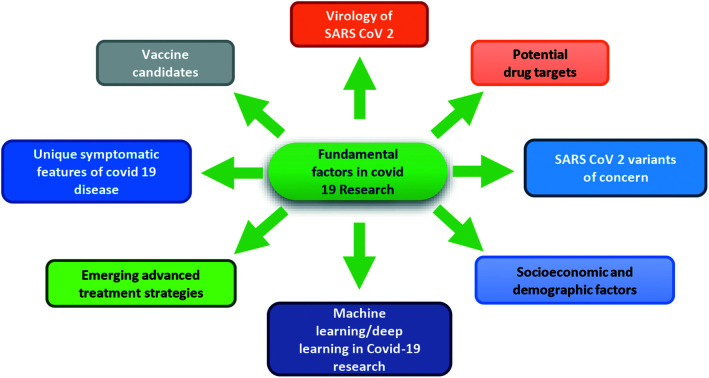
Abstract
SARS-CoV-2, the virus responsible for the COVID-19 pandemic, has been confirmed to be a new coronavirus having 79% and 50% similarity with SARS-CoV and MERS-CoV, respectively. For a better understanding of the features of the new virus SARS-CoV-2, we have discussed a possible correlation between some unique features of the genome of SARS-CoV-2 in relation to pathogenesis. We have also reviewed structural druggable viral and host targets for possible clinical application if any, as cases of reinfection and compromised protection have been noticed due to the emergence of new variants with increased infectivity even after vaccination. We have also discussed the types of vaccines that are being developed against SARS-CoV-2. In this review, we have tried to give a brief overview of the fundamental factors of COVID-19 research like basic virology, virus variants and the newly emerging techniques that can be applied to develop advanced treatment strategies for the management of COVID-19 disease.
1. Introduction
In the last twenty years, coronaviruses have caused several viral pandemics like severe acute respiratory syndrome (SARS) caused by SARS-CoV in 2002–2003 and Middle East respiratory syndrome (MERS) caused by MERS-CoV in 2012.1
In December 2019, several cases of some unknown type of pneumonia were first observed in patients who were admitted to a hospital of Wuhan city in Hubei province of China.2 Later it was diagnosed that an unknown virus of the coronavirus family was responsible for the disease.3 Further analysis revealed that the virus is 79% identical to SARS CoV and 50% identical to MERS CoV.5 In February 2020, the coronavirus study group (CSG) of the International Committee on Taxonomy of Viruses checked the novelty of the virus and preliminarily named it as 2019 nCov. Finally, after the further study based on taxonomy and phylogeny, the novel coronavirus got its official name as SARS-CoV-2.6
The symptoms of COVID-19 or coronavirus disease 2019 are similar to the symptoms of common flu like sore throat, fever, cough and fatigue. In some cases, these symptoms are coupled with diarrhoea and vomiting. Around 260 million people have been affected by this pandemic disease and more than 5 million deaths have been reported worldwide to date (https://www.worldometers.info/coronavirus/?utm_campaign=homeAdvegas1?%22%20%5Cl%22countries). Statistical data revealed that males are more prone to COVID-19 infection than females7 and people of older age groups (>55 years) with8 or without7 co-morbidities showed the highest infection fatality ratio. In addition to the common flu-like and respiratory distress symptoms, COVID-19 is now proven to induce cellular immune deficiency, coagulation activation, cardiac injury, hepatic injury, renal dysfunction and multiorgan failure.8 Europe, Italy and Spain were severely affected in early 2020. An epidemic peak started in the middle of February 2020, which rapidly evolved into a global pandemic.4 It has been reported that quite a large number of infected persons showed varied manifestations, whereas some showed no symptoms at all.6 A large majority of certain patients' sub-groups predominantly experienced severe respiratory syndrome with interstitial pneumonia in both lungs and acute respiratory distress.9,10 In such hospitalized conditions with respiratory failure states, patients required early and prolonged supply of mechanized ventilation.11 The second wave of COVID-19 has been already affecting most of the world. In India, the situation has become very harsh as compared to the first wave. Starting in February, India confirmed 10k to 20k new cases per day which have risen exponentially to 300k to 400k cases per day (https://www.worldometers.info/coronavirus/country/india/).
2. Taxonomy of SARS-CoV-2 and disease spectrum
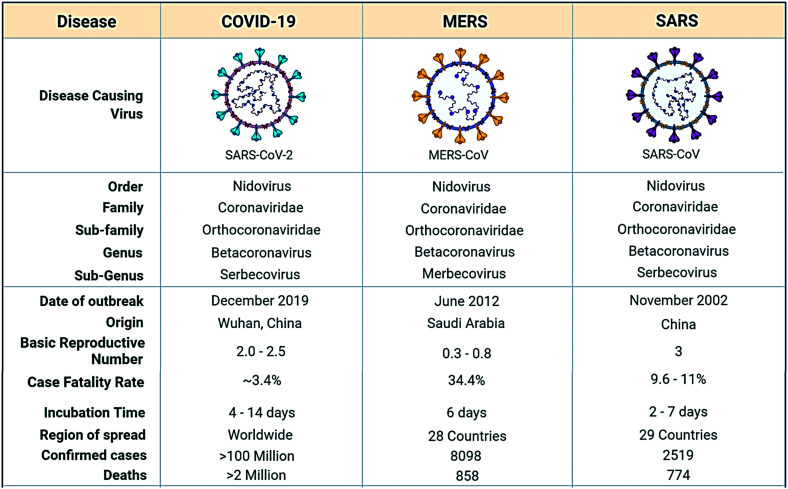
The coronavirus study group has identified the SARS-CoV-2 virus on the basis of phylogeny and taxonomy and confirmed the virus as a descendent of the severe acute respiratory syndrome coronavirus. (The taxonomic classification of SARS-CoV-2 being the order: Nidovirus, family: Coronaviridae, sub-family: Orthocoronaviridae, genus: Betacoronavirus and sub-genus: Serbecovirus).12
SARS-CoV-2 has an incubation period of about 14 days with a median of 4–5 days,13 whereas SARS-CoV has an incubation period of 2 to 7 days with a median incubation period of 4 days14 (Fig. 1).
Fig. 1. Taxonomical and epidemiological comparison of SARS-CoV-2, MERS CoV and SARS CoV.
The impact of an epidemic relies on some factors like the number of infected persons, transmissibility of the infection and spectrum of clinical severity. To control an epidemic or pandemic, some of the factors need to be taken into consideration: [i] the full spectrum of disease severity which ranges from asymptomatic to symptomatic-but-mild, to severe requiring hospitalization and to fatal, [ii] transmission of the virus, [iii] identification of infectors, age, illness severity, and risk of transmission to others particularly the role played by the asymptomatic or pre-symptomatic individual in transmission and [iv] the duration of virus presence in respiratory secretions, with severe illness and death.15
3. Structure of SARS-CoV-2
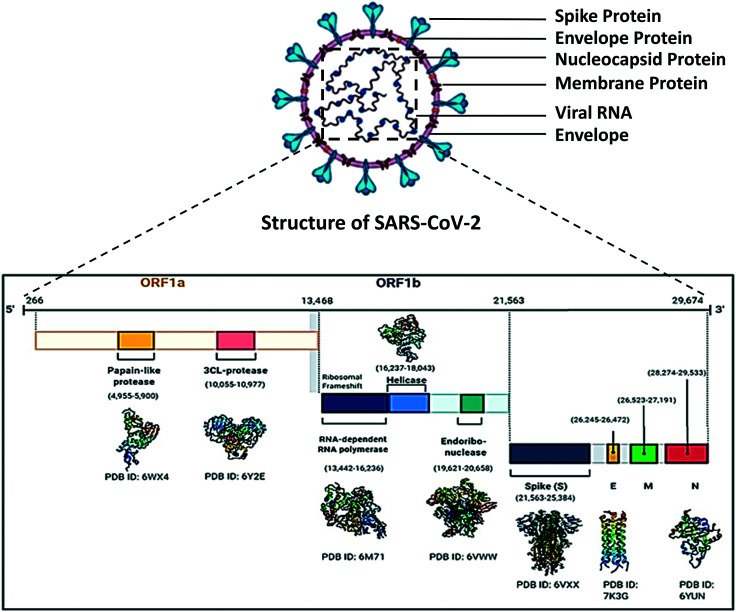
In 1965, June Almeida and David Tyrrell first used the name ‘corona’ to name coronaviruses, which means ‘crown’ or ‘wreath’ in Latin due to the presence of crown-shaped spike protein. Like other coronaviruses, SARS-CoV-2 viral particles are spherical in shape ranging from 50 to 200 nm in diameter.16,17 Coronaviruses are protected by a lipid bilayer envelope, to which the envelope proteins, membrane proteins and spike proteins are anchored,18 whereas the nucleocapsid proteins (SARS-CoV-2) bind with the virion19 RNA (Fig. 2).
Fig. 2. Structure of SARS-CoV-2 and organisation of the important genes in its genome.
3.1. Genome
Coronaviruses have the largest genomes of all known RNA viruses that incorporate ∼30 kb genomes inside the enveloped capsid. These viruses have variations among the genome due to significant alterations in the structure and morphology of the nucleocapsids of virions.16 RNA viruses are those viruses that use RNA (ribonucleic acid) as their genetic material.19 The genetic material of RNA viruses is mostly single-stranded RNA (ssRNA) but in some cases, double-stranded RNA (dsRNA) is also present.20 According to sense or polarity, RNA viruses are mainly of two types, negative-sense and positive-sense. The viral RNA of positive-sense RNA viruses is similar to mRNA and thus can be easily translated by the host cell, whereas the viral RNA of negative-sense RNA viruses is complementary to mRNA and for translation, the negative-sense RNA firstly needs to be converted to positive-sense RNA by the RNA-dependent RNA polymerase (RdRp). Ambisense RNA viruses contain at least one ambisense segment in their RNA and can translate genes from both the positive and negative strand.21
SARS-CoV-2 is a positive-sense single-stranded RNA virus22 of subgenus sarbecovirus (beta-CoV lineage B).23 Among the known beta coronaviruses, SARS-CoV-2 is unique in the aspect of the presence of a polybasic cleavage site, which is responsible for its severe transmissibility.24,25 From all the available data, it is now confirmed that the genome size of SARS-CoV-2 varies from 29.8 kb to 29.9 kb and its genome structure is similar to the specific gene characteristics of known CoVs; the 5′ region contains 5′ cap, leader sequence, UTR, a replicase gene which is more than two-thirds of the genome comprising ORF1ab that encodes ORF1ab polyprotein, while the 3′ possesses genes for structural proteins including spike (S), envelope (E), membrane (M), and nucleocapsid N proteins and accessory protein, 3′ UTR, and a poly-A tail26 (Fig. 2). Replicase polyprotein 1ab is a multifunctional protein, which is involved in the transcription as well as replication of viral RNAs. It contains the proteinases responsible for the cleavages of the polyprotein.
3.2. Proteins
Viral proteins are usually classified into 3 groups according to their functions i.e., structural proteins, non-structural proteins, regulatory proteins and/or accessory proteins.27
3.2.1. Structural proteins
The viral structural proteins make up the envelope or the protection of the virus. Most of the viruses have a protein capsid protecting the nucleic acid genome, but some viruses have an additional outer layer with glycoprotein spikes.
3.2.1.1. Spike glycoprotein
SARS-CoV-2 spike protein has two subunits, S1 and S2, that are non-covalently joined and form surface homotrimers. Subunit S1 binds to the host cell membrane by interacting with human ACE2 and CLEC4M/DC-SIGNR receptor.28 S2 mediates the viral entry by fusion of the viral membrane and host cell membrane by acting as a class I viral fusion protein.29 The most important characteristic of spike protein is that it has a functional polybasic (furin) cleavage site at the S1–S2 by the incorporation of 12 nucleotides.25 Spike protein forms three conformational states during host cell entry: native state, hairpin intermediate state, and post-fusion hairpin state. Spike protein is cleaved and activated by TMPRSS2, which is a transmembrane serine protease present in the host cell.30 Spike glycoproteins of SARS-CoV-2 possess some new glycosylation sites such as NGTK, NFTI, NLTT, and NTSN compared to SARS CoV. They also found that the SARS CoV and SARS-CoV-2 spike glycoproteins also share some common glycosylation sites such as NITN, NGTI, NITN, NFSQ, NESL, NCTF and NNTV.30 The spike protein of both strains of coronaviruses has 346 amino acid residues and 76% sequence identity.31 Glycan parts on S protein have been reported to help in escaping from the host immune response by masking “nonself” viral peptides with “self-glycans”, and glycosylation of S protein is also important for virus–receptor interactions, and in antibody production, as well as indications for vaccine development.32
3.2.1.2. Envelope protein
Envelope proteins of CoVs play multiple roles in virus pathogenesis,33 assembly,34,35 and viral release.36 It establishes an ion transport channel by self-assembling in host membranes and forming pentameric protein-lipid pores.37 SARS CoV and SARS CoV-2 envelope proteins have a sequence identity of 94.7%.31
3.2.1.3. Membrane protein
The main importance of viral membrane protein is its active participation in viral assembly.38 The M protein interacts with many viral proteins like the N protein to encapsulate the RNA genome.39 The SARS-CoV-2 M protein has 222 amino acids which have one amino acid more compared to the SARS CoV M protein. Both proteins have 90.5% sequence identity.31
3.2.1.4. Nucleocapsid proteins
Coronavirus nucleocapsid (N) proteins directly bind to the viral RNA. The N protein of coronaviruses plays a fundamental role in virion assembly by interacting with the viral genome and the membrane protein.40 An N protein plays an important role in packaging the viral RNA into a helical ribonucleocapsid protein (RNP) and enhances the efficiency of viral genomic replication. The nucleocapsid protein of SARS CoV was also found to inhibit the activity of the cyclin-dependent kinase (CDK) complex.41 SARS-CoV-2 N protein has 419 amino acid residues which are three amino acids less than the N protein of SARS CoV. The N proteins from SARS CoV and SARS CoV-2 have 90.5% sequence identity.31
3.2.2. Non-structural proteins
Recently, efforts have been made to consider non-structural proteins of SARS-CoV-2 as targets through repurposing drugs.42 Apart from the structural proteins, the SARS-CoV-2 genome expresses several nsps (non-structural proteins) (Fig. 2). Viral nsps are encoded by the genome of the virus but are not assembled in the virion; they are expressed in the infected cells. The nsps of SARS-CoV-2 play various vital roles in the life cycle of SARS-CoV-2.32 The nsps include various enzymes and transcription factors that the virus uses to replicate itself, such as a viral protease (Mpro or nsp 5), RNA, or other template-directed polymerases (RdRp or nsp 12). Other nsps like nsp1 interact with the 40S ribosome to degrade host mRNA, whereas nsp2 interacts with host prohibitin 1 (PHB1) and prohibitin 2 (PHB2) (prohibitin 1 and prohibitin 2 are highly expressed in mitochondrial function dependent cells). These two proteins assemble at the mitochondrial inner membrane to form a supra-macromolecular structure that regulates mitochondrial metabolism finally determining the lifespan of a cell43 and disrupts the host cell environment by playing a role in host cell survival signaling. Some of these non-structural proteins like nsp3 or PLpro, nsp4 and nsp6 are known to play a key role in the assembly process of membrane vesicles that are essential for viral replication. Several nsps like nsp7, nsp8 and nsp12 or RdRp form a complex that is responsible for nucleotide polymerization in SARS-CoV-2. Another important nsp i.e., nsp 13 possesses helicase activity that is essential for translation and splicing of mRNA. nsps also function in immunomodulation and transactivation of encoding genes of viral structural protein.26
3.2.3. Accessory proteins
Viral regulatory and accessory proteins are a broad category of viral proteins that indirectly affects the function, biological processes and activities of a virus.27 Many of the proteins in this category serve multiple functions like regulating the expression of viral genes and modification of host cell functions. The SARS-CoV-2 genome (Fig. 2) expresses 9 accessory proteins, encoded by the ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8, ORF9b, ORF14, and ORF10 gene. In a recent study, it has been observed that ORF6, ORF8 and ORF3b can inhibit the IFN β and NF-κB interferon signaling pathway, which benefits the viral replication in the early stages of SARS-CoV-2 infection by delaying the IFN release.44 ORF 3a of SARS-CoV-2 has been found to induce apoptosis in the host.45
4. SARS CoV-2 variants
It is already evident that a mutation, i.e., D614G, increases the ability of the SARS-CoV-2 virus to spread more quickly than the wild-type one.46,47 The D614G mutation, i.e., aspartic acid to glycine substitution at the 614th position within the spike protein, disrupts the hydrogen bond interaction with T859, which not only weakens the stability of the spike protein trimer but also shortens the distance between the backbone amino acid residue at the 614th position and the backbone carbonyl group of the residue at the 647th position that stabilizes the C terminal domain of the protein. This change leads to an open conformation of the spike protein and increases the availability of the spike trimer in the conformation that promotes the efficiency of the spike protein to bind with ACE 2.48 Recent studies confirmed that the D614G mutation has become dominant in the COVID-19 pandemic since Aug 2020.46,48,49 The WHO Virus Evolution Working Group (VEWG) has detected potential SARS-CoV-2 variants and marked them as variants of concern (VOC) that are already found to decrease the effectiveness of available diagnostics, therapeutics, vaccines, etc. and variants of interest (VOI) that are suspected to be responsible for community transmission or multiple COVID-19 cases/clusters or have been spread to multiple countries (tps://www.who.int/docs/default-source/coronaviruse/risk-comms-updates/update60_nomenclature-variants.pdf?sfvrsn=27fc6fa_4).
4.1. Alpha variant or B.1.1.7
This variant first emerged in the UK in September 2020 and has already invaded the USA, Canada and some other countries (https://www.who.int/csr/don/21-december-2020-sars-cov2-variant-united-kingdom/en). The variant has eight novel mutations i.e., HV 69-70del, Y144del, N501Y, A570D, P681H, T7161, S982A, and D1118H. The 69/70 deletion leads to a conformational change in spike protein. The P681H mutation near the furin cleavage site may have an impact on its enhanced transmissibility. Besides all these mutations, another mutation in the 27th position of ORF8 i.e., Q27stop was also observed in this variant.50 The variant showed increased, more efficient and rapid transmission than the wild type although there is no evidence of any impact on the severity of the disease or vaccine efficacy.51 However, in a recent study, Wu et al. showed that sera from the phase 1 participants of the mRNA-1273 vaccine were able to neutralize the B.1.1.7 variant to the same level as the D614G variant.52 Reduction in the neutralization titers was observed against B.1.1.7 by convalescent sera generated in early variants of SARS-CoV-2 and sera from Pfizer–BioNTech and Oxford-AstraZeneca vaccinated individuals.53
4.2. Beta variant or B.1.351
This variant was first identified at Nelson Mandela Bay, South Africa in October 2020 and became the predominant variant till now.54 Like the B.1.1.7 variant, this variant also possesses N501Y mutation but not 69/70 deletion. The eight mutations that the variant carries are L18F, D80A, D215G, R246I, K417N, E484K, N501Y, and A701V, in addition to the D614G mutation.55 It has been observed that the E484K mutation may affect the neutralization by some polyclonal and monoclonal antibodies56,57 and showed a 5-to-10-fold reduction in neutralization with sera from NHPs, immunized by mRNA-1273.58 In another study, Edara et al. observed that sera from previously infected and convalescent COVID-19 patients can reduce by 3-fold the binding affinity of antibody titers to the B.1.351 variant receptor-binding domain of the spike protein and can also reduce by 3.5-fold the neutralization of antibody titers against the SARS-CoV-2 B.1.351 variant compared to the B.1 variant.59 B.1.351 was found to be more resistant to neutralization by convalescent plasma (9.4-fold) and sera from vaccinated individuals (10.3–12.4-fold).60
4.3. Gamma variant or P.1
The SARS-CoV-2 epidemic in Brazil is dominated by two lineages, designated as B.1.1.33 and B.1.1.28. But a new variant that has recently circulated in Brazil is B.1.1.248 or P.1 which is a sub-variant of the B.1.1.28 lineage61 and is being known as the or gamma variant or Brazilian variant. The gamma variant has 17 unique mutations, 10 of which are in the spike protein (L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, H655Y, and T1027I).61 A recent study showed that this variant is capable of reinfecting the individuals of Manaus city in the Amazon region and Salvador, Bahia state of northeast Brazil, who were already recovered from COVID-19 pre-infection. This case of re-infection is raising the concern about the neutralizing ability of already developed vaccines against this variant.62 A lower neutralizing capacity (8·6 times) with plasma from individuals previously infected with SARS-CoV-2 and inefficient neutralization with plasma samples collected from CoronaVac vaccinated individuals against the P.1 isolates were observed.
4.4. Epsilon variant or CAL.20C or B.1.429 and B.1.427
The SARS-CoV-2 epsilon variant or CAL.20C or B.1.429 and B.1.427 was first identified in July 2020, in Southern California contemporaneously with the local surge in cases. This variant is defined by 3 mutations in the S protein unlike the clade 20G, which is the main reported clade in North America. This variant is defined by 5 mutations, 2 in ORFs (I4205V at ORF1a and D1183Y at ORF1b) and 3 in spike protein (S13I, W152C, and L452R). The prevalence of this variant has increased in the state of California. The L452R mutation in S protein is within a known receptor binding domain that has been found to be resistant to certain spike (S) protein monoclonal antibodies. Reduced neutralization by convalescent sera and BNT2b2, mRNA1273 vaccine-elicited sera was observed when compared to non-VOC/VOI variants.63
4.5. Delta variant or B.1.617.2 and Delta+ variant or B.1.617.2.1
The delta variant was first found in India and has T19R, (G142D), 156del, 157del, R158G, L452R, T478K, D614G, P681R, and D950N mutations in spike protein. The variant has now spread in 54 countries including the UK and USA. B.1.617.2 has now become the most dominant VOC of SARS-CoV-2 in England, having a presence of over 90% of all new cases. This variant has shown a 64% increase in transmissibility as compared to variant B.1.1.7.64,65 This VOC also showed reduced neutralization by post-vaccination sera and monoclonal antibody treatments.66
The Delta Plus, which first emerged in India, has now spread to more than 20 countries including the United States, England and Japan. This variant has unique mutation profiles, compared to the delta variant. Besides the signature mutations in spike (G142D, A222V, and T95I), three mutations in spike (K417N, V70F, and W258L) were exclusively found in the Delta Plus variant. Structural analyses showed that another five key mutations (T95I, A222V, G142D, R158G, and K417N) are more frequent in the Delta Plus variant. These mutations may alter the interactions with antibodies and strengthen the structural conformation of the virus. The delta variant is found to be less sensitive to sera from naturally immunized individuals, whereas humoral immune response above the threshold of neutralization was observed with sera from vaccinated pre-infected individuals. Vaccines from Pfizer and AstraZeneca showed three to five-fold lower potency compared to the alpha variant.66
4.6. Omicron variant or B.1.1.529
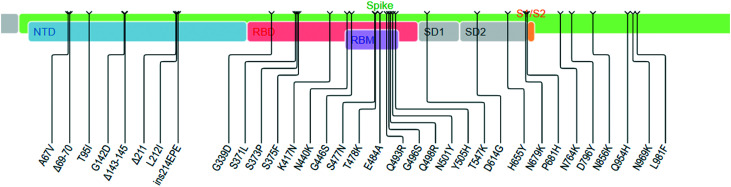
Very recently, another most divergent variant has changed the trajectory of the pandemic after its first identification on 24th November 2021. The WHO designated the new variant as Omicron (or B.1.1.529), on the advice of the WHO's Technical Advisory Group on Virus Evolution. The variant possesses a very high number of mutations in the spike protein resulting in 30 amino acid changes in the spike protein and out of which, 15 are located in the RBD domain. In addition, there are three deletions and three insertions in the N-terminal domain of the spike protein. There is considerable uncertainty about its transmission ability, pathogenesis and the efficacy of the existing vaccines in place. Based on the mutation profile, it is not unlikely that the Omicron may be linked to immune escape (Fig. 3) (https://covdb.stanford.edu/page/mutation-viewer/#omicron). The original variant of SARS-Cov-2 has a R0 of 2.5, while the same for the delta variant (B1.617.2) is around 7 and Omicron's R0 could be as high as 10 (R0 is a mathematical term that indicates how contagious an infectious disease is and is also referred to as the reproduction number). R0 describes the average number of people (who were previously free of infection and have not been vaccinated) who are going to contract a contagious disease from one person with that disease. As Omicron has a R0 value of 10, a person who has the disease will transmit the disease to an average of 10 other people.67 While there is still much to be observed about the transmissibility of Omicron, available data suggest that this variant is significantly more transmissible than the delta variant and also capable of significant immune evasion. However, in a recent study, Zhao et al. found that the replication and fusion activity of the Omicron variant is much less dependent on TMPRSS2, compared to the delta variant which suggests that the Omicron variant may have poor replication activity in the lungs.68 In addition to multiple mutations in the receptor-binding domain (RBD), Omicron also exhibits mutations at the N-terminal domain that are associated with increased infectivity, more efficient cell entry and immune evasion. In the Omicron variant, the SAg motif of SARS-COV-2 has two mutations, N679K and P681H (Fig. 3). Thus the sequence of Omicron-SAg becomes (678)TKSHRRARSVASQ(690). This would significantly alter the polar characteristics and plausibly its interactions with others. But to scrutinize its possible impact on the SAg characteristics, more work in this direction would be required. Although the information is still emerging, it now appears certain that Omicron maybe 2 to 3 times more transmissible than delta.69 Recent studies on the ability of RBD neutralizing antibodies (NAbs) to escape the mutation profile of 247 human anti-RBD Nabs have shown that 85% of the tested NAbs were escaped by Omicron.70 The World Health Organization (WHO) has said that this variant has now spread in 57 countries. The WHO is coordinating with researchers around the world to assess its transmissibility, the severity of infection (including symptoms), the performance of vaccines and diagnostic tests, and the effectiveness of the treatments.
Fig. 3. Mutations in the spike glycoprotein, Omicron variant B.1.1.529 (Source: Stanford University Coronavirus Database).
4.7. Other variants of interest
In addition to these variants, there are some other variants of SARS-CoV-2 that are circulating, have some unique mutations and have increased transmissibility compared to the wild-type variant of SARS-CoV-2. The WHO labeled these variants as zeta or P.2, eta or B.1.525, iota or B.1.526, kappa or B.1.617.1 and Mu or B.1.621 and B.1.621.1 (Table 1).
SARS-CoV-2 variants and their characteristics.
| WHO label of variants | Pango lineage | First reported site | First reported time | Notable major mutation site | Transmissibility compared to non-VOC/VOI | Neutralizing antibody activity compared to non-VOC/VOI |
|---|---|---|---|---|---|---|
| Alpha | B.1.1.7, B.1.1.7 with E484K mutation and Q lineages | UK | September 2020 | D614G, 69–70 del, N501Y, P681H and E484K | +29% | Reduction in the neutralization titers against B.1.1.7 by convalescent sera generated in early strains of SARS-CoV-2 and sera from Pfizer–BioNtech, Moderna, and Oxford-AstraZeneca vaccines |
| Beta | B.1.351 | South Africa | December 2020 | K417N, E484K, N501Y | +25% | B.1.351 is markedly more resistant to neutralization by convalescent plasma (9.4-fold) and sera from individuals who have been vaccinated (10.3–12.4-fold) |
| Gamma | P.1 | Brazil | December 2020 | K417T, E484K, N501Y | +38% | Plasma from individuals previously infected with SARS-CoV-2 had an 8·6 times lower neutralizing capacity against the P.1 isolates and inefficient neutralization of P.1 isolates was seen with plasma samples collected from individuals vaccinated with doses of CoronaVac |
| Delta and Delta+ | B.1.617.2 (Delta) | India | October 2020 | L452R, T478K, P681R | +97% | Less sensitive to sera from naturally immunized individuals, whereas sera from vaccinated pre-infected individuals boosted the humoral immune response to well above the threshold of neutralization. Pfizer and AstraZeneca vaccines showed three- to five-fold lower potency compared to the alpha variant |
| B.1.617.2.1 or AY.1 (Delta+) | K417N, V70F, L452R, T478K and W258L | |||||
| Epsilon | B.1.429 and B.1.427 | California, USA | July 2020 | S13I, W152C, L452R | +19–+24% | Reduced neutralization by convalescent sera and BNT2b2, mRNA1273 vaccine-elicited sera |
| Zeta | P.2 | Rio de Janeiro, Brazil | April and November 2020 | E484K, but not the N501Y and K417T | Not defined | Decreased neutralisation of 5.8-fold for Pfizer–BioNTech COVID-19 vaccine (BNT162b2) and 2.9-fold for Moderna COVID-19 vaccine (mRNA-1273) |
| Eta | B.1.525 | UK | December 2020 | E484K and F888L | +29% | BNT162b2 vaccine-elicited antibodies showed a similar neutralization effect on B.1.1.7 |
| Iota | B.1.526 | New York, USA | November 2020 | E484K, S477N, N501Y | +35% | This variant is partially or completely resistant to two therapeutic monoclonal antibodies in clinical use and less susceptible to neutralization by convalescent plasma or vaccine sera |
| Kappa | B.1.617.1 | India | October 2020 | L452R, D614G, P681R, E484Q | +48% | Showed similar neutralization with the alpha variant against B.1.617.1 with sera of vaccinated individuals by Covaxin or Covid 19 recovered cases |
| Mu | B.1.621 and B.1.621.1 | Colombo, Sri Lanka | January 2021 | E484K, N501Y, P681H, K417, R346K and D950N | Not defined | Mu variant is highly resistant to sera from COVID-19 convalescents and BNT162b2-vaccinated individuals |
| Omicron | B.1.1.529 | South Africa | November 2021 | E484A, Q493R, T478k, N501Y, Q498R | 2–3 times more transmissible than other variants | Recent studies on the ability of RBD neutralizing antibodies (NAbs) of 247 human anti-RBD NAbs have shown that 85% of the tested NAbs were escaped by Omicron |
5. Viral entry
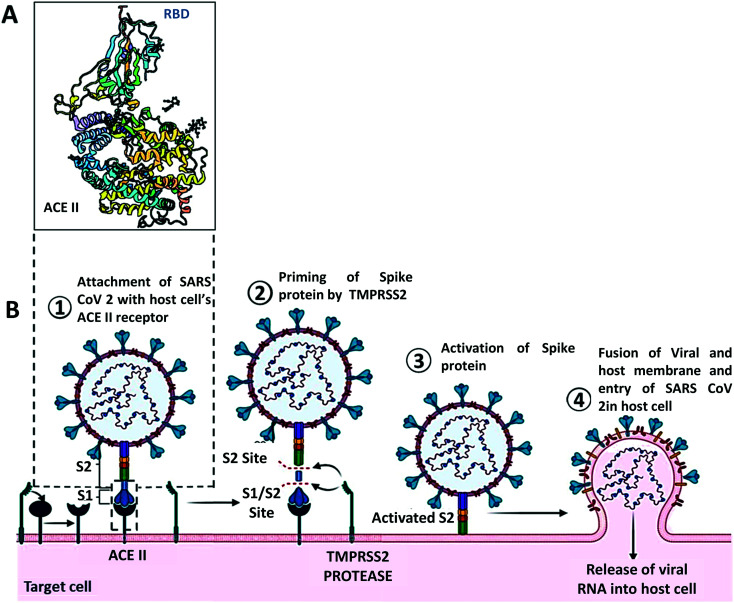
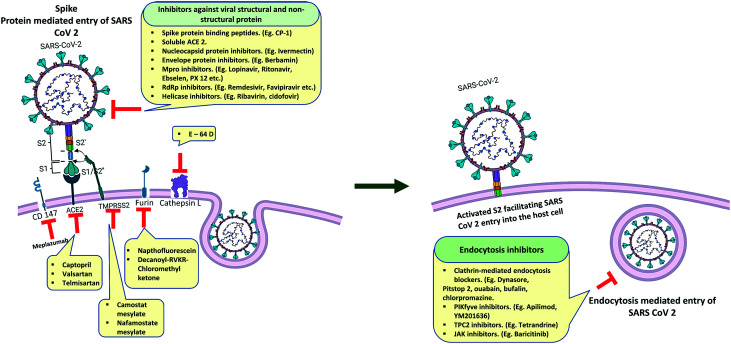
The entry of SARS-CoV-2 is mediated by its spike protein. Like many other coronaviruses, the S glycoprotein of SARS-CoV-2 is cleaved into two subunits S1 and S2 but only when they reach the host cell, whereas the entry glycoprotein of some viruses like HIV-1 and influenza is cleaved into two subunits during their biosynthesis or before the release of the newly formed virus from the host cell. The spike protein of SARS-CoV-2 consists of two subunits, S1 and S2. The S1 subunit binds to ACE 2, whereas the S2 subunit attaches the spike protein to the membrane. Viral entry by glycoproteins engaging receptors needs additional processes that bring some conformational changes in the glycoprotein which helps to form fusion pores by bringing the viral and cellular membranes together. In the case of SARS-CoV-2, the ACE 2 and S protein engagement facilitates the cleavage of an additional site i.e., the S2′ site is internalized to the S2 subunit of S protein71 (Fig. 4). The cleavage of the S2′ site is mediated by TMPRSS2 at the cell surface or cathepsin L in the endosomal compartment ensuring the ACE 2 mediated endocytosis, followed by the release of the fusion peptide composed of 20–25 residues initiating the fusion pore formation. As this pore forms and expands, the viral genome can access the host cell cytoplasm to initiate the process to replicate further.72 Recent studies have shown that there are several additional molecules other than ACE 2 which have been suggested to serve as alternative receptors for SARS-CoV-2. CD147, a transmembrane glycoprotein that is expressed in epithelial and immune cells, has been proposed to be an alternative receptor for SARS-CoV-2 infection.73 Another factor neuropilin 1 (NRP1) has been shown to promote S1 shedding through the furin-cleavage site and expose the S2′ site to TMPRSS2.74 BOAT 1, a natural amino acid transporter, has also shown some possibility to contribute to SARS-CoV-2 infection, although additional studies are needed to confirm its role.75 Other factors like C-type lectins, CD209 (DC-SIGN) and CD209 L (L SIGN) also promote the entry of SARS-CoV-2 in the host cell.76,77
Fig. 4. Structural features and host cell entry mechanism of SARS-CoV-2. A. Interaction between the ACE II receptor and SARS-CoV-2 receptor binding domain. B. Schematic representation of the mechanism of SARS-CoV-2 cell entry.
6. Pathogenesis of host immune response in COVID-19 patients
6.1. Super antigenic entity and cytokine storm
A hyperinflammatory syndrome reminiscent of toxic shock syndrome (TSS) was observed in severe COVID-19 patients and Multisystem Inflammatory Syndrome in children (MIS-C). TSS is typically caused by pathogenic superantigens,78,79 stimulating excessive activation of the adaptive immune system. There are two types of superantigens that can be observed, bacterial or viral. Bacterial superantigens like staphylococcal enterotoxins B and H can bind to major histocompatibility complex (MHC) class II (MHCII) molecules and T cell receptors (TCRs) of both CD4+ and CD8+ T cells separately or together. The characteristics of nonspecific binding of the superantigens with TCRs enable large-scale T cell activation and proliferation which results in huge production of IFNγ, TNFα, and IL-2 from T cells, and IL-1 and TNFα from antigen-presenting cells, leading to a cytokine storm and toxic shock.83,86 It has been reported that SARS-CoV-2 spike(s) glycoprotein contains a sequence and structure motifs highly similar to those of bacterial superantigens Staphylococcus enterotoxins and direct T cell receptors. This interaction between the virus and human T cells could be strengthened by a rare mutation (D839Y/N/E) obtained from a European strain of SARS-CoV. It was hypothesized that the skewed T cell receptor repertoire in COVID-19 patients with severe hyper inflammation may be associated with such a super antigenic effect. Notably, the super antigenic motif is not present in other SARS coronavirus families, which may explain the unique potential of SARS-CoV-2 to cause both MIS-C and the cytokine storm observed in adult COVID-19 patients with an important implication for the development of therapeutic approaches.
6.2. Search for an additional super antigenic motif
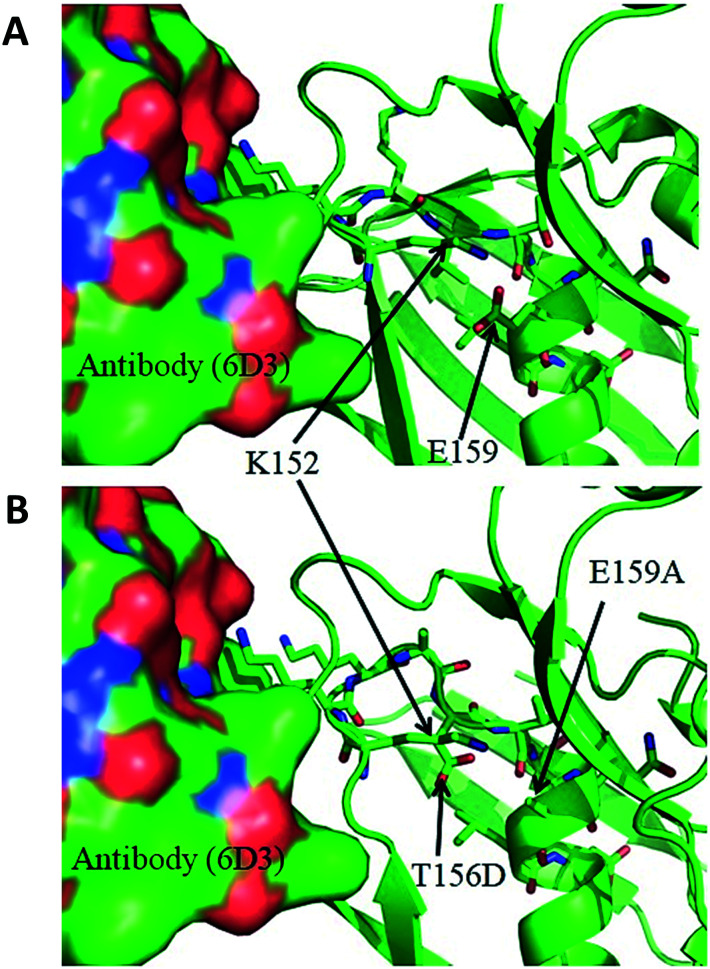
Cheng et al.79 identified the superantigenic motif (SAg) in the spike protein of SARS-CoV-2 as a stretch of residues that has substantial similarity with the superantigenic motif of staphylococcal enterotoxin B (SEB) (Fig. 5). An alignment of the primary sequences of the two motifs, available in the article by Cheng et al.,79 is as follows:SEB SAg: (150)TN-KKKATVQELD(161)SARS-CoV-2 spike: (678)TNSPRRARSVASQ(690)In connection with this, another report by Cheng et al. also suggested that 6D3 (an antibody that neutralizes the superantigenic bacterial toxin SEB) may also be repurposed as a mAb against SARS-COV-2 spike protein, as 6D3 is capable of binding to the similar sequence motifs shared by SEB and SARS CoV2 spike;80 the proposal was substantiated by the available molecular structures and modelling. At the same time, it raises the question, whether similar motifs are also present elsewhere, in the entire set of proteins of SARS-CoV-2 and if such an insert is available elsewhere that could also serve as a backup tool for the virus.
Fig. 5. A. Interaction between 6D3 (in surface representation) and SEB (in ribbon representation), with the side-chains of the SAg motif in sticks, as obtained from the PDB (4RGN). B. The complex after mutating the SAg motif residues to map the NPH residues (373 to 382) into 4RGN.
In pursuit of getting a clue towards this possibility, we carried out a few protein-blast searches81https://blast.ncbi.nlm.nih.gov/Blast.cgi), selecting the non-redundant protein sequences and limiting the search only for COVID-19, while keeping all other search criteria in standard settings. The searches were carried out using two query sequences separately; the first one was the SAg motif of SARS-CoV-2 itself and the second was the motif of the SEB SAg that was used by Cheng et al.79 The search with the motif of the spike protein did not identify any candidate with a significant match, except the spike protein itself and thus confirmed the absence of the same or similar motif elsewhere in SARS-CoV-2 and the same fact was also reported earlier.79 But the search with a subset of the sequence of the SEB SAg79 distilled out the following match with a motiff of the nucleocapsid phosphoprotein (NPH) (GenBank ref: QTE05854.1) of SARS-CoV-2, with 70% identity:SEB Ag = (152)KKKATV-QEL(160)NPHs = (373)KKKADVTQAL(382)The NPH is a structural protein of SARS-CoV-2 and it protects and packs the RNA into the virus82 and is likely to be a lucrative drug target. Obviously, a structural model of the SARS-CoV-2 NPH with or without SAg neutralizing antibodies would bring some more insight, but so far, the authors are aware that there is no such structure available in the PDB; neither is there any suitable template available for constructing a reliable homology model of the NPH. To intuitively create such a structural model, the available coordinates (PDB ID 4RGN)83 of the SEB-6D3 complex were suitably edited to graft the K(373)KKADVTQAL(382) motif of the NPH by replacing the equivalent stretch of residues of SEB. The resulting molecular model (Fig. 5) does not show any steric conflict between residues and the impact of the replacement of the motif seems to be comfortably absorbed. However, this modelling only hints at the possibility of NPH-6D3 (or similar) interactions based on an analogy with the earlier report.80 A quantitative prediction would require more work in this direction.
6.3. Macrophage activation syndrome and cytokine storm
However, the current studies state that most patients become critically ill and die due to activation of macrophages resulting in macrophage activation syndrome which causes the release of a high amount of pro-inflammatory cytokines such as interleukins (IL) leading to a cytokine storm, interferons (IFN), lymphokines, chemokines, tumour necrosis factors (TNF), and several other mediators.84–86 Severely infected patients with acute respiratory distress syndrome (ARDS) have elevated serum levels of IL-1B, IL-IRA, IL-6, IL-7, IL-8, IL-17, IL-9, IL-10, granulocyte-macrophage colony-stimulating factor (GM-CSF), fibroblast growth factor (FGF), interferon-ϒ-inducible protein (IP10), macrophage inflammatory protein-1α (MIP1α), monocyte chemoattractant protein (MCPI), platelet-derived growth factor (PDGF), tumor necrosis factor (TNFα) and vascular endothelial growth factor (VEGF) (Table 2). Hojyo et al. showed that ARDS with cytokine storms is the main cause of death due to COVID-19. Notably, intravascular coagulation is one of the major causes of multiorgan failure, which is mainly mediated by inflammatory cytokines, in particular, IL-6.87 A list of common inflammatory cytokines and their epidemiology has been shown in patients with SARS-CoV-2, compared to SARS-CoV and MERS-CoV.
Common inflammatory cytokines in patients with SARS-CoV-2, compared to SARS-CoV and MERS-CoV.
| Inflammatory cytokines | Expression | ||
|---|---|---|---|
| Covid 19 | MERS | SARS | |
| TNF α | High | High | High |
| IFN | High | Low | High |
| MCPI | High | High | Unknown |
| CRP | High | Not significant | Not significant |
| IL6 | High | High | High |
| IL1 | High | Unknown | High |
| IL17 | High | High | High |
| IL10 | High | High | High |
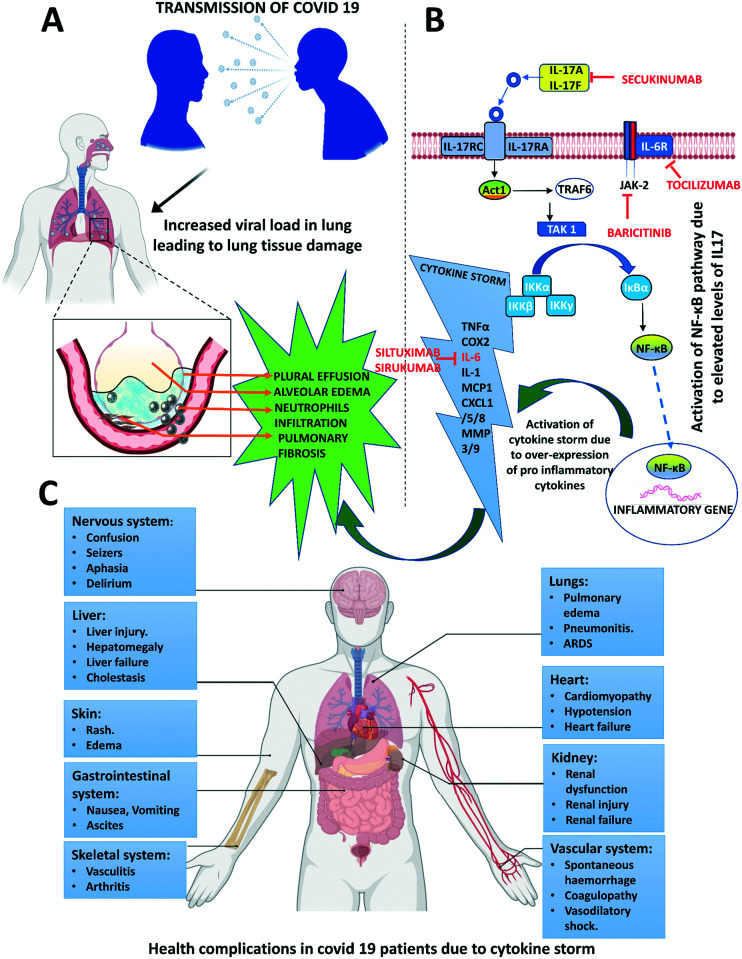
This physical process of a superfluity of cytokines is termed as “cytokine storm” or hypercytokinemia which brings havoc to the body such as septic shock, hemorrhage, heart and lung failure and even multi-system organ failure.88 Cytokines are a set of small extra-cellular signaling polypeptides released by various immune cells including macrophages, lymphocytes and mast cells, as well as other cell types such as endothelial cells for regulating a large number of biological processes such as pro and anti-inflammatory actions, cell proliferation, and innate and acquired immune responses via cell surface receptors.89 During viral infections, immune cells sense the viral particles and stimulate our immune system, releasing various cytokines to fight off the virus, removing the infected cells and repairing the injured tissue so that it does not spread to other cells and this is a normal protective mechanism.88,90 However, when too many immune cells are recruited to do such an activity, then cytokines become too abundant and overproduced resulting in them attacking their own cells and tissues instead of eliminating the virus. Once this process called cytokine storm starts, it is very difficult to switch off, leading to death of the infected person by affecting different parts of the body, attacking healthy cells and tissues, eating up red and white blood cells and damaging the liver, while blood vessels get leaky, the lungs get filled with fluid and blood pressure decreases; blood clots are also observed in the body, finally choking blood flow resulting in septic shock, hemorrhage, heart and lung failure and even multi-system organ failure (Fig. 6).88–90
Fig. 6. IL-17A-mediated activation of different immunopathological factors. A. Covid-19 infection leading to pleural effusion, alveolar oedema, and pulmonary fibrosis. B. Signalling transduction of IL-17A at alveolar epithelial cells of the lung. IL-17 induces inflammation by acting in synergy with IL-6, IL-1 and TNF and by expressing chemokines such as CXCL1, CXCL5, and MMPs as it is not a potent inducer of inflammatory response. Along with other chemokines and cytokines IL 17A generates a powerful inflammatory signal that results in a cytokine storm. C. Complications due to the cytokine storm in Covid 19 patients.
In the case of COVID-19, SARS-CoV-2 enters the host cells, starts replicating which results in the formation of progeny viruses and finally disintegrates the host cells to spread to other cells resulting in the release of a large number of cytokines in the lungs by alveolar macrophages and epithelial cells. These cytokines form a feedback loop to trigger the production of more pro-inflammatory cytokines by inducing macrophages, monocytes, and T cells. In the lung alveoli, IL-17A, IL-17F, IL-21 and IL-22 are produced as signature cytokines by Th17 cells in response to polarizing cytokines secondary to the presence of viral infection (Fig. 6).
6.4. Coagulopathy
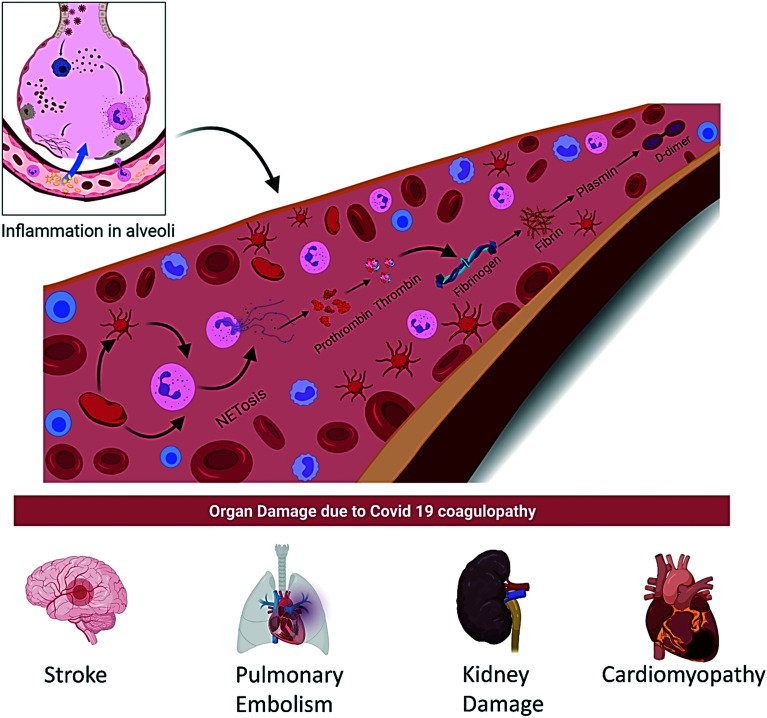
The incidence of thromboembolic disease is highly observed in COVID-19 patients ranging from cutaneous to pulmonary embolism and coronary to cerebral thrombosis. D-dimer and prothrombin have emerged as the most important biomarkers which have been analyzed at the time of hospital admission due to COVID-19. A schematic illustration of the implication of the COVID-19 coagulopathy cascade is shown in Fig. 7.
Fig. 7. Mechanism of coagulopathy disorder in Covid 19 patients and organ damage in Covid 19 patients.
Very recently, certain studies suggested that the effects of supplementation with a biological response modifier (BRM) may help to reduce the clinical severity and mortality due to coagulopathy especially in vulnerable populations namely Caucasian, African American and Hispanic elderly with co-morbidities.91
A study of 178 patients from the Wuhan Huoshenshan Hospital severe disease group showed a significantly lower platelet count but a high D dimer value as compared to the non-severe group. Similarly, the severe group also had highly abnormal other coagulation parameters compared to the non-severe group like prothrombin time (PT), disseminated intravascular coagulation (DIC) rate, etc. In severe COVID-19 patients, the DIC rate has been increased to 6.1% from 0 in the non-severe COVID-19 patients' group. The mechanism of activation of the coagulopathy cascade in COVID-19 patients remains unfathomable.94 However, some hypothesis has emerged about the development of thrombocytopenia in COVID-19 patients: (i) increased platelet destruction and platelet consumption due to intravascular coagulation disturbance; (ii) the decrease in platelet production which may be due to the cytokine storm following the virus infection that leads to the destruction of bone marrow progenitor cells; (iii) platelet aggregation in the lungs, resulting in microthrombi and platelet consumption. Finally, the low platelet count resulted in an increased risk of DIC, severe disease manifestation and also increased mortality rate in COVID-19 patients.92,93
In some cases, it has been observed that the abnormal coagulation parameters in COVID-19 patients increased the risk of gastrointestinal and intracranial hemorrhage that may occur due to the increased usage of anticoagulants.94,95
6.5. Specific race/ethnicity-based risk of coagulopathy
Studies suggest that Caucasians have a higher thrombotic risk compared to other Asian populations. However, studies on the USA population showed that the risk is even higher in African-American and Hispanic patients. Very recently, certain studies reported the effects of supplementation with a biological response modifier (BRM) on the development of clinical severity and reduction of mortality against the background of coagulopathy especially in vulnerable populations namely Caucasian, African American and Hispanic elderly with predisposing factors.96
6.6. Genome-wide association study (GWAS) revealing genes associated with the severity of COVID-19 disease
Genome-wide association study (GWAS) of severe COVID-19 was done to identify potential genetic factors in Italy and Spain, with 835 patients, 1235 control from Italy and 775 patients, 950 control from Spain. A meta-analysis was conducted on the basis of 8 582 968 single-nucleotide polymorphisms.97 The investigating team detected a cross-replicating association with rs11385942 at locus 3p21.31 and rs657152 at locus 9q34.2 in COVID-19 patients with respiratory failure, which was significant at the genome-wide level. Locus 3p21.31 is composed of six genes, i.e., SLC6A20, LZTFL1, CCR9, FYCO1, CXCR6, and XCR1. The insertion–deletion GA or G variant rs11385942 is associated with reduced expression of chemokine receptor gene CXCR6 and increased expression of SLC6A20 (encodes a protein that interacts with ACE II) and LZTFL1 (encodes Bardet–Biedl syndrome (BBS) proteins also known as the BBSome protein complex). The BBSome complex is known to function for sorting a complex of specific membrane proteins to the primary cilia and is also responsible for the regulation of ciliary trafficking of the hedgehog signal transducer.98 The complex also acts as a cargo adapter that recognizes signaling proteins such as GPCRs and links them to the intraflagellar transport machinery.99 The LZTFL1 gene is strongly expressed in human lung cells.100 The association signal at locus 9q34.2 found to be susceptible to the ABO blood group locus showing a higher risk in blood group A than in other blood groups but a protective effect was found in blood group O.97
6.7. Coronavirus disease (COVID-19) associated mucormycosis (CAM)
COVID-19-associated mucormycosis is a fungus infection causing nasal or eye infection in COVID-19 patients.101,102 This is a very rare condition. The most common risk factor among patients was diabetes. Most of the patients developed the condition after hospitalization. Critically ill patients or patients having a longer duration of hospital stays were more likely to develop fungal co-infections.103 Some studies suggested that the extensive use of steroids in COVID-19 management allowed opportunistic fungal infections to colonize due to suppressed immunity.104 Amphotericin B, the most common antifungal drug for the treatment of any kind of mycosis,105 may be used for the treatment of coronavirus disease (COVID-19) associated mucormycosis (CAM). Surgery may also be needed in some cases.
7. Potential therapeutic targets
The therapeutic targets of SARS-CoV-2 (Fig. 8) can be classified into two categories, ‘viral targets’ that are involved in different pathways of the life cycle of the virus, and ‘host targets’ that play some essential role in the viral life cycle and factors that are involved in host immune response.
Fig. 8. Viral and host targets and their inhibitors against SARS-CoV-2 entry in the host cell.
7.1. Viral targets
Most of the proteins of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are potential drug targets but some of them are more important in the aspect of their principal roles in the viral lifecycle and the absence of human protein homologs like the spike protein, papain-like protease (PLpro), chymotrypsin-like main protease (Mpro), and RNA-dependent RNA polymerase (RdRp).106
7.1.1. Structural proteins as targets
SARS-CoV-2 showed a different interaction pattern with ACE 2 compared with SARS CoV.107 A total of 18 residues of the SARS-CoV-2 S receptor-binding domain (RBD) instead of 15 residues of the SARS CoV S RBD were found to interact with the 20 residues of ACE II. Some unique interactions like Q 493 of the S-receptor binding domain and E35 of ACE II and K417 of the S-RBM (receptor binding motif) and D 30 of ACE II allowed better stabilization of the complex compared to the SARS CoV S-RBD and ACE II complex.108
These differences in the interaction may be responsible for the severe transmissible and pandemic nature of the virus. Several monoclonal antibodies (mAbs) that are previously found to be effective against SARS CoV have been investigated to check their activity against the SARS-CoV-2 RBD. But unfortunately, the majority of them were unable to bind to the SARS-CoV-2 RBD.109,110 Meanwhile, a SARS CoV-specific human mAb, CR3022, binds potently with SARS-CoV-2.111 Another mAb targeting the S1 subunit of spike protein inhibited both SARS-CoV-2 and SARS-CoV infections.112
Wang et al. proposed a novel route for virus entry that involves an immunoglobulin-like host-protein CD147 that is previously known for its role in the entry process of Plasmodium falciparum which causes malaria113 which may correlate with the activity of hydroxychloroquine though contradictory against SARS-CoV-2. An anti-CD147 humanized antibody, meplazumab, has preliminarily shown activity for inhibiting virus entry.113 A peptide named CP-1 has been found to block the virus–cell fusion process by binding with S1.114 Lipopeptides EK1C4 and IPB02 which have been designed as fusion inhibitors of pan-coronavirus were found to efficiently inhibit SARS-CoV-2 entry115 (Fig. 8).
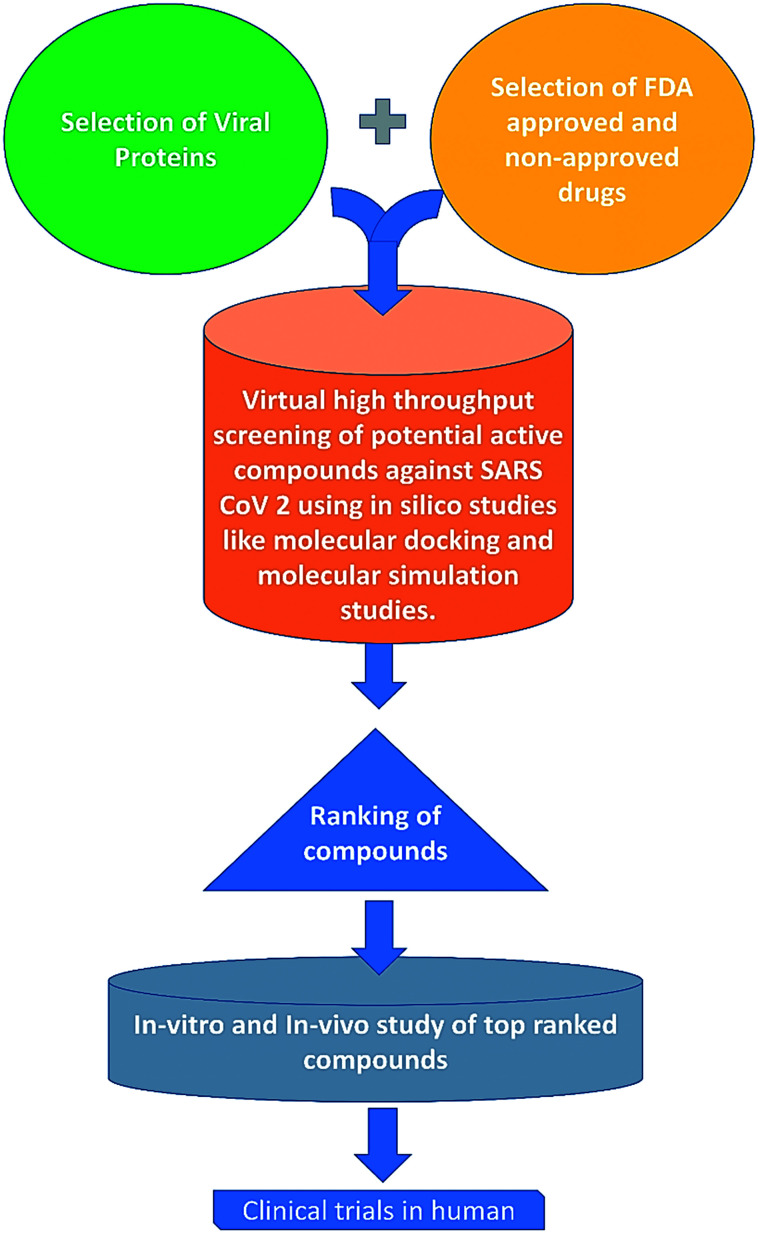
The nucleocapsid protein of SARS-CoV-2 can be a crucial drug target due to its critical function in the viral life cycle.116 In a recent study, Caly et al. showed that the FDA approved drug ivermectin is capable of inhibiting the N protein of SARS-CoV-2. They hypothesized that ivermectin may inhibit the viral replication of SARS-CoV-2 through inhibition of IMPα/β1-mediated nuclear import of viral proteins.117 Some in silico studies also showed that several natural compounds may significantly bind with SARS-CoV-2 nucleocapsid protein.118,119 In the process, top-ranked compounds are not yet tested in humans but docking a repurposing library, with billions of compounds, can result in the discovery of a useful deployable new antiviral compound on cells and in animals (Fig. 9).
Fig. 9. Schematic representation of the probable cascade of drug development against Covid 19. Drug development with an in silico study like docking a repurposing library with billions of compounds can recognize several natural compounds that significantly bind with SARS-CoV-2 proteins and the top-ranked compounds can be tested in humans after successive in vitro and in vivo studies, which can finally lead to the discovery of useful new antiviral compounds against SARS-CoV-2.
The SARS-CoV-2 envelope protein is the most neglected protein among all the structural proteins. In a recent study, SARS-CoV-2 E protein has been shown to form a cation channel that is lethal to host cells. Some natural compounds (BE12 or berbamin, BE30, BE31, BE32, BE33) were also assessed to check their inhibition efficacy on the envelope protein of the virus.120 Brom Ac, a combination therapy of bromelain & acetylcysteine, has also shown a disintegrating effect on the envelope as well as spike protein of SARS-CoV-2.121 In an in silico study, belachinal, macaflavanone E, and vibsanols have also shown an inhibitory effect on SARS-CoV-2 envelope protein.64
Till now no such study has been reported on the membrane protein of SARS-CoV-2, although it can be a good target for vaccine production.
7.1.2. Non-structural proteins as targets
Non-structural proteins (nsps) of SARS-CoV-2 present different crucial functions in the viral life cycle just like other coronaviruses. In this article, we will focus on Mpro, PLpro, RNA dependent RNA polymerase (RdRp) and helicase due to their essential role in the viral life cycle and infectivity.
7.1.2.1. Mpro and PLpro
Yang et al. have already determined the crystal structure of SARS-CoV-2 Mpro and performed structure-based virtual screening of different chemical compounds that are already FDA approved and other active compounds. They have identified eight compounds that can inhibit Mpro.122 Some FDA-approved drugs, ebselen, shikonin, tideglusib, and PX-12, are currently in clinical trials or preclinical studies123 to use against 3CLpro. Lopinavir–ritonavir is already recommended for use against the viral protein.124 In another study, Zhang et al. have designed a range of α-ketoamide inhibitors of SARS-CoV-2 3CLpro. Several in silico studies have also suggested a lot of potential natural compounds and FDA-approved drugs that can bind and inhibit SARS-CoV-2 3CLpro.125–129
Paxlovid™ (PF-07321332 + ritonavir), a new antiviral by Pfizer against Mpro, showed robust antiviral activity even against the current VOC omicron and received EUA (emergency use authorization) from the FDA. PF-07321332 is a novel SARS-CoV-2 MPRO inhibitor with potent oral bioavailability which alone showed promising results in preclinical studies with significantly low toxicity. In order to achieve maximal potency in clinical trials, PF-07321332 was combined with ritonavir, an anti-HIV drug, which slowed the metabolism of PF-07321332 by inhibiting cytochrome 450 enzymes. The interim data of a clinical trial with paxlovid showed significantly reduced COVID-19 related hospitalization requirements. Several other clinical trials with paxlovid are being conducted around the world and the outcomes could suggest full implementation of paxlovid on infected patients.130–132
No in vitro or in vivo study against PLpro has been found during the preparation of this manuscript but several in silico studies have been performed in search of potential compounds against PLpro.133–136
7.1.2.2. RdRp
RdRp is the most crucial enzyme in the RNA virus family as it mediates the transcription and replication of the RNA genome during infection which facilitates the chance to use it as a drug target. Several adenosine analogs like avifavir, remdesivir and favipiravir are already recommended to treat COVID-19 patients.137–140 A molecular study suggested that nucleotide analogs like sofosbuvir, alovudine, tenofovir alafenamide, AZT, abacavir, lamivudine, and emtricitabine can inhibit SARS-CoV-2 RdRp.141 Recently another molecule, molnupiravir (MK-4482, EIDD-2801) or N-hydroxy-5′-O-(2-methyl propanol)-3,4-dihydrocytidine that received EUA from the FDA, showed promising results in prevention of severe SARS-CoV-2 infections when initiated within 5 days of the onset of symptoms and reduced the event of hospitalization and death through day 29 (molnupiravir for oral treatment of COVID-19 in non-hospitalized patients); similar to other nucleoside analogs, this molecule targets SARS-CoV-2 RdRp which in turn inhibits proper replication and transcription.142,143 This antiviral drug candidate hinders viral propagation by inducing lethal mutations in the genome of the virus. In contrast to the established nucleoside analog 5-fluorouracil, molnupiravir is resistant to exoribonuclease-mediated proofreading activity.144 This makes the molecule an attractive drug candidate against COVID-19. Gordon et al. and Kabinger et al. separately investigated the selective incorporation of molnupiravir triphosphate (MTP) in the viral transcript. They found that MTP competes most effectively with CTP for incorporation in the viral RNA.145,146
7.1.2.3. Helicase
Like RdRp, the helicase of SARS-CoV-2 is one of the important enzymes that is involved in viral replication. An in silico drug repurposing study showed that cangrelor, pemetrexed, fludarabine, cidofovir and ribavirin can bind with the ATP binding site of the protein.147 Some other in silico studies also suggested some potential helicase inhibitors against SARS-CoV-2 helicase.147–150
7.2. Host targets
7.2.1. ACE II
Like SARS CoV, SARS-CoV-2 also utilizes ACE II (Fig. 4) as its main receptor to enter the host cell.151 ACE catalyzes the formation of Ang II from Ang I by proteolytic cleavage of Ang I which is an important process in the renin–angiotensin system (RAS) which is involved in the maintenance of blood pressure in mammals.152 ACE 2 is expressed in vascular endothelia, gastrointestinal system, heart, and kidney.153 Several strategies have been hypothesized and devoted to inhibiting the SARS-CoV-2 spike–ACE II interaction. In that sense, though controversial, angiotensin receptor blockers like captopril, valsartan, losartan and telmisartan can be used for COVID-19 therapy.154 The “brace corona clinical trial” on ACE II inhibitors has already suggested that the use of these ACE II blockers is safe for COVID-19 patients.155 Another hypothesis has been proposed, that is, the delivery of soluble recombinant ACE II can be effective for neutralizing the virus–receptor interaction. The proposed mechanism is that the soluble human enzyme ACE II or APN01 will imitate the ACE II receptors present in the organs of the human body which will help to eliminate the virus from the body as a complex with recombinant ACE II receptors.156,157
7.2.2. TMPRSS2
TMPRSS2 is mainly expressed in the gastrointestinal tract, but also in lower levels in several other tissues like in the prostate, colon, stomach, salivary gland, urogenital, and respiratory tracts.158,159 It has been previously known that TMPRSS2 promotes viral spread and pathogenesis by neutralizing antibodies and priming SARS CoV spike protein for virus–cell receptor interaction.153,160 It is now confirmed that SARS-CoV-2 entry is dependent on TMPRSS2 that can be interfered with by the protease inhibitor camostat mesylate.151 Another study from the same researcher group revealed that nafamostat mesylate can block SARS-CoV-2 spike protein activation more efficiently than camostat mesylate.161 Viracept or nelfinavir mesylate, another protease inhibitor prescribed for AIDS, suppresses SARS-CoV-2 entry which may be through inhibition of TMPRSS2.162 Maggio et al. suggested repurposing of the TMPRSS2 protease inhibitor due to the advantage of its pre-established clinical use and low cost compared to camostat mesylate.163 Due to the dependency of TMPRSS2 on androgens, it has been also hypothesized that androgen receptor–inhibitory therapies may reduce the susceptibility to COVID-19. The expression modulation of TMPRSS2 by estrogens and androgens may also correlate with the high infection rate in males.164
7.2.3. Cathepsin L
Cathepsin L is one of the important cysteine lysosomal endopeptidase enzymes of the peptidase C1 family which is involved in the initiation process of protein denaturation.165 Cysteine peptidase inhibitors can be peptide molecules such as pro-peptides and monoclonal antibodies or compounds like aldehydes, ketones, α-keto amides, α-keto-β-aldehydes, α-ketoacids, α-ketoesters, nitriles, azapeptide nitriles, and thiosemicarbazones.166–169 It was previously known that like TMPRSS2, cathepsin L also cleaves SARS-CoV spike protein during the post-receptor-binding stage followed by the virus entry into the host cell.170 In one of the earliest studies on SARS-CoV-2, Hoffmann et al. showed that a CatB/L inhibitor, E-64d, can efficiently block SARS-CoV-2 entry individually in the TMPRSS2− cell line and in combination with camostat mesylate in the TMPRSS2+ cell line.151 In another study, it has been observed that a cathepsin L selective inhibitor, SID26681509, can also reduce the entry of SARS-CoV-2 by more than 76%. Another interesting observation was, SARS-CoV-2 entry is dependent on cathepsin L but not on cathepsin B.171 Some cysteine inhibitors like iCP (Napsul-Ile-Trp-CHO),172 KGP94 (ref. 173) and a tetrahydroquinoline oxocarbazate derivative (CID 23631927)174 are yet to be evaluated against SARS-CoV-2.175
7.2.4. Furin
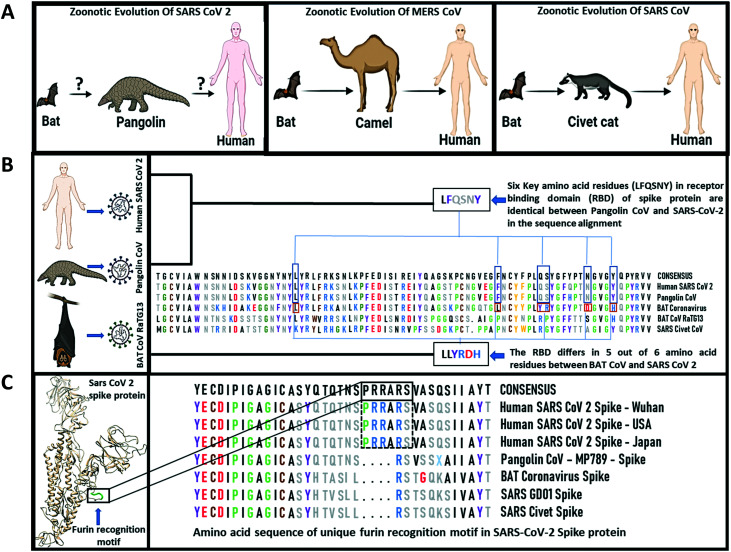
It is now known from some recent studies that the high pathogenicity of SARS-CoV-2 is related to the presence of a “furin-like cleavage site” (FCS) in the S protein25 that makes it unique among the other coronaviruses. Furin, a type 1 membrane-bound protease expressed in multiple tissues, belongs to the subtilisin-like proprotein convertase family.176 This family includes proteases with specific roles in the secretory pathway. The insertion of such cleavage sites in other CoVs, such as the infectious bronchitis virus, increased the pathogenicity, including neural symptoms in infected chickens.177 As furin is highly expressed in the lungs, it is very likely to be involved in SARS-CoV-2 infection, increasing its pathogenicity over other sarbecoviruses, as they lack this cleavage site.178 Recently, it has been proposed that the FCS may be an important site of coronavirus evolution. Mutations that appeared near the FCS (F1–2) region in samples isolated from mild COVID-19 patients from Zhejiang Province, China, showed that it can affect the electrostatic distribution of the S protein surface and its structure, finally reducing its ability to bind to furin. Experimental results in samples from those patients showed that furin had low protein expression levels in the lungs compared with other tissues, such as colon, glands, liver, and kidney.179 Comparison of the S1/S2 cleavage site sequence from pangolin CoV and SARS-CoV-2 shows insertion of the furin recognition motif which contains a sequence (PRRARSV) that mediates fusion of the viral and cell membranes (Fig. 10). The spike protein is cleaved by cell proteases to enable exposure of the fusion sequences. This indicates a distinct mechanism for entry of the viral genome into the host cytoplasm for replication.180 The FCS may contribute to SARS-CoV-2 infection of these organs. Inhibition of furin with peptides and, more recently, with small molecules is a strategy pursued to arrest tumor growth, inflammation, and some viral and bacterial infections.181 However, due to the pleiotropic role of furin-like enzymes in a large number of cellular processes, side effects are a concern.182 Determination of the crystal structure of furin will aid the design of specific small molecules.183 A recent study demonstrated that furin inhibitors like decanoyl-RVKR-chloromethyl ketone (CMK) and naphthofluorescein have antiviral activity on SARS-CoV-2-infected cells by decreasing the viral production load and other cytopathic effects. CMK can block virus entry by suppressing the cleavage of spike proteins, whereas naphthofluorescein primarily suppresses the viral RNA transcription.184
Fig. 10. The origin and features of the spike protein receptor binding domain of SARS-CoV-2. A. Zoonotic evolution of SARS-CoV-2, SARS CoV and MERS CoV. B. Sequence alignment of the spike protein of SARS-CoV-2, bat and pangolin CoV. C. Structure and amino acid sequence of the furin recognition motif observed only in human SARS-CoV-2 spike protein.
7.2.5. Clathrins
Clathrins are triskelion shaped scaffold proteins, composed of three heavy and three light chains.185–187 Clathrin accumulates around a mature vesicle and forms a coat to form clathrin-coated vesicles.188 Clathrin-coated vesicles (CCVs) are involved in the selective transport of membrane-bound proteins by receptor-mediated endocytosis that is essential for several pathways of the intracellular membrane transport system at the trans-Golgi network (TGN).189 Accumulation of phosphatidylinositol-4,5-bisphosphate initiates clathrin complex formation.190,191 The heterotetrameric adaptors, AP1 and AP2, are one of the important components of the clathrin complex that uses cytoplasmic domains to incorporate transmembrane molecules into CCVs. These adaptors are responsible for the selection of cargo through CCVs.192 CCVs also require BAR (bin/amphiphysin/Rvs) domain proteins and act1 to grow.193 In a previous study, it has been observed that SARS-CoV binds to ACE2 in a clathrin-dependent manner.194 Meanwhile, another study showed that SARS CoV enters the host cells through a clathrin-independent endocytic pathway.195 However, in a recent study Bayati et al. showed that SARS-CoV-2 uses clathrin-mediated endocytosis (CME) to enter the host cells. They observed that two previously known (CME) blockers, dynasore and pitstop 2, could reduce the entry of SARS-CoV-2 S protein in the host cell.196 Some FDA-approved drugs like ouabain, bufalin and chlorpromazine are also known to block CME which can be studied further against SARS-CoV-2.197
7.2.6. Phosphatidylinositol 3-phosphate 5-kinase (PIKfyve)
Phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) is the main enzyme that synthesizes phosphatidylinositol 3-phosphate, a phospholipid that regulates the dynamic process of endosome maturation in late endosomes.198 Apilimod199 and YM201636,200 two potent inhibitors of PIKfyve35, were found to significantly suppress the entry mechanism of SARS-CoV-2 in host cells through early endosomes in a dose-dependent manner. A major downstream effector of phosphatidylinositol 3-phosphate i.e., two-pore channel subtype 2 (TPC2)201 is also found to be an important factor for SARS-CoV-2 entry and can be inhibited by tetrandrine (an inhibitor of TPC2).108
7.2.7. Other members of the kinase family
Studies on other kinase (number associated kinase: serine/threonine kinase 16: NAK) family members namely tyrosine kinase inhibitors showed good antiviral activity in vitro.202 JAK (Janus-kinase) inhibitors, i.e., baricitinib, ruxolitinib and fedratinib were shown to inhibit NAK, also limiting systematic inflammatory response and cytokine production through inhibition of the canonical JAK-STAT (signal transducer & activator of transcription) pathway.203 However, baricitinib is the only JAK inhibitor that achieved sufficient plasma concentration to inhibit NAK members at therapeutic and well-tolerated doses.204 Out of the 3 drugs, only ruxolitinib (ChiCTR2000029580) has been recently studied and the recipients showed improved clinical outcomes and faster recovery.205
8. Role of monoclonal antibodies in COVID-19 treatment
Monoclonal antibodies (mAbs) are being used to treat different diseases like infectious diseases, autoimmune diseases and different types of cancers. These antibodies are laboratory-produced peptide molecules that are designed to serve as substitute antibodies that can mimic, modify and even enhance the body's immune response on cells that aren't wanted, such as viruses and cancer cells. The benefits of using these antibodies are they are well characterized and possess strong specificity and binding affinity.206 In the case of COVID-19, in addition to preventive function, mAbs have a beneficial role over vaccines in managing critical patient populations even after exposure to SARS-CoV-2.207 REGN-COV2, a combination of two neutralizing IgG1 mAbs, i.e., basiliximab and imdevimab, that bind two different, non-overlapping sites on the RBD of SARS-CoV-2 spike protein along with placebo showed that this combination is capable of neutralizing SARS-CoV-2 variants with all known S protein mutations. Clinical data showed that the casirivimab and imdevimab combination reduced the high risk of progression to severe COVID-19 and/or hospitalization (3% versus 9%) when compared with placebo (https://investor.regeneron.com/news-releases/news-release-details/regenerons-regn-cov2-antibody-cocktail-reduced-viral-levels-and/). Another IgG mAb i.e., bamlanivimab alone or in combination with another IgG mAb, etesevimab, also decreased the viral load when compared with placebo within day 3 to day 11.208 mAbs against cytokines are also considered in clinical trials to control the uncontrolled way of the response of cytokines in COVID-19 patients. More than hundreds of clinical trials for the repurposing of mAbs against cytokines are reported for the treatment of COVID-19 infected patients from all over the world. The majority of the trials are registered in the USA, France, and Italy.209 Tocilizumab recently received the US FDA approval for a phase III trial (Table 3). Another anti-IL6R antibody, sarilumab, has also been tested in Denmark and the results were similar to those for tocilizumab.210,211
Summary of clinical trials of different monoclonal antibodies on COVID-19 patients.
| Monoclonal antibodies | NCT No. | Title | Clinical trial status | Sample | Study area | Phase |
|---|---|---|---|---|---|---|
| Sarilumab (IL-6 receptors (sIL-6R and mIL-6R) inhibitor) | NCT04315298 | Evaluation of the efficacy and safety of sarilumab in hospitalized patients with COVID-19 | Active, not recruiting | 1912 | USA | III |
| https://clinicaltrials.gov/ct2/show/NCT04315298 | ||||||
| NCT04324073 | Cohort multiple randomized controlled trials open-label of immune modulatory drugs and other treatments in COVID-19 patients – sarilumab trial – CORIMUNO-19 – SARI (CORIMUNO-SARI) | Active, not recruiting | France | III | ||
| https://clinicaltrials.gov/ct2/show/NCT04324073 | ||||||
| NCT04327388 | Sarilumab COVID-19 | Active, not recruiting | Canada | III | ||
| https://clinicaltrials.gov/ct2/show/NCT04327388 | ||||||
| NCT02735707 | Randomized, embedded, multifactorial adaptive platform trial for community-acquired pneumonia (REMAP-CAP) | Recruiting | Australia | IV | ||
| https://clinicaltrials.gov/ct2/show/NCT02735707 | ||||||
| NCT04345289 | Efficacy and safety of novel treatment options for adults with COVID-19 pneumonia (CCAP) | Recruiting | Denmark | III | ||
| https://clinicaltrials.gov/ct2/show/NCT04345289 | ||||||
| Olokizumab | NCT04380519 | Study of the efficacy and safety of a single administration of olokizumab and RPH-104 with standard therapy in patients with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection (COVID-19) | Recruiting | 372 | Russian Federation | III |
| https://clinicaltrials.gov/ct2/show/NCT04380519 | ||||||
| Levilimab | NCT04397562 | A clinical trial of the efficacy and safety of levilimab (BCD-089) in patients with severe COVID-19 (CORONA) | Active, not recruiting | 206 | Russian Federation | III |
| https://clinicaltrials.gov/ct2/show/NCT04397562 | ||||||
| Lenzilumab (GM-CSF inhibitor) | NCT04351152 | Phase 3 study to evaluate the efficacy and safety of lenzilumab in hospitalized patients with COVID-19 pneumonia | Recruiting | 520 | USA | III |
| https://clinicaltrials.gov/ct2/show/NCT04351152 | ||||||
| Mavrilimumab | NCT04447469 | Study of mavrilimumab (KPL-301) in participants hospitalized with severe corona virus disease 2019 (COVID-19) pneumonia and hyper-inflammation | Recruiting | 588 | USA | III |
| https://clinicaltrials.gov/ct2/show/NCT04447469 | ||||||
| 37Canakinumab (IL-1β blocker) | NCT04362813 | Study of the efficacy and safety of canakinumab treatment for CRS in participants with COVID-19-induced pneumonia (CAN-COVID) | Recruiting | 451 | USA | III |
| https://clinicaltrials.gov/ct2/show/NCT04362813 | ||||||
REGN10933  REGN10987 combination therapy REGN10987 combination therapy |
NCT04452318 | Study assessing the efficacy and safety of anti-spike SARS CoV-2 monoclonal antibodies for prevention of SARS CoV-2 | Recruiting | 3750 | USA | III |
| Infection asymptomatic in healthy adults who are household contacts to an individual with a positive SARSCoV-2 RT-PCR assay | ||||||
| https://clinicaltrials.gov/ct2/show/NCT04452318 | ||||||
| Anakinra | NCT04324021 | Efficacy and safety of emapalumab and anakinra in reducing hyperinflammation and respiratory distress in patients with COVID-19 infection | Recruiting | 16 | Italy | III |
| https://clinicaltrials.gov/ct2/show/NCT04324021 | ||||||
| Emapalumab | NCT04324021 | Efficacy and safety of emapalumab and anakinra in reducing hyperinflammation and respiratory distress in patients with COVID-19 infection | Recruiting | 16 | Italy | III |
| https://clinicaltrials.gov/ct2/show/NCT04324021 | ||||||
| Ravulizumab | NCT04390464 | mulTi-arm therapeutic study in pre-ICU patients admitted with Covid-19 – repurposed drugs (TACTIC-R) (TACTIC-R) | Recruiting | 1167 | UK | III |
| https://clinicaltrials.gov/ct2/show/NCT04390464 | ||||||
| Bamlanivimab | NCT04701658 | A real world study of bamlanivimab in participants with mild-to-moderate coronavirus disease 2019 (COVID-19) (BLAZE-5) | Completed | 109 | USA | II |
| Casirivimab + imdevimab | NCT05092581 | COVID-19 study of pharmacokinetics, safety, tolerability, and efficacy of intravenous anti-spike(s) SARS-CoV-2 monoclonal antibodies (casirivimab + imdevimab) for the treatment of pediatric patients hospitalized due to COVID-19 | Active, not recruiting | 40 | USA | I |
| Bamlanivimab and etesevimab or LY3832479 (LY-CoV016) | NCT04497987 | A study of LY3819253 (LY-CoV555) and LY3832479 (LY-CoV016) in preventing SARS-CoV-2 infection and COVID-19 in nursing home residents and staff (BLAZE-2) | Completed | 1374 | USA | III |
9. Vaccines
9.1. Vaccines and the SARS-CoV-2 pandemic
The use of vaccines against different diseases like smallpox, rabies, plague, cholera, typhoid, and hepatitis has improved global health historically and saved several lives during different pandemics. It not only saved animal and human lives but also raised the quality of lives with low treatment costs.212 The goal of vaccine development against SARS-CoV-2 infection is to diminish the effects of the virus on public health, as well as the economy and society. The SARS-CoV-2 pandemic has devastated the most vulnerable in our society, i.e., adults of 65 years of age or older and economically deprived people. The time within which the SARS-CoV-2 mRNA vaccines are developed,213 counting from the publication of the first SARS-CoV-2 sequences through phase 1 in 6 months, is remarkable as compared to a typical timeline of 4 to 10 years. A total of 102 vaccines are in clinical development and 184 vaccine candidates are in the pre-clinical stage.
The Bacillus Calmette–Guérin (BCG) vaccine has been in use since 1921. In some recent studies, an interesting correlation between the BCG vaccinated population and the morbidity & mortality of COVID-19 patients has been shown. It has been found that countries having universal BCG policies like Africa and India showed a good survival rate against COVID-19 compared to countries without universal policies of BCG vaccination like Italy and USA.214,215 Countries with a high BCG vaccinated population also showed a lower incidence of COVID-19, as well as milder illness as compared to countries with a BCG non-vaccinated population.216,217 It has been hypothesized that the cause of the observed “off-target” protection against COVID-19 may be due to the non-specific boosting of innate immunity in BCG-vaccinated individuals.218 However, Hensel et al. showed that there is a correlation between the “current” universal BCG vaccination policy and the lower spread rate and mortality of COVID-19 but the claim has been also contradicted when several other important factors were also taken into consideration.219 Recently Escobar et al. also showed that the BCG vaccine provides protection from the severity of the disease. They provided evidence that the SARS-CoV-2 testing rate is the major confounding factor between the BCG vaccination policy and COVID-19. Additional factors were also included for the study like population density, urban population, smoking rates, the prevalence of diabetes and cardiovascular disease (CVD) with both BCG vaccination policies. The result indicated that the cardiovascular death rate along with the low testing rate was independently associated with high COVID-19 spread rates.220 Hence, there is a warranted need for clinical trials on the BCG vaccine to check its efficacy against SARS-CoV-2.
9.2. COVID-19 vaccine candidates
9.2.1. DNA and RNA vaccines
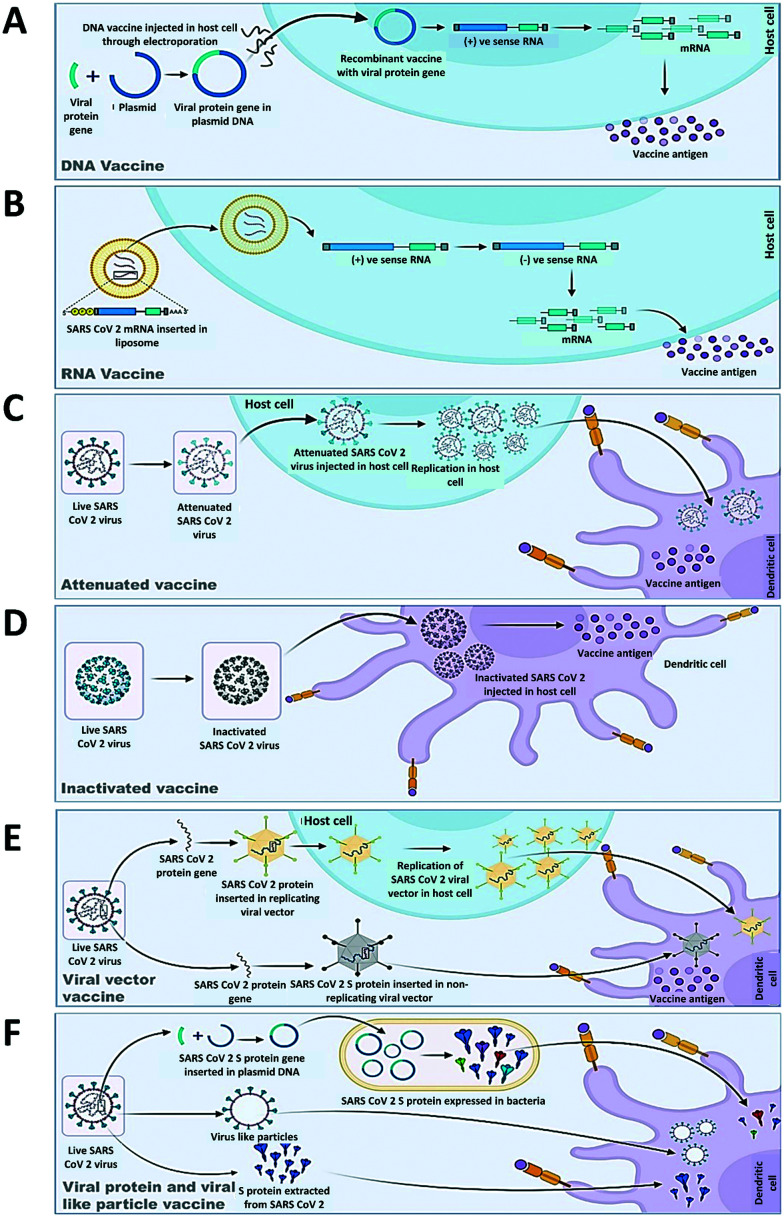
DNA and RNA vaccines (Fig. 11) are generated from the genetic material of viruses. The main advantage of developing these vaccines is that they can be produced on a large scale within a short time. DNA vaccines that can be produced in bacteria are based on plasmid DNA which contains expression promoters specific to mammalian cells and the gene of viral protein like spike or RBD in the case of SARS-CoV-2. Most of the generated RNA vaccines are either modified mRNA or self-replicating RNA. DNA and RNA vaccines both require some delivery agents like bacteria221,222 in the case of DNA and nanoparticles223,224 in the case of RNA vaccines. The disadvantages of these vaccines are that they produce low immunogenicity which ultimately results in multiple doses. There are ten DNA and sixteen RNA vaccines in clinical trials. In phase III, there are two RNA vaccines, in phase II two RNA and three DNA vaccines and two RNA and two DNA vaccines are in the clinical phase till now (Table 4) (https://www.who.int/publications/m/item/draft-landscape-of-COVID-19-candidate-vaccines).
Fig. 11. Mechanism of antigenic response of different vaccines under development against SARS-CoV-2. A. DNA vaccine. B. RNA vaccine. C. Attenuated vaccine. D. Inactivated vaccine. E. Viral vector vaccine. F. Viral protein and viral-like particle vaccine.
Covid-19 vaccine candidates in different clinical trial phases.
| Sl no | Vaccine name | Developer | Vaccine type | Number of doses with schedule and route of administration | Current status | Clinical trial report |
|---|---|---|---|---|---|---|
| 1 | CoronaVac | Sinovac Life Sciences, Beijing, China | Inactivated virus | 2 doses at 14 day interval via intra muscular route | Phase 3 | From phase 2 clinical trial, it was deduced that 3 μg dose of CoronaVac is suggested for assessment in phase 3 trial |
| https://doi.org/10.1016/S1473-3099(20)30843-4 | ||||||
| 2 | ChAdOx1 nCoV-19 vaccine (AZD1222) | AstraZeneca + University of Oxford | Viral vector (non-replicating) | 2 doses at 28 day interval via intra muscular route | Phase 4 | With an overall efficacy of 70.4% in phase 3 clinical trials ChAdOx1 nCoV-19 has been signalled for mass administration and phase 4 trials |
| DOI: https://doi.org/10.1016/S0140-6736(20)32661-1 | ||||||
| 3 | Recombinant novel coronavirus vaccine (adenovirus type 5 vector) | CanSino Biological Inc./Beijing Institute of Biotechnology | Viral vector (non-replicating) | Single dose | Phase 3 | With the administration of this vaccine at 2 different doses (1 × 1011 and 5 × 1010 viral particles) it was found that both doses could induce neutralizing antibody in response to live SARS-CoV-2. The safe dose of 5 × 1010 viral particles induced significant immune response |
| DOI: https://doi.org/10.1016/S0140-6736(20)31605-6 | ||||||
| 4 | Gam-COVID-Vac (Sputnik V) | “Gamaleya Research Institute; Health Ministry of the Russian Federation” | Viral vector (non-replicating) | 2 doses at 21 day interval via intra muscular route | Phase 3 | The interim analysis of a phase 3 clinical trial showed that Sputnik V was 91.6% effective against COVID-19 and was well tolerated in a large cohort |
| https://doi.org/10.1016/S0140-6736(21)00234-8 | ||||||
| 5 | mRNA-1273 | Moderna + National Institute of Allergy and Infectious Diseases (NIAID) | mRNA vaccine | 2 doses at 28 day interval via intra muscular route | Phase 4 | The mRNA-1273 vaccine was capable of preventing severe disease with an overall efficacy of 94.1%. Apart from transient local and systemic reaction it did not elicit any major problems |
| DOI: 10.1056/NEJMoa2035389 | ||||||
| 6 | BNT162b2 (3 LNP-mRNAs), also known as “Comirnaty” | Pfizer/BioNTech + Fosun Pharma | RNA based vaccine | 2 doses at 21 day interval via intra muscular route | Phase 4 | BNT162b2 conferred 95% protection against Covid-19 in persons who are 16 years or older |
| 10.1056/NEJMoa2034577 | ||||||
| 7 | Recombinant SARS-CoV-2 vaccine (CHO cell) | Anhui Zhifei Longcom Biopharmaceutical + Institute of Microbiology, Chinese Academy of Sciences | Protein subunit vaccine | 2 doses at 28 day interval or 3 doses at 28 day interval via intra muscular route | Phase 3 | This protein subunit vaccine was found to be well tolerated and significantly immunogenic. The 25 μg dose in a 3 dose schedule is being studied for its safety and efficacy on a broad scale |
| DOI: https://doi.org/10.1016/S1473-3099(21)00127-4 |
9.2.2. Recombinant protein vaccines (virus-like particle delivering saRNA)
The development of recombinant vaccines (Fig. 11) against SARS-CoV-2 is advantageous over other platforms as they can be made without dealing with this deadly virus and can be expressed in different expression systems like yeast, bacteria, plant and mammalian cells.225–227 Recombinant vaccines that are being developed are based on protein fragments that safely generate an immune response but no disease. There are mainly three types of recombinant vaccines for SARS-CoV-2 that are getting developed namely spike protein-based vaccines, virus-like particle (VLP)-based vaccines and spike protein receptor binding domain-based vaccines. Out of a total of 104 protein subunit vaccines, 71 are in the preclinical stage and 33 are in the clinical phase, out of which two are in phase 3 clinical trials (Table 4) (https://www.who.int/publications/m/item/draft-landscape-of-COVID-19-candidate-vaccines).
9.2.3. Viral vector vaccines (in vitro transcribed saRNA)
There are two types of viral vector-based vaccines (Fig. 11) that are getting developed for SARS-CoV-2, one is replication-competent vector vaccines and another one is replication-incompetent vector vaccines. Replication incompetent vector vaccines are based on another virus like adeno-associated virus (AdV) vectors, modified vaccinia Ankara (MVA) vectors, human parainfluenza virus vectors, influenza virus vectors, etc., which are basically engineered to express the SARS-CoV-2 spike protein and compromised in replication by the deletion of parts from its genome. Replication-competent vectors are typically derived from the attenuated or vaccine strains of engineered viruses that express a transgene-like spike protein. In some cases, animal viruses that do not replicate efficiently and cause no disease in humans are used as well. This technique can induce a more robust immune response as the vector can propagate to some extent in the vaccinated individual and hence can trigger a strong innate immune response. There are six replication-incompetent vector vaccine candidates against SARS-CoV-2 which have progressed far in clinical development, out of which ChAdOx1 nCoV-19 mutually developed by the University of Oxford, AstraZeneca and the Serum Institute of India showed the best efficiency (Table 4) (https://www.who.int/publications/m/item/draft-landscape-of-COVID-19-candidate-vaccines).
9.2.4. Self-amplifying RNA vaccine
This provides amplified and durable production of antigens in vivo, coupled with potent inherent immune-stimulating properties (Fig. 11). This may also require dose-sparing, i.e., to obtain the same immune responses with smaller doses of vaccine. With regard to manufacturability, scale-up production can be a challenge for very long RNA transcripts, such as self-amplifying RNAs, whereas the trans-amplifying approach permits the shorter length of RNA, albeit with two potential drawbacks: the requirement to manufacture two RNAs and the complexity due to the need for efficient in vivo delivery of both RNAs into the same cell. Finally new strategies in mRNA technology228 like nucleoside modifications, stabilization of sequences, and optimization of the codon of the entire replicon gene should be implemented. When the replicase genes were optimized for translational efficiency (and lack of flanking regions), enhanced immunogenicity was observed.
9.2.5. Live attenuated vaccines
Live attenuated vaccines (Fig. 11) are produced by a weakened version of the virus that can be generated by growing the virus under limited conditions like lower temperature and in non-human cells or by genetic modification of the virus-like deleting genes that are responsible for the host innate immune response.229 These vaccines can be given intranasally, which can induce immune responses but no disease. The major disadvantage of these vaccines is that the development of vaccines is very time-consuming. There are three SARS-CoV-2 attenuated vaccines in the developmental stage. Out of which, only one has entered the phase I clinical trial (https://www.who.int/publications/m/item/draft-landscape-of-COVID-19-candidate-vaccines). Inactivated or weakened virus vaccines can be produced by growing the virus in cell culture and then disrupting them with chemicals, heat, detergent, etc.230,231 Their productions are relatively easy and fast as compared to live attenuated viruses, but their yield could be limited depending on the conditions of the cell culture. There are sixteen inactivated vaccine candidates that have entered clinical trials, out of which two are in phase III now (Table 4) (https://www.who.int/publications/m/item/draft-landscape-of-COVID-19-candidate-vaccines).
10. Conclusions
The genetic features of SARS-CoV-2 discussed in this study provide a possible correlation between some unique features of the genome of SARS-CoV-2 as well as the pathogenesis and also the transmission of the disease in humans. We have also reviewed structural druggable viral and host targets that may be used against SARS-CoV-2. The 16 nsps of SARS-CoV-2 have special relevance for viral replication and function which makes them relevant drug targets specially the protease (3CL pro and PL pro), RdRp and helicase. Several studies also pointed the key host proteins related to SARS CoV entry like ACE II TMPRSS2, furin, or proteins that are involved in the regulation of intercellular viral trafficking during endocytic entry which can also be effective drug targets to inhibit SARS-CoV-2 in the host cell including the viral structural or non-structural proteins. A fast approach on research of potential vaccine candidates in this hour of need from all over the world provided several candidates, out of which 17 vaccine candidates (three RNA vaccines from Pfizer–BioNTech, Moderna and Takeda, seven conventional inactivated vaccines i.e., BBIBP-CorV, CoronaVac, Covaxin, WIBP-CorV, CoviVac, Minhai-Kangtai and QazVac, five viral vector vaccines i.e., Sputnik Light, Sputnik V, ChAdOx1, Convidecia, and Ad26.COV2.S, and two protein subunit vaccines i.e., EpiVacCorona and ZIFIVAX) are authorized by national authorities from around the globe for mass vaccination (https://vac-lshtm.shinyapps.io/ncov_vaccine_landscape/). But the key question is how long vaccine immunity will persist as the incidence of reinfection and finally death even after completion of the doses of vaccination is getting reported from all over the world. At this time, as the virus is continuously evolving it is also not clear whether vaccine-induced immune responses can give protection for a long time or not. The emergence of highly virulent and infective variants of SARS-CoV-2 has created queries on the effectiveness of the approved vaccines. Most of the approved vaccines showed inhibiting activity against the variants, but for how long? However, a reduction in immunity over longer periods of time may not be a major obstacle if booster doses of COVID-19 vaccines are given every few years. Another fact that is also not clear is how well older individuals, who are the most at risk from COVID-19, will respond to the vaccines. Due to these uncertainties, focus on different drug targets is needed as they may provide additional therapeutic options besides the COVID-19 vaccines. Our study provides an overview of options in the context of potent drugs and new treatment strategies relating to the management of COVID-19. For a better approach in COVID-19 therapy, we also discussed the importance of machine learning in the management of COVID-19 disease. In addition to the highly transmissible nature of the virus, some socioeconomic and demographic factors have also contributed to the spread of the disease. We have discussed some statistical tools that can also contribute to the better management of the disease.
Abbreviations
- ACE2
Angiotensin-converting enzyme 2
- AdV
Adeno-associated virus
- ARDS
Acute respiratory distress syndrome
- BCG
Bacillus Calmette–Guérin
- BRM
Biological response modifier
- CCV
Clathrin-coated vesicles
- CDC
Centres for Disease Control and Prevention
- CDK
Cyclin dependent kinase
- CME
Clathrin-mediated endocytosis
- CMK
Chloromethylketone
- CoV
Coronavirus
- COVID
Coronavirus disease
- CVD
Cardiovascular disease
- DIC
Disseminated intravascular coagulation
- DNA
Deoxyribonucleic acid
- dsRNA
Double stranded ribonucleic acid
- FCS
Furin-like cleavage site
- GISAID
Global initiative on sharing avian influenza data
- GMP
Guanosine mono phosphate
- IFN
Interferon
- IL
Interleukin
- JAK–STAT
Janus-kinase – signal transducer & activator of transcription
- kb
Kilobase
- kDa
Kilodalton
- mAb
Monoclonal antibody
- MCPI
Monocyte chemoattractant protein
- MERS
Middle East respiratory syndrome
- MHC
Major histocompatibility complex
- MIP1α
Macrophage inflammatory protein-1α
- MPRO(3CLPRO)
Main protease (3C-like protease)
- mRNA
Messenger ribonucleic acid
- MVA
Modified vaccinia Ankara
- NAK
Number associated kinase
- NF-kB
Nuclear factor kappa light chain enhancer of activated B cells
- NHP
Non-human primates
- nm
Nanometre
- NSP
Non-structural proteins
- ORF
Open reading frame
- PDGF
Platelet-derived growth factor
- PHB1 and PHB2
Prohibitin
- PIKfyve
Phosphatidylinositol 3-phosphate 5-kinase
- PLPRO
Papain-like protease
- PRRSV
Porcine reproductive and respiratory syndrome virus
- PT
Prothrombin time
- RdRp
RNA dependent RNA polymerase
- RNA
Ribonucleic acid
- RNP
Ribonucleocapsid
- SAg
Superantigenic motif
- SARS
Severe acute respiratory syndrome
- ssRNA
Single stranded ribonucleic acid
- TCR
T cell receptors
- TGN
trans-Golgi network
- TMPRSS2
Transmembrane protease serine 2
- TNF
Tumor necrosis factor
- TNFα
Tumour necrosis factor
- US FDA
United States Food and Drug Administration
- UTR
Untranslated region
- VEGF
Vascular endothelial growth factor
- VLP
Virus like particle
- VOC
Variants of concern
- WHO
World Health Organization
Conflicts of interest
There is no conflict of interest to declare.
Supplementary Material
Acknowledgments
We would like to acknowledge the Indian Council of Medical Research for sponsoring Emeritus Professorship to Dr. Syamal Roy and the Department of Pharmaceuticals, Ministry of Fertilizer, Government of India for providing the fellowship to Mr. Rudra Chakravarti and Mr. Rajveer Singh. We would also like to acknowledge the DBT, Govt of India [grant number BT/PR26301/GET/119/258/2017], Govt of West Bengal Department of Biotechnology WBDBT [grant number–BT/P/Budget/RD-74/2017] for their support and funding.
References
- Petrosillo N. Viceconte G. Ergonul O. Ippolito G. Petersen E. Clin. Microbiol. Infect. 2020;26:729–734. doi: 10.1016/j.cmi.2020.03.026. doi: 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. Horby P. W. Hayden F. G. Gao G. F. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. Hu B. Hu C. Zhu F. Liu X. Zhang J. Wang B. Xiang H. Cheng Z. Xiong Y. Zhao Y. Li Y. Wang X. Peng Z. JAMA, J. Am. Med. Assoc. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N. Zhang D. Wang W. Li X. Yang B. Song J. Zhao X. Huang B. Shi W. Lu R. Niu P. Zhan F. Ma X. Wang D. Xu W. Wu G. Gao G. F. Tan W. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C. Li X. Liu S. Sang Y. Gao S. J. Gao F. Emerging Microbes Infect. 2020;9:378–381. doi: 10.1080/22221751.2020.1727299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E. Baker S. C. Baric R. S. de Groot R. J. Drosten C. Gulyaeva A. A. Haagmans B. L. Lauber C. Leontovich A. M. Neuman B. W. Penzar D. Perlman S. Poon L. L. M. Samborskiy D. Sidorov I. A. Sola I. Ziebuhr J. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward H., Atchison C., Whitaker M., Ainslie K. E. C., Elliott J., Okell L., Redd R., Ashby D., Donnelly C. A., Barclay W., Darzi A., Cooke G., Riley S. and Elliott P., medRxiv, 2020, 10.1101/2020.08.12.20173690 [DOI] [PMC free article] [PubMed]
- Chen N. Zhou M. Dong X. Qu J. Gong F. Han Y. Qiu Y. Wang J. Liu Y. Wei Y. Xia J. Yu T. Zhang X. Zhang L. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V. M. Lienau J. Witzenrath M. Internist. 2019;60:1136–1145. doi: 10.1007/s00108-019-00671-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin D. A. Gulick R. M. Martinez F. J. N. Engl. J. Med. 2020;383:2451–2460. doi: 10.1056/NEJMcp2009575. [DOI] [PubMed] [Google Scholar]
- Marini J. J. Gattinoni L. JAMA, J. Am. Med. Assoc. 2020;323:2329–2330. doi: 10.1001/jama.2020.6825. [DOI] [PubMed] [Google Scholar]
- Zhou P. Yang X. L. Wang X. G. Hu B. Zhang L. Zhang W. Si H. R. Zhu Y. Li B. Huang C. L. Chen H. D. Chen J. Luo Y. Guo H. Jiang R. D. Liu M. Q. Chen Y. Shen X. R. Wang X. Zheng X. S. Zhao K. Chen Q. J. Deng F. Liu L. L. Yan B. Zhan F. X. Wang Y. Y. Xiao G. F. Shi Z. L. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder A. Arora M. Bharadiya V. Berry P. Agarwal M. Behera P. Shewade H. D. Lohiya A. Gupta M. Rao A. Parameswaran G. G. F1000Research. 2020;9:315. doi: 10.12688/f1000research.23496.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z. Lian X. Su X. Wu W. Marraro G. A. Zeng Y. Respir. Res. 2020;21:1–14. doi: 10.1186/s12931-020-01479-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M. Swerdlow D. L. Finelli L. N. Engl. J. Med. 2020;382:1194–1196. doi: 10.1056/NEJMp2002125. [DOI] [PubMed] [Google Scholar]
- Almeida J. D. Berry D. M. Cunningham C. H. Hamre D. Hofstad M. S. Mallucci L. McIntosh K. Nature. 1968;220:650. [Google Scholar]
- Lalchhandama K. Sci. Vis. 2020;20:78–92. [Google Scholar]
- Lai M. M. Cavanagh D. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D. Cagliani R. Clerici M. Sironi M. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton J. T., Segmented double-stranded RNA viruses: structure and molecular biology, Horizon Scientific Press, 2008 [Google Scholar]
- Nguyen M. Haenni A. L. Virus Res. 2003;93:141–150. doi: 10.1016/S0168-1702(03)00094-7. [DOI] [PubMed] [Google Scholar]
- Machhi J. Herskovitz J. Senan A. M. Dutta D. Nath B. Oleynikov M. D. Blomberg W. R. Meigs D. D. Hasan M. Patel M. Kline P. Chang R. C. C. Chang L. Gendelman H. E. Kevadiya B. D. J. Neuroimmune Pharmacol. 2020;15:359–386. doi: 10.1007/s11481-020-09944-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A. C. P. Li X. Lau S. K. P. Woo P. C. Y. Viruses. 2019:11. doi: 10.3390/v11020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A. C. Park Y. J. Tortorici M. A. Wall A. McGuire A. T. Veesler D. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B. Valle C. de Lamballerie X. Canard B. Seidah N. G. Decroly E. Antiviral Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi A. A. T. Fatima K. Mohammad T. Fatima U. Singh I. K. Singh A. Atif S. M. Hariprasad G. Hasan G. M. Hassan M. I. Biochim. Biophys. Acta, Mol. Basis Dis. 2020;1866:165878. doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky V. N. and Longhi S., Flexible Viruses: Structural Disorder in Viral Proteins, John Wiley and Sons, 2011 [Google Scholar]
- Sørensen B. Susrud A. Dalgleish A. G. Q. Rev. Biophys. Discov. 2020;1:e6. doi: 10.1017/qrd.2020.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S. Millet J. K. Licitra B. N. Whittaker G. R. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. Maurya V. K. Prasad A. K. Bhatt M. L. B. Saxena S. K. Virusdisease. 2020;31:13–21. doi: 10.1007/s13337-020-00571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto F. K. Protein J. 2020;39:198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. Chen H. Wang H. Front. Mol. Biosci. 2021;8:629873. doi: 10.3389/fmolb.2021.629873. doi: 10.3389/fmolb.2021.629873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres J. L. DeDiego M. L. Verdiá-Báguena C. Jimenez-Guardeño J. M. Regla-Nava J. A. Fernandez-Delgado R. Castaño-Rodriguez C. Alcaraz A. Torres J. Aguilella V. M. Enjuanes L. PLoS Pathog. 2015;425:330–339. doi: 10.1371/journal.ppat.1004077. [DOI] [Google Scholar]
- Lim K. P. Liu D. X. J. Biol. Chem. 2001;276:17515–17523. doi: 10.1074/jbc.M009731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruch T. R. Machamer C. E. Viruses. 2012;4:363–382. doi: 10.3390/v4030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerbeck J. W. Machamer C. E. J. Virol. 2019;93:e00015-19. doi: 10.1128/jvi.00015-19. doi: 10.1128/jvi.00015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdiá-Báguena C. Nieto-Torres J. L. Alcaraz A. DeDiego M. L. Torres J. Aguilella V. M. Enjuanes L. Virology. 2012;432:485–494. doi: 10.1016/j.virol.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B. W. Kiss G. Kunding A. H. Bhella D. Baksh M. F. Connelly S. Droese B. Klaus J. P. Makino S. Sawicki S. G. Siddell S. G. Stamou D. G. Wilson I. A. Kuhn P. Buchmeier M. J. J. Struct. Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu Y. L. Teoh K. T. Lo J. Chan C. M. Kien F. Escriou N. Tsao S. W. Nicholls J. M. Altmeyer R. Peiris J. S. M. Bruzzone R. Nal B. J. Virol. 2008;82:11318–11330. doi: 10.1128/JVI.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald M. E. Fehr A. R. Athmer J. Perlman S. Virology. 2018;517:62–68. doi: 10.1016/j.virol.2017.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M. Liu B. Chow V. T. K. Lal S. K. J. Biol. Chem. 2006;281:10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeski E. T. Moradi S. Raoufi R. Shahlaei M. Ali M. Janlou M. Zolghadri S. J. Biomol. Struct. Dyn. 2021;39:4633–4646. doi: 10.1080/07391102.2020.1779133. doi: 10.1080/07391102.2020.1779133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signorile A. Sgaramella G. Bellomo F. De Rasmo D. Cell. 2019;8:71. doi: 10.3390/cells8010071. doi: 10.3390/cells8010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Y. Liao C. H. Wang Q. Tan Y. J. Luo R. Qiu Y. Ge X. Y. Virus Res. 2020;286:198074. doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y. Shu T. Wu D. Mu J. Wang C. Huang M. Han Y. Zhang X.-Y. Zhou W. Qiu Y. Zhou X. Cell. Mol. Immunol. 2020;17:881–883. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y. J. Chiba S. Halfmann P. Ehre C. Kuroda M. Dinnon K. H. Leist S. R. Schäfer A. Nakajima N. Takahashi K. Lee R. E. Mascenik T. M. Graham R. Edwards C. E. Tse L. V. Okuda K. Markmann A. J. Bartelt L. De Silva A. Margolis D. M. Boucher R. C. Randell S. H. Suzuki T. Gralinski L. E. Kawaoka Y. Baric R. S. Science. 2021;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves D. C. Rowland-Jones S. L. Angyal A. Biochem. Biophys. Res. Commun. 2021;538:104–107. doi: 10.1016/j.bbrc.2020.10.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L. Wang X. Pascal K. E. Tomkins-Tinch C. Nyalile T. Wang Y. Baum A. Diehl W. E. Dauphin A. Carbone C. Veinotte K. Egri S. B. Schaffner S. F. Lemieux J. E. Munro J. Rafique A. Barve A. Sabeti P. C. Kyratsous C. A. Dudkina N. Shen K. Luban J. Cell. 2020;183:739–751. doi: 10.1016/j.cell.2020.09.032. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J. A. Liu Y. Liu J. Xia H. Johnson B. A. Lokugamage K. G. Zhang X. Muruato A. E. Zou J. Fontes-Garfias C. R. Mirchandani D. Scharton D. Bilello J. P. Ku Z. An Z. Kalveram B. Freiberg A. N. Menachery V. D. Xie X. Plante K. S. Weaver S. C. Shi P. Y. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Loman N., Pybus O., Barclay W., Barrett J., Carabelli A., Connor T., Peacock T., Robertson D. and Volz E., Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations - SARS-CoV-2 coronavirus/nCoV-2019 Genomic Epidemiology - Virological, https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-SARS-CoV-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563, (accessed 20 May 2021)
- Harrington D. Kele B. Pereira S. Couto-Parada X. Riddell A. Forbes S. Dobbie H. Cutino-Moguel T. Clin. Infect. Dis. 2021;73:1946–1947. doi: 10.1093/cid/ciab014. doi: 10.1093/cid/ciab014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Werner A. P., Moliva J. I., Koch M., Choi A., Stewart-Jones G. B. E., Bennett H., Boyoglu-Barnum S., Shi W., Graham B. S., Carfi A., Corbett K. S., Seder R. A. and Edwards D. K., bioRxiv Prepr. Serv. Biol., 2021, vol. 73, pp. 1946–1947, 10.1101/2021.01.25.427948 [DOI]
- Supasa P. Zhou D. Dejnirattisai W. Liu C. Mentzer A. J. Ginn H. M. Zhao Y. Duyvesteyn H. M. Nutalai R. Tuekprakhon A. Wang B. Cell. 2021;184:2201–2211. doi: 10.1016/j.cell.2021.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horby P., Huntley C., Davies N., Edmunds J., Ferguson N., Medley G. and Semple C., NERVTAG https//assets. Publ. Serv. gov. uk/government/uploads/system/uploads/attachment_data/file/955239/NERVTAG_paper_on_variant_of_concern__VOC__B
- Zhou D. Dejnirattisai W. Supasa P. Liu C. Mentzer A. J. Ginn H. M. Zhao Y. Duyvesteyn H. M. E. Tuekprakhon A. Nutalai R. Wang B. Paesen G. C. Lopez-Camacho C. Slon-Campos J. Hallis B. Coombes N. Bewley K. Charlton S. Walter T. S. Skelly D. Lumley S. F. Dold C. Levin R. Dong T. Pollard A. J. Knight J. C. Crook D. Lambe T. Clutterbuck E. Bibi S. Flaxman A. Bittaye M. Belij-Rammerstorfer S. Gilbert S. James W. Carroll M. W. Klenerman P. Barnes E. Dunachie S. J. Fry E. E. Mongkolsapaya J. Ren J. Stuart D. I. Screaton G. R. Cell. 2021;184:2348–2361.e6. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P. D., Graham B. S., Mascola J. R., Chang J. Y., Yin M. T., Sobieszczyk M., Kyratsous C. A., Shapiro L., Sheng Z., Nair M. S., Huang Y. and Ho D. D., bioRxiv Prepr. Serv. Biol., 2021, 10.1101/2021.01.25.428137 [DOI]
- Greaney A. J. Loes A. N. Crawford K. H. D. Starr T. N. Malone K. D. Chu H. Y. Bloom J. D. Cell Host Microbe. 2021;29:463–476.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. Schmidt F. Weisblum Y. Muecksch F. Barnes C. O. Finkin S. Schaefer-Babajew D. Cipolla M. Gaebler C. Lieberman J. A. Oliveira T. Y. Yang Z. Abernathy M. E. Huey-Tubman K. E. Hurley A. Turroja M. West K. A. Gordon K. Millard K. G. Ramos V. Da Silva J. Xu J. Colbert R. A. Patel R. Dizon J. Unson-O'Brien C. Shimeliovich I. Gazumyan A. Caskey M. Bjorkman P. J. Casellas R. Hatziioannou T. Bieniasz P. D. Nussenzweig M. C. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edara V. V. Norwood C. Floyd K. Lai L. Davis-Gardner M. E. Hudson W. H. Mantus G. Nyhoff L. E. Adelman M. W. Fineman R. Patel S. Cell Host Microbe. 2021;29:516–521. doi: 10.1016/j.chom.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. Nair M. S. Liu L. Iketani S. Luo Y. Guo Y. Wang M. Yu J. Zhang B. E. Kwong P. D. Graham B. S. Mascola J. R. Chang J. Y. Yin M. T. Sobieszczyk M. Kyratsous C. A. Shapiro L. Sheng Z. Huang Y. Ho D. D. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Voloch C. M., da Silva Francisco R., de Almeida L. G. P., Cardoso C. C., Brustolini O. J., Gerber A. L., Guimarães A. P. D. C., Mariani D., da Costa R. M., Ferreira O. C., Cavalcanti A. C., Frauches T. S., de Mello C. M. B., Galliez R. M., Faffe D. S., Castiñeiras T. M. P. P., Tanuri A., de Vasconcelos A. T. R., Chieppe A. O., Ribeiro M. S., de Carvalho Cardoso S. C., de Farias Figueiredo J., De Souza L. M., de Nogueira V. G., Nascimento F. F., Filho J. P. M., de Carvalho Leitão I., Castiñeiras A. C. P., Ferreira R. C., da Silva B. O., de Oliveira H. D. A., de Lira G. S. A., Scheid H. T., Renault L. Z., Correa L. A., dos Santos Durães M., Coelho R. F., Ota V. A., Bastos V. C., Medronho R., da Silva L. D. R., de Senna Costa C. A., Pansera C., dos Santos Rodrigues C., Costa A. R., de Souza W. R., de Azevedo M. C. V. M., Costa L. J., Herlinger A. L., Gonçalves C. C. A., Policarpo C., dos Santos Nascimento É. R., dos Santos F. L., Moreira F. R. R., Schiffler F. B., de Almeida J. M., Fortuna J. T. S., Boullosa L. T., de Paula Tôrres M. C., da Costa M. D. A. F., da Paz P. H. C., da Cruz T. F. C., Salviano R. J. B., Ciapina L. P., Souza R. C., dos Santos Correa É., da Silva B. Z., da Fonseca G. C., Klôh V. P., Wagner E. and de Faria Cavalcante L. T., medRxiv, 2020, 10.1101/2020.12.23.20248598 [DOI]
- Nonaka C. K. V. Franco M. M. Gräf T. de Lorenzo Barcia C. A. de Ávila Mendonça R. N. de Sousa K. A. F. Neiva L. M. C. Fosenca V. Mendes A. V. A. de Aguiar R. S. Giovanetti M. de Freitas Souza B. S. Emerging Infect. Dis. 2021;27:1522–1524. doi: 10.3201/eid2705.210191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm A. Toptan T. Pallas C. Wolf T. Goetsch U. Gottschalk R. Vehreschild M. J. Ciesek S. Widera M. Viruses. 2021;13:1693. doi: 10.3390/v13091693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav P. D., Sapkal G. N., Abraham P., Ella R., Deshpande G., Patil D. Y., Nyayanit D. A., Gupta N., Sahay R. R., Shete A. M., Panda S., Bhargava B. and Mohan V. K., bioRxiv, 2021, 10.1101/2021.04.23.441101 [DOI]
- Hester Allen A., Vusirikala A., Flannagan J., Twohig K. A., Zaidi A., Harris R., Charlett A., Dabrera G., Kall M. and Author M. affiliations, Increased household transmission of COVID-19 cases associated with SARS-CoV-2 Variant of Concern B.1.617.2: a national case-control study
- Planas D. Veyer D. Baidaliuk A. Staropoli I. Guivel-Benhassine F. Rajah M. M. Planchais C. Porrot F. Robillard N. Puech J. Prot M. Gallais F. Gantner P. Velay A. Le Guen J. Kassis-Chikhani N. Edriss D. Belec L. Seve A. Courtellemont L. Péré H. Hocqueloux L. Fafi-Kremer S. Prazuck T. Mouquet H. Bruel T. Simon-Lorière E. Rey F. A. Schwartz O. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Burki T. K. Lancet Respir. Med. 2021;2213–2600(21):00559. doi: 10.1016/S2213-2600(21)00559-2. [DOI] [Google Scholar]
- Zhao H. Lu L. Peng Z. Chen L. L. Meng X. Zhang C. Ip J. D. Chan W. M. Chu A. W. Chan K. H. Jin D. Y. Chen H. Yuen K. Y. To K. K. Emerging Microbes Infect. 2021:1–18. doi: 10.1080/22221751.2021.2023329. [DOI] [PMC free article] [PubMed] [Google Scholar]; , Epub ahead of print. PMID: 34951565
- del Rio C. Omer S. B. Malani P. N. JAMA, J. Am. Med. Assoc. 2022;327:319–320. doi: 10.1001/jama.2021.24315. doi: 10.1001/jama.2021.24315. [DOI] [PubMed] [Google Scholar]
- Cao Y. Wang J. Jian F. Xiao T. Song W. Yisimayi A. Huang W. Li Q. Wang P. An R. Wang Y. Nature. 2021 doi: 10.1038/s41586-021-04385-3. [DOI] [Google Scholar]
- Hoffmann M. Kleine-Weber H. Pöhlmann S. Mol. Cell. 2020;78:779–784. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J. Wan Y. Luo C. Gang Y. Geng Q. Auerbach A. Li F. Proc. Natl. Acad. Sci. U. S. A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. Mi L. Xu J. Yu J. Wang X. Jiang J. Xing J. Shang P. Qian A. Li Y. Shaw P. X. Wang J. Duan S. Ding J. Fan C. Zhang Y. Yang Y. Yu X. Feng Q. Li B. Yao X. Zhang Z. Li L. Xue X. Zhu P. J. Infect. Dis. 2005;191:755–760. doi: 10.1086/427811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L. Ojha R. Pedro L. D. Djannatian M. Franz J. Kuivanen S. van der Meer F. Kallio K. Kaya T. Anastasina M. Smura T. Levanov L. Szirovicza L. Tobi A. Kallio-Kokko H. Österlund P. Joensuu M. Meunier F. A. Butcher S. J. Winkler M. S. Mollenhauer B. Helenius A. Gokce O. Teesalu T. Hepojoki J. Vapalahti O. Stadelmann C. Balistreri G. Simons M. Science. 2020;370:856. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo S. M. R. Singer D. Makrides V. Huggel K. Pos K. M. Wagner C. A. Kuba K. Danilczyk U. Skovby F. Kleta R. Penninger J. M. Verrey F. Gastroenterology. 2009;136:872. doi: 10.1053/j.gastro.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers S. A. Tusell S. M. Gillim-Ross L. Hemmila E. M. Achenbach J. E. Babcock G. J. Thomas W. D. Thackray L. B. Young M. D. Mason R. J. Ambrosino D. M. Wentworth D. E. DeMartini J. C. Holmes K. V. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.-Y. Huang Y. Ganesh L. Leung K. Kong W.-P. Schwartz O. Subbarao K. Nabel G. J. J. Virol. 2004;78:5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouo T. Chaisawangwong W. J. Clin. Invest. 2021;131:e149327. doi: 10.1172/JCI149327. doi: 10.1172/JCI149327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M. H. Zhang S. Porritt R. A. Rivas M. N. Paschold L. Willscher E. Binder M. Arditi M. Bahar I. Proc. Natl. Acad. Sci. U. S. A. 2020;117:25254–25262. doi: 10.1073/pnas.2010722117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M. H. Porritt R. A. Rivas M. N. Krieger J. M. Ozdemir A. B. Garcia G. Arumugaswami V. Fries B. C. Arditi M. Bahar I. Structure. 2021;29:951–962.e3. doi: 10.1016/j.str.2021.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F. Madden T. L. Schäffer A. A. Zhang J. Zhang Z. Miller W. Lipman D. J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. Ye Q. Singh D. Cao Y. Diedrich J. K. Yates J. R. Villa E. Cleveland D. W. Corbett K. D. Nat. Commun. 2021;12:502. doi: 10.1038/s41467-020-20768-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta K. Varshney A. K. Franklin M. C. Goger M. Wang X. Fries B. C. J. Biol. Chem. 2015;290:6715. doi: 10.1074/jbc.M114.630715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C. Zhou L. Hu Z. Zhang S. Yang S. Tao Y. Xie C. Ma K. Shang K. Wang W. Tian D. S. Clin. Infect. Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakauer T. Toxins. 2019:11. [Google Scholar]
- Burgos-Blasco B. Güemes-Villahoz N. Santiago J. L. Fernandez-Vigo J. I. Espino-Paisán L. Sarriá B. García-Feijoo J. Martinez-de-la-Casa J. M. Exp. Eye Res. 2020;200:108253. doi: 10.1016/j.exer.2020.108253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojyo S. Uchida M. Tanaka K. Hasebe R. Tanaka Y. Murakami M. Hirano T. Inflammation Regener. 2020;40:37. doi: 10.1186/s41232-020-00146-3. doi: 10.1186/s41232-020-00146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L. Lu L. Cao W. Li T. Emerging Microbes Infect. 2020;9:727–732. doi: 10.1080/22221751.2020.1746199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costela-Ruiz V. J. Illescas-Montes R. Puerta-Puerta J. M. Ruiz C. Melguizo-Rodríguez L. Cytokine Growth Factor Rev. 2020;54:62–75. doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. Wang Y. Li X. Ren L. Zhao J. Hu Y. Zhang L. Fan G. Xu J. Gu X. Cheng Z. Yu T. Xia J. Wei Y. Wu W. Xie X. Yin W. Li H. Liu M. Xiao Y. Gao H. Guo L. Xie J. Wang G. Jiang R. Gao Z. Jin Q. Wang J. Cao B. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikewaki N. Rao K. S. Archibold A. D. Iwasaki M. Senthilkumar R. Preethy S. Katoh S. Abraham S. J. K. Thromb. J. 2020;18:1–8. doi: 10.1186/s12959-020-00239-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao C. Tao X. Cui W. Yi B. Pan T. Young K. H. Qian W. Exp. Hematol. Oncol. 2020;9:1–8. doi: 10.1186/s40164-019-0157-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P. Zhou Q. Xu J. Ann. Hematol. 2020;99:1205–1208. doi: 10.1007/s00277-020-04019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti C. B. Henchi S. Coppeta G. P. Testa S. Grassia R. Eur. J. Intern. Med. 2020;77:147–149. doi: 10.1016/j.ejim.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T. A. Wan D. W. Hajifathalian K. Tewani S. Shah S. L. Mehta A. Kaplan A. Ghosh G. Choi A. J. Krisko T. I. Fortune B. E. Crawford C. V. Sharaiha R. Z. Am. J. Gastroenterol. 2020;115:1609–1616. doi: 10.14309/ajg.0000000000000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikewaki N. Rao K. S. Archibold A. D. Iwasaki M. Senthilkumar R. Preethy S. Katoh S. Abraham S. J. K. Thromb. J. 2020;18:1–8. doi: 10.1186/s12959-020-00239-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- S. C.-19 G. Groupe N. Engl. J. Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S. Zhang Q. Bugge K. Breslow D. K. Searby C. C. Nachury M. V. Sheffield V. C. PLoS Genet. 2011;7:1002358. doi: 10.1371/journal.pgen.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink B. U. Gatsogiannis C. Hofnagel O. Wittinghofer A. Raunser S. eLife. 2020;9:e53910. doi: 10.7554/eLife.53910. doi: 10.7554/eLife.53910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q. Chen Z. H. Wang L. Zhang T. Duan L. Behrens C. Wistuba I. I. Minna J. D. Gao B. Luo J. H. Liu Z. P. Oncogene. 2016;35:2655–2663. doi: 10.1038/onc.2015.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg D. Muthu V. Sehgal I. S. Ramachandran R. Kaur H. Bhalla A. Puri G. D. Chakrabarti A. Agarwal R. Mycopathologia. 2021;186:289–298. doi: 10.1007/s11046-021-00528-2. doi: 10.1007/s11046-021-00528-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S. Gokhale T. Choudhury S. Deb A. Indian J. Ophthalmol. 2021;69:1002–1004. doi: 10.4103/ijo.IJO_3763_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. Yu Y. Xu J. Shu H. Xia J. Liu H. Wu Y. Zhang L. Yu Z. Fang M. Yu T. Wang Y. Pan S. Zou X. Yuan S. Shang Y. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangneux J. P. Bougnoux M. E. Dannaoui E. Cornet M. Zahar J. R. J. Mycol. Med. 2020;30:100971. doi: 10.1016/j.mycmed.2020.100971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza A. C. O. Amaral A. C. Front. Microbiol. 2017:8. doi: 10.3389/fmicb.2017.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks J. M. Smith J. C. N. Engl. J. Med. 2020;382:2261–2264. doi: 10.1056/NEJMcibr2007042. [DOI] [PubMed] [Google Scholar]
- Lan J. Ge J. Yu J. Shan S. Zhou H. Fan S. Zhang Q. Shi X. Wang Q. Zhang L. Wang X. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Gil C. Ginex T. Maestro I. Nozal V. Barrado-Gil L. Cuesta-Geijo M. Á. Urquiza J. Ramírez D. Alonso C. Campillo N. E. Martinez A. J. Med. Chem. 2020;63:12359–12386. doi: 10.1021/acs.jmedchem.0c00606. [DOI] [PubMed] [Google Scholar]
- Wang C. Li W. Drabek D. Okba N. M. A. van Haperen R. Osterhaus A. D. M. E. van Kuppeveld F. J. M. Haagmans B. L. Grosveld F. Bosch B. J. Nat. Commun. 2020;11:1–6. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. Zhang Y. Wu L. Niu S. Song C. Zhang Z. Lu G. Qiao C. Hu Y. Yuen K. Y. Wang Q. Zhou H. Yan J. Qi J. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X. Li C. Huang A. Xia S. Lu S. Shi Z. Lu L. Jiang S. Yang Z. Wu Y. Ying T. Emerging Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. Li R. Pan Z. Qian C. Yang Y. You R. Zhao J. Liu P. Gao L. Li Z. Huang Q. Xu L. Tang J. Tian Q. Yao W. Hu L. Yan X. Zhou X. Wu Y. Deng K. Zhang Z. Qian Z. Chen Y. Ye L. Cell. Mol. Immunol. 2020;17:647–649. doi: 10.1038/s41423-020-0426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Chen W., Zhou Y.-S., Lian J.-Q., Zhang Z., Du P., Gong L., Zhang Y., Cui H.-Y., Geng J.-J., Wang B., Sun X.-X., Wang C.-F., Yang X., Lin P., Deng Y.-Q., Wei D., Yang X.-M., Zhu Y.-M., Zhang K., Zheng Z.-H., Miao J.-L., Guo T., Shi Y., Zhang J., Fu L., Wang Q.-Y., Bian H., Zhu P. and Chen Z.-N., bioRxiv, 2020, 10.1101/2020.03.14.988345 [DOI]
- Liu S. Xiao G. Chen Y. He Y. Niu J. Escalante C. R. Xiong H. Farmar J. Debnath A. K. Tien P. Jiang S. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S. Liu M. Wang C. Xu W. Lan Q. Feng S. Qi F. Bao L. Du L. Liu S. Qin C. Sun F. Shi Z. Zhu Y. Jiang S. Lu L. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S. Yang M. Hong Z. Zhang L. Huang Z. Chen X. He S. Zhou Z. Zhou Z. Chen Q. Yan Y. Zhang C. Shan H. Chen S. Acta Pharm. Sin. B. 2020;10:1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly L. Druce J. D. Catton M. G. Jans D. A. Wagstaff K. M. Antiviral Res. 2020;178:104787. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolta R. Yadav R. Salaria D. Trivedi S. Imran M. Sourirajan A. Baumler D. J. Dev K. J. Biomol. Struct. Dyn. 2020:1. doi: 10.1080/07391102.2020.1850364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández J. F. Lavecchia M. J. J. Biomol. Struct. Dyn. 2020;28:1–8. doi: 10.1080/07391102.2020.1860828. doi: 10.1080/07391102.2020.1860828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B. Shen X. He Y. Pan X. Wang Y. Yang F. Fang S. Wu Y. Zuo X. Xie Z. Jiang X. Chi H. Meng Q. Zhou H. Zhou Y. Cheng X. Chen T. Xin X. Jiang H. Xiao G. Zhao Q. Zhang L.-K. Shen J. Li J. Gao Z. Cell Res. 2021;31:847–860. doi: 10.1101/2020.06.27.174953. doi: 10.1101/2020.06.27.174953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhter J., Pillai K., Badar S., Mekkawy A., Valle S. J. and Morris D. L., bioRxiv, 2020, 10.1101/2020.09.07.286906 [DOI]
- Jin Z. Du X. Xu Y. Deng Y. Liu M. Zhao Y. Zhang B. Li X. Zhang L. Peng C. Duan Y. Yu J. Wang L. Yang K. Liu F. Jiang R. Yang X. You T. Liu X. Yang X. Bai F. Liu H. Liu X. Guddat L. W. Xu W. Xiao G. Qin C. Shi Z. Jiang H. Rao Z. Yang H. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Nutho B. Mahalapbutr P. Hengphasatporn K. Pattaranggoon N. C. Simanon N. Shigeta Y. Hannongbua S. Rungrotmongkol T. Biochemistry. 2020;59:1769–1779. doi: 10.1021/acs.biochem.0c00160. [DOI] [PubMed] [Google Scholar]
- Wu C. Liu Y. Yang Y. Zhang P. Zhong W. Wang Y. Wang Q. Xu Y. Li M. Li X. Zheng M. Chen L. Li H. Acta Pharm. Sin. B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. Lin D. Kusov Y. Nian Y. Ma Q. Wang J. Von Brunn A. Leyssen P. Lanko K. Neyts J. De Wilde A. Snijder E. J. Liu H. Hilgenfeld R. J. Med. Chem. 2020;63:4562–4578. doi: 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- Singh R. Gautam A. Chandel S. Sharma V. Ghosh A. Dey D. Roy S. Ravichandiran V. Ghosh D. In Silico Pharmacol. 2021;9:27. doi: 10.1007/s40203-021-00089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth D. S. N. B. K. Murahari M. Chandramohan V. Panda S. P. Atmakuri L. R. Guntupalli C. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2021.1965027. [DOI] [PubMed] [Google Scholar]
- Singh R. Gautam A. Chandel S. Ghosh A. Dey D. Roy S. Ravichandiran V. Ghosh D. Molecules. 2020;25:4604. doi: 10.3390/molecules25204604. doi: 10.3390/molecules25204604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S. Sarmah S. Lyndem S. Singha Roy A. J. Biomol. Struct. Dyn. 2021;39:3347–3357. doi: 10.1080/07391102.2020.1763201. doi: 10.1080/07391102.2020.1763201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D. R. Allerton C. M. Anderson A. S. Aschenbrenner L. Avery M. Berritt S. Boras B. Cardin R. D. Carlo A. Coffman K. J. Dantonio A. Science. 2021;374:1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- Cully M. Nat. Rev. Drug Discovery. 2021;21:3–5. doi: 10.1038/d41573-021-00202-8. doi: 10.1038/d41573-021-00202-8. [DOI] [PubMed] [Google Scholar]
- Pfizer's novel COVID-19 oral antiviral treatment candidate reduced risk of hospitalization or death by 89% in interim analysis of phase 2/3 EPIC-HR study. Accessed December 30, 2021. https://www.pfizer.com/news/press-release/press-release-detail/pfizers-novel-covid-19-oral-antiviral-treatment-candidate
- Naidoo D. Roy A. Kar P. Mutanda T. Anandraj A. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1794972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya R., Das A., Prashar V. and Kumar M., ChemRxiv, 2020, 10.26434/chemrxiv.11860011.v2 [DOI]
- Amin S. A. Ghosh K. Gayen S. Jha T. J. Biomol. Struct. Dyn. 2020:1. doi: 10.1080/07391102.2020.1780946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza M. U. Ahmad S. Abdullah I. Froeyen M. Comput. Biol. Chem. 2020;89:107376. doi: 10.1016/j.compbiolchem.2020.107376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Zhang D. Du G. Du R. Zhao J. Jin Y. Fu S. Gao L. Cheng Z. Lu Q. Hu Y. Luo G. Wang K. Lu Y. Li H. Wang S. Ruan S. Yang C. Mei C. Wang Y. Ding D. Wu F. Tang X. Ye X. Ye Y. Liu B. Yang J. Yin W. Wang A. Fan G. Zhou F. Liu Z. Gu X. Xu J. Shang L. Zhang Y. Cao L. Guo T. Wan Y. Qin H. Jiang Y. Jaki T. Hayden F. G. Horby P. W. Cao B. Wang C. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal U. Raju R. Udwadia Z. F. Med. J. Armed Forces India. 2020;76:370–376. doi: 10.1016/j.mjafi.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashchenko A. A. Dmitriev K. A. Vostokova N. V. Azarova V. N. Blinow A. A. Egorova A. N. Gordeev I. G. Ilin A. P. Karapetian R. N. Kravchenko D. V. Lomakin N. V. Merkulova E. A. Papazova N. A. Pavlikova E. P. Savchuk N. P. Simakina E. N. Sitdekov T. A. Smolyarchuk E. A. Tikhomolova E. G. Yakubova E. V. Ivachtchenko A. V. Clin. Infect. Dis. 2021;73:531–534. doi: 10.1093/cid/ciaa1176. doi: 10.1093/cid/ciaa1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A. Arunachalam R. Elango A. J. Pharmacol. Pharmacother. 2020;11:1–7. doi: 10.4103/jpp.JPP_105_20. [DOI] [Google Scholar]
- Chen W. H. Tao X. Agrawal A. S. Algaissi A. Peng B. H. Pollet J. Strych U. Bottazzi M. E. Hotez P. J. Lustigman S. Du L. Jiang S. Tseng C. T. K. Vaccine. 2020;38:7533–7541. doi: 10.1016/j.vaccine.2020.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T. P. Sims A. C. Zhou S. Graham R. L. Pruijssers A. J. Agostini M. L. Leist S. R. Schäfer A. Dinnon 3rd K. H. Stevens L. J. Chappell J. D. Lu X. Hughes T. M. George A. S. Hill C. S. Montgomery S. A. Brown A. J. Bluemling G. R. Natchus M. G. Saindane M. Kolykhalov A. A. Painter G. Harcourt J. Tamin A. Thornburg N. J. Swanstrom R. Denison M. R. Baric R. S. Sci. Transl. Med. 2020;12:eabb5883. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl A. Gralinski L. E. Johnson C. E. Yao W. Kovarova M. Dinnon III K. H. Liu H. Madden V. J. Krzystek H. M. De C. White K. K. Gully K. Schäfer A. Zaman T. Leist S. R. Grant P. O. Bluemling G. R. Kolykhalov A. A. Natchus M. G. Askin F. B. Painter G. Browne E. P. Jones C. D. Pickles R. J. Baric R. S. Garcia J. V. Nature. 2021;591:451–457. doi: 10.1038/s41586-021-03312-w. doi: 10.1038/s41586-021-03312-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M. L. Pruijssers A. J. Chappell J. D. Gribble J. Lu X. Andres E. L. Bluemling G. R. Lockwood M. A. Sheahan T. P. Sims A. C. Natchus M. G. Saindane M. Kolykhalov A. A. Painter G. R. Baric R. S. Denison M. R. J. Virol. 2019;93:e01348. doi: 10.1128/JVI.01348-19. doi: 10.1128/JVI.01348-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C. J. Tchesnokov E. P. Schinazi R. F. Götte M. J. Biol. Chem. 2021;297:100770. doi: 10.1016/j.jbc.2021.100770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabinger F. Stiller C. Schmitzová J. Dienemann C. Kokic G. Hillen H. S. Höbartner C. Cramer P. Nat. Struct. Mol. Biol. 2021;28:740–746. doi: 10.1038/s41594-021-00651-0. doi: 10.1038/s41594-021-00651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugurel O. M. Mutlu O. Sariyer E. Kocer S. Ugurel E. Inci T. G. Ata O. Turgut-Balik D. Int. J. Biol. Macromol. 2020;163:1687–1696. doi: 10.1016/j.ijbiomac.2020.09.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza M. U. Froeyen M. J. Pharm. Anal. 2020;10:320–328. doi: 10.1016/j.jpha.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung A. B. Gene Rep. 2020;21:100860. doi: 10.1016/j.genrep.2020.100860. doi: 10.1016/j.genrep.2020.100860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis Borgio J. Alsuwat H. S. Al Otaibi W. M. Ibrahim A. M. Almandil N. B. Al Asoom L. I. Salahuddin M. Kamaraj B. AbdulAzeez S. Arch. Med. Sci. 2020;16:508–518. doi: 10.5114/aoms.2020.94567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M. Kleine-Weber H. Schroeder S. Krüger N. Herrler T. Erichsen S. Schiergens T. S. Herrler G. Wu N. H. Nitsche A. Müller M. A. Drosten C. Pöhlmann S. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K. Imai Y. Ohto-Nakanishi T. Penninger J. M. Pharmacol. Ther. 2010;128:119–128. doi: 10.1016/j.pharmthera.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A. B. Verma A. JAMA, J. Am. Med. Assoc. 2020;323:1769–1770. doi: 10.1001/jama.2020.8946. [DOI] [PubMed] [Google Scholar]
- Rothlin R. P. Vetulli H. M. Duarte M. Pelorosso F. G. Drug Dev. Res. 2020;81:768–770. doi: 10.1002/ddr.21679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes R. D. Macedo A. V. S. de Barros e Silva P. G. M. Moll-Bernardes R. J. Feldman A. D'Andréa Saba Arruda G. de Souza A. S. de Albuquerque D. C. Mazza L. Santos M. F. Salvador N. Z. Gibson C. M. Granger C. B. Alexander J. H. de Souza O. F. Am. Heart J. 2020;226:49–59. doi: 10.1016/j.ahj.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haschke M. Schuster M. Poglitsch M. Loibner H. Salzberg M. Bruggisser M. Penninger J. Krähenbühl S. Clin. Pharmacokinet. 2013;52:783–792. doi: 10.1007/s40262-013-0072-7. [DOI] [PubMed] [Google Scholar]
- Monteil V. Kwon H. Prado P. Hagelkrüys A. Wimmer R. A. Stahl M. Leopoldi A. Garreta E. Hurtado del Pozo C. Prosper F. Romero J. P. Wirnsberger G. Zhang H. Slutsky A. S. Conder R. Montserrat N. Mirazimi A. Penninger J. M. Cell. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y. Atlas Genet. Cytogenet. Oncol. Haematol. 2010;14:1163–1165. doi: 10.4267/2042/44922. [DOI] [Google Scholar]
- Vaarala M. H. Porvari K. S. Kellokumpu S. Kyllönen A. P. Vihko P. T. J. Pathol. 2001;193:134–140. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH743>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Glowacka I. Bertram S. Muller M. A. Allen P. Soilleux E. Pfefferle S. Steffen I. Tsegaye T. S. He Y. Gnirss K. Niemeyer D. Schneider H. Drosten C. Pohlmann S. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M. Schroeder S. Kleine-Weber H. Müller M. A. Drosten C. Pöhlmann S. Antimicrob. Agents Chemother. 2020;64:e00754-20. doi: 10.1128/AAC.00754-20. doi: 10.1128/AAC.00754-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musarrat F. Chouljenko V. Dahal A. Nabi R. Chouljenko T. Jois S. D. Kousoulas K. G. J. Med. Virol. 2020;92:2087–2095. doi: 10.1002/jmv.25985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio R. Corsini G. U. Pharmacol. Res. 2020:157. doi: 10.1016/j.phrs.2020.104837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopsack K. H. Mucci L. A. Antonarakis E. S. Nelson P. S. Kantoff P. W. Cancer Discovery. 2020;10:779–782. doi: 10.1158/2159-8290.CD-20-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett A. J. Buttle D. J. Mason R. W. ISI Atlas Sci.: Biochem. 1988;1:256–260. [Google Scholar]
- Kos J. Mitrović A. Mirković B. Future Med. Chem. 2014;6:1355–1371. doi: 10.4155/fmc.14.73. [DOI] [PubMed] [Google Scholar]
- Pogorzelska A. Żołnowska B. Bartoszewski R. Biochimie. 2018;151:85–106. doi: 10.1016/j.biochi.2018.05.023. [DOI] [PubMed] [Google Scholar]
- Dana D. Pathak S. K. Molecules. 2020:25. doi: 10.3390/molecules25030698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaket T. P. Kwon T. K. Kang S. C. Pharmacol. Ther. 2019;198:1–19. doi: 10.1016/j.pharmthera.2019.02.003. [DOI] [PubMed] [Google Scholar]
- Bosch B. J. Bartelink W. Rottier P. J. M. J. Virol. 2008;82:8887–8890. doi: 10.1128/JVI.00415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X. Liu Y. Lei X. Li P. Mi D. Ren L. Guo L. Guo R. Chen T. Hu J. Xiang Z. Mu Z. Chen X. Chen J. Hu K. Jin Q. Wang J. Qian Z. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbaa A. Chu F. Sudha T. Gallati C. Dier U. Dyskin E. Yalcin M. Bianchini C. Shaker O. Mousa S. A. Anticancer Res. 2009;29:4473–4481. [PubMed] [Google Scholar]
- Sudhan D. R. Siemann D. W. Clin. Exp. Metastasis. 2013;30:891–902. doi: 10.1007/s10585-013-9590-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P. P. Wang T. Kaletsky R. L. Myers M. C. Purvis J. E. Jing H. Huryn D. M. Greenbaum D. C. Smith A. B. Bates P. Diamond S. L. Mol. Pharmacol. 2010;78:319–324. doi: 10.1124/mol.110.064261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pišlar A. Mitrovic A. Sabotič J. Fonovic U. P. Nanut M. P. Jakoš T. Senjor E. Kos J. PLoS Pathog. 2020:16. doi: 10.1371/journal.ppat.1009013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K. Biochem. J. 1997;327:625–635. doi: 10.1042/bj3270625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J. Zhao Y. Hu Y. Zhao J. Xue J. Zhang G. J. Virol. 2021;95:e02447-20. doi: 10.1128/jvi.02447-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segreto R. Deigin Y. McCairn K. Sousa A. Sirotkin D. Sirotkin K. Couey J. J. Jones A. Zhang D. Environ. Chem. Lett. 2021:1–15. doi: 10.1007/s10311-021-01211-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X. Xu K. Jiang P. Lian J. Hao S. Yao H. Jia H. Zhang Y. Zheng L. Zheng N. Chen D. Yao J. Hu J. Gao J. Wen L. Shen J. Ren Y. Yu G. Wang X. Lu Y. Yu X. Yu L. Xiang D. Wu N. Lu X. Cheng L. Liu F. Wu H. Jin C. Yang X. Qian P. Qiu Y. Sheng J. Liang T. Li L. Yang Y. Emerging Microbes Infect. 2020;9:1474–1488. doi: 10.1080/22221751.2020.1781551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K. G. Rambaut A. Lipkin W. I. Holmes E. C. Garry R. F. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture F. Kwiatkowska A. Dory Y. L. Day R. Expert Opin. Ther. Pat. 2015;25:379–396. doi: 10.1517/13543776.2014.1000303. [DOI] [PubMed] [Google Scholar]
- Ivanova T. Hardes K. Kallis S. Dahms S. O. Than M. E. Künzel S. Böttcher-Friebertshäuser E. Lindberg I. Jiao G. S. Bartenschlager R. Steinmetzer T. ChemMedChem. 2017;12:1953–1968. doi: 10.1002/cmdc.201700596. [DOI] [PubMed] [Google Scholar]
- Dahms S. O. Hardes K. Becker G. L. Steinmetzer T. Brandstetter H. Than M. E. ACS Chem. Biol. 2014;9:1113–1118. doi: 10.1021/cb500087x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y. W. Chao T. L. Li C. L. Chiu M. F. Kao H. C. Wang S. H. Pang Y. H. Lin C. H. Tsai Y. M. Lee W. H. Tao M. H. Ho T. C. Wu P. Y. Jang L. T. Chen P. J. Chang S. Y. Yeh S. H. Cell Rep. 2020;33:108254. doi: 10.1016/j.celrep.2020.108254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T. Trends Cell Biol. 2009;19:596–605. doi: 10.1016/j.tcb.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter Haar E. Musacchio A. Harrison S. C. Kirchhausen T. Cell. 1998;95:563–573. doi: 10.1016/S0092-8674(00)81623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edeling M. A. Smith C. Owen D. Nat. Rev. Mol. Cell Biol. 2006;7:32–44. doi: 10.1038/nrm1786. [DOI] [PubMed] [Google Scholar]
- McMahon H. T. Boucrot E. Nat. Rev. Mol. Cell Biol. 2011;12:517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- Brodsky F. M., Chen C. Y., Knuehl C., Towler M. C. and Wakeharn D. E., in Annual Review of Cell and Developmental Biology, Annu Rev Cell Dev Biol, 2001, vol. 17, pp. 517–568 [DOI] [PubMed] [Google Scholar]
- Mousavi S. A. Malerød L. Berg T. Kjeken R. Biochem. J. 2004;377:1–16. doi: 10.1042/bj20031000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost M. Simpson F. Kavran J. M. Lemmon M. A. Schmid S. L. Curr. Biol. 1998;8:1399–1404. doi: 10.1016/S0960-9822(98)00022-0. [DOI] [PubMed] [Google Scholar]
- Haucke V., in Biochemical Society Transactions, Biochem Soc Trans, 2005, vol. 33, pp. 1285–1289 [DOI] [PubMed] [Google Scholar]
- Kumari S. Mg S. Mayor S. Cell Res. 2010;20:256–275. doi: 10.1038/cr.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y. Tanaka N. Tanaka Y. Inoue S. Morita K. Zhuang M. Hattori T. Sugamura K. J. Virol. 2007;81:8722–8729. doi: 10.1128/JVI.00253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. Yang P. Liu K. Guo F. Zhang Y. Zhang G. Jiang C. Cell Res. 2008;18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayati A., Kumar R., Francis V. and McPherson P., bioRxiv, 2020, 10.1101/2020.07.13.201509 [DOI]
- Yang N. Shen H. M. Int. J. Biol. Sci. 2020;16:1724–1731. doi: 10.7150/ijbs.45498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K. Marsland R. Upadhyayula S. Song E. Dang S. Capraro B. R. Wang W. Skillern W. Gaudin R. Ma M. Kirchhausen T. Nature. 2017;552:410–414. doi: 10.1038/nature25146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson E. A. Dyall J. Hoenen T. Barnes A. B. Zhou H. Liang J. Y. Michelotti J. Dewey W. H. DeWald L. E. Bennett R. S. Morris P. J. Guha R. Klumpp-Thomas C. McKnight C. Chen Y. C. Xu X. Wang A. Hughes E. Martin S. Thomas C. Jahrling P. B. Hensley L. E. Olinger G. G. White J. M. PLoS Neglected Trop. Dis. 2017;11:e0005540. doi: 10.1371/journal.pntd.0005540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta-Geijo M. A. Galindo I. Hernáez B. Quetglas J. I. Dalmau-Mena I. Alonso C. PLoS One. 2012;7:48853. doi: 10.1371/journal.pone.0048853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintzer A. F. Stroud R. M. FEBS J. 2018;285:233–243. doi: 10.1111/febs.14154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg E. Parent A. Yang P. L. Sattler M. Liu Q. Liu Q. Wang J. Meng C. Buhrlage S. J. Gray N. Griffin J. D. Pharm. Res. 2020;37:1–29. doi: 10.1007/s11095-019-2719-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchino B. Di Franco M. Conti F. Rheumatol. 2020;59:1200–1203. doi: 10.1093/rheumatology/keaa194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P. Griffin I. Tucker C. Smith D. Oechsle O. Phelan A. Stebbing J. Lancet. 2020;395:e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y. Wei J. Zou L. Jiang T. Wang G. Chen L. Huang L. Meng F. Huang L. Wang N. Zhou X. Luo H. Mao Z. Chen X. Xie J. Liu J. Cheng H. Zhao J. Huang G. Wang W. Zhou J. J. Allergy Clin. Immunol. 2020;146:137–146.e3. doi: 10.1016/j.jaci.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khawli L. A. Goswami S. Hutchinson R. Kwong Z. W. Yang J. Wang X. Yao Z. Sreedhara A. Cano T. Tesar D. Nijem I. Allison D. E. Wong P. Y. Kao Y. H. Quan C. Joshi A. Harris R. J. Motchnik P. mAbs. 2010;2:613. doi: 10.4161/mabs.2.6.13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P. C. Adams A. C. Hufford M. M. de la Torre I. Winthrop K. Gottlieb R. L. Nat. Rev. Immunol. 2021;21:382–393. doi: 10.1038/s41577-021-00542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb R. L. Nirula A. Chen P. Boscia J. Heller B. Morris J. Huhn G. Cardona J. Mocherla B. Stosor V. Shawa I. Kumar P. Adams A. C. Van Naarden J. Custer K. L. Durante M. Oakley G. Schade A. E. Holzer T. R. Ebert P. J. Higgs R. E. Kallewaard N. L. Sabo J. Patel D. R. Klekotka P. Shen L. Skovronsky D. M. JAMA, J. Am. Med. Assoc. 2021;325:632–644. doi: 10.1001/jama.2021.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S. Saxena B. Mehta P. Heliyon. 2021:7. doi: 10.1016/j.heliyon.2021.e06158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X. Han M. Li T. Sun W. Wang D. Fu B. Zhou Y. Zheng X. Yang Y. Li X. Zhang X. Pan A. Wei H. Proc. Natl. Acad. Sci. U. S. A. 2020;117:10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K. J. Su I. J. Theron M. Wu Y. C. Lai S. K. Liu C. C. Lei H. Y. J. Med. Virol. 2005;75:185–194. doi: 10.1002/jmv.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oli A. N. Obialor W. O. Ifeanyichukwu M. O. Odimegwu D. C. Okoyeh J. N. Emechebe G. O. Adejumo S. A. Ibeanu G. C. ImmunoTargets Ther. 2020;9:13–30. doi: 10.2147/ITT.S241064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L. A. Anderson E. J. Rouphael N. G. Roberts P. C. Makhene M. Coler R. N. McCullough M. P. Chappell J. D. Denison M. R. Stevens L. J. Pruijssers A. J. McDermott A. Flach B. Doria-Rose N. A. Corbett K. S. Morabito K. M. O'Dell S. Schmidt S. D. Swanson P. A. Padilla M. Mascola J. R. Neuzil K. M. Bennett H. Sun W. Peters E. Makowski M. Albert J. Cross K. Buchanan W. Pikaart-Tautges R. Ledgerwood J. E. Graham B. S. Beigel J. H. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. L. Morris J. Pollock T. M. Br. Med. J. 1984;288:564. doi: 10.1136/bmj.288.6416.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otu A. Ebenso B. Labonte R. Yaya S. J. Glob. Health. 2020:10. doi: 10.7189/jogh.10.010339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan M. Pahuja S. Mohan A. Pandey R. M. Madan K. Hadda V. Tiwari P. Guleria R. Mittal S. Public Health. 2020;185:91–92. doi: 10.1016/j.puhe.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir C. Kucuksezer U. C. Tamay Z. U. Allergy. 2020;75:1824–1827. doi: 10.1111/all.14344. [DOI] [PubMed] [Google Scholar]
- Mohapatra P. R. Mishra B. Behera B. Indian J. Tuberc. 2021;68:119–124. doi: 10.1016/j.ijtb.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel J. McAndrews K. M. McGrail D. J. Dowlatshahi D. P. LeBleu V. S. Kalluri R. Sci. Rep. 2020;10:18377. doi: 10.1038/s41598-020-75491-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar L. E. Cruze A. M. Mury C. B. Proc. Natl. Acad. Sci. U. S. A. 2020;117:17720–17726. doi: 10.1073/pnas.2008410117. doi: 10.1073/pnas.2008410117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurina V. Med. Sci. 2018;6:27. doi: 10.3390/medsci6020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoen C. Stritzker J. Goebel W. Pilgrim S. Int. J. Med. Microbiol. 2004;294:319–335. doi: 10.1016/j.ijmm.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Reichmuth A. M. Oberli M. A. Jeklenec A. Langer R. Blankschtein D. Ther. Delivery. 2016;7:319–334. doi: 10.4155/tde-2016-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin M. D. Shukla S. Chung Y. H. Beiss V. Chan S. K. Ortega-Rivera O. A. Wirth D. M. Chen A. Sack M. Pokorski J. K. Steinmetz N. F. Nat. Nanotechnol. 2020;15:646–655. doi: 10.1038/s41565-020-0737-y. [DOI] [PubMed] [Google Scholar]
- Merlin M. Gecchele E. Capaldi S. Pezzotti M. Avesani L. BioMed Res. Int. 2014:2014. doi: 10.1155/2014/136419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R. Kumar P. FEMS Yeast Res. 2019;19:7. [Google Scholar]
- Chen J. Miao L. Li J. M. Li Y. Y. Zhu Q. Y. Zhou C. L. Fang H. Q. Chen H. P. World J. Gastroenterol. 2005;11:6159–6164. doi: 10.3748/wjg.v11.i39.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. Maruggi G. Shan H. Li J. Front. Immunol. 2019;10:594. doi: 10.3389/fimmu.2019.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupprecht C. and Nagarajan T., Current Laboratory Techniques in Rabies Diagnosis, Research and Prevention, Academic Press, 2015, vol. 2 [Google Scholar]
- Sanders B., Koldijk M. and Schuitemaker H., in Vaccine Analysis: Strategies, Principles, and Control, Springer, Berlin Heidelberg, 2015, pp. 45–80 [Google Scholar]
- Amanat F. Stadlbauer D. Strohmeier S. Nguyen T. H. O. Chromikova V. McMahon M. Jiang K. Arunkumar G. A. Jurczyszak D. Polanco J. Bermudez-Gonzalez M. Kleiner G. Aydillo T. Miorin L. Fierer D. S. Lugo L. A. Kojic E. M. Stoever J. Liu S. T. H. Cunningham-Rundles C. Felgner P. L. Moran T. García-Sastre A. Caplivski D. Cheng A. C. Kedzierska K. Vapalahti O. Hepojoki J. M. Simon V. Krammer F. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]