Abstract
Contemporary microbial community analysis frequently involves PCR-amplified sequences of the 16S rRNA gene (rDNA). However, this technology carries the inherent problem of heterogeneity between copies of the 16S rDNA in many species. As an alternative to 16S rDNA sequences in community analysis, we employed the gene for the RNA polymerase beta subunit (rpoB), which appears to exist in one copy only in bacteria. In the present study, the frequency of 16S rDNA heterogeneity in bacteria isolated from the marine environment was assessed using bacterial isolates from the red alga Delisea pulchra and from the surface of a marine rock. Ten strains commonly used in our laboratory were also assessed for the degree of heterogeneity between the copies of 16S rDNA and were used to illustrate the effect of this heterogeneity on microbial community pattern analysis. The rock isolates and the laboratory strains were also used to confirm nonheterogeneity of rpoB, as well as to investigate the versatility of the primers. In addition, a comparison between 16S rDNA and rpoB PCR-DGGE (denaturing gradient gel electrophoresis)-based community analyses was performed using a DNA mixture of nine isolates from D. pulchra. Eight out of 14 isolates from D. pulchra, all rock isolates, and 6 of 10 laboratory strains displayed multiple bands for 16S rDNA when analyzed by DGGE. There was no indication of heterogeneity for either the rock isolates or the laboratory strains when rpoB was used for PCR-DGGE analysis. Microbial community pattern analysis using 16S rDNA PCR-DGGE showed an overestimation of the number of laboratory strains in the sample, while some strains were not represented. Therefore, the 16S rDNA PCR-DGGE-based community analysis was proven to be severely limited by 16S rDNA heterogeneity. The mixture of isolates from D. pulchra proved to be more accurately described using rpoB, compared to the 16S rDNA-based PCR-DGGE.
Community analysis of bacteria using molecular methods such as PCR amplification of the 16S rRNA gene (rDNA) in combination with denaturing or temperature gradient gel electrophoresis (DGGE or TGGE) is commonly performed in microbial ecology (15). Molecule-based community analysis is also increasingly employed in related fields such as ecotoxicology (23, 25). These methodologies have provided a new insight into microbial diversity and allow a more rapid, high-resolution description of microbial communities than that provided by the traditional approach of isolation of microorganisms. The use of 16S rDNA and PCR-DGGE or PCR-TGGE has often been combined with sequencing and subsequent identification of the species present in a sample (7, 16, 21). This is made possible through the extensive database of 16S rDNA sequences that has accumulated.
Increasingly, the banding pattern of the DGGE or TGGE gel is being used for community analysis by correlating the number of bands with environmental factors (22, 28) or by calculating different indices to trace changes in community structure with changes in environmental conditions (2, 18, 27). Due to the heterogeneity of 16S rDNA, recently reviewed by Fogel et al. (6), several bands per species will be seen in high-resolution PCR-DGGE analysis. However, little information has been published on the frequency of 16S rDNA heterogeneity in species isolated from the environment.
The intraspecies heterogeneity observed in a DGGE banding pattern is the result of the presence of multiple copies of the ribosomal genes and the fact that the gene copies have evolved differently (26). The amplified fragment of 16S rDNA will therefore appear as several bands on a DGGE or TGGE gel, instead of a single band that is representative of that particular species. The implications of such heterogeneity for community analysis have been discussed but not resolved (12, 17). The occurrence of heterogeneity in gene sequences in the GenBank database was investigated by Clayton et al. (1). These authors showed that between 48 and 82% of the species with two published sequences displayed heterogeneity that could not be explained by sequencing mistakes. We suggest that microbial community pattern analysis using 16S rDNA-based PCR-DGGE is significantly limited by its inherent heterogeneity.
A solution to the problem of 16S rDNA heterogeneity which can still capitalize on the advantages of PCR-DGGE is offered by the analysis of a gene that exists in a single copy. The gene should possess the same key attributes as 16S rDNA, namely, that it is common to all bacteria, that it has conserved as well as variable regions, and that it functions as an evolutionary clock. The gene for the RNA polymerase beta subunit, rpoB, is suggested to fulfill these criteria and can be used as an alternative to 16S rDNA in species identification (13, 14, 19). Therefore, a PCR product of this gene will result in a single band on a DGGE gel, making it possible to distinguish different species using DGGE or TGGE without having to sequence the individual bands. Furthermore, correlation and diversity measurements can thereby be applied directly to the DGGE or TGGE pattern.
This study was aimed at determining the frequency of intraspecies heterogeneity of 16S rDNA in strains isolated from the marine environment, as well as confirming the nonheterogeneity of rpoB and comparing 16S rDNA and rpoB community pattern analysis. The results presented in this study suggest that 16S rDNA-based PCR-DGGE community analysis is not suitable for microbial community analysis based on PCR-DGGE banding patterns. This study also shows that an alternative gene, such as rpoB, can be used successfully.
MATERIALS AND METHODS
Bacterial strains.
Fourteen randomly sampled bacterial isolates from Delisea pulchra and 14 randomly sampled isolates from a marine rock were used to investigate the frequency of 16S rDNA heterogeneity in environmental bacteria. Ten strains frequently used in our laboratory, hereafter called laboratory strains, (Pseudoalteromonas tunicata D2, Vibrio angustum S14, Bacillus subtilis, Staphylococcus epidermidis, Escherichia coli, Vibrio harveyi, V. fischeri, Serratia liquefaciens MG1, Staphylococcus aureus, and Helicobacter pylori) were also assessed for heterogeneity and for the effect of heterogeneity on microbial community pattern analysis. To confirm the nonheterogeneity of rpoB, DNA from the rock isolates and the laboratory strains was amplified using the rpoB primers and analyzed with DGGE. A mixture of DNAs from nine isolates from D. pulchra was used to compare 16S rDNA and rpoB microbial community pattern analyses. The nine isolates from D. pulchra represented the phylogenetic diversity of bacteria from the midsection of the plant and are here given with their tentative identification based on sequence comparison of approximately 500 bp from 16S rDNA (γ-proteobacterial strain HTB111, V. rumoiensis, B. licheniformis, Ferrimonas balearica, Pseudomonas luteola, Microbulbifer hydrolyticus, Prionitis lanceolata gall symbiont, B. cohnii, and Agrobacterium atlanticum).
Construction of rpoB primers.
The sequences for rpoB from E. coli, B. subtilis, S. aureus, and H. pylori (accession numbers AE000472, 2632267, 677848, and AE000625) were compared, and two regions containing conserved sequences were used to construct primers. The primer regions for the three species were not 100% identical. However, degenerate primers could not be used in conjunction with DGGE since they themselves gave rise to multiple products (data not shown). Mismatches in the primers therefore had to be accepted. Primers that gave PCR products for the 10 type strains were constructed and subsequently used for all of the bacteria in this study.
Isolation of bacteria from D. pulchra and a marine rock.
D. pulchra and the marine rock were sampled from Botany Bay, Sydney, New South Wales, Australia, in March 1999. The bacteria from D. pulchra and the rock were isolated by vortexing the sample in sterile seawater for 5 min and thereafter spreading 0.1 ml of the sample on plates containing Oxoid marine agar 2216. All colonies that visibly differed from each other in morphology and color were further isolated.
DNA extraction.
One milliliter of an overnight liquid culture of the individual bacteria was spun down, and the supernatant was discarded. One gram of silica zirconium beads and ∼1.5 ml of XS buffer (1 g of sodium xanthogenate, 20 ml of 4 M ammonium acetate, 10 ml of 1 M Tris (pH 7.4), 4 ml of 0.45 M EDTA [pH 8] per 100 ml) were added, and the cells were lysed in a Bio 101 Fastprep bead beater for 30 s at 5.5 m s−1. The samples were put on ice for 30 min and then spun for 30 min at 21,000 × g. One milliliter of supernatant was transferred, and 100 μl of 3 M sodium acetate (pH 5.2) and 0.9 ml isopropanol were added. The samples were left overnight at −20°C, and the DNA was collected through centrifugation at 4°C. The DNA pellet was washed in 70% ethanol and dissolved in Tris-EDTA buffer.
PCR conditions.
The 16S rDNA primers used were 27F (5′-GAGTTTGATCCTGGCTCAG-3′) and 536R (5′-GTATTACCGCGGCTGCTG-3′). These 16S rDNA primers are not degenerate, as were those described by Suzuki and Giovannoni (24), since degenerate primers give rise to multiple products with different melting profiles for the same strain (data not shown). The rpoB primers used were rpoB1698f (5′-AACATCGGTTTGATCAAC-3′; corresponding to E. coli position 1643) and rpoB2041r (5′-CGTTGCATGTTGGTACCCAT-3′; corresponding to E. coli position 2041). A GC clamp (5′-CGCCCCCCGCGCCCCGCGCCCGGCCCGCCGCCCCCGCCCC-3′) was added to the forward primers.
A 2.5-μl template sample (∼100 ng of DNA) was added to a 47.5-μl PCR mixture containing 5 μl of Sigma REDtaq buffer, 2.5 mM each deoxynucleoside triphosphate, 25 pmol of each primer, 20 μg of bovine serum albumin, sterile filtered milliQ water, and 1 μl Taq polymerase (Sigma REDTaq). The PCR protocol consisted of a denaturing step of 94°C for 5 min, followed by 25 cycles of denaturing for 30 s at 94°C, annealing for 1.5 min at 50°C, and a 1.5-min extension at 72°C. A final extension step of 72°C for 10 min was then performed. The same PCR mixture was used for the rpoB-based PCR but with the primers changed to rpoB1698f and rpoB2041r. The MgCl2 concentration was also raised to 2.6 mM. The cycling conditions were the same as for the 16S rDNA amplification, with the exception that certain strains (F. balearica, γ-proteobacterium strain HTB111, S. epidermidis, and S. aureus) had an annealing temperature of 40°C for the first 6 cycles, followed by 19 cycles with a 50°C annealing temperature. The same cycling conditions also applied to the mixtures containing these strains. The PCR products were run on a 2% agarose gel containing ethidium bromide and analyzed using the Bio-Rad Gel Doc imaging system.
DGGE conditions.
The PCR products were run on a 6% polyacrylamide gel in a 45 to 65% denaturing gradient using the Bio-Rad DCode system. The running conditions were 75 V at 60°C for 15 to 17 h. The DGGE gel was stained with ethidium bromide and analyzed using the Bio-Rad Gel Doc imaging system.
RESULTS
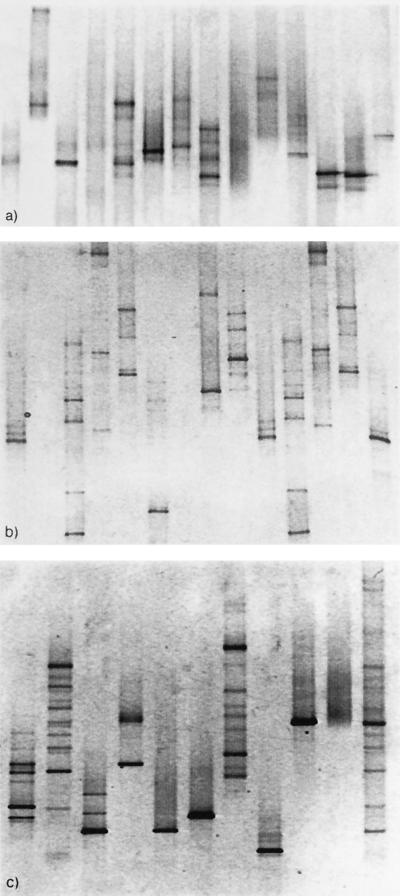
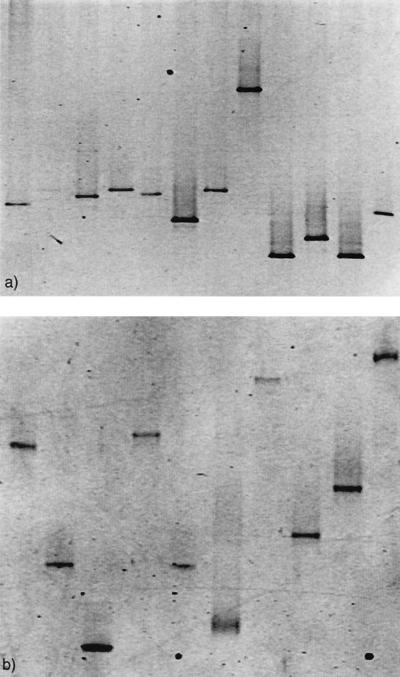
Eight of the 14 isolates from D. pulchra and 12 of the isolates from the marine rock showed multiple bands for the 16S rDNA PCR amplification, indicating intraspecies heterogeneity (Fig. 1a and b). For two of the marine rock isolates, the 16S rDNA could not be amplified and these were therefore excluded from further analysis (Fig. 1b, lanes 2 and 7). Of the 10 laboratory strains, 6 displayed intraspecies 16S rDNA heterogeneity (Fig. 1c, lanes 1 to 9). In the DGGE pattern for the mixture of the 10 laboratory strains, at least 12 bands could easily be detected but only some of the bands could be directly related to a single species. This indicates that 16S rDNA intraspecies heterogeneity severely hampers community pattern analysis. When the 12 remaining marine rock isolates, as well as the 10 laboratory strains, were amplified using rpoB, only one band per species could be seen (Fig. 2a and b), indicating a single copy of rpoB.
FIG. 1.
DGGE banding pattern of 16S rDNA PCR amplification of 14 D. pulchra isolates (a), 14 marine rock isolates (b), and 10 laboratory strains (c). Lanes: 1, P. tunicata; 2, V. angustum; 3, B. subtilis; 4, S. epidermidis; 5, E. coli; 6, V. harveyi; 7, V. fischeri; 8, S. liquefaciens; 9, S. aureus; 10, H. pylori; 11, a mixture of all 10 strains.
FIG. 2.
DGGE banding pattern of rpoB PCR amplification of 12 marine rock isolates (a) and 10 laboratory species (b). Lanes: 1, P. tunicata; 2, V. angustum; 3, B. subtilis; 4, S. epidermidis; 5, E. coli; 6, V. harveyi; 7, V. fischeri; 8, S. liquefaciens; 9, S. aureus; 10, H. pylori.
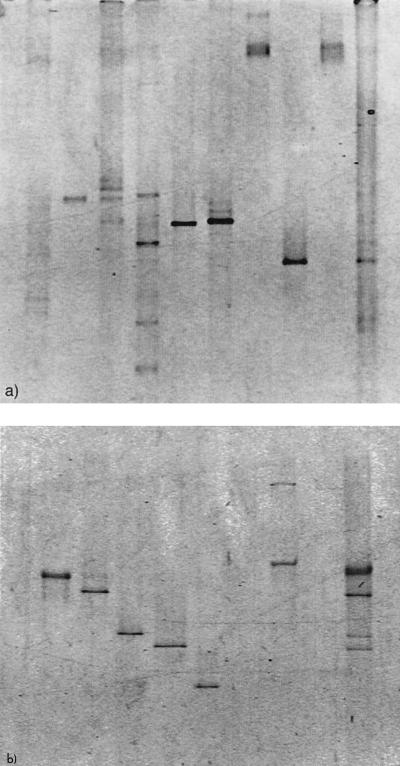
Nine bacterial isolates from D. pulchra were selected for a comparison between 16S rDNA and rpoB for PCR-DGGE or PCR-TGGE community analysis. DNA from two of the isolates could not be amplified with either set of primers (Fig. 3a and b, lanes 7 and 9). DNA from one further isolate was weakly amplified using the 16S rDNA primers but not at all using the rpoB primers (Fig. 3a and b, lane 1). We therefore assumed that DNA from seven isolates could be amplified in the mixture using 16S rDNA primers and DNA from six isolates could be amplified using the rpoB primers. We found that the 16S rDNA banding pattern of the mixture of isolates from D. pulchra revealed at least eight bands (Fig. 3a, lane 10). Moreover, some of the bands could not be related to individual isolates. When the mixture was amplified using rpoB, a clear pattern emerged (Fig. 3b). Five bands representing six possible isolates could be detected using rpoB, and these bands could easily be related to the individual isolates (Fig. 3b). A larger by-product could be seen when some isolates were amplified individually (Fig. 3b, lane 8), but these bands could not be seen when DNAs from the isolates were amplified in a mixture.
FIG. 3.
Comparison of DGGE banding pattern between 16S rDNA (a) and rpoB (b) PCR amplifications of a mixture of nine bacterial isolates from D. pulchra. Lanes: 1, γ-proteobacterial strain HTB111; 2, V. rumoiensis; 3, B. lichenformis; 4, F. balearica; 5, P. luteola; 6, M. hydrolyticus; 7, Prionitis lanceolata gall symbiont; 8, B. cohnii; 9, A. atlanticum; 10, a mixture of all nine isolates.
A BLAST search of the National Center for Biotechnology Information database using the primer sequences, and allowing for mismatches, resulted in a large number of hits of rpoB sequences from bacteria. This is in agreement with the findings in the present study, in which the rpoB primers constructed successfully amplified rpoB fragments from a variety of bacteria. It was, however, necessary to lower the annealing temperature to 40°C for some bacterial DNAs (S. epidermidis, S. aureus, y-proteobacterial strain HTB111, and F. balearica).
DISCUSSION
The results obtained in this study suggest that 16S rDNA heterogeneity is typical of bacteria isolated from the environment (Fig. 1a and b). This implies that the appearance of different bands in the analysis of 16S rDNA PCR products from mixed communities is not a measure of species diversity, as can be seen in the banding pattern for the mixture of 10 laboratory strains (Fig. 1c). Rather, it is sequence diversity, including that of intraspecies variation, that is reflected in the 16S rDNA banding pattern (27). Obviously, a comparison of sequence diversity does not provide the same information as a comparison of species diversity. Sequence diversity will change depending on how many bands the individual species of the community give rise to and will therefore not necessarily reflect the true changes in species diversity. It is clear that diversity indices, and correlations, based on the banding patterns from 16S rDNA PCR-DGGE are not suitable for the comparison of changes in microbial communities.
It is sometimes argued that the 16S rDNA-based DGGE patterns obtained are highly reproducible and therefore accurately reflect the community that is present (8). Both the PCR and DGGE steps in this study were performed numerous times and proved to give reproducible results for the 16S rDNA banding pattern. This demonstrates that the heterogeneity is not an artifact of the PCR step and that reproducibility is not related, in this or in any other case, to accuracy.
The results from this study demonstrate the need to employ a gene other than the 16S rRNA gene in PCR-DGGE microbial community analysis. Based on the results obtained, it is suggested that rpoB is one such suitable gene for PCR-DGGE community analysis, since only one band was observed for each bacterial isolate and the DGGE banding pattern from the mixture could clearly be related to single isolates (Fig. 3b).
A comparison of rpoB and 16S rDNA for the identification of members of the family Enterobacteriaceae, performed by Mollet et al. (13), showed that in 85 of 91 cases rpoB demonstrated higher resolution than did the 16S rDNA sequences in distinguishing between strains. They further demonstrated that rpoB exhibited between 1 and 15.4% more variability than the 16S rRNA gene in 82 strains. Kim et al. (10) identified 44 mycobacterial species and 107 clinical isolates based on rpoB amplification and sequencing. All strains appeared as separate entities in the rpoB phylogenetic tree, including one species (Mycobacterium kansasii) that cannot be separated from M. gastri using 16S rRNA gene comparison. These studies indicate that the rpoB gene is also highly suitable for species identification through sequencing.
It is noteworthy that the heterogeneity of the 16S rDNA copies makes species identification through sequencing difficult. The sequences of different copies of 16S rDNA within a species can differ by as much as 6.5% (26). Earlier work by Clayton et al. (1) showed that 48 to 82% of the species with more than one sequence in the GenBank database had intraspecies differences (1). Nübel et al. reported heterogeneity in Paenibacillus polymyxa and discussed its implications for species identification and community analysis (17). To date, however, neither of these articles has had much impact on studies of 16S rDNA-based community analysis. In fact, relatively few publications have attempted to address the problem related to the heterogeneity of 16S rDNA (4, 5).
Two different reviews of PCR-based microbial community analysis have been published in the past 2 years (15, 29). Both of these discussed the problems with heterogeneity, heteroduplex formation, and other biases during PCR. The problem with heterogeneity can be solved, as we have shown here, but the possibility of heteroduplex formation still exists. This is a problem that can occur in PCR products of any gene when a mixed-species template is used. The occurrence of heteroduplexes increases with increased similarity between sequences (30). This implies that intraspecies heteroduplex formation due to heterogeneous genes is more common than interspecies heteroduplex formation. Recommendations on how to minimize heteroduplex formation have been given, such as increasing the primer concentration and decreasing the number of cycles during PCR, but are not often considered. Recognition of heteroduplexes through sequencing and computer analysis can be carried out to a certain extent (11), but if community analysis using PCR-DGGE or PCR-TGGE is to be a rapid tool, this is not a lasting solution. The results described in this paper suggest that a nonheterogeneous gene may form the basis for future microbial community analysis. Use of a nonheterogeneous gene should also make it more feasible to understand and find solutions to heteroduplex formation through modified PCR conditions. Interestingly, we note that heteroduplex formation has so far not been observed for rpoB amplification.
The need to amplify at 40°C in order to detect certain bacteria can be a result of the mismatches to the primer sites or to regions flanking the primer site which affect annealing differently for different species (9). A larger by-product could also be seen for the bacterial DNA amplified at 40°C using rpoB primers (Fig. 3b, lane 8). However, this product was not present when these strains were amplified together in the mixture (Fig. 3b, lane 10). Preferential amplification during PCR (3, 19) was found to be the case for the rpoB genes since roughly the same amount of template DNA from the different strains produced different amounts of PCR product. This was, however, also the case for the 16S rDNA primers. The problem of preferential amplification, as well as relatively low annealing temperature, when the rpoB primers reported here are used may be addressed when a larger set of rpoB gene sequences becomes available. We are presently sequencing a 1,000-bp stretch of the gene in the species isolated in this study in order to generate an extended library for rpoB sequence information on environmental bacteria.
We suggest that introduction of a new gene for PCR-DGGE or PCR-TGGE microbial community analysis, such as rpoB, will provide considerably more reliable data and hence allow much-improved understanding of microbial diversity and community composition in environmental habitats.
ACKNOWLEDGMENTS
Strains isolated from a rock were kindly donated by Sharon Longford, University of New South Wales, Sydney, New South Wales, Australia.
Contributions to Ingela Dahllöf by Jubileumsfonden at Göteborg University are gratefully acknowledged. This work was supported by Australian Research Council funds to Staffan Kjelleberg and by the Centre for Marine Biofouling and Bio-Innovation at the University of New South Wales.
REFERENCES
- 1.Clayton R A, Sutton G, Hinkle P S, Bult C, Fields C. Intraspecific variation in small-subunit rRNA sequences in Genbank—why single sequences may not adequately represent prokaryotic taxa. Int J Syst Bacteriol. 1995;45:595–599. doi: 10.1099/00207713-45-3-595. [DOI] [PubMed] [Google Scholar]
- 2.El Fantroussi S, Verschuere L, Verstraete W, Top E M. Effect of phenylurea herbicides on soil microbial communities estimated by analysis of 16S rRNA gene fingerprints and community-level physiological profiles. Appl Environ Microbiol. 1999;65:982–988. doi: 10.1128/aem.65.3.982-988.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farrelly V, Rainey F A, Stackebrandt E. Effect of genome size and rRNA gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Felske A, Vancanneyt M, Kersters K, Akkermans A D L. Application of temperature-gradient gel electrophoresis in taxonomy of coryneform bacteria. Int J Syst Bacteriol. 1999;49:113–121. doi: 10.1099/00207713-49-1-113. [DOI] [PubMed] [Google Scholar]
- 5.Ferris M J, Ward D M. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1997;63:1375–1381. doi: 10.1128/aem.63.4.1375-1381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fogel G B, Collins C R, Li J, Brunk C F. Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb Ecol. 1999;38:93–113. doi: 10.1007/s002489900162. [DOI] [PubMed] [Google Scholar]
- 7.Fuhrman J A, Davies A A. Widespread Archaea and novel Bacteria from the deep sea as shown by 16S rRNA gene sequences. Mar Ecol Prog Ser. 1997;150:275–285. [Google Scholar]
- 8.Gelsomino A, Keijzer-Wolters A C, Cacco G, van Elsas J D. Assessment of bacterial community structure in soil by polymerase chain reaction and denaturing gradient gel electrophoresis. J Microbiol Methods. 1999;38:1–15. doi: 10.1016/s0167-7012(99)00054-8. [DOI] [PubMed] [Google Scholar]
- 9.Hansen M C, Tolkernielsen T, Givskov M, Molin S. Biased 16S rDNA PCR amplification caused by interference from DNA flanking the template region. FEMS Microbiol Ecol. 1998;26:141–149. [Google Scholar]
- 10.Kim B J, Lee S H, Lyu M A, Kim S J, Bai G H, Chae G T, Kim E C, Cha C Y, Kook Y H. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB) J Clin Microbiol. 1999;37:1714–1720. doi: 10.1128/jcm.37.6.1714-1720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kopczynski E D, Bateson M M, Ward D M. Recognition of chimeric small-subunit ribosomal DNAs composed of genes from uncultivated microorganisms. Appl Environ Microbiol. 1994;60:746–748. doi: 10.1128/aem.60.2.746-748.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez-Murcia A J, Anton A I, Rodriguez-Valera F. Patterns of sequence variation in two regions of the 16S rRNA multigene family of Escherichia coli. Int J Syst Bacteriol. 1999;49:601–610. doi: 10.1099/00207713-49-2-601. [DOI] [PubMed] [Google Scholar]
- 13.Mollet C, Drancourt M, Raoult D. rpoB sequence analysis as a novel basis for bacterial identification. Mol Microbiol. 1997;26:1005–1011. doi: 10.1046/j.1365-2958.1997.6382009.x. [DOI] [PubMed] [Google Scholar]
- 14.Morse R, Collins M D, Ohanlon K, Wallbanks S, Richardson P T. Analysis of the beta subunit of DNA-dependent RNA polymerase does not support the hypothesis inferred from 16S rRNA analysis that Oenococcus oeni (formerly Leuconostoc oenos) is a tachytelic (fast-evolving) bacterium. Int J Syst Bacteriol. 1996;46:1004–1009. doi: 10.1099/00207713-46-4-1004. [DOI] [PubMed] [Google Scholar]
- 15.Muyzer G, Smalla K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie van Leeuwenhoek. 1998;73:127–141. doi: 10.1023/a:1000669317571. [DOI] [PubMed] [Google Scholar]
- 16.Nielsen A T, Liu W T, Filipe C, Grady L, Molin S, Stahl D A. Identification of a novel group of bacteria in sludge from a deteriorated biological phosphorus removal reactor. Appl Environ Microbiol. 1999;65:1251–1258. doi: 10.1128/aem.65.3.1251-1258.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nübel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann R I, Ludwig W, Backhaus H. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol. 1996;178:5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nübel U, Garcia-Pichel F, Kuhl M, Muyzer G. Quantifying microbial diversity: morphotypes, 16S rRNA genes, and carotenoids of oxygenic phototrophs in microbial mats. Appl Environ Microbiol. 1999;65:422–430. doi: 10.1128/aem.65.2.422-430.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palenik B. Polymerase evolution and organism evolution. Curr Opin Gen Dev. 1992;2:931–936. doi: 10.1016/s0959-437x(05)80118-2. [DOI] [PubMed] [Google Scholar]
- 20.Polz M F, Cavanaugh C M. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol. 1998;64:3724–3730. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rölleke S S, Gurtner C, Drewello U, Drewello R, Lubitz W, Weissmann R. Analysis of bacterial communities on historical glass by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. J Microbiol Methods. 1999;36:107–114. doi: 10.1016/s0167-7012(99)00015-9. [DOI] [PubMed] [Google Scholar]
- 22.Sievert S M, Brinkhoff T, Muyzer G, Ziebis V, Kuever J. Spatial heterogeneity of bacterial populations along an environmental gradient at a shallow submarine hydrothermal vent near Milos Island (Greece) Appl Environ Microbiol. 1999;65:3834–3842. doi: 10.1128/aem.65.9.3834-3842.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smit E, Leeflang P, Wernars K. Detection of shifts in microbial community structure and diversity in soil caused by copper contamination using amplified ribosomal DNA restriction analysis. FEMS Microbiol Ecol. 1997;23:249–261. [Google Scholar]
- 24.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torsvik V, Daae F L, Sandaa R A, Øvreas L. Novel techniques for analysing microbial diversity in natural and perturbed environments. J Biotechnol. 1998;64:53–62. doi: 10.1016/s0168-1656(98)00103-5. [DOI] [PubMed] [Google Scholar]
- 26.Ueda K, Seki T, Kudo T, Yoshida T, Kataoka M. Two distinct mechanisms cause heterogeneity of 16S rRNA. J Bacteriol. 1999;181:78–82. doi: 10.1128/jb.181.1.78-82.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Hannen E J, Mooij W, van Agterveld M P, Gons H J, Laanbroek H J. Detritus-dependent development of the microbial community in an experimental system: qualitative analysis by denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1999;65:2478–2484. doi: 10.1128/aem.65.6.2478-2484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Hannen E J, Zwart G, van Agterveld M P, Gons H J, Ebert J, Laanbroek H J. Changes in bacterial and eukaryotic community structure after mass lysis of filamentous cyanobacteria associated with viruses. Appl Environ Microbiol. 1999;65:795–801. doi: 10.1128/aem.65.2.795-801.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Wintzingerode F, Göbel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 30.Wang G C Y, Wang Y. Frequency of formation of chimeric molecules is a consequence of PCR coamplification of 16S rRNA genes frommixed bacterial genomes. Appl Environ Microbiol. 1997;63:4645–4650. doi: 10.1128/aem.63.12.4645-4650.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]