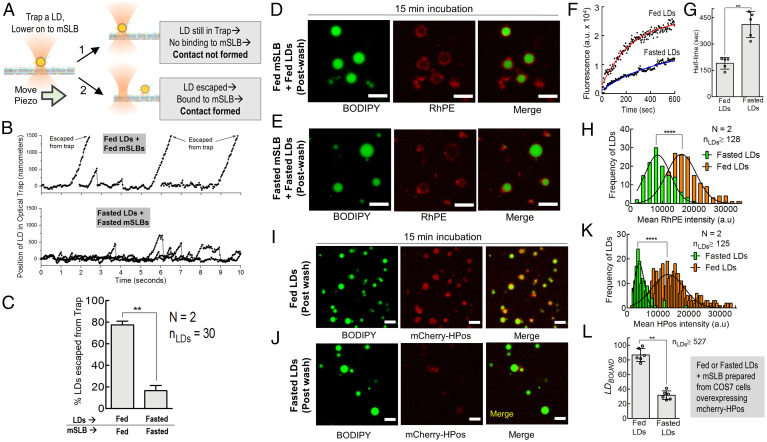
Visual proximity of two organelles in fluorescence images does not guarantee that a physical tether is actually forming at the contact. Furthermore, the strength of individual tethers has not been measured and may provide valuable information to understand the energy scales that control the biophysics of organelle-to-organelle contacts (31). A recent study (32) used hypotonicity to form large vesicles from the ER, permitting ER–LD contacts to be imaged in cells. The study revealed possible ordering of lipid domains at these contacts (32), and the authors suggested the use of optical tweezers to measure the strength of such contacts. However, calibrating the optical tweezer inside cells for an organelle whose size is not precisely known becomes difficult (33). To address this problem, we took advantage of the LD–mSLB assay. We held individual LDs in an optical trap and moved the mSLB underneath using a piezo stage, expecting that LD to mSLB tether formation would displace the LD from the trap center (see the schematic in Fig. 4A). Fig. 4B shows such experiments for fed-LDs on fed-mSLBs, or for fasted-LDs on fasted-mSLBs. In the fed case, most LDs attached to the moving mSLB within 20 s and escaped from the trap (displacement > 1 μm; Movie S1). However, in the fasted case, most LDs exhibited transient attachments and detachments without escapes, revealing weaker LD–mSLB interactions (Fig. 4B and Movie S2). We found that approximately 80% of fed-LDs escaped from the trap (Fig. 4C), in excellent agreement with the finding that LDBOUND was approximately 80% for fed-LDs flowed over fed-mSLB (Fig. 1D). Therefore, approximately 80% of fed-LDs form stable physical contacts with fed-mSLBs. Furthermore, only approximately 20% of fasted-LDs trapped above fasted-mSLBs could escape the trap by forming stable contacts (Fig. 4C). This low value is close to the LDBOUND value of approximately 30% observed for this combination in the flow-cell assay (Fig. 1D) and supports that stable LD–mSLB contacts do not pertain to LD biogenesis (induced by fasting) in the liver.
Fig. 4.
Optical trapping to measure individual ER–LD tether strength and exchange of lipids and proteins across LD–mSLB contact sites. A) Schematic representing ER–mSLB contact formation assay using an optical trap. A single LD (yellow circle) is optically trapped (focused laser beam) and brought in contact with the mSLB adhered to a flow cell. The flow cell is displaced (keeping the trap position invariant) using a piezo stage with a constant velocity of 1 μm/s for a given duration (20 s). If the LD does not bind strongly to the mSLB surface, then it remains in the trap. If the LD binds strongly to the mSLB surface, then it escapes from the trap. B) Position-time plot for fed-LDs and fasted-LDs held in an optical trap on respective mSLB surfaces. Displacement of LDs as a function of time was obtained by video tracking while moving the mSLB with a piezo stage. C) Percentage of LDs escaped from the optical trap. **P < 0.01, Mann–Whitney test. D) Representative postwash image of fed-LDs incubated for 15 min with fed-mSLB. LDs were labeled with BODIPY (green). The mSLB is doped with Rh-PE (red). Scale bar, 10 μm. E) Representative postwash image of fasted-LDs incubated with fasted-mSLB for 15 min. Scale bar, 10 μm. F) Change in Rh-PE fluorescence with time on a single LD of the indicated metabolic state (fed or fasted) bound to mSLB. G) Half-time for LD fusion with mSLB (as detected by Rh-PE fluorescence acquisition on LDs). Data represent mean ± SD. **P = 0.0079, Mann–Whitney test. The average is obtained across five LDs that were used in each case. H) Frequency distribution of Rh-PE-fluorescence intensity on fed (orange bars; mean ± SD, 16,005 ± 4,657 a.u) and fasted LDs (green bars; mean ± SD, 8,525± 3,996 a.u) after incubation with mSLBs doped with Rh-PE. Black trace represents a Gaussian fit. ****P < 0.0001, Student’s t test. I) Representative image of fed-LDs (green) incubated for 15 min with mSLB (red) made from COS7 cells overexpressing mCherry-HPos. J) Representative images of fasted-LDs (green) incubated for 15 min with mSLB (red) made from mCherry-HPos overexpressing COS7 cells. K) Frequency distribution of mCherry-HPos fluorescence intensity on fed (orange bars; mean ± SD, 13,258 ± 5,175), and fasted LDs (green bars; mean ± SD, 3,926 ± 1,563) after incubation with mSLBs prepared from COS7 cells overexpressing mCherry-HPos. Black trace represents Gaussian fit. ****P < 0.0001, Student’s t test. L) LDBOUND for fed-LDs and fasted-LDs over mCherry-HPos–labeled mSLB prepared from COS7 cells. Each point on the graph represents the LDBOUND calculated from a single field of view (area, 18,090 µm2) containing multiple LDs across independent experiments. Data represent mean ± SD. **P < 0.01, Mann–Whitney test. a.u., arbitrary units; N, the number of independent pair of animals used in different experiments; nLDs, total number of LDs observed.
The optical trap works as a spring of certain stiffness. We have developed a method for calibrating this stiffness for LDs of unknown size in vitro, wherein a maximum force of approximately20 pN could be exerted on LDs by the optical trap under our experimental conditions (20). Therefore, the stable fed-LD to fed-mSLB contacts that remain after washing off consist of a physical tether with strength exceeding 20 pN. Fewer stable tethers form between fasted-LDs on fasted-mSLBs, but the ones that do form also have a strength exceeding 20 pN, suggesting that their molecular nature may be similar to the one that is operative in the fed case. Taken together, the optical-trap studies show a binding strength of >20 pN for stable LD–mSLB contacts in fed and fasted states, but stable contacts are significantly reduced in the fasted state.
Stable LD–mSLB Contacts Support Efficient Protein and Lipid Trafficking.
ER–LD contacts that support LD catabolism should allow trafficking of proteins (e.g., lipases) and lipids across the contact site. To test if the LD–mSLB contacts also support exchange of lipids and proteins, a trace amount of rhodamine-PE (Rh-PE) was incorporated into the mSLB by mixing microsomes with Rh-PE-containing liposomes prior to mSLB preparation.
Note that unlike BODIPY-C12, which enters the hydrophobic LD core, Rh-PE would only incorporate into the peripheral phospholipid membrane of bilayer and monolayer vesicles. Rh-PE incorporation was similar for fed and fasted mSLBs (SI Appendix, Fig. S3D). Next, we added fed-LDs or fasted-LDs, respectively, to Rh-PE–labeled fed-mSLBs or fasted-mSLBs. Live imaging showed that fed-LDs acquired rhodamine fluorescence much more rapidly than fasted-LDs (Fig. 4 D–G and Movies S3 and S4). Accordingly, Rh-PE intensity after 15 min of incubation was higher on fed-LDs than fasted-LDs (Fig. 4H). Protein transfer has been observed across ER–LD contacts using the model peptide HPos (18). We therefore prepared mSLBs using microsomes from COS7 cells overexpressing mCherry-HPos, incubated the microsomes with fed-LDs or fasted-LDs, and imaged the LDs after 15 min of incubation (Fig. 4 I and J). Significantly more mCherry-HPos fluorescence could be detected on fed-LDs than on fasted-LDs (Fig. 4K), thus demonstrating efficient transfer of HPos to fed-LDs. Contact formation was also more efficient for fed-LDs on mSLBs prepared from COS7 cells, as evidenced from a higher LDBOUND value (Fig. 4L). Note that Fig. 4 D–L shows lipid and protein exchange only for postwash LDs that are stably bound to the mSLB.
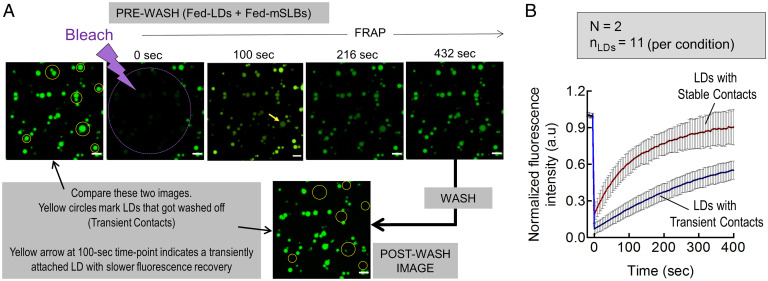
Because LD–mSLB contacts support lipid exchange, we next asked if stable contacts differ from transient contacts in their ability to traffic lipids. Microsomes prepared from liver of fed rat was doped with a trace amount of fluorescent BODIPY-C12 before preparing mSLBs. Fed-LDs were added to this fluorescent mSLB, incubated for 15 min, and then a field of view was bleached using a laser (Fig. 5A). FRAP data of all LDs in this field were recorded until full fluorescence recovery. The flow cell was then washed, causing approximately 20% of LDs to get washed off, as expected (because LDBOUND was ∼80% for fed-LDs flowed over HEK-microsomes; Fig. 2C). A postwash image of the same field of view was then compared with the prewash image to identify LDs that got washed off (Fig. 5A; yellow circles). The prewash FRAP data were now divided into two groups and analyzed: LDs that did not get washed off (stable contacts) and LDs that were washed off (transient contacts). While some LD-to-LD heterogeneity existed, significantly faster recovery was seen in the first group on average (Fig. 5B), suggesting more efficient lipid transfer of BODIPY-C12 from mSLB to LDs across stable contacts, followed by partitioning of BODIPY-C12 into the LD core. We also observed that Rh-PE labeled the membrane of a large fraction of fed-LDs (presumably stable contacts), but a smaller fraction did not acquire detectable amounts of Rh-PE (presumably transient contacts). Representative images of such LDs are shown in SI Appendix, Fig. S4 A–C, and real-time transfer of Rh-PE to fed-LDs can be seen in Movie S5. Therefore, the stable LD–mSLB contacts seen here are functionally different from the transient LD–mSLB contacts that possibly form due to nonspecific interactions.
Fig. 5.
Stable LD–mSLB contacts support efficient trafficking. A) Prewash FRAP of fed-LDs on BODIPY-C12–labeled fed-mSLB. FRAP measurement is followed by a wash. Yellow circles mark LDs that got washed off (compare prebleaching image with postwash image). The arrow in the 100-s image shows a transiently bound LD with slower fluorescence recovery. Scale bar, 10 μm. B) Prewash fluorescence recovery of fed-LDs divided into two groups 1) LDs that got washed off, assumed to have transient contacts’ and 2) LDs that did not get washed off, assumed to have stable contacts. Data are fitted to a recovery equation [blue and red trace; Y=(F0+Fi*(X/Thalf))/(1+(X/Thalf))]. Black dots represent mean and error bars represent SD. Data are the average of 19 different LDs. a.u., arbitrary units.
Artificial LDs and Conventional SLBs Confirm a Role for PA in Contact Formation.
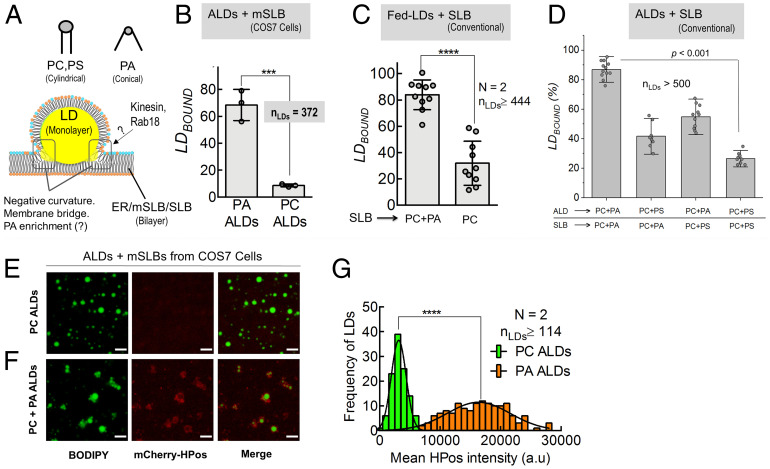
PA is a conical phospholipid (34) that can stabilize the negative curvature needed for forming lipidic bridges between two organelles (see schematic in Fig. 6A). Indeed, increasing PA in cells causes LDs to fuse together (35) and phospholipase-D, an enzyme that converts PC to PA, is enriched at LD–LD contacts (15). A role for PA in monolayer-to-monolayer (LD-to-LD) contacts is therefore expected. However, can PA serve the same role at monolayer-to-bilayer contacts, as depicted in Fig. 6A? This question is interesting because PA is highly enriched on LDs in the liver in fed state, and these LDs likely form contacts with the ER membrane to facilitate LD catabolism (15). Such a function of PA is supported by observing that PA-bound Spo-20p blocks LD–mSLB contacts (Fig. 3C). To test for a function of PA in monolayer-to-bilayer contacts, we prepared ALDs with a phospholipid membrane consisting of only PC or PC+PA (10 mol%). Both kinds of ALDs had similar size (SI Appendix, Fig. S4D). ALDs were then introduced over mSLBs prepared from COS7 cells to find significantly enhanced LDBOUND for PA–ALDs (Fig. 6B). PA is a bioactive phospholipid present on diverse cellular membranes (34). It is therefore possible that PA on the ER membrane also facilitates ER–LD contacts (in addition to PA on LDs). We therefore tested if addition of PA to conventional SLBs assists LD–SLB contacts. Conventional SLBs containing only PC or PC+PA (10 mol%) were deposited onto a glass coverslip. Fed-LDs from liver were then introduced to again find significantly higher LDBOUND on PA–SLBs (Fig. 6C).
Fig. 6.
Role of PA in LD-bilayer contact using ALDs and conventional SLB. A) Upper: Schematic showing lipids with no intrinsic curvature (e.g., PC, PS) or with intrinsic negative curvature (PA; a conical lipid is shown). Lower: - Schematic of a LD-bilayer interface demonstrating the negative-curvature region that could be stabilized by PA. Proteins such as kinesin-1 and Rab18 could be recruited to this interface by PA (speculation). B) LDBOUND for PA+PC ALDs or PC-only ALDs incubated with mSLB prepared from COS7 cells. Each point on the graph represents the LDBOUND calculated from a single field of view (area, 18,090 μm2) containing multiple LDs. More than 500 LDs were observed in these experiments. Data represent mean ± SD. ***P = 0.0009, Mann–Whitney test. C) LDBOUND for fed-LDs on conventional SLBs of the indicated types. Each data point on the graph represents a field of view (area, 18,090 µm2) with ∼100 LDs across multiple experiments. Data represent mean ± SD. ****P < 0.0001, **P = 0.003, Mann–Whitney test. D) LDBOUND for ALDs on conventional SLBs prepared using the indicated lipid types. PA or PS was added to ALDs or SLBs at 10 mol% (PC is 90 mol%). Each data point on the graph represents a field of view (area, 18090 μm2) with ∼100 LDs across multiple experiments. Data represent mean ± SD. E) Representative image of PC–ALDs (green) incubated for 15 min with mCherry-HPos mSLB (red). Scale bar, 10 μm. F) Representative image of PC+PA ALDs (green) incubated for 15 min with mCherry-HPos mSLB (red). Scale bar, 10 μm. G) Frequency distribution of mCherry-HPos-fluorescence intensity on PA (orange bars; mean ± SD, 16,378 ± 5,221 a.u) and PC–ALDs (green bars; mean ± SD, 3,161 ± 1,227 a.u) after incubation with mCherry-HPos mSLB. Black line represents a Gaussian fit. ****P < 0.0001, Student’s t test. a.u., arbitrary units; N, the number of independent pair of animals used in different experiments; nLDs, total number of LDs observed.
Next, we investigated requirements for the conical shape and also the negative headgroup charge of PA in engineering monolayer–bilayer contacts. We introduced ALDs over conventional SLBs, with both membranes having defined lipid composition consisting of (PC + 10 mol% PA) or (PC + 10 mol% phosphatidyl-serine [PS]). PS has a negatively charged headgroup like PA, but unlike PA, PS is a cylindrical phospholipid (31). LDBOUND was highest when both membranes contained PA and was lowest when PA was replaced by PS in both membranes (Fig. 6D). A similar observation was made (SI Appendix, Fig. S4E) when PA was replaced by phosphatidylethanolamine, a phospholipid that is conical like PA but has a zwitterionic head group, unlike the negatively charged PA. Therefore, the ability of PA to engineer monolayer–bilayer contacts even in the absence of proteins (Fig. 6C) likely derives from its conical shape as well as the negative charge in the headgroup. This is an interesting observation because PA also uses the aforesaid combination of biophysical properties for recruiting proteins to membranes with great specificity (34). If PA promotes ER–LD contacts, then does PA also enhance trafficking across these contacts? To test this, we prepared PC or (PC+PA)–ALDs as described earlier and introduced them over mSLBs prepared from COS7 cells overexpressing mCherry-HPos. Significantly more mCherry-HPos was transferred from mSLBs to PA–ALDs after a 15-min incubation (Fig. 6 E–G). We also verified that lipid transfer between LDs and mSLB is bidirectional, by incubating Rh-PE–labeled ALDs (PC or PC+PA) with unlabeled mSLB. Significantly more fluorescence was transferred to the mSLB from PA-containing ALDs (SI Appendix, Fig. S5).
Discussion
Perturbation in LD–organelle contacts causes lipodystrophy and lipid-storage diseases (6, 7). Several researchers have visualized these contacts after loading fatty acids into cells, thus focusing on ER–LD contacts in the context of LD biogenesis (1, 11). However, other pathways including LD catabolism and immune activation could also require LDs to interact with the ER (3, 5, 15). It is not clear if the molecular composition and biophysical properties of ER–LD contacts operative in these pathways are similar to LD biogenesis at the ER. Synthetic droplet–emulsified vesicles revealed interesting biophysical aspects of ER–LD contacts (36), but these cannot incorporate the complex (and often unknown) protein and lipid machinery that sustains diverse kinds of ER–LD contacts inside cells. The LD–mSLB assay developed here provides a first analysis of native-like contacts in vitro. Multiple contacts are observed simultaneously in a single focal plane, making this a high-throughput assay. This planar geometry may also permit future incorporation of microfluidics and total internal reflection fluorescence imaging to dissect the molecular architecture of ER–LD contacts and also to screen rapidly for molecules that can target these contacts.
ER–LD contacts are unique in the sense that the monolayer membrane can be continuous with the bilayer (Fig. 6A). Such membrane bridges (also called membrane contact sites), mostly discussed in the context of LD biogenesis, can support significant protein and lipid exchange, unlike organelle-to-organelle protein tethers (4, 5). A membrane bridge is likely formed at stable LD–mSLB contacts because these stable contacts can sustain efficient mSLB to LD transfer of lipids and proteins, unlike transient contacts (Fig. 5). Notably, LD–mSLB contacts were minimal when LDs and mSLB were prepared from the liver of fasted rat, where LD biogenesis is induced (Fig. 1B). Furthermore, inhibiting DGAT2 or Seipin knockdown had no effect on LD–mSLB formation (Fig. 2C). A possible reason is that we are selecting for mature, triglyceride-rich, buoyant LDs that are highly enriched with perilipin-2 when we purify LDs by centrifugation. We found that purified fed-LDs move rapidly over a long distance along microtubules (16, 20). Therefore, these LDs may have budded off from the ER (1, 11), and the ER-associated molecular machinery that gave birth to the LDs may have been lost or inactivated. It is also possible that the “triglyceride lenses” that appear in the ER membrane to drive LD biogenesis cannot be reproduced in the mSLB.
Nevertheless, and precisely because of the aforementioned reasons, the LD–mSLB assay provides an opportunity to understand novel kinds of contacts that must form between the ER and mature LDs in response to other cellular and physiological requirements. Our findings suggest two common molecular players (Rab18 and PA) that engineer LD–ER contacts for two very different pathways in the liver: feeding induced LD catabolism and immune activation. Rab GTPases are master regulators of vesicle trafficking, with Rab18 consistently detected on LDs. The presence of Rab18 at ER–LD contacts is important for dengue and hepatitis C virus replication (7). Rab18 associates with the NRZ-SNARE complex at ER–LD contacts, but the function of Rab18 in LD biology appears controversial, because some authors suggest that Rab18 aids LD growth (24), whereas others suggest Rab18 functions in lipolysis and LD catabolism (25, 26), and has lesser roles in LD biogenesis (27). Our in vitro results support the latter viewpoint, also pointing toward an interesting lipid (i.e., PA) that may partner Rab18 at ER–LD contacts.
PA is a bioactive signaling lipid vital for membrane remodeling in diverse processes such as vesicle trafficking, membrane fission and fusion, and cell signaling (37). PA is also a conical lipid that promotes negative curvature at organelle-to-organelle contacts (Fig. 6A). Our results suggest that PA may have a central biophysical role in stabilizing monolayer-to-bilayer contacts because it could enhance ALD–SLB contacts in a simplified protein-free system (Fig. 6D). Choudhary et al. (38) calculated the energy of a bilayer membrane harboring an LD, emphasizing how negative-curvature phospholipids such as diacylglycerol and phosphatidylethanolamine favor LD embedding into the bilayer during LD biogenesis. Surprisingly, we find no mention of PA in the context of ER–LD contacts and LD biogenesis. An intriguing possibility is that PA is generated at the ER–LD interface in the liver specifically to aid LD catabolism (e.g., after feeding), but not for LD biogenesis (after fasting). How might this be possible? We found that insulin activates phospholipase-D in the liver in the fed state, thus converting PC into PA. This elevates PA by approximately twofold on fed-LDs, following which kinesin-1 binds to PA and transports LDs to contact the ER (15). The PA on these PA-rich LDs may diffuse along the LD membrane to localize at the ER–LD interface, where PA stabilizes the negative-curvature zone (Fig. 6A). This possibility is supported by increased ALD–SLB contacts after addition of PA (Fig. 6D). It is intriguing how PA, kinesin-1 (15), and Rab18 (Fig. 3A) show a similar enrichment on fed-LDs. PA recruits proteins to membranes with exquisite specificity through a combination of curvature, electrostatics, hydrogen bonding, and hydrophobicity (34). PA can also increase the α-helical content of proteins bound to it (39), a conformational change that could stabilize the protein on a LD membrane (40). It is possible that PA is enriched at the ER–LD interface to stabilize membrane fusion, whereafter PA recruits specific factors (e.g., Rab18) to impart subtle functions to the ER–LD interface that control downstream physiological responses (e.g., LD catabolism, immune response).
We point out that the mSLB assay has limitations because the mSLB–coverslip interface could perturb membrane function. Furthermore, certain asymmetries within the bilayer membrane of the ER may not be reproduced on mSLBs. Nevertheless, we could reproduce certain aspects of native-like ER–LD interactions that were observed in response to fed/fasted transition and immune activation in animals or inside cells (3, 15, 16). Furthermore, blocking the factors (i.e., PA and Rab18) that are important for LD catabolism or immune activation also inhibited LD–mSLB contacts. Because PA can recruit proteins (including motors) with great specificity and can stabilize such proteins at organelle-to-organelle interfaces, it is imperative that we understand how this simplest phospholipid executes such functions downstream of diverse physiological pathways (34, 37). It may be possible to adapt the mSLB assay to interrogate in vitro contacts between mitochondria, peroxisomes, endosomes, ER, and other cellular membranes, providing much-needed molecular insight into neurological, metabolic, and pathogenic diseases (12). Such efforts are in progress in our laboratory.
Methods
Animal Procedures.
Sprague–Dawley rats were bred and maintained by the animal house staff at Tata Institute of Fundamental Research, Mumbai, India. All animal protocols were approved by the Institutional Animal Ethics Committee formulated by the Committee for the Purpose of Control and Supervision of Experiments on Animals in India. Male Sprague–Dawley rats (2–3 mo old) were used. Rats from the same litter were used for a fed-fasted pair. Rats were maintained in a regular 12-h light/dark cycle and fed a standard laboratory chow diet. Fed-group rats had ad libitum access to food and water. Fasted-group rats were fasted for 16 h and had ad libitum access to water. LPS/saline treatment was performed as described (3). Briefly, rats were intraperitoneally injected with 300 μL of saline buffer (control) or 6 mg/kg LPS (final dose, dissolved in saline; L2639, Sigma-Aldrich) and fasted overnight (16 h).
Isolation of Microsomes.
Total rat liver microsomes were isolated from rat liver using a previously standardized protocol, and purity of microsomes was confirmed (SI Appendix, Fig. S1C) as described (41). Sprague–Dawley rats were used according to the following categories: category 1, fed; category 2, fasted for 16 h; or category 3, LPS-injected and saline-injected (as control) and then fasted for 16 h. Rats were anesthetized using sodium thiopentane at 40 mg/kg body weight. Rats were killed and the liver was perfused with 50 mL of cold PBS (1× PBS) through the hepatic portal vein. Liver tissue was weighed in 1× PBS and 9 g of liver tissue was placed in three volumes of 0.25 M sucrose solution having 4 mM dithiothreitol, 8 μg/μL pepstatin, 4 mM phenylmethylsulfonyl fluoride (PMSF) and Roche protease mixture inhibitor. Mincing and homogenization of tissue were carried out in the cold room. The minced tissue was transferred into a 50-mL Potter-Elvehjem tissue grinder (kept on ice) and homogenized using a ribbed Teflon pestle for up to 20 strokes. The homogenate was filtered through cotton mesh (2 layers) using a glass funnel. The homogenates were centrifuged at 8,700g at 4 °C for 15 min to obtain a postnuclear supernatant (PNS). The PNS was spun at 43,000g (Beckman Coulter ultracentrifuge, Type 70 Ti rotor) at 4 °C for 7 min to pellet out mitochondria. The microsomes were pelleted at 110,000g at 4 °C for 60 min. Microsomes were resuspended in 1× PBS, flash-frozen, and stored at −80 °C for further experiments. Microsomes were prepared from cultured cells, following similar methods.
LD Isolation.
LDs from rat liver were isolated using a previously described protocol (15, 16, 20). Briefly, male Sprague–Dawley rats (2–3 mo old and in the aforementioned categories 1–3) were anesthetized (with sodium thiopentone, 40 mg/kg, via intraperitoneal injection). The abdominal cavity was cut open to perfuse the liver through the hepatic portal vein with cold PBS. The perfused liver was dissected, washed, and weighed. The liver was minced and resuspended in 1.5 times weight per volume 0.9 M sucrose containing MEPS buffer and homogenized using Dounce homogenizer at 4 °C. MEPS buffer is composed of 35 mM PIPES (piperazine-N,N′-bis(2-ethanesulfonic acid), 5 mM ethylene glycol tetraacetic acid, and 5 mM MgSO4 at pH 7.1, supplemented with protease inhibitor mixture (Roche), 4 mM PMSF (Sigma), 8 μg/mL pepstatin A (Sigma), and 4 mM dithiothreitol (Sigma). We centrifuged the homogenate at 1,800g for 10 min at 4 °C to obtain PNS. The PNS thus obtained was mixed with 1.5 times volume for volume (vol/vol) 2.5 M sucrose containing MEPS buffer (without PMSF) and was loaded as the bottom layer of sucrose-density gradient. This layer was overlaid with 5 mL (each) of 1.4 M, 1.2 M, 0.5 M, and 0 M sucrose in MEPS buffer. This gradient was centrifuged at 120,000g at 4 °C for 1 h to obtain LDs (the top-most whitish layer). LDs were collected using an 18G needle, flash-frozen, and stored in liquid nitrogen.
Preparation of Liposomes and Small Unilamellar Vesicles.
A mixture of dioleoylphosphatidylcholine (DOPC), egg PA, dioleoylphosphatidylserine, and Rh-PE (Avanti Polar Lipids) in the ratios of 89.5:10:0:0.5 mol% for PA SLB, 99.5:0:0:0.5 mol% for PC SLB, and 89.5:0:10:0.5 mol% for PS SLB lipids was aliquoted in a glass test tube (final concentration, 3 mM) and mixed gently. This chloroform-dissolved lipid mix was dried rapidly under a nitrogen stream and vacuum desiccated for 60 min. The dried lipid film was hydrated in 1× PBS at 50 °C for 30 min. After incubation, the hydrated liposomes were vigorously vortexed for 5 min. The hydrated liposomes were sonicated using a probe sonicator for 5 min at 4 °C to make small unilamellar vesicles (SUVs; Branson SFX250 Sonifier; 15% amplitude, 3-s on and 2-s off cycle). The sample was centrifuged at 20,000g for 10 min, 4 °C to remove large unilamellar vesicles and titanium particles (eroded from the probe).
Conventional SLB and mSLB Preparation.
The procedure for producing conventional SLBs was adapted from previous protocols (17). A glass coverslip was cleaned by piranha (6:4 vol/vol concentrated H2SO4:H2O2) treatment or by oxygen plasma cleaning (Harrick plasma PDC-32G). The coverslip was then assembled in a flow cell using double-sided tape. SUVs, prepared as described in the preceding section, were introduced into the flow cell and allowed to fuse with the hydrophilic surface of the coverslip for 45 min inside a humid chamber. After incubation, excess unbound liposomes were washed off using 1× PBS (200 μL × three washes).
To produce mSLBs, the flow cell was prepared as described earlier in Methods. Total rat liver microsomes (protein concentration, 5 mg/mL) were sonicated using a probe sonicator (Branson SFX250 Sonifier; 15% amplitude, 3-s on and 2-s off cycle) for 5 min at 4 °C in an ice bath. The sample was centrifuged at 20,000g for 10 min to remove aggregated proteins. These sonicated microsomes (protein concentration, 1.5 mg/mL) were labeled with BODIPY-C12 (Invitrogen, catalog no. D3822; 8 μM final probe concentration). Alternatively, microsomes were mixed with sonicated SUVs made from DOPC:Rh-PE liposomes for labeling (DOPC:Rh-PE, 99.5:0.5 mol%, 0.2 mM final concentration). Microsomes were then introduced into the flow cell and allowed to deposit on the cleaned coverslip for 45 min in a humid chamber. After incubation, excess unbound microsomes were washed off using 1× PBS (200 μL × three washes) and the sample was imaged.
LD–mSLB Interaction Assay.
LDs across different preparations were normalized to equal optical density in MEPS buffer. Then, we flowed 200 μL of this solution containing normalized LDs into a flow cell containing an mSLB-coated coverslip stuck to a cover glass by double-stick tape of approximately 90-µm thickness (the height of the flow cell). The buffer was then wicked to establish flow. Care was taken to avoid introduction of bubbles when flowing LDs and washing the flow cell. The flow cell was then incubated with the mSLB side on top (Fig. 1) to allow buoyant LDs to float up and come in contact with the mSLB. This incubation was done for 15 min by keeping the flow cell inside a humid chamber at room temperature. The 15-min incubation allows most LDs to float up across the flow chamber and come in contact with the mSLB. The flow-cell assembly was imaged in an upright confocal microscope to first observe LDs before the wash. Unbound LDs were then washed off using 200 μL 1× PBS, and the assembly was imaged again to observe LDs stably bound to the mSLB.
Preparation of ALDs.
ALDs were prepared by using a freeze-thaw technique as previously described (15). Briefly, 70 μL of glyceryl trioleate and 0.5 μmol egg PC were mixed in acid-washed, clean glass tubes to prepare 1 mL of ALDs. We made PA–ALDs by supplementing this reaction mix with 25 nmol of egg-PA. This mix was also supplemented with trace amounts of Rh-PE (1.6 nmol) to aid imaging and detect lipid exchange. This reaction mixture was dried under nitrogen gas stream for 30 min, followed by vacuum desiccation for 1 h to remove trace amounts of chloroform. After desiccation, 930 μL of HKM buffer (50 mM HEPES-KOH, 120 mM potassium acetate, and 1 mM MgCl2, at pH 7.4) was added to the reaction mix. This reaction mixture was vigorously mixed by vortexing for 10 min to aid emulsification. The whitish emulsion was subjected to five freeze-thaw cycles of flash freezing in liquid nitrogen and thawing at 55 °C. Each cycle was accompanied by vigorous vortexing for 10 min. ALDs were stored in liquid nitrogen and thawed prior to experiments.
Antibody Inhibition Experiment.
mSLBs were prepared as described earlier in Methods and incubated with relevant antibody for 30 min in a humid chamber. After incubation, excess antibody was washed off from mSLBs, LDs were introduced and incubated for 15 min. After incubation, the unbound or weakly bound LDs were washed off and stable contacts LD–mSLB contacts were imaged.
Fluorescent Recovery after Photobleaching.
FRAP analysis was performed with BODIPY-C12. The total scan area (2,025 μm2) and bleached region (6-μm radius) was restricted to obtain high temporal resolution. Images were acquired at 1× optical magnification. An argon laser (488 nm) at 8% power and 150 iterations at 100% transmission were used for photobleaching. Standardization of imaging parameters was performed to minimize photobleaching during acquisition. The mean postbleached intensity was normalized to the mean prebleach intensity. The recovery kinetics (characteristic diffusion time) was analyzed by fitting data to the recovery equation Y = (F0+Fi*(X/thalf))/(1+(X/thalf)), as described previously (42).
Plasmids.
GST-Spo20p (PABD) was a gift from Nicolas Vitale, Université de Strasbourg, Strasbourg, France. HPos overexpression construct was a gift from Albert Pol, Universitat de Barcelona, Barcelona, Spain. GFP-Rab18 (Q67L) and GFP-Rab18 (S22N) were a gift from R. G. Parton, University of Queensland, St. Lucia, Queensland, Australia .
Antibodies.
ADRP antibody (651102) was purchased from PROGEN Biotechnik. Hsp60 antibody (ab46798) was purchased from Abcam. PDI antibody (612117) was purchased from BD Bioscience. Anti-FLAG antibody (F1804) was purchased from Sigma. Rab18 antibody (sc393168) and Calnexin (sc 11397) antibody were purchased from Santa Cruz Biotechnology. S6 ribosomal protein antibody (2317) was brought from Cell Signaling Technologies. Alexa-594–labeled anti-mouse and anti-rabbit secondary antibodies were purchased from Invitrogen. Anti-BSCL2/seipin antibody was purchased from Abcam (ab106793). Anti–histone H3 antibody was purchased from Abcam (ab1791).
Cell Culture, Transfection, and Knockdown.
HEK293T (ATCC catalog no. CRL-3216), and COS-7 (ATCC catalog no. CRL-1651) were used in this study. All types of cells used in this study were maintained in Dulbecco's Modified Eagle Medium (DMEM; Sigma, D7777) containing 10% fetal bovine serum (FBS; Himedia). All cells were maintained in a humidified atmosphere of 5% CO2 in air at 37 °C. Lipofectamine 2000 (Invitrogen) was used for transient plasmid and shRNA transfections per the manufacturer’s instructions.
Seipin/BSCL2 shRNA.
shRNA against human BSCL2 was procured from ShRNA Resource Center at the Division of Biological Science, IISc.
Oleic Acid Loading.
Oleic acid was purchased from Sigma (O1383). HEK293T cells were grown in 6 × 10 cm plates and loaded with 400 μM oleic acid conjugated with fatty acid–free bovine serum albumin (at a molar ratio of 6:1) in complete medium 12 h before collection.
DGAT2 Inhibitor Treatment.
DGAT2 inhibitor (PF-06424439) was purchased from Sigma (catalog no. PZ0233). To block TAG synthesis, HEK293T cells in 6 × 10 cm plates were starved by treatment with low-glucose DMEM (Sigma, catalog no. D5523) without FBS, supplemented with 10 µM DGAT2 inhibitor PF-06424439 for 24 h before collection.
Optical Trapping.
The instrument and techniques for optical trapping have been described in detail (20). Briefly, LDs were observed at room temperature in a custom-developed DIC microscope (Nikon TE2000-U) using a ×100, 1.4 numerical aperture oil objective. Images were acquired at video rate (30 frames/sec) with a Cohu 4910 camera. Individual LDs were captured in the optical trap and lowered to be brought in contact with the mSLB surface. A piezo stage was then used to move the mSLB below the trapped LD at a constant velocity of 1 μm/s for 20 s. The position of LDs was tracked frame by frame using custom software.
Proteinase K Treatment.
Proteinase K was purchased from MP Biomedicals (catalog no. 193981). mSLB was prepared and proteinase K solution was flowed in the chamber with the final concentration of 500 µg/mL. The flow cell was kept in a humid chamber and incubated at 37 °C for 90 min. After the treatment, the solution was washed off with three washes with 100 µL 1× PBS each. For control, instead of proteinase K solution, 1× PBS was flowed in a control mSLB and incubation was done in the same humid chamber.
Purification of GST-Tagged Spo20p.
The GST fusion proteins were expressed in bacteria (Escherichia coli; BL21) and purified by glutathione-Sepharose (catalog no. 27–4574-01, Qiagen) per the manufacturer’s instructions. The purified proteins were dialyzed against 1× PBS. The samples were checked for quality by running on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie brilliant blue staining.
Thin-Layer Chromatography.
Lipids from LDs were extracted using chloroform extraction (13, 16). Briefly, 0.8 mL of aqueous sample containing LDs was mixed with 2 mL of methanol and 1 mL of chloroform followed by vortexing; 1 mL each of chloroform and water was added, which resulted in phase separation. The lower organic phase was transferred to a new glass tube, dried under a nitrogen stream, and resuspended in 20 μL of chloroform. The silica thin-layer chromatography plates (Merck) were precleaned using chloroform, followed by air drying. The sample was then spotted onto these plates using a glass capillary. The solvent system used was according to Wilfling and Thiam et al. (43) with minor modifications. The first solvent system, containing a mixture of n-hexane/diethyl ether/acetic acid (70:30:1), was run halfway and air dried. The plate was then run in a solvent mixture of n-hexene/diethyl ether (59:1). The plate was dried and visualized by spraying with 10% CuSO4 in 8% H3PO4, followed by baking in the oven above 150 °C for 15–20 min.
Antibody Staining of mSLBs.
mSLBs were incubated with an equal concentration of respective primary antibodies and incubated overnight at 4 °C in a humid chamber. After incubation, primary antibody was washed off using 1× PBS, and mSLB was incubated with fluorescently labeled secondary antibody (1:200 dilution) for 1 h. The excess unbound secondary antibody was then washed off and the sample was imaged.
Microscopy.
mSLBs and LDs were imaged using a Zeiss LSM 880 upright laser scanning microscope equipped with a ×63, 1.4 numerical aperture oil-immersion objective. The sequential excitation of fluorophores was achieved by using 488-nm and 561-nm lasers. Spectral bandpass emission filters were used for the acquisition of confocal images with high-sensitivity detectors. Imaging parameters were kept constant across the compared experiments.
Immunoblotting.
Equal concentrations of protein samples were resolved on a 10% SDS-PAGE gel transferred to polyvinylidene fluoride membrane for immunoblotting. The blot was blocked for 1 h at room temperature with 5% nonfat dry milk in Tris-buffered saline with 0.1% Tween-20 (blocking buffer). The membrane was incubated with primary antibody diluted in the blocking buffer for 1 h at room temperature or overnight at 4 °C. The membrane was washed with 0.1% TBST three times for 15 min each. The membrane was incubated with horseradish peroxidase–conjugated secondary antibody diluted in the blocking buffer for 1 h at room temperature. The membrane was washed with 0.1% TBST three times for 15 min each and developed using an ECL kit (Millipore). The blots were imaged on an Amersham Imager 600 and band intensity was quantified using Image J software.
Image and Statistical analyses.
Image analysis was carried out using ImageJ-Fiji software. Statistical analysis was performed using GraphPad Prism (version 5.0a).
Supplementary Material
Acknowledgments
We thank S. Ojha, V. Soppina, and A. Meghna for help with experiments. N. Vitale for the GST-Spo20p construct, A. Pol for the HPos construct, and R. Parton for the Rab18 constructs. We acknowledge the ShRNA Resource Center at the Division of Biological Science, IISc, Bangalore (funded by the Department of Biotechnolgy, Government of India : Grant BT/PR4982/AGR/36/718/2012) for human gene–specific shRNAs. S.T. Suryavanshi and Tata Institute of Fundamental Research are thanked for maintaining and providing animals. R.M. acknowledges the Tata Institute of Fundamental Research for access to some experimental facilities. This work was supported by an Augmented Seed Grant of the Indian Institute of Technology Bombay, the Department of Atomic Energy Government of India, and from Department of Biotechnology, Wellcome Trust India Alliance Senior Fellowships to R.M. (Grants IA/S/11/2500255 and IA/S/19/2/504634).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2200513119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Pol A., Gross S. P., Parton R. G., Review: Biogenesis of the multifunctional lipid droplet: Lipids, proteins, and sites. J. Cell Biol. 204, 635–646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cermelli S., Guo Y., Gross S. P., Welte M. A., The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr. Biol. 16, 1783–1795 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Bosch M., et al. , Mammalian lipid droplets are innate immune hubs integrating cell metabolism and host defense. Science 370, eaay8085 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Wu H., Carvalho P., Voeltz G. K., Here, there, and everywhere: The importance of ER membrane contact sites. Science 361, eaan5835 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thiam A. R., Dugail I., Lipid droplet-membrane contact sites - From protein binding to function. J. Cell Sci. 132, jcs230169 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Krahmer N., Farese R. V., Walther T. C., Balancing the fat: Lipid droplets and human disease. EMBO Mol. Med. 5, 973–983 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herker E., Vieyres G., Beller M., Krahmer N., Bohnert M., Lipid droplet contact sites in health and disease. Trends Cell Biol. 31, 345–358 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Valm A. M., et al. , Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546, 162–167 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilfling F., et al. , Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell 24, 384–399 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H., et al. , Seipin is required for converting nascent to mature lipid droplets. eLife 5, e16582 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walther T. C., Chung J., Farese R. V. Jr., Lipid droplet biogenesis. Annu. Rev. Cell Dev. Biol. 33, 491–510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salo V. T., Ikonen E., Moving out but keeping in touch: Contacts between endoplasmic reticulum and lipid droplets. Curr. Opin. Cell Biol. 57, 64–70 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Salo V. T., et al. , Seipin regulates ER-lipid droplet contacts and cargo delivery. EMBO J. 35, 2699–2716 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datta S., Liu Y., Hariri H., Bowerman J., Henne W. M., Cerebellar ataxia disease-associated Snx14 promotes lipid droplet growth at ER-droplet contacts. J. Cell Biol. 218, 1335–1351 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar M., et al. , Insulin activates intracellular transport of lipid droplets to release triglycerides from the liver. J. Cell Biol. 218, 3697–3713 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rai P., et al. , Kinesin-dependent mechanism for controlling triglyceride secretion from the liver. Proc. Natl. Acad. Sci. U.S.A. 114, 12958–12963 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamm L. K., McConnell H. M., Supported phospholipid bilayers. Biophys. J. 47, 105–113 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kassan A., et al. , Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J. Cell Biol. 203, 985–1001 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dar S., Kamerkar S. C., Pucadyil T. J., A high-throughput platform for real-time analysis of membrane fission reactions reveals dynamin function. Nat. Cell Biol. 17, 1588–1596 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Barak P., Rai A., Rai P., Mallik R., Quantitative optical trapping on single organelles in cell extract. Nat. Methods 10, 68–70 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadh K., Rai P., Mallik R., Feeding-fasting dependent recruitment of membrane microdomain proteins to lipid droplets purified from the liver. PLoS One 12, e0183022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye J., et al. , Cideb, an ER- and lipid droplet-associated protein, mediates VLDL lipidation and maturation by interacting with apolipoprotein B. Cell Metab. 9, 177–190 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Barak P., Rai A., Dubey A. K., Rai P., Mallik R., Reconstitution of microtubule-dependent organelle transport. Methods Enzymol. 540, 231–248 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Xu D., et al. , Rab18 promotes lipid droplet (LD) growth by tethering the ER to LDs through SNARE and NRZ interactions. J. Cell Biol. 217, 975–995 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin S., Driessen K., Nixon S. J., Zerial M., Parton R. G., Regulated localization of Rab18 to lipid droplets: Effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J. Biol. Chem. 280, 42325–42335 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Bekbulat F., et al. , RAB18 loss interferes with lipid droplet catabolism and provokes autophagy network adaptations. J. Mol. Biol. 432, 1216–1234 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Jayson C. B. K., et al. , Rab18 is not necessary for lipid droplet biogenesis or turnover in human mammary carcinoma cells. Mol. Biol. Cell 29, 2045–2054 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozeki S., et al. , Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J. Cell Sci. 118, 2601–2611 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Kassas N., et al. , Comparative characterization of phosphatidic acid sensors and their localization during frustrated phagocytosis. J. Biol. Chem. 292, 4266–4279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kassas N., et al. , Genetically encoded probes for phosphatidic acid. Methods Cell Biol. 108, 445–459 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Thiam A. R., Farese R. V. Jr., Walther T. C., The biophysics and cell biology of lipid droplets. Nat. Rev. Mol. Cell Biol. 14, 775–786 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King C., Sengupta P., Seo A. Y., Lippincott-Schwartz J., ER membranes exhibit phase behavior at sites of organelle contact. Proc. Natl. Acad. Sci. U.S.A. 117, 7225–7235 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rai A. K. A., Rai A., Ramaiya A. J., Jha R., Mallik R., Molecular adaptations allow dynein to generate large collective forces inside cells. Cell 152, 172–182 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Kooijman E. E., et al. , An electrostatic/hydrogen bond switch as the basis for the specific interaction of phosphatidic acid with proteins. J. Biol. Chem. 282, 11356–11364 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Fei W., et al. , A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 7, e1002201 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thiam A. R., et al. , COPI buds 60-nm lipid droplets from reconstituted water-phospholipid-triacylglyceride interfaces, suggesting a tension clamp function. Proc. Natl. Acad. Sci. U.S.A. 110, 13244–13249 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhukovsky M. A., Filograna A., Luini A., Corda D., Valente C., Phosphatidic acid in membrane rearrangements. FEBS Lett. 593, 2428–2451 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Choudhary V., et al. , Architecture of lipid droplets in endoplasmic reticulum is determined by phospholipid intrinsic curvature. Curr. Biol. 28, 915–926.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahankali M., Farkaly T., Bedi S., Hostetler H. A., Gomez-Cambronero J., Phosphatidic acid (PA) can displace PPARα/LXRα binding to the EGFR promoter causing its transrepression in luminal cancer cells. Sci. Rep. 5, 15379 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prévost C., et al. , Mechanism and determinants of amphipathic helix-containing protein targeting to lipid droplets. Dev. Cell 44, 73–86.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paiement J., Young R., Roy L., Bergeron J. J. M., Isolation of rough and smooth membrane domains of the endoplasmic reticulum from rat liver. Cell Biology. Four -Volume Set 2, 41–44 (2006). [Google Scholar]
- 42.Soumpasis D. M., Theoretical analysis of fluorescence photobleaching recovery experiments. Biophys. J. 41, 95–97 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilfling F., et al. , Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. eLife 3, e01607 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.