Abstract
Background and Objective: Incomplete Kawasaki disease (KD) has often been misdiagnosed due to a lack of the clinical manifestations of classic KD. However, it is associated with a markedly higher prevalence of coronary artery lesions. Identifying coronary artery lesions by echocardiography is important for the timely diagnosis of and favorable outcomes in KD. Moreover, similar to KD, coronavirus disease 2019, currently causing a worldwide pandemic, also manifests with fever; therefore, it is crucial at this moment that KD should be distinguished clearly among the febrile diseases in children. In this study, we aimed to validate a deep learning algorithm for classification of KD and other acute febrile diseases. Methods: We obtained coronary artery images by echocardiography of children (n = 138 for KD; n = 65 for pneumonia). We trained six deep learning networks (VGG19, Xception, ResNet50, ResNext50, SE-ResNet50, and SE-ResNext50) using the collected data. Results: SE-ResNext50 showed the best performance in terms of accuracy, specificity, and precision in the classification. SE-ResNext50 offered a precision of 81.12%, a sensitivity of 84.06%, and a specificity of 58.46%. Conclusions: The results of our study suggested that deep learning algorithms have similar performance to an experienced cardiologist in detecting coronary artery lesions to facilitate the diagnosis of KD.
Keywords: Explanable AI, Deep learning, Kawasaki disease, Coronary artery lesion, Ultrasound Image
Abbreviations: Kawasaki disease, KD; Class activation map, CAM; Global average pooling, GAP; Coronavirus disease 2019, COVID-19; Areas under the precision-recall curve, AUPRC; Adaptive Moment Estimation, ADAM
1. Introduction
Kawasaki disease (KD) is the most common acquired heart disease in childhood. It was first described by Dr. Kawasaki in 1967 [1]. KD mostly occurs in children aged less than 5 years and has a high prevalence in countries of Northeast Asia, particularly Japan, South Korea, and Taiwan [2], [3].
The main symptoms of KD are unexplained high fever, diffuse erythematous polymorphous rash, bilateral conjunctival injection, cervical lymphadenopathy, oral mucosal changes, and extremity changes, sometimes with perineal desquamation, or reactivation of the bacillus Calmette-Gurin injection site [4], [5], [6], [7]. While the widely used diagnostic criteria for KD are useful, incomplete KD in infants or children aged 10 years or older can often be problematic, causing misdiagnosis due to a lack of manifestation of the full clinical criteria of classic KD. Nevertheless, this disease has a much higher prevalence of coronary artery lesions [4], [7].
Given the difficulty in diagnosing incomplete or atypical KD by clinical features alone, identifying coronary artery findings by echocardiography, along with evaluation of various biomarkers by blood tests, becomes more significant for ensuring an appropriate diagnosis [4]. Even though the choice of treatment with a high dose of intravenous immunoglobulin infusion decreases the risk of coronary artery complications, about 5% of treated children and 15–25% of untreated children have a risk of coronary artery aneurysms or ectasia. Certainly, one of the fatal complications of untreated KD is coronary artery aneurysm. Accordingly, the role of echocardiography in recognizing coronary artery lesions is substantial to ensure timely diagnosis and favorable outcome [8].
For the proper diagnosis of incomplete KD, the pediatric cardiologists need to perform echocardiography to investigate the patient’s coronary arteries. The most important therapeutic goal of KD is the prevention of coronary artery aneurysm formation. When an aneurysm is noticed, it is critical to prevent development of a giant aneurysm or formation of a thrombus [9].
Unfortunately, without an experienced pediatric cardiologist and a KD expert, it is challenging to diagnose incomplete KD because the fever patterns of many acute febrile diseases in children appear similar to KD, particularly the initial high grade of fever. In 2020, reports of severely ill pediatric cases have shown that KD and coronavirus disease 2019 (COVID-19) presented similar symptoms [10], [11]. Moreover, the incidence of KD suddenly increased in Europe and the USA during the COVID-19 pandemic. In addition, KD-like multi-systemic inflammatory syndrome in children affected many children in Europe and in the USA. This is of particular concern, as it can result in missed or delayed KD diagnosis and treatment [12].
Several studies on computer-aided diagnosis based on deep learning algorithms have shown good performance. Deep learning-based approaches for the diagnosis of cancer or lesions have been shown to perform well and have already surpassed human doctors in some categories [13]. Additionally, deep learning algorithms have shown better performance in the medical vision field than conventional methods [14]. Several deep learning algorithms have been proposed to diagnose various diseases, such as breast cancer [15], liver cancer [16], and thyroid nodules [17] on ultrasound images. These medical deep learning algorithms have been proposed for computer-aided enhancement of diagnostic performance. However, deep learning algorithms have not yet been applied to KD diagnosis.
The purpose of this study was to assess whether explainable deep learning algorithms could be used to identify coronary artery lesions on echocardiographic images for the timely diagnosis of KD. We also evaluated the performance of these algorithms in distinguishing KD from another acute febrile disease, pneumonia.
2. Methods
2.1. Data acquisition
To investigate whether it would be possible to distinguish between KD and another similar acute febrile disease, we selected pneumonia as an alternative representative acute febrile disease. Pneumonia is one of the most common febrile diseases in children.
For this study, echocardiographic imaging data from January 2016 to August 2019 were acquired from Yonsei University Gangnam Severance Hospital. Giant coronary aneurysm cases were excluded from this study. Echocardiographic images of 138 children with incomplete KD and 65 children with pneumonia (203 in total) were acquired and labeled as KD and non-KD by an experienced cardiologist. 2D echocardiographic coronary artery short axis view images were obtained for the appropriate diagnosis when the children initially presented with high grade fever. We cropped the echocardiographic images to pixels.
2.2. Deep learning algorithm
In this study, to distinguish incomplete KD from non-KD using echocardiographic images, we applied six deep learning architectures: VGG-19 [18], Xception [19], ResNet-50 [20], ResNext-50 [21], SE-ResNet-50, and SE-ResNext-50 [22].
VGG [18] is the most basic network with a simple structure for classification and good performance. Therefore, it is still widely used as a comparison architecture. Xception [19] is a linear stack of depth-wise separable convolutions with residual connections and shows higher performance than the baseline Inception architecture [23]. ResNet [20] is a deep learning network which is stacked more deeply using a skip connection and is being used in various fields. ResNext [21] has shown higher performance while reducing the computation cost compared to the existing ResNet by using a group convolution. SE-ResNet and SE-ResNext [22] are networks in which squeeze-and-excitation blocks are added to ResNet and ResNext, respectively [24].
Since the data in our study were limited to training the models, only networks with 50 or fewer convolution layers were used in this experiment. Thus, we used VGG-19, Xception, ResNet-50, ResNext-50, SE-ResNet50, and SE-ResNext50 in this experiment. We then evaluated the capability of the deep learning algorithms to distinguish between KD and non-KD.
For the training, we used a stochastic optimization method, adaptive moment estimation (ADAM) optimizer, [25] with parameter , , and . The initial learning rate was , and it decreased by 1/10 every 30 epochs. We trained each network for a total of 120 epochs. Training batch sizes consisted of 32 patches. We used a binary cross-entropy loss function to train each network. We used the pretrained weights of each network on ImageNet to achieve better performance [24].
The deep learning framework used for training and testing the deep learning algorithms was PyTorch [26]. We trained and tested all networks using a 2-GHz Intel Xeon E5-2620 processor and an NVIDIA TITAN RTX graphics card (24 GB).
2.3. Class activation map
To explain deep learning algorithms, we used the class activation map (CAM) [27]. Previously, it was not possible to know which salient parts of a medical image would be highlighted by a deep learning algorithm for classification. The CAM has been proposed to solve this issue and to be able to explain deep learning algorithms. Most deep learning algorithms use a fully connected layer to classify the values obtained by applying a global average pooling (GAP) to the feature maps from the last convolution layer. A linear transform with a class number of filters is then applied to the weight obtained through GAP.
Here, to obtain the CAM, the weight of the linear transform for each class was multiplied by the feature map obtained from the last convolutional layer. The CAM at class , , can be calculated as follows:
| (1) |
where is a feature map from the last convolution layer for a unit k, is the weight of linear transformation corresponding to class for the unit , and and are the spatial information of and , respectively. The class activation map, , indicates a class-specific highlight map at a spatial grid . Therefore, through the CAM, it is possible to understand which parts of the image are considered when the deep learning algorithm proceeds with classification. Hence, the CAM is an excellent tool for analyzing medical image deep learning algorithms [28], [29], as in this study.
2.4. Ethics statement
This study was approved by the Yonsei University College of Medicine Institutional Review Board and the Research Ethics Committee of Severance Hospital (study approval number: 2020-1127-001). All research was performed in accordance with relevant guidelines and regulations. The requirement for written informed consent was waived by the Institutional Review Board due to the retrospective study design.
3. Results
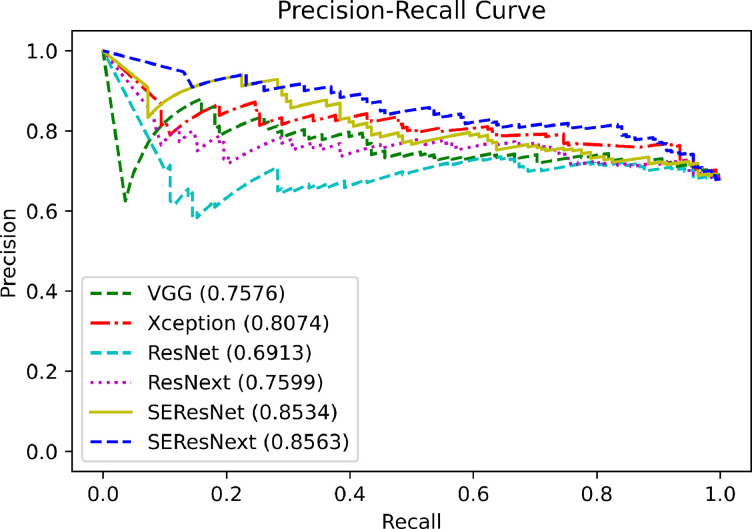
Since we conducted the experiments using 10-fold cross validation, there were 10 outcomes. Each subset had 182 or 183 echocardiographic images for training and 21 or 20 echocardiographic images for testing. The test images in each subset included 14 or 15 echocardiographic images labeled as KD and 6 or 7 echocardiographic images labeled as non-KD (pneumonia) by an echocardiographic specialist. Table 1 shows the diagnostic performance of each network trained with each subset for the test dataset. The accuracies of VGG19, Xception, ResNet50, ResNext50, SE-ResNet50, and SE-ResNext50 were 67.49%, 73.40%, 65.03%, 64.04%, 67.00%, and 75.86% for the classification of KD and non-KD, respectively. The F1 score of VGG19, Xception, ResNet50, ResNext50, SE-ResNet50, and SE-ResNext50 were 77.55%, 81.88%, 75.43%, 74.02%, 75.81%, and 82.56% for the classification of KD and non-KD, respectively. In contrast, the sensitivities of VGG19, Xception, ResNet50, ResNext50, SE-ResNet50, and SE-ResNext50 were 82.61%, 88.41%, 78.99%, 75.36%, 76.09%, and 84.06%, respectively. The specificities of VGG19, Xception, ResNet50, ResNext50, SE-ResNet50, and SE-ResNext50 were 35.38%, 41.54%, 35.38%, 40.00%, 47.69%, and 58.46%, respectively. The precision (positive predictive value; PPV) of VGG19, Xception, ResNet50, ResNext50, SE-ResNet50, and SE-ResNext50 were 73.08%, 76.25%, 72.19%, 72.73%, 75.54%, and 81.12%, respectively. The negative predictive value (NPV) of VGG19, Xception, ResNet50, ResNext50, SE-ResNet50, and SE-ResNext50 were 48.94%, 62.79%, 44.23%, 43.33%, 48.44%, and 63.33%, respectively. In these results, SE-ResNext50 showed the best performance in terms of accuracy, F1 score, sensivitivity, precision, and NPV for the distinction of KD and non-KD. It identified 154 true-positive diagnoses among 203 images. Fig. 2 shows the precision-recall curve of the deep learning algorithms used for the classification between KD and pneumonia. The areas under the precision-recall curve (AUPRC) of VGG19, Xception, ResNet50, ResNext50, SE-ResNet50, and SE-ResNext50 were 0.7576, 0.8074, 0.6913, 0.7599, 0.8534, and 0.8563, respectively.
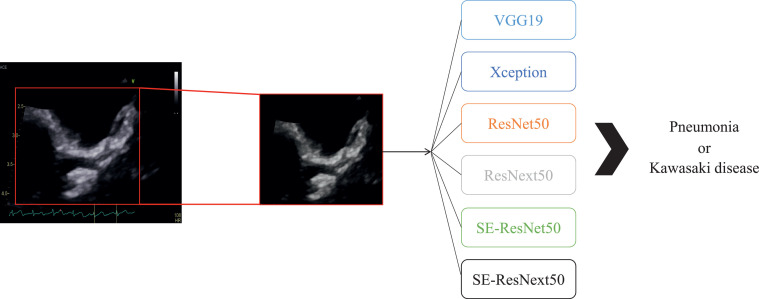
Fig. 1.
Echocardiography analysis process using deep learning algorithms in this study.
Table 1.
Diagnostic performance of deep learning algorithms. The best performance is in bold, and the second best performance is in underlined.
| Networks | VGG19 | Xception | ResNet50 | ResNext50 | SE-ResNet50 | SE-ResNext50 |
|---|---|---|---|---|---|---|
| Accuracy | 67.49 | 73.40 | 65.03 | 64.04 | 67.00 | 75.86 |
| F1 score | 77.55 | 81.88 | 75.43 | 74.02 | 75.81 | 82.56 |
| Sensitivity | 82.61 | 88.41 | 78.99 | 75.36 | 76.09 | 84.06 |
| Specificity | 35.38 | 41.54 | 35.38 | 40.00 | 47.69 | 58.46 |
| Precision (PPV) | 73.08 | 76.25 | 72.19 | 72.73 | 75.54 | 81.12 |
| NPV | 48.94 | 62.79 | 44.23 | 43.33 | 48.44 | 63.33 |
Fig. 2.
The precision-recall curves of each deep learning algorithm for KD. Areas under the precision-recall curve of VGG19, Xception, ResNet50, ResNext50, SE-ResNet50, and SE-ResNext50 were 0.7576, 0.8074, 0.6913, 0.7599, 0.8534, and 0.8563, respectively.
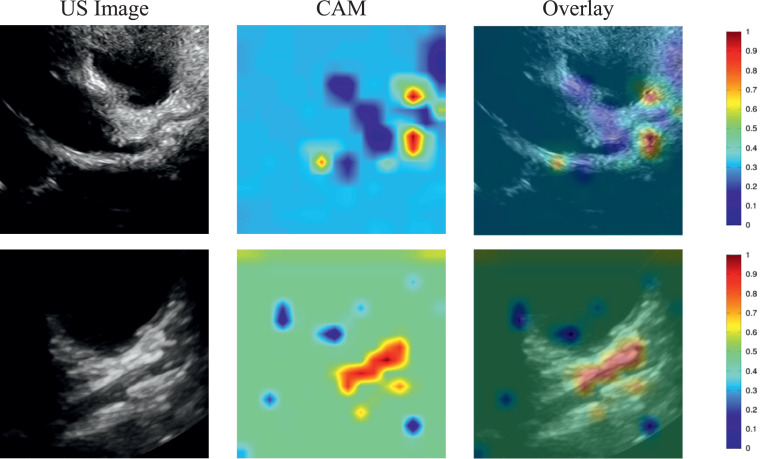
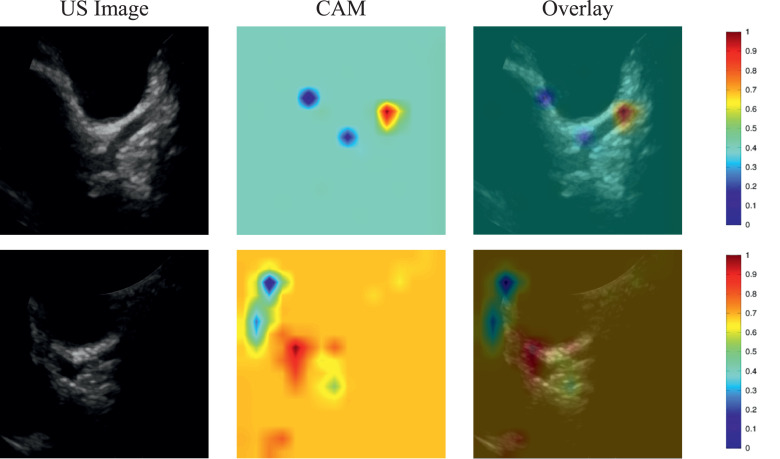
For non-KD, the echocardiographic images of pneumonia and its corresponding CAM images are illustrated in Fig. 3 . These images were correctly identified as non-KD by SE-ResNext50. The images in the first row represent the pneumonia image correctly recognized as non-KD by SE-ResNext50, whereas the second row shows the pneumonia image incorrectly classified as KD by SE-ResNext50.
Fig. 3.
Illustration of a class activation map of SE-ResNext50 for non-KD: the first column shows the echocardiographic images of non-KD, the second column shows the CAMs, and the third column shows the overlay of the echocardiographic image and the CAM.
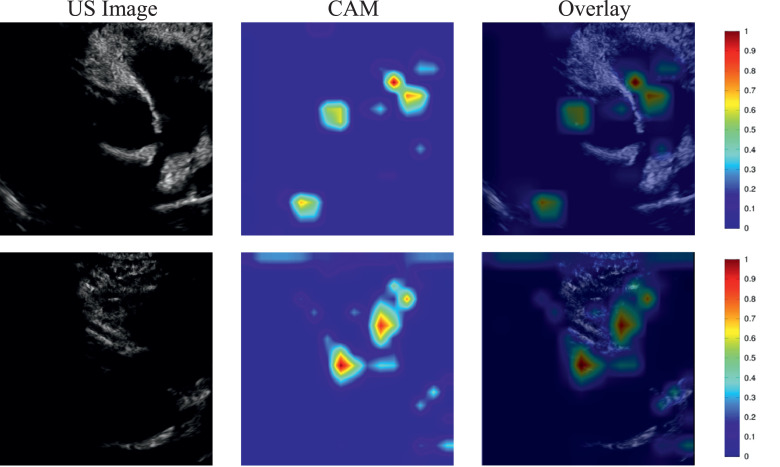
For incomplete KD, the echocardiographic image of KD and its corresponding CAM image are demonstrated in Fig. 4 . The images in the first row were correctly identified as KD by SE-ResNext50, whereas the images in the second row were incorrectly classified as non-KD by SE-ResNext50. The thicker red regions indicate the parts of images focused on by the deep learning algorithm during the process of classification as KD and non-KD.
Fig. 5.
Illustration of a class activation map of failure case of SE-ResNext50 for KD: the first row shows the echocardiographic images of non-KD, but the SE-ResNext50 predicts a KD. In contrast, the second row shows the echocardiographic images of KD, but the SE-ResNext50 predicts a non-KD.
Fig. 4.
Illustration of a class activation map of SE-ResNext50 for KD: the first column shows the echocardiographic images of KD, the second column shows the CAMs, the third column shows the overlays of the echocardiographic images and the CAM.
4. Discussion
The goal of this study was to investigate the potential of explainable deep learning algorithms to identify and differentiate KD from acute febrile diseases. We therefore selected several well-known deep learning algorithms (VGG19, Xception, ResNet50, ResNext50, SE-ResNet50, and SE-ResNext50) to distinguish incomplete KD from other acute febrile diseases. We selected pneumonia as a representative of other acute febrile diseases because it is the most common febrile disease in children. KD and pneumonia show similar fever patterns before the occurrence of respiratory symptoms in pneumonia. Despite the small training dataset, the results in our study demonstrated that the deep learning algorithms show excellent performance for the identification of the KD. Nevertheless, as the performance of a deep learning algorithm depends on the quantity of training data [30], [31], the deep learning algorithm for KD diagnosis should be extended.
Figs. 3 and 4 show the parts of the echocardiographic images that are considered important by the deep learning algorithm to distinguish between KD and non-KD. These figures indicate that the explainable deep learning algorithms identified KD by using the features of the coronary arteries. This is comparable to how pediatric cardiologists diagnose and differentiate the diseases. Clinical reports have mentioned that coronary artery imaging could be key to the appropriate diagnosis of KD, particularly incomplete KD [3]. Through this analysis, our results revealed that deep learning algorithms can identify KD among KD and non-KD, as cardiologists do, which suggests that deep learning algorithms could be applied in a clinical setting to recognize incomplete KD among various acute febrile diseases in children. Thus, these results indicate that explainable deep learning algorithms might be used to diagnose KD at a general hospital without a KD expert. In Korea, an experienced pediatric cardiologist is not always available at each hospital, owing to a lack of human resources. Nevertheless, timely diagnosis of KD is essential for proper treatment, to prevent poor outcomes of coronary artery lesions.
The SE-ResNext and SE-ResNet have several squeeze-and-excitation blocks allow using global information from an ultrasound image by using the global average pooling. Several works demonstrated that the performance of deep learning algorithms on different scale objects is affected by a receptive field. To distinguish incomplete KD and pneumonia in echocardiographic images, it is important to use the difference in the size of the coronary artery on the images. To fully discriminate the difference in the size of the coronal artery for the two diseases, it is beneficial to have a large receptive field or to use global spatial information. Thus, the SE-ResNext and SE-ResNet, which could use global spatial information, show better performance than ResNext and ResNet in distinguishing the incomplete KD and pneumonia diseases.
Previous studies have analyzed echocardiographic images using deep learning to perform classification of myocardial disease [32], detect hypertrophic cardiomyopathy, cardiac amyloid, and pulmonary arterial hypertension [33], and evaluate chamber segmentation [34] and wall motion abnormalities [35], [36]. However, there has been no study to date on diagnosis of incomplete KD by echocardiographic images of coronary artery lesions using deep learning, as we have done here. This study indicates that explainable deep learning has potential to diagnose KD among acute febrile diseases. In the global COVID-19 pandemic in particular, there is a risk that KD might be misdiagnosed, as WHO stated that COVID-19 has a similar febrile clinical presentation as KD [10], [11]. Therefore, now more than ever, it is important to distinguish KD from other febrile diseases in children; this may be possible by using an explainable deep learning algorithm.
In this study, we used 138 KD and 65 pneumonia images for evaluation of the performance of the deep learning models. Although the number of datasets was limited for training and testing the deep learning models to be translated into the clinics, we conducted cross-validation to confirm the performance of deep learning models in this study. It was found that the sensitivity of SE-ResNext was 82.64% which is comparable to that of an experienced cardiologist, which typically offers a sensitivity of more than 85%. However, the specificity of SE-ResNext was 58.46% which is 10% less than that of an experienced cardiologist, which usually offers a specificity of almost 70% [8]. Although the specificity of deep learning algorithms is lower than that of an experienced cardiologist, the deep learning models can be useful for the diagnosis of incomplete KD in the clinics that do not have experienced cardiologists when they are further improved. Note that the class activation maps from the deep learning models clearly [Figs. 3 and 4] showed that the coronary artery regions were salient for incomplete KD, but not clearly for pneumonia as experienced cardiologists do for the diagnosis of incomplete KD. Therefore, the results, shown in this study, demonstrate that the deep learning models have the potential for discriminating between incomplete KD and other febrile diseases such as pneumonia with echocardiographic images.
Many research has proven that the performance of deep learning algorithms depends on the amount of training data. The specificity of the deep learning algorithms in the discrimination between the incomplete KD and other febrile diseases like pneumonia in this study was relatively low because of the imbalanced ultrasound image dataset. To solve this issue, it may be necessary to acquire more KD and pneumonia images which have the similar number. However, it has been known that it is very difficult to acquire a large amount of medical data. Therefore, various deep learning models such as few-shot or zero-shot learning techniques have been studied to overcome this issue caused by the limited number of medical datasets [37]. Also, the problem from the imbalanced datasets of medical datasets has been tried to be solved by using various techniques such as focal loss which offers to focus training on a sparse dataset and simultaneously and prevent the vast number during training [38], [39]. The deep learning techniques can be applied to discriminate between incomplete KD and pneumonia using ultrasound imaging. The related study remains future work.
5. Conclusions
We have shown the feasibility of using an explainable deep learning approach for detection of KD based on echocardiography images. The AUPRCs of the deep learning algorithms, including VGG19, Xception, ResNet50, ResNext50, SE-ResNet50, and SE-ResNext50, were found to be 0.7576, 0.8074, 0.6913, 0.7599, 0.8534, and 0.8563, respectively, for discrimination between KD and non-KD. In particular, the SE-ResNext50 offered the best performance among the deep learning algorithms with an accuracy of 75.86% and an AUPRC of 0.8563. The explainable deep learning algorithms highlighted salient features of coronary arteries, similar to how an experienced pediatric cardiologist would examine coronary artery regions for the detection of KD. Although the specificity of deep learning algorithms was still lower than that of highly experienced clinicians for the discrimination between incomplete KD and non-KD, the deep learning algorithms used in this study were promising in terms of sensitivity and precision. Therefore, deep learning algorithms may assist clinicians in reducing the probability of misdiagnosing KD in clinical practice. The abilities of deep learning algorithms should be further developed to be comparable to the performance of highly experienced clinicians in order to translate this approach to application in the clinic.
Statements of ethical approval
The study was approved by the Ethics Committee of Severance Hospital. All the participants provided their written informed consent to participate in this study.
Declaration of Competing Interest
There are no conflicts of interest to disclose for publication of this paper.
Acknowledgements
This study was supported by a new faculty research seed money grant of Yonsei University College of Medicine for 202 (2020-32-0035), the Korea Medical Device Development Fund grant funded by the Korea goverment (RS-2020-KD000125, 9991006798), and the 2022 Joint Research Project of Institutes of Science and Technology (2460871).
References
- 1.Kuo H.-C. Preventing coronary artery lesions in Kawasaki disease. Biomed. J. 2017;40(3):141–146. doi: 10.1016/j.bj.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Makino N., Nakamura Y., Yashiro M., Ae R., Tsuboi S., Aoyama Y., Kojo T., Uehara R., Kotani K., Yanagawa H. Descriptive epidemiology of Kawasaki disease in japan, 2011–2012: from the results of the 22nd nationwide survey. J. Epidemiol. 2015:JE20140089. doi: 10.2188/jea.JE20140089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim G.B., Park S., Eun L.Y., Han J.W., Lee S.Y., Yoon K.L., Yu J.J., Choi J.-W., Lee K.-Y. Epidemiology and clinical features of Kawasaki disease in south korea, 2012–2014. Pediatr. Infect. Dis. J. 2017;36(5):482–485. doi: 10.1097/INF.0000000000001474. [DOI] [PubMed] [Google Scholar]
- 4.McCrindle B., Rowley A., Newburger J., Burns J., Bolger A., Gewitz M., Baker A., Jackson M., Takahashi M., Shah P., et al. American heart association rheumatic fever, endocarditis, and Kawasaki disease committee of the council on cardiovascular disease in the young; council on cardiovascular and stroke nursing; council on cardiovascular surgery and anesthesia; and council on epidemiology and prevention. diagnosis, treatment, and long-term management of Kawasaki disease: a scientific statement for health professionals from the american heart association. Circulation. 2017;135(17):e927–e999. doi: 10.1161/CIR.0000000000000484. [DOI] [PubMed] [Google Scholar]
- 5.Dietz S., Van Stijn D., Burgner D., Levin M., Kuipers I., Hutten B., Kuijpers T. Dissecting Kawasaki disease: a state-of-the-art review. Eur. J. Pediatr. 2017;176(8):995–1009. doi: 10.1007/s00431-017-2937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newburger J.W., Takahashi M., Burns J.C. Kawasaki disease. J. Am. Coll. Cardiol. 2016;67(14):1738–1749. doi: 10.1016/j.jacc.2015.12.073. [DOI] [PubMed] [Google Scholar]
- 7.Singh S., Jindal A.K., Pilania R.K. Diagnosis of Kawasaki disease. Int. J. Rheum. Dis. 2018;21(1):36–44. doi: 10.1111/1756-185X.13224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Na J.-H., Kim S., Eun L.Y. Utilization of coronary artery to aorta for the early detection of Kawasaki disease. Pediatr. Cardiol. 2019;40(3):461–467. doi: 10.1007/s00246-018-1985-6. [DOI] [PubMed] [Google Scholar]
- 9.Rowley A.H., Duffy C.E., Shulman S.T. Prevention of giant coronary artery aneurysms in Kawasaki disease by intravenous gamma globulin therapy. J. Pediatr. 1988;113(2):290–294. doi: 10.1016/s0022-3476(88)80267-1. [DOI] [PubMed] [Google Scholar]
- 10.Jones V.G., Mills M., Suarez D., Hogan C.A., Yeh D., Segal J.B., Nguyen E.L., Barsh G.R., Maskatia S., Mathew R. Covid-19 and Kawasaki disease: novel virus and novel case. Hosp. Pediatr. 2020;10(6):537–540. doi: 10.1542/hpeds.2020-0123. [DOI] [PubMed] [Google Scholar]
- 11.Viner R.M., Whittaker E. Kawasaki-like disease: emerging complication during the covid-19 pandemic. Lancet. 2020;395(10239):1741–1743. doi: 10.1016/S0140-6736(20)31129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan W.-j., Ni Z.-y., Hu Y., Liang W.-h., Ou C.-q., He J.-x., Liu L., Shan H., Lei C.-l., Hui D.S., et al. Clinical characteristics of coronavirus disease 2019 in china. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esteva A., Kuprel B., Novoa R.A., Ko J., Swetter S.M., Blau H.M., Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu S., Wang Y., Yang X., Lei B., Liu L., Li S.X., Ni D., Wang T. Deep learning in medical ultrasound analysis: a review. Engineering. 2019;5(2):261–275. [Google Scholar]
- 15.Lee H., Park J., Hwang J.Y. Channel attention module with multiscale grid average pooling for breast cancer segmentation in an ultrasound image. IEEE Trans. Ultrason. Ferroelectr. Freq. Control. 2020;67(7):1344–1353. doi: 10.1109/TUFFC.2020.2972573. [DOI] [PubMed] [Google Scholar]
- 16.Wu K., Chen X., Ding M. Deep learning based classification of focal liver lesions with contrast-enhanced ultrasound. Optik. 2014;125(15):4057–4063. [Google Scholar]
- 17.Ma J., Wu F., Zhu J., Xu D., Kong D. A pre-trained convolutional neural network based method for thyroid nodule diagnosis. Ultrasonics. 2017;73:221–230. doi: 10.1016/j.ultras.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 18.K. Simonyan, A. Zisserman, Very deep convolutional networks for large-scale image recognition, arXiv:1409.1556 (2014).
- 19.Chollet F. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Xception: deep learning with depthwise separable convolutions; pp. 1251–1258. [Google Scholar]
- 20.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 21.Xie S., Girshick R., Dollár P., Tu Z., He K. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Aggregated residual transformations for deep neural networks; pp. 1492–1500. [Google Scholar]
- 22.Hu J., Shen L., Sun G. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2018. Squeeze-and-excitation networks; pp. 7132–7141. [Google Scholar]
- 23.Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Rethinking the inception architecture for computer vision; pp. 2818–2826. [Google Scholar]
- 24.Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., Huang Z., Karpathy A., Khosla A., Bernstein M., et al. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 2015;115(3):211–252. [Google Scholar]
- 25.D.P. Kingma, J. Ba, Adam: a method for stochastic optimization, arXiv:1412.6980 (2014).
- 26.Paszke A., Gross S., Chintala S., Chanan G., Yang E., DeVito Z., Lin Z., Desmaison A., Antiga L., Lerer A. NIPS-W. 2017. Automatic differentiation in pytorch; pp. 1–4. [Google Scholar]
- 27.Zhou B., Khosla A., Lapedriza A., Oliva A., Torralba A. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Learning deep features for discriminative localization; pp. 2921–2929. [Google Scholar]
- 28.Ma X., Ji Z., Niu S., Leng T., Rubin D.L., Chen Q. Ms-cam: multi-scale class activation maps for weakly-supervised segmentation of geographic atrophy lesions in sd-oct images. IEEE J. Biomed. Health Inform. 2020;24(12):3443–3455. doi: 10.1109/JBHI.2020.2999588. [DOI] [PubMed] [Google Scholar]
- 29.Qiao N., Song M., Ye Z., He W., Ma Z., Wang Y., Zhang Y., Shou X. Deep learning for automatically visual evoked potential classification during surgical decompression of sellar region tumors. Transl. Vision Sci. Technol. 2019;8(6) doi: 10.1167/tvst.8.6.21. [DOI] [PMC free article] [PubMed] [Google Scholar]; 21–21.
- 30.Sun C., Shrivastava A., Singh S., Gupta A. Proceedings of the IEEE International Conference on Computer Vision. 2017. Revisiting unreasonable effectiveness of data in deep learning era; pp. 843–852. [Google Scholar]
- 31.D. Rolnick, A. Veit, S. Belongie, N. Shavit, Deep learning is robust to massive label noise, arXiv:1705.10694 (2017).
- 32.Zhang J., Gajjala S., Agrawal P., Tison G.H., Hallock L.A., Beussink-Nelson L., Lassen M.H., Fan E., Aras M.A., Jordan C., et al. Fully automated echocardiogram interpretation in clinical practice: feasibility and diagnostic accuracy. Circulation. 2018;138(16):1623–1635. doi: 10.1161/CIRCULATIONAHA.118.034338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vidal M.L.B., Diller G.-P., Kempny A., Li W., Dimopoulos K., Wort S.J., Gatzoulis M. Utility of deep learning algorithms in diagnosing and automatic prognostication of pulmonary arterial hypertension based on routine echocardiographic imaging. J. Am. Coll. Cardiol. 2021;77(18_Supplement_1) [Google Scholar]; 1670–1670.
- 34.Leclerc S., Smistad E., Pedrosa J., Østvik A., Cervenansky F., Espinosa F., Espeland T., Berg E.A.R., Jodoin P.-M., Grenier T., et al. Deep learning for segmentation using an open large-scale dataset in 2d echocardiography. IEEE Trans. Med. Imaging. 2019;38(9):2198–2210. doi: 10.1109/TMI.2019.2900516. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez-Martinez S., Duchateau N., Erdei T., Kunszt G., Aakhus S., Degiovanni A., Marino P., Carluccio E., Piella G., Fraser A.G., et al. Machine learning analysis of left ventricular function to characterize heart failure with preserved ejection fraction. Circulation. 2018;11(4):e007138. doi: 10.1161/CIRCIMAGING.117.007138. [DOI] [PubMed] [Google Scholar]
- 36.Omar H.A., Domingos J.S., Patra A., Upton R., Leeson P., Noble J.A. 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) IEEE; 2018. Quantification of cardiac bull’s-eye map based on principal strain analysis for myocardial wall motion assessment in stress echocardiography; pp. 1195–1198. [Google Scholar]
- 37.Feng R., Zheng X., Gao T., Chen J., Wang W., Chen D.Z., Wu J. Interactive few-shot learning: limited supervision, better medical image segmentation. IEEE Trans. Med. Imaging. 2021;40(10):2575–2588. doi: 10.1109/TMI.2021.3060551. [DOI] [PubMed] [Google Scholar]
- 38.Lee K., Kim J.Y., Lee M.H., Choi C.-H., Hwang J.Y. Imbalanced loss-integrated deep-learning-based ultrasound image analysis for diagnosis of rotator-cuff tear. Sensors. 2021;21(6):2214. doi: 10.3390/s21062214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang H., Lin L., Tong R., Hu H., Zhang Q., Iwamoto Y., Han X., Chen Y.-W., Wu J. ICASSP 2020-2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) IEEE; 2020. Unet 3+: a full-scale connected unet for medical image segmentation; pp. 1055–1059. [Google Scholar]