Significance
Proteins that undergo phase separation promote biomolecular condensate formation and play a significant role in many biological processes. We divided these proteins into two categories according to their underlying driving force when forming condensates: self-assembling proteins, which interact with the same protein species, and partner-dependent proteins, which interact with different biomolecule species. Most of the current computational tools preferentially predict self-assembling proteins and perform poorly in screening partner-dependent proteins. We thus built machine-learning models to predict the two protein categories separately. Further validation on the condensate proteome revealed that partner-dependent proteins are widespread in cells. We also developed a web server that integrates multiple phase-separation predictors, providing a convenient way for biologists to discover candidate phase-separating proteins.
Keywords: phase separation, metapredictor, self-assembly, partner-dependent, phosphorylation
Abstract
Protein self-assembly is one of the formation mechanisms of biomolecular condensates. However, most phase-separating systems (PS) demand multiple partners in biological conditions. In this study, we divided PS proteins into two groups according to the mechanism by which they undergo PS: PS-Self proteins can self-assemble spontaneously to form droplets, while PS-Part proteins interact with partners to undergo PS. Analysis of the amino acid composition revealed differences in the sequence pattern between the two protein groups. Existing PS predictors, when evaluated on two test protein sets, preferentially predicted self-assembling proteins. Thus, a comprehensive predictor is required. Herein, we propose that properties other than sequence composition can provide crucial information in screening PS proteins. By incorporating phosphorylation frequencies and immunofluorescence image-based droplet-forming propensity with other PS-related features, we built two independent machine-learning models to separately predict the two protein categories. Results of independent testing suggested the superiority of integrating multimodal features. We performed experimental verification on the top-scored proteins DHX9, Ki-67, and NIFK. Their PS behavior in vitro revealed the effectiveness of our models in PS prediction. Further validation on the proteome of membraneless organelles confirmed the ability of our models to identify PS-Part proteins. We implemented a web server named PhaSePred (http://predict.phasep.pro/) that incorporates our two models together with representative PS predictors. PhaSePred displays proteome-level quantiles of different features, thus profiling PS propensity and providing crucial information for identification of candidate proteins.
Phase separation (PS) is one of the mechanisms mediating the formation of membraneless compartments from macromolecules, such as proteins and nucleic acids (1). Multivalent weak interactions between these molecules are the driving force of PS. The interactions can generally be classified into two categories: one mediated by intrinsically disordered regions (IDRs) and the other mediated by multiple modular domains or motifs (2, 3). Proteins with high IDR content can interconvert between a range of different low-energy states. A single species can undergo IDR-mediated PS. In contrast, multivalent interactions mediated by multiple modular domains are more specific and usually require two or more different protein species to participate in PS (2). For example, the IDRs of Ddx4 self-assemble spontaneously to form membraneless compartments in living cells and in vitro (4), while single proteins within the LAT–Grb2–Sos1 PS system cannot undergo liquid–liquid PS (LLPS) (5). Herein, we characterize proteins that can self-assemble to form condensates as self-assembling PS (PS-Self) proteins, and we define proteins whose PS behaviors are regulated by partner components (proteins or nucleic acids) as partner-dependent PS (PS-Part) proteins.
Many bioinformatics tools have been developed to predict PS-related features and aid in screening PS proteins. Representative tools include PScore (6), PLAAC (7), catGRANULE (8), LARKS (9), ZipperDB (10), and the recently published Fuzdrop (11) and DeePhase (12). Among these tools, PLAAC and ZipperDB were not originally developed to screen PS proteins. Instead, they predict prion-like domains (PLDs) and fibril-forming segments, respectively. Although trained on the yeast proteome, PLAAC was later extended to screen human proteins and displayed exemplary performance in predicting PS proteins. PScore, catGRANULE, and LARKS were first-generation PS predictors. However, the different training samples of these methods lead to differences in their predictive behavior. For example, PScore and LARKS learned PS sequence patterns from proteins that have self-assembling behaviors, while catGRANULE was trained on granule participants. Compared with the first-generation predictors, FuzDrop and DeePhase were developed on proteins collected from PS databases. Using a larger number of training samples allows them to provide a broader perspective for screening PS proteins.
Recently, we performed a comprehensive analysis of 6 PS predictors on 278 PS proteins. The results showed that although these tools perform well in predicting PS proteins, they all prefer proteins with high IDR content (13). In addition, these first-generation predictors recognize vastly different kinds of proteins because they were developed to screen various sequence features (14). This calls for the development of a comprehensive metapredictor. There is currently no computational tool that can identify partner-dependent PS proteins. However, most PS systems involve multiple partners in biological conditions (14), as observed for RNA-binding proteins (15) and signaling complexes (5). The annotations collected from PhaSepDB (16) display similar patterns, with more PS-Part than PS-Self proteins (Dataset S1). Therefore, it is necessary to develop a predictor for screening potential partner-dependent proteins.
Here, we propose that the different amino acid patterns of self-assembling and partner-dependent proteins allow us to establish independent models to separately screen the two PS protein categories. We found that sequence-based features can distinguish PS proteins from non-PS proteins, and demonstrated that properties other than sequence composition, such as posttranslational modification (PTM) frequencies and immunofluorescence (IF) images, can provide crucial information. By incorporating multimodal features, we trained machine-learning models on experimentally validated proteins. Results of independent testing showed that our models outperform other tools in predicting the two categories of PS proteins. We then performed in vitro experiments on the top-scored candidates DHX9, Ki-67, and NIFK. Their PS behaviors prove the accuracy of our methods. Further evaluation on a high-quality proteome from membraneless organelles (MLOs) revealed the superiority of our model in screening partner-dependent proteins. With benchmark datasets provided by PhaSepDB, our method incorporates multimodal features for prediction of PS-Self and PS-Part proteins, and provides a metapredictor for identification of potential PS proteins.
Results
Datasets of PS-Self Proteins, PS-Part Proteins, and Non-PS Proteins.
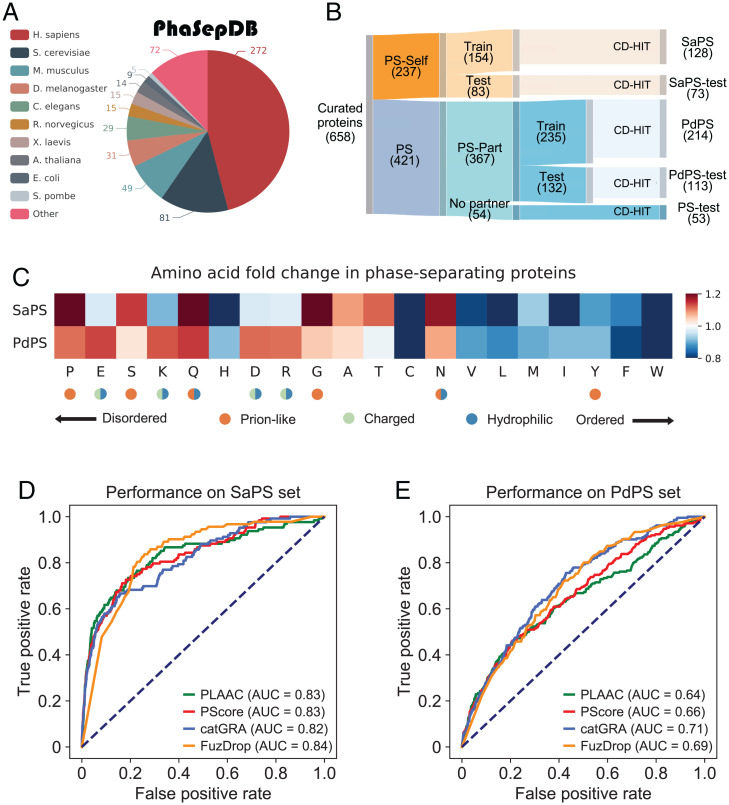
We collected 658 experimentally validated PS proteins from PhaSepDB (16), LLPSDB (17), and PhaSePro (18) (Dataset S1). The latest version of PhaSepDB provides comprehensive annotations of 592 nonredundant proteins (Fig. 1A), from which we collected 203 PS-Self and 335 PS-Part proteins; we used the remaining 54 PS proteins as an additional test set to measure model performance (Materials and Methods). We divided the PS-Self and PS-Part sets into training and independent testing sets according to the version number, then applied the CD-HIT algorithm (19) to all these sets with a sequence identity cutoff of 0.4 to reduce sequence similarity (Materials and Methods). We adopted the same classification criteria for two protein sets collected from LLPSDB and PhaSePro. Twenty-nine PS-Self and 28 PS-Part proteins were, respectively, selected to supplement the independent testing set (Materials and Methods). Finally, we grouped 658 proteins into five sets: the training and independent testing sets of PS-Self proteins (Fig. 1B, [SaPS, 128 proteins] Dataset S2, and [SaPS-test, 73 proteins] Dataset S3), the training and independent testing sets of PS-Part proteins (Fig. 1B, [PdPS, 214 proteins] Dataset S2, and [PdPS-test, 113 proteins] Dataset S3), and the independent PS test set (Fig. 1B and [PS-test, 53 proteins] Dataset S3). Proteins without PS reports were defined as non-PS proteins: 60,251 proteins collected from 10 representative species were used as non-PS proteins (Materials and Methods), from which we sampled 80% for training (NoPS, 48,187 proteins) (Dataset S2), and the remaining 20% were used for independent testing (NoPS-test, 12,064 proteins) (Dataset S3).
Fig. 1.
PS-Self and PS-Part proteins possess different amino acid patterns. (A) Species distribution of 592 nonredundant proteins that collected from PhaSepDB. (B) PS proteins collected from PhaSepDB, LLPSDB, and PhaSePro are divided into five nonoverlapping sets. (C) Amino acid frequency fold-changes of the SaPS set and the PdPS set are calculated against the NoPS set. Amino acids are ranked by their propensity to form IDRs. (D) To measure the performance of four representative PS predictors in screening self-assembling proteins, we plotted the ROC curve for each predictor by scoring proteins in the SaPS and NoPS sets. (E) ROC curves of four predictors are plotted for the PdPS and NoPS sets. The AUCs show the poor performance of these tools in screening partner-dependent proteins.
Together, we constructed three datasets for training: the positive sets SaPS and PdPS, and the negative set NoPS (Fig. 1B and SI Appendix, Dataset S2). We also defined four datasets as independent test sets: the positive sets SaPS-test, PdPS-test and PS-test, and the negative set NoPS-test (Fig. 1B and SI Appendix, Dataset S3).
Previous Tools Prefer PS-Self Rather than PS-Part Proteins.
Interactions like electrostatic, hydrophobic, π–π stacking and cation–π stacking drive biological macromolecules to aggregate and undergo PS (20, 21). Specific physical properties of different amino acids elicit specific interactions. For example, charged residues play a role in forming electrostatic interactions, aromatic residues and nonaromatic amino acids with π bonds in their side chains contribute to π–π stacking (2). Therefore, the amino acid composition of a PS protein may reflect the underlying driving force to some extent.
We compared the amino acid composition of self-assembling and partner-dependent proteins by calculating the fold-changes of amino acid frequency against the non-PS proteins (SaPS, PdPS, NoPS) (Materials and Methods and Dataset S2). After ranking the amino acids by their propensity to form disordered regions (22), we found that both PS protein groups possess high proportions of such residues (Fig. 1C). Prions can switch from nonaggregated states to self-templating highly ordered aggregates (7). We found that PLD-promoting amino acids were more common in PS-Self proteins than in PS-Part proteins. We also found a decreased frequency of charged residues in the SaPS set and an increased frequency of charged and hydrophilic residues in the PdPS set (Fig. 1C). To further verify the results, we performed the same statistical analysis on the human proteome (hSaPS, hPdPS, hNoPS) (Dataset S2). Similar results were obtained (SI Appendix, Fig. S1A).
Due to amino acid composition differences, a PS predictor may have different performances when distinguishing the two types of PS proteins from non-PS proteins. We chose PScore (6), PLAAC (7), catGRANULE (8), and FuzDrop (11), which have batch prediction interfaces or provide predicted results, as representative tools to compare their prediction performances. By scoring proteins in the SaPS, PdPS, and NoPS sets, the receiver operating characteristic (ROC) curve was plotted for each tool. The area under the curves (AUCs) showed that these predictors are excellent at predicting self-assembling proteins (Fig. 1D), but are unsatisfactory when screening PS-Part proteins (Fig. 1E). We performed the same analysis on the human proteome and achieved similar results (SI Appendix, Fig. S1 B and C).
Together, these results showed that current tools perform poorly in predicting PS-Part proteins compared to PS-Self proteins, yet both kinds of proteins are enriched in disorder-associated residues. Our previous work found that the scores of PS predictors are significantly correlated with the IDR scores (13). The two protein categories likely possess different IDR patterns, leading to different performances of current PS predictors in distinguishing them from non-PS proteins.
Multimodal Features Provide Information for Identifying PS-Self and PS-Part Proteins.
Next, we compared the distribution of PS-related features between the two PS protein sets and the non-PS protein set. As mentioned above, sequence properties such as disorder are closely related to protein PS. We used the PLAAC score to indicate prion-like propensity (7), the catGRANULE score to indicate granule-formation propensity (8), the IDR score from the ESpritz algorithm (23), and the low-complexity region (LCR) score from the SEG algorithm (24) to reflect a protein’s propensity to form IDRs and LCRs (Materials and Methods). Non-IDR interacting elements like coiled-coil (CC) structures can also drive PS (25, 26). We used the DeepCoil algorithm to detect potential CC structures (27) (Materials and Methods). In addition, we used the hydropathy score, the fraction of charged residues (FCR), and PScore to illustrate multivalent interactions including hydrophobicity, electrostatic interactions, and π–π stacking (6, 28) (Materials and Methods). Apart from amino acid sequence composition, features such as PTM frequencies and IF images can also provide information in screening PS proteins. PTMs play an extensive role in PS by regulating the reversibility of a binding reaction or altering a protein’s physical properties (3). Our previous studies showed that the PTM frequencies of PS proteins are significantly higher than those of background proteins (13). Here we chose phosphorylation (Phos) as the representative PTM type and calculated the Phos frequency for human proteins recorded in PhosphoSitePlus (29) (Materials and Methods). PS proteins usually appear as spherical-shaped structures in IF images. Trained on proteome-level IF images, the DeepPhase algorithm estimates the probability of proteins to be droplet-forming (30). We collected DeepPhase scores of 11,982 human proteins and used them for further comparison (Materials and Methods).
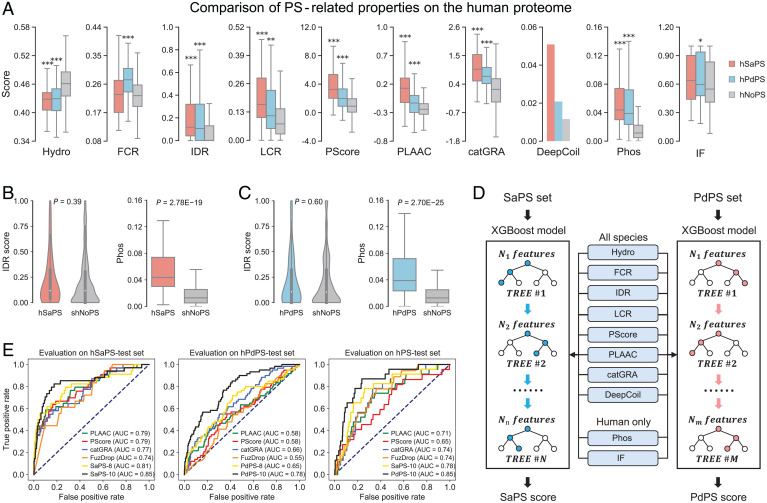
To test whether these features can discriminate the two protein categories from non-PS proteins, we used corresponding tools (Dataset S4) to score proteins in positive and negative datasets. We first compared the different sequence-based features in the datasets from the human proteome (hSaPS, hPdPS, hNoPS) (Dataset S2). As shown in Fig. 2A, most features are significantly different between the two PS protein sets and the non-PS set. Proteins in the hSaPS set possess higher IDR, LCR, PScore, PLAAC, and catGRANULE scores than those in the hPdPS set. The hPdPS set, but not the hSaPS set, has significantly higher levels of FCR than the hNoPS set. About 5% of the PS-Self proteins were predicted to contain a CC structure, compared to 2% of PS-Part proteins and only 1% of non-PS proteins (Fig. 2A). We then compared these features for all species data and found similar patterns (SaPS, PdPS, NoPS) (Fig. S2A and Dataset S2). When focusing on sequence-irrelevant features, we found that both PS sets have high levels of Phos frequency and IF-based droplet-forming propensity (Fig. 2A). Since PTM sites are enriched in IDRs, we downsampled the hNoPS set with similar IDR distributions of the hSaPS and the hPdPS sets (Materials and Methods and Dataset S5). The significance of the P value suggests that the enrichment of Phos frequency in self-assembling and partner-dependent proteins is IDR-independent (Fig. 2 B and C).
Fig. 2.
Constructing self-assembling and partner-dependent protein predictors with PS-related features. (A) Comparison of 10 PS-related features between the two PS protein sets and the non-PS set. P value is calculated through the two-sided Mann–Whitney U test (*P < 0.05; **P < 0.01; ***P < 0.001). (B) The hNoPS set is downsampled according to the IDR distribution in the hSaPS set. The Phos frequency of the hSaPS set is still significantly higher than that of the sampled hNoPS set. (C) The Phos frequency of the hPdPS set is significantly higher than that of the sampled hNoPS set with similar IDR distribution. (D) Schematic view of the SaPS and PdPS models. (E) Evaluating model performance using the independent test sets of self-assembling (hSaPS-test, Left), partner-dependent (hPdPS-test, Center), and PS protein sets (hPS-test, Right).
Together, these results show that our selected properties can provide information to distinguish PS proteins from non-PS proteins.
Models Integrating Multimodal Features Outperform Current Tools.
Using the properties mentioned above, we constructed two independent machine-learning models to separately predict self-assembling and partner-dependent proteins (31) (Fig. 2D). We first tested the learning effects of incorporating eight sequence-based features (hydropathy, FCR, IDR, LCR, PScore, PLAAC, catGRANULE, DeepCoil) on all-species data (SaPS, PdPS, NoPS) (Dataset S2). To increase the generalizability of our trained models, we adopted a cross-validation strategy for both positive and negative samples (Materials and Methods). Average AUCs on the validation sets revealed that the sequence-based features can work together to provide insights into the prediction of PS proteins (Table 1). To further compare the performance between our models and four PS predictors, we trained final models with all the positive samples (Materials and Methods). Evaluation on the independent test sets of self-assembling and partner-dependent proteins showed that our models have more stable prediction performance when faced with different PS protein categories (SaPS-test, PdPS-test, NoPS-test) (Fig. S2 B and C and Dataset S3). When scoring PS proteins without Self or Partner annotations, we found that the AUC value of PScore is relatively low (PS-test) (Fig. S2D and Dataset S3). This indicates that features other than π-contacts participate in the PS process of these proteins.
Table 1.
AUCs of our models with a fivefold cross-validation training strategy
| Model category | Species | Feature no. | Partition 1 | Partition 2 | Partition 3 | Partition 4 | Partition 5 | Average |
|---|---|---|---|---|---|---|---|---|
| SaPS | All species | 8 | 0.881 | 0.853 | 0.856 | 0.865 | 0.855 | 0.862 |
| hSaPS | Human | 8 | 0.921 | 0.88 | 0.871 | 0.859 | 0.858 | 0.878 |
| hSaPS | Human | 10 | 0.988 | 0.95 | 0.898 | 0.896 | 0.889 | 0.924 |
| PdPS | All species | 8 | 0.762 | 0.739 | 0.732 | 0.732 | 0.732 | 0.739 |
| hPdPS | Human | 8 | 0.765 | 0.786 | 0.777 | 0.762 | 0.765 | 0.771 |
| hPdPS | Human | 10 | 0.828 | 0.834 | 0.815 | 0.83 | 0.828 | 0.827 |
We next built and tested models with 8 and 10 features (the 8 features described above plus Phos frequency and DeepPhase) on the human proteome (hSaPS, hPdPS, hNoPS) (Dataset S2). The averaged AUCs of cross-validation indicated the significance of increasing the number of incorporated features, with a 5% increase on the hSaPS set and a 6% increase on the hPdPS set when comparing the 10-feature model to the 8-feature model (Table 1). We then evaluated the performance of the 8- and 10-feature models on the independent test sets (hSaPS-test, hPdPS-test, hNoPS-test) (Materials and Methods and Dataset S3). Although current PS predictors already perform well in screening self-assembling proteins, AUCs from the hSaPS-test set revealed that our model understands this type of protein better than any of the existing predictors (Fig. 2 E, Left). Evaluation on the hPdPS-test set showed that the PdPS model incorporating Phos frequency and DeepPhase score has an outstanding performance compared to the existing methods, with a 12%, 20%, and 20% increase compared with catGRANULE, PLAAC, and PScore, respectively (Fig. 2 E, Center). We also evaluated our 10-feature models on the hPS-test set (Dataset S3). The high AUC of the PdPS model suggests that these proteins are more likely to phase separate through interaction with other components than through self-assembly (Fig. 2 E, Right).
Taking these results together, we conclude that even though the current PS predictors perform well in predicting self-assembling proteins, the increased number of sequence-based features in our method may provide additional information for model decisions. Furthermore, constructing an independent model for predicting partner-dependent proteins is essential, and incorporating multimodal features like Phos level and IF images is of great importance in improving prediction performance.
The PdPS Model Performs Better in Screening MLO Participants.
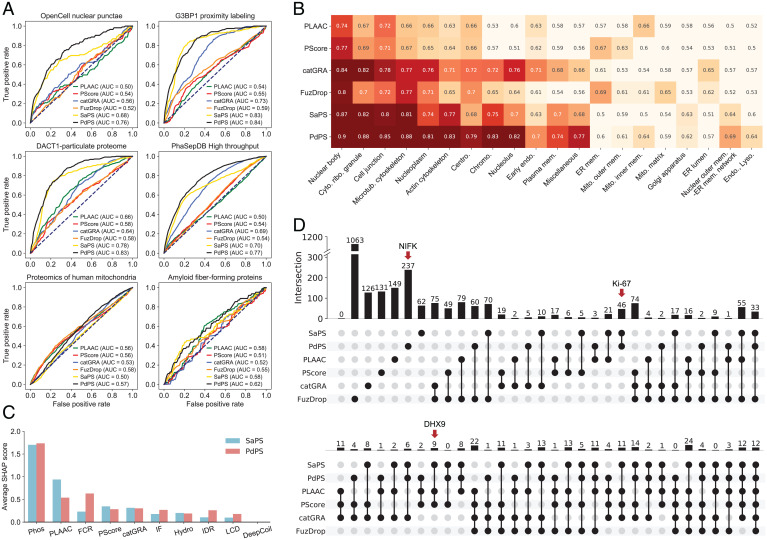
As mentioned before, PS biomolecular condensates usually contain multiple proteins (5, 14, 15), which we refer to as MLO participants. We tested the ability of the current tools to screen these proteins. We applied our 10-feature models and the other four PS predictors to estimate the proteins capable of undergoing PS in four human MLO participant datasets: the OpenCell nuclear punctae set (32), the DACT1-particulate proteome set (33), the G3BP1 proximity labeling set (34), and the PhaSepDB high-throughput set (16) (Materials and Methods and SDataset S6). To prevent self-validation, we removed the proteins that were included in the training sets. We then used the remaining proteins in these datasets and the human NoPS-test set as positive and negative samples, respectively, to calculate AUC values (hNoPS-test) (Dataset S3). As shown in Fig. 3A, our two methods have the highest confidence in predicting PS proteins in all four datasets. Significantly, the PdPS model has excellent ability in screening these MLO participants. Since catGRANULE screens for granule-localized proteins, it performs well in analyzing the G3BP1 proximity-labeling proteome and the PhaSepDB high-throughput dataset (Fig. 3A). We also created datasets for the human mitochondrial proteome and the amyloid fiber-forming proteome, most of which have not yet been assessed for undergoing PS (35, 36) (Materials and Methods and Dataset S6). We applied the same analytical strategy for these two sets. The AUCs showed that none of the tools had high confidence in predicting these proteins as PS proteins, which indicates that amyloid fiber-forming proteins and mitochondrial proteins differ significantly from PS proteins in the properties compared above (Fig. 3A).
Fig. 3.
Comparing the SaPS and PdPS models with another four representative PS predictors. (A) Comparison of six PS predictors on four datasets of proteins that participate in MLOs and two datasets of proteins located in membrane-bound organelles. AUC values are calculated by, respectively, using proteins in these datasets as positive samples and proteins in the human NoPS-test set as negative samples. (B) Comparison of 6 PS predictors on a BioID interactome with 20 intracellular locations. The value and the color of each block corresponds to the AUC value, which is calculated by, respectively, using proteins in these datasets as positive samples and proteins in the human NoPS-test set as negative samples. (C) The averaged SHAP values of the SaPS and PdPS models are calculated on the OpenCell nuclear punctae set. Phos frequency has the highest weight among the 10 incorporated features. (D) Overlap of top-scored proteins from six PS predictors. Only 12 of the 2,667 collected proteins are predicted as PS proteins by all predictors. The red arrows indicate the location of candidate proteins DHX9, Ki-67, and NIFK.
To further verify the results, we collected a BioID interactome that localizes 4,424 proteins to 20 intracellular locations (37) (Materials and Methods and Dataset S7). Using these annotated proteins as positive samples, we adopted the same strategy as in Fig. 3A to calculate AUC values for each method, then we ranked the 20 compartments according to the total prediction performance of these methods. Results showed that all the six tools performed well in predicting MLO participants and ignored proteins within membrane-bound organelles, such as the Golgi apparatus and lysosomes (Fig. 3B). Among the high-ranking MLOs, microtubules and the actin cytoskeleton have been reported previously to be constructed regionally with the help of a large pool of interacting proteins (2). Therefore, proteins within these two MLOs are more likely to phase separate in a partner-dependent manner. For example, short actin filaments form spindle-like tactoids in vitro through PS in the presence of Filamin (38). We found that the PdPS model better screens cytoskeleton-related proteins and outperforms existing PS predictors when distinguishing possible PS-Part proteins.
To evaluate how each of the incorporated properties contributes to the predictive ability of our methods, we adopted the model interpreter SHAP on proteins in the OpenCell nuclear punctae set to measure the importance of each feature to the model decision (39) (Materials and Methods). The averaged absolute value of the SHAP score indicates that the Phos frequency is more important than other features (Fig. 3C), suggesting a significant role of Phos in distinguishing PS proteins. We also found that the PLAAC score has a higher SHAP value in the SaPS model than in the PdPS model, while the FCR score is more critical in the PdPS than in the SaPS model (Fig. 3C). This result is consistent with the amino acid composition analysis (Fig. 1C and SI Appendix, Fig. S1A), which shows that PS-Self proteins are enriched in prion-like amino acids, while PS-Part proteins are enriched in charged amino acids. As shown in Fig. 2A, IDRs and LCRs are more significant discriminating properties than IF image for PS proteins. However, these two features displayed lower importance for model decisions than IF image (Fig. 3C). One possible reason is that these two features are strongly correlated with PLAAC and PScore (SI Appendix, Fig. S3), thus their information is redundant with PLAAC and PScore ranked second and fourth for model decision. Nevertheless, IF images can provide orthogonal information besides PLAAC and PScore.
We next compared the differences of the six methods by overlapping their top-scored proteins (Materials and Methods). Considering the impact of the training samples on protein scoring, we removed the proteins included in the positive training sets for each method (Dataset S8). Among 2,667 selected proteins, only 12 were predicted as PS proteins by all predictors (Fig. 3D). In contrast, most of these top-scored proteins are identified by only one method. Although the PLAAC, PScore, and catGRANULE scores are integrated into the SaPS and PdPS models, the high weight given to Phos frequency makes our models distinct from these three methods (Fig. 3C).
In summary, using protein annotations provided by high-throughput technology, we verified the ability of PS predictors to distinguish proteins in MLOs from those in membrane-bound organelles. Specifically, the PdPS model was excellent at identifying MLO participants. Using the model interpreter SHAP, we found differences in the contribution of incorporated features to model decisions. However, the Phos frequency has outstanding importance in both the SaPS and PdPS models, leading to better predictive performance.
Experimental Validation of DHX9, Ki-67, and NIFK Show the Effectiveness of the SaPS and PdPS Models.
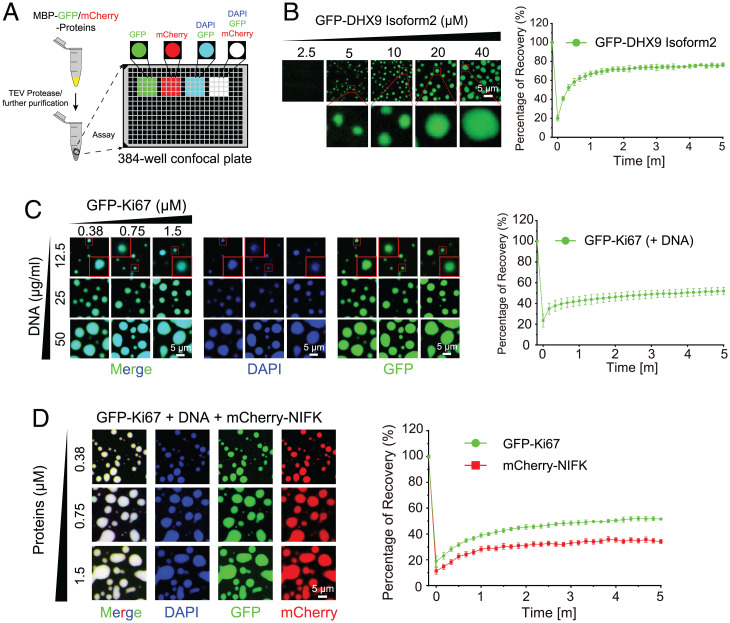
We performed in vitro PS experiments to test the effectiveness of our methods (Fig. 4A). We first selected protein DHX9 from the nine candidates that were predicted as PS proteins in both SaPS model, PLAAC, and PScore to test the ability of our model in screening PS-Self proteins (Fig. 3D and Dataset S8). DHX9 contains 1,270 amino acids, which is too long to ensure high-quality purification of the protein. We therefore used its short isoform (isoform2), which contains C-terminal RGG and disordered regions to perform PS experiments (Materials and Methods and SI Appendix, Fig. S4 A and B). The phase diagram showed that GFP–DHX9 isoform2 form green puncta, and the quantitative fluorescence recovery after photobleaching (FRAP) analysis indicated the dynamicity of formed puncta (Materials and Methods and Fig. 4B). Together, these results suggest that DHX9 isoform2 can self-assemble to undergo PS in vitro.
Fig. 4.
Experimental validation of DHX9 isoform2, Ki-67 truncation, and NIFK. (A) Schematic diagram of in vitro PS assay to illustrate the PS capacity of GFP or mCherry fused proteins after MBP removal. N-terminal MBP tags of MBP–GFP–DHX9 Isoform2, MBP–GFP–Ki-67 truncation and MBP–mCherry–NIFK were cleaved before droplet assembly with TEV protease overnight. Further droplet assembly for these proteins was performed on 384-well confocal plate. (B) Phase diagrams with blow-up images of GFP–DHX9 Isoform2 (Left). Quantitative results for FRAP analyses of the average recovery traces of GFP–DHX9 Isoform2 (Right). (C) Phase diagrams with blow-up images of GFP–Ki-67 truncation with DNA (Left). Quantitative results for FRAP analyses of the average recovery traces of GFP–Ki-67 truncation (Right). (D) Phase diagrams of GFP–Ki-67 truncation with DNA and mCherry–NIFK (Left). Quantitative results for FRAP analyses of the average recovery traces of GFP–Ki-67 truncation and mCherry–NIFK (Right).
We next selected another protein, Ki-67, from 46 candidates that ranked top-500 only in our SaPS and PdPS models (Fig. 3D and Dataset S8). Previous research has reported that Ki-67 promotes cell proliferation through its interaction with NIFK (40), which is also present among the top-ranking proteins by our PdPS model (Fig. 3D and Dataset S8). A recently published article showed that Ki-67 acts as a scaffold for mitotic chromosome proteins, and NIFK formed aggregates in Ki-67 knockout cells (41). Therefore, we chose these two proteins as candidates to verify their ability to undergo PS (Materials and Methods and SI Appendix, Fig. S4 C and D). The full length of Ki-67 is 3,256 amino acids, which makes Ki-67 difficult to purify. Since the deletion of Ki-67 repeats did not affect the distribution of Ki-67 into nucleolar heterochromatin during the interphase (42, 43), we performed PS experiments using Ki-67 truncation. As is shown in SI Appendix, Fig. S4 E and F, both GFP–Ki-67 and mCherry–NIFK can only form puncta with slow recovery after bleaching in the presence of PEG8000. However, Ki-67 phase separates at a lower concentration when mixed with DNA, suggesting its partner-dependent PS ability in vitro (Fig. 4C). Since NIFK and Ki-67 interact with each other, we further mixed NIFK, Ki-67, and DNA and found that NIFK can incorporate into the condensate of DNA and Ki-67 (Fig. 4D). NIFK does not possess a DNA binding domain, the interactions between NIFK and Ki-67 should be the driving force for its partition into the condensates.
Besides the above top-ranking proteins, we chose MBP and SUMO, whose SaPS scores were lower than 0.1, as the negative controls. Results show that both proteins cannot undergo PS (SI Appendix, Fig. S4 G and H). To conclude, the results above demonstrate that our methods can help to identify potential PS proteins.
Enriched Pathways and Domains Reveal the Functions of Two PS Protein Categories.
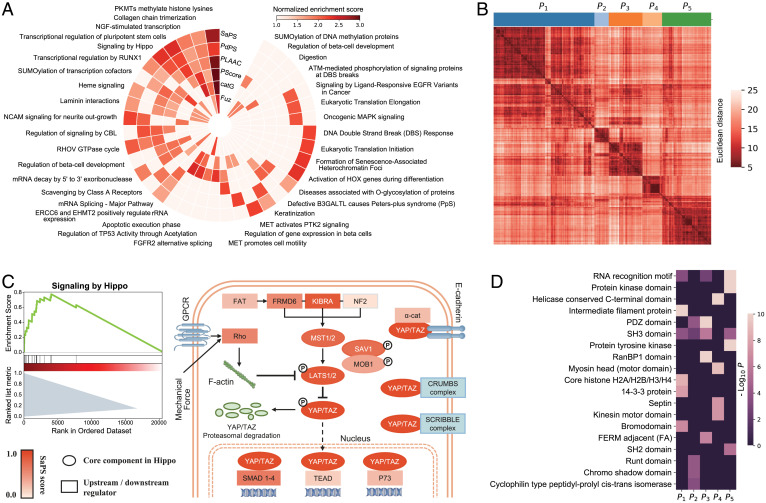
Next, we analyzed the pathways and domains that are enriched among the high-scoring proteins of our SaPS and PdPS models. To compare the enriched pathways generated by our two methods and the other four PS predictors, we used the single-sample gene set enrichment analysis (GSEA) on the Reactome database (44). Nine representative pathways were selected for each method to comprise a nonredundant set with 37 pathways (Materials and Methods and Fig. 5A). We found that Phos plays an essential role in these top-ranking pathways, including the transcriptional activity of RUNX1 and the signaling activity of PTK2. In addition, PTMs such as SUMOylation and O-glycosylation are also enriched in regulating these top-ranking pathways (Fig. 5A). When examining individual pathways, we found that Hippo signaling was enriched in both the SaPS and PdPS models. The Hippo pathway regulates several biological processes through the main effectors, YAP and TAZ (45). Although YAP and TAZ were not used to train the PdPS model, they were predicted to have high PS-Part scores, which is in line with a report that TAZ forms PS droplets with interacting proteins in cells, and YAP forms droplets in the presence of specific crowding agents (46). A recently published article indicated that LATS1, another core component of the Hippo pathway, can self-assemble through its PLD and also interact with partners like small nucleolar RNA host gene 9 (SNHG9) and phosphatidic acids to undergo PS (47). LATS1 was not used for training, but scored highly in both SaPS and PdPS models (Fig. 5B).
Fig. 5.
Functional analysis of self-assembling and partner-dependent candidates in the human proteome. (A) Single-sample GSEA of SaPS, PdPS, and another four representative PS predictors in the human proteome. Thirty-seven representative pathways are shown. If a method enriches any of the 37 pathways, the corresponding block would be colored according to its NES. (B) GSEA plot of Hippo pathway according to the scores of SaPS in the human proteome (Left). Schematic view of the Hippo pathway, in which the core components are shown as an ellipse, and the other regulators are shown as a rectangle. All components are colored according to their SaPS score (Right). (C) Clustering of 1,609 proteins with PdPS score greater than 0.8 into five sets according to the similarity of embedded protein sequences. The distance between clusters is measured by Ward’s minimum variance method. (D) Enriched domains in the five clustered sets of PdPS candidates. Nineteen representative domains are shown. If a cluster enriches any of the 19 domains, the corresponding block would be colored by −log10 P value.
Multivalent interactions mediated by modular domains are the main driving force of PS for PS-Part proteins. Therefore, we checked which domains were enriched and how they were distributed in 1,609 proteins with PdPS score greater than 0.8. Using a sequence-embedding method, we encoded each protein sequence to a 3,705-dimension vector (48). We then clustered these proteins into five groups according to vector similarity and analyzed the domain enrichment with the DAVID web server (49) (Materials and Methods and Fig. 5C). As is shown in Fig. 5D, the RNA recognition motif was the most enriched domain, suggesting the significance of RNA-binding proteins in partner-dependent PS. We also found enrichment of the PDZ, the SH2, and the SH3 domains, which are well-studied PS-promoting domains (3, 5, 50–52).
In summary, our methods can uncover pathways that are enriched in PS proteins and find domains that facilitate PS. Systematic analysis of the human proteome may suggest the involvement of PS-Self or PS-Part proteins in multiple biological processes, with profound implications for future studies.
Discussion
Multivalent interactions are the driving force of protein PS (3). However, these interactions are extremely complex. For example, the hydrophobicity, electrostatic interactions, and cation–π interactions are insufficient by themselves to rationalize the PS behavior of Ddx4 (21). In this study, we divided PS proteins into two categories, self-assembling proteins and partner-dependent proteins, based on their corresponding multivalent interactors. Unlike the driver/client theory in which clients are recruited into the condensates formed by drivers, partner-dependent proteins can also drive the droplet-forming process, such as LAT, Sos1, and Grb2 in the T cell receptor PS system. Using the collected datasets, we constructed separate predictors for the two protein categories. Independent testing indicated the excellent performances of the SaPS and PdPS models, and experimental validation of the top-scored proteins DHX9, Ki-67, and NIFK suggested the accuracy of our methods.
Many IDR-containing proteins can self-assemble to undergo PS. Although first-generation PS predictors are sufficient to screen such proteins (13), a comprehensive method that incorporates these sequence-based features yields better performance. Moreover, the involvement of multimodal features, such as Phos frequency and IF image-based droplet-forming propensity, can further improve prediction accuracy. However, the detected Phos sites may be affected by protein abundance. Considering the high weight of Phos frequency in the model decision, proteins with low abundance may rank low in our 10-feature model. Therefore, careful consideration of the scores from our 8-feature and 10-feature models may be helpful.
The development of high-quality proximity labeling and image-based subcellular localization technology provides insights in screening potential PS proteins. Based on AUCs, the PdPS model outperforms other tools in predicting MLO participants. We assume that these predicted MLO participants might act in a partner-dependent manner to undergo PS. However, our understanding of the regulatory relationship between these participants is limited, even though the underlying relationship is essential in explaining the PS behavior (5). Therefore, information like protein–protein interaction networks should be considered to find potential PS regulatory relationships in future studies.
We performed in vitro PS and FRAP experiments for all three candidate proteins (Fig. 4 B–D and SI Appendix, Fig. S4 E and F). FRAP assays are measured immediately after droplet formation, and all three proteins can recover partially in vitro. The recovery percentage indicates that the assemblies formed by three proteins, or at least Ki-67 and NIFK, are more gel-like rather than liquid-like. In fact, of the 342 PS proteins used to train the SaPS and PdPS models, 297 have their material state recorded in PhaSepDB, of which 94 could undergo hydrogel-like or solid-like PS, including well-studied PS protein TDP43 (53), HP1α (54), NUP98 (55), and NUP153 (56). Since we did not distinguish between the different material states of PS proteins when training, some proteins that rank high in our method may possess a gel-like rather than a liquid-like state when undergoing PS.
We compared the top-scored proteins from our methods and four representative PS predictors. Low overlap ratios between different candidate sets and enriched pathways suggest the variety of different tools. Therefore, a gallery that displays the scores of multiple PS predictors may provide convenience for biologists when screening candidate proteins or specific regions. In this study, we implemented a comprehensive web server named PhaSePred (predict.phasep.pro) (SI Appendix, Fig. S5), which incorporates residue-level scores of several PS predictors and PS-related features. The radar chart shows the proteome-level quantiles of different features, profiling the propensity for protein PS.
In conclusion, our study provides methods to predict self-assembling and partner-dependent proteins, and the web server PhaSePred may act as a metapredictor for researchers to systematically identify potential PS proteins.
Materials and Methods
Collecting PS and Background Proteins.
Datasets of proteins representing two droplet-forming mechanisms.
The newly released version of PhaSepDB (db.phasep.pro/) provides detailed information about experimentally validated PS proteins; it contains 592 nonredundant proteins localized in 59 different organelles (16). The annotated proteins were divided into two groups: PS-self and PS-other. “PS-self” refers to those proteins that can undergo self-assembling PS in vitro. “PS-other” refers to those proteins contributing to the formation of biomolecular condensates. If a protein participates in an MLO with partner components, its partners will be recorded in the “Partner” column. Since proteins may rely on interaction with different kinds of biomacromolecules to form MLOs, we only selected those with protein or nucleic acid partners as our defined partner-dependent proteins. Using these criteria, we collected 203 PS-Self and 380 PS-Part proteins, constituting a nonredundant set with 538 proteins. Forty-five proteins possess annotations of both PS categories. We used them as PS-Self proteins in downstream analysis (203 PS-Self proteins and 335 PS-Part proteins) (Dataset S1). We further divided the two protein sets according to the version number: those labeled “v1” and “v2_1” were used for training, and the remaining proteins were used for independent testing. For some proteins in PhaSepDB, it is unknown whether they undergo PS through self-assembly or through partner proteins. We did not utilize these proteins for training but used them as an additional test set to verify the prediction effect (54 PS proteins) (Dataset S1).
We applied the CD-HIT algorithm to the five protein sets with a sequence identity cutoff of 0.4 as a quality-control process (19). This yielded the following final sets: the training and independent testing sets of PS-Self proteins [SaPS (Dataset S2), SaPS-test (Dataset S3)], the training and independent testing sets of PS-Part proteins [PdPS (Dataset S2), PdPS-test (Dataset S3)], and the independent testing set of PS proteins [PS-test (Dataset S3)]. In addition to PhaSepDB, two other databases provide annotations of both in vivo and in vitro PS experiments: LLPSDB (17) (bio-comp.org.cn/llpsdb/) and PhaSePro (18) (https://phasepro.elte.hu/). LLPSDB collects annotations of 1,192 entries from 295 independent proteins, which provides various in vitro experimental conditions and indicates whether the protein can undergo PS under this condition. Since LLPSDB groups experimental information according to the number of components involved in MLOs, we selected proteins from one-component droplets with in vitro experimental annotations and defined them as self-assembling proteins. We then selected proteins from multicomponent droplets with in vivo or in vitro experiments and defined them as partner-dependent proteins. PhaSePro contains only 121 PS proteins, but it provides a wide range of information on the biophysical driving forces and the regulation of these molecular systems, such as PTM regulation and the interaction partner. Using these annotations, we defined the proteins labeled “partner dependent” as partner-dependent proteins. We merged the protein groups from the two databases and excluded proteins involved in our PS dataset (Dataset S1). We then applied the CD-HIT algorithm to the remaining proteins to remove similar sequences. Finally, we collected 29 PS-Self proteins and 28 PS-Part proteins and grouped them into our independent test set (SaPS-test, PdPS-test) (Dataset S3).
Altogether, we collected 201 self-assembling proteins, of which 128 were used for training and 73 were used for independent testing (Fig. 1B; SaPS, see Dataset S2; SaPS-test, see Dataset S3); we also collected 327 partner-dependent proteins, of which 214 were used for training and 113 were used for independent testing (Fig. 1B; PdPS, see Dataset S2; PdPS-test, see Dataset S3).
Datasets of non-PS proteins.
Since our PS proteins were retrieved from 49 organisms, we collected the corresponding proteomes from the Swiss-Prot database as the background proteins. To reduce data redundancy, only 10 organisms with 5 or more records of PS proteins in our datasets and with protein numbers greater than 3,000 were retained for further usage (Homo sapiens, Saccharomyces cerevisiae, Mus musculus, Drosophila melanogaster, Caenorhabditis elegans, Rattus norvegicus, Xenopus laevis, Arabidosis thaliana, Escherichia coli, Schizosaccharomyces pombe). We then removed the protein sequences recorded in the three PS databases (PhaSepDB, LLPSDB, PhaSePro). The remaining proteins were submitted to the CD-HIT algorithm with a sequence identity cutoff of 0.4 to reduce sequence similarity (19). Finally, 60,251 proteins that passed the quality-control process were used as the non-PS protein set. Since we collected additional PS proteins as positive samples in the independent test set, we randomly sampled 20% of the proteins from the non-PS set and used them as the negative samples of the independent test set (NoPS-test) (Dataset S3). The remaining 80% of the proteins were used for training (NoPS) (Dataset S2).
Fold-Changes of Amino Acid Frequencies.
The amino acid frequency was defined as the proportion of a certain amino acid type of all amino acids in the sequence. Therefore, the frequency of all amino acid types in a protein sequence sum to 1. For a certain amino acid type, we calculated the averaged amino acid frequency for proteins in the positive and negative sets, then divided the frequency of the positive set by the frequency of the negative set to get the fold-change value.
Calculating PS-Related Properties at the Proteome Level.
We used the tools introduced in Dataset S4 to calculate sequence-based PS-related features. The hydropathy and FCR score of a protein was calculated by localCIDER using the default parameter (28). The hydropathy score was defined as the average hydropathy of each residue from a normalized Kyte–Doolittle hydrophobicity scale (57), and the FCR score was calculated by dividing the total number of D, E, R, and K residues by the sequence length. We used the ESpritz DisProt program with the decision threshold set at a 5% false-positive rate to predict potential disordered regions (23), and we used the SEG local package with default parameters to detect LCRs within a given protein sequence (24). The fraction of IDR or LCR was defined as the number of amino acids in the corresponding domain divided by the sequence length. Each protein’s PScore, PLAAC, and catGRANULE score was calculated using the corresponding tools under the default parameters (6–8). However, PLAAC provides three summary scores for a given sequence, including LLR, CORE, and PRD. Since the LLR score is more appropriate in whole-proteome screening, we used the normalized LLR score to represent the PLD-forming propensity. The Python package DeepCoil was used to detect potential CC structures (27). We used 0.82 as the threshold for CC structure detection and changed the score to 0 and 1 to indicate whether a protein contains the predicted CC structure.
Other PS-related features were integrated to help model decisions, such as Phos frequency and IF images. To calculate the Phos frequency, we downloaded the Phos sites of human proteins from PhosphoSitePlus (29) (retrieved 8 September 2020). The Phos frequency was defined as the number of Phos sites divided by the protein sequence length. IF image-based droplet-forming propensities were collected from supplementary table 2 of ref. 30, DeepPhase. We submitted 12,073 human Ensembl gene IDs provided by DeepPhase to UniProtKB and retrieved 11,982 UniProt entries for further analysis.
Downsampling the hNoPS Set to Eliminate the Effect of IDR Distribution on Phos Frequency.
In order to eliminate the influence of IDRs on Phos frequency, we downsampled the hNoPS set according to the IDR distributions of the hSaPS set and the hPdPS set. Since the IDR scores generated by ESpritz range from 0 to 1, we divided proteins in the negative and positive sets into 10 groups with a step size of 0.1, respectively. For each of the 10 groups, we can divide the number of proteins in the negative set by the number of proteins in the corresponding positive set and define this value as the N/P ratio. Then, except for the group with the smallest N/P ratio, we randomly sampled proteins in the negative set of remaining groups to make the N/P ratios of the 10 groups identical. Finally, we combined proteins in the 10 sampled negative sets to get the downsampled hNoPS set with a similar IDR distribution to the positive set.
Constructing Machine-Learning Models for Predicting Two PS Protein Categories.
XGBoost classification model.
Our models were constructed using the Python package XGBoost, a tree-based machine-learning algorithm with high efficiency and exemplary performance in handling tabular data (31). Since the different tools introduced in Dataset S4 have different restrictions on the input data, there are some missing values when calculating features for the protein sequences. Fortunately, the XGBoost algorithm provides a strategy to deal with these missing values. Therefore, our models have a higher tolerance for the input data.
Data used for training and testing.
As introduced above, we have 128 PS-Self proteins, 214 PS-Part proteins, and 45,484 non-PS proteins for training (SaPS, PdPS, NoPS) (Dataset S2). In addition, we have 73 PS-Self proteins, 113 PS-Part proteins, 53 PS proteins, and 12,064 non-PS proteins for independent testing (SaPS-test, PdPS-test, PS-test, NoPS-test) (Dataset S3).
Model training.
We first adopted the fivefold cross-validation strategy to test the performance of the XGBoost model in distinguishing two PS protein sets from the non-PS set. The training process of the SaPS model and the PdPS model are separate, and both involved five rounds of training. For each training round, the positive training, positive validation, and negative validation sets were fixed, then 10 models were generated with 10 different negative training sets to prevent the overuse of negative samples. The negative validation set and 10 negative training sets were randomly sampled from the NoPS set. These generated sets contained twice the number of proteins as the positive validation set and the positive training set, respectively. Using the 10 trained models, we defined the prediction score of a single round as the average prediction score of these models. Due to utilization of a fivefold cross-validation strategy, the final prediction score was averaged on the five-round training. AUCs for validation sets indicate good performance of the SaPS and PdPS models, and increasing the number of features can further improve the predictive performance (Table 1).
Since we collected additional PS proteins as the independent test set, we did not use the cross-validation strategy in the final training. Therefore, the final model was determined with all the positive samples as the training set. To prevent the overuse of negative samples, 10 different subsets of negative samples were randomly sampled from the NoPS set with twice the number of proteins in the positive set to train the 10 models. The parameters of all models were set as default to prevent model overfitting on the training set, and the averaged prediction scores of the 10 trained models were used as the final prediction score.
Collecting Annotations of MLO Participants and Control Proteins.
We collected four MLO participant datasets from previously published databases and articles (Dataset S6). OpenCell is a human protein localization resource generated from 1,311 CRISPR-edited cell lines harboring fluorescent tags. We collected 140 proteins annotated with “nuclear punctae” and defined them as the “OpenCell nuclear punctae” set (32). A recently published article (33) reveals that DACT1 forms PS proteinaceous cytoplasmic bodies to repress Wnt signaling. The authors performed liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) and quantified the DACT1-particulate proteome. We collected proteins with relative abundance greater than 0.01 compared to DACT1 and defined them as the “DACT1-particulate proteome” set (33). Yang et al. (34) reported that the G3BP1-centered protein–RNA interaction network drives stress granule (SG) formation. They analyzed the proximity proteomics of SGs using APEX2-labeled G3BP1. We collected proteins with relative abundance greater than 0.01 compared to G3BP1 and defined them as the “G3BP1 proximity labeling” set. PhaSepDB provides collections of PS-associated proteins that can be identified with a high-throughput method, including organelle purification, proximity labeling, IF image-based screening, and affinity purification. We defined them as the “PhaSepDB high-throughput” set (16).
In addition to the MLO participant datasets, we collected two control sets for comparison (Dataset S6). The proteome of human mitochondria was collected from the Human Protein Atlas. Among 1,156 experimentally detected proteins, 1,126 are recorded by UniProt and were used for further comparison (35). Amyloid fiber-forming proteins were collected from the AmyPro database, which contains 162 entries of validated amyloid precursor proteins and prions. These entries make up a nonredundant set of 154 proteins, 68 of which were from humans. Since the performance of the 6 PS predictors was evaluated on the human proteome, we used these 68 human proteins for evaluation (36).
We also collected the intracellular location annotations of 4,424 proteins from the BioID interactomes of 192 subcellular markers (Dataset S7). The application of nonnegative matrix factorization localized these proteins to 20 compartments, including membrane-bound organelles, like the Golgi apparatus, and MLOs, like nuclear bodies (37).
To prevent self-validation, we removed the training proteins included in above datasets before using these datasets to calculate the AUC values (Datasets S6 and S7).
Selecting Top-Scored Proteins.
We selected top-scored 500 proteins from SaPS, PdPS, PLAAC, PScore, and catGRANULE with the corresponding positive training samples removed, respectively. There are 1,733 proteins with a FuzDrop score of 1 after training-data removal, we kept all these proteins for comparison (Dataset S8). Together, these proteins make up a nonredundant set with 2,667 proteins, from which we selected DHX9, Ki-67, and NIFK for experimental verification.
Model Explanation Using SHAP Value.
SHAP is a game-theoretic approach to explain the output of the machine-learning model. It provides an interface for tree models and ensembles of tree models (39). The SHAP values can reflect how features work together to push the model output from the base value to the model output for a single sample. Therefore, the absolute value of SHAP measures the importance of each feature for model decisions. We used proteins in the OpenCell nuclear punctae set that are supported by microscopy images as candidates, then we selected the top-scored proteins of the SaPS model and the PdPS model by setting the false-positive rate at 0.1. To get a global model explanation, we calculated the SHAP value for all the features of each sample with our trained models. The averaged absolute value on 10 models was used as the final score of a feature’s importance.
Experimental Materials.
Construction of recombinant plasmids.
cDNA encoding for Human DHX9 isoform2 (235 amino acids, with 1 to 1,035 residues missing while C-terminal RGG and disordered regions remaining), human Ki-67 truncation (821 amino acids, with a deletion of the Ki-67 repeat domain in 494 to 2,928 residues), and human NIFK (293 amino acids, full length) were synthesized (Genewiz) and cloned into a modified pET11 expression vector (a solubility MBP tag followed by a tobacco etch virus (TEV) cleavage site and a GFP or mCherry at the N terminus, and a noncleavable 6×His tag located at the C terminus) (Novagen) for expression and purification, respectively. cDNA encoding for SUMO protein was synthesized (Genewiz) and cloned into a pET28a vector (SUMO at the N terminus, 6×His at the C terminus) for expression and purification.
Protein purification.
Three modified pET11-based plasmids and the pET28a-SUMO plasmid were transferred into BL21 (DE3) bacteria cells (Tiangen). Transferred cells were cultured to OD600 = 0.8 at 37 °C in LB media with ampicillin (Inalco) or kanamycin (Inalco) and induced in the presence of 0.1 mM isopropyl β-d-1-thiogalactopyranoside (Inalco) at 16 °C overnight. Then, bacteria cells were harvested and resuspended in lysis buffer (20 mM Hepes, pH 7.4, 500 mM NaCl, 1 mM PMSF). The cells were next lysed using a high-pressure homogenizer (ATS Engineering) and centrifuged. The supernatants were loaded onto Ni-NTA affinity columns (GenScript), washed sufficiently with lysis buffer supplemented with 20 mM imidazole, and then eluted using reaction buffer (20 mM Hepes, pH 7.4, 100 mM NaCl) supplemented with 200 mM imidazole. The eluted solutions were collected, concentrated, and applied to gel-filtration for further purifications.
All proteins were further purified by a HiTrap Heparin HP (5 mL; Cytiva) to remove nucleic acid impurity. MBP–GFP–DHX9, MBP–GFP–Ki-67, and MBP–mCherry–NIFK were digested by 6×His-TEV protease at 4 °C overnight. Cleaved MBP tags were removed from Ki-67 and NIFK proteins by Ni-NTA affinity columns (GenScript). The removed MBP tag was collected for further in vitro PS experiments. MBP–GFP–DHX9 solution became turbid after TEV digestion even in a high-salt buffer (2 M NaCl). The cleaved MBP tag cannot be removed by either Ni-NTA affinity columns or MBPTrap HP (5 mL; Cytiva), thus the PS experiments of DHX9 were conducted after TEV digestion without further purification.
Preparation of DNA template.
DNA template of mono 177-bp of the Widom 601 sequence was cloned and purified as previously described (58). The sequence for the 177-bp DNA sequence is listed as following with 601 DNA sequence:
GAGCATCCGGATCCCCTGGAGAATCCCGGTGCCGAGGCCGCTCAATTGGTCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCACATATATACATCCTGTTCCAGTGCCGGACCC
In vitro PS assay.
Proteins concentration was measured after purification. PS in vitro assays of GFP–DHX9 was performed after TEV protease digestion, while that of GFP–Ki-67 and mCherry–NIFK were performed after removing the N-terminal MBP tags via mixing proteins with/without DNA. DNA was labeled with DAPI. In vitro PS assays were performed in reaction buffer (20 mM Hepes, pH 7.4, 100 mM NaCl), with various protein or DNA concentrations on 384 low-binding multiwell 0.17-mm microscopy plates (In Vitro Scientific) and sealed with optically clear adhesive film.
Imaging or in vitro FRAP experiments were conducted with a NIKON A1 microscope equipped with a 100× oil immersion objective. NIS-Elements AR Analysis was used to analyze these images.
Droplets were bleached with the corresponding laser pulse (three repeats, 20% intensity, dwell time 1.9 s). Recovery from photobleaching was recorded for the indicated time.
Finding Enriched Pathways with GSEA.
We performed the single-sample GSEA on 1,214 human pathways collected from the Reactome database (44). P values for each pathway were calculated with a 1,000-round permutation. We selected pathways with P value less than 0.05, then ranked them according to the normalized enrichment score (NES). If the overlap of proteins in two pathways exceeds 50%, the pathway with lower NES will be removed. The 9 most-enriched pathways in each method were selected to comprise a nonredundant set with 37 pathways.
Sequence Embedding.
We embedded protein sequences with a pretrained language model, which was developed on structural information including pairwise residue contact maps within individual proteins and global structural similarity between proteins (48). This model can map every amino acid of a protein into a 3,705-dimensional vector. Therefore, this model returns a 3,705 × N matrix (if a matrix has m rows and n columns, it is an m × n matrix) for a protein sequence with N amino acids. We then averaged the matrix along the axis of sequence length to get a 3,705 × 1 vector.
Protein Clustering and Domain Enrichment Analysis.
We performed hierarchical clustering for embedded vectors of 1,609 top-scored PS-Part candidates. We adopted Ward’s minimum variance method to calculate the distance between clusters, then grouped these proteins into five clusters. For each cluster, domain items retrieved from DAVID were ranked by −log10 P value, and the top-ranked four items were chosen for further analysis. These items together constitute a nonredundant set with 19 domains.
Developing a Comprehensive Web Server that Integrates Current PS Predictors.
We implemented a metapredictor named PhaSePred (http://predict.phasep.pro/) to access residue-level predictions of multiple PS predictors and PS-related features, including PLAAC for PLD detection (7), PScore for π-contact prediction (6), catGRANULE for granule-formation propensity prediction (8), ESPritz for IDR detection (23), SEG for LCR detection (24), CIDER for hydropathy prediction (28), DeepCoil for CC detection (27), and InterProScan for modular domain prediction (59) (SI Appendix, Fig. S5A). As of August 2021, integrated predictions for 116,806 sequences from 18 species are available (SI Appendix, Fig. S5B).
Users can search a protein’s name or the UniProt entry in the “Home” page. The query results are presented as a responsive table. The UniProt entry in the “Query result” page can be clicked to navigate to the detailed page. Take the protein DHX9 as an example (http://predict.phasep.pro/detail/Q08211/), the detailed page includes four sections. 1) Protein information: This includes the gene name and organism (SI Appendix, Fig. S5C, green dotted box). 2) PhaSePred and related scores: This includes predictions made by the 8- and 10-feature SaPS and PdPS models and other PS predictors. The ranks for these scores in the corresponding organism are displayed in a radar chart to provide the PS profile. For the immunofluorescence image-based method DeepPhase, the IF image from The Human Protein Atlas is shown (SI Appendix, Fig. S5C, red-dotted box). 3) Protein feature viewer: The PS-related predictions with residue-level scores are displayed in an interactive and scalable interface created by the neXtProt feature viewer. Residue-level annotations shown in the blue-dotted box of SI Appendix, Fig. S5C indicate that the functional region associated with PS is located at the C terminus of DHX9, which includes the low-complexity region predicted by SEG, prion-like domain predicted by PLAAC, granule-forming region predicted by catGRANULE, and π-contact region predicted by PScore. 4) Protein sequence viewer: The amino acid sequence for the protein is shown, and the regions with higher PS-related scores are highlighted (SI Appendix, Fig. S5C, orange-dotted box).
Detailed instructions and a data summary are described in the PhaSePred “Guide” and “About” pages, respectively. All data in PhaSePred can be freely downloaded in the “Download” page.
Supplementary Material
Acknowledgments
This work was supported by the National Key Research and Development Program of China (Grants 2021YFF1200900, 2018YFA0507504, and 2019YFA0508403); the National Natural Science Foundation of China (Grants 32070666, 31871443, 32150023, and 32100417); the Clinical Medicine Plus X-Young Scholars Project of Peking University (Grant PKU2021LCXQ012); and the Fundamental Research Funds for the Central Universities.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2115369119/-/DCSupplemental.
Data Availability
All study data are included in the main text and supporting information.
References
- 1.Alberti S., Gladfelter A., Mittag T., Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176, 419–434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang H., et al. , Liquid-liquid phase separation in biology: Mechanisms, physiological functions and human diseases. Sci. China Life Sci. 63, 953–985 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Li P., et al. , Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nott T. J., et al. , Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su X., et al. , Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352, 595–599 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vernon R. M., et al. , Pi-Pi contacts are an overlooked protein feature relevant to phase separation. eLife 7, e31486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lancaster A. K., Nutter-Upham A., Lindquist S., King O. D., PLAAC: A web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics 30, 2501–2502 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolognesi B., et al. , A concentration-dependent liquid phase separation can cause toxicity upon increased protein expression. Cell Rep. 16, 222–231 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes M. P., et al. , Atomic structures of low-complexity protein segments reveal kinked β sheets that assemble networks. Science 359, 698–701 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stanislawski J., Kotulska M., Unold O., Machine learning methods can replace 3D profile method in classification of amyloidogenic hexapeptides. BMC Bioinformatics 14, 21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardenberg M., Horvath A., Ambrus V., Fuxreiter M., Vendruscolo M., Widespread occurrence of the droplet state of proteins in the human proteome. Proc. Natl. Acad. Sci. U.S.A. 117, 33254–33262 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saar K. L., et al. , Learning the molecular grammar of protein condensates from sequence determinants and embeddings. Proc. Natl. Acad. Sci. U.S.A. 118, e2019053118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen B., et al. , Computational screening of phase-separating proteins. Genomics Proteomics Bioinformatics 19, 13–24 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vernon R. M., Forman-Kay J. D., First-generation predictors of biological protein phase separation. Curr. Opin. Struct. Biol. 58, 88–96 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Mittag T., Parker R., Multiple modes of protein-protein interactions promote RNP granule assembly. J. Mol. Biol. 430, 4636–4649 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.You K., et al. , PhaSepDB: A database of liquid-liquid phase separation related proteins. Nucleic Acids Res. 48 (D1), D354–D359 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Q., et al. , LLPSDB: A database of proteins undergoing liquid-liquid phase separation in vitro. Nucleic Acids Res. 48 (D1), D320–D327 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mészáros B., et al. , PhaSePro: The database of proteins driving liquid-liquid phase separation. Nucleic Acids Res. 48 (D1), D360–D367 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu L., Niu B., Zhu Z., Wu S., Li W., CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murthy A. C., et al. , Molecular interactions underlying liquid-liquid phase separation of the FUS low-complexity domain. Nat. Struct. Mol. Biol. 26, 637–648 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das S., Lin Y. H., Vernon R. M., Forman-Kay J. D., Chan H. S., Comparative roles of charge, π, and hydrophobic interactions in sequence-dependent phase separation of intrinsically disordered proteins. Proc. Natl. Acad. Sci. U.S.A. 117, 28795–28805 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campen A., et al. , TOP-IDP-scale: A new amino acid scale measuring propensity for intrinsic disorder. Protein Pept. Lett. 15, 956–963 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walsh I., Martin A. J., Di Domenico T., Tosatto S. C., ESpritz: Accurate and fast prediction of protein disorder. Bioinformatics 28, 503–509 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Wootton J. C., Federhen S., Statistics of local complexity in amino acid sequences and sequence databases. Comput. Chem. 17, 149–163 (1993). [Google Scholar]
- 25.Rebane A. A., et al. , Liquid-liquid phase separation of the Golgi matrix protein GM130. FEBS Lett. 594, 1132–1144 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang X., et al. , Arabidopsis FLL2 promotes liquid-liquid phase separation of polyadenylation complexes. Nature 569, 265–269 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludwiczak J., Winski A., Szczepaniak K., Alva V., Dunin-Horkawicz S., DeepCoil-a fast and accurate prediction of coiled-coil domains in protein sequences. Bioinformatics 35, 2790–2795 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Holehouse A. S., Das R. K., Ahad J. N., Richardson M. O., Pappu R. V., CIDER: Resources to analyze sequence-ensemble relationships of intrinsically disordered proteins. Biophys. J. 112, 16–21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hornbeck P. V., et al. , PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu C., et al. , Proteome-scale analysis of phase-separated proteins in immunofluorescence images. Brief. Bioinform. 22, bbaa187 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Chen T., Guestrin C., XGBoost: A scalable tree boosting system. in Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, August 2016, (Association for Computing Machinery, 2016) pp 785–794. [Google Scholar]
- 32.Cho N. H., et al. , OpenCell: Endogenous tagging for the cartography of human cellular organization. Science 375, eabi6983 (2022). [DOI] [PMC free article] [PubMed]
- 33.Esposito M., et al. , TGF-β-induced DACT1 biomolecular condensates repress Wnt signalling to promote bone metastasis. Nat. Cell Biol. 23, 257–267 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang P., et al. , G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181, 325–345.e28 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thul P. J., et al. , A subcellular map of the human proteome. Science 356, eaal3321 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Varadi M., De Baets G., Vranken W. F., Tompa P., Pancsa R., AmyPro: A database of proteins with validated amyloidogenic regions. Nucleic Acids Res. 46 (D1), D387–D392 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Go C. D., et al. , A proximity-dependent biotinylation map of a human cell. Nature 595, 120–124 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Weirich K. L., et al. , Liquid behavior of cross-linked actin bundles. Proc. Natl. Acad. Sci. U.S.A. 114, 2131–2136 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lundberg S. M., Lee S.-I., “A unified approach to interpreting model predictions” in 31st Conference on Neural Information Processing Systems (NIPS, Long Beach, CA, 2017). [Google Scholar]
- 40.Lin T. C., et al. , The nucleolar protein NIFK promotes cancer progression via CK1α/β-catenin in metastasis and Ki-67-dependent cell proliferation. eLife 5, e11288 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stenström L., et al. , Mapping the nucleolar proteome reveals a spatiotemporal organization related to intrinsic protein disorder. Mol. Syst. Biol. 16, e9469 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saiwaki T., Kotera I., Sasaki M., Takagi M., Yoneda Y., In vivo dynamics and kinetics of pKi-67: Transition from a mobile to an immobile form at the onset of anaphase. Exp. Cell Res. 308, 123–134 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Kill I. R., Localisation of the Ki-67 antigen within the nucleolus. Evidence for a fibrillarin-deficient region of the dense fibrillar component. J. Cell Sci. 109, 1253–1263 (1996). [DOI] [PubMed] [Google Scholar]
- 44.Jassal B., et al. , The reactome pathway knowledgebase. Nucleic Acids Res. 48 (D1), D498–D503 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng Y., Pan D., The Hippo signaling pathway in development and disease. Dev. Cell 50, 264–282 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu Y., et al. , Phase separation of TAZ compartmentalizes the transcription machinery to promote gene expression. Nat. Cell Biol. 22, 453–464 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li R. H., et al. , A phosphatidic acid-binding lncRNA SNHG9 facilitates LATS1 liquid-liquid phase separation to promote oncogenic YAP signaling. Cell Res. 31, 1088–1105 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bepler T., Berger B., “Learning protein sequence embeddings using information from structure” in 7th International Conference on Learning Representations (ICLR, New Orleans, LA, 2019).
- 49.Jiao X., et al. , DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics 28, 1805–1806 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dionne U., et al. , Protein context shapes the specificity of SH3 domain-mediated interactions in vivo. Nat. Commun. 12, 1597 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beutel O., Maraspini R., Pombo-García K., Martin-Lemaitre C., Honigmann A., Phase separation of zonula occludens proteins drives formation of tight junctions. Cell 179, 923–936.e11 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Araki Y., et al. , SynGAP isoforms differentially regulate synaptic plasticity and dendritic development. eLife 9, e56273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gasset-Rosa F., et al. , Cytoplasmic TDP-43 de-mixing independent of stress granules drives inhibition of nuclear import, loss of nuclear TDP-43, and cell death. Neuron 102, 339–357.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ackermann B. E., Debelouchina G. T., Heterochromatin protein HP1α gelation dynamics revealed by solid-state NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 58, 6300–6305 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmidt H. B., Görlich D., Nup98 FG domains from diverse species spontaneously phase-separate into particles with nuclear pore-like permselectivity. eLife 4, e04251 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Milles S., Lemke E. A., Single molecule study of the intrinsically disordered FG-repeat nucleoporin 153. Biophys. J. 101, 1710–1719 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kyte J., Doolittle R. F., A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 (1982). [DOI] [PubMed] [Google Scholar]
- 58.Luger K., Rechsteiner T. J., Richmond T. J., Preparation of nucleosome core particle from recombinant histones Methods Enzymol. 304, 3–19 (1999). [DOI] [PubMed] [Google Scholar]
- 59.Jones P., et al. , InterProScan 5: Genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and supporting information.