Abstract
Restriction fragment length polymorphism and DNA sequence analysis discern two main types of Cryptosporidium parvum. We present a survey of length polymorphism at several microsatellite loci for type 1 and type 2 isolates. A total of 14 microsatellite loci were identified from C. parvum DNA sequences deposited in public databases. All repeats were mono-, di-, and trinucleotide repeats of A, AT, and AAT, reflecting the high AT content of the C. parvum genome. Several of these loci showed significant length polymorphism, with as many as seven alleles identified for a single locus. Differences between alleles ranged from 1 to 27 bp. Karyotype analysis using probes flanking three microsatellites localized each marker to an individual chromosomal band, suggesting that these markers are single copy. In a sample of 19 isolates for which at least three microsatellites were typed, a majority of isolates displayed a unique multilocus fingerprint. Microsatellite analysis of isolates passaged between different host species identified genotypic changes consistent with changes in parasite populations.
Genotypically, most isolates of Cryptosporidium parvum fall into two subgroups, designated type 1 and type 2 (13, 24, 29). The taxonomic status of these subgroups, as well as of other host-associated genotypes (13, 16), has not been resolved. The absence of recombinants between type 1 and type 2 C. parvum (2, 18) suggests that some genotypes may be reproductively isolated. Type 1 and type 2 are distributed worldwide but differ in host specificity. The majority of human infections excrete oocysts of type 1 (14, 28), which is not found in naturally infected animals. This observation has led to the hypothesis that C. parvum is transmitted via two routes, one anthroponotic and the other zoonotic. Humans excreting type 2 oocysts presumably contracted the infection from animal reservoirs.
Most genotypic surveys of C. parvum have been based on restriction fragment length polymorphism (RFLP) (6, 16–19, 24) or direct DNA sequence analysis of the ribosomal region and selected protein-coding regions (4, 8, 13, 15). RFLP markers are limited in resolution: typically only two restriction profiles are identified. Consequently, most of these markers do not differentiate among isolates belonging to the same genotype, even when they originate from widely separate geographic origins or different host species.
Microsatellites, or simple sequence repeats, constitute a rich source of polymorphisms and have been used extensively for high-resolution genotyping and mapping (25). In the taxonomically related malaria parasite, Plasmodium falciparum, numerous microsatellites of 1- to 5-nucleotide repeat units were described (21) and applied to the study of meiotic recombination (23) and genome mapping (22). Aiello et al. (1) analyzed six microsatellites in C. parvum. At one locus, four alleles were identified; the remaining five loci showed sequence differences between human and bovine isolates but no polymorphism within isolates from the same host species. Recently, Cacciò et al. (5) identified a polymorphic C. parvum microsatellite located within what appears to be a protein coding sequence.
Until recently little sequence information from C. parvum was available. RFLP markers used for genotypic analyses were identified in the few available sequences, which were either ribosomal genes or open reading frames. Genomic sequence surveys have generated new sequence information which is not restricted to coding regions and is therefore a potential source of microsatellite or sequence polymorphism.
We describe here the identification of 14 microsatellites in noncoding regions of the genome of C. parvum. Oocysts of known genotype were typed using these markers in order to assess the extent of polymorphism at these loci. For several isolates, unique multilocus profiles were obtained, emphasizing the utility of microsatellite analysis for genetic and epidemiological studies.
MATERIALS AND METHODS
C. parvum isolates.
The following isolates originated from calves: GCH1 (27), IOWA, OHIO (18), ICP, UCP (6), TAMU, CISD, and MD. All originate from the United States, except the MD (Moredun) isolate, which was originally isolated in Scotland. Three isolates (0676I, 2066K, and 0583K) were obtained directly from AIDS patients in the United States (30). NEMC1 was isolated from an AIDS patient also residing in the United States and was subsequently propagated in piglets (32). H39 and P12 (17) originated from HIV-negative individuals in Australia and Wales, respectively. Isolates UG350, UG502, UG408, UG489, and UG405 were obtained from Ugandan children with diarrhea. Oocysts were purified from feces by a salt flotation step, followed by either centrifugation on a 15%/30% (wt/vol) Nycodenz (Sigma, St. Louis, Mo.) step gradient (29) or immunomagnetic separation using Dynabeads anti-Cryptosporidium (Dynal, Lake Success, N.Y.) or Crypto-Scan IMS (ImmuCell, Portland, Maine). DNA extraction from oocysts was performed essentially as described previously (5). Oocysts were subjected to three cycles of freeze-thawing in STE buffer (120 mM NaCl, 25 mM Tris base, 1 mM EDTA, and 1% sodium dodecyl sulfate) followed by incubation overnight in 0.2 mg of proteinase K/liter (0.2 mg/ml) at 45°C. DNA was then extracted with phenol-chloroform and purified using GeneClean (Bio101, Vista, Calif.). All isolates were genotyped using a combination of published RFLPs (18).
Mouse infections.
Gamma interferon knockout (GKO) mice (26) were orally infected with 10 oocysts of isolate UCP, GCH1, TAMU, or MD of calf origin. Oocysts were recovered from mouse stool by immunomagnetic separation.
Identification of microsatellites.
A total of 1,150 sequences from the Stanford University-University of California—San Francisco C. parvum genome sequencing project (http://sequence-www.stanford.edu), now deposited in GenBank, were screened. Fifty-four microsatellites identified in this manner had enough flanking sequences to design primers. Twenty-one primer sets encompassing an equal number of noncoding microsatellite sequences were synthesized. Fourteen of 21 primer sets amplified microsatellite loci.
PCR amplification of microsatellites and gel analysis.
PCR amplifications were performed in 25-μl volumes containing 1× PCR buffer (4 mM Tris-HCl [pH 8.3], 20 mM KCl, 0.004% gelatin), 2.0 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 10 pmol of forward PCR primer, 2 pmol of 32P-labeled reverse primer, 0.5 U of Taq polymerase (Promega, Madison, Wis.), and an estimated 10 ng of template DNA. The reaction mixtures were denatured at 94°C for 3 min and cycled 36 times at 94°C for 15 s, at the respective annealing temperature (Table 1) for 25 s and at 72°C for 40 s in an MJ Research (Watertown, Mass.) thermal cycler. The reaction was terminated with a 7-min incubation at 72°C. An alternative labeling method consisted of using a 1-μl portion of unlabeled PCR mixture as a template for a second 15-cycle amplification in the presence of 2 pmol of 5′ 32P-labeled reverse PCR primer. The primers were 5′ end labeled with polynucleotide kinase in the presence of [γ-32P]ATP.
TABLE 1.
Microsatellites included in this study
| Accession no. | Locus | Repeat | Primer F | Primer R | Fragc | Tmd | Allelesb
|
||
|---|---|---|---|---|---|---|---|---|---|
| Type 1 | Type 1 and 2 | Type 2 | |||||||
| AQ449854 | 5B12 | TA × 19 | TTCCCCATATTACTCTATTTG | AGGAGGAGGAGAAAAATAG | 152 | 55 | + | + | + |
| AQ449550 | 1G09 | T × 18 | CATTCGGATGTGTGGAAAG | TAGAAGGTTTGTATACTTATTG | 110 | 52 | + | + | + |
| AQ449537 | 1F07 | TA × 15 | TAGCTTACCAAAGCTTCCTG | TCTTTGATTGAATGTTGAGATC | 197 | 53 | + | + | + |
| AQ988782 | 7E1C | TA × 16 | TCCATCCAAAAGGCATGAG | TTTAGGATGAATATTTGGTGG | 77 | 52 | + | + | + |
| AQ450211 | 9B10 | TTA × 7 | TGGGGAAATGCTCATTATAG | ATAAAACAATAAAAGCGGCAG | 99 | 55 | + | + | − |
| AQ449632 | 2G04 | TA × 16 | TGCGGGATAAATCGCATATT | CACTTCCTTCATTATTACGC | 134 | 54 | + | + | − |
| AQ449938 | 6B03 | TA × 13 | TTGCATGAATCGATATTTCAAG | TCAATGAATATTATAGTTGTAG | 149 | 52 | + | + | − |
| AQ449877 | 5D11 | T × 20 | AAATTCCACATTCAAAATTAAG | TCAAATCACTCAGTTATTTAAC | 128 | 52 | + | + | − |
| AQ855193 | CP193 | TAA × 8 | AGAGAGTTTAAGATTGGACGG | TCGATACCTTTAATAGGTACC | 104 | 52 | − | + | − |
| AQ450484a | 12C07 | TA × 19 | TGCATGTATTACCCCTATAAC | CGGGTAGCACAATTTATGA | 197 | 55 | − | − | + |
| A × 17 | |||||||||
| AQ449797 | 4E 12 | TA × 13 | TGCATGAATCGATATTTCAAG | ATCAATGAATATTATAGTTGTAG | 141 | 55 | − | − | + |
| AQ855273 | CP273 | TA × 21 | ATAATTCCTCTTCATTCTTCC | TTAGTAGCTAATAAACTCGTG | 156 | 52 | − | − | + |
| AQ450221 | 9C09 | TA × 14 | TTTCTCTATTTTCTTACCAATC | TATATCAGTGTCTATTAACTAC | 137 | 55 | − | − | − |
| AQ449992 | 6F11 | A × 19 | ATCCGTTTACGCACGCTT | TAGCCACCGTTTAAAGGC | 80 | 55 | − | − | − |
Labeled PCR products were mixed 1:1 with loading buffer (10 mM NaOH, 95% formamide, 0.05% bromophenol blue, and 0.05% xylene cyanol), and 2-μl samples were fractionated on 8% polyacrylamide urea sequencing gels electrophoresed at 2,000 V and 36 W in 1× Tris-borate-EDTA and 8 M urea for 2 to 3 h. The gels were dried for 30 min and exposed to Kodak XAR-5 film.
Karyotype analysis.
The chromosomal locations of microsatellites 1G09, 1F07, and 5B12 were determined by Southern blotting of contour-clamped homogeneous electrical field electrophoresis (CHEF) gels of KSU-1 isolate DNA followed by Southern blotting as described previously (10). The probes were 105-, 144-, and 317-bp PCR fragments amplified from regions immediately downstream (1G09) or upstream (1F07 and 5B12) of the repeat. The actual repeat was excluded from the probe.
Data analysis.
Isolates which were typed with a minimum of three microsatellite loci were included. Pairwise genetic distances between isolates were calculated using Jaccard's index [D = 1 − C/(2N − C), where C is the number of loci with a shared allele and N is the total number of loci included in the comparison] (9).
RESULTS
Searches in GenBank for simple repeats identified 54 C. parvum sequences. Of these, 20 contained (TA)10, 11 contained (TAA)6, and 23 contained T15. Searches for (TG)10, (TC)10, (TTG)6, (TGG)6, (TCC)6, (GGCC)6, (TTTA)6, (TTTG)6, or (TTTTA)5 did not identify any matches. This distribution is consistent with recently published surveys of the C. parvum genome (11, 20, 32). From the original selection of microsatellites, 40 sequences were excluded because they were located within coding regions, because of insufficient flanking sequence, or because the flanking sequences were unsuitable for primer design, usually due to low G+C content. Table 1 lists 14 C. parvum microsatellites selected for this study. In the table, microsatellites are listed approximately in order of decreasing variability, starting with those which discriminate within type 1 and within type 2, as well as between the genotypes. Next, loci differentiating among genotype 1 isolates but not among genotype 2 isolates are listed, followed by less polymorphic loci and finally by two monomorphic loci.
Southern hybridization of CHEF-separated oocyst DNA using probes for 1G09, 1F07, and 5B12 was performed to assess the possible existence of multiple copies of these markers in the genome of C. parvum. The three probes hybridized to individual chromosome bands. 1G09 and 1F07 hybridized to a band corresponding to chromosomes 3, 4, and 5 (3, 10), whereas 5B12 hybridized to chromosome band 2.
The number of repeat units in the microsatellites included in this study ranged from 7 (9B10) to 21 (CP273). There was no apparent correlation between the number of repeats and the degree of polymorphism. The average number of repeats in the most polymorphic group, i.e., those loci differentiating both within type 1 and type 2 isolates (5B12, 1G09, 1F07, and 7E1C), was 17 compared to 16.5 for the two monomorphic loci, 9C09 and 6F11.
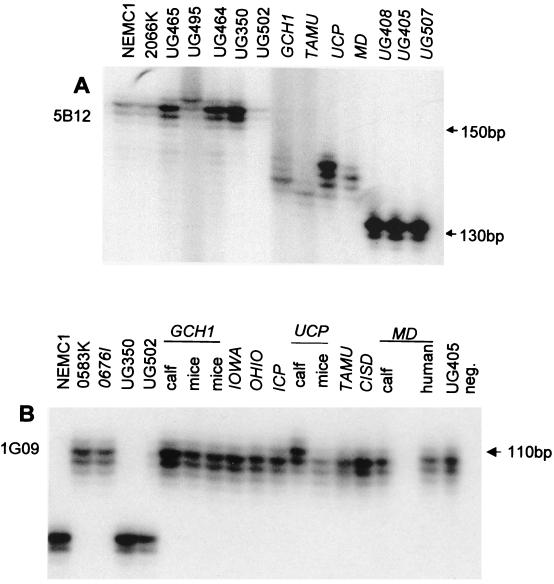
Figure 1 illustrates the microsatellite polymorphism at two loci, 5B12 and 1G09. Figure 1A illustrates the occurrence of seven alleles at the 5B12 locus, the most polymorphic microsatellite encountered in this study. Four alleles were observed at the 1G09 locus, of which three are shown in Fig. 1B: a unique allele for UCP originating from a calf, one allele for the remaining type 2 isolates, and one for the type 1 isolates. The presence of different alleles in UCP oocysts originating from different hosts is discussed further below.
FIG. 1.
Electrophoretic analysis of 5B12 and 1G09 microsatellites. DNA was amplified from the isolates shown in the presence of 5′ 32P-labeled reverse primer, resolved on a sequencing gel, and visualized by autoradiography. (A) 5B12 locus; (B) 1G09 locus. Genotype 2 isolates are italicized. The host origins of GCH1, MD, and UCP oocysts are indicated in panel B. The positions of selected markers from a 10-bp ladder are shown at the right. neg., negative PCR control.
To confirm the electrophoretic profiles of three microsatellite loci, cloned PCR products from the 1F07, 5B12, and 12C07 loci originating from five, four, and two isolates, respectively, were sequenced. As expected, the alignment of these sequences revealed differences in the number of repeats (Fig. 2). Alignment of five sequences from the 1F07 locus showed four alleles differing in the number of AT repeats. The 5B12 sequences obtained from three genotype 2 isolates (GCH1, IOWA, and UG405) and one genotype 1 isolate (UG464) also revealed different numbers of AT repeats. The sequences are consistent with the electrophoretic profiles shown in Fig. 1 in showing the largest deletion in UG405 and the longest repeat sequence in UG464. However, a discrepancy between the electrophoretic profile of UG464 (Fig. 1) and its sequence (Fig. 2) was noticed. The 5B12 amplicon size on the gel was 10 to 15 bp larger than the sequenced insert. A possible explanation for this difference is that each sequence was obtained from a randomly selected PCR clone, which may not be identical to the predominant PCR band. The sequence comparison of four 12C07 alleles, including two from different genome surveys of isolate IOWA (accession numbers AQ083613 and AQ450484) and one each from GCH1 and MD identified a 27-bp deletion in an AAT repeat in GCH1 and in one sequence from IOWA. The sequences are consistent with the electrophoretic profiles obtained for these isolates (not shown). Two additional short deletions were identified in two poly(A) and poly(T) stretches located downstream of the 12C07 AAT repeat. Accession numbers AQ083613 and AQ450484 and the gel analysis of 12C07 suggest that isolate IOWA is polymorphic at this locus. Interestingly, several other isolates amplified two 12C07 bands with the same 27-bp size difference, but no alleles with intermediate mobility were identified.
FIG. 2.
Sequence analysis of cloned PCR products originating from 1F07 and 5B12 loci. One clone was picked randomly for each isolate and sequenced in both orientations. The locations of forward (F) and reverse (R) primers are shown with arrows. Genotype 2 isolates are italicized. Periods (.), deletions; tildas (∼), not sequenced.
Table 2 summarizes the microsatellite analysis of 19 C. parvum isolates with up to 12 polymorphic loci. The two monomorphic loci, 9C09 and 6F11, are excluded. Only CP193 did not reveal any polymorphism in addition to the type 1-type 2 subdivision, although only six isolates were typed with this marker. Four loci differentiated among type 1 but not among type 2 isolates, whereas three loci differentiated among type 2 but not among type 1 isolates. Using the data from Table 2, a distance matrix was constructed from all pairwise comparisons between isolates. From this matrix, the mean genetic heterogeneity within each type and between type 1 and type 2 was calculated (Table 3). Consistent with previous RFLP analyses, the mean distances between pairs of isolates within the same type were smaller than the distances between isolates belonging to different types. Due to the presence of unique alleles in samples originating from Uganda, the mean genetic distances were smaller when these isolates were excluded. This analysis also suggests that the degrees of heterogeneity within the types are similar.
TABLE 2.
Summary of microsatellite polymorphismsa
| Microsatellite | Alleleb
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1
|
?c
|
2
|
|||||||||||||||||
| NEMC1 | 2066K | H39 | UG350 | UG502 | P12 | 0583K | 0676I | UG405 | UG408 | UG489 | GCH1 | IOWA | OHIO | ICP | UCP | TAMU | CISD | MD | |
| 5B12d | 2 | 2 | 2 | 2 | 2 | 5 | 5 | 5 | 7 | 7 | 7 | 5 | 6 | 5 | 5 | 5 | 6 | 5 | 4 |
| 1G09 | 3 | 3 | 3 | 3 | 3 | 4 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 |
| 1F07 | 2 | 2 | 2 | 1 | 3 | 2 | 4 | 5 | 7 | 7 | 7 | 5 | 4 | 4 | 4 | 5 | 4 | 4 | 4 |
| 7 E1C | 2 | 3 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | ||||||||
| 9B10 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |||||||
| 2G04 | 1 | 1 | 3 | 4 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |||||||
| 6B03 | 2 | 1 | 3 | 3 | 3 | 3 | 3 | ||||||||||||
| 5D11 | 1 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | ||||||
| Cp193 | 2 | 1 | 1 | 1 | 1 | 1 | |||||||||||||
| 12C07e | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 4 | 1 | |||||||
| 4 E12 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |||||||
| Cp273 | 1 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | |||||||
TABLE 3.
Arithmetic mean of pairwise genetic distances within C. parvum type 1, within type 2, and between type 1 and type 2a
| Comparison | All isolates
|
Uganda excluded
|
||
|---|---|---|---|---|
| nb | Mean distance | nb | Mean distance | |
| Within type 1 | 15 | 0.50 | 7 | 0.43 |
| Within type 2 | 65 | 0.61 | 36 | 0.44 |
| Between 1 and 2 | 72 | 0.97 | 36 | 0.93 |
Genetic distances were derived from microsatellite data (Table 2) using Jaccard's index as described in Materials and Methods.
Number of pairwise comparisons.
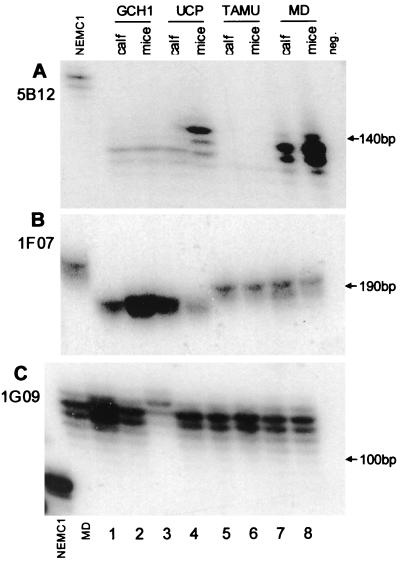
Four isolates of calf origin were passaged to GKO mice with the aim of studying the stability of microsatellite markers following transmission between different host species. Of the isolates tested, only isolate UCP showed a change in profile (Fig. 3). Two loci, 5B12 and 1G09, displayed a profile for the calf-derived oocysts different from that for the oocysts recovered from mice (Fig. 3, lanes 3 and 4). A third locus, 1F07, showed no differences between UCP oocysts obtained from calves and mice.
FIG. 3.
Genotypic profile of four genotype 2 isolates passaged from calves to GKO mice. Oocysts originating from calves (lanes 1, 3, 5, and 7) and GKO mice (lanes 2, 4, 6, and 8) were genotyped in parallel. Notice the change in the 5B12 and 1G09 profiles in UCP recovered from mice. NEMC1 samples and one MD sample in 1G09 were included as PCR controls. neg., negative PCR control.
DISCUSSION
Because of its sensitivity and simplicity, PCR-based RFLP typing has been extensively applied to the characterization of C. parvum isolates from clinical and epidemiological studies. The limited resolution offered by this method emphasizes the need for techniques capable of discriminating among different isolates, even if they belong to the same type. Microsatellite analysis provides improved resolution due to the extensive length polymorphism found at simple repeat loci. The microsatellites we used were chosen because they are located outside coding regions and are therefore more likely to mutate than protein-coding sequences. In contrast to random-amplification techniques, such as random-amplified polymorphic DNA (12) and amplified fragment length polymorphism analyses, microsatellite fingerprints are not affected by exogenous DNA and may therefore be more suitable for analyzing environmental samples.
Although shared alleles in type 1 and type 2 isolates were identified at several loci, genetic fingerprints combining multiple microsatellites and numerical analysis of these data support the type 1-type 2 classification obtained previously with RFLP and sequence analysis. This observation is consistent with our hypothesis that C. parvum includes at least two reproductively separate genotypes.
The availability of several isolates originating from Uganda enabled us to identify for the first time a correlation between geographic origin and genotype. In fact, the three type 2 isolates originating from that country shared at the 5B12 and 1F07 loci unique alleles not encountered in other isolates.
As observed with an RFLP marker located within the poly(T)-GP900 gene (7), transmitting C. parvum populations between different host species may affect their genetic makeup. Observations with four type 2 isolates reported here confirm this observation and suggest that isolates may consist of heterogeneous parasite populations, as previously documented by sequence analysis of the β-tubulin locus (31). It is our assumption that the observed changes reflect a shift in population makeup following transmission to a different host species. It is unclear why this phenomenon was only observed in isolate UCP and not in GCH1, TAMU, and MD. However, based on the appearance of a unique allele in UCP oocysts recovered from mice (Fig. 1 and 3), we can exclude the possibility that laboratory contaminations caused the observed changes in microsatellite profile. Under the assumption that C. parvum populations undergo sexual reproduction, the observed changes in two of three loci examined following calf-to-mouse passage suggests a link between these loci and genes conferring a selective advantage in mice. Since the two loci displaying an allele replacement following host change (5B12 and 1G09) reside on different chromosome bands, we assume that such a selective mechanism affects multiple loci throughout the genome.
An interesting observation related to the heterogeneous structure of C. parvum isolates is the identification in GenBank of two sequences originating from locus 12C07 of isolate IOWA which differ by 27 bp in an AAT repeat. The possible frequent occurrence of heterogeneous parasite populations implies that direct gel analysis of microsatellite polymorphisms may be superior to sequencing of cloned PCR products. The former method generates a fingerprint of the entire population, whereas cloned PCR products are selected from a potentially heterogeneous collection of alleles. An illustration of the difference between these methods is provided by our data on the 5B12 locus, where the predominant allele from isolate UG464 was estimated at >150 bp (Fig. 1) but was not encountered among four plasmids we sequenced. This could indicate that longer repeat sequences are inefficiently replicated in the bacterial host and are therefore less likely to be sequenced.
In summary, the availability of polymorphic microsatellite markers allows the differentiation of isolates with identical RFLP profiles, a significant improvement over the current RFLP method. Genotypic characterization using such markers could find application in outbreak investigations and in epidemiological studies. For instance, microsatellite fingerprints obtained from oocysts recovered from finished or surface water could be compared with those from potential sources of oocysts within the watershed. Identity between fingerprints would point to a putative source of contaminating oocysts, whereas different fingerprints would exclude a particular source. However, surveys based on these markers need to consider the potential for microsatellite fingerprints to be affected by the host species. A practical advantage of the microsatellite polymorphisms is the reliance on conventional PCR methods and the potential for automation when fluorescently labeled PCR products are used instead of radioactively labeled primers. The use of multiple markers will generate multilocus fingerprints capable of discriminating between individual isolates.
ACKNOWLEDGMENTS
This study was supported by USDA NRI grant 9800919.
Our thanks to Sylvie LeBlancq (Columbia University School of Public Health) for performing CHEF analyses, to Mike Buckholt, Julia Dilo, Kim Deary, and Michelle Beauchemin for logistical help and technical assistance, and to Cynthia Chappell (University of Texas), Joe Crabb (ImmuCell), and Una Morgan (Murdoch University) for providing C. parvum samples.
REFERENCES
- 1.Aiello A E, Xiao L, Limor J R, Liu C, Abrahamsen M S, Lal A A. Microsatellite analysis of the human and bovine genotypes of Cryptosporidium parvum. J Eukaryot Microbiol. 1999;46:46S–47S. [PubMed] [Google Scholar]
- 2.Awad-El-Kariem F M, Robinson H A, Petry F, McDonald V, Evans D, Casemore D. Differentiation between human and animal isolates of Cryptosporidium parvum using molecular and biological markers. Parasitol Res. 1998;84:297–301. doi: 10.1007/s004360050399. [DOI] [PubMed] [Google Scholar]
- 3.Blunt D S, Khramtsov N V, Upton S J, Montelone B A. Molecular karyotype analysis of Cryptosporidium parvum: evidence for eight chromosomes and a low-molecular-size molecule. Clin Diagn Lab Immunol. 1997;4:11–13. doi: 10.1128/cdli.4.1.11-13.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cacciò S, Homan W, van Dijk K, Pozio E. Genetic polymorphism at the β-tubulin locus among human and animal isolates of Cryptosporidium parvum. FEMS Microbiol Lett. 1999;170:173–179. doi: 10.1111/j.1574-6968.1999.tb13371.x. [DOI] [PubMed] [Google Scholar]
- 5.Cacciò S, Homan W, Camilli R, Traldi G, Koterbeek T, Pozio E. A microsatellite marker reveals population heterogeneity within human and animal genotypes of Cryptosporidium parvum. Parasitology. 2000;120:237–244. doi: 10.1017/s0031182099005508. [DOI] [PubMed] [Google Scholar]
- 6.Carraway M, Tzipori S, Widmer G. Identification of genetic heterogeneity in the Cryptosporidium parvum ribosomal repeat. Appl Environ Microbiol. 1996;62:712–716. doi: 10.1128/aem.62.2.712-716.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carraway M, Tzipori S, Widmer G. New restriction fragment length polymorphism marker in Cryptosporidium parvum identifies mixed parasite populations and genotypic instability in response to host change. Infect Immun. 1997;65:3958–3960. doi: 10.1128/iai.65.9.3958-3960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiGiovanni G D, Hashemi F H, Shaw N J, Abrams F A, LeChevallier M W, Abbaszadegan M. Detection of Cryptosporidium parvum oocysts in surface and filter backwash water samples by immunomagnetic separation and integrated cell culture-PCR. Appl Environ Microbiol. 1999;65:3427–3432. doi: 10.1128/aem.65.8.3427-3432.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaccard P. Nouvelles recherches sur la distribution florale. Bull Soc Vaudoise Sci Nat. 1908;44:223–270. [Google Scholar]
- 10.LeBlancq S M, Khramtsov N, Zamani F, Upton S J, Wu T W. Ribosomal RNA gene organization in Cryptosporidium parvum. Mol Biochem Parasitol. 1997;90:463–478. doi: 10.1016/s0166-6851(97)00181-3. [DOI] [PubMed] [Google Scholar]
- 11.Liu C, Vigdorovich V, Kapur V, Abrahamsen M S. A random survey of the Cryptosporidium parvum genome. Infect Immun. 1999;67:3960–3969. doi: 10.1128/iai.67.8.3960-3969.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan U M, Constantine C C, O'Donoghue P, Meloni B P, O'Brien P A, Thompson R C A. Molecular characterization of Cryptosporidium isolates from humans and other animals using random amplified polymorphic DNA analysis. Am J Trop Med Hyg. 1995;52:559–564. doi: 10.4269/ajtmh.1995.52.559. [DOI] [PubMed] [Google Scholar]
- 13.Morgan U M, Deplazes P, Forbes D A, Spano F, Hertzberg H, Sargent K D, Elliot A, Thompson R C A. Sequence and PCR-RFLP analysis of the internal transcribed spacers of the rDNA repeat unit in isolates of Cryptosporidium from different hosts. Parasitology. 1999;118:49–58. doi: 10.1017/s0031182098003412. [DOI] [PubMed] [Google Scholar]
- 14.Patel S, Pedraza-Diaz S, McLauchlin J, Casemore D P. Molecular characterization of Cryptosporidium parvum from two large suspected waterborne outbreaks. Comm Dis Public Health. 1998;1:231–233. [PubMed] [Google Scholar]
- 15.Peng M M, Xiao L, Freeman A R, Arrowood M J, Escalante A A, Weltman A C, Ong C S L, MacKenzie W R, Lal A A, Beard C B. Genetic polymorphism among Cryptosporidium parvum isolates: evidence for two distinct human transmission cycles. Emerg Infect Dis. 1997;3:567–573. doi: 10.3201/eid0304.970423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pieniazek N J, Bornay-Llinares F J, Slemenda S B, da Silva A J, Moura I N S, Arrowood M J, Ditrich O, Adiss D G. New Cryptosporidium genotypes in HIV-infected persons. Emerg Infect Dis. 1999;5:444–449. doi: 10.3201/eid0503.990318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spano F, Putignani L, McLauchlin J, Casemore D P, Crisanti A. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol Lett. 1997;150:209–217. doi: 10.1016/s0378-1097(97)00115-8. [DOI] [PubMed] [Google Scholar]
- 18.Spano F, Putignani L, Crisanti A, Sallicandro A, Morgan U M, Le Blancq S M, Tchack L, Tzipori S, Widmer G. A multilocus genotypic analysis of Cryptosporidium parvum from different hosts and geographical origins. J Clin Microbiol. 1998;36:3255–3259. doi: 10.1128/jcm.36.11.3255-3259.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spano F, Putignani L, Guida S, Crisanti A. Cryptosporidium parvum: PCR-RFLP analysis of the TRAP-C1 (thrombospondin-related adhesive protein of Cryptosporidium-1) gene discriminates between two alleles differentially associated with parasite isolates of animal and human origin. Exp Parasitol. 1998;90:195–198. doi: 10.1006/expr.1998.4324. [DOI] [PubMed] [Google Scholar]
- 20.Strong W B, Nelson R G. Preliminary profile of the Cryptosporidium parvum genome: an expressed sequence tag and genome survey sequence analysis. Mol Biochem Parasitol. 2000;107:1–32. doi: 10.1016/s0166-6851(99)00225-x. [DOI] [PubMed] [Google Scholar]
- 21.Su X, Wellems T. Towards a high resolution Plasmodium falciparum linkage map: polymorphic markers from hundreds of simple sequence repeats. Genomics. 1996;33:430–444. doi: 10.1006/geno.1996.0218. [DOI] [PubMed] [Google Scholar]
- 22.Su X, Kirkman L A, Fujioka H, Wellems T E. Complex polymorphisms in an ∼330 kDa protein are linked to chloroquine-resistant P. falciparum in Southeast Asia and Africa. Cell. 1997;91:593–603. doi: 10.1016/s0092-8674(00)80447-x. [DOI] [PubMed] [Google Scholar]
- 23.Su X, Ferdig M T, Huang Y, Huynh C Q, Liu A, You J, Wotton J C, Wellems T E. A genetic map and recombination parameters of the human malaria parasite Plasmodium falciparum. Science. 1999;286:1351–1353. doi: 10.1126/science.286.5443.1351. [DOI] [PubMed] [Google Scholar]
- 24.Sulaiman I M, Xiao L, Yang C, Escalante L, Moore A, Beard C B, Arrowood M J, Lal A A. Differentiating human from animal isolates of Cryptosporidium parvum. Emerg Infect Dis. 1998;4:681–685. doi: 10.3201/eid0404.980424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tautz D, Schloeterer C. Simple sequences. Curr Opin Genet Dev. 1994;4:832–837. doi: 10.1016/0959-437x(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 26.Theodos C, Sullivan K, Griffiths J, Tzipori S. The profile of healing and non-healing Cryptosporidium parvum infection in mice with functional B and T lymphocytes: the extent of gamma interferon modulation determines infection outcome. Infect Immun. 1997;65:4761–4769. doi: 10.1128/iai.65.11.4761-4769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tzipori S, Rand W, Griffiths J, Widmer G, Crabb J. Evaluation of an animal model system for cryptosporidiosis: therapeutic efficacy of paromomycin and hyperimmune bovine colostrum-immunoglobulin. Clin Diagn Lab Immunol. 1994;1:450–463. doi: 10.1128/cdli.1.4.450-463.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tzipori, S., and G. Widmer. In F. Petri (ed.), The biology of Cryptosporidium, in press. Cryptosporidosis and microsporidiosis. Karger, Basel, Switzerland.
- 29.Widmer G. Genetic heterogeneity and PCR detection of Cryptosporidium parvum. Adv Parasitol. 1998;40:224–241. doi: 10.1016/s0065-308x(08)60122-0. [DOI] [PubMed] [Google Scholar]
- 30.Widmer G, Tzipori S, Fichtenbaum C J, Griffiths J K. Genotypic and phenotypic characterization of Cryptosporidium parvum isolates from people with AIDS. J Infect Dis. 1998;178:834–840. doi: 10.1086/515373. [DOI] [PubMed] [Google Scholar]
- 31.Widmer G, Tchack L, Chappell C L, Tzipori S. Sequence polymorphism in the β-tubulin gene reveals heterogeneous and variable population structure in Cryptosporidium parvum. Appl Environ Microbiol. 1998;64:4477–4481. doi: 10.1128/aem.64.11.4477-4481.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Widmer G, Akiyoshi D, Buckholt M A, Feng X, Rich S M, Deary K M, Bowman C A, Xu P, Wang Y, Wang X, Buck G A, Tzipori S. Animal propagation and genomic survey of a genotype 1 isolate of Cryptosporidium parvum. Mol Biochem Parasitol. 2000;108:187–197. doi: 10.1016/s0166-6851(00)00211-5. [DOI] [PubMed] [Google Scholar]