Abstract
We synthesized a Yarrowia lipolytica strain overproducing lipase for industrial applications by using long terminal repeat (ζ) of the Y. lipolytica retrotransposon Ylt1 and an allele of URA3 with a promoter deletion to construct JMP3. JMP3 is a derivative of plasmid pHSS6 carrying a NotI-NotI cassette which contains a defective URA3 allele, a polylinker sequence, and the ζ region for targeting to multiple sites in the genome of the recipient. We inserted the LIP2 gene (encoding extracellular lipase) under the control of the strong POX2 promoter into JMP3 to generate JMP6. The pHSS6 region was removed by NotI digestion prior to transformation. Two Y. lipolytica strains transformed with the JMP6 LIP2 cassette had a mean of 10 integrated copies devoid of the Escherichia coli region, corresponding to an autocloning event. The copy number in the transformants was stable even after 120 generations in nonselective and lipase-inducing conditions. The resulting strains could produce 0.5 g of active lipase per liter in the supernatant, 40 times more than the single-copy strain with the LIP2 promoter. This work provides a new expression system in Y. lipolytica that results in strains devoid of bacterial DNA and in strains producing a high level of lipase for industrial uses, waste treatment, and pancreatic insufficiency therapy.
Some yeasts can use fat as the sole carbon source. One such yeast, Yarrowia lipolytica, is considered a promising host for protein production because it naturally and efficiently secretes large amounts of a variety of proteins (for a review, see reference 1). Integrative and replicative vectors are available for this yeast, but the replicative vectors are present at low copy number (28). We have previously used defective URA3 markers (Y. lipolytica URA3 gene with promoter deletions) to construct multicopy integrative plasmids. These plasmids, pINA764 through pINA773, contain the XPR2 gene (coding for the alkaline extracellular protease [AEP]) as a reporter gene and a fragment of ribosomal DNA (rDNA) to target integration (20). Transformed cells contain one integrated copy if the nondefective allele ura3d1 is used. If the defective allele ura3d4 is used, then 12 to 60 integrated copies are found if the plasmids were targeted to the rDNA loci (20) and about 30 integrated copies are found if the plasmids were targeted to XPR2 (1). These integrations occur in tandem at one or two sites. Strains containing XPR2 integrated in tandem at the rDNA locus are stable under nonselective conditions and in media in which XPR2 is not expressed. However, in conditions in which XPR2 was induced, the overproduction of AEP poisoned the culture, resulting in the rapid selection of deamplified cell lines. Nevertheless, in some transformants, AEP production was 10 to 12 times higher than in the wild type (20).
Repetitive elements often are used as target sites for integration of plasmids carrying genes to be amplified. The rDNA locus is commonly used in Y. lipolytica, Kluyveromyces lactis, Saccharomyces cerevisiae, Candida utilis, Schizosaccharomyces pombe, and Phaffia rhodozyma (20, 22, 35, 39, 41, 46), although other repetitive elements, e.g., Ty, also have been used (5). The Ylt1 retrotransposon is a repetitive element that has recently been characterized in Y. lipolytica (38) and is present at ca. 35 copies per genome. An additional 30 copies of long terminal repeat (LTR) solo (ζ) also are present. Thus, this region provides at least 65 potential target sites per genome and could be useful in developing multicopy transformants.
Derivatives of the pINA764 to pINA773 vectors were constructed by exchanging the rDNA region for the ζ region (1). With both types of plasmid, the complete vector was integrated, including the expression cassette and the bacterial part (Ori and ampicillin resistance gene). The bacterial DNA is a disadvantage if the resulting strain is to be used for industrial protein production, since current European regulations classify strains containing bacterial DNA as genetically modified organisms (10).
Lipases are secreted by many bacteria and fungi. The biotechnological potential of these enzymes is steadily increasing with various applications such as in the oleochemical, detergent, and food industries, surfactant production, organic chemistry, and fat-containing waste effluent treatment (for reviews see references 15 and 40). Lipases also may be used for human therapy of pancreatic deficiency (48). Y. lipolytica secretes several lipases and esterases. We recently identified the extracellular lipase, a triacylglycerol acylhydrolase (EC 3.1.1.3), encoded by the LIP2 gene (33). This extracellular 38.5-kDa lipase was used in oleochemistry (8) and Y. lipolytica strains in waste treatment (7, 43). This lipase is acid resistant and not inhibited by biliary salt and may be used in human therapy (32).
Our objectives in this study were to adapt our single- and multicopy vectors to use ζ regions as targeting sites and to obtain integrative cassettes free of bacterial DNA and to use this technology to construct Y. lipolytica strains that overexpressed the extracellular lipase Lip2p.
MATERIALS AND METHODS
Strains, media, and induction conditions.
Escherichia coli DH5α used for plasmid preparation was grown in Luria-Bertani (LB) medium (36). The Y. lipolytica strains used were PO1d (MatA leu2-270 ura3-302 xpr2-322; strain CLIB139 from the Collection de Levures d'Interet Biotechnologique [CLIB], Thiverval-Grignon, France and E150 (MatB leu2-270 his1 ura3-302 xpr2-322; CLIB122). The yeast media YPD, YNB, and YNBcas have been described elsewhere (45). The induction media YPH and YNBcasH are YPD and YNBcas, respectively, with glucose replaced by olive oil (10 g/liter) (Sigma-Aldrich, St. Quentin Fallavier, France). For solid media, 1.5% agar was added. The yeast was transformed using the lithium acetate procedure (47), and clones were selected on YNBcas. Tributyrin plates (YNBT) were used for lipase detection; they contained YNB supplemented with tributyrin (Sigma-Aldrich) (5 g/liter).
DNA manipulation.
Recombinant DNA techniques were carried out by standard methods (36). The expression cassettes used for yeast transformation were recovered as NotI or SalI-EcoRI DNA fragments from agarose gels and purified with the GeneCleanII extraction kit (Ozyme, St. Quentin Yvelines, France). Total genomic DNA was prepared and Southern hybridization was performed as previously described (20). Membranes were scanned using a STORM 860 PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). Copy number was determined using Molecular Dynamics ImageQuaNT software.
Northern blot analysis.
Samples of total RNA (30 μg) were subjected to electrophoresis in a 1.2% agarose–formaldehyde gel, transferred to a Hybond-N+ nylon membrane (Amersham-Pharmacia, Orsay, France), and UV-cross-linked to the membrane. The membrane was hybridized with a 32P-labeled probe at 45°C in 50% formamide–5× SSPE (0.75 M NaCl, 0.05 M NaH2PO4, 0.005 M EDTA [pH 7.7])–5× Denhardt's solution–0.5% sodium dodecyl sulfate (SDS)–100 μg of salmon sperm DNA per ml for 16 h. Washing and exposure conditions were identical to those for Southern blotting. mRNA levels were quantified using Molecular Dynamics ImageQuaNT software.
Construction of basal expression vectors.
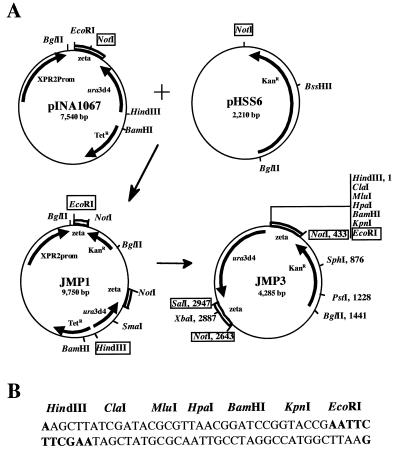
JMP3 (Fig. 1A) contains the defective ura3d4 marker and a polylinker, flanked by the ζ region, inserted at the NotI site of the pHSS6 (14) vector. First, pINA1067 (also known as pINA970 [1]) and pHSS6 were digested with NotI and ligated. The resulting plasmid, JMP1, was selected on tetracycline (10 μg/ml) and kanamycin (40 μg/ml). The pBR322 and XPR2 promoter regions were eliminated by HindIII-EcoRI digestion of JMP1 and replaced with an HindIII-EcoRI adaptor containing several unique restriction sites (Fig. 1B), giving rise to plasmid JMP3. For the nondefective vector JMP5, the 1,398-bp HindIII-XbaI fragment containing ura3d4 was replaced with a 1,484-bp HindIII-XbaI fragment containing the ura3d1 allele.
FIG. 1.
Schematic diagram of JMP3 construction. (A) pHSS6 was introduced into pINA1067 at the NotI site, giving rise to JMP1. pBR322 and the XPR2 promoter were replaced by a polylinker, resulting in the defective JMP3 vector. (B) The sequences of the polylinker and the unique cloning sites present are shown. The expression cassette is liberated by NotI or SalI-EcoRI digestion (boxed). Genes conferring tetracycline (TetR) and kanamycin (KanR) resistance are indicated by an arrow. ura3d4 is a modified version of the Y. lipolytica URA3 gene (20). Arrows indicate direction of transcription. Zeta regions are indicated by an open box.
Construction of the expression vectors.
Vectors containing the POX2 promoter (45)-LIP2 expression cassette were constructed by inserting a 3,229-bp ClaI-EcoRI fragment into the ClaI and EcoRI sites of JMP3 and JMP5 to give JMP6 (defective) and JMP7 (nondefective), respectively. Digestion of the plasmids with NotI yields two fragments. The first fragment represents the pHSS6 moiety, and the second represents the targeting cassette, consisting of the URA3 marker and the expression cassette flanked by the ζ region (Fig. 1). This targeting cassette was purified in an agarose gel prior to transformation.
We used the Y. lipolytica LIP2 gene, which codes for the 38.5-kDa extracellular lipase Lip2p (33). This lipase has potential applications in the food industry and waste treatment (8, 9). We fused it under the control of the strong and inducible POX2 promoter. POX2 codes for the long-chain acyl-coenzyme A oxidase, which is induced by fatty acids (45). The POX2p-LIP2 expression cassette is released from the vectors with NotI digestion (expression cassette with ζ region) or SalI and EcoRI digestion (expression cassette without the ζ region). The expression cassettes are free of bacterial DNA.
Stability of the transformants.
Strains were grown in YPD and YPDH for 2 weeks to keep the cells in the exponential phase of growth throughout the experiment. Cells were cultured in 50 ml of medium in 200-ml baffled flasks with shaking. Every morning, fresh cultures were inoculated at an initial A600 of 0.5 and grown for 8 h. In the evening, fresh cultures were inoculated at an initial A600 of 0.2 for 16 h. Samples of the previous culture were used to inoculate the new culture. Under our growth conditions, one cell generation corresponded to 2.5 h.
General protein techniques.
Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) with a 12% polyacrylamide resolving gel and a 4% polyacrylamide stacking gel (19). Proteins were stained with Coomassie brilliant blue. Gels were dried with DryEase (Novex, San Diego, Calif.) on cellulose-acetate films (Novex). Protein concentration was determined using the bicinchoninic (BCA) protein assay (Pierce, Rockford, Ill.) with bovine serum albumin as a reference. Lipase activity was routinely determined by titrimetric assay with olive oil (Sigma) as the substrate emulsion. Supernatant (5 to 20 μl) was added to 5 ml of the emulsion and 2 ml of 50 mM phosphate buffer (pH 6.8). Samples were incubated for 20 min at 37°C with shaking (200 rpm). The reaction was stopped by adding 4 ml of acetone-ethanol (50:50, vol/vol) containing 0.09% thymolphthalein indicator. Enzymatic activity was determined by titration of the released fatty acid with 50 mM sodium hydroxide. One unit of lipase activity corresponds to the quantity of enzyme that liberates 1 μmol of fatty acid per min.
RESULTS
Transformation and production of lipase by transformants.
Ylt1 and the ζ solo element are not present in all wild-type Y. lipolytica isolates. The repetitive element was absent from PO1d but present in E150. These two yeasts were used as recipients for transformation. Typically, we obtained high transformation frequencies (104 transformants per μg of DNA) with nondefective vectors (JMP5 and JMP7). In contrast, we obtained 10 to 30 transformants per μg of DNA with PO1d and 50 to 150 transformants per μg of DNA with E150 when we used defective vectors (JMP3 and JMP6).
Transformants appeared on YNBcas after 5 to 15 days with the defective vector and after 2 to 3 days with the nondefective vector. Transformation frequencies and the appearance of the clones were very similar to those of our first-generation plasmids (1), indicating that the ura3d4 allele remained defective in the new vectors independent of the location and orientation of the selection marker in the plasmid. Single-colony isolates were named according to strain, plasmid and clone number as follows: PO1d-6-1 represents strain PO1d, plasmid JMP6, and clone number 1.
Ten transformants obtained with the nondefective vector and 40 transformants obtained with the defective vector were selected from each strain and maintained on YNBcas. Their lipase production on solid YNBT and in liquid YNBH medium was assessed. Transformants obtained with the nondefective vector JMP7 contained one integrated copy of POX2-LIP2 and the chromosomal copy of LIP2 and showed no significant increase in lipase production (data not shown).
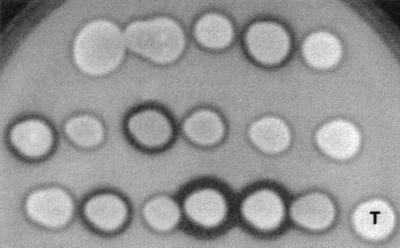
Three of the 40 PO1d clones transformed with JMP6 (PO1d-6-1 through PO1d-6-40) produced halos similar to those of the wild type, corresponding to URA3 conversion events, although only 27 and 568 bp (promoter and terminator regions, respectively) are shared between the genomic ura3-302 allele (ura3::SUC2 deletion [27]) and the ura3d1 allele. The conversion frequency was 5 to 20% depending on the transformation experiment. Such conversion events also were observed in E150. For 17 PO1d transformants, we observed differences in halo size on tributyrin plates, indicating clone heterogeneity for lipase production (Fig. 2A). These results were confirmed by determining lipase activity in YNBH after 48 and 96 h of induction. We observed 2 to 16 times higher levels of lipase production than those of the wild type, suggesting that multiple copies of the expression cassette were integrated. Similar results were obtained with E150-6-1 through E150-6-40 (data not shown).
FIG. 2.
Lipase production by Ura+ transformants. Lipase detection was done on YNBT plates. Seventeen random Ura+ transformants were grown in liquid YNBcas medium, and 3 μl of each culture was spotted onto YNBT. Plates were incubated for 2 days at 28°C. The size of the clear zone around the colonies, which reflects lipase production, was compared with that of the wild-type, PO1d (T).
Copy number and integration event analysis in multicopy integrants.
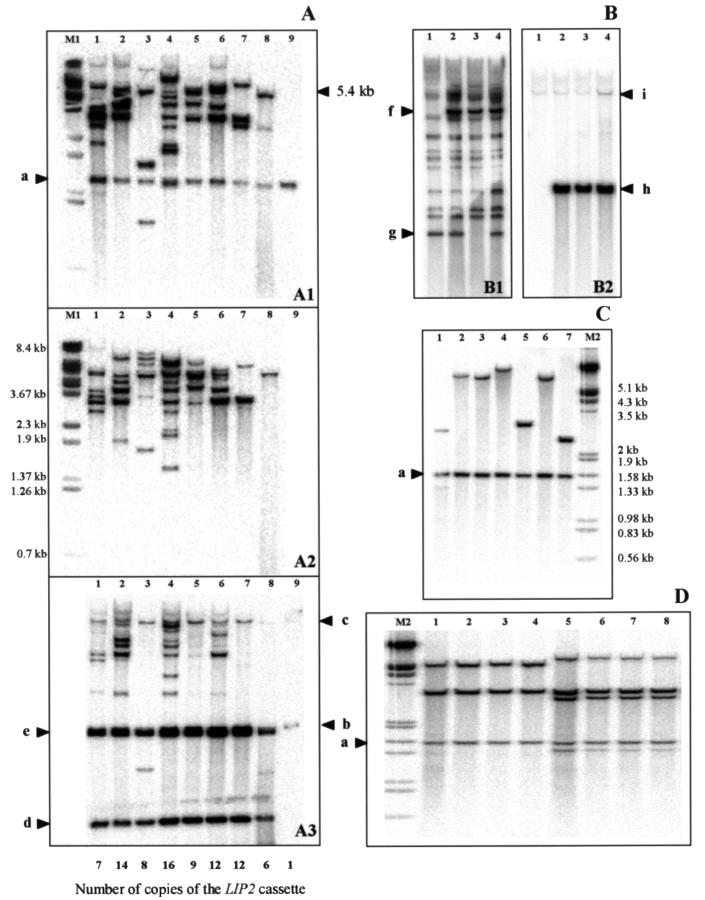
We analyzed the JMP6 (defective) transformants of E150 and PO1d giving the largest halos and the highest levels of lipase production. Total DNA was prepared from eight PO1d transformants (PO1d-6-2, -4, -8, -13, -14, -15, -16, and -17). Southern blot analysis was performed with the LIP2 gene ζ, and POX2 promoter probes. Multiple signals were detected (Fig. 3A). For the wild-type strain PO1d, we expected a 1.6-kb HindIII band with the LIP2 probe (Fig. 3, panel A1, lane 9, band a, chromosomal LIP2 gene), no band with the ζ probe (Fig. 3, panel A2, lane 9), and 1.5- and 8-kb bands with the POX2 promoter probe (Fig. 3, panel A3, lane 9, bands b and c, chromosomal POX2 locus). For the expression cassette, we expected to detect different fragments with the LIP2 and ζ probes if integration occurred at different loci or a single 5.4-kb band corresponding to the full-length expression cassette if integration occurred in tandem. With the POX2 probe, HindIII digestion should give bands d and e from the expression cassette (0.6 and 1.58 kb). For PO1d transformants probed with LIP2 (Fig. 3A1), the 1.6-kb HindIII chromosomal band was detected in all recombinant strains together with other bands of various sizes and numbers, with no strong signal at 5.4 kb. Thus, the expression cassette was integrated not in tandem but in a dispersed manner, with the number of bands reflecting the number of integration events. We confirmed the dispersed integration by detecting the same bands with the ζ probe. We observed from 2 to more than 15 different bands. The two POX2 bands were very intense (Fig. 3, panel A3). For each clone, the level of radioactivity in each of the various bands was determined using the PhosphorImager. We used either the 1.5-kb chromosomal POX2 band or the 1.6-kb chromosomal LIP2 gene as an indicator of the intensity signal for one copy (Fig. 3A). We found an average of 10 integrated copies (range, 6 to 16).
FIG. 3.
Southern blot analysis of JMP6 transformants. (A) PO1d transformed with the defective JMP6 expression cassette released by NotI. Genomic DNA from eight transformants (lanes 1 to 8) was digested with HindIII and hybridized with radiolabeled probes: the LIP2 gene (A1), ζ (A2), and the POX2 promoter (A3). PO1d was used as a control (lane 9). Specific bands are indicated by arrows and correspond to the LIP2 genomic gene (band a), the POX2 promoter (bands b and c), and the POX2 promoter of the expression cassette (bands d and e). The copy number of the LIP2 cassette in each transformant (relative to the chromosomal LIP2 signal) is indicated at the bottom of panel A. (B) Analysis of three E150 transformants obtained with the NotI JMP6 expression cassette. Genomic DNA was digested with EcoRI (B1) and EcoRI-KpnI (B2) and probed with ζ (B1) and LIP2 (B2). Strains used were E150 as a control (lane 1) and E150-6-1 through E150-6-3 (lanes 2 to 4, respectively). (B1) The strong hybridization signal at 5.4 kb (band f) corresponds to the expression cassette. The disappearance of a ζ signal was observed (band g). (B2) The strong hybridization signal at 1.6 kb (band h) corresponds to the expression cassette (part of POX2 promoter and LIP2), and the weak signal corresponds to the genomic LIP2 locus (band i). (C) Analysis of PO1d transformant obtained with the EcoRI-SalI expression cassette (without the ζ region). Genomic DNA was digested and probed as in panel A1. (D) Stability of two PO1d-JMP6 transformants. Genomic DNA from clones PO1d-6-17 (lanes 1 to 4) and -16 (lanes 5 to 8) after 5 (1 and 5), 50 (2 and 6), 90 (3 and 7), and 120 (4 and 8) generations cultured in normal growth and lipase induction conditions, respectively, for each lane pair. The 1.6-kb chromosomal lipase gene is also detected (band a). Lambda DNA digested with BstEII (M1) or with HindIII-EcoRI (M2) was used as molecular size standards. Total DNA from the various recombinant strains was extracted, blotted, and hybridized as previously described (20).
In the E150 transformants probed with ζ, we detected a strong signal corresponding to the size of the expression cassette (Fig. 3, panel B1, lanes 2 to 4, band f), which is absent from the parental E150 strain (lane 1). We also observed several bands that were present in all strains (Fig. 3, panel B1, lanes 1 to 4). We interpret these results to mean that E150 contains a large number of Ylt1 and/or ζ loci and that integration occurs primarily in tandem. For E150-6-2, integration was targeted to the ζ site, giving a 1-kb EcoRI fragment (disappearance of this band in this transformant; Fig. 3, panel B1, lane 3, band g). With the LIP2 probe, all transformants had a single strong 1.6-kb EcoRI-KpnI band (Fig. 3, panel B2, lanes 2 to 4, band h) compared to the weak band corresponding to the genomic LIP2 locus (Fig. 3, panel B2, lanes 1 to 4, band i). Copy numbers were estimated as for PO1d transformants and found to be in the same range (6 to 16 copies).
The ζ region and gene amplification.
PO1d was transformed with the expression cassette devoid of the ζ region (SalI and EcoRI digestion, Fig. 1). Few transformants were obtained, and their profile on Southern blots was different from that of transformants obtained with the expression cassette containing the ζ region (Fig. 3C). When the transformants were probed with LIP2, we detected the 1.6-kb genomic band of the LIP2 locus and one or two additional bands corresponding to the integrated cassette. In this context, integration occurred at random, and the extent of LIP2 amplification was significantly less than with the cassette with the ζ region.
Transformants are stable.
We tested the stability of the recombinant strains under inducing and noninducing conditions by culturing PO1d-6-15 and PO1d-6-17 separately in YPD and YPDH for 2 weeks (approximately 120 generations). No change was detected in the patterns obtained in YPD (data not shown) or YPDH (Fig. 3D), indicating that the selected transformants were stable even after 120 generations of induction conditions.
Increase in lipase production.
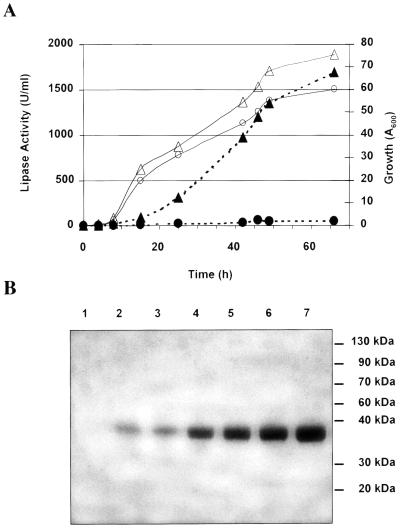
We compared the lipase secretion of the PO1d strain and the amplified transformant PO1d-6-15 (JMY184). In minimal medium (YNBH), the two strains grew similarly. Lipase activity at 52 h after induction was 20 U/ml in the supernatant for PO1d and 150 U/ml for the transformant, JMY184. In the rich YPDH medium, JMY184 grew faster and to a higher cell density (Fig. 4A). Lipase activity at 60 h after induction was 50 U/ml for PO1d and 1,500 U/ml for JMY184. In YPDH, we observed the accumulation over time of a single protein band in the cell supernatant (Fig. 4B). Conventional protein quantification methods could not be used due to the presence of olive oil, so protein production was estimated from SDS-PAGE gels stained with Coomassie blue and found to be about 0.5 g/liter. For the end points, when all the olive oil had disappeared, this estimated protein concentration was confirmed by the BCA titration method, which gave 0.556 g of protein per liter in the supernatant.
FIG. 4.
Lipase production by JMY184. (A) Growth and lipase activity in rich YPDH medium. Cell growth and lipolytic activity in the supernatant of the cell cultures are shown by open and solid symbols, respectively, with circles and triangles indicating the PO1d strain and the PO1d-6-15 transformant, respectively. (B) Extracellular lipase accumulation in YPDH medium. Samples (5 μl of crude supernatant) were resolved by SDS-PAGE (12%). Collection times were 0, 18, 25, 42, 46, 49, and 69 h (lanes 1 to 7, respectively). The gel was stained with Coomassie brilliant blue R-250. BenchMark prestained protein ladder (Life Technologies) was used as size standards. Sizes are indicated on the right-hand side.
Lipase activity and LIP2 mRNA levels in JMY184.
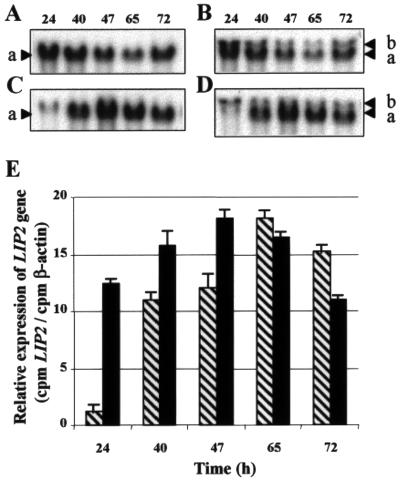
We analyzed LIP2 expression by JMY184 grown in different media by Northern blotting (Fig. 5). A Y. lipolytica β-actin probe was used to measure the amount of RNA. The amount of the LIP2 transcript (1.6 kb) was maximal at 47 h in YPH medium containing only olive oil (2%) and at 65 h in YPDH medium containing both olive oil (2%) and glucose (1%). The POX2 promoter driving expression of the LIP2 gene is repressed by glucose and induced when glucose is completely consumed (24 h) (data not shown and Fig. 5E). At the same time (24 h), in the absence of glucose, the level of LIP2 mRNA was two-thirds of the maximum. These differences in transcript level in the presence of olive oil as the carbon source and inducer and in the presence and absence of glucose correlate well with the lipase activity in the cell supernatant in the same growth conditions (Fig. 4B and data not shown) and are consistent with the hypothesis that regulation occurs at the transcriptional level.
FIG. 5.
(A to D) Northern blot analysis of LIP2 and β-actin transcripts in JMY184 grown in two different media. Total RNA was extracted from a JMY184 transformant grown in YPH (A) or in YPDH (C) at various time points. The probes used were the HindIII-EcoRI fragment containing the complete LIP2 open reading frame (A to D) and the 1-kb ScaI-XbaI fragment of Y. lipolytica β-actin (B and D). Band a corresponds to the LIP2 transcript signal, and band b corresponds to the β-actin transcript signal, indicated by an arrow. (E) Relative levels of LIP2 gene expression in JMY184 in YPH (solid bars) and YPDH (hatched bars) at 24, 40, 47, 65, and 72 h, normalized to the β-actin signal used as a control.
DISCUSSION
Previous plasmids used in Y. lipolytica contained fragments of E. coli DNA that restrict the utility of the plasmids in industrial applications. We constructed a series of vectors that release the expression cassette from the bacterial DNA prior transformation. These vectors are the first of their kind for Y. lipolytica and are the third example of this type. Such integrative cassettes devoid of bacterial DNA which could be amplified have also been used for the expression of Aspergillus oryzae α-amylase in an industrial baker's yeast strain (29) and for the expression of monellin in Candida utilis (17).
Amplification is usually based on drug resistance markers (18) or a defective marker such as poorly expressed heterologous auxotrophic markers (5, 11) and promoter-defective alleles (ura3d, leu2d, and trp1d) (3, 20, 22, 29). For promoter-defective alleles, amplification depends upon localization of the marker to the integrative cassette (29).
Culture stability is a major problem with the genetically modified organisms used for industrial protein production, as the large-scale production of protein often requires long-term culture. Integrating the expression cassette into the yeast genome increases stability (34), but the presence of tandem repeat copies at a genomic site can lead to mitotic or meiotic instability of the integrated plasmid (16, 44). To circumvent this problem, we targeted our new plasmids to the ζ sequence, a sequence widely dispersed in the yeast genome (38).
The integration pattern depended upon the strain used. In E150, which has ζ targets in its genome, integration was targeted to one ζ site (Fig. 3B, lane 3), and multicopy integrations occurred essentially in tandem. The PO1d strain does not have a genomic ζ region, and integration was dispersed throughout the genome (Fig. 3A). The ζ region at the end of the expression cassette is required for multiple and dispersed integration (Fig. 3C), although the mechanism underlying this phenomenon remains to be elucidated (4). This is the first description of a dispersed multiple integration event in a yeast other than Saccharomyces cerevisiae (5). Only dispersed single integration was obtained previously by restriction enzyme-mediated integration (37). We found no evidence that the LTR increases movement of the cassettes following integration. Scattered integrations may account for the high stability of the two transformants of PO1d. The small size of the expression cassette (5.4 kb) also may be important in mitotic stability (21), and the nature of the expressed gene: LIP2 dispersed multicopy integrants are more stable than XPR2 tandem multicopy integrants, which tend to deamplify under induction conditions (20).
The copy number was 6 to 16 in the various transformants that we obtained. This is lower than the 60 copies previously described when rDNA was used as the target (20). This difference may be due to the target sequence or to the reporter gene. For most of the transformants analyzed, we observed a strong correlation between copy number and lipase production, similar to that seen for AEP production when the XPR2 copy number was low (10 copies) (20, 26). The lipase activity in the supernatant of some transformants was 40 times higher than that of the recipient strain.
We also wanted to determine if the PO1d host, when combined with our vectors, reaches the biological limits of the expression system or whether production could be increased further. We are now developing vectors based on the JPM3 technology with the LIP2 expression cassette but carrying a defective version of LEU2. However, increasing gene dosage does not necessarily improve productivity (34), as shown in Pichia pastoris for the tetanus toxin fragment C). Alternatively, changing the POX2 promoter to another inducible promoter such as XPR2, ICL1, or RPS7 or to a constitutive promoter such as TEF or hp4d (2, 23–25, 30) might also increase productivity.
Y. lipolytica has previously produced up to 1,200 U of lipase per ml in a fermentor (9). After two rounds of chemical mutagenesis, some of the mutants secreted 25 times more lipase than the wild-type strain. Some of our transformed strains secrete at least equivalent amounts of active lipase in flask culture (1,500 U/ml), suggesting that molecular biological methods are at least as useful for strain development as are the traditional methods. Good production processes can greatly improve total production capacity (6, 12). Attempts to optimize batch production conditions in a fermentor with JMY184 have already resulted in an increase in lipase activity up to 10,000 U/ml (data not shown). This confirms the great capacity of Y. lipolytica for homologous (AEP and lipase) (9, 20) and heterologous (rice α-amylase, prochymosin, factor XIIIa, and hepatitis B virus middle surface antigen) (13, 28, 31, 42) protein secretion.
In summary, we have developed an amplification system that allows multiple integration of an expression cassette devoid of bacterial DNA, either in tandem or dispersed depending on the strain used. We obtained strains with as many as 16 integrated copies that are stable for at least 120 generations and that produced high levels of lipase. This technology may be a general one for the production of proteins in Y. lipolytica for commercial use.
ACKNOWLEDGMENTS
This work was supported in part by Mayoly-Spindler Laboratories, by the Institut National de la Recherche Agronomique, and by the Centre National de la Recherche Scientifique. Strains and plasmids are available upon request to the CLIB under specific INRA agreement.
G. Pignède and H.-J. Wang contributed equally to this work.
REFERENCES
- 1.Barth G, Gaillardin C. Yarrowia lipolytica. In: Wolf W K, editor. Nonconventional yeasts in biotechnology. Vol. 1. Berlin, Germany: Springer-Verlag; 1996. pp. 313–318. [Google Scholar]
- 2.Barth G, Scheuber T. Cloning of the isocitrate lyase gene (ICL1) from Yarrowia lipolytica and characterization of the deduced protein. Mol Gen Genet. 1993;241:422–430. doi: 10.1007/BF00284696. [DOI] [PubMed] [Google Scholar]
- 3.Bergkamp R J, Kool I M, Geerse R H, Planta R J. Multiple-copy integration of the alpha-galactosidase gene from Cyamopsis tetragonoloba into the ribosomal DNA of Kluyveromyces lactis. Curr Genet. 1992;21:365–370. doi: 10.1007/BF00351696. [DOI] [PubMed] [Google Scholar]
- 4.Boeke J, Sandmeyer S. Yeast transposable elements. In: Broach J, Pringle J, Jones E, editors. The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis, and energetics. I. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1991. pp. 193–261. [Google Scholar]
- 5.Boeke J D, Xu H, Fink G R. A general method for the chromosomal amplification of genes in yeast. Science. 1988;239:280–282. doi: 10.1126/science.2827308. [DOI] [PubMed] [Google Scholar]
- 6.Chang C C, Ryu D D, Park C S, Kim J Y, Ogrydziak D M. Recombinant bioprocess optimization for heterologous protein production using two-stage, cyclic fed-batch culture. Appl Microbiol Biotechnol. 1998;49:531–537. doi: 10.1007/s002530051209. [DOI] [PubMed] [Google Scholar]
- 7.De Felice B, Pontecorvo G, Carfagna M. Degradation of waste waters from olive oil mills by Yarrowia lipolytica ATCC 20255 and Pseudomonas putida. Acta Biotechnol. 1997;3:231–239. [Google Scholar]
- 8.Destain J. Ph.D. thesis. Gembloux, Belgium: Faculté universitaire des sciences agronomiques de Gembloux; 1998. [Google Scholar]
- 9.Destain J, Roblain D, Thonart P. Improvement of lipase production from Yarrowia lipolytica. Biotechnol Lett. 1997;19:105–107. [Google Scholar]
- 10.European Commission. Council directive 90/219/EEC. Off. J. Eur. Commun. L117 of 5 August 1990. 1990. pp. 124–133. [Google Scholar]
- 11.Gilbert S C, van Urk H, Greenfield A J, McAvoy M J, Denton K A, Coghlan D, Jones G D, Mead D J. Increase in copy number of an integrated vector during continuous culture of Hansenula polymorpha expressing functional human haemoglobin. Yeast. 1994;10:1569–1580. doi: 10.1002/yea.320101206. [DOI] [PubMed] [Google Scholar]
- 12.Gordillo M A, Montesinos J L, Casas C, Valero F, Lafuente J, Sola C. Improving lipase production from Candida rugosa by a biochemical engineering approach. Chem Phys Lipids. 1998;93:131–142. doi: 10.1016/s0009-3084(98)00037-1. [DOI] [PubMed] [Google Scholar]
- 13.Hamsa P V, Chattoo B B. Cloning and growth-regulated expression of the gene encoding the hepatitis B virus middle surface antigen in Yarrowia lipolytica. Gene. 1994;143:165–170. doi: 10.1016/0378-1119(94)90092-2. [DOI] [PubMed] [Google Scholar]
- 14.Hoekstra M F, Seifert H S, Nickoloff J, Heffron F. Shuttle mutagenesis: bacterial transposons for genetic manipulations in yeast. Methods Enzymol. 1991;194:329–342. doi: 10.1016/0076-6879(91)94025-8. [DOI] [PubMed] [Google Scholar]
- 15.Jaeger K E, Reetz M T. Microbial lipases form versatile tools for biotechnology. Trends Biotechnol. 1998;16:396–403. doi: 10.1016/s0167-7799(98)01195-0. [DOI] [PubMed] [Google Scholar]
- 16.Keller N P, Bergstrom G C, Yoder O C. Mitotic stability of transforming DNA is determined by its chromosomal configuration in the fungus Cochliobulus heterosporum. Curr Genet. 1991;25:227–233. [Google Scholar]
- 17.Kondo K, Miura Y, Sone H, Kobayashi K, Iijima H. High-level expression of a sweet protein, monellin, in the food yeast Candida utilis. Nat Biotechnol. 1997;15:453–457. doi: 10.1038/nbt0597-453. [DOI] [PubMed] [Google Scholar]
- 18.Kondo K, Saito T, Kajiwara S, Takagi M, Misawa N. A transformation system for the yeast Candida utilis: use of a modified endogenous ribosomal protein gene as a drug-resistant marker and ribosomal DNA as an integration target for vector DNA. J Bacteriol. 1995;177:7171–7177. doi: 10.1128/jb.177.24.7171-7177.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Le Dall M T, Nicaud J M, Gaillardin C. Multiple-copy integration in the yeast Yarrowia lipolytica. Curr Genet. 1994;26:38–44. doi: 10.1007/BF00326302. [DOI] [PubMed] [Google Scholar]
- 21.Lopes T S, de Wijs I J, Steenhauer S I, Verbakel J, Planta R J. Factors affecting the mitotic stability of high-copy-number integration into the ribosomal DNA of Saccharomyces cerevisiae. Yeast. 1996;12:467–477. doi: 10.1002/(sici)1097-0061(199604)12:5<467::aid-yea933>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 22.Lopes T S, Klootwijk J, Veenstra A E, van der Aar P C, van Heerikhuizen H, Raue H A, Planta R J. High-copy-number integration into the ribosomal DNA of Saccharomyces cerevisiae: a new vector for high-level expression. Gene. 1989;79:199–206. doi: 10.1016/0378-1119(89)90202-3. [DOI] [PubMed] [Google Scholar]
- 23.Madzak C, Blanchin-Roland S, Cordero Otero R R, Gaillardin C. Functional analysis of upstream regulating regions from the Yarrowia lipolytica XPR2 promoter. Microbiology. 1999;145:75–87. doi: 10.1099/13500872-145-1-75. [DOI] [PubMed] [Google Scholar]
- 24.Madzak C, Treton B, Blanchin-Roland S. Strong hybrid promoters and integrative expression/secretion vectors for quasi-constitutive expression of heterologous proteins in the yeast Yarrowia lipolytica. Mol Microbiol Biotechnol. 2000;2:207–216. [PubMed] [Google Scholar]
- 25.Muller S, Sandal T, Kamp-Hansen P, Dalboge H. Comparison of expression systems in the yeasts Saccharomyces cerevisiae, Hansenula polymorpha, Klyveromyces lactis, Schizosaccharomyces pombe and Yarrowia lipolytica: cloning of two novel promoters from Yarrowia lipolytica. Yeast. 1998;14:1267–1283. doi: 10.1002/(SICI)1097-0061(1998100)14:14<1267::AID-YEA327>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 26.Nicaud J M, Fabre E, Beckerich J M, Fournier P, Gaillardin C. Cloning, sequencing and amplification of the alkaline protease (XPR2) gene of the yeast Yarrowia lipolytica. J Biotechnol. 1989;12:285–298. [Google Scholar]
- 27.Nicaud J M, Fabre E, Gaillardin C. Expression of invertase activity in Yarrowia lipolytica and its use as a selective marker. Curr Genet. 1989;16:253–260. doi: 10.1007/BF00422111. [DOI] [PubMed] [Google Scholar]
- 28.Nicaud J M, Fournier P, La Bonnadiere C, Chasles M, Gaillardin C. Use of ars18 based vectors to increase protein production in Yarrowia lipolytica. J Biotechnol. 1991;19:259–270. doi: 10.1016/0168-1656(91)90063-2. [DOI] [PubMed] [Google Scholar]
- 29.Nieto A, Prieto J A, Sanz P. Stable high-copy-number integration of Aspergillus oryzae alpha-amylase cDNA in an industrial baker's yeast strain. Biotechnol Prog. 1999;15:459–466. doi: 10.1021/bp9900256. [DOI] [PubMed] [Google Scholar]
- 30.Ogrydziak D M, Demain A L, Tannenbaum S R. Regulation of extracellular protease production in Candida lipolytica. Biochim Biophys Acta. 1977;497:525–538. doi: 10.1016/0304-4165(77)90209-4. [DOI] [PubMed] [Google Scholar]
- 31.Park C S, Chang C C, Kim J Y, Ogrydziak D M, Ryu D D. Expression, secretion, and processing of rice alpha-amylase in the yeast Yarrowia lipolytica. J Biol Chem. 1997;272:6876–6881. doi: 10.1074/jbc.272.11.6876. [DOI] [PubMed] [Google Scholar]
- 32.Pignède G, Fudalej F, Nicaud J-M, Gaillardin C, Seman M. Clonage et expression de lipases extracellulaires acido resistantes de levures. French patent 98,899. September 1998. [Google Scholar]
- 33.Pignède G, Wang H J, Fudalej F, Gaillardin C, Seman M, Nicaud J-M. Characterization of an extracellular lipase encoded by LIP2 in Yarrowia lipolytica. J Bacteriol. 2000;182:2802–2810. doi: 10.1128/jb.182.10.2802-2810.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romanos M A, Scorer C A, Clare J J. Foreign gene expression in yeast: a review. Yeast. 1992;8:423–488. doi: 10.1002/yea.320080602. [DOI] [PubMed] [Google Scholar]
- 35.Rossolini G M, Riccio M L, Gallo E, Galeotti C L. Kluyveromyces lactis rDNA as a target for multiple integration by homologous recombination. Gene. 1992;119:75–81. doi: 10.1016/0378-1119(92)90068-z. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Maniatis T, Fritsch E F. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Schiestl R H, Petes T D. Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:7585–7589. doi: 10.1073/pnas.88.17.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmid-Berger N, Schmid B, Barth G. Ylt1, a highly repetitive retrotransposon in the genome of the dimorphic fungus Yarrowia lipolytica. J Bacteriol. 1994;176:2477–2482. doi: 10.1128/jb.176.9.2477-2482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smerdon G R, Walton E F, Aves S J. Stable production of human gastric lipase by chromosomal integration in the fission yeast Schizosaccharomyces pombe. Appl Microbiol Biotechnol. 1998;49:45–50. doi: 10.1007/s002530051135. [DOI] [PubMed] [Google Scholar]
- 40.Stead D. Microbial lipases: their characteristics, role in food spoilage and industrial uses. J Dairy Res. 1986;53:481–505. doi: 10.1017/s0022029900025103. [DOI] [PubMed] [Google Scholar]
- 41.Szostak J W, Wu R. Insertion of a genetic marker into the ribosomal DNA of yeast. Plasmid. 1979;2:536–554. doi: 10.1016/0147-619x(79)90053-2. [DOI] [PubMed] [Google Scholar]
- 42.Tharaud C, Ribet A M, Costes C, Gaillardin C. Secretion of human blood coagulation factor XIIIa by the yeast Yarrowia lipolytica. Gene. 1992;121:111–119. doi: 10.1016/0378-1119(92)90168-o. [DOI] [PubMed] [Google Scholar]
- 43.Thonart P, Destain J, Zgouli S, Antoine P, Godefroid J, Evrard P. Les bacs à graisse—une solution aux problèmes des matières grasses pour les PME. Trib Eau. 1997;50:41–46. [Google Scholar]
- 44.Tooley P W, Leung H, Leong S A. Meiotic and mitotic stability of transforming DNA in the phytopathogenic fungus Magnaporthe grisea. Curr Genet. 1992;21:55–60. [Google Scholar]
- 45.Wang H J, Le Dall M T, Wach Y, Laroche C, Belin J M, Gaillardin C, Nicaud J M. Evaluation of acyl coenzyme A oxidase (Aox) isozyme function in the n-alkane-assimilating yeast Yarrowia lipolytica. J Bacteriol. 1999;181:5140–5148. doi: 10.1128/jb.181.17.5140-5148.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wery J, Gutker D, Renniers A C, Verdoes J C, van Ooyen A J. High copy number integration into the ribosomal DNA of the yeast Phaffia rhodozyma. Gene. 1997;184:89–97. doi: 10.1016/s0378-1119(96)00579-3. [DOI] [PubMed] [Google Scholar]
- 47.Xuan J W, Fournier P, Declerck N, Chasles M, Gaillardin C. Overlapping reading frames at the LYS5 locus in the yeast Yarrowia lipolytica. Mol Cell Biol. 1990;10:4795–4806. doi: 10.1128/mcb.10.9.4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zentler-Munro P L, Assoufi B A, Balasubramanian K, Cornell S, Benoliel D, Northfield T C, Hodson M E. Therapeutic potential and clinical efficacy of acid-resistant fungal lipase in the treatment of pancreatic steatorrhoea due to cystic fibrosis. Pancreas. 1992;7:311–319. doi: 10.1097/00006676-199205000-00007. [DOI] [PubMed] [Google Scholar]