ABSTRACT
Vertebrates harbor hundreds of endogenous retroviral (ERV) sequences in their genomes, which are considered signs of past infections that occurred during evolution. On rare occasions, ERV genes like env are maintained and coopted by hosts for physiological functions, but they also participate in recombination events with exogenous retroviruses to generate rearranged viruses with novel tropisms. In domestic cats, feline leukemia virus type D (FeLV-D) has been described as a recombinant virus between the infectious FeLV-A and likely the ERV-DC14 env gene that resulted in an extended tropism due to the usage of a new uncharacterized retroviral receptor. Here, we report the identification of SLC31A1 encoding the copper transporter 1 (CTR1) as a susceptibility gene for ERV-DC14 infection. Expression of human CTR1 into nonpermissive cells was sufficient to confer sensitivity to ERV-DC14 pseudotype infection and to increase the binding of an ERV-DC14 Env ligand. Moreover, inactivation of CTR1 by genome editing or cell surface downmodulation of CTR1 by a high dose of copper dramatically decreased ERV-DC14 infection and binding, while magnesium treatment had no effect. We also investigated the role of CTR1 in the nonpermissivity of feline and hamster cells. While feline CTR1 was fully functional for ERV-DC14, we found that binding was strongly reduced upon treatment with conditioned medium of feline cells, suggesting that the observed resistance to infection was a consequence of CTR1 saturation. In contrast, hamster CTR1 was inactive due to the presence of a N-linked glycosylation site at position 27, which is absent in the human ortholog. These results provide evidence that CTR1 is a receptor for ERV-DC14. Along with chimpanzee endogenous retrovirus type 2, ERV-DC14 is the second family of endogenous retrovirus known to have used CTR1 during past infections of vertebrates.
IMPORTANCE Receptor usage is an important determinant of diseases induced by pathogenic retroviruses. In the case of feline leukemia viruses, three subgroups (A, B, and C) based on their ability to recognize different cell host receptors, respectively, the thiamine transporter THTR1, the phosphate transporter PiT1, and the heme exporter FLVCR1, are associated with distinct feline diseases. FeLV-A is horizontally transmitted and found in all naturally infected cats, while FeLV-B and FeLV-C have emerged from FeLV-A, respectively, by recombination with endogenous retroviral env sequences or by mutations in the FeLV-A env gene, both leading to a switch in receptor usage and in subsequent in vivo tropism. Here, we set up a genetic screen to identify the retroviral receptor of ERV-DC14, a feline endogenous provirus whose env gene has been captured by infectious FeLV-A to give rise to FeLV-D in a process similar to FeLV-B. Our results reveal that the copper transporter CTR1 was such a receptor and provide new insights into the acquisition of an expanded tropism by FeLV-D.
KEYWORDS: ERV-DC, FeLV-D, endogenous retrovirus, host receptor, CTR1, feline leukemia virus, retroviral receptor
INTRODUCTION
Retroviral envelope glycoproteins (Env) anchored at the surface of retroviral particles are essential players of infection. They ensure viral entry function by interacting with specific cell surface receptors and by mediating fusion of viral and cellular membranes (1). This is rendered possible by the conformational changes of Env triggered after receptor contact, which unmask a fusion peptide leading to membrane fusion and subsequent release of the viral nucleocapsid core. The expression profile of cellular receptors and their specific recognition by Env determine in part the host range of retroviruses and their classification. When cells become infected, they express viral Env from newly established proviruses, which can interact with their cognate receptor and prevent further infections in a process called interference to superinfection. This viral restriction by receptor interference is the basis of virus classification according to their receptor usage (2–4).
Env are also considered to be host proteins. They can be expressed from endogenous retroviral (ERV) env genes with reading frames that have been maintained open during evolution and coopted by hosts for physiological functions. This is exemplified by syncytins among mammals for their role in fusion of syncytiotrophoblast membranes during placentation (5–7). Coopted env genes have also been described as resistant genes to exogenous retroviral infections. They have been identified in various animals, including mice and cats, and encode either full-length Env or Env-truncated receptor-binding domains (RBD), referred to as restriction factors, that can block virus entry at the receptor level by saturation (8–12). Endogenous env can also be donors of ERV sequences in recombination events that occur during infection with exogenous retroviruses, such as the ecotropic murine leukemia viruses (E-MLV) in mice and feline leukemia virus type A (FeLV-A) in cats (12, 13). The resulting viruses always harbor a rearranged env sequence, display an extended tropism due to receptor switching and often lead to exacerbated pathogenicity.
Gammaretroviral Env interact with their host receptor through their RBD located at the N terminus (14, 15). The structure of RBD is highly conserved among retroviral Env with a compact core composed of antiparallel beta-sheets on which discreet variable loops conferring receptor specificity are connected (16–18). RBD fold autonomously and can be found in hosts as a natural soluble factor with conserved receptor-binding capacities (10, 19). They can also be engineered as RBD ligands for the monitoring of plasma membrane expression of host receptors (15, 20–24). To date, all of the identified host receptors used by gammaretroviruses and deltaretroviruses are nutrient transporters belonging to the solute carrier (SLC) family (25). Since the discovery of the first retroviral receptor for E-MLV (26) and of its physiological function as a transporter of cationic amino acids (27, 28), all of the newly identified receptors were shown to share a multipass-transmembrane topology and to be transporters of solutes, including inorganic phosphate, myoinositol, neutral amino acids, heme, riboflavin, thiamine, folate, glucose, or lactate (11, 29–41). The targeted selection of SLC by gammaretroviruses and deltaretroviruses for infection is currently unknown.
Superinfection interference analysis of FeLV strains has led to the identification of four interference groups in domestic cats. These includes FeLV-A, -B, -C, and -D, each entering host cells through distinct cell surface receptors (4, 42, 43). FeLV-A is found in all infected cats, is transmitted horizontally, and required the thiamine transporter SLC19A2/THTR1 for cell entry (38, 44). FeLV-A causes slow immunosuppressive and lymphoma diseases but can become highly pathogenic following recombination events with endogenous inherited retroviral sequences or mutations in the env gene, giving rise to FeLV-B or -C, respectively (13). These two FeLV have broader host ranges due to the use of new receptors. The FeLV-B receptor was identified as the sodium-dependent phosphate transporter SLC20A1/PiT1 (45), while FeLV-C requires the heme exporter SLC49A1/FLVCR1 (36, 46). FeLV-T is another FeLV variant with T-cell tropism, described as a fusion-defective virus requiring PiT1 as receptor in a FeLIX cofactor-dependent manner (19, 47). More recently, FeLV-D has been described as a novel recombinant virus between FeLV-A and a unique endogenous sequence belonging to the newly described ERV family of domestic cats called ERV-DC (4, 48, 49). ERV-DC comprise 13 proviral loci classified in 3 genotype groups. FeLV-D originated from genotype group I, of which ERV-DC14 is the prototype (48). Like FeLV-B, FeLV-D has acquired an extended host range with the capacity to replicate in many cell line of different species, suggesting the use of a new receptor. FeLV-D falls in an interference group different from those of FeLV-A, FeLV-B, and FeLV-C, likely meaning that FeLV-D and its ERV-DC relatives from genotype group I utilize a receptor different than THTR1, PiT1, and FLVCR1 (43). Here, we have employed a cDNA library screen strategy and identified the copper transporter 1 (CTR1/SLC31A1) as a receptor used by ERV-DC14. Interestingly, after CERV2, ERV-DC14 is the second family of endogenous retroviruses that requires CTR1 as a host receptor for infection (50).
RESULTS
The ERV-DC14 receptor is widely expressed.
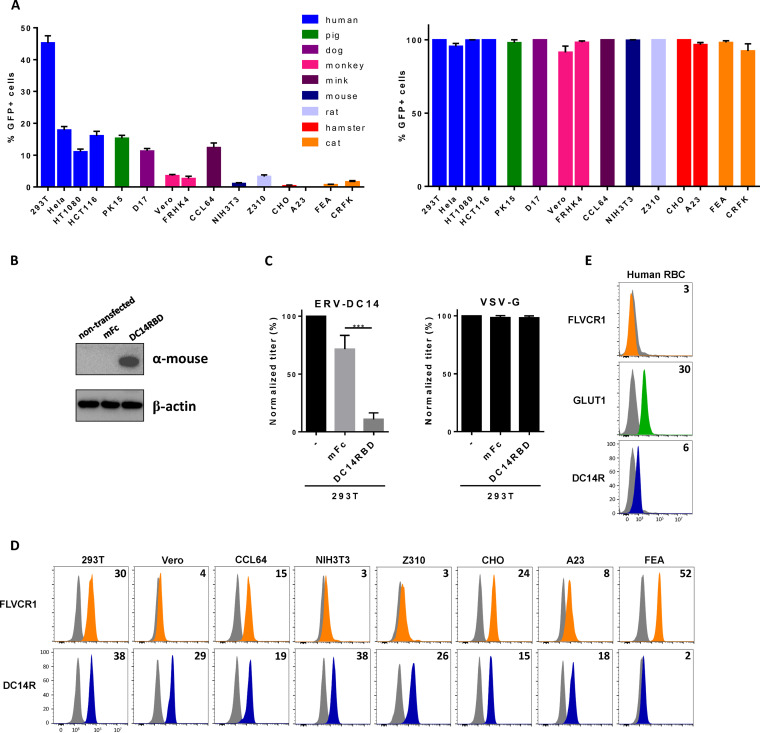
To assess the presence of the ERV-DC14 receptor at the cell surface, we first constructed an ERV-DC14 Env expression vector in order to produce enhanced green fluorescent protein (EGFP) lentiviral vector particles pseudotyped with the ERV-DC14 Env. These particles were shown to be infectious and could infect more than 40% of human 293T cells (Fig. 1A), demonstrating the presence of a functional cell surface ERV-DC14 receptor (DC14R) on human cells as reported (4). Further infection analyses of cells from various species revealed a large host range of ERV-DC14 pseudotypes. Of note, rodent cells appeared to be poorly infectible, with hamster A23 and CHO cells fully resistant to infection (Fig. 1A), although they are readily infectible by vesicular stomatitis virus G (VSV-G) pseudotypes. We also derived an ERV-DC14 immunoadhesin comprising the retroviral Env RBD fused to a mouse IgG1 Fc fragment for receptor detection by flow cytometry. To verify that this RBD ligand was able to interact with the cellular receptor used by ERV-DC14 pseudotypes, we developed an interference assay. DC14RBD was stably introduced in 293T cells by transfection (Fig. 1B), and cells were challenged with pseudotyped virions. Figure 1C shows that the presence of DC14RBD in cells conferred resistance to ERV-DC14 pseudotype infection compared to that of an empty vector control but had no effect on lentivirus infection pseudotyped with the VSV-G protein. Therefore, DC14RBD is a specific ligand for cell surface detection of DC14R. We next used this RBD ligand to assess the presence of DC14R at the surface of cell lines from different species. As shown in Fig. 1D, binding was detected at various levels for each of the cell lines tested, including hamster cells that were resistant to ERV-DC14 pseudotype infection. Thus, no strict correlation could be observed between DC14R binding and infection. We also assessed the expression of DC14R on human red blood cells (RBC). As expected, HTLV2 RBD binding to the glucose transporter GLUT1 was detected on human RBC (Fig. 1E) (40, 51). Similarly, DC14RBD binding was observed albeit at a lower level than HTLV2 RBD, suggesting the presence of DC14R at the surface of human RBC. In contrast, binding of an FeLV-C Env RBD, which recognizes the FLVCR1 heme exporter, was undetectable on human RBC.
FIG 1.
ERV-DC14 receptor is widely expressed. (A) Sensitivity of various cell lines from different species to infection by EGFP lentiviral vector pseudotyped with the ERV-DC14-specific Env or the VSV-G protein. Data are means ± SEM from n = 4 experiments. (B) Representative immunoblot of DC14RBD in cell lysates from 293T cells stably transduced with the MLV-based retroviral vectors carrying either the ERV-DC14 RBD cDNA fused to mouse IgG1 Fc fragment or the mouse Fc (mFc) alone. (C) Cells from panel B were evaluated for their sensitivity to infection by EGFP retroviral vectors pseudotyped with the ERV-DC14-specific Env or the VSV-G protein. Data are means ± SEM from n = 3 experiments. One-way analysis of variance (ANOVA) with Dunnett’s multiple-comparison test; ***, P ≤ 0.001. (D) Evaluation of cell surface expression of ERV-DC14 receptor (DC14R) or FLVCR1 (heme exporter) on various cell lines by flow cytometry using RBD ligands from ERV-DC4 and FeLV-C Env, respectively. Numbers (× 103) indicate the delta mean fluorescence intensity of a representative experiment (n = 3) (orange, green, blue), compared with nonspecific staining with the secondary antibody (gray). (E) Evaluation of cell surface expression of FLVCR1, GLUT1, or DC14R on human red blood cells (RBC) by flow cytometry using RBD ligands from ERV-DC14, HTLV2, and FeLV-C Env, respectively. Numbers (× 103) indicate the delta mean fluorescence intensity of a representative experiment (n = 3).
Identification of CTR1 as a functional receptor for ERV-DC14.
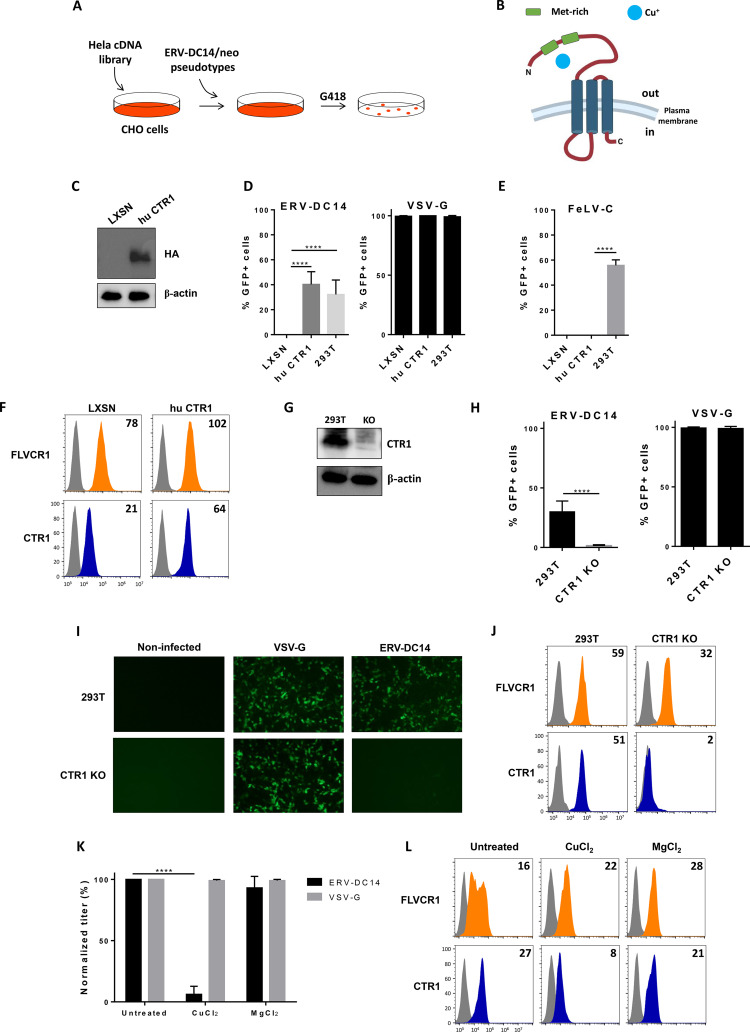
Hamster CHO cells were shown to be resistant to ERV-DC14 pseudotype infection due to the absence of a functional receptor for viral entry, although it can interact with DC14RBD. Since human HeLa cells express a functional DC14R, we introduced a HeLa cDNA library expressed from a retrovirus vector (52) into CHO cells by transduction and screened cells for their capacity to become susceptible to ERV-DC14 infection (Fig. 2A). Transduced cells were then challenged with ERV-DC14-enveloped LXSN vectors (53) carrying the neo gene and subjected 2 days later to G418 selection. About one hundred independent G418-resistant clones were obtained after 10 days of selection and pooled before genomic DNA extraction. PCR amplification and sequencing of cDNAs contained in the library-expressing vector identified multiple copies of SLC31A1, which encode the copper transporter CTR1 (Fig. 2B). Reintroduction of human SLC31A1 cDNA into CHO cells, in order to express a hemagglutinin (HA)-tagged CTR1 (Fig. 2C), conferred sensitivity to ERV-DC14 pseudotype infection as efficiently as infection on 293T cells (Fig. 2D) but did not affect VSV-G pseudotype infection or confer permissivity of CHO cells to FeLV-C virions (Fig. 2E). Human CTR1 also facilitated the binding of DC14RBD to CHO cells (Fig. 2F). Thus, our results suggest that CTR1 is a potent and specific receptor for both attachment and infection of ERV-DC14. Surprisingly, FeLV-C RBD binding was slightly increased, suggesting that copper transport and heme export may be linked in a common copper-iron metabolic pathway. We next asked whether CTR1 was the sole receptor for ERV-DC14 in human cells. For that purpose, we inactivated the CTR1 gene in 293T cells by genome editing (Fig. 2G). Challenge of CTR1 KO cells by ERV-DC14 pseudotypes revealed a total absence of infection (Fig. 2H and I) and a dramatic loss in DC14RBD binding (Fig. 2J). In contrast, absence of CTR1 did not affect infection by VSV-G-enveloped virions and slightly decreased the expression of the FLVCR heme exporter. Another mean of downmodulating CTR1 expression is to treat cells with a high dose of copper. Indeed, although copper is an essential cation for biological processes like the mitochondrial electron chain transfer, its excess is toxic for cells that can adapt to this situation by decreasing the expression of the CTR1 copper transporter from the plasma membrane (54). Figure 2K and L show that overnight treatment of human 293T cells with 100 μM copper chloride specifically inhibit ERV-DC14 infection and binding, while magnesium chloride had no effect. Overall, these data indicated that CTR1 is a specific and the sole receptor for ERV-DC14 Env binding and viral infection in human cells.
FIG 2.
CTR1 is a functional receptor for ERV-DC14 pseudotype infection and binding. (A) Schematic representation of human cDNA screen used to identify ERV-DC14 receptor. (B) Representation of CTR1 transmembrane receptor. (C) Representative immunoblot of HA-tagged CTR1 in cell lysates from CHO cells stably transduced with MLV-based LXSN retroviral vector either empty or carrying the CTR1 cDNA from human (hu CTR1). Cells from panel C were evaluated for their sensitivity to retroviral infection by EGFP lentiviral vectors pseudotyped with either the ERV-DC14-specific Env or the VSV-G protein (D) or with either the FeLV-C-specific Env (E). Infection of 293T control cells is shown. Data are means ± SEM from n = 3 experiments. One-way ANOVA with Dunnett’s multiple-comparison test; ****, P ≤ 0.0001. (F) CHO cells from panel C were evaluated for FLVCR1 or CTR1 cell surface expression by flow cytometry using RBD ligands from FeLV-C and ERV-DC14 Env, respectively. Numbers (× 103) indicate the delta mean fluorescence intensity of a representative experiment (n = 3). (G) Representative immunoblot of CTR1 in cell lysates of 293T control and CTR1 KO (invalidated by CRISPR/Cas9 technology). (H) 293T CTR1 KO and control cells were evaluated for their sensitivity to infection by EGFP lentiviral vectors pseudotyped with ERV-DC14-specific Env or the VSV-G protein. Data are means ± SEM from n = 3 experiments. Student unpaired t test; ****, P ≤ 0.0001. (I) Visualization of infected cells (green) from panel G with a Nikon fluorescence microscope. (J) 293T cells from panel G were evaluated for FLVCR1 or CTR1 cell surface expression by flow cytometry using RBD ligands. Numbers (× 103) indicate the delta mean fluorescence intensity of a representative experiment (n = 3). (K) 293T cells grown overnight in DMEM supplemented with 100 μM CuCl2 or MgCl2 were evaluated for their sensitivity to infection by EGFP lentiviral vector pseudotyped with either the VSV-G protein or the ERV-DC14-specific Env. Data are means ± SEM from n = 3 experiments. Student unpaired t test; ****, P ≤ 0.0001. (L) 293T cells from panel K were evaluated for cell surface expression of FLVCR1 and CTR1 by flow cytometry using RBD ligands. Numbers (× 103) indicate the delta mean fluorescence intensity of a representative experiment (n = 3).
Species specificity of CTR1 to ERV-DC14 infection.
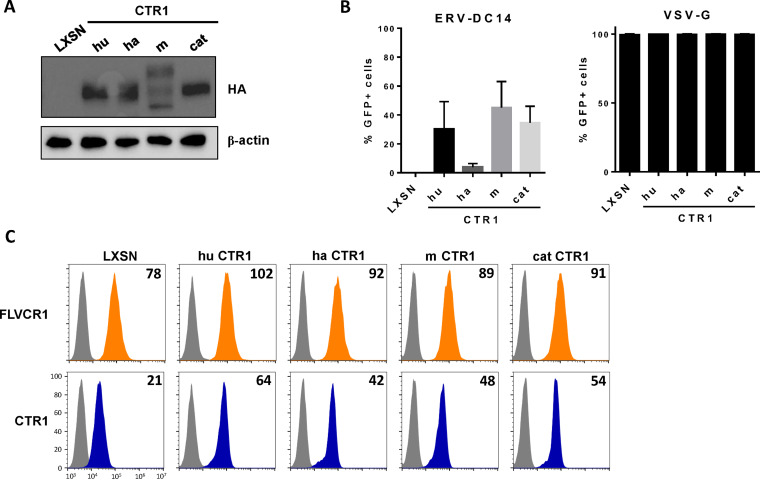
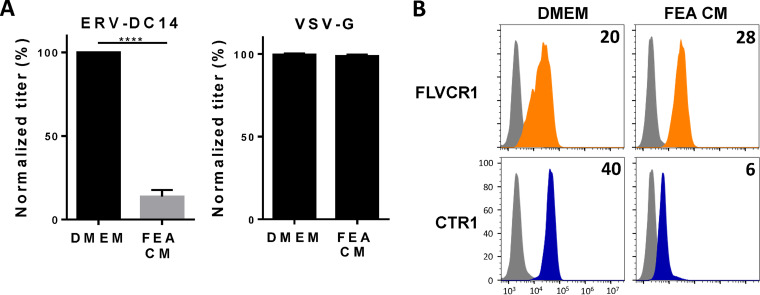
Although ERV-DC14 can efficiently infect human cells, mouse cells are weakly infectible and hamster and feline cells are fully resistant (Fig. 1A). To investigate the role of CTR1 in this ERV-DC14 host range, CTR1 cDNAs were amplified from mouse NIH3T3, hamster CHO, and cat FEA cells, introduced in LXSN-based retroviral vector, expressed as HA-tagged proteins in CHO cells, and compared to the human ortholog (Fig. 3A). With the exception of hamster CTR1, we found that all of the CTR1 orthologs conferred the sensitivity of CHO cells to ERV-DC14 pseudotype infection but did not affect VSV-G pseudotype infection, demonstrating functional and specific receptor activity (Fig. 3B). Moreover, all of the CTR1 proteins including the hamster one increased the binding of DC14RBD (Fig. 3C). Therefore, CTR1 exhibited no species specificity for Env binding, which did not correlate with infection for the hamster CTR1. Thus, the resistance of hamster cells appears to be supported by a nonfunctional CTR1 at a postbinding step. Interestingly, the feline CTR1 was a potent retroviral receptor for ERV-DC14, while all feline cell lines are described as resistant (4). This is most likely due to the presence of the inhibitory activity of Refrex1, a pair of soluble RBD-like Env derived from the ERV-DC7 and ERV-DC16 loci (10, 55). We confirmed that pretreatment of 293T cells with conditioned medium of FEA cells (FEA-CM) resulted in a strong decrease in both ERV-DC14 infection (Fig. 4A) and DC14RBD binding (Fig. 4B), while infectivity of VSV-G-pseudotyped particles was unchanged as well as FeLV-C RBD binding. By performing reverse transcriptase PCR (RT-PCR) on FEA total RNAs, we confirmed that FEA cells, like many feline cells, express Refrex1 transcripts, although we detected only the presence of ERV-DC16 transcripts by direct sequencing of the PCR product (data not shown). Hence, resistance of feline cells is not due to the inability of feline CTR1 to serve as an ERV-DC14 receptor but is likely due to the presence of the ERV-DC7/16 env gene product Refrex1, which is able to interact with and block CTR1 receptor function.
FIG 3.
CTR1 requirement for ERV-DC14 in different species. (A) Representative immunoblot of HA-tagged CTR1 in cell lysates from CHO cells stably transduced with MLV-based LXSN retroviral vector either empty or carrying the CTR1 cDNA from human (hu), hamster (ha), mouse (m), or cat. (B) Cells from panel A were evaluated for their sensitivity to retroviral infection by EGFP lentiviral vectors pseudotyped with either the ERV-DC14-specific Env or the VSV-G protein. Data are means ± SEM from n = 3 experiments. (C) CHO cells from panel A were evaluated for FLVCR1 and CTR1 or cell surface expression by flow cytometry using RBD ligands. Numbers (× 103) indicate the delta mean fluorescence intensity of a representative experiment (n = 3).
FIG 4.
Conditioned medium of feline cells inhibits ERV-DC14 infection and binding. (A) 293T cells incubated for 5 h in conditioned medium (CM) from feline FEA cells or complete DMEM were evaluated for their sensitivity to infection by EGFP retroviral vectors pseudotyped with the ERV-DC14-specific Env or the VSV-G protein. Data are means ± SEM from n = 3 experiments. Student unpaired t test; ****, P ≤ 0.0001. (B) 293T cells from panel A were evaluated for FLVCR1 or CTR1 cell surface expression by flow cytometry using RBD ligands. Numbers (× 103) indicate the specific delta mean fluorescence intensity of a representative experiment (n = 3).
CTR1 determinants for receptor function.
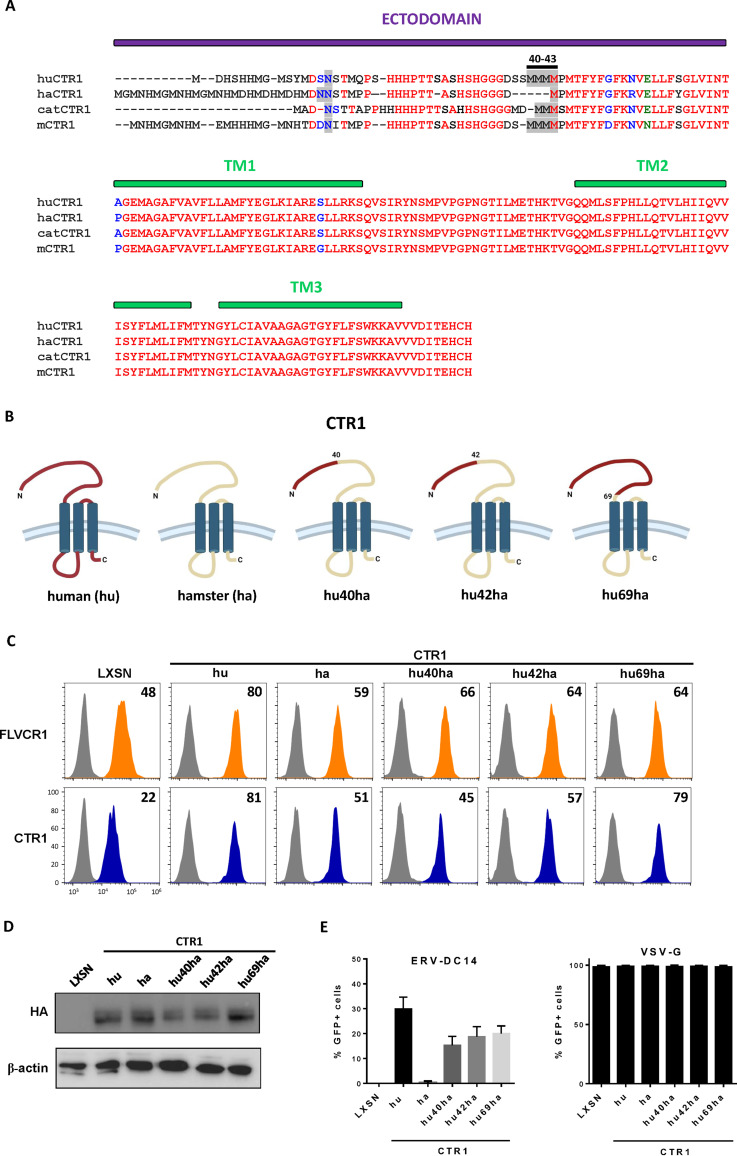
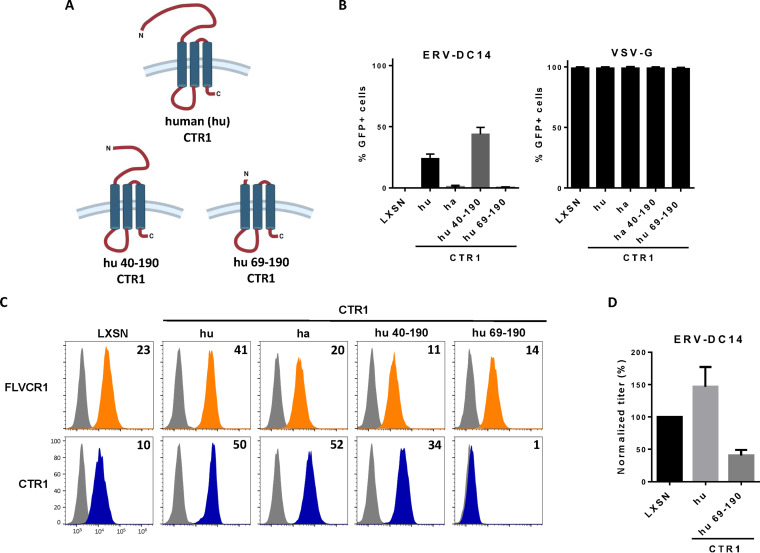
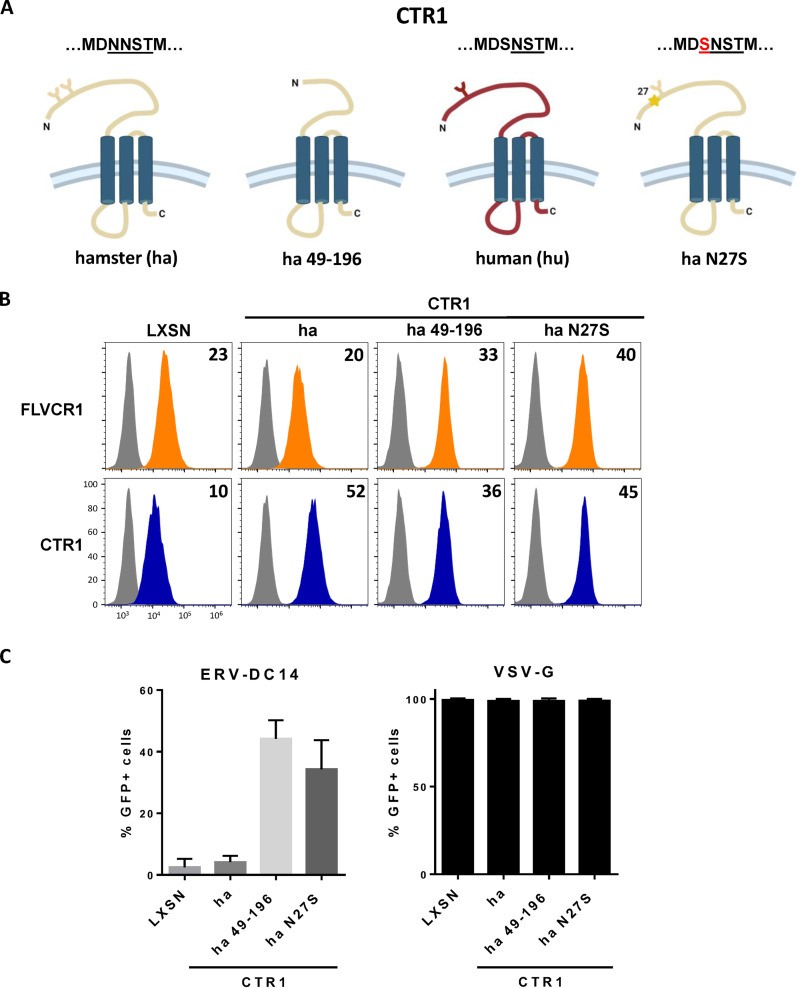
As shown in Fig. 1A, CHO cells are resistant to ERV-DC14 infection, although they can interact with ERV-DC14 Env, suggesting that the endogenous CTR1 is expressed at the cell surface but is incompetent for virus entry and subsequent infection. We, therefore, sought to identify the determinants in hamster CTR1 responsible for this entry defect. When compared to the amino acid sequence of the functional human CTR1, variations of hamster CTR1 in sequence and length are mostly located in the N-terminal ectodomain before the first transmembrane (Fig. 5A). To test whether viral entry determinants could be contained in this domain, we constructed a set of chimeras between human and hamster CTR1 as well as truncated mutants and evaluated their capacity to promote ERV-DC14 infection. We first exchanged the hamster CTR1 ectodomain with its human counterpart either partially until a quadruplet of methionine (amino acids 40 to 43 in human CTR1) at positions 40 (hu40ha) and 42 (hu42ha) or totally just before the first transmembrane at position 69 (hu69ha). The hu40ha construct maintained the four methionines like in human CTR1, while only one methionine was present in hu42ha as in hamster CTR1 (Fig. 5B). All of these CTR1 chimeras were expressed in CHO cells and present at the cell surface as assayed by flow cytometry (Fig. 5C and D). In addition, they also rendered CHO cells susceptible to ERV-DC14 infection and remained permissive to VSV-G pseudotype infection (Fig. 5E), suggesting that the first 40 amino acids of the ectodomain contained the determinants required for ERV-DC14 entry and infection. Surprisingly, deletion of this fragment in human CTR1 still conferred ERV-DC14 susceptibility and Env binding, while a larger deletion until amino acid 69 did not (Fig. 6A to C). Moreover, human CTR1 deleted of its first 68 residues was not detected at the cell surface and likely trapped endogenous CTR1 inside cells, as suggested by the decrease in DC14RBD binding (Fig. 6C) and in ERV-DC14 pseudotype infection when expressed in 293T cells (Fig. 6D). Since the 1 to 40 ectodomain was not required for retroviral receptor function, we tested the possibility that the 1 to 48 corresponding fragment in hamster CTR1 could impede virus entry function without affecting DC14RBD binding (Fig. 7A). As observed in Fig. 7B, the deletion of this fragment did not affect DC14RBD binding compared to wild-type hamster CTR1 and indeed allowed CHO cell permissivity to ERV-DC14 virions (Fig. 7C). A closer examination of hamster CTR1 ectodomain sequence revealed the presence of an additional consensus N-linked glycosylation site at position N27 that is absent in human CTR1 (Fig. 5A and 7A). Because extra-glycosylation of retroviral receptors has been shown to exert a protective effect of hamster cells toward infection (56), we removed this glycosylation site by site-directed mutagenesis (N27S mutant) and tested its role in viral receptor function. Expression of the N27S hamster CTR1 in CHO cells maintained DC14RBD binding (Fig. 7B) and conferred ERV-DC14 sensitivity compared to wild-type hamster CTR1 (Fig. 7C). Therefore, the hamster CTR1 contains all of the determinants required for productive infection, but its receptor activity is inhibited by the presence of carbohydrate moieties at position N27.
FIG 5.
Functional analysis of human/hamster CTR1 chimeras. (A) Amino acid sequences of human, hamster, cat, and mouse CTR1 were aligned using ClustalW. The ectodomain and the 3 transmembranes (TM) are indicated in purple and green boxes, respectively. The N-linked glycosylation sites and the quadruplet of methionines at positions 40 to 43 of the human sequence are highlighted in gray. Amino acid color code is as follows: red, identical; green, strongly similar; blue, weakly similar; black, different. Deletions are indicated with dashes. (B) Schematic representations of chimeric constructs between human and hamster CTR1. (C) CHO cells stably transduced with LXSN retroviral vector either empty or carrying the CTR1 cDNA from human (hu), hamster (ha), or chimeras (hu40ha, hu42ha, and hu69ha) were evaluated for CTR1 or FLVCR1 cell surface expression by flow cytometry using the RBD ligands. Numbers (× 103) indicate the delta mean fluorescence intensity of a representative experiment (n = 3). (D) Representative immunoblot of HA-tagged CTR1 in cell lysates from panel C. (E) Cells from panel C were evaluated for their sensitivity to infection by EGFP retroviral vectors pseudotyped with either the ERV-DC14-specific Env or the VSV-G protein. Data are means ± SEM from n = 3 experiments.
FIG 6.
Ectodomain of CTR1 is not required for ERV-DC14 infection and binding. (A) Schematic representations of truncated CTR1 constructs. (B) CHO cells stably transduced with LXSN retroviral vector either empty or carrying the CTR1 cDNA from human (hu) or truncated human CTR1 (hu 40 to 190 and hu 69 to 190) were evaluated for their sensitivity to infection by EGFP retroviral vectors pseudotyped with either the ERV-DC14-specific Env or the VSV-G protein. Data are means ± SEM from n = 3 experiments. (C) Cells from panel B were evaluated for FLVCR1 or CTR1 cell surface expression by flow cytometry using RBD ligands. Numbers (× 103) indicate the delta mean fluorescence intensity of a representative experiment (n = 3). (D) 293T cells transduced with retroviral LXSN either empty or carrying the CTR1 cDNA from human (hu) or truncated (hu 69 to 190) were evaluated for their sensitivity to infection by EGFP retroviral vectors pseudotyped with the ERV-DC14-specific Env. Data are means ± SEM from n = 3 experiments.
FIG 7.
N-glycosylation of hamster CTR1 prevents ERV-DC14 infection. (A) Schematic representations of human and hamster CTR1 constructs. Y forms represent carbohydrate chains. (B) CHO cells stably transduced with LXSN retroviral vector either empty or carrying the CTR1 cDNA from hamster (ha), truncated hamster (ha 49 to 196), or mutated hamster CTR1 (N-linked glycosylation mutant N27S) were evaluated for FLVCR1 or CTR1 cell surface expression by flow cytometry using the RBD ligands. Numbers (× 103) indicate the delta mean fluorescence intensity of a representative experiment (n = 3). (C) Cells from panel B were evaluated for their sensitivity to infection by EGFP retroviral vectors pseudotyped with either the ERV-DC14-specific Env or the VSV-G protein. Data are means ± SEM from n = 3 experiments.
DISCUSSION
Using a human expression library, we identified a cDNA that confers susceptibility to ERV-DC14 infection. Sequence analysis revealed the presence of SLC31A1, which encodes CTR1, the main copper transporter in vertebrates. Expression of human SLC31A1 into nonpermissive CHO cells rendered cells susceptible to ERV-DC14 pseudotype infection and conferred increased binding to a RBD ligand generated from ERV-DC14 Env. Moreover, inactivation of SLC31A1 in human cells by CRISPR/Cas9 technology abolished infectivity of ERV-DC14 pseudotypes and RBD binding, suggesting that CTR1 is the sole functional host receptor of ERV-DC14. Because FeLV-D and ERV-DC14 Env cross-interfere with each other (10) and because FeLV-D is a recombinant virus with an env gene originating from the ERV-DC14 genotype group I, it is likely that CTR1 is also a receptor for FeLV-D.
It has been suggested that ERV-DC invaded cat genomes a million years ago on three independent occasions, giving rise to three distinct genotype groups of ERV-DC based on phylogenetic analysis of their env gene (4, 10). Our present study suggests that infection of cat germ cells by two of these ERV-DC genotype groups occurred through the copper transporter CTR1 as the main entry receptor, namely, genotype groups I and II with ERV-DC14 and ERV-DC7/16 as prototypes, respectively. Interestingly, chimpanzee endogenous retrovirus-2 (CERV2) present in the genome of several Old Word primates but absent in humans likely used CTR1 during primate invasion as well (50). However, no sequence homology was found in their Env SU, suggesting that CERV2 and ERV-DC14/7/16 do not share a recent common ancestor despite similar receptor usage (10, 50). Surprisingly, CERV2 virions could readily infect feline CRFK cells (50), suggesting that CTR1 was fully functional in these cells. The inability of ERV-DC14 to infect feline cells was therefore not due to the absence of CTR1 at the plasma membrane or to a defective CTR1 as evidenced by the susceptibility of hamster cells to ERV-DC14 infection when expressing feline CTR1 (Fig. 3B). Instead, feline cells express the ERV-DC7 and 16 env gene product Refrex1, which has been described as an inhibitory factor of ERV-DC14 virions and its FeLV-D derivative (10, 55), likely by CTR1 competition. The recognition of different determinants on CTR1 probably explains why CERV2 is insensitive toward Refrex1 in CRFK cells. This suggests that ERV-DC14/FeLV-D may not cross-interfere with CERV2 despite the use of a common CTR1 receptor. This also suggests that the underlying mechanism of Refrex1 restriction does not involve CTR1 retention inside cells but rather CTR1 saturation at the plasma membrane by soluble Refrex1 proteins released by feline cells.
In addition to feline cells, hamster cells are also nonpermissive to ERV-DC14 infection and mouse cells are slightly permissive (Fig. 1A). While mouse Ctr1 appeared fully functional when overexpressed in CHO cells, hamster CTR1 remained defective. Low endogenous expression of Ctr1 or low affinity between Ctr1 and DC14RBD could explain why mouse NIH3T3 cells were less readily infected than human cells. However, binding of DC14RBD was comparable for both cell types. Other parameters like absence or weak expression of a Ctr1 cofactor could explain this discrepancy. Consistent with this hypothesis, we found that overexpression of human CTR1 in mouse NIH3T3 cells resulted in significantly increased levels of DC14RBD binding but modestly increased cell susceptibility to ERV-DC14 infection (not shown). CTR1 orthologs present high sequence homology except in their N-terminal extracellular ectodomain (50). This domain has a high content of methionine and histidine residues, known to capture cuprous Cu+ ions for CTR1-mediated transport (57). Human CTR1 ectodomain also contains one consensus N-linked glycosylation site at N15 (58), which is conserved in hamster CTR1 at N28. We showed that a second N-linked glycosylation site at N27 in hamster CTR1 was responsible for the inability of CTR1 to function as a retroviral receptor. This additional glycosylation moiety compared to the human CTR1 did not prevent plasma membrane expression of hamster CTR1 since its presence was detected by flow cytometry using the DC14RBD. Preventing glycosylation by site-directed mutagenesis or by deleting the first 48 residues of the ectodomain restored the capacity of hamster CTR1 to function as an ERV-DC14 receptor. Like for the E-MLV receptor CAT1 (SLC7A1), the amphotropic-MLV receptor PiT2 (SLC20A2), and the xenotropic-MLV receptor XPR1 (SLC53A1) expressed in hamster cells (56, 59), hamster CTR1 is rendered nonfunctional by addition of extra sugar moieties during synthesis. A similar glycosylation-inactivated form of mouse CTR1, which also contains 2 consensus glycosylation sites in the extracellular ectodomain, may explain the low sensitivity of NIH3T3 to ERV-DC14 infection.
So far, all receptors used by gammaretroviruses and deltaretroviruses are solute carriers with a topology consisting of multiple transmembrane domains. It is not known whether all of the members of the SLC family have the potential to serve as retroviral receptors for virus entry. Although they are recognized by Env RBD harboring very similar three-dimensional structures, these SLC have limited sequence homology and possess a variable number of transmembranes, with CTR1 having only three of them. One common feature of these SLC is their ability to shuttle between plasma membrane and lysosomes upon solute interactions, which is likely to be crucial for viral entry. In this regard, most of the SLC found to be retroviral receptors are cargos of the SNX27-retromer, a major hub retrieving transmembrane proteins from lysosome degradation and recycling them to the plasma membrane (60). CTR1 is also subject to the SNX27-retomer regulation (61) since elevated copper levels trigger its endocytosis from the plasma membrane, preventing excessive copper uptake and toxic accumulation in cells (54). Another common feature is the fact that they transport solutes involved directly or indirectly in energy metabolism. Glucose and lactate are the initial and end products of glycolysis, inorganic phosphate and myoinositol participate in high-energy molecule synthesis like ATP, and inositol pyrophosphates, thiamine, and riboflavin are vitamin B metabolites participating, respectively, in the citric acid cycle and in the mitochondrial electron transport chain, as does copper. Thus, cell surface expression of these metabolite transporters reflects the high metabolic activity of cells, which is required for proper gammaretroviral infections.
MATERIALS AND METHODS
Cell culture.
Human embryonic kidney (HEK) 293T, HeLa, HT1080, HCT116, pig PK15, dog D17, simian Vero and FRHK4, mink CCL64, mouse NIH3T3, rat Z310, Chinese hamster ovary (CHO) and A23, cat feline embryonic fibroblast (FEA), and Crandell feline kidney (CRFK) cell lines were maintained in complete Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, penicillin/streptomycin, and nonessential amino acids. Cells were maintained at 37°C with 5% CO2. When indicated, 293T cells were incubated in filtered FEA cell supernatant or incubated overnight in the presence of 100 μM CuCl2 (307483; Sigma) or MgCl2 (208367; Sigma).
For CRISPR/Cas9-mediated gene disruption, 293T cells were cotransfected with the pX458 vector carrying the Cas9 protease fused to GFP and a Sanger lentiviral CRISPR vector (Merck) carrying the blue fluorescent protein (BFP) and a single guide RNA (sgRNA) against human CTR1 under the control of the U6 promoter (sgRNA, 5′-TACTAGCAATGTTCTATGAAGG-3′). A BD FACSAria was used for GFP/BFP double-positive cell sorting 48 h later. Cells were cloned by limiting dilution in a 96-well plate.
Plasmids, viral productions, and infections.
pLXSN vector (53) pseudotyped with the VSV-G protein were produced by cotransfecting 293T cells with the LXSN vectors carrying CTR1 cDNAs, the MLV Gag-Pol expression vector pC57GPBEB (62), and the VSV-G Env expression vector pCSI-G (52). Viral supernatants were harvested 48 h after transfection, filtered, and stored at −80°C or used directly for CHO cell transduction. CHO cells expressing CTR1 cDNA were generated by transduction with pLXSN retroviral vector pseudotyped with vesicular stomatitis virus G protein (VSV-G), followed by G418 selection. Lentiviral CSGW vectors expressing the enhanced green fluorescent protein (EGFP) were produced as previously described (23). CSGW pseudotype vectors bearing envelope glycoproteins (Env) from VSV (VSV-G) and feline leukemia virus (ERV-DC14 or FeLV-C) were used to infect for 48 h 1 × 104 cells/well seeded the day before in a 96-well plate. Cells were then resuspended in 50 μL trypsin and 100 μL PBA (phosphate-buffered saline [PBS] with 2% fetal bovine serum [FBS]) and analyzed on a NovoCyte flow cytometer (ACEA Biosciences, Inc). Data analyses measuring the percentage of EGFP-positive infected cells were performed using FlowJo software.
cDNA library screen for identification of ERV-DC14 receptor.
The previously described HeLa cell cDNA library expressed from an MLV-based vector was used to produce viral supernatants and introduced into hamster CHO cells by transduction as previously described (52). The library-containing cells were then screened for their susceptibility to ERV-DC14 pseudotype vectors. Briefly, the CHO cell library was transduced with an MLV vector carrying the neo gene and pseudotyped with the ERV-DC14 Env and selected 48 h later in the presence of 2 mg/mL of G418. G418-resistant clones were pooled and harvested and genomic DNA was extracted with QIAamp DNA minikit (Qiagen) and quantified by NanoDrop. cDNA from the library was amplified by PCR and sequenced as described (52).
RNA extraction, 5′ RACE, and PCR.
CHO and FEA cells were harvested with trypsin and washed in PBS. Total RNA was extracted with GenElute mammalian total RNA miniprep kit (Sigma) and quantified by NanoDrop. cDNAs were obtained by 5′ RACE (rapid amplification cDNA ends) from FEA cell total RNA using the SMARTer RACE cDNA amplification kit (Clontech). The full-length cDNA from hamster CTR1 was obtained from RT-PCR amplification of CHO cell RNA. All PCR products were cloned in the pLXSN retroviral vector. All CTR1 cDNA were fused in frame at their C-terminal end with two copies of the HA tag. pLhumanCTR1SN (hu CTR1), pLhamsterCTR1SN (ha CTR1), pLmouseCTR1SN (m CTR1), and pLcatCTR1SN (cat CTR1) were carriers of human, hamster, mouse, and cat CTR1, respectively. Chimera between human and hamster CTR1 were obtained by recombinant PCR as follows: pLH40SN (hu40ha CTR1), pLH42SN (hu42ha CTR1), and pLH69SN (hu69ha CTR1). Residues 40, 42 and 69 are the position of the recombination points. Truncated CTR1 from human: pLH69-190SN (hu 69-190 CTR1), pLH69-190SN (hu 40-190 CTR1) and hamster: pLH49-196SN (ha 49-196 CTR1) were generated by PCR (details upon request). Site-directed mutagenesis of N-linked glycosylation site was used to introduce asparagine to serine at position 27 in hamster cDNA (N27S CTR1).
Monitoring retroviral receptors cell surface expression.
Detection of CTR1, FLVCR1, or GLUT1 at the plasma membrane was performed by flow cytometry using soluble receptor-binding domains (RBD) derived from ERV-DC14, FeLV-C, or HTLV2 retroviral Env fused to a mouse IgG1 Fc domain (40). RBD ligands were produced and used as previously described (23). Briefly, 105 cells were harvested with trypsin, resuspended in 100 μL PBA (PBS with 2% FBS) containing the different ligands, and incubated at 37°C for 30 min. Cells were then washed twice with cold PBA and labeled for 20 min on ice with Alexa 488-conjugated anti-mouse IgG1 antibodies (1:400; Thermo Fisher reference A21121). Cells were then washed in PBA and analyzed on a NovoCyte flow cytometer (ACEA Biosciences, Inc). Data analysis was performed using FlowJo software.
Immunoblotting.
Cells were lysed in lysis buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM MgCl2, 1% Triton X-100) and protease inhibitors (Complete; Roche), resolved by SDS-PAGE, and analyzed by immunoblotting using HA antibodies (3F10; Roche; 11867423001, 1:5,000) followed by horseradish peroxidase (HRP)-conjugated anti-rat antibodies (Sigma; A9037; 1:10,000) or CTR1 monoclonal antibody (Proteintech; 67221-1; 1:1,000) followed by horseradish peroxidase (HRP)-conjugated anti-mouse antibodies (Jackson; 715-035-150; 1:10,000) or anti β-actin-HRP (A3854; Sigma; 1:100,000) antibodies. Signals were visualized using the Luminata Forte detection reagent (Merck Millipore). A Bio-Rad ChemiDoc imager was used.
Statistical analysis.
Statistical analyses were performed using GraphPad Prism 6 software. The data were expressed as the mean ± standard error of the mean (SEM). The results were considered statistically significant at P values of <0.05 (*), <0.01 (**), <0.001 (***), and <0.0001 (****). Statistical tests and number of independent experiments are indicated in the figure legends.
ACKNOWLEDGMENTS
The authors are grateful to Andrea Cimarelli (CIRI, Lyon, France) for providing CRFK cells. We also thank all of the members of our teams for technical help and advice. We acknowledge the Montpellier Rio Imagine facility, member of the national infrastructure France-BioImaging supported by the French National Research Agency (ANR-10-INBS-04) for cell sorting by flow cytometry.
This work was supported by the French National Research agency (ANR CALCIPHOS, ANR-17-CE14-0008 to J.-L.B.), by the French SIDACTION (19-2-AEQ-12560 to J.-L.B.), by the French medical research foundation FRM (DBI201312285579), and by the Labex GR-Ex (ANR-11-LABX-0051). S.T. was supported by the ANR CALCIPHOS, D.G. by FRM, S.I. by CNRS and by the METAFORA Biosystems company, V.C. by CNRS, and J.-L.B. by INSERM.
Conceived the project, J.-L.B.; designed the experiments and analyzed the data, S.T., D.G., S.I., V.C., and J.-L.B.; performed the experiments, S.T., D.G., S.I., J.T., V.C., and J.-L.B.; wrote the original manuscript, S.T. and J.-L.B.; edited the manuscript, S.T., D.G., S.I., V.C., and J.-L.B.
We declare no competing interests.
Contributor Information
Jean-Luc Battini, Email: jean-luc.battini@irim.cnrs.fr.
Frank Kirchhoff, Ulm University Medical Center.
REFERENCES
- 1.Weiss RA. 1993. Cellular receptors and viral glycoproteins involved in retroviral entry, p 1–108. In Levy J (ed), The Retroviridae. Plenum Press, New York, NY. [Google Scholar]
- 2.Miller AD, Wolgamot G. 1997. Murine retroviruses use at least six different receptors for entry into Mus dunni cells. J Virol 71:4531–4535. 10.1128/JVI.71.6.4531-4535.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeuchi Y, Patience C, Magre S, Weiss RA, Banerjee PT, Le Tissier P, Stoye JP. 1998. Host range and interference studies of three classes of pig endogenous retrovirus. J Virol 72:9986–9991. 10.1128/JVI.72.12.9986-9991.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anai Y, Ochi H, Watanabe S, Nakagawa S, Kawamura M, Gojobori T, Nishigaki K. 2012. Infectious endogenous retroviruses in cats and emergence of recombinant viruses. J Virol 86:8634–8644. 10.1128/JVI.00280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, Keith JC, McCoy JM. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785–789. 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 6.Blaise S, de Parseval N, Bénit L, Heidmann T. 2003. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci USA 100:13013–13018. 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupressoir A, Lavialle C, Heidmann T. 2012. From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta 33:663–671. 10.1016/j.placenta.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Buller RS, Sitbon M, Portis AL. 1988. The endogenous mink cell focus-forming (MCF) gp70 linked to the Rmcf gene restricts MCF virus replication in vivo and provides partial resistance to erythroleukemia induced by Friend murine leukemia virus. J Exp Med 167:1535–1546. 10.1084/jem.167.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikeda H, Laigret F, Martin MA, Repaske R. 1985. Characterization of a molecularly cloned retroviral sequence associated with Fv-4 resistance. J Virol 55:768–777. 10.1128/JVI.55.3.768-777.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito J, Watanabe S, Hiratsuka T, Kuse K, Odahara Y, Ochi H, Kawamura M, Nishigaki K. 2013. Refrex-1, a soluble restriction factor against feline endogenous and exogenous retroviruses. J Virol 87:12029–12040. 10.1128/JVI.01267-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanco-Melo D, Gifford RJ, Bieniasz PD. 2017. Co-option of an endogenous retrovirus envelope for host defense in hominid ancestors. Elife 6:e22519. 10.7554/eLife.22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozak CA. 2015. Origins of the endogenous and infectious laboratory mouse gammaretroviruses. Viruses 7:1–26. 10.3390/v7010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiu ES, Hoover EA, VandeWoude S. 2018. A retrospective examination of feline leukemia subgroup characterization: viral interference assays to deep sequencing. Viruses 10:29. 10.3390/v10010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Battini JL, Heard JM, Danos O. 1992. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J Virol 66:1468–1475. 10.1128/JVI.66.3.1468-1475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Battini JL, Danos O, Heard JM. 1995. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J Virol 69:713–719. 10.1128/JVI.69.2.713-719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fass D, Davey RA, Hamson CA, Kim PS, Cunningham JM, Berger JM. 1997. Structure of a murine leukemia virus receptor-binding glycoprotein 2.0 angstrom resolution. Science 277:1662–1666. 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- 17.Barnett AL, Wensel DL, Li W, Fass D, Cunningham JM. 2003. Structure and mechanism of a coreceptor for infection by a pathogenic feline retrovirus. J Virol 77:2717–2729. 10.1128/jvi.77.4.2717-2729.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarthy KR, Timpona JL, Jenni S, Bloyet LM, Brusic V, Johnson WE, Whelan SPJ, Robinson-Mccarthy LR. 2020. Structure of the receptor binding domain of EnvP(B)1, an endogenous retroviral envelope protein expressed in human tissues. mBio 11:e02772-20. 10.1128/mBio.02772-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson MM, Lauring AS, Burns CC, Overbaugh J. 2000. Identification of a cellular cofactor required for infection by feline leukemia virus. Science 287:1828–1830. 10.1126/science.287.5459.1828. [DOI] [PubMed] [Google Scholar]
- 20.Kim FJ, Manel N, Garrido EN, Valle C, Sitbon M, Battini JL. 2004. HTLV-1 and -2 envelope SU subdomains and critical determinants in receptor binding. Retrovirology 1:41. 10.1186/1742-4690-1-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavanya M, Kinet S, Montel-Hagen A, Mongellaz C, Battini J-L, Sitbon M, Taylor N. 2008. Cell surface expression of the bovine leukemia virus-binding receptor on B and T lymphocytes is induced by receptor engagement. J Immunol 181:891–898. 10.4049/jimmunol.181.2.891. [DOI] [PubMed] [Google Scholar]
- 22.Lagrue E, Abe H, Lavanya M, Touhami J, Bodard S, Chalon S, Battini JL, Sitbon M, Castelnau P. 2010. Regional characterization of energy metabolism in the brain of normal and MPTP-intoxicated mice using new markers of glucose and phosphate transport. J Biomed Sci 17:91. 10.1186/1423-0127-17-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López-Sánchez U, Tury S, Nicolas G, Wilson MS, Jurici S, Ayrignac X, Courgnaud V, Saiardi A, Sitbon M, Battini JL. 2020. Interplay between primary familial brain calcification-associated SLC20A2 and XPR1 phosphate transporters requires inositol polyphosphates for control of cellular phosphate homeostasis. J Biol Chem 295:9366–9378. 10.1074/jbc.RA119.011376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peric D, Barragan I, Giraud-Triboult K, Egesipe AL, Meyniel-Schicklin L, Cousin C, Lotteau V, Petit V, Touhami J, Battini JL, Sitbon M, Pinset C, Ingelman-Sundberg M, Laustriat D, Peschanski M. 2015. Cytostatic effect of repeated exposure to simvastatin: a mechanism for chronic myotoxicity revealed by the use of mesodermal progenitors derived from human pluripotent stem cells. Stem Cells 33:2936–2948. 10.1002/stem.2107. [DOI] [PubMed] [Google Scholar]
- 25.Overbaugh J, Miller AD, Eiden MV. 2001. Receptors and entry cofactors for retroviruses include single and multiple transmembrane-spanning proteins as well as newly described glycophosphatidylinositol-anchored and secreted proteins. Microbiol Mol Biol Rev 65:371–389. 10.1128/MMBR.65.3.371-389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albritton LM, Tseng L, Scadden D, Cunningham JM. 1989. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57:659–666. 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- 27.Kim JW, Closs EI, Albritton LM, Cunningham JM. 1991. Transport of cationic amino acids by the mouse ecotropic retrovirus receptor. Nature 352:725–728. 10.1038/352725a0. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Kavanaugh MP, North RA, Kabat D. 1991. Cell-surface receptor for ecotropic murine retroviruses is a basic amino-acid transporter. Nature 352:729–731. 10.1038/352729a0. [DOI] [PubMed] [Google Scholar]
- 29.Kavanaugh MP, Miller DG, Zhang W, Law W, Kozak SL, Kabat D, Miller AD. 1994. Cell-surface receptors for gibbon ape leukemia virus and amphotropic murine retrovirus are inducible sodium-dependent phosphate symporters. Proc Natl Acad Sci USA 91:7071–7075. 10.1073/pnas.91.15.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olah Z, Lehel C, Anderson WB, Eiden MV, Wilson CA. 1994. The cellular receptor for gibbon ape leukemia virus is a novel high affinity sodium-dependent phosphate transporter. J Biol Chem 269:25426–25431. 10.1016/S0021-9258(18)47267-5. [DOI] [PubMed] [Google Scholar]
- 31.Giovannini D, Touhami J, Charnet P, Sitbon M, Battini JL. 2013. Inorganic phosphate export by the retrovirus receptor XPR1 in metazoans. Cell Rep 3:1866–1873. 10.1016/j.celrep.2013.05.035. [DOI] [PubMed] [Google Scholar]
- 32.Hein S, Prassolov V, Zhang Y, Ivanov D, Löhler J, Ross SR, Stocking C. 2003. Sodium-dependent myo-inositol transporter 1 is a cellular receptor for Mus cervicolor M813 murine leukemia virus. J Virol 77:5926–5932. 10.1128/jvi.77.10.5926-5932.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tipper CH, Cingoz O, Coffin JM. 2012. Mus spicilegus endogenous retrovirus HEMV uses murine sodium-dependent myo-inositol transporter 1 as a receptor. J Virol 86:6341–6344. 10.1128/JVI.00083-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tailor CS, Nouri A, Zhao Y, Takeuchi Y, Kabat D. 1999. A sodium-dependent neutral-amino-acid transporter mediates infections of feline and baboon endogenous retroviruses and simian type D retroviruses. J Virol 73:4470–4474. 10.1128/JVI.73.5.4470-4474.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rasko JEJ, Battini JL, Gottschalk RJ, Mazo I, Miller AD. 1999. The RD114/simian type D retrovirus receptor is a neutral amino acid transporter. Proc Natl Acad Sci USA 96:2129–2134. 10.1073/pnas.96.5.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quigley JG, Yang Z, Worthington MT, Phillips JD, Sabo KM, Sabath DE, Berg CL, Sassa S, Wood BL, Abkowitz JL. 2004. Identification of a human heme exporter that is essential for erythropoiesis. Cell 118:757–766. 10.1016/j.cell.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 37.Yonezawa A, Masuda S, Katsura T, Inui KI. 2008. Identification and functional characterization of a novel human and rat riboflavin transporter, RFT1. Am J Physiol Cell Physiol 295:632–641. 10.1152/ajpcell.00019.2008. [DOI] [PubMed] [Google Scholar]
- 38.Mendoza R, Anderson MM, Overbaugh J. 2006. A putative thiamine transport protein is a receptor for feline leukemia virus subgroup A. J Virol 80:3378–3385. 10.1128/JVI.80.7.3378-3385.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsang J, Ribet D, Heidmann T, Dewannieux M. 2019. Identification of the receptor used by the ecotropic mouse GLN endogenous retrovirus. J Virol 93:e01125-18. 10.1128/JVI.01125-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manel N, Kim FJ, Kinet S, Taylor N, Sitbon M, Battini JL. 2003. The ubiquitous glucose transporter GLUT-1 is a receptor for HTLV. Cell 115:449–459. 10.1016/s0092-8674(03)00881-x. [DOI] [PubMed] [Google Scholar]
- 41.Miyake A, Kawasaki J, Ngo H, Makundi I, Muto Y, Khan AH, Smith DJ, Nishigaki K. 2019. Reduced folate carrier: an entry receptor for a novel feline leukemia virus variant. J Virol 93:e00269-19. 10.1128/JVI.00269-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarma PS, Log T. 1973. Subgroup classification of feline leukemia and sarcoma viruses by viral interference and neutralization tests. Virology 54:160–169. 10.1016/0042-6822(73)90125-6. [DOI] [PubMed] [Google Scholar]
- 43.Miyake A, Watanabe S, Hiratsuka T, Ito J, Ngo MH, Makundi I, Kawasaki J, Endo Y, Tsujimoto H, Nishigaki K. 2016. Novel feline leukemia virus interference group based on the env gene. J Virol 90:4832–4837. 10.1128/JVI.03229-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hardy WD, Old LJ, Hess PW, Essex M, Cotter S. 1973. Horizontal transmission of feline leukaemia virus. Nature 244:266–269. 10.1038/244266a0. [DOI] [PubMed] [Google Scholar]
- 45.Takeuchi Y, Vile RG, Simpson G, O'Hara B, Collins MK, Weiss RA. 1992. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J Virol 66:1219–1222. 10.1128/JVI.66.2.1219-1222.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quigley JG, Burns CC, Anderson MM, Lynch ED, Sabo KM, Overbaugh J, Abkowitz JL. 2000. Erratum: cloning of the cellular receptor for feline leukemia virus subgroup C (FeLV-C), a retrovirus that induces red cell aplasia. Blood 95:1093–1099. 10.1182/blood.V95.3.1093.003k01_1093_1099. [DOI] [PubMed] [Google Scholar]
- 47.Lauring AS, Anderson MM, Overbaugh J. 2001. Specificity in receptor usage by T-cell-tropic feline leukemia viruses: implications for the in vivo tropism of immunodeficiency-inducing variants. J Virol 75:8888–8898. 10.1128/JVI.75.19.8888-8898.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawasaki J, Nishigaki K. 2018. Tracking the continuous evolutionary processes of an endogenous retrovirus of the domestic cat: ERV-DC. Viruses 10:179. 10.3390/v10040179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ngo MH, Arnal M, Sumi R, Kawasaki J, Miyake A, Grant CK, Otoi T, Fernández de Luco D, Nishigaki K. 2019. Tracking the fate of endogenous retrovirus segregation in wild and domestic cats. J Virol 93:e01324-19. 10.1128/JVI.01324-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soll SJ, Neil SJD, Bieniasz PD. 2010. Identification of a receptor for an extinct virus. Proc Natl Acad Sci USA 107:19496–19501. 10.1073/pnas.1012344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Montel-Hagen A, Kinet S, Manel N, Mongellaz C, Prohaska R, Battini JL, Delaunay J, Sitbon M, Taylor N. 2008. Erythrocyte Glut1 triggers dehydroascorbic acid uptake in mammals unable to synthesize vitamin C. Cell 132:1039–1048. 10.1016/j.cell.2008.01.042. [DOI] [PubMed] [Google Scholar]
- 52.Battini JL, Rasko JEJ, Miller AD. 1999. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proc Natl Acad Sci USA 96:1385–1390. 10.1073/pnas.96.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller AD, Rosman GJ. 1989. Improved retroviral vectors for gene transfer and expression. Biotechniques 7:980–990. [PMC free article] [PubMed] [Google Scholar]
- 54.Petris MJ, Smith K, Lee J, Thiele DJ. 2003. Copper-stimulated endocytosis and degradation of the human copper transporter, hCtr1. J Biol Chem 278:9639–9646. 10.1074/jbc.M209455200. [DOI] [PubMed] [Google Scholar]
- 55.Ito J, Baba T, Kawasaki J, Nishigaki K. 2016. Ancestral mutations acquired in refrex-1, a restriction factor against feline retroviruses, during its cooption and domestication. J Virol 90:1470–1485. 10.1128/JVI.01904-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller DG, Miller AD. 1992. Tunicamycin treatment of CHO cells abrogates multiple blocks to retrovirus infection, one of which is due to a secreted inhibitor. J Virol 66:78–84. 10.1128/JVI.66.1.78-84.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Öhrvik H, Thiele DJ. 2014. How copper traverses cellular membranes through the mammalian copper transporter 1, Ctr1. Ann N Y Acad Sci 1314:32–41. 10.1111/nyas.12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maryon EB, Molloy SA, Kaplan JH. 2007. O-linked glycosylation at threonine 27 protects the copper transporter hCTR1 from proteolytic cleavage in mammalian cells. J Biol Chem 282:20376–20387. 10.1074/jbc.M701806200. [DOI] [PubMed] [Google Scholar]
- 59.Miller DG, Miller AD. 1993. Inhibitors of retrovirus infection are secreted by several hamster cell lines and are also present in hamster sera. J Virol 67:5346–5352. 10.1128/JVI.67.9.5346-5352.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Steinberg F, Gallon M, Winfield M, Thomas EC, Bell AJ, Heesom KJ, Tavaré JM, Cullen PJ. 2013. A global analysis of SNX27-retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nat Cell Biol 15:461–471. 10.1038/ncb2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Curnock R, Cullen PJ. 2020. Mammalian copper homeostasis requires retromer-dependent recycling of the high-affinity copper transporter 1. J Cell Sci 133:jcs249201. 10.1242/jcs.249201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lassaux A, Sitbon M, Battini J-L. 2005. Residues in the murine leukemia virus capsid that differentially govern resistance to mouse Fv1 and human Ref1 restrictions. J Virol 79:6560–6564. 10.1128/JVI.79.10.6560-6564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]