Significance
We study microvilli of Caenorhabditis elegans larvae and mouse intestinal tissues by combining high-pressure freezing, cryo-focused ion-beam milling, cryo-electron tomography, and subtomogram averaging. We find that many radial nanometer bristles, referred to as nanobristles, project from the lateral surface of nematode and mouse microvilli. The C. elegans nanobristles are 37.5 nm long. We show that nanobristle formation requires a protocadherin family protein, CDH-8, in C. elegans. The loss of nanobristles in cdh-8 mutants slows down animal growth and ectopically increases the number of Y-shaped microvilli, the putative intermediate structures if microvilli split from their tips. Our results reveal a potential role of nanobristles in separating microvilli and suggest that microvilli division may help generate nascent microvilli with uniformity.
Keywords: nanobristles, cryo-FIB, cryo-ET, intestinal apical surface, microvilli
Abstract
Microvilli are actin-bundle-supported membrane protrusions essential for absorption, secretion, and sensation. Microvilli defects cause gastrointestinal disorders; however, mechanisms controlling microvilli formation and organization remain unresolved. Here, we study microvilli by vitrifying the Caenorhabditis elegans larvae and mouse intestinal tissues with high-pressure freezing, thinning them with cryo-focused ion-beam milling, followed by cryo-electron tomography and subtomogram averaging. We find that many radial nanometer bristles referred to as nanobristles project from the lateral surface of nematode and mouse microvilli. The C. elegans nanobristles are 37.5 nm long and 4.5 nm wide. Nanobristle formation requires a protocadherin family protein, CDH-8, in C. elegans. The loss of nanobristles in cdh-8 mutants slows down animal growth and ectopically increases the number of Y-shaped microvilli, the putative intermediate structures if microvilli split from tips. Our results reveal a potential role of nanobristles in separating microvilli and suggest that microvilli division may help generate nascent microvilli with uniformity.
Microvilli are membrane-bound cell-surface protrusions that contain a core bundle of actin filaments enveloped in the plasma membrane (1–3). Many epithelial cells develop microvilli above their apical surface to enhance functional capacity for a range of physiological tasks, including nutrient absorption in the intestine (4), solute uptake in the renal tubules (5), mechanosensation in sensory stereocilia of the inner ear (6), and chemosensation in the gut, lung, and urogenital tracts (7–9). Abnormal microvillar structure and function lead to human disorders, such as life-threatening nutrient malabsorption, osmotic imbalances, and inherited deafness in Usher syndrome (1, 3, 4, 10).
An intestinal absorptive cell enterocyte develops up to 1,000 densely packed microvilli in an array known as the brush border. These fingerlike outward projections enhance the functional surface area for nutrient absorption and provide the barrier for host defense against pathogens and toxins (1, 3, 4). Because the gut epithelium undergoes constant regenerative renewal, microvillus assembly is a process that continues throughout our lifetime (1, 3, 4, 11). The long-standing questions regarding microvillus formation are how microvilli are formed with striking uniformity in sizes and how these protrusions are maximally packed in a hexagonal pattern.
The tip of microvilli is known to be decorated by additional filamentous structures. The glycoprotein-rich glycocalyx localizes between the apical tip of microvilli and the luminal space (Fig. 1A), provides a barrier for pathogens, and serves as the interface for nutrient digestion (3, 4). The protocadherin-based adhesion tip links localize between adjacent microvilli (Fig. 1A) (12). The mammalian cadherin superfamily members, including CDHR2 and CDHR5, play essential roles in packing microvilli, increasing surface density, and controlling microvilli length (13). Other cadherins, specifically CDH23 and PCDH15, have been implicated in organizing the exaggerated microvilli found on inner-ear hair cells (14, 15).
Fig. 1.
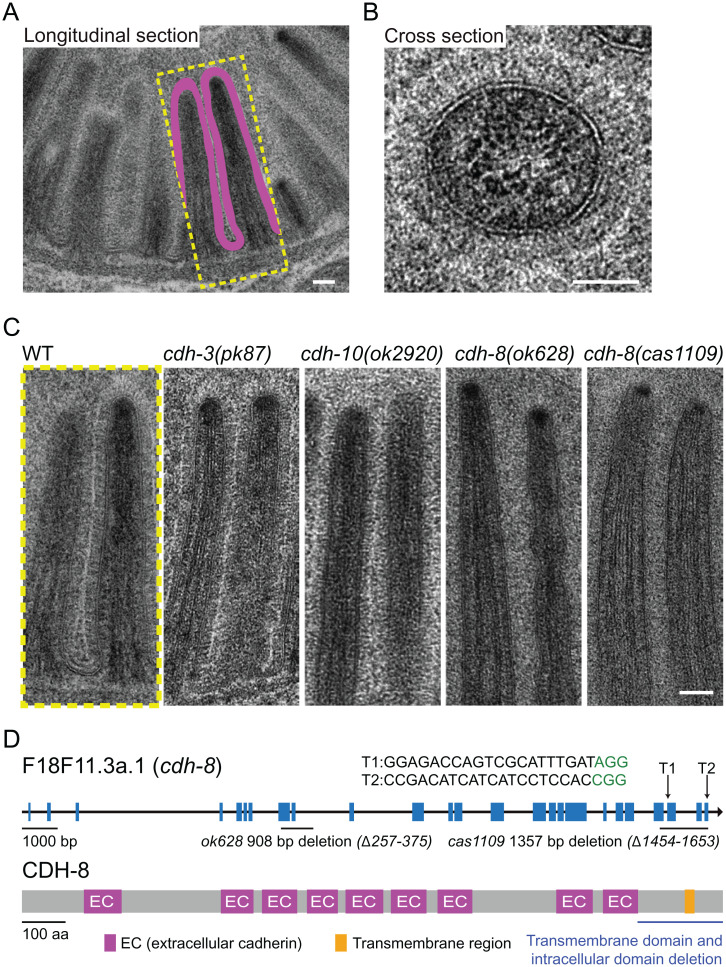
In situ cryo-ET of the C. elegans intestinal brush border reveals nanobristles on the lateral surface of microvilli. (A) A schematic diagram of an intestinal epithelial cell (Left) and two microvilli (Right) from the dotted box in Left. The glycocalyx and the protocadherin tip link are the characterized cell-surface structure at microvillar tips. This work shows that numerous nanobristles (magenta) decorate the lateral surface of microvilli. (B, Left and Center) Representative cryo-SEM images of the C. elegans L1 larvae before and after FIB milling. (Scale bars, 10 μm.) (B, Right) Representative FIB image of the ∼200-nm-thick cryo-lamella. (Scale bar, 5 μm.) (C) A 3D rendering of the C. elegans intestinal brush border showing various macromolecules and structures. Magenta, nanobristles; cyan, membrane; yellow, actin; beige, ribosome; green, mitochondria; orange, ER) Nanobristles and ribosomes were mapped back in the tomogram with the computed location and orientation. (D) A selected microvillus from E magnified for visualization. (E and F) Cryo-ET tomogram slices of microvilli (E, top view; F, side view). (Scale bars in C–F, 50 nm.)
An individual microvillus is only 0.1 μm in diameter and 1 to ∼2 μm in height (Fig. 1A), the tiny dimension of which, along with their high density and lumen localization, becomes a technical hurdle for in situ structural investigations at high resolution (1–3). Despite our understanding of microvillar tip decorations, it is unclear whether any structure projects from the lateral surface of microvilli. Recent methodology advance of cryo-electron tomography (cryo-ET) makes the platform well suited to address the challenges of studying microvillus structure in animals (16, 17). Here, we used cryo-ET to reveal a previously unrecognized nanobristle structure on the lateral surface of microvilli. We provide evidence that nanobristle formation depends on a protocadherin family protein, CDH-8, and that nanobristles regulate microvilli separation.
Results
Cryo-ET of C. elegans Larvae.
We initiated the cryo-ET study of intestinal microvilli in the nematode Caenorhabditis elegans. Classical transmission electron microscopy (TEM) described the anatomy and molecular organization of the C. elegans intestine and microvilli, which highly resemble those in mammals, demonstrating that microvilli are evolutionarily conserved organelles (Fig. 1A) (18, 19). Studies of microvilli in other species require tissue dissection or in vitro cell-culture systems. In contrast, the thickness of the C. elegans L1 larvae, which is ∼12.5 μm in diameter (SI Appendix, Fig. S1A) (18, 19), allows high-pressure freezing (HPF) of an entire animal, enabling structural analysis of microvilli in their close-to-native conditions.
We employed cryo-focused ion-beam milling (cryo-FIB) to produce thin lamellas from the frozen-hydrated vitrified worms (16). By taking advantage of the C. elegans gut development and anatomy invariance (18, 19), we focused on the intestine from the newly hatched first-stage juvenile (L1 larva, 6 h after hatching). The larval intestine runs ∼70% of body length between the anterior pharynx and the posterior hindgut (18, 19). We thinned the intestine’s central region, about 150 μm posterior from the animal’s nose, corresponding to the intestinal cells from Int4 to Int7 (Fig. 1B, SI Appendix, Fig. S1A, and Movie S1). Cryo-FIB milling removed the biological material above and below the area, generating a 150- to 200-nm-thick lamella traversing the whole animal with high consistency and reproducibility (Fig. 1B).
We applied cryo-ET to image the cryo-lamella, which facilitates the three-dimensional (3D) visualization of microvilli at unprecedented molecular resolution. Each lamella provided a suitable imaging area of ∼10 × 10 µm2, from which we obtained approximately two to three cryo-ET tilt series. In agreement with the early TEM observations (18, 19), our tomograms of C. elegans microvilli showed that the plasma membrane surrounds the actin bundles in the core (Fig. 1C and Movie S1). The plasma membrane appears as two smooth and parallel black lines, and actin bundles from the cross-section were resolved as individual black dots (Fig. 1D and Movie S1). The excellent contrast and rich structural details from the reconstructed tomogram demonstrate the high quality of prepared samples (Fig. 1D and Movie S1).
Nanobristles Project from C. elegans Intestinal Microvilli.
We found that many bristle-like structures project from the lateral surface of each microvillus (Fig. 1 C and F and Movie S2). These structures are highly uniform, with a constant diameter of 4.5 ± 0.5 nm (mean ± SD; n = 4,983). From the surface of the microvilli, we found 9,195 ± 351 (mean ± SD) nanobristles per µm2 on the microvillus membrane. They appear to be the transmembrane (TM) structure whose extracellular length is 37.5 ± 0.2 nm (mean ± SD). Each tomographic slice contains 50 ± 5 (mean ± SD; from 12 focal planes) of the bristle-like structures (Fig. 1 D and E), highlighting their abundance. These structures distribute along the lateral surface, but not the tip of microvilli, which are distinct from adhesion tip links between adjacent microvillar tips (Fig. 1A and SI Appendix, Fig. S1B) (12). Our cryo-ET did not detect fibrous meshwork at the tip of microvilli (SI Appendix, Fig. S1B), which is consistent with the notion that the glycocalyx, a highly-hydrated fibrous meshwork that consists of sulfate mucopolysaccharides, can be visualized at the microvillar tip by TEM only by additional straining with heavy-metal ions (1–3). Microvilli are micrometer appendages decorating the epithelium surface, and nanometer bristle-like structures cover the lateral surface of microvilli (SI Appendix, Fig. S1B). We termed these structures nanobristles, whose dimension and location distinguish them from the known bristles in other cell types, such as millimeter-long Drosophila mechanosensory bristles (20). While this study focused on nanobristles on the surface of microvilli, our cryo-ET images also reveal a circumferential granular structure between the plasma membrane and actin filaments, which are likely formed by the actin-membrane cross-linking ERM (Ezrin/Radixin/Moesin family) proteins or myosin-I or other molecules (Fig. 1D, SI Appendix, Fig. S1C, and Movies S3 and S4) (3).
We wondered why the previous TEM studies did not describe nanobristles. The TEM sample preparation involves chemical fixation, dehydration, and heavy-metal staining, which are incompatible with high-resolution structural studies. Although TEM cannot resolve an individual nanobristle, this technique may detect an electron-dense layer that contains numerous nanobristles at the lateral surface of microvilli. To test this idea, we performed TEM of HPF, freeze-substituted C. elegans. We determined an excellent-quality TEM if the lipid bilayer of the plasma membrane is resolved as two parallel black lines. From such microvilli, we found an electron-dense area along the microvilli’s lateral surface from either longitudinal or cross-sections (Fig. 2 A and B). The thickness of these layers is 37.3 ± 0.5 nm (51 nanobristle layers from three animals; mean ± SD) (Fig. 2A), comparable to that of cryo-ET tomograms. We noticed a thin band with no electron density localized between two adjacent nanobristle layers (Fig. 2A), indicating that nanobristles from two neighboring microvilli are not connected.
Fig. 2.
C. elegans microvilli in WT and cdh-8 mutant animals. (A) TEM of the longitudinal view of the C. elegans intestinal brush border. (Scale bar, 100 nm.) (B) High-magnification cross-section image of the C. elegans microvilli. (Scale bar, 50 nm.) (C) TEM images of microvilli in WT, cdh-3(pk87), cdh-10(ok2920), cdh-8(ok628), and cdh-8(cas1109) mutant animals. (Scale bar, 100 nm.) (D, Upper) Gene structure of cdh-8. CRISPR-Cas9 generated the TM and intracellular domain deletion allele cas1109 of cdh-8. Blue boxes represent exons. Arrow, sgRNA; green, protospacer adjacent motif sequence. (D, Lower) Schematics of the C. elegans CDH-8 protein. Purple, cadherin extracellular domain; orange, TM domain.
Nanobristle Formation Requires CDH-8.
We sought to identify the molecular composition of nanobristles. We performed single-cell messenger RNA (mRNA) sequencing of C. elegans larvae to gain insights into the intestine transcriptome (SI Appendix, Fig. S2 A and B and Dataset S1; see details in Materials and Methods). A previous study described the proteome of the mouse brush border (21), and we suspected that protocadherins might be involved and used their dataset to identify candidates. Among the overlapped hits between the C. elegans intestinal transcriptome and mouse brush-border proteome, the intriguing candidates include the protocadherin family proteins (SI Appendix, Fig. S2C).
We asked whether a protocadherin might regulate nanobristle formation. The protocadherins encoded by cdh-3, cdh-8, and cdh-10 might have the suitable size and number of repeats of nanobristles (SI Appendix, Fig. S3A), and we thus obtained their mutant alleles to probe this possibility. Because TEM offers sufficient throughput to screen for the nanobristle layer and electron-transparent intermicrovillar band, we quickly used this method to assess nanobristles in multiple mutants. In comparison to wild-type (WT) animals, we did not find apparent defects of the nanobristle layers in cdh-3 or cdh-10 deletion mutants (Fig. 2C; >200 examined microvilli from three animals of each genotype). By contrast, the nanobristle layer and the electron transparent band become undetectable in cdh-8(ok628) mutant animals (530 examined microvilli from five animals), despite excellent preservation of the microvillar membrane (Fig. 2C). Our cryo-ET tomograms confirmed the loss of nanobristles in cdh-8(ok628) animals (SI Appendix, Fig. S3 B and C and Movie S5).
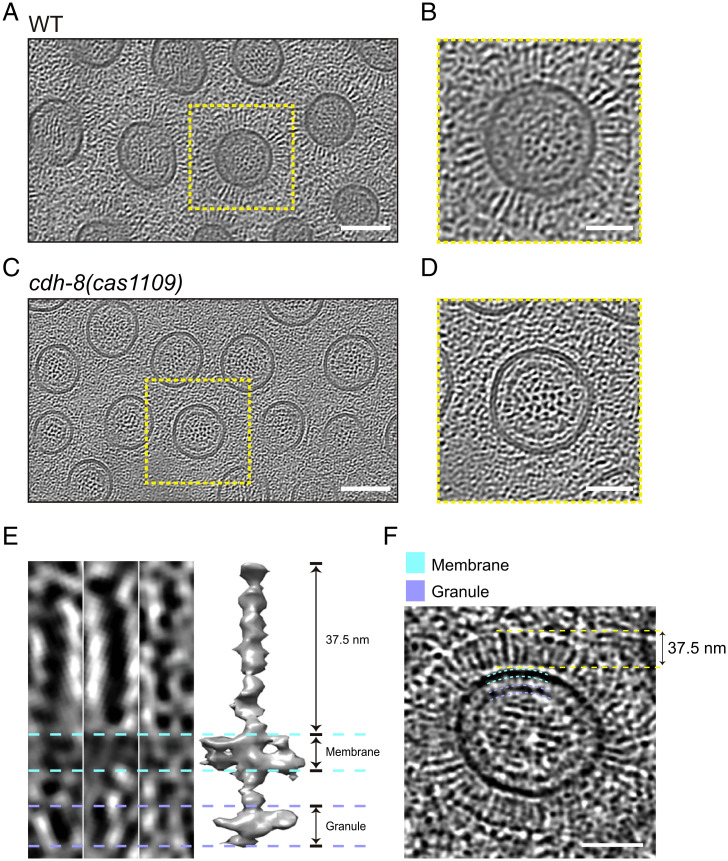
Protocadherin genes are well-known for their alternative splicing (22), and the cdh-8(ok628) allele has an in-frame deletion that removes 119 residues in its extracellular domain (Fig. 2D). We detected a layer of fibrous appearance perpendicular to the membrane surrounding the microvilli in the cdh-8(ok628) mutant (SI Appendix, Fig. S3 B, D, and E). We suspect that the cdh-8(ok628) allele may not entirely abolish the surface features, although it prevented the formation of typical nanobristles, making them invisible in HPF samples. To rule out the effects of a large variety of protocadherin isoforms, we implemented the CRISPR-Cas9–based homologous recombination strategy to precisely remove the TM domain, along with the entire intracellular domain of CDH-8, which are shared by all the CDH-8 isoforms, generating a cdh-8(cas1109) mutant allele (Fig. 2D). Our TEM datasets did not detect the nanobristle layer or the electron transparent band along the microvilli in the TM-domain-removed cdh-8(cas1109) mutant animals (Fig. 2C; 365 examined microvilli from four animals). We found that the absence of nanobristles did not affect the length of microvilli (SI Appendix, Fig. S4A). Cryo-ET results further confirmed that cdh-8(cas1109) animals lost nanobristles on the lateral surface of microvilli (Fig. 3 A–D and Movies S6 and S7), demonstrating an essential and specific role of CDH-8 in nanobristle formation. From all the examined seven sets of cryo-ET images of cdh-8(cas1109) mutant microvilli, we did not detect the extra fibrous structures observed in the cdh-8(ok628) mutant (SI Appendix, Fig. S3 D and E), indicating that a severe loss of CDH-8 does not produce these extra structures. Our quantification of the intermicrovillus distance between WT and cdh-8(cas1109) showed that the intermicrovillus distance was significantly reduced in cdh-8(cas1109) mutant animals (SI Appendix, Fig. S4B), which agrees with the nanobristle’s roles in separating microvilli.
Fig. 3.
The CDH-8 protein and nanobristles. (A–D) Cryo-ET of the C. elegans intestinal brush border in WT (A and B) or cdh-8(cas1109) (C and D). Microvilli were enlarged in the yellow boxes. (Scale bars, 100 nm [A and C]; 50 nm [B and D].) (E and F) Structural analysis of nanobristles from the tomogram slice view (E, Left and F) and subtomogram averaging model (E, Right). (Scale bar, 50 nm.)
In line with these genetic and cryo-ET results, the GFP-based transcriptional reporter showed that the cdh-8 gene was expressed in the C. elegans intestinal cells (SI Appendix, Fig. S4C), validating our single-cell sequencing data. By constructing a red fluorescence protein mScarlet knock-in animal to label an established microvillus marker ERM-1 (2, 3), we found that the GFP-tagged CDH-8 full-length protein localized to the intestinal lumen, which was marked with ERM-1::mScarlet (SI Appendix, Fig. S4D).
To define the molecular architecture of nanobristles, we performed subtomogram averaging of nanobristles from cryo-ET datasets. A total of 4,983 nanobristles were hand-picked from 11 tomograms, aligned, and averaged to produce a low-resolution average structure (Fig. 3 E and F). The density map shows clear features of a single nanobristle embedded in the membrane, including the extracellular domain with ∼37.5-nm length, the TM region, and the intracellular domain (Fig. 3E), which matches well the observation on the raw micrographs and the tomograms (Fig. 3F). CDH-8 is predicted to consist of nine extracellular cadherin domains, a single TM helix, and an intracellular domain. While the resolved structural features do not conflict with the CHD-8 structure, the accurate docking of CHD-8 in the reconstructed map is still not available under such resolution. Future cryo-ET analyses with an atomic resolution are needed to determine how the CDH-8 protein forms nanobristles.
Loss of Nanobristles Slows Down Animal Growth.
Next, we used the cdh-8 mutant animals to dissect the function of nanobristles. We did not notice any apparent defects in the overall animal morphology or the number of progeny; however, our quantifications of the body length at each developmental stage revealed reduced animal growth in cdh-8 mutant animals (SI Appendix, Fig. S5). Because the eggshell prevents embryos from accessing the bacterial food, our measurements started from the newly hatched larvae, when the worm body lengths are indistinguishable between cdh-8 mutants and WT. With the progression of animal growth in the presence of the same food, the body lengths of cdh-8 mutant alleles were shorter than those of WT animals at the various larval stages (SI Appendix, Fig. S5). At 122 h after hatching, when C. elegans already developed into the day-2 adults, cdh-8 deletion animals reached the WT body length (SI Appendix, Fig. S5). Notably, the GFP-tagged cdh-8 gene expression using an intestinal-specific promoter Pges-1 fully rescued the slow-growth phenotype in cdh-8 mutants (SI Appendix, Fig. S5), demonstrating a cell-autonomous function of cdh-8 in promoting animal growth. We suggest that the growth delay might result from the less efficient food update and that the slow, but continuous, growth eventually restored the cdh-8 mutant body length to the WT level.
Y-Shaped Microvilli in cdh-8 Mutants.
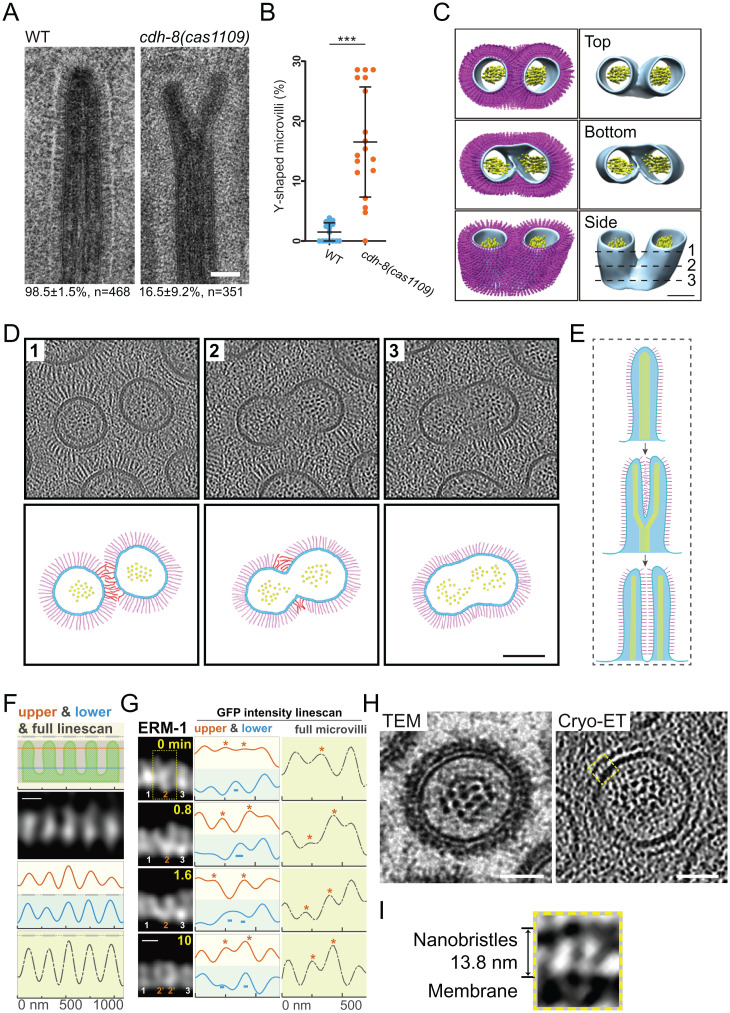
To examine if nanobristles contribute to microvilli assembly and organization, we compared microvilli structure in cdh-8 mutant and WT animals. Our TEM analyses detected the forked or branched (collectively called Y-shaped) microvilli in cdh-8(cas1109) mutant animals with 17% penetrance (Fig. 4 A and B; 351 microvilli from four animals). The Y-shaped microvilli were rarely detectable in WT animals (Fig. 4 A and B; 468 microvilli from five animals). Curiously, half a century ago, the Y-shaped microvilli were documented during the salamander microvilli regeneration (SI Appendix, Fig. S4E) (23). Using hydrostatic pressure to eliminate microvilli on the small intestine, Tilney and Cardell (23) observed the Y-shaped regenerating microvilli 4 min after pressure release, but not later on, suggesting that Y-shaped microvilli might represent an intermediate structure during microvilli regeneration.
Fig. 4.
CDH-8 regulates microvillus assembly and a proposed model. (A) TEM images of microvilli in WT and the Y-shaped microvilli in cdh-8(1109) mutants. (Scale bar, 100 nm.) Below are percentages of the observed bristle-like microvilli or Y-shaped microvilli. (B) Quantification of Y-shaped microvilli in WT and cdh-8(1109) mutant animals. (C) The 3D reconstruction of the representative Y-shaped microvilli in WT animals. (Scale bar, 100 nm.) (D) Cryo-ET tomogram slices of the Y-shape microvilli. Lines 1 to 3 in C indicate the localization for tomographic slices in D, Upper. D, Lower shows the schematic. (Scale bar, 100 nm.) (E) A proposed role of nanobristles in microvilli division. Nanobristles, magenta; actin in microvilli, yellow; plasma membrane, cyan. (F) Schematics of high-resolution live-cell imaging of microvilli in a developing C. elegans larva using the Airyscan confocal microscopy. In the first row, the orange line (40 nm below the top of microvilli), the blue line (40 nm above the bottom of microvilli), and the gray bar (the entire microvilli) indicate the line scan for measuring the fluorescence intensity from microvilli images (ERM-1::GFP) in the second row or G. (Scale bar, 200 nm.) (G) Time-lapse fluorescence images of a Y-shaped microvillus separation into two nascent microvilli. The yellow boxes, asterisks, and − show the no. 2 microvillus separation, whereas other microvilli remained unchanged. (Scale bar, 200 nm.) (H) Cross-section TEM (Left) or cryo-ET image (Right) of mouse microvilli. A high-magnification view of the dotted yellow box in I. (Scale bars, 50 nm.) (I) The yellow boxed region shows nanobristle-like structures on the surface of mouse microvilli. *P < 0.05; ***P < 0.001.
In a developing WT C. elegans larva, one of the cryo-ET tomograms showed a Y-shaped microvillus with two separated tips, but a single base (Fig. 4 C and D, SI Appendix, Fig. S4F, and Movie S8). Consistent with our TEM results, we only detected one Y-shaped microvillus from more than 100 examined ones, which indicates the occurrence of such a structure during animal development and suggests that a Y-shaped microvillus might be transient or unusual. In cdh-8 mutant animals, the Y-shaped microvilli may be an arrested status of a splitting microvillus, and the lack of nanobristles failed to space two daughter microvilli efficiently, thereby enriching Y-shaped microvilli (Fig. 4A). To explain the observed Y-shaped microvilli, we propose that an existing microvillus is divided from the tip to the base, generating two nascent microvilli and nanobristles inserted into the newly split microvilli to promote their separation (Fig. 4 D and E and SI Appendix, Fig. S4F). By counting the number of actin filaments from the tip to the base of Y-shaped microvilli, we found that the two tip regions contain 22 and 27 actin filaments, respectively, whereas the base has 49 actin filaments (SI Appendix, Fig. S6), which is consistent with our model that microvilli can split from the tip to the base. Alternatively, two adjacent microvilli may fuse from their bases to form Y-shaped microvilli; however, previous studies have never detected microvilli whose diameters are twice more prominent than others, which a microvilli fusion model does not easily explain.
Live Imaging of Microvilli Division.
To test the microvillus division model, we followed the microvilli-assembly process in C. elegans. Although fluorescence live-cell imaging recorded microvilli dynamics in cell cultures (24, 25), time-lapse visualization of microvilli in a living organism has been challenging. The microvillus membrane-to-membrane spacing is ∼88 nm, not allowing conventional confocal microscopy to resolve neighboring microvilli. Neither could the superresolution imaging platforms be applied because their limited working spaces prevent determining the intestinal lumen structures, at least 5 μm away from the C. elegans cuticle. Using the Airyscan technology in a GFP::ERM-1 KI animal, we successfully resolved individual microvilli along the intestinal lumen in developing C. elegans larvae (Fig. 4F). Line scan from the microvillar top or bottom or the whole microvilli revealed separate fluorescence peaks, and the average distance between the peaks of adjacent microvilli is 201.0 ± 9.1 nm (mean ± SD, n = 44; Fig. 4F), comparable to the measurements from the cryo-ET tomogram slice (205.8 ± 28.0 nm, mean ± SD, n = 49). Time-lapse recording started from a Y-shaped microvillus (Fig. 4 G, yellow dotted box), and its two tips gradually separated, forming a V-shaped microvillus (0.8 and 1.6 min in Fig. 4G). Eventually, two nascent microvilli were isolated (Fig. 4G). Another microvillus underwent a similar process to assemble two new ones, and the Y- or V-shaped microvilli were intermediate structures when a mother microvillus split into two daughter microvilli (SI Appendix, Fig. S7A and more examples in SI Appendix, Fig. S7). Microvilli are dynamic, changing their orientation and position over time in the culture systems, raising alternative explanations for the images and shifting intensities shown in Fig. 4 F and G. While we cannot exclude other possibilities, many static neighboring microvilli around dividing microvilli validate the stability of our imaging system and indicate that most microvilli in these imaging areas remain still during the recording time (SI Appendix, Fig. S8A). These results support the microvilli division model in which two nascent microvilli may be generated by dividing an old microvillus.
Nanobristles on Mouse Microvilli.
We addressed whether mammals develop a similar nanobristle structure on their intestinal microvilli. Using the HPF, cryo-FIB milling, and cryo-ET platform to examine mouse intestinal tissues, we found that 13.8 ± 0.8-nm-long bristle-like structures decorate the lateral surface of mouse microvilli (64 mouse nanobristles from four tomograms; mean ± SD) (Fig. 4 H and I and SI Appendix, Fig. S8B). Despite the difference in length, the morphology and location of these bristle-like structures suggest that nanobristles, like microvilli, might be widespread from nematodes to mammals.
Discussion
This study shows that nanobristles decorate the lateral surface of microvilli and that a protocadherin protein, CDH-8, is essential for nanobristle formation. Our results indicate that nanobristles are involved in microvillus separation and suggest that microvilli may divide to form nascent microvilli of the exact sizes, providing insights into microvilli uniformity.
The idea that mother microvilli give rise to new daughter microvilli has been recently described by using an in vitro-cultured system (26). Live-cell imaging results from Gaeta et al. (26) showed that the formation of new microvilli involved the growth of new protrusions from the lateral or base of preexisting ones. While it remains untested whether the exact mechanism applies to the live mammalian intestine, in situ cryo-ET and live-imaging results from developing worm larvae support the possibility that one mother microvillus generates daughter microvilli of the identical dimension in live C. elegans. Thus, microvillus formation involves multiple pathways, including de novo formation, elongation from the lateral or base of a preexisting one, or perhaps dividing a mother microvillus. The microvilli division model addresses how to assemble microvilli from existing ones during tissue homeostasis or late development, complementing a microvillar elongation strategy for de novo microvilli formation (1–3, 25). Together, these models address how microvilli are assembled at different developmental stages or in various animal species.
Future studies will improve the resolution of nanobristle structure in C. elegans to determine whether CDH-8 is its component more directly. Our observation of nanobristles on the lateral surface of microvilli offers a glimpse of microvilli spacing. In the absence of the nanobristles in the cdh-8 mutants, other molecules probably separate two microvilli. Nanobristles may promote the separation, and without them, the separation is less efficient, increasing the chance of detecting Y-shaped microvilli. Protocadherin family proteins have been generally considered to form homophilic interactions between adjacent plasma membranes; however, our study detected a space that separates nanobristles from adjacent microvillar membranes. We speculate that CDH-8 may function with other molecules on the microvilli surface to generate such a space, and, alternatively, protocadherins may play roles beyond homophilic interactions.
In addition to microvilli separation, the decoration of nanobristles on microvilli can have other advantages, such as host defense against microbe infection. On the other hand, nanobristle decoration appears to be a paradox to microvilli’s role in increasing surface area for nutrient uptake. Numerous nanobristles occupy microvilli’s lateral surface, where food absorption occurs (3, 4). One possibility could be that nanobristles act as a food filter, selecting the predigested food particles whose sizes are smaller than the internanobristle distance to contact the microvillar membrane. The food particles of various sizes may occupy the membrane without such a filter, but cannot be absorbed efficiently, explaining the slow growth phenotype in cdh-8 mutant larvae (SI Appendix, Fig. S5).
Microvilli are widespread across species (5, 12, 14, 27), and, likewise, other animals may develop a similar nanobristle structure on their microvillar lateral surface. The length of the mouse nanobristles is only ∼15 nm, significantly shorter than the 37.5-nm-long worm nanobristles. We suggest that another protocadherin family protein with a smaller extracellular domain or a protein other than a protocadherin family protein may generate the mouse nanobristles. Therefore, the nanobristle structures on the lateral surface of microvilli are likely built up by distinct proteins across species.
Despite decades of ultrastructural studies by TEM, our research indicates that anatomical structures can be more complex and fascinating than is currently appreciated; given the power of the cryo-ET platform, perhaps nanobristles are the “tip of the iceberg,” and additional biological structures will be uncovered in situ.
Materials and Methods
The C. elegans Strains and Culture.
SI Appendix, Table S1 lists the C. elegans strains used in this study. Using the standard protocol (28), we cultivated C. elegans on nematode growth medium agar plates seeded with the Escherichia coli OP50 at 20 °C. The WT strain was Bristol N2. Some strains were provided by the Caenorhabditis Genetics Center (CGC), funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
Molecular Biology, Genome Editing, and Transgenesis.
We performed the genome editing of the cdh-8 and erm-1 genes in C. elegans following the established protocols (29, 30). We used the CRISPR design tool to select the target sites. The single-guide RNA (sgRNA) sequences (SI Appendix, Table S2) were inserted into the pDD162 vector (Addgene catalog [cat.] no. 47549) by linearizing this vector with 15-bp overlapped primers using Phusion high-fidelity DNA polymerase (New England Biolabs, cat. no. MO531L). PCR products were digested with Dpn I (Takara, cat. no. 1235A) for 2 h at 37 °C and transformed into Trans5α bacterial chemically competent cells (TransGen Biotech, cat. no. CD201-01). The linearized PCR products with 15-bp overlapping ends were cyclized to generate plasmids by spontaneous recombination in bacteria. Homology recombination templates were constructed by cloning the 1.5-kb upstream and downstream homology arms into pPD95.77 plasmids using the In-Fusion Advantage PCR cloning kit (Clontech, cat. no. 639621). A fluorescence tag (e.g., GFP) was inserted into the constructs with a flexible linker before the stop codons. Synonymous mutations were introduced to Cas9 target sites to avoid the cleavage of the homologous repair template by Cas9. The plasmids are listed in SI Appendix, Table S3. GFP-tagged CHD-8 was generated by using a PCR slicing-by-overlap-extension method (31) that fused a 2,844-bp promoter, plus the coding region of cdh-8 with gfp::unc-54 3′ untranslated region DNA fragments. The primers are listed in SI Appendix, Table S3.
To generate a knock-in strain, we purified the sgRNA construct and the repair template plasmids using the PureLink Quick PCR Purification Kit (Invitrogen, cat. no. K310001) and coinjected them into N2 animals with the pRF4 [rol-6 (su1006)] coinjection marker. F1 transgenic progenies were singled and screened by PCR and Sanger sequencing. Transgenic animals were generated by the germ-line microinjection of DNA plasmids or PCR products into N2 with the Podr-1::gfp plasmid as a coinjection marker. We maintained at least two independent transgenic lines with a constant transmission rate (>50%). Concentrations of DNA constructs used for generating knock-in or overexpression were 50 ng/µL or 20 ng/µL, respectively.
HPF and TEM of C. elegans.
We performed HPF experiments using an early protocol with minor modifications (16). C. elegans L1 larvae were washed off plates with M9 buffer. An ∼2-μL droplet of worm suspension was applied directly onto the glow-discharged cryo-EM grid. The extra bulk of buffer was blotted with filter paper. The cryo-EM grids were immediately transferred into the 100-μm-deep cavity of 6-mm aluminum carriers (Beijing Wulundes Biotech Ltd.), which were filled with 2-methyl pentane (Sigma, cat. no. M65807). The carriers were quickly covered with 0.12-mm-thick sapphire discs. The sandwiched assembly was transferred into the HPM100 HPF machine (Leica Microsystems) using a 6-mm correlative light EM holder. The assembly was transferred into the FC6 cryo-ultramicrotome chamber (Leica Microsystems) at −150 °C. The assemblies with the intact sapphire discs were kept in the chamber for 10 min to sublime the extra 2-methyl pentane. The cryo-EM grids were then picked out from the assemblies and stored in liquid nitrogen.
For TEM experiments, C. elegans strains were transferred to a 50-μm-deep specimen carrier with a pipette and frozen rapidly by a Leica EM HPM100 HPF system. Carriers were immersed in fixative (acetone solution of 1% osmium tetroxide and 0.1% uranyl acetate) in a 2-mL microcentrifuge tube. The tubes were processed in a Leica EM AFS2 machine by using a standard substitution and fixation program: −90 °C for 48 h, −60 °C for 24 h, −30 °C for 18 h, and, finally, to 4 °C. After fixation, specimens were washed three times with pure acetone, infiltrated with SPI-PON 812 resin for 2 d, embedded in a flat mold, and polymerized at 60 °C. Then, 90-nm ultrathin sections were generated by using a Leica EM UC7 Ultramicrotome and picked on 200 mesh copper grids. Sections were poststained with 2% uranyl acetate and Reynold’s lead citrate to enhance contrast and imaged by using the FEI Tecnai G2 Spirit (120 kV) electron microscope.
Cryo-FIB Milling of C. elegans Larvae.
Helios NanoLab G3 UC dual-beam microscope (Thermo Fisher Scientific Co.) equipped with the Quorum PP3010T cryo-transfer system was used for cryo-FIB milling experiments. We used the homemade cryo-FIB shuttle adapted to the PP3010T system to mount and transfer the grid samples. The shuttle keeps two frozen specimens on a slope of 30°, including the naked grids and packaged autogrids. A protective shutter (32, 33) covers the samples to avoid ice contamination during transport. The shutter was opened or closed by the manipulation knife within the PP3010T preparation chamber. The HPF samples were loaded on the cryo-FIB shuttle and transferred to the cryo-stage of the PP3010T preparation chamber. To eliminate the residual cryoprotectant, we kept the samples in the chamber for 30 to ∼60 min. Sputter coating of a platinum (Pt) layer was performed in the PP3010T preparation chamber, with a parameter of 10 mA and 60 s. The Pt layer improves sample conductivity and decreases the charge-induced movement during FIB milling.
SEM images were acquired at the accelerating voltage of 2 kV and beam current of 0.2 nA by an Everhart–Thornley Detector (34). A gas-injection system was used to do protective Pt coating before FIB milling. The stage was tilted to 18°. Rough milling of the parallel pattern from two sides was performed by using an ion-beam current of 2.5 to ∼0.23 nA until the lamella thickness was ∼1 to 2 μm. An ion-beam current of 80 to ∼24 pA was used for fine polishing until the lamella’s thickness was about 150 to 200 nm. The grids were transferred into grid boxes within a liquid-nitrogen tank for storage.
Cryo-ET Data Collection.
We collected the cryo-ET data using the Titan Krios Microscopy (Thermo Fisher Scientific Co.) operated at a voltage of 300 KV and equipped with a Cs corrector, GIF Quantum energy filter (Gatan), and K2 Summit direct electron detector (Gatan). All the tilt series were recorded from 60° to −60° 2ith the SerialEM software (35). The recording state was at a nominal magnification of 33,000× in counting mode with a pixel size of 3.421 Å⋅pixel−1. Each stack was exposed for 2.4 s with an exposure time of 0.3 s per frame and recorded as a movie of eight frames, resulting in the total dose rate of ∼1.927 electrons per Å2 for each stack. GIF was set to a slit width of 20 eV. The defocus ranged from −3 to −7 μm. The MotionCor2 program was used to correct the beam-induced motion (36). For the C. elegans cdh-8(cas1109) mutant samples, the tilt series were collected with a Volta phase plate, and the defocus was about −1 μm.
Cryo-ET Reconstruction and Subtomogram Averaging.
Tilt series were aligned with the patch-tracking method in IMOD software (37). The tomograms were reconstructed with TOMO3D scripts (38). The binning four tomograms (pixel size, 13.684 Å) reconstructed with the Simultaneous Iterative Reconstruction Technique were used for particle picking manually. Tomograms reconstructed with weighted-back projection were used for further subtomogram averaging analysis. A total of 4,983 nanobristles were manually picked from the reconstructed tomograms with IMOD software. The protomo/i3 software (39) package was used for subtomogram analysis, including alignment and classification. All the nanobristles were used to generate an initial model. With the initial model as the reference, 10 rounds of 3D classification were performed to gather the homogenous 3D particles. In total, 3,688 nanobristle particles were selected to perform further for used alignment to generate the final 3D map.
Membrane Segmentation Atomic Model Fitting.
To analyze the arrangement of nanobristles in the microvilli membrane, we remapped the nanobristle subtomogram averaging map back into the tomograms with the refined positions and orientations. The segmentation of the microvilli, endoplasmic reticulum (ER), and mitochondria was performed by using the Amira software (Thermo Fisher). Ribosome particles were manually picked and aligned with i3 software to generate the orientation angles. The cryo-ET map of the yeast 80S ribosome (Electron Microscopy Data Bank code 8799) was filtered to 25 Å and remapped back into the tomogram. The rendering images were made with UCSF Chimera software. The tomograms were processed to increase contrast using Cryo-CARE software (40).
Single-Cell mRNA Sequencing in C. elegans.
To identify nanobristle components, we applied SPLiT-seq (split-pool ligation-based transcriptome sequencing), a single-cell RNA-sequencing method that labels the cellular origin of RNA through combinatorial barcoding (41), to profile intestinal cell transcriptomes in C. elegans larvae. Three rounds of combinatorial barcoding yielded 18,432 barcode combinations [three rounds of barcoding processed in 4-well, 96-well, and 48-well plates (41)]. After removing low-quality reads and transcriptomes, we obtained 5,054 single-cell transcriptomes, including transcripts from 18,790 genes. Among them, 10 transcriptomes were assigned to the intestinal cell on the intestinal cell-specific expression of the ifb-2 or pgp-1 gene (19). Comparison of transcribed genes between the intestinal cell and the epithelial seam cells that express ceh-16, nhr-74, or nhr-73 resulted in 2,261 transcripts that were only detected in the intestine, but not epithelia. We performed gene-set enrichment analysis for gene ontology cellular component and found 95 plasma-membrane protein-coding genes. Gene ontology molecular function analysis of these plasma membrane-related genes exhibited a significant enrichment of genes categorized as calcium-ion binding, including several CDH (CaDHerin) family genes. Considering that nanobristle proteins might be conserved across species, we searched 2,261 intestine-specifically transcribed genes for those coding mouse homologs that reside in the intestinal epithelial cell brush border (21). Our cross-species analysis generated a list of 94 genes, also including members of the cadherin family. Below are the detailed methods.
We prepared the single C. elegans cells as described (42). Sodium dodecyl sulfate–DTT–pronase E treatment and mechanical disruption of synchronized larvae yielded cell suspension with debris. Single cells were separated from doublets, debris, and undigested larvae by filtering and were then fixed and penetrated.
To perform single-cell mRNA sequencing, we used the SPLiT-seq method as described (41) with modifications. Transcriptomes of single cells were labeled with three rounds of split-pool barcoding and released from cells for amplification (SPLiT-seq). In each split-pool round, fixed cells were randomly distributed into wells, and transcripts were labeled with well-specific barcodes. Segmentation of complementary DNA amplicons by Tn5 transposons and PCR amplification of fragments were applied during sequencing library preparation. The TruePrep DNA Library Prep Kit V2 constructed the sequencing library for Illumina (Vazyme Biotech, cat. no. TD502), according to the manufacturer’s instructions. The library was sequenced on HiSeq systems (Illumina) using 150-nt kits and paired-end sequencing. According to the results of FastQC, adaptors or low-quality nucleotides were trimmed by Trim Galore (version [v] 0.5.2) using default parameters. For each paired-end sequencing read, a 10-bp Unique Molecular Identifier (UMI) sequence and a 24-bp cell barcode were extracted from the Read 2 file by the tool “preprocess_splitseq.pl” of zUMIs (v0.0.6). The Read 1 was split by different cell barcodes in Read 2 and mapped to the C. elegans genome (WS263) by zUMIs (v0.0.6) and STAR (v2.6.0c). We kept unique mapping reads. The duplicated reads from the same transcript were excluded based on the UMI information in Read 2.
The raw and processed data are provided at the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE167859).
C. elegans Body-Length Measurement.
To measure the C. elegans body length, we synchronized the worms by allowing day-1 young adults to lay eggs on fresh plates for 1 h at 20 °C. We photographed 16, 40, 64, 88, and 112 h after these eggs hatched. Worms were mounted on 3% agarose pads, paralyzed with a drop of 10 mM sodium azide, and photographed with a Zeiss Axio Observer Z1 microscope. All the length measurements were performed from the worm nose to the tail tip using the Java image-processing program ImageJ.
Live-Cell Imaging.
Live-cell imaging of the worm was performed as described (43). Worms were anesthetized with 1 mg/mL levamisole and mounted on 3% agarose pads at 20 °C. The immobilized larvae were subsequently imaged with an Axio Observer Z1 microscope (Carl Zeiss MicroImaging, Inc.) and the 488 nm and 568 nm lines of a Sapphire CW CDRH USB Laser System attached to the spinning disk confocal scan head (Yokogawa CSU-X1 Spinning Disk Unit). We conducted live-cell imaging to follow microvilli dynamics at a high spatial resolution using a Zeiss LSM 900 confocal microscope with AiryScan superresolution module equipped with highly sensitive gallium arsenide phosphide detectors using a 63x Zeiss objective (1.4 numerical aperture) 20 °C.
Statistical Analysis.
The quantitative data were analyzed in GraphPad Prism v8. Independent Student’s t tests were performed to compare the mean values between the two groups. Statistical significance was designated as *P < 0.05, **P < 0.01, and ***P < 0.001.
Supplementary Material
Acknowledgments
We thank Drs. Mingjie Zhang, Piali Sengupta, Jianlin Lei, Hongwei Wang, Li Yu, Ye-Guang Chen, and Mark Mooseker for discussions; the CGC for providing some strains; and the Tsinghua University Cryo-EM Facility of China National Center for Protein Sciences (Beijing) for HPF, cryo-FIB milling, and tomography data collection. This work was supported by the National Key R&D Program of China Grant 2019YFA0508401; and National Natural Science Foundation China Grants 31991190, 31730052, 31525015, 31861143042, 31561130153, 31671444, and 31871352.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. J.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2122249119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information. The raw and processed data are provided at the GEO database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE167859).
References
- 1.Crawley S. W., Mooseker M. S., Tyska M. J., Shaping the intestinal brush border. J. Cell Biol. 207, 441–451 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pelaseyed T., Bretscher A., Regulation of actin-based apical structures on epithelial cells. J. Cell Sci. 131, jcs221853 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sauvanet C., Wayt J., Pelaseyed T., Bretscher A., Structure, regulation, and functional diversity of microvilli on the apical domain of epithelial cells. Annu. Rev. Cell Dev. Biol. 31, 593–621 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Delacour D., Salomon J., Robine S., Louvard D., Plasticity of the brush border—The yin and yang of intestinal homeostasis. Nat. Rev. Gastroenterol. Hepatol. 13, 161–174 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Coudrier E., Kerjaschki D., Louvard D., Cytoskeleton organization and submembranous interactions in intestinal and renal brush borders. Kidney Int. 34, 309–320 (1988). [DOI] [PubMed] [Google Scholar]
- 6.Schwander M., Kachar B., Müller U., Review series: The cell biology of hearing. J. Cell Biol. 190, 9–20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerbe F., Jay P., Intestinal tuft cells: Epithelial sentinels linking luminal cues to the immune system. Mucosal Immunol. 9, 1353–1359 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Krasteva G., Kummer W., “Tasting” the airway lining fluid. Histochem. Cell Biol. 138, 365–383 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Kummer W., Deckmann K., Brush cells, the newly identified gatekeepers of the urinary tract. Curr. Opin. Urol. 27, 85–92 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Schneeberger K., Roth S., Nieuwenhuis E. E. S., Middendorp S., Intestinal epithelial cell polarity defects in disease: Lessons from microvillus inclusion disease. Dis. Model. Mech. 11, dmm031088 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gehart H., Clevers H., Tales from the crypt: New insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 16, 19–34 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Crawley S. W., et al. , Intestinal brush border assembly driven by protocadherin-based intermicrovillar adhesion. Cell 157, 433–446 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weck M. L., Crawley S. W., Tyska M. J., A heterologous in-cell assay for investigating intermicrovillar adhesion complex interactions reveals a novel protrusion length-matching mechanism. J. Biol. Chem. 295, 16191–16206 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narui Y., Sotomayor M., Tuning inner-ear tip-link affinity through alternatively spliced variants of protocadherin-15. Biochemistry 57, 1702–1710 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richardson G. P., Petit C., Hair-bundle links: Genetics as the gateway to function. Cold Spring Harb. Perspect. Med. 9, a033142 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harapin J., et al. , Structural analysis of multicellular organisms with cryo-electron tomography. Nat. Methods 12, 634–636 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Mahamid J., et al. , Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 351, 969–972 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Dimov I., Maduro M. F., The C. elegans intestine: Organogenesis, digestion, and physiology. Cell Tissue Res. 377, 383–396 (2019). [DOI] [PubMed] [Google Scholar]
- 19.McGhee J. D., The C. elegans intestine. WormBook, 1–36 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bitan A., Guild G., Abdu U., The highly elongated Drosophila mechanosensory bristle—A new model for studying polarized microtubule function. Fly (Austin) 4, 246–248 (2010). [DOI] [PubMed] [Google Scholar]
- 21.McConnell R. E., Benesh A. E., Mao S., Tabb D. L., Tyska M. J., Proteomic analysis of the enterocyte brush border. Am. J. Physiol. Gastrointest. Liver Physiol. 300, G914–G926 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zipursky S. L., Grueber W. B., The molecular basis of self-avoidance. Annu. Rev. Neurosci. 36, 547–568 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Tilney L. G., Cardell R. R., Factors controlling the reassembly of the microvillous border of the small intestine of the salamander. J. Cell Biol. 47, 408–422 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Postema M. M., Grega-Larson N. E., Neininger A. C., Tyska M. J., IRTKS (BAIAP2L1) elongates epithelial microvilli using EPS8-dependent and independent mechanisms. Curr. Biol. 28, 2876–2888.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meenderink L. M., et al. , Actin dynamics drive microvillar motility and clustering during brush border assembly. Dev. Cell 50, 545–556.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaeta I. M., Meenderink L. M., Postema M. M., Cencer C. S., Tyska M. J., Direct visualization of epithelial microvilli biogenesis. Curr. Biol. 31, 2561–2575.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Araya-Secchi R., Neel B. L., Sotomayor M., An elastic element in the protocadherin-15 tip link of the inner ear. Nat. Commun. 7, 13458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brenner S., The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10, 1028–1034 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedland A. E., et al. , Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10, 741–743 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hobert O., PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32, 728–730 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Rigort A., et al. , Micromachining tools and correlative approaches for cellular cryo-electron tomography. J. Struct. Biol. 172, 169–179 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Zhang J., Ji G., Huang X., Xu W., Sun F., An improved cryo-FIB method for fabrication of frozen hydrated lamella. J. Struct. Biol. 194, 218–223 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Schaffer M., et al. , Optimized cryo-focused ion beam sample preparation aimed at in situ structural studies of membrane proteins. J. Struct. Biol. 197, 73–82 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kremer J. R., Mastronarde D. N., McIntosh J. R., Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Agulleiro J. I., Fernandez J. J., Tomo3D 2.0—Exploitation of advanced vector extensions (AVX) for 3D reconstruction. J. Struct. Biol. 189, 147–152 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Winkler H., et al. , Tomographic subvolume alignment and subvolume classification applied to myosin V and SIV envelope spikes. J. Struct. Biol. 165, 64–77 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buchholz T., Jordan M., Pigino G., Jug F., “Cryo-CARE: Content-aware image restoration for cryo-transmission electron microscopy data” in 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019) Fiona Pattison, Ed. (IEEE, Piscataway, NJ, 2019), pp. 502–506.
- 41.Rosenberg A. B., et al. , Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang S., Banerjee D., Kuhn J. R., Isolation and culture of larval cells from C. elegans. PLoS One 6, e19505 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chai Y., et al. , Live imaging of cellular dynamics during Caenorhabditis elegans postembryonic development. Nat. Protoc. 7, 2090–2102 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information. The raw and processed data are provided at the GEO database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE167859).