Abstract
Cryptosporidium species has been identified as an important pediatric diarrheal pathogen in resource-limited countries, particularly in very young children (0–24 months). However, the only available drug (nitazoxanide) has limited efficacy and can only be prescribed in a medical setting to children older than one year. Many drug development projects have started to investigate new therapeutic avenues. Cryptosporidium’s unique biology is challenging for the traditional drug discovery pipeline and requires novel drug screening approaches. Notably, in recent years, new methods of oocyst generation, in vitro processing, and continuous three-dimensional cultivation capacities have been developed. This has enabled more physiologically pertinent research assays for inhibitor discovery. In a short time, many great strides have been made in the development of anti-Cryptosporidium drugs. These are expected to eventually turn into clinical candidates for cryptosporidiosis treatment in the future. This review describes the latest development in Cryptosporidium biology, genomics, transcriptomics of the parasite, assay development, and new drug discovery.
Keywords: Cryptosporidium, Genomics, Virulence factors, Drug targets, Therapeutics
Introduction
Cryptosporidium is second only to rotavirus as a leading global cause of moderate-to-severe diarrhea in children younger than two years (Kotloff et al. 2019, 2013). Apart from being an unfortunate contributor to childhood mortality, Cryptosporidium can also cause persistent infection, cognitive issues, malnutrition, and stunted growth (Khalil et al. 2018; Platts-Mills et al. 2015). These adverse effects are probably mediated by environmental enteric disruption, leading to local chronic inflammation and barrier disruption. Disruption of barriers might lead to recurrent infection and hence nutrient malabsorption (Checkley et al. 2015). Considering the severity of these outcomes and the unavailability of safe drugs or vaccines, the pathogen has emerged as of utmost public health importance.
Cryptosporidium is a protozoan apicomplexan parasite. First observed by Ernest Edward Tyzzer in 1907 (Tyzzer 1907), Cryptosporidium was initially considered harmless until the 1970s, when it got its reputation as a causative agent of diarrhea (Nime et al. 1976). After the early reports of cryptosporidiosis in humans in 1976 (Tzipori et al. 1980), it was later associated with HIV/AIDS, causing life-threatening cryptosporidiosis in hepatobiliary, respiratory, and gastrointestinal tract infections (Cama et al. 2007; Ma and Soave 1983). Of the various Cryptosporidium species, in humans, Cryptosporidium hominis and Cryptosporidium parvum are responsible for over 90% of cases (Sow et al. 2016). Cryptosporidium spreads specifically through the fecal–oral route, primarily via contaminated food or water (Efstratiou et al. 2017). It has also been documented to spread by person-to-person routes. By invading the microvillus layer of the gastrointestinal epithelium, it infects a large range of vertebrate hosts (Checkley et al. 2015; Deng et al. 2004a, b; O’Handley and Olson 2006). Various in vivo studies now point towards altered tight junction protein expressions during Cryptosporidium infections aiding the barrier disruption (Kumar et al. 2018). The parasite often spends most of its life within a specific “parasitophorous vacuole,” which also helps the pathogen evade the host immune defense system (Lendner and Daugschies 2014; Schmid-Hempel 2009). Parasite-induced pattern recognition receptors and inflammasome activation have also been described (Laurent and Lacroix-Lamandé 2017; Sateriale et al. 2021). Though infections are not always associated with clinical symptoms in most animals and are often considered a self-limiting illness (Checkley et al. 1997; Osman et al. 2016), they take a lethal turn in severely immunocompromised individuals with a risk of recurrent infections (Sparks et al. 2015). Infected individuals show a wide range of clinical presentations, such as watery diarrhea, vomiting, nausea, abdominal cramps, and a low-grade fever (Hunter and Nichols 2002). Children may have multiple stages or phases of cryptosporidiosis, indicating short-lived or incomplete acquired immunity from this infection. The Global Enteric Multicenter Study (GEMS) indicates that in seven countries of sub-Saharan Africa and South Asia, Cryptosporidium is a major contributor to infant diarrheal incidents (Kotloff et al. 2013). In many developed countries, water-borne or food-borne cryptosporidiosis may also occur in adults. In addition, because of the minimally invasive feature of the pathogen, i.e., it does not penetrate deep into the host mucosal layer, the infection can spread into different parts in the case of immunocompromised individuals (López-Vélez et al. 1995; Miyamoto and Eckmann 2015). Moreover, asymptomatic fecal shedding of C. parvum oocysts has also been depicted elsewhere (Eibach et al. 2015; Zambriski et al. 2013). Since several studies have depicted a poor clinical outcome with currently approved therapeutics in malnourished and immunocompromised individuals (Amadi et al. 2009; Ashigbie et al. 2021), a new array of drugs with enhanced efficacy should be developed by properly understanding the biological processes of the parasite.
Biology of Cryptosporidium
Considering the problems with the parasite and the public health hazard status, seeking answers to the fundamental questions about infection pattern, virulence, genomics, and proteomics, is necessary for developing successful drugs or vaccine candidates against the pathogen.
Cryptosporidium life cycle
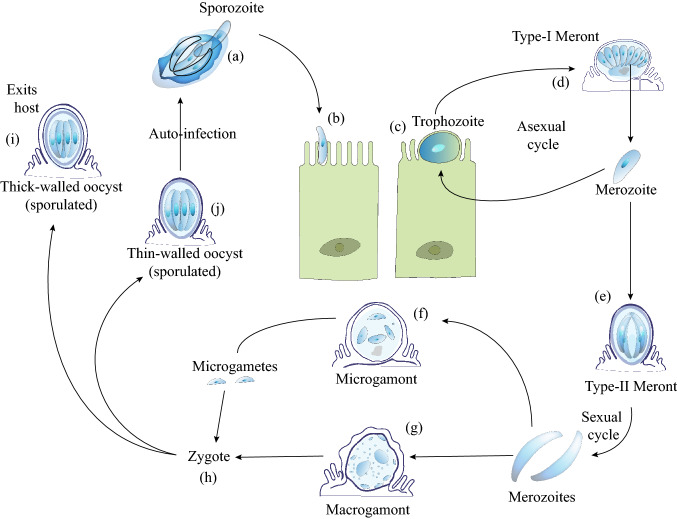
Based on the developmental phases, the life cycle of Cryptosporidium can be sub-divided into six groups. The first stage is the excystation phase that releases infectious sporozoites, leading to the second stage of asexual proliferation within the host cell, called merogony. The third stage of gametogony refers to the formation of the micro-and macro-gametes, followed by the fourth phase of fertilization of these gametes. The fifth stage constitutes the formation of oocyst walls to create an environmentally tolerant stage for the transition of the infection from one host to the next. The sixth and final stage of sporogony refers to the formation of infectious sporozoites (Current and Garcia 1991; Dumaine et al. 2020; Miyamoto and Eckmann 2015). The parasite stays within a closed compartment named “parasitophorous vacuole” composed of the host cell plasmalemma and acquires nutrients from the host cell through an apicomplexan-specific feeder organelle (Thompson et al. 2005; Tzipori and Ward 2002). Cryptosporidium has a monoxenous life cycle. Sporulated oocysts, once ingested or inhaled by the susceptible host, come into contact with the reducing environment of the human digestive tract where a combination of pH, pancreatic enzymes, and salts induces the process of excystation (Fayer et al. 1984; Tzipori and Ward 2002), marking the beginning of the six-phased life cycle. The stages of the parasite life cycle are depicted in Fig. 1.
Fig. 1.
Cryptosporidium parvum life cycle. a The excystation of a single oocyst releases four infective sporozoites. Using gliding motility as a means of locomotion, the sporozoites ultimately reach the microvilli of the intestinal epithelial cells. b The parasite remains in the microvillar region inside a parasitophorous vacuole in the plasma membrane of the host. c The sporozoites develop into spherical trophozoites. d Trophozoites undergo merogony, to form Type-I meront, consisting of 8 merozoites. Meront ruptures and infective merozoites are released to infect other nearby cells. e Type-II meront formed from a type-I meront contains four merozoites, but instead of continuing the infection cycle, each merozoite now undergoes gametogony, giving rise to either a (f) microgamont or a (g) macrogamont. h Each micro or macro gamont ultimately gets fertilized to produce a zygote. The zygote, after undergoing sporogony, produces an oocyst containing four sporozoites. It is covered with either a thick or a thin wall. i The thick-walled oocyst is released into the intestinal lumen eventually being excreted out, ready to infect a new host (j) The thin-walled oocyte, on the other hand, can re-infect the same host in a process called autoinfection. (Adapted with modification from CDC, Atlanta, GA, USA. https://www.cdc.gov/dpdx/cryptosporidiosis/index.html). The figure was created with the help of Adobe Illustrator 2020 software
Genomics of Cryptosporidium species
Tyzzer first acknowledged the existence of multiple species forms within the Cryptosporidium genus. He thoroughly illustrated the C. muris species from the gastric glands of laboratory mice by publishing a comprehensive life cycle report and later also describing another Cryptosporidium species (Tyzzer 1910; Tzipori and Widmer 2008). Cryptosporidium was used in early whole-genome sequencing (WGS) projects as a pathogen of interest to public health with two reference strains as members of the genome: C. hominis TU502 and C. parvum Iowa, and has been reannotated (Isaza et al. 2015). Their genome sequences were seen with almost similar genome sizes of 9.16 Mb and 9.11 Mb with reported sequence divergences of 3–5%, respectively (Abrahamsen et al. 2004; Xu et al. 2004). It has also been reported that Cryptosporidium retains certain genes for mitochondrial biosynthesis but lacks an apicoplast, unlike other apicomplexans (LaGier et al. 2003). Unusual nuclear protein-dependent degenerate mitochondria and mitochondrial-derived compartments have puzzled researchers ever since their discovery (Henriquez et al. 2005; Keithly et al. 2005; Xu et al. 2004). Further, mitosomes have also been recently reported to contain the Fe-S biosynthesis machinery in the parasite (Miller et al. 2018). The genomic studies revealed highly streamlined pathways, a lack of some cell structures, and metabolic pathways seen in other apicomplexans (Barta and Thompson 2006; Bouzid et al. 2013). Energy metabolism is primarily via glycolysis, with the availability of both aerobic and anaerobic components, thus providing versatility for the parasite (Barta and Thompson 2006). After the availability of WGS data of C. muris, biological differences among different species were realized (Wang et al. 2012). C. muris, a gastric species of rodent, was found to have a genome size of 9.2 Mb and is equipped with all enzymes of the TCA cycle and a conventional respiratory chain, unlike the other two Cryptosporidium species (Mogi and Kita 2010). Due to advanced scientific approaches, unlike in previous WGS analyses, now the oocysts can directly be isolated from stool specimens, purified, and whole genome amplification of the genomic DNA can easily be performed (Guo et al. 2015a; Morris et al. 2019). Illumina and Roche 454 high-throughput sequencing technologies have been used in WGS analyses of field and clinical isolates (Andersson et al. 2015; Feng et al. 2017; Guo et al. 2015b; Hadfield et al. 2015; Troell et al. 2016). High genomic diversity and high recombination rates were also observed by WGS of C. hominis samples collected from Bangladesh (Gilchrist et al. 2018). There are differences between Cryptosporidium species in terms of total length, number of genes with introns, GC content, number of tRNAs, proteins with transmembrane domains, and so on, and such comparative genomic analysis of Cryptosporidium have been extensively reviewed elsewhere (Fan et al. 2019; Khan et al. 2018; Xu et al. 2019).
Transcriptomics study of Cryptosporidium species
Despite challenges regarding parasite cell culture and the presence of multiple life cycle stages, thanks to recent advancements, transcriptome analysis from different stages of the parasite have advanced our understanding of the host-parasite interactions. Considering the minimalistic metabolism pathways of the parasite, the survival of oocysts in open environments has puzzled the researchers. Zhang et al. developed a specific microarray chip for C. parvum for transcriptome analysis and found around 2000 upregulated genes, of which more than 50% of the genes were hypothetical (Zhang et al. 2012). Other upregulated genes were found associated with transcription, RNA synthesis, metabolism, and modification. The authors found an active expression of genes involved in ubiquitin/proteasome-mediated protein degradation and post-translational modification, implying the recycling of proteins to overcome the parasite's inability to endogenously synthesize amino acids (Zhang et al. 2012). The highest level of energy metabolism-associated gene expression was identified in the lactate dehydrogenase (LDH) gene in sporozoites (Matos et al. 2019). Since LDH is often associated with the parasitophorous vacuole, drug targets against the enzyme could be of therapeutic advantage, which will be discussed in subsequent sections. RNA-seq analysis has revealed stark transcriptomic differences between the intracellular and extracellular stages of the parasite (Lippuner et al. 2018). Genes associated with biosynthetic processes such as mucin (gp40/15 and gp900) were found upregulated in intracellular stages. There are also specific genes that are expressed throughout all stages of the parasite, such as calcium-dependent protein kinases (CDPKs), which could be potential drug targets against the parasite (Hulverson et al. 2017a). However, this approach might be challenging as the parasite has five different CDPKs and they vary in their expression profiles in different parasite stages and biological importance.
After in vitro parasite infection of the human ileocecal adenocarcinoma (HCT-8) cells, RT-PCR was used to assess the temporal gene expression patterns. Two hours post-infection at the early trophozoite stage, sugar-nucleotide transporter genes, transcription, and DNA-associated genes were upregulated, which indicates that the parasite obtains nutrition from the host (Matos et al. 2019). At the late trophozoite stage at 6 h post-infection, an increase in genes encoding for translation, protein folding, transport, and proteasome was observed, indicating the formation of meronts. At 12 h, with the appearance of early type-I meronts, genes encoding for cytoskeletal proteins and micronemes/rhoptries get upregulated, signifying merozoite travel to adjacent cells (Mauzy et al. 2012). After 24 h of infection, the appearance of matured type-I meront is observed, along with the upregulation of genes encoding for translation and other metabolic pathways. Depending on the cell type, a slight difference in gene expression patterns can be observed (Relat and O’Connor 2020). After 36 h of infection, high gene expression pertaining to Rab-GDP dissociation inhibitor is observed, which indicates nutritional transport to the newly formed microgamonts (Mauzy et al. 2012; Relat and O’Connor 2020). At 48 h post-infection, sexual stages begin to take over the culture setting, and enrichment of genes coding for meiosis-related proteins and oocyst wall formation was observed, implying the formation of micro- and macrogametes (Tandel et al. 2019). After 72 h of infection, genes encoding for pyruvate decarboxylase and other necessary wall-producing proteins were found overexpressed (Heo et al. 2018).
The parasite can also affect the host transcriptome, and studies have shown an upregulation in cell cytoskeleton arrangement-related proliferation, inflammation, and apoptosis-related genes (Deng et al. 2004a, b). However, later RNA-seq data of infected pig intestinal monolayer cells suggested no changes to stress or apoptosis-associated host genes, indicating the host was not much affected by the parasite infection (Mirhashemi et al. 2018). Moreover, a recent microarray study on long non-coding RNAs (lncRNAs) depicted an upregulation of inflammatory factors (TNF and interleukins), cell proliferation factors, Wnt-signaling pathway, hedgehog pathway, and tight junction-associated proteins (Liu et al. 2018). Recent studies have demonstrated the production and transport of lncRNAs into the host cell nucleus during infection. One such transcript of C. parvum, Cdg7_FLc_0990, interacts with the host cell chaperone heat shock protein 70 (hsp70) to get delivered into the host nucleus (Wang, et al. 2017a). Such transcripts affect the host cell transcriptome by upregulating pro-inflammatory genes and hijacking the host histone regulation system (Wang et al. 2017b). Other putative lncRNAs were also reported, projecting lncRNAs as a mechanism of parasite-induced host transcriptome regulation (Li et al. 2021; Ming et al. 2018; Zhao et al. 2018).
CryptoDB (https://cryptodb.org), a robust genome database, acts as a public repository for sequences of the Cryptosporidium genome (Heiges et al. 2006). This database provides access to various tools that allow genes to be retrieved on the basis of text, motif queries, and sequence similarity (Warrenfeltz and Kissinger 2020). Researchers have demonstrated that Cryptosporidium and other species share more than 150 ancestral apicomplexan proteins, predominantly involved in eukaryotic host cell interactions and apical complex biogenesis (Gordon and Sibley 2005; Templeton et al. 2004). The Cryptosporidium comparative transcriptomics and genomics fields are still at an early stage in comparison to other apicomplexan pathogens such as Plasmodium. Therefore, there is a need to obtain additional WGS data from diverse pathogenic and zoonotic Cryptosporidium spp. to have a clear view of the genetic basis of virulence, host infection, and transmissibility (Nader et al. 2019; Widmer et al. 2012). In the wake of new bioinformatics genotyping tools developed for quality control, classification, and analysis of sequence chromatograms, it would be possible to initiate drug candidate searches against the parasite by understanding its genome biology (Yanta et al. 2021).
Cryptosporidium virulence factors and host immune system intervention
Several virulence factors are known to initiate processes of infection and disease maintenance by the parasite. Host factors also play a significant role in the outcomes of the pathogen-host interactions. A few reports have classified the factors responsible for initiating, developing, and perpetuating Cryptosporidium infection. Usually, Cryptosporidium does not induce systemic inflammation or reach into deep tissues; rather, the parasite is localized on the apical surface of the intestinal epithelium in membrane-bound compartments (Okhuysen and Chappell 2002; Valigurová et al. 2008). Hence, the absence of some apicomplexan-conserved cellular invasion components in Cryptosporidium is not surprising (Valigurová et al. 2008). Evolutionary analyses have also indicated that Cryptosporidium has a unique combination of features from Plasmodium and Toxoplasma as well as from gut-infecting gregarine parasites (Aldeyarbi and Karanis 2016; Barta and Thompson 2006). However, it does cause significant defects in the absorptive and secretory functions of the gut. These defects may result from direct damage to the epithelial cells of the host or may be mediated by indirect pathways connected to them (Smith et al. 2005). To date, it has been difficult for researchers to effectively characterize Cryptosporidium-specific virulence factors to establish their roles as damage-causing agents or to prove that those factors can be modified to reduce the severity of the disease. In contrast to other apicomplexan parasites (Plasmodium and Toxoplasma), in the case of Cryptosporidium, it remains difficult to use reverse genetic engineering techniques in in-vivo or in-vitro culture settings. Difficulty in knocking down or knocking out specific genes to investigate virulence has limited the crypto-specific drug research field (Vinayak et al. 2015). In recent years, several advanced gene manipulation techniques like CRISPR-Cas9 have been employed in the case of Cryptosporidium to overcome the hurdle of novel anti-parasitic drug discovery (Vinayak et al. 2015). Various novel in vivo animal models for parasite infection has been depicted elsewhere (Costa et al. 2012; Lee et al. 2019; Sateriale et al. 2019). A recent review describes a controlled human model named CHIM (controlled human infection model) in volunteers for accelerated and robust drug discovery (Jumani et al. 2021). Despite initial success in animal model-related studies, host specificity of the parasite, different symptoms, and diverse infection patterns should be kept in mind while addressing the issue in humans. Cryptosporidial factors of putative virulence were established as genes involved in the initial interactions with host epithelial cells of Cryptosporidium oocysts and sporozoites. It also includes the formation of the parasitophorous vacuole, attachment, excystation, invasion, intracellular maintenance, membrane lysis, gliding motility, and damage to host cells (Bouzid et al. 2013).
From an immunological response point of view, the release of pro-inflammatory cytokines/chemokines (IL-8, CXCL10, and CCL2) from the immune cells and up-regulated antimicrobial peptide (β-defensin) production has been demonstrated after Cryptosporidium infection (Chen et al. 2001; Zaalouk et al. 2004). Cryptosporidium parvum infection activates the cellular toll-like receptors (TLRs) and the TLR2 and TLR4-mediated NF-κB pathway, accounting for anti-apoptotic properties, thereby helping the parasite to propagate (Chen et al. 2005, 2001; Yang et al. 2015). Researchers have also demonstrated that C. parvum infection reduces the expression of key cellular micro-RNA let-7i, which is associated with the upregulation of TLR4 in cholangiocytes responsible for epithelial cellular defense (Chen et al. 2007). Further, down-regulation of let-7i during C. parvum infection is also associated with the overexpression of SNAP23 and increased TLR4-mediated exosome release from the epithelial cells. Released apical exosomes were observed to contain antimicrobial peptides dedicated to reducing infection levels (Hu et al. 2013). By using genetically tractable mouse models, the role of interferon-gamma (IFN-γ) during the early stage of Cryptosporidium infection and T-cell-mediated parasite clearance has been demonstrated (Sateriale et al. 2019). During an infection, IFN-γ, which is secreted by macrophages, NK cells, and dendritic cells, coordinates both the innate and adaptive immune responses. Furthermore, IL-18 may protect against C. parvum infection by increasing the secretion of IFN-γ and other anti-microbial peptides (Bedi et al. 2015; Choudhry et al. 2012). Moreover, an immunocompetent rat model has recently been described by the Tzipori group, where rats are protected against secondary infection after a primary intra-tracheal infection by sporozoites. This model can be used for future vaccine development screening trials against C. parvum and C. hominis (Dayao et al. 2020).
The role of the adaptive immune system during Cryptosporidium infection is also poorly understood. Insightful use of CD8 + or CD4 + T-lymphocytes has been recognized to protect against Cryptosporidium infection in immunodeficient mice (Kváč et al. 2011). Studies have found an association between infection history and anti-Cryptosporidium antibodies in humans (Wanyiri et al. 2014). Significantly, higher levels of CD4 + T cells, circulating IgG, and fecal IgA were observed in HIV-infected patients with cryptosporidiosis (Wanyiri et al. 2014). Increased levels of IgG, IgM, and IgA were also observed in response to acute and asymptomatic Cryptosporidium infections (Ajjampur et al. 2011). Future studies related to immune system intervention should particularly focus on the correlative role of inflammasomes and the humoral immune system in asymptomatic and persistent parasite infections.
Therapeutic avenues for cryptosporidiosis
Anti-parasitic treatments for cryptosporidiosis in a patient who is suboptimal or immunocompromised along with enhancement of cellular immune function should be the key goal in the management of cryptosporidiosis (e.g., combined antiretroviral therapy for AIDS). During initial treatment, avoiding mortality and malnutrition should be the aim. Different drugs have been identified in animal models and patients with action against Cryptosporidium in vitro.
Anti-parasitic drugs repurposing
Nitazoxanide (NTZ) is a nitrothiazole benzamide broad-spectrum anti-parasitic compound and is the only FDA-approved drug for the treatment of cryptosporidiosis (Checkley et al. 2015). In a study in Egypt, about 80% of NTZ patients reported cessation of diarrhea compared with 41% of the placebo group (Rossignol et al. 2001). Similarly, NTZ reduced mortality in malnourished children infected with cryptosporidiosis in comparison to placebo (Amadi et al. 2002). NTZ is postulated to inhibit the parasitic enzyme pyruvate-ferredoxin oxidoreductase (PFOR), thereby hampering the anaerobic energy transfer reactions (Fox and Saravolatz 2005). Though NTZ was reported to be effective in non-HIV patients, depicting enhanced parasitic clearance, and reduced oocyst shedding (Rossignol et al. 2006), however, it was found ineffective in the case of HIV/AIDS patients, even at prolonged higher doses (Amadi et al. 2002, 2009). The precise mechanism of reduced efficacy of NTZ in immunocompromised individuals is still unknown and needs further research. The side effects associated with higher doses of NTZ have also been reported elsewhere (Lee et al. 2017).
NTZ has also been used with other antimicrobial drugs and has shown promising results. NTZ, along with antibiotic azithromycin (AZR), has been used in a piglet model of C. hominis (Lee et al. 2017). Though NTZ alone reduced oocyst shedding and other symptoms, AZR alone or AZR + NTZ didn’t have any significant effect on early oocyst shedding or mucosal injury reduction (Lee et al. 2017). Paromomycin is an amino-glycoside that has effectiveness in AIDS patients with cryptosporidiosis. A comparative analysis between NTZ and paromomycin revealed that both the drugs were effective against the parasitic infection in a neonatal mouse model (Blagburn et al. 1998). Clofazimine (CFZ), a lipophilic riminophenazine drug used to treat leprosy and multidrug-resistant tuberculosis, was found effective against Cryptosporidium in an in vitro approach (Love et al. 2017). However, it was later found to be unsuitable in a phase-2 human trial involving severely immunocompromised HIV patients (Iroh Tam et al. 2020), due to poor absorbance of CFZ in crypto-infected HIV patients.
Other antimicrobial drugs, such as clarithromycin, rifabutin, rifaximin, and roxithromycin, may also be effective in controlling Cryptosporidium growth and outcome (Amenta et al. 1999; Fichtenbaum et al. 2000; Holmberg et al. 1998; Uip et al. 1998). One study reported that the Halogeno-Thiazolides were more effective in inhibiting Cryptosporidium growth and oocyst shedding (95%) in immunosuppressed Mongolian gerbils compared with NTZ (47%) (Gargala et al. 2013). Other studies have also indicated the effectiveness of natural product extracts such as Citrus maxima and pomegranate (Punica granatum) peel against C. parvum in murine models of infection (Al-Mathal and Alsalem 2013; Hafez and Hamed 2021). However, extended in vivo research is necessary to establish the proper therapeutic effect of such treatments.
Apart from AIDS patients, Cryptosporidium infection also appears to be problematic for organ transplant recipients. A French multicenter study in organ transplant recipients reported cryptosporidiosis as a post-transplantation infection that can be eradicated without an absolute reduction in immunosuppression (Lanternier et al. 2017), with the administration of NTZ and immunosuppressive agents (Bhadauria et al. 2015). Several reports show that in organ transplant recipients, Cryptosporidium infection should routinely be tested owing to the parasite’s seasonal pattern, and long-term therapy with NTZ is often found effective (Krause et al. 2012; Legrand et al. 2011).
Recently, we found that KDU731 is a lead chemical, and has been shown to exhibit substantial in-vitro activity against C. parvum and C. hominis (Love and Choy 2021). It is also effective against mouse and calf models of cryptosporidiosis, along with rat toxicity investigation (Manjunatha et al. 2017). Further in-vitro activity profiling revealed that a similar analog, KDU691, was parasiticidal at its EC90 and its MoA (mechanisms of action) was a hindrance to merozoite formation (Funkhouser-Jones et al. 2020). This is most likely because of lipid membrane processing impairment caused by CpPI (4) K inhibition. Interestingly, KDU731 pharmacokinetics in C. parvum-infected calves revealed low systemic exposure, indicating that it may not be necessary for clinical effectiveness in this cohort.
The limited efficacy of these repurposed drugs against the parasite in vulnerable and malnourished hosts has paved the way for research into novel therapeutic agents. However, some technical difficulties regarding the genetic manipulation of the parasite and suitable in vivo experimental models remain unresolved. Further, a lack of knowledge regarding the host immune system’s interaction with the parasite is also a major hurdle in developing novel therapeutic targets. In the next section, putative therapeutic candidates harnessing the benefits of a bioinformatics-based discovery approach will be discussed.
Novel drug targets
Despite the challenges of animal model-based research and in vitro culture, the availability of complete genome sequences of Cryptosporidium species has opened up new avenues for developing novel drug candidates. The streamlined metabolic pathways of the parasite allow greater opportunities for selective drug therapy (Striepen and Kissinger 2004). Various in silico approaches have been employed using comparative genomics analysis, homology modeling, prioritization parameters, epitope prediction, virtual screening, molecular docking, and simulation studies (Dhal et al. 2019; Panda and Mahapatra 2018). In the life cycle of apicomplexan parasites, the regulation of Ca2+ binding by calcium-dependent protein kinases (CDPKs) is necessary for parasite secretion, motility, and growth (Etzold et al. 2014). Therefore, CDPKs are thought to be potential therapeutic targets against the parasites (Su et al. 2022). After understanding the crystal structures of CDPKs having a gatekeeper glycine residue, specific bumped-kinase inhibitors (BKIs) were developed that inhibit CDPK1 functions through the hydrophobic pocket that opens up next to the glycine residue (Huang et al. 2017; Hulverson, et al. 2017a, b; Van Voorhis et al. 2021). Calcium-dependent protein kinase 6 (CpCDPK6) (cgd4_3330) is a hypothetical protein of the CDPK family that regulates sporozoite invasion, gliding, and parasite egress (Billker et al. 2009; Wernimont et al. 2010; Zhang et al. 2021). An in silico study that evaluated potential inhibitors against the CpCDPK6 reported the Tres Cantos Antimalarial Set (TCAMS) _11730 as potential inhibitors (Dhal et al. 2020).
Similarly, inhibitors like pyrazolopyridines and imidazopyrazines have been employed against phosphatidylinositol-4-OH kinase (PI(4)K) of Cryptosporidium. The imidazopyrazines work by inhibiting PI(4)K's ATP-binding pocket, hence changing the intracellular distribution of phosphatidylinositol-4-phosphate (Manjunatha et al. 2017; McNamara et al. 2013). The genome sequencing of C. parvum shows that it neither has a functioning mitochondrion, or active Krebs cycle, nor has a cytochrome-based respiration chain, but encodes all glycolytic enzymes (Keithly et al. 2005; Yu et al. 2014). The parasite relies primarily on anaerobic glucose oxidation for energy metabolism, and enzymes to generate ATP in the glycolytic pathways. The lactate dehydrogenase and isomerase glucose-6-phosphate in Cryptosporidium (CpLDH and CpGPI) have been found to be associated with the parasitophorous vacuole membrane formation and energy dynamics (Madern et al. 2004; H. Zhang et al. 2015). Therefore, LDH, GPI, and other glycolysis pathway members could be potential therapeutic targets (Dhal et al. 2018; Eltahan et al. 2018).
Further, clan-CA cysteine proteases, expressed during the sporozoite stage, are vital for surface protein processing, hemoglobin hydrolysis, host cell invasion, nutrition uptake, and are structurally different from their analogous human counterparts (Kim 2004; Na et al. 2009). Cysteine protease inhibitors effectively block the host cell invasion, suggesting that cysteine proteases could be an appropriate new chemotherapeutic target (Kang et al. 2012; Siqueira-Neto et al. 2018). N-methyl-piperazine-Phe-homoPhe-vinyl sulfone phenyl (K11777) has been used in an in vitro setting as well as in immunocompromised mice to inhibit the activity of cathepsin L-like or papain-like cysteine proteases, thereby reducing the parasite growth (Ndao et al. 2013).
Another putative drug target could be the oxidoreductase inosine 5′-monophosphate dehydrogenase (IMPDH) essential for guanine synthesis in Cryptosporidium. Various inhibitors for CpIMPDH have been reported (Kirubakaran et al. 2012; Gorla et al. 2014; Sun et al. 2014; Shigetomi et al. 2019; Asmare et al. 2022). Similarly, another study reported the use of Phylomer® designed peptides as an inhibitor of CpIMPDH, effectively reducing Cryptosporidium growth (Jefferies et al. 2015). Oleylphosphocholine- C23H48NO4P (OlPC) is structurally similar to the antiparasitic drug miltefosine, and interferes with the integrity of parasitic lipid biosynthesis, thereby inducing parasite apoptosis (Hernández et al. 2014). OIPC successfully inhibited C. parvum growth in infected HCT-8 cells and was able to cure infected mice (Sonzogni-Desautels et al. 2015). In contrast to miltefosine, OIPC demonstrated a better toxicity profile for HCT-8 cells (Fortin et al. 2014; Sinkala et al. 2011; Sonzogni-Desautels et al. 2015). Considering the importance of acyl-coenzyme-A synthetase (ACS) enzymes in parasite fatty acid metabolism (Chattopadhyay and Mahapatra 2019), inhibitors like triacsin-C has been employed to reduce the development of parasite oocysts (about 88%) in IL-12-KO mice (Guo et al. 2014). The usefulness of elongation factor 2 (EF-2) protein as a therapeutic target has also been described elsewhere (Dhal et al. 2022).
In an activity-centered drug repurposing screen approach, out of a library of 727 FDA-approved drugs tested in HCT-8 cells, 3-hydroxy-3-methyl-glutaryl-coenzyme A (HMG-CoA) reductase was found to be a suitable target (Bessoff et al. 2013). A group of anti-inflammatory lipid-lowering agents known as statins (HMG-CoA reductase inhibitors) has been repurposed for handling Cryptosporidium infections both in an in vitro and in vivo setup by inhibiting the intermediates of the mevalonate pathway (Parihar et al. 2019) (Madbouly Taha et al. 2017). Putative drug candidates against the parasite discussed in this section and their structures are provided in Fig. 2. In another experimental setup, an existing drug library of 400 compounds against the apicomplexan Plasmodium falciparum from the Malaria Box (MMV) was tested against C. parvum. The screen yielded about 19 compounds with activity against C. parvum growth (Bessoff et al. 2014). Other studies have also reported piperazine-based compounds (MMV665917) from the Malaria Box as potential drug candidates (Jumani et al. 2018). Moreover, the ReFRAME library with 12,000 compounds has also been evaluated for their efficacy against Cryptosporidium (Janes et al. 2018). Several other novel inhibitors have been described against Cryptosporidium containing specific new chemical entities (NCEs), i.e., lysine tRNA synthetase (KRS) inhibitors (Baragaña et al. 2019), phenylalanyl-tRNA synthetase inhibitors (bicyclic azetidines) (Vinayak et al. 2020), benzoxaboroles (AN7973) (Lunde et al. 2019), and cleavage and polyadenylation specificity factor-3 (CPSF3) inhibitors (Swale et al. 2019). A comprehensive list of drugs and putative drug targets against the parasite is provided in Table 1.
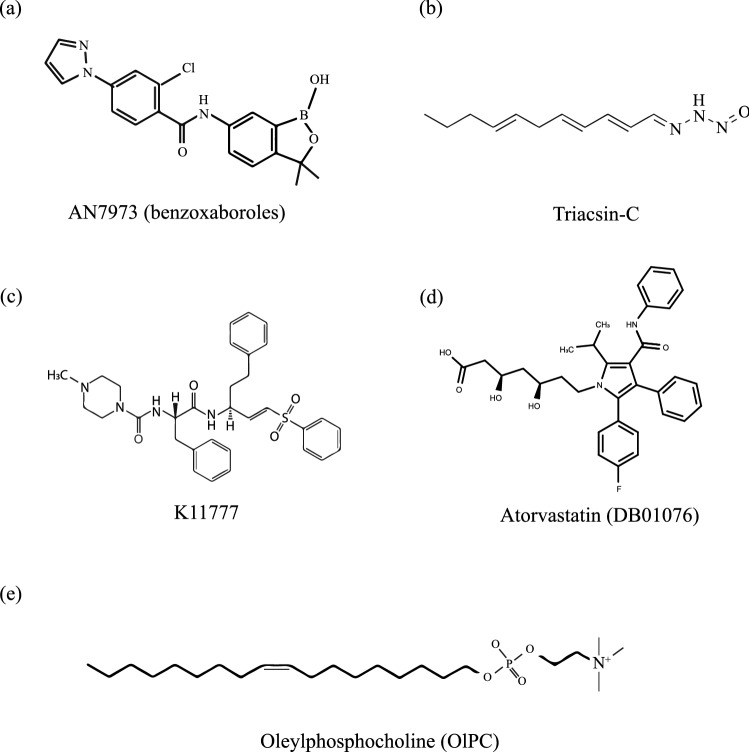
Fig. 2.
Novel therapeutics against Cryptosporidium. a Structures of AN7973, a benzoxaborole (Lunde et al. 2019), and b triacsin-C targeting acyl-coenzyme-A synthetases have been employed against the parasite (Guo et al. 2014). c The structure of compound K11777 targeting the cysteine proteases has also been employed against the parasite (Ndao et al. 2013). d The structure of atorvastatin, a statin compound, having DrugBank database ID: DB01076, in combination with nitazoxanide (NTZ), has been tested for a synergistic approach against the parasite-infected mice (Madbouly Taha et al. 2017). e Oleylphosphocholine (OlPC), an alkylphosphocholine, has also been employed against the parasite-infected immunocompromised mice (Sonzogni-Desautels et al. 2015)
Table 1.
Comprehensive list of drugs and putative drug targets against Cryptosporidium effective here implies drugs reducing the oocyst shedding, enhancing parasitic clearance, and alleviating symptoms)
| Drugs/drug targets | Outcome of the parasite | Reference(s) |
|---|---|---|
| Nitazoxanide (NTZ) |
FDA-approved Limited efficacy in reducing parasite burden in non-AIDS patients Non-effective in AIDS patients |
Amadi et al. (2002, 2009), Rossignol et al. (2001, 2006) |
| Clofazimine (CFZ) |
Effective in an in vitro approach Found ineffective in phase-2 human trial |
Iroh Tam et al. (2020), Love et al. (2017) |
| Azithromycin (AZR) | AZR alone or AZR + NTZ having no significant effect | Lee et al. (2017) |
| Paromomycin | Effective against the parasite in a mouse model | Blagburn et al. (1998) |
| Halogeno-Thiazolides | Found effective in immunosuppressed Mongolian gerbils | Gargala et al. (2013) |
| Rifabutin, Rifaximin, Clarithromycin, Roxithromycin | Effective in vitro and in vivo (lack of extended research) | Amenta et al. (1999), Fichtenbaum et al. (2000), Holmberg et al. (1998), Uip et al. (1998) |
| Drugs against the calcium-dependent protein kinases (CDPKs) |
Effective in an in silico, in vitro, and in vivo setting Not entered human trials |
Huang et al. (2017; Hulverson et al. (2017a, b), Van Voorhis et al. (2021) |
| Phosphatidylinositol-4-OH kinase (PI(4)K) and lactate dehydrogenase inhibitors | Deemed effective in an in silico and in vitro setup | Dhal et al. (2018), Eltahan et al. (2018) Manjunatha et al. (2017), McNamara et al. (2013), Zhang et al. (2015) |
| K11777 (Inhibitors of clan-CA cysteine proteases) | In vitro and in vivo efficacy | Ndao et al. (2013), Siqueira-Neto et al. (2018) |
| Statin (HMG-CoA reductase inhibitors) | Atorvastatin found effective in immunocompromised infected mice | Bessoff et al. (2013), Kang et al. (2012), Madbouly Taha et al. (2017), Parihar et al. (2019) |
| Inhibitors against IMPDH enzyme | Effective in a mouse model and other in vitro approaches | Gorla et al. (2014), Jefferies et al. (2015), Shigetomi et al., (2019) |
| Alkylphosphocholines (Oleylphosphocholine-OIPC) | Effective in HCT-8 cell model and mice | Sonzogni-Desautels et al. (2015) |
| Triacsin-C (inhibitor of acyl-coenzyme-A synthetases) | Effective in immunocompromised mice | Guo et al. (2014) |
| MMV665917 (a piperazine-based compound repurposed from the Malaria Box) | In vitro efficacy | Jumani et al. (2018) |
| Benzoxaboroles (AN7973) and bicyclic azetidines | Observed in an in silico study, extended research warranted | Lunde et al. (2019), Swale et al. (2019), Vinayak et al. (2020) |
Most of these NCEs have depicted superior in vitro anti-cryptosporidial activity and in vivo efficacy in reducing fecal oocyst burden. However, extended evaluation of in vivo safety, efficacy, and pharmacological characterization are required for these compounds to enter human trials. The active compound or the repurposed drugs should be deemed effective in malnourished children and immunocompromised individuals. The drug should also be affordable and be available in heat-stable conditions so as to be useful in low-resource settings (Ashigbie et al. 2021; Manjunatha et al. 2016; Sow et al. 2016).
Advanced methods of oocysts generation, in vitro processing, and drug testing
A powerful method to quantify the parasite load by real-time PCR is the key to the in vitro evaluation of anti-cryptosporidial drug efficacy. However, high-throughput screening (HTS) of drugs against C. parvum was deemed impractical by the labor-intensive traditional assays. A simplified quantitative RT-PCR assay suitable for HTS of compounds and for evaluating drug efficacy against the growth of C. parvum in vitro has been described (Zhang and Zhu 2020). Parasite load assessment by qRT-PCR provides several advantages when compared with qPCR. The RNA used in qRT-PCR, due to its quick degradation, better indicates cell viability than DNA. Total RNA isolation is also easier by directly allowing the samples to be used in a high-throughput analysis (Zhang and Zhu 2020). Furthermore, hollow fiber technology is a powerful tool for the culture of difficult-to-grow cells. Cryptosporidium parvum has a multistage sexual and asexual life cycle that has proved difficult to culture by conventional in vitro culture methods (Baydoun et al. 2017; Morada et al. 2016). This novel method enables the evaluation of potential therapeutic compounds under in vitro conditions that emulate the dynamic processes of the host, facilitating preliminary pharmacokinetics and pharmacodynamics data to be assessed (Yarlett et al. 2020).
Challenges involving in vitro and in vivo models during the development of therapeutics for Cryptosporidium are extensively reviewed elsewhere (Jumani et al. 2021; Manjunatha et al. 2016). Briefly, using the mouse stem cell-derived intestinal epithelial monolayers, long-term C. parvum growth can be supported. Monolayers are usually grown as spheroids and plated onto transwells, allowing for separate basolateral and apical compartments. In the apical chamber, the cell growth medium is removed to create a mixed interface, enhancing host cell differentiation and long-term C. parvum growth. Such stem-cell-derived setups could serve as useful in vitro models of the natural host’s intestinal niche (Heo et al. 2018; Wilke et al. 2019). The complete life cycle of C. parvum has been grown in human organoids (Heo et al. 2018). Human intestinal enteroids (HIEs) are cultures containing stem cells isolated from human biopsies. Such multi-cellular cultures can be differentiated into different intestinal cells. In a three-dimensional matrix, HIEs spatially emulate the native host intestinal epithelium. These scaffold-based models can be used to grow Cryptosporidium and can also be used to evaluate drug efficacy and pharmacokinetics (Bhalchandra et al. 2020; Chen et al. 2017).
Conclusion
The drug discovery strategies for Cryptosporidium are largely unexplored in relation to other neglected tropical diseases. However, despite the many challenges unique to Cryptosporidium biology, great deals of promising advancements have been made in a comparatively short period of time. The high risk of parasite resistance to existing therapies illustrates the critical need for substitutes, as well as new targets and new chemotypes for which resistance has not yet been developed. The most promising drug therapies are expected to have these. Several developments in screening techniques to classify new anti-cryptosporidium compounds and developments in Cryptosporidium culture cultivation have resulted in the development of new phenotypic assays to profile and prioritize drug compounds. Robust and diverse pipelines for potential cryptosporidiosis therapeutics have been established. Novel compounds will need to be evaluated by these pipelines and prioritized by their molecular mechanism of action as compounds eventually occur in compound progression. Phenotypic screening is prioritized based on its performance in related fields, especially in pathogens with homologs in Cryptosporidium. A wealth of new chemical tools with various objectives and mechanisms of action have been created by the phenotypic screening of targeted or repositioned chemical collections. As an underestimated disease affecting mostly developing countries, cryptosporidiosis attracts low funding opportunities and limited commercial interest, keeping it in the category of neglected diseases. In such a scenario, in silico methods of drug development could generally boon due to the reduction of expenses and time required for developing new therapeutic candidates and treatment regimens.
Acknowledgements
The authors would like to acknowledge the bioinformatics lab facility of the School of Biotechnology, KIIT Deemed to be University during the course of work.
Abbreviations
- C. hominis
Cryptosporidium hominis
- C. parvum
Cryptosporidium parvum
- WGS
Whole-genome sequencing
- GEMS
Global enteric multicenter study
- CDPKs
Calcium-dependent protein kinases
- CHIM
Controlled human infection model
- NTZ
Nitazoxanide
- AZR
Azithromycin
- CFZ
Clofazimine
- CpLDH
C. parvum Lactate dehydrogenase
- CpGPI
C. parvum Isomerase glucose-6-phosphate
- IMPDH
Inosine 5′-monophosphate dehydrogenase
- CpCDPK6
C. parvum Calcium-dependent protein kinase 6
- BKIs
Bumped-kinase inhibitors
- NCEs
New chemical entities
- HTS
High-throughput screening
Author contributions
AKD and CP wrote the manuscript and made the diagrams. RKM and SI Yun supervised the study and corrected the manuscript.
Funding
The work is supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF-2021R1A2C1094316).
Declarations
Conflict of interest
None declared.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ajit Kumar Dhal and Chinmaya Panda are contributed equally.
Contributor Information
Soon-IL Yun, Email: siyun@jbnu.ac.kr.
Rajani Kanta Mahapatra, Email: rmahapatra@kiitbiotech.ac.in.
References
- Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, et al. Complete genome sequence of the apicomplexan, Cryptosporidium Parvum. Science. 2004;304(5669):441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- Ajjampur SSR, Sarkar R, Allison G, Banda K, Kane A, Muliyil J, et al. Serum IgG response to cryptosporidium immunodominant antigen gp15 and polymorphic antigen gp40 in children with cryptosporidiosis in South India. Clin Vaccine Immunol. 2011;18(4):633–639. doi: 10.1128/CVI.00464-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldeyarbi HM, Karanis P. The ultra-structural similarities between Cryptosporidium parvum and the gregarines. J Eukaryot Microbiol. 2016;63(1):79–85. doi: 10.1111/jeu.12250. [DOI] [PubMed] [Google Scholar]
- Al-Mathal EM, Alsalem AA. Pomegranate (Punica granatum) peel is effective in a murine model of experimental Cryptosporidium parvum ultrastructural studies of the ileum. Exp Parasitol. 2013;134(4):482–494. doi: 10.1016/j.exppara.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Amadi B, Mwiya M, Musuku J, Watuka A, Sianongo S, Ayoub A, Kelly P. Effect of nitazoxanide on morbidity and mortality in Zambian children with cryptosporidiosis: a randomised controlled trial. The Lancet. 2002;360(9343):1375–1380. doi: 10.1016/S0140-6736(02)11401-2. [DOI] [PubMed] [Google Scholar]
- Amadi B, Mwiya M, Sianongo S, Payne L, Watuka A, Katubulushi M, Kelly P. High dose prolonged treatment with nitazoxanide is not effective for cryptosporidiosis in HIV positive Zambian children: a randomised controlled trial. BMC Infect Dis. 2009;9(1):195. doi: 10.1186/1471-2334-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amenta M, Nogare ERD, Colomba C, Prestileo TS, Lorenzo FD, Fundarò S (1999) Intestinal Protozoa in HIV-Infected Patients: Effect of Rifaximin in Cryptosporidium parvum and Blastocystis hominis Infections. J Chemother 11(5), 391–395. 10.1179/joc.1999.11.5.391 [DOI] [PubMed]
- Andersson S, Sikora P, Karlberg ML, Winiecka-Krusnell J, Alm E, Beser J, Arrighi RBG. It’s a dirty job—a robust method for the purification and de novo genome assembly of Cryptosporidium from clinical material. J Microbiol Methods. 2015;113:10–12. doi: 10.1016/j.mimet.2015.03.018. [DOI] [PubMed] [Google Scholar]
- Ashigbie PG, Shepherd S, Steiner KL, Amadi B, Aziz N, Manjunatha UH, et al. Use-case scenarios for an anti-Cryptosporidium therapeutic. PLoS Negl Trop Dis. 2021;15(3):e0009057. doi: 10.1371/journal.pntd.0009057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asmare MM, Nitin N, Yun SI, Mahapatra RK. QSAR and deep learning model for virtual screening of potential inhibitors against Inosine 5’Monophosphate dehydrogenase (IMPDH) of Cryptosporidium parvum. J Mol Graph Model. 2022;111:108108. doi: 10.1016/j.jmgm.2021.108108. [DOI] [PubMed] [Google Scholar]
- Baragaña B, Forte B, Choi R, Nakazawa Hewitt S, Bueren-Calabuig JA, Pisco JP, et al. Lysyl-tRNA synthetase as a drug target in malaria and cryptosporidiosis. Proc Natl Acad Sci. 2019;116(14):7015–7020. doi: 10.1073/pnas.1814685116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barta JR, Thompson RCA. What is Cryptosporidium? Reappraising its biology and phylogenetic affinities. Trends Parasitol. 2006;22(10):463–468. doi: 10.1016/j.pt.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Baydoun M, Vanneste SB, Creusy C, Guyot K, Gantois N, Chabe M, et al. Three-dimensional (3D) culture of adult murine colon as an in vitro model of cryptosporidiosis: Proof of concept. Sci Rep. 2017;7(1):17288. doi: 10.1038/s41598-017-17304-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedi B, McNair NN, Förster I, Mead JR. IL-18 Cytokine levels modulate innate immune responses and cryptosporidiosis in mice. J Eukaryot Microbiol. 2015;62(1):44–50. doi: 10.1111/jeu.12164. [DOI] [PubMed] [Google Scholar]
- Bessoff K, Sateriale A, Lee KK, Huston CD. Drug repurposing screen reveals FDA-approved inhibitors of human HMG-CoA reductase and isoprenoid synthesis that block Cryptosporidium parvum growth. Antimicrob Agents Chemother. 2013;57(4):1804–1814. doi: 10.1128/AAC.02460-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessoff K, Spangenberg T, Foderaro JE, Jumani RS, Ward GE, Huston CD. Identification of Cryptosporidium parvum active chemical series by repurposing the open access malaria box. Antimicrob Agents Chemother. 2014;58(5):2731–2739. doi: 10.1128/AAC.02641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadauria D, Goel A, Kaul A, Sharma RK, Gupta A, Ruhela V, et al. Cryptosporidium infection after renal transplantation in an endemic area. Transpl Infect Dis. 2015;17(1):48–55. doi: 10.1111/tid.12336. [DOI] [PubMed] [Google Scholar]
- Bhalchandra S, Lamisere H, Ward H. Intestinal organoid/enteroid-based models for Cryptosporidium. Curr Opin Microbiol. 2020;58:124–129. doi: 10.1016/j.mib.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billker O, Lourido S, Sibley LD. Calcium-dependent signaling and kinases in apicomplexan parasites. Cell Host Microbe. 2009;5(6):612–622. doi: 10.1016/j.chom.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagburn BL, Drain KL, Land TM, Kinard RG, Hutton MP, Lindsay DS, et al. Comparative efficacy evaluation of dicationic carbazole compounds, nitazoxanide, and paromomycin againstCryptosporidium parvum infections in a neonatal mouse model. Antimicrob Agents Chemother. 1998;42(11):2877–2882. doi: 10.1128/AAC.42.11.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzid M, Hunter P, Chalmers R, Tyler K. Cryptosporidium pathogenicity and virulence. Clin Microbiol Rev. 2013;26:115–134. doi: 10.1128/CMR.00076-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cama VA, Ross JM, Crawford S, Kawai V, Chavez-Valdez R, Vargas D, et al. Differences in clinical manifestations among cryptosporidium species and subtypes in HIV-infected persons. J Infect Dis. 2007;196(5):684–691. doi: 10.1086/519842. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Mahapatra RK. Identification of adaptive inhibitors of Cryptosporidium parvum fatty acyl-coenzyme A synthetase isoforms by virtual screening. Parasitol Res. 2019;118(11):3159–3171. doi: 10.1007/s00436-019-06445-0. [DOI] [PubMed] [Google Scholar]
- Checkley W, Gilman RH, Epstein LD, Suarez M, Diaz JF, Cabrera L, et al. Asymptomatic and symptomatic cryptosporidiosis: their acute effect on weight gain in Peruvian children. Am J Epidemiol. 1997;145(2):156–163. doi: 10.1093/oxfordjournals.aje.a009086. [DOI] [PubMed] [Google Scholar]
- Checkley W, White AC, Jaganath D, Arrowood MJ, Chalmers RM, Chen X-M, et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect Dis. 2015;15(1):85–94. doi: 10.1016/S1473-3099(14)70772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Levine SA, Splinter PL, Tietz PS, Ganong AL, Jobin C, et al. Cryptosporidium parvum activates nuclear factor κB in biliary epithelia preventing epithelial cell apoptosis. Gastroenterology. 2001;120(7):1774–1783. doi: 10.1053/gast.2001.24850. [DOI] [PubMed] [Google Scholar]
- Chen X-M, O’Hara SP, Nelson JB, Splinter PL, Small AJ, Tietz PS, et al. Multiple TLRs are expressed in human cholangiocytes and mediate host epithelial defense responses to Cryptosporidium parvum via activation of NF-κB. J Immunol. 2005;175(11):7447–7456. doi: 10.4049/jimmunol.175.11.7447. [DOI] [PubMed] [Google Scholar]
- Chen X-M, Splinter PL, O’Hara SP, LaRusso NF. A Cellular Micro-RNA, let-7i, regulates toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum Infection*. J Biol Chem. 2007;282(39):28929–28938. doi: 10.1074/jbc.M702633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhou W, Roh T, Estes MK, Kaplan DL. In vitro enteroid-derived three-dimensional tissue model of human small intestinal epithelium with innate immune responses. PLoS ONE. 2017;12(11):e0187880. doi: 10.1371/journal.pone.0187880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhry N, Petry F, van Rooijen N, McDonald V. A protective role for interleukin 18 in interferon γ–mediated innate immunity to Cryptosporidium parvum that is independent of natural killer cells. J Infect Dis. 2012;206(1):117–124. doi: 10.1093/infdis/jis300. [DOI] [PubMed] [Google Scholar]
- Costa LB, Noronha FJ, Roche JK, Sevilleja JE, Warren CA, Oriá R, et al. Novel in vitro and in vivo models and potential new therapeutics to break the vicious cycle of cryptosporidium infection and malnutrition. J Infect Dis. 2012;205(9):1464–1471. doi: 10.1093/infdis/jis216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Current WL, Garcia LS. Cryptosporidiosis. Clin Microbiol Rev. 1991;4(3):325–358. doi: 10.1128/CMR.4.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayao DA, Sheoran A, Carvalho A, Xu H, Beamer G, Widmer G, Tzipori S. An immunocompetent rat model of infection with Cryptosporidium hominis and Cryptosporidium parvum. Int J Parasitol. 2020;50(1):19–22. doi: 10.1016/j.ijpara.2019.10.002. [DOI] [PubMed] [Google Scholar]
- Deng M, Lancto CA, Abrahamsen MS. Cryptosporidium parvum regulation of human epithelial cell gene expression. Int J Parasitol. 2004;34(1):73–82. doi: 10.1016/j.ijpara.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Deng M, Rutherford MS, Abrahamsen MS. Host intestinal epithelial response to Cryptosporidium parvum. Adv Drug Deliv Rev. 2004;56(6):869–884. doi: 10.1016/j.addr.2003.10.034. [DOI] [PubMed] [Google Scholar]
- Dhal AK, Pani A, Mahapatra RK, Yun S-I. In-silico screening of small molecule inhibitors against Lactate Dehydrogenase (LDH) of Cryptosporidium parvum. Comput Biol Chem. 2018;77:44–51. doi: 10.1016/j.compbiolchem.2018.09.002. [DOI] [PubMed] [Google Scholar]
- Dhal AK, Pani A, Mahapatra RK, Yun S-I. An immunoinformatics approach for design and validation of multi-subunit vaccine against Cryptosporidium parvum. Immunobiology. 2019;224(6):747–757. doi: 10.1016/j.imbio.2019.09.001. [DOI] [PubMed] [Google Scholar]
- Dhal AK, Pani A, Yun S-I, Mahapatra RK. In-silico analysis of Calcium Dependent Protein Kinase 6 of Cryptosporidium parvum through molecular modeling, docking, and dynamics simulation study. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1790036. [DOI] [PubMed] [Google Scholar]
- Dhal AK, Young GM, Yun SIL, Mahapatra RK. Computational analysis of elongation factor 2 (EF-2) of Cryptosporidium parvum for identification of therapeutics. Biologia. 2022 doi: 10.1007/s11756-022-01030-w. [DOI] [Google Scholar]
- Dumaine JE, Tandel J, Striepen B. Cryptosporidium parvum . Trends Parasitol. 2020;36(5):485–486. doi: 10.1016/j.pt.2019.11.003. [DOI] [PubMed] [Google Scholar]
- Efstratiou A, Ongerth JE, Karanis P. Waterborne transmission of protozoan parasites: review of worldwide outbreaks—an update 2011–2016. Water Res. 2017;114:14–22. doi: 10.1016/j.watres.2017.01.036. [DOI] [PubMed] [Google Scholar]
- Eibach D, Krumkamp R, Al-Emran HM, Sarpong N, Hagen RM, Adu-Sarkodie Y, et al. Molecular Characterization of Cryptosporidium spp. among Children in Rural Ghana. PLoS Negl Trop Dis. 2015;9(3):e0003551. doi: 10.1371/journal.pntd.0003551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltahan R, Guo F, Zhang H, Xiang L, Zhu G. Discovery of ebselen as an inhibitor of Cryptosporidium parvum glucose-6-phosphate isomerase (CpGPI) by high-throughput screening of existing drugs. Int J Parasitol Drugs Drug Resist. 2018;8(1):43–49. doi: 10.1016/j.ijpddr.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzold M, Lendner M, Daugschies A, Dyachenko V. CDPKs of Cryptosporidium parvum—stage-specific expression in vitro. Parasitol Res. 2014;113(7):2525–2533. doi: 10.1007/s00436-014-3902-0. [DOI] [PubMed] [Google Scholar]
- Fan Y, Feng Y, Xiao L. Comparative genomics: how has it advanced our knowledge of cryptosporidiosis epidemiology? Parasitol Res. 2019;118(12):3195–3204. doi: 10.1007/s00436-019-06537-x. [DOI] [PubMed] [Google Scholar]
- Fayer R, Leek GR. The effects of reducing conditions, medium, pH, temperature, and time on in vitro excystation of cryptosporidium. J Protozool. 1984;31(4):567–569. doi: 10.1111/j.1550-7408.1984.tb05504.x. [DOI] [PubMed] [Google Scholar]
- Feng Y, Li N, Roellig DM, Kelley A, Liu G, Amer S, et al. Comparative genomic analysis of the IId subtype family of Cryptosporidium parvum. Int J Parasitol. 2017;47(5):281–290. doi: 10.1016/j.ijpara.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichtenbaum CJ, Zackin R, Feinberg J, Benson C, Griffiths JK. Rifabutin but not clarithromycin prevents cryptosporidiosis in persons with advanced HIV infection. AIDS. 2000 doi: 10.1097/00002030-200012220-00010. [DOI] [PubMed] [Google Scholar]
- Fortin A, Caridha DP, Leed S, Ngundam F, Sena J, Bosschaerts T, et al. Direct comparison of the efficacy and safety of oral treatments with oleylphosphocholine (OlPC) and Miltefosine in a Mouse Model of L. major Cutaneous Leishmaniasis. PLoS Negl Trop Dis. 2014;8(9):e3144. doi: 10.1371/journal.pntd.0003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox LM, Saravolatz LD. Nitazoxanide: a new thiazolide antiparasitic agent. Clin Infect Dis. 2005;40(8):1173–1180. doi: 10.1086/428839. [DOI] [PubMed] [Google Scholar]
- Funkhouser-Jones LJ, Ravindran S, Sibley LD. Defining stage-specific activity of potent new inhibitors of Cryptosporidium parvum growth in vitro. Mbio. 2020;11(2):e00052–e120. doi: 10.1128/mBio.00052-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargala G, François A, Favennec L, Rossignol J-F. Activity of Halogeno-Thiazolides against Cryptosporidium parvum in Experimentally infected immunosuppressed gerbils (Merio) Antimicrob Agents Chemother. 2013;57(6):2821–2823. doi: 10.1128/AAC.01538-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist CA, Cotton JA, Burkey C, Arju T, Gilmartin A, Lin Y, et al. Genetic diversity of cryptosporidium hominis in a bangladeshi community as revealed by whole-genome sequencing. J Infect Dis. 2018;218(2):259–264. doi: 10.1093/infdis/jiy121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JL, Sibley LD. Comparative genome analysis reveals a conserved family of actin-like proteins in apicomplexan parasites. BMC Genom. 2005;6(1):179. doi: 10.1186/1471-2164-6-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorla SK, McNair NN, Yang G, Gao S, Hu M, Jala VR, et al. Validation of IMP dehydrogenase inhibitors in a mouse model of cryptosporidiosis. Antimicrob Agents Chemother. 2014;58(3):1603–1614. doi: 10.1128/AAC.02075-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Zhang H, Fritzler JM, Rider SD, Jr, Xiang L, McNair NN, et al. Amelioration of Cryptosporidium parvum infection in vitro and in vivo by targeting parasite fatty acyl-coenzyme a synthetases. J Infect Dis. 2014;209(8):1279–1287. doi: 10.1093/infdis/jit645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Li N, Lysén C, Frace M, Tang K, Sammons S, et al. Isolation and enrichment of cryptosporidium DNA and verification of DNA purity for whole-genome sequencing. J Clin Microbiol. 2015;53(2):641–647. doi: 10.1128/JCM.02962-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Tang K, Rowe LA, Li N, Roellig DM, Knipe K, et al. Comparative genomic analysis reveals occurrence of genetic recombination in virulent Cryptosporidium hominis subtypes and telomeric gene duplications in Cryptosporidium parvum. BMC Genomics. 2015;16(1):320. doi: 10.1186/s12864-015-1517-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield SJ, Pachebat JA, Swain MT, Robinson G, Cameron SJS, Alexander J, et al. Generation of whole genome sequences of new Cryptosporidium hominis and Cryptosporidium parvum isolates directly from stool samples. BMC Genomics. 2015;16(1):650. doi: 10.1186/s12864-015-1805-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafez EN, El Hamed WFA. The efficacy of citrus maxima peels aqueous extract against cryptosporidiosis in immunecompromised mice. Acta Parasitol. 2021;66(2):638–653. doi: 10.1007/s11686-020-00315-x. [DOI] [PubMed] [Google Scholar]
- Heiges M, Wang H, Robinson E, Aurrecoechea C, Gao X, Kaluskar N, et al. CryptoDB: a Cryptosporidium bioinformatics resource update. Nucleic Acids Res. 2006;34(1):D419–D422. doi: 10.1093/nar/gkj078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriquez FL, Richards TA, Roberts F, McLeod R, Roberts CW. The unusual mitochondrial compartment of Cryptosporidium parvum. Trends Parasitol. 2005;21(2):68–74. doi: 10.1016/j.pt.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Heo I, Dutta D, Schaefer DA, Iakobachvili N, Artegiani B, Sachs N, et al. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat Microbiol. 2018;3(7):814–823. doi: 10.1038/s41564-018-0177-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández L, Gálvez R, Montoya A, Checa R, Bello A, Bosschaerts T, et al. First study on efficacy and tolerability of a new alkylphosphocholine molecule (oleylphosphocholine—OlPC) in the treatment of canine leishmaniosis due to Leishmania infantum. Parasitol Res. 2014;113(1):157–164. doi: 10.1007/s00436-013-3638-2. [DOI] [PubMed] [Google Scholar]
- Holmberg SD, Moorman AC, Von Bargen JC, Palella FJ, Loveless MO, Ward DJ, et al. Possible effectiveness of clarithromycin and Rifabutin for cryptosporidiosis chemoprophylaxis in HIV disease. JAMA. 1998;279(5):384–386. doi: 10.1001/jama.279.5.384. [DOI] [PubMed] [Google Scholar]
- Hu G, Gong A-Y, Roth AL, Huang BQ, Ward HD, Zhu G, et al. Release of luminal exosomes contributes to TLR4-mediated epithelial antimicrobial defense. PLoS Pathog. 2013;9(4):e1003261. doi: 10.1371/journal.ppat.1003261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Choi R, Hulverson MA, Zhang Z, McCloskey MC, Schaefer DA, et al. 5-aminopyrazole-4-carboxamide-based compounds prevent the growth of Cryptosporidium parvum. Antimicrob Agents Chemother. 2017;61(8):e00020–e117. doi: 10.1128/AAC.00020-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulverson MA, Choi R, Arnold SLM, Schaefer DA, Hemphill A, McCloskey MC, et al. Advances in bumped kinase inhibitors for human and animal therapy for cryptosporidiosis. Int J Parasitol. 2017;47(12):753–763. doi: 10.1016/j.ijpara.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulverson MA, Vinayak S, Choi R, Schaefer DA, Castellanos-Gonzalez A, Vidadala RSR, et al. Bumped-kinase inhibitors for cryptosporidiosis therapy. J Infect Dis. 2017;215(8):1275–1284. doi: 10.1093/infdis/jix120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter PR, Nichols G. Epidemiology and clinical features of Cryptosporidium infection in immunocompromised patients. Clin Microbiol Rev. 2002;15(1):145–154. doi: 10.1128/CMR.15.1.145-154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iroh Tam P, Arnold SLM, Barrett LK, Chen CR, Conrad TM, Douglas E, et al. Clofazimine for treatment of cryptosporidiosis in human immunodeficiency virus infected adults: an experimental medicine, randomized, double-blind, placebo-controlled phase 2a trial. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaza JP, Galván AL, Polanco V, Huang B, Matveyev AV, Serrano MG, et al. Revisiting the reference genomes of human pathogenic Cryptosporidium species: reannotation of C. parvum Iowa and a new C. hominis reference. Sci Rep. 2015;5(1):16324. doi: 10.1038/srep16324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes J, Young ME, Chen E, Rogers NH, Burgstaller-Muehlbacher S, Hughes LD, et al. The ReFRAME library as a comprehensive drug repurposing library and its application to the treatment of cryptosporidiosis. Proc Natl Acad Sci. 2018;115(42):10750–10755. doi: 10.1073/pnas.1810137115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies R, Yang R, Woh CK, Weldt T, Milech N, Estcourt A, et al. Target validation of the inosine monophosphate dehydrogenase (IMPDH) gene in Cryptosporidium using Phylomer® peptides. Exp Parasitol. 2015;148:40–48. doi: 10.1016/j.exppara.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Jumani RS, Bessoff K, Love MS, Miller P, Stebbins EE, Teixeira JE, et al. A novel piperazine-based drug lead for cryptosporidiosis from the medicines for malaria venture open-access malaria box. Antimicrob Agents Chemother. 2018;62(4):e01505–e1517. doi: 10.1128/AAC.01505-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumani RS, Blais J, Tillmann H-C, Segal F, Wetty D, Ostermeier C, et al. Opportunities and challenges in developing a cryptosporidium controlled human infection model for testing antiparasitic agents. ACS Infectious Diseases. 2021;7(5):959–968. doi: 10.1021/acsinfecdis.1c00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J-M, Ju H-L, Yu J-R, Sohn W-M, Na B-K. Cryptostatin, a chagasin-family cysteine protease inhibitor of Cryptosporidium parvum. Parasitology. 2012;139(8):1029–1037. doi: 10.1017/S0031182012000297. [DOI] [PubMed] [Google Scholar]
- Keithly JS, Langreth SG, Buttle KF, Mannella CA. Electron tomographic and ultrastructural analysis of the Cryptosporidium parvum relict mitochondrion, its associated membranes, and organelles. J Eukaryot Microbiol. 2005;52(2):132–140. doi: 10.1111/j.1550-7408.2005.04-3317.x. [DOI] [PubMed] [Google Scholar]
- Khalil IA, Troeger C, Rao PC, Blacker BF, Brown A, Brewer TG, et al. Morbidity, mortality, and long-term consequences associated with diarrhoea from Cryptosporidium infection in children younger than 5 years: a meta-analyses study. Lancet Glob Health. 2018;6(7):e758–e768. doi: 10.1016/S2214-109X(18)30283-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Shaik JS, Grigg ME. Genomics and molecular epidemiology of Cryptosporidium species. Acta Trop. 2018;184:1–14. doi: 10.1016/j.actatropica.2017.10.023. [DOI] [PubMed] [Google Scholar]
- Kim K. Role of proteases in host cell invasion by Toxoplasma gondii and other Apicomplexa. Acta Trop. 2004;91(1):69–81. doi: 10.1016/j.actatropica.2003.11.016. [DOI] [PubMed] [Google Scholar]
- Kirubakaran S, Gorla SK, Sharling L, Zhang M, Liu X, Ray SS, et al. Structure–activity relationship study of selective benzimidazole-based inhibitors of Cryptosporidium parvum IMPDH. Bioorg Med Chem Lett. 2012;22(5):1985–1988. doi: 10.1016/j.bmcl.2012.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. The Lancet. 2013;382(9888):209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- Kotloff KL, Nasrin D, Blackwelder WC, Wu Y, Farag T, Panchalingham S, et al. The incidence, aetiology, and adverse clinical consequences of less severe diarrhoeal episodes among infants and children residing in low-income and middle-income countries: a 12-month case-control study as a follow-on to the Global Enteric Multicenter St. Lancet Glob Health. 2019;7(5):e568–e584. doi: 10.1016/S2214-109X(19)30076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause I, Amir J, Cleper R, Dagan A, Behor J, Samra Z, Davidovits M. Cryptosporidiosis in children following solid organ transplantation. Pediatr Infect Dis J. 2012;31(11):1135–1138. doi: 10.1097/INF.0b013e31826780f7. [DOI] [PubMed] [Google Scholar]
- Kumar A, Chatterjee I, Anbazhagan AN, Jayawardena D, Priyamvada S, Alrefai WA, et al. Cryptosporidium parvum disrupts intestinal epithelial barrier function via altering expression of key tight junction and adherens junction proteins. Cell Microbiol. 2018;20(6):e12830. doi: 10.1111/cmi.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kváč M, Kodádková A, Sak B, Květoňová D, Jalovecká M, Rost M, Salát J. Activated CD8+ T cells contribute to clearance of gastric Cryptosporidium muris infections. Parasite Immunol. 2011;33(4):210–216. doi: 10.1111/j.1365-3024.2010.01271.x. [DOI] [PubMed] [Google Scholar]
- LaGier MJ, Tachezy J, Stejskal F, Kutisova K, Keithly JS. Mitochondrial-type iron–sulfur cluster biosynthesis genes (IscS and IscU) in the apicomplexan Cryptosporidium parvum. Microbiology. 2003;149(12):3519–3530. doi: 10.1099/mic.0.26365-0. [DOI] [PubMed] [Google Scholar]
- Lanternier F, Amazzough K, Favennec L, Mamzer-Bruneel M-F, Abdoul H, Tourret J, et al. Cryptosporidium spp infection in solid organ transplantation: the nationwide “TRANSCRYPTO” study. Transplantation. 2017;101(4):826. doi: 10.1097/TP.0000000000001503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent F, Lacroix-Lamandé S. Innate immune responses play a key role in controlling infection of the intestinal epithelium by Cryptosporidium. Int J Parasitol. 2017;47(12):711–721. doi: 10.1016/j.ijpara.2017.08.001. [DOI] [PubMed] [Google Scholar]
- Lee S, Harwood M, Girouard D, Meyers MJ, Campbell MA, Beamer G, Tzipori S. The therapeutic efficacy of azithromycin and nitazoxanide in the acute pig model of Cryptosporidium hominis. PLoS ONE. 2017;12(10):e0185906. doi: 10.1371/journal.pone.0185906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Beamer G, Tzipori S. The piglet acute diarrhea model for evaluating efficacy of treatment and control of cryptosporidiosis. Hum Vaccin Immunother. 2019;15(6):1445–1452. doi: 10.1080/21645515.2018.1498436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand F, Grenouillet F, Larosa F, Dalle F, Saas P, Millon L, et al. Diagnosis and treatment of digestive cryptosporidiosis in allogeneic haematopoietic stem cell transplant recipients: a prospective single centre study. Bone Marrow Transplant. 2011;46(6):858–862. doi: 10.1038/bmt.2010.200. [DOI] [PubMed] [Google Scholar]
- Lendner M, Daugschies A. Cryptosporidium infections: molecular advances. Parasitology. 2014;141(11):1511–1532. doi: 10.1017/S0031182014000237. [DOI] [PubMed] [Google Scholar]
- Li Y, Baptista RP, Sateriale A, Striepen B, Kissinger JC. Analysis of long non-coding RNA in Cryptosporidium parvum reveals significant stage-specific antisense transcription. Front Cell Infect Microbiol. 2021 doi: 10.3389/fcimb.2020.608298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippuner C, Ramakrishnan C, Basso WU, Schmid MW, Okoniewski M, Smith NC, et al. RNA-Seq analysis during the life cycle of Cryptosporidium parvum reveals significant differential gene expression between proliferating stages in the intestine and infectious sporozoites. Int J Parasitol. 2018;48(6):413–422. doi: 10.1016/j.ijpara.2017.10.007. [DOI] [PubMed] [Google Scholar]
- Liu T-L, Fan X-C, Li Y-H, Yuan Y-J, Yin Y-L, Wang X-T, et al. Expression profiles of mRNA and lncRNA in HCT-8 cells infected with Cryptosporidium parvum IId subtype. Front Microbiol. 2018 doi: 10.3389/fmicb.2018.01409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Vélez R, Tarazona R, Camacho AG, Gomez-Mampaso E, Guerrero A, Moreira V, Villanueva R. Intestinal and extraintestinal cryptosporidiosis in AIDS patients. Eur J Clin Microbiol Infect Dis. 1995;14(8):677–681. doi: 10.1007/BF01690873. [DOI] [PubMed] [Google Scholar]
- Love MS, Choy RKM. Emerging treatment options for cryptosporidiosis. Curr Opin Infect Dis. 2021;34(5):455. doi: 10.1097/2FQCO.0000000000000761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MS, Beasley FC, Jumani RS, Wright TM, Chatterjee AK, Huston CD, et al. A high-throughput phenotypic screen identifies clofazimine as a potential treatment for cryptosporidiosis. PLoS Negl Trop Dis. 2017;11(2):e0005373. doi: 10.1371/journal.pntd.0005373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde CS, Stebbins EE, Jumani RS, Hasan MM, Miller P, Barlow J, et al. Identification of a potent benzoxaborole drug candidate for treating cryptosporidiosis. Nat Commun. 2019;10(1):2816. doi: 10.1038/s41467-019-10687-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P, Soave R. Three-step stool examination for cryptosporidiosis in 10 homosexual men with protracted watery diarrhea. J Infect Dis. 1983;147(5):824–828. doi: 10.1093/infdis/147.5.824. [DOI] [PubMed] [Google Scholar]
- Madbouly Taha N, Salah A, Yousof H-A, El-Sayed SH, Younis AI, Ismail Negm MS. Atorvastatin repurposing for the treatment of cryptosporidiosis in experimentally immunosuppressed mice. Exp Parasitol. 2017;181:57–69. doi: 10.1016/j.exppara.2017.07.010. [DOI] [PubMed] [Google Scholar]
- Madern D, Cai X, Abrahamsen MS, Zhu G. Evolution of Cryptosporidium parvum lactate dehydrogenase from malate dehydrogenase by a very recent event of gene duplication. Mol Biol Evol. 2004;21(3):489–497. doi: 10.1093/molbev/msh042. [DOI] [PubMed] [Google Scholar]
- Manjunatha UH, Chao AT, Leong FJ, Diagana TT. Cryptosporidiosis drug discovery: opportunities and challenges. ACS Infect Dis. 2016;2(8):530–537. doi: 10.1021/acsinfecdis.6b00094. [DOI] [PubMed] [Google Scholar]
- Manjunatha UH, Vinayak S, Zambriski JA, Chao AT, Sy T, Noble CG, et al. A Cryptosporidium PI(4)K inhibitor is a drug candidate for cryptosporidiosis. Nature. 2017;546(7658):376–380. doi: 10.1038/nature22337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos LVS, McEvoy J, Tzipori S, Bresciani KDS, Widmer G. The transcriptome of Cryptosporidium oocysts and intracellular stages. Sci Rep. 2019;9(1):7856. doi: 10.1038/s41598-019-44289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauzy MJ, Enomoto S, Lancto CA, Abrahamsen MS, Rutherford MS. The Cryptosporidium parvum transcriptome during in vitro development. PLoS ONE. 2012;7(3):e31715. doi: 10.1371/journal.pone.0031715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara CW, Lee MCS, Lim CS, Lim SH, Roland J, Nagle A, et al. Targeting Plasmodium PI(4)K to eliminate malaria. Nature. 2013;504(7479):248–253. doi: 10.1038/nature12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CN, Jossé L, Tsaousis AD. Localization of Fe-S biosynthesis machinery in Cryptosporidium parvum mitosome. J Eukaryot Microbiol. 2018;65(6):913–922. doi: 10.1111/jeu.12663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming Z, Gong A-Y, Wang Y, Zhang X-T, Li M, Dolata CE, Chen X-M. Trans-suppression of defense DEFB1 gene in intestinal epithelial cells following Cryptosporidium parvum infection is associated with host delivery of parasite Cdg7_FLc_1000 RNA. Parasitol Res. 2018;117(3):831–840. doi: 10.1007/s00436-018-5759-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirhashemi ME, Noubary F, Chapman-Bonofiglio S, Tzipori S, Huggins GS, Widmer G. Transcriptome analysis of pig intestinal cell monolayers infected with Cryptosporidium parvum asexual stages. Parasit Vectors. 2018;11(1):176. doi: 10.1186/s13071-018-2754-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y, Eckmann L. Drug development against the major diarrhea-causing parasites of the small intestine, Cryptosporidium and Giardia. Front Microbiol. 2015;5:4. doi: 10.3389/fmicb.2015.01208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogi T, Kita K. Diversity in mitochondrial metabolic pathways in parasitic protists Plasmodium and Cryptosporidium. Parasitol Int. 2010;59(3):305–312. doi: 10.1016/j.parint.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Morada M, Lee S, Gunther-Cummins L, Weiss LM, Widmer G, Tzipori S, Yarlett N. Continuous culture of Cryptosporidium parvum using hollow fiber technology. Int J Parasitol. 2016;46(1):21–29. doi: 10.1016/j.ijpara.2015.07.006. [DOI] [PubMed] [Google Scholar]
- Morris A, Robinson G, Swain MT, Chalmers RM. Direct sequencing of cryptosporidium in stool samples for public health. Front Public Health. 2019 doi: 10.3389/fpubh.2019.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na B-K, Kang J-M, Cheun H-I, Cho S-H, Moon S-U, Kim T-S, Sohn W-M. Cryptopain-1, a cysteine protease of Cryptosporidium parvum, does not require the pro-domain for folding. Parasitology. 2009;136(2):149–157. doi: 10.1017/S0031182008005350. [DOI] [PubMed] [Google Scholar]
- Nader JL, Mathers TC, Ward BJ, Pachebat JA, Swain MT, Robinson G, et al. Evolutionary genomics of anthroponosis in Cryptosporidium. Nat Microbiol. 2019;4(5):826–836. doi: 10.1038/s41564-019-0377-x. [DOI] [PubMed] [Google Scholar]
- Ndao M, Nath-Chowdhury M, Sajid M, Marcus V, Mashiyama ST, Sakanari J, et al. A cysteine protease inhibitor rescues mice from a lethal Cryptosporidium parvum Infection. Antimicrob Agents Chemother. 2013;57(12):6063–6073. doi: 10.1128/AAC.00734-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nime FA, Burek JD, Page DL, Holscher MA, Yardley JH. Acute enterocolitis in a human being infected with the protozoan cryptosporidium. Gastroenterology. 1976;70(4):592–598. doi: 10.1016/S0016-5085(76)80503-3. [DOI] [PubMed] [Google Scholar]
- O’Handley RM, Olson ME. Giardiasis and cryptosporidiosis in ruminants. Vet Clin Food Anim Pract. 2006;22(3):623–643. doi: 10.1016/j.cvfa.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Okhuysen PC, Chappell CL. Cryptosporidium virulence determinants—Are we there yet? Int J Parasitol. 2002;32(5):517–525. doi: 10.1016/S0020-7519(01)00356-3. [DOI] [PubMed] [Google Scholar]
- Osman M, El Safadi D, Cian A, Benamrouz S, Nourrisson C, Poirier P, et al. Prevalence and risk factors for intestinal protozoan infections with Cryptosporidium, Giardia, Blastocystis and Dientamoeba among Schoolchildren in Tripoli, Lebanon. PLOS Negl Trop Dis. 2016;10(3):e0004496. doi: 10.1371/journal.pntd.0004496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda C, Mahapatra RK. Identification of novel therapeutic candidates in Cryptosporidium parvum: an in silico approach. Parasitology. 2018;145(14):1907–1916. doi: 10.1017/S0031182018000677. [DOI] [PubMed] [Google Scholar]
- Parihar SP, Guler R, Brombacher F. Statins: a viable candidate for host-directed therapy against infectious diseases. Nat Rev Immunol. 2019;19(2):104–117. doi: 10.1038/s41577-018-0094-3. [DOI] [PubMed] [Google Scholar]
- Platts-Mills JA, Babji S, Bodhidatta L, Gratz J, Haque R, Havt A, et al. Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED) Lancet Glob Health. 2015;3(9):e564–e575. doi: 10.1016/S2214-109X(15)00151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relat RMB, O’Connor RM. Cryptosporidium: host and parasite transcriptome in infection. Curr Opin Microbiol. 2020;58:138–145. doi: 10.1016/j.mib.2020.09.012. [DOI] [PubMed] [Google Scholar]
- Rossignol J-FA, Ayoub A, Ayers MS. Treatment of diarrhea caused by Cryptosporidium parvum: A prospective randomized, double-blind, placebo-controlled study of nitazoxanide. J Infect Dis. 2001;184(1):103–106. doi: 10.1086/321008. [DOI] [PubMed] [Google Scholar]
- Rossignol J, Kabil SM, El-GoharyYounis YAM. Effect of nitazoxanide in diarrhea and enteritis caused by cryptosporidium species. Clin Gastroenterol Hepatol. 2006;4(3):320–324. doi: 10.1016/j.cgh.2005.12.020. [DOI] [PubMed] [Google Scholar]
- Sateriale A, Šlapeta J, Baptista R, Engiles JB, Gullicksrud JA, Herbert GT, et al. A genetically tractable, natural mouse model of cryptosporidiosis offers insights into host protective immunity. Cell Host Microbe. 2019;26(1):135–146. doi: 10.1016/j.chom.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sateriale A, Gullicksrud JA, Engiles JB, McLeod BI, Kugler EM, Henao-Mejia J, et al. The intestinal parasite Cryptosporidium is controlled by an enterocyte intrinsic inflammasome that depends on NLRP6. Proc Natl Acad Sci. 2021;118(2):e2007807118. doi: 10.1073/pnas.2007807118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Hempel P. Immune defence, parasite evasion strategies and their relevance for ‘macroscopic phenomena’ such as virulence. Philos Trans R Soc B Biol Sci. 2009;364(1513):85–98. doi: 10.1098/rstb.2008.0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigetomi K, Sarwono AEY, Ichikawa S, Ubukata M. Novel adenosine-derived inhibitors of Cryptosporidium parvum inosine 5′-monophosphate dehydrogenase. J Antibiot. 2019;72(12):934–942. doi: 10.1038/s41429-019-0199-3. [DOI] [PubMed] [Google Scholar]
- Sinkala E, Katubulushi M, Sianongo S, Obwaller A, Kelly P. In a trial of the use of miltefosine to treat HIV-related cryptosporidiosis in Zambian adults, extreme metabolic disturbances contribute to high mortality. Ann Trop Med Parasitol. 2011;105(2):129–134. doi: 10.1179/136485911X12899838683160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siqueira-Neto JL, Debnath A, McCall L-I, Bernatchez JA, Ndao M, Reed SL, Rosenthal PJ. Cysteine proteases in protozoan parasites. PLoS Negl Trop Dis. 2018;12(8):e0006512. doi: 10.1371/journal.pntd.0006512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HV, Nichols RAB, Grimason AM. Cryptosporidium excystation and invasion: getting to the guts of the matter. Trends Parasitol. 2005;21(3):133–142. doi: 10.1016/j.pt.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Sonzogni-Desautels K, Renteria Flores A, Vasquez Camargo F, Di Lenardo T, Mikhail A, Arrowood M, et al. Oleylphosphocholine (OlPC) arrests Cryptosporidium parvum growth in vitro and prevents lethal infection in interferon gamma receptor knock-out mice. Front Microbiol. 2015 doi: 10.3389/fmicb.2015.00973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sow SO, Muhsen K, Nasrin D, Blackwelder WC, Wu Y, Farag TH, et al. The burden of cryptosporidium diarrheal disease among children < 24 Months of Age in moderate/high mortality regions of Sub-Saharan Africa and South Asia, Utilizing Data from the Global Enteric Multicenter Study (GEMS) PLoS Negl Trop Dis. 2016;10(5):e0004729. doi: 10.1371/journal.pntd.0004729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks H, Nair G, Castellanos-Gonzalez A, White AC. Treatment of Cryptosporidium: what we know, gaps, and the way forward. Curr Trop Med Rep. 2015;2(3):181–187. doi: 10.1007/s40475-015-0056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striepen B, Kissinger JC. Genomics meets transgenics in search of the elusive Cryptosporidium drug target. Trends Parasitol. 2004;20(8):355–358. doi: 10.1016/j.pt.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Su J, Shen Y, Li N, Li Y, Zhang Z, Xiao L, et al. Comparative characterization of CpCDPK1 and CpCDPK9, two potential drug targets against cryptosporidiosis. Microorganisms. 2022;10(2):333. doi: 10.3390/microorganisms10020333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Khan J, Makowska-Grzyska M, Zhang M, Cho JH, Suebsuwong C, et al. Synthesis, in vitro evaluation and cocrystal structure of 4-oxo-[1]benzopyrano[4,3-c]pyrazole cryptosporidium parvum inosine 5′-monophosphate dehydrogenase (CpIMPDH) Inhibitors. J Med Chem. 2014;57(24):10544–10550. doi: 10.1021/jm501527z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swale C, Bougdour A, Gnahoui-David A, Tottey J, Georgeault S, Laurent F, et al. Metal-captured inhibition of pre-mRNA processing activity by CPSF3 controls Cryptosporidium infection. Sci Translat Med. 2019;11(517):eaax761. doi: 10.1126/scitranslmed.aax7161. [DOI] [PubMed] [Google Scholar]
- Tandel J, English ED, Sateriale A, Gullicksrud JA, Beiting DP, Sullivan MC, et al. Life cycle progression and sexual development of the apicomplexan parasite Cryptosporidium parvum. Nat Microbiol. 2019;4(12):2226–2236. doi: 10.1038/s41564-019-0539-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton TJ, Iyer LM, Anantharaman V, Enomoto S, Abrahante JE, Subramanian GM, et al. Comparative analysis of apicomplexa and genomic diversity in eukaryotes. Genome Res. 2004;14(9):1686–1695. doi: 10.1101/gr.2615304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RCA, Olson ME, Zhu G, Enomoto S, Abrahamsen MS, Hijjawi NS (2005). Cryptosporidium and Cryptosporidiosis. In: Baker JR, Muller R, D B. T.-A. in P. Rollinson (Eds), Vol 59, pp 77–158). Academic Press. 10.1016/S0065-308X(05)59002-X [DOI] [PubMed]
- Troell K, Hallström B, Divne A-M, Alsmark C, Arrighi R, Huss M, et al. Cryptosporidium as a testbed for single cell genome characterization of unicellular eukaryotes. BMC Genom. 2016;17(1):471. doi: 10.1186/s12864-016-2815-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyzzer EE. A sporozoan found in the peptic glands of the common mouse. Proc Soc Exp Biol Med. 1907;5(1):12–13. doi: 10.3181/00379727-5-5. [DOI] [Google Scholar]
- Tyzzer, E. E. (1910). An extracellular Coccidium, Cryptosporidium Muris (Gen. Et Sp. Nov.), of the gastric Glands of the Common Mouse. J Med Res 23(3): 487–510. [PMC free article] [PubMed]
- Tzipori S, Ward H. Cryptosporidiosis: biology, pathogenesis and disease. Microbes Infect. 2002;4(10):1047–1058. doi: 10.1016/S1286-4579(02)01629-5. [DOI] [PubMed] [Google Scholar]
- Tzipori S, Widmer G. A hundred-year retrospective on cryptosporidiosis. Trends Parasitol. 2008;24(4):184–189. doi: 10.1016/j.pt.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzipori S, Angus K, Gray E, Campbell I. Vomiting and diarrhea associated with cryptosporidial infection. N Engl J Med. 1980;303(14):818. doi: 10.1056/NEJM198010023031416. [DOI] [PubMed] [Google Scholar]
- Uip DE, Lima AL, Amato VS, Boulos M, Neto VA, Bem David D. Roxithromycin treatment for diarrhoea caused by Cryptosporidium spp in patients with AIDS. J Antimicrob Chemother. 1998;41(2):93–97. doi: 10.1093/jac/41.suppl_2.93. [DOI] [PubMed] [Google Scholar]
- Valigurová A, Jirků M, Koudela B, Gelnar M, Modrý D, Šlapeta J. Cryptosporidia: epicellular parasites embraced by the host cell membrane. Int J Parasitol. 2008;38(8):913–922. doi: 10.1016/j.ijpara.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Van Voorhis WC, Hulverson MA, Choi R, Huang W, Arnold SLM, Schaefer DA, et al. One health therapeutics: Target-Based drug development for cryptosporidiosis and other apicomplexa diseases. Vet Parasitol. 2021;289:109336. doi: 10.1016/j.vetpar.2020.109336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayak S, Pawlowic MC, Sateriale A, Brooks CF, Studstill CJ, Bar-Peled Y, et al. Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum. Nature. 2015;523(7561):477–480. doi: 10.1038/nature14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayak S, Jumani RS, Miller P, Hasan MM, McLeod BI, Tandel J, et al. Bicyclic azetidines kill the diarrheal pathogen Cryptosporidium in mice by inhibiting parasite phenylalanyl-tRNA synthetase. Sci Translat Med. 2020;12(563):8412. doi: 10.1126/scitranslmed.aba8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Jian F, Zhang L, Ning C, Liu A, Zhao J, et al. Multilocus sequence subtyping and genetic structure of cryptosporidium muris and cryptosporidium andersoni. PLoS ONE. 2012;7(8):e43782. doi: 10.1371/journal.pone.0043782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gong A-Y, Ma S, Chen X, Li Y, Su C-J, et al. Delivery of Parasite RNA Transcripts Into Infected Epithelial Cells During Cryptosporidium Infection and Its Potential Impact on Host Gene Transcription. J Infect Dis. 2017;215(4):636–643. doi: 10.1093/infdis/jiw607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gong A-Y, Ma S, Chen X, Strauss-Soukup JK, Chen X-M. Delivery of parasite Cdg7_Flc_0990 RNA transcript into intestinal epithelial cells during Cryptosporidium parvum infection suppresses host cell gene transcription through epigenetic mechanisms. Cell Microbiol. 2017;19(11):e12760. doi: 10.1111/cmi.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanyiri JW, Kanyi H, Maina S, Wang DE, Steen A, Ngugi P, et al. Cryptosporidiosis in HIV/AIDS patients in Kenya: clinical features, epidemiology, molecular characterization and antibody responses. Am Soc Trop Med Hygiene. 2014;91(2):319–328. doi: 10.4269/ajtmh.13-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrenfeltz S, Kissinger JC (2020) Accessing Cryptosporidium Omic and Isolate Data via CryptoDB.org. In J. R. Mead & M. J. Arrowood (Eds.), Cryptosporidium. Methods in molecular biology (vol 2052, pp 139–192). Humana, New York, NY, United States. 10.1007/978-1-4939-9748-0_10 [DOI] [PubMed]
- Wernimont AK, Artz JD, Finerty P, Lin Y-H, Amani M, Allali-Hassani A, et al. Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium. Nat Struct Mol Biol. 2010;17(5):596–601. doi: 10.1038/nsmb.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmer G, Lee Y, Hunt P, Martinelli A, Tolkoff M, Bodi K. Comparative genome analysis of two Cryptosporidium parvum isolates with different host range. Infect Genet Evol. 2012;12(6):1213–1221. doi: 10.1016/j.meegid.2012.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke G, Funkhouser-Jones LJ, Wang Y, Ravindran S, Wang Q, Beatty WL, et al. A stem-cell-derived platform enables complete Cryptosporidium development in vitro and genetic tractability. Cell Host Microbe. 2019;26(1):123–134. doi: 10.1016/j.chom.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Widmer G, Wang Y, Ozaki LS, Alves JM, Serrano MG, et al. The genome of cryptosporidium hominis. Nature. 2004;431(7012):1107–1112. doi: 10.1038/nature02977. [DOI] [PubMed] [Google Scholar]
- Xu Z, Guo Y, Roellig DM, Feng Y, Xiao L. Comparative analysis reveals conservation in genome organization among intestinal Cryptosporidium species and sequence divergence in potential secreted pathogenesis determinants among major human-infecting species. BMC Genom. 2019;20(1):406. doi: 10.1186/s12864-019-5788-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Fu Y, Gong P, Zheng J, Liu L, Yu Y, et al. Bovine TLR2 and TLR4 mediate Cryptosporidium parvum recognition in bovine intestinal epithelial cells. Microb Pathog. 2015;85:29–34. doi: 10.1016/j.micpath.2015.05.009. [DOI] [PubMed] [Google Scholar]
- Yanta CA, Bessonov K, Robinson G, Troell K, Guy RA. CryptoGenotyper: A new bioinformatics tool for rapid Cryptosporidium identification. Food and Waterborne Parasitology. 2021;23:e00115. doi: 10.1016/j.fawpar.2021.e00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarlett N, Morada M, Gobin M, Van Voorhis W, Arnold S (2020) In vitro culture of Cryptosporidium parvum using hollow fiber bioreactor: applications for simultaneous pharmacokinetic and pharmacodynamic evaluation of test compounds. In: Cryptosporidium. Springer, pp 335–350. 10.1007/978-1-4939-9748-0_19 [DOI] [PubMed]
- Yu Y, Zhang H, Guo F, Sun M, Zhu G. A Unique Hexokinase in Cryptosporidium parvum, an apicomplexan pathogen lacking the krebs cycle and oxidative phosphorylation. Protist. 2014;165(5):701–714. doi: 10.1016/j.protis.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaalouk TK, Bajaj-Elliott M, George JT, McDonald V (2004) Differential Regulation of β-Defensin Gene Expression during Cryptosporidium parvum Infection. Infect Immun 72(5): 2772 LP–2779. 10.1128/IAI.72.5.2772-2779.2004 [DOI] [PMC free article] [PubMed]
- Zambriski JA, Nydam DV, Bowman DD, Bellosa ML, Burton AJ, Linden TC, et al. Description of fecal shedding of Cryptosporidium parvum oocysts in experimentally challenged dairy calves. Parasitol Res. 2013;112(3):1247–1254. doi: 10.1007/s00436-012-3258-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Guo F, Zhou H, Zhu G. Transcriptome analysis reveals unique metabolic features in the Cryptosporidium parvum oocysts associated with environmental survival and stresses. BMC Genomics. 2012;13(1):1–15. doi: 10.1186/1471-2164-13-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Guo F, Zhu G. Cryptosporidium lactate dehydrogenase is associated with the parasitophorous vacuole membrane and is a potential target for developing therapeutics. PLoS Pathog. 2015;11(11):e1005250. doi: 10.1371/journal.ppat.1005250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Shao Q, Guo Y, Li N, Li Y, Su J, et al. Characterization of three calcium-dependent protein kinases of Cryptosporidium parvum. Front Microbiol. 2021 doi: 10.3389/fmicb.2020.622203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhu G (2020) High-throughput screening of drugs against the growth of Cryptosporidium parvum in vitro by qRT-PCR. In: Cryptosporidium. Springer, pp 319–334. 10.1007/978-1-4939-9748-0_18 [DOI] [PubMed]
- Zhao G-H, Gong A-Y, Wang Y, Zhang X-T, Li M, Mathy NW, Chen X-M. Nuclear delivery of parasite Cdg2_FLc_0220 RNA transcript to epithelial cells during Cryptosporidium parvum infection modulates host gene transcription. Vet Parasitol. 2018;251:27–33. doi: 10.1016/j.vetpar.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]