Abstract
Lactobacillus plantarum NC8 contains a pdc gene coding for p-coumaric acid decarboxylase activity (PDC). A food grade mutant, designated LPD1, in which the chromosomal pdc gene was replaced with the deleted pdc gene copy, was obtained by a two-step homologous recombination process using an unstable replicative vector. The LPD1 mutant strain remained able to weakly metabolize p-coumaric and ferulic acids into vinyl derivatives or into substituted phenyl propionic acids. We have shown that L. plantarum has a second acid phenol decarboxylase enzyme, better induced with ferulic acid than with p-coumaric acid, which also displays inducible acid phenol reductase activity that is mostly active when glucose is added. Those two enzymatic activities are in competition for p-coumaric and ferulic acid degradation, and the ratio of the corresponding derivatives depends on induction conditions. Moreover, PDC appeared to decarboxylate ferulic acid in vitro with a specific activity of about 10 nmol · min−1 · mg−1 in the presence of ammonium sulfate. Finally, PDC activity was shown to confer a selective advantage on LPNC8 grown in acidic media supplemented with p-coumaric acid, compared to the LPD1 mutant devoid of PDC activity.
Substituted hydroxycinnamic acids (principally ferulic acid and p-coumaric acid), also called phenolic acids, are abundant molecules that bind the complex lignin polymer to the hemicellulose and cellulose in plant cell walls (16). Various bacteria and fungi produce a wide range of hemicellulases in order to degrade these cell wall polymers (17) and to release phenolic acids, which are biologically important molecules. First, they serve as signals for the plant-associated Agrobacterium tumefaciens and induce vir gene expression through a two-component system consisting of the VirA and VirG proteins (28). The Vir-inducing properties of ferulic acid in A. tumefaciens were shown to be strain dependent (29), while ferulic acid could be O-demethylated into caffeic acid by the VirH2 protein in order to turn off vir gene expression (26).
Phenolic acid catabolism is also essential in the biodegradation of plant wastes. Several bacteria, such as Pseudomonas spp. and Acinetobacter calcoaceticus, are able to grow on these compounds as the sole source of carbon. In the first step, they convert ferulic and p-coumaric acids into vanillic and p-hydroxybenzoic acids, respectively, which are then transformed into protocatechuic acid and integrated into the tricarboxylic acid cycle via the β-ketoadipate pathway (33, 38, 41). Ferulic acid can also be degraded into vanillin by a two-step process involving either a coenzyme A (CoA) ligase followed by side chain cleavage in Pseudomonas fluorescens (21) and Pseudomonas sp. strain HR199 (34) or a propionic acid chain cleavage followed by a reductase in the white-rot fungus Pycnoporus cinnabarinus (2, 31). In other microbial systems, phenolic acids are metabolized into volatile phenols by two different pathways. Most often, they are first decarboxylated into 4-vinyl derivatives and then reduced to 4-ethyl derivatives. Phenolic acid decarboxylases (PAD) have been characterized in yeast and bacteria (7, 19). Four genes coding for PAD enzymes, pad1 from Saccharomyces cerevisiae (13), fdc from Bacillus pumilus (44), pdc from Lactobacillus plantarum (7), and pad from Bacillus subtilis (9) have been cloned and expressed in Escherichia coli. Recently, several gene products sharing significant homology with PAD1 from S. cerevisiae have been discovered through genome sequencing; these are encoded by pad1 from Archaeoglobus fulgidus (27), pad1 from E. coli (accession no., AJ006210), vdcB from Streptomyces sp. strain D7 (12), yclB from B. subtilis (43), and MJ0102 from Methanococcus jannaschii (4), although the corresponding enzymatic activities have not been demonstrated so far. A second pathway has been proposed for Lactobacillus pastorianus where caffeic and p-coumaric acids are first reduced into substituted phenyl propionic acids and then decarboxylated into 4-ethyl derivatives (42).
The interest in improving our understanding of phenolic acid biodegradation is multiple. First, as was shown for S. cerevisiae, PAD activity may confer a selective advantage upon microorganisms during growth on plants, where PAD expression could constitute a stress response induced by phenolic acid (13, 23). Second, phenol derivatives are valuable intermediates in the biotechnological production of new flavor and fragrance chemicals (25). Third, they are regarded as sources of phenolic off-flavors in many beers and wines, due to their characteristic aroma and their low threshold of detection (20, 39).
L. plantarum is now considered a model for ubiquitous lactic acid bacteria in the plant kingdom. Moreover, it is used as a malolactic starter in some wines and as a lactic starter for many vegetable fermentations which contain phenolic acids. L. plantarum displays substrate-inducible PAD activity encoded by the pdc gene, which is transcriptionally regulated by p-coumaric, ferulic, and caffeic acids (7). Under the conditions tested, the purified p-coumaric acid decarboxylase (PDC) exhibits high specific activity on p-coumaric acid (0.6 μmol · min−1 · mg−1) but not detectable activity on ferulic acid (8).
We have constructed an L. plantarum mutant strain deficient in PDC activity in order to investigate alternate pathways for phenolic acid degradation and to study the stress response. Kinetic studies of the pdc mutant revealed the existence of a second PDC activity, called PDC2, as well as an alternate phenolic acid reduction pathway, in L. plantarum. Our results also suggest that the synthesis of PDC in L. plantarum could constitute a stress response induced by phenolic acid toxicity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. L. plantarum LPNC8 (kindly provided by Lars Axelsson), LPCHL2, and LPD1 were grown at 37°C in MRS medium (15) with glucose (20 g/liter) as the source of carbon. E. coli TG1 was grown at 37°C on Luria-Bertani (LB) medium (3). Antibiotics were used in the following concentrations: erythromycin at 200 mg/liter for E. coli and 5 mg/liter for L. plantarum and ampicillin at 200 mg/liter for E. coli.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli TGI | supE hsdΔ5thi Δ(lac-proAB)F′ [traD36 proAB+ lacqlacZΔM15] | 22 |
| L. plantarum CHL2 | Wild-type, gram-positive, ubiquitous homolactic acid bacterium | Charles Hansen's BioSystems, Horsholm, Denmark |
| L. plantarum NC8 | Plasmid-free strain | L. Axelsson, presented at the Fifth Symposium on Lactic Acid Bacteria, Veldhoven, The Netherlands, 1996. |
| Plasmids | ||
| pBluescript SK(−) | Apr ΔlacZ | Stratagene |
| pGID023 | Shuttle vector for E. coli and L. plantarum; derivative of pJDC9 containing the pE194 replication functions; used as an unstable integration vector; Emr | 24 |
| pBSK−FA | Apr; pBluescript SK(−) containing the 537-bp fragment (FA) of L. plantarum amplified by PCR with primers D1 and D2 | This study |
| pBSK−FAC | Apr; pBluescript SK(−) containing FA and the 497-bp (FC) fragment of L. plantarum amplified by PCR with primers D3 and D4 | This study |
| pGFAC | Emr; pGID023 containing the 1,034-bp FAC fragment | This study |
| pJDC9 | Emr; ΔlacZ | 11 |
| PJPDC1 | pJDC9 containing the 2.3-kb Sau3A fragment of L. plantarum CHL2 with the pdc gene | 7 |
PCR amplification of DNA.
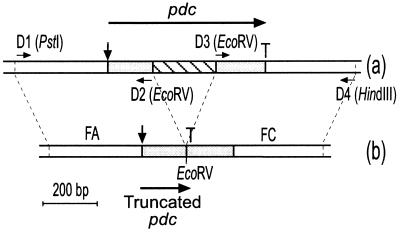
To construct the L. plantarum pdc mutant, the plasmid-free strain LPNC8 was preferred to LPCHL2, which carries two plasmids, in order to avoid incompatibility between any of these plasmids and the unstable autoreplicative vector pGID023. To verify that the pdc gene locus was identical in LPCHL2 and LPNC8, Southern blotting and DNA sequencing of the pdc gene region were carried out in both strains. L. plantarum NC8 chromosomal DNA was used to perform PCR amplification. Primers D1 (5′ AGCCTGCAGACCGACACTGATCCACTC 3′) and D2 (5′ AGCGATATCGACCCAACGACCGGCACC 3′), which include PstI and EcoRV sites, respectively, (underlined nucleotides), were used to amplify the FA region, which corresponds to a 537-bp DNA fragment of the pdc locus encompassing the putative promoter region and the first 152 nucleotides of the open reading frame. Primers D3 (5′ AGCGATATCGTCTCGTGAAAAGTATGCC3′) and D4 (5′ GGCAAGCTTGCAGAGCAAGGTAAG 3′), which include EcoRV and HindIII sites, respectively (underlined nucleotides), were used to amplify the FC region, which corresponds to a 497-bp DNA fragment containing the last 174 nucleotides of the pdc open reading frame followed by a putative transcriptional terminator (Fig. 1). A 160-bp pdc internal fragment, located within the deleted region, was amplified with primers R1 (5′ GCTGACATCGTCATGTTG 3′) and R2 (5′ GTTTCCATTAAATCGATG 3′). PCR amplification was performed using 0.1 μg of DNA template with 0.5 U of Taq DNA polymerase (Appligene) under standard conditions in an automatic DNA thermocycler (Hybaid, Ltd., Teddington, United Kingdom).
FIG. 1.
Physical map of the pdc locus in the wild-type strain LPNC8 (a) and the mutant strain LPD1 (b). Long horizontal arrows represent the two ORFs and their orientations. The start sites are indicated by vertical arrows and the stop codons by T. The positions and orientations of the primers are indicated by short horizontal arrows, and restriction sites that were created are noted between brackets.
DNA manipulation and transformation procedures.
Standard procedures described by Sambrook et al. (36) were used for DNA manipulation. L. plantarum chromosomal DNA was prepared by the method described by Posno et al. (35). PCR products were purified with the Jet Pur Kit (Q.BIOgene, Illkirch, France) and sequenced by the dideoxy chain termination method (37) with the Thermosequenase radiolabeled terminator cycle sequencing kit (Amersham Life Science, Inc., Cleveland, Ohio) in accordance with the recommendations of the manufacturer. Southern blotting was performed as previously described (7). The 160-bp PCR product was labeled with 10 μCi of [α-32P]dATP (NEN, Boston, Mass.) by 10 PCR amplification cycles with primers R1 and R2 to generate a highly radiolabeled double-stranded probe. E. coli and L. plantarum strains were transformed by electroporation as described by Dower et al. (18) and Aukrust and Nes (1), respectively.
Preparation of whole-cell suspensions and cell extracts.
Cells of L. plantarum grown in MRS medium and E. coli grown in LB medium were harvested by centrifugation, washed twice with 25 mM potassium phosphate buffer (pH 6.0), and resuspended in the same buffer. For low-activity detection (a few nanomoles per minute per milligram), concentrated resting cells (5 g/liter [dry weight]) were used in kinetic reactions. For high-activity detection, diluted resting cells (0.2 g/liter [dry weight]) were used. For cell extract preparation, cells were harvested as described above and disrupted with a French press at 1.2 × 108 Pa. The total protein concentration in the cell extract was determined by using a protein assay kit (Bio-Rad, Richmond, Calif.) with bovine serum albumin as the standard. The specific activity was expressed as micromoles or nanomoles of substrate degraded per minute per milligram of protein. For whole cells, the protein concentration was deduced from the dry biomass in the cell suspension (1 g of dry biomass per liter corresponds to 0.5 g of total protein per liter).
Assay of phenolic acid degradation.
Phenolic acid degradation and derivative production were monitored by UV spectrophotometry (using quartz cuvettes in a Beckman DU600 spectrophotometer). The products identified by UV spectrophotometry (phenolic acids and phenolic derivatives) have been previously identified by gas chromatographic analysis (6) and also by high-pressure liquid chromatographic (HPLC) analysis (14). UV spectrophotometry has since been used to study phenolic acid metabolism, taking advantage of the rapidity, sensitivity, and reliability of this method (7, 9, 14). p-coumaric acid has a main absorption peak at 285 nm and a second, lower peak at 305 nm. Ferulic acid has two absorption peaks, at 285 and 300 nm. Decarboxylation and reduction of phenolic acids lead to a hypsochrome UV spectrum displacement, as was shown for the p-coumaric acid derivatives 4-vinyl phenol, 4-ethyl phenol, and phloretic acid (Fig. 2). Analysis of phenolic acid metabolism could be performed using this method in different reaction mixtures. In MRS medium, the sample has to be centrifuged first and the supernatant is diluted 100-fold prior to UV analysis. Ferulic and p-coumaric acid degradation could be detected at 300 and 305 nm, respectively, as these two acids have a second absorption peaks at these wavelengths. Detection of the phenolic acid derivatives is not possible because components of MRS medium interfered with the UV spectra in a range of 255 to 280 nm. In order to identify the phenolic acid degradation products, analyses were carried out with cell extracts or with whole cells in phosphate buffer. The absence of UV absorbance of the phosphate buffer and Stop buffer (20 mM Tris-HCl–0.3% sodium dodecyl sulfate [SDS] to stop activity; pH 6.0) used in these experiments was first checked. Moreover, the stability of phenolic acids in the phosphate buffer was also verified in the time of the experiments. Whole cells of L. plantarum were incubated in phosphate buffer to check that this bacterium did not produce extracellular proteins which could interfere with UV analysis. Kinetic reactions with cell extracts (about 5 to 50 mg of proteins/liter) in phosphate buffer were started by adding 0.6 mM substrate. During incubation, samples were taken and diluted 50-fold in Stop buffer prior to UV analysis. Nevertheless, low-specific-activity detection by UV spectrophotometry was critical in cell extracts, since high protein concentrations have to be incubated with relatively small amounts of substrate (0.06 to 0.3 mM). Under these conditions, UV spectra are disturbed by UV absorbance of soluble proteins, and the appearance of 4-vinyl phenol or other derivatives may remain undetected. Since the uptake of phenolic acids into bacterial cells is not a limiting step for their metabolism, the problem of low-specific-activity detection was solved by running kinetic experiments in phosphate buffer using highly concentrated whole-cell suspensions. In this case, whole-cell suspensions were prepared as described above. Reactions were started by adding 0.6 mM substrates. Samples were taken and were immediately centrifuged (at 4°C, for 15 min, at 12,000 × g) to eliminate the cells. The supernatant was diluted 50-fold in Stop buffer before UV spectrophotometry analysis.
FIG. 2.
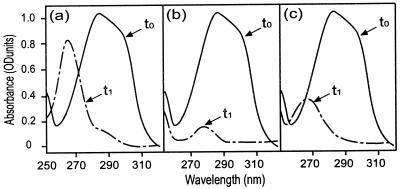
UV spectra of p-coumaric acid and metabolic derivatives: 1, 60 μM p-coumaric acid; 2, 60 μM 4-vinyl phenol; 3, 60 μM phloretic acid; 4, 60 μM 4-ethyl phenol; 5, 30 μM 4-vinyl phenol with 30 μM phloretic acid. Arrows point to the maximum absorbance for each compound.
UV spectrophotometry analysis is suitable for detecting low phenolic compound concentrations (1 to 10 μM). As kinetic reactions are most often performed with 1.2 or 0.6 mM phenolic acids, the dilution of the sample before UV analysis strongly reduces the UV absorbance of the mixture reaction components when necessary (for samples from MRS medium). In all cases, the UV spectra obtained under these conditions have no background signals.
RESULTS
Cloning of a deleted copy of the pdc gene.
The FA and FC regions of the pdc gene (Fig. 1) were amplified separately by PCR and used for the construction of plasmid pGFAC. The FA fragment was cloned into the pBluescript SK(−) vector between the PstI and EcoRV sites. The FC fragment was then ligated into pBSK−FA between EcoRV and HindIII in the same transcriptional orientation. The resulting plasmid, named pBSK−FAC, bears a copy of the pdc gene lacking 199 bp of its internal region. A frameshift in the deleted gene copy downstream from the EcoRV site creates a stop codon, which causes the synthesis of a truncated peptide of 51 amino acids (Fig. 1). The 1,034-bp PstI/HindIII fragment from pBSK−FAC was cloned into the shuttle vector pGID023 to give plasmid pGFAC, which was transformed into E. coli TG1. Whole cells and corresponding cell extracts of E. coli TG1(pGFAC) were prepared as described in Materials and Methods and were tested for their ability to metabolize p-coumaric and ferulic acids (data not shown). No PDC activity was detected on either substrate, even after 24 h of incubation, while whole cells and cell extracts of E. coli TG1(pJPDC1), containing the wild-type pdc gene, exhibited PDC activity of 8.5 μmol · min−1 · mg−1 on p-coumaric acid (7). These results indicated that plasmid pGFAC was suitable for generating a pdc mutant strain of L. plantarum by gene replacement.
Disruption of the pdc gene in LPNC8.
Plasmid pGFAC was introduced in LPNC8 by electroporation, and transformants were selected for erythromycin resistance. The primary recombination event between pGFAC and the chromosomal pdc locus led to integration of the whole plasmid and thus conferred erythromycin resistance. The second recombination event, between the chromosomal pdc gene and its deleted copy, followed by segregational loss of the excised vector, produced erythromycin-sensitive clones. Since the excision step can generate either the wild-type PDC phenotype or a deletion in the chromosomal pdc gene, PCR analyses on total DNA of erythromycin-sensitive candidates (Ems) were carried out to screen for clones bearing the deleted pdc gene copy. Two primers located on each side of the deleted region were used, which should result in amplification of a 540-bp fragment for the wild-type gene and of a 342-bp fragment for the deleted gene. Among 10 Ems colonies tested, four clones, designated D1, D2, D8, and D10, yielded the shorter PCR fragment, while all others, as well as the wild-type control LPNC8, gave the 540-bp fragment of the full-length gene. Sequencing of the 342-bp PCR fragment confirmed the deletion (data not shown). Southern blotting was performed on total DNA of D1, D2, D8, and D10, which was then digested by EcoRI/HindIII and hybridized with a 160-bp labeled probe located in the deleted region. Positive hybridization was obtained with the wild-type LPNC8 strain, while no hybridization could be detected with clones D1, D2, D8, and D10 (data not shown). These results further confirmed that clones D1, D2, D8, and D10 carried a 199-bp internal deletion in the pdc gene. One of the latter clones, designated LPD1, was retained for further studies.
Ability of an LPD1 mutant to metabolize p-coumaric acid.
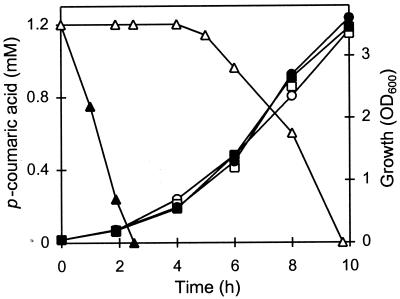
Wild-type LPNC8 and the LPD1 pdc mutant were grown in MRS medium alone or supplemented with 1.2 mM p-coumaric acid. Samples were taken during the growth cycle to measure the p-coumaric acid concentration by UV spectrophotometry at 305 nm, in order to avoid interference with MRS components. The two strains displayed similar growth curves with and without p-coumaric acid but showed different rates of p-coumaric acid degradation (Fig. 3). LPNC8 degraded 100% of available p-coumaric acid within the first 2.5 h of growth, while no p-coumaric acid appeared to be degraded during the same period in the LPD1 culture. However, the LPD1 mutant started to weakly metabolize p-coumaric acid after 5 h, and all available p-coumaric acid had disappeared from the medium after 10 h. This indicated that LPD1 still produced one or more enzymes, distinct from PDC, responsible for p-coumaric acid utilization. Phenolic acid pathway analysis was performed using whole cells and cell extracts in phosphate buffer to eliminate absorption in the UV range by MRS components. LPNC8 and LPD1 were grown in MRS medium and divided in two subcultures. One subculture was used as a control and was incubated for 2 h at 37°C with no addition, while the other was supplemented with 3 mM ferulic acid and incubated under the same conditions. Whole cells and cell extracts prepared from these subcultures were tested for p-coumaric acid degradation activity. No acid degradation was detected in the uninduced whole cells and cell extracts of either strain. In LPNC8, the wild-type strain, p-coumaric acid was decarboxylated by whole induced cells and corresponding cell extracts with an activity of 500 nmol · min−1 · mg−1. In addition, p-coumaric acid was also decarboxylated into 4-vinyl phenol by the LPD1 mutant, but at a lower rate of about 5 nmol · min−1 · mg−1, indicating that a second PAD was synthesized by LPD1. The PDC activity of LPD1 was 100-fold lower than that of LPNC8, the wild-type strain, and was for this reason totally concealed by the high PDC activity conferred by the pdc gene. In the LPD1 cell extract, this second PAD activity was not detected. Studies of phenolic acid metabolism were therefore carried out with whole cells of LPNC8 and LPD1.
FIG. 3.
Growth of strain LPNC8 (squares) and the LPD1 mutant (circles) supplemented (filled symbols) or not (open symbols) with 1.2 mM p-coumaric acid at pH 6.5. Residual p-coumaric acid concentrations in LPNC8 (filled triangles) and in LPD1 (open triangles) were measured by UV spectrophotometry (see Materials and Methods).
Evidence of a second PDC enzyme and a phenolic acid reductase in L. plantarum.
In order to characterize the second PAD activity, growing cultures of LPD1 and LPNC8 were divided into four samples and induced with 1.2 or 3 mM p-coumaric acid or ferulic acid. Whole resting cell suspensions were prepared in 25 mM phosphate buffer (pH 6.0) in order to identify p-coumaric acid degradation products (Table 2). In the wild-type strain, LPNC8, UV spectra indicate that p-coumaric acid was strongly decarboxylated into 4-vinyl phenol with a specific activity of 500 nmol · min−1 · mg−1 (Fig. 4a). In LPD1 mutant, p-coumaric acid metabolism was dependent on the nature and concentration of the inducer. Cells induced with ferulic acid (1.2 or 3 mM) displayed p-coumaric acid decarboxylation activity of about 8 nmol · min−1 · mg−1. In cells induced with 1.2 mM p-coumaric acid, p-coumaric acid was metabolized but 4-vinyl phenol was not the product of the reaction. Instead, phloretic acid or 4-ethyl phenol appeared to be produced, based on the UV spectrum (Fig. 4b). For cells induced with 3 mM p-coumaric acid, UV spectra indicate that p-coumaric acid was degraded into a mixture of 4-vinyl phenol and phloretic acid or 4-ethyl phenol (Fig. 4c). The production of 4-ethyl derivatives from phenolic acids requires the prior formation of 4-vinyl derivatives or substituted phenyl propionic acids (7). As 4-vinyl derivatives were never detected during kinetic experiments with LPD1 cells, phenolic acids were likely reduced into substituted phenyl propionic acids and subsequently decarboxylated into 4-ethyl derivatives. These results suggested that LPD1 could produce a phenolic acid reductase, which metabolized p-coumaric acid into phloretic acid, as was shown for L. pastorianus (42). Since a reduced cofactor is generally required for enzymatic reduction (30, 32), similar experiments were done to confirm the hypothesis of reduction, using whole cells incubated for 15 min with 20 mM glucose at 30°C prior to starting the kinetics, in order to stimulate glycolysis and increase the pool of reduced cofactors. No difference was detected in the wild-type strain, while the addition of glucose in whole cells of LPD1 strongly stimulated the transformation of p-coumaric acid into a phenol derivative which was not 4-vinyl phenol but was phloretic acid and/or 4-ethyl phenol, for cells induced by p-coumaric acid (1.2 and 3 mM) and 1.2 mM ferulic acid (Table 2). Only LPD1 cells induced with 3 mM ferulic acid still produced a mixture of 4-vinyl phenol and phloretic acid and/or 4-ethyl phenol (Table 2). Taken together, our results indicate the presence, in both wild-type and LPD1 strains, of a second PAD (named PDC2), highly induced by ferulic acid, and of a putative phenolic acid reductase activity (named PAR) induced by p-coumaric and ferulic acids in the presence of glucose. These two enzymes displayed low specific activities (about 10 nmol · min−1 · mg−1 for p-coumaric acid) compared to PDC.
TABLE 2.
Derivatives accumulated upon biotransformation of p-coumaric acid by whole cells of LPNC8 and the LPD1 mutant, induced with p-coumaric or ferulic acid
| Strain | Derivative(s) present after inductiona by:
|
|||||||
|---|---|---|---|---|---|---|---|---|
|
p-Coumaric acid
|
Ferulic acid
|
|||||||
| 1.2 mM
|
3 mM
|
1.2 mM
|
3 mM
|
|||||
| − | + | − | + | − | + | − | + | |
| LPNC8 | 4VP | 4VP | 4VP | 4VP | 4VP | 4VP | 4VP | 4VP |
| LPD1 | RD | RD | 4VP/RD | RD | 4VP | RD | 4VP | 4VP/RD |
Whole cells were incubated for 15 min at 30°C with (+) or without (−) 20 mM glucose before kinetic reactions. 4VP, 4-vinyl phenol; RD, reduced derivatives, i.e., phloretic acid and/or 4-ethyl phenol; 4VP/RD, mixture of 4-vinyl phenol and reduced derivatives.
FIG. 4.
UV spectra resulting from the conversion of 0.6 mM p-coumaric acid by induced whole cells of LPNC8 (0.2 g/liter) or LPD1 (5 g/liter) incubated 1 h without glucose in 25 mM phosphate buffer. t0, UV spectra corresponding to the sample taken at the start of kinetic reaction. t1, UV spectra corresponding to the sample taken after 1 h of kinetic reaction. (a) LPNC8 induced with p-coumaric or ferulic acid (1.2 or 3 mM); (b) LPD1 induced with 1.2 mM p-coumaric acid; (c) LPD1 induced with 3 mM p-coumaric acid.
The PDC enzyme displays weak decarboxylase activity on ferulic acid.
Whole resting cell suspensions of LPD1 and LPNC8 induced with p-coumaric or ferulic acid were prepared as previously described and tested for ferulic acid metabolism. Ferulic acid derivatives produced by whole cells of LPD1 were different from those produced by whole cells of LPNC8. Ferulic acid was partially or totally reduced, depending on the nature and the concentration of the inducers, by the LPD1 mutant, but was preferentially decarboxylated in the wild-type strain, LPNC8 (Table 3). This indicates that the PDC enzyme is involved in ferulic acid decarboxylation in whole cells. To confirm this hypothesis, which is contradictory to the results obtained previously with the purified PDC (8), ferulic acid metabolism was tested in whole cells of the recombinant strain E. coli TG1(pJPDC1) overexpressing the PDC enzyme (7) and on the corresponding cell extract. In whole cells, ferulic acid was decarboxylated at a rate of about 40 nmol · min−1 · mg−1, while the corresponding cell extract displayed no detectable activity on ferulic acid in phosphate buffer. Reaction conditions were then modified by varying the pH and temperature independently and by adding glycerol or salts. Ferulic acid decarboxylase activity was stimulated in cell extracts supplemented with 20% ammonium sulfate or 20% NaCl, with an optimum activity of about 30 nmol · min−1 · mg−1, indicating that PDC displays a low ferulic acid decarboxylase activity under those conditions.
TABLE 3.
Derivatives from the biotransformation of ferulic acid by whole cells of LPNC8 and the LPD1 mutant, induced with p-coumaric or ferulic acid
| Strains | Derivative(s) present after inductiona by:
|
|||||||
|---|---|---|---|---|---|---|---|---|
|
p-Coumaric acid
|
Ferulic acid
|
|||||||
| 1.2 mM
|
3 mM
|
1.2 mM
|
3 mM
|
|||||
| − | + | − | + | − | + | − | + | |
| LPNC8 | 4VG | 4VG/RD | 4VG | 4VG/RD | 4VG | 4VG/RD | 4VG | 4VG |
| LPD1 | RD | RD | 4VG/RD | RD | 4VG | RD | 4VG | 4VG/RD |
Whole cells were incubated for 15 min at 30°C with (+) or without (−) 20 mM glucose before kinetic reactions. 4VG, 4-vinyl gaïacol; RD, reduced derivatives, i.e., dihydroferulic acid and/or 4-ethyl gaïacol; 4VG/RD, mixture of 4-vinyl gaïacol and reduced derivatives.
Influence of the p-coumaric acid concentration on the growth of the wild-type strain, LPNC8, and the mutant LPD1 strain at different pHs.
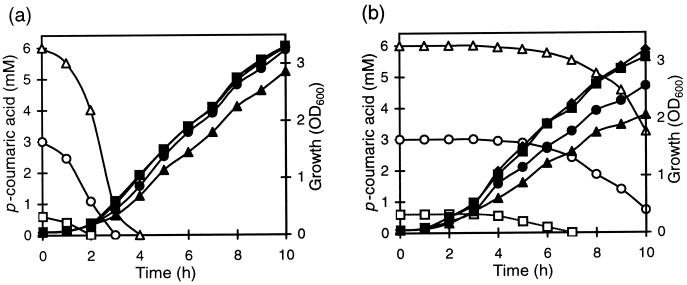
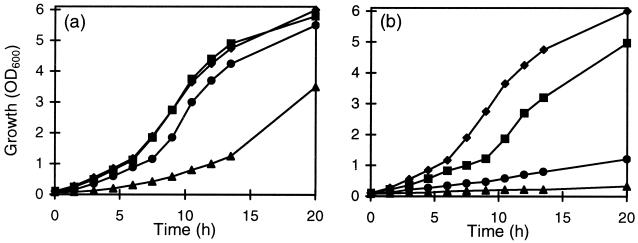
Three concentrations of p-coumaric acid (0.6, 3, and 6 mM) were tested on the growth of the LPNC8 and LPD1 strains in MRS broth at pH 6.5. The residual p-coumaric acid concentration was measured during growth. For the wild-type strain, LPNC8, addition of 0.6 or 3 mM p-coumaric acid in the culture medium had no apparent effect on growth (Fig. 5a). The totality of this acid was degraded in, respectively, 2 or 3 h. Addition of 6 mM p-coumaric increased the latency period, but when all the available p-coumaric acid was metabolized, the final biomass was the same as that of the control culture without p-coumaric acid (data not shown). In contrast to results for the wild-type strain, p-coumaric acid was not degraded during the first 5 h of LPD1 mutant growth (Fig. 5b). With 0.6 mM p-coumaric acid, LPD1 metabolized 100% of this acid after 7 h of growth and its growth was not inhibited. For cultures with 3 and 6 mM p-coumaric acid, only 80 and 90%, respectively, was metabolized in the same period. LPD1 growth was significantly decreased, and the final biomass reached only 78 and 57% compared to that of the control without acid (data not shown). These results seem to indicate that p-coumaric acid is toxic for L. plantarum. The entrance by diffusion of weak acids into bacterial cells increases with a decrease in external pH (45). In order to observe the influence of different rates of acid uptake on the growth, cultures were performed at different initial pHs (5.5, 4.5, and 3.5) in MRS medium and at three p-coumaric acid concentrations (1.2, 3, and 6 mM). The most significant results were obtained at pH 4.5. The growth of the wild-type strain, LPNC8, was not modified by 1.2 or 3 mM p-coumaric acid and was only reduced with 6 mM (Fig. 6a). The growth of the LPD1 mutant strain was strongly inhibited in the presence of 1.2 or 3 mM p-coumaric acid and was totally inhibited with 6 mM p-coumaric acid (Fig. 6b). These results indicate that p-coumaric acid toxicity was higher at a low initial pH of the growth medium.
FIG. 5.
Growth of (filled symbols) and degradation of p-coumaric acid by (open symbols) LPNC8 (a) and LPD1 (b) at different p-coumaric acid concentrations (⧫, 0 mM; ■, 0.6 mM; ●, 3 mM; ▴, 6 mM) at pH 6.5. Samples were taken during growth to determine the biomass (OD600) and p-coumaric acid degradation using UV spectrophotometry (see Materials and Methods).
FIG. 6.
Growth of LPNC8 (a) and LPD1 (b) at different p-coumaric acid concentrations (⧫, 0 mM; ■, 1.2 mM; ●, 3 mM; ▴, 6 mM) at pH 4.5.
DISCUSSION
Inactivation of the pdc gene revealed some interesting features of phenolic acid metabolism in L. plantarum. Most notable was a low residual PAD activity in the LPD1 mutant, attributed to an alternate PDC2 activity that weakly decarboxylates p-coumaric and ferulic acids into 4-vinyl derivatives and is better induced with ferulic acid. This is, to our knowledge, the first report which demonstrates the existence of two distinct and functional acid phenol decarboxylases in a single microorganism.
Our study also reveals that L. plantarum converts phenolic acids into substituted phenyl propionic acids. Therefore, L. plantarum appears to have p-coumaric and ferulic acid reductase activities (PAR), induced by both substrates and mostly active when glucose is added (Tables 2 and 3). Reduction of p-coumaric and caffeic acid has been previously demonstrated in L. pastorianus (42). In L. plantarum, PAR and PDC2 activities are in competition for p-coumaric and ferulic acid degradation, and the ratio of the corresponding derivatives depends on induction conditions. The stable food grade mutant LPD1 strain, which was obtained by a two-step homologous recombination event, is devoid of its major PAD activity. However, it is still able to metabolize phenolic acids, and it is not suitable for producing fermented food or beverages without phenolic acid derivatives.
In a previous work (8), the existence of the second PAD, PDC2, was supported by the facts that ferulic acid was decarboxylated by whole cells of L. plantarum and that the purified PDC from L. plantarum did not display activity on ferulic acid under the conditions tested. The present results nevertheless indicate that PDC exhibits a weak decarboxylase activity in vitro on ferulic acid when ammonium sulfate or NaCl are added in the reaction buffer. This constitutes the major difference between L. plantarum PDC and both ferulic acid decarboxylase (FDC) from B. pumilus (44) and PAD from B. subtilis (9), which metabolize p-coumaric and ferulic acids in vivo and in vitro at similar rates, without salt addition. The FDC and PAD enzymes display strong amino acid sequence identity (88%), while PDC shares only 62 and 66% identity with FDC and PAD, respectively, with the most diverging domains being located in the N- and C-terminal protein regions. Construction of chimeric proteins is currently in progress in order to correlate the protein domain(s) with the substrate specificity and metabolic characteristics of these enzymes.
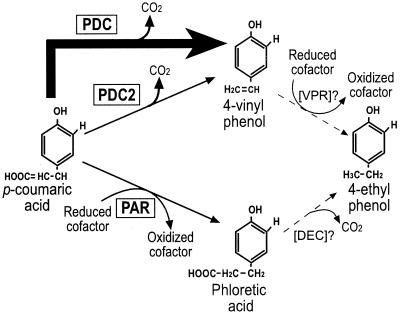
We have demonstrated that L. plantarum has three inducible activities for the degradation of p-coumaric and ferulic acids (Fig. 7), which we think may be involved in the stress response induced by phenolic acids. The functional PDC enzyme clearly confers a selective advantage on the wild-type strain, LPNC8, for growth in the presence of p-coumaric acid, while the growth of the LPD1 mutant at acidic pHs is strongly inhibited by p-coumaric acid. These results are consistent with those obtained recently for E. coli growth with inhibitory concentrations of ferulic or vanillic acid (45). Therefore, PDC synthesis in L. plantarum appears to be the most efficient cellular response to quickly convert p-coumaric acid into a less toxic derivative. The low PDC2 and PAR activities are much less efficient as detoxification systems, and their biological significance remains to be established. Two mechanisms are known to be involved in phenolic acid toxicity under acidic growth conditions: the dissipation of the ΔpH and a specific toxicity of phenolic acids. At pHs lower than their pKa (about 4.5), phenolic acids are essentially found in the protonated form. They can enter the bacterial cell by diffusion, while the released H+ protons depress the internal pH, thereby inhibiting bacterial growth. Recent studies have shown that in S. cerevisiae, the H+-ATPase which pumps protons out of the cell is induced by cinnamic acid at inhibitory concentrations (10). This proton pump could be activated in order to counteract dissipation of the transmembrane proton motive force and to restore the ΔpH (5). Such a response is probably the primary mechanism to overcome phenolic acid toxicity, since S. cerevisiae displays a very weak PAD activity (13), 5,000-fold weaker than the L. plantarum PDC. Similarly, the corresponding H+-ATPase of L. plantarum (40) could counteract the dissipation of ΔpH upon the entrance of phenolic acids. Proton pump activation uses ATP molecules, which are no longer available for biosynthesis purposes, explaining the lower growth rate and lower final biomass in medium supplemented with p-coumaric acid. The specific molecular toxicity of p-coumaric acid, which is evident in the LPD1 mutant, is considerably reduced by the high PDC activity in the wild-type strain, LPNC8.
FIG. 7.
Proposed pathway for the degradation of p-coumaric acid in L. plantarum. The arrow thickness represents the relative intensity of enzymatic activity. PDC, p-coumaric acid decarboxylase; PDC2: phenolic acid decarboxylase; PAR, phenolic acid reductase; VPR, putative 4-vinyl phenol reductase; DEC, putative phloretic acid decarboxylase.
In conclusion, knockout of the pdc gene from L. plantarum reveals the existence of two other inducible enzymatic activities involved in phenolic acid metabolism. However, these activities are about 100-fold lower than that of PDC. Therefore, PDC activity appears to be one of the major components of the stress response caused by phenolic acids, particularly in acidic media, which are the natural habitats of lactic acid bacteria. Our mutant provides a convenient model for studying mechanisms of the stress response which involve specific phenolic acid-dependent regulation systems. Such regulation systems are currently under investigation in our laboratory.
ACKNOWLEDGMENTS
We are grateful to Véronique Dartois (Microgenomics, San Diego, Calif.) for critical review of the manuscript, to Lars Axelsson (MATFORSK, Norwegian Food Research Institute, Osloveien, Norway) for the gift of strain LPNC8, and to Christine Bernard-Rojas for technical assistance.
This study was supported by the “Ministère de l'Education Nationale, de la Recherche et de la Technologie” and the “Conseil Régional de Bourgogne.”
REFERENCES
- 1.Aukrust T, Nes I F. Transformation of Lactobacillus plantarum with the plasmid pTV1 by electroporation. FEMS Microbiol Lett. 1988;52:127–132. [Google Scholar]
- 2.Bernard O, Bastin G, Stentelaire C, Lesage-Meessen L, Asther M. Mass balance modeling of vanillin production from vanillic acid by cultures of the fungus Pycnoporus cinnabarinus in bioreactors. Biotechnol Bioeng. 1999;65:558–571. doi: 10.1002/(sici)1097-0290(19991205)65:5<558::aid-bit9>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 3.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;60:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bult C J, White O, Olsen G J, Zhou L, Fleishmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kervalage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;23:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 5.Carmelo V, Santos H, Sa-Correia I. Effect of extracellular acidification on the activity of plasma membrane ATPase and on the cytosolic and vacuolar pH of Saccharomyces cerevisiae. Biochim Biophys Acta. 1997;1325:63–70. doi: 10.1016/s0005-2736(96)00245-3. [DOI] [PubMed] [Google Scholar]
- 6.Cavin J-F, Andioc V, Etievant P X, Diviès C. Ability of wine lactic acid bacteria to metabolize phenol carboxylic acids. Am J Enol Vitic. 1993;44:76–80. [Google Scholar]
- 7.Cavin J-F, Barthelmebs L, Diviès C. Molecular characterization of an inducible p-coumaric acid decarboxylase from Lactobacillus plantarum: gene cloning, transcriptional analysis, overexpression in Escherichia coli, purification and characterization. Appl Environ Microbiol. 1997;63:1939–1944. doi: 10.1128/aem.63.5.1939-1944.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavin J-F, Barthelmebs L, Guzzo J, Van Beeumen J, Samyn B, Travers J-F, Diviès C. Purification and characterization of an inducible p-coumaric acid decarboxylase from Lactobacillus plantarum. FEMS Microbiol Lett. 1997;147:291–295. doi: 10.1128/aem.63.5.1939-1944.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavin J-F, Dartois V, Diviès C. Gene cloning, transcriptional analysis, purification and characterization of phenolic acid decarboxylase from Bacillus subtilis. Appl Environ Microbiol. 1998;64:1466–1471. doi: 10.1128/aem.64.4.1466-1471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chambel A, Viegas C A, Sa-Correia I. Effect of cinnamic acid on growth and on plasma membrane H+-ATPase activity of Saccharomyces cerevisiae. Int J Food Microbiol. 1999;50:173–179. [Google Scholar]
- 11.Chen J D, Morrison D A. Construction and properties of a new insertion vector, pJDC9, that is protected by transcriptional terminators and useful for cloning of DNA from Streptococcus pneumoniae. Gene. 1988;64:155–164. doi: 10.1016/0378-1119(88)90489-1. [DOI] [PubMed] [Google Scholar]
- 12.Chow K T, Pope M K, Davies J. Characterization of a vanillic acid non-oxidative decarboxylation gene cluster from Streptomyces sp. D7. Microbiology. 1999;145:2393–2403. doi: 10.1099/00221287-145-9-2393. [DOI] [PubMed] [Google Scholar]
- 13.Clausen M, Lamb C J, Megnet R, Doerner P W. PAD1 encodes phenylacrylic acid decarboxylase which confers resistance to cinnamic acid in Saccharomyces cerevisiae. Gene. 1994;142:107–112. doi: 10.1016/0378-1119(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 14.Degrassi G, Polverino de Laureto P, Bruschi C V. Purification and characterization of ferulate and p-coumarate decarboxylase from Bacillus pumilus. Appl Environ Microbiol. 1995;61:326–332. doi: 10.1128/aem.61.1.326-332.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Man P J, Rogosa M, Sharpe M. A medium for the cultivation of Lactobacilli. J Appl Bacteriol. 1960;23:130–135. [Google Scholar]
- 16.De Vries R P, Michelsen B, Poulsen C H, Kroon P A, Van Den Heuvel R H H, Faulds C B, Williamson G, Van Den Hombergh J P T W, Visser J. The faeA genes from Aspergillus niger and Aspergillus tubingensis encode ferulic acid esterases involved in degradation of complex cell wall polysaccharides. Appl Environ Microbiol. 1997;63:4638–4644. doi: 10.1128/aem.63.12.4638-4644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Vries R P, Poulsen C H, Madrid S, Visser J. aguA, the gene encoding an extracellular α-glucuronidase from Aspergillus tubingensis, is specifically induced on xylose and not on glucuronic acid. J Bacteriol. 1998;180:243–249. doi: 10.1128/jb.180.2.243-249.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dower W J, Miller F, Ragsdale C W. High efficiency transformation of Escherichia coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edlin D A N, narbad A, Gasson M J, Dickinson J R, Lloyd D. Purification and characterization of hydroxycinnamate decarboxylase from Brettanomyces anomalus. Enzyme Microb Technol. 1998;22:232–239. [Google Scholar]
- 20.Etiévant P X, Issanchou S N, Marie S, Ducruet V, Flanzy C. Sensory impact of volatile phenols on red wine aroma: influence of carbonic maceration and time of storage. Sci Aliment. 1989;9:19–33. [Google Scholar]
- 21.Gasson M J, Kitamura Y, McLauchlan W R, Narbad A, Parr A J, Parsons E L H, Payne J, Rhodes M J C, Walton N J. Metabolism of ferulic acid to vanillin. J Biol Chem. 1998;273:4163–4170. doi: 10.1074/jbc.273.7.4163. [DOI] [PubMed] [Google Scholar]
- 22.Gibson T J. Studies on the Epstein-Barr virus genome. Ph.D. thesis. Cambridge, England: Cambridge University; 1984. [Google Scholar]
- 23.Goodey A R, Tubb R S. Genetic and biochemical analysis of the ability of Saccharomyces cerevisiae to decarboxylate cinnamic acids. J Gen Microbiol. 1982;128:2615–2620. [Google Scholar]
- 24.Hols P, Ferain T, Garmyn D, Bernard N, Delcour J. Use of homologous expression-secretion signals and vector-free stable chromosomal integration in engineering of Lactobacillus plantarum for α-amylase and levanase expression. Appl Environ Microbiol. 1994;60:1401–1413. doi: 10.1128/aem.60.5.1401-1413.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Z, Dostal L, Rosazza J P N. Microbial transformation of ferulic acid by Saccharomyces cerevisiae and Pseudomonas fluorescens. Appl Environ Microbiol. 1993;59:2244–2250. doi: 10.1128/aem.59.7.2244-2250.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalogeraki V S, Zhu J, Eberhard A, Madsen E L, Winans S. The phenolic vir gene inducer ferulic acid O-demethylated by the VirH2 protein of an Agrobacterium tumefaciens Ti plasmid. Mol Microbiol. 1999;34:512–522. doi: 10.1046/j.1365-2958.1999.01617.x. [DOI] [PubMed] [Google Scholar]
- 27.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Venter J C. The complete genome sequence of the hyperthermophilic sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;27:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 28.Lee Y W, Jin S, Sim W S, Nester E W. Genetic evidence for direct sensing of phenolic compounds by the VirA protein of Agrobacterium tumefaciens. Proc Natl Acad Sci USA. 1995;92:12245–12249. doi: 10.1073/pnas.92.26.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee Y W, Jin S, Sim W S, Nester E W. The sensing of plant signal molecules by Agrobacterium: genetic evidence for direct recognition of phenolic inducers by the VirA protein. Gene. 1996;179:83–88. doi: 10.1016/s0378-1119(96)00328-9. [DOI] [PubMed] [Google Scholar]
- 30.Leonardo M R, Dailly Y, Clark D P. Role of NAD in regulating the adhE gene of Escherichia coli. J Bacteriol. 1996;178:6013–6018. doi: 10.1128/jb.178.20.6013-6018.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lomascolo A, Stentelaire C, Asther M, Lesage-Meessen L. Basidiomycetes as new biotechnological tools to generate natural aromatic flavours for the food industry. Trends Biotechnol. 1999;17:282–289. doi: 10.1016/s0167-7799(99)01313-x. [DOI] [PubMed] [Google Scholar]
- 32.Lopez De Felipe F, Kleerebezem M, De Vos W M, Hugenholtz J. Cofactor engineering: a novel approach to metabolic engineering in Lactococcus lactis by controlled expression of NADH oxidase. J Bacteriol. 1998;180:3804–3808. doi: 10.1128/jb.180.15.3804-3808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narbad A, Gasson M J. Metabolism of ferulic acid via vanillin using a novel CoA-dependent pathway in a newly isolated strain of Pseudomonas fluorescens. Microbiology. 1998;144:1397–1405. doi: 10.1099/00221287-144-5-1397. [DOI] [PubMed] [Google Scholar]
- 34.Overhage J, Priefert H, Steinbüchel A. Biochemical and genetic analyses of ferulic acid catabolism in Pseudomonas sp. strain HR199. Appl Environ Microbiol. 1999;65:4837–4847. doi: 10.1128/aem.65.11.4837-4847.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Posno M, Leer R J, Van Luik N, Van Giezen M J F, Heuvelmans P T H M, Lokman B C, Pouwels P H. Incompatibility of Lactobacillus vectors with replicons derived from small cryptic Lactobacillus plasmids and segregational instability of the introduced vectors. Appl Environ Microbiol. 1991;57:1822–1828. doi: 10.1128/aem.57.6.1822-1828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segura A, Bünz P V, D'Argenio D A, Ornston L N. Genetic analysis of a chromosomal region containing vanA and vanB, genes required for conversion of either ferulate or vanillate to protocatechuate in Acinetobacter. J Bacteriol. 1999;181:3494–3504. doi: 10.1128/jb.181.11.3494-3504.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thurston P A, Tubb R S. Screening yeast strains for their ability to produce phenolic off-flavours: a simple method for determining phenols in wort and beer. J Inst Brew. 1981;87:177–179. [Google Scholar]
- 40.Tseng C-P, Jya-Ly T, Montville T J. Bioenergetic consequences of catabolic shifts by Lactobacillus plantarum in response to shifts in environmental oxygen and pH in chemostat cultures. J Bacteriol. 1991;173:4411–4416. doi: 10.1128/jb.173.14.4411-4416.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Venturi V, Zennaro F, Degrassi G, Okeke B C, Bruschi C V. Genetics of ferulic acid bioconversion to protocatechuic acid in plant-growth-promoting Pseudomonas putida WCS358. Microbiology. 1998;144:965–973. doi: 10.1099/00221287-144-4-965. [DOI] [PubMed] [Google Scholar]
- 42.Whiting G C, Carr J G. Metabolism of cinnamic acid and hydroxy-cinnamic acids by Lactobacillus pastorianus var. quinicus. Nature. 1959;184:1427–1428. doi: 10.1038/1841427a0. [DOI] [PubMed] [Google Scholar]
- 43.Yamane K, Kumano M, Kurita K. The 25 degrees-36 degrees region of the Bacillus subtilis chromosome: determination of the sequence of a 146-kb segment and identification of 113 genes. Microbiology. 1996;142:3047–3056. doi: 10.1099/13500872-142-11-3047. [DOI] [PubMed] [Google Scholar]
- 44.Zago A, Degrassi G, Bruschi C V. Cloning, sequencing, and expression in Escherichia coli of the Bacillus pumilus gene for ferulic acid degradation. Appl Environ Microbiol. 1995;61:4484–4486. doi: 10.1128/aem.61.12.4484-4486.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zaldivar J, Ingram L O. Effect of organic acids on the growth and fermentation of ethanologenic Escherichia coli LY01. Biotechnol Bioeng. 1999;66:203–210. doi: 10.1002/(sici)1097-0290(1999)66:4<203::aid-bit1>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]