ABSTRACT
The immediate early viral protein replication and transcription activator (RTA) of Kaposi’s sarcoma-associated herpesvirus (KSHV) is essential for activating the lytic cycle of KSHV. RTA induces the KSHV lytic cycle by several mechanisms, acting as a viral transcription factor that directly induces viral and host genes and acting as a viral E3 ubiquitin ligase by degrading host proteins that block viral lytic replication. Recently, we have characterized the global gene expression changes in primary effusion lymphoma (PEL) upon lytic reactivation of KSHV, which also led to the identification of rapidly downregulated genes such as ID2, an inhibitor of basic helix-loop-helix transcription factors. Here, we demonstrate that ID2 overexpression in PEL ablates KSHV lytic reactivation, indicating that ID2 inhibits the KSHV lytic cycle. Furthermore, we show that while ID2 is highly expressed during latency, its protein level is rapidly reduced by 4 h postinduction during lytic reactivation. Our results indicate that RTA binds to ID2 and induces its degradation during the KSHV lytic cycle by N-terminal ubiquitination through the ubiquitin-proteasome pathway. Importantly, we found that not only KSHV RTA but also its Epstein-Barr virus (EBV) and murine gammaherpesvirus 68 (MHV68) homologs interact with ID2, and they can induce the degradation of all four members of the ID protein family, suggesting an evolutionarily conserved interplay between gammaherpesvirus RTAs and ID proteins. Taken together, we propose that ID2 acts as a repressor of the KSHV lytic cycle, which is counteracted by its RTA-mediated degradation. We also predict that ID proteins may act as restriction factors of the lytic phase of the other gammaherpesviruses as well.
IMPORTANCE In addition to its transcription regulatory role, RTA is also known to have an E3 ubiquitin ligase activity, which RTA utilizes for inducing protein degradation. However, it is still largely unknown what host factors are downregulated during KSHV lytic reactivation by RTA-mediated protein degradation and what the biological significance of the degradation of these host factors is. In this study, we discovered that RTA employs N-terminal ubiquitination to induce degradation of ID2, a potent transcription repressor of host genes, via the ubiquitin-proteasome pathway to promote KSHV lytic reactivation in PEL cells. Furthermore, we found that not only KSHV RTA but also RTA of EBV and MHV68 gammaherpesviruses can induce the degradation of all four human ID proteins, indicating that the interplay between gammaherpesvirus RTAs and ID proteins is evolutionarily conserved.
KEYWORDS: ID proteins, ID2, Kaposi's sarcoma-associated herpesvirus, PEL, RTA, lytic cycle, reactivation, ubiquitination
INTRODUCTION
Kaposi’s sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8 (HHV-8), is the etiological agent of the endothelial neoplasm Kaposi’s sarcoma, which is frequently found in AIDS patients (1). KSHV also causes KSHV-associated inflammatory syndrome (KICS) and two lymphoproliferative diseases, primary effusion lymphoma (PEL) and multicentric Castleman’s disease (2, 3). Analogous to other herpesviruses, KSHV has a biphasic life cycle, including a lytic and a latent phase (4). During latency, the majority of lytic genes are repressed, and there is no virus production. The lytic cycle can be triggered in latently infected cells by various environmental and physiological stimuli (5). Of the KSHV-encoded viral factors, the replication and transcription activator (RTA) is the essential viral transcription factor that is responsible for inducing KSHV from latency to lytic replication (6, 7). RTA initiates the cascade-like expression of lytic genes categorized as immediate early (IE), early (E), and late (L) genes, which allows the replication of viral genome and, eventually, virus production (8). Although it is well established that the transcriptional activity of RTA is essential for viral gene regulation and virus production, RTA has also been shown to be capable of inducing protein degradation, preferentially of sumoylated proteins, which can contribute to RTA-induced lytic reactivation (9, 10). Previous studies demonstrated that RTA possesses an E3 ubiquitin ligase activity whereby RTA can polyubiquitinate and induce protein degradation, or RTA hijacks host E3 ubiquitin ligases for targeted protein degradation (10–16). However, so far, only a few host targets have been identified for RTA-induced protein degradation that affects the progress of the KSHV lytic cycle and viral transmission. By identifying the key host factors targeted by RTA for degradation and revealing their biological importance in infected cells, we can better understand the RTA-driven mechanisms responsible for KSHV replication, transmission, and viral pathogenesis.
Recently, our lab has determined the global host gene expression changes during the lytic cycle of KSHV in PEL cells, which revealed a gradual reprogramming of the host transcriptome upon RTA-induced lytic reactivation (17). One of the host genes rapidly downregulated during KSHV lytic reactivation was the inhibitor of DNA binding protein 2 (ID2). ID2 is a helix-loop-helix (HLH) cellular protein that is one of the four members of the ID protein family (18). The ID proteins have no DNA binding properties but are able to interact with basic HLH (bHLH) transcription factors and prevent them from binding to promoters, thereby inhibiting their transactivation function (19). The expression of the ID protein family is tightly regulated due to their potent dominant negative effect on bHLH proteins, as well as other transcription factors that are crucial for embryonic development, cell cycle progression, and immune system development (20, 21). The ID genes are expressed well in tissues undergoing proliferation such as in early development, while their expression is usually restricted in mature, differentiated tissues (22). A previous study revealed that the expression levels of ID1, ID2, and ID3 are upregulated upon KSHV latent infection, and, in addition, they can contribute to KSHV-induced tumorigenesis (23). However, the impact of ID proteins on the KSHV lytic cycle has not been investigated. Because ID2 is expressed during latency while it is downregulated during KSHV lytic cycle, we hypothesized that ID2 may function as a repressor of lytic reactivation and the downregulation of ID2 possibly benefits the lytic cycle of KSHV.
Here, we have discovered that RTA can induce the swift degradation of ID2 through N-terminal ubiquitination during lytic reactivation of KSHV in PEL cells. We found that if ID2 is overexpressed during KSHV reactivation in PEL, every step of the lytic cycle is repressed, from lytic gene induction to virus replication and virus production. In addition, we also show that not only KSHV RTA but also RTA proteins from Epstein-Barr virus (EBV) and murine gammaherpesvirus 68 (MHV68) can induce the degradation of all four ID proteins, indicating an evolutionarily conserved interplay between gammaherpesvirus RTAs and ID proteins. Taken together, we propose that ID2 acts as a repressor of the KSHV lytic cycle; however, RTA can overcome this barrier by inducing ID2 degradation. This function of RTA may be evolutionarily conserved among a subset of gammaherpesviruses and ID proteins.
RESULTS
ID2 is rapidly downregulated during KSHV lytic reactivation.
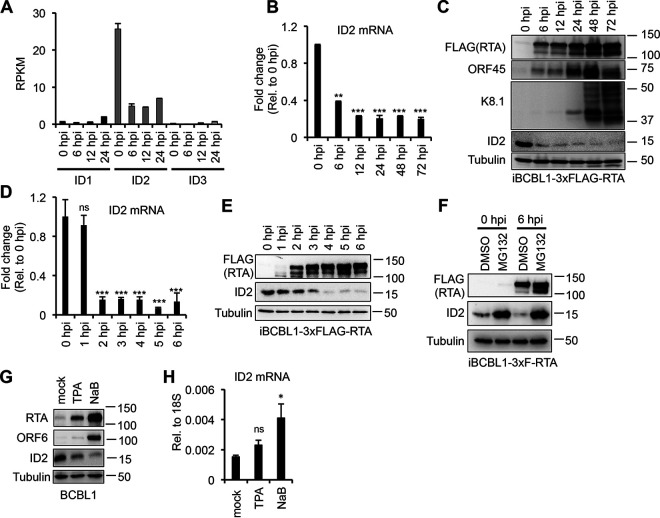
Analysis of our published transcriptome data derived from iBCBL1-3xFLAG-RTA cells indicates that, of the four ID genes, ID2 expression is the highest in BCBL1 during KSHV latency, which is greatly reduced during KSHV lytic reactivation triggered by doxycycline (Dox)-inducible 3xFLAG-RTA (Fig. 1A) (17). Independent reverse transcriptase quantitative PCR (RT-qPCR) and immunoblot analyses further confirmed that ID2 is significantly downregulated in iBCBL1-3xFLAG-RTA by 6 h postinduction (hpi) (Fig. 1B and C). To determine when ID2 expression was reduced at the mRNA and protein levels within the first 6 h of KSHV lytic reactivation, we sampled at each hour from 0 hpi (latency) to 6 hpi (Fig. 1D and E). We found that by 2 hpi, the mRNA abundance of ID2 was dramatically decreased, and by 4 hpi, ID2 protein was nearly undetectable (Fig. 1D and E). Like all ID proteins, the half-life of ID2 is very short, around 15 to 20 min (24, 25). It has been suggested that the half-life of ID proteins can extend when they are actively bound to another factor (26, 27). Since the reduction of ID2 at the protein level was delayed by 2 h relative to its transcript level, we hypothesized that ID2 may be bound to a factor during KSHV latency, thus extending its half-life, but it gets degraded after 4 hpi. In fact, proteasome inhibitor MG132 treatment of iBCBL1-3xFLAG-RTA restored ID2 protein level, indicating that ID2 does undergo proteasomal degradation during KSHV lytic reactivation (Fig. 1F). We speculate that the reduction of the ID2 mRNA level prior to protein reduction in iBCBL1-3xFLAG-RTA is caused by RTA overexpression, which may deregulate host factors involved in ID2 transcription. We also tested how the mRNA and protein levels of ID2 change if KSHV lytic reactivation is induced in BCBL1, using different drugs that are known KSHV inducers. We treated BCBL1 cells for 24 h with either the protein kinase C activator 12-O-tetradecanoylphorbol-13-acetate (TPA) or the histone deacetylase inhibitor sodium butyrate (NaB). We found that ID2 was reduced at the protein level but not at the RNA level in the reactivated cells (Fig. 1G and H). These data show that regardless of how KSHV lytic reactivation is induced, the ID2 protein level is always reduced, while its mRNA level can remain unchanged. Altogether, these results suggest that ID2 can be actively degraded during KSHV lytic cycle.
FIG 1.
ID2 is rapidly downregulated during KSHV lytic reactivation. (A) Transcriptome sequencing (RNA-seq) analysis of the expression of ID genes during KSHV lytic reactivation at various time points in iBCBL1-3xFLAG-RTA cells. The average of three biological replicates is shown. (B) ID2 gene expression was measured by RT-qPCR in iBCBL1-3xFLAG-RTA during latency and at various time points of lytic reactivation. (C) Immunoblot analysis of ID2 and lytic viral protein expression during KSHV lytic reactivation in iBCBL1-3xFLAG-RTA cells. (D) RT-qPCR analysis of ID2 gene expression during KSHV latency (0 hpi) up to 6 hpi in iBCBL1-3xFLAG-RTA cells. (E) Western blot analysis of FLAG-RTA and ID2 expression in reactivated iBCBL1-3xFLAG-RTA cells. (F) Immunoblot analysis of 3xFLAG-RTA and ID2 protein expression during MG132 or DMSO (vehicle control) treatment of reactivated iBCBL1-3xF-RTA cells at 0 (latency) and 6 hpi. (G) Immunoblot analysis of lytic viral proteins and ID2 expression at 24 hpi in BCBL1 in which lytic reactivation was induced with TPA or NaB. (H) RT-qPCR testing of ID2 gene expression during TPA- and NaB-mediated KSHV reactivation in BCBL1 cells at 24 hpi. t tests were performed between GFP-OE and ID2-OE (sample of n = 3), and degree of statistical significance is indicated as *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; and ns, not significant.
KSHV RTA induces polyubiquitination and degradation of ID2 through the ubiquitin-proteasome pathway.
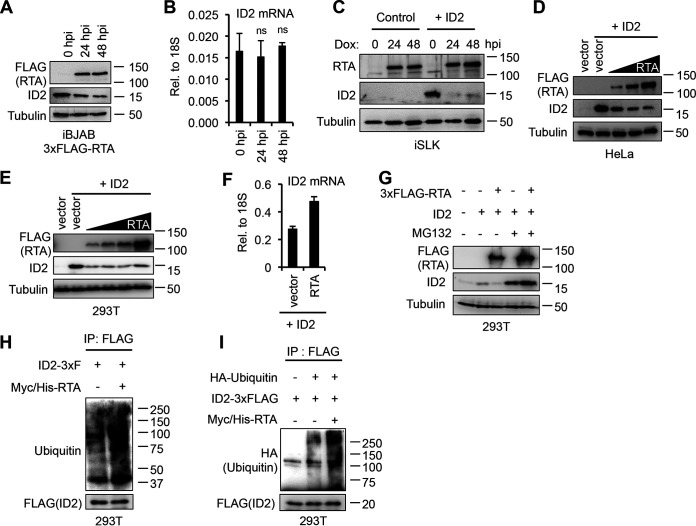
In addition to being a potent gene transcription activator, RTA has also been shown to have intrinsic E3 ubiquitin ligase activity that allows it to induce the degradation of cellular and viral proteins (10, 12–14, 16). We hypothesized that since the downregulation of ID2 occurred within the first few hours of KSHV reactivation, the immediate early viral factor RTA could be causing the rapid degradation of ID2. To test this, we analyzed ID2 expression in a KSHV-free transgenic B cell lymphoma cell line encoding a Dox-inducible 3xFLAG-RTA (iBJAB-3xFLAG-RTA). We found that upon RTA expression, ID2 was reduced at the protein level but not at the mRNA level (Fig. 2A and B). We also confirmed RTA-mediated downregulation of ID2 in three different epithelial cell lines (Fig. 2C to F). Since iSLK, HeLa, and 293T cell lines have a barely detectable amount of ID2, we expressed ID2 using lentiviral transduction in iSLK and plasmid transfection in HeLa and 293T cells. We found that the Dox-inducible RTA dramatically reduced the ID2 protein level in iSLK (Fig. 2C). To investigate if ID2 was downregulated by RTA in a dose-dependent manner, we cotransfected HeLa and 293T cells with ID2 and an increasing amount of RTA (Fig. 2D and E). ID2 immunoblot analysis showed an RTA-mediated dose-dependent reduction of ID2 in HeLa cells (Fig. 2D), while we could observe robust ID2 downregulation even at the lowest concentration of RTA in 293T cells (Fig. 2E). To check that ID2 downregulation was not due to RTA repressing its transcription, we performed RT-qPCR and detected no reduction of ID2 mRNA level (Fig. 2F).
FIG 2.
KSHV RTA induces the polyubiquitination and degradation of ID2 through the ubiquitin-proteasome pathway. (A) Immunoblot detection of 3xFLAG-RTA and ID2 in Dox-induced iBJAB-3xFLAG-RTA cells. (B) RT-qPCR analysis of ID2 transcript abundance in iBJAB-3xFLAG-RTA cells. t tests were performed between 0 h postinduction (hpi) and 24 or 48 hpi (ns, not significant). (C) Immunoblot analysis of lenti-GFP and lenti-ID2-transduced iSLK cells expressing Dox-inducible RTA. (D) Western blot analysis of HeLa cells cotransfected with ID2 and either vector control or an increasing amount of 3xFLAG-RTA. (E) Immunoblot detection of 3xFLAG-RTA and ID2 in 293T cells that were cotransfected with ID2 and either vector control or increasing amounts of 3xFLAG-RTA. (F) RT-qPCR analysis of ID2 in the experiment performed in panel E when control vector or the largest amount of RTA was cotransfected with ID2. (G) Western blot analysis of 293T cells that were cotransfected with ID2 and vector or 3xFLAG-RTA, and treated with DMSO (control) or 40 μM MG132. (H) FLAG immunoprecipitation was performed using 293T cells that were cotransfected with ID2-3xFLAG and either a vector control or Myc/His-tagged RTA (M/H-RTA). FLAG IP was subjected to ubiquitin Western blotting. (I) 293T cells were cotransfected with HA-ubiquitin, ID2-3xFLAG, and Myc/His-RTA (M/H-RTA). After FLAG immunoprecipitation (IP) was performed, the IP samples were subjected to immunoblot analysis.
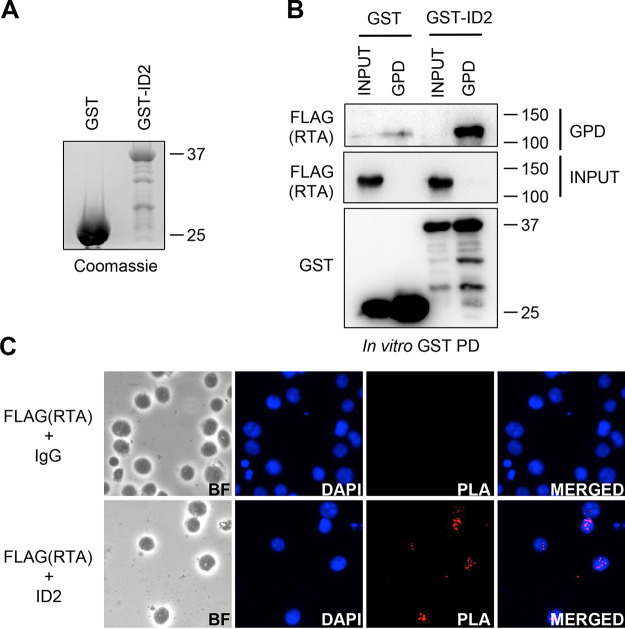
Next, we investigated whether RTA was inducing ID2 degradation through the ubiquitin-proteasome pathway. For this, we cotransfected 293T cells with ID2 and RTA in the presence of either dimethyl sulfoxide (DMSO) (control) or the proteasome inhibitor MG132. We observed an increase in ID2 level with MG132 treatment alone, which is in line with previous studies showing that ID2 degradation, which is normally mediated by the anaphase-promoting complex (APC/CCdh1) in cells, can be blocked by MG132 (26). Importantly, we also observed a complete rescue of ID2 protein expression in the presence of RTA with MG132 (Fig. 2G). Ubiquitin immunoblot of immunoprecipitated ID2-3xFLAG showed increased ubiquitination of ID2 in the presence of Myc/His-tagged RTA (M/H-RTA) during MG132 treatment indicating that RTA induces polyubiquitination of ID2, which can ultimately lead to its degradation (Fig. 2H and I). Furthermore, using an in vitro glutathione S-transferase (GST) pulldown and a proximity ligation assay (PLA), we found that RTA directly binds to ID2 (Fig. 3), which is in agreement with other studies showing that RTA interacts with its substrate proteins targeted for degradation (12, 13). Taken together, these data demonstrate that RTA interacts with ID2 and induces polyubiquitination and subsequent degradation of ID2 through the ubiquitin-proteasome pathway.
FIG 3.
KSHV RTA and ID2 interact. (A) Coomassie blue staining of GST (control) and GST-ID2 protein purified from E. coli. (B) Immunoblot analysis of in vitro GST pulldown (GST-PD) using GST (control) and GST-ID2 with cell extracts of 293T expressing 3xFLAG-RTA. (C) iBCBL1-3xFLAG-RTA cells were reactivated and treated with MG132 for 6 h and then fixed and permeabilized before the proximity ligation assay was performed. Cells were probed with anti-FLAG and anti-IgG control (top) or with anti-FLAG and anti-ID2 antibodies (bottom).
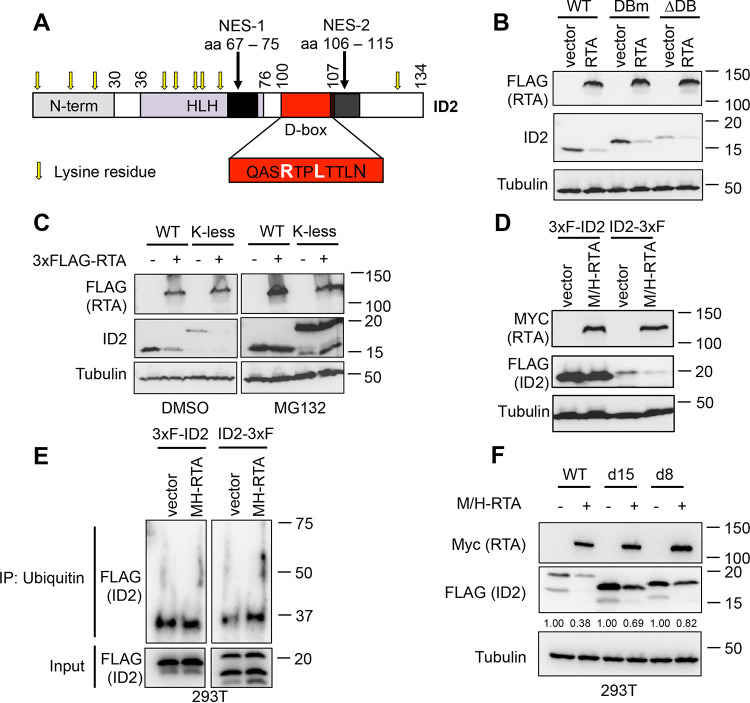
RTA induces ID2 degradation through its N-terminal ubiquitination.
Lasorella and her colleagues have shown that the ID2 protein level is tightly controlled by APC/CCdh1, which binds to ID2 through its C-terminal D-box motif that leads to polyubiquitination and degradation of ID2 (25). To determine whether RTA was also using the ID2’s D-box to target it for degradation, we cotransfected 293T cells with 3xFLAG-RTA and wild-type (WT) ID2 or ID2 mutants, such as a D-box point mutant (DBm), where the arginine and leucine were mutated to a glycine and valine (depicted in Fig. 4A) or a D-box deletion (ΔDB) mutant. We note that mutation of D-box causes an abnormal mobility shift of ID2 in protein gel electrophoresis (Fig. 4B), which has also been observed in previous studies (25). We found that RTA targets ID2 for protein degradation through a D-box-independent mechanism (Fig. 4B).
FIG 4.
KSHV RTA induces ID2 degradation through the ubiquitin-proteasome pathway by N-terminal ubiquitination. (A) Domain map of the full-length (WT) ID2. (B) Immunoblot analysis of ID2 WT, D-box point mutant (DBm), or D-box deletion mutant (ΔDB) cotransfected with a vector control or 3xFLAG-tagged RTA. (C) Western blot analysis of WT or lysine less mutant (K-less) of ID2 cotransfected with a vector control or 3xFLAG-RTA for 2 days before being treated with DMSO (vehicle control) or MG132 (40 μM) for 12 h. (D) Immunoblot detection of N- and C-terminal 3xFLAG-tagged ID2 cotransfected with vector control or Myc/His-tagged RTA in 293T cells. (E) Ubiquitin immunoprecipitation was performed using 293T cells cotransfected with Myc/His-RTA (M/H-RTA) and 3xFLAG-ID2 or ID2-3xFLAG. After ubiquitin immunoprecipitation was performed, the samples were subjected to FLAG immunoblot analysis. (F) Immunoblot analysis of 293T cells that were cotransfected with Myc/His-RTA (M/H-RTA) and WT ID2-3xFLAG or its N-terminal truncation mutants. Band intensities of ID2-3xFLAG were quantified relative to tubulin (loading control) and presented as a ratio relative to the vector control of each condition in panel F.
Most E3 ubiquitin ligases add polyubiquitin chains to their substrate proteins via an internal lysine residue (28). This led us to test whether RTA was using this mechanism to induce ID2 degradation by mutating all 9 lysine residues of ID2 to alanine to make a lysine-less (K-less) ID2 mutant (Fig. 4A). We cotransfected 293T cells with 3xFLAG-RTA and either the WT or K-less ID2 and discovered that RTA mediates ID2 degradation in a lysine-independent manner, which could be rescued by MG132 treatment (Fig. 4C). In addition to internal lysine residues, polyubiquitination of a subset of short-lived cellular and viral proteins can also occur by N-terminal ubiquitination, which means that the ubiquitin chain is attached to the free N-terminal residue of the substrate protein (29–33). Importantly, the N-terminal epitope tagging of these proteins can prevent them from degradation by the ubiquitin-proteasome pathway (30, 32). To test if RTA mediates N-terminal ubiquitination of ID2, we epitope tagged either the N or C terminus of ID2 with 3xFLAG. We cotransfected 293T cells with M/H-RTA and the 3xFLAG epitope-tagged ID2 clones (Fig. 4D). We observed that the N-terminal-tagged ID2 was stabilized in the presence of RTA, while ID2-3xFLAG was still undergoing degradation by RTA (Fig. 4D).
To further support that RTA can mediate N-terminal ubiquitination of ID2, we compared the abundance of differentially epitope tagged ID2 in ubiquitin (Ub) immunoprecipitation (IP) in the absence and in the presence of RTA (Fig. 4E). 3xFLAG-ID2 or ID2-3xFLAG was expressed alone or coexpressed with Myc/His-RTA in 293T cells. After MG132 treatment, Ub IP was performed, which was followed by FLAG Western blotting for the tagged ID2 proteins. When 3xFLAG-ID2 was used, we found a comparable amount of ID2 in the Ub IPs without and with RTA. In contrast, there was more ID2 in the Ub IP when ID2-3xFLAG was coexpressed with RTA than ID2-3xFLAG expression alone. Our interpretation is that while ID2 can undergo ubiquitination likely via its lysine residues regardless of how ID2 was epitope tagged, RTA can increase the ubiquitination of ID2-3xFLAG via its N-terminal ubiquitination but not that of 3xFLAG-ID2. That is why more ID2 can be detected in the Ub IP with ID2-3xFLAG in the presence of RTA than in the absence of RTA.
Previous studies have shown that the deletion of a portion of the N terminus of proteins undergoing N-terminal ubiquitination can prevent them from degradation (30, 32, 34). To further confirm that RTA targets ID2 for degradation in an N-terminal-dependent manner, we made two N-terminal truncation mutants of ID2-3xFLAG that were missing the first 8 or 15 amino acids (d8 or d15) (Fig. 4F). We found that the N-terminal truncation mutants of ID2 were indeed stabilized to some degree in the presence of RTA in comparison to WT ID2 (Fig. 4F). Since the protein level of the d15 mutant was not completely restored, we hypothesize that a larger part of the N terminus or other parts of ID2 are also involved in its RTA-mediated degradation. Taken together, these results indicate that RTA induces ID2 degradation through its N-terminal ubiquitination. This is a novel finding since KSHV RTA has not yet been shown to utilize N-terminal ubiquitination to induce degradation of any other substrate proteins.
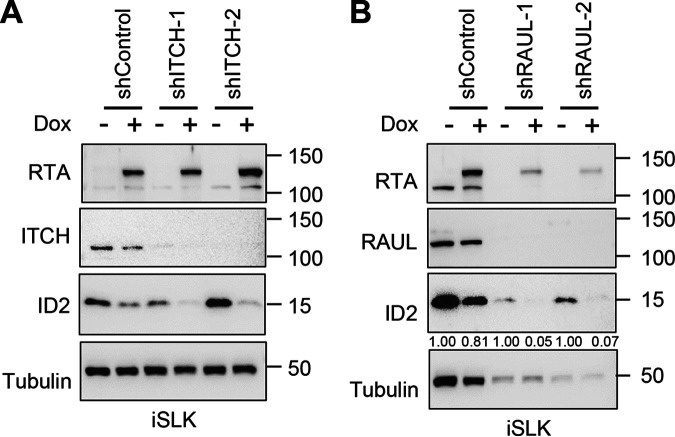
The host E3 ubiquitin ligases ITCH and RAUL are not needed, but the first 530 amino acids of RTA are required for RTA-meditated degradation of ID2.
RTA has been demonstrated to induce degradation of its substrate proteins either through chaperoning a host E3 ubiquitin ligase to polyubiquitinate and degrade its substrate, or RTA itself ubiquitinates its substrate using its RING-like domain. Previous studies showed that RTA can utilize the host E3 ligases ITCH or RAUL (aka UBE3C) for RTA-mediated protein degradation (11, 15). To assess the role of ITCH and RAUL in RTA-mediated degradation of ID2, we treated iSLK cells with a lentivirus expressing ID2 along with lentiviral ITCH or RAUL short hairpin RNA (shRNA) for 2 days before inducing RTA expression with Dox (Fig. 5A and B). We found that neither ITCH nor RAUL is involved in RTA-mediated degradation of ID2 (Fig. 5A and B). We note that although shRNA with RAUL (shRAUL) affected the viability of iSLK cells as seen by the reduced tubulin expression, we could determine that shRAUL did not abrogate RTA-mediated ID2 degradation (Fig. 5B).
FIG 5.
RTA-mediated degradation of ID2 does not require host E3 ubiquitin ligases ITCH or RAUL. (A) Immunoblot analysis of iSLK cells that were transduced with lenti-ID2 and lenti-shControl or lenti-shITCH for 3 days and then treated with 1 μg/mL of Dox to induce RTA expression for 24 h. (B) Western blot analysis of iSLK cells that were treated with lenti-ID2 and lenti-shControl or lenti-shRAUL for 3 days and then induced with 1 μg/mL of Dox to express RTA for 24 h. Band intensities of ID2 were quantified relative to tubulin (loading control) and presented as a ratio relative to the Dox- negative control of each condition in panel B.
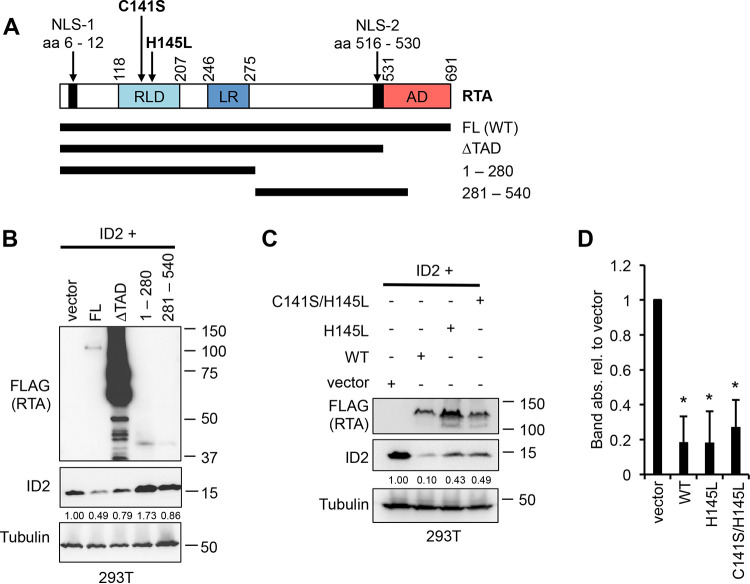
Next, we tested what region of RTA was necessary to induce ID2 degradation by using different RTA truncation mutants (Fig. 6A). We cotransfected 293T cells with ID2 along with full-length (FL) RTA or an RTA mutant such as the transactivation domain deletion mutant (amino acids [aa] 1 to 530 or ΔTAD), aa 1 to 280, or aa 281 to 540 (Fig. 6B). We identified that the first 530 amino acids of RTA are sufficient to induce ID2 degradation, although they are less efficient than full-length RTA (Fig. 6B). Since the RING-like domain (RLD) of RTA, which contains its E3 ubiquitin ligase activity, is in aa 1 to 530, we also tested if the E3 ubiquitin ligase activity of RTA is necessary for ID2 degradation (10). Previous studies showed that a point mutation in RLD at aa 145, changing histidine to leucine (H145L) and/or changing cysteine to serine at aa141 (C141S), is sufficient to abrogate the E3 ligase activity of RTA (10, 12, 13). After cotransfecting 293T cells with ID2-HA along with either WT or the H145L or C141S/H145L mutant of RTA, we found that the RTA mutants were not able to reduce the ID2 protein level as efficiently as WT RTA (Fig. 6C). Altogether, these results show that the first 530 amino acids of RTA, which involves its RING-like domain, are required but not sufficient for the robust ID2 degradation, but it is likely that other parts of RTA are also required for the efficient downregulation of ID2.
FIG 6.
The first 530 amino acids of RTA are sufficient to target ID2 for degradation. (A) Model representation of the full-length (FL) KSHV RTA and its truncation mutants. (B) Immunoblot detection of ID2 and 3xFLAG-tagged RTA FL and mutants following a 2-day 293T cotransfection experiment. Band intensities of ID2 were quantified relative to tubulin (loading control) and presented as a ratio relative to ID2 cotransfected with vector. (C) 293T was cotransfected with ID2 and 3xFLAG-tagged WT, H145L mutant of RTA, or the C141S/H145L double mutant of RTA, and immunoblot analysis was performed 2 days after transfection. (D) Band intensities of ID2 in three independent experiments, which were done exactly as in panel C, were quantified relative to tubulin (loading control) and presented as a ratio relative to ID2 cotransfected with vector. The degree of statistical significance calculated by t tests is indicated as *, P ≤ 0.001 (sample of n = 3).
ID2 represses the lytic cycle of KSHV.
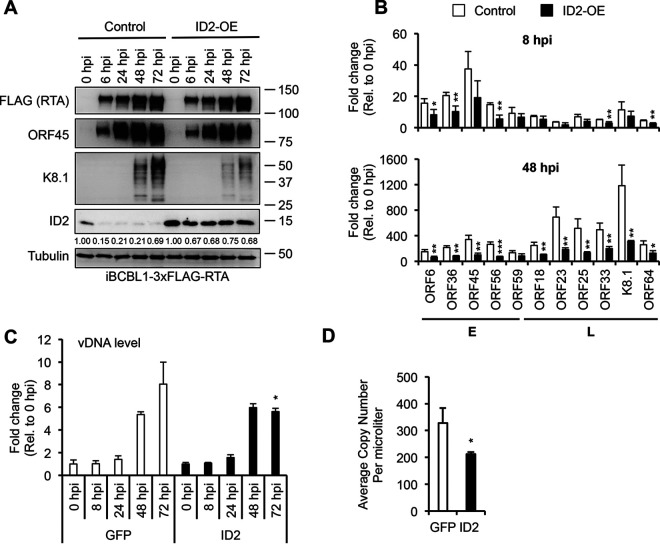
Because of how quickly ID2 is degraded during the KSHV lytic cycle, it prompted us to test whether ID2 acts as a suppressor of the viral lytic phase. To address this question, we overexpressed ID2 (ID2-OE) in iBCBL1-3xFLAG-RTA cells using lentiviral transduction, while lytic reactivation was induced by Dox-induced 3xFLAG-RTA. Lenti-GFP (GFP-OE) was used as a control (Fig. 7). Immunoblot analysis in Fig. 7A shows that while the expression of the lytic cycle inducer 3xFLAG-RTA was not affected by ID2-OE, we observed slight reduction of early viral protein expression (ORF45) and strong abrogation of late viral protein expression (K8.1). Viral gene expression analysis by RT-qPCR showed that ID2-OE could reduce early and late lytic gene expression as early as 8 hpi, but the repression of viral gene expression was more pronounced and significant at 48 hpi (Fig. 7B). We also found diminished viral DNA replication and virus production upon ID2-OE (Fig. 7C and D).
FIG 7.
ID2 negatively regulates KSHV lytic cycle in iBCBL1-3xFLAG-RTA cells. (A) Immunoblot analysis of ID2 and viral protein expression in lenti-GFP and lenti-ID2-transduced iBCBL13xFLAG-RTA cells during KSHV reactivation. Band intensities of ID2 were quantified relative to tubulin (loading control) and presented as a ratio relative to ID2 at 0 hpi with lenti-GFP transduction. (B) RTA-qPCR of viral gene expression at 8 hpi and 48 hpi in samples shown in panel A. (C) Viral DNA replication was measured by qPCR during KSHV reactivation in samples shown in panel A. (D) Quantification of virus production at 72 hpi by qPCR based on virion-associated KSHV DNA in supernatants of iBCBL13xFLAG-RTA cells treated with lenti-GFP or lenti-ID2. t tests were performed between GFP-OE and ID2-OE, and degree of statistical significance is indicated as *, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001 (sample of n = 3).
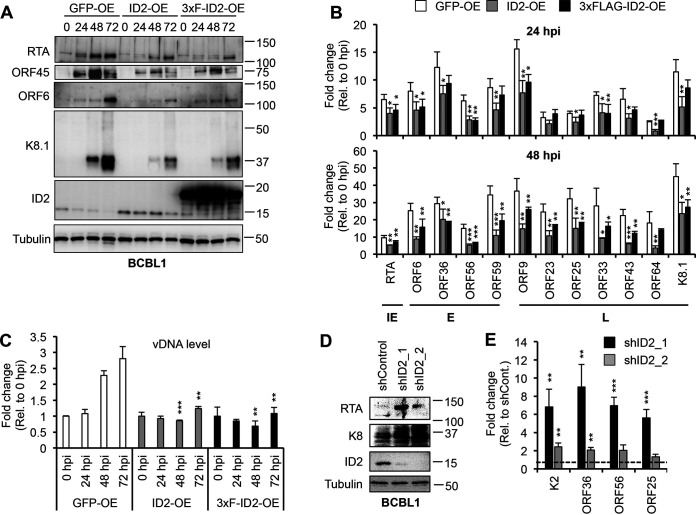
Because lytic reactivation in the iBCBL1-3xFLAG-RTA cells is triggered by Dox-inducible 3xFLAG-RTA, it is not possible to determine if ID2 can also block the viral lytic cycle by downregulating the activation of KSHV-encoded RTA gene itself. To determine whether ID2 can inhibit RTA gene expression, thereby impeding the KSHV lytic cycle, untagged ID2 or 3xFLAG-ID2 was overexpressed in BCBL1 by lentiviral transduction, and lytic reactivation was triggered by TPA, which initiates the lytic cycle through inducing RTA gene expression (Fig. 8). Immunoblot analysis in Fig. 8A shows that in lenti-GFP (control) BCBL1 cells, endogenous ID2 was rapidly downregulated, while the expression of lytic viral proteins (RTA, ORF45, ORF6, and K8.1) was increased, but in ID2-OE and 3xFLAG-ID2-OE BCBL1 cells, ID2 expression remained high, and the expression of lytic viral proteins, including RTA, was greatly reduced. RT-qPCR analysis confirmed that ID2 overexpression significantly reduced lytic viral gene expression (Fig. 8B), while qPCR test of viral DNA level showed that viral DNA replication was abolished (Fig. 8C). In agreement with ID2 being a repressor of the KSHV lytic cycle, we found that shRNA knockdown of ID2 in BCBL1 increased KSHV lytic gene expression (Fig. 8D and E). Taken together, these results indicate that ID2 is a repressor of the KSHV lytic cycle, and we hypothesize that is why ID2 expression must be downregulated to allow robust lytic gene expression during lytic reactivation.
FIG 8.
ID2 diminishes TPA-induced lytic reactivation of KSHV in BCBL1 cells. (A) Immunoblot analysis of ID2 and viral proteins in lenti-GFP-, lenti-ID2-, and lenti-3xFLAG-ID2-transduced BCBL1 cells during TPA-induced KSHV reactivation. (B) RT-qPCR measurement of viral gene expression at 24 hpi and 48 hpi in samples shown in panel A. (C) Viral DNA replication measured by qPCR during KSHV reactivation in samples described in panel A. (D) Immunoblot detection of lytic viral proteins and ID2 in ID2 lenti-shRNA-transduced BCBL1 cells at 3 days postransduction. (E) RT-qPCR analysis of lytic viral gene expression in samples shown in panel D. t tests were performed between GFP-OE and ID2-OE or shControl and shID2, and degree of statistical significance is indicated as *, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001 (sample of n = 3).
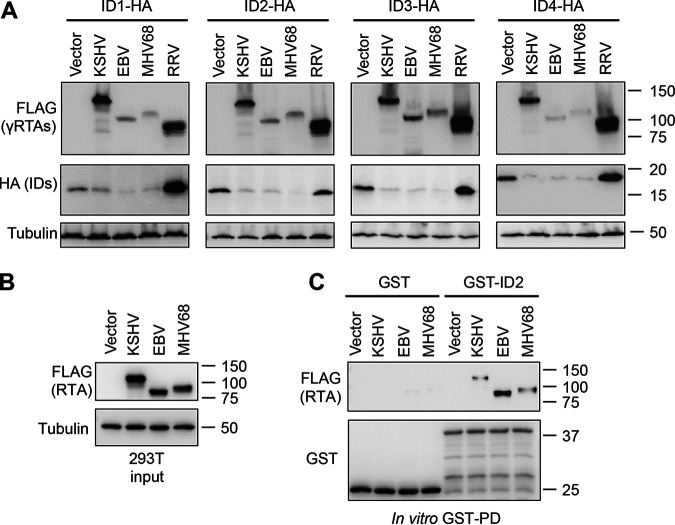
KSHV, EBV, and MHV68 RTAs can induce degradation of ID proteins.
RTA in the gammaherpesvirus subfamily is highly conserved in its transactivation function, while less is known about their protein degradation activities and whether they have evolutionary conserved host targets (35). It is also known that the four ID proteins are very similar to each other in humans, and furthermore, they are also highly conserved across species (36). Therefore, we aimed to determine whether gammaherpesvirus RTAs from KSHV, Epstein-Barr virus (EBV), rhesus murine gammaherpesvirus 68 (MHV68), and macaque rhadinovirus (RRV) can modulate the expression of any of the ID proteins (Fig. 9). To test this, 293T cells were cotransfected with C-terminally HA-tagged ID1, ID2, ID3, or ID4 in conjunction with a KSHV, EBV, MHV68, or RRV RTA, and then, we analyzed ID protein expression (Fig. 9A). Interestingly, the expression of each of the ID proteins could be downregulated by KSHV, EBV, and MHV68 RTAs, but not by RRV RTA, despite it being robustly expressed (Fig. 9A). In addition, in vitro GST pulldown revealed that both EBV and MHV68 RTAs interact with GST-ID2 similarly to KSHV RTA. These results indicate that the modulation of the abundance of ID proteins by gammaherpesvirus RTAs is evolutionarily conserved among gammaherpesviruses, which can be critical in the regulation of gammaherpesvirus lytic replication.
FIG 9.
Degradation of ID proteins by gammaherpesviral RTAs. (A) Immunoblot analysis of 293T cells after being cotransfected for 2 days with C-terminally HA-tagged ID1-ID4 and either a vector control or a 3xFLAG-tagged RTA derived from KSHV, EBV, MHV68 or RRV. (B) 293T cells were transfected with a vector control or 3xFLAG-tagged RTA from KSHV, EBV, and MHV68 for 2 days. Immunoblot detection of input fraction of cell lysates that were used for in vitro GST pulldown (GST-PD) in panel C. (C) Immunoblot analysis of in vitro GST-PD samples.
DISCUSSION
ID2 is a well-known dominant negative regulator of bHLH and other non-HLH transcription factors, thereby acting as a potent modulator of cellular gene expression (37). In this study, we discovered that ID2 acts as a repressor of KSHV viral lytic gene expression, diminishing viral DNA replication and virus production in PEL cells. We found that RTA mediates N-terminal ubiquitination and the rapid degradation of ID2 during KSHV reactivation, which favors robust lytic gene expression. We also discovered that in addition to KSHV RTA, its EBV and MHV68 homologs can also interact with ID2 and can block the expression of each of the ID proteins. Our findings suggest that ID proteins may play a repressive role in the lytic cycle of gammaherpesviruses, which is counteracted by gammaherpesviral RTAs to facilitate the lytic replication of viruses.
While the transactivation function of KSHV RTA is known to be essential to drive lytic viral gene expression, we still have limited knowledge about the biological significance of its protein degradation function during the KSHV lytic cycle. RTA has been shown to mediate the degradation of a few cellular and viral factors that affect the progress of the KSHV lytic cycle. RTA can polyubiquitinate and induce degradation of its substrate proteins through two different mechanisms. RTA either uses its own RING-like domain for ubiquitination and degradation of proteins such as MyD88 and Hey1, or it hijacks a host E3 ubiquitin ligase for targeted protein degradation, like in the case of IRF3, IRF7, and KSHV vFLIP (10–15). Since RTA interacts with the host E3 ubiquitin ligases ITCH and RAUL, we first tested if they play a role in RTA-mediated ID2 degradation. Even though we did not find any evidence for ITCH or RAUL being involved in degrading ID2, it is possible that RTA can be chaperoning, stabilizing, or upregulating the expression of another host E3 ligase to target ID2 for degradation. Since mutating the D-box of ID2 did not cause its stabilization in the presence of RTA, we also excluded that the anaphase-promoting complex (APC/CCdh1) is involved in the RTA-mediated degradation of ID2 (25). Next, we tested if RTA utilizes its own E3 ubiquitin ligase function in ID2 degradation. We found that the H145L and C141S mutations in the RING-like domain of RTA, which were demonstrated by others to fully abrogate its E3 ubiquitin ligase function (10, 12), reduced, but did not fully abolish, RTA’s protein degradation function. It is possible that the function of RING-like domain of RTA was not fully abrogated by the point mutations, which allowed for partial degradation of ID2. Interestingly, the RTA mutant lacking the C-terminal transactivation domain (aa 1 to 530) can still downregulate ID2 protein expression but less efficiently than the full-length RTA. This indicates that the first 530 residues of RTA are required but not sufficient for robust ID2 degradation. It is possible that the complete degradation of diverse substrates of RTA requires different E3 ubiquitin ligase activity from RTA or other parts of RTA than the RING-like domain. Nevertheless, based on our findings that RTA directly interacts with ID2 and RTA alone can induce polyubiquitination and degradation of ID2, which requires the N terminus of RTA, and MG132 treatment can block RTA-mediated ID2 degradation, we concluded that RTA induces ID2 degradation through the ubiquitin-proteasome pathway.
In general, proteins destined to be degraded through the proteasome pathway get polyubiquitinated via internal lysine residues located between the amino and carboxyl termini of proteins. Surprisingly, we found that RTA can induce the degradation of a lysine-less mutant of ID2. However, the N-terminal epitope tagging of ID2 prevented its RTA-mediated degradation. These results indicate that ID2 degradation by RTA does not need any lysine residues within ID2, but it does require the free N terminus of ID2. Importantly, several studies have shown that a subset of short-lived cellular and viral proteins, such as p21, MyoD, and human papillomavirus 16 E7 oncoprotein, can be targeted for protein degradation by an alternative mechanism of polyubiquitination, which is called N-terminal ubiquitination (29–33). This differs from the N-end rule of ubiquitination where the E3 ubiquitin ligase binds to a degron motif in the N terminus of a substrate protein to polymerize a ubiquitin chain from an internal lysine residue (38). N-terminal ubiquitination does not require a protein to contain internal lysine residues. Instead, the initial ubiquitin moiety is attached to the free N-terminal residue of the substrate protein, resulting in the subsequent polymerization of a Lys-48 ubiquitin chain (33). Although an earlier study has identified the type of polyubiquitin chain polymerized on the substrate protein of RTA, it has not been shown where in the substrate protein the ubiquitin chain is anchored (11). To the best of our knowledge, this is the first study demonstrating that RTA uses N-terminal ubiquitination to target a host protein for degradation. Further studies are required to determine whether RTA utilizes N-terminal ubiquitination for all its substrate proteins or just a small subset to target them for 26S proteasome-mediated degradation.
Based on the biological functions of proteins that are currently known to be degraded by RTA, they have one of the three following characteristics: (i) they possess the ability to induce antiviral immune responses (10, 11, 13), (ii) they dampen RTA gene expression, or (iii) they reduce the transcriptional activity of RTA (12, 14, 16, 39). Therefore, degradation of any of these host factors by RTA can evidently facilitate lytic viral gene expression. We identified ID2 as a novel suppressor of the KSHV lytic cycle, which can inhibit lytic replication through downregulating RTA expression or reducing lytic viral gene induction and viral DNA replication downstream of RTA. However, the question remains of how ID2 represses the viral lytic cycle. The primary target of ID proteins is the E protein family of bHLH transcription factors that have diverse functions in the regulation of development, cell growth, and cell survival (18). However, in addition to bHLH proteins, ID2 can also interact with other classes of transcription factors such as Rb, Pax, and the ternary complex factor (TCF) subfamily of ETS domain transcription factors (22, 40, 41). Importantly, TCF is a major activator of gene expression of c-Fos, which is one of the subunits of the AP-1 transcription complex (42, 43). TPA was shown to activate the KSHV lytic cycle by upregulating the expression of RTA and other lytic genes by inducing AP-1 activity (44). Since we found that ID2 overexpression in BCBL1 inhibits TPA-mediated induction of RTA expression, it is possible that ID2 may exert its repressive function through inhibiting TCF and TCF-mediated AP-1 expression. Further studies will be required to establish the specific mechanism of how ID2 suppresses RTA and RTA-mediated lytic gene expression.
While there is abundant information about the functions of ID proteins in development and cell cycle regulation, we still know very little about the role of ID proteins in the regulation of viral infections. Recent studies identified ID2 as an antiviral restriction factor of dengue virus, while ID1 and ID3 can repress the replication of foot-and-mouth disease virus, but the mechanism of how these ID proteins inhibit viral replication is still unknown (45, 46). In addition, ID1 and ID3 have been shown to be upregulated by viral factors of oncogenic viruses such as the EBV latent membrane protein 1 (LMP-1) and E6 of several different human papillomavirus (HPV) types, while KSHV latent protein LANA upregulates ID1, ID2, and ID3 in latently infected endothelial cells (23, 47–49). In the case of KSHV, upregulation of ID1-3 has been implicated to play a role in KSHV-induced oncogenesis, which is associated with latently infected cells, while their role in the lytic cycle was not addressed (23). Importantly, our study revealed that ID2 can also function in the repression of the lytic cycle, which promotes the maintenance of the pool of latently infected cells whose hyperproliferation can contribute to KSHV-associated tumorigenesis.
Furthermore, we also discovered that in addition to KSHV RTA, EBV and MHV68 RTAs can also bind to ID2, and they can downregulate the expression of all four ID proteins. These data indicate that not only the promoter transactivation role but also the protein degradation activity of gammaherpesvirus RTAs are evolutionarily conserved. In line with our observation, MHV68 RTA has also been shown to possess E3 ubiquitin ligase activity that is associated with its protein degradation function, while EBV RTA can induce protein degradation but without having its own E3 ubiquitin ligase activity (50, 51). Future studies are warranted to test how individual ID proteins affect the lytic cycle of different gammaherpesviruses to delineate their role in regulating latent-lytic switch of gammaherpesviruses.
MATERIALS AND METHODS
Cell lines.
293T (ATCC), HeLa (NIH AIDS Reagent Program), and iSLK cells (obtained from Jae Jung at the University of Southern California) were maintained in Dulbecco’s modified Eagle medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin (P/S). BCBL1 (NIH AIDS Reagent program) and iBCBL1-3xFLAG-RTA (also known as TREx-BCBL1-3xFLAG-RTA; see reference 17) are KSHV-positive PEL cell lines, while iBJAB-3xFLAG-RTA (also known as TREx-BJAB-3xFLAG-RTA; see reference 17) is a KSHV-free B cell lymphoma cell line. BCBL1 was maintained in RPMI supplemented with 10% FBS and P/S. The iBCBL1-3xFLAG-RTA and iBJAB-3xFLAG-RTA cell lines were cultured in RPMI containing 10% Tet system-approved FBS (TaKaRa), P/S, and 20 μg/mL hygromycin B.
Antibodies, inducers, and inhibitors.
The following primary antibodies were used in the study: anti-RTA (a gift from Yoshihiro Izumiya, UC Davis), anti-ORF6 (gift from Gary S. Hayward, Johns Hopkins University), anti-ORF45 (Santa Cruz; catalog no. sc-53883), anti-K8 (Santa Cruz; catalog no. sc-57889), anti-K8.1 (Santa Cruz; catalog no. sc-65446), anti-ID2 (Santa Cruz; catalog no. sc-489; Cell Signaling; catalog no. 3431S), anti-UBE3C/RAUL (Bethyl Lab; catalog no. A304-122A-T), anti-Itch (Bethyl Laboratories; catalog no. A301-992A-T), anti-ubiquitin (Cell Signaling; catalog no. 3933S), anti-tubulin (Sigma; catalog no. T5326), anti-HA (BioLegend; catalog no. 901501), anti-FLAG (Sigma; catalog no. F1804), anti-GST (Santa Cruz; catalog no. sc-138), and anti-Myc (BioLegend; catalog no. 626802). To induce the KSHV lytic cycle in BCBL1, TPA (12-O-tetradecanoylphorbol-13-acetate) and NaB (sodium butyrate) from Sigma were used at 20 ng/mL and 1 mM concentration, respectively. For inhibiting protein synthesis, cells were treated with 40 μM MG132 (https://www.selleckchem.com/) for 12 h before harvesting the cells for immunoblot analysis.
Plasmids and DNA transfection.
We used pCDH-CMV-MCS-EF1-puro vector to express RTA and ID2 in cells such as 3xFLAG-RTA of KSHV, EBV, MHV68, and RRV; mutants of KSHV RTA; untagged wild-type ID2 or its mutants; N- or C-terminal 3xFLAG-tagged ID2; and C-terminal HA-tagged ID1 to ID4. RTA and ID2 mutants were generated by PCR and In-Fusion cloning. For ID2 shRNA expression, the pLKO.1 lentiviral vector was used. GST and GST-ID2, which are expressed from a pGEX-2T plasmid, were gifts from Antonio Iavarone (Columbia University). To clone 3xFLAG-tagged EBV, MHV68, and RRV RTAs into pCDH-CMV-3xFLAG-MCS-EF1-puro vector, they were PCR amplified from different plasmids that were received from Sumita Bhaduri-McIntosh and Scott Tibbetts (University of Florida) and Dirk Dittmer (University of North Carolina at Chapel Hill), respectively. Plasmid transfection of 293T and HeLa cells was performed with polyethyleneimine (PEI; Polysciences) and Lipofectamine 2000 (Thermo Fisher Scientific) transfection reagents, respectively.
Lentivirus production and lentiviral transduction.
To produce lentivirus, the ID2-, shID2-, shITCH-, and shRAUL-expressing lentiviral vectors were cotransfected with the third-generation lentivirus packaging plasmids pmDC gag/pol, pRSV rev, and pmDK VSVg into 293T cells. Sixty hours after DNA transfection, the cell culture media were harvested, passed through 0.45-μm syringe filters, and concentrated by ultracentrifugation (24,000 rpm, 2 h, 10°C). PEL cells and iSLK cells were transduced by lentiviruses in the presence of 8 μg/mL polybrene by using spinning infection (2,000 rpm, 20 min, 30°C). Cells were infected with lentiviruses for 2 days before inducing the KSHV lytic cycle or RTA expression. The target sequences of shRNAs are the following: shID2-1, GCCTACTGAATGCTGTGTATA; shID2-2, GAGCCTGCTATACAACATGAA; shITCH-1, GCCGACAAATACAAATACAAA; shITCH-2, CCCAAGAATCAGAGGTTATAT; shRAUL-1, CCAGACATTACTACTTCCTA; and shRAUL-2, GCAGATAAGCAAGAAGTTCAA.
Total RNA isolation and RT-qPCR analysis.
Total RNA was purified from cells using TRIzol reagent (Invitrogen), and cDNA synthesis was performed with iScript cDNA synthesis kit (Bio-Rad) by using 1 μg of total RNA, which was followed by qPCR analysis as previously published (52). The qPCR was performed using SYBR green supermix (Bio-Rad) in a CFX96 real-time PCR machine. Gene expression changes were calculated either by using the threshold cycle (2−ΔCT) method, where gene expression was determined relative to the expression level of 18S rRNA, or using 2−ΔΔCT, which calculates fold change relative to a control sample. The gene expression graphs are based on three biological replicates. The sequences of RT-qPCR primers are shown in 5′ to 3′ orientation in Table 1. The primers were designed based on the BAC16 DNA sequence (GenBank accession no. GQ994935). Significance was determined through a two-tailed Student's t test, and the degree of significance was signified as ns, not significant; *, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001.
TABLE 1.
List of primers used in this study
| Gene | Forward sequence (5′–3′) | Reverse sequence (5′–3′) |
|---|---|---|
| AMP | TCGGTCCTCCGATCGTTGTCA | GTTGAGTACTCACCAGTCACA |
| HS1 | TTCCTATTTGCCAAGGCAGT | CTCTTCAGCCATCCCAAGAC |
| ID1 | ACGAGCAGCAGGTAAACGTGC | TCTCCACCTTGCTCACCTTGCGGT |
| ID2 | CACGGATATCAGCATCCTGTC | CACACAGTGCTTTGCTGTCAT |
| ID3 | ACTACATTCTCGACCTGCAGGTAG | GTTGGAGATGACAAGTTCCGGAGT |
| ID4 | TGCCTGCAGTGCGATATGAACGAC | TAACGTGCTGCAGGATCTCCACTT |
| 18S | TTCGAACGTCTGCCCTATCAA | GATGTGGTAGCCGTTTCTCAGG |
| RTA | CAAGGTGTGCCGTGTAGAGAT | GGTCAAAGCCTTACGCTTCTT |
| ORF6 | GTTCAAGATACCCTTGTATGACGA | CTTAGAGCCTGTGCTATTCCAGT |
| ORF9 | TGCCTAAACATAGCGGAGACCGT | GTCTACTGGCCTCCGCAGGAGA |
| ORF11 | GGCACCCATACAGCTTCTACGA | CGTTTACTACTGCACACTGCA |
| ORF18 | TGTGGAAGCTCGTGTACGAT | TATCGTTCAAGTGCATCCAG |
| ORF23 | GCACGCTCCATGATGGTAAC | CGTGTCACTTAACACAGACA |
| ORF25 | ACAGTTTATGGCACGCATAGTG | GGTTCTCTGAATCTCGTCGTGT |
| ORF33 | ATCATGTACGTCGTAAGCCA | GAAATCTCAGTCCGGATGCT |
| ORF36 | ATTGCCAACGACCTGATGCA | ACTCCAGTCCAGCTGCAGCA |
| ORF43 | GAGGACTCGTATGTCCTCCAGTC | GTCAGAGGTCAGACTCTACACTG |
| ORF45 | CCATACAGCGACCCTGATGA | CCGATTCTCTGACTCAATACT |
| ORF56 | CACAGATTCCCGTCAATACAAA | GTATCTTCAGTAGGCGGCAGAG |
| ORF59 | AACCGCAGTTCGTCAGGACCACCA | CCTTAGCCACTTAAGTAGGAATG |
| ORF64 | CTTCCTCGAGGGCATCATATAC | TATACGGTGATGGACTTGATGG |
| K2 | TCACTGCGGGTTAATAGGATTT | CATGACGTCCACGTTTATCACT |
| K8.1 | ATCTCCGTCGAGTGAGTATCCAA | GTGTCATAAAAGTCACGTGGGAG |
Quantification of KSHV replication and virus production.
Total DNA was purified from latent uninduced and lytic-induced PELs by lysing cells in radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, pH 8.0, 140 mM NaCl, 0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 1× protease inhibitor cocktail) followed by sonication and then phenol-chloroform extraction. Viral DNA replication was measured by qPCR using ORF11 (viral DNA)- and HS1 (host DNA)-specific primers and calculated by the 2−ΔΔCT method as described previously (52). To quantify virus production, virion-associated viral DNA was purified from supernatants of induced PEL cells at 72 hpi. Supernatants were first treated with DNase I (Sigma), which was followed by proteinase K (Invitrogen) treatment at 60°C for 1 h. Afterward, the supernatants were spiked with 1 pg of plasmid DNA carrying an ampicillin (AMP) gene, and DNA was purified from supernatants with phenol-chloroform extraction. ORF11- and AMP-specific primers were used to quantify viral DNA and plasmid DNA, respectively. AMP qPCR was used for normalization. Viral DNA copies per milliliter were calculated by using a standard curve based on serial dilution of purified KSHV BAC16 DNA. The qPCR graphs show the average of three biological replicates.
In vitro GST pulldown assay.
GST and GST-ID2 were expressed in Escherichia coli BL21(DE) strain for the GST pulldown assay. Overnight bacterial cultures were diluted 1:5 in LB medium supplemented with 100 μg/mL ampicillin and grown until an optical density at 600 nm (OD600) was between 0.4 and 0.6. Overexpression of GST and GST-ID2 was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h at 37°C. Afterward, the cells were collected, washed with cold Dulbecco's phosphate-buffered saline (DPBS), and lysed in lysis buffer (10 mM Tris-HCl, pH 8.0, 120 mM NaCl, 1 mM EDTA, 1 mM PMSF, and 500 μg/mL lysozyme) for 15 min on ice, which was followed by sonication. After centrifugation of the lysates, supernatants were precleared with protein A Sepharose for 1 h at 4°C and then incubated with glutathione agarose (Thermo Scientific) for 2 h at 4°C. The pelleted glutathione agarose was washed with immunoprecipitation buffer (50 mM Tris, pH 7.5, 150 mM NaCl, and 0.5% NP-40) three times and then incubated with 293T lysates for 2 h at 4°C. After centrifugation, the GST pulldown assays were washed with IP buffer three times, resuspended in 2× Laemmli buffer (Bio-Rad) supplemented with 2-mercaptoethanol, and boiled for 5 min at 95°C, and the samples were analyzed by immunoblotting.
Immunoprecipitation.
293T cells were cotransfected with 3xFLAG-tagged ID2 and Myc/His-RTA expression plasmids, and after 24 h, the cells were treated with 40 μM MG132 for 12 additional hours. Cells were harvested, washed with cold dPBS, and lysed in NP-40 lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, and 0.5% NP-40) containing 1 mM PMSF and 1× protease inhibitor cocktail. The lysates were passed through a 23-gauge needle 8 to 10 times, and then, they were incubated on ice for 30 min. After centrifugation of the lysates, supernatants were precleared with protein A Sepharose at 4°C overnight. The following day, the precleared lysate was incubated with primary antibodies for 3 h at 4°C and then protein A/G Plus-agarose (Santa Cruz) was added to the lysates and incubated for 2 h at 4°C. The immunoprecipitants (IPs) were washed three times for 10 min with rotating at 4°C before being resuspended in 2× Laemmli buffer. The IPs and inputs were analyzed by immunoblotting.
Proximity ligation assay.
The iBCBL1-3xFLAG-RTA cell line was induced with 1 μg/mL of Dox and treated with 40 μM MG132 immediately before being plated onto poly-l-lysine (Sigma)-coated coverslips at a density of 2.5 × 105 cells through centrifugation at 2,000 rpm for 20 min at 30°C. The cells were then left to incubate for 6 h at 37°C before being fixed with 4% paraformaldehyde for 10 min at room temperature (RT), permeabilized with 0.5% Triton-X for 5 min at RT, and washed 3 times with PBS containing 0.2% Tween 20. PLA was performed on the cells according to the manufacturer’s protocol (Sigma-Aldrich; catalog no. DUO92101). The primary antibodies used were all diluted 1:500 in antibody diluent provided by the PLA kit. Cells were either incubated with anti-IgG rabbit polyclonal antibody (Cell Signaling Technology; catalog no. 2729S) and anti-FLAG (Sigma; catalog no. F1804) as a negative control or with anti-ID2 (Santa Cruz Biotechnology; catalog no. sc-489) and anti-FLAG for 1 h at 37°C.
ACKNOWLEDGMENTS
We thank Antonio Iavarone (Columbia University), Jae U. Jung (University of Southern California), Gary S. Hayward (Johns Hopkins University), and Yoshihiro Izumiya (University of California, Davis) for providing reagents. We also thank the members of the Toth and Papp laboratories for the helpful discussions during the project and Julia Chen for the technical assistance.
This work was supported by NIH grants R01AI132554 and R01DE028331 as well as the American Cancer Society Research Scholar grant RSG-18-221-01-MPC. L.R.C. was supported by NSF Bridge to Doctorate fellowship (number 1500579) and NIH-NIDCR training grant T90DE021990. L.M.S. was supported by NIH-NIDCR training grant T90DE021990.
Contributor Information
Zsolt Toth, Email: ztoth@dental.ufl.edu.
Felicia Goodrum, University of Arizona.
REFERENCES
- 1.Mesri EA, Cesarman E, Boshoff C. 2010. Kaposi's sarcoma and its associated herpesvirus. Nat Rev Cancer 10:707–719. 10.1038/nrc2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cesarman E. 2011. Gammaherpesvirus and lymphoproliferative disorders in immunocompromised patients. Cancer Lett 305:163–174. 10.1016/j.canlet.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polizzotto MN, Uldrick TS, Hu D, Yarchoan R. 2012. Clinical manifestations of Kaposi sarcoma herpesvirus lytic activation: multicentric Castleman disease (KSHV-MCD) and the KSHV inflammatory cytokine syndrome. Front Microbiol 3:73. 10.3389/fmicb.2012.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Purushothaman P, Uppal T, Verma SC. 2015. Molecular biology of KSHV lytic reactivation. Viruses 7:116–153. 10.3390/v7010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aneja KK, Yuan Y. 2017. Reactivation and lytic replication of Kaposi's sarcoma-associated herpesvirus: an update. Front Microbiol 8:613. 10.3389/fmicb.2017.00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lukac DM, Kirshner JR, Ganem D. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J Virol 73:9348–9361. 10.1128/JVI.73.11.9348-9361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun R, Lin SF, Gradoville L, Yuan Y, Zhu F, Miller G. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc Natl Acad Sci USA 95:10866–10871. 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun R, Lin SF, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J Virol 73:2232–2242. 10.1128/JVI.73.3.2232-2242.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Izumiya Y, Kobayashi K, Kim KY, Pochampalli M, Izumiya C, Shevchenko B, Wang DH, Huerta SB, Martinez A, Campbell M, Kung HJ. 2013. Kaposi's sarcoma-associated herpesvirus K-Rta exhibits SUMO-targeting ubiquitin ligase (STUbL) like activity and is essential for viral reactivation. PLoS Pathog 9:e1003506. 10.1371/journal.ppat.1003506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu Y, Wang SE, Hayward GS. 2005. The KSHV immediate-early transcription factor RTA encodes ubiquitin E3 ligase activity that targets IRF7 for proteosome-mediated degradation. Immunity 22:59–70. 10.1016/j.immuni.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Yu Y, Hayward GS. 2010. The ubiquitin E3 ligase RAUL negatively regulates type i interferon through ubiquitination of the transcription factors IRF7 and IRF3. Immunity 33:863–877. 10.1016/j.immuni.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gould F, Harrison SM, Hewitt EW, Whitehouse A. 2009. Kaposi's sarcoma-associated herpesvirus RTA promotes degradation of the Hey1 repressor protein through the ubiquitin proteasome pathway. J Virol 83:6727–6738. 10.1128/JVI.00351-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Q, Liang D, Sun R, Jia B, Xia T, Xiao H, Lan K. 2015. Kaposi's sarcoma-associated herpesvirus-encoded replication and transcription activator impairs innate immunity via ubiquitin-mediated degradation of myeloid differentiation factor 88. J Virol 89:415–427. 10.1128/JVI.02591-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Z, Yan Z, Wood C. 2008. Kaposi's sarcoma-associated herpesvirus transactivator RTA promotes degradation of the repressors to regulate viral lytic replication. J Virol 82:3590–3603. 10.1128/JVI.02229-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chmura JC, Herold K, Ruffin A, Atuobi T, Fabiyi Y, Mitchell AE, Choi YB, Ehrlich ES. 2017. The Itch ubiquitin ligase is required for KSHV RTA induced vFLIP degradation. Virology 501:119–126. 10.1016/j.virol.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyu Y, Nakano K, Davis RR, Tepper CG, Campbell M, Izumiya Y. 2017. ZIC2 is essential for maintenance of latency and is a target of an immediate early protein during Kaposi's sarcoma-associated herpesvirus lytic reactivation. J Virol 91:e00980-17. 10.1128/JVI.00980-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papp B, Motlagh N, Smindak RJ, Jin Jang S, Sharma A, Alonso JD, Toth Z. 2019. Genome-wide identification of direct RTA targets reveals key host factors for Kaposi's sarcoma-associated herpesvirus lytic reactivation. J Virol 93:e01978-18. 10.1128/JVI.01978-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang LH, Baker NE. 2015. E proteins and ID proteins: helix-loop-helix partners in development and disease. Dev Cell 35:269–280. 10.1016/j.devcel.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lasorella A, Benezra R, Iavarone A. 2014. The ID proteins: master regulators of cancer stem cells and tumour aggressiveness. Nat Rev Cancer 14:77–91. 10.1038/nrc3638. [DOI] [PubMed] [Google Scholar]
- 20.Lasorella A, Uo T, Iavarone A. 2001. Id proteins at the cross-road of development and cancer. Oncogene 20:8326–8333. 10.1038/sj.onc.1205093. [DOI] [PubMed] [Google Scholar]
- 21.Verykokakis M, Zook EC, Kee BL. 2014. ID'ing innate and innate-like lymphoid cells. Immunol Rev 261:177–197. 10.1111/imr.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iavarone A, Garg P, Lasorella A, Hsu J, Israel MA. 1994. The helix-loop-helix protein Id-2 enhances cell proliferation and binds to the retinoblastoma protein. Genes Dev 8:1270–1284. 10.1101/gad.8.11.1270. [DOI] [PubMed] [Google Scholar]
- 23.Liang D, Hu H, Li S, Dong J, Wang X, Wang Y, He L, He Z, Gao Y, Gao SJ, Lan K. 2014. Oncogenic herpesvirus KSHV Hijacks BMP-Smad1-Id signaling to promote tumorigenesis. PLoS Pathog 10:e1004253. 10.1371/journal.ppat.1004253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sullivan JM, Havrda MC, Kettenbach AN, Paolella BR, Zhang Z, Gerber SA, Israel MA. 2016. Phosphorylation regulates Id2 degradation and mediates the proliferation of neural precursor cells. Stem Cells 34:1321–1331. 10.1002/stem.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lasorella A, Stegmuller J, Guardavaccaro D, Liu G, Carro MS, Rothschild G, de la Torre-Ubieta L, Pagano M, Bonni A, Iavarone A. 2006. Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature 442:471–474. 10.1038/nature04895. [DOI] [PubMed] [Google Scholar]
- 26.Trausch-Azar JS, Lingbeck J, Ciechanover A, Schwartz AL. 2004. Ubiquitin-proteasome-mediated degradation of Id1 is modulated by MyoD. J Biol Chem 279:32614–32619. 10.1074/jbc.M403794200. [DOI] [PubMed] [Google Scholar]
- 27.Deed RW, Armitage S, Norton JD. 1996. Nuclear localization and regulation of Id protein through an E protein-mediated chaperone mechanism. J Biol Chem 271:23603–23606. 10.1074/jbc.271.39.23603. [DOI] [PubMed] [Google Scholar]
- 28.Glickman MH, Ciechanover A. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82:373–428. 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 29.Sadeh R, Breitschopf K, Bercovich B, Zoabi M, Kravtsova-Ivantsiv Y, Kornitzer D, Schwartz A, Ciechanover A. 2008. The N-terminal domain of MyoD is necessary and sufficient for its nuclear localization-dependent degradation by the ubiquitin system. Proc Natl Acad Sci USA 105:15690–15695. 10.1073/pnas.0808373105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fajerman I, Schwartz AL, Ciechanover A. 2004. Degradation of the Id2 developmental regulator: targeting via N-terminal ubiquitination. Biochem Biophys Res Commun 314:505–512. 10.1016/j.bbrc.2003.12.116. [DOI] [PubMed] [Google Scholar]
- 31.Bloom J, Amador V, Bartolini F, DeMartino G, Pagano M. 2003. Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell 115:71–82. 10.1016/s0092-8674(03)00755-4. [DOI] [PubMed] [Google Scholar]
- 32.Reinstein E, Scheffner M, Oren M, Ciechanover A, Schwartz A. 2000. Degradation of the E7 human papillomavirus oncoprotein by the ubiquitin-proteasome system: targeting via ubiquitination of the N-terminal residue. Oncogene 19:5944–5950. 10.1038/sj.onc.1203989. [DOI] [PubMed] [Google Scholar]
- 33.Breitschopf K, Bengal E, Ziv T, Admon A, Ciechanover A. 1998. A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J 17:5964–5973. 10.1093/emboj/17.20.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aviel S, Winberg G, Massucci M, Ciechanover A. 2000. Degradation of the Epstein-Barr virus latent membrane protein 1 (LMP1) by the ubiquitin-proteasome pathway. Targeting via ubiquitination of the N-terminal residue. J Biol Chem 275:23491–23499. 10.1074/jbc.M002052200. [DOI] [PubMed] [Google Scholar]
- 35.Damania B, Jeong JH, Bowser BS, DeWire SM, Staudt MR, Dittmer DP. 2004. Comparison of the Rta/Orf50 transactivator proteins of gamma-2-herpesviruses. J Virol 78:5491–5499. 10.1128/jvi.78.10.5491-5499.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roschger C, Cabrele C. 2017. The Id-protein family in developmental and cancer-associated pathways. Cell Commun Signal 15:7. 10.1186/s12964-016-0161-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. 1990. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 61:49–59. 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 38.Varshavsky A. 1996. The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci USA 93:12142–12149. 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yada K, Do E, Sakakibara S, Ohsaki E, Ito E, Watanabe S, Ueda K. 2006. KSHV RTA induces a transcriptional repressor, HEY1 that represses rta promoter. Biochem Biophys Res Commun 345:410–418. 10.1016/j.bbrc.2006.04.092. [DOI] [PubMed] [Google Scholar]
- 40.Stinson J, Inoue T, Yates P, Clancy A, Norton JD, Sharrocks AD. 2003. Regulation of TCF ETS-domain transcription factors by helix-loop-helix motifs. Nucleic Acids Res 31:4717–4728. 10.1093/nar/gkg689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts EC, Deed RW, Inoue T, Norton JD, Sharrocks AD. 2001. Id helix-loop-helix proteins antagonize pax transcription factor activity by inhibiting DNA binding. Mol Cell Biol 21:524–533. 10.1128/MCB.21.2.524-533.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cavigelli M, Dolfi F, Claret FX, Karin M. 1995. Induction of c-fos expression through JNK-mediated TCF/Elk-1 phosphorylation. EMBO J 14:5957–5964. 10.1002/j.1460-2075.1995.tb00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Halazonetis TD, Georgopoulos K, Greenberg ME, Leder P. 1988. c-Jun dimerizes with itself and with c-Fos, forming complexes of different DNA binding affinities. Cell 55:917–924. 10.1016/0092-8674(88)90147-x. [DOI] [PubMed] [Google Scholar]
- 44.Cohen A, Brodie C, Sarid R. 2006. An essential role of ERK signalling in TPA-induced reactivation of Kaposi's sarcoma-associated herpesvirus. J Gen Virol 87:795–802. 10.1099/vir.0.81619-0. [DOI] [PubMed] [Google Scholar]
- 45.Luo Y, Wang G, Ren T, Zhang T, Chen H, Li Y, Yin X, Zhang Z, Sun Y. 2021. Screening of host genes regulated by ID1 and ID3 proteins during foot-and-mouth disease virus infection. Virus Res 306:198597. 10.1016/j.virusres.2021.198597. [DOI] [PubMed] [Google Scholar]
- 46.Zanini F, Pu SY, Bekerman E, Einav S, Quake SR. 2018. Single-cell transcriptional dynamics of flavivirus infection. Elife 7:e32942. 10.7554/eLife.32942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ikeda JI, Wada N, Nojima S, Tahara S, Tsuruta Y, Oya K, Morii E. 2016. ID1 upregulation and FoxO3a downregulation by Epstein-Barr virus-encoded LMP1 in Hodgkin's lymphoma. Mol Clin Oncol 5:562–566. 10.3892/mco.2016.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lo AK, Dawson CW, Lo KW, Yu Y, Young LS. 2010. Upregulation of Id1 by Epstein-Barr virus-encoded LMP1 confers resistance to TGFbeta-mediated growth inhibition. Mol Cancer 9:155. 10.1186/1476-4598-9-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akil N, Yasmeen A, Kassab A, Ghabreau L, Darnel AD, Al Moustafa AE. 2008. High-risk human papillomavirus infections in breast cancer in Syrian women and their association with Id-1 expression: a tissue microarray study. Br J Cancer 99:404–407. 10.1038/sj.bjc.6604503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De La Cruz-Herrera CF, Shire K, Siddiqi UZ, Frappier L. 2018. A genome-wide screen of Epstein-Barr virus proteins that modulate host SUMOylation identifies a SUMO E3 ligase conserved in herpesviruses. PLoS Pathog 14:e1007176. 10.1371/journal.ppat.1007176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dong X, He Z, Durakoglugil D, Arneson L, Shen Y, Feng P. 2012. Murine gammaherpesvirus 68 evades host cytokine production via replication transactivator-induced RelA degradation. J Virol 86:1930–1941. 10.1128/JVI.06127-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toth Z, Smindak RJ, Papp B. 2017. Inhibition of the lytic cycle of Kaposi's sarcoma-associated herpesvirus by cohesin factors following de novo infection. Virology 512:25–33. 10.1016/j.virol.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]