Abstract
Bacterial communities in the large intestines of pigs were compared using terminal restriction fragment length polymorphism (T-RFLP) analysis targeting the 16S ribosomal DNA. The pigs were fed different experimental diets based on either modified standard feed or cooked rice supplemented with dietary fibers. After feeding of the animals with the experimental diets for 2 weeks, differences in the bacterial community structure in the spiral colon were detected in the form of different profiles of terminal restriction fragments (T-RFs). Some of the T-RFs were universally distributed, i.e., they were found in all samples, while others varied in distribution and were related to specific diets. The reproducibility of the T-RFLP profiles between individual animals within the diet groups was high. In the control group, the profiles remained unchanged throughout the experiment and were similar between two independent but identical experiments. When the animals were experimentally infected with Brachyspira hyodysenteriae, causing swine dysentery, many of the T-RFs fluctuated, suggesting a destabilization of the microbial community.
The gastrointestinal tract of pigs is densely populated with bacteria, and the intestinal microbiota has important influence on animal health and growth performance. The lumenal contents of the colon support between 1010 and 1011 culturable bacteria per g (wet weight) (1, 6, 21). The majority of the culturable bacteria in the pig colon are gram-positive, strict anaerobic streptococci, lactobacilli, eubacteria, clostridia, and peptostreptococci (21, 27, 28). The gram-negative organisms comprise about 10% of the total culturable bacteria. Most isolates belong to the Bacteroides and Prevotella groups (27). The gastrointestinal tract bacterial community structure is susceptible to changes in the diet of the animal. For example, the bacterial community will adapt to the introduction of high levels of dietary fibers by increased growth of bacteria with cellulolytic and xylanolytic activities (8, 21, 32). Other dietary treatments that are known to affect the intestinal bacterial community are the addition of organic acids to the feed (25) and prefermentation of commercial dry feeds (26). However, the effects of these treatments on the structure of the bacterial community are unknown. Pathogenic bacteria and bacteria that are part of the indigenous microbiota in the pig intestine may interact. Resistance to colonization by pathogens is well known (4), while synergistic relations with one or more indigenous intestinal bacterial species are a prerequisite for the pathogenicity of Brachyspira hyodysenteriae (34) and Lawsonia intracellularis (16). Both species cause severe intestinal disorders. On the other hand, it has been shown previously that the onset of swine dysentery causes a dramatic disturbance of the intestinal bacterial community (24). All these studies are based on culturing of the bacteria. However, comparisons with direct microscopic counts have shown that only part of the intestinal bacteria can be accounted for by colony counting (27, 28). Moreover, in most studies the phenotypic characterization of the bacterial isolates is left at the genus level or even higher taxonomic levels, resulting in an inadequate description of the bacterial community that can detect only profound changes.
The coherent phylogeny of the bacteria based on small subunit (16S) rRNA sequence analysis (35) and the implementation of molecular methods have provided microbial ecologists with a set of tools for analyzing and a framework for understanding complex microbial communities. This rRNA approach is based on the direct analysis of the rRNA or the genes coding for it (rDNA) after an initial PCR amplification with universal primers. Sequence analysis of libraries of cloned rDNA from various environments including the pig intestine has demonstrated a species richness undetectable by culturing methods (23). Even in a well-studied microbial system such as human feces, a so-far-unknown bacterial diversity was discovered when molecular methods were applied (12, 30). However, the cloning-sequencing procedure is time-consuming and may limit the number of samples that can be processed. Alternatively, gel-based typing methods such as terminal restriction fragment length polymorphism (T-RFLP) analysis can be used for fingerprinting or profiling microbial communities. The T-RFLP method, originally developed for the identification of bacteria (3), has been implemented for the characterization of microbial diversity in various environments (7, 14, 15, 19). Here we report on the use of T-RFLP to characterize and compare the bacterial communities in the colons of pigs fed different experimental diets and after infection with B. hyodysenteriae, the agent of swine dysentery.
MATERIALS AND METHODS
Animals.
One hundred twelve healthy pigs (Danish Landrace/Yorkshire × Duroc) with an average weight of 23.5 kg (range, 18 to 29.3 kg) were purchased from a high-health herd, free of B. hyodysenteriae.
The animals were negative for swine dysentery as determined by bacteriological culture. The animals were divided into groups and were housed in separate pens with raised mesh floors.
Diets.
The composition of the diets for the different groups is shown in Table 1. All diets were supplemented with the essential vitamins and minerals. The pigs were fed twice daily. The fermented liquid feed was prepared in an 80-liter closed tank with continuous stirring as previously described (17). A standard Danish pig diet (wheat, 33%; barley, 33%; soybeans, 11%; fish meal, 11%) was mixed with water at a ratio of 2.75/1 (wt/wt). The feed was allowed to preferment 4 to 5 days before the onset of the experiment. At feeding, 18.75 liters of the content was removed from the tank. The rice diets were prepared by mixing parboiled rice with water at a 1:1 (vol/vol) ratio and boiling the mixture in an autoclave at 121°C for 20 min. After cooling, the rice was thoroughly mixed with the remaining ingredients. The rice diets were stored at −20°C and thawed before they were fed to the pigs.
TABLE 1.
Composition of the experimental diets
| Diet | Composition | Content (% [wt/wt])
|
||
|---|---|---|---|---|
| RS | Total NSP | sNSP | ||
| K | Wheat (33%), barley (33%), soybeans (11%), fish meal (11%) | 20 | 5 | |
| B | CPa | 2 | 1.2 | |
| C | CP and 10% potato starch | 10 | 2 | 1.2 |
| D | CP and 20% wheat bran | 10 | 1.2 | |
| E | CP and 12% sugar beet pulp | 10 | 6.2 | |
| F | Fermented liquid diet K (pH 4) | —b | —b | |
| G | Diet K with 2% lactic acid (pH 4) | 20 | 5 | |
CP, cooked rice and animal protein.
—, unknown due to microbial fermentation.
Sampling from pigs.
Two independent but identical experiments were done. At the beginning of each experiment, the pigs were divided into seven groups, each group having eight pigs. The animal groups were each fed one of the experimental diets. After 2 weeks, two pigs in each group were sacrificed. Each animal was autopsied immediately, and a 10-cm section of the intestine from the top of the spiral colon was tied off and stored at −80°C until further processing. The remaining pigs were experimentally infected with B. hyodysenteriae, as previously described (10), except in the control group (K). Four weeks after infection, two pigs from each diet group were killed and sampled. The remaining pigs were killed and sampled another 2 weeks later or earlier if severe symptoms of dysentery developed. Monitoring of swine dysentery was done by feces scoring: ++, watery mucoid and blood-tinged feces to semiliquid mucoid feces; +, semisolid feces.
Extraction and purification of DNA.
For DNA extraction, 200 mg of intestinal content was suspended in 600 μl of phosphate-buffered saline. The samples were kept on ice throughout the procedure. The samples were vortexed thoroughly and then centrifuged for 2 min at 200 × g in a microcentrifuge. The supernatant was transferred to a new microcentrifuge tube and centrifuged at 12,000 × g for 5 min. The supernatant was discarded, and the pellet was resuspended in 570 μl of TE (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). The suspension was transferred to 2-ml screw-cap tubes to which 350 to 400 mg of 100-μm zirconia-silica beads (Biospec Products Inc., Bartlesville, Okla.) and 30 μl of 10% sodium dodecyl sulfate had been added, and the bacterial cells were lysed by being shaken for 4 min on a minibead beater (Biospec Products Inc.) on high speed. After a brief spin in a microcentrifuge, the samples were transferred to a microcentrifuge tube and the DNA was purified by the cetyltrimethylammonium bromide method (2). DNA was finally dissolved in 50 μl of TE and stored at −21°C.
The DNA concentrations were measured on a GeneQuant RNA-DNA calculator (Pharmacia LKB Biochrom Ltd., Cambridge, United Kingdom) and adjusted to a concentration of 5 μg of DNA ml−1 before the PCR.
PCR conditions.
Four replicate 50-μl PCR mixtures were made from each sample on a PTC-200 thermal cycler (MJ Research, Watertown, Mass.). Reaction conditions were as follows: 5 μl PCR buffer (HT Biotechnology Ltd., Cambridge, United Kingdom); 200 μM (each) deoxynucleoside triphosphates, 0.1 μM forward primer S-D-Bact-0008-a-S-20 (5′-AGAGTTTGATCMTGGCTCAG-3′), 0.1 μM reverse primer S-D-Bact-0926-a-A-20 (5′-CCGTCAATTCCTTTRAGTTT-3′), and 1.25 U of DNA polymerase (SuperTaq; HT Biotechnology Ltd.) in a 50-μl reaction. Primer S-D-Bact-0008-a-S-20 was 5′ FAM (carboxyfluorescein-N-hydroxysuccinimide ester-dimethyl sulfoxide) labeled. Two-microliter aliquots of extracted DNA (5 μg ml−1) were amplified. PCR cycling consisted of an initial denaturation at 94°C for 6 min; followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 57°C for 45 s, and extension at 72°C for 2 min; and a final extension at 72°C for 3 min. Amplified DNA was verified by electrophoresis on 1.5% agarose gels.
Restriction digests.
The fluorescently labeled PCR products were purified on QIAquick PCR purification kit columns (Qiagen GmbH, Hilden, Germany) and eluted in a final volume of 50 μl of double-distilled water. The concentrations of the purified PCR products were measured, and 200 ng of purified PCR products was digested overnight at 37°C with 20 U of CfoI (Boehringer, Mannheim, Germany) in 20-μl reaction mixtures.
T-RFLP analysis.
The fluorescently labeled terminal restriction fragments (T-RFs) were analyzed by electrophoresis on an automatic sequence analyzer (ABI PRISM 373 DNA Sequencer; PE Biosystems, Foster City, Calif.) in GeneScan mode. Aliquots (2 μl) of T-RFs were mixed with 2 μl of deionized formamide, 0.4 μl of loading buffer (PE Biosystems), and 0.6 μl of DNA fragment length standard. The standard size marker was a 1:1 mixture of the size standards GS-500 ROX and GS-1000 ROX (PE Biosystems). The T-RF mixture was denatured at 94°C for 2 min and chilled on ice prior to electrophoresis. Five-microliter aliquots of the mixture were loaded on a 36-cm, 6% denaturing polyacrylamide gel. Electrophoresis settings were 2,500 V and 40 mA for 10 h, using the B filter set.
Data analysis.
After electrophoresis, the lengths of the T-RFs were determined by comparison with the internal size standard using GeneScan software (PE Biosystems).
For pairwise comparisons of the profiles, Dice coefficients (SD) were calculated as
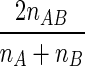 |
where nAB is the number of bands common for samples A and B, nA is the total number of bands in sample A, and nB is the total number of bands in sample B.
RESULTS
T-RFs were determined in the range of 30 to 674 bp, which was the range of the size marker that could be determined reliably under the applied electrophoresis conditions (Fig. 1).
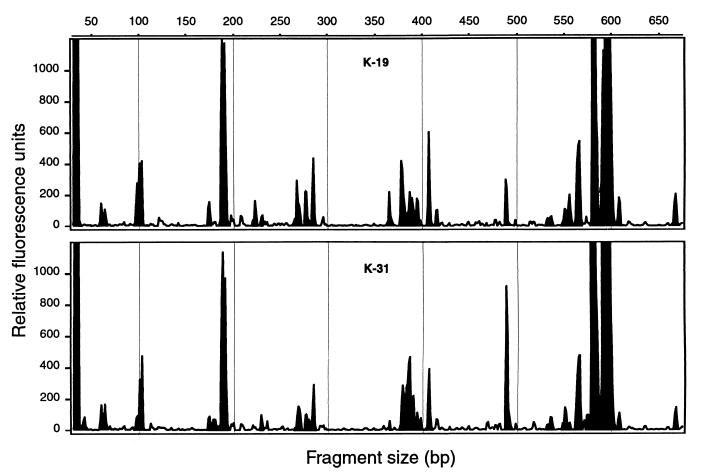
FIG. 1.
T-RFLP profiles from the colon lumenal contents of two pigs in the control group from experiment 1. K-31 was sampled after the animals had been fed the standard diet for 2 weeks; K-19 was sampled 4 weeks later. Thirty-nine peaks were detected in the K-31 sample (solid fill); 41 peaks were detected in the K-19 sample. The Dice coefficient of the two samples was 0.975.
Due to inadequate automatic peak calling by the GeneScan software, peaks were determined manually. T-RFLP profiles from replicate samples (in each group: two animals, same sampling day) were aligned, and only those peaks occurring in both profiles were included in the analysis. The Dice coefficients for pairwise comparisons of the T-RFLP profiles within the diet groups averaged 0.974 before the infection (Table 2); thus, the variation between replicate animals was small. The size of the peaks was ignored during peak calling, and fragments were determined to be either present or not. The fragments included in the analysis are shown in Fig. 2.
TABLE 2.
Dice coefficients for pairwise comparisons of T-RFs from colons of pigs fed experimental diets before experimental infection
| Diet | Coefficient within group | Coefficient between diet groupsa
|
|||||
|---|---|---|---|---|---|---|---|
| K | B | C | D | E | F | ||
| B | 0.988 | 0.625 | |||||
| C | 0.967 | 0.630 | 0.750 | ||||
| D | —b | 0.636 | 0.714 | 0.833 | |||
| E | 0.990 | 0.729 | 0.761 | 0.846 | 0.840 | ||
| F | 0.964 | 0.719 | 0.659 | 0.557 | 0.559 | 0.653 | |
| G | 0.974 | 0.813 | 0.609 | 0.731 | 0.700 | 0.815 | 0.733 |
Refer to Table 1 for definitions of the diets.
—, not determined; only one sample available.
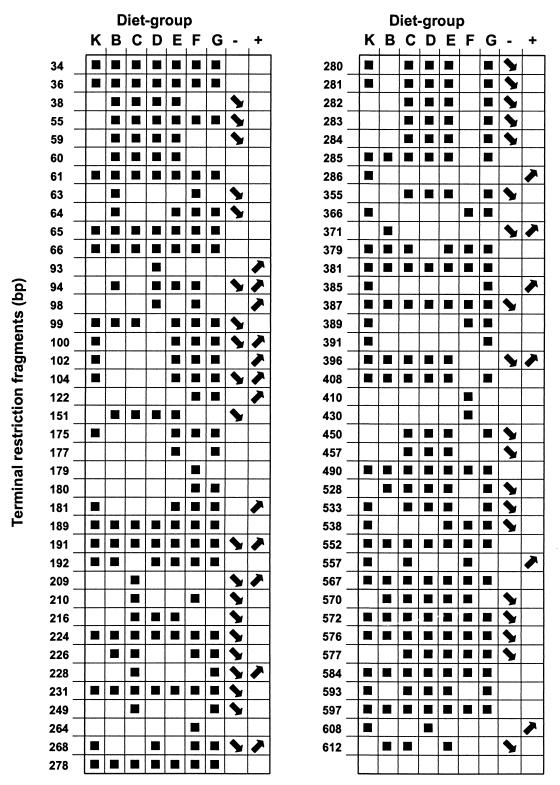
FIG. 2.
T-RFs included in the analysis. The sizes of the fragments are in base pairs. Refer to Table 1 for an explanation of the diets (K to G). Solid squares show fragments that were present in samples from the diet groups after 2 weeks of feeding the experimental diets to the animals and before the infection with B. hyodysenteriae. Arrows pointing downward in the “−” column indicate that the fragment disappeared in one or more diet groups after infection. Arrows pointing upward in the “+” column indicate fragments that were first detected in a new diet group after the infection.
A total of 77 fragments were included in the analysis. In the control group (K), 42 fragments were detected; in the B group, 38 fragments were; in the C group, 50 fragments were; in the D group, 47 fragments were; in the E group, 54 fragments were; in the F group, 47 fragments were; and in the G group, 54 fragments were.
The mean of the Dice coefficients for pairwise comparisons of all the samples from the control group (K) was 0.959 (n = 8; sampled after 2 and 4 weeks of feeding in two identical experiments), indicating highly similar microbial communities in all the control animals irrespective of the sampling day and the experiment.
Nineteen fragments had a universal distribution, i.e., they were found in all the diet groups (Fig. 2).
Effects of the different diets on the intestinal bacterial community could be seen after 2 weeks. The Dice coefficients for pairwise comparisons of the T-RFLP profiles prior to infection are shown in Table 2. The control group (standard diet; K) had moderate coefficients (0.625 to 0.636) for the animals fed the cooked rice diet (group B) and the rice diets supplemented with either resistant starch (RS; group C) or high concentrations of total nonstarch polysaccharides (NSP) but low soluble NSP (l-sNSP; group D). The coefficient was somewhat higher between the control group and the group receiving a rice-based diet with high sNSP supplement (group E; 0.729) or the group that was fed the fermented standard feed (group F; 0.719). The control group had the highest Dice coefficient for the animals that got the standard diet with lactic acid supplement (group G). All the rice groups had high Dice coefficients for each other, but the highest coefficients were found among the rice groups with RS or dietary fiber supplement (0.833 to 0.846). The fermented feed group (F) differed most from the rice groups with RS (group C; 0.557), or low sNSP (group D; 0.559), while the coefficient between this group and both the group fed unsupplemented cooked rice (group B; 0.659) and the group fed cooked rice with high sNSP (group E; 0.653) was moderate. The coefficient between the F group and the group fed a standard diet with lactic acid supplement was moderate (group G; 0.733).
Among the variable fragments, five patterns of distribution could be recognized (Fig. 2). Four fragments (38, 59, 60, and 151 bp) were found only in animals receiving a rice-based diet (groups B, C, D, and E). Two fragments (216 and 457 bp) were found in the animals fed the cooked rice plus potato starch or dietary fibers (groups C, D, and E), and five fragments (282, 283, 284, 355, and 450 bp) were found in these groups plus the group receiving the standard diet supplemented with lactic acid (groups C, D, and E plus G). Five fragments (100, 102, 104, 181, and 538 bp) were found in the standard diet group (K) plus groups E, F, and G. Four fragments (280, 281, 533, and 593 bp) were present in pigs from all groups except those fed cooked rice and fed fermented feed (groups K plus C, D, and E plus G). Four fragments (179, 264, 410, and 430 bp) were found exclusively in diet group F.
The fragments of 102 and 104 bp match the size of the theoretical T-RFs of all species in the Prevotella genus derived from the Ribosomal Database Project, alignment version 7.1 (http://www.cme.msu.edu/rdp/html/analyses.html). The only other bacteria in the database matching these terminal fragment sizes are Bacteroides fragilis, Bacteroides uniformis, and Bacteroides ovatus. Thus, it is reasonable to conclude that the absence of these two fragments in the B, C, and D groups implies that no prevotellas could be detected in the animals fed the cooked rice diet or the cooked rice diets with the potato starch or wheat bran supplement.
The remaining fragments were distributed in a more scattered fashion, with no systematic trends relating to the diet groups.
Following the infection with B. hyodysenteriae, severe clinical signs of swine dysentery with mucoid to mucohemorrhagic feces were found in all diet groups, except in the uninfected control group and the group receiving the fermented diet (group F).
With the onset of swine dysentery, the microbial communities in the colon destabilized. Thirty-nine fragments disappeared from one or more diet groups where they had previously been detected, while 18 fragments appeared in groups where they had not been detected before the infection (Table 2). Nine fragments both disappeared in some groups and appeared for the first time in others. The mean of the Dice coefficients for pairwise comparisons of the T-RFLP profiles decreased from 0.977 (standard error of the mean = 0.005) before the infection to 0.822 (standard error of the mean = 0.043) after the infection in the groups showing clinical signs. No changes occurred in the F group, and the similarity remained high after the infection (SD = 1.000).
Small peaks of 666 bp, corresponding to the size of the fragment characteristic of B. hyodysenteriae, were seen in all groups including the control group and were found randomly on all sampling days.
DISCUSSION
The susceptibility of the bacterial community in the colon of pigs to animal dietary changes and infection with an intestinal pathogen was investigated. The experimental diets were chosen in an attempt to minimize the frequency of clinical infections with B. hyodysenteriae. Diets based on cooked white rice have been shown to protect pigs against swine dysentery, whereas increasing amounts of dietary fibers compound the incidents of swine dysentery (22, 29). A proposed mechanism is that the amount of fermentable substrate in the large intestine affects components of the intestinal microbiota that are synergistic with B. hyodysenteriae (8). Fermented feed and feed supplemented with organic acid were also included, as these treatments may also reduce the incidence of swine dysentery.
We used the T-RFLP analysis to characterize the bacterial community in the colon of the pigs. T-RFLP is a sensitive method for demonstrating differences between microbial communities in location and time (7, 14, 15, 19). The enzymatic digestion of amplified, 5′-fluorescently labeled community 16S rDNA produces T-RFs that vary in length due to restriction site polymorphism and insertions or deletions (5). Microbial communities with different species compositions produce different and characteristic T-RFLP profiles. Each peak of the profile may originate from a single species or from a group of species sharing a common location of the restriction site. Thus, interpretation of the profiles, in terms of which bacterial species may be present in the community, is often ambiguous. T-RFLP has proven more powerful than denaturing gradient gel electrophoresis due to its greater sensitivity, which facilitates the detection of more and smaller populations (15, 19).
Only one set of PCR primers and one restriction enzyme were used for the analysis. The primer-enzyme combination that produced the largest number of unique T-RFs based on computer simulations of the sequences in the Ribosomal Database Project small-subunit database was selected (14). Clement and colleagues (7) recommend the use of several restriction enzyme digests to increase the information derived from the community. However, as the CfoI digestion resulted in a large number of T-RFs, which provided sufficient resolution to demonstrate differences in the microbial communities between the diet groups, only this enzyme was used in the analysis.
Four replicate amplification reactions were combined to provide enough rDNA for the analysis. Another advantage of combining multiple PCRs is to reduce the significance of possible tube-to-tube variation of individual amplification reactions. The PCR was run for 30 cycles to produce sufficient T-RFs for detection. Thirty cycles of amplification may introduce a bias toward certain 16S rDNA types; however, Suzuki and Giovannoni (31) state that this bias is likely to be small from samples of highly diverse templates. Moreover, Zoetendal and coworkers (36) did not find any differences in band patterns after 20 or 35 cycles of amplification when applying temperature gradient gel electrophoresis to environmental samples. The size of the peaks was ignored in the analysis because a proportionality between bacterial abundance in the sample community and the size of the peak in the complex profile remains to be documented.
The total number of T-RFs included in the analysis was 77, and the highest number of T-RFs in a single diet group was 54. The culturable bacterial community in the human colon has been estimated to be composed of 400 to 500 phenotypes (20), although this diversity has never been confirmed by actually culturing all those species. Thus, assuming that the pig intestinal microbiota is made up of an equally large number of species, the T-RFLP grossly underestimated the species richness in our samples. Underestimation of the diversity may result from preferential amplification of certain 16S rDNA species or from multiple rDNA species competing during amplification, causing insufficient amplification of the minority rDNAs for detection (13). However, the most significant factor is probably the identity of T-RFs from different bacterial species. Computer simulations have shown that dispersed phylogenetic groups of bacteria may produce T-RFs of identical size and that this reduces the resolution of the T-RFLP analysis from the level of species to that of higher-order groups (14). Consequently, the number of T-RFs in a profile is not a measure of the species richness in the community.
A small 666-bp peak was seen in many of the samples. This peak matches the size of the B. hyodysenteriae T-RF. However, since this fragment was detected prior to the infection and was also found in the uninfected control group, which was culture negative for B. hyodysenteriae throughout the experiment, the 666-bp peak most likely comprises T-RFs from species other than B. hyodysenteriae. Thus, this peak is not specific and cannot be used for identification of B. hyodysenteriae.
The bacterial community in the colon of pigs in the uninfected control group was stable throughout the entire first experiment, and the T-RFLP profiles were identical between the two experiments. No systematic variation in the T-RFs was detected, although peaks present in only one of the replicate samples were sometimes found. These T-RFs may represent animal-to-animal variation in the distribution and abundance of certain populations or groups of populations. This variation was small as reflected by a Dice coefficient of 0.959 for all the control samples. However, for clarity, peaks that were not present in all replicate samples were omitted in the subsequent analysis, and a consensus profile was constructed by comparing the control group samples from all the sampling days and from both experiments. The Dice coefficients for pairwise comparisons of the replicate samples from each of the diet groups before the infection were high (Table 2; mean = 0.974), and consensus profiles of the fragments occurring in both replicate samples could be constructed. Large variation in the abundance of the bacterial groups between individual pigs on the same diet has been found by culturing methods (1, 28). Our finding of a core of consistent T-RFs in each diet group supports the results of Durmic and colleagues (8) that the major culturable components of the large intestinal microbiota were identical in groups of pigs fed the same diet.
The type of diet greatly influenced the intestinal microbiota as expressed by the T-RFLP profiles after 2 weeks. We did not sample the animals earlier in the experiment and cannot draw any conclusions on how fast the bacterial community responded to the shift to the experimental diets. Varel and coworkers (32) found a growth response of xylanolytic and cellulytic bacteria, which increased almost an order of magnitude in numbers, 12 days after pigs were shifted to a high-fiber diet. Nineteen of the 77 T-RFs were found in all diet groups, i.e., these fragments originated from bacterial populations or groups of populations that were members of the community independent of the diet. Unique communities for each individuals but also some dominant bacteria present in all individuals have been found using temperature gradient gel electrophoresis to characterize the bacterial communities in human fecal samples (36).
The rice-based diets resulted in five characteristic peaks, suggesting the presence of at least five bacterial populations in the pig gut being favored by the cooked rice compared to the other diets. Other patterns recognized were related to the amount and type of fiber in the feed. These T-RFs may represent bacteria that utilize the dietary fibers as a substrate for fermentation. A substantial microbial fermentation of NSP and dietary fibers takes place in the cecum and colon of monogastric animals, and the fatty acids produced may contribute up to 30% of the maintenance energy needs of growing pigs (33). An interesting finding was the lack of the T-RFs corresponding to the Prevotella group in the pigs fed the cooked rice or the cooked rice supplemented with either 10% potato starch or 20% wheat bran. These diets had low concentrations of sNSP.
The Dice coefficient between the control group and the group fed the lactic acid-amended standard diet (group G) was the highest observed (0.813), suggesting that the addition of lactic acid had little effect on the bacterial community in the colon. The fermented standard diet had a moderate Dice coefficient for the control group, while the lowest observed coefficients were those between the group given fermented feed and the groups given cooked rice diets with high RS and insoluble NSP. Thus, these diets supported different microbial populations in the pig colon.
The 102- and 104-bp T-RFs match the sizes of fragments from B. uniformis, B. fragilis, and Prevotella melaninogenica, which are known to be synergistic with B. hyodysenteriae in producing dysentery in mice (9, 11). However, although no peaks matching the T-RFs from these species were detected in the low-sNSP diet groups, the pigs developed dysentery. Other bacteria have synergistic effects in pigs, including Fusobacterium nucleatum and Fusobacterium necrophorum (34), but T-RFs of sizes corresponding to these species were not detected in any of the groups. It is likely that there are many more bacterial species that are synergistic with B. hyodysenteriae.
Infection of the pigs with B. hyodysenteriae tended to destabilize the colon bacterial community. In contrast to the stable T-RFLP profile found throughout the experiment in the control group, many T-RFs fluctuated with the onset of swine dysentery. The fluctuations were not systematic, although the overall tendency was to reduce the number of T-RFs. Eighteen T-RFs appeared in diet groups where they had not been detected before the infection. These T-RFs probably originated from small bacterial populations that were already present in the animals before the infection. The mean of the Dice coefficients within the diet groups decreased to 0.822, and consensus profiles could not reasonably be constructed. A shift in the colonic epithelial microbiota from predominating nonmotile gram-positive bacteria to motile gram-negative bacteria after infection with B. hyodysenteriae has previously been reported (24). Assuming that Bacteroides and Prevotella organisms constitute the main portion of the gram-negative intestinal bacteria (27), we did not detect such a shift after the infection. Severe clinical signs of dysentery were not observed in the group fed the fermented standard feed, and the T-RFLP profiles remained unchanged in this group, suggesting that the destabilization in the groups having dysentery was due to the changed conditions imposed by the diarrhea.
In conclusion, the T-RFLP analysis was useful for demonstrating changes in the bacterial community structure in pigs fed different experimental diets. However, the inability to link the T-RFs to bacterial species is a serious drawback of the method. To address this problem, extensive sequencing of 16S rRNA from the environment under investigation is needed. By comparison of the theoretical T-RFs derived from clone 16S rRNA libraries, it should be possible to generate tentative bacterial names for the T-RFs or at least to determine their phylogenetic affiliation. Our study has shown that the pig intestinal microbial ecosystem responds fast and dynamically to perturbations such as dietary changes or infection with intestinal pathogens and that it is a useful model ecosystem for basic microbial ecological studies.
ACKNOWLEDGMENTS
This work was supported by the Research Secretariat of the Ministry of Food, Agriculture and Fisheries (project no. Alt-98-1).
We acknowledge Sebastian Vous for his excellent technical assistance.
REFERENCES
- 1.Allison M J, Robinson I M, Bucklin J A, Booth G D. Comparison of bacterial populations of the pig cecum and colon based upon enumeration with specific energy sources. Appl Environ Microbiol. 1979;37:1142–1151. doi: 10.1128/aem.37.6.1142-1151.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1988. pp. 2.4.1–2.4.5. [Google Scholar]
- 3.Avaniss-Aghajani E, Jones K, Chapman D, Brunk C. A molecular technique for identification of bacteria using small subunit ribosomal RNA sequences. BioTechniques. 1994;17:144–149. [PubMed] [Google Scholar]
- 4.Bourlioux P. What is currently known about the molecular mechanisms of colonisation resistance? Anaerobe. 1997;3:179–184. doi: 10.1006/anae.1997.0098. [DOI] [PubMed] [Google Scholar]
- 5.Brunk C F, Avaniss-Aghajani E, Brunk C A. A computer analysis of primer and probe hybridization potential with bacterial small-subunit rRNA sequences. Appl Environ Microbiol. 1996;62:872–879. doi: 10.1128/aem.62.3.872-879.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butine T J, Leedle J A Z. Enumeration of selected anaerobic bacterial groups in cecal and colonic contents of growing-finishing pigs. Appl Environ Microbiol. 1989;55:1112–1116. doi: 10.1128/aem.55.5.1112-1116.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clement B G, Kehl L E, DeBord K L, Kitts C L. Terminal restriction fragment patterns (TRFPs), a rapid, PCR-based method for the comparison of complex bacterial communities. J Microbiol Methods. 1998;31:135–142. [Google Scholar]
- 8.Durmic Z, Pethick D W, Pluske J R, Hampson D J. Changes in bacterial populations in the colon of pigs fed different sources of dietary fibre, and the development of swine dysentery after experimental infection. J Appl Microbiol. 1998;85:574–582. doi: 10.1046/j.1365-2672.1998.853539.x. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi T, Suenega I, Komeda T, Yamazaki T. Role of Bacteroides uniformis in susceptibility of Ta:CF#1 mice to infection with Treponema hyodysenteriae. Zentbl Bakteriol. 1990;274:118–125. doi: 10.1016/s0934-8840(11)80981-3. [DOI] [PubMed] [Google Scholar]
- 10.Jensen T K, Boye M, Møller K, Leser T D, Jorsal S E. Association of Serpulina hyodysenteriae with the colonic mucosa in experimental swine dysentery studied by fluorescent in situ hybridization. APMIS. 1998;106:1061–1068. [PubMed] [Google Scholar]
- 11.Joens L A, Robinson I M, Glock R D, Matthews P J. Production of lesions in gnotobiotic mice by inoculation with Treponema hyodysenteriae. Infect Immun. 1981;31:504–506. doi: 10.1128/iai.31.1.504-506.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langendijk P S, Schut F, Jansen G J, Raangs G C, Kamphuis G R, Wilkinson M H F, Welling G W. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl Environ Microbiol. 1995;61:3069–3075. doi: 10.1128/aem.61.8.3069-3075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leser T D. Quantitation of Pseudomonas sp. strain B13(FR1) in the marine environment by competitive polymerase chain reaction. J Microbiol Methods. 1995;22:249–262. [Google Scholar]
- 14.Liu W-T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marsh T L, Liu W-T, Forney L J, Cheng H. Beginning a molecular analysis of the eucaryal community in activated sludge. Water Sci Technol. 1998;37:455–460. [Google Scholar]
- 16.McOrist S, Jasni S, Mackie R A, MacIntyre N, Neef N, Lawson G H K. Reproduction of porcine proliferative enteropathy with pure cultures of ileal symbiont intracellularis. Infect Immun. 1993;61:4286–4292. doi: 10.1128/iai.61.10.4286-4292.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mikkelsen L M, Jensen B B. Performance and microbial activity in the gastrointestinal tract of pigs fed fermented liquid feed at weaning. J Anim Feed Sci. 1998;7:211–215. [Google Scholar]
- 18.Miyazaki K, Martin J C, Marinsek-Logar R, Flint H J. Degradation and utilization of xylans by the rumen anaerobe Prevotella bryantii (formerly P. ruminicola subsp. brevis) B14. Anaerobe. 1997;3:373–381. doi: 10.1006/anae.1997.0125. [DOI] [PubMed] [Google Scholar]
- 19.Moeseneder M M, Arrieta J M, Muyzer G, Winter C, Herndl G J. Optimization of terminal-restriction fragment length polymorphism analysis for complex marine bacterioplankton communities and comparison with denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1999;65:3518–3525. doi: 10.1128/aem.65.8.3518-3525.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore W E C, Holdeman L V. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl Microbiol. 1974;27:961–979. doi: 10.1128/am.27.5.961-979.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore W E C, Moore L V H, Cato E P, Wilkins T D, Kornegay E T. Effect of high-fiber and high-oil diets on the fecal flora of swine. Appl Environ Microbiol. 1987;53:1638–1644. doi: 10.1128/aem.53.7.1638-1644.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pluske J R, Siba P M, Pethick D W, Durmic Z, Mullan B P, Hampson D J. The incidence of swine dysentery in pigs can be reduced by feeding diets that limit the amount of fermentable substrate entering the large intestine. J Nutr. 1996;126:2920–2933. doi: 10.1093/jn/126.11.2920. [DOI] [PubMed] [Google Scholar]
- 23.Pryde S E, Richardson A J, Stewart C S, Flint H J. Molecular analysis of the microbial diversity present in the colonic wall, colonic lumen, and cecal lumen of a pig. Appl Environ Microbiol. 1999;65:5372–5377. doi: 10.1128/aem.65.12.5372-5377.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson I M, Whipp S C, Bucklin J A, Allison M J. Characterization of predominant bacteria from the colons of normal and dysenteric pigs. Appl Environ Microbiol. 1984;48:964–969. doi: 10.1128/aem.48.5.964-969.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roth F X, Kirchgessner M. Organic acids as feed additives for young pigs: nutritional and gastrointestinal effects. J Anim Feed Sci. 1998;7(Suppl. 1):25–33. [Google Scholar]
- 26.Russel P J, Geary T M, Brooks P H, Campbell A. Performance, water use effluent output of weaner pigs fed ad libitum with either dry pellets or liquid feed and their role of microbial activity in the liquid feed. J Sci Food Agric. 1996;72:8–16. [Google Scholar]
- 27.Russell E G. Types and distribution of anaerobic bacteria in the large intestine of pigs. Appl Environ Microbiol. 1979;37:187–193. doi: 10.1128/aem.37.2.187-193.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salanitro J P, Blake I G, Muirhead P A. Isolation and identification of fecal bacteria from adult swine. Appl Environ Microbiol. 1977;33:79–84. doi: 10.1128/aem.33.1.79-84.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siba P M, Pethick D W, Hampson D J. Pigs experimentally infected with Serpulina hyodysenteriae can be protected from developing swine dysentery by feeding them a highly digestible diet. Epidemiol Infect. 1996;116:207–216. doi: 10.1017/s0950268800052456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suau A, Bonnet R, Sutren M, Godon J-J, Gibson G R, Collins M D, Doré J. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999;65:4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varel V H, Robinson I M, Jung H-J G. Influence of dietary fiber on xylanolytic and cellulytic bacteria of adult pigs. Appl Environ Microbiol. 1987;53:22–26. doi: 10.1128/aem.53.1.22-26.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varel V H, Yen J T. Microbial perspective on fiber utilization by swine. J Anim Sci. 1997;75:2715–2722. doi: 10.2527/1997.75102715x. [DOI] [PubMed] [Google Scholar]
- 34.Whipp S C, Robinson I M, Harris D L, Glock R D, Matthews P J, Alexander T J L. Pathogenic synergism between Treponema hyodysenteriae and other selected anaerobes in gnotobiotic pigs. Infect Immun. 1979;26:1042–1047. doi: 10.1128/iai.26.3.1042-1047.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zoetendal E G, Akkermans A D L, deVos W M. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol. 1998;64:3854–3859. doi: 10.1128/aem.64.10.3854-3859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]