Abstract
Microbes interact in natural communities in a spatially structured manner, particularly in biofilms and polymicrobial infections. While next generation sequencing approaches provide powerful insights into diversity, metabolic capacity, and mutational profiles of these communities, they generally fail to recover in situ spatial proximity between distinct genotypes in the interactome. Hi‐C is a promising method that has assisted in analysing complex microbiomes, by creating chromatin cross‐links in cells, that aid in identifying adjacent DNA, to improve de novo assembly. This study explored a modified Hi‐C approach involving an initial lysis phase prior to DNA cross‐linking, to test whether adjacent cell chromatin can be cross‐linked, anticipating that this could provide a new avenue for study of spatial‐mutational dynamics in structured microbial communities. An artificial polymicrobial mixture of Pseudomonas aeruginosa, Staphylococcus aureus, and Escherichia coli was lysed for 1–18 h, then prepared for Hi‐C. A murine biofilm infection model was treated with sonication, mechanical lysis, or chemical lysis before Hi‐C. Bioinformatic analyses of resulting Hi‐C interspecies chromatin links showed that while microbial species differed from one another, generally lysis significantly increased links between species and increased the distance of Hi‐C links within species, while also increasing novel plasmid‐chromosome links. The success of this modified lysis‐Hi‐C protocol in creating extracellular DNA links is a promising first step toward a new lysis‐Hi‐C based method to recover genotypic microgeography in polymicrobial communities, with potential future applications in diseases with localized resistance, such as cystic fibrosis lung infections and chronic diabetic ulcers.
Keywords: biofilm structure, DNA cross‐linking, genomics, Hi‐C, polymicrobial infections
1. INTRODUCTION
Microbial communities in chronic infections and biofilms tend to diversify genetically in a spatially structured manner (Burmølle et al., 2014; Cullen & McClean, 2015; Hotterbeekx et al., 2017; Jorth et al., 2015; Whiteson et al., 2014; Winstanley et al., 2016), posing serious challenges to researchers and clinicians. For example, in cystic fibrosis (CF) lung infections, localized cell clusters undergo mutational changes and develop distinct infection niches (Cullen & McClean, 2015; Hauser et al., 2011; Jorth et al., 2015; Oliver & Mena, 2010; Whiteson et al., 2014; Winstanley et al., 2016). Similarly, in chronic diabetic ulcers and wounds, biofilm communities form resistant, structured polymicrobial complexes (Burmølle et al., 2014; DeLeon et al., 2014; Kirketerp‐Møller et al., 2008; Misic et al., 2014). More broadly, spatial structure is a key aspect in other biofilms, such as those causing problems in water system piping, medical devices, and living tissues, as microcolonies diverge while embedded in their extracellular polymeric substance supported by a lattice of extracellular DNA (eDNA) (Hall‐Stoodley et al., 2004; Percival et al., 2014). However, despite the importance of spatial dynamics and lattice eDNA in polymicrobial infections and other biofilms, current genomic approaches fail to capture these data.
Next‐generation sequencing (NGS) approaches can characterize genotypes and community DNA, but they do not recover spatial microgeography of infection genotypes or matrix eDNA. We propose that exposing this missing microgeography could reveal intriguing dynamics in these infections and uncover important mechanisms of antibiotic resistance evolution and biofilm resistance to treatment. For example, in natural microbial communities spatial dynamics may promote exchange metabolites by cross‐feeding (Seth & Taga, 2014; Wintermute and Silver, 2010) which can promote synergistic interactions between microbes such as complementary auxotrophies that worsen infections and virulence (Baishya & Wakeman, 2019; Burmølle et al., 2014; Hotterbeekx et al., 2017). Importantly, spatially proximity in biofilm and microbial communities may promote interactions that select for localized antimicrobial resistance (Bisht & Wakeman, 2019; Calhoun et al., 2008; Oliver, 2010; Oliver & Mena, 2010) or exchanges of mobile DNA and plasmids (Burmølle et al., 2014; Davey & O’toole, 2000; López‐Collazo et al., 2015) that confer antibiotic resistance. Finally, eDNA structure and interactions critically support biofilms (Devaraj et al., 2019; Eisenbeis et al., 2018; Pakkulnan et al., 2019).
New NGS approaches, for the first time, may offer promise to simultaneously recover distinct local genotypes, eDNA interactions, and the spatial context of these elements. For example, microfluidics‐based linked‐read approaches or read cloud approaches can yield long‐distance genome sequence contexts by linking barcodes onto contiguous chromatin prior to short‐read sequencing (Kang et al., 2019; Zheng et al., 2016). Alternatively, single‐cell approaches using flow cytometry and whole genome amplification can associate chromosomes with intracellular plasmids (Conlan et al., 2014; Koren & Phillippy, 2015). An even more promising set of approaches include the proximity ligation or Hi‐C‐based methods. Hi‐C uses simple formaldehyde crosslinking followed by restriction digestion and novel ligation of neighbouring chromatin prior to sequencing to trace long‐distance chromatin interactions (Belaghzal et al., 2017; van Berkum et al., 2010). For microbial communities, Hi‐C has been used to improve the grouping of sequences to species, thereby assisting in microbiome de novo assembly (DeMaere and Darling, 2018; Demaere & Darling, 2019; Marbouty et al., 2014; Yaffe & Relman, 2020) and improving assignment of plasmids or prophage to their proper hosts (Kent et al., 2020; Marbouty et al., 2017; Ramani et al., 2017; Stalder et al., 2019). However, all these approaches fail to reveal DNA proximity between adjacent cells and have not been studied in the context of eDNA in biofilms, because the standard method is to barcode or crosslink and ligate DNA within intact cells.
Therefore, here we explore a novel approach to retrieve cellular and eDNA microgeography along with community genome sequencing using a modified application of Hi‐C. The approach seeks to lyse cells in situ, thus allowing neighbouring cell DNA to interact prior to crosslinking the DNA during Hi‐C library preparation. Then released crosslinked DNA is precipitated and retained, rather than being washed away as in conventional Hi‐C, and eDNA present in the extracellular matrix may also be retained and sequenced. Here, we test the effectiveness of the first steps in this process, comparing different lysis conditions in multispecies cell mixtures which have been washed to remove eDNA, and in mouse wound biofilms. In both cases, we focus on microbes that are well‐studied laboratory models that also form biofilms and are commonly isolated from CF lungs or chronic wounds, Pseudomonas aeruginosa, Staphylococcus aureus, and Escherichia coli (Hauser et al., 2011; Kirketerp‐Møller et al., 2008; Waters & Smyth, 2015). Results showed that initial in situ lysis prior to Hi‐C crosslinking increased cell‐cell chimeric link associations between species, demonstrating interacting eDNA crosslinks can be recovered, a promising first step toward a new lysis‐Hi‐C based method to recover genotypic microgeography in polymicrobial communities.
2. MATERIALS AND METHODS
2.1. Cell culturing and artificial polymicrobial mixture preparation
Pseudomonas aeruginosa (PA14), Staphylococcus aureus (Newman), and Escherichia coli (K‐12 MG1655) containing the pUPC18 plasmid, were prepared by inoculation separately on tryptic soy agar plates. A single colony from each species was transferred to Luria‐Bertani (LB) broth and shaken at 220 rpm overnight at 37°C to obtain cultures with optical densities of 0.5 in 1:10 dilution of phosphate buffered saline (PBS). Cell suspensions were stored at 4°C briefly, then washed to remove aqueous phase including possible extracellular DNA (eDNA) from dead/lysed cells by centrifuging at 10,000 g for 3 min, discarding the supernatant. The three species were combined in a sterile 1.5 ml tube, gently vortexed for 1 min, pelleted and supernatant was removed to remove further eDNA, and then cells were resuspended. The final mixture was spread into thin smears on the side of sterile 1.5 ml microcentrifuge tubes and allowed to air dry, open for 30 min so they formed a 1 mm thin layer of cells concentrated and adhered to the tube, serving as in vitro communities (Figure 1).
FIGURE 1.
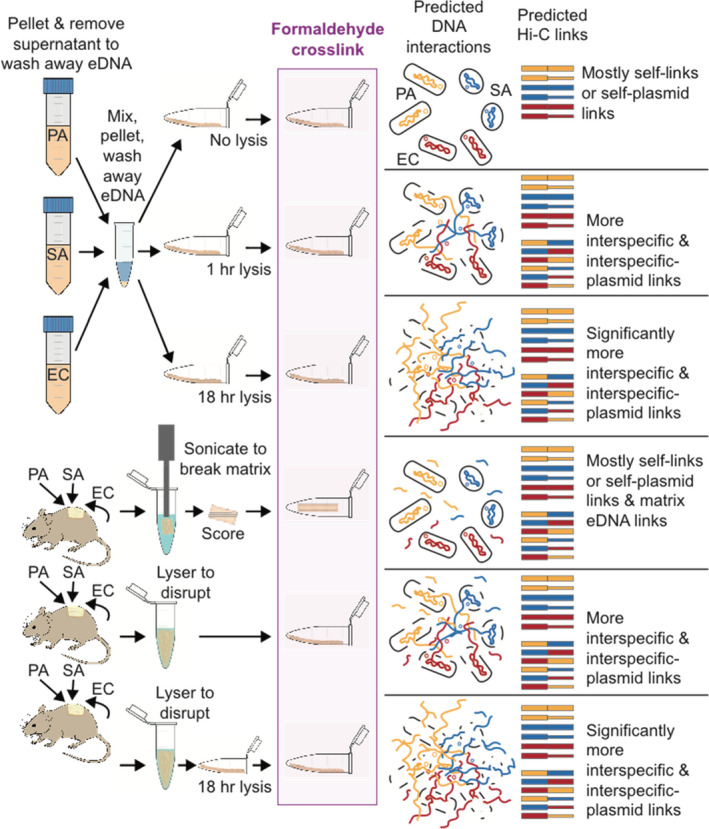
Sample treatments and predicted DNA interactions during formaldehyde crosslinking with predicted Hi‐C link outcomes expected. EC, Escherichia coli (DNA in red); PA, Pseudomonas aeruginosa (DNA in yellow); SA, Staphylococcus aureus (DNA in blue). Plasmid DNA is shown as small circles and as thinner lines in Hi‐C links
2.2. Mouse wound biofilm in vivo model growth and preparation
Mouse wound models were used for in vivo biofilm samples following protocols described previously (DeLeon et al., 2014). Six female Swiss Webster non‐SPF, non‐germ free mice (Charles River Laboratory) of similar body size (18–20 g), raised in cohoused conditions were shaved, anesthetized and received 1.5 x 1.5 cm surgical excisions dorsally, then wounds were covered with transparent, semipermeable polyurethane dressing (OPSITE, Smith & Nephew) which allowed for topical application of bacteria onto wounds and protection from contamination (Watters et al., 2013). Pseudomonas aeruginosa (PAO1) and S. aureus (SA31) were grown overnight in LB broth cultures and subcultured to new LB, then grown at 37°C for 4 h to obtain an optical density of 0.9 at 600 nm, as described previously (Watters et al., 2013). Aliquots of 100 µl were pelleted and washed with PBS then serially diluted tenfold to obtain 10−3 dilutions. Both species were applied topically to wounds by injection under the dressing at approximately 104 CFUs per 100 µl. Mice were euthanized at 5 days post treatment and wound biofilms were collected. Escherichia coli from these biofilms (see Result) were probably present through natural murine passage from housing and grooming. Mice housing and study procedures followed protocols approved by the Institutional Animal Care and Use Committee in the animal facility of Texas Tech University Health Sciences Center. Although P. aeruginosa and S. aureus strains differed between our cell and mouse experiments, these differences should have little to no impact on our study because the genomes and chromatin within each species are known to be similar and therefore would be relatively similar in DNA crosslinking patterns during Hi‐C.
2.3. Biofilm disruption
Initial trials, showing no yield of biotin‐captured products during Hi‐C, suggested that mouse wound biofilm matrix disruption was necessary to increase formaldehyde crosslinking effectiveness. To achieve this, biofilms from separate mice were sliced to 5 mm by 1 cm pieces and transferred to separate 2 ml centrifuge tubes. Samples were treated to disrupt the biofilm matrix while leaving cells intact in situ in the biofilm using a short, low‐power sonication protocol developed in our laboratory based on published studies (Joyce et al., 2011; Mandakhalikar et al., 2018). Briefly this involved sonicating in 1 ml of tris buffered saline with TritonX‐100 (TBS‐T) for 1 min (20% amplitude, pulse 5 s, duration 30 s, achieving ~24 Watts) using a Qsonica Q500 Sonicator (Fischer Scientific). After sonication, the biofilm sample was inspected and scored on all sides under a dissection microscope on an cold plate with sterile forceps before being transferred to a new tube for formaldehyde cross‐linking. Remaining biofilm samples were disrupted by shaking in the presence of a 7 mm stainless steel bead and 1 ml of tris buffered saline with TritonX‐100 (TBS‐T) cooled to 4°C. Samples were shaken at 50 Hz on the TissueLyserLT (Qiagen) and placed on ice every few minutes to cool until tissue (skin and biofilm) appeared broken down to a slurry. These biofilm slurries were transferred to separate 1.5 ml tubes and spread along the side of the tube and allowed to air‐dry to concentrate cells and adhere them to the tube for 1 h prior to downstream lysis (or no lysis) and crosslinking treatments (Figure 1). These mechanical disruptions would undoubtedly change community spatial structure, and therefore are not proposed as a part of future applications of this method. However, in the present study we sought to compare factors among treatments differing in few variables, thus, we included a biofilm close to its original state (sonication/scoring treatment) and treatments of disrupted/lysed biofilms closely resembling mixed cells (described below).
2.4. Cell lysis and formaldehyde crosslinking
Prior to formaldehyde crosslinking, samples were subjected to one of six treatments (see Figure 1 and Table S1). For cultured cell mixtures spread thinly to mimic artificial polymicrobial mixtures or “communities,” treatments were: (1) no lysis, (2) short chemical lysis (1 h), and (3) long chemical lysis (18 h). For mouse wound biofilms, treatments were: (4) no lysis on sonicated and scored intact biofilm, (5) short mechanical disruption (as described above), and (6) short mechanical disruption followed by long chemical lysis (18 h). Chemical lysis was performed by addition of 40 µl of Igepal lysis buffer (5 mM Tris‐HCl, 10 mM NaCl, and 0.02% Igepal CA‐630 detergent) to the surface of adhered cell suspensions or biofilm slurries followed by incubation for 1 h (short lysis) or 18 h (long lysis). All samples were then crosslinked by saturation of the surface layer with 40 µl of 1% formaldehyde for 20 min at room temperature. Formaldehyde cross‐linking was quenched with 50 µl of 2.5 M glycine on ice for 10 min to prevent over cross‐linking. Samples were then centrifuged to pellet cellular material, and then 2.5× volume of 100% ethanol and 1/10 volume of 3 M sodium acetate was added to each tube before storage at −20°C overnight to precipitate any free crosslinked DNA. After DNA precipitation, samples were centrifuged for 30 min at 4°C, at 14,000× g, supernatant was removed, pellets were washed 3 times with 1 ml of 80% ethanol.
2.5. Hi‐C library preparation and Illumina sequencing
Hi‐C libraries were prepared with the Dovetail Hi‐C Library Preparation Kit (Dovetail Genomics) with a modified protocol to optimize for bacterial targets (Crémazy et al., 2018) and additional steps to conserve free DNA that tends not to precipitate by centrifugation but instead is precipitated in ethanol or isolated by binding on AMPure beads (see detailed protocol in Supporting Information Materials Appendix S1). Finished lysis‐Hi‐C libraries were checked for quality on the Agilent TapeStation 2200, and normalized, and pooled for sequencing. Illumina multiplexed libraries were sequenced on the HiSeq platform with 150 bp paired end reads, performed at Genewiz Inc.
2.6. Read filtering and HiC‐Pro reference creation
Initial total read analysis was performed using read mapping analyses in bwa mem v1.9–63 (Li, 2013) and results parsing in SAMtools (Li et al., 2009) using reference genomes for Pseudomonas aeruginosa (PAO1 and PA14), Staphylococcus aureus (Newman/SA31), and Escherichia coli (K‐12 MG1655), along with sequences from their known plasmids, pUCP18 used in E. coli and pG527, a common natural IncP‐1‐type plasmid in Proteobacteria (Sen et al., 2013) were downloaded from NCBI (Table S2).
Then, to prepare raw reads for mapping using Bowtie2 (Langmead & Salzberg, 2012) the HiC‐Pro (Servant et al., 2015) pipeline, we used an in house script to merge read overlaps in Pear v0.9.11 (Zhang et al., 2014), trim and quality filter reads in Trimmomatic v.0.38 (Bolger et al., 2014), and split merged overlapped reads again produce nonoverlapping paired files for Bowtie2.
2.7. HiC‐Pro analysis and statistical analysis of interchromosome links
HiC‐Pro 2.11.1 (Servant et al., 2015) was run using a singularity container as recommended in the user manual. First, references were indexed in Bowtie2 for estimated insert size ranging from 50–800 bp. Then DpnII restriction enzyme sites were calculated for the reference files using the ligation site sequence (GATCGATC) and python utilities for in silico digestion, outputting a bed format file of restriction sites. A chromosome sizes file was prepared using a simple awk command, then a HiC‐Pro configuration file was prepared. HiC‐Pro analysis was then performed in the following steps. First, reads were aligned end to end to the reference genomes in Bowtie2, then chimers created by formaldehyde crosslinking were detected using ligation site information and mapping. Valid Hi‐C interactions were detected by determining if the mapped reads were of the appropriate insert size, near a restriction site, and spanned two different restriction fragments when mapped. Valid interactions were binned into equal sized groups across the genomes with map counts tallied per each bin. Interchromosomal links were analysed from HiC‐Pro 20,000 bp bin matrix files, normalized per bin and per count for each genome within each sample, to control for differences in numbers of cells of each species and number of reads and valid interactions from each sample.
2.8. Statistical analyses, HiTC contact maps, and Circos links display
From output non‐normalized HiC‐Pro 20,000 matrices and bed files, links were annotated based on mapping position in a spreadsheet, then link distances from the origin of replication were calculated and median distance from the origin was compared using the one‐tailed Mann‐Whitney U test. Differences in link types were compared using the two‐proportion z‐test, and error bars were calculated to indicate 95% confidence intervals around the proportions using the Wilson score method without continuity correction (Newcombe, 1998). Contact maps were computed from annotated matrices in R using the HiTC BioC package (Servant et al., 2012). Circular link diagrams were generated in Circos (Krzywinski et al., 2009).
3. RESULTS
3.1. Hi‐C read statistics, read abundance, and coverage for species and plasmids
Total reads obtained per sample ranged from 48.3 to 255.4 million (Table S3). Of these reads, for cells 75.4% to 93.8% of reads mapped to microbial genomes and plasmids, whereas for mouse samples only 0.52% to 1.88% of reads mapped to microbes and plasmids. In all samples, including both in vitro cell polymicrobial mixtures and in vivo mouse wound biofilm samples, the greatest percent of properly paired reads mapped to the pUCP18 plasmid (Figure S1 and Table S3), with the next highest rates of read mapping to S. aureus (SA), followed by E. coli (EC), with fewest reads mapping to P. aeruginosa (PA) and the pG527 plasmid. Genome and plasmid sequencing coverage levels were well over 30x for cells, but were lower than 30× for mouse samples, and particularly low for PA. For all samples, the percentages of Hi‐C interactions assessed as valid by the HiC‐Pro software were similar, with most invalid interactions being dangling end pairs (Figure S2).
3.2. Within‐species Hi‐C link distance changes associated with lysis treatments
Hi‐C links within species, which may include links within the same cell and between neighbouring cells of the same species were the most common category of link in all treatments, except in E. coli (EC) short and long lysis treatments, in which links with other species were more common. Analysis of link distances within species (measured from the origin) showed significant increase in the distance of links in response to lysis treatments for E. coli (EC) for cells, and for P. aeruginosa (PA) in both cell polymicrobial mixtures and mouse wound biofilms (Figure 2). Statistical tests for difference in median within‐species link differences in 20kb units: Mann‐Whitney U tests (one‐tailed) of difference in medians (e.g., for EC, CNL 1 vs. CSL 39: z‐score = −16.0584, p‐value = 2.4955 x 10−58; for EC, CSL 39 vs. CLL 9: z‐score = –9.3285, p‐value = 5.365 x 10−21; for PA, CNL 35 vs. CSL 72: z‐score = −15.1748, p‐value = 2.5933 x 10−52; for PA, CNL 35 vs. CLL 69: z‐score = –46.629274, p‐value = 0; for PA, MS 0 vs. MLL 24.5: z‐score = −8.1279, p‐value = 2.1831 x 10−16; for PA, MML 37 vs. MLL 24.5: z‐score = –1.4135, p‐value = 0.07875, i.e., not significant). However, unlike EC and PA, for all samples including cell polymicrobial mixtures and mouse wound biofilms S. aureus (SA) did not show median self‐link distance differences in response to lysis and instead showed similar link numbers for all distances (i.e., a straight slope in Figure 2). Inspection of SA‐SA self‐link read mapping density (Figure S3) showed a largely uniform distribution of read density across link distances from 0 to 72 x 20 kb regions, with many higher read‐density regions with 250–500 reads mapped per link, dispersed throughout the genome. However, the proportion of SA self‐link reads mapping to <20 kb distance (i.e., links of “0”) was significantly decreased by lysis treatments for both cells and mouse wound biofilm samples (e.g., percent of SA‐SA reads with <20 kb distance for CNL 16.9% vs. CSL 6.53% vs. CLL 7.32%, all statistically significant, and for MS 7.59% vs. MML 5.55% vs. MLL 5.70%; see p‐values in Table S4).
FIGURE 2.
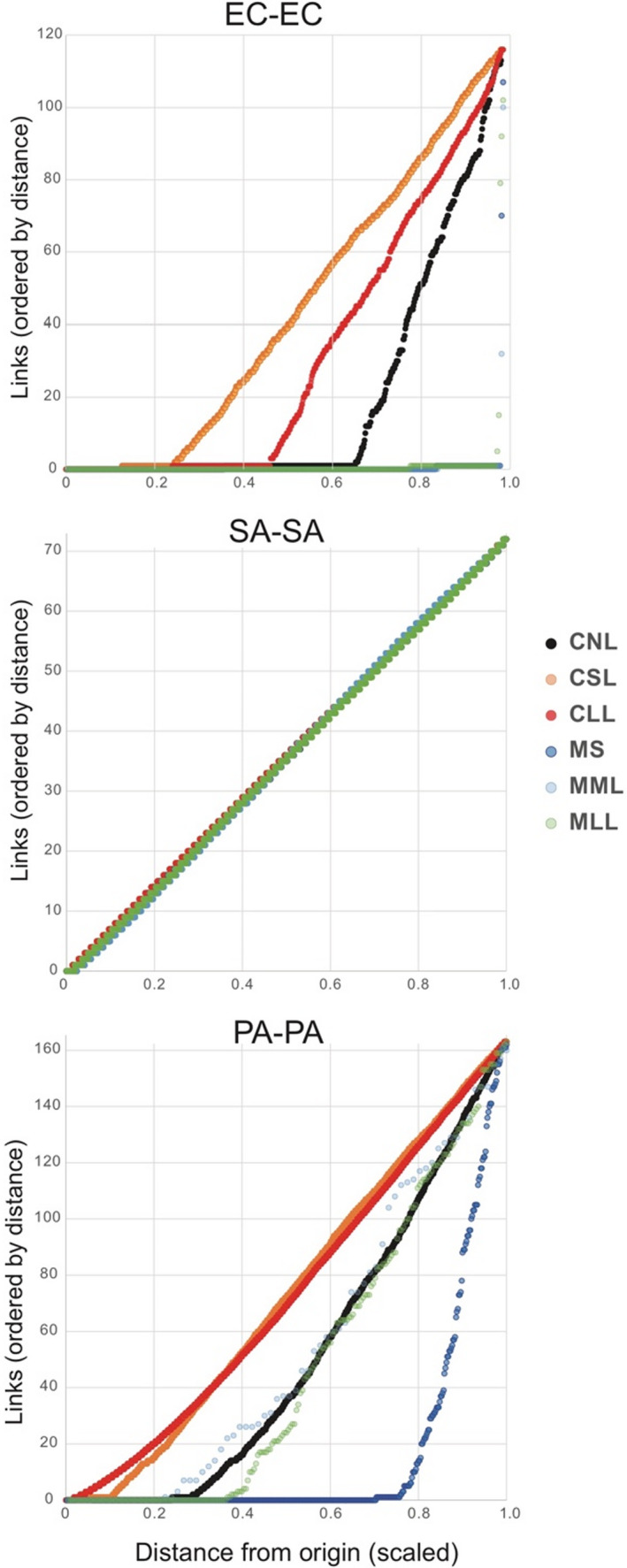
Distance of Hi‐C links within species (i.e., self‐links) measured from origin of replication, normalized between species (i.e., link distance from origin was scaled to a value of 1). CLL, cells long lysis; CNL, cells no lysis; CSL, cells short lysis; EC, Escherichia coli; MLL, mouse long lysis; MML, mouse mechanical lysis; MS, mouse sonicated; PA, Pseudomonas aeruginosa; SA, Staphylococcus aureus. Note: in the SA–SA plot, all six treatments are depicted, but points are difficult to distinguish because they fall on the same line
3.3. Effects of lysis on interspecies Hi‐C links
Raw intra‐ and interspecies Hi‐C links depicted in non‐normalized contact maps (Figure S4) showed differences between species and treatments in the link sites formed. The proportion of Hi‐C links that were interspecies, rather than intraspecies (i.e., self‐links) was generally higher for cell polymicrobial mixtures (e.g., 4.3% to 67.2%) than for mouse wound biofilms (e.g., 0.031% to 10.1%). Read density mapping to the links >20 kb distance showed a similar pattern between cells and mouse biofilms, although percent values were usually lower (Figure 3).
FIGURE 3.
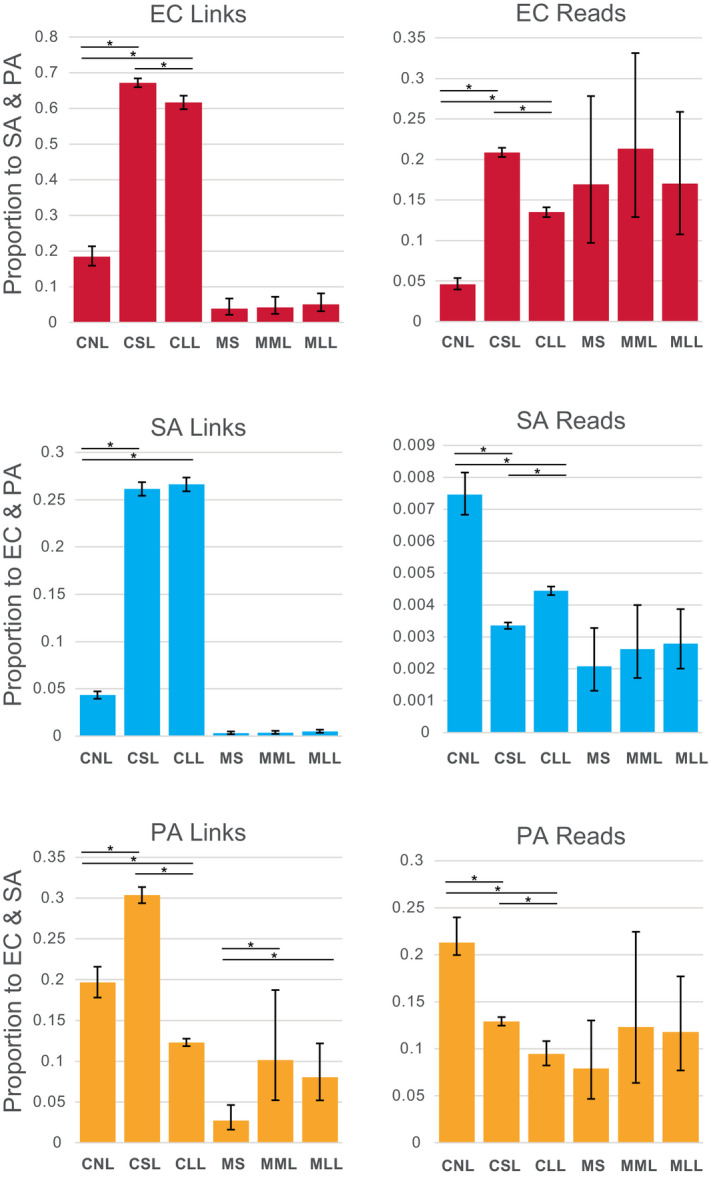
Proportion of Hi‐C links and reads occurring between species plotted for cell polymicrobial “communities” and mouse wound biofilms. Asterisks show statistically significant differences (two‐proportion z‐test) and error bars show 95% confidence intervals calculated by the Wilson score method without continuity correction (Newcombe, 1998). CLL, cells long lysis; CNL, cells no lysis; CSL, cells short lysis; EC, Escherichia coli; MLL, mouse long lysis; MML, mouse mechanical lysis; MS, mouse sonicated; PA, Pseudomonas aeruginosa; SA, Staphylococcus aureus
The proportion of interspecies Hi‐C links within species for cell mixtures increased following short lysis for all three species (e.g., for EC, CNL 18.5% vs. CSL 67.2%: two‐proportion z‐test significant z‐score: −26.270901, p‐value < .00000001; for SA, CNL 4.33% vs. CSL 26.1%: z‐score: −45.464189, p‐value < .00000001; and for PA, CNL 19.6% vs. CSL 30.3%: z‐score: –8, Table S5). Interspecies Hi‐C links also increased significantly compared to no lysis controls for cell polymicrobial mixtures following long lysis for EC and SA (e.g., for EC, CNL 18.5% vs. CLL 61.7%: two‐proportion z‐test significant z‐score: –21.1853919, p‐value < .00000001; for SA, CNL 4.33% vs. CLL 26.6%: z‐score: –46.1893673, p‐value < .00000001). However, longer lysis significantly reduced the proportion of interspecies Hi‐C links for PA (e.g., CNL 19.6% vs. CLL 9.45%: z‐score: 8.667221, p‐value < .00000001). The same significant increase in interspecies Hi‐C links following lysis treatments of cells was evident in read density to links for EC (e.g., CNL 4.62% vs. CSL 20.9%: z‐score: −22.751932, p‐value < .00000001; CNL 4.62% vs. CLL 13.5%: z‐score −14.4369787, p‐value < .00000001) (Figure 3). However, lysis treatments significantly reduced relative read density of interspecies links compared to intra‐species links for SA and PA (e.g., for SA, CNL 0.746% vs. CSL 0.335%: z‐score: 17.2520165, p‐value < .00000001; CNL 0.746% vs. CLL 0.444%: z‐score: 11.008855, p‐value < .00000001; and for PA, CNL 21.3% vs. CSL 12.9%: z‐score: 9.54093336, p‐value < .00000001; CNL 21.3% vs. CLL 9.45%: z‐score 15.467366, p‐value < .00000001).
For mouse wound biofilms proportions of interspecies Hi‐C links and interspecies read densities to these links tended to increase with lysis, however, these patterns were generally not statistically significant (Figure 3 and Table S5). The exception was in PA in mice wound biofilms, for which both mechanical and Igepal lysis showed significantly greater interspecies Hi‐C links (e.g., MS 2.74% vs. MML 10.13%: z‐score: −3.17899263, p‐value 0.0007389; MS 2.74% vs. MLL 8.02%: z‐score: –3.19779, p‐value: 0.0006924).
Lysis duration (1 h vs. 18 h treatment with Igepal) had less impact on the magnitude of interspecies links than the presence or absence of lysis steps for EC and SA, but not PA, in which longer lysis produced significantly fewer interspecies links than other treatments (as described above). This pattern of lower magnitude difference between lysis methods for mouse wound biofilms, comparing mechanical lysis to mechanical and 18 h Igepal, was similar for SA and PA (with statistical support in PA as described above), but not for EC (Figure 3).
3.4. Effects of lysis on plasmid‐chromosome Hi‐C links
The proportion of Hi‐C links between chromosomes and plasmids compared to self‐links differed between species (Figure 4), was highest levels in EC cell polymicrobial mixtures (e.g., 13.6% to 27.2%) and mouse wound biofilms (e.g., 6.76% to 11.1%). Lower levels of plasmid links occurred in SA (e.g., 0.379% to 2.05%) and PA (e.g., 0.860% to 4.73%). Similar patterns occurred for levels of read densities for chromosome‐plasmid links, with EC having higher rates of read density for plasmid links than SA and PA (Figure 4). Lysis treatments had significant effect on plasmid links for all cell polymicrobial mixtures in all species, and in mouse wound biofilms, for some comparisons. The number of links between EC and plasmids decreased significantly following lysis in cell mixtures (e.g., CNL 27.2% vs. CSL 13.6%: z‐score: 8.89626892, p‐value < .00000001; CNL 27.2% vs. CLL 20.8%: z‐score: 3.4447239, p‐value: .0002858; Figure 4 and Table S5). A similar pattern was observed for EC links to plasmids for read densities (e.g., CNL 33.4% vs. CSL 28.1%: z‐score: 7.35328577, p‐value < .00000001; CNL 33.4% vs. CLL 30.4%: z‐score: 3.8872897, p‐value: .0000507). Conversely, the number of links between SA and PA and plasmids significantly increased following lysis for cell mixtures. For example, in SA, lysis increased links to plasmids in both lysis treatments (e.g., CNL 1.30% vs. CSL 2.05%: z‐score: −4.2524199, p‐value: .0000106; CNL 1.30% vs. CLL 1.75%: z‐score: −2.66190574, p‐value: .0038850). In PA, lysis increased links for short lysis treatment (e.g., CNL 2.29% vs. CSL 4.73%: z‐score: –4.0545417, p‐value: .0000251), but decreased links for long lysis treatment (e.g., CNL 2.29% vs. CLL 1.23%: z‐score: 3.3485574, p‐value: .0004062). In PA cell “communities,” plasmid link read densities followed the same pattern as described above for link numbers, with short lysis increasing plasmid links, although this was not significant, and long lysis significantly decreasing plasmid links (e.g., CNL 2.87% vs. CLL 1.48%: z‐score: 3.9949629, p‐value: .0000324). Conversely, SA plasmid link read densities were significantly decreased for short lysis (e.g., CNL 0.672% vs. CSL 0.479%: z‐score: 6.89141075, p‐value < .00000001), but not long lysis (Figure 4 and Table S5). For mouse wound biofilms, lysis increased the plasmid link numbers significantly in several cases, for example, in EC (e.g., MS 6.76% vs. MML 11.1%: z‐score –1.85647694, p‐value: .0316928), in SA (MML 0.379% vs. MLL 0.608%: z‐score: –1.79246863, p‐value: 0.0365290), and in PA (MS 0.860% vs. MLL 1.80%: z‐score: −4.955125, p‐value: 0.0000004). Plasmid link read densities were only significantly increased in EC (e.g., MS 28% vs. MML 42.9%: z‐score: −1.95014374, p‐value: .0255795).
FIGURE 4.
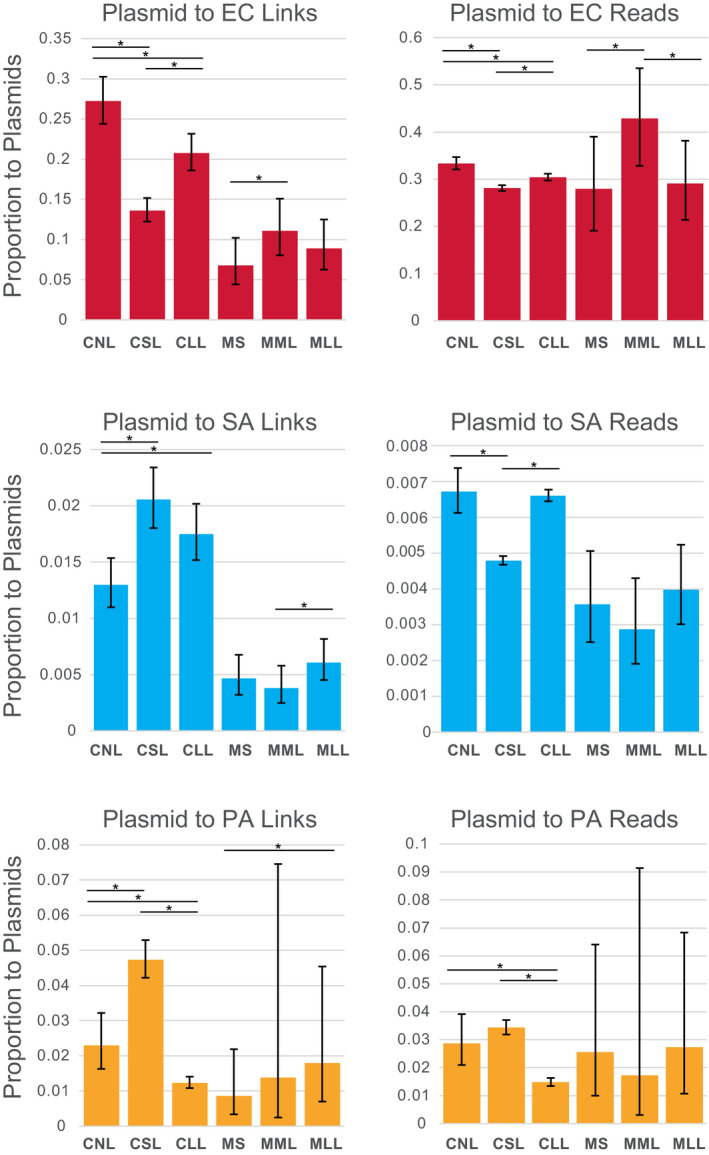
Proportion of Hi‐C links and reads occurring between cells and plasmids (pG527 and pUPC18) plotted for cell polymicrobial “communities” and mouse wound biofilms. Asterisks show statistically significant differences (two‐proportion z‐test) and error bars show 95% confidence intervals calculated by the Wilson score method without continuity correction (Newcombe, 1998). CLL, cells long lysis; CNL, cells no lysis; CSL, cells short lysis; EC, Escherichia coli; MLL, mouse long lysis; MML, mouse mechanical lysis; MS, mouse sonicated; PA, Pseudomonas aeruginosa; SA, Staphylococcus aureus
Lysis duration (1 h vs. 18 h treatment with Igepal for cells or mechanical lysis to mechanical and 18 h Igepal for mouse wound biofilms) had a variable impact on the magnitude of plasmid links in all species (Figure 4), with generally similar effects as described for interspecies links described above.
3.5. Effects of lysis on Hi‐C link positions
Interspecies Hi‐C link positions were dispersed broadly and variably across genomes, regardless of treatment or species (Figure 5 and Figure S5). Similarly, plasmid link positions for pUPC18 and pG527 were scattered across genomes, without any notable pattern (Figure 5).
FIGURE 5.
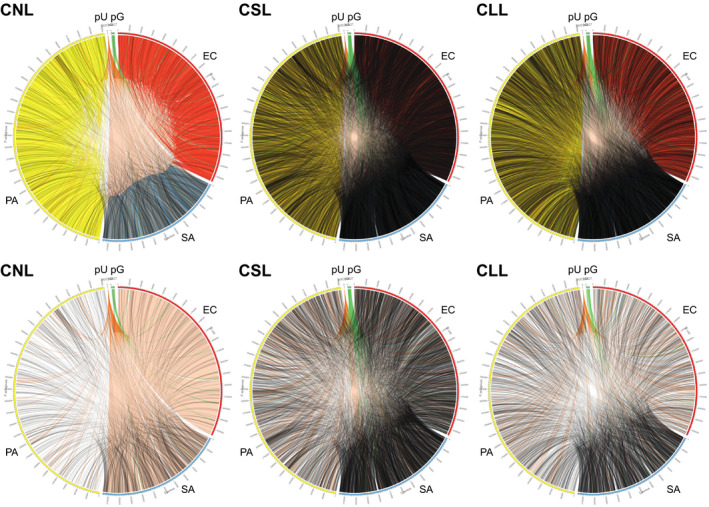
Hi‐C link positions within and between cells and plasmids for cell mixtures (3‐species artificial polymicrobial “communities”), drawn in Circos. Upper row shows plots for all links (not normalized); lower row shows only interspecies or chromosome‐to‐plasmid links, normalized to the same number of links between samples. CLL, cells long lysis; CNL, cells no lysis; CSL, cells short lysis; EC, Escherichia coli; PA, Pseudomonas aeruginosa; pU, pUCP18 plasmid; pG, pG527 plasmid; SA, Staphylococcus aureus
4. DISCUSSION
Our results showed that in situ lysis prior to Hi‐C formaldehyde cross‐linking significantly increased interspecies Hi‐C cross‐links. Specifically, we found that short lysis treatment to the surface of artificial mixed species “communities” increased the proportion of interspecies Hi‐C links between Escherichia coli, Staphylococcus aureus, and Pseudomonas aeruginosa from 1.5‐ to 6‐fold, up to levels of one quarter to two thirds of sequenced inter‐chromosomal Hi‐C links. These findings support the potential feasibility of lysis‐Hi‐C as an approach for assessing microgeography and localized microevolution in cell communities. Such an approach could improve upon existing NGS methods which fail to capture mutational spatial dynamics and nuanced complexities of resistant communities arising in natural biofilms and chronic infections (Calhoun et al., 2008; López‐Collazo et al., 2015; Oliver, 2010; Oliver & Mena, 2010; Waters & Smyth, 2015). For example, sequencing of neighbouring species in polymicrobial biofilms of chronic wounds and CF lung infections could help identify enhanced virulence due to synergistic and evolved interactions between microbes (Burmølle et al., 2014; Hauser et al., 2011; Hotterbeekx et al., 2017; Jorth et al., 2015; Seth & Taga, 2014; Wintermute and Silver, 2010; Winstanley et al., 2016;).
These data also revealed differences between interspecies links depending on bacterial species and lysis method. Cells of E. coli formed the highest level of interspecies links after lysis (up to 67%), whereas S. aureus showed the highest increase in interspecies links following lysis (up to 6‐fold), whereas P. aeruginosa was the most sensitive to lysis duration, with a decrease in interspecies links in the long lysis treatment. It is not clear why interspecies links for P. aeruginosa decreased following the 18 h lysis, but we speculate this might have been a result of a combination of exposure to native DNases and a lower overall abundance of P. aeruginosa Hi‐C captured DNA in these samples, based on our read mapping analyses. Lower abundance of P. aeruginosa Hi‐C links may be due to self‐encapsulation with matrix polysaccharides (alginate, Pel, present in PA14) that may inhibit formaldehyde and enzymatic steps (Franklin et al., 2011; Limoli et al., 2017). Other differences between interspecies link levels among these species could derive from a combination of factors including differences in initial species abundance and differences in cell permeability to formaldehyde treatment, or amenability to downstream Hi‐C procedures such as frequency of DpnII cut sites or accessibility of chromatin to enzymatic steps. Although DpnII is one of the most popular restriction enzymes in bacterial Hi‐C experiments (Marbouty et al., 2014; Yaffe & Relman, 2020) due to the high frequency of GATC cut sites, this methyl inhibited type II endonuclease may perform differently for different bacterial species in response to varying in vitro conditions (Barnes et al., 2014). DpnII may respond to different levels of methylation which can also vary based on growth conditions, as described for P. aeruginosa for which methylation ranges from 65%–85% at target sites (Doberenz et al., 2017).
We found consistent differences among species in the rate of and type of Hi‐C link. Specifically, S. aureus Hi‐C captured reads were more abundant than reads from E. coli and these species consistently produced more reads than P. aeruginosa. These results are consistent with unequal species proportions in source material, however, whereas we attempted to create artificial polymicrobial mixtures with equal ratios of three species, we made no attempt to control for species abundance in the mouse biofilms, yet the final mapped Hi‐C link ratios were similar across all samples, including mouse wound biofilms. These similar patterns in cell cultures and biofilms suggest there may be species‐specific differences in these species' cellular permeability or chromatin features described above. However, it is critical to note that if species differ in abundance of Hi‐C links, regardless of the reason, this will affect the expected rate of interspecies links. Notably, both lower‐abundance species or lower link‐forming species would be expected to form more interspecies links relative to intraspecies links, while higher‐abundance species or higher link‐forming species might be expected to form higher proportions of links to the same species (i.e., self‐links) due to proximity of same‐species cells or same‐species linkable DNA fragments. Mathematical modeling of predicted propensities for inter‐ and intraspecies links, based on species' relative abundance, density, and propensity to form links is beyond the scope of the current study, but would be worthy of future study if this method becomes broadly used. Our data emphasize the need to perform statistical corrections for differences between species’ link patterns due to differences in histone‐like protein structure (Anuchin et al., 2011; Stojkova et al., 2019) as part of the assessment of spatial proximity following lysis‐Hi‐C.
A major finding was the high rate of plasmid‐to‐host genome Hi‐C links, both for artificial polymicrobial mixtures and mouse wound biofilms. Our data showed that after lysis, the E. coli plasmid formed fewer links with its host and more links with other species, indicating that lysis promotes new plasmid associations in Hi‐C. Specifically, we found high levels of E. coli genome‐to‐pUCP18 links in intact (not lysed) cells, with one third as many E. coli‐pUCP18 links as E. coli‐to‐E. coli links (i.e., self‐links), suggesting formaldehyde cross‐linking was highly effective when plasmids were in close proximity to host DNA in intact cells. This result is consistent with past studies (Kent et al., 2020; Marbouty et al., 2017; Ramani et al., 2017; Stalder et al., 2019), showing the utility of conventional Hi‐C for assignment of extrachromosomal DNA. Conversely, plasmid links to alternate hosts S. aureus and P. aeruginosa increased by approximately 50% to over twofold following lysis, suggesting successful ligation of extracellular plasmid DNA and extruded neighbouring cell DNA. To date, the effects of cell lysis on novel plasmid Hi‐C links are poorly understood and has not been systematically explored. Arguably, our finding that Hi‐C plasmid links are affected by lysis should be a consideration in analysing plasmid‐host associations in Hi‐C studies of natural microbiomes or biofilms. Given that natural cell lysis may produce significant extracellular DNA and extracellular plasmids, these novel links could introduce error in interpreting plasmid‐host identities.
Our results on plasmid links in mouse wound biofilms showed that lysis increased the plasmid to alternate host link numbers significantly, specifically for S. aureus between mechanical lysis and long Igepal lysis, and for P. aeruginosa between sonicated and long Igepal lysis treatments. However, for E. coli in these biofilms, chromosome‐to‐plasmid links were highest in mechanically lysed specimens, suggesting either that sonication and Igepal treatments were more effective at dispersing plasmid DNA than mechanical lysis. This may be due to additional inhibitory effects of biofilm matrix polysaccharides that are disrupted by sonication and Igepal, or due to high plasmid abundance in the intercellular spaces in these biofilms compared to the cell artificial “communities.” Assessing the identity and microgeography of biofilm matrix eDNA (Kavanaugh et al., 2019; Tahrioui et al., 2019) will be critical in future studies that consider application of Hi‐C to evaluate the role of eDNA in this context (Boháčová et al., 2019), particularly of interest as a source of eDNA‐recombination (Devaraj et al., 2019).
While the main goal in this study was to examine how lysis affects Hi‐C links, especially interspecies and plasmid links, we also examined changes in link distance within species, reasoning that extracellular DNA (eDNA) from lysed cells will often cross‐link with neighbours of the same species. We found, as expected, that lysis increased average distance between intraspecific Hi‐C links in E. coli and P. aeruginosa, with median self‐link distance increased up to 38‐ or 37‐fold, respectively, following lysis. This suggests lysis had significant effects on the formation of new chromatin links, either through forming new within‐genome links resulting from disrupted chromatin packing, or through intercellular links with neighbouring cells. In mouse wound biofilms, median self‐link distance in P. aeruginosa also increased 37‐fold, suggesting that a similar transformation in chromatin microgeography occurs both in cultured cells and in biofilms. In contrast, for S. aureus median self‐link distance was not affected by lysis. Instead, lysis of S. aureus significantly increased the proportion of distant links (i.e., links <20 kb distance were significantly decreased by lysis), both for artificial polymicrobial mixtures and for mouse wound biofilms. Self‐links should decay with linear distance along the chromosome (Marbouty et al., 2014; Servant et al., 2015), except where chromatin packaging occurs. The consistent absence of this pattern in S. aureus in all samples in our study was surprising. We propose two possible explanations. Either a large proportion of S. aureus cells may have inadvertently lysed including within the “no lysis” controls during sample preparation, perhaps during the short period of drying or during formaldehyde cross‐linking, causing the observed linear relationship of link distance to link number. Given the cell cluster morphology of S. aureus, this could then lead to high rates of links across genome distances from lysed neighbour cells. Or, a second explanation could be that S. aureus has highly‐linked DNA due to intrinsic nucleoid‐associated histone‐like proteins or secreted DNA‐binding proteins (Anuchin et al., 2011; Stojkova et al., 2019). Noting that we found that self‐links seemed to be random in their location across the genome, even for close links (<20 kb) in S. aureus, these data do not support regularly distributed nucleoid loops anchored by histone‐like proteins (Anuchin et al., 2011). Nevertheless, differences in self links between species could be due to differences in frequency of histone‐like proteins and chromatin architecture. Clearly, further experiments will be needed understand these potential factors.
While mouse wound biofilms produced fewer Hi‐C links than artificial polymicrobial cell mixtures, interspecies links increased following lysis. Specifically, interspecies links tended to increase following both mechanical lysis and mechanical + Igepal lysis, albeit with statistical support only in the case of P. aeruginosa. We found most Hi‐C links in the mouse material were between mouse chromosomes (data not shown), due to inclusion of mouse skin in these preparations. Repeated attempts to directly apply Igepal lysis reagents on these biofilm surfaces failed, producing extremely low Hi‐C‐captured (biotin‐streptavidin) products, suggesting significant inhibition of reagent permeation in these samples. For this reason, for a “no lysis” control, we used a sonication and scoring method on whole biofilms, and to facilitate lysis we broke the matrix with mechanical lysis. Based on these challenges with abundant non‐target mouse DNA and challenges of reagent permeation into the biofilm, we suggest that future optimization protocols for biopsied skin biofilms should incorporate mammalian DNA depletion or should mechanically reduce skin material with a microtome or scraping approach.
Ultimately, these experiments provided new impetus for developing and refining a lysis‐Hi‐C method that will be useful in identifying spatial‐mutational dynamics in polymicrobial communities. Our procedures herein for mouse biofilms were optimized for comparative purposes with mixed cells and therefore were not optimized to recover genotypic microgeography. However, our outcomes show that a lysis‐Hi‐C strategy has promise to recover genotypes and eDNA microgeography. In such applications, it will be essential to use non‐disruptive approaches, such as mild enzymatic treatment of the matrix prior to in situ formaldehyde crosslinking. Our lysis‐Hi‐C methods for cells met with the quality criteria for well‐prepared Hi‐C libraries, with up to 90% of our Hi‐C reads mapping to our references (Servant et al., 2015). These data showed the most common type of invalid read was typical of many experiments: dangling ends, comprising incomplete biotinylated dNTP overhang fill‐ins on one side of the cross‐link (Belaghzal et al., 2017; Servant et al., 2015). In future, this method could be improved at this step by extending end fill‐in (e.g., from 25 to 45 min) or fill‐in temperature could be increased to improve enzyme efficiency (Belaghzal et al., 2017; van Berkum et al., 2010). Further experiments could explore alternative lysis in the range of our short lysis, which was generally more effective, or alternatively, DNase inhibitors could be added to reduce potential undesired endogenous nuclease activity during lysis. We followed standard formaldehyde application steps, but our results suggest possible variance in effectiveness between species. We recommend that this cross‐linking step be explored systematically in future studies. Finally, one of the most interesting findings and critical aspects of our methods was the apparent presence of intercellular (extracellular) DNA even in the “no lysis” controls. Standard Hi‐C protocols for bacteria (Crémazy et al., 2018; Marbouty et al., 2014; Yaffe & Relman, 2020) involve successive cell precipitation and aqueous wash steps before and after formaldehyde fixation, which will tend to remove eDNA. In contrast, our methods not only applied lysis and cross‐linking reagents to undisturbed cell‐film of biofilm surfaces, but also painstakingly sought to preserve free DNA at all steps of the protocol, using ethanol precipitation or AMPure bead binding. In doing this, we were able to detect high background levels of intercellular links even in non‐lysed treatments, suggesting a wealth of hidden eDNA to be discovered by this method.
Polymicrobial infections and biofilms pose serious challenges to researchers and clinicians, which necessitates novel technological advancements to study them (Burmølle et al., 2014; Hall‐Stoodley et al., 2004; Hughes & Webber, 2017; Misic et al., 2014), particularly in the context of cross‐feeding and evolved cooperation contributing to resistance (Cullen & McClean, 2015; Seth & Taga, 2014). Currently, NGS studies of in situ biofilms improve upon classical microbiology methods by gathering broad taxonomic community data, but no NGS method has been available to investigate spatial‐mutational dynamics. By demonstrating the feasibility of lysis‐Hi‐C in creating and sequencing links between extracellular DNA, chromosomes and plasmids, and between different species, this study opens the door to new methods for exploring the dynamics of polymicrobial microgeography and evolution.
AUTHOR CONTRIBUTIONS
Amanda M. V. Brown designed the initial study with assistance from Catherine A. Wakeman. Experiments were led by Bravada M. Hill with assistance from Karishma Bisht for cell culture preparation and Kendra P. Rumbaugh for mouse biofilm preparation. Georgia Rae Atkins and Amy A. Gomez assisted with experimental optimization. Bravada M. Hill led the initial bioinformatic analysis with assistance from Amanda M. V. Brown. Final analyses and statistics were performed by Amanda M. V. Brown. The manuscript was written by Bravada M. Hill and Amanda M. V. Brown and all coauthors assisted with editing of the final manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interests.
Supporting information
Appendix S1
ACKNOWLEDGEMENTS
We thank several past undergraduates in the Brown Laboratory for assistance with initial experiments. We thank technicians at Dovetail for helpful discussions on adapting the kit for our protocol. Graduate funding support to B.M.H. was through the Texas Tech Association of Biologists, the Helen DeVitt Jones Fellowship, Tech ASM, and the ASM Capstone Fellowship. Graduate funding support to K.B. was through the Texas Tech University Doctoral Dissertation Competition Fellowship. Undergraduate research support to G.R.A. and A.A.G. was provided by the Center for the Integration of STEM Education & Research (CISER) program. Support for mouse biofilm materials to K.P.R. was through grants from the National Institutes of Health (R21 AI137462‐01A1) and the Ted Nash Long Life Foundation. Support for polymicrobial culture preparation to C.A.W. was through NIH/NIGMS R15GM128072. Support for sequencing, experiments, and analysis to A.M.V.B. were through startup funding from the Department of Biological Sciences at Texas Tech.
Hill, B. M. , Bisht, K. , Atkins, G. R. , Gomez, A. A. , Rumbaugh, K. P. , Wakeman, C. A. , & Brown, A. M. V. (2022). Lysis‐Hi‐C as a method to study polymicrobial communities and eDNA. Molecular Ecology Resources, 22, 1029–1042. 10.1111/1755-0998.13535
DATA AVAILABILITY STATEMENT
Raw DNA sequences have been deposited in the NCBI Sequence Read Archives under BioProjectID PRJNA762485, BioSamples SAMN21396244‐SAMN21396249. HiC‐Pro link data files and bioinformatic code are deposited in Dryad https://doi.org/10.5061/dryad.tmpg4f507.
REFERENCES
- Anuchin, A. M. , Goncharenko, A. V. , Demidenok, O. I. , & Kaprelyants, A. S. (2011). Histone‐like proteins of bacteria (review). Applied Biochemistry and Microbiology, 47(6), 580–585. 10.1134/S0003683811060020 [DOI] [PubMed] [Google Scholar]
- Baishya, J. , & Wakeman, C. A. (2019). Selective pressures during chronic infection drive microbial competition and cooperation. NPJ Biofilms and Microbiomes, 5(1), 1–9. 10.1038/s41522-019-0089-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, H. E. , Liu, G. , Weston, C. Q. , King, P. , Pham, L. K. , Waltz, S. , Helzer, K. T. , Day, L. , Sphar, D. , Yamamoto, R. T. , & Forsyth, R. A. (2014). Selective microbial genomic DNA isolation using restriction endonucleases. PLoS One, 9(10), e109061. 10.1371/journal.pone.0109061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaghzal, H. , Dekker, J. , & Gibcus, J. H. (2017). Hi‐C 2.0: An optimized Hi‐C procedure for high‐resolution genome‐wide mapping of chromosome conformation. Methods, 123, 56–65. 10.1016/j.ymeth.2017.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisht, K. , & Wakeman, C. A. (2019). Discovery and therapeutic targeting of differentiated biofilm subpopulations. Frontiers in Microbiology, 10, 1908. 10.3389/fmicb.2019.01908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boháčová, M. , Pazlarová, J. , Fuchsová, V. , Švehláková, T. , & Demnerová, K. (2019). Quantitative evaluation of biofilm extracellular DNA by fluorescence‐based techniques. Folia Microbiologica, 64(4), 567–577. 10.1007/s12223-019-00681-8 [DOI] [PubMed] [Google Scholar]
- Bolger, A. M. , Lohse, M. , & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30(15), 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmølle, M. , Ren, D. , Bjarnsholt, T. , & Sørensen, S. J. (2014). Interactions in multispecies biofilms: Do they actually matter? Trends in Microbiology, 22(2), 84–91. 10.1016/j.tim.2013.12.004 [DOI] [PubMed] [Google Scholar]
- Calhoun, J. H. , Murray, C. K. , & Manring, M. M. (2008). Multidrug‐resistant organisms in military wounds from Iraq and Afghanistan. Clinical Orthopaedics and Related Research, 466(6), 1356–1362. 10.1007/s11999-008-0212-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlan, S. , Thomas, P. J. , Deming, C. , Park, M. , Lau, A. F. , Dekker, J. P. , Snitkin, E. S. , Clark, T. A. , Luong, K. , Song, Y. I. , Tsai, Y.‐C. , Boitano, M. , Dayal, J. , Brooks, S. Y. , Schmidt, B. , Young, A. C. , Thomas, J. W. , Bouffard, G. G. , Blakesley, R. W. , … Segre, J. A. (2014). Single molecule sequencing to track plasmid diversity of hospital‐associated carbapenemase‐producing Enterobacteriaceae. Science Translational Medicine, 6(254), 254ra126. 10.1126/scitranslmed.3009845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crémazy, F. G. , Rashid, F.‐Z.‐ M. , Haycocks, J. R. , Lamberte, L. E. , Grainger, D. C. , & Dame, R. T. (2018). Determination of the 3D genome organization of bacteria using Hi‐C. In Dame R. T. (Ed.), Bacterial Chromatin (pp. 3–18). Humana Press. 10.1007/978-1-4939-8675-0_1 [DOI] [PubMed] [Google Scholar]
- Cullen, L. , & McClean, S. (2015). Bacterial adaptation during chronic respiratory infections. Pathogens (Basel, Switzerland), 4(1), 66–89. 10.3390/pathogens4010066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey, M. E. , & O'toole, G. A. (2000). microbial biofilms: From ecology to molecular genetics. Microbiology and Molecular Biology Reviews, 64(4), 847–867. 10.1128/MMBR.64.4.847-867.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeon, S. , Clinton, A. , Fowler, H. , Everett, J. , Horswill, A. R. , & Rumbaugh, K. P. (2014). Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an In vitro wound model. Infection and Immunity, 82(11), 4718–4728. 10.1128/IAI.02198-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMaere, M. Z. , & Darling, A. E. (2018). Sim3C: Simulation of Hi‐C and Meta3C proximity ligation sequencing technologies. GigaScience, 7(2), 1–12. 10.1093/gigascience/gix103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaere, M. Z. , & Darling, A. E. (2019). Bin3C: Exploiting Hi‐C sequencing data to accurately resolve metagenome‐assembled genomes. Genome Biology, 20(1), 1–16. 10.1186/s13059-019-1643-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj, A. , Buzzo, J. R. , Mashburn‐Warren, L. , Gloag, E. S. , Novotny, L. A. , Stoodley, P. , Bakaletz, L. O. , & Goodman, S. D. (2019). The extracellular DNA lattice of bacterial biofilms is structurally related to Holliday junction recombination intermediates. Proceedings of the National Academy of Sciences of the United States of America, 116(50), 25068–25077. 10.1073/pnas.1909017116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doberenz, S. , Eckweiler, D. , Reichert, O. , Jensen, V. , Bunk, B. , Sproer, C. , Kordes, A. , Frangipani, E. , Luong, K. , Korlach, J. , Heeb, S. , Overmann, J. , Kaever, V. , & Häussler, S. (2017). Identification of a Pseudomonas aeruginosa PAO1 DNA methyltransferase, its targets, and physiological roles. MBio, 8(1), 1–14. 10.1128/mBio.02312-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbeis, J. , Saffarzadeh, M. , Peisker, H. , Jung, P. , Thewes, N. , Preissner, K. T. , Herrmann, M. , Molle, V. , Geisbrecht, B. V. , Jacobs, K. , & Bischoff, M. (2018). The staphylococcus aureus extracellular adherence protein Eap Is a DNA binding protein capable of blocking neutrophil extracellular trap formation. Frontiers in Cellular and Infection Microbiology, 8, 1–12. 10.3389/fcimb.2018.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin, M. J. , Nivens, D. E. , Weadge, J. T. , & Lynne Howell, P. (2011). Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Frontiers in Microbiology, 2, 1–16. 10.3389/fmicb.2011.00167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall‐Stoodley, L. , Costerton, J. W. , & Stoodley, P. (2004). Bacterial biofilms: From the natural environment to infectious diseases. Nature Reviews Microbiology, 2(2), 95–108. 10.1038/nrmicro821 [DOI] [PubMed] [Google Scholar]
- Hauser, A. R. , Jain, M. , Bar‐Meir, M. , & McColley, S. A. (2011). Clinical significance of microbial infection and adaptation in cystic fibrosis. Clinical Microbiology Reviews, 24(1), 29–70. 10.1128/CMR.00036-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotterbeekx, A. , Kumar‐Singh, S. , Goossens, H. , & Malhotra‐Kumar, S. (2017). In vivo and In vitro Interactions between Pseudomonas aeruginosa and Staphylococcus spp. Frontiers in Cellular and Infection Microbiology, 7, 1–13. 10.3389/fcimb.2017.00106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, G. , & Webber, M. A. (2017). Novel approaches to the treatment of bacterial biofilm infections. British Journal of Pharmacology, 174(14), 2237–2246. 10.1111/bph.13706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorth, P. , Staudinger, B. J. , Wu, X. , Hisert, K. B. , Hayden, H. , Garudathri, J. , Harding, C. L. , Radey, M. C. , Rezayat, A. , Bautista, G. , Berrington, W. R. , Goddard, A. F. , Zheng, C. , Angermeyer, A. , Brittnacher, M. J. , Kitzman, J. , Shendure, J. , Fligner, C. L. , Mittler, J. , … Singh, P. K. (2015). Regional isolation drives bacterial diversification within cystic fibrosis lungs. Cell Host and Microbe, 18(3), 307–319. 10.1016/j.chom.2015.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce, E. , Al‐Hashimi, A. , & Mason, T. J. (2011). Assessing the effect of different ultrasonic frequencies on bacterial viability using flow cytometry. Journal of Applied Microbiology, 110(4), 862–870. 10.1111/j.1365-2672.2011.04923.x [DOI] [PubMed] [Google Scholar]
- Kang, J. B. , Siranosian, B. A. , Moss, E. L. , Banaei, N. , Andermann, T. M. , & Bhatt, A. S. (2019). Intestinal microbiota domination under extreme selective pressures characterized by metagenomic read cloud sequencing and assembly. BMC Bioinformatics, 20(Suppl 16), 1–13. 10.1186/s12859-019-3073-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh, J. S. , Flack, C. E. , Lister, J. , Ricker, E. B. , Ibberson, C. B. , Jenul, C. , Moormeier, D. E. , Delmain, E. A. , Bayles, K. W. , & Horswill, A. R. (2019). Identification of extracellular DNA‐binding proteins in the biofilm matrix. MBio, 10(3), 1–30. 10.1128/mBio.01137-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, A. G. , Vill, A. C. , Shi, Q. , Satlin, M. J. , & Brito, I. L. (2020). Widespread transfer of mobile antibiotic resistance genes within individual gut microbiomes revealed through bacterial Hi‐C. Nature Communications, 11(1), 1–9. 10.1038/s41467-020-18164-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirketerp‐Møller, K. , Jensen, P. , Fazli, M. , Madsen, K. G. , Pedersen, J. , Moser, C. , Tolker‐Nielsen, T. , Høiby, N. , Givskov, M. , & Bjarnsholt, T. (2008). Distribution, organization, and ecology of bacteria in chronic wounds. Journal of Clinical Microbiology, 46(8), 2717–2722. 10.1128/JCM.00501-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren, S. , & Phillippy, A. M. (2015). One chromosome, one contig: Complete microbial genomes from long‐read sequencing and assembly. Current Opinion in Microbiology, 23, 110–120. 10.1016/j.mib.2014.11.014 [DOI] [PubMed] [Google Scholar]
- Krzywinski, M. , Schein, J. , Birol, I. , Connors, J. , Gascoyne, R. , Horsman, D. , Jones, S. J. , & Marra, M. A. (2009). Circos: An information aesthetic for comparative genomics. Genome Research, 19(9), 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B. , & Salzberg, S. L. (2012). Fast gapped‐read alignment with Bowtie 2. Nature Methods, 9(4), 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA‐MEM. ArXiv, 1–3. Preprint arXiv:1303.3997v2 [Google Scholar]
- Li, H. , Handsaker, B. , Wysoker, A. , Fennell, T. , Ruan, J. , Homer, N. , Marth, G. , Abecasis, G. , Durbin, R. & 1000 Genome Project Data Processing Subgroup . (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25(16), 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limoli, D. H. , Whitfield, G. B. , Kitao, T. , Ivey, M. L. , Davis, M. R. , Grahl, N. , Hogan, D. A. , Rahme, L. G. , Howell, P. L. , O’Toole, G. A. , Goldberg, J. B. , O’Toole, G. A. , & Goldberg, J. B. (2017). Pseudomonas aeruginosa alginate overproduction promotes coexistence with Staphylococcus aureus in a model of cystic fibrosis respiratory infection. MBio, 8(2), 1–18. 10.1128/mBio.00186-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Collazo, E. , Jurado, T. , De Dios Caballero, J. , Pérez‐Vázquez, M. , Vindel, A. , Hernández‐Jiménez, E. , Tamames, J. , Cubillos‐Zapata, C. , Manrique, M. , Tobes, R. , Máiz, L. , Cantón, R. , Baquero, F. , & Del Campo, R. (2015). In vivo attenuation and genetic evolution of a ST247‐SCCmecI MRSA clone after 13 years of pathogenic bronchopulmonary colonization in a patient with cystic fibrosis: Implications of the innate immune response. Mucosal Immunology, 8(2), 362–371. 10.1038/mi.2014.73 [DOI] [PubMed] [Google Scholar]
- Mandakhalikar, K. D. , Rahmat, J. N. , Chiong, E. , Neoh, K. G. , Shen, L. , & Tambyah, P. A. (2018). Extraction and quantification of biofilm bacteria: Method optimized for urinary catheters. Scientific Reports, 8(1), 1–9. 10.1038/s41598-018-26342-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbouty, M. , Baudry, L. , Cournac, A. , & Koszul, R. (2017). Scaffolding bacterial genomes and probing host‐virus interactions in gut microbiome by proximity ligation (chromosome capture) assay. Science Advances, 3(2), e1602105. 10.1126/sciadv.1602105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbouty, M. , Cournac, A. , Flot, J. F. , Marie‐Nelly, H. , Mozziconacci, J. , & Koszul, R. (2014). Metagenomic chromosome conformation capture (meta3C) unveils the diversity of chromosome organization in microorganisms. ELife, 3, e03318. 10.7554/eLife.03318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misic, A. M. , Gardner, S. E. , & Grice, E. A. (2014). The wound microbiome: Modern approaches to examining the role of microorganisms in impaired chronic wound healing. Advances in Wound Care, 3(7), 502–510. 10.1089/wound.2012.0397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcombe, R. G. (1998). Two‐sided confidence intervals for the single proportion: Comparison of seven methods. Statistics in Medicine, 872(17), 857–872. [DOI] [PubMed] [Google Scholar]
- Oliver, A. (2010). Mutators in cystic fibrosis chronic lung infection: Prevalence, mechanisms, and consequences for antimicrobial therapy. International Journal of Medical Microbiology, 300(8), 563–572. 10.1016/j.ijmm.2010.08.009 [DOI] [PubMed] [Google Scholar]
- Oliver, A. , & Mena, A. (2010). Bacterial hypermutation in cystic fibrosis, not only for antibiotic resistance. Clinical Microbiology and Infection, 16(7), 798–808. 10.1111/j.1469-0691.2010.03250.x [DOI] [PubMed] [Google Scholar]
- Pakkulnan, R. , Anutrakunchai, C. , Kanthawong, S. , Taweechaisupapong, S. , Chareonsudjai, P. , & Chareonsudjai, S. (2019). Extracellular DNA facilitates bacterial adhesion during Burkholderia pseudomallei biofilm formation. PLoS One, 14(3), 1–19. 10.1371/journal.pone.0213288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival, S. L. , McCarty, S. M. , & Lipsky, B. (2014). Biofilms and wounds: An overview of the evidence. Advances in Wound Care, 4(7), 373–381. 10.1089/wound.2014.0557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani, V. , Deng, X. , Qiu, R. , Gunderson, K. L. , Steemers, F. J. , Disteche, C. M. , Noble, W. S. , Duan, Z. , & Shendure, J. (2017). Massively multiplex single‐cell Hi‐C. Nature Methods, 14(3), 263–266. 10.1038/nmeth.4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen, D. , Brown, C. J. , Top, E. M. , & Sullivan, J. (2013). Inferring the evolutionary history of IncP‐1 plasmids despite incongruence among backbone gene trees. Molecular Biology and Evolution, 30(1), 154–166. 10.1093/molbev/mss210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant, N. , Lajoie, B. R. , Nora, P. , Giorgetti, L. , Chen, C. , Heard, E. , Dekker, J. , & Barillot, E. (2012). HiTC: exploration of high‐throughput ‘C’ experiments. Bioinformatics, 28(21), 2843–2844. 10.1093/bioinformatics/bts521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant, N. , Varoquaux, N. , Lajoie, B. R. , Viara, E. , Chen, C. J. , Vert, J. P. , Heard, E. , Dekker, J. , & Barillot, E. (2015). HiC‐Pro: An optimized and flexible pipeline for Hi‐C data processing. Genome Biology, 16(1), 1–11. 10.1186/s13059-015-0831-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth, E. C. , & Taga, M. E. (2014). Nutrient cross‐feeding in the microbial world. Frontiers in Microbiology, 5, 350. 10.3389/fmicb.2014.00350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder, T. , Press, M. O. , Sullivan, S. , Liachko, I. , & Top, E. M. (2019). Linking the resistome and plasmidome to the microbiome. ISME Journal, 13(10), 2437–2446. 10.1038/s41396-019-0446-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojkova, P. , Spidlova, P. , & Stulik, J. (2019). Nucleoid‐associated protein Hu: A lilliputian in gene regulation of bacterial virulence. Frontiers in Cellular and Infection Microbiology, 9, 1–7. 10.3389/fcimb.2019.00159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahrioui, A. , Duchesne, R. , Bouffartigues, E. , Rodrigues, S. , Maillot, O. , Tortuel, D. , Hardouin, J. , Taupin, L. , Groleau, M. C. , Dufour, A. , Déziel, E. , Brenner‐Weiss, G. , Feuilloley, M. , Orange, N. , Lesouhaitier, O. , Cornelis, P. , & Chevalier, S. (2019). Extracellular DNA release, quorum sensing, and PrrF1/F2 small RNAs are key players in Pseudomonas aeruginosa tobramycin‐enhanced biofilm formation. Npj Biofilms and Microbiomes, 5(1), 15. 10.1038/s41522-019-0088-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berkum, N. L. , Lieberman‐Aiden, E. , Williams, L. , Imakaev, M. , Gnirke, A. , Mirny, L. A. , Dekker, J. , & Lander, E. S. (2010). Hi‐C: A method to study the three‐dimensional architecture of genomes. Journal of Visualized Experiments, 39, 1869. 10.3791/1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, V. , & Smyth, A. (2015). Cystic fibrosis microbiology: Advances in antimicrobial therapy. Journal of Cystic Fibrosis, 14(5), 551–560. 10.1016/j.jcf.2015.02.005 [DOI] [PubMed] [Google Scholar]
- Watters, C. , Deleon, K. , Trivedi, U. , Griswold, J. A. , Lyte, M. , Hampel, K. J. , Wargo, M. J. , & Rumbaugh, K. P. (2013). Pseudomonas aeruginosa biofilms perturb wound resolution and antibiotic tolerance in diabetic mice. Medical Microbiology and Immunology, 202(2), 131–141. 10.1007/s00430-012-0277-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteson, K. L. , Bailey, B. , Bergkessel, M. , Conrad, D. , Delhaes, L. , Felts, B. , Harris, J. K. , Hunter, R. , Lim, Y. W. , Maughan, H. , Quinn, R. , Salamon, P. , Sullivan, J. , Wagner, B. D. , & Rainey, P. B. (2014). The upper respiratory tract as a microbial source for pulmonary infections in cystic fibrosis: Parallels from island biogeography. American Journal of Respiratory and Critical Care Medicine, 189(11), 1309–1315. 10.1164/rccm.201312-2129PP [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winstanley, C. , O’Brien, S. , & Brockhurst, M. A. (2016). Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends in Microbiology, 24(5), 327–337. 10.1016/j.tim.2016.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermute, E. H. & Silver, P. A. (2010). Emergent cooperation in microbial metabolism. Molecular Systems Biology, 6(1), 407. 10.1038/msb.2010.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe, E. , & Relman, D. A. (2020). Tracking microbial evolution in the human gut using Hi‐C reveals extensive horizontal gene transfer, persistence, and adaptation. Nature Microbiology, 5(2), 343–353. 10.1038/s41564-019-0625-0.Tracking [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Kobert, K. , Flouri, T. , & Stamatakis, A. (2014). PEAR: A fast and accurate Illumina Paired‐End reAd mergeR. Bioinformatics, 30(5), 614–620. 10.1093/bioinformatics/btt593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, G. X. Y. , Lau, B. T. , Schnall‐Levin, M. , Jarosz, M. , Bell, J. M. , Hindson, C. M. , Kyriazopoulou‐Panagiotopoulou, S. , Masquelier, D. A. , Merrill, L. , Terry, J. M. , Mudivarti, P. A. , Wyatt, P. W. , Bharadwaj, R. , Makarewicz, A. J. , Li, Y. , Belgrader, P. , Price, A. D. , Lowe, A. J. , Marks, P. , … Ji, H. P. (2016). Haplotyping germline and cancer genomes with high‐throughput linked‐read sequencing. Nature Biotechnology, 34(3), 303–311. 10.1038/nbt.3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1
Data Availability Statement
Raw DNA sequences have been deposited in the NCBI Sequence Read Archives under BioProjectID PRJNA762485, BioSamples SAMN21396244‐SAMN21396249. HiC‐Pro link data files and bioinformatic code are deposited in Dryad https://doi.org/10.5061/dryad.tmpg4f507.


