Abstract
SARS-CoV-2 is the coronavirus causing the ongoing pandemic with > 460 millions of infections and > 6 millions of deaths. SARS-CoV-2 nucleocapsid (N) is the only structural protein which plays essential roles in almost all key steps of the viral life cycle with its diverse functions depending on liquid–liquid phase separation (LLPS) driven by interacting with various nucleic acids. The 419-residue N protein is highly conserved in all variants including delta and omicron, and composed of both folded N-/C-terminal domains (NTD/CTD) as well as three long intrinsically disordered regions (IDRs). Recent results have suggested that its CTD and IDRs are also cryptic nucleic acid–binding domains. In this context, any small molecules capable of interfering in its interaction with nucleic acids are anticipated to modulate its LLPS and associated functions. Indeed, ATP, the energy currency existing at very high concentrations (2–12 mM) in all living cells but absent in viruses, modulates LLPS of N protein, and consequently appears to be evolutionarily hijacked by SARS-CoV-2 to promote its life cycle. Hydroxychloroquine (HCQ) has been also shown to specifically bind NTD and CTD to inhibit their interactions with nucleic acids, as well as to disrupt LLPS. Particularly, the unique structure of the HCQ-CTD complex offers a promising strategy for further design of anti-SARS-CoV-2 drugs with better affinity and specificity. The finding may indicate that LLPS is indeed druggable by small molecules, thus opening up a promising direction for drug discovery/design by targeting LLPS in general.
Keywords: Adenosine triphosphate (ATP), Hydroxychloroquine (HCQ): SARS-CoV-2, Nucleocapsid (N) protein, Liquid–liquid phase separation (LLPS), NMR spectroscopy
Liquid–liquid phase separation (LLPS) in the life cycle of SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) caused the ongoing pandemic (Wu et al. 2020), which already led to > 460 millions of infections and > 6 millions of deaths. SARS-CoV-2 is a member of a large family of positive-stranded RNA coronaviruses with ∼30 kb genomic RNA (gRNA) packaged by nucleocapsid (N) protein in a membrane-enveloped virion. SARS-CoV-2 contains four structural proteins: namely the spike (S) protein that recognizes the host cell receptors angiotensin–converting enzyme-2 (ACE2), membrane (M), membrane-associated envelope (E), and N proteins.
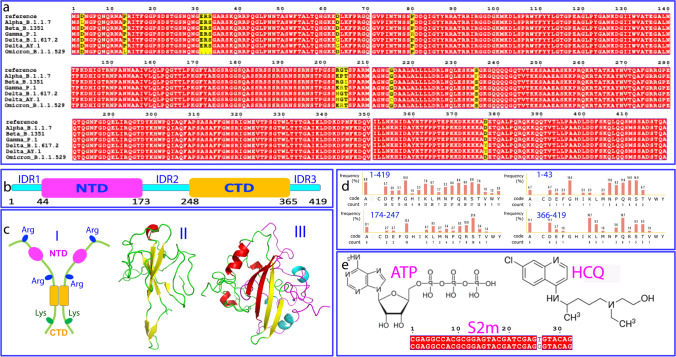
SARS-CoV-2 N protein is a 419-residue multifunctional protein whose sequences are highly conserved in all variants of concern (VOCs) including delta and omicron (Fig. 1a). It is composed of the folded N-terminal domain (NTD) and C-terminal domain (CTD) (Fig. 1b and 1c), as well as three long intrinsically disordered regions (IDRs) respectively over 1–43 with 6 Arg and 1 Lys, 174–247 with 8 Arg and 1 Lys, 366–419 with 1 Arg and 9 Lys (Fig. 1d). Previous studies indicate that its NTD is an RNA-binding domain (RBD) functioning to bind a large assay of viral and host cell nucleic acids including RNA and DNA (Dinesh et al. 2020), while CTD acts to dimerize/oligomerize for forming high-order structures (Zinzula et al. 2021). However, our recent NMR studies unveiled that CTD is a cryptic domain for binding nucleic acids with the affinity even higher than that of NTD in binding the 32-mer stem-loop II motif (S2m) derived from SARS-CoV-2 gRNA (Fig. 1e) (Dang and Song 2022).
Fig. 1.
SARS-CoV-2 nucleocapsid protein, ATP and HCQ. a Sequence alignment of N protein of variants of concern (VOCs) of SARS-CoV-2 according to WHO: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants. b Domain organization of SARS-CoV-2 N protein. c (I) Cartoon diagram of the dimeric N protein with three IDRs rich in Arg or Lys. Three-dimensional structures of NTD (II) and dimeric CTD (III). d Amino acid compositions of the full-length N protein and three IDRs respectively. e Chemical structures of ATP and HCQ, as well as sequences of 32-mer stem-loop II motif (S2m) of SARS-CoV-1 (upper) and SARS-CoV-2 (lower)
Coronavirus N proteins have two major categories of functions: while their primary role is to assemble the gRNA to form the gRNA-Nprotein (vRNP) complex into the new virions at the final stage of the life cycle, they also appear to suppress the immune system of the host cell and to hijack cellular machineries to achieve the replication by forming stress granules (SGs) and localizing gRNA onto the replicase-transcriptase complexes (RTCs) (Carlson et al. 2020; Dang et al. 2021a, b, c; Dang and Song 2021, 2022; Iserman et al. 2020; Lu et al. 2021; Perdikari et al. 2020; Savastano et al. 2020; Cascarina and Ross 2022).
Very recently, liquid–liquid phase separation (LLPS), the emerging principle for commonly underlying the formation of the membrane-less organelles (MLOs) critical for cellular physiology and pathology (Hyman et al. 2014; Patel et al. 2017; Shin and Brangwynne 2017; Kang et al. 2019a, b; Dang et al. 2021a), has been found to be the key mechanism for the diverse functions of SARS-CoV-2 N protein (Carlson et al. 2020; Dang et al. 2021a, b, c; Iserman et al. 2020; Lu et al. 2021; Perdikari et al. 2020; Savastano et al. 2020; Cascarina and Ross 2022). Strikingly, the functions of N protein appear to be dependent on its binding to various nucleic acids of both specific and non-specific sequences. Indeed, LLPS of N protein appears to be mainly driven by its multivalent and dynamic interactions with nucleic acids, because the pure N protein with nucleic acids completely removed is largely lacking of the intrinsic capacity in LLPS (Carlson et al. 2020; Dang et al. 2021a, b, c).
As seen in Fig. 1d, although three long IDRs with a total of ~ 170 residues contain a number of Arg/Lys residues, they only have two Phe residues. Therefore, unlike the 512-residue human FUS protein, which is rich in both Arg/Lys and aromatic residues particularly Tyr within its IDRs, and consequently able to establish intermolecular π–π and π-cation interactions for driving LLPS even without needing the presence of nucleic acids (Kang et al. 2019a, b; Song 2021), for SARS-CoV-2 N protein, nucleic acids appear to be essential for driving LLPS by binding both NTD and CTD, as well as by providing aromatic rings to establish π–π or/and π-cation interactions with Arg/Lys residues within IDRs of N protein. In particular, although the exact mechanism still remains elusive, the package of the gRNA into the new virions requires the complex but precise interaction between gRNA and N protein, which must be extremely challenging for SARS-CoV-2 with such a large genome (~ 30 kb). As such, N protein needs to be highly conserved in SARS-CoV-2 variants, because only the variants which have the N proteins correctly functional in binding various nucleic acids can survive and effectively spread in evolution. In this context, any small molecule capable of intervening in the interaction of N protein with nucleic acids is expected to modulate most, if not all, key steps of the life cycle of SARS-CoV-2, some of which may manifest the anti-SARS-CoV-2 activity.
ATP is hijacked by SARS-CoV-2 to promote its life cycle
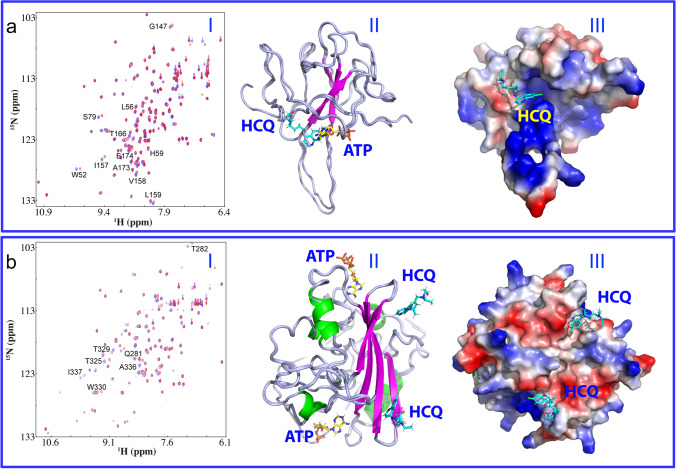
Mysteriously, ATP, the universal “biological fuel” for all living cells (Fig. 1e), has very high concentrations from 2 to 12 mM depending on the types of cells (Patel et al. 2017; Kang et al. 2019a, b; Song 2021). By contrast, all viruses have no ability to generate ATP (Wessner 2010). Very unexpectedly, ATP has been recently shown to specifically bind a pocket on NTD of SARS-CoV-2 N protein (Fig. 2a), which is located within the conserved surface for binding various nucleic acids (Dinesh et al. 2020). Very recently, as shown in Fig. 2b, the dimeric CTD was also unveiled to own two binding pockets for ATP (Dang and Song 2022). Moreover, ATP is not only able to act as a bivalent binder to biphasically modulate LLPS of SARS-CoV-2 N protein in the absence of nucleic acids: namely induction at low and dissolution at high ATP concentrations, but also to monotonically dissolve LLPS induced by the presence of nucleic acids (Dang et al. 2021a, b, c; Dang and Song 2021). Mechanistically, the results suggest that ATP can compete with nucleic acids not only in binding the folded NTD/CTD but also Arg/Lys-rich IDRs (Dang et al. 2021a, b, c; Song 2021; Dang and Song 2022).
Fig. 2.
ATP and HCQ specifically bind NTD and CTD. a (I) Superimposition of HSQC spectra of NTD in the free state (blue) and in the presence of HCQ at 1:15 (NTD:HCQ) (red). The significantly perturbed residues are labeled. (II) Superimposition of the structures of the ATP-NTD and HCQ-NTD complexes with ATP and HCQ displayed in stick. (III) the structure of the HCQ-NTD complex with HCQ in stick and NTD in the electrostatic potential surface. b (I) Superimposition of HSQC spectra of CTD in the free state (blue) and in the presence of HCQ at 1:7.5 (CTD:HCQ) (red). The significantly-perturbed residues are labeled. (II) Superimposition of the structures of the ATP-CTD and HCQ-CTD complexes with ATP and HCQ in stick. (III) the structure of the HCQ-CTD complex with HCQ in stick and CTD in the electrostatic potential surface
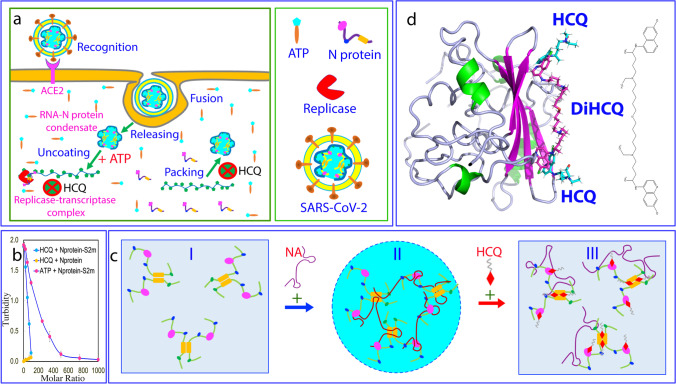
Therefore, ATP appears to be evolutionarily hijacked by SARS-CoV-2 to facilitate its life cycle. Briefly, as illustrated in Fig. 3a, upon entry into the cell, SARS-CoV-2 will release its gRNA-Nprotein condensate. As at this stage, the ratio between ATP and Nprotein/gRNA condensate is very high, ATP thus serves to facilitate the uncoating of gRNA by dissolving the vRNP condensate. Later after the new copies of the viral RNA or/and N protein are synthesized by the host cell machinery, the ratio will become reduced. Consequently, ATP may even act to enhance LLPS of the viral gRNA and N protein together with the host cell replicases and related factors to form replicase-transcriptase complexes. Finally, once the synthesis of all viral components has been completed, the ratio between ATP and Nprotein/gRNA becomes further reduced, and consequently a large population of N protein molecules will become unbound with ATP. As such, the ATP-unbound NTD/CTD as well as IDRs of N protein will become available for recognizing and binding the specific sites of the viral gRNA to initiate the package of gRNA and N proteins to form new virions.
Fig. 3.
HCQ disrupts LLPS of SARS-CoV-2 N protein induced by nucleic acid. a A scheme to illustrate the role of LLPS of SARS-CoV-2 N protein in the viral cycle which is critically driven by the multivalent interaction with various nucleic acids but can be disrupted by HCQ. In this context, ATP appears to be hijacked by SARS-CoV-2 to promote its life cycle including the initial uncoating of the gRNA-Nprotein condensate, subsequent localizing to forming replicase-transcriptase complex, and final package of gRNA and N protein. By contrast, HCQ appears to manifest the anti-SARS-CoV-2 activity by disrupting the interaction of N protein with nucleic acids and LLPS at least at two key steps: formation of replicase-transcriptase complexes, as well as final package of gRNA and N protein into new virions. b Turbidity (absorption at 600 nm) curves of N protein without and with the pre-existence of S2m at 1:0.75 upon addition of ATP and HCQ at different ratios. c Speculative model for LLPS of N protein induced by nucleic acids which can be dissolved by HCQ due to the displacement of nucleic acids from being bound with NTD/CTD of N protein. d The docking structure of the dimeric CTD in complex with HCQ and DiHCQ by covalently linking two individual HCQ molecules
HCQ disrupts the interaction of SARS-CoV-2 N protein with nucleic acids and LLPS
Hydroxychloroquine (HCQ) (Fig. 1e) has been proposed for clinically combating the SARS-CoV-2 pandemic (Roldan et al. 2020; Satarker et al. 2020). Very recently, a clinical study in Singapore demonstrated that oral HCQ could indeed prevent the SARS-CoV-2 infection in the high transmission environments (Seet et al. 2921). On the other hand, the mechanisms for its anti-SARS-CoV-2 activity remain poorly understood and previously no viral protein has been experimentally identified to interact with HCQ. Indeed, all the actions of HCQ have been proposed to target the sites on the host cells including the interference in the endocytic pathway, blockade of sialic acid receptors, restriction of pH-mediated S protein cleavage at the ACE2-binding site, and prevention of cytokine storm.
Very unexpectedly, our NMR study recently decrypted that HCQ could in fact specifically bind both NTD (I of Fig. 2a) and CTD (I of Fig. 2b) with dissociation constants (Kds) of 112.1 and 57.1 μM respectively, which consequently inhibited the interactions of N protein with nucleic acids as well as dissolved its LLPS induced by nucleic acids (Dang and Song 2021; Song 2021). Moreover, with NMR-derived constraints, the structures of the HCQ-NTD and HCQ-CTD complexes have been successfully constructed. In the complexes, while ATP and HCQ bind the pockets of NTD with an overlap over their aromatic rings (II and III of Fig. 2a), the distinctive pockets were identified on the dimeric CTD to respectively bind ATP and HCQ (II and III of Fig. 2b). Noticeably in the HCQ-CTD complex, two HCQ molecules are bound with two distinctive pockets but within a cleft on the same side of the dimeric CTD structure, and this binding appears to be mainly driven by the insertion of the aromatic ring of HCQ into the CTD pockets.
On the other hand, unlike ATP which acts as a bivalent binder capable of biphasically modulating LLPS of SARS-CoV-2 N protein (Dang et al. 2021a, b, c), HCQ has no ability to induce LLPS of N protein in the absence of nucleic acids (Fig. 3b), indicating that HCQ cannot behave as a bivalent binder. Nevertheless, HCQ has the capacity much higher than that of ATP in dissolving LLPS of SARS-CoV-2 N protein induced by both specific and non-specific nucleic acids (Dang and Song 2021; Song 2021), which include 24-mer poly(dA) (A24) and S2m (Fig. 1d and Fig. 3b). Mechanistically, as illustrated by Fig. 3c, upon introduction of nucleic acids, SARS-CoV-2 N protein existing as homogenous solution (I of Fig. 3c) will undergoes LLPS into forming dynamic liquid droplets (II of Fig. 3c) by establishing dynamic and multivalent interactions between nucleic acids and NTD/CTD as well as Arg/Lys residues within IDRs. In this context, the ability of HCQ which is much stronger than that of ATP to displace nucleic acids from NTD/CTD appears to be sufficient to disrupt LLPS (III of Fig. 3c) because the binding affinities between nucleic acids and folded NTD/CTD are much higher than those between nucleic acids and Arg/Lys residues within IDRs (Song 2021). This also indicates that the interaction of the folded NTD and CTD with nucleic acids is the major driving force for LLPS of SARS-CoV-2 N protein.
Therefore, in addition to acting on the multiple sites of the host cell as extensively proposed (Roldan et al. 2020; Satarker et al. 2020), HCQ can also manifest its anti-SARS-CoV-2 activity by directly targeting SARS-CoV-2 N protein to at least block two key steps of the viral life cycle: the formation of the replicase-transcriptase complex and final package of gRNA and N protein (Fig. 3a). Intriguingly, HCQ was shown to inhibit the maturation of SARS-CoV-2 virions, which was previously proposed to result from the HCQ-induced changes of host cell structures/conditions such as pH (Roldan et al. 2020; Satarker et al. 2020). Our newly discovered mechanism suggests that the capacity of HCQ to specifically inhibit the binding of N protein with nucleic acids and to disrupt LLPS may at least partly contribute to the inhibition of the maturation of SARS-CoV-2 virions.
Summary and outlook
Currently, great efforts have been directed to developing the spike-based vaccines to combat the pandemic. Nevertheless, challenges still remain to terminate the pandemic, which include rapidly emerging antibody-resistance variants such as omicron (Wang et al. 2021) and the potential adverse effects (Lei et al. 2021; Hoepel et al. 2021; Olofsson et al. 2022). Therefore, small molecule drugs by directly targeting SARS-CoV-2 proteins are extremely valuable and urgently needed to finally terminate the pandemic.
Out of four SARS-CoV-2 structural proteins, N protein is the only one which plays the essential roles in almost all key steps of the viral life cycle. With our recent discovery (Dang and Song 2022), SARS-CoV-2 N protein has thus been established to be a multivalent nucleic acid–binding protein, whose folded NTD and CTD as well as Arg/Lys residues within IDRs are all capable of binding nucleic acids of the extreme diversity in types and sequences. Most importantly, almost all functions of N protein including LLPS appear to depend on its capacity in interacting with nucleic acids and as a result its sequences are highly conserved in all variants. In this context, SARS-CoV-2 N protein represents a top target for discovery/design of anti-SARS-CoV-2 drugs by disrupting its interaction with nucleic acids. Nevertheless, this task appeared to be very challenging as the interaction interfaces of N protein with nucleic acids are very large. Therefore, it is very unexpected to find that ATP, the universal energy currency in all living cells but absent in viruses, has the ability to generally compete with nucleic acids by binding various folded and intrinsically disordered nucleic acid–binding domains not only of human proteins such as FUS, TDP-34, and hnRNPA1 (Kang et al. 2019a, b; Kang et al. 2019a; Dang et al. 2021a, 2021b; Song 2021), but also SARS-CoV-2 N protein (Dang et al. 2021a, b, c; Dang and Song 2022). In the biological context, ATP appears to be emerging as a cellular factor which has been hijacked by SARS-CoV-2 in evolution to facilitate its life cycle. As all coronaviruses have the highly conserved N proteins and share the conserved life cycle mechanisms, likely ATP is also exploited by other coronaviruses to facilitate their life cycles.
On the other hand, despite its small size, HCQ, a safe drug recommended by WHO to treat other diseases (Roldan et al. 2020; Satarker et al. 2020), has been now shown to specifically bind NTD/CTD to inhibit the interactions of SARS-CoV-2 N protein with nucleic acids as well as to dissolve LLPS. This finding not only provides an acting mechanism for HCQ to manifest the anti-SARS-CoV-2 activity, but also unveils that the large interaction interfaces of SARS-CoV-2 N protein with nucleic acids as well as the associated LLPS are in fact druggable by small molecules, thus suggesting a promising strategy for further development of anti-SARS-CoV-2 drugs. Furthermore, HCQ appears to be the first existing drug which has been found to target LLPS, thus bearing immediate implications for further design of drugs in general by modulating LLPS of other protein–nucleic acid systems, whose roles in underlying various human diseases are just starting to be recognized.
At the fundamental level, the core mechanisms of LLPS induced by nucleic acids appear to be highly conserved (Dang et al. 2021a, b, c; Kang et al. 2019a, b; Song 2021). Consequently, HCQ which acts to target LLPS of N proteins is anticipated to be effective to most, if not all, variants of SARS-CoV-2. This may explain why African countries where HCQ is regularly in-taken to prevent malaria, have a low incidence of SARS-CoV-2 although they have very low vaccination rates (BBC news 2020). On the other hand, accumulating evidence suggests that the nucleic acid–induced LLPS is not only critical for the life cycles of coronaviruses, but might be also essential for other virus host interactions. This might thus rationalize the puzzling observations that HCQ was not only effective for SARS-CoV-2 and SARS-CoV-1, but also fot Dengue/Zika infections (Roldan et al. 2020; Satarker et al. 2020).
The relatively low binding affinities of HCQ to NTD/CTD of SARS-CoV-2 N protein (Kd of 112.1 and 57.1 μM respectively) rationalize the reports that HCQ is highly effective in preventing the infection as well as in treating the infection at the early stage (Roldan et al. 2020; Satarker et al. 2020; Seet et al. 2921). However, as shown in Fig. 2b, in the HCQ-CTD complex, two HCQ molecules utilize the aromatic rings to insert into the binding pockets within a cleft on the same side of the dimeric CTD structure. This unique HCQ-CTD structure might offer a promising strategy for further design of anti-SARS-CoV-2 drugs which are expected to own better affinity and specificity (Dang and Song 2021). Briefly, by engineering the proper groups to link the aromatic rings of two HCQ molecules, the bivalent or even multivalent binders (DiHCQ) might be obtained (Fig. 3d). For such molecules, their Kd values are the time of the Kd values of individual groups (Song and Ni 1998). Furthermore, the bivalent/multivalent binders might also have the high specificity because the probability should be extremely low for human proteins to own such unique two HCQ-binding pockets observed on the dimeric CTD structure.
Here, we further propose to integrate the combinatorial chemistry and biophysical methods including experimental such as NMR and computational such as MD simulations (Shi et al. 2011; Dang and Song, 2021; Dang et al. 2022) to facilitate the high through-put design of bivalent or even multivalent small molecules by starting from HCQ in order to obtain more efficient anti-SARS-CoV-2 drugs, which are capable of binding the dimeric CTD of SARS-CoV-2 N protein not only with its two aromatic rings, but also with its linker groups.
Author contribution
Conceived the research: J.S. Performed research and analyzed data: M.D and J.S; acquired funding: J.S; wrote manuscript: J.S.
Funding
This study is supported by Ministry of Education of Singapore (MOE) Tier 1 Grants R-154–000-B92-114 to Jianxing Song.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- BBC news (2020) Coronavirus in Africa: five reasons why Covid-19 has been less deadly than elsewhere. https://www.bbc.com/news/world-africa-54418613
- Carlson CR, Asfaha JB, Ghent CM et al (2020) Phosphoregulation of phase separation by the SARS-CoV-2 N protein suggests a biophysical basis for its dual functions. Mol Cell 17:1092-1103.e4. 10.1016/j.molcel.2020.11.025 [DOI] [PMC free article] [PubMed]
- Cascarina SM, Ross ED (2022) Phase separation by the SARS-CoV-2 nucleocapsid protein: consensus and open questions. J Biol Chem 298:101677. 10.1016/j.jbc.2022.101677 [DOI] [PMC free article] [PubMed]
- Dang M, Song J (2021) Structural basis of anti-SARS-CoV-2 activity of HCQ: specific binding to N protein to disrupt its interaction with nucleic acids and LLPS. QRB Discovery 2:1–9. 10.1017/qrd.2021.12 [DOI] [PMC free article] [PubMed]
- Dang M, Song J (2022) CTD of SARS-CoV-2 N protein is a cryptic domain for binding ATP and nucleic acid that interplay in modulating phase separation. Protein Sci 31:345–356. 10.1002/pro.4221 [DOI] [PMC free article] [PubMed]
- Dang M, Li Y, Song J (2021) ATP biphasically modulates LLPS of SARS-CoV-2 nucleocapsid protein and specifically binds its RNA-binding domain. Biochem Biophys Res Commun 19:50–55. 10.1016/j.bbrc.2021.01.018 [DOI] [PMC free article] [PubMed]
- Dang M, Lim L, Kang J, Song J (2021) ATP biphasically modulates LLPS of TDP-43 PLD by specifically binding arginine residues. Commun Biol 4:714. 10.1038/s42003-021-02247-2 [DOI] [PMC free article] [PubMed]
- Dang M, Li Y, Song J (2021) Tethering-induced destabilization and ATP-binding for tandem RRM domains of ALS-causing TDP-43 and hnRNPA1. Sci Rep 11:1034. 10.1038/s41598-020-80524-6 [DOI] [PMC free article] [PubMed]
- Dang M, Lim L, Roy A, Song J (2022) Myricetin allosterically inhibits the dengue NS2B-NS3 protease by disrupting the active and locking the inactive conformations. ACS Omega 7:2798–2808. 10.1021/acsomega.1c05569 [DOI] [PMC free article] [PubMed]
- Dinesh DC, Chalupska D, Silhan J et al (2020) Structural basis of RNA recognition by the SARS-CoV-2 nucleocapsid phosphoprotein. PLoS Pathog 16:e1009100–e1009100. 10.1371/journal.ppat.1009100 [DOI] [PMC free article] [PubMed]
- Hoepel W, Chen HJ, Geyer1 CE et al (2021) High titers and low fucosylation of early human anti-SARS-CoV-2 IgG promote inflammation by alveolar macrophages. Sci Transl Med. 10.1126/scitranslmed.abf8654 [DOI] [PMC free article] [PubMed]
- Hyman AA, Weber CA, Julicher F (2014) Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol 30:39–58. 10.1146/annurev-cellbio-100913-013325 [DOI] [PubMed]
- Iserman C, Roden CA, Boerneke MA et al (2020) Genomic RNA elements drive phase separation of the SARS-CoV-2 Nucleocapsid. Mol Cell 80:1078–1091. 10.1016/j.molcel.2020.11.041 [DOI] [PMC free article] [PubMed]
- Kang J, Lim L, Lu Y et al (2019) A unified mechanism for LLPS of ALS/FTLD-causing FUS as well as its modulation by ATP and oligonucleic acids. PLoS Biol 17:e3000327. 10.1371/journal.pbio.3000327 [DOI] [PMC free article] [PubMed]
- Kang J, Lim L, Song J (2019) ATP binds and inhibits the neurodegeneration-associated fibrillization of the FUS RRM domain. Commun Biol 20:223. 10.1038/s42003-019-0463-x [DOI] [PMC free article] [PubMed]
- Lei Y, Zhang J, Schiavon CR et al (2021) SARS-CoV-2 spike protein impairs endothelial function via downregulation of ACE 2. Circ Res 128:1323–1326. 10.1161/CIRCRESAHA.121.318902 [DOI] [PMC free article] [PubMed]
- Lu S, Ye Q, Singh D et al (2021) The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nat Commun 12:502. 10.1038/s41467-020-20768-y [DOI] [PMC free article] [PubMed]
- Olofsson AM, Falla F, Yang D et al (2022) Intracellular reverse transcription of Pfizer BioNTech COVID-19 mRNA vaccine BNT162b2 in vitro in human liver cell line. Curr Issues Mol Biol 44:1115–1126. 10.3390/cimb44030073 [DOI] [PMC free article] [PubMed]
- Patel A, Malinovska L, Saha S et al (2017) ATP as a biological hydrotrope. Science 356:753–756. 10.1126/science.aaf6846 [DOI] [PubMed]
- Perdikari TM, Murthy AC, Ryan VH et al (2020) SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs. EMBO J 17:e106478. 10.15252/embj.2020106478 [DOI] [PMC free article] [PubMed]
- Roldan EQ, Biasiotto G, Magro P et al (2020) The possible mechanisms of action of 4-aminoquinolines (chloroquine/hydroxychloroquine) against Sars-Cov-2 infection (COVID-19): a role for iron homeostasis? Pharmacol Res 158:104904. 10.1016/j.phrs.2020.104904 [DOI] [PMC free article] [PubMed]
- Satarker S, Ahuja T, al Banerjeeet M (2020) Hydroxychloroquine in COVID-19: potential mechanism of action against SARS-CoV-2. Curr Pharmacol Rep 24:1–9. 10.1007/s40495-020-00231-8 [DOI] [PMC free article] [PubMed]
- Savastano A, de Opakua AI, Rankovic M et al (2020) Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat Commun 11:6041. 10.1038/s41467-020-19843-1 [DOI] [PMC free article] [PubMed]
- Seet RCS, Quek AML, Ooi DSQ et al (2921) Positive impact of oral hydroxychloroquine and povidone-iodine throat spray for COVID-19 prophylaxis: an open-label randomized trial. Int J Infect Dis 106:314–322. 10.1016/j.ijid.2021.04.035 [DOI] [PMC free article] [PubMed]
- Shi J, Han N, Lim L et al (2011) Dynamically-driven inactivation of the catalytic machinery of the SARS 3C-Like protease by the N214A mutation on the extra domain. PLoS Comput Biol 7:e1001084. 10.1371/journal.pcbi.1001084 [DOI] [PMC free article] [PubMed]
- Shin Y, Brangwynne CP (2017) Liquid phase condensation in cell physiology and disease. Sci 357:6357. 10.1126/science.aaf4382 [DOI] [PubMed]
- Song J (2021) Adenosine triphosphate energy-independently controls protein homeostasis with unique structure and diverse mechanisms. Protein Sci Apr 7. 10.1002/pro.4079 [DOI] [PMC free article] [PubMed]
- Song J, Ni F (1998) NMR for the design of functional mimetics of protein-protein interactions: one key is in the building of bridges. Biochem Cell Biol 76:177–188. 10.1139/bcb-76-2-3-177 [DOI] [PubMed]
- Wang P, Nair MS, Liu L, et al. (2021) Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. Mar 8. 10.1038/s41586-021-03398-2 [DOI] [PubMed]
- Wessner DR. The origins of viruses. Nat Educ. 2010;3:37. [Google Scholar]
- Wu F, Zhao S, Yu B et al (2020) New coronavirus associated with human respiratory disease in China. Nature 579:265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed]
- Zinzula L, Basquin J, Bohn S et al (2021) High-resolution structure and biophysical characterization of the nucleocapsid phosphoprotein dimerization domain from the Covid-19 severe acute respiratory syndrome coronavirus 2. Biochem Biophys Res Commun 538:54–62. 10.1016/j.bbrc.2020.09.13 [DOI] [PMC free article] [PubMed]