Summary
The analysis of morphological features of neurons derived from human pluripotent stem cells (hPSCs) is important to describe neuronal phenotypes and changes observed throughout development. Using free and easily accessible tools, we describe a protocol for the morphometric quantification of hPSCs-derived neurons in two- and three-dimensions in vitro cultures. We detail the analysis of soma area and main and secondary dendrites lengths of GFP-transfected neurons and the measurement of area and perimeter of immunostained neurospheres.
Subject areas: Cell Biology, Microscopy, Molecular Biology, Neuroscience, Stem Cells, Cell Differentiation
Graphical abstract
Highlights
-
•
Transfection of neurons derived from human pluripotent stem cells (hPSCs)
-
•
Generation of neurospheres from neural stem cells (NSCs) derived from hPSCs
-
•
Morphometric analysis of soma area and dendrites length of GFP-transfected neurons
-
•
Measurement of area and perimeter of immunostained neurospheres
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
The analysis of morphological features of neurons derived from human pluripotent stem cells (hPSCs) is important to describe neuronal phenotypes and changes observed throughout development. Using free and easily accessible tools, we describe a protocol for the morphometric quantification of hPSCs-derived neurons in two- and three-dimensions in vitro cultures. We detail the analysis of soma area and main and secondary dendrites lengths of GFP-transfected neurons and the measurement of area and perimeter of immunostained neurospheres.
Before you begin
Human pluripotent stem cells (hPSCs), which include embryonic and induced pluripotent stem cells (hESCs and hiPSCs, respectively), can differentiate into a wide range of specialized cells, thus emerging as a valuable model to study human development and diseases in a dish.
Neurons derived from hPSCs have become a powerful tool for studying neural mechanisms, including homeostasis, development, and neuronal stress. The structural features of a neuron are important for the fulfillment of the different functions that neurons must carry out in each biological process and, for this reason; the generation of a morphometric quantification protocol is extremely helpful when studying these neurons under different conditions, either at different stages of differentiation or under conditions of neuronal stress.
There are available different published procedures for neuronal morphological quantification that mostly rely on mouse primary cultures or histological preparations (Billeci et al., 2013). In this protocol we combine the easy and simple quantification of morphometric changes of neurons differentiated from hPSCs by a reliable step-by-step procedure that can be performed and especially useful to any new to the field researcher. In particular, we detailed a 2-dimension (2D) differentiation protocol that allows the quantification of soma area and main and secondary dendrites lengths of the obtained neurons and a 3-dimension (3D) differentiation protocol, which permits the measurement of neurospheres area and perimeter.
Neural stem cells generation, maintenance and cryopreservation
Timing: 15 days
There are several available protocols to derive neural stem cells (NSCs) from hPSCs. In particular, we followed the protocol described by Yan (Yan et al., 2013), which allows an easily reproducible generation of NSCs from hPSCs, by using a commercial kit from Thermo Scientific (PSC Neural Induction Medium, Gibco). The NSCs were obtained and validated following manufacturer’s instructions. For this protocol it is mandatory to start from good quality hPSCs. The cells should be arranged in colonies with defined borders and must express high levels of the pluripotency markers OCT-4, NANOG, SSEA4 and TRA-1-80. This protocol should be started when hPSCs growing in mTeSR1 medium in a 37°/5% CO2 incubator chamber reach 70–80% confluency. Prior to use, warm mTeSR1 to 20°C–25°C. Do not leave mTeSR1 medium at 20°C–25°C for longer than 2 h per day and avoid exposure to light to avoid degradation of medium components.
Although we present a NSCs protocol workflow, we highly recommend accessing the manufacturer’s manual with all the specifications and troubleshooting (MAN0008031, Thermo Scientific, https://assets.thermofisher.com/TFS-Assets/LSG/manuals/MAN0008031.pdf).
Preparing hPSCs for the neural induction
-
1.
Remove old medium (1 mL of mTeSR1 medium) from each well of a 6-well cell culture plate.
-
2.
Wash hPSCs cells with 1 mL of warm (37°C) PBS 1×.
-
3.
Incubate hPSCs with 1 mL of Accutase 1× for 7 min at 37°C.
-
4.
Dilute Accutase 1× with 3 mL of PBS 1×, and gently detach and transfer hPSCs into a 15 mL conical tube using a 5- or 10-mL serological plastic pipette.
-
5.
Centrifuge hPSCs at 200 × g for 5 min and remove the supernatant.
-
6.
Determine cell number using your preferred method.
-
7.
Plate hPSCs at a density of 2.5 × 105–3 × 105 into each well of Geltrex-coated 6-well cell culture plates (add 1 mL of 1/100 Geltrex diluted in DMEM/F12 per well and incubate for 1 h at 37°C) in a final volume of 1 mL mTeSR1 medium per well containing 10 μM of ROCK inhibitor Y27632.
Neural induction of hPSCs
-
8.
Day 1 of the Neural Induction: remove mTeSR1 medium with ROCK inhibitor by aspiration and place 2.5 mL of pre-warmed (37°C) complete PSC Neural Induction Medium in each well of a 6-well cell culture well.
Note: Neural Induction Medium consists in Neurobasal medium containing 2% of Neural Induction Supplement provided by the manufacturer. This medium can be stored at 4°C protected from the light for up to 2 weeks. Do not warm the medium at 37°C longer than 10 minutes to preserve the biological activity of the morphogens present in the supplement.
-
9.
Incubate at 37°C/5% CO2 and change to 2.5 mL of fresh Neural Induction Medium every other day.
CRITICAL: It is very important to eliminate by aspiration cells that do not exhibit a neural phenotype throughout the protocol to obtain high-purity NSCs cultures.
NSCs expansion
-
10.
On day 7 of the protocol NSCs (Passage 0) are ready to be expanded.
-
11.
Wash NSCs with 1 mL of warm (37°C) DPBS 1× containing calcium and magnesium.
-
12.
Incubate NSCs with 1 mL of Accutase 1× for 7 min at 37°C.
-
13.
Dilute Accutase 1× with 3 mL of DPBS 1× with calcium and magnesium, and gently detach and transfer cells into a 15 mL conical tube using a 5- or 10-mL serological plastic pipette.
-
14.
Centrifuge NSCs at 300 × g for 5 min and remove the supernatant.
-
15.
Aspirate the supernatant and re-suspend NSCs with 1 mL of pre-warmed (37°C) complete Neural Expansion Medium.
Note: Neural Expansion Medium consists in 49% of Neurobasal medium and 49% Advanced DMEM-F12 containing 2% of Neural Induction Supplement provided by the manufacturer. This medium can be stored at 4°C protected from the light for up to 2 weeks. Do not warm the medium at 37°C longer than 10 minutes to preserve the biological activity of the growth factors present in the supplement.
-
16.
Determine cell number using your preferred method.
-
17.
Plate NSCs at a density of 0.5 × 105–1 × 105 cell/cm2 into each well of Geltrex-coated 6-well cell culture plates.
-
18.
Add ROCK inhibitor Y27632 to a final concentration of 5 μM to prevent cell death.
-
19.
Incubate at 37°C/5% CO2 and change to 2.5 mL of fresh Neural Expansion Medium every other day until NSCs culture reaches 100% of confluency.
CRITICAL: You need to confirm that NSCs were correctly generated. To evaluate the phenotype of the NSCs confirm the expression of the following markers: the SRY-like HMG-box 1 and 2 (SOX-1 and SOX-2) transcription factors, the class VI intermediate filament protein NESTIN and the paired box 6 (PAX-6) transcription factor by immunostaining. For a more information about how to validate the generated NSCs please refer to Yan et al. publication (Yan et al., 2013).
NSCs passage and maintenance
NSCs are grown as monolayers in NSCs medium at 37°C in an incubator with a humidified atmosphere of 5% CO2 in air. An optimal management of this culture is important in order to achieve good quality neuronal cultures upon 2D or 3D (neurospheres formation) neuronal differentiation.
-
20.
Remove 2.5 mL of medium from each well of a 6-well cell culture plate.
-
21.
Wash NSCs with 1 mL of warm (37°C) PBS 1×.
-
22.
Incubate NSCs with 500 μL of Accutase 1× for 7 min at 37°C.
-
23.
Dilute Accutase 1× with 1 mL of PBS 1×, and gently detach and transfer cells into a 15 mL conical tube using a 5- or 10-mL serological plastic pipette.
-
24.
Centrifuge NSCs at 200 × g for 5 min and remove the supernatant.
-
25.Plate cells.
-
a.For culture maintenance, plate NSCs at a density of 1 × 105 cells/cm2 into each well of a Geltrex-coated 6-well cell culture plate (add 1 mL of 1/100 Geltrex diluted in DMEM/F12 per well and incubate for 1 h at 37°C) in a final volume of 2.5 mL NSCs medium per well.
-
b.For 2D Neuronal differentiation or 3D Neurospheres obtention, look at the corresponding sections below.
-
a.
-
26.
Incubate at 37°C/5% CO2 and change to 2.5 mL fresh NSCs medium every other day and store NSC medium in the dark at 2°C–8°C.
NSCs cryopreservation
-
27.
Remove 2.5 mL of medium from each well of a 6-well cell culture plate.
-
28.
Wash NSCs with 1 mL of warm (37°C) PBS 1×.
-
29.
Incubate NSCs with 500 μL of Accutase 1× for 7 min at 37°C.
-
30.
Dilute Accutase 1× with 1 mL of PBS 1×, and gently detach and transfer cells into a 15 mL conical tube using a 5- or 10-mL serological plastic pipette.
-
31.
Centrifuge NSCs at 200 × g for 5 min and remove the supernatant.
-
32.
Determine the cell number using your preferred method.
-
33.
Cryopreserve NSCs in a concentration of 2 × 106–4 × 106 cells/mL. Final volume per cryotube is 1 mL of NSCs Medium containing 10% of DMSO and 5 μM of ROCK inhibitor Y27632.
-
34.
Place the cryotubes containing the cells in a freezing chamber filled with isopropanol and stored them at −80°C ON.
-
35.
Transfer the frozen cells into a liquid nitrogen tank for long-term storage.
Escherichia coli calcium chloride competent cell protocol
Timing: 1 day
To obtain high transformation efficiency, it is crucial that E. coli growth is in the mid-log phase at the time of harvest, which generally occurs at OD600 between 0.4 and 0.9, with the optimal value depending on the culture volume, strain, and protocol. In all steps, care must be taken to use sterile tools and labware, media, and reagents where appropriate or required.
-
36.
Inoculate a single colony into 25 mL LB in a 250 mL bottle in the morning.
-
37.
Shake at 37°C for approximately 4–6 h to reach mid-log phase (OD600 between 0.4 and 0.9).
-
38.
Put the E. coli cells on ice for 10 min (keep at 0°C–4°C from now on).
-
39.
Collect the cells by centrifugation for 3 min at 2,500 × g. Decant supernatant and gently resuspend on 10 mL cold 0.1 M CaCl2 (cells are susceptible to mechanical disruption, so handle them carefully).
-
40.
Incubate on ice for 20 min.
-
41.
Centrifuge as in 39.
-
42.
Discard supernatant and gently resuspend on 5 mL cold 0.1 M CaCl2/15% Glycerol.
-
43.
Dispense in microtubes (300 μL/tube). Freeze in - 80°C for up to 6 months.
Note: There are many different aspects of the biology of E. coli, which will provide important parameters when it comes to choosing the right E. coli strain for transformation. Information regarding strain specific characteristics can be found in Escherichia coli host strains (Casali, 2003).
Plasmid amplification and purification for neuronal transfection
Timing: 3 days
Morphometric analysis is performed in GFP-expressing neurons. Neurons are transfected with an eGFP-encoding plasmid, a fluorescent marker that allows the visualization of cell morphology under fluorescence microscope, and further perform the desired morphometric measurements. Plasmid # 6085-1 available from Clontech (https://www.addgene.org/vector-database/2491/) is used. Other alternative plasmids from Addgene are pcDNA3-EGFP (#13031), EGFP (#176015) or CMV-eGFP-Z1 (#135049). All these plasmids use a strong CMV promoter, which is suitable for a sustained expression of fluorescent reporters in neurons.
-
44.Day 1: Plasmid transformation into competent E. coli cells.
-
a.Thaw competent Escherichia coli on ice and cooling the 1.5 mL microtubes that will be used in transformation.
-
b.Spray down the benchtop with a 70% ethanol solution and wipe down with a paper towel.
-
c.Set up a Bunsen burner and work slowly, carefully, and deliberately within the sterile field area created by the updraft of the flame.
 CRITICAL: It is important to keep competent cells cold and minimize the time that cultures and growth media are open to the environment.
CRITICAL: It is important to keep competent cells cold and minimize the time that cultures and growth media are open to the environment. -
d.Add 10 ng of plasmid into 50 μL of competent E. coli in 1.5 mL ice cold microtubes and mix gently.
-
e.Incubate the mixture on ice for 30 min.
-
f.Heat shock at 42°C for 90 s.
-
g.Incubate the mixture on ice for 5 min.
-
h.Add 500 μL of LB medium to the mixture.
-
i.Incubate the mixture at 37°C for 30 min.
-
j.Pre-warm LB agar plates with ampicillin (100 μg/μL) at 20°C–25°C for 1 h.
-
k.Spread the transformed cells onto the LB agar with ampicillin plates.
-
l.Incubate the plates at 37°C ON (16–18 h).Note: We suggest a control for untransformed competent cells by plating on antibiotic-free LB-agar to evaluate bacterial viability.Alternatives: This protocol can be performed using different strains of E. coli such as STBL3, DH5⍺, TOP10, BL21.
-
a.
-
45.
Day 2: select a single colony and culture it in a flask containing 250 mL of LB with 100 μg/μL ampicillin.
-
46.
Grow it shaking at 200 rpm at 37°C ON (16–18 h).
Note: We suggest incubating an antibiotic control containing 5 mL of LB with 100 μg/μL ampicillin in a 15 mL conical tube with untransformed bacterial cells.
-
47.
Day 3: transfer the bacterial culture to 50 mL conical tubes (or a 400 mL wide neck bottle) and centrifuge them at 3,200 × g for 15 min.
-
48.
Discard the supernatant and keep the pellet.
Pause point: At this point, the protocol can be paused. Store pellets up to 1 week at −20°C or up to 1 month at −80°C to continue with the plasmid extraction later.
-
49.
Purify the amplified GFP-encoding plasmid using commercially available purification systems such as plasmid Wizard® Plus Midipreps DNA Purification System #A760, QIAGEN plasmid purification Midi Kit #12145 or Genopure Plasmid Midi Kit Roche #03143414001.
-
50.
Finally, quantify plasmid concentration with your preferred method such as NanoDrop, QUBIT or quantification by agarose gel electrophoresis.
Note: Plasmid concentration must be greater than 500 ng/μL because high concentrations of plasmids are required to transfect eukaryotic cells. Verify that 260/280 and 260/230 ratio of the plasmid is in the appropriate range for each ratio. The absorption at 260 nm correlates with the concentration of nucleotides. Absorption at 280 nm gives a measure of residual protein. The A260/A280 ratio is a measure of purification; it should fall in the range 1.8–2.0. The ratio of absorbance at 260 and 230 nm can be used as a secondary measure of DNA. Absorbance at 230 nm is accepted as being the result of unwanted contamination such as Trizol, phenol, Guanidine HCL and guanidine thiocyanate. The 260/230 values for “pure” nucleic acid are often higher than the respective 260/280 values. Expected 260/230 values are commonly in the range of 2.0–2.2.
Key resource table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Alexa Fluor 488 goat anti-mouse IgG (1:400) | Invitrogen | A11001 |
| Anti-Tubulin beta III (Tuj-1) (1:1000) | Covance | MMS-435P |
| Bacterial strains | ||
| One Shot Stbl3 Chemically Competent E. coli | Invitrogen | C7373-03 |
| Chemicals, peptides, and recombinant proteins | ||
| Agar Bacteriological | Oxoid | 1082469-02 |
| Ampicillin | Laboratorio DRAWER S.A. | 52308 |
| Anti-Adherence Rinsing Solution | STEMCELL Technologies | 07010 |
| B-27 Supplement (50×) | Gibco | 17504-001 |
| Bovine Serum Albumin (BSA) | Sigma-Aldrich | A7906-50G |
| Calcium Chloride Dihydrate | Anedra | 6456 |
| DMEM/F12 (1:1) (1×) | Gibco | 11330-032 |
| Advanced DMEM/F-12 | Gibco | 12634010 |
| DPBS (1×) calcium, magnesium | Gibco | 14040133 |
| Epidermal Growth Factor Recombinant Human Protein (EGF) | Gibco | PHG0313 |
| Geltrex | Gibco | A14132-02 |
| GlutaMAX (100×) | Gibco | 35050-061 |
| Glycerin | Anedra | 6754 |
| Hoechst 33342, trihydrochloride, trihydrate | Invitrogen | H3570 |
| Human Fibroblast Growth Factor-basic (bFGF) | Gibco | PHG0261 |
| L-Ascorbic Acid | Anedra | 6019 |
| Lipofectamine Stem Transfection Reagent | Invitrogen | STEM00015 |
| MEM Non-Essential Amino Acids (100×) | Gibco | 11140-050 |
| mTeSR1 Basal Medium | STEMCELL Technologies | 85850 |
| N-2 Supplement (100×) | Gibco | 17502-048 |
| Neubauer counting chamber | Marienfeld | 0610110 |
| Neurobasal Medium | Gibco | 21103-049 |
| Normal Goat Serum (NGS) | Sigma-Aldrich | S26-M |
| Paraformaldehyde | Sigma-Aldrich | P6148-1KG |
| Penicillin Streptomycin | Gibco | 15140-122 |
| Peptone from meat | Merck | 1.07214.1000 |
| Phosphate Buffered Saline (PBS) pH7.4 (1×) | Gibco | 10010-023 |
| Recombinant Human Brain-Derived Neurotrophic Factor (BDNF) | Gibco | PHC7074 |
| Recombinant Human Glial-Derived Neurotrophic Factor (GDNF) | Gibco | PHC7041 |
| Rotenone | Sigma-Aldrich | R8875-1G |
| Sodium chloride | Anedra | 7647-14-5 |
| StemPro Accutase | Gibco | A11105-01 |
| Triton X-100 | Sigma | T8787-100ML |
| Y-27632 dihydrochloride, Rho kinase inhibitor | Abcam | Ab120129 |
| Yeast extract granulated | Merck | 1.03753.0500 |
| Critical commercial assays | ||
| PSC Neural Induction Medium | Gibco | A1647801 |
| Wizard Plus Midipreps DNA Purification System | Promega | A7640 |
| Experimental models: Cell lines | ||
| FN2.1 hiPSCs (human, 46 XY, 4 years old, male) | Lab resource | n/a |
| H9 hESCs (human, 46 XX, blastocyst, female) | Lab resource | n/a |
| Software and algorithms | ||
| FIJI (FIJI Is Just ImageJ) | NIH | V. 2.1.0/1.53c |
| Eclipse NET | Laboratory imaging | V. 1.20.0 |
| Other | ||
| 1.5-mL microcentrifuge tube | Axygen | 311-08-051 |
| 12-well plate | Corning | 3513 |
| 15-mL conical tube | Tarsons | 546021 |
| 2.0 Cryogenic Vials | Corning | 430488 |
| 50-mL conical tube | Tarsons | 500041 |
| 48-well plate CELLSTAR | Greiner Bio-One | 677 180 |
| 6-well plate COSTAR | Corning | 3516 |
| 96-well conical V plate | Greiner CELLSTAR | M9561 |
| AccuBlock digital dry bath | Labnet International | D1100 |
| Heraeus rotor | Sorvall | 75006445 |
| Nikon Eclipse TE2000-S inverted microscope | Nikon | M314E |
| Petri dish | TNT | 704001 |
| Wide neck bottle 200 mL | Eppendorf | A24934 |
Materials and equipment
LB liquid medium
| Reagent | Final concentration | Amount |
|---|---|---|
| Peptone from meat | 1% | 1 g |
| Sodium chloride | 1% | 1 g |
| Yeast extract granulated | 0.5% | 0,5 g |
| ddH2O | n/a | up to 100 mL |
| Total | n/a | 100 mL |
LB agar medium
| Reagent | Final concentration | Amount |
|---|---|---|
| Peptone from meat | 1% | 1 g |
| Sodium chloride | 1% | 1 g |
| Yeast extract granulated | 0.5% | 0,5 g |
| Agar bacteriological | 1.5% | 1,5 g |
| ddH2O | n/a | up to 100 mL |
| Total | n/a | 100 mL |
Note: Autoclave sterilized LB mediums can be stored at a temperature between 20°C–25°C for up to a month. Before adding your antibiotic to LB mediums and LB-agar solutions, you should create a 1,000× stock. The antibiotic solution should not be prepared and stored in advance for an extended period. Antibiotics are sensitive to high temperatures, so add antibiotics after sterilization and before the agar is solidified. An optimal temperature to aim for is ∼50°C–55°C. Moreover, prepare the antibiotic-containing mediums right before usage to ensure their bioactivity.
NSCs medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM/F-12 | n/a | 46.98 mL |
| B27 (50×) | 1× | 1 mL |
| N2 (100×) | 1× | 500 μL |
| GlutaMAX (100×) | 1× | 500 μL |
| MEM NNEA (100×) | 1× | 500 μL |
| Pen/Strep (100×) | 1× | 500 μL |
| EGF (100 μg/mL) | 20 ng/mL | 10 μL |
| bFGF (100 μg/mL) | 20 ng/mL | 10 μL |
| Total | n/a | 50 mL |
CRITICAL: Prepare stocks of bFGF and EGF to 100 μg/mL in sterile 10 mM Tris-HCl pH7.6 /0.1% BSA. Make aliquots of 10 μL and frozen them at −20°C, once thawed do not refrozen. Once supplemented NSCs medium is stable for up to two weeks when stored in the dark at 2°C–8°C.
Neuronal differentiation medium
| Reagent | Final concentration | Amount |
|---|---|---|
| Neurobasal | n/a | 47.47 mL |
| B27 (50×) | 1× | 1 mL |
| GlutaMAX (100×) | 1× | 500 μL |
| MEM NNEA (100×) | 1× | 500 μL |
| Pen/Strep (100×) | 1× | 500 μL |
| Ascorbic Acid (200 μg/mL) | 35.2 ng/mL | 8.8 μL |
| GDNF (100 μg/mL) | 20 ng/mL | 10 μL |
| BDNF (100 μg/mL) | 20 ng/mL | 10 μL |
| Total | n/a | 50 mL |
CRITICAL: Prepare stocks of BDNF and GDNF to 100 μg/mL in sterile 10 mM Tris-HCl pH7.6 /0.1% BSA. Make aliquots of 10 μL and frozen them at −20°C, once thawed do not refrozen. Once supplemented neuronal differentiation medium is stable for up to two weeks when stored in the dark at 2°C–8°C.
Fixation immunostaining solution
| Reagent | Final concentration | Amount |
|---|---|---|
| DPBS (1×) calcium, magnesium | 1× | up to 500 mL |
| Bovine Serum Albumin | 0.1% | 0,5 g |
| Paraformaldehyde | 4% | 4 g |
| Total | n/a | 500 mL |
Permeabilization immunostaining solution
| Reagent | Final concentration | Amount |
|---|---|---|
| DPBS (1×) calcium, magnesium | 1× | up to 500 mL |
| Bovine Serum Albumin | 0.1% | 0,5 g |
| Normal Goat Serum | 10% | 50 mL |
| Triton | 0.1% | 500 μL |
| Total | n/a | 500 mL |
Washing immunostaining solution
| Reagent | Final concentration | Amount |
|---|---|---|
| DPBS (1×) calcium, magnesium | 1× | up to 500 mL |
| Bovine Serum Albumin | 0.1% | 0,5 g |
| Total | n/a | 500 mL |
Note: We keep sterile immunostaining solutions at 8°C for up to 6 months.
Step-by-step method details
Differentiation and transfection of neurons (2D NSCs-differentiation)
Timing: 15 days
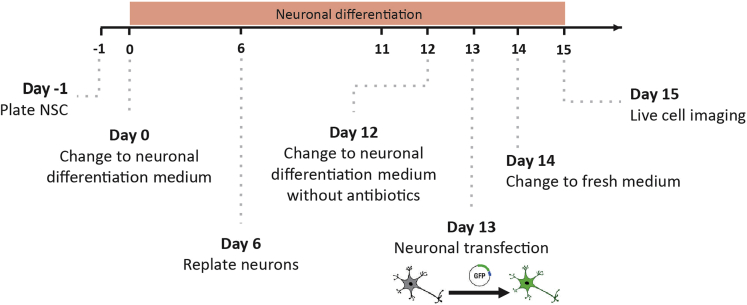
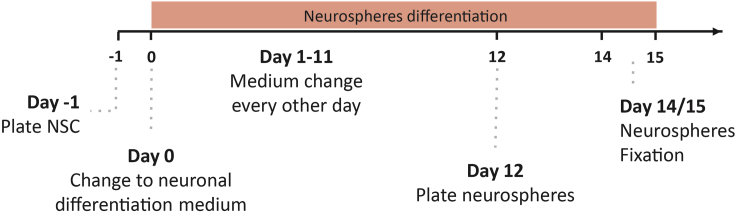
At the beginning of the protocol (Figure 1), NSCs should exhibit a typical triangular morphology, very short processes, and large cells. As the differentiation protocol progresses, cells begin to clump together, presenting a smaller cell body and longer processes typical of neurons. By day 7 of the neuronal differentiation protocol, phenotypic changes should be detectable by the untrained eye.
-
1.
Plate 3 × 104 NSCs per cm2 in NSCs medium in one well of a 6-well cell culture plate pretreated with 1 mL of 1/100 Geltrex at 1 h at 37°C.
-
2.Day 0: Start neuronal differentiation.
-
a.Remove 2.5 mL of NSCs medium from the well of 6-well plate.
-
b.Wash with a 1 mL of warm (37°C) PBS 1×.
-
c.Add 2.5 mL of Neuronal differentiation medium to the well.
-
a.
Note: Neuronal differentiation medium must be warmed at 20°C–25°C and stored the dark at 2°C–8°C.
CRITICAL: Add medium and buffers through the walls of the well avoiding the detachment of the cells.
-
3.
Day 2–4: Change to 2.5 mL of fresh medium every other day.
-
4.Day 6: Re-plate cells.
-
a.Remove 2.5 mL of Neuronal differentiation medium from the well.
-
b.Wash with a 1 mL of warm (37°C) DPBS 1× with calcium and magnesium.
-
c.Incubate cells with 500 μL of Accutase 1× for 7 min at 37°C.
-
d.Dilute Accutase 1× with 1 mL of DPBS 1× with calcium and magnesium and gently detach and transfer cells into a 15 mL conical tube using a 5- or 10-mL serological plastic pipette.
-
e.Centrifuge cells at 200 × g for 5 min and remove the supernatant.
-
f.Determine cell viability after dissociation by Trypan blue exclusion dye assay:
-
i.Prepare a 0.4% solution of Trypan blue in buffered isotonic salt solution, pH 7.2–7.3 (i.e., PBS).
-
ii.Centrifuge an aliquot of cell suspension being tested for viability 5 min at 200 × g and discard supernatant.
-
iii.Resuspend the cell pellet by pipetting up and down 10 times in 1 mL PBS.
-
iv.Add 0.1 mL of Trypan blue stock solution to 0.1 mL of cell suspension.
-
v.Pipette up and down to thoroughly mix the two solutions. Allow mixture to incubate ∼3 min at 20°C–25°C. Cells should be counted within 3–5 min of mixing with Trypan blue, as longer incubation periods will lead to cell death and reduced viability counts.
-
vi.Load a hemocytometer and examine immediately under a microscope at low magnification.
-
vii.Count the unstained (viable) and stained (non-viable) cells separately in the hemocytometer.
-
i.
-
g.Plate 7 × 105 cells per well in a 12-well cell culture plate pretreated with 1 mL Geltrex (1/100) for 1 h at 37°C.
-
h.Incubate at 37°C and 5% CO2.
-
a.
CRITICAL: Cell clumping reduces access to critical nutrients and, as a result, hinders overall cell growth. Gently pipet and thoroughly disaggregate cell clumps, to loosen monolayer into a single cell suspension, avoiding bubbles in the suspension when pipetting.
-
5.
Day 8–11: Change Neuronal differentiation medium to 2.5 mL of fresh medium every other day.
-
6.
Day 12: Change medium to a 2.5 mL of Neuronal differentiation medium without the addition of antibiotics.
-
7.Day 13: Neuronal transfection of GFP-encoding plasmid with Lipofectamine Stem Transfection Reagent in a 12-well cell culture plate.
-
a.Dilute 4 μL of Lipofectamine Stem Transfection Reagent in 50 μL of Neuronal differentiation medium without antibiotics.
-
b.Dilute 500 ng of DNA plasmid in 50 μL of Neuronal differentiation medium without antibiotics.
-
c.Add diluted DNA to the diluted lipofectamine and mix pipetting. Avoid bubble formation.
-
d.Incubate the mix for 10 min at 20°C–25°C.
-
e.Remove the medium from the well, add 900 μL of Neuronal differentiation medium without antibiotics and incubate at 37°C.
-
f.Mix gently by pipetting the DNA-lipofectamine complexes and add them to the wells.
-
g.Incubate 12–16 h at 37°C in 5% CO2.
-
a.
CRITICAL: The average neuronal transfection efficiency we observed is 0.4%. It is important to work with transfections efficiencies ranging 1% or less, to avoid the presence of overlapping transfected cells and misinterpretation of the results.
-
8.
Day 14: Change medium containing lipofectamine with 2.5 mL of fresh Neuronal differentiation medium.
Note: If high toxicity is observed, change the medium with lipofectamine for fresh medium after 4–6 hours of addition.
-
9.
Day 15: GFP expression will typically start around 6 h post-transfection and will reach to a pick at 48–72 h. Add Hoechst (1 μg/mL final concentration) and incubate for 15 min to visualize nuclei.
CRITICAL: In the case of not using Hoechst as a staining dye for nuclei, it is important to use another type of cell-permeable stain that marks living cells. We do not recommend using DAPI because it stains the nuclei of dead cells.
-
10.
Visualize and photograph transfected neurons using an inverted fluorescence microscope.
CRITICAL: You need to confirm that NSCs are correctly differentiated to cells with neuronal phenotype. Perform immunofluorescence staining for neuronal markers such as βIII-tubulin/TUJ-1, Microtubule-associated protein 2/MAP-2 and Microtubule-associated protein 5/MAP-5. For a more information about how to validate the differentiated neurons please refer to Yan et al. publication (Yan et al., 2013).
Figure 1.
2D Neuronal differentiation protocol outline
Timeline for 2D-neuronal differentiation protocol where all the essential steps are marked, from NSCs plate to live cell imaging.
Quantification of 2D neuronal NSC-derived differentiation protocol with FIJI software
Timing: 2 h
Live cell imaging was visualized with an Eclipse TE2000-S inverted microscope (Nikon) using E-Plan 10× and 20× objectives with a long working distance and a super high-pressure mercury lamp. Photographic images were acquired with a Nikon DXN1200F digital camera, which was controlled by the EclipseNet software (version 1.20.0 build 61). Open-source FIJI software was used for processing images (Schindelin et al., 2012).
-
11.
Open FIJI software, a window with different bars and menu should appear (Figure 2).
-
12.Calibrate the FIJI software with the measurements in micrometers for the photographs.
-
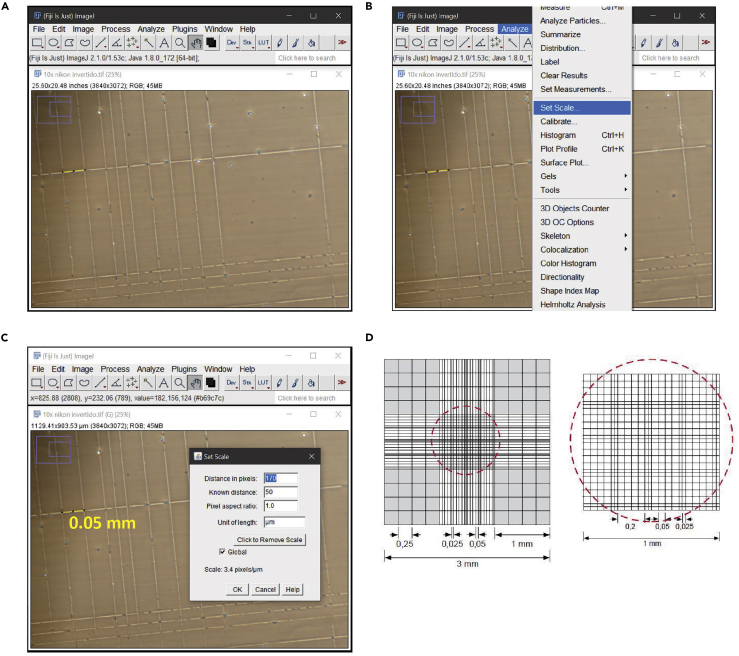
a.For this step it is necessary to photograph an element with a known distance with the same objective with which the neuron photographs will be taken (in this case a Neubauer chamber but can be a ruler) (Figure 3).
-
b.Load the Neubauer chamber photograph using the file button in the MENU BAR and then select “Open…” from the sub-menu (Figure 3A).
-
c.Trace a line using “straight” tool from the TOOL bar between two points (Figure 3B).
-
d.Select the Analyze button from MENU BAR and select Set Scale… from the sub-menu (Figure 3B).
-
e.Set the scale with the known distance in Set scale windows as you can see in Figure 3C. In this case we use 50 μm which corresponds to the width of the squares corresponding to the center of the counting chamber (Figure 3D). It is important to check the “global” box to keep the scale parameters in all the photos that will be uploaded.
-
a.
Note: In case the spatial information of the sample is not embedded in the metadata of the photo file. It is important to work with uncompressed file formats such as TIFF to have all the photo information available. This procedure must be done with each photograph captured with different objectives used and with files that have different pixel size (resolution).
-
13.
Load the GFP-transfected neurons and Hoechst-stained nuclei images using the file button in the MENU BAR and then select “Open…” from the sub-menu.
Note: The differentiation protocol detailed renders a high percentage (almost 100%) of neurons. The neurons display a very different morphology when compared to undifferentiated cells. Thus, only after a thorough observation of each neuron, cells would be included in the analysis. In case of having doubts, the analysis would not consider cells with doubtful morphology.
-
14.
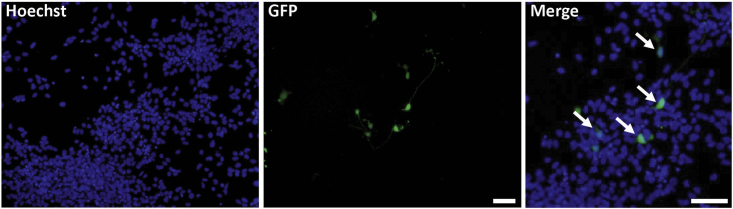
Use images showing Hoechst-stained blue nuclei to distinguish the soma of the neurons, and then work with images showing the expression of GFP in transfected cells neurons (Figure 4).
-
15.
Mark the area corresponding to the neural body with the polygon selection tool from the TOOLS MENU.
-
16.
Click Analyze from MENU BAR and select Measure from the sub-menu.
-
17.
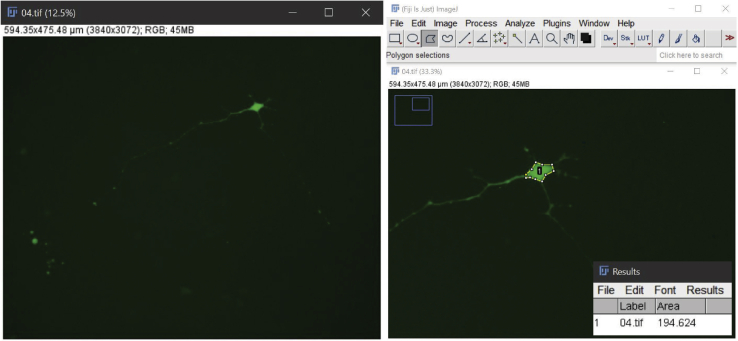
A new window would appear with the measurement in μm2 determining the area of the neuronal soma (Figure 5).
-
18.
Save this quantification in a file.
-
19.
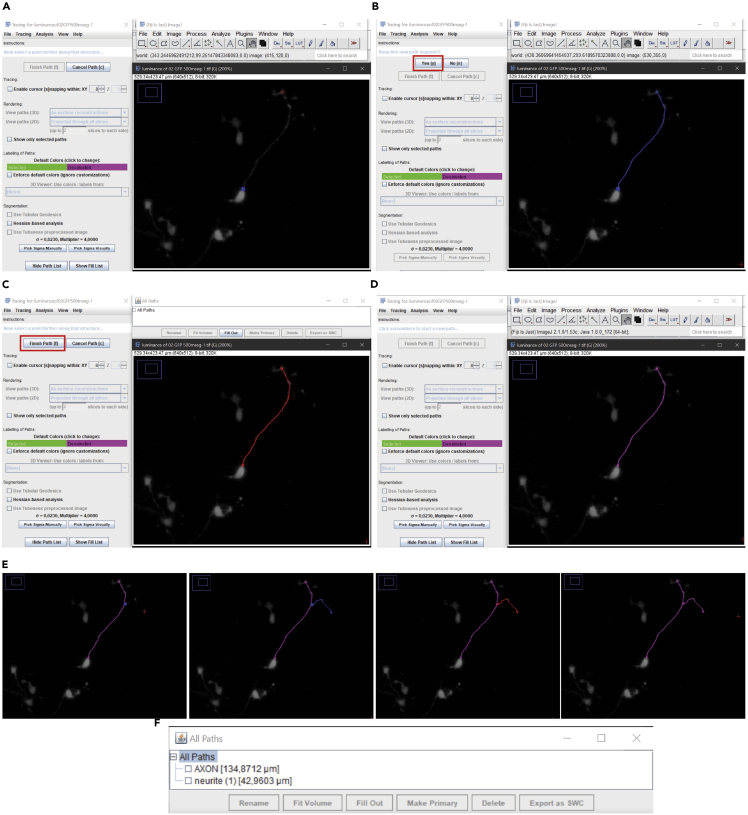
Open the Simple Neurite Tracer plugin from the Plugin button in MENU BAR to measure the neurites length.
-
20.
Mark the start and end of the main neurite and end the pathway to observe the length of the prolongation in micrometers (Figure 6A).
-
21.
Mark “yes” if the parts are well connected (Figure 6B).
-
22.
Click the “finish path” button if all the neurite is marked (Figures 6C and 6D).
-
23.
Make the same procedure for secondary neurites (Figure 6E).
-
24.
You can see the measurements in the path windows with all the quantification in μm (Figure 6F). Save this quantification in a file.
Note: It is possible to save the paths made on the neurites by accessing the "File" menu of the SNT plugin and then accessing the "Save all paths as SWC..." submenu. This will allow you to save all the traces made. Once these data are saved, to access them you must load the corresponding image in FIJI and open the file with the .swc extension in the SNT plugin from the "File" menu option and the "Load Traces/ (e)SWT" option. File..." from the submenu.
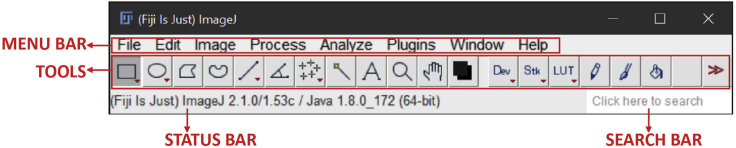
Figure 2.
FIJI software main parts for image processing
The FIJI software interface consists of 4 main parts. A MENU BAR where you can access file, edit, image, process, analyze, window and help submenus. A TOOLS bar where you can find selection shape as rectangle, oval, polygon, freehand, straight, angle, multi-point, wand, text, magnifying glass, scrolling, color picker tools, command finder, developer menu, paintbrush, flood fill, arrow and LUT menu. A STATUS BAR where you see information about TOOLS and FIJI version and finally a SEARCH BAR. For more information access the software page (https://imagej.net/tutorials/).
Figure 3.
Setting photographs scale
(A) Microphotograph of a Neubauer chamber obtained at the same magnification as the neuron images that will be quantified.
(B) Access to the menu to set the scale of the photograph.
(C) Window with scale settings.
(D) Diagram with the measurements of the counting chamber provided by the manufacturer.
Figure 4.
Locating the neuronal soma in neuronal culture
Microphotographs of 2D neuronal culture stained with Hoechst (blue) and expressing GFP (green). The bar represents 100 μm.
Figure 5.
Measuring the soma
Original image of a neuron together with the tools used and the result of the measurement of the neuronal body’s area.
Figure 6.
Measuring the axon and neurites length
(A–E) Sequence of photographs from (A) to (E) showing step-by-step the measurement of axon and neurites length.
(F) In section (F), an example of how the results measured by the plugin is shown.
3D NSCs differentiation protocol for the formation of neurospheres (Figure 7)
Figure 7.
3D neurospheres differentiation protocol outline
Timeline for 3D-neuronal differentiation protocol where all the essential steps are marked, from NSCs plate to neurospheres immunostaining.
Timing: 15 days
During 3D NSCs neuronal differentiation, it would be possible to observe shiny spheres, with abundance of large, healthy cells. After transferring the neurospheres to Geltrex-coated plates neuronal processes emanating from the neurospheres should be observed. The neurospheres remain as clumps and express the neuronal marker TUJ-1.
-
25.
Incubate a 96-well conical V bottom plate with Anti-Adherent Rising Solution (30 μL per well) at 20°C–25°C for 30 min.
CRITICAL: NSCs cultured in a 6-well plate must be maintained in optimal conditions and good morphology. It is necessary to adapt this protocol according to the cell culture area. All reagents should be warmed at 20°C–25°C before use.
Alternatives: Alternatively, PrimeSurface 3D culture Ultra-low Attachment plates 96-well, V bottom plates S-BIO technologies #MS-9096VZ can be used.
-
26.
Remove the 2.5 mL medium from the well of 6-well plate.
-
27.
Wash NSCs with 1 mL of warm (37°C) DPBS 1× with calcium and magnesium.
-
28.
Incubate NSCs with 500 μL of Accutase 1× for 7 min at 37°C.
-
29.
Dilute Accutase 1× with 1 mL of DPBS 1× with calcium and magnesium and gently detach and transfer cells into a 15 mL conical tube using a 5- or 10-mL serological plastic pipette.
-
30.
Centrifuge at 200 × g for 5 min and remove the supernatant.
-
31.
Resuspend the pellet in 1 mL of NSCs medium and count cell number with your preferred method.
-
32.
Plate 8000 NSCs in each well of a 96-well plate in a final volume of 100 μL.
-
33.
Centrifuge the 96-well plate at 200 × g for 5 min.
-
34.
Incubate at 37°C and 5% CO2 for 24 h.
-
35.
Remove NSCs Medium and wash carefully with 150 μL of warm (37°C) DPBS 1× with calcium and magnesium.
-
36.
Add 150 μL of Neuronal differentiation medium.
-
37.
Change the medium every other day (1–2 days) by removing 130 μL of medium and placing 130 μL of fresh medium.
CRITICAL: When removing the medium, tilt the plate at a 45-degree angle toward us and slowly remove it touching the wall of the well without reaching the bottom where the neurosphere is placed. The neurospheres are stressed when are without medium, therefore it is important to leave a small amount so that they are covered (20 μL). Change medium in turns alternating with 37°C incubations (i.e.: if we have a 96-well plate with neurospheres we will change in 24 wells per turn in a total of 4 times). The use of vacuum is not recommended; instead, we used a p200 with a 200 μL tip with a 10 μL tip at its end for greater precision when removing the medium.
-
38.On day 11 of the neuronal differentiation plate neurospheres into a 48-well cell culture plate pretreated with Geltrex at 37°C for 1 h.
-
a.Add 300 μL of Neuronal differentiation medium per well of the 48-well plate and warm at 37°C.
-
b.Remove medium from the 96-well plate and add 50 μL of fresh medium.
-
c.Use a sterile cut P1000 tip to transfer the neurospheres carefully from the 96-well conical V bottom plate to each well of the 48-well cell culture plate without damaging them.
-
d.Plate 1–2 neurospheres per well.
-
a.
Note: If 2 co-cultured neurospheres stick together they should be excluded from the analysis, only individual neurosphere must be included.
-
39.
Change the medium every other day to a final volume of 400 μL per each well of the 48-well cell culture plate until day 14 of the neuronal differentiation protocol. It is important to leave a small amount of medium.
CRITICAL: We recommend marking the location where the neurospheres were adhered to the plate with a marker. This allows us to know its location in the well and avoid manipulating the area when changing media or proceeding with the neurospheres staining. The use of vacuum is not recommended; instead, we used a p200 with a 200 μL tip with a 10 μL tip at its end for greater precision when removing the medium.
Neurospheres immunostaining
Timing: 2 days
It is important to be careful when performing the steps of the technique so that the neurospheres do not detach. For this reason, we recommend placing and removing the solutions slowly and never on the neurosphere but against the walls of the well. All buffers used in this section should contain calcium and magnesium to prevent neurospheres detaching from the plate.
-
40.
Remove the 400 μL of Neuronal differentiation medium from each well of 48-well plate.
-
41.
Wash with 200 μL warm (37°C) DPBS 1× containing magnesium and calcium.
-
42.
Fix cells with 200 μL Fixation Immunostaining Solution for 15 min at 20°C–25°C.
Pause point: Once neurospheres are fixed, they can be stored at 4°C for up to 2 weeks in 400 μL of Washing Immunostaining Solution per well of 48-well plate. We still recommend performing this technique as soon as possible after fixing.
-
43.
Wash with 200 μL Washing Immunostaining Solution for 5 min.
-
44.
Repeat the previous step twice.
-
45.
Permeate for 45 min with 200 μL Permeabilization Immunostaining Solution.
-
46.
Incubate with 50 μL Anti-tubulin ꞵ3 (TUJ-1) primary antibody (1:1000 in Permeabilization Immunostaining Solution) 12–16 h at 4°C without agitation or shacking.
-
47.
Wash with 200 μL Washing Immunostaining Solution for 5 min.
-
48.
Repeat the previous step twice.
-
49.
Incubate with 50 μL of a secondary antibody recognizing primary antibody heavy chain (Alexa 488, 1:400 in Permeabilization Immunostaining Solution) for 45 min at 20°C–25°C protected from light.
-
50.
Wash with 200 μL Washing Immunostaining Solution for 5 min.
-
51.
Repeat the previous step twice.
-
52.
Place 400 μL of Washing Immunostaining Solution per well of 48-well plate.
-
53.
Observe under the fluorescence microscope.
Pause point: This plate can be stored at 4°C protected from light for up to 2 weeks.
Quantification of 3D NSCs differentiation protocol with FIJI software
Timing: 1 h
We visualized immunostained neurospheres with an Eclipse TE2000-S inverted microscope (Nikon) using a 4× E-Plan objective and a super high-pressure mercury lamp. We adquired photographic images with a Nikon DXN1200F digital camera, which was controlled by the EclipseNet software (version 1.20.0 build 61). We used open-source FIJI software for processing images (Schindelin et al., 2012).
-
54.
Open FIJI software and repeat steps 10 and 11 of “quantification of 2D neuronal NSC-derived differentiation protocol with FIJI software”.
-
55.
Load the images of TUJ-1 immunostained neurospheres using the file button in the MENU BAR and then select “Open…” from the sub-menu.
-
56.
Select the area corresponding to the neurosphere with the polygon selection tool from the TOOLS MENU.
-
57.
Choose Analyze from MENU BAR and select “Set measurements…” from the sub-menu to make sure to tick area and perimeter boxes.
-
58.
Then select Analyze from MENU BAR and click Measure from the sub-menu.
-
59.
A new window would appear with the measurement in μm2 determining the area of the neurosphere and the measurement in μm for the perimeter (Figure 8).
-
60.
Save this quantification in a file.
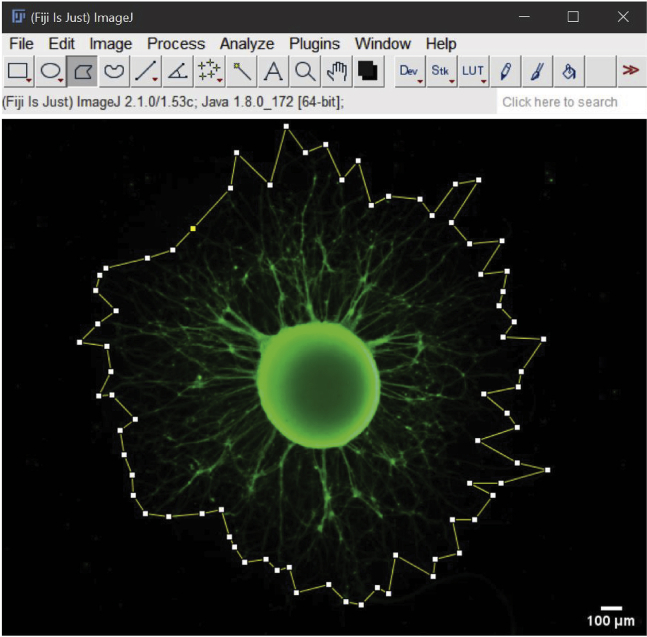
Figure 8.
Measuring the area and perimeter of a neurosphere
For the quantification of the area and perimeter of a neurosphere, the polygon tool was used and we delimited the edge of the neurosphere through the junction of the neurites.
Expected outcomes
We studied neuronal stress in neurons derived from hPSCs. For this purpose, we use a 24 h treatment with Rotenone at a concentration of 1 μM as a stressful stimulus, concentration that is not cytotoxic to the neurons (Pei et al., 2016 and data not shown) but induces changes in mitochondrial membrane potential (data not shown). Neurons were derived from NSCs generated with the PSC Neural Induction Medium kit (Gibco, Thermo Fisher Scientific) from the hESCs line WA09 (H9) (Thomson 1998) and hiPSCs line FN2.1 (Questa et al., 2016) lines. To evaluate the effects caused by Rotenone it is relevant to analyze and quantify morphometric features in a minimum of 30 neurons or neurospheres per condition.
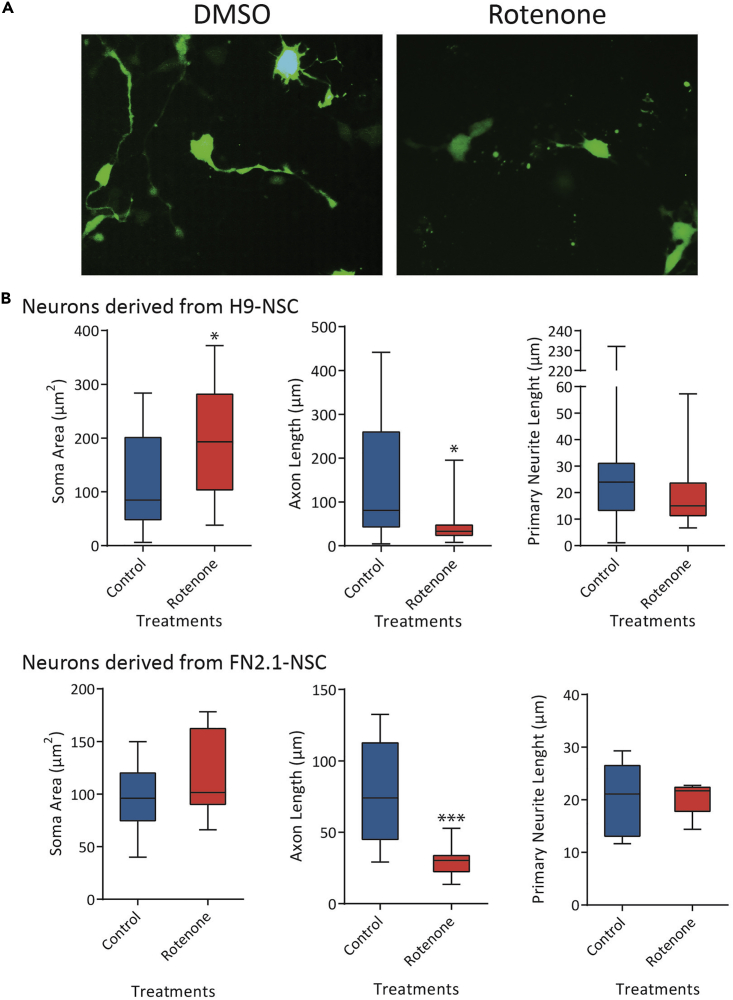
We performed a 14 days-2D neuronal differentiation protocol from hPSCs-derived NSCs and then treated the obtained neurons with Rotenone for 24 h to observe and quantify the morphometric changes. We were able to observe: (1) an increase in the area of the soma, (2) a considerable reduction in axonal length, (3) a reduction in the length of primary neurites, and (4) a decrease in the presence of secondary neurites upon Rotenone treatment (Figure 9).
Figure 9.
Neuronal morphological changes upon stressful Rotenone treatment
Neurons derived from H9 hESCs and FN2.1 hiPSCs were transfected with a GFP-encoding plasmid and then treated with Rotenone (1 μM) for 24 h.
(A) Fluorescent images of control (DMSO) and Rotenone-treated neuronal cultures.
(B) Quantifications of the neuronal morphometric changes (soma area and axon, primary and secondary neurites length) upon Rotenone treatment. Media + SEM is graphed. Student’s t test: ∗=p<0.05; ∗∗=p<0.01; ∗∗∗=p<0.001 vs. Ctrl (Control: DMSO).
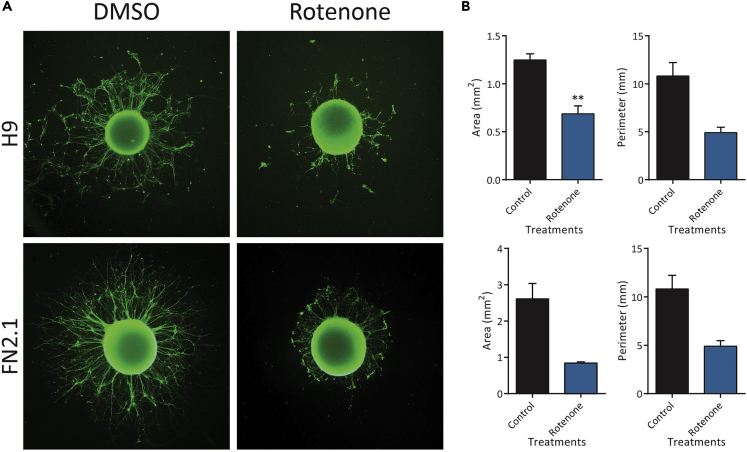
We also performed a 14 days-3D neuronal differentiation protocol (neurospheres formation) with H9 and FN2.1 hPSCs-derived NSCs and then added Rotenone for 24 h to quantify axonal and primary neurites retraction. We observed changes at the level of the area and perimeter of the neurospheres upon Rotenone treatment, which indicates that the stressful stimulus effectively affects the length of the neurites (Figure 10).
Figure 10.
Changes in area and perimeter of neurospheres treated with Rotenone
Neurospheres derived from H9 hESCs and FN2.1 hiPSCs were treated or not with Rotenone (1 μM) for 24 h and then fixed and stained against TUJ-1 neuron-specific marker.
(A) Images of control (DMSO) and Rotenone-treated neurospheres.
(B) Quantifications of neurospheres area and perimeter upon Rotenone treatment. Media + SEM are graphed. Student’s t test: ∗=p<0.05; ∗∗=p<0.01; ∗∗∗=p<0.001 vs. Ctrl (Control: DMSO).
Limitations
We describe this protocol to be used in any laboratory facility without the needs of special programming/machine learning skills or expensive equipment or software such as cellSens Dimension (Olympus Corporation, Japan) or ImagePro plus (Media Cybernetics, USA). Therefore, the following limitations are related to the processing time of the images and the possibility of detecting and quantifying small neuronal structures.
In the 2D-neuronal NSC-derived differentiation protocol the morphometric quantifications are difficult to automate and, therefore, the measurement of structures is very time-consuming. Furthermore, when working with 10× and 20× objectives, smaller structures such as dendritic spines and smaller neurites cannot be distinguished.
Regarding the 3D-neuronal differentiation protocol, neurosphere quantifications are made at the level of the whole neuronal population without any possibility of analyzing individual neurons. In consequence the changes that can be studied are limited to the retraction or elongation of the longest axons or primary neurites.
Troubleshooting
2D NSCs-neuronal differentiation protocol
Problem 1
NSCs do not adhere to the plate or very high cell death rate is observed.
Potential solution
Check the quality of NSCs from which the 2D neuronal differentiation protocol is started. The QC criteria used consists in checking viability by a Trypan blue exclusion dye assay, proliferation by immunocytochemistry using Ki-67 antibody, and validation of NSCs identity by immunofluorescence staining using specific antibodies against SOX-1, SOX-2, NESTIN and PAX-6.
Check that culture media is at 20°C–25°C and not cold.
Check that the media are stored at a temperature of 4°C protected from light and do not have a date greater than 2 weeks. The medium should not be warmed in a bath at 37°C, because the recombinant proteins can be degraded.
Check that the culture is not long out of the 37°C incubator.
Check that during the incubation of the Geltrex coating the plate has not dried. This circumstance will directly affect the adhesion of the cells and their survival.
Check that when detaching the cells, the transfer is carried out with a serological pipette. There are cell lines that suffer mechanical stress when passing through the small hole of a 200- or 1000-μL tips.
Try using NSCs of lower passages.
Problem 2
hPSCs and NSCs do not detach when incubate them with Accutase under the conditions described in the protocol.
Potential solution
Sometimes when cultures are 100% confluent, hPSCs and NSCs may take longer to detach. If this happens, we recommend using a cell scraper to lift them mechanically. This must be done with great care to stress the cells as little as possible.
Check that the Accutase is at 37°C (not cold) so that it can exert its biological activity.
Problem 3
hPSCs grow very fast or very slow.
Potential solution
Each cell line presents metabolic characteristics that are inherent to each line. In this protocol we use two lines that have been cultured and studied in our laboratory for years and we know their characteristics. In case of observing very different growth rates, we recommend adapting the protocol to the lines with which experiments are carried out.
Problem 4
Individual GFP-transfected neurons cannot be distinguished from each other, impeding satisfactory quantification.
Potential solution
It is possible that neuron culture confluence was higher than 70% at the time of transfection and/or that transfection efficiency was too high. We work with transfection efficiencies of less than 1%.
Be sure that the neurons reached 40–60% confluence at the time of transfection.
We recommend performing dose-response curves to establish optimal experimental conditions (plasmid, lipofectamine) for each tested cell line concomitantly with cell viability verification by a Trypan blue exclusion dye assay.
Problem 5
Not sufficient GFP-transfected neurons in culture.
Potential solution
Be sure that the neurons reached 40–60% confluence at the time of transfection.
We recommend performing dose-response curves to establish optimal experimental conditions (plasmid, lipofectamine) for each tested cell line concomitantly with cell viability verification by a Trypan blue exclusion dye assay.
Corroborate that the plasmid is not degraded by means of an electrophoretic run in 0.8% agarose.
Problem 6
I cannot access the SNT plugin, because it does not allow me to click it.
Potential solution
Some versions of FIJI do not allow clicking on the menu where the plugin name appears. This can be very easily solved by forcing the software to start the plugin by pressing the “alt” button on the keyboard while clicking the plugin name in the menu.
3D NSCs-neuronal differentiation protocol (Neurospheres)
Problem 7
It is difficult to observe neurospheres at the microscope when they are growing in the 96 well conical plates.
Potential solution
To observe the neurosphere in this condition use the micrometer dial of the microscope to observe the mass of cells in the center of the well at different planes. We recommend comparing it with an empty well until the operator gets used to seeing the neurosphere in this type of conical plate.
Confirm that the microscope is in good working conditions.
Problem 8
Neurospheres are not formed.
Potential solution
Confirm that the neurospheres are not being aspirated when changing the medium. For this reason, we discourage the use of aspiration and opt for manual medium change with a pipette.
Check the quality of NSCs from which the neurosphere protocol is started as described in problem 1. Additionally, verify the lack of more differentiated lineage markers including TUJ-1 for neurons, GFAP for astrocytes and O1 for oligodendrocytes, and the absence of pluripotency markers OCT-4 and NANOG by immunofluorescence staining.
Try using NSCs of lower passages.
Problem 9
Neurospheres do not adhere to the extracellular matrix (Geltrex) coating.
Potential solution
If the body of the neurosphere is dark, it is most likely that many cells are dead. We recommend minimizing the time outside the incubator and confirming that they do not dry out during medium change.
Check the quality of NSCs from which the neurosphere protocol is started as mentioned above.
Geltrex coating condition can be cell line specific. Try making a concentration curve to find the optimal Geltrex concentration needed for the proper neurospheres adhesion.
Try using NSCs of lower passages.
We recommend performing immunofluorescence staining for NSCs markers.
Change the branch of the extracellular matrix used. You can try using Matrigel as an example.
Problem 10
Neurosphere prolongations cannot be observed.
Potential solution
If the body of the neurosphere is dark, it is most likely that many cells are dead. We recommend minimizing the time outside the incubator and confirming that they do not dry out during medium change.
This can be caused because p1000 tips used to transfer neurospheres were not properly cut and caused neurospheres damage or by rough handling (step 15c). You need to work very gently on this step.
In addition, we recommend the use of NSCs of low passages, and to perform immunofluorescence staining for NSCs markers to check NSCs quality as previously mentioned.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Leonardo Romorini (lromorini@fleni.org.ar).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
This work was supported by grants from ANPCyT PICT-2016-2621 and Fundación FLENI.
Author contributions
S.M.: involved in the conception of the research idea, laboratory work, interpretation of the results, preparation and editing of manuscript, M.S.R.V.: cell culture, laboratory work, L.I. and S.L.F.L.: laboratory work, G.E.S., M.E.S., and L.R.: involved in the conception of the research idea, data analysis, result interpretation, manuscript preparation and editing. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Sofía Mucci, Email: sofiamucci27@gmail.com.
Leonardo Romorini., Email: lromorini@fleni.org.ar.
Data and code availability
This study did not generate any datasets or code.
References
- Billeci L., Magliaro C., Pioggia G., Ahluwalia A. NEuronMOrphological analysis tool: open-source software for quantitative morphometrics. Front. Neuroinf. 2013;7:2. doi: 10.3389/fninf.2013.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casali N. In: Casali N., Preston A., editors. Vol. 235. Humana Press; 2003. Escherichia coli host strains. (E. coli Plasmid Vectors. Methods in Molecular Biology). [DOI] [PubMed] [Google Scholar]
- Pei Y., Peng J., Behl M., Sipes N.S., Shockley K.R., Rao M.S., Tice R.R., Zeng X. Comparative neurotoxicity screening in human iPSC-derived neural stem cells, neurons and astrocytes. Brain Res. 2016;1638:57–73. doi: 10.1016/j.brainres.2015.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Questa M., Romorini L., Blüguermann C., Solari C.M., Neiman G., Luzzani C., Scassa M.É., Sevlever G.E., Guberman A.S., Miriuka S.G. Generation of iPSC line iPSC-FH2.1 in hypoxic conditions from human foreskin fibroblasts. Stem Cell Res. 2016;16:300–303. doi: 10.1016/j.scr.2015.12.026. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Yan Y., Shin S., Jha B.S., Liu Q., Sheng J., Li F., Davis J., Bharti K., Zeng X., Rao M., et al. Efficient and rapid derivation of primitive neural stem cells and generation of brain subtype neurons from human pluripotent stem cells. Stem Cells Transl. Med. 2013;2:862–870. doi: 10.5966/sctm.2013-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate any datasets or code.