Significance
While ZBP1 is well documented to drive cell death in response to viruses, its role in the context of Toll-like receptor (TLR)–mediated immune activation remains less defined. Here, we show that ZBP1 promotes inflammation in response to bacterial lipopolysaccharide (LPS) or double-stranded RNA (dsRNA). In a dose dependent manner, ZBP1 promotes the recruitment of RIPK1 to the TLR3/4 adaptor TRIF, activating downstream inflammatory signaling. ZBP1 plays a crucial role in TRIF-dependent responses in vivo, as Zbp1−/− mice exhibited resistance to LPS-induced hypothermia and attenuated inflammatory responses to dsRNA. Our findings suggest that ZBP1 plays a synergistic role in the immune response by driving inflammation downstream of TLRs in response to bacterial and viral components.
Keywords: ZBP1, RIPK1, endotoxic shock, inflammation
Abstract
ZBP1 is widely recognized as a mediator of cell death for its role in initiating necroptotic, apoptotic, and pyroptotic cell death pathways in response to diverse pathogenic infection. Herein, we characterize an unanticipated role for ZBP1 in promoting inflammatory responses to bacterial lipopolysaccharide (LPS) or double-stranded RNA (dsRNA). In response to both stimuli, ZBP1 promotes the timely delivery of RIPK1 to the Toll-like receptor (TLR)3/4 adaptor TRIF and M1-ubiquitination of RIPK1, which sustains activation of inflammatory signaling cascades downstream of RIPK1. Strikingly, ZBP1-mediated regulation of these pathways is important in vivo, as Zbp1−/− mice exhibited resistance to LPS-induced septic shock, revealed by prolonged survival and delayed onset of hypothermia due to decreased inflammatory responses and subsequent cell death. Further findings revealed that ZBP1 promotes sustained inflammatory responses by mediating the kinetics of proinflammatory “TRIFosome” complex formation, thus having a profound impact downstream of TLR activation. Given the well-characterized role of ZBP1 as a viral sensor, our results exemplify previously unappreciated crosstalk between the pathways that regulate host responses to bacteria and viruses, with ZBP1 acting as a crucial bridge between the two.
ZBP1 (Z-DNA-binding protein 1) is a cytosolic nucleic acid sensor and RHIM (receptor interacting protein [RIP] homotypic interaction motif) domain–containing protein that has been studied extensively in the context of RIPK3 (RIP kinase)-dependent necroptosis (1–4). As such, ZBP1 uses its two Z-DNA-binding domains to recognize viruses such as murine cytomegalovirus and influenza A virus (IAV) (2, 4–6), driving RHIM-mediated interactions between ZBP1 and RIPK3 and the activation of mixed lineage kinase domain like pseudokinase. This activation cascade ultimately induces necroptosis, as well as the assembly of a RIPK1/Fas receptor-associated death domain protein (FADD)/caspase-8 (CASP8)-containing complex that drives apoptosis in response to viral infection (1). In response to IAV infection, ZBP1 also promotes RIPK3-independent apoptosis, dependent on the direct recruitment of RIPK1 and activation of FADD and CASP8 (1, 7). ZBP1-mediated induction of necroptosis can also be unleashed during mammalian development if RIPK1 is deleted or mutated. Indeed, mutation of the RHIM domain of RIPK1 results in embryonic lethality in mice that can be rescued by the additional deletion of ZBP1 (8, 9).
Recently, we reported that when the host inflammatory response is inhibited, ZBP1 initiates CASP8-mediated, RIPK1-dependent pyroptosis in response to bacterial lipopolysaccharide (LPS) (10–12). That is, ZBP1 promoted formation of a prodeath complex (or TRIFosome) downstream of the RHIM domain-containing protein and Toll-like receptor 4 (TLR4) adaptor TRIF (TIR domain containing adaptor protein-inducing interferon [IFN] β), suggesting that ZBP1 might also regulate LPS-induced inflammatory responses downstream of TRIF. Herein, we demonstrate that ZBP1 is important for the core functions of the TLR4 and TLR3 pathways by promoting the production of proinflammatory cytokines in response to LPS or polyinosine-polycytidylic acid (poly(I:C)) in vitro and in vivo. Via RHIM-dependent interactions, ZBP1 tunes the timing and magnitude of the inflammatory response by regulating the kinetics of proinflammatory complex formation and activation of mitogen activated protein kinase (MAPK), nuclear factor-κappa B (NF-κB), and IRF3-mediated signaling cascades. Importantly, deficiency in ZBP1 promotes resistance to LPS-induced septic shock in vivo by damping serum and tissue-specific inflammatory responses and cell death, mirroring the decrease and delay in the inflammatory response observed in vitro in the absence of ZBP1. Together with our previously reported role for ZBP1 in the regulation of the prodeath TRIFosome (10) complex, these data suggest that a similar complex is assembled in the proinflammatory context, thus presenting the TRIFosome as a universal regulator of cell death and inflammatory responses.
Results
ZBP1 Specifically Promotes TRIF-Mediated Inflammatory Responses.
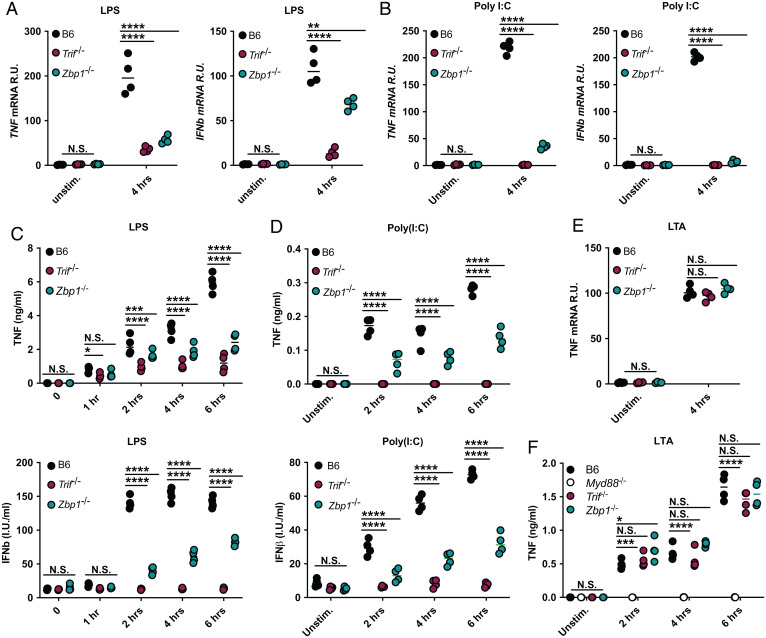
We previously reported that ZBP1 interacts with the TLR3/4 adaptor protein TRIF to promote RIPK1-mediated cell death (10). Since RIPK1 also plays an integral role in TRIF-mediated inflammatory responses (13), we hypothesized that ZBP1 may be involved in the inflammatory responses initiated downstream of TLR4 and TLR3. Accordingly, treatment of bone marrow–derived macrophages (BMDMs) with the TLR4 agonist LPS or the TLR3 agonist poly(I:C) resulted in robust tumor necrosis factor (TNF) and IFNβ messenger RNA (mRNA) and protein production that was decreased in the absence of ZBP1 and nearly abolished by loss of TRIF, indicating that ZBP1 played an essential role in TRIF-mediated proinflammatory signaling (Fig. 1 A–D). To investigate whether the role for ZBP1 was confined to TRIF-mediated immune responses, we used lipoteichoic acid (LTA) to activate the TLR2/MyD88-dependent, TRIF-independent pathway (Fig. 1 E and F). Unlike in response to LPS or poly(I:C), LTA-mediated inflammatory responses were not dependent on ZBP1, suggesting that ZBP1 specifically promotes TRIF-mediated inflammatory responses (Fig. 1 E and F).
Fig. 1.
ZBP1 specifically promotes TRIF-mediated inflammatory responses. (A and B) TNF and IFNβ mRNA levels after 4 h of (A) LPS or (B) poly(I:C) treatment in B6, Trif−/−, and Zbp1−/− macrophages. (C and D) TNF and IFNβ protein levels at indicated timepoints in B6, Trif−/−, and Zbp1−/− macrophages stimulated with (C) LPS or (D) poly(I:C). (E and F) TNF (E) mRNA and (F) protein levels at indicated timepoints after stimulation with LTA in indicated macrophages. For ELISA and qPCR data, data points indicate the mean from triplicate wells of four biologically independent experiments. ANOVA was used for comparison between groups: ns, nonsignificant (P > 0.05); *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
ZBP1 Enhances TRIF-Mediated MAPK, NF-κB, and IRF3 Activation.
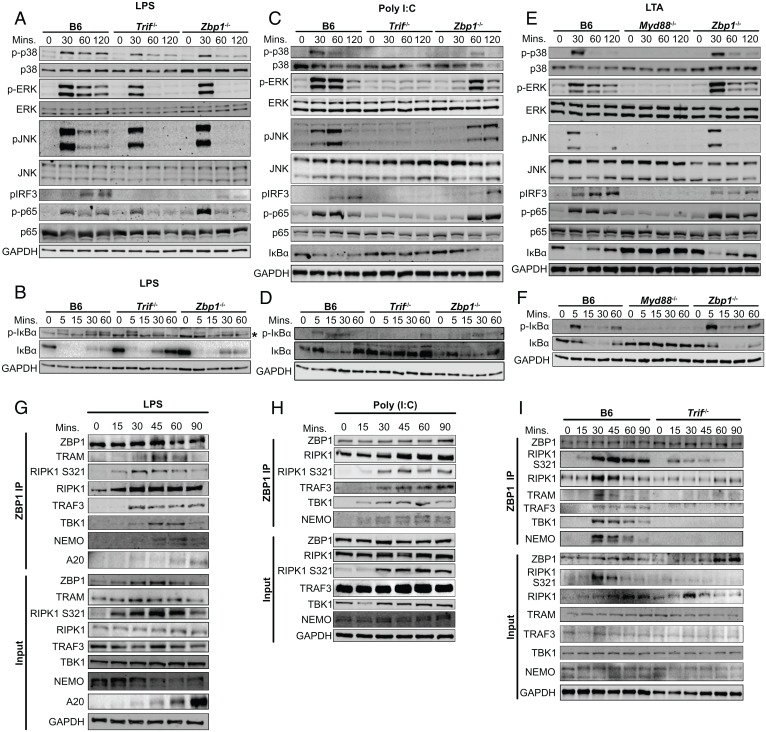
To determine the level at which ZBP1 regulates inflammation in response to LPS, we analyzed MAPK, NF-κB and IRF3 activation in response to LPS in Zbp1−/− BMDMs. We found that ZBP1 is critical for sustained, but not early, phosphorylation of p38, extracellular signal-regulated kinase (ERK), and c-Jun terminal kinase (JNK) in response to LPS (Fig. 2A). Similar loss of sustained MAPK activation was observed in Trif−/− cells, in agreement with current models of early induction of inflammatory signaling mediated at the plasma membrane through the MyD88-dependent pathway and sustained MAPK activation at later timepoints occurring through the TRIF-dependent pathway (14). Similar, albeit attenuated, dependence on TRIF and ZBP1 at late, rather than early, timepoints was observed for NF-κB activation as measured by p65 phosphorylation (Fig. 2A). In contrast, strong IκBα phosphorylation in B6 BMDMs was abrogated at 30 and 60 min in the absence of ZBP1 or TRIF (Fig. 2B). This loss of IκBα phosphorylation correlated with robust replenishment of IkBα in Trif−/− BMDMs and a slight increase in total IκBα levels in Zbp1−/− BMDMs (Fig. 2B). From these data, we conclude that ZBP1 deficiency largely phenocopies TRIF deficiency in its effect on the MAPK activation pathway, but to a lesser extent on the NF-κB pathway. However, given the interplay between NF-κB-mediated production and degradation of IκBα, changes in IκBα levels may be difficult to interpret at this these later timepoints. Finally, IRF3 phosphorylation, which drives IFN induction downstream of TRIF (15), was significantly decreased in Zbp1−/− BMDMs and completely absent in Trif −/− BMDMs (Fig. 2A). In further support of a role for ZBP1 in TRIF-dependent signaling, deficiency in ZBP1 significantly delayed MAPK, NF-κB, and IRF3 activation in response to TLR3-activation with poly(I:C) but had no impact on MyD88-dependent, LTA-induced inflammatory responses downstream of TLR2 (Fig. 2 C–F). These findings identify a key role for ZBP1 in mediating TRIF-dependent inflammatory responses downstream of TLR3/4 and support previous overexpression-based studies that demonstrated that ZBP1 can modulate proinflammatory cytokine production via interactions with RIPK1 and induction of NF-κB signaling (16–18). Altogether, these results show that ZBP1 plays a nonredundant role in the activation of multiple inflammatory pathways, likely via interactions downstream of TRIF.
Fig. 2.
ZBP1 enhances TRIF-mediated MAPK, NF-κB, and IRF3 activation (A and B) Levels of total and phosphorylated (A) p38, ERK and JNK, IRF3, p65, and (B) IκBα in B6, Trif−/−, and Zbp1−/− macrophages after treatment with LPS for indicated time. Asterisk indicates nonspecific bottom band. (C and D) Levels of total and phosphorylated (C) p38, ERK and JNK, IRF3, p65, and (D) IκBα in B6, Trif−/−, and Zbp1−/− macrophages after treatment with poly(I:C) for indicated time. (E and F) Levels of total and phosphorylated (E) p38, ERK and JNK, IRF3, p65, and (F) IκBα in B6, Myd88−/−, and Zbp1−/− macrophages after treatment with LTA for indicated time. (G and H) ZBP1 immunoprecipitation in B6 BMDMs stimulated with (G) LPS or (H) poly(I:C) and probed for proinflammatory complex components as indicated. (I) ZBP1 immunoprecipitation in B6 and Trif−/− BMDMs stimulated with LPS and probed for proinflammatory complex components as indicated. Data from immunoblots are representative of three or more biologically independent experiments. Unless otherwise indicated, blots were singly or doubly probed to ensure highest quality. All blots from a specific panel are run from a single set of lysates that were identically handled.
To identify mediators of the inflammatory response that might interact with ZBP1, we considered that ZBP1 promotes RHIM-mediated interactions between TRIF and RIPK1 that drive prodeath complex formation and CASP8-mediated pyroptosis (10). Since RIPK1 also serves as a scaffold to promote inflammatory responses downstream of TRIF (13, 19), we hypothesized that ZBP1 may similarly promote TRIF-RIPK1 interactions in response to LPS or poly(I:C) to drive TLR3/4-mediated cytokine production. In agreement with this idea, RIPK1-deficiency attenuated inflammatory responses downstream of TRIF (13, 19) (SI Appendix, Fig. S1A), suggesting that deficiency in ZBP1 may attenuate TRIF-mediated inflammation by perturbing the scaffolding functions of RIPK1. To test this hypothesis, we activated macrophages with LPS or poly(I:C), performed ZBP1-specific immunoprecipitations, and probed for proinflammatory mediators that might interact with ZBP1 based on their involvement in TRIF signaling (Fig. 2 G and H). As we have seen previously (10), RIPK1 was constitutively bound to ZBP1, and RIPK1-ZBP1 binding was enhanced in response to both LPS and poly(I:C) (Fig. 2 G and H). We observed S321 phosphorylated RIPK1 bound to ZBP1 at early timepoints in LPS- and poly(I:C)-treated cells. Phosphorylation of RIPK1 at S321 is mediated by MAPKs and is known to promote RIPK1 kinase-independent proinflammatory complex stability (20, 21), supporting a role for ZBP1 within the proinflammatory complex induced downstream of TRIF. This was further supported by the binding of inflammatory signaling components such as TRAF3, TBK1, and NF-kappa-B-essential modulator (NEMO) to ZBP1, the latter providing evidence of NF-κB component binding and activation downstream of TRIF (Fig. 2 G and H). Although reliable antibodies capable of detecting TRIF by immunoblot are not available, the TRIF-associated adaptor Toll/interleukin-1 receptor adaptor molecule 2 (TRAM) is similarly required for TRIF-mediated inflammatory responses to LPS, but not poly(I:C) (22, 23), and can be detected by immunoblot (SI Appendix, Fig. S1B and Fig. 2G). Using TRAM as a proxy for TRIF, we observed association between ZBP1 and TRAM, strongly indicating that ZBP1 interacts with the signaling components of the TRIF-mediated proinflammatory complex, or proinflammatory TRIFosome (Fig. 2G).
In further support of a TRIF-specific function for ZBP1, TRIF deficiency abrogated binding of inflammatory signaling components to ZBP1 in response to LPS (Fig. 2I). While the constitutive binding between RIPK1 and ZBP1 was unaltered by TRIF deficiency, the presence of S321 phosphorylated RIPK1 bound to ZBP1 was decreased in the absence of TRIF, highlighting the importance of TRIF in the induction of this proinflammatory phosphorylation event (Fig. 2I). Although some S321 phosphorylated RIPK1 bound to ZBP1 was detectable in the absence of TRIF, this phosphorylation can likely be attributed to induction of the MyD88-dependent pathway in response to LPS. This is supported by stronger S321 phosphorylation at early timepoints that is decreased at later timepoints in the absence of TRIF. Binding interactions within the ZBP1/TRIF proinflammatory complex were transient and mostly terminated within 90 min of activation, as evidenced by the appearance of A20, a deubiquitinase, which resolves inflammatory activation (Fig. 2G). Overall, these data demonstrate that ZBP1 interacts with numerous proinflammatory TRIFosome components.
ZBP1 Promotes Proinflammatory Complex Formation and TRIF-RIPK1 Colocalization.
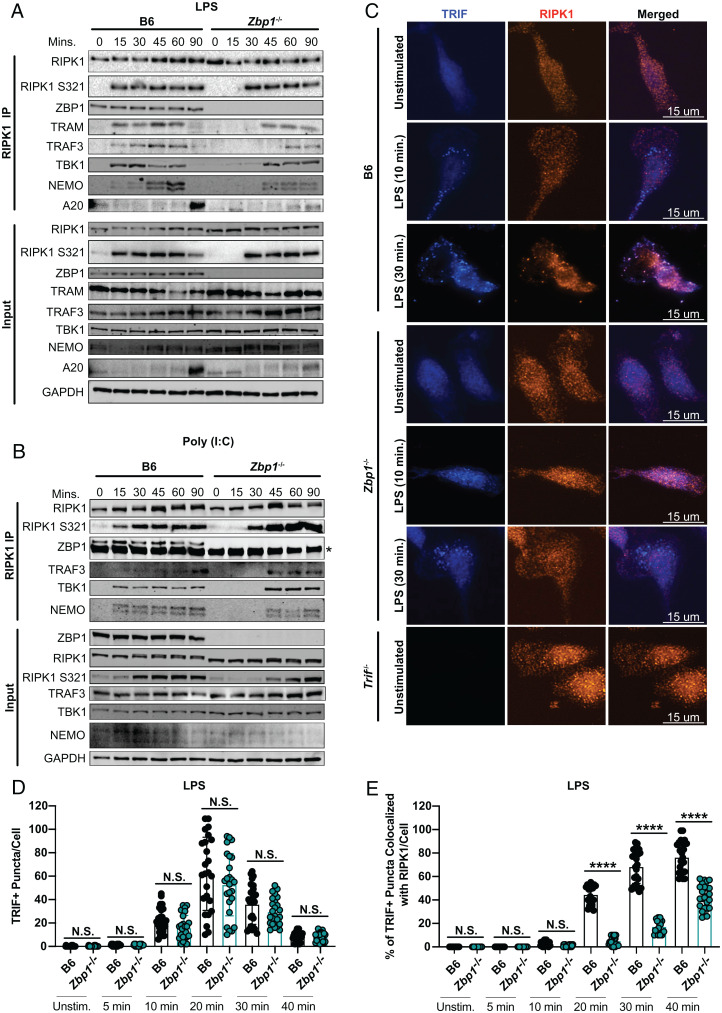
In addition to revealing a unique role for ZBP1 in TRIF-mediated inflammation, ZBP1-specific immunoprecipitations provided a tool to investigate which interactions in the TRIF-dependent complex rely on ZBP1. To this end, we compared the binding of RIPK1 to proinflammatory mediators in ZBP1-deficient and sufficient BMDMs activated with LPS or poly(I:C) (Fig. 3 A and B). In support of ZBP1 promoting proinflammatory TRIFosome formation, deficiency in ZBP1 delayed RIPK1 S321 phosphorylation and recruitment of TRAF3, TBK1, and NEMO to RIPK1 in response to LPS and poly(I:C) (Fig. 3 A and B). Importantly, ZBP1 deficiency also delayed RIPK1 recruitment to TRAM in response to LPS, supporting a role for ZBP1 in promoting RIPK1 recruitment to TRIF (Fig. 3A).
Fig. 3.
ZBP1 promotes proinflammatory complex formation and TRIF-RIPK1 colocalization. (A and B) RIPK1 immunoprecipitation in B6 and Zbp1−/− BMDMs stimulated with (A) LPS or (B) poly(I:C) and probed for proinflammatory complex components as indicated. Asterisk indicates band from antibody heavy chain. (C) Representative 60× images of TRIF and RIPK1 staining in B6, Zbp1−/−, and Trif−/− macrophages stimulated with LPS as indicated. (D) Quantification of the number of TRIF+ puncta/cell in B6 and Zbp1−/− macrophages stimulated with LPS at indicated timepoints. (E) Quantification of the percentage of TRIF+ puncta colocalized with RIPK1/cell in B6 and Zbp1−/− macrophages stimulated with LPS at indicated timepoints. Data points indicate individual cells, n = 25 cells/group imaged across four or five fields of view. Data from immunoblots and colocalization experiments are representative of three or more biologically independent experiments. Unless otherwise indicated, blots were singly or doubly probed to ensure highest quality. All blots from a specific panel are run from a single set of lysates that were identically handled. ANOVA was used for comparison between groups: ns, nonsignificant (P > 0.05); *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Delayed binding of RIPK1 to TRAM in the absence of ZBP1 supports a model in which ZBP1 initiates proinflammatory complex formation by promoting TRIF-RIPK1 interactions (Fig. 3A). In the absence of antibodies to TRIF validated for immunoblot, we resorted to specific detection of TRIF by immunofluorescence (Fig. 3C). Strikingly, as early as 10 min after LPS treatment of B6 macrophages, TRIF puncta were detectable (Fig. 3 C and D). While these puncta did not initially appear to colocalize with RIPK1, after 30 min of LPS treatment, nearly 80% of TRIF puncta were colocalized with RIPK1 puncta (Fig. 3 C and E). Supporting the role for ZBP1 in recruiting RIPK1 to TRIF, ZBP1 deficiency did not affect TRIF puncta formation but dramatically decreased the colocalization of TRIF- and RIPK1-positive puncta (Fig. 3 C–E).
ZBP1 Mediates RIPK1 M1-Ubiquitination to Promote Interactions within the Proinflammatory Complex.
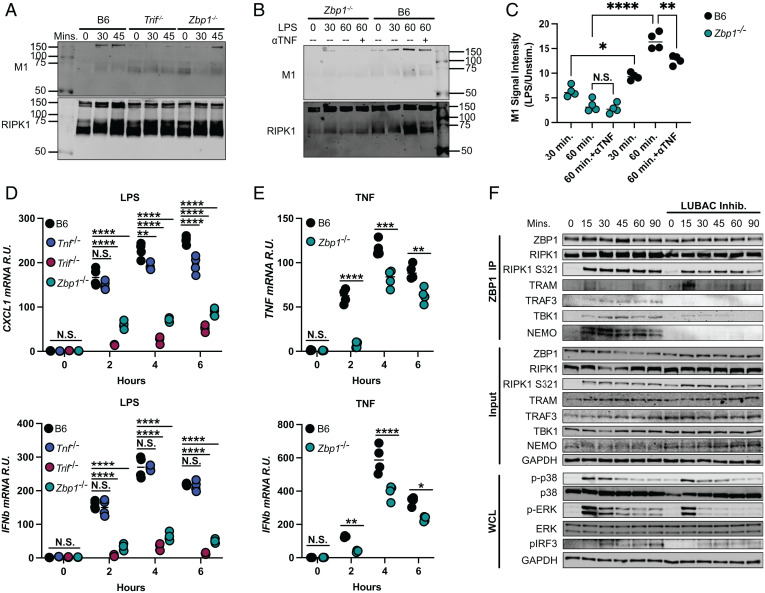
TBK1 and NEMO are known to bind RIPK1 and drive inflammatory signaling downstream of TNF receptor (TNFR) in addition to TLRs (24, 25). Based on the fact that TBK1:ZBP1 and NEMO:ZBP1 interactions relied on TRIF (Fig. 2I) and that the binding of TBK1 and NEMO to RIPK1 is mediated by M1-ubiquitination of RIPK1 (26), we investigated whether ZBP1 promotes the addition of M1-ubiquitin chains on RIPK1, thus facilitating binding of TBK1 and NEMO to RIPK1. Strikingly, ZBP1 and TRIF deficiency reduced M1-ubiquitination of RIPK1 at 30 min after treatment with LPS (Fig. 4A). While M1-ubiquitination of RIPK1 was inhibited at later timepoints in the absence of TRIF, it was restored at 45 min in Zbp1−/− (Fig. 4A). These results support our finding that ZBP1 delays, but does not entirely prevent, proinflammatory complex formation downstream of TRIF (Fig. 3A). Because LPS induces autocrine and paracrine TNF signaling at later timepoints, which may contribute to M1-ubiquitination of RIPK1 downstream of TNFR as opposed to TRIF, we next used TNF-neutralizing antibody (ɑTNF) (Fig. 4 B and C) to isolate TRIF-dependent ubiquitination events in LPS-stimulated B6 and Zbp1−/− BMDMs. ɑTNF appeared to slightly reduce M1-ubiquitination of RIPK1 in B6 cells at 60 min, indicating a potential contribution of TNF signaling to this RIPK1 modification in response to LPS (Fig. 4 B and C). However, M1-ubiquitination in Zbp1−/− cells was unaffected by ɑTNF, highlighting the importance of ZBP1 in the addition of linear ubiquitin chains to RIPK1 in response to LPS. (Fig. 4 B and C). The fact that ɑTNF treatment did not further attenuate M1-ubiquitination in Zbp1−/− cells suggests that ZBP1 may additionally regulate M1-ubiquitination of RIPK1 downstream of TNFR.
Fig. 4.
ZBP1 mediates RIPK1 M1-ubiquitination to promote interactions within the proinflammatory complex. (A and B) RIPK1-specific immunoprecipitation in (A) B6, Trif−/−, and Zbp1−/− macrophages treated with LPS for 30 or 45 min or (B) B6 and Zbp1−/− macrophages treated with LPS for 30 or 60 min ± ɑTNF and probed for M1-ubiquitin and RIPK1. (C) Quantification of M1-ubiquitination in LPS-activated B6 and Zbp1−/− macrophages/unstimulated ± ɑTNF. (D and E) mRNA levels in indicated cells stimulated with (D) LPS or (E) TNF. (F) ZBP1-specific immunoprecipitation in B6 BMDMs stimulated with LPS for indicated timepoints ± LUBAC inhibition and probed for proinflammatory complex components. Data from immunoblots are representative of three or more biologically independent experiments. Unless otherwise indicated, blots were singly or doubly probed to ensure highest quality. All blots from a specific panel are run from a single set of lysates that were identically handled. For qPCR data, data points indicate the mean from triplicate wells of four biologically independent experiments. ANOVA was used for comparison between groups: ns, nonsignificant (P > 0.05); *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To rule out the possibility that ZBP1 regulates LPS-induced inflammatory signaling downstream of autocrine/paracrine TNF signaling instead of via direct interactions with TRIF, we compared inflammatory cytokine production in TNF- and ZBP1-deficient macrophages (Fig. 4D). While TNF deficiency did reduce CXCL-1 mRNA levels in response to LPS, particularly at later timepoints, Tnf−/− cells did not recapitulate the reduction in cytokine production observed in Zbp1−/− (Fig. 4D). Although these results indicate that ZBP1 does not regulate LPS-inflammatory responses solely downstream of autocrine/paracrine TNF, it does not rule out the possibility that ZBP1 contributes to inflammatory responses downstream of TNFR. Indeed, we found that ZBP1 deficiency reduced inflammatory cytokine production in response to TNF, suggesting a potential additional role for ZBP1 in TNF signaling, potentially via regulation of interactions with RIPK1 (Fig. 4E).
M1-ubiquitination of RIPK1 is mediated by the E3 ligase linear ubiquitin chain assembly complex (LUBAC) (27, 28), which has been characterized primarily downstream of TNFR activation. To see if LUBAC exerts its effect at the level of ZBP1, we investigated whether inhibition of LUBAC affects the recruitment of proinflammatory complex components to ZBP1 in response to LPS (Fig. 4F). Strikingly, LUBAC inhibition decreased binding of TRAF3, TBK1, and NEMO to ZBP1 but had no impact on S321 phosphorylation of RIPK1 or ZBP1 binding to TRAM (Fig. 4F). LUBAC inhibition also decreased MAPK and IRF3 activation at later timepoints after treatment with LPS, in agreement with a role for LUBAC in promoting TRIF/RIPK1-dependent inflammatory responses (Fig. 4F). These findings supported the model that ZBP1 enhances recruitment of RIPK1 to TRIF, promoting LUBAC-mediated binding of proinflammatory complex components.
ZBP1 Levels Modulate RHIM-Mediated Inflammatory Responses.
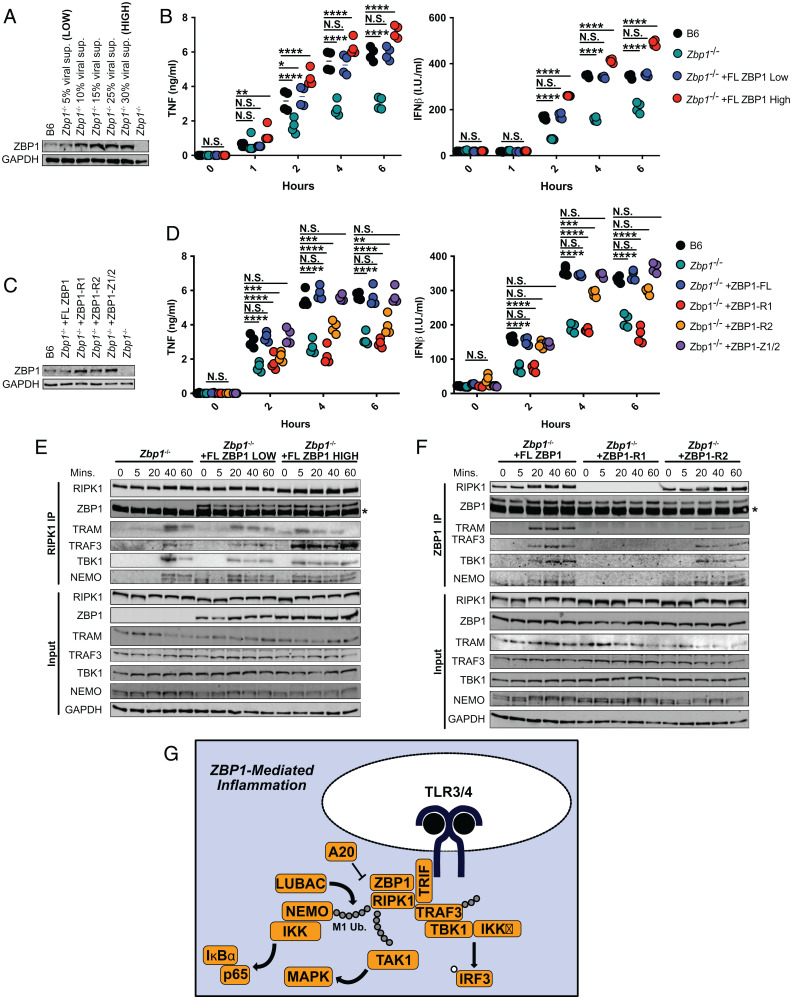
The delay, rather than the abrogation, of the RIPK1-specific proinflammatory complex formation in the absence of ZBP1 (Fig. 3 A–E) suggested a quantitative contribution of ZBP1 to inflammatory responses to LPS. To investigate if the levels of ZBP1 might modulate the extent of ZBP1-dependent activation, we generated cells with HIGH and LOW levels of FLAG-tagged ZBP1 (Fig. 5A and SI Appendix, Fig. S2 A and B). The levels of ZBP1 in ZBP1 LOW cells closely recapitulated endogenous wild-type ZBP1 levels, while ZBP1 HIGH cells expressed 2 to 3× more ZBP1 (Fig. 5A and SI Appendix, Fig. S2 A and B). Reconstitution of Zbp1−/− macrophages with low levels of ZBP1 recapitulated the TNF and IFNβ production observed in B6 (Fig. 5 A and B). However, reconstitution with high levels of ZBP1 enhanced inflammatory cytokine production significantly, compared to B6, indicating that ZBP1 levels tightly mediate the extent of the inflammatory response to LPS (Fig. 5 A and B). This is of particular interest, since constitutive and induced IFNs regulate ZBP1 levels (10, 29), providing physiologically relevant contexts in which differential ZBP1 expression may modulate inflammatory responses across cell types and tissues.
Fig. 5.
ZBP1 levels modulate RHIM-mediated inflammatory responses. (A) ZBP1 levels in B6 and Zbp1−/− macrophages transduced with indicated concentrations of viral supernatant for ZBP1. (B) TNF and IFNβ protein levels in B6, Zbp1−/−, and Zbp1−/− macrophages reconstituted with low or high levels of ZBP1. (C) ZBP1 levels in Zbp1−/− macrophages transduced with mutant ZBP1 constructs lacking RHIM1 (ZBP1-R1), RHIM2 (ZBP1-R2), or both Zɑ domains. (D) TNF and IFNβ protein levels in B6, Zbp1−/−, and Zbp1−/− macrophages reconstituted with RHIM or Zɑ mutant ZBP1 constructs. (E) RIPK1 immunoprecipitation in Zbp1−/− macrophages reconstituted with low and high levels of ZBP1 and probed for proinflammatory complex components. Asterisk indicates band from antibody heavy chain. (F) ZBP1 immunoprecipitation in Zbp1−/− macrophages reconstituted with full-length or RHIM-mutant ZBP1 and probed for proinflammatory complex components. Asterisk indicates band from antibody heavy chain. (G) Model of ZBP1-regulated complex formation leading to inflammation and cell death in response to LPS. Data from immunoblots are representative of three or more biologically independent experiments. Unless otherwise indicated, blots were singly or doubly probed to ensure highest quality. All blots from a specific panel are run from a single set of lysates that were identically handled. For ELISA data, data points indicate the mean from triplicate wells of four biologically independent experiments. ANOVA was used for comparison between groups: ns, nonsignificant (P > 0.05); *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To determine if ZBP1 levels define the magnitude of the inflammatory response to LPS by modulating the timing of inflammatory complex formation, we performed a RIPK1-specific immunoprecipitation in Zbp1−/− cells reconstituted with low and high levels of ZBP1 and stimulated with LPS (Fig. 5E). Indeed, reconstitution with low levels of ZBP1 accelerated complex formation compared to Zbp1−/−, with inflammatory complex component recruitment occurring 20 min after stimulation, compared to 40 min in the absence of ZBP1 (Fig. 5E). High levels of ZBP1 enhanced inflammatory complex formation further, with binding of TRAM, TRAF3, TBK1, and NEMO detectable as early as 5 min after stimulation with LPS (Fig. 5E). These results support our hypothesis that the levels of ZBP1 modulate LPS-induced inflammation by tightly controlling the timing of inflammatory complex formation.
It is important to note that overexpression of full-length ZBP1 often results in extensive cell death during transduction (10). To rule out the possibility that differential cell death contributes to the phenotypes we observed in our ZBP1 HIGH and ZBP1 LOW cells, we confirmed that no appreciable death occurred in response to LPS in these cells (SI Appendix, Fig. S2C). However, when cell death was induced via addition of TAK1 inhibitor 5z7 to LPS, reconstitution of Zbp1−/− cells with low levels of ZBP1 recapitulated B6, while ZBP1 HIGH cells exhibited accelerated and enhanced cell death compared to wild type (SI Appendix, Fig. S2D).
To determine the potential differential roles for the RHIM and Zɑ domains in ZBP1-mediated inflammation, we generated FLAG-tagged mutant ZBP1 constructs lacking RHIM1 (ZBP1-R1), RHIM2 (ZBP1-R2), or both Zɑ domains (Fig. 5C). As before, we titrated the levels of the ZBP1 mutants to recapitulate the endogenous levels of ZBP1 observed in wild-type cells (Fig. 5C and SI Appendix, Fig. S2 A and B). Reconstitution of Zbp1−/− cells with full-length ZBP1 rescued TNF and IFNβ production (Fig. 5D). However, reconstitution with ZBP1 lacking either RHIM1 (ZBP1-R1) or RHIM2 (ZBP1-R2) resulted in attenuated inflammatory cytokine production, indicating that both RHIM domains are important for ZBP1-mediated regulation of inflammation (Fig. 5D). The effect of RHIM1 deletion appeared to have a more significant impact on inflammatory cytokine production than RHIM2 deletion, particularly at later timepoints (Fig. 5D). Even though the Zɑ domains of ZBP1 have proven to play an important role in the context of viral infection (1, 5, 29), the Zɑ domains were not required for ZBP1-mediated inflammation in response to LPS (Fig. 5D).
To investigate the potential roles for the individual RHIM domains of ZBP1 further, we performed a ZBP1-specific immunoprecipitation in Zbp1−/− macrophages reconstituted with full-length and RHIM-mutant ZBP1 (Fig. 5F). As expected, lack of RHIM1 abrogated ZBP1-RIPK1 binding but also appeared to completely inhibit recruitment of inflammatory complex components to ZBP1, highlighting the importance of RIPK1 in the ZBP1-mediated inflammatory response to LPS (Fig. 5F). While a lack of RHIM2 appeared to only slightly decrease the recruitment of TRAF3, TBK1, and NEMO to ZBP1, TRAM binding was significantly reduced, suggesting that the second RHIM of ZBP1 may enhance or stabilize interactions with TRIF (Fig. 5F).
In light of these findings, we present a model in which in macrophages, ZBP1 is constitutively bound to RIPK1 via RHIM1-mediated interactions. The RHIM domains of ZBP1 further mediate interactions between TRIF and RIPK1 upon LPS induction. When ZBP1 levels are high, these interactions are accelerated and enhanced, and when ZBP1 is absent, interactions between TRIF and RIPK1 are substantially delayed. Recruitment of RIPK1 to TRIF induces ubiquitination of RIPK1 by LUBAC, subsequently nucleating the recruitment of TAK1, NEMO, and the IkappaB kinase (IKK) complex to drive MAPK and NF-κB activation as well as TRAF3 and TBK1 to promote induction of IFN signaling downstream of TRIF (Fig. 5G).
ZBP1 Promotes LPS-Induced Septic Shock and Poly(I:C)-Induced Inflammation.
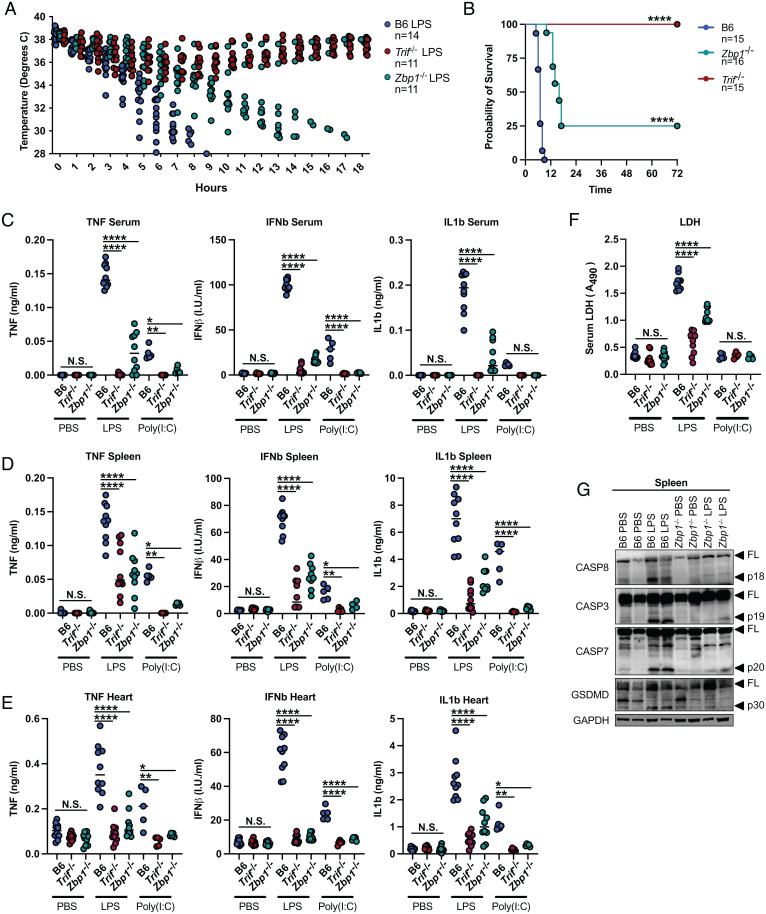
To determine the physiological relevance of ZBP1 in the regulation of LPS responses, we injected Zbp1−/− and Trif−/− mice intraperitoneally (IP) with LPS to induce septic shock. While Zbp1−/− mice were not as resistant to LPS as Trif−/−, they exhibited significantly delayed onset of hypothermia (Fig. 6A). To this end, while B6 mice succumbed to septic shock within 5 to 9 h of injection, 75% of Zbp1−/− mice did not succumb until 12 to 20 h after injection, and 25% survived for over 72 h (Fig. 6B). These results mirror our in vitro data in which ZBP1 deficiency delays, but does not entirely prevent, inflammatory complex formation and inflammatory signaling cascade activation in response to LPS. Furthermore, these results indicate that ZBP1 plays a critical role in the pathology of septic shock, potentially by promoting inflammatory responses and cell death in response to LPS.
Fig. 6.
ZBP1 promotes LPS-induced septic shock and poly(I:C)-induced inflammation. (A and B) Mouse body temperature (A) and survival (B) over time in B6, Trif−/−, and Zbp1−/− mice after IP injection with 25 mg/kg LPS. (C–E) TNF, IFNβ, and IL-1β protein levels in the (C) serum, (D) spleen, and (E) heart of B6, Trif−/−, and Zbp1−/− mice injected IP with 25 mg/kg LPS or i.v. with 15 mg/kg poly (I:C) 4 h after injection. (F) Relative LDH levels in the serum of B6, Trif−/−, and Zbp1−/− mice injected IP with 25 mg/kg LPS or i.v. with 15 mg/kg poly (I:C) 4 h after injection. For ELISA and LDH assays, data points indicate individual mice tested (B6 PBS, n = 10; Trif−/− PBS, n = 10; Zbp1−/− PBS, n = 10; B6 LPS, n = 10; Trif−/− LPS, n = 10; Zbp1−/− LPS, n = 10; B6 poly(I:C), n = 5; Trif−/− poly(I:C), n = 5; Zbp1−/− poly(I:C), n = 5). (G) Levels of full-length and cleaved CASP8, CASP3, CASP7, and GSDMD in the spleen of B6 and Zbp1−/− mice injected IP with 25 mg/kg LPS or PBS. Data from immunoblots are representative of three or more independent experiments. Unless otherwise indicated, blots were singly or doubly probed to ensure highest quality. All blots from a specific panel are run from a single set of lysates that were identically handled. Log-rank (Mantel-Cox) test was used for comparison of survival curves. ANOVA was used for comparison between groups: ns, nonsignificant (P > 0.05); *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To determine if ZBP1 confers resistance to LPS shock by dampening TRIF-mediated inflammatory responses in vivo, we compared serum and tissue cytokine levels in B6, Zbp1−/−, and Trif−/− mice 4 h after intraperitoneal injection with LPS or intravenous (i.v.) injection of poly(I:C). Poly(I:C) injection does not readily induce acute hypothermia and lethality in mice and is more commonly used to promote protective immunity to viral and bacterial infections (30, 31). However, both LPS and poly(I:C) injection induced robust TNF production which was strikingly reduced in the serum and spleen of Zbp1−/− and Trif−/− mice (Fig. 6 C and D). Similarly, IFNβ and interleukin (IL)-1β levels in the serum and spleen were significantly reduced in the absence of ZBP1 compared to wild-type B6 control mice, suggesting a role for ZBP1 in the induction of inflammatory responses to LPS and poly(I:C) in vivo (Fig. 6 C and D). In further support, levels of TNF, IFNβ, and IL-1β in the heart were significantly reduced by ZBP1 deficiency (Fig. 6E), which was of particular interest since the impaired myocardial contractile function and elevated cardiomyocyte proinflammatory cytokine production induced in the context of septic shock contribute significantly to pathogenesis (32, 33). ZBP1-dependent IL-1β secretion was indicative of inflammatory cell death and suggested a potential role for ZBP1 in the regulation of cell death in response to LPS. In support of ZBP1-dependent cell death, serum lactate dehydrogenase (LDH) levels indicative of lytic cell death were attenuated in Zbp1−/− mice compared to B6 in response to LPS (Fig. 6F). However, in agreement with the low serum IL-1β levels observed in response to poly(I:C) (Fig. 6C), poly(I:C) injection did not induce LDH release, likely due to a lack of lytic cell death (Fig. 6F). Cleavage of CASP8, CASP3, CASP7, and the effector of pyroptosis gasdermin-D (GSDMD) was abrogated in the spleen of LPS-injected Zbp1−/− mice, indicating decreased apoptotic and pyroptotic cell death within the tissue (Fig. 6G). These results reveal a critical role for ZBP1 in driving TRIF-mediated inflammatory and cell death responses in vivo.
Discussion
These findings further extend the role of ZBP1 in the TRIF-dependent complex termed the TRIFosome, previously characterized in the context of pyroptotic cell death (10). Here, we show that a similar complex assembles in conditions that favor the proinflammatory response rather than cell death and that the formation of the TRIFosome is dependent on the levels of ZBP1. These data also demonstrate an unanticipated and key role for ZBP1, typically considered to be a sensor of viral infections, in TRIF-mediated inflammation and cell death downstream of TLR3/4. These findings support a model in which ZBP1, by functioning as a RHIM-containing scaffolding protein, promotes nucleation of TRIF-RIPK1 complexes downstream of TLR3/4. In this role, ZBP1 modulates TRIF-mediated inflammation, with slight changes in ZBP1 levels tuning the onset, kinetics, and magnitude of the proinflammatory response to LPS. This model implicates the ubiquitinase complex LUBAC, previously characterized primarily in the context of TNFR ligation, in the assembly of the TRIFosome. Specifically, recruitment of TRAF3, TBK1, and NEMO to the TRIFosome was abrogated upon inhibition of LUBAC activity, while ZBP1-dependent recruitment of RIPK1 to TRAM was unaffected. These findings suggest there may be additional crossover between TNFR and TLR signaling pathways in their mechanism of activation. The discovery that LUBAC and ZBP1 are involved in both TLR- and TNFR-mediated inflammatory signaling demonstrates the versatility and redundancy of the mechanisms of pathway activation. As such a common mediator, ZBP1 facilitates not only the intersection of viral and bacterial responses, but also the junction of TLR- and TNFR1-mediated responses.
This function has crucial consequences to the outcome of the LPS-mediated response, best exemplified by the striking discovery that mice lacking ZBP1 exhibit delayed hypothermia and 25% survival in response to LPS-induced septic shock. This finding also has important implications in the context of RIPK1 biology, as it positions RIPK1 as an important regulator of septic shock. However, it is difficult to assess the role of RIPK1 in vivo as mice lacking RIPK1 die perinatally, and various RIPK1 conditional knockouts result in autoimmune phenotypes (34–36), LPS-induced lethality has been shown to be independent of RIPK1 kinase activity (37). However, this role for ZBP1 in LPS-induced septic shock suggests that RIPK1 scaffolding functions as opposed to kinase activity may drive the inflammatory responses that contribute to lethality. Additionally, slight changes in the kinetics of inflammatory complex formation mediated by ZBP1 have a significant impact on the magnitude of the inflammatory response. More broadly, our results hint at the possibility of widespread crosstalk between the various RHIM-driven pathways, wherein RHIM-containing members of one pathway mediate significant effects as rheostatic regulators of other pathways, with profound physiological outcomes.
Materials and Methods
Mice and Macrophages.
C57BL/6 (B6) and Ticam1−/− (Trif−/−) mice were obtained from The Jackson Laboratory. Mice were housed according to protocols approved by the Tufts University Medical School Animal Care and Use Committees. Femurs from Ticam2−/− (Tram−/−) mice were generously donated by L. Li. Zbp1−/− mice were provided by S. Balachandran. Zbp1−/− mice were backcrossed to C57BL/6 for more than 11 generations before being intercrossed to eliminate any potential contribution from the 129 genetic background on which the knockout was initially derived. To generate the BMDMs used in this study, BM was isolated from the long bones of mice, propagated in Roswell Park Memorial Institute (RPMI) containing 20% fetal bovine serum (FBS), 2% Pen-Strep, and 30% L cell supernatant on nontissue-culture-treated Petri dishes for 7 d. Once differentiated, BMDMs were plated for experiments at a density of 1 × 106 cm2 in RPMI containing 20% FBS and 2% Pen-Strep. Ripk1−/− mouse fetal liver macrophages were generated and donated by K. Fitzgerald.
Injection of LPS or Poly(I:C) and Preparation of In Vivo Samples.
Prior to induction of the septic shock or poly(I:C)-induced inflammation models, mice were cohoused for at least 2 wk. The 11-wk-old male and female C57BL/6J, and Zbp1−/− mice were injected IP with 25 mg/kg LPS in sterile phosphate buffered saline (PBS) (500 μL) or i.v. with 15 mg/kg poly(I:C) (CAS #31852-29-6) in sterile PBS (100 μL). Temperature was monitored regularly by rectal thermometer. At indicated timepoints after administration of LPS or poly(I:C), or when mouse body temperature reached <30 °C, mice were euthanized by CO2 asphyxiation. Spleen, heart, and blood were harvested for quantification of cytokine levels by enzyme-linked immunosorbent assay (ELISA) and death effector cleavage by immunoblot. All experiments were performed in accordance with regulations and approval of the Tufts University Institutional Animal Care and Use Committee.
Reagents.
LPS S. minnesota R5 (10 ng/mL), LTA S. aureus (2 μg/mL), poly(I:C) (CAS #31852-29-6; 20 ug/mL), and 5z7 (125 nM) were purchased from Sigma. Recombinant murine TNF-ɑ (50ng/mL) was purchased from Peprotech (Cat. 315-01A). The LUBAC inhibitor HOIPIN-8 (30 μM) was purchased from Axon Medchem.
RNA Isolation and Analysis.
The 5 × 105 BMDMs were plated on 24-well tissue-culture-treated plates. Cells were lysed with TRIzol (Invitrogen), and RNA extraction was carried out according to the manufacturer’s instructions. Reverse transcription was performed using M-MuLV reverse transcriptase, RNase inhibitor, random primers 9, and deoxyribonucleotide triphosphate mix (New England BioLabs) to synthesize complementary DNA (cDNA). cDNA was analyzed for relative mRNA levels using SYBR Green (Applied Biosystems) and intron spanning primers. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used to normalize mRNA levels. Postamplification melting curve analysis was performed to confirm primer specificity.
Quantitative PCR Primers.
TNF: (F) 5′-CTGTAGCCCACGTCGTAGC-3′,
(R) 5′-TTGAGATCCATGCCGTTG-3′
CXCL-1: (F) 5′-TGAGCTGCGCTGTCAGTG-3′,
(R) 5′-AGAAGCCAGCGTTCACCAGA-3′
IFNβ: (F) 5′-CAGCTCCAAGAAAGGACGAAC-3′,
(R) 5′-GGCAGTGTAACTCTTCTGCAT-3′
GAPDH: (F) 5′-GGAGAGTGTTTCCTCGTCCC-3′,
(R) 5′-TTCCCATTCTCGGCCTTGAC-3′.
ELISA.
At indicated timepoints after treatment with LPS, cell supernatants were collected, and cytokine secretion was measured by ELISA. Murine TNF (DY410) and CXCL-1 (DY453) and IL-1β (DY401) DuoSet ELISA kits were used according to the manufacturer’s instructions. For IFNβ ELISA, cell supernatants were added to 384-well ELISA plates coated overnight at 4 °C with monoclonal rat anti-mouse IFNβ antibody (Santa Cruz sc-57201, 1:500 dilution in 0.1 M carbonate buffer) and blocked with 10% FBS in PBS for 2 h at 37 °C. Supernatants were incubated on plates overnight at 4 °C before washing with 0.005% polysorbate 20 in PBS and adding polyclonal rabbit anti-mouse IFNβ antibody (R&D Systems 32400-1, 1:2,000 dilution in 10% FBS in PBS) overnight at 4 °C. After washing, goat anti-rabbit-HRP antibody (Cell Signaling Technologies, 7074, 1:2,000 dilution in 10% FBS in PBS) was added for 2 to 3 h at room temperature. Tetramethylbenzidine substrate was added, and the reaction was stopped with 2N H2SO4.
Immunoblotting.
After indicated treatments, cells were lysed in 1X Laemmli Buffer containing 5% β-mercaptoethanol, boiled for 15 min, and incubated on ice for 15 min. Primary antibodies against p38 (9212), p-p38 (4511), ERK1/2 (4696), p-ERK1/2 (4370), JNK (9255), p-JNK (9255), pIRF3 (29047), IκBα (4814), p-IκBα (9246), full-length CASP8 (4790), cleaved CASP8 (8592), CASP3 (9665), CASP7 (9492), RIPK1 (3493), pRIPK1 (Ser166) (31122), pRIPK1 (Ser321) (83613), TRAF3 (4729), TBK1 (3504), NEMO (2685), A20 (5630), p65 (8242), p-p65 (3033), and GAPDH (2118) were purchased from Cell Signaling Technologies. ZBP1 antibody (AG-20B-0010-C100) was purchased from Adipogen. GSDMD (ab209845) was purchased from Abcam. FADD antibody (05-486) was purchased from Millipore Sigma. TRAM antibody was purchased from Santa Cruz (sc-376076). M1-ubiquitin antibody (MABS451) was purchased from Sigma. Secondary antibodies, anti-rabbit IgG (H+L) (DyLight 800 4X PEG Conjugate) (5151), and anti-mouse IgG (H+L) (DyLight 800 4X PEG Conjugate) (5257) were purchased from Cell Signaling Technologies.
ZBP1 and RIPK1 Immunoprecipitations.
Indicated BMDMs were plated on 6-well tissue-culture-treated plates, stimulated as indicated, and harvested in immunoprecipitation lysis buffer (0.5% Triton X, 50 mM Tris Base [pH 7.4], 150 mM NaCl, 2 mM ethylenediaminetetraacetic acid, 2 mM ethylene glycol tetraacetic acid, 1X protease inhibitor mixture). Lysed cells were rotated for 60 min at 4 °C with intermittent vortexin and centrifuged at 5,000 X g for 5 min, and the supernatant was incubated with α-RIPK1 (BD Biosciences 610459) or α-ZBP1 (AG-20B-0010-C100) antibody-conjugated protein G agarose beads (Cell Signaling Technology 37478). Samples were washed three times in immunoprecipitation lysis buffer, and protein complexes were eluted with 1X Laemmli buffer containing 5% β-mercaptoethanol at 90 °C for 15 min.
High-Magnification TRIF and RIPK1 Imaging.
To observe TRIF and RIPK1 localization, macrophages were seeded at a density of 1 × 106 cm2 in RPMI on 1.17-mm-thick glass bottom imaging plates. Cells were stimulated with LPS for indicated times, fixed in 4% paraformaldehyde for 15 min, blocked in 1X PBS (5% FBS, 0.2% Triton X-100), and incubated overnight with anti-TRIF (abcam ab13810) or anti-RIPK1 (BD Biosciences 610459) antibody followed by a 2-h incubation in the presence of Alexa Fluor 555 conjugated goat anti-mouse (ab150114) and Alexa Fluor 405 conjugated goat anti-rabbit (ab175652) antibodies. The Lionheart automated microscope was used to image cells at 60× magnification to capture ∼5 cells/field of view in quadruplicate. Gen5 3.10 imaging software was used to quantify positive staining for each protein, and a mask was generated to calculate puncta/cell.
Lentiviral Constructs and Transduction.
For ZBP1 reconstitution experiments, full-length and mutagenized versions of ZBP1 sequences were ligated into a pLEX lentiviral vector. ZBP1 sequences lacking RHIM 1 were generated using 5′-tgtggtgagccaggtcc-3′, 5′-ggcaatggagatgtggc-3′ primers. Sequences lacking RHIM 2 were generated using 5′-gcagggaagcacccc-3′, 5′-ctgagctccgtgggc-3′ primers. Sequences lacking Z-DNA-binding domains 1 and 2 (Z1/2) were generated sequentially using 5′-tctggagatggggctcc-3′, 5′-aggagcttctgccatggt-3′ followed by 5′-caggaaggccaagacatagc-3′, 5′-agggattgcaggagcc-3′ primers. Lentiviral particles were generated by transfection with packaging vector psPAX (plasmid 12260; Addgene) and the VSV-G pseudotyping vector pMD2.G (plasmid 12259; Addgene) into the 293T cell line. Generated viral supernatant containing lentiviral particles was used to transduce Zbp1−/− macrophages at indicated concentrations on day 4 of differentiation, followed by puromycin selection (3 ug/mL, 48 h) starting on day 6.
Quantification and Statistical Analysis.
For ELISA and qPCR data, data points indicate the mean from triplicate wells of four biologically independent experiments. Immunoblots are representative of three or more independent experiments. Significance was determined using a one-way or two-way ANOVA as appropriate: ns (nonsignificant), P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 . Log-rank (Mantel-Cox) test was used for comparison of survival curves.
Supplementary Material
Acknowledgments
We thank Dr. K. Fitzgerald and Dr. L. Li for sharing various mouse strains and cell lines for this study. We thank the Tufts University Genomics Core and A. Tai for help with RNA-sequencing data analysis. This work was supported by NIH grants R01 AI167245 and R01 AI056234 to A.P.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2113872119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Thapa R. J., et al. , DAI senses influenza A virus genomic RNA and activates RIPK3-dependent cell death. Cell Host Microbe 20, 674–681 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Upton J. W., Kaiser W. J., Mocarski E. S., DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host Microbe 11, 290–297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kesavardhana S., et al. , ZBP1/DAI ubiquitination and sensing of influenza vRNPs activate programmed cell death. J. Exp. Med. 214, 2217–2229 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuriakose T., et al. , ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci. Immunol. 1, 1–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang T., et al. , Influenza virus Z-RNAs induce ZBP1-mediated necroptosis. Cell 180, 1115–1129.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Upton J. W., Kaiser W. J., Mocarski E. S., Virus inhibition of RIP3-dependent necrosis. Cell Host Microbe 7, 302–313 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nogusa S., et al. , RIPK3 activates parallel pathways of MLKL-driven necroptosis and FADD- mediated apoptosis to protect against influenza A virus. Cell Host Microbe 20, 13–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin J., et al. , RIPK1 counteracts ZBP1-mediated necroptosis to inhibit inflammation. Nature 540, 124–128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newton K., et al. , RIPK1 inhibits ZBP1-driven necroptosis during development. Nature 540, 129–133 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Muendlein H. I., et al. , ZBP1 promotes LPS-induced cell death and IL-1β release via RHIM-mediated interactions with RIPK1. Nat. Commun. 12, 86 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muendlein H. I., et al. , cFLIPL protects macrophages from LPS induced pyroptosis via inhibition of complex II formation. Science 367, 1379–1384 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarhan J., et al. , Caspase-8 induces cleavage of gasdermin D to elicit pyroptosis during Yersinia infection. Proc. Natl. Acad. Sci. U.S.A. 115, E10888–E10897 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meylan E., et al. , RIP1 is an essential mediator of Toll-like receptor 3-induced NF-κ B activation. Nat. Immunol. 5, 503–507 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Covert M. W., Leung T. H., Gaston J. E., Baltimore D., Achieving stability of lipopolysaccharide-induced NF-kB activation. Science 309, 1854–1858 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Fitzgerald K. A., et al. , IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4, 491–496 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Rebsamen M., et al. , DAI/ZBP1 recruits RIP1 and RIP3 through RIP homotypic interaction motifs to activate NF-kappaB. EMBO Rep. 10, 916–922 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaiser W. J., Upton J. W., Mocarski E. S., Receptor-interacting protein homotypic interaction motif-dependent control of NF-κ B activation via the DNA-dependent activator of IFN regulatory factors. J. Immunol. 181, 6427–6434 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takaoka A., et al. , DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448, 501–505 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Cusson-Hermance N., Khurana S., Lee T. H., Fitzgerald K. A., Kelliher M. A., Rip1 mediates the Trif-dependent toll-like receptor 3- and 4-induced NF-kappaB activation but does not contribute to interferon regulatory factor 3 activation. J. Biol. Chem. 280, 36560–36566 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Geng J., et al. , Regulation of RIPK1 activation by TAK1-mediated phosphorylation dictates apoptosis and necroptosis. Nat. Commun. 8, 359 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dondelinger Y., et al. , MK2 phosphorylation of RIPK1 regulates TNF-mediated cell death. Nat. Cell Biol. 19, 1237–1247 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Kagan J. C., et al. , TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat. Immunol. 9, 361–368 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto M., et al. , TRAM is specifically involved in the Toll-like receptor 4-mediated MyD88-independent signaling pathway. Nat. Immunol. 4, 1144–1150 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Degterev A., Ofengeim D., Yuan J., Targeting RIPK1 for the treatment of human diseases. Proc. Natl. Acad. Sci. U.S.A. 116, 9714–9722 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mifflin L., Ofengeim D., Yuan J., Receptor-interacting protein kinase 1 (RIPK1) as a therapeutic target. Nat. Rev. Drug Discov. 19, 553–571 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ofengeim D., Yuan J., Regulation of RIP1 kinase signalling at the crossroads of inflammation and cell death. Nat. Rev. Mol. Cell Biol. 14, 727–736 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Zinngrebe J., et al. , LUBAC deficiency perturbs TLR3 signaling to cause immunodeficiency and autoinflammation. J. Exp. Med. 213, 2671–2689 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oikawa D., et al. , Molecular bases for HOIPINs-mediated inhibition of LUBAC and innate immune responses. Commun. Biol. 3, 163 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu Y., et al. , Cloning of DLM-1, a novel gene that is up-regulated in activated macrophages, using RNA differential display. Gene 240, 157–163 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Gowen B. B., et al. , TLR3 is essential for the induction of protective immunity against Punta Toro Virus infection by the double-stranded RNA (dsRNA), poly(I:C12U), but not Poly(I:C): Differential recognition of synthetic dsRNA molecules. J. Immunol. 178, 5200–5208 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Homan E. R., Zendzian R. P., Schott L. D., Levy H. B., Adamson R. H., Studies on poly I:C toxicity in experimental animals. Toxicol. Appl. Pharmacol. 23, 579–588 (1972). [DOI] [PubMed] [Google Scholar]
- 32.Boyd J. H., Mathur S., Wang Y., Bateman R. M., Walley K. R., Toll-like receptor stimulation in cardiomyoctes decreases contractility and initiates an NF-kappaB dependent inflammatory response. Cardiovasc. Res. 72, 384–393 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Binck B. W., et al. , Bone marrow-derived cells contribute to contractile dysfunction in endotoxic shock. Am. J. Physiol. Heart Circ. Physiol. 288, H577–H583 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Kelliher M. A., et al. , The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity 8, 297–303 (1998). [DOI] [PubMed] [Google Scholar]
- 35.O’Donnell J. A., et al. , Dendritic cell RIPK1 maintains immune homeostasis by preventing inflammation and autoimmunity. J. Immunol. 200, 737–748 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dannappel M., et al. , RIPK1 maintains epithelial homeostasis by inhibiting apoptosis and necroptosis. Nature 513, 90–94 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newton K., et al. , RIPK3 deficiency or catalytically inactive RIPK1 provides greater benefit than MLKL deficiency in mouse models of inflammation and tissue injury. Cell Death Differ. 23, 1565–1576 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.