Abstract
Transfer of the 2,4-dichlorophenoxyacetic acid (2,4-D) degradation plasmids pEMT1 and pJP4 from an introduced donor strain, Pseudomonas putida UWC3, to the indigenous bacteria of two different horizons (A horizon, depth of 0 to 30 cm; B horizon, depth of 30 to 60 cm) of a 2,4-D-contaminated soil was investigated as a means of bioaugmentation. When the soil was amended with nutrients, plasmid transfer and enhanced degradation of 2,4-D were observed. These findings were most striking in the B horizon, where the indigenous bacteria were unable to degrade any of the 2,4-D (100 mg/kg of soil) during at least 22 days but where inoculation with either of the two plasmid donors resulted in complete 2,4-D degradation within 14 days. In contrast, in soils not amended with nutrients, inoculation of donors in the A horizon and subsequent formation of transconjugants (105 CFU/g of soil) could not increase the 2,4-D degradation rate compared to that of the noninoculated soil. However, donor inoculation in the nonamended B-horizon soil resulted in complete degradation of 2,4-D within 19 days, while no degradation at all was observed in noninoculated soil during 89 days. With plasmid pEMT1, this enhanced degradation seemed to be due only to transconjugants (105 CFU/g of soil), since the donor was already undetectable when degradation started. Denaturing gradient gel electrophoresis (DGGE) of 16S rRNA genes showed that inoculation of the donors was followed by a shift in the microbial community structure of the nonamended B-horizon soils. The new 16S rRNA gene fragments in the DGGE profile corresponded with the 16S rRNA genes of 2,4-D-degrading transconjugant colonies isolated on agar plates. This result indicates that the observed change in the community was due to proliferation of transconjugants formed in soil. Overall, this work clearly demonstrates that bioaugmentation can constitute an effective strategy for cleanup of soils which are poor in nutrients and microbial activity, such as those of the B horizon.
The highly diverse microbial communities present in fresh and marine waters, sewage, and soils are able to transform a wide range of organic chemicals. However, many synthetic organic compounds, although biodegradable, may persist in nature for a long time because the required catabolic capacity is not present or because the populations of microorganisms bringing about their destruction are not large or active enough. One way to enhance breakdown of these chemicals is bioaugmentation by inoculation of a habitat with microorganisms that either are known to readily metabolize the chemicals or have been equipped with the necessary degradative genes in the laboratory (25, 46). This bioaugmentation of polluted soils has already been studied by different authors (6, 10, 11, 12, 21, 33). However, in many cases the introduced bacteria fail to degrade the pollutants due to their poor survival or low activity in the environment, caused by abiotic and biotic stresses which are not encountered in the laboratory environment (1, 2, 25, 46, 58, 59). An alternative approach involves the introduction of appropriate plasmid-borne catabolic genes into well-established and competitive indigenous bacterial populations. Here, the survival of the introduced donor strain is no longer needed once the catabolic genes are transferred to the indigenous bacteria. In addition, the plasmid transfer may result in vertical movement of the catabolic genes through the soil, resulting in the dissemination of the desired catabolic activity into deeper soil layers. This dissemination is especially important since the indigenous metabolic activity in these subsurface soils is lower, which may result in contamination of groundwater (24). Also, the high costs of mixing the soil layers to bring the bacteria in close proximity of the pollutant might be avoided. To date, a large number of studies have reported the occurrence of conjugative gene transfer between bacteria in soil (17, 31); however, there is only little information about transfer of catabolic plasmids as a means of bioaugmentation (8, 14, 15, 42, 54, 55). Research in our laboratory has shown before that this approach accelerates the degradation of biphenyl or 2,4-dichlorophenoxyacetic acid (2,4-D) in soil, although the introduced donor strains survived only between 3 and 14 days (14, 54, 55). Other groups have shown a less pronounced effect of 2,4-D degradation, using the same strategy (15, 42).
A model compound for the study of plasmid-borne catabolic genes is the herbicide 2,4-D, which is a commercially used chlorinated aromatic compound. Genes encoding 2,4-D degradation are often located on conjugative plasmids (7, 16, 36, 52, 53) but have recently been found to be also chromosomally located (37, 50). The most extensively studied plasmid is pJP4 from Ralstonia eutropha JMP134 (16), which has become a model for the study of 2,4-D degradation. Plasmid pEMT1 (52) carries virtually the same degradative genes in an organization similar to that of plasmid pJP4 but probably does not belong to the same incompatibility group as pJP4 (IncPβ) (27, 52). Nothing is known about the differences in their transfer frequencies and host ranges in soil.
In this study we have investigated the transfer of the 2,4-D degradation plasmids pEMT1 and pJP4, from an introduced donor strain, Pseudomonas putida UWC3, to the indigenous bacteria of the A and B horizons of a 2,4-D-contaminated sandy-loam soil. Compared to the soil from the A horizon (depth, 0 to 30 cm), the soil from the B horizon (depth, 30 to 60 cm) has a different texture, a different organic-matter content, and probably also a different microbial community, with a different metabolic activity. The impact of these differences on the 2,4-D degradation capacities of the soils, the plasmid transfer rates, and the subsequent accelerated 2,4-D degradation was investigated. In addition, we examined if the formation of indigenous transconjugants in soil had an impact on the overall microbial community structure, as revealed by the culture-independent technique denaturing gradient gel electrophoresis (DGGE) of 16S rRNA genes.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. The 2,4-D degradation plasmids pEMT1 and pJP4 were tagged with a mini-transposon, mini-Tn5 KmlacZ (13), as previously described (53) and were designated pEMT1::lacZ and pJP4::lacZ. In brief, a triparental conjugation was performed with Escherichia coli CC118 λpir (pUT-mini-Tn5 KmlacZ) as the donor strain (29), with E. coli HB101 (pRK2013) as the helper strain (23), and with R. eutropha JMP134, which contains plasmid pJP4 (16), or R. eutropha JMP228 (pEMT1) as the recipient. The mating mixture was resuspended in MMO mineral medium (48) with 1,000 mg of 2,4-D per liter, 50 mg of kanamycin per liter, and 20 mg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) per liter, the latter as a chromogenic substrate for detection of β-galactosidase activity. Cultures were shaken at 150 rpm at 28°C. Tagged plasmids were subsequently transferred to E. coli DH5α (28) and then to P. putida UWC3, which was used as a donor in the transfer experiments. Marking of the plasmids had no measurable effect on their transfer frequencies or on the 2,4-D degradation capacity of the host strain, R. eutropha JMP228 (data not shown). We chose P. putida UWC3 as a donor strain because P. putida UWC3 (pEMT1) has been shown to degrade 2,4-D poorly or not at all in mineral medium with or without an additional C source (55). Additional degradation tests with autoclaved soil showed that P. putida UWC3 (pEMT1::lacZ) degraded 100 mg of 2,4-D per kg of soil in 15 days in nutrient-amended soil but did not degrade 2,4-D at all, even after 80 days, in nonamended soil. This poor degradation capacity of P. putida UWC3 (pEMT1::lacZ) seemed to be due to the instability of the plasmid. P. putida UWC3 (pJP4::lacZ), on the contrary, degraded 100 mg of 2,4-D per kg both in the presence (5 days) and in the absence (21 days) of nutrients (data not shown).
TABLE 1.
Most important bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| Pseudomonas putida UWC3 | Rifr ILV− | 30 |
| Ralstonia eutropha JMP228 | Rifr | 16 |
| Ralstonia eutropha JMP228n | Rifr Nxr | 52 |
| Plasmids | ||
| pUT-mini-Tn5 KmlacZ | Kmr LacZ+ | 13 |
| pEMT1 | Tfd+ | 52 |
| pJP4 | Tfd+ 3-CBA+ Hgr | 16 |
| pEMT1::lacZ | Tfd+ Kmr LacZ+ | This study |
| pJP4::lacZ | Tfd+ 3-CBA+ Hgr Kmr LacZ+ | This study |
Rifr, Nxr, Kmr, and Hgr, resistance to rifampin, nalidixic acid, kanamycin, and mercury, respectively; ILV−, auxotrophic for isoleucine, leucine, and valine; Tfd+ and 3-CBA+, abilities to degrade 2,4-D and 3-chlorobenzoate, respectively; LacZ+, β-galactosidase activity.
Media and culture conditions.
P. putida UWC3 was grown on MKB agar (47) with 100 mg of rifampin per liter. The MKB medium for the donors P. putida UWC3 (pEMT1::lacZ) and P. putida UWC3 (pJP4::lacZ) contained, besides 100 mg of rifampin per liter, 50 mg of kanamycin per liter to provide selection pressure for the plasmids and 20 mg of X-Gal per liter. R. eutropha JMP228 (pEMT1) and R. eutropha JMP228 (pJP4) were maintained on MMO mineral medium supplemented with 500 mg of 2,4-D (52) per liter and 40 mg of bromothymol blue, a pH indicator (35), per liter. All other strains were grown on Luria-Bertani (LB) (47) medium with an appropriate antibiotic (100 mg of rifampin per liter, 200 mg of nalidixic acid per liter, or 50 mg of kanamycin per liter) and 20 mg of X-Gal per liter. All plates were incubated at 28°C.
Soils.
The soils used in this study were collected from a farm (Pittem, Belgium) and had not previously been exposed to 2,4-D. The A-horizon soil was sampled at a depth of 0 to 30 cm and was characterized as a sandy-loam soil (7.3% clay, 16.8% loam, 75.9% sand). This soil has an organic-matter content of 1.38%, a pH of 5.6 (KCl), and a moisture content at field capacity of 15.9% ± 0.9% on dry soil (57). The microbial activity in the A horizon was 1.97 mg of CO2-C · kg of soil−1 · day−1 as determined by CO2 respiration measurements (44). The B horizon (depth, 30 to 60 cm) belongs to the sandy-texture class (7.6% clay, 9.2% loam, 83.2% sand) with an organic-matter content of 0.38%, a pH of 5.3 (KCl), a moisture content at field capacity of 11.8% ± 0.8% on dry soil (57), and a microbial activity of 0.52 mg of CO2-C · kg−1 · day−1. The heterotrophic plate counts (R2A agar; Difco, Detroit, Mich.) of both the A- and the B-horizon soils varied between ca. 107 and 108 CFU/g (fresh weight) of soil. Soils were stored at room temperature.
Microcosm design and treatments.
Transfer experiments were performed with 500-ml glass microcosms which contained 100 g of the A- or B-horizon soil. An overnight culture of the donors P. putida UWC3 (pEMT1::lacZ) and P. putida UWC3 (pJP4::lacZ) in LB medium with 50 mg of kanamycin per liter was washed twice in 0.85% sterile saline. When transfer experiments were performed in the presence of nutrients, the pellet was resuspended in 5 ml of 5× LB medium; otherwise, the pellet was suspended in 5 ml of saline. One milliliter of this bacterial suspension was mixed thoroughly with 100 mg of 2,4-D per kg in 100 g of the A- or the B-horizon soil. To noninoculated control soils only 2,4-D and either 5× LB medium or saline, but no donor cultures, were added. After all amendments were made, the moisture content was adjusted with water to 75% of the water-holding capacity. Soil microcosms were incubated at room temperature in a plastic container covered with aluminum foil in which the relative humidity was kept high by means of an open beaker filled with tap water. All treatments were performed in duplicate using independent microcosms.
The survival of the donor strain in the soil was monitored as a function of time by plating 0.1-ml samples of serial 10-fold dilutions of 1 g of soil in saline on MKB agar that contained 100 mg of rifampin per liter and 200 mg of cycloheximide per liter (MKBRC), a medium on which the donor is fluorescent under UV light. The survival of the donor with the plasmid was monitored by plating on the previous medium, to which 50 mg of kanamycin per liter had been added (MKBRCK) to select for the tagged plasmids. Transconjugants were detected as blue colonies on MMO mineral medium with 500 mg of 2,4-D per liter as the sole C source, 20 mg of X-Gal per liter, 50 mg of kanamycin per liter, and 200 mg of cycloheximide per liter (MXK2,4-D). Since the P. putida UWC3 donors require isoleucine, leucine, and valine, they cannot grow on this medium. The detection limit for donors and transconjugants was 102 CFU/g of soil. All plates were incubated at 28°C, and plate counts (CFU per gram of soil) are expressed per gram (fresh weight) of soil.
2,4-D was extracted from 1 g of soil with 2 ml of sterile demineralized water. Its concentration was determined by high-performance liquid chromatography analysis (Kontron) using an Alltima C18 reversed-phase column (inside diameter, 250 by 8 mm; particle size, 5 μm; Alltech, Deerfield, Ill.), a methanol–0.1% aqueous phosphoric acid (85:15) mixture as an eluent at a flow rate of 0.8 ml/min, and a Kontron diode array detector, with the wavelength set at 230 nm for detection and quantification of 2,4-D (54).
Confirmation of the potential transconjugants.
Blue colonies which developed from the highest dilution on MXK2,4-D plates were transferred to LB medium, MKBRCK, and MXK2,4-D plates by replica plating to compare the colony morphologies of the transconjugants and to eliminate false positives or potential donor cells forming colonies on MXK2,4-D. Putative transconjugants with different morphologies were mated with R. eutropha JMP228n (39) with selection on LB medium plates containing 50 mg of kanamycin per liter, 200 mg of nalidixic acid per liter, and 20 mg of X-Gal per liter. The presence of the plasmids was confirmed by a modified Kado and Liu plasmid extraction procedure (32, 51, 52). Plasmids were visualized on a 0.8% agarose gel (47) and compared with the plasmids extracted from the donors. This procedure also suggested that transconjugants formed in soil were able to act as donors of the plasmids. We also confirmed that the pJP4 and pEMT1 plasmid-borne tfdA gene was present in unique isolates by performing a PCR with the primers TVU and TVL as previously described (56). The 2,4-D degradation capacities of some of the positive transconjugants were compared with those of the donors, R. eutropha JMP228 (pEMT1::lacZ) and R. eutropha JMP228 (pJP4::lacZ) (positive controls), and of P. putida UWC3 and R. eutropha JMP228n (negative controls) by transferring one colony of each strain to 5 ml of liquid MMO mineral medium with 100 mg of 2,4-D per liter and, when appropriate, 50 mg of kanamycin per liter. After 3 days of shaking at 150 rpm at 28°C, the remaining amount of 2,4-D was quantified by high-performance liquid chromatography analysis (54).
DNA extraction and DGGE.
For pure cultures, the PCR template was obtained by boiling a colony for 10 min in 200 μl of MilliQ-water. For DGGE analysis of colony mixtures resuspended from plates (referred to as plate DNAs), colonies were scraped off with a loop, after which the plates were washed with 2 ml of water. Two hundred microliters of this suspension was boiled for 10 min and used as the template in the PCR. For soil analyses, samples (2 g) were taken at regular intervals from one single microcosm per treatment. Total DNA (referred to as soil DNA) was extracted based on the protocol described previously (20, 22), followed by purification with the Wizard DNA Clean-Up System (Promega, Madison, Wis.). In all cases 1 μl of the template was used for PCR with the bacterium-specific 16S rRNA forward primer P63f and the reverse primer P518r, based on a universally conserved region, as previously described (22). The PCR product contains a GC clamp of 40 bases, added to the forward primer, and has a total length of 531 bp (based on the length of the reference strain E. coli K-12). PCR products were subjected to DGGE based on the protocols of Muyzer et al. (40) and El Fantroussi et al. (22). In brief, PCR samples were run for 17 h at 50 V on a 6% (wt/vol) polyacrylamide gel with a denaturing gradient ranging from 50 to 65% (where a 100% denaturant contains 7 M urea and 40% formamide). After electrophoresis the gels were stained with SYBR Green I nucleic acid gel stain and photographed. The pictures presented in this paper are negative images.
Some bands were excised from the gel and incubated overnight at 4°C in 20 μl of MilliQ-water. Subsequently, 1 μl of this solution was reamplified in 25 μl of a PCR mix and the amplified products were cloned into the PCR-TOPO 2.1 cloning vector (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. Recombinant (white) colonies were screened by using a two-stage procedure to ensure recovery of the DGGE band of interest. First, plasmid inserts (eight for each band) were reamplified by PCR with vector-specific primers (M13 primer set; Invitrogen Corp.). The PCR products were immediately reamplified with the 16S rRNA-specific PCR primers described above (22) and subjected to DGGE analysis. Sequences that comigrated with the original band of interest were selected and sequenced by Eurogentec (Liège, Belgium). The partial sequences, of approximately 400 bp, were aligned to 16S rRNA sequences obtained from the National Centre for Biotechnology Information database by using the BLAST, version 2.0, search program (3).
Nucleotide sequence accession numbers.
The nucleotide sequences for bands 1 to 5 have been deposited in the GenBank database under accession no. AF247780 to AF247784.
RESULTS
Horizontal transfer of the mini-Tn5 KmlacZ-tagged plasmids pEMT1 and pJP4 from an introduced donor strain, P. putida UWC3, to indigenous bacteria of the A and B horizons of a sandy-loam soil was studied. The effect of this transfer on the degradation of 2,4-D and on the microbial-community structure was investigated.
Transfer of plasmids and degradation of 2,4-D in nutrient-amended soil.
Approximately 105 CFU of P. putida UWC3 (pEMT1::lacZ) per g of soil and 106 CFU of P. putida UWC3 (pJP4::lacZ) per g of soil were inoculated separately into duplicate independent microcosms containing either the nutrient-amended A- or B-horizon soil spiked with 2,4-D (100 mg/kg). The microcosms were sampled on day 0 and 2, 6, 14, 22, and 50 days after inoculation. In the A-horizon soil, 2,4-D-degrading transconjugants with plasmids pEMT1::lacZ and pJP4::lacZ were first detected after 6 days (4.2 × 104 and 3.0 × 103 CFU/g of soil, respectively), and their numbers remained quite stable throughout the experiment. On day 0 and day 2 counts of putative transconjugant colonies on plates did not exceed those of the background growth of noninoculated control soil (ca. 103 CFU/g of soil) (data not shown). Randomly picked transconjugants from day 6 and subsequent days contained a plasmid of the same size as pEMT1::lacZ or pJP4::lacZ, gave a positive PCR signal with the tfdA primers, and degraded 100 mg of 2,4-D per liter in liquid mineral medium in 3 days, while no degradation at all was observed during this period for the donors.
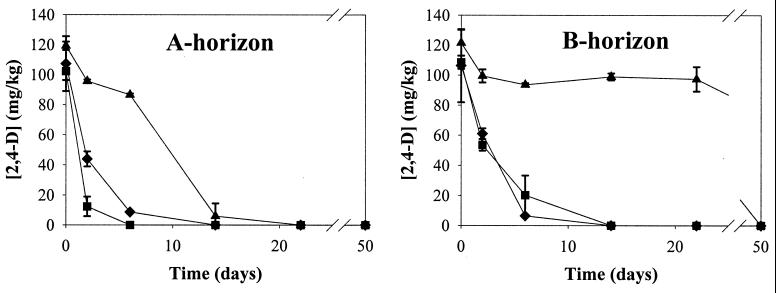
In noninoculated nutrient-amended soil from the A horizon, indigenous bacteria degraded 100 mg of 2,4-D per kg in 14 days, a period that was shortened when P. putida UWC3 (pEMT1::lacZ) and P. putida UWC3 (pJP4::lacZ) were added (Fig. 1). This result shows that inoculation of the donors enhanced the degradation of 2,4-D.
FIG. 1.
Effect of donor inoculation and subsequent plasmid transfer on the degradation of 2,4-D in the nutrient-amended A- and B-horizon soil. The data points and error bars show the means and standard deviations based on data from duplicate microcosms. ▴, noninoculated soil plus 2,4-D; ⧫, soil plus 2,4-D plus pEMT1::lacZ; ■, soil plus 2,4-D plus pJP4::lacZ.
In the nutrient-amended inoculated soil from the B horizon, transconjugants were also first detected on day 6, but in slightly higher numbers than in the A-horizon soil (8.3 × 105 CFU/g of soil for pEMT1::lacZ and 2.9 × 105 CFU/g of soil for pJP4::lacZ), and their numbers also remained constant for at least 50 days. The background on transconjugant-selective plates was the same as in the A-horizon soil (data not shown). Randomly isolated and purified transconjugants were confirmed to be true transconjugants as described above.
Interestingly, indigenous bacteria of the B horizon started to degrade the 2,4-D only after 22 days, while in the inoculated soils 80 to 90% of the 2,4-D was degraded in 6 days and 100% was degraded between day 6 and day 14 (Fig. 1). Hence, the inoculation of donors in the B horizon enhanced the degradation of 2,4-D in an even more pronounced way than in the A horizon.
Transfer of plasmids and degradation of 2,4-D in soil without nutrients.
In order to mimic more closely the natural soil conditions, the same experiment was performed as described above but without addition of nutrients to the soils (nonamended soil).
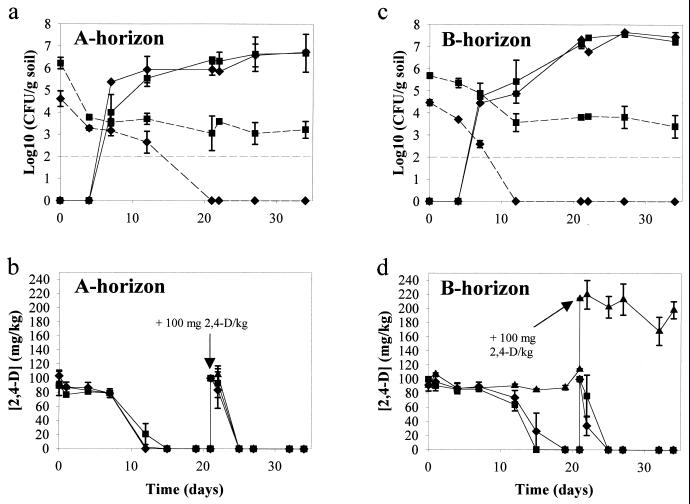
In the A-horizon soil, spiked with 2,4-D, transconjugants with either pEMT1::lacZ or pJP4::lacZ were first observed on day 7 and slightly increased in numbers until day 34 (Fig. 2a). On day 0 and day 4 counts of transconjugants on plates did not exceed those of the background growth of noninoculated control soil (ca. 102 CFU/g of soil). For each plasmid donor 50 putative transconjugants were picked from the transconjugant-selective plates of days 7, 12, 22, and 34. Of all colony types, a representative was selected (22 for pEMT1::lacZ, 14 for pJP4::lacZ) and confirmed to be true transconjugants, as described above.
FIG. 2.
Transfer of plasmids pEMT1::lacZ and pJP4::lacZ in the nonamended A- and B-horizon soil. (a and c) Survival of donors (dotted lines) and formation of transconjugants (solid lines). (b and d) Effect of donor inoculation and subsequent plasmid transfer on the degradation of 2,4-D. The data points and error bars show the means and standard deviations based on data from duplicate microcosms. ▴, noninoculated soil plus 2,4-D; ⧫, soil plus 2,4-D plus pEMT1::lacZ; ■, soil plus 2,4-D plus pJP4::lacZ. The broken straight line represents the detection limit of plate counts. The arrow indicates the second amendment with 100 mg of 2,4-D/kg of soil on day 21.
In the A horizon the number of plasmid-containing donors declined rapidly, especially for P. putida UWC3 (pEMT1::lacZ) (Fig. 2a). This seemed to be due to the instability of the plasmid, since counts of P. putida UWC3 on plates without selection for the plasmid (kanamycin) remained quite constant (104 CFU/g of soil) (data not shown).
In this nonamended A-horizon soil, the addition of donors and formation of transconjugants did not result in enhanced 2,4-D degradation. The 100 mg of 2,4-D per kg was degraded as fast in the noninoculated control soil as in the inoculated soils (Fig. 2b). When 100 mg of 2,4-D per kg was added to the soil for a second time at day 21, it was completely degraded in less than 4 days in the A-horizon soil both with and without the donors.
In the B-horizon soil spiked with 2,4-D and inoculated with approximately the same numbers of donors as was the A-horizon soil, transconjugants with either plasmid were first detected on day 7 (Fig. 2c) and further increased by 2 log units. Background on transconjugant-selective plates was the same as in the A horizon. For each donor 50 transconjugants were picked from the selective plates on days 7, 12, 22, and 34. A selection of these transconjugants was confirmed to contain the corresponding plasmid and to degrade 2,4-D (36 transconjugants for pEMT1::lacZ, 14 transconjugants for pJP4::lacZ).
As in the A horizon, poor survival of the plasmid-containing donors was observed, especially for P. putida UWC3 (pEMT1::lacZ). On day 12 the number of this donor had already decreased below the detection limit (Fig. 2c).
Interestingly, unlike the indigenous bacteria in the nonamended A-horizon soil, those in the nonamended B-horizon control soil were not able to degrade any of the 100 mg of 2,4-D per kg over the first 21 days. However, in the B-horizon soils inoculated with P. putida UWC3 (pEMT1::lacZ) and P. putida UWC3 (pJP4::lacZ), the 2,4-D was completely degraded by 19 and 15 days, respectively (Fig. 2d). A second addition of 100 mg of 2,4-D per kg on day 21 was completely degraded in less than 4 days in the soils inoculated with donors, but still no degradation occurred in the noninoculated B-horizon soil (Fig. 2d). Even 68 days after the second amendment of 2,4-D there was still 174 of the initial 200 mg of 2,4-D per kg present in this control soil (data not shown). These results demonstrate a clear success of bioaugmentation in the B horizon.
Revelation of transconjugants by DGGE.
To study the effect of the inoculation of the P. putida donors on the structure of the microbial community, DGGE of 16S rRNA genes was used, based on total soil DNA. More specifically, the aim was to investigate if the transconjugants, detected in high numbers by selective plating (Fig. 2a and c), would also be revealed as members of the numerically dominant populations in soil using a noncultivation-based approach. Since the effect of donor inoculation and subsequent plasmid transfer on 2,4-D degradation was most profound in the nonamended B-horizon soil, this experiment was selected to be studied in more detail.
In the noninoculated nonamended B-horizon soil, the addition of 100 mg of 2,4-D per kg did not cause clear changes in the DGGE profiles (Fig. 3). When the P. putida donors were added to this soil, four bands corresponding to the donor, as shown in the DGGE patterns of strain UWC3, were always very dominant (data not shown). The intensities of these four bands diminished gradually from day 21, and this change was in accordance with the appearance of new dominant bands, as shown for soil with P. putida UWC3 (pEMT1::lacZ) in Fig. 3. These bands remained clearly dominant throughout the experiment, and most were not visible in the patterns of the noninoculated soil.
FIG. 3.
Comparison of the DGGE profiles on days 0, 4, 7, 12, 21, 26, and 34 of 16S rRNA gene fragments amplified from DNAs of the nonamended B horizon (soil DNA) treated with 2,4-D and with or without the donor P. putida UWC3 (pEMT1::lacZ) and from DNAs extracted from a mixture of colonies resuspended from transconjugant selective plates (plate DNA). Bands 4 and 5 are present in the DGGE profile of soil DNA but not in that of plate DNA. Lanes 1 and 24, P. putida UWC3 (pEMT1::lacZ); lanes 2 and 9, native B-horizon soil without 2,4-D or a donor at days 0 and 89, respectively; lanes 3 through 8, soil DNA from noninoculated B-horizon soil plus 2,4-D; lanes 10 through 16, soil DNA from B-horizon soil plus 2,4-D plus pEMT1::lacZ; lanes 17 through 23, plate DNA from transconjugant selective plates. B, B-horizon soil.
To investigate if the new bands observed in the DGGE pattern of the nonamended B-horizon soil, inoculated with P. putida UWC3 (pEMT1::lacZ), corresponded to transconjugants, the DGGE patterns from total soil DNA extracted on days 0, 4, 7, 12, 21, 27, and 34 (soil DNA) were first compared to the DGGE patterns of the mixture of transconjugant colonies resuspended from the transconjugant-selective agar plates (plate DNA) (Fig. 2c). Figure 3 shows that the different DGGE profiles of soil and colony suspensions matched very well. Interestingly, the bands that corresponded with putative transconjugants were detected earlier in the DGGE profiles of the colony suspension than in the DGGE profile from total soil DNA (day 7 compared to day 21). The bands in the DGGE pattern of plate DNA of day 0 and day 4 correspond to false negatives, since these two bands are also visible in the DGGE pattern of soil inoculated with 2,4-D but without the donor strain (Fig. 3), and since on day 0 and day 4 counts of transconjugants on plates did not exceed those of the background of noninoculated control soil (Fig. 2c).
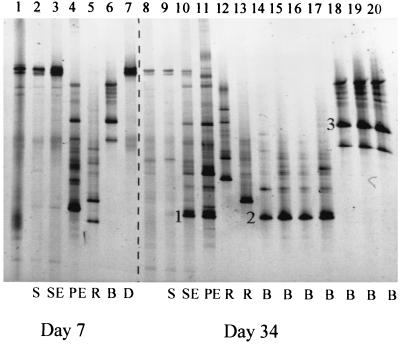
To confirm that the new bands in the DGGE patterns of soil and plate DNA were indeed derived from real transconjugants, the partial 16S rRNA genes of a few purified and confirmed transconjugant colonies with different colony morphologies isolated on days 7 and 34 were loaded on the same DGGE gel (Fig. 4). Most of the DGGE bands in the patterns of soil and plate DNAs corresponded with bands of purified transconjugants. The DNA sequences of two bands from a different origin (soil DNA, lane 10; transconjugant colony, lane 14) but with the same position in the DGGE pattern (see bands 1 and 2 in Fig. 4) were almost identical (99.8% similarity) and had a high nucleotide sequence similarity (96%) with Burkholderia graminis. This clearly indicates that the lowest band in the DGGE profile from soil corresponds with a real transconjugant.
FIG. 4.
Comparison of the DGGE patterns of soil DNA of the B-horizon soil with 2,4-D, with or without the donor P. putida UWC3 (pEMT1::lacZ) (soil DNA), and with DNAs of a mixture of colonies resuspended from transconjugant selective plates (plate DNA) and of single colonies of purified and confirmed transconjugants on days 7 and 34. Bands 1 and 2 are very similar, and their sequences are most related to B. graminis (96%). The sequence of band 3 matched most closely Burkholderia caribensis (96.6%). Lanes 1 and 8, native B-horizon soil without 2,4-D or the donor at days 0 and 89, respectively; lanes 2 and 9, soil DNA from noninoculated B-horizon soil plus 2,4-D on days 7 and 34, respectively (S); lanes 3 and 10, soil DNA from B-horizon soil plus 2,4-D plus pEMT1::lacZ on days 7 and 34, respectively (SE); lanes 4 and 11, plate DNA from transconjugant-selective plates on days 7 and 34, respectively (PE); lanes 5, 6, and 12 through 20, single colonies of purified transconjugants on days 7 and 34, respectively (B, Burkholderia species; R, R. eutropha-like organism); lane 7, P. putida UWC3 (pEMT1::lacZ) (D).
As can be seen in Fig. 4, there were at least four major groups of transconjugants on day 34 based on the position of the brightest band in the DGGE gel (lanes 12, 13, 14 through 17, and 18 through 20). In a separate study, these transconjugants were identified as R. eutropha-like (lanes 12 and 13), B. graminis-like (lanes 14 through 17), and Burkholderia caribensis-like (lanes 18 through 20) organisms (J. Goris, W. Dejonghe, B. Geeraerts, E. De Clerck, B. Hoste, E. M. Top, P. Vandamme, and P. de Vos, unpublished data). These findings were confirmed by sequence analysis of the DGGE bands of two of these transconjugants (Fig. 4, band 2 was 96% similar to B. graminis and band 3 was 96.6% similar to B. caribensis). These results indicate that the plasmid transferred to different species within at least two different genera.
Two additional bands were detected in the DGGE pattern of the soil DNA that were absent in the pattern of plate DNA and also invisible in the pattern of noninoculated soil (bands 4 and 5 in Fig. 3). The sequence of band 4 matched most closely (98%) with Bradyrhizobium sp., while band 5 showed high sequence similarity to a grassland soil clone (98%) (38) and to a Sphingomonas sp. (97%). This may suggest that some transconjugants which may not be detected by plating can be observed by DGGE analysis.
The DGGE patterns of soil and plate DNAs from the nonamended B horizon with P. putida UWC3 (pEMT1::lacZ) were compared with those of the same soil inoculated with the other donor, P. putida UWC3 (pJP4::lacZ). It was clear that the DGGE patterns on day 34 were almost identical for both plasmids and that also with pJP4::lacZ, the DGGE profile of soil DNA contained two additional bands that were not visible in the pattern of the plate DNA (data not shown).
DISCUSSION
Plasmid transfer and a clearly increased rate of 2,4-D degradation were observed when P. putida UWC3 was added as the donor of the 2,4-D degradation plasmids pEMT1::lacZ and pJP4::lacZ to nutrient-amended A- and B-horizon soil (Fig. 1). However, the enhanced 2,4-D degradation was probably mainly due to the activities of the donor strains and not to those of the transconjugants, since by day 2, ca. 50% or more of the added 2,4-D was already degraded, while no transconjugants could yet be detected at that point in time and the donors were still present at ca. 106 CFU/g of soil (data not shown). These findings correspond with the degradation results with sterile nutrient-amended soil, where the two donor strains could degrade 50% of the added 2,4-D in 2 days (data not shown). However, we cannot totally exclude the involvement of the transconjugants in 2,4-D degradation, since their numbers rose drastically between day 2 and day 6, during which the remaining 50 mg of 2,4-D per kg was almost completely degraded.
In contrast, in the nonamended B-horizon soil inoculated with P. putida UWC3 (pEMT1::lacZ), the observed 2,4-D degradation must have been due to transconjugants, since they were already detected when the 2,4-D degradation started at day 7 and since most of the P. putida UWC3 donor cells had already lost their plasmid by then (Fig. 2c and d). This conclusion is supported by the results obtained in sterile-soil experiments, where P. putida UWC3 (pEMT1::lacZ) was not able to degrade 2,4-D due to the instability of its plasmid (data not shown). With plasmid pJP4::lacZ, the observed disappearance of 2,4-D in the nonamended B-horizon soil was probably due to both transconjugants and donors, since P. putida UWC3 (pJP4::lacZ) was still present at 104 CFU/g of soil when 2,4-D degradation started and was able to degrade 2,4-D in sterile soil (100 mg/kg in 21 days). Since the numbers of transconjugants of both plasmids increased dramatically while the 2,4-D was degraded, they were probably involved in the degradation in both cases.
The more striking effect of bioaugmentation in the B horizon compared to that in the A horizon was due to the poor 2,4-D degradation capacity of the indigenous bacteria in this deeper soil layer (Fig. 1 and 2d). A similar decrease in 2,4-D degradation activity with depth was observed by Veeh et al. (60) in a silt loam and silt clay soil. This was explained by the fact that microbial plate counts, which are positively correlated with the soil organic carbon content, declined as a function of depth. In our study, the total plate count of the B-horizon soil was not different from that of the A-horizon soil, but its microbial activity, as shown by the respiration rate (1.97 mg of CO2-C · kg of soil−1 · day−1 compared to 0.52 mg of CO2-C · kg−1 · day−1), was clearly lower. The microbial capacity to degrade 2,4-D seemed to be present in the B horizon of our soil since 2,4-D was degraded in the nutrient-amended soil after a lag period of more than 22 days. Audus (5) and Veeh et al. (60) also observed this lag phase, which increased with depth. They characterized it as a period of adaptation during which the enzymes needed for decomposition of the substrate and its metabolites are synthesized.
The transfer of the 2,4-D degradation plasmid pEMT1 in nutrient-amended soil has been shown before by our group, first with E. coli XL1 Blue (pEMT1k) as a donor strain (54). Transconjugants were detected at numbers similar to those found in this study, and the transfer also resulted in enhanced degradation of 2,4-D. A more recent study showed transfer of plasmid pJP4 from an E. coli strain to different indigenous bacteria in some but not all tested soils and in the presence of 500 or 1,000 mg of 2,4-D per kg. The effect of transfer on accelerated 2,4-D degradation was very small and shown in only one of the four soils studied (42). Since E. coli is not a suitable strain for bioremediation purposes, Top et al. (55) extended their earlier study (54) using the same soil without nutrients and with P. putida UWC3 as the donor of plasmid pEMT1k. They found a transfer rate that was similar to that found in the present study and also observed an enhanced degradation of 2,4-D. However, since the donor strain was still present when degradation started (104 CFU/g of soil), its involvement in the degradation of 2,4-D could not be totally excluded. As far as we know, the present study is the first one to demonstrate so clearly that bioaugmentation can be effective in soils with low microbial activity, such as those of the B horizon.
It was clear that the transfer rate of the plasmids was independent of the kind of plasmid (pEMT1 or pJP4) and soil (A or B horizon) used and that the addition of nutrients to the soil did not increase the number of transconjugants. The latter observation is in contrast with the results of many studies, such as those of Top et al. (51) and Götz and Smalla (26), who showed that addition of nutrients to soil enhanced the number of transconjugants per gram of soil. A possible explanation why the transconjugant numbers in the nutrient-amended soil were not higher than in the nonamended soil is that 2,4-D was degraded much faster in the nutrient-amended soil. Due to this, the stimulating effect of 2,4-D on the transconjugants was diminished. Top et al. (54) showed that few or no transconjugants with plasmids pEMT1k and pEMT3k were detected in soil without 2,4-D, compared to the high numbers in 2,4-D-contaminated soil. Also, Neilson et al. (41) and Newby et al. (42) could detect the transfer of plasmid pJP4 from an introduced donor to the indigenous bacteria of nonsterile soil only when 2,4-D was added.
From the comparison of DGGE patterns and the sequence information it can be concluded that the new bands in the DGGE pattern of the B-horizon soil, first visible on day 21, most probably correspond to transconjugants. The fact that the formation of transconjugants could be revealed by DGGE analysis means that these transconjugants formed very dominant populations in this soil. This was also clear by plating, since the transconjugant numbers at day 21 were ca. 107 CFU/g of soil, as high as the total plate count. Interestingly, transconjugants were not yet detected in the DGGE profile of soil DNA on day 7, when ca. 105 CFU/g of soil was counted by plating. Since the DGGE pattern of soil DNA represents a profile of the numerically dominant populations in the soil, transconjugants can be visualized only if they are present in sufficiently high numbers. These high numbers were due rather to growth of transconjugants than to additional plasmid transfer into new species, since no change occurred in the DGGE patterns of soil and plate DNAs as a function of time.
The heterogeneity of the 16S rRNA genes, observed in the P. putida UWC3 strain and in some transconjugants, has been reported previously for other species (9, 43, 45). Also, the clear visibility of the DGGE bands of the P. putida UWC3 inoculum corresponds with the results of other studies that were able to track inocula in soil, aquifers, and sludge (18, 19, 49) by their prominent DGGE band(s). The strong DGGE signal of the donor could be explained by the fact that the initial donor concentration represented ca. 10% of the total heterotrophic plate count of the B-horizon soil. Over time, the number of donors decreased while those of transconjugants increased, resulting in dominant transconjugant bands and faint-to-invisible donor bands in the DGGE pattern. Also, Stephen et al. (49) found that DGGE analysis of amplified 16S rRNA gene fragments from soil was a useful method of tracking the survival of introduced bacteria even though they provided only 2% of the viable biomass.
The possibility that some transconjugants which may not be detected by plating can be observed by DGGE analysis of 16S rRNA is in agreement with the conclusions of other authors who found that certain bacteria which do not grow on synthetic media can be visualized by 16S rRNA analysis (4). The strain most related to Bradyrhizobium sp. may indeed have acquired the 2,4-D degradation plasmids pEMT1 and pJP4 since Kinkle et al. (34) have demonstrated transfer of plasmid pJP4 between B. japonicum USDA438 and several Bradyrhizobium sp. strains in nonsterile soil.
The very similar DGGE profiles of the B-horizon soil inoculated with either pEMT1::lacZ or pJP4::lacZ indicate that the numerically dominant transconjugants of each plasmid belonged to the same species. This finding is in agreement with the identification of several transconjugants isolated on agar plates, which revealed that most transconjugants were Burkholderia species and R. eutropha-like organisms, when both pEMT1::lacZ and pJP4::lacZ were used (J. Goris, W. Dejonghe, B. Geeraerts, E. De Clerck, B. Hoste, E. M. Top, P. Vandamme, and P. de Vos, unpublished data). This suggests that the host ranges for transfer and expression of the degradative genes in soil are very similar for both plasmids. Transconjugants of plasmids pEMT1 and pJP4 obtained in other transfer studies of surface soils (A horizon) were also identified as Ralstonia or Burkholderia species (15, 42, 54, 55). Since the transfer range of pJP4 is a lot broader than these two genera (16), the limited diversity of transconjugants found in soil, when growth on 2,4-D as the sole C source was selected for, must be due to the limited abilities of several transconjugants to mineralize 2,4-D.
In conclusion, bioaugmentation of a 2,4-D-contaminated soil was very successful in soil from the B horizon, where the indigenous community could not, or could only very slowly, degrade the herbicide. With plasmid pEMT1::lacZ, this success must be due to the transfer of the plasmid to the indigenous bacteria. To our knowledge, such a clear effect of bioaugmentation through the spread of natural catabolic genes in a soil community has not yet been reported in the literature. In addition, DGGE analysis of the soil 16S rRNA gene pool was used to show that the in situ formation and subsequent proliferation of high numbers of 2,4-D-degrading transconjugants caused clear changes in the soil microbial community structure.
ACKNOWLEDGMENTS
This work was supported by a research grant of the Flemish Fund for Scientific Research (FWO-Vlaanderen), by a project G.O.A. (1997 to 2002) of the Ministerie van de Vlaamse Gemeenschap, Bestuur Wetenschappelijk Onderzoek, and by the EU-concerted action MECBAD. E. M. Top and P. De Vos are indebted to the FWO-Vlaanderen for the positions of Research Associate and Research Director, respectively.
We thank S. De Neve for providing the soils and their characteristics, I. De Laere for technical assistance, and N. Boon, R. Kozak, J. Robbens, and R. Wouters for their useful suggestions on the manuscript.
REFERENCES
- 1.Akkermans A D L. Application of bacteria in soils: problems and pitfalls. FEMS Microbiol Rev. 1994;15:185–194. [Google Scholar]
- 2.Alexander M. Inoculation. In: Alexander M, editor. Biodegradation and bioremediation. San Diego, Calif: Academic Press; 1994. pp. 226–247. [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Audus L J. Microbiological breakdown of herbicides in soils. In: Woodford E K, Sagar G R, editors. Herbicides and the soil. Oxford, United Kingdom: Blackwell; 1960. pp. 1–19. [Google Scholar]
- 6.Barles R W, Daughton C G, Hsieh P H. Accelerated parathion degradation in soil inoculated with acclimated bacteria under field conditions. Arch Environ Contam Toxicol. 1979;8:647–660. doi: 10.1007/BF01054867. [DOI] [PubMed] [Google Scholar]
- 7.Bhat M A, Tsuda M, Horiike K, Nozaki M, Vaidyanathan C S, Nakazawa T. Identification and characterization of a new plasmid carrying genes for degradation of 2,4-dichlorophenoxyacetate from Pseudomonas cepacia CSV90. Appl Environ Microbiol. 1994;60:307–312. doi: 10.1128/aem.60.1.307-312.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brokamp A, Schmidt F R J. Survival of Alcaligenes xylosoxidans degrading 2,2-dichloropropionate and horizontal transfer of its halidohydrolase gene in a soil microcosm. Curr Microbiol. 1991;22:299–306. [Google Scholar]
- 9.Cilia V, Lafay B, Christen R. Sequence heterogeneities among 16S ribosomal RNA sequences, and their effect on phylogenetic analyses at the species level. Mol Biol Evol. 1996;13:451–461. doi: 10.1093/oxfordjournals.molbev.a025606. [DOI] [PubMed] [Google Scholar]
- 10.Clark C G, Wright S J L. Detoxification of isopropyl N-phenylcarbamate (IPC) and isopropyl N-3-chlorophenylcarbamate (CIPC) in soil, and isolation of IPC-metabolizing bacteria. Soil Biol Biochem. 1970;2:19–27. [Google Scholar]
- 11.Crowley D E, Brennerova M V, Irwin C, Brenner V, Focht D D. Rhizosphere effects on biodegradation of 2,5-dichlorobenzoate by a bioluminescent strain of root-colonizing Pseudomonas fluorescens. FEMS Microbiol Ecol. 1996;20:79–89. [Google Scholar]
- 12.Daane L L, Häggblom M M. Earthworm egg capsules as vectors for the environmental introduction of biodegradative bacteria. Appl Environ Microbiol. 1999;65:2376–2381. doi: 10.1128/aem.65.6.2376-2381.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Rore H, Demolder K, De Wilde K, Top E, Houwen F, Verstraete W. Transfer of the catabolic plasmid RP4::Tn4371 to indigenous soil bacteria and its effect on respiration and biphenyl breakdown. FEMS Microbiol Ecol. 1994;15:71–78. [Google Scholar]
- 15.DiGiovanni G D, Neilson J W, Pepper I L, Sinclair N A. Gene transfer of Alcaligenes eutrophus JMP134 plasmid pJP4 to indigenous soil recipients. Appl Environ Microbiol. 1996;62:2521–2526. doi: 10.1128/aem.62.7.2521-2526.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Don R H, Pemberton J M. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J Bacteriol. 1981;145:681–686. doi: 10.1128/jb.145.2.681-686.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dröge M, Pühler A, Selbitschka W. Horizontal gene transfer among bacteria in terrestrial and aquatic habitats as assessed by microcosm and field studies. Biol Fertil Soils. 1999;29:221–245. [Google Scholar]
- 18.Dybas M J, Barcelona M, Bezborodnikov S, Davies S, Forney L, Heuer H, Kawka O, Mayotte T, Sepulveda-Torres L, Smalla K, Sneathen M, Tiedje J, Voice T, Wiggert D C, Witt M E, Criddle C S. Pilot-scale evaluation of bioaugmentation for in-situ remediation of a carbon tetrachloride-contaminated aquifer. Environ Sci Technol. 1998;32:3598–3611. [Google Scholar]
- 19.Eichner C A, Erb R W, Timmis K N, Wagner-Döbler I. Thermal gradient gel electrophoresis analysis of bioprotection from pollutant shocks in the activated sludge microbial community. Appl Environ Microbiol. 1999;65:102–109. doi: 10.1128/aem.65.1.102-109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El Fantroussi S, Mahillon J, Naveau H, Agathos S N. Introduction and PCR detection of Desulfomonile tiedjei in soil microcosms. Biodegradation. 1997;8:125–133. doi: 10.1023/a:1008262426800. [DOI] [PubMed] [Google Scholar]
- 21.El Fantroussi S, Belkacemi M, Top E M, Mahillon J, Naveau H, Agathos S N. Bioaugmentation of a soil bioreactor designed for pilot-scale anaerobic bioremediation studies. Environ Sci Technol. 1999;33:2992–3001. [Google Scholar]
- 22.El Fantroussi S, Verschuere L, Verstraete W, Top E M. Effect of phenylurea herbicides on soil microbial communities estimated by analysis of 16S rRNA gene fingerprints and community-level physiological profiles. Appl Environ Microbiol. 1999;65:982–988. doi: 10.1128/aem.65.3.982-988.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Figurski D H, Helinski D R. Replication of an origin containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fomsgaard I S. Degradation of pesticides in subsurface soils, unsaturated zone—a review of methods and results. Int J Environ Anal Chem. 1995;58:231–245. [Google Scholar]
- 25.Goldstein R M, Mallory L M, Alexander M. Reasons for possible failure of inoculation to enhance biodegradation. Appl Environ Microbiol. 1985;50:977–983. doi: 10.1128/aem.50.4.977-983.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Götz A, Smalla K. Manure enhances plasmid mobilization and survival of Pseudomonas putida introduced into field soil. Appl Environ Microbiol. 1997;63:1980–1986. doi: 10.1128/aem.63.5.1980-1986.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gstalder M E, Faelen M, Vuye A, Couturier M. First Symposium of the EU-Concerted Action on “Mobile Elements' Contribution to Bacterial Adaptability and Diversity” (MECBAD). Magdeburg, Germany: Scriptum Verlag Magdeburg GmbH; 1999. Spread of IncP plasmids in environment: a reality? p. 33. [Google Scholar]
- 28.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Microb Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 29.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hill K E, Weightman A J, Fry J C. Isolation and screening of plasmids from the epilithon which mobilize recombinant plasmid pD10. Appl Environ Microbiol. 1992;28:1292–1300. doi: 10.1128/aem.58.4.1292-1300.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill K E, Top E M. Gene transfer in soil systems using microcosms. FEMS Microbiol Ecol. 1998;25:319–329. [Google Scholar]
- 32.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kilbane J J, Chatterjee D K, Chakrabarty A M. Detoxification of 2,4,5-trichlorophenoxyacetic acid from contaminated soil by Pseudomonas cepacia. Appl Environ Microbiol. 1983;45:1697–1700. doi: 10.1128/aem.45.5.1697-1700.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinkle B K, Sadowsky M J, Schmidt E L, Koskinen W C. Plasmids pJP4 and r68.45 can be transferred between populations of Bradyrhizobia in nonsterile soil. Appl Environ Microbiol. 1993;59:1762–1766. doi: 10.1128/aem.59.6.1762-1766.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loos M A. Indicator media for microorganisms degrading chlorinated pesticides. Can J Microbiol. 1975;21:104–107. doi: 10.1139/m75-016. [DOI] [PubMed] [Google Scholar]
- 36.Maë A A, Marits R O, Ausmees N R, Kôiv V M, Heinaru A L. Characterization of a new 2,4-dichlorophenoxyacetic-acid degrading plasmid pEST4011: physical map and localization of catabolic genes. J Gen Microbiol. 1993;139:3165–3170. [Google Scholar]
- 37.Matheson V G, Forney L J, Suwa Y, Nakatsu C H, Sextone A J, Holben W E. Evidence for acquisition in nature of a chromosomal 2,4-dichlorophenoxyacetic acid/α-ketoglutarate dioxygenase gene by different Burkholderia spp. Appl Environ Microbiol. 1996;62:2457–2463. doi: 10.1128/aem.62.7.2457-2463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCaig A E, Glover L A, Prosser J I. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pasture. Appl Environ Microbiol. 1999;65:1721–1730. doi: 10.1128/aem.65.4.1721-1730.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mergeay M, Springael D. Conjugation-mediated gene transfer in bacterial strains to be used for bioremediation. Methods Biotechnol. 1997;2:153–167. [Google Scholar]
- 40.Muyzer G, de Waal E C, Uitterlinden A. Profiling of complex microbial populations using denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neilson J W, Josephson K L, Pepper I L, Arnold R B, DiGiovanni G D, Sinclair N A. Frequency of horizontal gene transfer of a large catabolic plasmid (pJP4) in soil. Appl Environ Microbiol. 1994;60:4053–4058. doi: 10.1128/aem.60.11.4053-4058.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newby D T, Josephson K L, Pepper I L. Detection and characterization of plasmid pJP4 transfer to indigenous soil bacteria. Appl Environ Microbiol. 2000;66:290–296. doi: 10.1128/aem.66.1.290-296.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nübel U, Engelen B, Felske A, Snaider J, Wieshuber A, Amann R, Ludwig W, Backhaus H. Sequence heterogeneities of genes encoding 16S RNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol. 1996;178:5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paul E A, Clark F E. Methods for studying soil microorganisms. In: Paul E A, Clark F E, editors. Soil microbiology and biochemistry. San Diego, Calif: Academic Press; 1989. pp. 62–63. [Google Scholar]
- 45.Rainey F A, Ward-Rainey N L, Janssen P H, Hippe H, Stackebrandt E. Clostridium paradoxum DSM 7308T contains multiple 16S rRNA genes with heterogeneous intervening sequences. Microbiology. 1996;142:2087–2095. doi: 10.1099/13500872-142-8-2087. [DOI] [PubMed] [Google Scholar]
- 46.Ramadan M A, El-Tayeb O M, Alexander M. Inoculum size as a factor limiting success of inoculation for biodegradation. Appl Environ Microbiol. 1990;56:1392–1396. doi: 10.1128/aem.56.5.1392-1396.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 48.Stanier R Y, Palleroni N J, Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966;43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 49.Stephen J R, Chang Y-J, MacNaughton S J, Whitaker S L, Hicks C L, Leung K T, Flemming C A, White D C. Fate of a metal-resistant inoculum in contaminated and pristine soils assessed by denaturing gradient gel electrophoresis. Environ Toxicol Chem. 1999;18:1118–1123. [Google Scholar]
- 50.Suwa Y, Wright A D, Fukimore F, Nummy K A, Hausinger R P, Holben W E, Forney L J. Characterization of a chromosomally encoded 2,4-dichlorophenoxyacetic acid/α-ketoglutarate dioxygenase from Burkholderia sp. strain RASC. Appl Environ Microbiol. 1996;62:2462–2469. doi: 10.1128/aem.62.7.2464-2469.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Top E, Mergeay M, Springael D, Verstraete W. Gene escape model: transfer of heavy metal resistance genes from Escherichia coli to Alcaligenes eutrophus on agar plates and in soil samples. Appl Environ Microbiol. 1990;56:2471–2479. doi: 10.1128/aem.56.8.2471-2479.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Top E M, Holben W E, Forney L J. Characterization of diverse 2,4-dichlorophenoxyacetic acid-degradative plasmids isolated from soil by complementation. Appl Environ Microbiol. 1995;61:1691–1698. doi: 10.1128/aem.61.5.1691-1698.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Top E M, Maltseva O V, Forney L J. Capture of a catabolic plasmid that only encodes a 2,4-dichlorophenoxyacetic acid:α-ketoglutaric acid dioxygenase (TfdA) by genetic complementation. Appl Environ Microbiol. 1996;62:1470–1476. doi: 10.1128/aem.62.7.2470-2476.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Top E M, Van Daele P, De Saeyer N, Forney L J. Enhancement of 2,4-dichlorophenoxyacetic acid (2,4-D) degradation in soil by dissemination of catabolic plasmids. Antonie Leeuwenhoek Int J G. 1998;73:87–94. doi: 10.1023/a:1000663619522. [DOI] [PubMed] [Google Scholar]
- 55.Top E M, Maila M P, Clerinx M, Goris J, De Vos P, Verstraete W. Methane oxidation as a method to evaluate the removal of 2,4-dichlorophenoxyacetic acid (2,4-D) from soil by plasmid mediated bioaugmentation. FEMS Microbiol Ecol. 1999;28:203–213. [Google Scholar]
- 56.Vallaeys T, Fulthorpe R R, Wright A M, Soulas G. The metabolic pathway of 2,4-dichlorophenoxyacetic acid degradation involves different families of tfdA and tfdB genes according to PCR-RFLP analysis. FEMS Microbiol Ecol. 1996;20:163–172. [Google Scholar]
- 57.Van De Steene J. Het volgen van de NO3− N-dynamiek in bodems via time domain reflectrometry (TDR). Engineer's thesis. Ghent, Belgium: Ghent University; 1998. [Google Scholar]
- 58.van Elsas J D, Heijnen C E. Methods for the introduction of bacteria in soil: a review. Biol Fertil Soils. 1990;10:127–133. [Google Scholar]
- 59.van Veen J A, van Overbeek L S, van Elsas J D. Fate and activity of microorganisms introduced into soil. Microbiol Mol Biol Rev. 1997;61:121–135. doi: 10.1128/mmbr.61.2.121-135.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Veeh R H, Inskeep W P, Camper A K. Soil depth and temperature effects on microbial degradation of 2,4-D. J Environ Qual. 1996;25:5–12. [Google Scholar]