Abstract
Two constructs derived from the α-amylase gene (amyA) of Lactobacillus amylovorus were expressed in Lactobacillus plantarum, and their expression products were purified, characterized, and compared. These products correspond to the complete (AmyA) and truncated (AmyAΔ) forms of α-amylase; AmyAΔ lacks the 66-kDa carboxyl-terminal direct-repeating-unit region. AmyA and AmyAΔ exhibit similar amylase activities towards a range of soluble substrates (amylose, amylopectin and α-cyclodextrin, and soluble starch). The specific activities of the enzymes towards soluble starch are similar, but the KM and Vmax values of AmyAΔ were slightly higher than those of AmyA, whereas the thermal stability of AmyAΔ was lower than that of AmyA. In contrast to AmyA, AmyAΔ is unable to bind to β-cyclodextrin and is only weakly active towards glycogen. More striking is the fact that AmyAΔ cannot bind or hydrolyze raw starch, demonstrating that the carboxyl-terminal repeating-unit domain of AmyA is required for raw-starch binding activity.
Raw-starch-degrading amylases are commercially important enzymes in the beverage, food, and textile industries (13). Starch is commonly used as a renewable raw material for the industrial production of lactic acid (6). The amylolytic lactic acid bacteria are attractive alternatives for this type of process, since they can directly produce lactic acid from starch (37, 38). Various Lactobacillus strains exhibit amylase activity: Lactobacillus cellobiosus (32), Lactobacillus amylovorus (26), Lactobacillus amylophilus (27), Lactobacillus plantarum (10, 30), Lactobacillus manihotivorans (24), and Lactobacillus amylolyticus (2). The α-amylase genes of L. plantarum A6 (12), L. amylovorus (12), and L. manihotivorans (J. Morlon-Guyot, I. Jacobé, de Haut, J. Boniface, and J. P. Guyot, Abstr. Aff. VI4, 9ème Colloque Club Bact. Lact., 1998) have been sequenced. These lactobacillus amyA genes have more than 98% nucleotide sequence identity and a similar level of identity for the deduced amino acid sequence (Morlon-Guyot et al., Abstr. Aff. VI4, 9ème Colloque Club Bact. Lact., 1998). With such high homologies, it is assumed that the three enzymes possess similar structure-function relationships. It has been demonstrated that the first 410 amino acids of L. amylovorus are sufficient to transfer an amylolytic activity to a nonamylolytic strain of L. plantarum (17), suggesting that the N-terminal parts of these enzymes contains the active site. The 3′-terminal halves of the three genes exhibit a special tandem repeated-unit structure (12; Morlon-Guyot et al., Abstr. Aff., VI4, 9ème Colloque Club Bact. Lact., 1998) whose role is still unknown. Giraud and Cuny (12) suggested that the region of repeated sequences might be responsible for raw-starch binding. A recent study showed that some proteolytic fragments of the L. plantarum A6 amylase lose the raw-starch-digesting ability (9), but their positions in the primary structure were not determined. In the present work, the functional role of the carboxyl-terminal repeat sequences of the L. amylovorus amylase (AmyA) is investigated using clones encoding either the entire AmyA or the truncated amylase AmyAΔ, which has the repeated sequences deleted (17). Comparison of the enzymatic properties of the two amylases reveals that the C-terminal part of the enzyme containing the repeated sequences is involved in the ability of this enzyme to bind to raw starch.
MATERIALS AND METHODS
Material.
Soluble potato starch, calcium carbonate, dinitrosalicylic acid (DNS), and glucose were from Prolabo, Paris, France. Raw corn starch, pullulan maltose, amylose (type III from potato), amylopectin (from corn), glycogen (from mussels), and α-, β-, and γ-cyclodextrins were from Sigma Chemical Co. (St. Louis, Mo.). Sepharose was from Pharmacia Biotech (Uppsala, Sweden).
Bacterial strains, plasmids, and culture conditions.
L. plantarum LMG9211 was from the Laboratorium voor Microbiologie Universiteit Gent (Ghent, Belgium) culture collection. Plasmids pLPCR2-3 carrying the 5.5-kb amyA gene from L. amylovorus and pLPCR2-3ΔB/X carrying the 4.0-kb truncated amyA gene were constructed as described previously (17). MRS medium (7) was used to grow the lactobacillus strains at 30°C. This medium was first made without glucose, and, depending on the experiments, the carbon source added was either 1% (wt/vol) glucose, maltose, or soluble or raw starch, with or without 4% calcium carbonate. For enzyme preparation, cultures were grown in a 2-liter bioreactor (Biolafitte, Poissy, France) at 30°C and agitated at 300 rpm. The pH was maintained at 6.0 by automatic addition of 5 N NaOH.
Transformation of L. plantarum and selection of transformants.
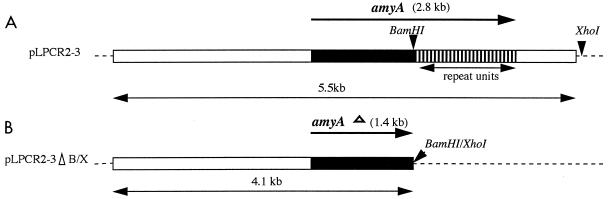
Plasmids pLPCR2-3 and pLPCR2-3ΔB/X (Fig. 1) were introduced into L. plantarum LMG9211 by electroporation, as described previously (32). The transformants were selected by adding erythromycin (2.5 μg/ml) to the solid culture medium. The production of α-amylase by the recombinant clones was detected by visualization of halos due to starch degradation on 1% starch-containing plates after staining them with iodine vapors.
FIG. 1.
Restriction map of pLPCR2-3 and pLPCR2-3ΔB/X plasmids. (A) pLPCR2-3 plasmid, carrying the 2.8-kb amyA gene of L. amylovorus, ligated at the BglII site of the pLPCR2 plasmid (dashed line). (B) pLPCR2-3ΔB/X plasmid derived from pLPCR2-3 after deletion of the 2-kb BamHI-XhoI fragment.
Enzyme purification.
Following an 18-h batch culture, the fermentation broth was collected and centrifuged at 9,000 × g for 15 min at 4°C.
The intact L. amylovorus α-amylase was purified from L. plantarum(pLPCR2-3) supernatant by affinity chromatography as described previously (34), using a β-cyclodextrin–epoxy-activated Sepharose 6B column (16 by 35 mm). After being washed with 0.1 M citrate-phosphate buffer, pH 5.5, the bound amylase was eluted with β-cyclodextrin (8 mM) in the same buffer at a flow rate of 0.5 ml · min−1 for 400 min.
The truncated amylase from L. amylovorus was purified from L. plantarum(pLPCR2-3ΔB/X) supernatant. (NH4)2SO4 was added to the clarified culture to 70% saturation, and the precipitate was collected by centrifugation (15,000 × g for 15 min at 4°C), resuspended in 50 ml of 0.05 M citrate-phosphate buffer (pH 5.5), concentrated by ultrafiltration (10,000-Da cutoff [Amicon; Millipore]) and loaded onto a Protein Pak 200SW gel exclusion column (8 by 300 mm; Waters). The enzyme-containing fractions were pooled and concentrated (Microcon centrifugal device YM-10 [Amicon; Millipore]).
Electrophoresis analyses.
Sodium dodecyl sulfate–7.5% polyacrylamide gel electrophoresis (SDS–7.5% PAGE) was performed according to the method of Laemmli (19). Proteins were visualized by Coomassie blue staining as described by Blakesly and Boezi (1). Activity staining was performed in the gel after renaturation of the enzymes, using the method described by Lacks and Springhorn (18).
Enzyme activity assay.
Routine α-amylase activity assay was used to determine the activity of α-amylase in the fractions collected throughout the purification. The activity of α-amylase towards soluble potato starch was determined by measurement of its iodine-complexing ability, using the protocol described by Giraud et al. (11) with minor modifications: enzyme incubation was performed at pH 5.0 and 63°C. One enzyme unit is defined as the amount of enzyme that permits the hydrolysis of 10 mg of soluble starch over 30 min.
For enzymatic characterization of the recombinant enzymes, amylase activity was determined by measuring the increase of reducing sugar formed by the enzymatic hydrolysis of 1% soluble potato starch or other substrates (α-, β-, and γ-cyclodextrins, amylopectin, amylose, glycogen, maltose, pullulan, and raw corn starch). The reducing sugars were quantified by the DNS method using glucose as a standard (22) at pH 5.0 and 63°C. One unit of amylase activity was defined as the amount of enzyme which liberated 1 μmol of reducing sugars per min. The protein concentration was estimated by the method of Bradford (4), using bovine serum albumin as a standard (Bio-Rad [Richmond, Calif.] protein assay).
The apparent Michaelis constant (KM) of each enzyme was determined at 10 different concentrations of soluble starch (from 0 to 20 g/liter) at their temperature and pH optima. Kinetic parameters were calculated by fitting initial velocities and substrate concentrations to the Michaelis-Menten equation using the quasi-Newton minimization method (Microsoft Excel, version 5).
Effects of pH and temperature.
The amylase activity was determined at various pH values (ranging from 3 to 7) and at various temperatures (from 45 to 75°C) in 0.1 M citrate-phosphate buffer. A second-order factorial design was used (3, 25) in order to study the combined effect of pH and temperature on amylase activity.
To determine the thermostabilities of the recombinant enzymes, samples were preincubated in 0.1 M citrate-phosphate buffer at optimum pH and at several temperatures (from 35 to 75°C) for various times (from 0 to 2 h). The samples were then chilled on ice for at least 30 min, and the residual activity was determined at pH 5.0 and 63°C, using the DNS method as described above.
Adsorption of α-amylase on raw starch.
Various amounts (from 0 to 140 μg/ml) of recombinant enzyme (AmyA or AmyAΔ) were added to a raw-corn-starch suspension (10 mg/ml) in 0.1 M citrate-phosphate buffer, pH 5, to a final volume of 60 μl. The mixture was incubated at 4°C for 30 min under gentle shaking (6 rpm) and centrifuged at 13,000 × g for 5 min. The protein concentration in the supernatant was assayed, and the amount of adsorbed enzyme was calculated by subtraction (35). Adsorption constants (Kad [in milliliters per milligram of starch]) were calculated from the slopes obtained from the linear adsorption using 10 initial concentrations of purified enzyme (5).
SEM.
Samples were prepared from L. plantarum(pLPCR2-3) and L. plantarum(pLPCR2-3ΔB/X) cultures incubated for various times (up to 48 h) with 1% raw starch. The samples were then centrifuged, and the resulting pellets were lyophilized and submitted to homogenous gold metallization. SEM examination was performed using a JEOL JSM-6300F microscope (Montpellier).
RESULTS
Amylase production.
α-Amylase constructs (Fig. 1) were transferred by electroporation into L. plantarum LMG9211, and the resulting transformants, L. plantarum(pLPCR2-3) and L. plantarum(pLPCR2-3ΔB/X), were tested for the production of α-amylase in the medium. Recombinant clones from both transformants formed halos on starch-containing plates, which indicate secretion of active α-amylase (data not shown). In order to select transformants with the best amylase secretion efficiencies, L. plantarum transformants were grown for 24 h on various carbon sources, using L. plantarum LMG9211 as a negative control. The recombinant transformants displayed comparable growth rates in all media tested. The cultures were harvested in the late logarithmic growth phase (optical density at 600 nm, 2), and amylolytic activities on soluble starch were measured in the supernatant. Amylase produced by the L. plantarum(pLPCR2-3ΔB/X) recombinant strain was found to be secreted at a much higher yield (from 2- to 15-fold, depending on the carbon source) than that produced by L. plantarum(pLPCR2-3) (Table 1). The carbon source had an effect on the amylase production of L. plantarum(pLPCR2-3): the highest amylolytic activity was obtained using soluble starch as a carbon source, while glucose decreased amylase production (from 1.5- to 8.5-fold, depending on the carbon source to which it is compared). Such an effect was not observed with the L. plantarum(pLPCR2-3ΔB/X) transformant, which exhibited the same level of α-amylase activity independent of the carbon source. For both recombinant strains, the presence of calcium carbonate in the growth medium slightly increased amylase production, except for the transformants grown on soluble starch, where the effect was negligible.
TABLE 1.
Levels of α-amylase activity in the culture supernatants of L. plantarum strains in response to medium compositiona
| Carbon source (1% [wt/vol]) | Amylase activity (U/ml)
|
|||||
|---|---|---|---|---|---|---|
|
L. plantarum LMG9211
|
L. plantarum(pLPCR2-3)
|
L. plantarum(pLPCR2-3ΔB/X)
|
||||
| − CaCO3 | + CaCO3 | − CaCO3 | + CaCO3 | − CaCO3 | + CaCO3 | |
| Glucose | 0 | 0 | 1 | 2 | 13 | 15 |
| Maltose | 0 | 0 | 4 | 5 | 10 | 17 |
| Soluble starch | 0 | 0 | 7 | 7 | 16 | 16 |
| Raw starch | 0 | 0 | 2 | 4 | 14 | 15 |
With (+) and without (−) CaCO3.
Purification of the recombinant AmyA and AmyAΔ amylases.
The amylase (AmyA) produced by the L. plantarum(pLPCR2-3) transformant (grown on soluble starch) was purified from the culture supernatant by affinity chromatography on β-cyclodextrin–Sepharose. The elution pattern showed a unique peak of protein which was superimposable on the amylase activity of the enzyme. The recombinant enzyme, purified to homogeneity, migrated as a single band on SDS-PAGE which was active on the zymogram (data not shown). Thus, using a single chromatography step, about 75-fold purification was obtained with a yield of 69% (Table 2).
TABLE 2.
Purification steps of recombinant α-amylases secreted by L. plantarum strainsa
| α-Amylase-producing strain | Purification step | Vol (ml) | Total protein (mg) | Total activity (U) | Sp act (U · mg−1) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|---|---|
| L. plantarum(pLPCR2-3) | Culture supernatant | 150 | 24 | 1,345 | 56 | 1 | 100 |
| Affinity chromatography | 5 | 0.2 | 930 | 4,076 | 72 | 69 | |
| L. plantarum(pLPCR2-3ΔB/X) | Culture supernatant | 1,000 | 168 | 9,864 | 58 | 1 | 100 |
| (NH4)2SO4 precipitate and ultrafiltration | 4 | 31 | 6,003 | 192 | 3 | 61 | |
| Gel filtration | 2 | 0.5 | 409 | 766 | 13 | 4 | |
| Ultrafiltration and concentration | 0.1 | 0.1 | 299 | 3,511 | 60 | 3 |
Data are from 1.5 liters of L. plantarum culture supernatant. Activity was determined as described in Materials and Methods, using soluble starch as a substrate. Protein concentrations were determined with the Bio-Rad protein assay.
Since the amylase (AmyAΔ) produced by L. plantarum(pLPCR2-3ΔB/X) was not retained on β-cyclodextrin–Sepharose, a different purification strategy was undertaken. Following ammonium sulfate precipitation and concentration by ultrafiltration, the enzyme preparation was separated by gel filtration chromatography (Table 2). Using this procedure, the enzyme was purified about 60-fold, with a yield of 3%, and showed a single band on SDS-PAGE and the zymogram (data not shown).
The apparent molecular mass of the purified amylases deduced from the gel analysis was 127 kDa, and it was 59 kDa for AmyA and AmyAΔ.
The specific activities of the enzymes on soluble starch were similar: 4,076 U · mg−1 for AmyA and 3,511 U · mg−1 for AmyAΔ.
Effects of temperature and pH.
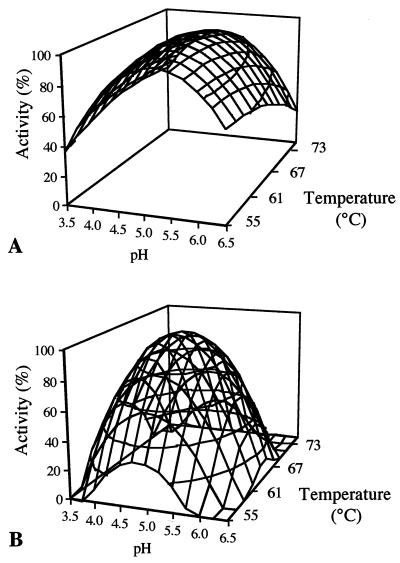
Response surfaces of AmyA and AmyAΔ amylase activities as a function of pH and temperature are shown in Fig. 2. AmyA showed optimal activity at pH 5.0 and 62°C, whereas AmyAΔ had optimal activity at pH 4.8 and 64°C. A given percentage of AmyA activity was maintained over a broader range of temperatures than was AmyAΔ activity; as an example, a minimum of 80% of optimum activity was obtained between 47 and 75°C for AmyA, whereas for AmyAΔ, the same percentage was observed between 59 and 69°C.
FIG. 2.
Effect of pH and temperature on the amylase activities of AmyA (A) and AmyAΔ (B).
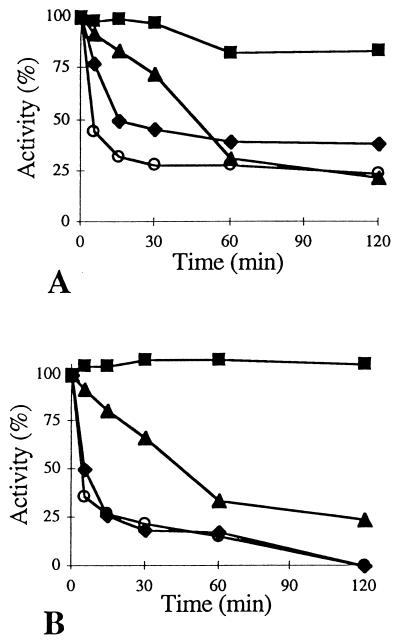
The thermostabilities of the enzymes were similar at temperatures below 65°C. However, at 65°C, the activity of AmyAΔ was totally lost after 120 min, whereas AmyA still exhibited about 40% of its optimum activity (Fig. 3). At higher temperatures, AmyA activity decreased less rapidly than that of AmyAΔ. AmyA thus exhibited higher thermostability than AmyAΔ.
FIG. 3.
Thermostabilities of AmyA (A) and AmyAΔ (B) at 75 (○), 65 (⧫), 55 (▴), and 45°C (■).
Substrate specificity.
AmyA and AmyAΔ relative activities towards various polysaccharide substrates were tested (Table 3). For both enzymes, the best hydrolyzing substrates were found to be amylopectin, soluble starch, and α-cyclodextrin, while amylose was hydrolyzed at a lower rate. Both enzymes were unable to hydrolyze either pullulan or β- and γ-cyclodextrins. With glycogen, a relatively high activity was observed with AmyA, whereas it was very low with AmyAΔ. AmyA was active towards raw starch, while AmyAΔ was unable to hydrolyze it.
TABLE 3.
Relative activity of purified amylases towards different substrates
| Substrate | Relative activity (%)
|
|
|---|---|---|
| AmyA | AmyAΔ | |
| Soluble starch (potato) | 100a | 100a |
| Raw starch (corn) | 20 | 0 |
| Amylose (potato; type III) | 69 | 73 |
| Amylopectin (corn) | 137 | 112 |
| Glycogen | 69 | 9 |
| Pullulan | 0 | 0 |
| α-Cyclodextrin | 92 | 114 |
| β-Cyclodextrin | 0 | 0 |
| γ-Cyclodextrin | 0 | 0 |
Soluble potato starch was taken as the reference, giving 100% of enzyme activity.
The affinities of both enzymes for soluble starch were determined at their pH and temperature optima. Both amylases followed a typical Michaelis-type kinetic. The apparent KM and Vmax values for AmyA were 3 mg · ml−1 and 0.13 μmol · ml−1 · s−1 (equivalent to a kcat of 5,652 s−1), respectively, while AmyAΔ had a KM of 4 mg · ml−1 and a Vmax of 0.20 μmol · ml−1 · s−1 (equivalent to a kcat of 3,878 s−1).
Ability to bind to raw starch.
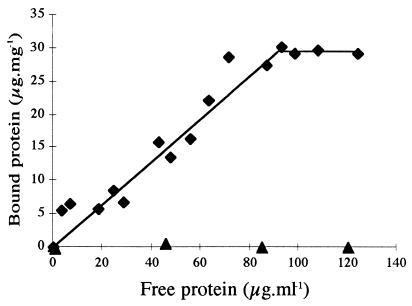
Adsorption of AmyA and AmyAΔ to raw-starch granules was assayed at various protein concentrations (5). As shown in Fig. 4, AmyA was adsorbed to raw starch whereas AmyAΔ was unable to bind to it. For AmyA, the Kad (see Material and Methods) was 0.26 ml of protein suspension per mg of raw starch, and the curve reached a plateau when 30 μg of protein was bound to 1 mg of raw starch, indicating limited availability or accessibility of raw starch for the enzyme. This maximum is reached when the protein forms a monolayer on the starch surface (35).
FIG. 4.
Adsorption of AmyA (⧫) and AmyAΔ (▴) on raw starch. The linear adsorption isotherms indicate the apparent equilibrium distribution of enzymes between the solid phase (bound protein) and the liquid phase (unbound protein) at various protein concentrations.
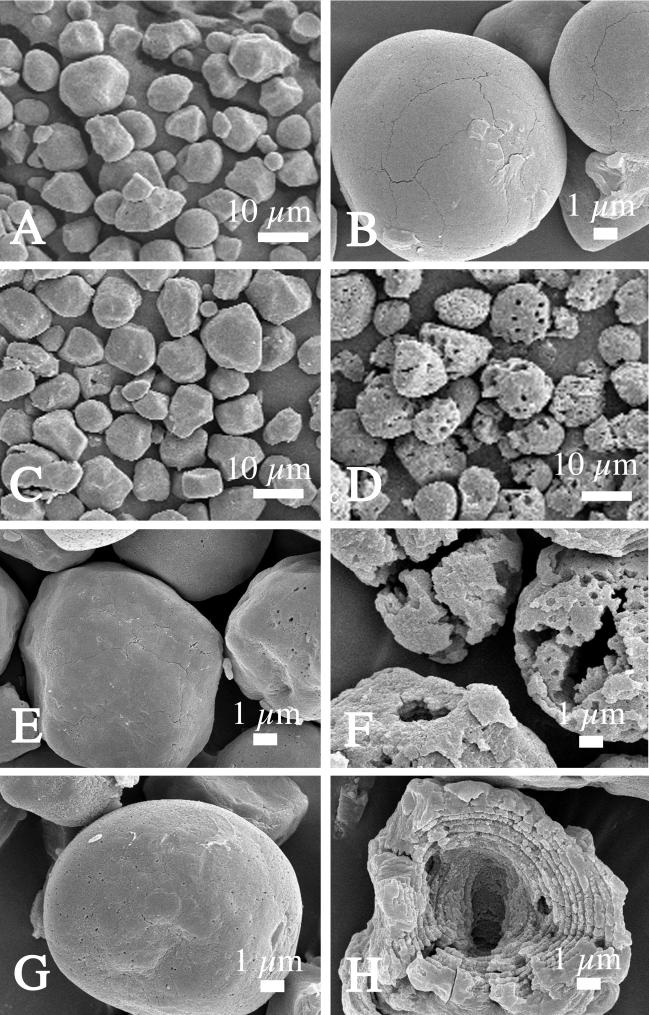
SEM observation of raw-starch granules after fermentation with the recombinant L. plantarum strains.
Enzymatic attack on raw-starch granules was observed by SEM during fermentation. In each fermentation sample, the aspects of starch granules were relatively homogenous (Fig. 5A, C, and D). When the fermentation was carried out with L. plantarum(pLPCR2-3ΔB/X), the initially smooth granules (Fig. 5A and B) did not show any measurable degradation after 24 (Fig. 5C and E) or 48 (Fig. 5G) h. In contrast, the starch granules became rougher and perforated after 24 h when fermentation was carried out with L. plantarum(pLPCR2-3) (Fig. 5D and F). After 48 h of fermentation with this transformant, many starch granules displayed large cavities, and their lamellar organization could be observed (Fig. 5H).
FIG. 5.
Scanning electron micrographs showing the effects of α-amylases produced by recombinant L. plantarum strains on raw corn starch granules after fermentation. Shown are native raw starch (A and B) and raw-starch granules after 24 (C and E) or 48 (G) h of fermentation with L. plantarum(pLPCR2-3ΔB/X) and after 24 (D and F) or 48 (H) h of fermentation with L. plantarum(pLPCR2-3).
DISCUSSION
Expression and secretion.
Since L. plantarum is easily electroporable (31) and had already been used for efficient L. amylovorus α-amylase gene expression and secretion (8, 17), it was chosen as the host for transformation of the two α-amylase constructs, pLPCR2-3 and pLPCR2-3ΔB/X. Despite identical promoter and signal sequences, AmyAΔ was expressed at a higher level of amylase than AmyA. This variation in amylase production could be due to a difference in plasmid copy number among the transformants or to the folding of the recombinant protein.
One form of catabolite repression in the gram-positive bacteria is mediated by the cis-acting catabolite responsive element (cre) and a trans-acting repressor protein (14). A putative cre sequence has been identified in the three Lactobacillus amyA genes which have been sequenced so far (12; Morlon-Guyot et al., Abstr. Aff. VI4 9ème Colloque Club Bact. Lact., 1998), and it is therefore postulated that the carbon source should have an effect on the amylase production of these strains. Despite the presence of this sequence in both AmyA and AmyAΔ constructs, the carbon source had an effect only on amylase produced by L. plantarum(pLPCR2-3) and not on amylase produced by L. plantarum(pLPCR2-3ΔB/X). As suggested above, if the copy number of the plasmid pLPCR2-3ΔB/X is high, the number of cre sequences would be higher than the number of available repressor molecules and might therefore prevent their titration.
Enzymatic activities.
AmyAΔ specific activity was very similar to that of AmyA on all substrates tested. These data are in agreement with those obtained previously with AmyA and AmyAΔ on soluble starch (17), suggesting that the C-terminal part of the amylase is not involved in the activity of the enzyme. These data also suggest that the truncated amylase folds correctly, as already shown for Bacillus stearothermophilus and Bacillus subtilis C-terminally truncated amylases (28, 33). However, AmyAΔ showed lower activity for soluble starch, with one-third-fold increase in KM and one-third decrease in kcat compared to AmyA, indicating altered substrate binding in AmyAΔ. A similar effect of C-terminal truncation was observed with the B. stearothermophilus amylase (33).
AmyA was found to be slightly more thermostable than AmyAΔ. It is therefore suggested that the C terminus of AmyA plays a positive role in the thermostability of the enzyme by maintaining the intact conformation of the enzyme. These results are in agreement with those previously observed with the amylases from Cryptococcus sp. strain S-2 (15) but are different from those obtained with the amylases from Bacillus (28, 33), for which the carboxyl-terminally truncated enzymes were more thermostable than the entire enzymes.
Surprisingly, AmyAΔ was almost unable to hydrolyze glycogen but was active towards amylopectin, whose structure is similar to that of glycogen. These data could be explained by the fact that in contrast to glycogen, amylopectin is often contaminated with small oligosaccharides, which were used as substrates for AmyAΔ.
Binding to raw starch.
Only weak amylolytic activity was obtained in the culture supernatant of L. plantarum(pLPCR2-3) grown in the presence of raw starch compared to that obtained with L. plantarum(pLPCR2-3ΔB/X) on the same medium. These data can be explained by the fact that the intact amylase (AmyA) was adsorbed on raw starch whereas AmyAΔ was not retained on raw starch and was thus totally recovered in the supernatant.
As has been demonstrated by Leloup et al. (20), enzyme adsorption is the limiting parameter of hydrolysis. The method used in this work for measuring the adsorption of the enzyme to raw starch clearly separates raw-starch binding ability from enzymatic activity. Under these conditions, AmyA bound raw starch whereas AmyAΔ could not bind to it. Taken together, these results demonstrate the existence in AmyA of a C-terminal region affecting binding to raw starch. Some amylases have been shown to contain a starch binding domain located at the C terminus of the enzyme (for a review, see reference 16), but structurally different from the repeat unit structure characteristic of the Lactobacillus amylases. The role of the C-terminal repeat unit regions in ligand binding has also been demonstrated in other proteins (for reviews, see references 21, 23, 29, and 36), but this is the first report of such a role in the amylase family.
ACKNOWLEDGMENT
R. Rodriguez was supported by a personal grant from the Conacyt (Consejo Nacional de Ciencia y Technologia) Mexico.
REFERENCES
- 1.Blakesly R W, Boezi J A. A new staining technique for protein in polyacrylamide gels using Coomassie Brilliant Blue G-250. Anal Biochem. 1977;82:580–582. doi: 10.1016/0003-2697(77)90197-x. [DOI] [PubMed] [Google Scholar]
- 2.Bohak I, Back W, Richter L, Eirman M, Ludwig W, Schleifer K H. Lactobacillus amylolyticus sp. nov., isolated from beer malt and beer wort. Syst Appl Microbiol. 1998;21:360–364. doi: 10.1016/S0723-2020(98)80045-3. [DOI] [PubMed] [Google Scholar]
- 3.Box G E P, Hunter W G, Hunter J S. Statistics for experimenters: an introduction to design, data analysis, and model building. New York, N.Y: John Wiley and Sons, Inc.; 1978. [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Chen L, Coutinho P M, Nikolov Z, Ford C. Deletions analysis of the starch-binding domain of Aspergillus glucoamylase. Protein Eng. 1995;8:1049–1055. doi: 10.1093/protein/8.10.1049. [DOI] [PubMed] [Google Scholar]
- 6.Cheng P, Mueller R E, Jaeger S, Bajpai R, Iannotti E L. Lactic acid production from enzyme-thinned corn starch using Lactobacillus amylovorus. J Ind Microbiol. 1991;7:27–34. [Google Scholar]
- 7.De Man J C, Rogosa M, Sharpe M E. A medium for the cultivation of lactobacilli. J Appl Bacteriol. 1960;23:130–135. [Google Scholar]
- 8.Fitzsimmons A, Hols P, Jore J, Leer R J, O'Connell M, Delcour J. Development of an amylolytic Lactobacillus plantarum silage strain expressing the Lactobacillus amylovorus α-amylase gene. Appl Environ Microbiol. 1994;60:3529–3535. doi: 10.1128/aem.60.10.3529-3535.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Florencio, J. A., M. Raimbault, J. P. Guyot, D. R. Eiras-Stofella, C. R. Soccol, and J. D. Fontana.Lactobacillus plantarum amylase acting on crude starch granules: native isoforms and activity changes after limited proteolysis. Appl. Biochem. Biotechnol., in press. [DOI] [PubMed]
- 10.Giraud E, Brauman A, Keleke S, Lelong B, Raimbault M. Isolation and physiological study of an amylolytic strain of Lactobacillus plantarum. Appl Microbiol Biotechnol. 1991;36:379–383. [Google Scholar]
- 11.Giraud E, Gosselin L, Marin B, Parada J L, Raimbault M. Purification and characterization of an extracellular amylase activity from Lactobacillus plantarum strain A6. J Appl Bacteriol. 1993;75:276–282. [Google Scholar]
- 12.Giraud E, Cuny G. Molecular characterization of the α-amylase genes of Lactobacillus plantarum A6 and Lactobacillus amylovorus reveals an unusual 3′ end structure with direct tandem repeats and suggests a common evolutionary origin. Gene. 1997;198:149–157. doi: 10.1016/s0378-1119(97)00309-0. [DOI] [PubMed] [Google Scholar]
- 13.Glazer A N, Nikado H. Microbial Biotechnology. W. H. New York, N.Y: Freeman and Co.; 1994. [Google Scholar]
- 14.Hueck C J, Hillen W. Catabolite repression in Bacillus subtilis, a global regulatory mechanism for the gram-positive bacteria? Mol Microbiol. 1995;15:395–401. doi: 10.1111/j.1365-2958.1995.tb02252.x. [DOI] [PubMed] [Google Scholar]
- 15.Iefuji H, Chino M, Kato M, Iimura Y. Raw-starch-digesting and thermostable α-amylase from the yeast Cryptococcus sp. S-2: purification, characterization, cloning and sequencing. Biochem J. 1996;318:989–996. doi: 10.1042/bj3180989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janecek S, Sevcik J. The evolution of starch-binding domain. FEBS Lett. 1999;456:119–125. doi: 10.1016/s0014-5793(99)00919-9. [DOI] [PubMed] [Google Scholar]
- 17.Jore J P M, De Parasis J. Studies on the α-amylase of Lactobacillus amylovorus as a model for heterologous protein secretion by lactobacilli. FEMS Microbiol Rev. 1993;12:26. [Google Scholar]
- 18.Lacks S A, Springhorn S S. Renaturation of enzymes after polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate. J Biol Chem. 1980;225:7467–7473. [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Leloup V M, Colonna P, Ring S G. α-Amylase adsorption on starch crystallites. Biotechnol Bioeng. 1991;38:127–134. doi: 10.1002/bit.260380204. [DOI] [PubMed] [Google Scholar]
- 21.Lemaire M, Beguin P. Nucleotide sequence of the celG gene of Clostridium thermocellum and characterization of its product, endoglucanase CelG. J Bacteriol. 1993;175:3353–3356. doi: 10.1128/jb.175.11.3353-3360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller G L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428. [Google Scholar]
- 23.Monchois V, Reverte A, Remaud-Simeon M, Monsan P, Willemot R M. Effect of Leuconostoc mesenteroides NRRL B-512F dextransucrase carboxy-terminal deletions on dextran and oligosaccharide synthesis. Appl Environ Microbiol. 1998;64:1644–1649. doi: 10.1128/aem.64.5.1644-1649.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morlon-Guyot J, Guyot J P, Pot B, Jacobe de Haut I, Raimbault M. Lactobacillus manihotivorans sp. nov., a new starch-hydrolyzing lactic acid bacterium isolated from cassava sour starch fermentation. Int J Syst Bacteriol. 1998;48:1101–1109. doi: 10.1099/00207713-48-4-1101. [DOI] [PubMed] [Google Scholar]
- 25.Murado M A, Siso I G, Gonzalez P, Montemayor I, Pastrana L, Pintado J. Characterization of microbial biomasses and amylolytic preparations obtained from mussel processing waste treatment. Bioresour Technol. 1993;43:117–125. [Google Scholar]
- 26.Nakamura L K. Lactobacillus amylovorus, a new starch-hydrolyzing species from cattle waste corn fermentation. Int J Syst Bacteriol. 1981;31:56–63. [Google Scholar]
- 27.Nakamura L K, Crowell C D. Lactobacillus amylovorus, a new starch-hydrolysing species from swine waste-corn fermentation. Dev Ind Microbiol. 1979;20:535–540. [Google Scholar]
- 28.Ohdan K, Kuriki T, Kaneko H, Shimada J, Takada T, Fujimoto Z, Mizuno H, Okada S. Characteristics of two forms of α-amylases and structural implication. Appl Environ Microbiol. 1999;65:4652–4658. doi: 10.1128/aem.65.10.4652-4658.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohura T, Kasuya K I, Doi Y. Cloning and characterization of the polyhydroxybutyrate depolymerase gene of Pseudomonas stutzeri and analysis of the function of substrate binding domains. Appl Environ Microbiol. 1999;65:189–197. doi: 10.1128/aem.65.1.189-197.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olympia M, Fukuda H, Ono H, Kaneko Y, Takano M. Characterisation of starch hydrolyzing Lactic Acid Bacteria isolated from a fermented fish and rice food «Burong Isda» and its amylolytic enzyme. J Ferment Bioeng. 1995;80:124–130. [Google Scholar]
- 31.Rodriguez S R, Morlon-Guyot J, Guyot J P. Electrotransformation of Lactobacillus manihotivorans LMG 18010T and LMG 18011. J Appl Microbiol. 1999;87:99–107. [Google Scholar]
- 32.Sen S, Chakrabarty S L. Amylase from Lactobacillus cellobiosus D-39 isolated from vegetable wastes; purification and characterisation. J Appl Bacteriol. 1986;60:419–423. [Google Scholar]
- 33.Vihinen M, Peltonen T, Litia A, Suominen I, Mantsala P. C-terminal truncations of thermostable Bacillus stearothermophilus α-amylase. Protein Eng. 1994;7:1255–1259. doi: 10.1093/protein/7.10.1255. [DOI] [PubMed] [Google Scholar]
- 34.Vretblad P. Immobilization of ligands for biospecific affinity chromatography via their hydroxyl groups. Their cyclo-hexaamylose-β-amylase system. FEBS Lett. 1974;47:86–89. doi: 10.1016/0014-5793(74)80431-x. [DOI] [PubMed] [Google Scholar]
- 35.Williamson G, Belshaw N J, Williamson M P. O-Glycosylation in Aspergillus glucoamylase. Biochem J. 1992;282:423–428. doi: 10.1042/bj2820423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wren B W. A family of clostridial and streptococcal ligand-binding proteins with conserved C-terminal repeat sequences. Mol Microbiol. 1991;5:797–803. doi: 10.1111/j.1365-2958.1991.tb00752.x. [DOI] [PubMed] [Google Scholar]
- 37.Xiaodong W, Xuan G, Rakhit S K. Direct fermentative production of lactic acid on cassava and other starch substrates. Biotechnol Lett. 1997;19:841–843. [Google Scholar]
- 38.Zhang D X, Cheryan M. Starch to lactic acid in a continuous membrane bioreactor. Processes Biochem. 1994;29:145–150. [Google Scholar]