ABSTRACT
Gammaherpesviruses (GHVs) are lymphotropic tumor viruses with a biphasic infectious cycle. Lytic replication at the primary site of infection is necessary for GHVs to spread throughout the host and establish latency in distal sites. Dissemination is mediated by infected B cells that traffic hematogenously from draining lymph nodes to peripheral lymphoid organs, such as the spleen. B cells serve as the major reservoir for viral latency, and it is hypothesized that periodic reactivation from latently infected B cells contributes to maintaining long-term chronic infection. While fundamentally important to an understanding of GHV biology, aspects of B cell infection in latency establishment and maintenance are incompletely defined, especially roles for lytic replication and reactivation in this cell type. To address this knowledge gap and overcome limitations of replication-defective viruses, we generated a recombinant murine gammaherpesvirus 68 (MHV68) in which ORF50, the gene that encodes the essential immediate-early replication and transcription activator protein (RTA), was flanked by loxP sites to enable conditional ablation of lytic replication by ORF50 deletion in cells that express Cre recombinase. Following infection of mice that encode Cre in B cells with this virus, splenomegaly and viral reactivation from splenocytes were significantly reduced; however, the number of latently infected splenocytes was equivalent to WT MHV68. Despite ORF50 deletion, MHV68 latency was maintained over time in spleens of mice at levels approximating WT, reactivation-competent MHV68. Treatment of infected mice with lipopolysaccharide (LPS), which promotes B cell activation and MHV68 reactivation ex vivo, yielded equivalent increases in the number of latently infected cells for both ORF50-deleted and WT MHV68, even when mice were simultaneously treated with the antiviral drug cidofovir to prevent reactivation. Together, these data demonstrate that productive viral replication in B cells is not required for MHV68 latency establishment and support the hypothesis that B cell proliferation facilitates latency maintenance in vivo in the absence of reactivation.
IMPORTANCE Gammaherpesviruses establish lifelong chronic infections in cells of the immune system and place infected hosts at risk for developing lymphomas and other diseases. It is hypothesized that gammaherpesviruses must initiate acute infection in these cells to establish and maintain long-term infection, but this has not been directly tested. We report here the use of a viral genetic system that allows for cell-type-specific deletion of a viral gene that is essential for replication and reactivation. We employ this system in an in vivo model to reveal that viral replication is not required to initiate or maintain infection within B cells.
KEYWORDS: B cell, MHV68, ORF50, RTA, chronic infection, gammaherpesvirus, latency, lytic replication, reactivation
INTRODUCTION
Herpesviruses are large, enveloped viruses that contain a double-strand DNA genome within a protein capsid and establish lifelong chronic infections (1). The gammaherpesvirus (GHV) subfamily includes the human pathogens, Epstein-Barr virus (EBV) and Kaposi sarcoma-associated herpesvirus (KSHV), which infect a large percentage of the adult population worldwide. GHV infections do not typically result in severe disease in healthy individuals but can cause a variety of cancers, especially in immunocompromised patients (2–9). EBV is the causative agent of infectious mononucleosis and is associated with Burkitt lymphoma, nasopharyngeal carcinoma, post-transplant lymphoproliferative disorder, Hodgkin lymphoma, and other cancers in chronically infected persons (3, 8, 10–12). KSHV is the etiologic agent of Kaposi sarcoma, an AIDS-defining malignancy, as well as multicentric Castleman disease and primary effusion lymphoma (2, 4–6, 9, 13–15). These diseases are epidemiologically and economically impactful, and laboratory-based pathogenesis studies are important to understand how these viruses infect a host and cause disease.
Like all herpesvirus infections, the GHV infection cycle is biphasic. The productive replication phase, a process known as lytic replication, occurs following primary infection of a host. Here, viral gene products are expressed in a regulated cascade and infectious viral particles are produced (16). The second phase of infection is chronic and persists for the life of the infected host. This phase, known as latency, is characterized by minimal expression of viral genes as the virus maintains the genome as a circular episome within the host-cell nucleus (17–19). Proteins expressed by the virus in latency facilitate maintenance of the viral genome and promote cellular proliferation and survival (20). Upon receiving an appropriate stimulus, a latently infected cell can reactivate and reinitiate the lytic cycle.
Because chronic GHV infection can lead to severe disease, understanding mechanisms of viral infection and maintenance of chronic infection is fundamental. However, human GHVs exhibit a very narrow host range and do not readily infect laboratory animals. Murine gammaherpesvirus 68 (MHV68) is a natural pathogen of murid rodents that provides a highly tractable small-animal model for studying GHV infection and latency in vivo (21–24). MHV68 is a close genetic relative of the human GHVs; genes essential for viral replication are largely conserved among the GHVs, and there is overlap in the host RNAs targeted by EBV, KSHV, and MHV68 non-coding RNAs (25–27). Several MHV68 and KSHV latency gene products also are conserved and functionally interchangeable (28–31). MHV68 pathogenesis recapitulates key aspects of human GHV infections, targeting B cells as the preferred reservoir for latency and promoting development of lymphomas and lymphoproliferative diseases following infection (22, 27).
The MHV68 mouse model has shaped our understanding of GHV infection, dissemination, and persistence in a host. Following intranasal (IN) inoculation of mice, MHV68 undergoes lytic replication in the respiratory epithelium (32). This enables MHV68 to infect tissue-resident and infiltrating immune cells, cross the epithelial barrier, and traffic to draining lymph nodes (33–36). MHV68 is thought to expand in the lymph node prior to disseminating to distal sites, such as the spleen (33, 35, 37–39). In addition to B cells, MHV68 establishes latency in macrophages, dendritic cells, epithelial cells, and endothelial cells (40); however, several lines of evidence indicate that B cells play an indispensable role in MHV68 trafficking to the spleen.
For example, MHV68 does not establish latency in the spleen following intranasal inoculation of B cell-deficient μMT mice, but dissemination to the spleen is restored if B cells are adoptively transferred into these animals prior to infection (41, 42). Experiments in which latency-associated genes ORF73 and M2 were conditionally deleted in B cells, demonstrate that virus accumulates in draining lymph nodes after IN inoculation, but bloodborne infection (for ORF73) and latency in the spleen are drastically reduced, indicating the importance of the B cell population in viral dissemination (33, 39). Interestingly, replication-defective viruses also either fail to disseminate after IN inoculation or exhibit severe attenuation in the spleen (43–45), suggesting that lytic replication also plays a pivotal role in seeding distal organs with latent virus. Further, despite the clear importance of B cells and lytic replication in systemic MHV68 dissemination and latency, it is not known if lytic replication within the B cell compartment is required for trafficking to and establishing latency in distal cellular reservoirs.
Additionally, the mechanisms used by GHVs to maintain latency within specific cellular populations are not completely defined. Within the spleen, latently infected B cells are the predominant reactivation competent cell type in MHV68-infected mice, and will reactivate if explanted, especially if cells are treated with B cell-activating stimuli (46–48). Clearly, reactivation can provide a means for herpesviruses to “get out” and infect new hosts. However, it is also suggested that periodic “homeostatic reactivation” contributes to the long-term maintenance of latency within a host. This hypothesis is mostly based on observations suggesting that reactivation occurs in splenic B cells and macrophages during latency. For instance, adoptive transfer of B cells from mice latently infected with MHV68 into naive mice leads to infection of B cells within recipient mice (49). Further, treating infected mice with Toll-like receptor (TLR) agonists, such as lipopolysaccharide (LPS), stimulates GHV reactivation from B cells and increases the number of infected splenocytes in vivo (48). However, whether periodic reactivation, plays a major role in maintaining long-term latency in vivo, especially in immunocompetent animals, is not completely understood. Another viable hypothesis is that proliferation of latently infected B cells in response to other infections or B cell activating stimuli is sufficient to maintain the B cell latency reservoir. This has not been directly tested.
In experiments described here, we sought to determine whether GHVs require lytic replication within B cells in order to traffic systemically, establish, and maintain long-term latency. Traditional loss-of-function mutations in MHV68 lytic genes are valuable tools for defining functions of viral gene products during acute viral replication at the site of inoculation, but they are not suitable to answer questions pertaining to cell-type-specific requirements for essential lytic-cycle proteins or defining downstream roles for such proteins in viral pathogenesis (45, 50). To address this limitation, we generated an MHV68 recombinant virus that encodes loxP sites flanking a gene essential for lytic replication, ORF50. Utilizing cell-type-specific, Cre-mediated deletion of this gene, we define B cell-specific roles for lytic replication and reactivation in chronic GHV infection.
RESULTS
Development and validation of a conditional ORF50 mutant.
The findings that (i) adoptive transfer of B cells into B cell-deficient mice restores trafficking to and latency establishment in the spleen after IN inoculation, and (ii) that deletion of ORF73, the gene that encodes mLANA, specifically in B cells prevented trafficking from mediastinal LNs to the spleen demonstrate that B cells are critical for MHV68 to colonize the mouse spleen after IN inoculation (33, 41). Replication defective viruses, such as those with null mutations in ORF50 or ORF31, essential genes that respectively encode the replication and transcription activator (RTA) or a homolog of EBV BDLF4, fail to establish latency in the spleen following IN inoculation, suggesting the importance of lytic replication to latency establishment (43, 44, 51). However, whether lytic replication in the B cell compartment is necessary for viral dissemination to distal sites of latency is not known.
To address this question, we generated an MHV68 conditional mutant that can be rendered incapable of lytic replication by deletion of ORF50 in a tissue-specific manner. We used a recombineering approach to insert loxP sites flanking the second exon of the ORF50 gene in the MHV68 BAC to enable deletion of ORF50 by Cre recombinase (O50.loxP MHV68) (Fig. 1A). To verify that loxP-flanked (floxed) ORF50 was efficiently deleted from O50.loxP MHV68, we infected normal or Cre-expressing Vero cells with either wild-type MHV68 (WT MHV68) or O50.loxP and evaluated ORF50 deletion by PCR. While intact ORF50 was readily detected in Vero cells infected with O50.loxP, only the deletion product was detected in Vero-Cre cells infected with the conditional mutants (Fig. 1B). The intact locus was detected in both cell types following infection with WT MHV68. The ORF73 gene was examined by PCR for off-target effects and found to be unaffected in all viruses tested.
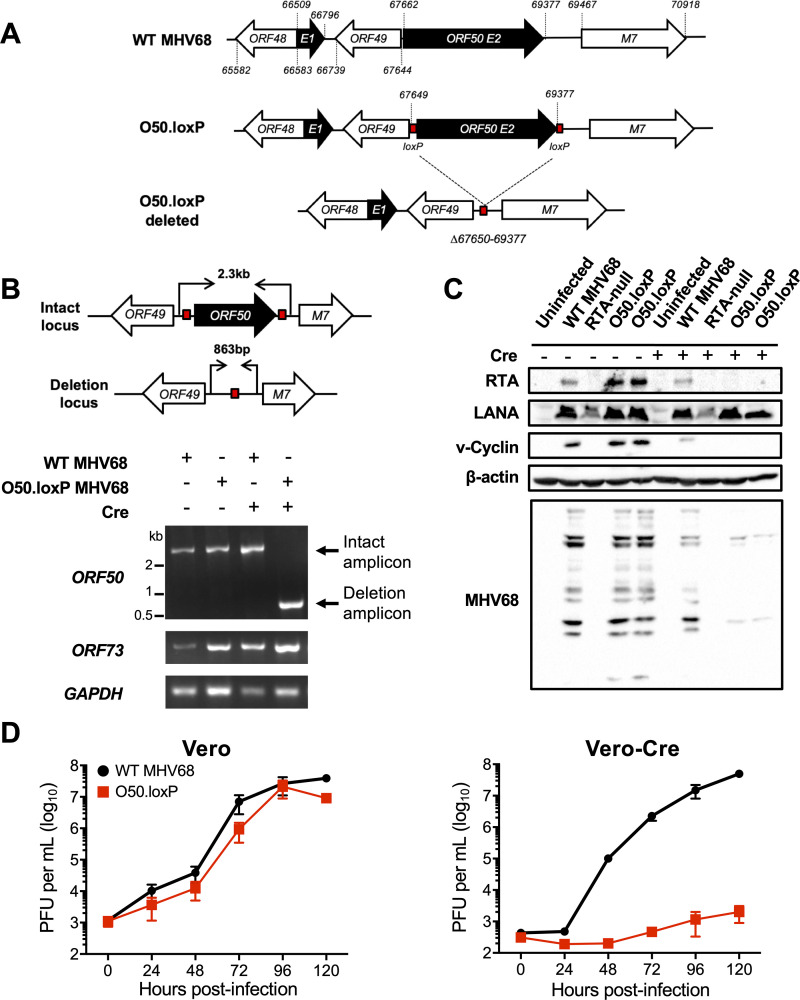
FIG 1.
Derivation and validation of O50.loxP MHV68. (A) Schematic depicting the insertion of loxP sites flanking ORF50 in the genome and its deletion after Cre-mediated recombination. loxP sites were inserted flanking ORF50 exon 2 (E2) after nucleotides 67649 and 69377 in the MHV68 genome. Cre-mediated recombination removes the ORF50 E2 coding region (Δ67650 to 69377). (B) Vero cells or Vero-Cre cells were infected with the indicated virus at an MOI of 5 PFU/cell. Total DNA was isolated at 24 h postinfection, and PCR was performed as illustrated in the schematic to detect the intact or deleted ORF50 locus, the distal ORF73 locus, and GAPDH as a control. (C) 3T3 fibroblasts that encode Cre-ERT2 were treated with vehicle (−) or 4-hydroxytamoxifen (+) to induce Cre activity 24 h prior to infection. Treated cells were infected with the indicated viruses at an MOI of 0.5 PFU/cell. Cells were lysed on day 4 postinfection, and proteins were resolved by SDS-PAGE. Immunoblot analyses were performed using antibodies to detect the indicated proteins. Cellular β-actin serves as a loading control. (D) Vero cells and Vero-Cre cells were infected with WT MHV68 or O50.loxP MHV68 at 0.05 PFU/cell. Viral titers were determined by plaque assay at the indicated times postinfection. Results are means of triplicate samples. Error bars represent standard deviations.
Because the RTA protein encoded by ORF50 is an essential immediate-early gene product that is required for early viral gene expression and ultimately viral replication, we evaluated whether ORF50 deletion correlated with a loss of viral protein production and inhibition of viral replication. Consistent with ORF50 deletion and the requirement for RTA in promoting viral gene expression, MHV68 proteins were not detected in immunoblot analyses using MHV68 antiserum on lysates from 3T3 cells expressing inducible-Cre (3T3-iCre) infected with O50.loxP (Fig. 1C). In contrast, MHV68 antigens were readily detected in lysates from non-induced 3T3-iCre cells infected with O50.loxP or WT MHV68. Expression of the immediate-early protein LANA was not affected by RTA ablation. The slight reduction in viral antigen detection observed in WT MHV68 infected 3T3-iCre cells is likely due to tamoxifen-related repression of viral entry and nuclear translocation (52, 53). Consistent with the essential role of RTA in viral replication, O50.loxP was severely defective for viral replication in Vero-Cre cells relative to WT MHV68 but replicated as efficiently as WT virus in Vero cells that did not express Cre (Fig. 1D). Together, these data demonstrate that Cre recombinase deletes a floxed ORF50 gene from the MHV68 genome during infection, thereby blocking downstream production of viral proteins and ultimately viral replication. Moreover, efficient O50.loxP replication in cells that did not express Cre suggests that the presence of loxP sites flanking ORF50 did not adversely impact progression through the lytic replication cycle.
We next evaluated O50.loxP MHV68 lytic replication, latency, and reactivation in vivo in WT mice that do not express Cre recombinase. On day 7 following IN inoculation of WT C57BL/6 mice, titers of both WT MHV68 and O50.loxP were equivalent in the lungs, strongly suggesting that O50.loxP lytic infection is not attenuated in vivo (Fig. 2A). Evaluating latency establishment and reactivation efficiency in spleens on days 16 to 18 after IN inoculation using limiting-dilution PCR analyses and cytopathic effect (CPE) assays, respectively, we found that O50.loxP established latency and reactivated from splenocytes as efficiently as WT MHV68 following infection of C57BL/6 mice (Fig. 2B and D; Table 1 and 2). Spleen weights, a correlate of virus-induced splenomegaly (54), also were comparable between WT MHV68 and O50.loxP infections (Fig. 2C). Together these data indicate that insertion of loxP sites flanking the ORF50 gene does not attenuate MHV68 infection of mice in the absence of Cre-recombinase.
FIG 2.
O50.loxP exhibits efficient acute replication, latency, and reactivation in C57BL/6 mice. C57BL/6 mice were infected IN with 1000 PFU of the indicated virus. (A) Mice were sacrificed on day 7 postinfection, and viral titers in lung homogenates were determined by plaque assay. (B to D) Mice were sacrificed on days 16–18 postinfection and spleens were collected. (B) Single-cell suspensions of spleen cells were serially diluted, and the frequencies of cells harboring MHV68 genomes were determined using a limiting-dilution PCR analysis. (C) Spleens were weighed as a measure of splenomegaly. The dashed line indicates the average mass of spleens from mock-infected mice. (D) Reactivation frequencies were determined by ex vivo plating of serially diluted cells on an indicator monolayer. Cytopathic effect was scored 2 to 3 weeks postplating. For A and C, each dot represents one mouse. For B and D, groups of three to five mice were pooled for each infection and analysis. Results are means of two to three independent infections. Error bars represent the standard error of the means. ns denotes P > 0.05 in a two-tailed Student’s t test.
TABLE 1.
Frequency of MHV68 latently infected cells
| Mouse strain | Virus | Route of infection | Cell population | Days postinfection | Latency frequency |
|---|---|---|---|---|---|
| C57BL/6 | WT MHV68 | IN | Spleen | 16 | 1/140 |
| 50.loxP | IN | Spleen | 16 | 1/75 | |
| CD19-Cre | WT MHV68 | IN | Spleen | 16 | 1/65 |
| 50.loxP | IN | Spleen | 16 | 1/100 | |
| WT MHV68 | IN | Spleen | 42 | 1/1,900 | |
| 50.loxP | IN | Spleen | 42 | 1/1,700 | |
| WT MHV68 | IP | Spleen | 16 | 1/140 | |
| 50.loxP | IP | Spleen | 16 | 1/550 | |
| WT MHV68 | IP | PEC | 16 | 1/500 | |
| 50.loxP | IP | PEC | 16 | 1/570 |
TABLE 2.
Frequency of reactivation competent cells
| Mouse strain | Virus | Route of infection | Cell population | Days postinfection | Reactivation frequencya |
|---|---|---|---|---|---|
| C57BL/6 | WT MHV68 | IN | Spleen | 16 | 1/25,000 |
| 50.loxP | IN | Spleen | 16 | 1/11,000 | |
| CD19-Cre | WT MHV68 | IN | Spleen | 16 | 1/7,800 |
| 50.loxP | IN | Spleen | 16 | BLD | |
| WT MHV68 | IP | Spleen | 16 | 1/4,500 | |
| 50.loxP | IP | Spleen | 16 | BLD | |
| WT MHV68 | IP | PEC | 16 | 1/1200 | |
| 50.loxP | IP | PEC | 16 | 1/2000 |
BLD, below the limit of detection of 1 in 100,000 cells.
Deletion of ORF50 in B cells does not prevent MHV68 latency establishment.
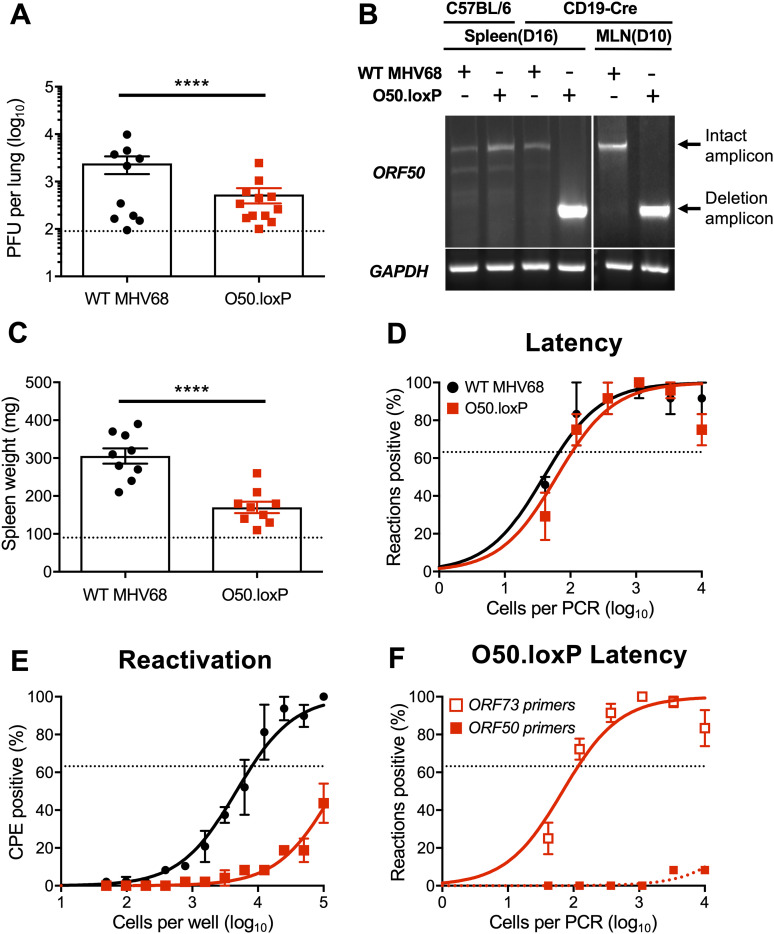
To test the hypothesis that productive viral replication in B cells influences MHV68 latency, we infected mice that encode Cre-recombinase under the control of the CD19 pan-B cell promoter (55), to allow for B cell-specific deletion of ORF50 in O50.loxP infected animals. Of note, we previously established this model in tests of viruses in which the ORF73 and M2 genes were floxed in the MHV68 genome (33, 39). On day 7 postinfection, O50.loxP titers were modestly lower than WT virus titers in lungs of CD19-Cre mice (Fig. 3A). A qualitative PCR analysis of the ORF50 locus in lymphoid tissues following infection by WT MHV68 and O50.loxP demonstrated that the floxed viral gene was deleted in both mediastinal lymph nodes (MLNs) and spleens on days 10 and 16 postinfection, respectively (Fig. 3B). Because several lines of evidence including our previous work with floxed ORF73 and M2 viruses indicate that MHV68 passes through draining lymph nodes such as the MLNs before systemic infection (33, 35–37), these observations suggest that ORF50 deletion occurs early during the MHV68 dissemination process in CD19-Cre mice. Compared with WT MHV68 infection, splenomegaly was significantly reduced in CD19-Cre mice infected with O50.loxP (Fig. 3C), suggesting that lytic replication in B cells contributes to the MHV68 IM-like syndrome (54). However, O50.loxP established latency in spleens of CD19-Cre mice at levels equivalent to WT MHV68 (Fig. 3D; Table 1). Consistent with the requirement for RTA in viral replication, reactivation by O50.loxP was severely compromised in CD19-Cre mice (Fig. 3E; Table 2). Importantly, a quantitative analysis of latently infected splenocytes using well-characterized primers specific to the ORF50 locus (56) revealed that essentially all viral genomes present in the spleen on day 16 postinfection no longer contained an intact ORF50 gene (Fig. 3F). Together, these data suggest that the presence of ORF50 and likely the RTA protein in B cells is critical for infection-associated splenomegaly and MHV68 reactivation from splenocytes but is not required for latency establishment in the spleen following IN inoculation.
FIG 3.
O50.loxP replicates efficiently and establishes latency, but reactivation is impaired in CD19-Cre mice. CD19-Cre mice were infected IN with 1,000 PFU of the indicated virus. (A) Mice were sacrificed on day 7 postinfection, and viral titers in lung homogenates were determined by plaque assay. Each dot represents one mouse. (B) Total DNA was isolated from spleens or lymph nodes of infected C57BL/6 or CD19-Cre mice on days 10 or 16 postinfection with the indicated virus. PCR was performed to evaluate the integrity of the ORF50 locus or GAPDH as a control. (C to F) Mice were sacrificed on day 16 postinfection. (C) Spleens were harvested and weighed as a measure of splenomegaly. The dashed line indicates the average mass of spleens from mock-infected mice. Each dot represents one mouse. (D) The frequency of cells harboring MHV68 genomes was determined by limiting-dilution PCR analysis. (E) Reactivation frequencies were determined by ex vivo plating of serially diluted cells on an indicator monolayer. Cytopathic effect was scored 2 to 3 weeks postplating. (F) ORF50 deletion was confirmed by comparing limiting-dilution PCR analyses performed using primers specific for either the ORF50 locus or ORF73 locus. Groups of three to five mice were pooled for each infection and analysis. Results are means of two to three independent infections. Error bars represent standard error of the means. **** denotes P < 0.0001 in a two-tailed Student’s t test.
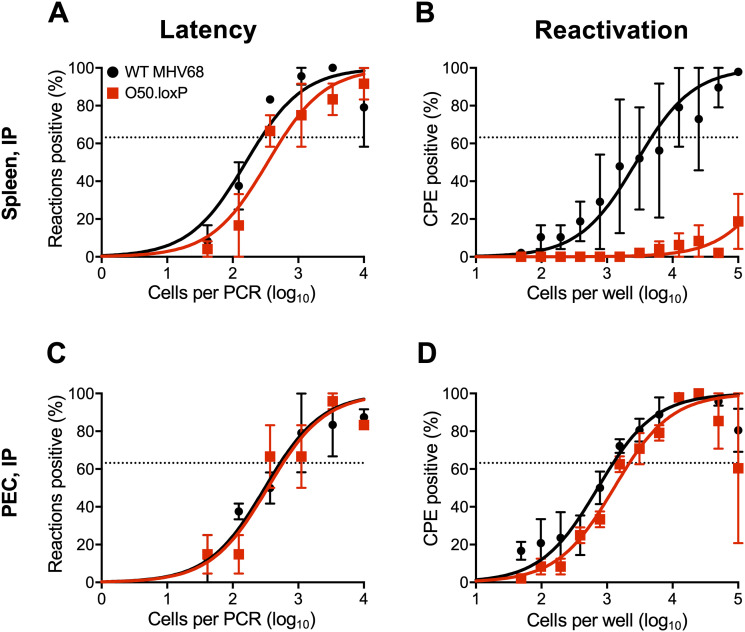
In addition to B cells, peritoneal macrophages also harbor latent MHV68, especially following intraperitoneal (IP) inoculation. To more completely understand how ORF50 deletion in B cells influences chronic MHV68 infection, we assessed latency and reactivation following IP inoculation of CD19-Cre mice. Consistent with results following IN infection, O50.loxP MHV68 established latency at frequencies comparable to WT MHV68 in splenocytes, while reactivation was substantially reduced (Fig. 4A and B; Table 1 and 2). In peritoneal exudate cells (PECs), O50.loxP established latency in CD19-Cre mice at levels equivalent to WT MHV68 and did not exhibit a defect in ex vivo reactivation, which agrees with the notion that macrophages, which are CD19- and should not express Cre-recombinase in these mice, are the major cell type in the peritoneal cavity infected by MHV68 (Fig. 4C and D; Table 1 and 2). These findings support the conclusion that reactivation from splenocytes, but not PECs, primarily involves B cells, and further suggests that MHV68 does not pass through a B cell prior to infecting the majority of PECs, following IP infection of mice.
FIG 4.
O50.loxP establishes latency in spleens of CD19-Cre mice, but does not reactivate, after IP infection. CD19-Cre mice were infected IP with 1,000 PFU of the indicated virus. Mice were sacrificed on days 16 to 18 postinfection and spleens (A and B) or peritoneal exudate cells (PECs, C and D) were isolated. (A and C) The frequency of cells harboring latent viral genomes was determined by limiting-dilution PCR analysis. (B and D) Reactivation frequencies were determined by ex vivo plating of serially diluted cells on an indicator monolayer. Cytopathic effect was scored 2 to 3 weeks postplating. Groups of three to five mice were pooled for each infection and analysis. Results are means of two to three independent infections. Error bars represent standard error of the means.
LPS stimulation promotes increased latency in the absence of reactivation.
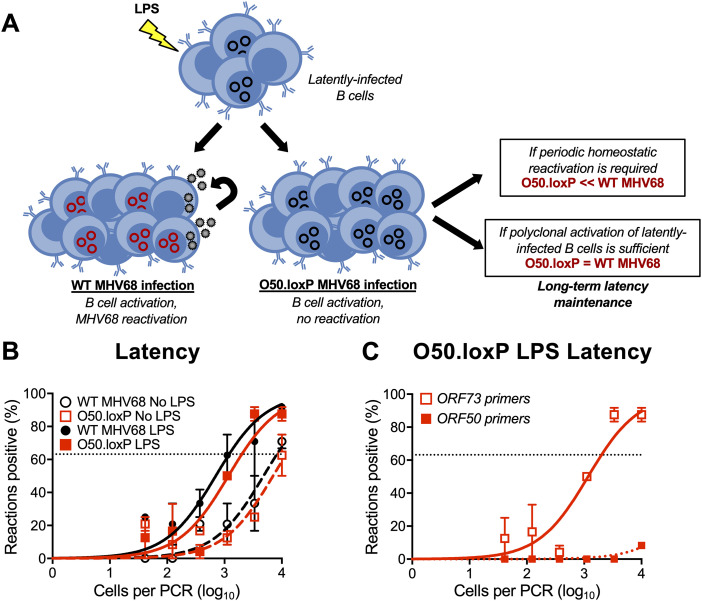
Reactivation is undoubtedly a major contributor to GHV shedding that allows spread from chronically infected to naive hosts. However, whether periodic reactivation occurs within a host already colonized by a GHV in a manner that contributes to the homeostatic maintenance of latent viral reservoirs is not yet clear. TLR agonists stimulate GHV reactivation ex vivo and promote expansion of the latent MHV68 reservoir in vivo when administered to mice (48, 57). This supports the hypothesis that reactivation facilitates reseeding of the latent pool. However, it is also possible that B cell proliferation driven by immune activation enables re-expansion of the latent reservoir without overt reactivation—a hypothesis that has not been directly tested.
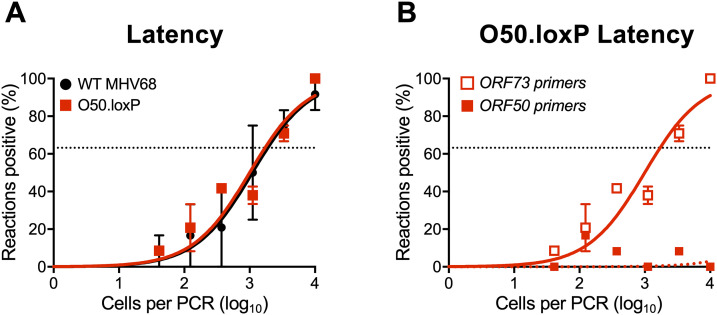
To begin addressing this question, we first evaluated O50.loxP maintenance over time relative to WT MHV68 in CD19-Cre mice. By day 42 postinfection, a time point at which lytic viral replication has cleared and immune activation has waned (20, 32, 58), we found that ORF50-deleted MHV68 was present at levels roughly equivalent to WT virus (Fig. 5A; Table 1). Control LD-PCR performed using primers specific to ORF50 indicated that the locus was, in fact, absent from viral genomes in splenocytes (Fig. 5B), which demonstrates that non-deleted WT virus has not expanded to reclaim the available space. While these observations do not directly address whether homeostatic reactivation is needed to seed naive cells in order to maintain a latency reservoir, they do indicate that O50.loxP MHV68 maintains latency essentially as well as WT virus in CD19-Cre splenocytes despite a severe defect in reactivation from B cells.
FIG 5.
O50.loxP latency is maintained over time despite ORF50 deletion. CD19-Cre mice were infected IN with 1000 PFU of the indicated virus. Mice were sacrificed on day 42 postinfection, and spleens were collected. (A) The frequency of cells harboring MHV68 genomes was determined by limiting-dilution PCR analysis. (B) ORF50 deletion was confirmed by comparing limiting-dilution PCR analyses performed using primers specific for either the ORF50 locus or ORF73 locus. Groups of three to five mice were pooled for each infection and analysis. Results are means of two to three independent infections. Error bars represent standard error of the means.
We previously demonstrated that lipopolysaccharide (LPS), a TLR4 agonist that stimulates polyclonal B cell activation in mammals, drives MHV68 reactivation from latency ex vivo and promotes re-expansion of the MHV68 latency reservoir in spleens of mice at late time points after infection (48). Viral recrudescence in lungs and a correlating virus-specific CD8+ T cell response support the hypothesis that reactivation promoted the increase in latently infected cells observed in these experiments (48), but it also is possible that cellular proliferation or undefined effects of LPS treatment contributed to the observed increase in MHV68 latency. Thus, to gain additional insight into how LPS stimulation leads to an increase in the number of latently infected splenocytes, we compared outcomes of LPS treatment of mice infected with WT or ORF50-deleted MHV68 (see schematic in Fig. 6A). Relative to mock-treated controls, both O50.loxP and WT MHV68 exhibited 5- to 10-fold increases in the number of latently infected cells per spleen 2 weeks after in vivo stimulation with LPS (Fig. 6B; Table 3). LD-PCR performed with primers specific to ORF50 indicated that this viral gene was deleted from the latent viral genomes present in O50.loxP infected spleens (Fig. 6C). These data strongly suggest that LPS-induced B cell activation promotes an increase in the number of latently infected cells despite deletion of ORF50.
FIG 6.
Stimulating B cell polyclonal activation increases O50.loxP genome frequencies. (A) Hypothesized outcomes following polyclonal B cell activation in the presence or absence of MHV68 reactivation from B cells. (B, C) CD19-Cre mice were infected IN with 1,000 PFU of the indicated virus. On day 42 postinfection mice were injected intraperitoneally with diluent (PBS) or 15 μg of LPS. Mice were sacrificed 14 days post-treatment (56 dpi), and spleens were harvested. (B) The frequency of cells harboring MHV68 genomes was determined by limiting-dilution PCR analysis. (C) ORF50 deletion was confirmed by comparing limiting-dilution PCR analyses performed using primers specific for either the ORF50 locus or ORF73 locus. The LPS-treated O50.loxP infection is shown. Groups of three to five mice were pooled for each infection and analysis. Results are means of two to three independent infections. Error bars represent standard error of the means.
TABLE 3.
Frequency of MHV68 latently infected cells following LPS treatment
| Mouse strain | Virus | Route of infection | Cell population | Days postinfection | Treatment | Latency frequencya |
|---|---|---|---|---|---|---|
| CD19-Cre | WT MHV68 | IN | Spleen | 58 | No LPS | 1/8,100 |
| 50.loxP | IN | Spleen | 58 | No LPS | BLD | |
| WT MHV68 | IN | Spleen | 58 | LPS | 1/1,200 | |
| 50.loxP | IN | Spleen | 58 | LPS | 1/2,000 |
BLD, below the limit of detection of 1 in 10,000 cells.
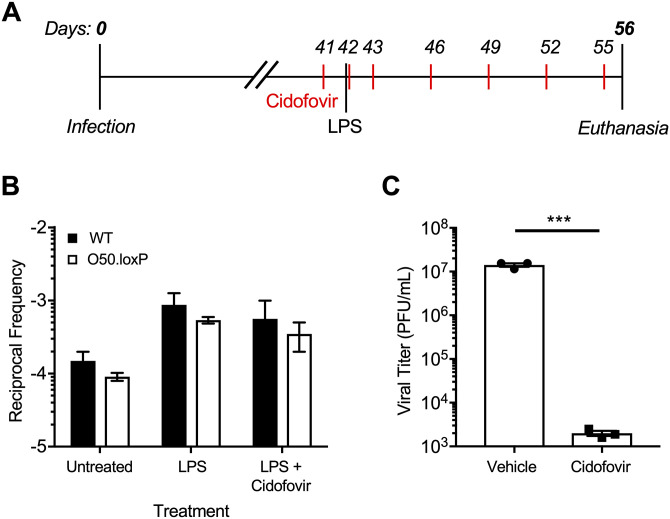
Although reactivation from B cells, the major long-term reservoir for MHV68, does not appear to influence re-expansion of the latent reservoir, a potential caveat to this interpretation is that latent O50.loxP MHV68 within non-Cre-expressing CD19- cells, such as macrophages, dendritic cells, or epithelial cells, could hypothetically reactivate and reseed latency in the spleen. To address this possibility, latently-infected mice were treated with the antiviral drug cidofovir over a 15-day dosing regimen beginning 1 day prior to LPS administration (Fig. 7A). Cidofovir is an inhibitor of the viral DNA polymerase that potently inhibits MHV68 reactivation and associated diseases in vivo, especially using the regimen described (45, 59). Two weeks after stimulation, the frequencies of cells harboring O50.loxP and WT MHV68 genomes were comparable following treatments with LPS alone or LPS in combination with cidofovir (Fig. 7B; Table 4). We confirmed in parallel experiments that the cidofovir used effectively blocked MHV68 replication (Fig. 7C). While there was a very slight reduction in the frequency of latently infected cells in the cidofovir and LPS treated mice compared with LPS alone (2.4- and 1.8-fold, respectively, for WT and O50.loxP infections), in both cases LPS stimulation correlated with an increase in the number of latently infected splenocytes compared with untreated controls. These data highlight the potential of a polyclonal B cell activating stimulus to increase the number of latently infected cells in vivo, even in the absence of contributions from reactivation.
FIG 7.
Blocking viral replication with antiviral drug Cidofovir does not prevent the LPS-driven increase in latency. (A) Experimental design depicting the LPS and antiviral treatment regimens. (B) CD19-Cre mice were infected IN with 1,000 PFU of the indicated virus. Mice were injected subcutaneously with Cidofovir on days 41, 42, 43, and every 3 days subsequently until sacrifice. On day 42 postinfection mice were injected intraperitoneally with diluent (PBS) or 15 μg of LPS. Mice were sacrificed 14 days post-treatment (56 dpi), and spleens were harvested. (B) The frequency of cells harboring MHV68 genomes was determined by limiting-dilution PCR analysis. Reciprocal frequencies of genome-positive cells are shown. Results are means of three independent infections. (C) To confirm Cidofovir potency, BHK21 cells were treated with vehicle (PBS) or cidofovir (50 μg/mL) prior to infection at an MOI of 5 PFU/cell. Cells were subjected to freeze-thaw lysis 24 h postinfection. Viral titers were determined by plaque assay. Error bars represent standard deviations. *** P < 0.001 in a two-tailed Student’s t test.
TABLE 4.
Frequency of MHV68 latently infected cells following LPS + Cidofovir treatment
| Mouse strain | Virus | Route of infection | Cell population | Days postinfection | Treatment | Latency frequency |
|---|---|---|---|---|---|---|
| CD19-Cre | WT MHV68 | IN | Spleen | 58 | No LPS | 1/7,500 |
| 50.loxP | IN | Spleen | 58 | No LPS | 1/7,550 | |
| WT MHV68 | IN | Spleen | 58 | LPS | 1/1,900 | |
| 50.loxP | IN | Spleen | 58 | LPS | 1/1,950 | |
| WT MHV68 | IN | Spleen | 58 | LPS + Cidofovir | 1/4,500 | |
| 50.loxP | IN | Spleen | 58 | LPS + Cidofovir | 1/3,500 |
DISCUSSION
It is well established that B cells represent a preferred cell type for gammaherpesvirus latency, offering a reservoir of infected cells with the capacity to proliferate and survive for the lifetime of the host (23, 41, 60–68). However, whether lytic replication and reactivation from infected B cells contribute to expansion and maintenance of latency is difficult to evaluate with traditional loss-of-function viral mutagenesis strategies. Here we report the use of cell-type-specific, Cre-lox-mediated deletion of ORF50, a gene that is essential for viral replication, to examine the importance of lytic replication in B cells in MHV68 latency. The conditional deletion of ORF50 differs from traditional loss-of-function ORF50 mutants which are replication-dead in all tissues from the start of the infection process. The results of this B cell-specific deletion approach better define steps in viral infection that facilitate viral dissemination and latency establishment and provide new evidence pertaining to mechanisms of long-term latency maintenance within an infected host.
Lytic replication in host colonization.
Following IN inoculation of mice, MHV68 undergoes lytic replication in the respiratory epithelium (32). This promotes infection of lung-resident and recruited antigen presenting cells that carry the virus to draining mediastinal lymph nodes (36, 38). In the draining lymph nodes, the virus is harbored in proliferating B cells before disseminating via hematogenous routes to the spleen (33, 35, 37, 60). Previous work using ORF50-null MHV68 or cidofovir treatment to block acute replication following WT MHV68 infection suggests that lytic replication after IN inoculation is critical for MHV68 dissemination and latency establishment in the spleen (43, 45, 59, 69). In contrast, lung-resident B cells seemingly take up and maintain MHV68 genomes (45). Because B cells facilitate trafficking from lungs to spleens (41, 42) and ORF50 deletion was evident in draining lymph nodes of CD19-Cre mice by day 10 postinfection with O50.loxP, our data suggest that lytic replication in the lung facilitates seeding of B cells in the draining lymph node. This event appears to be sufficient for latent colonization of B cells throughout the host.
Compared with other MHV68 genes we have conditionally deleted in B cells (33, 39), the observation that O50.loxP MHV68 establishes latency in the spleen at WT levels after IN inoculation is unique. Deletion of the M2 gene in both CD19+ and AID+ B cells resulted in a ca. 10-fold reduction in splenic latency (39). B cell-specific deletion of ORF73, which encodes the episome maintenance protein LANA, resulted in splenic latency that was below limits of detection (33). Similar to O50.loxP, however, both M2.loxP and O73.loxP viruses accumulate to levels similar to WT virus in MLNs after IN inoculation and in the spleen after IP infection. To reiterate, LANA and M2 are critically important latency gene products, while RTA is essential for lytic replication. Because ORF50 deletion in B cells did not prevent efficient latency establishment in the spleen after IN inoculation, while in contrast deletion of M2 or ORF73 restricted latency, these data support a model in which lytic replication facilitates escape across an epithelial barrier and spread to lymphoid organs. Our data suggest that once B cells within lymphoid tissues are infected, lytic replication is no longer necessary, and latency gene products take over. The latency gene expression program then facilitates B cell-mediated dissemination of infected cells to distal sites such as the spleen, thereby enabling viral colonization of lymphoid tissues throughout the host.
Contribution of periodic reactivation to latent persistence.
Our evaluations of long-term latency after ORF50 deletion and the impacts of in vivo stimulation with LPS offer unique insights into the contribution of reactivation from B cells to long-term latency maintenance. We initially suspected that B cell reactivation would play a critical role in long-term chronic infection, and as a result, the frequency of cells harboring ORF50-deleted MHV68 would more rapidly decay in the spleen than would frequencies of WT reactivation-competent virus. This hypothesis was based on the observations (i) that adoptive transfer of latently infected B cells leads to new infections of B cells within a naive congenic recipient mouse (49) and (ii) that TLR agonists such as LPS that trigger reactivation ex vivo also promote an increase in the percentage of latently infected cells when administered to infected mice (48). However, after observing relatively comparable levels of ORF50-deleted and WT virus in spleens at late time points after primary infection, especially after stimulation with LPS, we questioned whether mechanisms of maintenance that did not involve reactivation might also be at work, but we first considered whether reactivation from non-B cell latency reservoirs or non-deleted viruses might participate in maintaining latent viral infection. Because the LPS-driven increase in latently infected splenocytes was not blocked by the potent antiviral drug cidofovir, we concluded that our data strongly suggest that mechanisms other than reactivation play a critical role in maintaining MHV68 latency over time.
One particularly compelling hypothesis is that MHV68, and perhaps other GHVs as well, co-opt B cell proliferation driven by non-GHV stimulators as a critical means to maintain high levels of latency in the absence of reactivation. In support of this hypothesis, it is notable that a large percentage of MHV68 genomes reside in the highly proliferative germinal center B cell compartment for many weeks after infection (40, 61, 70, 71). TLR ligands like LPS are well known to promote polyclonal B cell activation and proliferation (72), a phenotype we confirmed in control experiments (S.M. Owens and J.C. Forrest, unpublished). We speculate that this particular mechanism could allow “passive” expansion of the number of latently infected cells in response to other infections while limiting the production of antigenic targets—both lytic and latent —that could promote immune detection and clearance of infected cells. Additional experiments are necessary to fully evaluate these hypotheses.
Comparison with similar studies and consideration of caveats.
Cre-lox recombination provides an elegant system for conditional deletion, but the deletion must be efficient and specific. We find it interesting that our results differed from those of a previous study (73), however nuances of the two studies may explain the apparently conflicting outcomes. Contrary to our findings, a virus in which both ORF49 and ORF50 were floxed (MHV-F50) was significantly attenuated following both IN and IP infection of CD19-Cre mice (73). This study employed the original MHV68 BAC system, which used loxP sites flanking the inserted BAC vector sequences to allow removal of the foreign bacterial origin of replication and antibiotic selection cassette from the viral genome during virus recovery in Cre-expressing cells (74). To employ Cre-loxP-mediated deletion of viral genes presents a complication, as recombination could occur between a floxed gene of interest and the other loxP sites in the genome, leading to deletion of large segments of viral sequences. To overcome this potential problem, we developed a BAC system that employed the Flp-Frt recombination system for BAC vector deletion and demonstrated its functionality in a previous study (33). We used this system here to ensure that ORF50 would not be inadvertently deleted while generating virus stocks. Because the MHV-F50 study utilized the original MHV68 BAC that contains additional loxP sites, it is possible that recombination led to a larger than expected deletion within the MHV68 genome in Cre-expressing mice. In addition to ORF50 (and ORF49), in vivo Cre-mediated recombination may have inadvertently deleted the left half of the viral genome.
It also is possible that the larger deletion of both ORF50 and ORF49 yields a much more severe attenuated phenotype than deletion of ORF50 alone. ORF49 encodes a protein that is essential for lytic infection, and viruses lacking ORF49 are attenuated for both lytic and latent infection (75, 76). Conditional deletion mutants of ORF49 have not been evaluated. Moreover, tiled array, RNA-seq, and long-read transcript analyses have demonstrated the incredibly complex nature of transcription within the MHV68 genome (25, 77–80). Although functional analyses of the transcripts are incomplete, recent work demonstrated the presence of numerous transcripts that overlap the ORF49 and ORF50 loci on both sense and anti-sense strands of the viral genome (80). It is conceivable that a larger deletion disrupts these (or other) transcripts in a manner that results in a more severe latency phenotype. It is worth noting that we have performed analogous experiments in which we floxed another essential immediate-early lytic gene, ORF57. Similar to O50.loxP, O57.loxP MHV68 disseminated to the spleen and established latency despite deletion of ORF57 from B cells (Owens, Gupta, and Forrest, manuscript in preparation). While the ORF57-encoded protein has a different function than RTA, this result offers independent confirmation and provides additional confidence in the conclusion that lytic replication in B cells is not necessary for viral dissemination and latency establishment. In conclusion, the data presented here highlight the utility of cell-type specific deletion of viral genes to foster an understanding of fundamental mechanisms of GHV infection and persistence in the host.
MATERIALS AND METHODS
Ethics statement.
Mouse experiments performed for this study were carried out in accordance with NIH, USDA, and UAMS Division of Laboratory Animal Medicine and Institutional Animal Care and Use Committee (IACUC) guidelines. The protocol supporting this study was approved by the UAMS IUCAC. Mice were anesthetized prior to inoculations and sacrificed humanely at the end of experiments.
Cells and viruses.
Vero (ATCC CCL-81) and Vero-Cre cells, NIH 3T12 (ATCC CCL-164), BHK21 (ATCC CCL-10), and Swiss albino 3T3 (ATCC CRL-1658) fibroblasts were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM l-glutamine (cDMEM). Cells were cultured at 37°C with 5% CO2 and ~99% humidity. Murine embryonic fibroblasts (MEFs) were harvested from C57BL/6 mouse embryos (47) and immortalized as previously described. Viruses used in this study included the FRT BAC-derived (WT MHV68) (33); the RTA deficient RTA-null MHV68, and the conditional ORF50 mutant (O50.loxP MHV68) (43, 45). WT and O50.loxP viruses were derived from MHV68 with FRT sites flanking the BAC cassette and were passaged in Flp-expressing 3T12 cells to remove the BAC cassette (33). Titers were quantified as described previously (81).
Generation of recombinant viruses.
The WT MHV68 BAC was created by en passant mutagenesis (82). To generate the O50.loxP, loxP sites were inserted adjacent to the 5′ and 3′ ends of the orf50 exon 2 (E2) in a FRT BAC template by two successive rounds of en passant mutagenesis (33) utilizing the following primers:
O50loxpUP_for 5’AGTCTGCAAGAAATAATAGCCTCCCACTTTTATGGAAATCATAACTTCGTATAGCATACATTATACGAAGTTATTGGTAGCTCCTCCTACATGATAGGGATAACAGGGTAATCGATT, O50loxpUP_rev 5’GGTTAATTGGTTGTAACACTGGCCTCCCACTTTTATGGAAATCATAACTTCGTATAGCATACATTATACGAAGTTATTGGTAGCTCCTCCTACATGAGTCATGAAATGTCCCTTCAA, O50loxpDWN_for 5’CATACTTAGTCCACTCGACCCAAACAGCCTGGAGTCATAAATAACTTCGTATAGCATACATTATACGAAGTTATACGGTGCCAAATACAAGACATTTAGGGATAACAGGGTAATCGATTT, O50loxpDWN_rev 5’GGTTAATTGGTTGTAACACTGGCCAAACAGCCTGGAGTCATAAATAACTTCGTATAGCATACATTATACGAAGTTATACGGTGCCAAATACAAGACATTTCTAAATCCTTAAGTATAA.
Viruses were passaged in Flp-expressing 3T12 fibroblasts to remove the BAC cassette, and titers were quantified as previously described (33).
Cell culture infections.
For immunoblot assays, 3T12 fibroblasts were infected at a multiplicity of infection (MOI) of 5 PFU/cell with WT MHV68, RTA-null, O50.loxP MHV68. Briefly, viral stocks were diluted in low-volume (200 μL) of cDMEM and added directly onto fibroblast cell monolayers seeded the previous day at a density of 2 × 105 cells/well in 6-well plates. Plates were incubated at 37°C and rocked gently every 15 min for 1 h. After 1 h, cDMEM was added back to the cultures.
For all other viral infections, 3T3 or 3T12 cells plated the previous day were inoculated with a low volume of virus diluted in cDMEM. Plates were rocked every 15 min for 1 h at 37°C. After low-volume adsorption, inocula were removed for high-multiplicity infections. After adsorption, cells were incubated in a normal culture volume of cDMEM for the indicated times at 37°C. For multistep growth curves, infected cells were frozen at −80°C at the indicated time points. Cells were subjected to freeze-thaw lysis to release progeny virions, and lysates were serially diluted for plaque assays as described (81).
Nucleic acid isolation and PCR.
Vero or Vero-Cre cells were mock infected or infected with MHV68 at a MOI of 5 PFU/cell. Nucleic acid was isolated from cells at 24 h or 48 h postinfection. Total DNA was isolated using a GenCatch blood and tissue genomic mini-prep kit (Epoch Life Science). PCR for the detection of the full-length ORF50 gene was performed using primers ORF50_del_for 5’GCTTCCTCGTCTACAGAGGTCAGG and ORF50_del_rev 5’GGCACCCATACTAAGTTGTGATTC. ORF73 and GAPDH genes were detected by PCR utilizing primers 73_IG_US and 73_IG_DS for ORF73 and GAPDHF and GAPDHR for GAPDH in GoTaq polymerase (Promega) as described (33).
Immunoblot analyses.
Immunoblot analyses were performed as previously described (33). Briefly, cells were lysed with radioimmunoprecipitation (RIPA) buffer (150 mM NaCl, 20 mM Tris, 2 mM EDTA, 1% NP-40, 0.25% deoxycholate supplemented with phosphatase and protease inhibitors), and protein samples were centrifuged at 16,000 × g to remove insoluble debris. Protein content for each sample was quantified using the Bio-Rad DC Protein Assay (Bio-Rad). Samples were diluted in 6x Laemmli sample buffer and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes (Thermo Scientific). Blots were probed with the indicated primary antibodies and with horseradish peroxidase (HRP) conjugated secondary antibodies (Jackson ImmunoResearch). Chemiluminescent signal was detected using a ChemiDoc MP Imaging System (Bio-Rad) on blots treated with Clarity ECL reagent (Bio-Rad).
Mouse infections and tissue harvests.
Male and female, C57BL/6, heterozygous CD19-Cre [B6.129P2(C)-Cd19tm1(cre)Cgn/J] mice were purchased from Jackson Laboratories or were bred and maintained in the animal housing facilities at the University of Arkansas for Medical Sciences (UAMS, Little Rock, AR). Note: Cre-encoding mice used were heterozygous for Cre expression. Mice were housed in sterile conditions and treated according to the guidelines of the Division of Laboratory Animal Medicine (DLAM) at UAMS. Eight- to 10-week-old mice were anesthetized using isoflurane and inoculated with 1,000 PFU of virus diluted in incomplete DMEM (20 μL) for intranasal inoculations or injected with 1,000 PFU of virus diluted in incomplete DMEM (200 μL) for intraperitoneal inoculations. Lungs were harvested 7 to 10 days postinfection. Serum, splenocytes, and peritoneal exudate cells (PECs) were harvested 16 to 18 days postinfection, or 42 and 90 days postinfection, as described previously (56). Cells from draining lymph nodes were isolated as previously described (33).
Splenocyte isolation and limiting-dilution analyses.
Spleens were homogenized using a Tenbroek tissue disrupter. Red blood cells were lysed by incubating tissue homogenate in 8.3 g/L ammonium chloride for 10 min at room temperature with shaking. Cells were filtered through 40-μM mesh to reduce clumping. Frequencies of cells harboring MHV68 genomes were determined using limiting-dilution (LD) PCR analysis with primers specific for an ORF73 target as previously described (33). We also performed the same experiments with primers specific for an ORF50 gene segment to confirm excision of floxed ORF50 in infected cells (33). Frequencies of latently infected cells capable of reactivating were determined using a limiting-dilution analysis for cytopathic effect induced on an indicator monolayer as previously described (47).
Plaque assays.
Plaque assays were performed as previously described (81) using BHK21 cells (2 × 105 cells/well). Briefly, infected cells were overlaid with 1.5% methylcellulose in DMEM supplemented with 2.5% calf serum, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM l-glutamine and incubated at 37°C for 4 to 6 days. Cell monolayers were stained with a solution of crystal violet in formalin for identification and quantification of plaques.
Antiviral treatment and in vivo stimulation of reactivation.
For in vitro analyses, BHK21 cells were infected at an MOI of 5 PFU/cell. Cells were treated with vehicle (PBS) or cidofovir (50 μg/mL) for 24 h. Cell lysates were isolated and viral titers were quantified by plaque assay.
For in vivo analyses, CD19-Cre mice were injected intraperitoneally with 15 μg of Escherichia coli lipopolysaccharide in 100 μL of sterile PBS 42 days postinfection to stimulate a TLR response (48). Where indicated, mice were subcutaneously injected with 25 mg/kg of cidofovir in sterile PBS on alternating flanks (60 to 100 μL inoculum volume) (83), on days 41, 42, 43, 46, 49, 52, and 55 postinfection. On day 56 postinfection (day 14 post-LPS, where indicated), mice were euthanized by CO2 asphyxiation and spleens were collected for analysis.
Antibodies and drug treatments.
Antibodies used in this study include rabbit polyclonal mLANA antiserum (84), mouse polyclonal MHV68 antiserum (81), anti-RTA (85), anti-v-cyclin (86), and mouse monoclonal anti-β-actin (Sigma-Aldrich, #A2228). For drug treatments, the 17β-estradiol agonist Z-4-hydroxytamoxifen (4-OHT; Alexis Biochemicals; #ALX-550-361-M001) was dissolved at a stock concentration of 2 mM. Inducible-Cre 3T3 fibroblasts plated the previous day were treated with either 0.2 μM 4-OHT to induce nuclear translocation of Cre or ethanol as a vehicle control for 24 h prior to infection.
Statistics.
All statistical analyses were performed using GraphPad Prism software (GraphPad Software, San Diego, CA). Statistical significance was determined by a two-tailed unpaired Student’s t test with a 95% confidence.
ACKNOWLEDGMENTS
This work was supported in part by grant R01CA167065 from the NIH National Cancer Institute and funds from the UAMS College of Medicine and Arkansas Biosciences Institute to J.C.F. The Flow Cytometry Core and the work described here were also supported in part by the Center for Microbial Pathogenesis and Host Inflammatory Responses award P20GM103625 from the NIH National Institute of General Medical Sciences Centers of Biomedical Research Excellence. S.M.O. was supported by a postdoctoral fellowship from Translational Research Institute (TRI) grant TL1 TR003109 through the NIH National Center for Advancing Translational Sciences. D.W.W. and D.G.O. were supported by the Gundersen Medical Foundation. The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
Contributor Information
J. Craig Forrest, Email: JCForrest@uams.edu.
Rozanne M. Sandri-Goldin, University of California, Irvine
REFERENCES
- 1.Davison AJ, Eberle R, Ehlers B, Hayward GS, McGeoch DJ, Minson AC, Pellett PE, Roizman B, Studdert MJ, Thiry E. 2009. The order Herpesvirales. Arch Virol 154:171–177. 10.1007/s00705-008-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackermann M. 2006. Pathogenesis of gammaherpesvirus infections. Vet Microbiol 113:211–222. 10.1016/j.vetmic.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 3.Ambinder RF. 2001. Epstein-Barr virus associated lymphoproliferations in the AIDS setting. Eur J Cancer 37:1209–1216. 10.1016/s0959-8049(01)00123-x. [DOI] [PubMed] [Google Scholar]
- 4.Boshoff C, Chang Y. 2001. Kaposi's sarcoma-associated herpesvirus: a new DNA tumor virus. Annu Rev Med 52:453–470. 10.1146/annurev.med.52.1.453. [DOI] [PubMed] [Google Scholar]
- 5.Geraminejad P, Memar O, Aronson I, Rady PL, Hengge U, Tyring SK. 2002. Kaposi's sarcoma and other manifestations of human herpesvirus 8. J Am Acad Dermatol 47:641–655. quiz 656-8. 10.1067/mjd.2002.128383. [DOI] [PubMed] [Google Scholar]
- 6.Goncalves PH, Ziegelbauer J, Uldrick TS, Yarchoan R. 2017. Kaposi sarcoma herpesvirus-associated cancers and related diseases. Curr Opin HIV AIDS 12:47–56. 10.1097/COH.0000000000000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinzone MR, Berretta M, Cacopardo B, Nunnari G. 2015. Epstein-Barr virus- and Kaposi sarcoma-associated herpesvirus-related malignancies in the setting of human immunodeficiency virus infection. Semin Oncol 42:258–271. 10.1053/j.seminoncol.2014.12.026. [DOI] [PubMed] [Google Scholar]
- 8.Wagner-Johnston ND, Ambinder RF. 2007. Epstein-Barr virus-related lymphoproliferative disorders. Curr Hematol Malig Rep 2:249–254. 10.1007/s11899-007-0034-y. [DOI] [PubMed] [Google Scholar]
- 9.Wong EL, Damania B. 2005. Linking KSHV to human cancer. Curr Oncol Rep 7:349–356. 10.1007/s11912-005-0061-6. [DOI] [PubMed] [Google Scholar]
- 10.Cesarman E. 2011. Gammaherpesvirus and lymphoproliferative disorders in immunocompromised patients. Cancer Lett 305:163–174. 10.1016/j.canlet.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohseni M, Boniface MP, Graham C. 2021. Mononucelosis. StatPearls, [Internet]. https://www.ncbi.nlm.nih.gov/books/NBK470387/. [PubMed] [Google Scholar]
- 12.Molloy M, Zhang W, Usherwood E. 2010. Mononucleosis and antigen-driven T cell responses have different requirements for interleukin-2 signaling in murine gammaherpesvirus infection. J Virol 84:10923–10927. 10.1128/JVI.00856-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abere B, Schulz TF. 2016. KSHV non-structural membrane proteins involved in the activation of intracellular signaling pathways and the pathogenesis of Kaposi's sarcoma. Curr Opin Virol 20:11–19. 10.1016/j.coviro.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Bishop BN, Lynch DT. 2021. Kaposi sarcoma. StatPearls, [Internet]. https://www.ncbi.nlm.nih.gov/books/NBK534839/. [PubMed] [Google Scholar]
- 15.Oksenhendler E, Boutboul D, Galicier L. 2019. Kaposi sarcoma-associated herpesvirus/human herpesvirus 8-associated lymphoproliferative disorders. Blood 133:1186–1190. 10.1182/blood-2018-11-852442. [DOI] [PubMed] [Google Scholar]
- 16.Honess RW, Roizman B. 1974. Regulation of herpesvirus macromolecular synthesis. cascade regulation of the synthesis of three groups of viral proteins. J Virol 14:8–19. 10.1128/JVI.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purushothaman P, Dabral P, Gupta N, Sarkar R, Verma SC. 2016. KSHV genome replication and maintenance. Front Microbiol 7:54. 10.3389/fmicb.2016.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang MS, Kieff E. 2015. Epstein-Barr virus latent genes. Exp Mol Med 47:e131. 10.1038/emm.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Leo A, Calderon A, Lieberman PM. 2020. Control of viral latency by episome maintenance proteins. Trends Microbiol 28:150–162. 10.1016/j.tim.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Speck SH, Ganem D. 2010. Viral latency and its regulation: lessons from the gamma-herpesviruses. Cell Host Microbe 8:100–115. 10.1016/j.chom.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blasdell K, McCracken C, Morris A, Nash AA, Begon M, Bennett M, Stewart JP. 2003. The wood mouse is a natural host for Murid herpesvirus 4. J Gen Virol 84:111–113. 10.1099/vir.0.18731-0. [DOI] [PubMed] [Google Scholar]
- 22.Dong S, Forrest JC, Liang X. 2017. Murine gammaherpesvirus 68: a small animal model for gammaherpesvirus-associated diseases. Adv Exp Med Biol 1018:225–236. 10.1007/978-981-10-5765-6_14. [DOI] [PubMed] [Google Scholar]
- 23.Fujiwara S. 2018. Animal models of human gammaherpesvirus infections. Adv Exp Med Biol 1045:413–436. 10.1007/978-981-10-7230-7_19. [DOI] [PubMed] [Google Scholar]
- 24.Mistríková J, Raslová H, Mrmusová M, Kúdelová M. 2000. A murine gammaherpesvirus. Acta Virol 44:211–226. [PubMed] [Google Scholar]
- 25.Ungerleider NA, Tibbetts SA, Renne R, Flemington EK. 2019. Gammaherpesvirus RNAs come full circle. mBio 10. 10.1128/mBio.00071-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Virgin HWt, Latreille P, Wamsley P, Hallsworth K, Weck KE, Dal Canto AJ, Speck SH. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol 71:5894–5904. 10.1128/JVI.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Tibbetts SA, Krug LT. 2021. Conquering the host: determinants of pathogenesis learned from murine Gammaherpesvirus 68. Annu Rev Virol 8:349–371. 10.1146/annurev-virology-011921-082615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta A, Oldenburg DG, Salinas E, White DW, Forrest JC. 2017. Murine Gammaherpesvirus 68 expressing Kaposi sarcoma-associated herpesvirus latency-associated nuclear antigen (LANA) reveals both functional conservation and divergence in LANA homologs. J Virol 91. 10.1128/JVI.00992-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, Zhu L, Lu X, Feldman ER, Keyes LR, Wang Y, Fan H, Feng H, Xia Z, Sun J, Jiang T, Gao SJ, Tibbetts SA, Feng P. 2015. Recombinant murine gamma herpesvirus 68 carrying KSHV G protein-coupled receptor induces angiogenic lesions in mice. PLoS Pathog 11:e1005001. 10.1371/journal.ppat.1005001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee KS, Suarez AL, Claypool DJ, Armstrong TK, Buckingham EM, van Dyk LF. 2012. Viral cyclins mediate separate phases of infection by integrating functions of distinct mammalian cyclins. PLoS Pathog 8:e1002496. 10.1371/journal.ppat.1002496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Habison AC, de Miranda MP, Beauchemin C, Tan M, Cerqueira SA, Correia B, Ponnusamy R, Usherwood EJ, McVey CE, Simas JP, Kaye KM. 2017. Cross-species conservation of episome maintenance provides a basis for in vivo investigation of Kaposi's sarcoma herpesvirus LANA. PLoS Pathog 13:e1006555. 10.1371/journal.ppat.1006555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sunil-Chandra NP, Efstathiou S, Arno J, Nash AA. 1992. Virological and pathological features of mice infected with murine gamma-herpesvirus 68. J Gen Virol 73:2347–2356. 10.1099/0022-1317-73-9-2347. [DOI] [PubMed] [Google Scholar]
- 33.Salinas E, Gupta A, Sifford JM, Oldenburg DG, White DW, Forrest JC. 2018. Conditional mutagenesis in vivo reveals cell type- and infection stage-specific requirements for LANA in chronic MHV68 infection. PLoS Pathog 14:e1006865. 10.1371/journal.ppat.1006865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frederico B, Chao B, Lawler C, May JS, Stevenson PG. 2015. Subcapsular sinus macrophages limit acute gammaherpesvirus dissemination. J Gen Virol 96:2314–2327. 10.1099/vir.0.000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frederico B, Chao B, May JS, Belz GT, Stevenson PG. 2014. A murid gamma-herpesviruses exploits normal splenic immune communication routes for systemic spread. Cell Host Microbe 15:457–470. 10.1016/j.chom.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 36.Frederico B, Milho R, May JS, Gillet L, Stevenson PG. 2012. Myeloid infection links epithelial and B cell tropisms of Murid Herpesvirus-4. PLoS Pathog 8:e1002935. 10.1371/journal.ppat.1002935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feldman ER, Kara M, Oko LM, Grau KR, Krueger BJ, Zhang J, Feng P, van Dyk LF, Renne R, Tibbetts SA. 2016. A gammaherpesvirus noncoding RNA is essential for hematogenous dissemination and establishment of peripheral latency. mSphere 1. 10.1128/mSphere.00105-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaspar M, May JS, Sukla S, Frederico B, Gill MB, Smith CM, Belz GT, Stevenson PG. 2011. Murid herpesvirus-4 exploits dendritic cells to infect B cells. PLoS Pathog 7:e1002346. 10.1371/journal.ppat.1002346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Owens SM, Oldenburg DG, White DW, Forrest JC. 2020. Deletion of murine gammaherpesvirus gene M2 in activation-induced cytidine deaminase-expressing B Cells impairs host colonization and viral reactivation. J Virol 95. 10.1128/JVI.01933-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flaño E, Husain SM, Sample JT, Woodland DL, Blackman MA. 2000. Latent murine gamma-herpesvirus infection is established in activated B cells, dendritic cells, and macrophages. J Immunol 165:1074–1081. 10.4049/jimmunol.165.2.1074. [DOI] [PubMed] [Google Scholar]
- 41.Stewart JP, Usherwood EJ, Ross A, Dyson H, Nash T. 1998. Lung epithelial cells are a major site of murine gammaherpesvirus persistence. J Exp Med 187:1941–1951. 10.1084/jem.187.12.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Usherwood EJ, Stewart JP, Robertson K, Allen DJ, Nash AA. 1996. Absence of splenic latency in murine gammaherpesvirus 68-infected B cell-deficient mice. J Gen Virol 77:2819–2825. 10.1099/0022-1317-77-11-2819. [DOI] [PubMed] [Google Scholar]
- 43.Pavlova IV, Virgin HWt, Speck SH. 2003. Disruption of gammaherpesvirus 68 gene 50 demonstrates that Rta is essential for virus replication. J Virol 77:5731–5739. 10.1128/jvi.77.10.5731-5739.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jia Q, Wu TT, Liao HI, Chernishof V, Sun R. 2004. Murine gammaherpesvirus 68 open reading frame 31 is required for viral replication. J Virol 78:6610–6620. 10.1128/JVI.78.12.6610-6620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moser JM, Farrell ML, Krug LT, Upton JW, Speck SH. 2006. A gammaherpesvirus 68 gene 50 null mutant establishes long-term latency in the lung but fails to vaccinate against a wild-type virus challenge. J Virol 80:1592–1598. 10.1128/JVI.80.3.1592-1598.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moser JM, Upton JW, Gray KS, Speck SH. 2005. Ex vivo stimulation of B cells latently infected with gammaherpesvirus 68 triggers reactivation from latency. J Virol 79:5227–5231. 10.1128/JVI.79.8.5227-5231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weck KE, Barkon ML, Yoo LI, Speck SH, Virgin HI. 1996. Mature B cells are required for acute splenic infection, but not for establishment of latency, by murine gammaherpesvirus 68. J Virol 70:6775–6780. 10.1128/JVI.70.10.6775-6780.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gargano LM, Forrest JC, Speck SH. 2009. Signaling through Toll-like receptors induces murine gammaherpesvirus 68 reactivation in vivo. J Virol 83:1474–1482. 10.1128/JVI.01717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freeman ML, Burkum CE, Yager EJ, Woodland DL, Blackman MA. 2011. De novo infection of B cells during murine gammaherpesvirus 68 latency. J Virol 85:10920–10925. 10.1128/JVI.05027-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Forrest JC, Paden CR, Allen RD, 3rd, Collins J, Speck SH. 2007. ORF73-null murine gammaherpesvirus 68 reveals roles for mLANA and p53 in virus replication. J Virol 81:11957–11971. 10.1128/JVI.00111-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watanabe T, Narita Y, Yoshida M, Sato Y, Goshima F, Kimura H, Murata T. 2015. The Epstein-Barr virus BDLF4 gene is required for efficient expression of viral late lytic genes. J Virol 89:10120–10124. 10.1128/JVI.01604-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng K, Chen M, Xiang Y, Ma K, Jin F, Wang X, Wang X, Wang S, Wang Y. 2014. Inhibition of herpes simplex virus type 1 entry by chloride channel inhibitors tamoxifen and NPPB. Biochem Biophys Res Commun 446:990–996. 10.1016/j.bbrc.2014.03.050. [DOI] [PubMed] [Google Scholar]
- 53.Drori A, Messerle M, Brune W, Tirosh B. 2014. Lack of XBP-1 impedes murine cytomegalovirus gene expression. PLoS One 9:e110942. 10.1371/journal.pone.0110942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flaño E, Woodland DL, Blackman MA. 2002. A mouse model for infectious mononucleosis. Immunol Res 25:201–217. 10.1385/IR:25:3:201. [DOI] [PubMed] [Google Scholar]
- 55.Rickert RC, Roes J, Rajewsky K. 1997. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res 25:1317–1318. 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weck KE, Kim SS, Virgin HI, Speck SH. 1999. Macrophages are the major reservoir of latent murine gammaherpesvirus 68 in peritoneal cells. J Virol 73:3273–3283. 10.1128/JVI.73.4.3273-3283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gargano LM, Moser JM, Speck SH. 2008. Role for MyD88 signaling in murine gammaherpesvirus 68 latency. J Virol 82:3853–3863. 10.1128/JVI.02577-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barton E, Mandal P, Speck SH. 2011. Pathogenesis and host control of gammaherpesviruses: lessons from the mouse. Annu Rev Immunol 29:351–397. 10.1146/annurev-immunol-072710-081639. [DOI] [PubMed] [Google Scholar]
- 59.Neyts J, De Clercq E. 1998. In vitro and in vivo inhibition of murine gamma herpesvirus 68 replication by selected antiviral agents. Antimicrob Agents Chemother 42:170–172. 10.1128/AAC.42.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Collins CM, Boss JM, Speck SH. 2009. Identification of infected B-cell populations by using a recombinant murine gammaherpesvirus 68 expressing a fluorescent protein. J Virol 83:6484–6493. 10.1128/JVI.00297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Collins CM, Speck SH. 2012. Tracking murine gammaherpesvirus 68 infection of germinal center B cells in vivo. PLoS One 7:e33230. 10.1371/journal.pone.0033230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dutia BM, Reid SJ, Drummond DD, Ligertwood Y, Bennet I, Rietberg W, Silvia O, Jarvis MA, Nash AA. 2009. A novel Cre recombinase imaging system for tracking lymphotropic virus infection in vivo. PLoS One 4:e6492. 10.1371/journal.pone.0006492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liang X, Collins CM, Mendel JB, Iwakoshi NN, Speck SH. 2009. Gammaherpesvirus-driven plasma cell differentiation regulates virus reactivation from latently infected B lymphocytes. PLoS Pathog 5:e1000677. 10.1371/journal.ppat.1000677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Siegel AM, Rangaswamy US, Napier RJ, Speck SH. 2010. Blimp-1-dependent plasma cell differentiation is required for efficient maintenance of murine gammaherpesvirus latency and antiviral antibody responses. J Virol 84:674–685. 10.1128/JVI.01306-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sunil-Chandra NP, Arno J, Fazakerley J, Nash AA. 1994. Lymphoproliferative disease in mice infected with murine gammaherpesvirus 68. Am J Pathol 145:818–826. [PMC free article] [PubMed] [Google Scholar]
- 66.Sunil-Chandra NP, Efstathiou S, Nash AA. 1992. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J Gen Virol 73:3275–3279. 10.1099/0022-1317-73-12-3275. [DOI] [PubMed] [Google Scholar]
- 67.van Dyk LF, Virgin HWt, Speck SH. 2003. Maintenance of gammaherpesvirus latency requires viral cyclin in the absence of B lymphocytes. J Virol 77:5118–5126. 10.1128/jvi.77.9.5118-5126.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weck KE, Kim SS, Virgin HI, Speck SH. 1999. B cells regulate murine gammaherpesvirus 68 latency. J Virol 73:4651–4661. 10.1128/JVI.73.6.4651-4661.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krug LT, Moser JM, Dickerson SM, Speck SH. 2007. Inhibition of NF-kappaB activation in vivo impairs establishment of gammaherpesvirus latency. PLoS Pathog 3:e11. 10.1371/journal.ppat.0030011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Flaño E, Kim IJ, Woodland DL, Blackman MA. 2002. Gamma-herpesvirus latency is preferentially maintained in splenic germinal center and memory B cells. J Exp Med 196:1363–1372. 10.1084/jem.20020890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nealy MS, Coleman CB, Li H, Tibbetts SA. 2010. Use of a virus-encoded enzymatic marker reveals that a stable fraction of memory B cells expresses latency-associated nuclear antigen throughout chronic gammaherpesvirus infection. J Virol 84:7523–7534. 10.1128/JVI.02572-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bekeredjian-Ding I, Jego G. 2009. Toll-like receptors–sentries in the B-cell response. Immunology 128:311–323. 10.1111/j.1365-2567.2009.03173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lawler C, de Miranda MP, May J, Wyer O, Simas JP, Stevenson PG. 2018. Gammaherpesvirus colonization of the spleen requires lytic replication in B cells. J Virol 92. 10.1128/JVI.02199-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adler H, Messerle M, Wagner M, Koszinowski UH. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J Virol 74:6964–6974. 10.1128/jvi.74.15.6964-6974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Noh CW, Cho HJ, Kang HR, Jin HY, Lee S, Deng H, Wu TT, Arumugaswami V, Sun R, Song MJ. 2012. The virion-associated open reading frame 49 of murine gammaherpesvirus 68 promotes viral replication both in vitro and in vivo as a derepressor of RTA. J Virol 86:1109–1118. 10.1128/JVI.05785-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee S, Cho HJ, Park JJ, Kim YS, Hwang S, Sun R, Song MJ. 2007. The ORF49 protein of murine gammaherpesvirus 68 cooperates with RTA in regulating virus replication. J Virol 81:9870–9877. 10.1128/JVI.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cheng BY, Zhi J, Santana A, Khan S, Salinas E, Forrest JC, Zheng Y, Jaggi S, Leatherwood J, Krug LT. 2012. Tiled microarray identification of novel viral transcript structures and distinct transcriptional profiles during two modes of productive murine gammaherpesvirus 68 infection. J Virol 86:4340–4357. 10.1128/JVI.05892-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Canny SP, Goel G, Reese TA, Zhang X, Xavier R, Virgin HW. 2014. Latent gammaherpesvirus 68 infection induces distinct transcriptional changes in different organs. J Virol 88:730–738. 10.1128/JVI.02708-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Johnson LS, Willert EK, Virgin HW. 2010. Redefining the genetics of murine gammaherpesvirus 68 via transcriptome-based annotation. Cell Host Microbe 7:516–526. 10.1016/j.chom.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.O'Grady T, Feswick A, Hoffman BA, Wang Y, Medina EM, Kara M, van Dyk LF, Flemington EK, Tibbetts SA. 2019. Genome-wide transcript structure resolution reveals abundant alternate isoform usage from murine gammaherpesvirus 68. Cell Rep 27:3988–4002.e5. 10.1016/j.celrep.2019.05.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stahl JA, Paden CR, Chavan SS, MacLeod V, Edmondson RD, Speck SH, Forrest JC. 2012. Amplification of JNK signaling is necessary to complete the murine gammaherpesvirus 68 lytic replication cycle. J Virol 86:13253–13262. 10.1128/JVI.01432-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol Biol 634:421–430. 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 83.Dal Canto AJ, Virgin HWt, Speck SH. 2000. Ongoing viral replication is required for gammaherpesvirus 68-induced vascular damage. J Virol 74:11304–11310. 10.1128/jvi.74.23.11304-11310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Paden CR, Forrest JC, Tibbetts SA, Speck SH. 2012. Unbiased mutagenesis of MHV68 LANA reveals a DNA-binding domain required for LANA function in vitro and in vivo. PLoS Pathog 8:e1002906. 10.1371/journal.ppat.1002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dong X, Feng H, Sun Q, Li H, Wu TT, Sun R, Tibbetts SA, Chen ZJ, Feng P. 2010. Murine gamma-herpesvirus 68 hijacks MAVS and IKKbeta to initiate lytic replication. PLoS Pathog 6:e1001001. 10.1371/journal.ppat.1001001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van Dyk LF, Hess JL, Katz JD, Jacoby M, Speck SH, Virgin HW. 1999. The murine gammaherpesvirus 68 v-cyclin gene is an oncogene that promotes cell cycle progression in primary lymphocytes. J Virol 73:5110–5122. 10.1128/JVI.73.6.5110-5122.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]