Abstract
Babesiosis among humans is on the rise in North America. Current diagnostic assays for the screening of babesiosis require blood collection by venipuncture, which is an invasive method. Urine on the other hand is a desirable biospecimen for biomarker analysis of Babesia microti infections because it can be collected periodically and non-invasively. Our group uses a new class of biomarker harvesting nanocage technology, which, when combined with mass spectrometry (MS), can determine the presence of parasite proteins shed in different bodily fluids of mammalian hosts, including urine. Using the hamster model of babesiosis, our nanoparticle-MS approach identified several B. microti proteins in erythrocytes, plasma, and urine samples. Surface and secreted antigens previously shown to elicit host immune responses against the parasite were particularly abundant in erythrocytes and plasma compared to other proteins. Two of these antigens, BmSA1 and BMR1_03g00947, showed different localization patterns by immunofluorescence of infected erythrocytes. Hamster urine samples from parasitemic animals harbored lower numbers of B. microti proteins compared to erythrocytes and plasma, with glycolytic enzymes, cytoskeletal components, and chaperones being the most frequently detected proteins. By applying novel nanoparticle-MS methods, a high level of analytical sensitivity can be achieved to detect multiple B. microti proteins in blood and urine. This is generally difficult to obtain with other techniques due to the masking of parasite biomarkers by the complex biomolecular matrix of bodily fluids from the host.
Keywords: Babesiosis, nanoparticle, mass spectrometry, proteomics, biomarker
INTRODUCTION
Babesiosis is caused by intraerythrocytic protozoan parasites of the genus Babesia. The incidence of the disease has been rising in recent years in the United States, with approximately 2,000 to 2,400 annual cases reported to the Centers for Disease Control and Prevention since 2015 (Gray et al. 2019). Babesia microti is the primary causative agent for the majority of human cases of babesiosis in the U.S. Transmission occurs primarily by Ixodes ticks in a similar fashion to Lyme disease, with other less common routes of transmission including pregnancy and blood transfusion (Lobo et al. 2013; Vannier et al. 2015). The latter is of concern since potential blood donors may harbor asymptomatic infections.
Human babesiosis is usually asymptomatic in immunocompetent populations, or results in mild symptoms that resolve within a few days. Immunocompromised individuals may experience severe symptoms of acute anemia, thrombocytopenia, organ failure, or even death (Krause 2019; Vannier et al. 2015). Diagnosis is primarily performed by microscopy of blood smears. Highly sensitive nucleic acid amplification tests have been developed for the detection of B. microti infections (Chan et al. 2013; Wang et al. 2015; Wilson et al. 2015), but these remain restricted to specialized laboratories with only two assays currently approved by the U.S. Food and Drug Administration (Tonnetti et al. 2019). An alternative approach to conventional tests for babesiosis is the direct identification of parasite antigens in bodily fluids by MS. However, some of the challenges of working in MS-based detection studies include a lack of optimal analytical sensitivity and the masking of pathogen biomarkers by highly abundant host proteins.
A recent report from our group described the use of hydrogel nanoparticles coupled with high affinity chemical baits to capture and concentrate B. microti proteins from whole blood of experimentally infected hamsters (Magni et al. 2019). Sequestered proteins were analyzed by MS, identification of tandem mass spectra was performed by searching against an annotated B. microti protein database (https://piroplasmadb.org/piro/), and the results provided a preliminary description of the parasite proteome during acute infection (Magni et al. 2019). The aim of the present study was to expand the applicability of our nanoparticle-MS approach to the direct identification of parasite proteins in erythrocytes, plasma, and urine of hamsters with acute infection. A comparison between the types of proteins detected in the different biological samples was performed with the ultimate goal of revealing novel candidates that could serve as potential diagnostic biomarkers of babesiosis in bodily fluids of the host.
MATERIALS AND METHODS
Babesia isolate
Babesia microti GI (BEI Resources NR-44070; ATCC® PRA-398™) was originally isolated from blood obtained from a human case of babesiosis in Nantucket, Massachusetts, USA, in 1983 (Gray et al. 2002; Piesman et al. 1986). The isolate was maintained by in vivo propagation in Syrian hamsters (Stock HsdHan:AURA, Envigo, Indianapolis, IN) according to published protocols (Cullen and Levine 1987; Oz and Hughes 1996) and procedures approved by the ATCC® IACUC.
Infection of animals
Four Syrian hamsters were inoculated with ~108 parasitized erythrocytes in 0.5 ml of blood. Blood samples were collected by the peri-orbital route following inhalational anesthesia with isoflurane and parasitemia was determined by microscopic examination of Giemsa-stained blood films. A minimum of 500 erythrocytes were counted to assess parasitemia in each sample. Parasitemia assessment included all parasitized cells regardless of intraerythrocytic stage or number of parasites per cell. After 30 days of infection, hamsters were anesthetized by ketamine injection (50 mg/kg) and 0.5 ml of blood was collected from each animal using 2 μg/ml of heparin solution as anticoagulant. Urine samples (0.1-0.2 ml) were collected directly from the bladders with a syringe during abdominal surgery and animals were subsequently euthanized using carbon dioxide inhalation.
Synthesis of hydrogel poly(N-isopropylacrylamide-co-allylammine) nanoparticles
Nanoparticles were synthesized as described previously (Douglas et al. 2011; Fredolini et al. 2008; Longo et al. 2009). Briefly, 9 g of N-isopropylacrylamide (NIPAm) and 0.4 g of N,N’-methylenebisacrylamide (BIS) were dissolved in 250 ml of Milli-Q water and filtered into a three neck round bottom flask under a vacuum. The filter was washed with an additional 100 ml of Milli-Q water and collected in the same flask. The suspension was purged with nitrogen for 30 min with stirring at medium speed. Allylammine (0.676 ml) was added to the suspension followed by purging for another 15 min with stirring at medium speed. The suspension was heated at 70 °C and subsequently treated with 5 ml of a 20 mg/ml solution of potassium persulfate (Sigma Aldrich, St. Louis, MO) in order to initiate polymerization. The reaction was heated at 70 °C for 6 h. Unreacted monomers were eliminated by washing the particles five times at room temperature in Milli-Q water using a centrifugation speed of 19,000 x g for 50 min. Lastly, particles were resuspended in a total volume of 300 ml of Milli-Q water. Dye functionalization and validation of hydrogel nanoparticles were performed as described previously (Magni et al. 2019).
Processing of biological samples with nanoparticles
Blood, plasma, and urine samples were heat-inactivated at 55 °C for 15 min. Blood samples were centrifuged at 2,000 x g for 10 min. A 0.1 ml suspension of nanoparticles (5 mg/ml) was diluted in 0.9 ml of deionized water and added to each erythrocyte pellet. The erythrocyte/nanoparticle suspensions were incubated at room temperature for 45 min, subjected to three freeze/thaw cycles at −80 °C, and centrifuged at 16,100 x g for 5 min. The resulting samples were centrifuged at 2,000 x g for 10 min to remove cellular debris. Plasma and urine samples (0.1 ml) were previously diluted 1:2 in 50 mM Tris-HCl (pH 7.0) before the addition of 0.1 ml nanoparticles and incubation at room temperature for 15 min. Nanoparticles were separated in all samples by high-speed centrifugation at 16,100 x g for 25 min and washed three times in ultrapure water (18 MΩ-cm). In order to elute captured proteins, 20 μl of 4% sodium dodecyl sulfate in 50 mM ammonium bicarbonate were added to the nanoparticle pellet and incubated for 20 min at room temperature. Samples were then centrifuged at 16,100 x g for 20 min. Supernatants were transferred into new tubes and mixed with 180 μl of 50 mM ammonium bicarbonate. Detergent residue was removed from the eluates using Pierce detergent removal spin columns (Thermo Scientific, Waltham, MA).
MS analysis
Eluates obtained from nanoparticle processing were reduced using 200 mM dithiothreitol at room temperature for 15 minutes and alkylated using 50 mM iodoacetamide at room temperature in the dark for 20 min. Enzymatic digestions were carried out overnight at 37 °C with 4 μl (0.5 μg/μl) of sequencing grade trypsin (Promega, Madison, WI) in 50 mM ammonium bicarbonate, pH 8.0. Digestions were stopped with 100% trifluoroacetic acid, samples were desalted using C18 spin columns (Pierce Biotechnology, Waltham, MA), and final eluates were dried using a Microvap 118 nitrogen evaporator (Organomation, Berlin, MA). Samples were then reconstituted in 10 μl of 0.1% formic acid with 100 fmol angiotensin-1 and analyzed with an Orbitrap Fusion™ Tribrid™ mass spectrometer coupled with a Nanospray EASY-nLC 1200 UHPLC system (Thermo Scientific). Reversed-phase chromatography separations of the peptide mixtures were performed using PepMap RSLC (75 μm i.d. × 15 cm long) with a 2 μm C18 resin LC column (Thermo Scientific). A 0.1% solution of formic acid was used as mobile phase A and 0.1% formic acid; 80% acetonitrile was used as mobile phase B. Peptides in the samples were eluted using a linear gradient starting from 5% mobile phase B/95% phase A to 50% mobile phase B/50% phase A for 90 min at 300 nl/min, then to 100% mobile phase B for an additional 2 min. The Orbitrap mass spectrometer was operated in a data-dependent mode in which each full MS scan was followed by TopN MS/MS scans of the most abundant molecular ions. Those ions with charge states from 2+ to 4+ were dynamically selected for collision induced dissociation (CID) using a normalized collision energy of 35%.
Identification of proteins
Tandem mass spectra obtained from MS analysis were searched against theoretical spectra derived from an annotated database of protein sequences from Babesia microti R1 (PiroplasmaDB.org) using the SEQUEST algorithm and tryptic cleavage constraints. High-confidence peptide identifications were obtained by applying the following filter criteria to the search results: 1) Xcorr > 2.0, > 3.0 and > 4.0 for 2+, 3+, 4+ precursor ions; 2) q-value < 0.05; and 3) probability of randomized identification: e0.01. Acceptable false discovery rate (FDR) based on forward-reverse decoy was < 1%. In addition, post-analysis filtering criteria were applied as follows: 1) peptide length >7 amino acids; 2) absence of carryover as determined by analyzing a blank sample with a 90 min gradient; 3) manual validation of each peptide spectra; and 4) rejection of peptides showing >90% similarity with any Mesocricetus auratus (Syrian hamster) protein and any other organism contained in the non-redundant database. The functional descriptions of the identified B. microti proteins were retrieved from UniProt (https://www.uniprot.org/).
Immunofluorescence
Rabbit polyclonal antibodies were raised against peptide antigens of the B. microti BmSA1 (BMR1_03g00785) and BMR1_03g00947 proteins. The selection of suitable peptide sequences for immunization, synthesis of peptide antigens, and the production of affinity purified antibodies were performed by GenScript, Piscataway, NJ. Peptide amino acid sequences are available upon request. A 0.5 ml aliquot of hamster blood with ~40% parasitemia was washed in 10 ml of PBS by centrifugation at 1,100 x g for 5 min. Erythrocytes were fixed in 4% paraformaldehyde and 0.0075% glutaraldehyde in PBS for 30 min as described (Tonkin et al. 2004). Fixed cells were washed in PBS as above and permeabilized with 0.1% Triton X-100/PBS for 10 min. Following another PBS wash, cells were blocked in 3% bovine serum albumin (BSA) in PBS for 1 h. Primary rabbit antibodies to BmSA1 or BMR1_03g00947 were added to the cell suspension at final dilutions of 1:500 and incubated for 1 h at room temperature. Cells were washed three times in PBS and incubated for 1 h with Alexa Fluor® 488-conjugated goat anti-rabbit secondary antibodies (Thermo Scientific) diluted 1:2000 in 3% BSA/PBS. Cells were suspended in mounting medium with DAPI nuclear counterstain and protein localization was visualized under 1000X magnification using a Zeiss Axioscope fluorescence microscope connected to a digital camera. Digital microscopic images were captured using Zen Imaging Software (Zeiss, Oberkochen, Germany).
RESULTS
Overview of Babesia proteins identified in erythrocytes, plasma, and urine
Four hamsters (H1 through H4) were inoculated with B. microti and parasitemia was monitored for 30 days. Only one of the hamsters (H2) did not develop significant parasitemias throughout infection (Fig. 1). Whole blood and urine samples were terminally collected at the 30-day time point for MS analysis. Parasitemias at this stage of infection reached 41% for H1, 1.5% for H2, 27% for H3, and 25% for H4 (Fig. 1 and Table 1). In general, a correlation between parasite burden and the total numbers of proteins identified in erythrocytes, plasma, and urine by MS was observed in our experiments (Table 1). Analysis of erythrocyte pellets showed 302 B. microti proteins in hamster H1, 22 in H2, 105 in H3, and 163 in H4. The highest numbers of parasite proteins identified in plasma and urine were found in H1 while no B. microti-derived proteins were detected in H2 urine, the animal with the lowest parasitemia in the group. H1 was the only animal with dark urine upon collection, indicative of hemoglobinuria (Table 1).
Fig. 1.
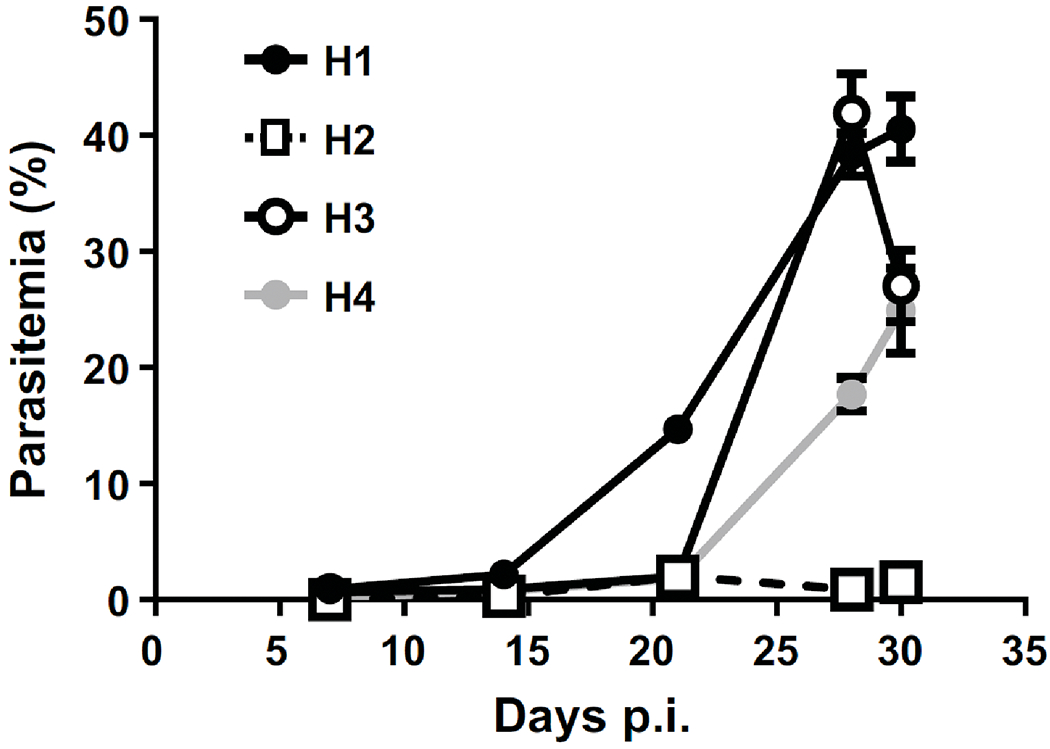
Parasitemia of hamsters infected with B. microti GI. Blood samples were collected at different times of infection and parasitemia was determined by microscopic examination of Giemsa-stained blood films as described in Materials and Methods. Data represent means ± SEM.
Table 1.
Summary of B. microti proteins identified in erythrocytes, plasma, and urine of experimentally infected hamsters
| Animal No. | Parasitemia (%)a | Sample | Total proteins | Housekeepingb | Hypotheticalc | Conserved Plasmodiumd | Seroreactive Antigense |
|---|---|---|---|---|---|---|---|
| H1 | 41 | Erythrocytes | 302 | 223 | 42 | 26 | 11 |
| Plasma | 25 | 17 | 4 | 1 | 3 | ||
| Urine | 23 | 21 | 2 | 0 | 0 | ||
|
| |||||||
| H2 | 1.5 | Erythrocytes | 22 | 14 | 4 | 0 | 4 |
| Plasma | 2 | 1 | 0 | 0 | 1 | ||
| Urine | 0 | 0 | 0 | 0 | 0 | ||
|
| |||||||
| H3 | 27 | Erythrocytes | 105 | 80 | 11 | 6 | 8 |
| Plasma | 20 | 14 | 3 | 0 | 3 | ||
| Urine | 8 | 8 | 0 | 0 | 0 | ||
|
| |||||||
| H4 | 25 | Erythrocytes | 163 | 126 | 17 | 13 | 7 |
| Plasma | 4 | 3 | 0 | 0 | 1 | ||
| Urine | 9 | 7 | 1 | 1 | 0 | ||
As determined at 30 days post-infection.
Proteins associated with common cellular and metabolic activities (i.e., ribosomal proteins, transporters, glycolytic enzymes, chaperones, etc.).
Proteins matching hypothetical genes in the B. microti genome.
Proteins of unknown function that are conserved in Plasmodium falciparum.
Proteins with reported immune responses in vivo (Silva et al., 2016).
In agreement with our previous study (Magni et al. 2019), the majority of parasite proteins detected in erythrocytes of all four animals (~70%) were associated with common cellular and metabolic activities (i.e., housekeeping functions). These included ribosomal proteins, transporters, glycolytic enzymes, chaperones, proteases, as well as proteins involved in trafficking, DNA and RNA metabolism, signaling, translation, lipid metabolism, motility, and invasion. Approximately 16 to 20% of all parasite proteins identified in erythrocytes of all four animals matched hypothetical genes in the B. microti genome. These included proteins of unknown function that are conserved in Plasmodium falciparum (Table 1). Complete MS data with descriptions of all proteins identified in the different biological samples of all four hamsters are shown in Supplemental Table S1. The results include the sums of the ion scores of all peptides that were identified (Sum PEP Scores), the percentage of the protein sequence covered by identified peptides, the total number of peptides identified for each protein, the numbers of peptide spectrum matches (PSMs), predicted amino acid lengths, and calculated molecular weights for each protein.
Babesia proteins identified in erythrocytes
To assess the relative abundance of B. microti proteins identified in erythrocytes of all four animals, we sorted the proteins listed in Table S1 based on the total number of distinct peptide sequences identified for each. Results from this analysis are depicted in Fig. 2A and Table S2 for each hamster examined in this study relative to H1, the animal with the highest parasitemia. A further selection of 38 proteins in H1 based on identifications by five or more peptides and their comparison with hamsters H2 to H4 is shown in Fig. 2B and C. Twenty four of the 38 proteins were among the top 10% most expressed genes in the B. microti genome as reported by Silva et al. (Silva et al. 2016). The majority of these were detected in at least two hamsters and included surface/secreted antigens (i.e., BmSA1, conserved protein BMR1_03g00947, and S1/P1 nuclease), several chaperones (i.e., heat shock proteins, peptidylprolyl isomerase/PPI), endoplasmic reticulum-resident calcium binding protein/ERC), and proteins involved in carbohydrate and energy metabolism (i.e., glycerol-3-phosphate dehydrogenase/GPDH and aconitate hydratase/CAN) (Fig. 2B–C and Table S2).
Fig. 2.
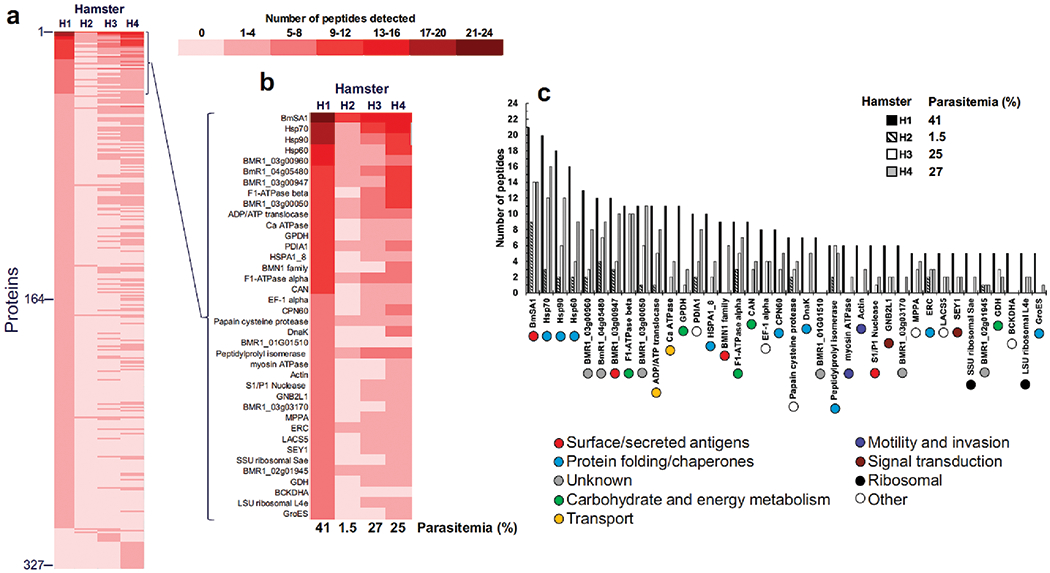
Relative abundance of B. microti proteins identified in erythrocytes of four hamsters. Proteins identified by MS in Supplemental Table S1 were sorted based on their total number of distinct peptide sequences detected and a comparison between experimental subjects was performed relative to H1, the animal with highest parasitemia (A). A selection of 38 proteins in hamster H1 based on identifications by five or more peptides and their comparison with hamsters H2 to H4 with ascribed biological functions of each protein is also shown (B and C).
Detection of surface/secreted antigens
The GPI-anchored BmSA1 protein (BMR1_03g00785) and the secreted conserved protein BMR1_03g00947 were detected in erythrocytes of all four animals (Figs. 2 and 3A, Table S2). Both proteins were previously reported to be among the top 20 reactive B. microti antigens to induce strong IgM/IgG responses in laboratory mice (Silva et al. 2016). Six additional top 20 seroreactive antigens were detected in the erythrocytes of at least two hamsters (Fig. 3A). These included the IgG inducers BmGPI10 (BMR1_02g04275) and N1-15 maltese-cross seroactive antigen (BmR1_04g07535), and the IgM/IgG inducers S1/P1 nuclease (BMR1_02g03140) and rhoptry neck protein RON2 (BMR1_03g04695). We verified the expression of BmSA1 and BMR1_03g00947 in infected erythrocytes by immunofluorescence (Fig. 3B). Using rabbit polyclonal antibodies raised against peptide antigens of each protein, we observed localization of BmSA1 primarily on ring forms of B. microti (Fig. 3B, top panel). Localization of BMR1_03g00947 on the other hand was distributed throughout the cytoplasm of infected erythrocytes (Fig. 3B, bottom panel). No immunostaining of parasites was observed with pre-immune rabbit sera (Data not shown).
Fig. 3.
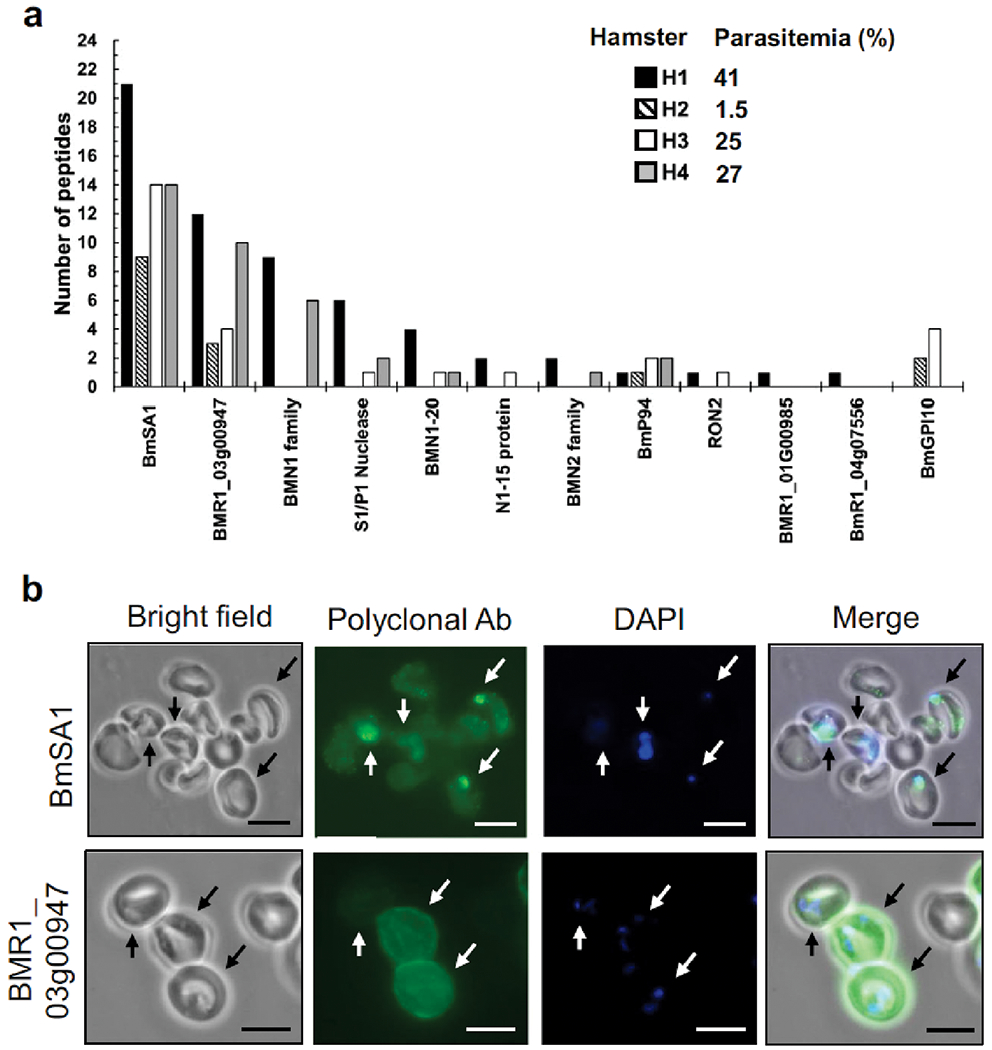
Detection of B. microti surface/secreted antigens in hamster erythrocytes. Twelve B. microti antigens were identified by MS analysis and their relative abundance was assessed in hamsters H1 through H4 by the numbers of distinct peptide sequences detected (A). Localization of B. microti BmSA1 and BMR1_3g00947 in infected erythrocytes was performed by immunofluorescence assays using rabbit polyclonal antibodies. Parasite nuclei in infected cells (arrows) were stained with DAPI (B).
Babesia proteins identified in plasma
The relative amounts of parasite proteins found in plasma as determined by the numbers of peptides identified for each are shown in Fig. 4 and Table S2. Out of the 34 proteins captured and identified by our nanoparticle-MS approach, 31 were also detected in erythrocytes and 14 of these were present in at least two hamsters with high parasitemias (Animals H1, H3, and H4 in Fig. 4). BmSA1 was the most abundant protein detected in plasma in a similar fashion to our results with erythrocytes. BMR1_03g00947 was also present, in addition to two other surface/secreted antigens, BMN1 family protein BMR1_03g00005 and BmP94 (BmR1_04g08155). Other noticeable proteins in the plasma of at least two animals included the glycolytic enzyme glyceraldehyde 3-phosphate dehydrogenase (GAPDH), the chaperones ERC and heat shock protein 1/8 (HSPA1_8), three proteins of unknown function (BMR1_03g00960, BmR1_04g05480, and BMR1_03g00050), and ribosomal proteins.
Fig. 4.
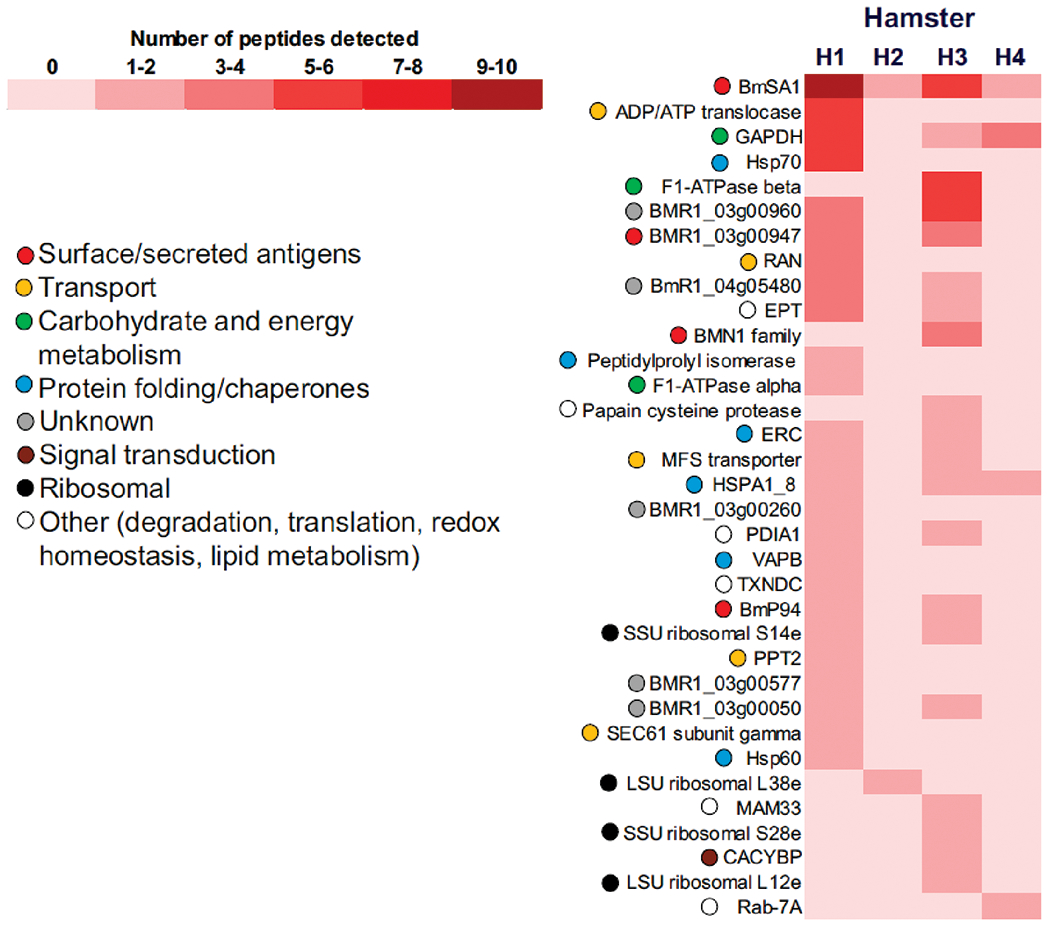
Relative abundance of B. microti proteins identified in plasma of four hamsters. Proteins identified by MS in Supplemental Table S1 were sorted based on their total number of distinct peptide sequences detected and a comparison between experimental subjects was performed relative to H1, the animal with highest parasitemia. Ascribed biological functions of each protein are also shown.
Babesia proteins identified in urine
Nanoparticles were useful in trapping >25 parasite proteins in urine samples of infected hamsters. The abundance of urinary proteins detected was dependent on the parasite burden with nearly 90% of the proteins identified belonging to animal H1 (Fig. 5). Of note, PPI was the only urinary protein found in erythrocytes and plasma, while four proteins were also present in erythrocytes: GPDH (BMR1_02g01010), hypothetical protein BmR1_04g09850, cofilin/actin-depolymerizing factor homolog 1 (ADF), and small GTP-binding protein BMR1_03g01795. The remaining 21 proteins were exclusively found in urine. These included proteins with functions in glycolysis (phosphoglycerate kinase/PGK and phosphoglycerate mutase/PGM), motility (profilin-like protein BMR1_02g02745), transport (nuclear transport factor 2/NUTF2), and trafficking (Rab family protein BMR1_02g02755).
Fig. 5.
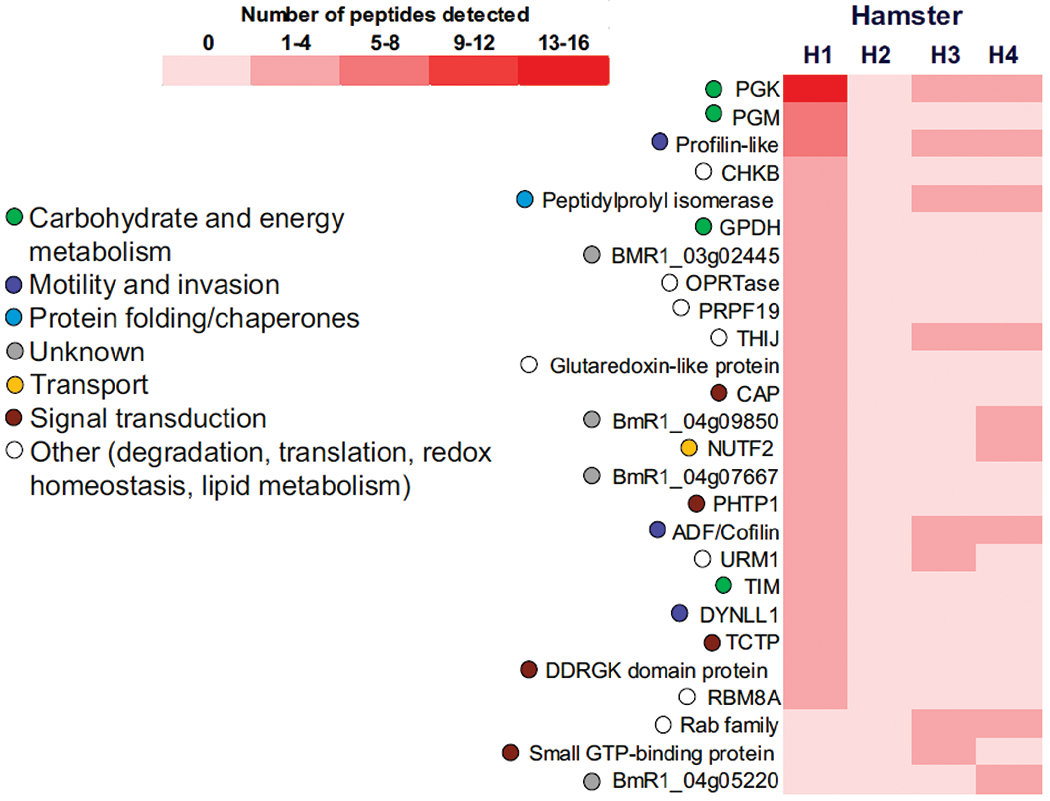
Relative abundance of B. microti proteins identified in urine of four hamsters. Proteins identified by MS in Supplemental Table S1 were sorted based on their total number of distinct peptide sequences detected and a comparison between experimental subjects was performed relative to H1, the animal with highest parasitemia. Ascribed biological functions of each protein are also shown.
DISCUSSION
The present study evaluated the application of hydrogel nanoparticle-MS technology in sequestering and identifying B. microti proteins in erythrocytes, plasma, and urine from hamsters with acute babesiosis. The use of this approach allowed us to compare the types of proteins detected in the different biological samples as we search for newer biomarkers of babesiosis in bodily fluids of the infected host. Similar to our previous report (Magni et al. 2019), the majority of proteins detected in erythrocytes (~70%) included ribosomal proteins, transporters, glycolytic enzymes, chaperones, proteases, and other factors commonly involved in housekeeping activities of the metabolically active trophozoite stage of the parasite. A subset of erythrocyte proteins was also found circulating in plasma of parasitemic hamsters, including the highly abundant BmSA1 surface antigen and three other surface/secreted antigens: BMR1_03g00947, BMN1 family protein BMR1_03g00005, and BmP94 (BmR1_04g08155). BmSA1 and BMR1_03g00947 differed in their expression in infected erythrocytes, with the former localizing on ring forms of B. microti and the latter showing a diffused distribution throughout the host cytoplasm. Proteins detected in urine were restricted to hamsters with high parasitemia (H1, H3, and H4) and included factors with functions in glycolysis, motility, and transport.
A limitation of this study was the small animal sample size used in the experiments. Although higher amounts of parasite proteins were detected in the three biological samples with parasitemias >25%, notable differences in the numbers and types of proteins were observed between H3 and H4, the two hamsters with comparable parasitemias. On the one hand it is not surprising to observe such differences as B. microti protein expression is likely to vary over time due to changes in the biological environment of infection at the time of analysis. On the other hand, differences among individuals with similar parasitemias may result from the technical challenge to detect extremely low concentrations of specific proteins in individual samples that fall below the 1 pg/ml limit of detection of the nanoparticle/MS workflow (Kim et al. 2018). Further work using a larger sample size combined with MS analysis of samples collected periodically is necessary to decipher the basis for the variability observed between individuals. Such studies will also help elucidate the parasitemia thresholds for detecting parasite urinary biomarkers and whether the different protein profiles observed between blood, plasma, and urine reflect changes in the B. microti proteome during infection.
Urine is a desirable biospecimen for biomarker analysis of Babesia microti infections because it can be collected periodically and non-invasively. Proteinuria as a result of kidney disease is often observed in human and animal cases of acute babesiosis (Blum et al. 2011; Filbin et al. 2001; Mylonakis 2001); however, proteomic studies of urinary proteins in the context of Babesia infections have not been reported. To our knowledge, this the first report demonstrating the detection of B. microti proteins in the urine of experimentally infected animals. The presence of urinary proteins correlated with the level of parasitemia with most identifications attributed to hamster H1, the animal showing evidence of hemoglobinuria. Most, if not all, of the proteins identified in urine samples of H1, H3, and H4 were likely derived from byproducts of erythrocyte lysis that became excreted by the kidneys. PPI was the only urinary protein also found in erythrocytes and plasma, while the great majority of proteins identified were exclusive to urine. These included PGK, PGM, and profilin-like protein BMR1_02g02745, which were highly abundant in H1. The notable absence in urine of parasite proteins such as BmSA1 and other surface antigens, heat shock proteins, F1-ATPases, and ribosomal proteins that were abundant in erythrocytes and plasma could be explained by: 1) a lower amount of groups of proteins in the urine due to different rates of glomerular filtration and tubular reabsorption in the kidneys; 2) an increased susceptibility of subsets of blood proteins to degradation in the kidneys or following excretion in the urine; or 3) differences in the affinities of nanoparticles to particular proteins present in blood or urine. Any of these scenarios may affect the detection limit of MS for proteins present at low concentrations in urine.
To date, there are limited FDA-approved assays for diagnosis of human babesiosis in the U.S. and many require blood collection by venipuncture. Results from this study show that the detection and identification of B. microti biomarkers in different bodily fluids is feasible using nanoparticle sequestration combined with mass spectrometry. The data also provide potential candidates that could be used in the development of a direct antigen detection test in urine, which in turn could complement or even replace current diagnostic tests for babesiosis. In the context of maintaining the safety of the blood supply, a sensitive and specific protein-based direct test for mass screening of potential donors would be of great benefit to rule out individuals with active disease. Future work is required to better examine temporal variations in the composition of peptides found in different bodily fluids of the host, their correlation with acute or chronic stages of babesiosis, and their impact in the severity of the disease.
Supplementary Material
ACKNOWLEDGMENTS
We thank Debra Adams-Fish from the ATCC® Specialty Laboratory Facilities for her assistance in the animal experiments. R.E. Molestina was supported by the ATCC® Internal Research and Development Program. A. Luchini, L. Liotta, and R. Magni were supported by grants W81XWH-17-1-0175 from the U.S. Army Medical Research Acquisition Activity, and R21HD097472, R01AR068436, and R21AI138135 from the U.S. National Institutes of Health. ATCC® is a registered trademark of the American Type Culture Collection.
REFERENCES
- Blum S, Gattringer R, Haschke E, Walochnik J, Tschurtschenthaler G, Lang F, Oberbauer R (2011) The case: hemolysis and acute renal failure. Babesiosis. Kidney Int 80(6):681–683. 10.1038/ki.2011.184 [DOI] [PubMed] [Google Scholar]
- Chan K, Marras SA, Parveen N (2013) Sensitive multiplex PCR assay to differentiate Lyme spirochetes and emerging pathogens Anaplasma phagocytophilum and Babesia microti. BMC Microbiol 13:295. 10.1186/1471-2180-13-295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen JM, Levine JF (1987) Pathology of experimental Babesia microti infection in the Syrian hamster. Lab Anim Sci 37(5):640–643 [PubMed] [Google Scholar]
- Douglas TA, Tamburro D, Fredolini C, Espina BH, Lepene BS, Ilag L, Espina V, Petricoin EF, Liotta LA, Luchini A (2011) The use of hydrogel microparticles to sequester and concentrate bacterial antigens in a urine test for Lyme disease. Biomaterials 32(4):1157–1166. 10.1016/j.biomaterials.2010.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filbin MR, Mylonakis EE, Callegari L, Legome E (2001) Babesiosis. J Emerg Med 20(1):21–24 [DOI] [PubMed] [Google Scholar]
- Fredolini C, Meani F, Reeder KA, Rucker S, Patanarut A, Botterell PJ, Bishop B, Longo C, Espina V, Petricoin EF, Liotta LA, Luchini A (2008) Concentration and preservation of very low abundance biomarkers in urine, such as human growth hormone (hGH), by Cibacron Blue F3G-A loaded hydrogel harticles. Nano Res 1(6):502–518. 10.1007/s12274-008-8054-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray EB, Niknafs AM, Herwaldt BL (2019) Surveillance for Babesiosis - United States, 2017. Annual Summary. https://www.cdc.gov/parasites/babesiosis/resources/babesiosis_surveillance_summary_2017b.pdf. Accessed 5 January 2020.
- Gray J, von Stedingk LV, Gurtelschmid M, Granstrom M (2002) Transmission studies of Babesia microti in Ixodes ricinus ticks and gerbils. J Clin Microbiol 40(4):1259–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Araujo R, Howard M, Magni R, Liotta LA, Luchini A (2018) Affinity enrichment for mass spectrometry: improving the yield of low abundance biomarkers. Expert Rev Proteomics 15(4):353–366. 10.1080/14789450.2018.1450631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause PJ (2019) Human babesiosis. Int J Parasitol 49(2):165–174. 10.1016/j.ijpara.2018.11.007 [DOI] [PubMed] [Google Scholar]
- Lobo CA, Cursino-Santos JR, Alhassan A, Rodrigues M (2013) Babesia: an emerging infectious threat in transfusion medicine. PLoS pathogens 9(7):e1003387. 10.1371/journal.ppat.1003387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo C, Patanarut A, George T, Bishop B, Zhou W, Fredolini C, Ross MM, Espina V, Pellacani G, Petricoin EF, Liotta LA (2009) Core-shell hydrogel particles harvest, concentrate and preserve labile low abundance biomarkers. PLoS One 4(3):e4763. 10.1371/journal.pone.0004763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magni R, Luchini A, Liotta L, Molestina RE (2019) Analysis of the Babesia microti proteome in infected red blood cells by a combination of nanotechnology and mass spectrometry. Int J Parasitol 49(2):139–144. 10.1016/j.ijpara.2018.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylonakis E (2001) When to suspect and how to monitor babesiosis. Am Fam Physician 63(10):1969–74 [PubMed] [Google Scholar]
- Oz HS, Hughes WT (1996) Acute fulminating babesiosis in hamsters infected with Babesia microti. Int J Parasitol 26(6):667–670 [DOI] [PubMed] [Google Scholar]
- Piesman J, Karakashian SJ, Lewengrub S, Rudzinska MA, Spielmank A (1986) Development of Babesia microti sporozoites in adult Ixodes dammini. Int J Parasitol 16(4):381–385 [DOI] [PubMed] [Google Scholar]
- Silva JC, Cornillot E, McCracken C, Usmani-Brown S, Dwivedi A, Ifeonu OO, Crabtree J, Gotia HT, Virji AZ, Reynes C, Colinge J, Kumar V, Lawres L, Pazzi JE, Pablo JV, Hung C, Brancato J, Kumari P, Orvis J, Tretina K, Chibucos M, Ott S, Sadzewicz L, Sengamalay N, Shetty AC, Su Q, Tallon L, Fraser CM, Frutos R, Molina DM, Krause PJ, Ben Mamoun C (2016) Genome-wide diversity and gene expression profiling of Babesia microti isolates identify polymorphic genes that mediate host-pathogen interactions. Sci Rep 6:35284 10.1038/srep35284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonnetti L, Young C, Kessler DA, Williamson PC, Reik R, Proctor MC, Bres V, Deisting B, Bakkour S, Schneider W, Diner S, Busch MP, Stramer SL, Linnen JM (2019) Transcription-mediated amplification blood donation screening for Babesia. Transfusion Dec 29 10.1111/trf.15630 [DOI] [PubMed] [Google Scholar]
- Tonkin CJ, van Dooren GG, Spurck TP, Struck NS, Good RT, Handman E, Cowman AF, McFadden GI (2004) Localization of organellar proteins in Plasmodium falciparum using a novel set of transfection vectors and a new immunofluorescence fixation method. Mol Biochem Parasitol 137(1):13–21. 10.1016/j.molbiopara.2004.05.009 [DOI] [PubMed] [Google Scholar]
- Vannier EG, Diuk-Wasser MA, Ben Mamoun C, Krause PJ (2015) Babesiosis. Infect Dis Clin North Am 29(2):357–370. 10.1016/j.idc.2015.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Villafuerte P, Zhuge J, Visintainer P, Wormser GP (2015) Comparison of a quantitative PCR assay with peripheral blood smear examination for detection and quantitation of Babesia microti infection in humans. Diagn Microbiol Infect Dis 82(2):109–113. 10.1016/j.diagmicrobio.2015.03.010 [DOI] [PubMed] [Google Scholar]
- Wilson M, Glaser KC, Adams-Fish D, Boley M, Mayda M, Molestina RE (2015) Development of droplet digital PCR for the detection of Babesia microti and Babesia duncani. Experimental parasitology 149:24–31 doi: 10.1016/j.exppara.2014.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


