Abstract
Polyamine composition in an aphid endosymbiotic bacterium, Buchnera sp., was determined by high-performance liquid chromatographic analysis. We found that Buchnera contained virtually only a single polyamine, spermidine. The spermidine content of Buchnera was considerably higher in young aphids and tended to decrease with the age of the host. Expression of speD and speE, whose gene products are key enzymes in the synthesis of spermidine, was analyzed by real-time quantitative reverse transcription-PCR. It was shown that the levels of their mRNAs fluctuated in line with the spermidine content.
Buchnera spp. are intracellular symbiotic bacteria harbored by aphid bacteriocytes, cells specifically differentiated for this purpose (1, 3, 15). The symbiotic association between Buchnera and aphids is mutualistic and obligate in that neither partner can reproduce in the absence of the other (11). This is partly because Buchnera produce essential amino acids (6, 7, 20, 26) and vitamins (21), which are utilized by the host aphid. Molecular phylogenetic studies of 16S rRNA genes suggested that Buchnera belong to the γ subdivision of the Proteobacteria and that they are closely related to Escherichia coli (32). However, there are significant differences between Buchnera and E. coli. Each Buchnera cell has more than 100 copies of the genome (17), whose size is about a seventh of that of the E. coli genome (4). This suggests that these genomic copies must be stabilized in a specific way in the Buchnera cell. In the meantime, Buchnera cells do not divide as frequently as free-living bacteria, suggesting that their proliferation is strictly controlled by the host bacteriocyte (14). Since polyamines are known to be important factors for DNA stabilization, DNA replication, and cell proliferation, we directed our attention to these polycationic compounds.
Polyamines are linear aliphatic compounds that are positively charged under physiological ionic and pH conditions. They are present in all prokaryotic and eukaryotic cells and account for the majority of intracellular cationic charge (29). Among several functions implicated, charge neutralization of intracellular polyanions, especially DNA, may be the most important physiological role of polyamines. The interaction of polyamines with DNA induces such conformational changes as transitions from B to A and Z forms (30), bending (8), and, at higher polyamine concentrations, condensation of DNA (9, 24, 25). These polyamine-induced conformational changes may affect DNA metabolism and modify the interactions of DNA with sequence-specific DNA-binding proteins (23).
As the first step to investigating the roles of polyamines in Buchnera, in this study, we determined the polyamine composition of Buchnera and further assessed the expression of genes involved in the biosynthesis of polyamines.
MATERIALS AND METHODS
Host aphids.
A long-established parthenogenetic clone of the pea aphid, Acyrthosiphon pisum Harris, was maintained on young broad bean plants, Vicia faba L., at 15°C in a long-day regimen of 16 h of light and 8 h of dark (13). Insects were collected within 24 h after larviposition by apterous mothers. These nymphs are described as 0-day aphids. Once the aphids reached adulthood, they were transferred twice a week to fresh plants in order to keep the nutritional conditions constant.
Isolation of Buchnera.
The aphids were dissected in a drop of buffer A (35 mM Tris-HCl [pH 7.5], 25 mM KCl, 10 mM MgCl2, 250 mM sucrose) (13) on a petri dish covered with 1% agarose gel. Bacteriocytes freed from the insect body were collected and gently crushed by pipetting. The homogenate was filtered through an isopore membrane filter (Millipore; pore size, 3 μm) to remove cell components of host origin. This filtration method was verified to give purer samples than other methods, such as the Percoll gradient method (26), and was applied to obtain DNA samples for Buchnera genome analysis (27). Shotgun sequencing of purified DNA detected no contaminant DNAs such as those of eukaryotic mitochondria or other bacteria (S. Shigenobu, personal communication), suggesting that this Buchnera sample was virtually free of contaminants.
Estimation of the volume of Buchnera cells used for HPLC analysis.
An aliquot of isolated Buchnera cells was used to estimate the volume of Buchnera applied for high-pressure liquid chromatography (HPLC) analysis. The number of Buchnera cells was determined using hemocytometers. The volume of Buchnera cells, treated as spheres, was calculated from the diameter, measured with a micrometer. The sum volume of Buchnera cells was calculated by multiplying the number by the average volume.
E. coli strain.
E. coli TOP10 cells were cultured overnight at 37°C in LB medium and collected by centrifugation at the stationary phase.
HPLC analysis.
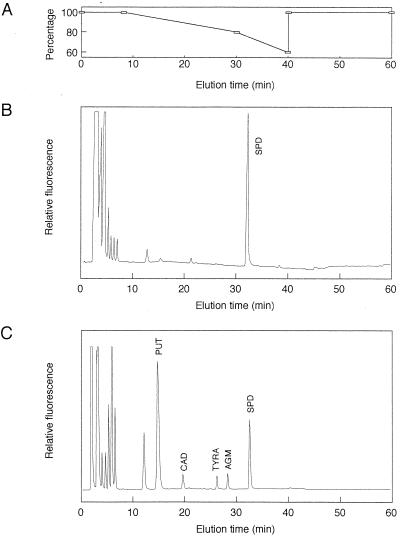
Buchnera and E. coli cells were homogenized in 5% perchloric acid (PCA), and the acid-soluble fractions were obtained by centrifugation at 18,000 × g for 5 min. Supernatants were analyzed in a JASCO HPLC system using a Crestpak C18S column (4.6 by 150 mm) heated to 40°C. Elution was done using a stepwise gradient with solvent A (0.1 M sodium acetate, 10 mM sodium 1-hexanesulfonate [pH 4.5]) and solvent B (methanol). The gradient parameters were as shown in Fig. 1A. The flow rate of the solvents was 1.0 ml/min. Polyamines were detected by fluorescence after mixing the column effluent with an o-phthalaldehyde solution containing 0.06% o-phthalaldehyde, 0.4% borate buffer (pH 10.5), 0.1% Brij 35, and 12 mM 2-mercaptoethanol at 40°C. Fluorescence was measured at an excitation wavelength of 365 nm and an emission wavelength of 455 nm. Quantification was achieved by determining the peak area of the fluorescence tracings and reference to standard curves prepared using polyamine standard solutions.
FIG. 1.
Analysis of polyamine composition by HPLC. (A) Elution from the reversed-phase column was done using a stepwise gradient with solvent A and solvent B. The gradient parameters are shown as percentages of solvent A. (B) Typical chromatogram of polyamines in Buchnera. (C) Typical chromatogram of polyamines in E. coli. PUT, putrescine; CAD, cadaverine; TYRA, tyramine; AGM, agmatine; SPD, spermidine.
Protein assay.
The PCA precipitates were dissolved in 0.1 N NaOH, and their protein contents were assayed using the bicinchoninic acid protein assay reagent (Pierce).
RNA preparation.
Total RNA extraction from bacteriocytes was performed using TRIzol reagent (Gibco-BRL). To remove chromosomal DNA contamination, RNA samples were treated with DNase I.
Real-time quantitative RT-PCR.
Total RNAs were prepared from bacteriocytes as described above, and the cDNAs were synthesized with 1 μg of total RNA, 1 μl of 200 mM dithiothreitol, and 1 μl of 0.2-μg/μl pd(N)6 primer using the First-Strand cDNA synthesis kit (Pharmacia). PCRs were carried out using reverse-transcribed (RT) samples from the preceding step, 0.2 μM (each) target-specific primers (Table 1) (these primers were designed on the basis of sequence data [GenBank accession number AP000398]), 3 mM MgCl2, and LightCycler-DNA Master SYBR Green I (Roche Diagnostics). To prevent the formation of primer dimers, TaqStart antibody (Clontech) was added to the PCR mixture for the hot start. This antibody keeps the DNA polymerase inactive until the temperature rises above 70°C and is inactivated by the same heating step that denatures the target DNA. A LightCycler (Boehringer Mannheim) instrument was used for real-time quantitative PCR. Temperature parameters for PCR amplification were 95°C for 0 s, 55°C for 5 s, and 72°C for 30 s for 40 cycles. The fluorometric intensity of SYBR Green I, a specific dye for double-stranded DNA, was measured at the end of each elongation phase, and a relative amount of each target cDNA was calculated by kinetic analysis (32). Fluorescence signals caused by primer dimers and nonspecific background were discriminated by melting-curve analysis, as recommended by the manufacturer.
TABLE 1.
Sequences of the primers used in quantitative RT-PCR
| Target gene | Primers | Size of PCR product (bp) |
|---|---|---|
| 16S rRNA | 5′-TGAGACACGGTCCAGACTCCTAC-3′ | 790 |
| 5′-GTTGCGCTCGTTGCGGGACTTAAC-3′ | ||
| speD | 5′-GCTATGCTAATACCAATGATTCGCG-3′ | 447 |
| 5′-TATCTCTAGTAAATCCCCGCACACG-3′ | ||
| speE | 5′-GATGATATTGTTCAAACAACTGAACGCG-3′ | 445 |
| 5′-GAAGAAAAAAAACACCGTTCTGTGCTAC-3′ |
RESULTS
Cell size of Buchnera.
Buchnera cells were isolated from about 20 individuals each of 10-, 20-, 30-, 40-, and 50-day-old adult aphids. The volume of Buchnera cells did not change significantly as the host aphids aged (Table 2).
TABLE 2.
Cell size of Buchneraa
| Age of aphid (days) | Cell diam (μm) | Vol (μm3) |
|---|---|---|
| 10 | 2.73 ± 0.11 | 12.7 ± 1.5 |
| 20 | 2.70 ± 0.10 | 11.6 ± 1.3 |
| 30 | 2.69 ± 0.07 | 11.0 ± 0.8 |
| 40 | 2.71 ± 0.07 | 11.4 ± 1.0 |
| 50 | 2.82 ± 0.06 | 12.3 ± 0.9 |
Values are means ± standard errors (n = 50).
Polyamine composition of Buchnera.
Irrespective of the age of the host aphids, Buchnera contained virtually only a single polyamine, spermidine (Fig. 1B). Although putrescine and cadaverine are major polyamines in many prokaryotic organisms, no significant amounts of these polyamines were detected in Buchnera. The polyamine composition of E. coli was also examined by the same method (Fig. 1C). In E. coli, considerable amounts of putrescine, spermidine, cadaverine, agmatine, and tyramine were detected, which was consistent with the previous reports (10, 16, 28).
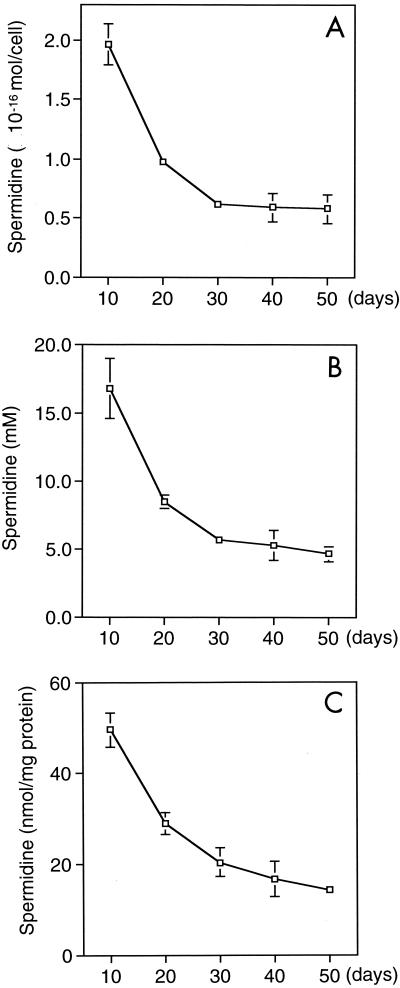
In Fig. 2, we present changes in the spermidine content in terms of the amount per cell (A), per volume (B), and per milligram of protein (C). In each case, spermidine was the most abundant in Buchnera isolated from 10-day-old aphids and amounted to (1.97 ± 0.17) × 10−16 mol/cell, 16.8 ± 3.2 mM, and 49.6 ± 3.7 nmol/mg of protein. According to the value expressed as the amount per cell, each Buchnera cell at day 10 contained about 1.2 × 108 molecules of spermidine, each of which has trivalent positive charges. In the meantime, it has been shown that a Buchnera cell in adult aphids contains about 6.4 × 107 bp of DNA (about 100 copies of the genome, whose size is about 640 kb) (17). This suggests that spermidine amounts sufficient to neutralize the total negative charges of phosphates due to the double-stranded DNA in a Buchnera cell of young aphids are present.
FIG. 2.
Spermidine content in Buchnera. Buchnera cells were isolated from about 20 individuals each of 10-, 20-, 30-, 40-, and 50-day-old adult aphids. Spermidine content is presented in terms of the amount per cell (A), per volume (B), and per milligram of protein (C). Each data point is the mean ± standard error of five replicate groups.
It is known that a single cell of E. coli contains 5.6 × 106 molecules of putrescine and 1.1 × 106 molecules of spermidine (22). As the host aphids grew older, the spermidine content in Buchnera decreased (Fig. 2A, B, and C), although the level was still higher than that detected in E. coli (1 to 2 mM).
Quantitative RT-PCR of mRNAs for speD and speE.
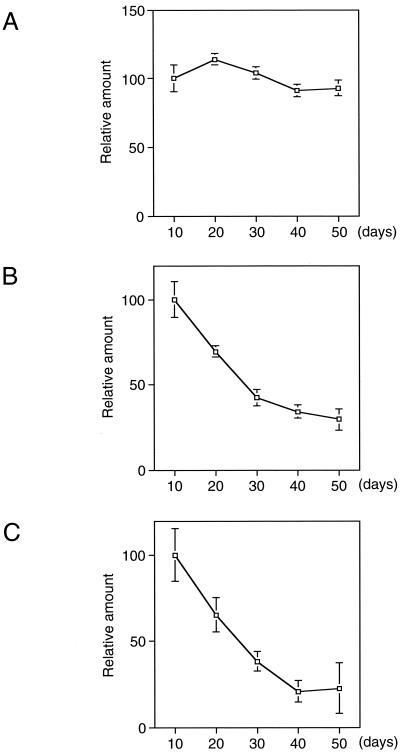
Total RNAs were extracted from bacteriocytes of 10-, 20-, 30-, 40-, and 50-day-old aphids and reverse transcribed. In the beginning, we quantified 16S rRNA in samples derived from the same amount of total RNAs (Fig. 3A). The amount of 16S rRNA in each age sample was used as an internal standard to calibrate the amounts of mRNAs for speD and speE (each value obtained was divided by the amount of 16S rRNA). Relative amounts of these mRNAs are shown in Fig. 3B and C. The mRNAs for speD and speE were the most abundant in the bacteriocytes isolated from 10-day-old aphids and decreased in amount with the age of the host. These results plausibly account for changes in the amount of spermidine estimated in Fig. 2.
FIG. 3.
Relative amounts of speD and speE mRNAs analyzed by quantitative RT-PCR. Total RNA was prepared from bacteriocytes of aphids at the indicated ages (days). cDNAs were synthesized with pd(N)6 primer using the First-Strand cDNA synthesis kit (Pharmacia) and quantified by PCR amplification with a LightCycler instrument (Boehringer Mannheim). Amounts of target cDNAs in each sample are expressed as a proportion of that in Buchnera from 10-day-old aphids. Each data point is the mean ± standard error of five replicate groups. (A) Relative amount of 16S rRNA. The value of each data point was used for calibration in panels B and C. (B) Relative amount of speD mRNA. (C) Relative amount of speE mRNA.
DISCUSSION
The present study revealed that Buchnera contained a large amount of only one polyamine, spermidine. This represented a marked difference in the polyamine composition between Buchnera and E. coli, a bacterium closely related to Buchnera (31). In E. coli, like most other prokaryotes, the most abundant polyamine is putrescine (10, 16, 28), a divalent amine. Spermidine is a trivalent amine with high affinity to DNA and thus, compared with putrescine, much higher activity to stabilize DNA molecules (5, 24, 25). In this context, it is important to consider the cell size and unique genome structure of Buchnera. The cell volume of Buchnera (ca. 10 μm3; Table 2) is about 10 times that of E. coli (0.5 to 1.0 μm3 [19]). Since Buchnera has more than 100 copies of the genome (17), whose size is about a seventh of that of the E. coli genome (4), the total amount of DNA molecules in a Buchnera cell is also about 10 times as great as that of an E. coli cell. Therefore, the DNA volume in Buchnera is roughly similar to that of E. coli, indicating that an extraordinarily large number of circular DNA molecules have to be stabilized in a large Buchnera cell. For this reason, it is conceivable that a unique mechanism for dealing with DNA molecules is needed by Buchnera. A high concentration of spermidine, which is an efficient stabilizer of DNA, in Buchnera can be involved in this mechanism. We also found that the spermidine content decreased with the age of the host aphid. It was demonstrated that the distribution of DNA in the Buchnera cell changes with the age of the host aphid (18). DNA molecules apparently spread uniformly throughout the cell that was isolated from young (18-day) aphids, while the Buchnera cells from middle-aged (30-day) or older (40-day) aphids showed heterogeneous distribution of DNA. These findings may support the hypothesis that spermidine is required to stabilize the large number of DNA molecules in Buchnera cells. However, this does not necessarily mean that all organisms containing large amounts of spermidine have many genomic copies. Spermidine is known as the major polyamine in Bacillus subtilis also, which is a gram-positive bacterium phylogenetically distant from Buchnera (12). In the case of B. subtilis, spermidine is essential for sporulation, which requires compaction of the genomic DNA into a small specialized cell, the spore.
DNA molecules stabilized by spermidine do not form tight aggregates but form highly fluid liquid crystal structures, which cannot be accomplished by inorganic cations or proteins (24). This fluidity enables DNA-binding proteins to get access to DNA molecules, which is prerequisite to gene expression and DNA replication, although it is uncertain whether all the copies of the Buchnera genome function actively. The concentration of spermidine in Buchnera was higher when the host aphids were young, suggesting that this polyamine also plays an important role in DNA replication in Buchnera, since the genomic copy number of Buchnera increases with time when aphids are young (18). This is consistent with the previous reports indicating that an increase in polyamine biosynthesis was required for DNA replication of many other prokaryotic and eukaryotic cells (5, 28).
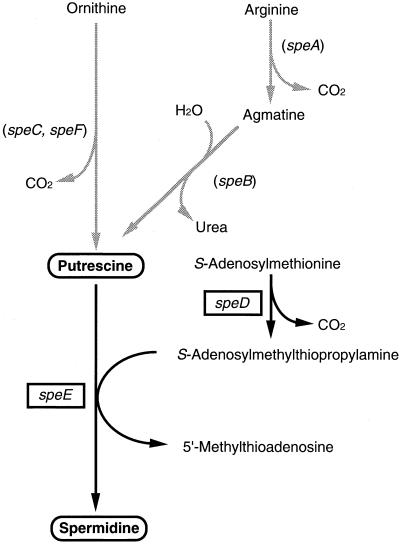
We examined the expression of the speD and speE genes in Buchnera, whose products are key enzymes in spermidine synthesis. The mRNAs for speD and speE were the most abundant in Buchnera isolated from 10-day-old aphids and decreased with age of the host, which was in line with the change in spermidine content. This finding suggests that spermidine detected in Buchnera (Fig. 2) is synthesized through Buchnera's own metabolism. Whole-genome analysis of Buchnera revealed that this bacterium has no other genes than speD and speE that are involved in the polyamine biosynthetic pathway (28), while E. coli has six of them, speA, speB, speC, speD, speE, and speF (2) (Fig. 4). In other words, Buchnera conserves genes that are essential to synthesize spermidine in spite of a drastic reduction in the genome size, suggesting that spermidine is an indispensable substance for Buchnera. However, it is yet to be answered how Buchnera produces spermidine without the ability to synthesize its precursors, such as agmatine and putrescine. The most probable scenario is that the host provides Buchnera with these precursors. It is already known that Buchnera and host aphids exchange amino acids to meet their metabolic requirements (26, 27). Therefore, it is not farfetched to suppose that host aphids affect the physiology of Buchnera through controlling the supply of polyamine precursors.
FIG. 4.
Biosynthetic pathway of polyamines. In Buchnera, the pathways to synthesize putrescine are absent. The enzymes encoded by the genes are as follows: speA, arginine decarboxylase (EC 4.1.1.19); speB, agmatinase (EC 3.5.3.11); speC, ornithine decarboxylase isozyme (EC 4.1.1.17); speD, S-adenosylmethionine decarboxylase (EC 4.1.1.50); speE, spermidine synthase=putrescine aminopropyl transferase (EC 2.5.1.16); speF, ornithine decarboxylase, inducible (EC 4.1.1.17). Genes present in the Buchnera genome are boxed, while those absent are in parentheses.
ACKNOWLEDGMENTS
This work was supported by Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists, a grant from the Program for Promotion of Basic Research Activities for Innovation Biosciences (ProBRAIN) of the Bio-oriented Technology Research Advancement Institution, and Grants-in-Aid for Scientific Research from the Japanese Ministry of Education, Science, Sports and Culture.
REFERENCES
- 1.Baumann P, Baumann L, Lai C-Y, Rouhbakhsh D, Moran N A, Clark M A. Genetics, physiology, and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Annu Rev Microbiol. 1995;49:55–94. doi: 10.1146/annurev.mi.49.100195.000415. [DOI] [PubMed] [Google Scholar]
- 2.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Buchner P. Endosymbiosis of animals with plant microorganisms. New York, N.Y: Interscience Publishers, Inc.; 1965. pp. 297–332. [Google Scholar]
- 4.Charles H, Ishikawa H. Physical and genetic map of the genome of Buchnera, the primary endosymbiont of the pea aphid Acyrthosiphon pisum. J Mol Evol. 1999;48:142–150. doi: 10.1007/pl00006452. [DOI] [PubMed] [Google Scholar]
- 5.Cohen S S. A guide to the polyamines. New York, N.Y: Oxford University Press, Inc.; 1998. [Google Scholar]
- 6.Douglas A E. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol. 1998;43:17–37. doi: 10.1146/annurev.ento.43.1.17. [DOI] [PubMed] [Google Scholar]
- 7.Febvay G, Liadouze I, Guillaud J, Bonnot G. Analysis of energetic amino acid metabolism in Acyrthosiphon pisum: a multidimensional approach to amino acid metabolism in aphids. Arch Insect Biochem Physiol. 1995;29:45–69. [Google Scholar]
- 8.Feuerstein B G, Pattabiraman N, Marton L J. Spermine-DNA interactions: a theoretical study. Proc Natl Acad Sci USA. 1986;83:5948–5952. doi: 10.1073/pnas.83.16.5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gosule L C, Schellmann J A. Compact form of DNA induced by spermidine. Nature. 1976;259:333–335. doi: 10.1038/259333a0. [DOI] [PubMed] [Google Scholar]
- 10.Hamana K. Distribution of diaminopropane and acetylspermidine in Enterobacteriaceae. Can J Microbiol. 1996;42:107–114. doi: 10.1139/m96-017. [DOI] [PubMed] [Google Scholar]
- 11.Houk E J, Griffiths G W. Intracellular symbiotes of the homoptera. Annu Rev Entomol. 1980;25:161–187. [Google Scholar]
- 12.Ishii I, Takada H, Terao K, Kakegawa T, Igarashi K, Hirose S. Decrease in spermidine content during logarithmic phase of cell growth delays spore formation of Bacillus subtilis. Cell Mol Biol. 1994;40:925–931. [PubMed] [Google Scholar]
- 13.Ishikawa H. Host-symbiont interactions in the protein synthesis in the pea aphid, Acyrthosiphon pisum. Insect Biochem. 1982;12:613–622. [Google Scholar]
- 14.Ishikawa H. Control of macromolecule synthesis in the aphid endosymbiont by the host insect. Comp Biochem Physiol. 1984;78B:51–57. [Google Scholar]
- 15.Ishikawa H. Biochemical and molecular aspects of endosymbiosis in insects. Int Rev Cytol. 1989;116:1–45. doi: 10.1016/s0074-7696(08)60637-3. [DOI] [PubMed] [Google Scholar]
- 16.Kashiwagi K, Igarashi K. Adjustment of polyamine contents in Escherichia coli. J Bacteriol. 1988;170:3131–3135. doi: 10.1128/jb.170.7.3131-3135.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komaki K, Ishikawa H. Intracellular bacterial symbionts of aphids possess many genomic copies per bacterium. J Mol Evol. 1999;48:717–722. doi: 10.1007/pl00006516. [DOI] [PubMed] [Google Scholar]
- 18.Komaki K, Ishikawa H. Genomic copy number of intracellular bacterial symbionts of aphids varies in response to developmental stage and morph of their host. Insect Biochem Mol Biol. 2000;30:253–258. doi: 10.1016/s0965-1748(99)00125-3. [DOI] [PubMed] [Google Scholar]
- 19.Mongold J A, Lenski R E. Experimental rejection of a nonadaptive explanation for increased cell size in Escherichia coli. J Bacteriol. 1996;178:5333–5334. doi: 10.1128/jb.178.17.5333-5334.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakabachi A, Ishikawa H. Differential display of mRNAs related to amino acid metabolism in the endosymbiotic system of aphids. Insect Biochem Mol Biol. 1997;27:1057–1062. doi: 10.1016/s0965-1748(97)00092-1. [DOI] [PubMed] [Google Scholar]
- 21.Nakabachi A, Ishikawa H. Provision of riboflavin to the host aphid, Acyrthosiphon pisum, by endosymbiotic bacteria, Buchnera. J Insect Physiol. 1999;45:1–6. doi: 10.1016/s0022-1910(98)00104-8. [DOI] [PubMed] [Google Scholar]
- 22.Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. [Google Scholar]
- 23.Panagiotidis C A, Artandi S, Calame K, Silverstein S J. Polyamines alter sequence-specific DNA-protein interactions. Nucleic Acids Res. 1995;23:1800–1809. doi: 10.1093/nar/23.10.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelta J, Livolant F, Sikorav J-L. DNA aggregation induced by polyamines and cobalthexamine. J Biol Chem. 1996;271:5656–5662. doi: 10.1074/jbc.271.10.5656. [DOI] [PubMed] [Google Scholar]
- 25.Saminathan M, Antony T, Shirahata A, Sigal L H, Thomas T, Thomas T J. Ionic and structural specificity effects of natural and synthetic polyamines on the aggregation and resolubilization of single-, double-, and triple-stranded DNA. Biochemistry. 1999;38:3821–3830. doi: 10.1021/bi9825753. [DOI] [PubMed] [Google Scholar]
- 26.Sasaki T, Ishikawa H. Production of essential amino acids from glutamate by mycetocyte symbionts of the pea aphid, Acyrthosiphon pisum. J Insect Physiol. 1995;41:41–46. [Google Scholar]
- 27.Shigenobu, S., H. Watanabe, M. Hattori, Y. Sakaki, and H. Ishikawa. Mutualism as revealed at the genomic level: the whole genome sequence of Buchnera sp. APS, an endocellular bacterial symbiont of aphids. Nature, in press. [DOI] [PubMed]
- 28.Tabor C W, Tabor H. Spermidine, spermine, and related amines. Pharmacol Rev. 1964;16:245–300. [PubMed] [Google Scholar]
- 29.Tabor C W, Tabor H. Polyamines. Annu Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- 30.Thomas T J, Gunnia U B, Thomas T. Polyamine-induced B-DNA to Z-DNA conformational transition of a plasmid DNA with (dG-dC)n insert. J Biol Chem. 1991;266:6137–6141. [PubMed] [Google Scholar]
- 31.Unterman B M, Baumann P, McLean D L. Pea aphid symbiont relationships established by analysis of 16S rRNAs. J Bacteriol. 1989;171:2970–2974. doi: 10.1128/jb.171.6.2970-2974.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wittwer C T, Herrmann M G, Moss A A, Rasmussen R P. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques. 1997;22:130–138. doi: 10.2144/97221bi01. [DOI] [PubMed] [Google Scholar]