Abstract
Microcystin, a hepatotoxin known to be the cause of animal and human deaths, is produced by the bloom-forming cyanobacterium Microcystis aeruginosa in freshwater bodies worldwide. The toxin is produced nonribosomally via a multifunctional enzyme complex, consisting of both peptide synthetase and polyketide synthase modules coded for by the mcy gene cluster. The recent identification of the mcy genes in the production of microcystin synthetase for the first time provides an avenue to study the regulation of microcystin production at a genetic level. In this study, M. aeruginosa PCC7806 was grown either under continuous light of various intensities or under low light with subsequent short-term exposure to different light intensities and qualities and various stress factors. RNase protection assays were employed to observe the level of mcyB and mcyD transcription under each condition. Both mcyB and mcyD transcript levels were increased under high light intensities and red light. Blue light and certain artificial stress factors (methylviologen and NaCl) led to reduced transcript amounts. There appeared to be two light thresholds, between dark and low light (16 μmol of photons m−2 s−1), and medium (31 μmol of photons m−2 s−1) and high light (68 μmol of photons m−2 s−1), at which a significant increase in transcription occurred. Our findings show that the effect of light on microcystin synthetase production is due to light quality and is initiated at certain threshold intensities, which are not necessarily reflected by observed intracellular toxin bioactivity.
Marine and freshwater cyanobacteria produce a wide range of bioactive, in part toxic, compounds, including non-ribosomally made peptides, polyketides, alkaloids, and lipopolysaccharides. Most intensively investigated have been those species which, under certain environmental conditions, tend to mass development, forming blooms. The hepatotoxins and neurotoxins (cyanotoxins) produced by bloom-forming cyanobacteria has been the cause of human and animal health hazards and even death (5, 14). Knowledge of the regulation of cyanotoxin biosynthesis could allow implementation of water management strategies to avoid environmental conditions that support toxin production and give clues to the as yet unknown functions of these substances.
One of the most common bloom-forming, hepatotoxin-producing species of cyanobacteria is Microcystis aeruginosa. While certain species can also produce neurotoxins, M. aeruginosa is most commonly known to produce the hepatotoxic heptapeptide microcystin in a variety of forms with varying toxicity (18, 28). Microcystin binds to the multispecific bile acid transport system, subsequently directing toxic effects to hepatocytes (29). Toxicity is exerted, perhaps not exclusively, by the inhibition of eukaryotic protein phosphatases PP2A and PP1 (10, 19, 29, 36), resulting in excessive phosphorylation of cytoskeletal filaments, loss of cellular support, and destruction of hepatic sinusoid endothelium (9, 11, 22).
Evaluation of the development of toxin concentrations in cyanobacterial populations during bloom events is important for the prediction of potential health hazards. Changing toxin concentrations in cyanobacterial blooms most probably reflect alterations in species and strain composition with various toxins and toxicities, as well as the regulation of toxin biosynthesis in specific strains under certain environmental conditions. Changes in toxin production due to variable laboratory conditions are usually lower than the observed differences in toxin levels between strains of a given species or that observed in natural blooms of M. aeruginosa (33). Nevertheless, several environmental factors have been described to influence the biosynthesis of cyanotoxins for several defined isolates.
A variety of studies have focused on the effects of nutrients, such as nitrogen and phosphorus (6, 24–26, 32, 43), trace metals (17, 38), temperature (32, 40, 43), pH (12, 41), and light (26, 32, 39, 43) on microcystin production. Several, but not all, studies have suggested that toxin production is highest under optimal growth conditions (33). Orr and Jones (25) concluded that the rate of microcystin production is directly proportional to the growth rate of the cyanobacterial population regardless of the environmental parameter tested. Increasing toxicity has been observed when light intensities were raised from approximately 7 to 40 μmol of photons m−2 s−1, depending on the study, with no further increases observed at higher light intensities (39, 40, 43). In contrast, microcystin concentrations in Anabaena and Oscillatoria strains were reduced at high light intensities (26, 32). Unfortunately, much of the published data seem controversial, as the individual studies are not readily comparable due to the various growth and toxicity assessment techniques employed.
More precise investigations of potential regulatory mechanisms of cyanotoxin biosynthesis require knowledge of the genes and enzymes involved. For the first time in the case of a cyanobacterial toxin, such studies are possible with the recent discovery of the genes and biosynthetic pathway required for the production of microcystins in M. aeruginosa (7). Microcystin is synthesized nonribosomally (3), catalyzed by a large multifunctional enzyme complex consisting of peptide synthetase and polyketide synthase modules (7). Individual peptide synthetase modules catalyze amino/hydroxy acid activation and thioester formation reactions in the same order in which their residues are incorporated into the growing heptapeptide chain (15, 20). The polyketide synthase is proposed to be involved in the production of the fatty acid side chain of the unique amino acid Adda, essential for toxicity of the microcystin peptide. Genes for the enzyme complex form a large gene cluster containing at least two operons, mcyABC (peptide synthetase) and mcyDE (hybrid polyketide-peptide synthetase) (35). The messages are transcribed in opposite directions by a central sequence of approximately 900 bp containing the promoter and putative regulatory cis elements.
In this study, mcyB and mcyD were chosen as representatives of the microcystin peptide synthetase and polyketide synthase genes, respectively, and investigated with respect to transcript accumulation in cells exposed to a variety of light and stress conditions. This is the first genetic study focusing on the effect of light intensity and quality and oxidative stress upon the transcriptional regulation of genes responsible for toxin synthesis in bloom-forming cyanobacteria.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
M. aeruginosa PCC7806 was grown in Z8 medium (26) as batch cultures, in glass vessels with a diameter of 4 cm, under continuous aeration at 23°C. The effect of light on mcyB and mcyD transcripts was tested in a series of experiments. In the first set, batch cultures (250 ml) were grown at low (16 μmol of photons m−2 s−1), medium (31 μmol of photons m−2 s−1), and high (68 μmol of photons m−2 s−1) light, from an optical density at 750 nm (OD750) of 0.15 to ∼1.6. During this growth period, OD750, chlorophyll-α (34), and total protein (Bio-Rad, Hercules, Calif.) were measured every day, starting on day 3. Samples for RNase protection assays (RPAs) (50 ml) were taken at early (OD750 = 0.4 to 0.7), middle (OD750 = 0.84 to 0.87), and late (OD750 = 1.6 to 2.0) growth phases, at which times cells were counted using a hemocytometer (Thoma, Bad Blankenburg, Germany). In the second set of experiments, a preculture of 1 liter was grown under low light (16 μmol of photons m−2 s−1) to an OD750 of 0.66. Thereafter, the culture was divided (100 ml/sample) and exposed to different light or stress conditions (see Fig. 4).
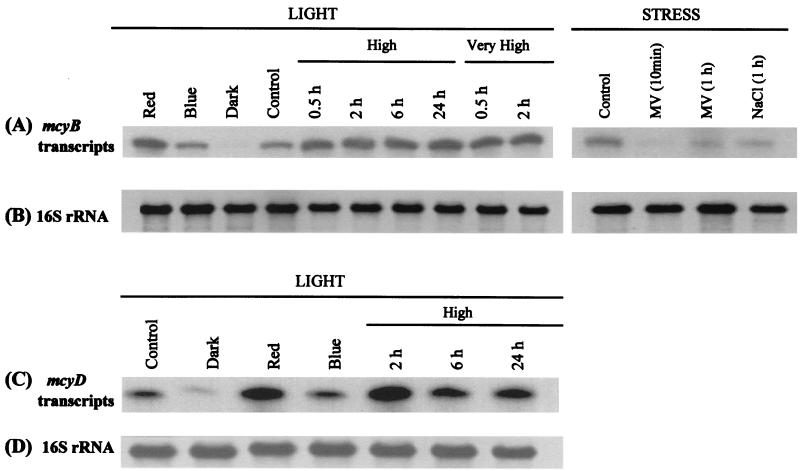
FIG. 4.
Transcript analysis of cells grown under low light to an OD750 of 0.66 and subsequently exposed to different light and stress conditions. (A) RPAs of mcyB transcripts. (B) Northern blot analysis of 16S rRNA from total RNA used for mcyB RPAs. (C) RPAs of mcyD transcripts. (D) Northern blot analysis of 16S rRNA from total RNA used for mcyD RPAs.
A light intensity of 68 μmol of photons m−2 s−1 (high) was provided by cool fluorescence globes. Low (16 μmol of photons m−2 s−1) and medium (31 μmol of photons m−2 s−1) light intensities were obtained by shading cultures with two and one grey filter (Lee Filters, Hampshire, England), respectively. Very high light (400 μmol of photons m−2 s−1) and low light (16 μmol of photons m−2 s−1) for light quality (red and blue light) experiments were produced by a Fibre Illuminator (FL-440; Walz, Effeltrich, Germany) in the dark. Red and blue light environments were generated using cut-off filter foils (Lee Filters) wrapped around the culture vessels. Blue filters allowed light transmission between 400 and 510 nm, peaking at 475 nm, and red light at 620 nm, with ∼1/2 at 666 nm. In all cases, light intensity was set and measured using a spherical (SPQA) light meter (LI-250; Walz) placed inside an empty culture vessel, surrounded with the appropriate filter foils, at the position of the test culture. Oxidative stress was induced by the addition of 1 mM methylviologen (MV) (C12H12N2 · 2HCl) (Serva, Feinbiochemica, Heidelberg, Germany) to low- and high-light cultures. A second stress factor was introduced by the addition of 250 mM sodium chloride to low-light cultures.
Sampling and RNA extraction.
During sampling, extreme care was taken to maintain light conditions similar to those tested. Twenty-five milliliters of culture was placed into 50-ml tubes, and the remaining volume was filled with ice prior to centrifugation at 5,500 × g for 10 min at 4°C. Cell pellets were frozen in liquid nitrogen and kept at −20°C until RNA extraction. Total RNA was extracted using Trizol reagent (Gibco BRL, Life Technologies, Rockville, Md.) following pretreatment of the cells. The frozen pellet was crushed in a precooled (with liquid nitrogen) mortar. This was filled three times with liquid nitrogen between each crushing of the cells. Still in a frozen state, Trizol reagent was added to the cells and combined with the cell suspension until the mixture was completely defrosted. Phenol extraction and precipitation were performed as per the manufacturer's instructions. RNA was further purified via columns (High Pure RNA isolation kit; Boehringer, Mannheim, Germany). Concentrations of purified RNA were measured in duplicate at OD260 (GeneQuant II RNA/DNA calculator; Pharmacia Biotech, Uppsala, Sweden).
Northern analysis.
One microgram of RNA from each sample was separated by electrophoresis on a 1.3% formaldehyde gel and blotted onto a charged nylon membrane (GeneScreen Plus hybridization transfer membrane; NEN Life Science Products, Boston, Mass.) as described by Sambrook et al. (30). Blots were probed with radioactively labeled (Readyprime; Amersham, Braunschweig, Germany) 16S rRNA gene PCR product. Primers for the 16S rRNA gene amplification were 16SF (5′-GCGTTATCCGGAATTATTGG-3′) and 16SR (5′-CCA CTAAGAACGAGGGTTGC-3′), and PCR conditions were as described previously (21). Hybridization and washing conditions were performed using standard procedures (30).
RPAs.
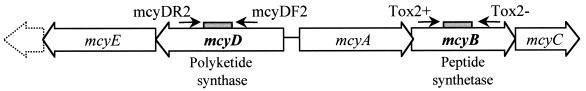
Oligonucleotides tox2+ (5′-AGGAACAAGTTGCACAGAATCCGCA-3′) and tox2− (5′-ACTAATCCCTATCTAAACACAGTAACTCA-3′) were used to amplify a 200-bp fragment within mcyB (peptide synthetase gene). Similarly, a 297-bp fragment of mcyD (microcystin polyketide synthase gene) was amplified with the oligonucleotide set mcyDF2 (5′-GGTTCGCCTGGTCAAAGTAA-3′) and mcyDR2 (5′-CCTCGCTAAAGAAGGGTTGA-3′) (Fig. 1). These fragments were ligated into the cloning vector pGEM (Promega) and checked for directional insertion via PCR and sequencing. After linearization of the vector, the probes for the RPAs were prepared via in vitro transcription and labeled with [α-32P]UTP (MaxiScript; Ambion, Austin, Tex.). The resulting probes were gel purified and eluted (0.5 M ammonium acetate, 1 mM EDTA, 0.1% sodium dodecyl sulfate) overnight. RPAs were carried out according to the manufacturer's instructions (Boehringer) with the coprecipitation of probe and RNA (exactly 8 μg of RNA used per sample) as the first step. Digestion products were subjected to polyacrylamide gel electrophoresis (5%, 19:1 cross-linking with 7 M urea in Tris-borate-EDTA) at 3 to 5 mA for 3 h (30). Gels were exposed to X-ray film for various time periods to obtain autoradiograms in the appropriate exposure ranges for photography.
FIG. 1.
Microcystin synthetase gene clusters mcyABC and mcyDE. Mutations within mcyABC or mcyDE result in loss of toxicity (7, 35). Relative positions of primers used to generate RPA probes within mcyB (peptide synthetase) and mcyD (polyketide synthase) are shown.
Toxin measurements.
Intracellular microcystin content was measured using the colorimetric PP2A inhibition assay (1). This assay has a 50% inhibitory concentration of 6.72 μg/liter for the microcystin-LR standard, using 0.5 mg of PP2a per ml, and a limit of detection of 0.033 μg/liter (33 pM). Prior to the assay, 1 ml of cell suspension in water (supernatant removed) was freeze-thawed three times and diluted 1:100 to 1:8,000. Samples were measured in duplicate in the same assay and also in repeated assays. After adjusting each assay to percentages of protein phosphatase activities, the concentration of microcystin was calculated from an average standard curve of all assays. PP2a inhibitor (microcystin) content was calculated for cells per milliliter of sample, resulting in values reported as picomoles of microcystin per cell.
RESULTS
Different light intensities had an effect on M. aeruginosa growth, chlorophyll-a and protein content, and cell size.
Batch cultures of M. aeruginosa PCC7806, set up with a starting OD750 of 0.15, were grown under various light intensities for 8 days. Within the first 3 days, cells under high light grew to an OD750 of 0.7, compared to an OD750 of 0.6 under medium light and 0.45 at low light. Increased growth under high light intensities continued until day 6, when the cells had reached an OD750 of between 1.1 and 1.3 (data not shown).
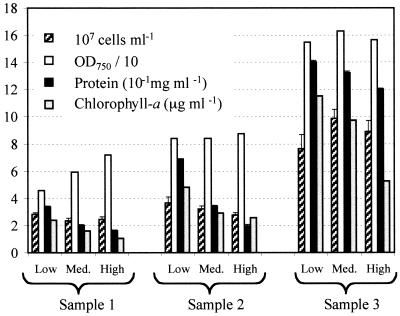
Cell counts and further analysis of cellular protein and chlorophyll-a concentrations were performed at early, middle, and late growth phases. Sample set 1 (sample 1) and sample set 3 (sample 3) were taken on days 3 and 8, respectively. Sample set 2 (sample 2) was taken when cells had reached an OD750 of between 0.84 and 0.87, which corresponded to day 5 for low-light cultures and day 4 for middle- and high-light cultures. At all three sampling times, a downward trend was observed for protein and chlorophyll-a concentrations from low-light to high-light cultures (Fig. 2). Interestingly, cell numbers were also slightly reduced from low to high light intensities whether at similar (sample 2) or increased (sample 1) optical densities. This trend was not observed during the late growth phase (sample 3) (Fig. 2).
FIG. 2.
Cell counts, optical density (10−1), protein concentration, and chlorophyll-a concentration in M. aeruginosa cultures grown under low (16 μmol of photons m−2 s−1), medium (31 μmol of photons m−2 s−1), and high (68 μmol of photons m−2 s−1) light intensities, measured from sample 1 (early growth phase), sample 2 (middle growth phase), and sample 3 (late growth phase).
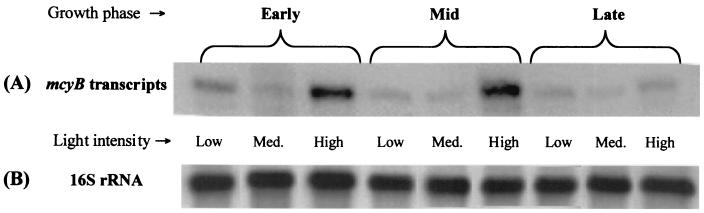
Transcript levels of mcyB from cells grown under different light intensities were higher at high light.
For each light intensity (low, medium, and high), total RNA was extracted from samples taken during early (sample 1), middle (sample 2), and late (sample 3) growth. Equal amounts of RNA for each sample were subjected to RPAs using the antisense probe for mcyB. Levels of mcyB transcripts increased when cells were grown under high light intensities, from samples taken at early and middle growth phases (Fig. 3A). Interestingly, this transcript response at high light was not seen in the late growth phase. To standardize these results, the same RNA was hybridized with a 16S rDNA probe. No differences were seen in 16S rRNA transcripts, relative to total RNA, under all light intensities and growth phases investigated (Fig. 3B). Comparison of mcyB transcripts from cells grown at low and medium light intensities and over growth showed no obvious differences (Fig. 3A).
FIG. 3.
Transcripts from cells grown under low, medium, and high light intensities from early, middle, and late growth phases. (A) mcyB transcript analysis by RPA. (B) Northern blot analysis of 16S rRNA from total RNA used for RPAs.
Transcript levels of mcyB and mcyD increased when cells were transferred from low to high light intensities and decreased when moved into the dark.
M. aeruginosa cells were grown to mid-exponential phase (OD750 = 0.66) under low light and then exposed to high and very high light for periods of 0.5 to 24 h. Transcripts of mcyB and mcyD increased within the first 0.5 h under high light, with no further increase thereafter (Fig. 4). While a difference in mcyB transcript amounts was seen from cells grown at low light (control) compared to high light, no obvious differences were observed when cells were exposed to high or very high light intensities (Fig. 4). Cells maintained in the dark for 2 h showed the lowest mcyB and mcyD transcript levels recorded in this study (Fig. 4).
Light quality affected mcyB and mcyD transcripts.
Responses to light intensities seen via transcript increases and decreases raised questions regarding the nature of the signal for gene transcription. When cells were moved from low white light to red light at the same intensity for 2 h, the amounts of mcyB and mcyD transcripts increased to a level comparable to that seen for high light (Fig. 4). Cells moved to the same intensity of blue light exhibited no change in transcript amounts compared to the control at low white light (Fig. 4).
Stress had a negative effect on mcyB transcription.
To find out if the increased transcript level under red light was due to oxidative stress, cultures were exposed to various stress-inducing factors. MV (1 mM), an agent causing oxidative stress by inhibiting the electron transfer between photosystem II (PSII) and PSI, was added to low- and high-light cultures. Only 10 min after the addition of MV to low-light cultures, the amounts of mcyB transcripts were reduced to levels experienced in cells kept in the dark (Fig. 4). Similar reductions in mcyB transcripts were seen in cells exposed to high light prior to addition of MV (data not shown). Cells under low light exposed to 250 mM NaCl for 1 h exhibited mcyB transcript decreases similar to those seen in cells exposed to MV (Fig. 4). Reduced transcripts seen for both stress factors do not correlate to the increased transcripts seen under red light.
There were no significant trends in cellular toxin content from cells grown under different light intensities.
Toxin content with respect to cell numbers was measured using the PP2A assay based on the inhibition of protein phosphatase activity. All samples exhibited toxin contents ranging from 0.95 ± 0.31 (standard deviation) to 3.43 ± 1.04 10−5 pmol/cell. The sensitivity of the assay led to high standard deviations between measurements of the same sample in repetitive assays. As a result, with the exception of cells at early growth under high light, which showed a lower toxin content (1.73 ± 0.38) than medium (2.82 ± 0.19) and high (3.43 ± 1.04) light-grown cells, there were no significant differences between any of the other samples.
DISCUSSION
The findings presented here show increases in microcystin peptide synthetase and polyketide synthase gene transcription as a result of high light intensities. For the first time, we were able to show that light quality (in the red light spectrum) was responsible for this effect on genes involved in microcystin biosynthesis. Furthermore, we observed that transcript levels were reduced under certain artificial stress conditions. It remains to be investigated whether any changes seen in transcript amounts are affected by transcript stability or instability under the conditions tested.
Increased transcript amounts of mcyB and mcyD seen in cells exposed to 68 and 400 μmol of photons m−2 s−1 were comparable to those seen for red light at 16 μmol of photons m−2 s−1. This intensity of red light is equivalent to the content of red light found during high-light exposure (68 μmol of photons m−2 s−1) and may thus lead to the observed high-light effect. However, the same intensity of blue light had no effect on the amount of transcripts.
At high light intensities, the saturation of the light-harvesting complex leads to the production of free radicals and susceptibility to photoinhibition (42). Thus, enhanced transcription under red and high light conditions may be due to a direct effect of light via a light receptor, an effect on the PS, or oxidative stress. Electron scavengers such as MV have commonly been used to create a situation of oxidative stress via inhibition of the PS (13, 45). Cells exposed to MV for only 10 min showed reduced mcyB and mcyD transcripts. If the high-light effect were due to the inhibition of PS or oxidative stress, transcripts should have increased under these conditions. The addition of a second stress factor, NaCl at a concentration comparable to that in seawater, also resulted in reduced transcript amounts. As NaCl does not inhibit PS, this result together with the response to MV may point to a more general negative effect of stress on the transcription of microcystin synthetase genes.
Comparison of transcript amounts in cells exposed to different light quantities revealed two approximate threshold intensities at which transcription may be initiated. The first increase in transcript amounts, identified in cells grown at low light compared to cells exposed to the dark for 2 h, was similar to the only effect of light reported for another Microcystis strain (23). The second quantum increase occurred between cells grown at 31 and 68 μmol of photons m−2 s−1, with no further transcript increases when cells were exposed to 400 μmol of photons m−2 s−1. Although not directly comparable, several ecological studies have observed a similar threshold when measuring toxin content in cells grown under different light conditions. Increasing microcystin content was observed when the light intensity was raised from approximately 2 to 40 μmol of photons m−2 s−1, depending on the study, with no further increases at higher light intensities (26, 39, 40, 43). This led to the conclusion that light intensities influencing the toxicity of M. aeruginosa are less than about 40 μmol of photons m−2 s−1 (39). Such light intensity is found at a depth of about 1 m during bloom conditions, with intensities of 400 μmol of photons m−2 s−1 measured at the surface (42). This correlates to the highest toxicity measured in the surface waters of a Microcystis bloom during periods of calm weather (2). Unfortunately, direct comparison of light intensities described by various studies is not possible due to different measuring techniques. In this study, a spherical globe measuring light from all angles was used instead of a flat light meter, which is likely to result in slightly lower measurements under diffused light (Li-Cor manual; Walz). Nevertheless, the notion of a light threshold for microcystin production is of interest in the investigation of putative roles for this substance.
Because of the numerous cellular processes affected by light, we cannot conclude that light is the single factor leading to these thresholds. Cell division is influenced by high light, which may have a pronounced effect on transcription and/or toxin production. Highest cell division rates may be expected at light saturation intensity, which for M. aeruginosa PCC7806 is about 32 μmol of photons m−2 s−1 (K. Hesse, E. Dittmann, W. Bleiss, T. Börner, and J.-G. Kohl, unpublished data), falling within the second threshold suggested. It has been reported as a summary of several previous studies that toxin production is limited by cell division rates regardless of the environmental factor under investigation (25). Short-term exposures to high light intensities in the present study revealed transcript increases within 2 h of the move from low to high light and decreases after the move from light into the dark. Cell division rates are also likely to increase and decrease under these conditions, respectively, and thus be coupled to transcription. However, cells of M. aeruginosa divide less than once a day, which suggests that the changes in transcript amount seen after just 2 h are due to a direct response to light and not entirely to cell division (25).
Constant transcript amounts were observed from early to late growth phase in samples from low and medium light intensities. Reduced transcripts visualized under high light during the late growth phase were probably due to increased culture density, reducing the amount of effective light able to penetrate the culture, thus producing a low-light effect on mcyB transcription (31).
Alternatively, the high-light response of transcripts may be repressed by a cell density response, similar to the quorum-sensing regulation of the biosynthesis of several other non-ribosomally produced peptides (8, 44). Preliminary data indicate that mcyD transcripts in an mcyB mutant of M. aeruginosa PCC7806 unable to produce microcystin (7) do not show the upregulated response to high light seen in the wild type (E. Dittmann, unpublished data). This may imply that microcystin plays some role in regulating its own biosynthesis and, taken with the putative effects of cell density, may also have a more general role as a signaling or quorum-sensing molecule.
Unlike the studies mentioned above, and with the exception of cells from sample 1 under high light, cellular toxin content between cells grown at low, medium, and high light intensities for up to 8 days did not change significantly. Toxin content was correlated on a per cell basis, as this parameter was less affected by light intensities than protein or chlorophyll-a contents. Total protein content is reduced with increasing light intensities, possibly due to the breakdown of PS components such as phycobilisomes (27, 42). Similarly, as Microcystis cells adapt to high light, carotenoid levels, providing protection against photooxidative damage, are increased while chlorophyll-a concentrations are reduced (K. Hesse, E. Dittmann, W. Bleiss, T. Börner, and J.-G. Kohl, unpublished data).
The negative trend of total proteins with increasing light may also have influenced microcystin content as reported by previous studies. Increasing microcystin concentrations identified under high light intensities, calculated with respect to total protein, may actually reflect decreasing protein concentrations at constant toxin levels and provide an explanation of varied results from this study and others (39). Similarly, decreasing toxin concentrations identified under high light conditions calculated with respect to dry weight (26, 32) may be a result of increasing cell mass due to the production of high-molecular-weight fatty acids and carbohydrate accumulation rather than toxin content.
Constant or decreasing (for sample 1) toxin contents found by our study do not correlate with our transcript analyses over increasing light intensities. However, toxin measurements detect the amount of microcystin in the cell at the time of sampling only and cannot distinguish whether the peptide is stored at some threshold amount or continuously synthesized or degraded as a result of some environmental factor. Furthermore, increases in toxicity as a result of changing the light intensity from 12 to 37 μmol of photons m−2 s−1 were shown to take longer than 5 days (39) and thus may not be observed in our toxin data. Hence, the identification of short-term regulatory factors upon toxin production is not possible via toxin analyses only. mRNA analysis showing increased transcription of mcyB and mcyD under high light may indicate higher toxin production under this condition. As this is not reflected by increased cellular PP2A inhibition, we speculate that the peptide is either altered to a derivative form not detected by the PP2A assay for microcystin toxicity, otherwise photodegraded, or released by the cell. While intracellular microcystin degradation mechanisms have not been identified, both sunlight irradiation and photolysis with UV light have been implicated in microcystin decomposition and isomerization to nontoxic forms (37).
Active release of microcystins from the cell has not been shown. Increased extracellular microcystin concentrations after exposure to high light intensities (26) have usually been attributed to cell lysis and subsequent leakage of the peptide (4, 16, 32). However, the recent identification of a putative ABC transporter gene (mcyH) located upstream of mcyE (35) may suggest the existence of a cytoplasmic or transmembrane microcystin transport system. Thus, it could be suggested, from the data presented here, that the toxin is constitutively produced under low and medium light intensities, but is exported only when a certain higher threshold intensity is reached. Low cellular toxin concentrations under high light at early growth may be a result of microcystin release before normal intracellular levels have been established. To offset these losses, the cell may engage in increased microcystin production, as shown by higher transcript levels of the biosynthesis genes. Toxin release under different light intensities requires further investigation, as does the possibility of posttranscriptional and posttranslational regulation of microcystin synthetase activity.
From our results, we conclude that light has a positive effect on mcyB and mcyD transcription and that this is not due to oxidative stress. Certain stress factors have a negative influence on transcription. We propose that the microcystin synthetase gene cluster is regulated by light quality, either directly or via another regulatory factor, and that transcription requires different thresholds of light intensity for initiation and upregulation. Cell division, density, and growth do not appear to influence transcription directly but may still be involved in a posttranscriptional regulation of microcystin synthetase gene expression. Alternatively, the lack of correlation between increasing mcyB and mcyD transcription and cellular toxin content may suggest microcystin release from the cell for a putative, as yet unknown, role of this peptide under high light conditions.
ACKNOWLEDGMENTS
This work was financially supported by funds from the DFG (BO1045/13-3) and the ARC, to T.B. and B.A.N., respectively. M.K. is a recipient of scholarships from the CRC for Water Quality and Treatment and the DAAD.
We thank Annegret Wilde and Michael Hisbergues for advice and assistance during this study.
REFERENCES
- 1.An J, Carmichael W W. Use of a colorimetric protein phosphatase inhibition assay and enzyme linked immunosorbent assay for the study of microcystins and nodularins. Toxicon. 1994;32:1495–1507. doi: 10.1016/0041-0101(94)90308-5. [DOI] [PubMed] [Google Scholar]
- 2.Annadotter H, Mattson R, Willen T. En ekologisk studie av blagronnalgen Microcystis toxinproduktion i Finjasjon samt undersokning av algtoxinets forekomst i vattenverkets olika steg. Uppsala, Sweden: Hassel-holms Kummuns Gatukontor and Statens Veterinaermediciniska anstalt och Limnologiska Institutionen i Uppsala; 1991. [Google Scholar]
- 3.Arment A R, Carmichael W W. Evidence that microcystin is a thiotemplate product. J Phycol. 1996;32:591–597. [Google Scholar]
- 4.Berg K, Skulberg O M, Skulberg R. Effects of decaying blue-green algae on the water quality—a laboratory study. Arch Hydrobiol. 1987;108:549–563. [Google Scholar]
- 5.Carmichael W W. The toxins of cyanobacteria. Sci Am. 1994;270:78–86. doi: 10.1038/scientificamerican0194-78. [DOI] [PubMed] [Google Scholar]
- 6.Codd G A, Poon G K. Cyanobacterial toxins. In: Rogers L F, Gallon J R, editors. Biochemistry of algae and cyanobacteria. Oxford, U.K: Clarendon Press; 1988. pp. 283–296. [Google Scholar]
- 7.Dittmann E, Neilan B A, Erhard M, von Döhren H, Börner T. Insertional mutagenesis of a peptide synthetase gene that is responsible for hepatotoxin production in the cyanobacterium Microcystis aeruginosa PCC 7806. Mol Microbiol. 1997;26:779–787. doi: 10.1046/j.1365-2958.1997.6131982.x. [DOI] [PubMed] [Google Scholar]
- 8.Dunny G M, Leonard B A B. Cell-cell communication in gram-positive bacteria. Annu Rev Microbiol. 1997;51:527–564. doi: 10.1146/annurev.micro.51.1.527. [DOI] [PubMed] [Google Scholar]
- 9.Eriksson J E, Paatero G I L, Meriluoto J A O, Codd G A, Kass G E N, Nicotera P, Orrenius S. Rapid microfilament reorganisation induced in isolated rat hepatocytes by microcystin-LR, a cyclic peptide toxin. Exp Cell Res. 1989;185:86–100. doi: 10.1016/0014-4827(89)90039-6. [DOI] [PubMed] [Google Scholar]
- 10.Eriksson J E, Toivola D, Meriluoto J A O, Karaki H, Han Y-G, Hartshorne D. Hepatocyte deformation induced by cyanobacterial toxins reflects inhibition of protein phosphatases. Biochem Biophys Res Commun. 1990;173:1347–1353. doi: 10.1016/s0006-291x(05)80936-2. [DOI] [PubMed] [Google Scholar]
- 11.Falconer E R, Yeung D S K. Cytoskeletal changes in hepatocytes induced by Microcystis toxins and their relation to hyperphosphorylation of cell proteins. Chem-Biol Interact. 1992;81:181–196. doi: 10.1016/0009-2797(92)90033-h. [DOI] [PubMed] [Google Scholar]
- 12.Gleason F K, Wood J M. Secondary metabolism in the cyanobacteria. In: Fay P, Van Baalen C, editors. The cyanobacteria. Amsterdam, The Netherlands: Elsevier; 1987. pp. 437–452. [Google Scholar]
- 13.Haertel H, Haseloff R F, Ebert B, Rank B. Free radical formation in chloroplasts: methyl viologen action. J Photochem Photobiol B Biol. 1992;12:375–385. [Google Scholar]
- 14.Jochimsen E M, Carmichael W W, An J S, Cardo D M, Cookson S T, Holmes C E M, Antunes M B D, Demelo D A, Lyra T M, Barreto V S T, Azevedo S M F O, Jarvis W R. Liver failure and death after exposure to microcystins at a haemodialysis centre in Brazil. N Engl J Med. 1998;338:873–878. doi: 10.1056/NEJM199803263381304. [DOI] [PubMed] [Google Scholar]
- 15.Kleinkauf H, von Döhren H. Nonribosomal biosynthesis of peptide antibiotics. Eur J Biochem. 1990;192:1–15. doi: 10.1111/j.1432-1033.1990.tb19188.x. [DOI] [PubMed] [Google Scholar]
- 16.Lehtimaeki K, Sivonen K, Luukkainen R, Niemelae S I. The effects of incubation time, temperature, light, salinity, and phosphorus on growth and heptotoxin production by Nodularia strains. Arch Hydrobiol. 1994;130:269–282. [Google Scholar]
- 17.Lukac M, Aegerter R. Influence of trace metals on growth and toxin production of Microcystis aeruginosa. Toxicon. 1993;31:293–305. doi: 10.1016/0041-0101(93)90147-b. [DOI] [PubMed] [Google Scholar]
- 18.Luukkainen R, Namikoshi M, Sivonen K, Rinehart K L, Niemelae S I. Isolation and identification of 12 microcystins from four strains and two bloom samples of Microcystis spp.: structure of a new hepatotoxin. Toxicon. 1994;32:133–139. doi: 10.1016/0041-0101(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 19.MacKintosh C, Beattie K A, Klumpp S, Cohen P, Codd G A. Cyanobacterial microcystin-LR is a potent and specific inhibitor of protein phosphatases 1 and 2A from both mammals and higher plants. FEBS Lett. 1990;264:187–192. doi: 10.1016/0014-5793(90)80245-e. [DOI] [PubMed] [Google Scholar]
- 20.Marahiel M A. Multidomain enzymes involved in peptide synthesis. FEBS Lett. 1992;307:40–43. doi: 10.1016/0014-5793(92)80898-q. [DOI] [PubMed] [Google Scholar]
- 21.Neilan B, Jacobs D, Del Dot T, Blackall L L, Hawkins P R, Cox P T, Goodman A E. rRNA sequences and evolutionary relationships among toxic and nontoxic cyanobacteria of the genus Microcystis. Int J Syst Bacteriol. 1997;47:693–697. doi: 10.1099/00207713-47-3-693. [DOI] [PubMed] [Google Scholar]
- 22.Nishiwaki-Matsushima R, Ohta T, Nishiwaki S, Suganuma M, Kohyama K, Ishikawa T, Carmichael W W, Fujiki H. Liver tumor promotion by the cyanobacterial cyclic peptide toxin microcystin-LR. J Cancer Res Clin Oncol. 1992;188:420–424. doi: 10.1007/BF01629424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishizawa T, Asayama M, Fujii K, Harada K, Shirai M. Genetic analysis of the peptide synthetase genes for a cyclic heptapeptide microcystin in Microcystis spp. J Biochem. 1999;126:520–526. doi: 10.1093/oxfordjournals.jbchem.a022481. [DOI] [PubMed] [Google Scholar]
- 24.Oh H-M, Lee S, Jang M-H, Yoon B-D. Microcystin production by Microcystis aeruginosa in a phosphorus-limited chemostat. Appl Environ Microbiol. 2000;66:176–179. doi: 10.1128/aem.66.1.176-179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orr P T, Jones G J. Relationship between microcystin production and cell division rates in nitrogen limited Microcystis aeruginosa cultures. Limnol Oceanogr. 1998;43:1604–1614. [Google Scholar]
- 26.Rapala J, Sivonen K, Lyra C, Niemelae S I. Variation of microcystins, cyanobacterial hepatotoxins, in Anabaena spp. as a function of growth stimuli. Appl Environ Microbiol. 1997;63:2206–2212. doi: 10.1128/aem.63.6.2206-2212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raps S, Kycia J H, Ledbetter M C, Siegelman H W. Light intensity adaptation and phycobilisome composition of Microcystis-aeruginosa. Plant Physiol. 1985;79:983–987. doi: 10.1104/pp.79.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rinehart K L, Namikoshi M, Choi B W. Structure and biosynthesis of toxins from blue-green algae (cyanobacteria) J Appl Phycol. 1994;6:159–176. [Google Scholar]
- 29.Runnegar M T, Kong S, Berndt N. Protein phosphatase inhibition and in vivo hepatotoxicity of microcystins. Am J Physiol. 1993;93:G224–G230. doi: 10.1152/ajpgi.1993.265.2.G224. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Schaefer M R, Golden S S. Differential expression of members of a cyanobacterial psbA gene family in response to light. J Bacteriol. 1989;171:3973–3981. doi: 10.1128/jb.171.7.3973-3981.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sivonen K. Effects of light, temperature, nitrate, orthophosphate, and bacteria on growth of and hepatotoxin production by Oscillatoria agardhii strains. Appl Environ Microbiol. 1990;56:2658–2666. doi: 10.1128/aem.56.9.2658-2666.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sivonen K, Jones G. Cyanobacterial toxins. In: Chorus I, Bartram J, editors. Toxic cyanobacteria in water: a guide to their public health consequences, monitoring and management. London, U.K: E & FN Spoon; 1999. pp. 41–111. [Google Scholar]
- 34.Talling J F, Driver D. Some problems in the estimation of chlorophyll a in phytoplankton. In: Doty E, editor. Proceedings of the Conference on Primary Productivity Measurement, Marine and Freshwater. Washington, D.C.: U.S. Atomic Energy Commission; 1963. p. 142. [Google Scholar]
- 35.Tillett D, Dittmann E, Erhard M, von Dören H, Börner T, Neilan B A. Structural organisation of microcystin biosynthesis in M. aeruginosa PCC7806: an integrated peptide-polyketide synthetase system. 2000. Chem. Biol., in press. [DOI] [PubMed] [Google Scholar]
- 36.Toivola D M, Eriksson J E, Brautigan D L. Identification of protein phosphatase 2A as the primary target for microcystin-LR in rat liver homogenates. FEBS Lett. 1994;344:175–180. doi: 10.1016/0014-5793(94)00382-3. [DOI] [PubMed] [Google Scholar]
- 37.Tsuji K, Watanuki T, Kondo F, Watanabe M F, Suzuki S, Nakazawa H, Suzuki M, Uchida J, Harada K I. Stability of microcystins from cyanobacteria. II. Effect of UV light on decomposition and isomerization. Toxicon. 1995;33:1619–1631. doi: 10.1016/0041-0101(95)00101-8. [DOI] [PubMed] [Google Scholar]
- 38.Utkilen H, Gjolme N. Iron-stimulated toxin production in Microcystis aeruginosa. Appl Environ Microbiol. 1995;61:797–800. doi: 10.1128/aem.61.2.797-800.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Utkilen H, Gjolme N. Toxin production by Microcystis aeruginosa as a function of light in continuous cultures and its ecological significance. Appl Environ Microbiol. 1992;58:1321–1325. doi: 10.1128/aem.58.4.1321-1325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Westhuizen A J, Eloff J N. Effect of temperature and light on the toxicity and growth of the blue-green alga Microcystis aeruginosa (UV-006) Planta. 1985;163:55–59. doi: 10.1007/BF00395897. [DOI] [PubMed] [Google Scholar]
- 41.van der Westhuizen A J, Eloff J N, Krueger G H J. Effect of culture age and pH of the culture medium on the composition of the toxin of the cyanobacterium Microcystis aeruginosa (UV-006) S-Afr Tydskr Plantkunde. 1988;54:372–374. [Google Scholar]
- 42.Walsh K, Jones G J, Dunstan R H. Effect of irradiance on fatty acid, carotenoid, total protein composition and growth of Microcystis aeruginosa. Phytochemistry. 1997;44:817–824. [Google Scholar]
- 43.Watanabe M F, Oishi S. Effects of environmental factors on toxicity of a cyanobacterium (Microcystis aeruginosa) under culture conditions. Appl Environ Microbiol. 1985;49:1342–1344. doi: 10.1128/aem.49.5.1342-1344.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whiteley M, Lee K M, Greenberg E P. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winterbourn C C. Production of hydroxyl radicals from paraquat radicals and H2O2. FEBS Lett. 1981;128:339–342. doi: 10.1016/0014-5793(81)80112-3. [DOI] [PubMed] [Google Scholar]