Abstract
Cyclin dependent kinase 9 (CDK9) plays an essential role in regulating transcriptional elongation. Aberrations in CDK9 activity have been observed in various cancers, which make CDK9 an attractive therapeutic target for cancers. This led to an intensive development of small-molecule CDK9 inhibitors or new emerging strategies, such as proteolysis targeting chimeras (PROTACs). Here, we review the CDK9 modulators in cancer not only for research purposes, but also for therapeutic applications.
This review outlines the representative CDK9 inhibitors in cancer research and application.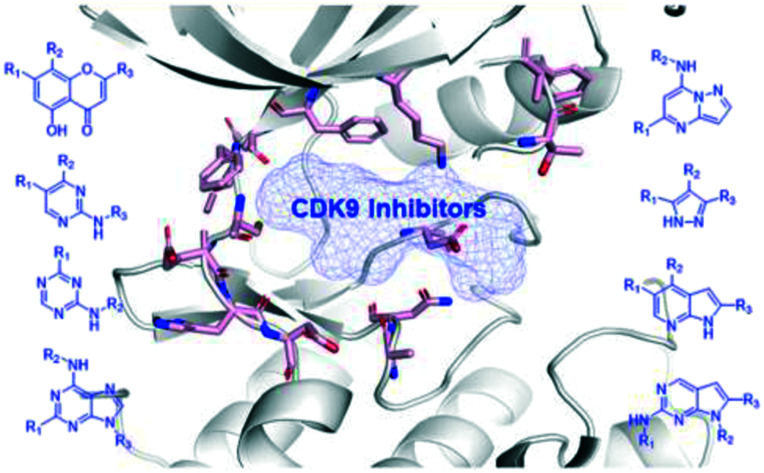
Introduction
Cyclin-dependent kinases (CDKs) belong to the serine/threonine kinase subfamily, and are involved in multiple functions, such as the cell division, apoptosis, transcription, and differentiation.1,2 Most CDKs form heterodimeric complexes with their partners called cyclins to perform their physiological function.3,4 CDKs are comprised of 21 hypo types that are divided into two groups based on their roles in cell cycle progression and transcription regulation.5,6 CDKs 1–6, 11 and 14–18 along with cyclins mainly regulate cell cycles and CDKs 7–13, 19 and 20 are associated with transcription control. CDKs 1–3 along with cyclins A, B, and E participate in S–G2, and M phases. CDKs 2, 4, and 6 associated with cyclins D and E are key players in the regulation of G0–G1 transition and the early phase of G1. CDK2 along with the cyclin A complex is also involved in the progression of the S phase. CDKs 7–13 along with cyclins C, H, L, K and T are associated with transcription regulation/RNA processing.7
Most CDKs have been implicated in human cancers. Abnormalities in CDK expression, activity and regulation have been found in human cancers and are considered as a hallmark of cancer development and progression.5,8 Due to their specialized roles in cell cycle progression and gene transcription regulation, CDKs present particularly promising drug targets for therapeutic interference for cancers.9,10 The first generation CDK inhibitors with multiple CDK inhibition have been explored for anticancer potential, such as flavopiridol11 and roscovitine.12 The second generation drugs were designed to have a smaller selective spectrum of CDKs, such as dinaciclib.13 Recently, drugs with more selectivity against CDKs have been developed. Three CDK4/6 inhibitors, palbociclib,14 ribociclib,15 and abemaciclib16 received the US Food and Drug Administration (FDA) approval for the treatment of HR+ breast cancer and other malignancies. During the past decade, attempts for discovering pharmacological CDK inhibitors have led to the realization that transcriptional CDKs are promising drug targets for therapeutic interference in human cancers.
Among the transcription-associated CDKs, CDK9 is the most extensively studied. CDK9 forms a functional complex with regulatory subunit cyclin T or cyclin K, which plays a critical role in regulating gene transcription elongation, messenger mRNA (mRNA) maturation and several physiologic processes.17 CDK9 with cyclin T forms the main component of the positive transcription elongation factor b (P-TEFb), which phosphorylates the carboxyl-terminal domain (CTD) of RNA polymerase II (RNAPII) to stimulate transcription elongation of most protein coding genes.18 The dysregulation of transcriptional programs has been observed in various human cancers, which makes CDK9 a prioritized target for cancer therapy across a range of tumor types.19–21 The importance of CDK9 has led to an intensive search for CDK9 inhibitors for therapeutic purposes. In this review, we provide an overview of CDK9 inhibitors in cancer research and therapy.
Cyclin dependent kinase 9 (CDK9)
CDK9 has two isoforms based on their respective molecular weight: a short form 42 kDa protein and a long-form 55 kDa protein. Compared to the 42 kDa isoform of CDK9, the 55 kDa isoform has an additional 117 amino acid residues at the amino terminus. The 42 kDa isoform of CDK9 is mainly distributed in the cytoplasm and nucleus and more highly expressed, while the 55 kDa isoform is mainly found in the nucleolar region.22 Both CDK9 isoforms form heterodimeric complexes with cyclin T. Cyclin T1 is the predominant CDK9-associated cyclin, while cyclin T2a and T2b are minor CDK9 partners.
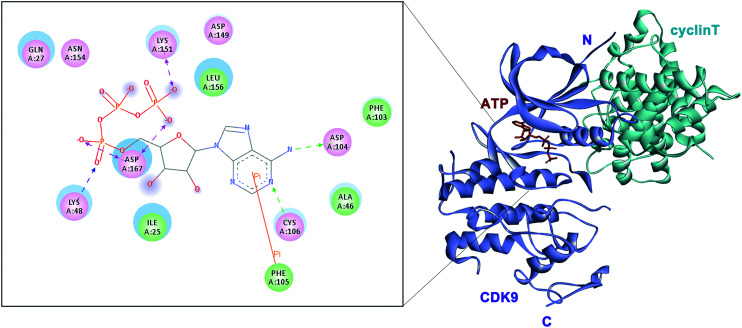
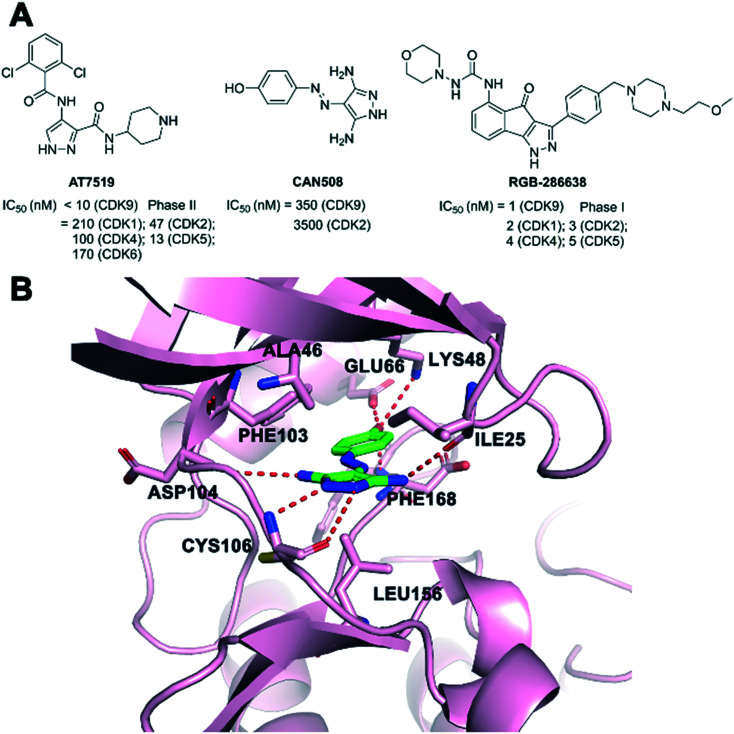
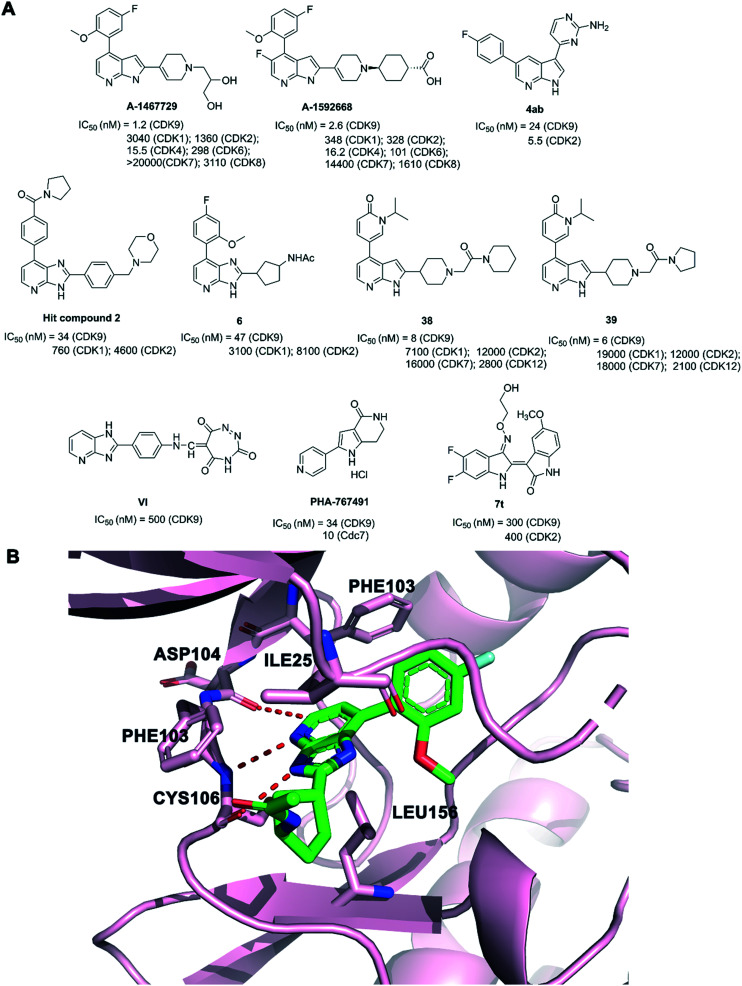
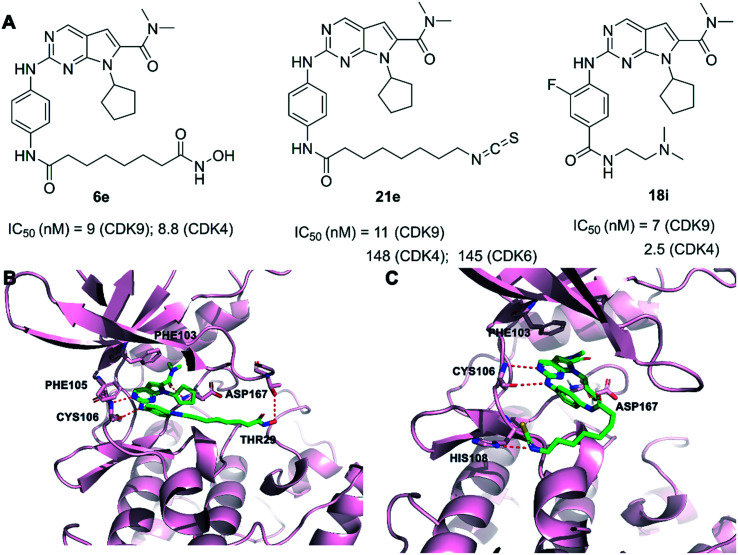
CDK9 consists of an N- and a C-terminal kinase lobe and a short C-terminal extension (Fig. 1). The ATP-binding site is sandwiched between the N- and C-terminal lobes. The adenine moiety of ATP forms hydrogen bonds with hinge residues ASP104 and CYS106. Hydrogen or ionic bonds are also formed between the phosphate group of ATP and the backbone residue of ASP167, LYS48 and LYS151. It is reported that CDK9 overexpressed in various tumor tissues and its high expression is associated with the poor survival of patients with cancer.17,23,24 Mechanistically, CDK9 inhibition blocks phosphorylation at Ser2 of the RNAPII CTD and induces downregulation of protein levels of oncogenes and anti-apoptotic proteins, leading to the inhibition of proliferation and the promotion of apoptosis, which are confirmed in various malignancies. Recent studies have shown that the prominent method of blocking the P-TEFb function is to inhibit the ATP-binding site of CDK9.
Fig. 1. Structure of CDK9/cyclin T (PDB: 3BLR).
During the past decade, attempts have been made toward the development of many CDK9 inhibitors for treatment of various malignancies.25,26 There are several pan-CDK inhibitors in clinical studies in various stages of their research and development with poor selectivity and high toxicity. Potent and selective CDK9 inhibitors have recently emerged.26 The high structural homology among the CDKs has presented challenge for the discovery of selective CDK inhibitors. CDK9 shows high sequence conservation with other CDK family members, thus the design of selective CDK9 inhibitor is particularly challenging. Representative pan-CDK inhibitors and selective CDK9 inhibitors are described in detail in this paper as follows.
CDK9 inhibitors for cancer therapy
CDK9 inhibitors with flavonoid cores
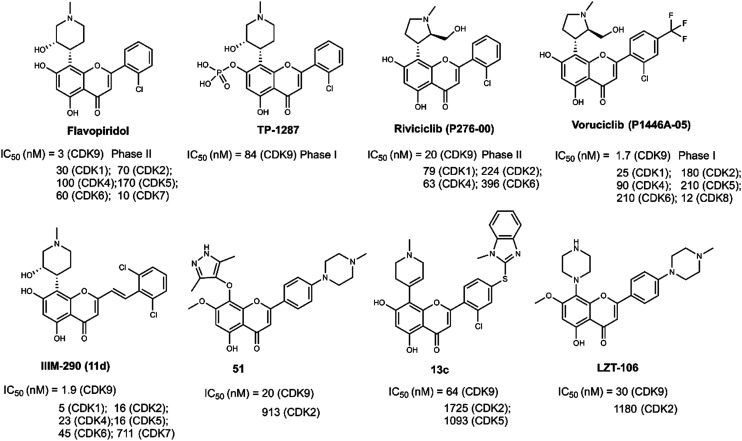
The first CDK9 inhibitor to be tested in clinical trials was flavopiridol (Fig. 2, alvocidib, [cis-5,7-dihydroxy-2-(2-chlorophenyl)-8-[4-(3-hydroxy-1-methyl)-piperidinyl]-1-benzopyran-4-one]), a flavonoid derived from an indigenous plant with multiple CDK inhibition.27 The Ki value of flavopiridol for CDK9/cyclin T was 10-fold lower than those for other CDKs.28 Preclinically, flavopiridol was an effective anti-cancer drug against various types of cancer, such as acute leukemia, breast cancer, lung cancer, prostate carcinoma, colon cancer and gastric cancer.29–33 Flavopiridol enhanced the radiosensitivity of carcinoma cells and displayed synergistic effects with chemotherapy drugs, such as taxanes, gemcitabine, cytarabine, topotecan and doxorubicin34–37 Flavopiridol inhibited CDKs that showed a clear blockade of cell cycle progression at the G1/S and G2/M boundaries and phosphorylated the CTD of RNAPII.38,39 The primary antitumor mechanism of flavopiridol in leukemia appeared to be the CDK9-mediated downregulation of transcription of antiapoptotic proteins.40 The crystal structure of flavopiridol with CDK9 (Fig. 3A) revealed that hydrogen bonds were formed from residues CYS106 and ASP104 to the hydroxyl of compound flavopiridol. The O hydroxyl of flavopiridol bonded with the ASP167 side chain. The chromenone group occupied a hydrophobic pocket formed by PHE103, PHE30, LYS48 and VAL33. Additionally, flavopiridol formed van der Waals contacts with the ILE25 and VAL33 residues of the glycine-rich loop (G-loop, residues 17–36). The PHE30 residue made van der Waals contacts (edge to face) with the piperidinyl group of flavopiridol.
Fig. 2. The development of CDK9 inhibitors with flavonoid cores.
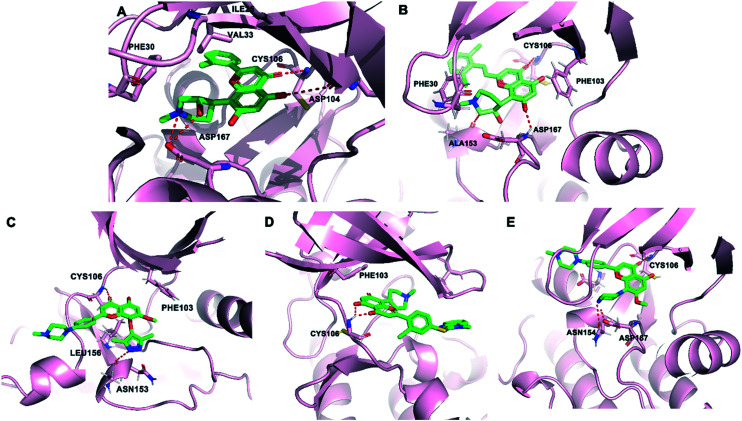
Fig. 3. (A) The crystal structure of flavopiridol (green) and CDK9 (pink, PDB: 3BLR). (B) The predicted binding mode of compound IIIM-290 (green) and CDK9 (pink, PDB: 3BLR). (C) The predicted binding mode of compound 51 (green) and CDK9 (pink, PDB: 3BLR). (D) The predicted binding mode of compound 13c (green) and CDK9 (pink, PDB: 3BLR). (E) The predicted binding mode of compound LZT-106 (green) and CDK9 (pink, PDB: 3BLR). The hydrogen bonds are depicted by red dotted lines.
TP-1287 (Fig. 2) was reported as a phosphate prodrug of flavopiridol with highly improved solubility under acidic, neutral, and basic conditions in 2017. Pharmacokinetic studies were conducted showing that TP-1287 has high oral bioavailability. TP-1287 strongly inhibited tumor growth, exhibiting 109.1% tumor growth inhibition at a dose of 7.5 mg kg−1.41 Phase I, the first-in-human study of oral TP-1287 (NCT03604783), has been launched recently in November 2021 and patients with advanced solid tumors have been enrolled.
Riviciclib (P276-00, Fig. 2) was obtained from Nicholas Piramal Ltd, Mumbai, India in 2007.42 Riviciclib inhibited CDK1, CDK4 and CDK9 with IC50 values at 79 nM, 63 nM and 20 nM, respectively.43 It modulated the cell cycle and induced apoptosis against mantle cell lymphoma (MCL), head and neck cancer. The anti-tumor activity and safety of riviciclib alone or with other agents have been evaluated in phase II clinical studies in patients. Single-agent treatment with riviciclib failed to produce objective responses among 13 patients with relapsed or refractory MCL in a phase II study.
Voruciclib (P1446A-05, P-1446) was obtained by introducing a trifluoromethyl group at the para position of the benzene ring in riviciclib in 2012 (Fig. 2).44 Voruciclib was a clinical stage oral CDK9 inhibitor, structurally similar to flavopiridol.45 The Ki value of voruciclib for CDK9/cyclin T2 was 0.626 nM. Structural differences of the methylpyrrolidin ring in voruciclib altered the target selectivity. Compared to flavopiridol, voruciclib was a CDK-selective inhibitor with potent activity against CDK9 and reduced activity against kinases outside of the CDK family. Voruciclib displayed effective CDK9 activity with repression of MCL-1.46 Voruciclib achieved synergistic anti-tumor efficacy with venetoclax and showed potential use in combination with conventional anti-cancer drugs.47,48
The 2,6-dichloro-styryl derivative IIIM-290 (11d, Fig. 2), a flavopiridol analogue, showed strong inhibition of CDK9 kinase (IC50 = 1.9 nM). It also exhibited inhibition of CDK1, CDK4 and CDK6 with IC50 values of 4.9 nM, 22.5 nM and 45 nM, respectively. IIIM-290 achieved 71% oral bioavailability with in vivo efficacy in pancreatic, colon, and leukemia xenografts at 50 mg kg−1, po.49 IIIM-290 enhanced the survival of animals with P388 and L1210 leukemia.50 The predicted binding mode of IIIM-290 with CDK9 was obtained (PDB: 3BLR). As shown in Fig. 3B, IIIM-290 could well embed in the ATP binding pocket of CDK9. The key interactions of IIIM-290 were hydrogen bonds and π–π interactions. The chromenone moiety formed an H-bond with the CYS106 residue. The protonated NH of the piperidine ring formed H-bonding interaction with the carbonyl oxygen of ALA153 and the oxygen formed an H-bond with the NH of ASP167. In addition, the ring connected with piperidine and the protonated NH of piperidine formed two important π–π interactions with PHE 103 and PHE 30, respectively.
In 2019, wogonin derivative compound 51 (Fig. 2) showed potent activity toward CDK9 (IC50 = 19.9 nM) and exhibited good selectivity toward other CDKs. Compound 51 displayed strong antiproliferative activity against leukemia cell lines and solid tumor cell lines. 51 inhibited the cell growth via inducing caspase-dependent apoptosis. Compound 51 showed significant antitumor efficacy without obvious toxic effects in vivo.51 As shown in Fig. 3C, compound 51 bound tightly to CDK9. The carbonyl group of 51 formed an important H-bonding interaction with the CYS106 residue. In addition, the pyrazole moiety of compound 51 formed another important H-bond with ALA153. Two H–π interactions with PHE103 and LEU156 were also observed. The benzomethylpiperazine moiety of 51 reached the solvent area.
Compound 13c (Fig. 2), a flavonoid derivative, showed significant nanomolar inhibition against CDK9 and GSK3β protein kinases with IC50 values of 64 nM and 59 nM, respectively. 13c exhibited high cytotoxic potency in tumor cell lines, such as ovarian cancer, gastric adenocarcinoma, breast cancer, prostate cancer, pancreatic cancer, colon carcinoma, and leukemia cell lines.52 As depicted in Fig. 3D, compound 13c formed key bidentate H-bonding interactions with the CYS106 residue of CDK9.
LZT-106 (Fig. 2) was a CDK9 kinase inhibitor with IC50 = 30 nM that could effectively downregulated the Mcl-1 protein via dual targeting of CDK9 and GSK-3β signaling.53 LZT-106 significantly reduced the tumor volume in a xenograft mouse model of colorectal cancer in in vivo experiments. The predicted binding mode study (Fig. 3E) was performed to explore the interactions of LZT-106 into the active site of CDK9 (PDB: 3BLR). LZT-106 is almost entirely buried in the CDK9 pocket. The carbonyl group of LZT-106 formed a hydrogen bond with the cysteine of CYS106 in the hinge region. The methylpiperazine group of LZT-106 formed hydrogen bonds with the residues ASP167 and ASN154 into the ATP binding site of CDK9. The chromenone moiety occupied the hydrophobic pocket at the back of the ATP binding site.
CDK9 inhibitors with aminopyrimidine cores
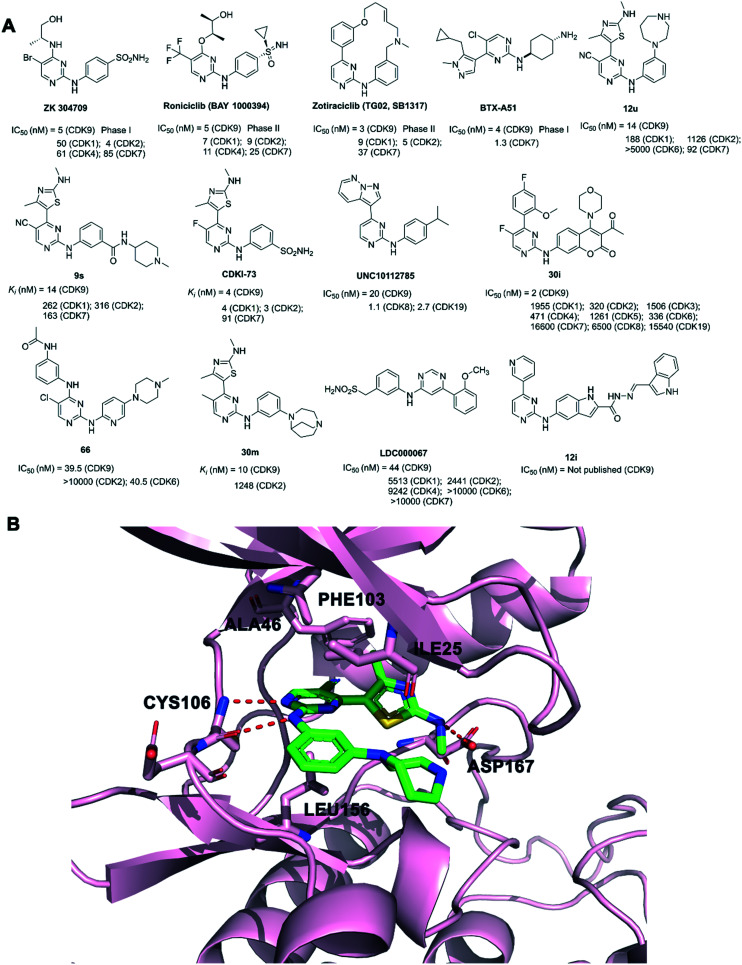
ZK 304709 (Fig. 4A) was an inhibitor of CDKs 1, 2, 4, 7, and 9, vascular endothelial growth factor receptor (VEGFR)1-3, platelet-derived growth factor receptor (PDGFR)-, and FMS-like tyrosine kinase (FLT)-3. ZK 304709 failed in phase I studies for patients with advanced solid tumors due to dose-limited absorption and high inter-patient variability.21
Fig. 4. (A) The development of CDK9 inhibitors with aminopyrimidine cores. (B) The crystal structure of 12u (green) bound to CDK9/cyclin T1 (pink, PDB: 4BCG).
Roniciclib (BAY 1000394, Fig. 4A) was derived from further lead optimization of ZK 304709.54 Roniciclib was a potent pan-CDK inhibitor and inhibited the activity of cell-cycle CDK1-4, and of transcriptional CDKs CDK7 and CDK9 with IC50 values in the range between 5 and 25 nM.55 It potently inhibited the growth of various human tumor xenografts. Furthermore, roniciclib showed more additive efficacy when combined with cisplatin and etoposide. Unfortunately, phase II clinical trials of roniciclib in extensive-disease small cell lung cancer, non-small cell lung cancer (NSCLC) and advanced breast cancer, were terminated due to the unfavorable risk–benefit profile in patients.56
Zotiraciclib (TG02, SB1317), a pyrimidine-based macrocycle compound, was an oral multi-kinase inhibitor of CDKs, JAK2 and FLT3 with potent anti-tumor properties (Fig. 4A).57 CDK9 was the most sensitive zotiraciclib target.58,59 Zotiraciclib was synergized with temozolomide representing a promising therapeutic strategy in glioblastoma.60 Zotiraciclib was also in combination with carfilzomib in patients with multiple myeloma.61 The combination of zotiraciclib and ibrutinib demonstrated moderate synergy.62 Zotiraciclib could induce cell-cycle arrest and lead to rapid commitment to apoptosis.63,64 It induced an effective blockade of both CDK and STAT signaling. The phase II study evaluating zotiraciclib plus temozolomide in recurrent anaplastic astrocytoma and glioblastoma (NCT02942264) and phase I studies of zotiraciclib in chronic lymphocytic leukemia (CLL) or small lymphocytic lymphoma (NCT01699152) and advanced hematological malignancies (NCT01204164) have been completed. Phase I studies of zotiraciclib in the treatment of recurrent/progressive high-grade glioma patients (NCT03904628) and in elderly newly diagnosed or adult relapsed patients with anaplastic astrocytoma or glioblastoma (NCT03224104) are in progress.
BTX-A51 (Fig. 4A) was a novel, oral, direct inhibitor of casein kinase 1α (CK1α), CDK7, and CDK9.65 BTX-A51 could increase p53 protein levels and downregulate negative regulators, Mdm2, enabling selective apoptosis of leukemia cells.66 Phase I studies of BTX-A51 in people with advanced solid tumor or non-Hodgkin lymphoma (NCT04872166) and relapsed or refractory acute myeloid leukemia or high-risk myelodysplastic syndrome (NCT04243785) are in progress.
Compound 12u (Fig. 4A), a 4-thiazol-2-anilinopyrimidine derivative, inhibited CDK9 with IC50 = 14 nM and showed over 80-fold selectivity for CDK9 versus CDK2.6712u exhibited potent anticancer activity in primary chronic lymphocytic leukemia cells and showed little toxicity in healthy normal cells. As shown in Fig. 4B, the aminopyrimidine moiety of 12u hydrogen-bonded with the residue CYS106 of the hinge regions in the crystal structure of CDK9 (PDB: 4BCG). In addition, the NH of the thiazole group formed a hydrogen bond with ASP167. At the back of the ATP binding site, the C-carbonitrile group in the hydrophobic region formed a favorable pair–π interaction with the gatekeeper residue PHE103. The pyrimidine ring of 12u was sandwiched between the hydrophobic side chains of ALA46 and LEU156 with van der Waals contacts.
Compound 9s (Fig. 4A), as a series of 2,4,5-trisubstituted pyrimidine derivative, inhibited CDK9 potently with Ki value as 14 nM.689s showed more than 20-fold selectivity for CDK9 over CDK1 and CDK2, and 10-fold selectivity over CDK7. 9s inhibited cellular CDK9 activity and induced cancer cell apoptosis.
CDKI-73 (Fig. 4A), a heterocyclic 3-(5-fluoro-4-(4-methyl-2-(methylamino)thiazol-5-yl)pyrimidin-2-ylamino)benzenesulfonamide, was a potent CDK9 inhibitor with IC50 = 4 nM.69,70 However, it also inhibited CDK1, CDK2 and CDK7 in the low nM range. CDKI-73 exhibited a favorable pharmacokinetic profile with an oral bioavailability of F = 56%. CDKI-73 significantly inhibited tumour growth, suppressed cellular CDK9 kinase activity and down-regulated the RNAPII phosphorylation in ovarian cancer, colorectal cancer and acute myeloid leukemia.20,71,72 CDKI-73 was also synergetic lethal with PARP inhibitor olaparib in BRCA1 wide-type ovarian cancer.73
UNC10112785 (Fig. 4A) potently inhibited the kinases of CDK8, CDK19, and CDK9, with IC50 values of 1.05, 2.67, and 19.9 nM, respectively, and about 10-fold selectivity over other kinases analyzed. Inhibition of CDK9 by UNC10112785 drove MYC oncoprotein loss in pancreatic cancer.74
Coumarin derivative 30i (Fig. 4A) was discovered as a CDK9 inhibitor with an IC50 value of 2 nM with high selectivity (160- to 8300-fold) over CDK1-8 and CDK 19. Compound 30i showed potent cellular antiproliferative activity in a panel of tumour cell lines, including leukaemia, pancreatic cancer, gastric cancer, melanoma, liver cancer, breast cancer, colon cancer and non-small-cell lung cancer. 30i displayed acceptable drug-like properties. In vivo, 30i significantly induced tumour growth inhibition in an acute myeloid leukaemia (AML) xenograft mouse model.75
Compound 66 (Fig. 4A) was an active dual CDK6 and CDK9 inhibitor (IC50 = 40.5 and 39.5 nM, respectively) and showed good selectivity over CDK2 (IC50 > 10 μM). Compound 66 inhibited cell proliferation by blocking cell cycle progression and inducing cellular apoptosis. 66 significantly inhibited tumor growth in a breast cancer xenograft mouse model.76
2,4,5-Trisubstituted pyrimidine 30m (Fig. 4A) was a potent and selective CDK9 inhibitor with an apparent inhibition constant (Ki) value of 10 nM, which showed 100-fold selectivity for CDK9 over CDK1 and CDK2. 30m was a potent anti-proliferative agent in both the ovarian cancer model A2780 and patient-derived CLL cells.77
LDC000067 (abbreviated to LDC067; WO 2008/129080) with a 2,4-aminopyrimidine scaffold inhibited CDK9 with an IC50 of 44 ± 10 nM (Fig. 4A).78 Its selectivity for CDK9 over other CDKs was in the range of 55-fold (vs. CDK2) to over 230-fold (vs. CDK6 and CDK7). Treatment with LDC000067 enhanced pausing of RNAPII on genes and induced apoptosis in cancer cells.
A novel potential CDK9 inhibitor, 12i (Fig. 4A), with a 5-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)-1H-indole scaffold showed good cytotoxicity against liver cancer, melanoma, breast cancer, and lung cancer cell lines.79
CDK9 inhibitors with pyridine (or triazine) cores
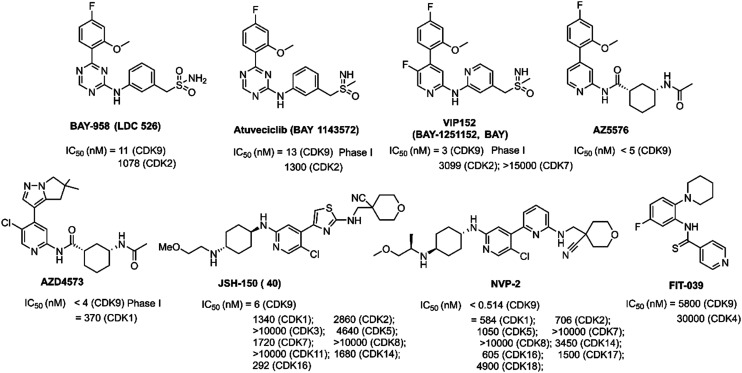
Triazine BAY-958 (LDC 526, Fig. 5) was a potent PTEFb/CDK9 inhibitor (IC50 = 11 nM). It displayed kinase selectivity, even within the CDK family. BAY-958 exhibited good antiproliferative activity in vitro. Daily oral administration of BAY-958 hydrochloride resulted in a marked inhibition of tumor growth. Nevertheless, BAY-958 had unfavorable physicochemical with a rather low aqueous solubility and DMPK properties. Lead optimization efforts of BAY-958 finally led to the identification of compound BAY-1143572.80
Fig. 5. The development of CDK9 inhibitors with pyridine (or triazine) cores.
Atuveciclib (BAY-1143572, Fig. 5), a sulfoximine compound, was a selective CDK9/PTEFb inhibitor currently being investigated in phase I clinical trials with advanced cancer and leukemia (NCT02345382, NCT01938638).81 It showed at least 50-fold selectivity against other CDKs in enzymatic assays.82,83 Atuveciclib possessed significant antitumor activity against AML, NK-cell leukemia/lymphoma.80,84 Atuveciclib also had an additive effect with the GSK-3β inhibitor, 9-ING-41 or p53-MDM2 interaction inhibitor, nutlin-3a, or the BCL2 inhibitor, venetoclax.85,86 Mechanism studies demonstrated that atuveciclib downregulated MDM4 and enhanced p53 activity induced by nutlin-3a in killing A375 melanoma cells.83 Atuveciclib was converted into PROTACs with enhanced the selectivity and activity.87
Extensive lead optimization efforts of atuveciclib led to identification of N-(pyridin-2-yl)-pyridin-2-amine VIP152 (previously BAY-1251152, BAY, Fig. 5). VIP152 exhibited potent CDK9 inhibitory activity (IC50 = 3 nM) and high selectivity against the structurally related kinases CDK2 (ratio of IC50 values: 1033) and CDK7 (ratio of IC50 values > 5000).88 VIP152 has progressed into phase I clinical trials for advanced cancers (NCT04978779, NCT02635672, NCT02745743).89 Compared with atuveciclib, VIP152 showed significantly increased biochemical CDK9 inhibition and cellular potency, increased selectivity against CDK2 as well as obvious efficacy upon i.v. application.90 The results of a phase I study showed that VIP152 displayed good tolerability and on-target activity, which indicated that VIP152 might be a good candidate for future combination therapies.91
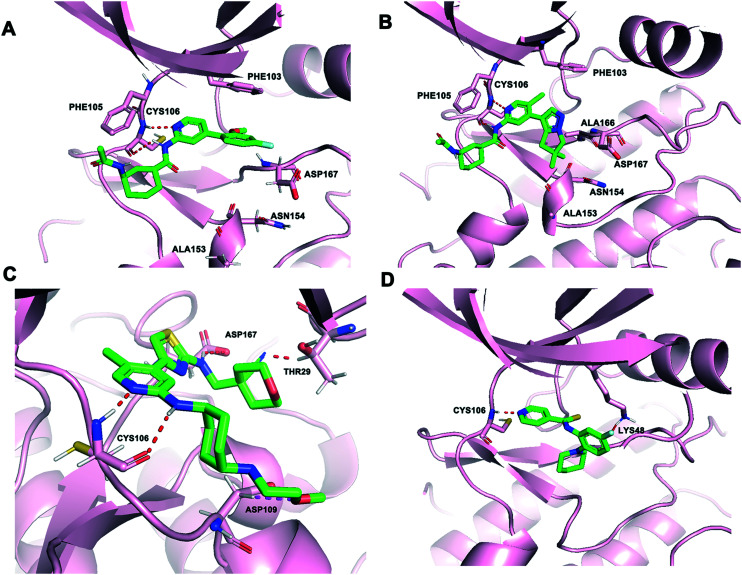
AZ5576 (Fig. 5) was a potent, selective, and orally bioavailable CDK9 inhibitor with an IC50 < 5 nM. AZ5576 demonstrated a selective effect on RNAPII phosphorylation at Ser2 in cells with an IC50 of 96 nM.92 AZ5576 exhibited potent efficacy in MYC-expressing diffuse large B-cell lymphoma (DLBCL). Treatment with AZ5576 inhibited growth of DLBCL cell lines in vitro and in vivo.93 The predicted binding mode of AZ5576 (PDB: 4BCF) is shown in Fig. 6A. CYS106 made two hydrogen-bonds with the pyridyl amide core of AZ5576. The cyclohexyl amide gave access to the solvent channel. The 4-fluoro-2-methoxyphenyl moiety filled the hydrophobic pocket formed near ASP167 of the activation loop. An important π–π interaction was also observed between 4-fluoro-2-methoxyphenyl group and the PHE103 residue. Further structure optimization of AZ5576 resulted in the identification of AZD4573.
Fig. 6. (A) The predicted binding mode of AZ5576 (green) and CDK9 (pink, PDB: 4BCF). (B) The crystal structure of AZD4573 (green) bound to CDK9 (pink, PDB: 6Z45). (C) The predicted binding mode of JSH-150 (green) and CDK9 (pink, PDB: 4BCG). (D) The predicted binding mode of FIT-039 (green) and CDK9 (pink, PDB: 3BLQ). The hydrogen bonds are depicted by red dotted lines.
AZD4573 (Fig. 5) was a potent and highly selective CDK9 inhibitor (IC50 < 4 nM) with suitable predicted human pharmacokinetic properties, resulting in activation of caspase 3/7 and cell apoptosis in a broad range of hematological cancer cell lines.94 AZD4573 enabled the indirect inhibition of cancer-promoting gene MCL-1, providing a therapeutic option for MCL-1 dependent diseases.95–97 AZD4573 was currently in phase I clinical trials for the treatment of hematological malignancies (NCT04630756, NCT03263637). The AZD4573 cocrystal structure bound to CDK9/cyclin T (PDB: 6Z45) is shown in Fig. 6B. Consistent with the predicted binding mode of AZ5576, AZD4573 interacted with the backbone atoms of CYS106 through the pyridine and the amidic NH. One of the gemdimethyl substituents of AZD4573 filled a small pocket formed near ASP167 of the activation loop, in close proximity to the side chains of ASN154 and ALA166.
JSH-150 (compound 40, Fig. 5), starting from the core scaffold of CDK7/9 inhibitor 12u, was a selective CDK9 inhibitor. JSH-150 exhibited an IC50 of 6 nM against CDK9 and achieved around 300–10 000-fold selectivity over other CDK kinase family members.98 It displayed high selectivity over other 468 kinases/mutants. JSH-150 displayed potent antiproliferative effects against a wide range of cancer cell lines including solid tumor and leukemia cells through arresting the cell cycle and inducing the apoptotic cell death. The predicted binding mode analysis of JSH-150-CDK9 complexes (PDB: 4BCG) revealed that aminopyridine of JSH-150 formed two hydrogen bonds with CYS106 (Fig. 6C). The CN group formed a hydrogen bond with THR29. In addition, the oxygen atom formed a hydrogen bond with ASP109. The aminothiazole group was embedded in the hydrophobic pocket, in which the NH formed a hydrogen bond with the oxygen atom of ASP167.
A structurally similar inhibitor to JSH-150, NVP-2 (Fig. 5), developed by Novartis, was a highly selective aminopyrimidine-derived CDK9 inhibitor.99 NVP-2 potently inhibited the kinase activity of CDK9 with an IC50 less than 0.514 nM and exhibited good kinome selectivity against 468 kinases. The only kinases that inhibited greater than 99% were CDK9 and DYRK1B with an IC50 of 350 nM, 700-fold higher than that observed for CDK9. NVP-2 exhibited anti-proliferative effects on leukemia cancer cell line, MOLT4.
CDK9 inhibitor FIT-039 (Fig. 5), N-[5-fluoro-2-(1-piperidinyl)phenyl]isonicotinthioamide, suppressed CKD9/cyclin T1 kinase activity with an IC50 value of 5.8 μM but did not have a marked inhibitory effect on other CDKs, such as CDK4, CDK2, CDK5, CDK6, or CDK7. FIT-039 suppressed viral oncogenes E6 and E7 and had a therapeutic effect on HPV-Induced neoplasia.100 The predicted binding mode of FIT-039 docked into the ATP binding site of the CDK9 crystal structure is shown in Fig. 6D. The fluoride group and nitrogen atom in pyridine of FIT-039 interacted with the LYS48 and CYS106 residues of CDK9 by forming hydrogen bonds, respectively. The pyridine group occupied a hydrophobic pocket formed by PHE103, ASP104, VAL79 and ALA46.
CDK9 inhibitors with purine cores
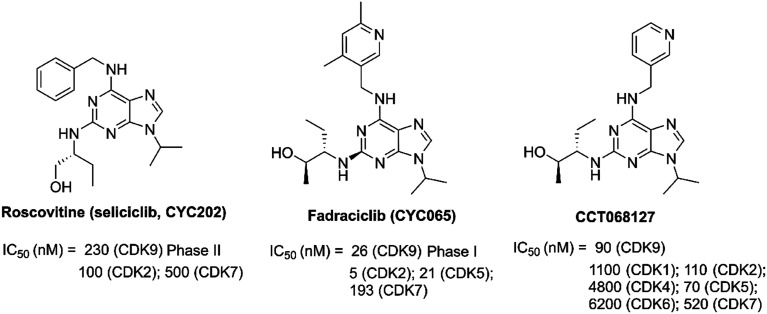
R-Roscovitine (seliciclib, CYC202, Fig. 7) was a synthetic CDK inhibitor with most potent activity against the CDK2, CDK7 and CDK9.101–103 It was the first CDK inhibitor to enter clinical trials as an anticancer agent.104–106R-Roscovitine has cytotoxic activity against a range of human cancers, such as lymphomas, multiple myeloma, lung cancer, colon cancer and so on.107–109 As reported, R-roscovitine caused cell cycle arrest and induced cell death through the inhibition of transcription and down-regulation of antiapoptotic proteins.101,107 A phase II clinical trial of R-roscovitine (NCT03774446) is currently being conducted to evaluate its safety. A phase I study (NCT00999401) of sequential administration of oral sapacitabine and oral seliciclib in patients with advanced solid tumors has been completed.
Fig. 7. The development of CDK9 inhibitors with purine cores.
Fadraciclib (CYC065, Fig. 7), a 2,6,9-trisubstituted purine analog, was an orally ATP competitive inhibitor of CDK9 and CDK2 over other CDKs and non-CDK enzymes against leukaemia, uterine serous carcinoma, breast cancer, lung cancer, and neuroblastoma.110–114 Treatment with fadraciclib alone or in combination with other agents induced a marked G1 arrest and gross apoptosis of tumor cells.115 The efficacy and mechanism of action of fadraciclib was identified and fadraciclib inhibited CDK9-dependent RNAPII phosphorylation, resulting in rapid induction of apoptosis. Fadraciclib could act synergistically with selected DNA damaging or targeted agents, such as trastuzumab and taselisib.116 Fadraciclib as a single agent trial is being examined in phase I clinical trials in advanced cancers (NCT02552953). Fadraciclib is a combined regimen with venetoclax for relapsed/refractory CLL (NCT04017546, NCT03739554).
CCT068127 (Fig. 7), a trisubstituted purine compound, was a CDK2 and CDK9 inhibitor derived from parental seliciclib.117 It has increased potency toward CDK2/cyclin E (22-fold) and CDK9/cyclin T (11-fold) kinase activities and selectivity toward CDK2 and 9 versus CDK4 and CDK7, compared to seliciclib. CT068127 treatment resulted in reduced phosphorylation of RNAPII, and cell cycle arrest and apoptosis of cancer cells. The combination of CCT068127 and ABT263, a BCL2 family inhibitor, showed potent inhibition activity for the treatment of human cancer.
CDK9 inhibitors with pyrazolo[1,5-a]pyrimidine cores
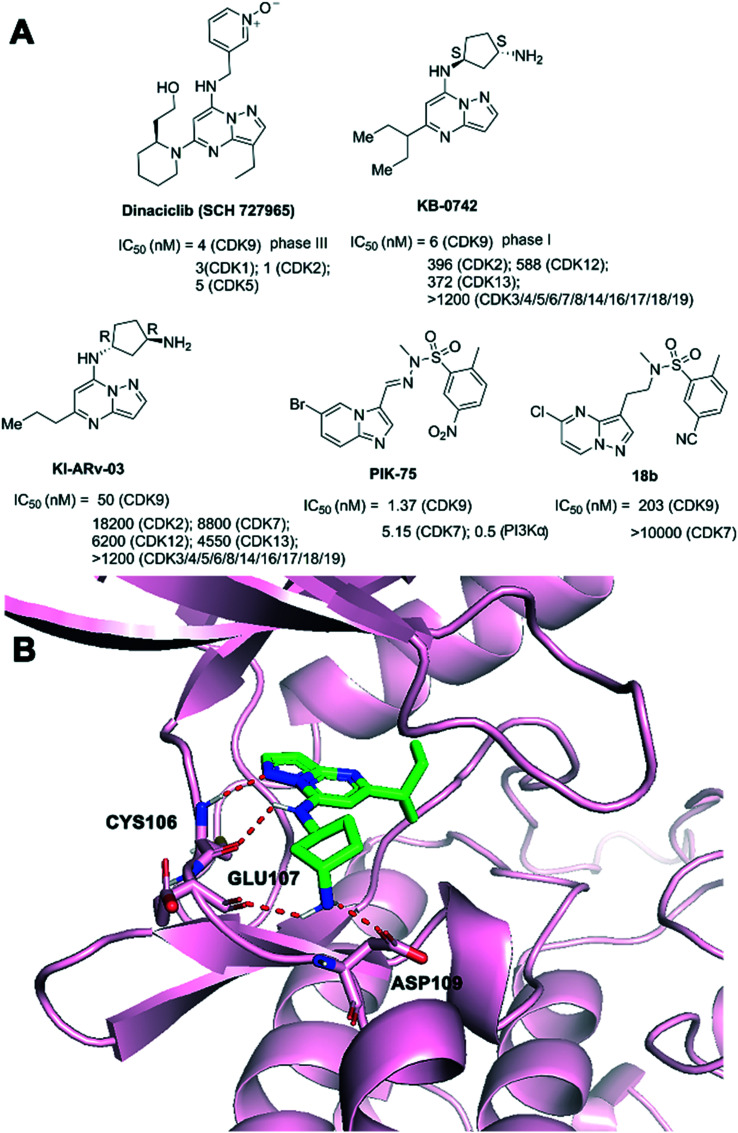
Dinaciclib (SCH 727965, Fig. 8A) inhibited CDK2, CDK5, CDK1, and CDK9 activity in vitro with IC50 values of 1, 1, 3, and 4 nM, that demonstrated single agent activity in solid tumor and myeloma.118–120 In addition, dinaciclib provided a novel strategy to overcome IFNG-triggered acquired resistance in pancreatic tumour immunity.121 Dinaciclib and anti-PD1 combination therapy promoted an antitumor immune response in patients with hematological malignancies or advanced breast cancer.122 Dinaciclib could result in cell-cycle arrest in many tumor cell lines. Apoptotic and antitumor effects of dinaciclib were demonstrated.123,124 Dinaciclib has been evaluated in clinical trials. A phase III study comparing dinaciclib versus of atumumab in patients with refractory CLL has been completed (NCT01580228). Phase II studies of dinaciclib monotherapy in patients with advanced breast and lung cancers (NCT00732810), relapsed or refractory multiple myeloma (NCT01096342) have been completed.
Fig. 8. (A) The development of CDK9 inhibitors with pyrazolo[1,5-a]pyrimidine cores. (B) The predicted binding mode of KB-0742 (green) and CDK9 (pink, PDB: 3MY1). The hydrogen bonds are depicted by red dotted lines.
KB-0742 (Fig. 8A) was an oral, potent, and selective CDK9 inhibitor (CDK9 inhibition IC50 = 6 nM at 10 mM ATP) with >50-fold selectivity over all CDKs profiled and >100-fold selectivity against cell-cycle CDKs (CDK1-6).125 KB-0742 was derived from KI-ARv-03, a selective inhibitor of CDK9 (IC50 = 0.15 mM at 45 mM ATP) over all other tested CDKs with a minimum of 130-fold selectivity. KB-0742 showed potent anti-tumor activity in castration-resistant prostate cancers (CRPCs) in vitro and in vivo. A clinical trial for KB-0742 was launched in participants with relapsed or refractory solid tumors or non-Hodgkin lymphoma (NCT04718675).21 As shown in Fig. 8B, the pyrazolopyrimidine core of KB-0742 made two hydrogen bonds to CYS106 (NH and CO) of the CDK9 hinge in the predicted binding mode. The terminal primary amine of KB-0742 interacted with GLU107 and ASP109 residues of CDK9. The 3-pentane extension of the propane alkyl chain formed van der Waals interactions with the G-loop.
Phillipson et al. reported the discovery of a new series of compounds based on a pyrazolo[1,5-a]pyrimidine nucleus. Compound 18b (Fig. 8A) was discovered as a novel CDK9 inhibitor with improved selectivity over PI3Ka based on PIK-75.126 PIK-75, a PI3Kα inhibitor (IC50 = 0.5 nM), showed significant activity against a number of kinases, such as CDK9 (IC50 = 1.37 nM) and CDK7 (IC50 = 5.15 nM). 18b inhibited CDK9 with an IC50 value of 203 nM and exhibited greater selectivity over PI3Kα.
CDK9 inhibitors with pyrazole cores
AT7519 (Fig. 9A) was a potent inhibitor of several CDK family members, which showed potent antiproliferative activity in a panel of human tumor cell lines.127,128 Global phosphoproteomics revealed AT7519 with CDK suppression as a vulnerability to KRAS addiction in pancreatic cancer. The mechanism of action showed that AT7519 caused cell cycle arrest followed by apoptosis via RNAPII inhibition in human tumor cells.129,130 Phase II studies of AT7519 in patients with relapsed and/or refractory CLL (NCT01627054) and with relapsed MCL (NCT01652144) have been completed. Effects of AT7519 alone and AT7519 plus bortezomib in a phase II clinical study have also been completed with multiple myeloma (NCT01183949). AT7519 in phase I clinical development treated patients with advanced or metastatic solid tumors or refractory non-Hodgkin's lymphoma (NCT00390117).
Fig. 9. (A) The development of CDK9 inhibitors with pyrazole cores. (B) The crystal structure of CAN508 (green) bound to CDK9 (pink, PDB: 3TN8). The hydrogen bonds are depicted by red dotted lines.
CAN508 (Fig. 9A) was an arylazopyrazole CDK9 inhibitor with an IC50 of 350 nM. It exhibited a 38-fold selectivity for CDK9/cyclin T over other CDKs.131 CAN508 has also been shown to inhibit the growth of various cancer cell lines and induced cell apoptosis. CAN508 treatment was identified to inhibit angiogenesis through a CDK9-dependent mechanism.132 The crystal structure (Fig. 9B) of CAN508 with CDK9/cyclin T was determined (PDB: 3TN8). CAN508 bound to the ATP binding site located between the N- and C-terminal lobes of CDK9. The diaminopyrazole ring of CAN508 made hydrogen bonds to the main chain nitrogen of CDK9 CYS106 and the chain oxygen of ASP104 in the hinge region. Additional hydrogen interaction between CAN508 and the carbonyl group of ILE25 of CDK9 was identified. The OH group of the CAN508 phenolic moiety engaged in a network of hydrogen bonds with residues GLU66, LYS48, and PHE168 of CDK9. CAN508 was sandwiched between the ALA46 in the N-terminal and LEU156 in the C-terminal lobe.
RGB-286638 (Fig. 9A) was an indenopyrazole-derived multi-targeted CDK inhibitor, which inhibited the kinase activity of CDK9/cyclin T1, CDK1/cyclin B1, CDK2/cyclin E, CDK4/cyclin D1, CDK3/cyclin E, and CDK5/p35 with IC50 values of 1, 2, 3, 4, and 5 nM, respectively.133 In addition, RGB-286638 also inhibited other tyrosine and serine/threonine non-CDK enzymes. CDK9 was the primary target of RGB-286638 that mediated its caspase-dependent apoptosis activity in multiple myeloma. A phase I study of RGB-286638 (NCT01168882) was performed to evaluate the pharmacokinetic and pharmacodynamic profiles in patients with solid tumors.134
CDK9 inhibitors with aminothiazole cores
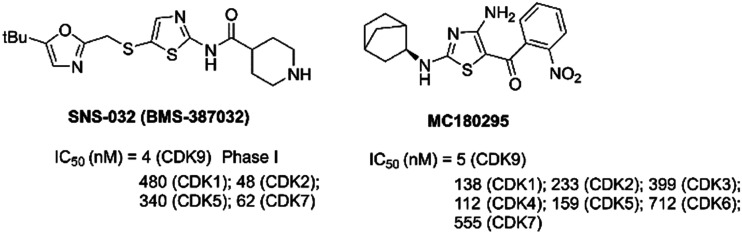
SNS-032 (BMS-387032, Fig. 10), an aminothiazole, was a highly efficacious and selective antitumor agent of CDKs 2, 7 and 9 (IC50 = 48 nM, 62 nM and 4 nM, respectively).135 SNS-032 prevented tumor cell-induced angiogenesis by inhibiting the vascular endothelial growth factor. SNS-032 showed single agent activity in malignant hematologic cells and solid tumors.136,137 Phase I clinical trials of SNS-032 in patients with advanced B-lymphoid malignancies (NCT00446342) and solid tumors (NCT00292864) have been completed.
Fig. 10. The development of CDK9 inhibitors with aminothiazole cores.
MC180295 (Fig. 10) was a novel potent and selective CDK9 inhibitor. MC180295 against a panel of 250 kinases at 1 μM was highly selective against CDKs within the human kinome. MC180295 had broad anti-cancer activity in vitro and was effective in in vivo cancer models.138
CDK9 inhibitors with azaindole cores or nitrogen-containing heterocycles
A-1592668 (Fig. 11A) was a potent small-molecule inhibitor of CDK9 (IC50 = 2.6 nM), with CDK selectivity profiles.139 A-1592668 was afforded by modification primarily to the right-hand portion of the pharmacophore from A-1467729 (Fig. 11A). A-1467729 was a potent CDK9 inhibitor (IC50 = 1.2 nM) with >1000× selectivity over CDK1, CDK2, CDK7 and CDK8. A-1592668 initiated apoptosis and tumor cell death in MCL-1-dependent hematologic tumors. A-1592668 demonstrated robust pharmacodynamic marker movement. It could be in combination with venetoclax for the treatment of patients with hematologic malignancies.
Fig. 11. (A) The development of CDK9 inhibitors with azaindole cores or nitrogen-containing heterocycles. (B) The crystal structure of compound 6 (green) with CDK9 (pink, PDB: 7NWK). The hydrogen bonds are depicted by red dotted lines.
Compound 4ab (Fig. 11A) inhibited CDK2 and CDK9 activity with nanomolar potency (IC50 = 5.5 and 24 nM, respectively). 4ab showed potent antiproliferative activities against a panel of tested tumor cell lines. In addition, 4ab demonstrated good aqueous solubility and an acceptable in vivo PK profile. 4ab showed good in vivo efficacy in a mouse mammary triple-negative breast cancer model.140
Azaindoles 38 and 39 (Fig. 11A) were obtained from the optimization of a hit compound 2 (CDK9 IC50 = 34 nM) and a co-crystal structure of the azabenzimidazole-based lead 6 (CDK9 IC50 = 47 nM) bound to CDK9. 38 and 39 were potent and highly selective CDK9 inhibitors, with an IC50 value of 8 and 6 nM, respectively. 38 and 39 showed no significant inhibition at 1 μM apart from CDK9 (99% inhibition), which exhibited high selectivity vs. CDK1, CDK2, CDK6, CDK7, and CDK12. 38 and 39 exhibited good physicochemical and pharmacokinetic properties.141 The crystal structure of 6 with CDK9-cyclin T1 is shown in Fig. 11B. The azabenzimidazole scaffold of compound 6 made hydrogen bonds to the CYS106 residue of the CDK9 hinge. The azabenzimidazole scaffold also formed a CH–O interaction to the backbone carbonyl oxygen of ASP104. The acetamido group oriented into the solvent and the 3H-imidazo[4,5-b]pyridine moiety made contact with the hydrophobic pocket formed by PHE103, PHE105, VAL79 and LEU156.
Ghanem et al. designed a series of novel imidazo[4,5-b]pyridine based compounds as potent anticancer agents with CDK9 inhibitory activity. The compounds had promising anticancer activity against either breast or colon cancer cell lines. Compound VI (Fig. 11A) was the most potent one exhibiting a submicromolar level against CDK9 enzyme assay (IC50 = 0.50 μM).142
PHA-767491, (Fig. 11A, 1,5,6,7-tetrahydro-2-(4-pyridinyl)-4H-pyrrolo[3,2-c]pyridin-4-one hydrochloride), was a dual inhibitor of the Cdc7/CDK9 inhibitor with IC50 values of 10 nM and 34 nM, respectively. PHA-767491 resulted in apoptotic cell death in multiple cancer cell types and tumor growth inhibition in cancer models.143 PHA-767491 was synergized with multiple epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs), such as lapatinib, erlotinib and gefitinib, to overcome resistance to EG.
7t (Fig. 11A), an indirubin-30-monoxime derivative, was a potent CDK inhibitor that showed significant inhibition against both CDK2/cyclin E1 and CDK9/cyclin T1. 7t exhibited IC50 values at the submicromolar level.144
CDK9 inhibitors with pyrrolo[2,3-d]pyrimidine cores
Our group has developed some CDK9 inhibitors based on pyrrolo[2,3-d]pyrimidine scaffolds. Compound 6e (Fig. 12A) showed CDK4/9 and HDAC1 inhibitory activity of IC50 = 8.8, 12, and 2.2 nM, respectively. 6e showed good selectivity in a kinase profiling assay against 375 kinases. In addition, 6e showed significant antitumor efficacy in vitro and in vivo.145 As shown in the predicted binding mode (Fig. 12B), two hydrogen bonds were formed between the aminopyrimidine of 6e and the backbone residue of CYS106 in CDK9. Compound 6e gained a hydrogen bonding interaction between the hydroxamic acid group and the THR29 residue of CDK9. The dimethylcarbamoyl moiety filled the hydrophobic pocket, which formed a hydrogen bond with ASP167 of the activation loop. Compound 21e (Fig. 12A) potently inhibited CDK9 with an IC50 value of 11 nM and suppressed the stemness properties of NSCLC effectively. 21e displayed good selectivity over the CDK family kinases and kinase profiling assay against 381 kinases. 21e efficiently inhibited the CDK9 signaling pathway and stemness both in vitro and in vivo.146 As shown in the predicted binding mode (Fig. 12C), the 2-aminopyrimidine of 21e hydrogen bonded with the CYS106 residue of the kinase hinge regions in CDK9. An additional hydrogen bond existed between the isothiocyanate group of compound 21e and the HIS108 residue. The pyrrolo-[2,3-d]-pyrimidine group exploited the hydrophobic region close to the gatekeeper residue PHE103 to form a π–π interaction in CDK9. The dimethylcarbamoyl moiety filled the hydrophobic pocket, forming a van der Waals contact with the ASP167 residue. Compound 18i (Fig. 12A) was discovered with potent activity against CDK4 and CDK9 at the nanomolar level with IC50 = 2.5 nM and 7 nM, respectively. 18i demonstrated only modest activity against 3 out of the 394 protein kinases. Oral administration of 18i showed significant therapeutic effects in xenograft mouse models of breast cancer.147
Fig. 12. (A) The development of CDK9 inhibitors with pyrrolo[2,3-d]pyrimidin cores. (B) The predicted binding mode of compound 6e (green) and CDK9 (pink, PDB: 4BCF). (C) The predicted binding mode of compound 21e (green) and CDK9 (pink, PDB: 4BCF). The hydrogen bonds are depicted by red dotted lines.
Selective CDK9 small molecule degraders (PROTACs)
The PROTAC strategy has been used to design selective CDK9 degraders. PROTACs are formed with one fragment that binds the target protein, another that binds a ubiquitin (E3) ligase, and a linker connecting these two fragments. Recent studies have reported the identification of selective CDK9 degraders.
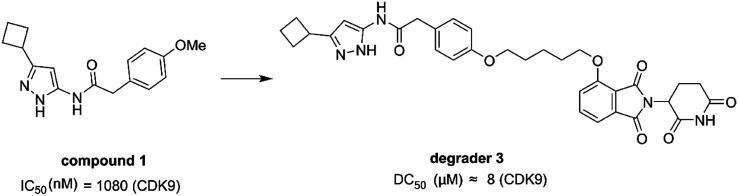
In 2017, Robb et al. reported the first example of a PROTAC that selectively degraded CDK9. The degrader 3 (Fig. 13) was composed of compound 1 and thalidomide binding to the E3 ubiquitin ligase Cereblon (CRBN), and an alkane chain. Compound 1 showed selectivity for CDK5 and CDK9 over CDK2 whereas degrader 3 was selective for CDK9. Degrader 3 degraded CDK9 with a DC50 about 8 μM (the value estimated from the reported data). In addition, 3 degraded CDK9 resulting in reduced phosphorylation of Ser2 on RPB1, a CDK9 substrate and reduced the levels of Mcl-1, a prosurvival protein, regulated by CDK9 activity.148
Fig. 13. Chemical structure of designing PROTAC degrader 3 based on compound 1.
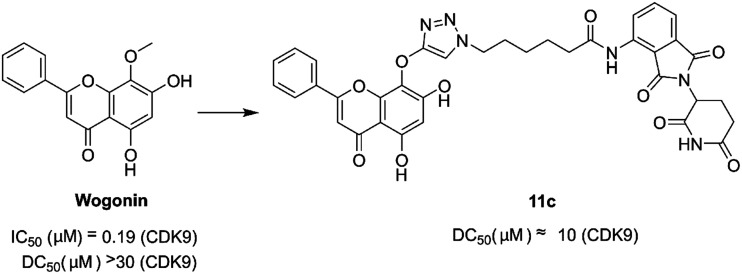
In 2018, Bian et al. disclosed the identification of a series of PROTACs targeting CDK9 based on the scaffold of wogonin. Compound 11c (Fig. 14) selectively promoted the proteasome-dependent and CRBN-dependent degradation of CDK9. The wogonin-based PROTAC 11c degraded CDK9 with moderate anti-proliferative effects in several cancer cell lines.149
Fig. 14. Chemical structure of designing PROTAC 11c based on wogonin.
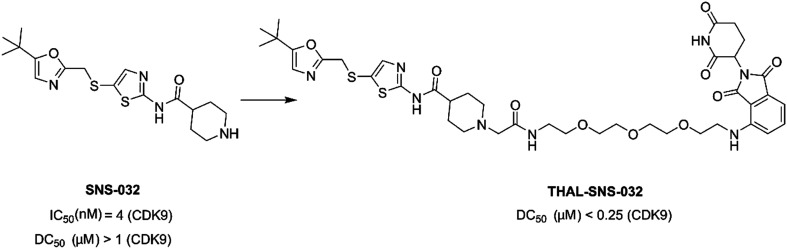
THAL-SNS-032 (Fig. 15) was a selective CDK9 degrader consisting of SNS-032, a thalidomide derivative which bound CRBN, and a 3-polyethylene glycol (PEG) linker. SNS-032 was a multi target inhibitor, while THAL-SNS-032 induced rapid and specific degradation of CDK9 with near complete degradation at <250 nM concentration.99
Fig. 15. Chemical structure of designing PROTAC THAL-SNS-032 based on SNS-032.
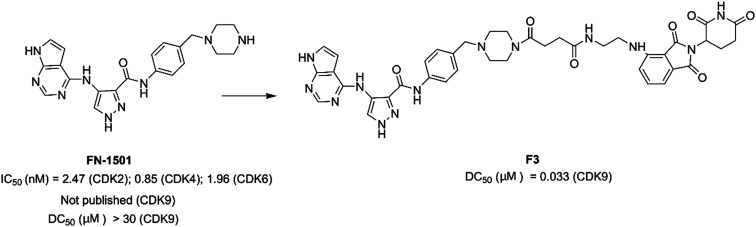
Zhou et al. designed and optimized a dual-degrader, compound F3 (Fig. 16), targeting multiple CDKs by using the PROTAC approach. Compound F3 potently induced degradation of both CDK2 (DC50 = 62 nM) and CDK9 (DC50 = 33 nM). In prostate cancer cells, compound F3 inhibited cell proliferation by arresting the cell cycle in S and G2/M phases.150
Fig. 16. Chemical structure of designing PROTAC F3 based on compound FN-1501.
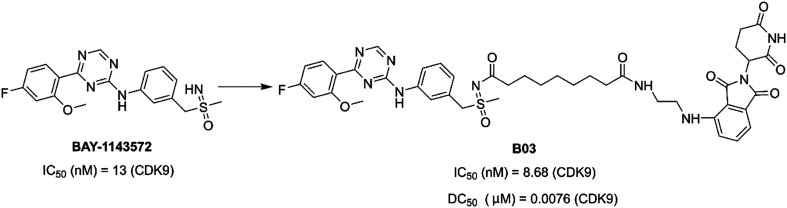
In 2021, Qiu et al. reported PROTAC B03 (Fig. 17) based on a selective CDK9 inhibitor BAY-1143572 and pomalidomide. B03 could inhibit cell growth more effectively than BAY-1143572 alone. Moreover, B03 induced the degradation of CDK9 in AML cells at a low nanomolar concentration, with little inhibition of other kinases.151
Fig. 17. Chemical structure of designing PROTAC B03 based on BAY-1143572.
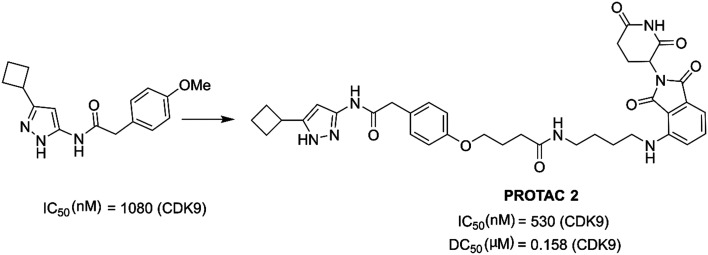
King et al. optimized and improved a previously reported144 aminopyrazole based CDK9 degrader. They explored aminopyrazole based PROTAC 2 (Fig. 18) that selectively degraded CDK9 (DC50 = 158 nM). PROTAC 2 selectively degraded CDK9 over other CDK family members. PROTAC 2 sensitized pancreatic cancer cells.152
Fig. 18. Chemical structure of designing PROTAC 2.
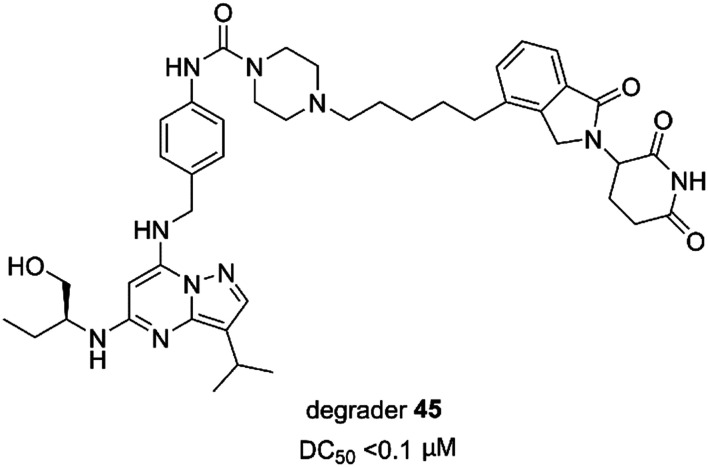
Degrader 45 (Fig. 19) was a heterobifunctional molecule and a selective and efficacious CDK9 degrader. Degrader 45 enabled potent inhibition of TNBC cell growth with an IC50 value of 4 nM. 45 could induce cell apoptosis in vitro and inhibited tumor growth in the TNBC xenograft model in vivo. Mechanistic investigation demonstrated that degrader 45 could effectively downregulate the downstream targets of CDK9 at the transcriptional level.153
Fig. 19. Chemical structure of designing degrader 45.
Conclusions
CDK9 plays an important role in the regulation of transcription. Deregulation of the CDK9 pathway or CDK9 overexpression was widespread in a variety of malignancies. Thus, blocking CDK9 signaling provided an appealing therapeutic approach to curb cancer development. During the past decade, many researchers have focused their efforts on the development of small molecule inhibitors CDK9 for anticancer potential. Several generations of CDK9 inhibitors have been discovered. Pan-CDK inhibitors with multiple CDK inhibition have been known for some time, and some involved in clinical trials. They displayed remarkable anti-tumor efficacy. However, due to their low specificity, pan-CDK inhibitors may come with toxicities, limiting their further applications. Several pan-CDK9 inhibitors, including flavopiridol, roniciclib, etc., have limited clinical application in cancer treatment due to their side effects. Subsequently, selective CDK9 inhibitors with improved selectivity have since emerged. Some CDK9 inhibitors, including atuveciclib, VIP152, AZD4573, JSH-150, NVP-2, KB-0742, AT7519, and so on, have been developed with better selectivity and less side effects. Selective CDK9 inhibitors may be beneficial to patients. Despite this, efforts and progress have been made. Notably, the development of PROTAC strategies in recent years has made it possible to reduce the toxicity and side effects.
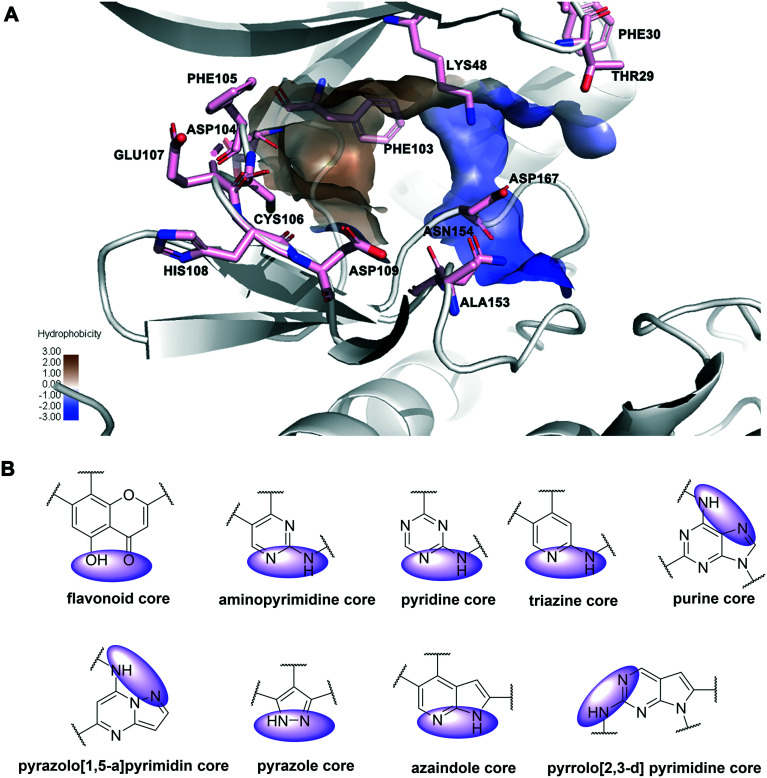
As depicted in Fig. 20A, there are some essential amino acid residues for the kinase inhibitory activity in the ATP-binding pocket of CDK9, such as CYS106, ASP167, ASP104, and THR29. Important interactions between inhibitors and CDK9 included hydrogen bonds, hydrophobic interactions, and the π–π stacking interaction. In particular, the residue CYS106 in the hinge region was important for hydrogen bonds. In addition, the shallow hydrophobic pocket was formed by the HIS108, PHE105, and PHE103 residues of CDK9. Moreover, the gatekeeper residue PHE103 at the back of the ATP binding site may form a favorable π–π interaction. Structural studies of CDK9 inhibitors reported indicated that, according to their core skeleton, CDK9 inhibitors included flavonoid, aminopyrimidine, pyridine (or triazine), purine, pyrazolo[1,5-a]pyrimidine, pyrazole, aminothiazole, azaindole and pyrrolo[2,3-d]pyrimidine cores (Fig. 20B). Nearly all the core skeletons (Fig. 20B) were anchored to CDK9 through hydrogen bonding interactions with the hinge region residues, similar to ATP mimetic inhibitors of other kinases.154
Fig. 20. (A) The main amino acid residues of CDK9 (PDB: 3BLR) that formed interactions with the inhibitors. The hydrophobicity ranges its intensity from 3.00 (maximum hydrophobic zones) to −3.00 (minimum hydrophobic zones). The minimum intensity areas are the blue shaded regions and the maximum intensity areas are denoted with dull brownish color. (B) Atoms of inhibitors participating in hydrogen bonds with the hinge region of CDK9 are shown in purple shadow.
Most of the kinase inhibitors reported could be classified into two main categories based on their mode of actions: ATP-competitive inhibitors and allosteric inhibitors.155 As above, ATP-competitive inhibitors formed one to three hydrogen bonds to the amino acids situated in the hinge region of the target kinase, similar to the binding mode of the adenine residue of the ATP.156 Allosteric inhibitors induced a conformational change to the kinase and modified its activity.157,158 Recently, several allosteric inhibitors for other protein kinases have been developed and reported with advantages of high selectivity and few adverse effects.159,160 To our knowledge, no CDK9 inhibitors targeting allosteric sites have been developed yet. Therefore, allosteric regulation may open a new avenue for CDK9 drug discovery.
From the perspective of the development of inhibitors, the design of drugs targeting CDK9 is a promising strategy for the development of antitumor drugs. Selectivity improvement is an important aspect for the development of CDK9 drugs.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81903099).
References
- Lim S. Kaldis P. Development. 2013;140:3079–3093. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- Peyressatre M. Prevel C. Pellerano M. Morris M. C. Cancers. 2015;7:179–237. doi: 10.3390/cancers7010179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z. P. Zhu Y. L. Ratner E. S. Front. Oncol. 2018;8:303. doi: 10.3389/fonc.2018.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra S. Joshi G. Munshi A. Kumar R. Eur. J. Med. Chem. 2017;142:424–458. doi: 10.1016/j.ejmech.2017.08.071. [DOI] [PubMed] [Google Scholar]
- Malumbres M. Barbacid M. Nat. Rev. Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- Thomas M. C. Chiang C. M. Crit. Rev. Biochem. Mol. Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- Asghar U. Witkiewicz A. K. Turner N. C. Knudsen E. S. Nat. Rev. Drug Discovery. 2015;14:130–146. doi: 10.1038/nrd4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicenas J. Valius M. J. Cancer Res. Clin. Oncol. 2011;137:1409–1418. doi: 10.1007/s00432-011-1039-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Fu L. Wu J. Liu M. Wang G. Liu B. Zhang L. Eur. J. Med. Chem. 2021;229:114056. doi: 10.1016/j.ejmech.2021.114056. [DOI] [PubMed] [Google Scholar]
- Miller D. M. Flaherty K. T. Pigm. Cell Melanoma Res. 2014;27:351–365. doi: 10.1111/pcmr.12211. [DOI] [PubMed] [Google Scholar]
- Cimini A. d'Angelo M. Benedetti E. D'Angelo B. Laurenti G. Antonosante A. Cristiano L. Di Mambro A. Barbarino M. Castelli V. Cinque B. Cifone M. G. Ippoliti R. Pentimalli F. Giordano A. J. Cell. Physiol. 2017;232:312–322. doi: 10.1002/jcp.25421. [DOI] [PubMed] [Google Scholar]
- Cicenas J. Kalyan K. Sorokinas A. Stankunas E. Levy J. Meskinyte I. Stankevicius V. Kaupinis A. Valius M. Ann. Transl. Med. 2015;3:135. doi: 10.3978/j.issn.2305-5839.2015.03.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry D. Guzi T. Shanahan F. Davis N. Prabhavalkar D. Wiswell D. Seghezzi W. Paruch K. Dwyer M. P. Doll R. Nomeir A. Windsor W. Fischmann T. Wang Y. Oft M. Chen T. Kirschmeier P. Lees E. M. Mol. Cancer Ther. 2010;9:2344–2353. doi: 10.1158/1535-7163.MCT-10-0324. [DOI] [PubMed] [Google Scholar]
- Beaver J. A. Amiri-Kordestani L. Charlab R. Chen W. Palmby T. Tilley A. Zirkelbach J. F. Yu J. Liu Q. Zhao L. Crich J. Chen X. H. Hughes M. Bloomquist E. Tang S. Sridhara R. Kluetz P. G. Kim G. Ibrahim A. Pazdur R. Cortazar P. Clin. Cancer Res. 2015;21:4760–4766. doi: 10.1158/1078-0432.CCR-15-1185. [DOI] [PubMed] [Google Scholar]
- Araki K. Miyoshi Y. N. Engl. J. Med. 2017;376:288. doi: 10.1056/NEJMc1615255. [DOI] [PubMed] [Google Scholar]
- Kim E. S. Drugs. 2017;77:2063–2070. doi: 10.1007/s40265-017-0840-z. [DOI] [PubMed] [Google Scholar]
- Mandal R. Becker S. Strebhardt K. Cancers. 2021;13:2181. doi: 10.3390/cancers13092181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassandri M. Fioravanti R. Pomella S. Valente S. Rotili D. Del Baldo G. De Angelis B. Rota R. Mai A. Front. Pharmacol. 2020;11:1230. doi: 10.3389/fphar.2020.01230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahaman M. H. Kumarasiri M. Mekonnen L. B. Yu M. Diab S. Albrecht H. Milne R. W. Wang S. Endocr.-Relat. Cancer. 2016;23:T211–T226. doi: 10.1530/ERC-16-0299. [DOI] [PubMed] [Google Scholar]
- Rahaman M. H. Lam F. Zhong L. Teo T. Adams J. Yu M. Milne R. W. Pepper C. Lokman N. A. Ricciardelli C. Oehler M. K. Wang S. Mol. Oncol. 2019;13:2178–2193. doi: 10.1002/1878-0261.12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan A. Pang Y. Butler M. Merchant M. Kim O. Yu G. Su Y. T. Gilbert M. R. Levens D. Wu J. Cancers. 2021;13:3039. doi: 10.3390/cancers13123039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales F. Giordano A. Cell Cycle. 2016;15:519–527. doi: 10.1080/15384101.2016.1138186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz A. L. Schaum M. Richter J. Kitzig E. F. Engler C. C. Leithauser F. Henne-Bruns D. Knippschild U. Lemke J. Tumor Biol. 2017;39:1010428317694304. doi: 10.1177/1010428317694304. [DOI] [PubMed] [Google Scholar]
- Wang J. Dean D. C. Hornicek F. J. Shi H. Duan Z. FASEB J. 2019;33:5990–6000. doi: 10.1096/fj.201801789RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffo S. Damato A. Alfano L. Giordano A. J. Exp. Clin. Cancer Res. 2018;37:36. doi: 10.1186/s13046-018-0704-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. Qin Z. Tian Y. Wang J. Xu C. Li Z. Bian J. J. Med. Chem. 2020;63:13228–13257. doi: 10.1021/acs.jmedchem.0c00744. [DOI] [PubMed] [Google Scholar]
- Senderowicz A. M. Invest. New Drugs. 1999;17:313–320. doi: 10.1023/a:1006353008903. [DOI] [PubMed] [Google Scholar]
- Baumli S. Lolli G. Lowe E. D. Troiani S. Rusconi L. Bullock A. N. Debreczeni J. E. Knapp S. Johnson L. N. EMBO J. 2008;27:1907–1918. doi: 10.1038/emboj.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi T. Karp J. E. Tan M. Doyle L. A. Peters T. Yang W. D. Wei D. Ross D. D. Clin. Cancer Res. 2003;9:3320–3328. [PubMed] [Google Scholar]
- Najmi S. Korah R. Chandra R. Abdellatif M. Wieder R. Clin. Cancer Res. 2005;11:2038–2046. doi: 10.1158/1078-0432.CCR-04-1083. [DOI] [PubMed] [Google Scholar]
- Litz J. Carlson P. Warshamana-Greene G. S. Grant S. Krystal G. W. Clin. Cancer Res. 2003;9:4586–4594. [PubMed] [Google Scholar]
- Jung C. Motwani M. Kortmansky J. Sirotnak F. M. She Y. H. Gonen M. Haimovitz-Friedman A. Schwartz G. K. Clin. Cancer Res. 2003;9:6052–6061. [PubMed] [Google Scholar]
- Soner B. C. Aktug H. Acikgoz E. Duzagac F. Guven U. Ayla S. Cal C. Oktem G. Int. J. Mol. Med. 2014;34:1249–1256. doi: 10.3892/ijmm.2014.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro G. I. Clin. Cancer Res. 2004;10:4270s–4275s. doi: 10.1158/1078-0432.CCR-040020. [DOI] [PubMed] [Google Scholar]
- Raju U. Nakata E. Mason K. A. Ang K. K. Milas L. Cancer Res. 2003;63:3263–3267. [PubMed] [Google Scholar]
- Mason K. A. Hunter N. R. Raju U. Ariga H. Husain A. Valdecanas D. Neal R. Ang K. K. Milas L. Int. J. Radiat. Oncol., Biol., Phys. 2004;59:1181–1189. doi: 10.1016/j.ijrobp.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Bible K. C. Kaufmann S. H. Cancer Res. 1997;57:3375–3380. [PubMed] [Google Scholar]
- Arguello F. Alexander M. Sterry J. A. Tudor G. Smith E. M. Kalavar N. T. Greene J. F. Koss W. Morgan C. D. Stinson S. F. Siford T. J. Alvord W. G. Klabansky R. L. Sausville E. A. Blood. 1998;91:2482–2490. [PubMed] [Google Scholar]
- Lapenna S. Giordano A. Nat. Rev. Drug Discovery. 2009;8:547–566. doi: 10.1038/nrd2907. [DOI] [PubMed] [Google Scholar]
- Chen R. Keating M. J. Gandhi V. Plunkett W. Blood. 2005;106:2513–2519. doi: 10.1182/blood-2005-04-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wontak Kim H. H. Peterson P. Whatcott C. J. Weitman S. Warner S. L. Bearss D. J. Siddiqui-Jain A. Cancer Res. 2017;77:5133. [Google Scholar]
- Joshi K. S. Rathos M. J. Joshi R. D. Sivakumar M. Mascarenhas M. Kamble S. Lal B. Sharma S. Mol. Cancer Ther. 2007;6:918–925. doi: 10.1158/1535-7163.MCT-06-0613. [DOI] [PubMed] [Google Scholar]
- Shirsath N. P. Manohar S. M. Joshi K. S. Mol. Cancer. 2012;11:77. doi: 10.1186/1476-4598-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalpana A. P. Joshi S. Rathos M. Wagh V. Manohar S. Bhatia D. Damre A. Sivakumar M. Sharma S. Cancer Res. 2012;72:3054. [Google Scholar]
- Roskoski R. Pharmacol. Res. 2019;139:471–488. doi: 10.1016/j.phrs.2018.11.035. [DOI] [PubMed] [Google Scholar]
- Dey J. Deckwerth T. L. Kerwin W. S. Casalini J. R. Merrell A. J. Grenley M. O. Burns C. Ditzler S. H. Dixon C. P. Beirne E. Gillespie K. C. Kleinman E. F. Klinghoffer R. A. Sci. Rep. 2017;7:18007. doi: 10.1038/s41598-017-18368-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedtke D. A. Su Y. Ma J. Li X. Buck S. A. Edwards H. Polin L. Kushner J. Dzinic S. H. White K. Lin H. Taub J. W. Ge Y. Signal Transduction Targeted Ther. 2020;5:17. doi: 10.1038/s41392-020-0112-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P. Zhang Y. K. Zhang X. Y. Wang Y. J. Lu K. W. Hall T. Peng R. Yang D. H. Xie N. Chen Z. S. Cell. Physiol. Biochem. 2018;45:1515–1528. doi: 10.1159/000487578. [DOI] [PubMed] [Google Scholar]
- Bharate S. B. Kumar V. Jain S. K. Mintoo M. J. Guru S. K. Nuthakki V. K. Sharma M. Bharate S. S. Gandhi S. G. Mondhe D. M. Bhushan S. Vishwakarma R. A. J. Med. Chem. 2018;61:1664–1687. doi: 10.1021/acs.jmedchem.7b01765. [DOI] [PubMed] [Google Scholar]
- Mintoo M. Khan S. Wani A. Malik S. Bhurta D. Bharate S. Malik F. Mondhe D. Mol. Carcinog. 2021;60:671–683. doi: 10.1002/mc.23332. [DOI] [PubMed] [Google Scholar]
- Wang J. Li T. Zhao T. Wu T. Liu C. Ding H. Li Z. Bian J. Eur. J. Med. Chem. 2019;178:782–801. doi: 10.1016/j.ejmech.2019.06.024. [DOI] [PubMed] [Google Scholar]
- Ibrahim N. Bonnet P. Brion J. D. Peyrat J. F. Bignon J. Levaique H. Josselin B. Robert T. Colas P. Bach S. Messaoudi S. Alami M. Hamze A. Eur. J. Med. Chem. 2020;199:112355. doi: 10.1016/j.ejmech.2020.112355. [DOI] [PubMed] [Google Scholar]
- Yu Z. Du J. Zhao Y. Gao Y. Li Y. Zhao K. Lu N. Cancer Lett. 2021;498:31–41. doi: 10.1016/j.canlet.2020.10.001. [DOI] [PubMed] [Google Scholar]
- Lucking U. Jautelat R. Kruger M. Brumby T. Lienau P. Schafer M. Briem H. Schulze J. Hillisch A. Reichel A. Wengner A. M. Siemeister G. ChemMedChem. 2013;8:1067–1085. doi: 10.1002/cmdc.201300096. [DOI] [PubMed] [Google Scholar]
- Siemeister G. Lucking U. Wengner A. M. Lienau P. Steinke W. Schatz C. Mumberg D. Ziegelbauer K. Mol. Cancer Ther. 2012;11:2265–2273. doi: 10.1158/1535-7163.MCT-12-0286. [DOI] [PubMed] [Google Scholar]
- Cho B. C. Dy G. K. Govindan R. Kim D. W. Pennell N. A. Zalcman G. Besse B. Kim J. H. Koca G. Rajagopalan P. Langer S. Ocker M. Nogai H. Barlesi F. Lung Cancer. 2018;123:14–21. doi: 10.1016/j.lungcan.2018.04.022. [DOI] [PubMed] [Google Scholar]
- William A. D. Lee A. C. H. Goh K. C. Blanchard S. Poulsen A. Teo E. L. Nagaraj H. Lee C. P. Wang H. S. Williams M. Sun E. T. Hu C. Y. Jayaraman R. Pasha M. K. Ethirajulu K. Wood J. M. Dymock B. W. J. Med. Chem. 2012;55:169–196. doi: 10.1021/jm201112g. [DOI] [PubMed] [Google Scholar]
- Goh K. C. Novotny-Diermayr V. Hart S. Ong L. C. Loh Y. K. Cheong A. Tan Y. C. Hu C. Jayaraman R. William A. D. Sun E. T. Dymock B. W. Ong K. H. Ethirajulu K. Burrows F. Wood J. M. Leukemia. 2012;26:236–243. doi: 10.1038/leu.2011.218. [DOI] [PubMed] [Google Scholar]
- Pallis M. Abdul-Aziz A. Burrows F. Seedhouse C. Grundy M. Russell N. Br. J. Haematol. 2012;159:191–203. doi: 10.1111/bjh.12018. [DOI] [PubMed] [Google Scholar]
- Su Y. T. Chen R. Wang H. R. Song H. Zhang Q. Chen L. Y. Lappin H. Vasconcelos G. Lita A. Maric D. Li A. G. Celiku O. Zhang W. Meetze K. Estok T. Larion M. Abu-Asab M. Zhuang Z. P. Yang C. Z. Gilbert M. R. Wu J. Clin. Cancer Res. 2018;24:1124–1137. doi: 10.1158/1078-0432.CCR-17-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder K. G. Matulis S. M. Hitosugi S. Gupta V. A. Sharp C. Burrows F. Nooka A. K. Kaufman J. L. Lonial S. Boise L. H. Cancer Biol. Ther. 2016;17:769–777. doi: 10.1080/15384047.2016.1192086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R. Tsai J. Thompson P. A. Chen Y. L. Xiong P. Liu C. M. Burrows F. Sivina M. Burger J. A. Keating M. J. Wierda W. G. Plunkett W. Blood Cancer J. 2021;11:57. doi: 10.1038/s41408-021-00436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Fernandez S. Ortiz-Ruiz M. J. Parrott T. Zaknoen S. Ocio E. M. San Miguel J. Burrows F. J. Esparis-Ogando A. Pandiella A. Clin. Cancer Res. 2013;19:2677–2687. doi: 10.1158/1078-0432.CCR-12-2118. [DOI] [PubMed] [Google Scholar]
- Le Rhun E. von Achenbach C. Lohmann R. Silginer M. Schneider H. Meetze K. Szabo E. Weller M. Int. J. Cancer. 2019;145:242–253. doi: 10.1002/ijc.32069. [DOI] [PubMed] [Google Scholar]
- Minzel W. Venkatachalam A. Fink A. Hung E. Brachya G. Burstain I. Shaham M. Rivlin A. Omer I. Zinger A. Elias S. Winter E. Erdman P. E. Sullivan R. W. Fung L. Mercurio F. Li D. Vacca J. Kaushansky N. Shlush L. Oren M. Levine R. Pikarsky E. Snir-Alkalay I. Ben-Neriah Y. Cell. 2018;175:171–185 e125. doi: 10.1016/j.cell.2018.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brian M. Ball J. Stein, MD A. S. Borthakur, MD G. Murray, RAC C. Kook, PhD K. Chan, PhD K. WH. Stein, MD E. M. Blood. 2020;136:18–19. [Google Scholar]
- Shao H. Shi S. H. Huang S. L. Hole A. J. Abbas A. Y. Baumli S. Liu X. R. Lam F. Foley D. W. Fischer P. M. Noble M. Endicott J. A. Pepper C. Wang S. D. J. Med. Chem. 2013;56:640–659. doi: 10.1021/jm301475f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H. Shi S. H. Foley D. W. Lam F. Abbas A. Y. Liu X. R. Huang S. L. Jiang X. R. Baharin N. Fischer P. M. Wang S. D. Eur. J. Med. Chem. 2013;70:447–455. doi: 10.1016/j.ejmech.2013.08.052. [DOI] [PubMed] [Google Scholar]
- Walsby E. Pratt G. Shao H. Abbas A. Y. Fischer P. M. Bradshaw T. D. Brennan P. Fegan C. Wang S. D. Pepper C. Oncotarget. 2014;5:375–385. doi: 10.18632/oncotarget.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hole A. J. Baumli S. Shao H. Shi S. H. Huang S. L. Pepper C. Fischer P. M. Wang S. D. Endicott J. A. Noble M. E. J. Med. Chem. 2013;56:660–670. doi: 10.1021/jm301495v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahaman M. H. Yu Y. Zhong L. Adams J. Lam F. Li P. Noll B. Milne R. Peng J. Wang S. Invest. New Drugs. 2019;37:625–635. doi: 10.1007/s10637-018-0661-2. [DOI] [PubMed] [Google Scholar]
- Lam F. Abbas A. Y. Shao H. Teo T. Adams J. Li P. Bradshaw T. D. Fischer P. M. Walsby E. Pepper C. Chen Y. Ding J. Wang S. D. Oncotarget. 2014;5:7691–7704. doi: 10.18632/oncotarget.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. J. Zhi X. L. Chen S. Y. Shen X. Q. Chen C. Yuan L. Guo J. Y. Meng D. Chen M. Yao L. Q. Am. J. Cancer Res. 2020;10:1140–1155. [PMC free article] [PubMed] [Google Scholar]
- Blake D. R. Vaseva A. V. Hodge R. G. Kline M. P. Gilbert T. S. K. Tyagi V. Huang D. W. Whiten G. C. Larson J. E. Wang X. D. Pearce K. H. Herring L. E. Graves L. M. Frye S. V. Emanuele M. J. Cox A. D. Der C. J. Sci. Signaling. 2019;12:eaav7259. doi: 10.1126/scisignal.aav7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. Li H. Wang X. Huang J. Li S. Liu C. Dong R. Zhu G. Duan C. Jiang F. Zhang Y. Zhu Y. Zhang T. Chen Y. Tang W. Lu T. Eur. J. Med. Chem. 2020;200:112424. doi: 10.1016/j.ejmech.2020.112424. [DOI] [PubMed] [Google Scholar]
- Wang Y. Chen X. Yan Y. Zhu X. Liu M. Liu X. J. Med. Chem. 2020;63:3327–3347. doi: 10.1021/acs.jmedchem.9b02121. [DOI] [PubMed] [Google Scholar]
- Shao H. Foley D. W. Huang S. Abbas A. Y. Lam F. Gershkovich P. Bradshaw T. D. Pepper C. Fischer P. M. Wang S. Eur. J. Med. Chem. 2021;214:113244. doi: 10.1016/j.ejmech.2021.113244. [DOI] [PubMed] [Google Scholar]
- Albert T. K. Rigault C. Eickhoff J. Baumgart K. Antrecht C. Klebl B. Mittler G. Meisterernst M. Br. J. Pharmacol. 2014;171:55–68. doi: 10.1111/bph.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H. Wu J. Ao M. Zhou X. Li B. Cui Z. Wu T. Wang L. Xue Y. Wu Z. Fang M. Bioorg. Chem. 2020;102:104064. doi: 10.1016/j.bioorg.2020.104064. [DOI] [PubMed] [Google Scholar]
- Lucking U. Scholz A. Lienau P. Siemeister G. Kosemund D. Bohlmann R. Briem H. Terebesi I. Meyer K. Prelle K. Denner K. Bomer U. Schafer M. Eis K. Valencia R. Ince S. von Nussbaum F. Mumberg D. Ziegelbauer K. Klebl B. Choidas A. Nussbaumer P. Baumann M. Schultz-Fademrecht C. Ruhter G. Eickhoff J. Brands M. ChemMedChem. 2017;12:1776–1793. doi: 10.1002/cmdc.201700447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y. Xing K. Zhang J. Tong T. Shi Y. T. Cao H. X. Yu H. Zhang Y. Liu D. Zhao L. X. Eur. J. Med. Chem. 2021;209:112885. doi: 10.1016/j.ejmech.2020.112885. [DOI] [PubMed] [Google Scholar]
- Arne Scholz U. L. Siemeister G. Lienau P. Boemer U. Ellinghaus P. Walter A. O. Valencia R. Ince S. von Nussbaum F. Mumberg D. Brands M. Ziegelbauer K. Cancer Res. 2015;75:AM2015-DDT2002-2002. [Google Scholar]
- Narita T. Ishida T. Ito A. Masaki A. Kinoshita S. Suzuki S. Takino H. Yoshida T. Ri M. Kusumoto S. Komatsu H. Imada K. Tanaka Y. Takaori-Kondo A. Inagaki H. Scholz A. Lienau P. Kuroda T. Ueda R. Iida S. Blood. 2017;130:1114–1124. doi: 10.1182/blood-2016-09-741983. [DOI] [PubMed] [Google Scholar]
- Kinoshita S. Ishida T. Ito A. Narita T. Masaki A. Suzuki S. Yoshida T. Ri M. Kusumoto S. Komatsu H. Shimizu N. Inagaki H. Kuroda T. Scholz A. Ueda R. Sanda T. Iida S. Haematologica. 2018;103:2059–2068. doi: 10.3324/haematol.2018.191395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson P. Dierichs L. Klein-Hitpass L. Bergmann A. K. Mollmann M. Menninger S. Habenberger P. Klebl B. Siveke J. T. Duhrsen U. Choidas A. Durig J. Ther. Adv. Hematol. 2020;11:1–15. doi: 10.1177/2040620720933761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetkova M. Growkova K. Fojtik P. Valcikova B. Palusova V. Verlande A. Jorda R. Krystof V. Hejret V. Alexiou P. Rotrekl V. Uldrijan S. Cell Death Discovery. 2020;11:754. doi: 10.1038/s41419-020-02971-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X. Q. Li Y. Q. Yu B. Ren J. Huang H. D. Wang M. Ding H. Li Z. Y. Wang J. B. Bian J. L. Eur. J. Med. Chem. 2021;211:113091. doi: 10.1016/j.ejmech.2020.113091. [DOI] [PubMed] [Google Scholar]
- Lucking U. Kosemund D. Bohnke N. Lienau P. Siemeister G. Denner K. Bohlmann R. Briem H. Terebesi I. Bomer U. Schafer M. Ince S. Mumberg D. Scholz A. Izumi R. Hwang S. von Nussbaum F. J. Med. Chem. 2021;64:11651–11674. doi: 10.1021/acs.jmedchem.1c01000. [DOI] [PubMed] [Google Scholar]
- Wu T. Z. Qin Z. Tian Y. C. Wang J. B. Xu C. X. Li Z. Y. Bian J. L. J. Med. Chem. 2020;63:13228–13257. doi: 10.1021/acs.jmedchem.0c00744. [DOI] [PubMed] [Google Scholar]
- Ulrich A. S. Luecking T. Kosemund D. Bohlmann R. Briem H. Lienau P. Siemeister G. Terebesi I. Meyer K. Prelle K. Valencia R. Ince S. von Nussbaum F. Mumberg D. Ziegelbauer K. Brands M. Cancer Res. 2017;77:AM2017–AM2984. [Google Scholar]
- Byrne M. G. F. M. Ottmann O. G. Mantzaris I. Wermke M. Lee D. J. Morillo D. Scholz, PhD A. Ince S. Valencia R. Souza F. Cordoba R. Blood. 2018;132:4055. [Google Scholar]
- Cidado M. S. J. Grondine M. Boiko S. Wang H. Borodovsky A. Mazzola A. M. Wu A. Lawson D. Ferguson D. Gao B. Cui A. D'Cruz C. Drew L. Cancer Res. 2016;76:3572. [Google Scholar]
- Hashiguchi T. Bruss N. Best S. Lam V. Danilova O. Paiva C. J. Wolf J. Gilbert E. W. Okada C. Y. Kaur P. Drew L. Cidado J. Hurlin P. Danilov A. V. Mol. Cancer Ther. 2019;18:1520–1532. doi: 10.1158/1535-7163.MCT-18-1023. [DOI] [PubMed] [Google Scholar]
- Barlaam B. Casella R. Cidado J. Cook C. De Savi C. Dishington A. Donald C. S. Drew L. Ferguson A. D. Ferguson D. Glossop S. Grebe T. Gu C. G. Hande S. Hawkins J. Hird A. W. Holmes J. Horstick J. Jiang Y. Lamb M. L. McGuire T. M. Moore J. E. O'Connell N. Pike A. Pike K. G. Proia T. Roberts B. San Martin M. Sarkar U. Shao W. L. Stead D. Sumner N. Thakur K. Vasbinder M. M. Varnes J. G. Wang J. Y. Wang L. Wu D. D. Wu L. W. Yang B. Yao T. G. J. Med. Chem. 2020;63:15564–15590. doi: 10.1021/acs.jmedchem.0c01754. [DOI] [PubMed] [Google Scholar]
- Alcon C. Manzano-Munoz A. Montero J. Clin. Cancer Res. 2020;26:761–763. doi: 10.1158/1078-0432.CCR-19-3670. [DOI] [PubMed] [Google Scholar]
- Cidado J. Boiko S. Proia T. Ferguson D. Criscione S. W. San Martin M. Pop-Damkov P. Su N. Franklin V. N. R. Chilamakuri C. S. R. D'Santos C. S. Shao W. L. Saeh J. C. Koch R. Weinstock D. M. Zinda M. Fawell S. E. Drew L. Clin. Cancer Res. 2020;26:922–934. doi: 10.1158/1078-0432.CCR-19-1853. [DOI] [PubMed] [Google Scholar]
- Carter B. Z. Mak P. Y. Tao W. Warmoes M. Lorenzi P. L. Mak D. Ruvolo V. Tan L. Cidado J. Drew L. Andreeff M. Haematologica. 2022;107:58–76. doi: 10.3324/haematol.2020.260331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B. L. Wu J. X. Wu Y. Chen C. Zou F. M. Wang A. L. Wu H. Hu Z. Q. Jiang Z. R. Liu Q. W. Wang W. Zhang Y. C. Liu F. Y. Zhao M. Hu J. Huang T. Ge J. Wang L. Ren T. Wang Y. X. Liu J. Liu Q. S. Eur. J. Med. Chem. 2018;158:896–916. doi: 10.1016/j.ejmech.2018.09.025. [DOI] [PubMed] [Google Scholar]
- Olson C. M. Jiang B. Erb M. A. Liang Y. Doctor Z. M. Zhang Z. Zhang T. Kwiatkowski N. Boukhali M. Green J. L. Haas W. Nomanbhoy T. Fischer E. S. Young R. A. Bradner J. E. Winter G. E. Gray N. S. Nat. Chem. Biol. 2018;14:163–170. doi: 10.1038/nchembio.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajiro M. Sakai H. Onogi H. Yamamoto M. Sumi E. Sawada T. Nomura T. Kabashima K. Hosoya T. Hagiwara M. Clin. Cancer Res. 2018;24:4518–4528. doi: 10.1158/1078-0432.CCR-17-3119. [DOI] [PubMed] [Google Scholar]
- Lacrima K. Valentini A. Lambertini C. Taborelli M. Rinaldi A. Zucca E. Catapano C. Cavalli F. Gianella-Borradori A. Maccallum D. E. Bertoni F. Ann. Oncol. 2005;16:1169–1176. doi: 10.1093/annonc/mdi217. [DOI] [PubMed] [Google Scholar]
- Raje N. Kumar S. Hideshima T. Roccaro A. Ishitsuka K. Yasui H. Shiraishi N. Chauhan D. Munshi N. C. Green S. R. Anderson K. C. Blood. 2005;106:1042–1047. doi: 10.1182/blood-2005-01-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvi A. J. Austen B. Weston V. J. Fegan C. MacCallum D. Gianella-Borradori A. Lane D. P. Hubank M. Powell J. E. Wei W. Taylor A. M. Moss P. A. Stankovic T. Blood. 2005;105:4484–4491. doi: 10.1182/blood-2004-07-2713. [DOI] [PubMed] [Google Scholar]
- Le Tourneau C. Faivre S. Laurence V. Delbaldo C. Vera K. Girre V. Chiao J. Armour S. Frame S. Green S. R. Gianella-Borradori A. Dieras V. Raymond E. Eur. J. Cancer. 2010;46:3243–3250. doi: 10.1016/j.ejca.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Benson C. White J. De Bono J. O'Donnell A. Raynaud F. Cruickshank C. McGrath H. Walton M. Workman P. Kaye S. Cassidy J. Gianella-Borradori A. Judson I. Twelves C. Br. J. Cancer. 2007;96:29–37. doi: 10.1038/sj.bjc.6603509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum D. E. Melville J. Frame S. Watt K. Anderson S. Gianella-Borradori A. Lane D. P. Green S. R. Cancer Res. 2005;65:5399–5407. doi: 10.1158/0008-5472.CAN-05-0233. [DOI] [PubMed] [Google Scholar]
- Dey A. Wong E. T. Cheok C. F. Tergaonkar V. Lane D. P. Cell Death Differ. 2008;15:263–273. doi: 10.1038/sj.cdd.4402257. [DOI] [PubMed] [Google Scholar]
- Raynaud F. I. Whittaker S. R. Fischer P. M. McClue S. Walton M. I. Barrie S. E. Garrett M. D. Rogers P. Clarke S. J. Kelland L. R. Valenti M. Brunton L. Eccles S. Lane D. P. Workman P. Clin. Cancer Res. 2005;11:4875–4887. doi: 10.1158/1078-0432.CCR-04-2264. [DOI] [PubMed] [Google Scholar]
- Whittaker S. R. Walton M. I. Garrett M. D. Workman P. Cancer Res. 2004;64:262–272. doi: 10.1158/0008-5472.can-03-0110. [DOI] [PubMed] [Google Scholar]
- Craig MacKay S. F. Saladino C. Pohler E. Zheleva D. Blake D. G. Mol. Cancer Ther. 2015;14:B182. [Google Scholar]
- Chiara Saladino S. F. Davis S. Blake D. Zheleva D. Cancer Res. 2015;75:1650–1650. [Google Scholar]
- Kawakami M. Mustachio L. M. Rodriguez-Canales J. Mino B. Roszik J. Tong P. Wang J. Lee J. J. Myung J. H. Heymach J. V. Johnson F. M. Hong S. Zheng L. Hu S. H. Villalobos P. A. Behrens C. Wistuba I. Freemantle S. Liu X. Dmitrovsky E. J. Natl. Cancer Inst. 2017;109:djw297. doi: 10.1093/jnci/djw297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A. L. Lind H. Hong A. Dokic D. Oppat K. Rosenthal E. Guo A. Thomas A. Hamden R. Jeruss J. S. Cell Cycle. 2017;16:1453–1464. doi: 10.1080/15384101.2017.1338988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon E. Liang T. Jamin Y. Walz S. Kwok C. Hakkert A. Barker K. Urban Z. Thway K. Zeid R. Hallsworth A. Box G. Ebus M. E. Licciardello M. P. Sbirkov Y. Lazaro G. Calton E. Costa B. M. Valenti M. Brandon A. D. Webber H. Tardif N. Almeida G. S. Christova R. Boysen G. Richards M. W. Barone G. Ford A. Bayliss R. Clarke P. A. De Bono J. Gray N. S. Blagg J. Robinson S. P. Eccles S. A. Zheleva D. Bradner J. E. Molenaar J. Vivanco I. Eilers M. Workman P. Lin C. Y. Chesler L. J. Clin. Invest. 2020;130:5875–5892. doi: 10.1172/JCI134132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaltriti M. Eichhorn P. J. Cortes J. Prudkin L. Aura C. Jimenez J. Chandarlapaty S. Serra V. Prat A. Ibrahim Y. H. Guzman M. Gili M. Rodriguez O. Rodriguez S. Perez J. Green S. R. Mai S. Rosen N. Hudis C. Baselga J. Proc. Natl. Acad. Sci. U. S. A. 2011;108:3761–3766. doi: 10.1073/pnas.1014835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocco E. Lopez S. Black J. Bellone S. Bonazzoli E. Predolini F. Ferrari F. Schwab C. L. Menderes G. Zammataro L. Buza N. Hui P. Wong S. Zhao S. M. Bai Y. L. Rimm D. L. Ratner E. Litkouhi B. Silasi D. A. Azodi M. Schwartz P. E. Santin A. D. Br. J. Cancer. 2016;115:303–311. doi: 10.1038/bjc.2016.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker S. R. Barlow C. Martin M. P. Mancusi C. Wagner S. Self A. Barrie E. Te Poele R. Sharp S. Brown N. Wilson S. Jackson W. Fischer P. M. Clarke P. A. Walton M. I. McDonald E. Blagg J. Noble M. Garrett M. D. Workman P. Mol. Oncol. 2018;12:287–304. doi: 10.1002/1878-0261.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry D. Guzi T. Shanahan F. Davis N. Prabhavalkar D. Wiswell D. Seghezzi W. Paruch K. Dwyer M. P. Doll R. Nomeir A. Windsor W. Fischmann T. Wang Y. L. Oft M. Chen T. Y. Kirschmeier P. Lees E. M. Mol. Cancer Ther. 2010;9:2344–2353. doi: 10.1158/1535-7163.MCT-10-0324. [DOI] [PubMed] [Google Scholar]
- Kumar S. K. LaPlant B. Chng W. J. Zonder J. Callander N. Fonseca R. Fruth B. Roy V. Erlichman C. Stewart A. K. Consortium M. P. Blood. 2015;125:443–448. doi: 10.1182/blood-2014-05-573741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. K. LaPlant B. R. Chng W. J. Zonder J. A. Callander N. Roy V. Furth B. Erlichman C. Stewart K. Blood. 2012;120:443–448. doi: 10.1182/blood-2014-05-573741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. Chen P. Liu K. Liu J. Zhou B. Wu R. Peng Q. Liu Z. X. Li C. Kroemer G. Lotze M. Zeh H. Kang R. Tang D. Gut. 2021;70:890–899. doi: 10.1136/gutjnl-2019-320441. [DOI] [PubMed] [Google Scholar]
- Hossain D. M. S. Javaid S. Cai M. M. Zhang C. S. Sawant A. Hinton M. Sathe M. Grein J. Blumenschein W. Pinheiro E. M. Chackerian A. J. Clin. Invest. 2018;128:644–654. doi: 10.1172/JCI94586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. J. Yeh Y. Y. Smith L. L. Wagner A. J. Hessler J. Gupta S. Flynn J. Jones J. Zhang X. Bannerji R. Grever M. R. Byrd J. C. Leukemia. 2012;26:2554–2557. doi: 10.1038/leu.2012.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker A. Gregory G. P. Verbrugge I. Kats L. Hilton J. J. Vidacs E. Lee E. M. Lock R. B. Zuber J. Shortt J. Johnstone R. W. Cancer Res. 2016;76:1158–1169. doi: 10.1158/0008-5472.CAN-15-1070. [DOI] [PubMed] [Google Scholar]
- Richters A. Doyle S. K. Freeman D. B. Lee C. Leifer B. S. Jagannathan S. Kabinger F. Koren J. V. Struntz N. B. Urgiles J. Stagg R. A. Curtin B. H. Chatterjee D. Mathea S. Mikochik P. J. Hopkins T. D. Gao H. Branch J. R. Xin H. Westover L. Bignan G. C. Rupnow B. A. Karlin K. L. Olson C. M. Westbrook T. F. Vacca J. Wilfong C. M. Trotter B. W. Saffran D. C. Bischofberger N. Knapp S. Russo J. W. Hickson I. Bischoff J. R. Gottardis M. M. Balk S. P. Lin C. Y. Pop M. S. Koehler A. N. Cell Chem. Biol. 2021;28:134–147 e114. doi: 10.1016/j.chembiol.2020.10.001. [DOI] [PubMed] [Google Scholar]
- Phillipson L. J. Segal D. H. Nero T. L. Parker M. W. Wan S. S. de Silva M. Guthridge M. A. Wei A. H. Burns C. J. Bioorg. Med. Chem. 2015;23:6280–6296. doi: 10.1016/j.bmc.2015.08.035. [DOI] [PubMed] [Google Scholar]
- Wyatt P. G. Woodhead A. J. Berdini V. Boulstridge J. A. Carr M. G. Cross D. M. Davis D. J. Devine L. A. Early T. R. Feltell R. E. Lewis E. J. McMenamin R. L. Navarro E. F. O'Brien M. A. O'Reilly M. Reule M. Saxty G. Seavers L. C. A. Smith D. M. Squires M. S. Trewartha G. Walker M. T. Woolford A. J. A. J. Med. Chem. 2008;51:4986–4999. doi: 10.1021/jm800382h. [DOI] [PubMed] [Google Scholar]
- Dolman M. E. M. Poon E. Ebus M. E. den Hartog I. J. M. van Noesel C. J. M. Jamin Y. Hallsworth A. Robinson S. P. Petrie K. Sparidans R. W. Kok R. J. Versteeg R. Caron H. N. Chesler L. Molenaar J. J. Clin. Cancer Res. 2015;21:5100–5109. doi: 10.1158/1078-0432.CCR-15-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires M. S. Feltell R. E. Wallis N. G. Lewis E. J. Smith D. M. Cross D. M. Lyons J. F. Thompson N. T. Mol. Cancer Ther. 2009;8:324–332. doi: 10.1158/1535-7163.MCT-08-0890. [DOI] [PubMed] [Google Scholar]
- Santo L. Vallet S. Hideshima T. Cirstea D. Ikeda H. Pozzi S. Patel K. Okawa Y. Gorgun G. Perrone G. Calabrese E. Yule M. Squires M. Ladetto M. Boccadoro M. Richardson P. G. Munshi N. C. Anderson K. C. Raje N. Oncogene. 2010;29:2325–2336. doi: 10.1038/onc.2009.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumli S. Hole A. J. Noble M. E. Endicott J. A. ACS Chem. Biol. 2012;7:811–816. doi: 10.1021/cb2004516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystof V. Rarova L. Liebl J. Zahler S. Jorda R. Voller J. Cankar P. Eur. J. Med. Chem. 2011;46:4289–4294. doi: 10.1016/j.ejmech.2011.06.035. [DOI] [PubMed] [Google Scholar]
- Cirstea D. Hideshima T. Santo L. Eda H. Mishima Y. Nemani N. Hu Y. Mimura N. Cottini F. Gorgun G. Ohguchi H. Suzuki R. Loferer H. Munshi N. C. Anderson K. C. Raje N. Leukemia. 2013;27:2366–2375. doi: 10.1038/leu.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biessen D. A. Burger H. de Bruijn P. Lamers C. H. Naus N. Loferer H. Wiemer E. A. Mathijssen R. H. de Jonge M. J. Clin. Cancer Res. 2014;20:4776–4783. doi: 10.1158/1078-0432.CCR-14-0325. [DOI] [PubMed] [Google Scholar]
- Misra R. N. Xiao H. Y. Kim K. S. Lu S. F. Han W. C. Barbosa S. A. Hunt J. T. Rawlins D. B. Shan W. F. Ahmed S. Z. Qian L. G. Chen B. C. Zhao R. L. Bednarz M. S. Kellar K. A. Mulheron J. G. Batorsky R. Roongta U. Kamath A. Marathe P. Ranadive S. A. Sack J. S. Tokarski J. S. Pavletich N. P. Lee F. Y. F. Webster K. R. Kimball S. D. J. Med. Chem. 2004;47:1719–1728. doi: 10.1021/jm0305568. [DOI] [PubMed] [Google Scholar]
- Wu Y. Chen C. Sun X. Shi X. Jin B. Ding K. Yeung S. C. Pan J. Clin. Cancer Res. 2012;18:1966–1978. doi: 10.1158/1078-0432.CCR-11-1971. [DOI] [PubMed] [Google Scholar]
- Walsby E. Lazenby M. Pepper C. Burnett A. K. Leukemia. 2011;25:411–419. doi: 10.1038/leu.2010.290. [DOI] [PubMed] [Google Scholar]
- Zhang H. Pandey S. Travers M. Sun H. Morton G. Madzo J. Chung W. Khowsathit J. Perez-Leal O. Barrero C. A. Merali C. Okamoto Y. Sato T. Pan J. Garriga J. Bhanu N. V. Simithy J. Patel B. Huang J. Raynal N. J. Garcia B. A. Jacobson M. A. Kadoch C. Merali S. Zhang Y. Childers W. Abou-Gharbia M. Karanicolas J. Baylin S. B. Zahnow C. A. Jelinek J. Grana X. Issa J. J. Cell. 2018;175:1244–1258 e1226. doi: 10.1016/j.cell.2018.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips D. C. Jin S. Gregory G. P. Zhang Q. Xue J. Zhao X. X. Chen J. Tong Y. S. Zhang H. C. Smith M. Tahir S. K. Clark R. F. Penning T. D. Devlin J. R. Shortt J. Hsi E. D. Albert D. H. Konopleva M. Johnstone R. W. Leverson J. D. Souers A. J. Leukemia. 2020;34:1646–1657. doi: 10.1038/s41375-019-0652-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh U. Chashoo G. Khan S. U. Mahajan P. Nargotra A. Mahajan G. Singh A. Sharma A. Mintoo M. J. Guru S. K. Aruri H. Thatikonda T. Sahu P. Chibber P. Kumar V. Mir S. A. Bharate S. S. Madishetti S. Nandi U. Singh G. Mondhe D. M. Bhushan S. Malik F. Mignani S. Vishwakarma R. A. Singh P. P. J. Med. Chem. 2017;60:9470–9489. doi: 10.1021/acs.jmedchem.7b00663. [DOI] [PubMed] [Google Scholar]
- Barlaam B. De Savi C. Dishington A. Drew L. Ferguson A. D. Ferguson D. Gu C. G. Hande S. Hassall L. Hawkins J. Hird A. W. Holmes J. Lamb M. L. Lister A. S. McGuire T. M. Moore J. E. O'Connell N. Patel A. Pike K. G. Sarkar U. Shao W. L. Stead D. Varnes J. G. Vasbinder M. M. Wang L. Wu L. W. Xue L. Yang B. Yao T. G. J. Med. Chem. 2021;64:15189–15213. doi: 10.1021/acs.jmedchem.1c01249. [DOI] [PubMed] [Google Scholar]
- Ghanem N. M. Farouk F. George R. F. Abbas S. E. S. El-Badry O. M. Bioorg. Chem. 2018;80:565–576. doi: 10.1016/j.bioorg.2018.07.006. [DOI] [PubMed] [Google Scholar]
- Montagnoli A. Valsasina B. Croci V. Menichincheri M. Rainoldi S. Marchesi V. Tibolla M. Tenca P. Brotherton D. Albanese C. Patton V. Alzani R. Ciavolella A. Sola F. Molinari A. Volpi D. Avanzi N. Fiorentini F. Cattoni M. Healy S. Ballinari D. Pesenti E. Isacchi A. Moll J. Bensimon A. Vanotti E. Santocanale C. Nat. Chem. Biol. 2008;4:357–365. doi: 10.1038/nchembio.90. [DOI] [PubMed] [Google Scholar]
- Yan L. Lai F. F. Chen X. G. Xiao Z. Y. Bioorg. Med. Chem. Lett. 2015;25:2447–2451. doi: 10.1016/j.bmcl.2015.03.066. [DOI] [PubMed] [Google Scholar]
- Li Y. Luo X. Guo Q. Nie Y. Wang T. Zhang C. Huang Z. Wang X. Liu Y. Chen Y. Zheng J. Yang S. Fan Y. Xiang R. J. Med. Chem. 2018;61:3166–3192. doi: 10.1021/acs.jmedchem.8b00209. [DOI] [PubMed] [Google Scholar]
- Wang X. Yu C. Wang C. Ma Y. Wang T. Li Y. Huang Z. Zhou M. Sun P. Zheng J. Yang S. Fan Y. Xiang R. Eur. J. Med. Chem. 2019;181:111535. doi: 10.1016/j.ejmech.2019.07.038. [DOI] [PubMed] [Google Scholar]
- Li Y. Du R. Nie Y. Wang T. Ma Y. Fan Y. Bioorg. Chem. 2021;109:104717. doi: 10.1016/j.bioorg.2021.104717. [DOI] [PubMed] [Google Scholar]
- Robb C. M. Contreras J. I. Kour S. Taylor M. A. Abid M. Sonawane Y. A. Zahid M. Murry D. J. Natarajan A. Rana S. Chem. Commun. 2017;53:7577–7580. doi: 10.1039/c7cc03879h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian J. Ren J. Li Y. Wang J. Xu X. Feng Y. Tang H. Wang Y. Li Z. Bioorg. Chem. 2018;81:373–381. doi: 10.1016/j.bioorg.2018.08.028. [DOI] [PubMed] [Google Scholar]
- Zhou F. Chen L. Cao C. Yu J. Luo X. Zhou P. Zhao L. Du W. Cheng J. Xie Y. Chen Y. Eur. J. Med. Chem. 2020;187:111952. doi: 10.1016/j.ejmech.2019.111952. [DOI] [PubMed] [Google Scholar]
- Qiu X. Li Y. Yu B. Ren J. Huang H. Wang M. Ding H. Li Z. Wang J. Bian J. Eur. J. Med. Chem. 2021;211:113091. doi: 10.1016/j.ejmech.2020.113091. [DOI] [PubMed] [Google Scholar]
- King H. M. Rana S. Kubica S. P. Mallareddy J. R. Kizhake S. Ezell E. L. Zahid M. Naldrett M. J. Alvarez S. Law H. C. Woods N. T. Natarajan A. Bioorg. Med. Chem. Lett. 2021;43:128061. doi: 10.1016/j.bmcl.2021.128061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D. Wang H. Zeng Q. Wang W. Hao B. Feng X. Wang P. Song N. Kan W. Huang G. Zhou X. Tan M. Zhou Y. Huang R. Li J. Chen X. H. J. Med. Chem. 2021;64:14822–14847. doi: 10.1021/acs.jmedchem.1c01350. [DOI] [PubMed] [Google Scholar]
- Attwood M. M. Fabbro D. Sokolov A. V. Knapp S. Schioth H. B. Nat. Rev. Drug Discovery. 2021;20:839–861. doi: 10.1038/s41573-021-00252-y. [DOI] [PubMed] [Google Scholar]
- Lu S. Zhang J. J. Med. Chem. 2019;62:24–45. doi: 10.1021/acs.jmedchem.7b01844. [DOI] [PubMed] [Google Scholar]
- Fan J. Liu Y. Kong R. Ni D. Yu Z. Lu S. Zhang J. J. Med. Chem. 2021;64:17728–17743. doi: 10.1021/acs.jmedchem.1c01695. [DOI] [PubMed] [Google Scholar]
- Lu S. He X. Yang Z. Chai Z. Zhou S. Wang J. Rehman A. U. Ni D. Pu J. Sun J. Zhang J. Nat. Commun. 2021;12:4721. doi: 10.1038/s41467-021-25020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. He X. Ni D. Zhang J. J. Med. Chem. 2019;62:6405–6421. doi: 10.1021/acs.jmedchem.8b01749. [DOI] [PubMed] [Google Scholar]
- Ni D. Wei J. He X. Rehman A. U. Li X. Qiu Y. Pu J. Lu S. Zhang J. Chem. Sci. 2020;12:464–476. doi: 10.1039/d0sc05131d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. Shen Q. Zhang J. Acc. Chem. Res. 2019;52:492–500. doi: 10.1021/acs.accounts.8b00570. [DOI] [PubMed] [Google Scholar]