Abstract
For ethanol production from lignocellulose, the fermentation of xylose is an economic necessity. Saccharomyces cerevisiae has been metabolically engineered with a xylose-utilizing pathway. However, the high ethanol yield and productivity seen with glucose have not yet been achieved. To quantitatively analyze metabolic fluxes in recombinant S. cerevisiae during metabolism of xylose-glucose mixtures, we constructed a stable xylose-utilizing recombinant strain, TMB 3001. The XYL1 and XYL2 genes from Pichia stipitis, encoding xylose reductase (XR) and xylitol dehydrogenase (XDH), respectively, and the endogenous XKS1 gene, encoding xylulokinase (XK), under control of the PGK1 promoter were integrated into the chromosomal HIS3 locus of S. cerevisiae CEN.PK 113-7A. The strain expressed XR, XDH, and XK activities of 0.4 to 0.5, 2.7 to 3.4, and 1.5 to 1.7 U/mg, respectively, and was stable for more than 40 generations in continuous fermentations. Anaerobic ethanol formation from xylose by recombinant S. cerevisiae was demonstrated for the first time. However, the strain grew on xylose only in the presence of oxygen. Ethanol yields of 0.45 to 0.50 mmol of C/mmol of C (0.35 to 0.38 g/g) and productivities of 9.7 to 13.2 mmol of C h−1 g (dry weight) of cells−1 (0.24 to 0.30 g h−1 g [dry weight] of cells−1) were obtained from xylose-glucose mixtures in anaerobic chemostat cultures, with a dilution rate of 0.06 h−1. The anaerobic ethanol yield on xylose was estimated at 0.27 mol of C/(mol of C of xylose) (0.21 g/g), assuming a constant ethanol yield on glucose. The xylose uptake rate increased with increasing xylose concentration in the feed, from 3.3 mmol of C h−1 g (dry weight) of cells−1 when the xylose-to-glucose ratio in the feed was 1:3 to 6.8 mmol of C h−1 g (dry weight) of cells−1 when the feed ratio was 3:1. With a feed content of 15 g of xylose/liter and 5 g of glucose/liter, the xylose flux was 2.2 times lower than the glucose flux, indicating that transport limits the xylose flux.
To obtain an economically feasible industrial process for ethanol production from lignocellulose, it is necessary to ferment all sugars present with high yields and productivities (53). The commonly used Saccharomyces cerevisiae has many advantages as an ethanol producer, such as fast sugar consumption, high ethanol yield from hexoses, and high resistance to inhibitory compounds that are present in the hydrolysates. However, a major drawback is that S. cerevisiae cannot utilize the pentose sugar xylose, only its isomer xylulose. In xylose-utilizing yeasts, the conversion from xylose to xylulose is a two-step process catalyzed by xylose reductase (XR) and xylitol dehydrogenase (XDH) (10), whereas bacteria perform the conversion in one step with xylose isomerase (XI) (23).
Xylose fermentation by recombinant S. cerevisiae carrying heterologous XYL1 and XYL2 genes from Pichia stipitis, which encode XR and XDH, respectively, has resulted mainly in xylitol formation (24, 44, 48). Similarly, if xylA from Thermus thermophilus, which encodes XI, is introduced into S. cerevisiae, then only limited xylose fermentation is observed (47). Limited xylose fermentation by recombinant S. cerevisiae has been ascribed to poor xylose uptake (9, 24, 25), a cofactor imbalance generated by the discrepancy in cofactor usage by XR and XDH (8, 24, 49), limitations in the pentose phosphate pathway (12, 24, 38, 48), and insufficient induction or activation of ethanologenic enzymes (5, 17, 20, 29). When homologous XKS1, which encodes xylulokinase (XK), was overexpressed in a Saccharomyces sp. strain carrying XYL1 and XYL2, the ethanol yield and the xylose uptake rate increased under oxygen-limited conditions, but xylitol was still a major by-product (22).
Although the shortcomings of xylose fermentation by recombinant S. cerevisiae have been investigated in several studies, data from anaerobic fermentations do not exist and quantitative data are sparse. Chemostat cultivations in which growth rate and concentrations of substrates and products are constant enable quantitative determinations of metabolic fluxes. Analysis of xylose fluxes is the first step towards identifying causes for by-product formation and low productivity during xylose fermentation. A stable recombinant strain was constructed by integration of the XYL1 and XYL2 genes from P. stipitis and the homologous XKS1 gene under control of the PGK1 promoter into the chromosomal HIS3 locus of S. cerevisiae. Metabolic fluxes were determined for different xylose-to-glucose ratios in chemostat cultivations under anaerobic conditions. Glucose was used as a cosubstrate since the recombinant strain was unable to grow on xylose in the absence of oxygen.
MATERIALS AND METHODS
Strains and plasmids.
Escherichia coli DH5α [F− Φ80 dlacZΔM15 Δ(lacZYA-argF) U169 deoR recA1 endA1 hsdR17(rK−, mK+) supE44 λ− thi-1 gyrA96 relA1] (GIBCO BRL, Gaithersburg, Md.) was used for subcloning. S. cerevisiae CEN.PK113-7A (MATa his3-Δ1 MAL2-8C SUC2) (18) was used as the recipient yeast strain for the integrating plasmid YIpXR/XDH/XK. All strains were stored frozen at −80°C. Agar plates streaked from the frozen stocks were used to inoculate the precultures.
Plasmids used for cloning of the XYL1 and XYL2, XKS1, and HIS3 genes were pY7 (46), pXks (B. Johansson, C. Christensson, T. Hobley, and B. Hahn-Hägerdal, submitted for publication), and YDp-H (3), respectively. YDp-H was obtained from Jörg Hauf (Scientific Research and Development GmbH, Oberursel, Germany).
Media.
S. cerevisiae CEN.PK PK113-7A was grown in YPD medium (40) for transformation. In all other experiments, a defined medium including vitamins and trace elements was used (50). l-Histidine was added at 50 mg per liter for strain CEN.PK PK113-7A. For the continuous cultivations, the medium was also supplemented with ergosterol and unsaturated fatty acids in the form of Tween 80 (Sigma, St. Louis, Mo.) (1, 2). Ergosterol and Tween 80 were dissolved in boiling 96% (vol/vol) ethanol to final concentrations of 0.01 and 0.42 g/liter, respectively. Bacterial strains were grown in Luria-Bertani medium (35). Transformants were selected by adding ampicillin (50 mg/liter). For growth on solid media, 20 g of agar per liter was added.
Nucleic acid manipulations.
Standard techniques for nucleic acid manipulations were used (35). Plasmids were prepared using the QIAGEN Maxi Plasmid Purification Kit (Qiagen GmbH, Hilden, Germany). Restriction enzymes and other modifying enzymes were purchased from Boehringer Mannheim Scandinavia AB (Bromma, Sweden). DNA fragments separated by agarose gel electrophoresis were purified with the QIAquick Gelextraction Kit (Qiagen GmbH).
Construction of integrating vector expressing XYL1, XYL2, and XKS1.
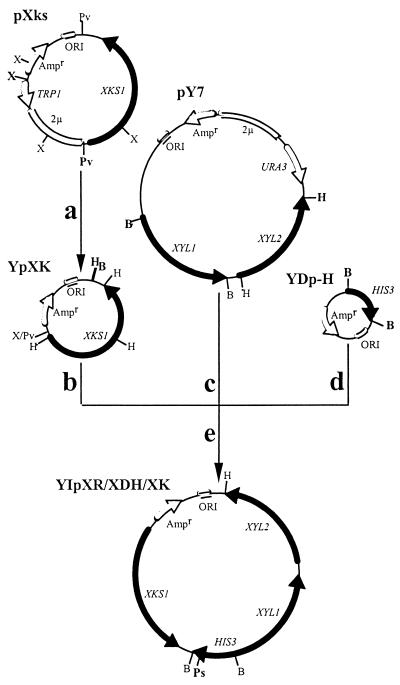
The 2μ origin of replication and the P-ribosyl-anthranilate isomerase (TRP1) gene were removed by partial digestion with XmnI and PvuII from plasmid pXks, a yeast episomal plasmid carrying the XKS1 gene under the control of the phosphoglycerate kinase (PGK) promoter and terminator (Johansson et al., submitted) (Fig. 1a). This construct has two base substitutions introduced in the XKS1 gene upstream from the start codon to maximize translational efficiency; furthermore, the codon for the N-terminal amino acid was altered to increase the protein stability of the gene product. The remaining fragment of 6,465 bp was recircularized by self-ligation and then partially digested with HindIII and BamHI, creating a fragment of 6,435 bp (Fig. 1b). The genes XYL1 and XYL2 under the control of alcohol dehydrogenase (ADH) and PGK promoters, respectively, were excised from pY7 (46) by partial digestion with HindIII and BamHI, yielding a fragment of 6,153 bp (Fig. 1c). The 1,150-bp HIS3 cassette was excised from YDp-H (3) by BamHI (Fig. 1d). Finally, the plasmid carrying XKS1 was ligated to the HIS3 cassette and the XYL1-XYL2 fragment, yielding YIpXR/XDH/XK, an integrating vector carrying XYL1, XYL2, and XKS1 and with HIS3 as a selection marker (Fig. 1e). Restriction enzyme digestions verified the map of the constructed vector.
FIG. 1.
Schematic flow sheet for the construction of the integrating vector YIpXR/XDH/XK harboring XYL1, XYL2, and XKS1. XYL1 is under control of the ADH promoter and terminator, whereas both XYL2 and XKS1 are under control of the PGK promoter and terminator. (a) Partial digestion of pXks with XmnI and PvuII to remove TRP1 and the 2μ origin of replication, followed by recircularization of the remaining fragment to create YpXK. (b) Partial digestion of YpXK with HindIII and BamHI. (c) Excision of XYL1 and XYL2 from pY7 by partial digestion with HindIII and BamHI. (d) Excision of the HIS3 cassette from YDp-H with BamHI. (e) Ligation of the three fragments to create YIpXR/XDH/XK. This integrating vector was linearized with PstI to target integration to HIS3 in the chromosome. The restriction sites are labeled as follows: B, BamHI; H, HindIII; Ps, PstI; Pv, PvuII; X, XmnI; and X/Pv, hybrid site of XmnI and PvuII. Only relevant restriction sites are shown for each step, and those that were used are in bold.
Yeast strain transformation.
The lithium acetate method was used for transformation (37).
Continuous cultivations.
Continuous fermentations were conducted anaerobically in computer-controlled glass bioreactors (Belach Bioteknik AB, Stockholm, Sweden) at 30°C with a stirring speed of 200 rpm. The working volume of the reactors was 600 ml, and the pH was adjusted to 5.5 with 3 M KOH. Anaerobic conditions were maintained by sparging with nitrogen containing less than 5 ppm of O2 (ADR class2 1A; AGA, Sundbyberg, Sweden) at a constant gas flow of 0.3 liter/min controlled by mass flow meters (El-Flow, Insat, Switzerland). The feed reservoir was degassed before connection to the fermentor. During the continuous cultivation, a rubber ball filled with nitrogen connected to the reservoir prevented a vacuum buildup when medium was pumped out. The off-gas condensers were cooled to 4°C. Antifoam (Dow Corning Antifoam RD Emulsion; BDH Laboratory Supplies, Poole, United Kingdom) was included in the feed at a concentration of 0.5 ml/liter of feed.
Precultures, 200 ml of medium in 1-liter baffled shake-flasks, were incubated overnight at 30°C and were shaken at 140 rpm in an orbital incubator (INR-200; Gallenkamp, Leicester, United Kingdom). Cells were harvested by centrifugation (5,000 × g, 5 min, 4°C) and were washed once with a solution containing 9 g of NaCl/liter before inoculation into the fermentors.
Fermentations were started as anaerobic batch cultures with glucose (5 g/liter) as the sole carbon source. When the batch fermentation was depleted of glucose, as detected by a rapid drop in the carbon dioxide evolution rate, a feed of fresh medium was started, yielding a dilution rate (D) of 0.06 h−1. The volume was kept at 600 ml by continuous removal of broth through a siphon. The total concentration of carbon source in the feed was 20 g/liter. Steady states with various ratios of xylose and glucose were analyzed. One set of fermentations was started with 20 g of glucose/liter, and after a steady state was reached, the feed was switched to 5 g of xylose/liter plus 15 g of glucose/liter, followed by 10 g of xylose/liter plus 10 g of glucose/liter and, finally, 15 g of xylose/liter plus 5 g of glucose/liter. A second set of fermentations was operated in the opposite mode and started with 15 g of xylose/liter plus 5 g of glucose/liter so that each steady state was performed in duplicate.
Analyses.
Substrates consumed and products formed were analyzed by column liquid chromatography. A Gilson (Villiers-le-bel, France) CLC system equipped with an ion-moderated partition chromatography column, Aminex HPX-87H (Bio-Rad, Richmond, Calif.), was used together with a Shimadzu (Kyoto, Japan) refractive index detector, RID-6A, with a mobile phase of 5 mM H2SO4. The flow rate was 0.6 ml/min, and the separation temperature was 45°C.
For determination of ethanol evaporation, the experimental setup was identical to the one used for the continuous cultivations. Ethanol was added to non-cell-containing medium to a concentration of 6 g/liter, and the ethanol concentration in the fermentor was assayed over time.
The composition of the outgoing gas was continuously monitored by a carbon dioxide and oxygen monitor (type 1308; Brüel&Kjœr, Copenhagen, Denmark) (11) using photoacoustic and magnetoacoustic detection for CO2 and O2, respectively.
The dry weight of the cells was determined by filtering a known volume of the culture broth through a 0.45-μm-pore-size Supor membrane (Gelman Sciences, Ann Arbor, Mich.). After being washed with 3 volumes of double-distilled water and dried in a microwave oven for 15 min, the filter was weighed. The dry weight of the cells was determined in triplicate for each steady state.
Enzymatic assays.
Cell extracts for enzyme assays were prepared using glass beads (0.5 mm in diameter). Cells were harvested by centrifugation and, after washing, were resuspended in a disintegration buffer, 0.1 M triethanolamine buffer (pH 7.0) containing 1 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol, and 0.5 mM EDTA. The suspension was vortexed for 5 min at 4°C, put on ice for 5 min, and then vortexed again. Cell debris and glass beads from the cell extract were separated by centrifugation (20,000 × g, 5 min, 4°C), and the remaining supernatant was used for enzyme determinations. Enzyme activities were measured with two different sample concentrations (each in duplicate) by using a U-2000 model spectrophotometer (Hitachi Ltd., Tokyo, Japan) operating at 340 nm and 30°C and with buffers and reagents according to Table 1. Assays were adapted from previously reported assays (4, 7, 32, 33, 39, 51). Specific activities are expressed as units per milligram of protein. Units are defined as micromoles of NADH reduced or oxidized per minute. For the XK assay, the reaction occurring before the addition of ATP (XDH in the reverse direction) was subtracted from the reaction observed in the presence of ATP. No reaction was observed in the absence of xylulose. Xylulose was produced as previously described (31).
TABLE 1.
Buffer and reagent concentrations for enzyme assaysa
| Enzyme (EC no.) | Buffer | Cofactor | Reagents | Start reagent (concn) |
|---|---|---|---|---|
| XR (EC 1.1.1.21) | Triethanolamine (100 mM, pH 7.0) | NADPH (0.2 mM) | Xylose (350 mM) | |
| XDH (EC 1.1.1.9) | Glycine (100 mM, pH 9.0), MgCl2 (50 mM) | NAD+ (3.0 mM) | Xylitol (300 mM) | |
| XK (EC 2.7.1.17) | Tris-HCl (50 mM, pH 7.5), MgCl2 (2.0 mM) | NADH (0.2 mM) | Xylulose (8.5 mM), phosphoenol pyruvate (0.2 mM), pyruvate kinase (10 U), lactate dehydrogenase (10 U) | ATP (2.0 mM) |
| PGK (EC 2.7.2.3) | Tris-HCl (100 mM, pH 8.0), EDTA (0.9 mM), MgCl2 (2 mM) | NADH (0.2 mM) | ATP (1 mM), glyceraldehyde-3-phosphate dehydrogenase (1 U) | 3-Phosphoglycerate (20 mM) |
| ADH (EC 1.1.1.1) | Glycine (100 mM, pH 9.0) | NAD+ (5.0 mM) | Ethanol (1.7 M) |
All concentrations refer to the final assay mixture. The final assay volume was 1 ml.
The protein content was assayed using Coomassie protein assay reagent (6) (Pierce, Rockford, Ill.) with bovine serum albumin as a standard.
Calculations.
Carbon balances and yields were calculated using single carbon unit equivalents (expressed as moles of carbon) (13) to allow comparison of hexose and pentose sugar metabolism. The elemental formula CH1.745O0.627N0.129S0.0025 was used for calculation of assimilated carbon converted to biomass (16).
RESULTS
Construction of a recombinant strain expressing XYL1, XYL2, and XKS1.
An integrating vector, YIpXR/XDH/XK, carrying XYL1, XYL2, and XKS1 and with HIS3 as selectable marker, was constructed (Fig. 1). HIS3 in S. cerevisiae CEN.PK 113-7A is deleted between the HindIII sites (37). The constructed plasmid was cut with PstI within the HIS3 gene, but outside the HindIII deletion in the recipient strain, to generate homologous ends to initiate integration. The resulting recombinant, S. cerevisiae TMB 3001, grew in the absence of histidine and expressed XR, XDH, and XK activities. The specific activities of XR, XDH, and XK in cell lysates of the parental strain CEN.PK 113-7A and the recombinant strain TMB 3001 grown on a xylose-glucose mixture in shaken-flask cultures were 0.03, 0.01, and 0.02 U/mg of protein and 0.21, 1.78, and 0.93 U/mg of protein, respectively.
Anaerobic fermentations of xylose and glucose.
Product formation by S. cerevisiae TMB 3001 was investigated in a high-performance bioreactor under anaerobic conditions. The recombinant strain carrying YIpXR/XDH/XK grew aerobically on xylose, but not anaerobically. We included glucose in all fermentations to enable anaerobic chemostat cultivation. The maximum growth rate anaerobically on glucose was 0.35 h−1 (data not shown). Four different steady states were established, three with mixtures of xylose and glucose, the xylose-to-glucose ratios in the feed being 3:1, 1:1, and 1:3, respectively, and one with glucose as the sole carbon source (Table 2). For one set of fermentations, the first steady state was established with the medium containing the highest xylose concentration; the following three steady states were conducted in the order of decreasing xylose concentration in the feed. A second set was conducted in the opposite mode, i.e., started with a feed containing glucose only. The total amount of carbon source in the feed was 667 mmol of C, equivalent to 20 g/liter, and the D was 0.06 h−1. A post hoc analysis (Scheffe, α = 0.05) showed no significant difference between replicates. Steady-state concentrations were independent of the order in which steady states were reached. The standard deviation was less than 5% of the consumption and production for mean values of all substances (Table 2).
TABLE 2.
Specific substrate consumption rates and product yields for xylose and glucose fermentation at different steady statesa
| Feed denomination (g/liter) | Substrate consumption rate (mmol of C h−1 g [dry wt] of cells−1)
|
Product yield (mmol of C of product/mmol of C of total carbohydrates)
|
% C recoverye | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Xylose | Glucose | EtOH | EtOHb (corrected) | EtOHc (on glucose) | XOH | XOHd (on xylose) | Acet | Glyc | Biomass | CO2 | ||
| 15 xylose + 5 glucose | −6.8 | −15.1 | 0.34 | 0.45 | 0.65 | 0.12 | 0.39 | 0.006 | 0.078 | 0.11 | 0.23 | 98.2 |
| 10 xylose + 10 glucose | −4.7 | −18.0 | 0.38 | 0.46 | 0.58 | 0.09 | 0.42 | 0.004 | 0.090 | 0.11 | 0.27 | 101.1 |
| 5 xylose + 15 glucose | −3.3 | −22.7 | 0.37 | 0.50 | 0.58 | 0.03 | 0.24 | 0.004 | 0.094 | 0.09 | 0.26 | 98.6 |
| 0 xylose + 20 glucose | 0.0 | −23.8 | 0.39 | 0.53 | 0.53 | 0.004 | 0.085 | 0.10 | 0.29 | 100.6 | ||
The total carbon concentration in the feed was 667 mmol of C/liter, or 20 g/liter. EtOH, ethanol; XOH, xylitol; Acet, acetate; Glyc, glycerol.
Ethanol yield was corrected by assuming that the missing percentage in the degree of reduction balance was due to evaporated ethanol.
Ethanol yield was calculated based on consumed glucose only.
Xylitol yield was calculated based on xylose only.
C recovery includes the product yields calculated for total carbohydrates.
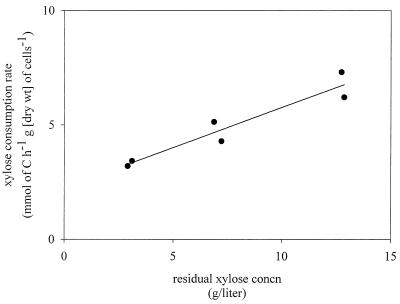
Xylose was coutilized with glucose under anaerobic conditions. The xylose uptake rate increased with increasing xylose concentration and decreasing glucose concentration in the feed, from 3.3 mmol of C h−1 g (dry weight) of cells−1 when the xylose-to-glucose ratio in the feed was 1:3 to 6.8 mmol of C h−1 g (dry weight) of cells−1 when the xylose-to-glucose ratio in the feed was 3:1 (Fig. 2; Table 2). However, even at the highest xylose concentration, only 12% of the xylose was consumed (Table 3).
FIG. 2.
Xylose uptake rate at different xylose concentrations in the feed during anaerobic fermentation of xylose-glucose mixtures with S. cerevisiae TMB 3001. The equation for the line is y = 2.266 + 0.349x, and the standard error of the estimate is 0.5298.
TABLE 3.
Concentrations of xylose and glucose in the different feeds and in the fermentor and biomass concentrations for the different steady states
| Feed denomination (g/liter) | Feed concn (g/liter)
|
Residual concn (g/liter)
|
Biomass concn (g/liter) | ||
|---|---|---|---|---|---|
| Xylose | Glucose | Xylose | Glucose | ||
| 15 xylose + 5 glucose | 15.04 | 5.08 | 12.80 | 0.074 | 0.64 |
| 10 xylose + 10 glucose | 9.56 | 9.67 | 7.05 | 0.066 | 1.09 |
| 5 xylose + 15 glucose | 5.23 | 15.27 | 3.01 | 0.049 | 1.35 |
| 0 xylose + 20 glucose | 21.02 | 0.032 | 1.78 | ||
Carbon balances at four steady states with increasing xylose concentration showed that the measured products accounted for 86.9, 84.8, 94.4, and 88.1% of consumed carbon (Table 2). Using a balance of the degree of reduction (34) of substrates and products and assuming that ethanol accounted for the missing percentage in the degree of reduction balances, the carbon balances were recalculated and closed to within 2% (Table 2). The resulting rates of ethanol evaporation calculated from measured values and degree of reduction balances were 0.04, 0.08, 0.11, and 0.14 g liter−1 h−1 for steady states with increasing glucose concentration. At an initial concentration of 6 g of ethanol/liter, the experimentally determined evaporation rate was 0.06 g liter−1 h−1, which is in agreement with the calculated values. The ethanol concentration differed at the four steady states, which may have influenced the evaporation rate. Furthermore, ethanol is highly volatile and the experimental error is large. To obtain as accurate a determination of the ethanol concentrations as possible, degree of reduction balances were used throughout the study. The corrected ethanol yield on total carbohydrates decreased with increasing xylose in the feed, from 0.53 to 0.45 mol of C/(mol of C of consumed carbohydrates) (0.41 to 0.35 g/g) (Table 2). However, the corrected ethanol yield calculated on consumed glucose increased, showing that xylose is converted to ethanol under anaerobic conditions. The corrected ethanol yield for the steady state with only glucose in the feed was 0.53 mol of C/(mol of C of glucose) (0.41 g/g), and with a xylose-to-glucose ratio in the feed of 3:1, the yield increased to 0.65 mol of C/(mol of C of glucose) (0.50 g/g). Hence, for the steady state with a xylose-to-glucose ratio in the feed of 3:1, the ethanol yield on xylose was estimated to be 0.27 mol of C/(mol of C of xylose) [(0.65 − 0.53) × 15.1/6.8 (Table 2)] (0.21 g/g), assuming the ethanol yield on glucose to be constant for the four different steady states.
The xylitol yield, calculated with total consumed carbohydrates, increased with increasing xylose uptake rate, from 0.03 to 0.12 mol of C/(mol of C of carbohydrates) (0.03 to 0.12 g/g), when the xylose concentration in the feed increased from 5 to 15 g/liter (Table 2) so that the xylose fraction excreted as xylitol increased from 24 to 40%. The glycerol yield increased slightly at the lowest xylose concentration. At the highest xylose concentration, the lowest glycerol yield and highest acetate yield were observed.
The biomass yield on consumed sugar was rather constant (Table 2), but the concentration of biomass increased from 0.64 to 1.78 g/liter with increasing glucose concentration (Table 3).
Enzyme activities and strain stability.
The specific activities of the enzymes XR, XDH, XK, PGK, and ADH were measured throughout the fermentations (Table 4). The recombinant S. cerevisiae strain TMB 3001 expressed XR, XDH, and XK with activities of 0.4 to 0.5, 2.7 to 3.4, and 1.5 to 1.7 U/mg of protein, respectively, corresponding to an approximate XR-to-XDH-to-XK ratio of 1:6:4. Furthermore, the strain exhibited stable recombinant enzyme activities throughout more than 4 weeks of continuous cultivation, equivalent to more than 40 generations. The PGK and ADH activities were 15 to 23 and 21 to 28 U/mg of protein, respectively (Table 4).
TABLE 4.
Specific enzyme activities in cell lysates of samples (four replicates) from different steady states of strain S. cerevisiae TMB 3001 in anaerobic, glucose-limited continuous cultures
| Feed denomination (g/liter) | Enzyme activity (U/mg of protein) ± SD
|
||||
|---|---|---|---|---|---|
| XR | XDH | XK | PGK | ADH | |
| 15 xylose + 5 glucose | 0.42 ± 0.05 | 2.74 ± 0.10 | 1.54 ± 0.07 | 21.9 ± 1.2 | 23.1 ± 1.2 |
| 10 xylose + 10 glucose | 0.52 ± 0.04 | 3.37 ± 0.09 | 1.71 ± 0.07 | 23.1 ± 1.30 | 28.4 ± 0.75 |
| 5 xylose + 15 glucose | 0.52 ± 0.05 | 3.14 ± 0.06 | 1.61 ± 0.03 | 15.4 ± 0.16 | 27.7 ± 0.54 |
| 0 xylose + 20 glucose | 0.43 ± 0.03 | 2.75 ± 0.11 | 1.56 ± 0.07 | 17.3 ± 1.21 | 21.2 ± 0.81 |
DISCUSSION
For the first time, anaerobic ethanol formation from xylose has been demonstrated for a recombinant xylose-utilizing strain of S. cerevisiae. The corrected ethanol yields (0.45 to 0.50 mmol of C/mmol of C; 0.35 to 0.38 g/g) and productivities (9.7 to 13.2 mmol of C h−1 g [dry weight] of cells−1; 0.24 to 0.30 g h−1 g [dry weight] of cells−1) obtained for cofermentation of xylose and glucose by TMB 3001 were slightly lower than those previously reported for Saccharomyces sp. strain 1400(LNH-ST) in oxygen-limited batch fermentation, 0.56 mmol of C/mmol of C and 14.3 mmol of C h−1 g [dry weight] of cells−1, respectively (21). The discrepancy could arise from differences in strain background as well as in the fermentation setup, i.e., absence or presence of oxygen and continuous or batch mode. For P. stipitis, the most efficient natural xylose-fermenting yeast, the highest ethanol yield on xylose, 0.63 mmol of C/mmol of C (0.48 g/g), was obtained in oxygen-limited continuous culture with a D of 0.06 h−1 (42). The productivity obtained under the same conditions was 8.7 mmol of C h−1 g (dry weight) of cells−1 (0.20 g h−1 g [dry weight] of cells−1). In contrast, both ethanol yield and productivity under anaerobic conditions were considerably lower (42). One reason for choosing S. cerevisiae as the host strain for development of a xylose-fermenting yeast is its rapid anaerobic growth on glucose (30, 52). However, anaerobic growth on xylose has not yet been demonstrated for recombinant xylose-utilizing S. cerevisiae. This lack of growth has been attributed to slow xylose metabolism (24), resulting in too little ATP formation to maintain growth. P. stipitis has recently been metabolically engineered for anaerobic growth (41). When S. cerevisiae URA1 was expressed in P. stipitis, the yeast grew anaerobically on glucose, but not on xylose, supporting the suggestion that anaerobic growth on xylose is limited by slow xylose metabolism.
The strategy for chromosomal integration of the genes encoding XR, XDH, and XK resulted in a stable recombinant strain which retained its physiological characteristics through more than 4 weeks of continuous cultivation without selection pressure. Translated to generation time, strain TMB 3001 was stable for more than 40 generations, which is considerably longer than the four to five generations of stability reported for strain 1400 (pLNH32), which carries the same genes on a 2μm-derived vector (22). Recombinant xylose-utilizing Saccharomyces strains carrying 2μm-based vectors have been stably maintained in batch cultivation (22, 24, 27, 44, 48), but in continuous cultivation they tend to be unstable (27, 28). The instability of strains carrying 2μm-based vectors may result from genetic instability at the plasmid level, i.e., spontaneous loss of the transformed phenotype and the plasmid (19, 26, 28) or high frequency of recombination, resulting in cells that still carry the selectable marker but have lost the cloned gene (15, 28). Consequently, strains carrying less foreign DNA usually dominate the culture since the high-copy-number plasmids and the high expression of the heterologous genes can reduce the growth rate and glycolytic flux (26–28, 43). For the construction of TMB 3001, a single target sequence rather than multiple integrations in ribosomal DNA or transposons was chosen to diminish the burden on the cells caused by expression of foreign DNA. The resulting XR, XDH, and XK activities for TMB 3001 were lower than those for a CEN.PK strain carrying the same genes on two multicopy plasmids (pY7 [46] and pXks [Johansson et al., submitted]) cultivated in batch, 0.4, 3, and 1.6 U/mg compared to 0.7, 18, and 36 U/mg, respectively (Johansson et al., submitted). Still, the ethanol yields of the two strains were similar, 0.45 and 0.42 mol of C/mol of C (0.35 and 0.32 g/g), respectively, implying that higher-level expression of XR, XDH, and XK does not increase the flux to ethanol.
The XYL1 gene, which encodes XR, that was expressed in S. cerevisiae originated from P. stipitis. This enzyme uses either NADPH or NADH as a cofactor, while XDH exclusively uses NAD+. Under anaerobic conditions, the fraction of xylose converted to xylitol by NADPH-dependent XR activity is not further converted to xylulose. This was demonstrated by increased xylitol excretion following the addition of the respiratory inhibitor antimycin A to an oxygen-limited culture of a recombinant xylose-utilizing strain of S. cerevisiae (24). Under the conditions applied in the present study, less than 50% of the consumed xylose was excreted as xylitol. The difference could result from the overexpression of XK, which increases ethanol production and lowers xylitol excretion (14, 17, 44), or from the simultaneous utilization of glucose, which may supply NAD+ by glycerol formation. The latter hypothesis is supported by the high glycerol yield at the steady state with the lowest xylose-to-glucose ratio, 1:3 (Table 2). The supplementary NAD+ might be used by XDH and reduce xylitol excretion. ATP is used in glycerol formation, but not in xylitol formation, and this result indicates that only when a surplus of glucose is available can the yeast use carbon for glycerol formation to supply XDH with NAD+.
In addition to redox constraints, xylose transport also may limit xylose utilization by recombinant S. cerevisiae. Xylose is transported by the facilitated glucose transport system in S. cerevisiae cells, which have a 200-fold lower affinity for xylose than for glucose (9, 24, 25). Increasing the xylose concentration in the feed enhanced the xylose flux. However, the xylose flux was still 2.2 times lower than the glucose flux when a feed consisting of 15 g of xylose/liter and 5 g of glucose/liter was utilized, suggesting that xylose transport to a large extent determines the xylose flux in recombinant S. cerevisiae TMB 3001.
ACKNOWLEDGMENTS
We thank Dace Leveika for technical assistance and Jörg Hauf for supplying the vector YDp-H.
This work was financially supported by EC contract BIO-CT95-0107, Energimyndigheten (Swedish National Energy Administration), and TEMPUS contract JEP-09273-95.
REFERENCES
- 1.Andreasen A A, Stier T J B. Anaerobic nutrition of Saccharomyces cerevisiae. I. Ergosterol requirement for growth in a defined medium. J Cell Comp Physiol. 1953;41:23–36. doi: 10.1002/jcp.1030410103. [DOI] [PubMed] [Google Scholar]
- 2.Andreasen A A, Stier T J B. Anaerobic nutrition of Saccharomyces cerevisiae. II. Unsaturated fatty acid requirement for growth in a defined medium. J Cell Comp Physiol. 1954;43:271–281. doi: 10.1002/jcp.1030430303. [DOI] [PubMed] [Google Scholar]
- 3.Berben G, Dumont J, Gilliquet V, Bolle P-A, Hilger F. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- 4.Bergmeyer H U, editor. Methods of enzymatic analysis. 2nd ed. Vol. 1. New York, N.Y: Academic Press; 1974. [Google Scholar]
- 5.Boles E, Heinisch J, Zimmermann F K. Different signals control the activation of glycolysis in the yeast Saccharomyces cerevisiae. Yeast. 1993;9:761–770. doi: 10.1002/yea.320090710. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Brooks S P J, Storey K B. Subcellular enzyme binding in glycolytic control: in vivo studies with fish muscle. Am J Physiol. 1988;255:R289–R294. doi: 10.1152/ajpregu.1988.255.2.R289. [DOI] [PubMed] [Google Scholar]
- 8.Bruinenberg P M, de Bot P H M, van Dijken J P, Scheffers W A. The role of redox balances in the anaerobic fermentation of xylose by yeasts. Eur J Appl Microbiol Biotechnol. 1983;18:287–292. [Google Scholar]
- 9.Busturia A, Lagunas R. Catabolite inactivation of the glucose transport system in Saccharomyces cerevisiae. J Gen Microbiol. 1986;132:379–385. doi: 10.1099/00221287-132-2-379. [DOI] [PubMed] [Google Scholar]
- 10.Chiang C, Knight S G. Metabolism of d-xylose by moulds. Nature. 1960;188:79–80. doi: 10.1038/188079a0. [DOI] [PubMed] [Google Scholar]
- 11.Christensen I H, Schulze U, Nielsen J, Villadsen J. Acoustic off-gas analyser for bioreactors: precision, accuracy and dynamics of detection. Chem Eng Sci. 1995;50:2601–2610. [Google Scholar]
- 12.Ciriacy M, Porep H. Conversion of pentoses to ethanol by baker's yeast. In: Magnien M, editor. Biomolecular engineering in the European Community. Dordrecht, The Netherlands: Martinus Nijhoff Publishers; 1986. pp. 675–681. [Google Scholar]
- 13.de Jong-Gubbels P, Vanrolleghem P, Heijnen S, van Dijken J P, Pronk J T. Regulation of carbon metabolism in chemostat cultures of Saccharomyces cerevisiae grown on mixtures of glucose and ethanol. Yeast. 1995;11:407–418. doi: 10.1002/yea.320110503. [DOI] [PubMed] [Google Scholar]
- 14.Deng X X, Ho N W Y. Xylulokinase activity in various yeasts including Saccharomyces cerevisiae containing the cloned xylulokinase gene. Appl Biochem Biotechnol. 1990;24/25:193–199. doi: 10.1007/BF02920245. [DOI] [PubMed] [Google Scholar]
- 15.Dobson M J, Futcher A B, Cox B S. Control of recombination within and between DNA plasmids of Saccharomyces cerevisiae. Curr Genet. 1980;2:193–200. doi: 10.1007/BF00435685. [DOI] [PubMed] [Google Scholar]
- 16.Duboc P, Schill N, Menoud L, van Gulik W, von Stockar U. Measurements of sulfur, phosphorus and other ions in microbial biomass: influence on correct determination of elemental composition and degree of reduction. J Biotechnol. 1995;43:145–158. doi: 10.1016/0168-1656(95)00135-0. [DOI] [PubMed] [Google Scholar]
- 17.Eliasson A, Boles E, Johansson B, Österberg M, Thevelein J M, Spencer-Martins I, Juhnke H, Hahn-Hägerdal B. Xylulose fermentation by mutant and wild-type strains of Zygosaccharomyces and Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2000;53:376–382. doi: 10.1007/s002530051629. [DOI] [PubMed] [Google Scholar]
- 18.Entian K-D, Koetter P. Yeast mutant and plasmid collections. In: Brown A J P, Tuite M F, editors. Yeast gene analysis. Vol. 26. London, United Kingdom: Academic Press; 1998. pp. 431–449. [Google Scholar]
- 19.Futcher A B, Cox B S. Copy number and the stability of 2-μm circle-based artificial plasmids of Saccharomyces cerevisiae. J Bacteriol. 1984;157:283–290. doi: 10.1128/jb.157.1.283-290.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn-Hägerdal B, Hallborn J, Jeppsson H, Meinander N, Walfridsson M, Ojamo H, Penttila M, Zimmermann F K. Redox balances in recombinant Saccharomyces cerevisiae. Ann N Y Acad Sci. 1996;782:286–296. doi: 10.1111/j.1749-6632.1996.tb40569.x. [DOI] [PubMed] [Google Scholar]
- 21.Ho, N. W. Y. 1997. World patent WO 97/42307.
- 22.Ho N W Y, Chen Z, Brainard A P. Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl Environ Microbiol. 1998;64:1852–1859. doi: 10.1128/aem.64.5.1852-1859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hochster R M, Watson R W. Enzymatic isomerization of D-xylose to D-xylulose. Arch Biochem. 1954;48:120–129. doi: 10.1016/0003-9861(54)90313-6. [DOI] [PubMed] [Google Scholar]
- 24.Kötter P, Ciriacy M. Xylose fermentation by Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 1993;38:776–783. [Google Scholar]
- 25.Kotyk A. Properties of the sugar carrier in baker's yeast. 2. Specificity of transport. Folia Microbiol. 1967;12:121–131. doi: 10.1007/BF02896872. [DOI] [PubMed] [Google Scholar]
- 26.Mason C A. Physiological aspects of growth and recombinant DNA stability in Saccharomyces cerevisiae. Antonie Leeuwenhoek. 1991;59:269–283. doi: 10.1007/BF00583680. [DOI] [PubMed] [Google Scholar]
- 27.Meinander N Q, Boels I, Hahn-Hägerdal B. Fermentation of xylose/glucose mixtures by metabolically engineered Saccharomyces cerevisiae strains expressing XYL1 and XYL2 from Pichia stipitis with and without overexpression of TAL1. Bioresour Technol. 1999;68:79–87. [Google Scholar]
- 28.Meinander N Q, Hahn-Hägerdal B. Fed-batch xylitol production with two recombinant Saccharomyces cerevisiae strains expressing XYL1 at different levels, using glucose as a cosubstrate: a comparison of production parameters and strain stability. Biotechnol Bioeng. 1997;54:391–399. doi: 10.1002/(SICI)1097-0290(19970520)54:4<391::AID-BIT12>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 29.Müller S, Boles E, May M, Zimmermann F K. Different internal metabolites trigger the induction of glycolytic gene expression in Saccharomyces cerevisiae. J Bacteriol. 1995;177:4517–4519. doi: 10.1128/jb.177.15.4517-4519.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagy M, Lacroute F, Thomas D. Divergent evolution of pyrimidine biosynthesis between anaerobic and aerobic yeasts. Proc Natl Acad Sci USA. 1992;89:8966–8970. doi: 10.1073/pnas.89.19.8966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olsson L, Lindén T, Hahn-Hägerdal B. A rapid chromatographic method for the production of preparative amounts of xylulose. Enzyme Microb Technol. 1994;16:388–394. [Google Scholar]
- 32.Rizzi M, Erlemann P, Bui-Thanh N-A, Dellweg H. Xylose fermentation by yeasts. 4. Purification and kinetic studies of xylose reductase from Pichia stipitis. Appl Microbiol Biotechnol. 1988;29:148–154. [Google Scholar]
- 33.Rizzi M, Harwart K, Erlemann P, Bui-Thahn N-A, Dellweg H. Purification and properties of the NAD+-xylitol-dehydrogenase from the yeast Pichia stipitis. J Ferment Bioeng. 1989;67:20–24. [Google Scholar]
- 34.Roels J A. Energetics and kinetics in biotechnology. 1st ed. Amsterdam, The Netherlands: Elsevier Biomedical Press BV; 1983. [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Scherer S, Davis R W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci USA. 1979;76:4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schiestl R H, Gietz R D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 38.Senac T, Hahn-Hägerdal B. Intermediary metabolite concentrations in xylulose- and glucose-fermenting Saccharomyces cerevisiae cells. Appl Environ Microbiol. 1990;56:120–126. doi: 10.1128/aem.56.1.120-126.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shamanna D K, Sanderson K E. Uptake and catabolism of d-xylose in Salmonella typhimurium LT2. J Bacteriol. 1979;139:64–70. doi: 10.1128/jb.139.1.64-70.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 41.Shi N-Q, Jeffries T W. Anaerobic growth and improved fermentation of Pichia stipitis bearing a URA1 gene from Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 1998;50:339–345. doi: 10.1007/s002530051301. [DOI] [PubMed] [Google Scholar]
- 42.Skoog K, Hahn-Hägerdal B. Effect of oxygenation on xylose fermentation by Pichia stipitis. Appl Environ Microbiol. 1990;56:3389–3394. doi: 10.1128/aem.56.11.3389-3394.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snoep J L, Yomano L P, Westerhoff H V, Ingram L O. Protein burden in Zymomonas mobilis: negative flux and growth control due to overproduction of glycolytic enzymes. Microbiology. 1995;141:2329–2337. [Google Scholar]
- 44.Tantirungkij M, Nakashima N, Seki T, Yoshida T. Construction of xylose-assimilating Saccharomyces cerevisiae. J Ferment Bioeng. 1993;75:83–88. [Google Scholar]
- 45.Toon S T, Philippidis G P, Ho N W Y, Chen Z, Brainard A, Lumpkin R E, Riley C J. Enhanced cofermentation of glucose and xylose by recombinant Saccharomyces yeast strains in batch and continuous operating modes. Appl Biochem Biotechnol. 1997;63–65:243–255. doi: 10.1007/BF02920428. [DOI] [PubMed] [Google Scholar]
- 46.Walfridsson M, Anderlund M, Bao X, Hahn-Hägerdal B. Expression of different levels of enzymes from the Pichia stipitis XYL1 and XYL2 genes in Saccharomyces cerevisiae and its effect on product formation during xylose utilisation. Appl Microbiol Biotechnol. 1997;48:218–224. doi: 10.1007/s002530051041. [DOI] [PubMed] [Google Scholar]
- 47.Walfridsson M, Bao X, Anderlund M, Lilius G, Bülow L, Hahn-Hägerdal B. Ethanolic fermentation of xylose with Saccharomyces cerevisiae harboring the Thermus thermophilus xylA gene, which expresses an active xylose (glucose) isomerase. Appl Environ Microbiol. 1996;62:4648–4651. doi: 10.1128/aem.62.12.4648-4651.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walfridsson M, Hallborn J, Penttila M, Keränen S, Hahn-Hägerdal B. Xylose-metabolizing Saccharomyces cerevisiae strains overexpressing the TKL1 and TAL1 genes encoding the pentose phosphate pathway enzymes transketolase and transaldolase. Appl Environ Microbiol. 1995;61:4184–4190. doi: 10.1128/aem.61.12.4184-4190.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Dijken J P, Scheffers W A. Redox balances in the metabolism of sugars by yeasts. FEMS Microbiol Rev. 1986;32:199–224. [Google Scholar]
- 50.Verduyn C, Postma E, Scheffers W A, van Dijken J P. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992;8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- 51.Verduyn C, van Kleef R, Frank J, Schreuder H, van Dijken J P, Scheffers W A. Properties of the NAD(P)H-dependent xylose reductase from the xylose-fermenting yeast Pichia stipitis. Biochem J. 1985;226:669–677. doi: 10.1042/bj2260669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Visser W, Scheffers W A, Batenburg-van der Vegte W H, van Dijken J P. Oxygen requirements of yeasts. Appl Environ Microbiol. 1990;56:3785–3792. doi: 10.1128/aem.56.12.3785-3792.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.von Sivers M, Zacchi G. A techno-economical comparison of three processes for the production of ethanol from pine. Bioresour Technol. 1995;51:43–52. [Google Scholar]