Abstract
Hexavalent chromium [Cr(VI)] is a common environmental carcinogen causing lung cancer in humans. This study investigates the mechanism of Cr(VI) carcinogenesis focusing on the role of the epitranscriptomic dysregulation. The epitranscriptomic effect of Cr(VI) was determined in Cr(VI)-transformed human bronchial epithelial cells, chromate-exposed mouse and human lungs. The epitranscriptomic effect and its role in Cr(VI)-induced cell transformation, cancer stem cell (CSC)-like property, and tumorigenesis were determined by microarray analysis, soft agar colony formation, suspension spheroid formation, and mouse xenograft tumorigenesis assays. It was found that chronic Cr(VI) exposure causes epitranscriptomic dysregulations as evidenced by the increased levels of total RNA N6-methyladenosine (m6A) modification and the RNA m6A methyltransferase like-3 (METTL3) in Cr(VI)-transformed cells and chromate exposure-caused mouse and human lung tumors. Knockdown of METTL3 expression in Cr(VI)-transformed cells significantly reduces their m6A levels and transformed phenotypes and tumorigenicity in mice. Moreover, knockdown of METTL3 expression in parental nontransformed cells significantly reduces the capability of chronic Cr(VI) exposure to induce cell transformation and CSC-like property. Together, this study reveals that chronic Cr(VI) exposure is capable of altering cellular epitranscriptome by increasing the m6A RNA modification via upregulating the RNA methyltransferase METTL3 expression, which plays an important role in Cr(VI)-induced cell transformation, CSC-like property, and tumorigenesis.
Keywords: hexavalent chromium, epitranscriptome, RNA modification, m6A, METTL3, cancer stem cell (CSC)-like property
Hexavalent chromium [Cr(VI)] is listed as one of the “Top 20 Hazardous Substances” by the U.S. Environmental Protection Agency and the Agency for Toxic Substances and Disease Registry (ATSDR, 2019). It is estimated that over a thousand hazardous waste sites on U.S. National Priority List contain high levels of Cr(VI) (ATSDR, 2012; ATSDR, 2019). Moreover, Cr(VI) is classified as a Group I carcinogen for humans by the International Agency for Research on Cancer (IARC, 1990). Chronic Cr(VI) exposure causes lung cancer and other types of cancer in humans (IARC, 1990). However, the mechanism of Cr(VI) carcinogenesis remains elusive (Chen et al., 2019; Holmes et al., 2008; Humphries et al., 2016; Wang and Yang, 2019; Wang et al., 2019a,b, 2021; Zhu and Costa, 2020).
The recent advances in demonstrating the dynamic and reversible feature of RNA modifications and their roles in regulating gene expression and many biological processes represent a breakthrough in our understanding of RNA biology and functions (Frye et al., 2018; Meyer and Jaffrey, 2014; Roundtree et al., 2017). The recognition of crucial biological functions of RNA modifications led to the birth of terms of “epitranscriptome” and “epitranscriptomics” (Saletore et al., 2012). RNA modifications refer to naturally occurring chemical modifications in nucleotides and ribose of RNA molecules. More than 150 types of chemical modifications in RNAs have been identified to date (Boccaletto and Bagiński, 2021; Boccaletto et al., 2018). Similar to DNA and histone protein modifications, RNA modifications are also dynamically regulated by 3 groups of proteins known as: (1) “Writers” that deposit various modifications to RNA molecules; (2) “Erasers” that remove modifications from RNA molecules; (3) “Readers” that recognize, bind to the modified RNA molecules and mediate the functional outcomes of RNA modifications (Shi et al., 2019; Zaccara et al., 2019). Although the roles of DNA and protein modifications in environmental carcinogenesis have been widely explored, the role of RNA modifications (the epitranscriptome) in environmental carcinogenesis especially metal carcinogenesis has been rarely studied (Yang, 2020).
Among over 150 types of chemical modifications in various RNA molecules, the N6-methyladenosine (m6A) modification is now known as the most prevalent internal modification in eukaryotic messenger RNAs (mRNAs) (Uddin et al., 2020, 2021). The installment of m6A in RNA molecules is accomplished by a multicomponent methyltransferase complex termed as the “m6A writer,” which catalyzes the transfer of a methyl group from S-adenosylmethionine to the N-6 position of adenosine (Uddin et al., 2021; Yang et al., 2018; Yue et al., 2015). The m6A writer complex includes several methyltransferases such as methyltransferase like-3 (METTL3) and METTL14, Wilms’ tumor 1-associating protein (WTAP), and other components. Although METTL3 is the major component that plays the catalytic function in the writer complex, the other components of the complex are also essential for achieving the m6A installation in RNAs (Yue et al., 2015). The erasers (demethylases) responsible for removing the m6A from RNA molecules are fat mass and obesity-associated protein (FTO) and AlkB homolog 5 (ALKBH5; Jia et al., 2011; Zheng et al., 2013). The reader proteins that determine the functional outcomes of the m6A modification include the YTH domain-containing family proteins 1–3 (YTHDF 1–3), YTH domain-containing protein1-2, and some others (Uddin et al., 2021; Yang et al., 2018; Yue et al., 2015). Although recent studies showed that dysregulations of m6A levels and its writers, erasers, and readers play important roles in cancer (Barbieri and Kouzarides, 2020; Huang et al., 2020; Liu et al., 2019; Uddin et al., 2021), it remains largely unexplored whether the m6A modification dysregulation plays a role in environmental carcinogenesis (Yang, 2020). The objective of this study is to investigate the mechanism of Cr(VI) carcinogenesis focusing on the effect of chronic Cr(VI) exposure on the epitranscriptome.
MATERIALS AND METHODS
Cell culture and cell transformation by chronic Cr(VI) exposure
Immortalized nontumorigenic human bronchial epithelial BEAS-2B cells were purchased from American Type Culture Collection (Manassas, Virginia). Immortalized nontumorigenic human bronchial epithelial 16HBE cells were generously provided by Dr Dieter C. Gruenert (University of California San Francisco, San Francisco, California). BEAS-2B cells were cultured in Dulbecco’s Modified Eagle Medium supplemented with 5% fetal bovine serum (FBS) and 16HBE cells were cultured in Minimum Essential Media supplemented with 10% FBS. BEAS-2B and 16HBE cell transformation processes by chronic Cr(VI) exposure were described in our recent publications (Wang et al., 2018, 2019a). Briefly, BEAS-2B and 16HBE cells were continuously exposed to a vehicle control or 0.25 μM of Cr(VI) (K2Cr2O7) for 20 and 40 weeks, respectively. Chronic Cr(VI) exposure-induced cell transformation was confirmed by soft agar colony formation assay, suspension serum-free culture sphere formation assay, and mouse tumorigenesis analysis as described in our recent publication (Wang et al., 2018).
Mouse lung tumorigenesis induced by chronic Cr(VI) exposure via oropharyngeal aspiration
Mouse chronic Cr(VI) exposure via oropharyngeal aspiration was performed in Dr Patti C Zeidler-Erdely’s laboratory at National Institute for Occupational Safety and Health (Morgantown, West Virginia). The detailed Cr(VI) exposure protocol and Cr(VI)-induced lung tumorigenesis in mice were reported in our recent publication (Zeidler-Erdely et al., 2020). Briefly, lung tumor susceptible male A/J mice, age 5–7 weeks, were purchased from Jackson Laboratories (Bar Harbor, Maine) and housed in an AAALAC-accredited, specific pathogen-free, environmentally controlled facility. Mice were housed 2 per cage in ventilated cages and provided HEPA-filtered air under a controlled light cycle (12-h light/12-h dark). Animals were acclimated to the animal facility for 1 week and allowed access to a conventional diet (6% Irradiated NIH-31 Diet, Harlan Teklad, Madison, Wisconsin) and filtered tap water ad libitum. All animal studies were approved by the Centers for Disease Control Morgantown Institutional Animal Care and Use Committee. The applicable international, national, and/or institutional guidelines for the care and use of animals were followed. After 1 week acclimation, 6- to 8-week-old male A/J mice were exposed to a vehicle control (PBS) or a suspension of calcium chromate (CaCrO4; 100 μg per mouse) via oropharyngeal aspiration once per week for 26 weeks. After 26 weeks vehicle control or chromate exposure, mice were maintained for additional 44 weeks and then euthanized. Vehicle control- and chromate-exposed mouse lungs were either freshly frozen for total RNA extraction for m6A level analysis or fixed in 10% buffered formalin, paraffin embedded and sectioned for pathological evaluations, and METLL3 immunofluorescence (IF) staining.
Human lung tissue sections from chronic chromate-exposed workers
The chromate-exposed workers’ lung tissue sections were processed and provided by Dr Kazuya Kondo from Tokushima University (Tokushima City, Japan). The proposal and protocol for using chromate-exposed workers’ lung tissue sections for METTL3 IF staining were reviewed and approved by the Institutional Review Board of University of Kentucky and Tokushima University. Chronic chromate exposure-induced lung cancer in chromate workers was characterized and reported in Dr Kazuya Kondo’s previous publications (Ali et al., 2011; Kondo et al., 2006; Tsuboi et al., 2020).
Human m6A RNA modification microarray analysis
Total RNAs from chronic Cr(VI) exposure-transformed BEAS-2B cells [BEAS-2B-Cr(VI)] and the passage-matched control BEAS-2B cells (BEAS-2B-Control) were submitted to Arraystar Inc. (Rockville, Maryland) for human m6A RNA modification microarray analysis. The microarray raw data were further analyzed by Arraystar Inc. and the microarray raw data were deposited to the National Center for Biotechnology Information’s data repository (GSE186605).
Nude mouse tumorigenesis assay
BEAS-2B-Cr(VI) with control shRNA [BEAS-2B-Cr(VI)-Control shRNA] or METLL3 stable knockdown shRNA [BEAS-2B-Cr(VI)-METTL3 shRNA] were used to test the effect of knocking down METTL3 expression on the tumor forming capability of Cr(VI)-transformed cells in nude mice. The animal study protocol was reviewed and approved by the University of Kentucky Animal Care and Use Committee. Briefly, BEAS-2B-Cr(VI)-Control shRNA and BEAS-2B-Cr(VI)-METTL3 shRNA cells (1.5 × 106 cells in 0.1 ml of 1:1 growth factor-reduced Matrigel and PBS) were injected subcutaneously into the flank areas of 7-week-old female nude mice (Nu/Nu, Charles River Laboratories). There were 7 mice in BEAS-2B-Cr(VI)-Control shRNA cell injection group and 8 mice in BEAS-2B-Cr(VI)-METTL3 shRNA cell injection group. Mice were maintained under specific pathogen-free conditions and monitored once per week before tumor formation. Once the subcutaneous xenograft tumor developed, mice were monitored twice per week. All mice were euthanized 12 weeks after the cell injection and the xenograft tumors were harvested and photographed.
Generation of vector control and METTL3 stable knockdown cells
Parental BEAS-2B cells, BEAS-2B-Cr(VI) cells, and 16HBE cells were used to generate vector control and METTL3 stable knockdown cells following the protocol described in our previous publications (Humphries et al., 2014; Zhao et al., 2011). Briefly, cells were transduced with nontargeting control shRNA lentiviral particles (pZIP-hCMV-ZsGreen-Puro-Control shRNA) or METTL3-specific targeting shRNA lentiviral particles (pZIP-hCMV-ZsGreen-Puro-METTL3 shRNA) obtained from Transomic Technologies (Huntsville, Alabama). Forty-eight hours after the lentiviral particle transduction, cells were selected with puromycin (1 μg/ml) following the procedures described in our previous studies (Humphries et al., 2014; Zhao et al., 2011). The METTL3 stable knockdown in parental BEAS-2B, Cr(VI)-transformed BEAS-2B [BEAS-2B-Cr(VI)] or Cr(VI)-transformed 16HBE [16HBE-Cr(VI)] cells were confirmed by Western blot analysis.
The m6A RNA modification ELISA-like colorimetric assay
Cellular and mouse lung tissue total RNA m6A levels were also measured using the EpiQuik m6A RNA Methylation Quantification Kit (Catalog No.: P-9005-96) from EpiGenTEK (Farmingdale, New York) following the manufacturer’s detailed instructions.
Western blot analysis
Cells were collected and washed with PBS and lysed using cell lysis buffer following our published protocol (Wang et al., 2012, 2013, 2014) and subjected to SDS–polyacrylamide gel electrophoresis (10–30 μg of protein/lane). Western blots were performed 3 times using cell pellets collected at different times. The following primary antibodies were used: anti-METTL3, anti-METTL14, anti-WTAP, FTO, anti-ALKBH5, and anti-YTHDF1–3 (Abcam, Cambridge, Massachusetts); anti-Oct4, anti-KLF4, anti-KLF5, and anti-NANOG (Cell Signaling Technology, Beverly, Massachusetts); and anti-β-actin (Millipore Sigma, St Louis, Missouri).
Soft agar colony formation assay
The soft agar colony formation assay was performed following our previous protocol (Li et al., 2019; Wang et al., 2020; Yang et al., 2005). Briefly, cells were harvested and suspended in their corresponding culture media containing 10% FBS at a concentration of 0.5 × 104 cells/ml. Normal melting point agar (5 ml of 0.6% agar in culture media) was placed into each 60-mm cell culture dish as the bottom agar. After solidification of the bottom agar, 4 ml of cell mixture consisting of 2 ml of cell suspension (0.5 × 104 cells/ml) and 2 ml of 0.8% lower melting point agar in culture media containing 10% FBS were poured over the bottom agar. After solidification of the upper agar, 3 ml of culture media containing 10% FBS were added, and dishes were incubated at 37°C in a humidified 5% CO2 atmosphere. For testing the effect of pharmacological inhibitors treatment on soft agar colony formation of Cr(VI) exposure-transformed cells, the inhibitors were added to upper agar and culture media. Soft agar colonies were stained with 0.003% crystal violet, photographed, and counted (if >100 μm) after 4-week incubation.
Serum-free suspension culture sphere formation assay
The serum-free suspension culture sphere formation assay was carried out to evaluate the cancer stem cell (CSC)-like property of Cr(VI)-transformed cells following the published protocol with minor modifications (Dontu et al., 2003; Wang et al., 2019b; Xiao et al., 2018). Briefly, single cells (2.5 × 103) were plated in ultralow attachment 24-well culture plates (Corning, Corning, New York). Cells were suspended in serum-free media supplemented with human recombinant basic fibroblast growth factor (20 ng/ml), human recombinant epidermal growth factor (20 ng/ml; R&D, Minneapolis, Minnesota), B27 (50 times diluted from the original 50× stock solution, Invitrogen, Carlsbad, California), and heparin (4 μg/ml, Sigma-Aldrich). For testing the effect of pharmacological inhibitors treatment on suspension culture sphere forming efficiency of Cr(VI)-transformed cells, inhibitors were added into the suspension media when cells were plated. Plates were incubated at 37°C in a humidified 5% CO2 atmosphere for 10 days. Spheres were viewed, photographed, and counted (if > 100 μm) under a phase-contrast microscope after 10-day culture.
Mouse and human lung tissue IF staining
The mouse and human lung tissue section IF staining was carried out following our previous procedures (Wang et al., 2018; Zhao et al., 2010). The presented IF staining pictures are the overlaid images of METTL3 staining in red fluorescence with nuclear 4′6-diamidino-2-phenylindole staining in blue fluorescence. The IF staining images were taken and overlaid using Nikon NIS-Elements software.
Statistical analysis
The statistical analyses for the significance of differences between different groups (mean ± standard deviation [SD]) were carried out by testing different treatment effects using 2-tailed t tests for a comparison of 2 data sets. A p <.05 was considered statistically significant. The significance of difference in nude mouse xenograft tumor incidence rate was tested using Fisher’s exact test. A p < .05 was considered statistically significant.
RESULTS
Chronic Cr(VI) Exposure Dysregulates the Epitranscriptome
To explore whether chronic Cr(VI) exposure dysregulates cells’ epitranscriptome, we first performed the m6A epitranscriptomic microarray analysis of total RNAs extracted from chronic low-dose Cr(VI) exposure-transformed human bronchial epithelial BEAS-2B cells [BEAS-2B-Cr(VI)] and the passage-matched control BEAS-2B cells (BEAS-2B-Control). The microarray results presented as the heatmap show that the total RNA m6A modification levels in Cr(VI)-transformed BEAS-2B cells are greatly increased compared with that in the passage-matched control BEAS-2B cells (Figure 1A). These microarray results are further confirmed by using an EpiQuik m6A RNA Methylation Quantification Assay directly measuring total RNA m6A levels. When compared with the passage-matched control BEAS-2B cells, the total RNA m6A levels in chronic BEAS-2B-Cr(VI) cells is about 3.5-fold higher (Figure 1B). In addition, a significantly higher level of total RNA m6A modification is also detected in chronic Cr(VI) exposure-transformed human bronchial epithelial cell line 16HBE cells, compared with their corresponding passage-matched control 16HBE cells (Supplementary Figure 1).
Figure 1.
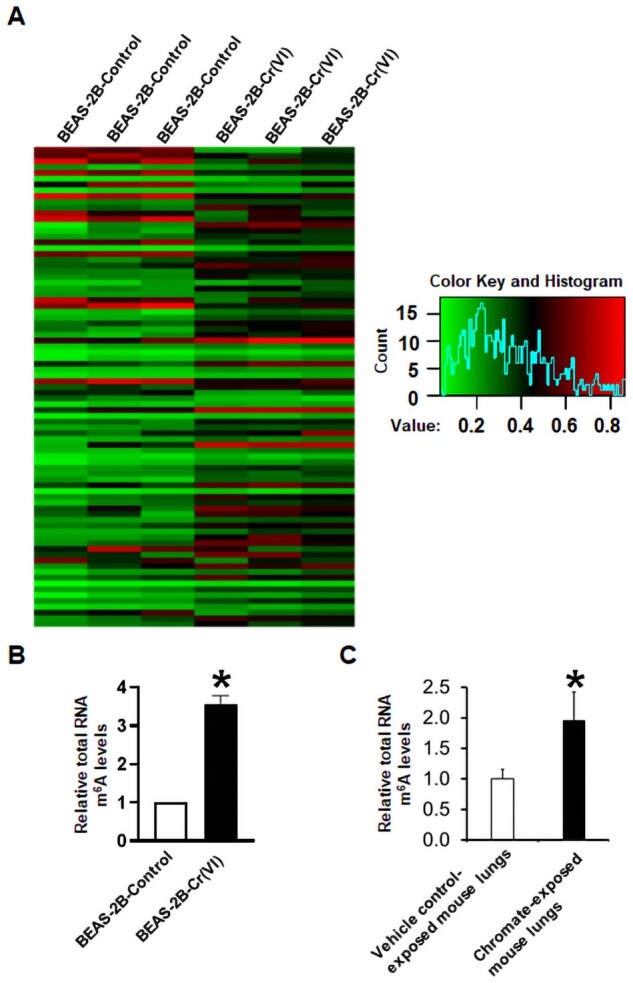
Total RNA N6-methyladenosine (m6A) modification levels are significantly increased in chronic hexavalent chromium [Cr(VI)] exposure-transformed human bronchial epithelial cells and chronic chromate-exposed mouse lung tissues. A, The heatmap from m6A microarray analysis showing the extent of total messenger RNA m6A methylation in passage-matched control cells (BEAS-2B-Control) and chronic Cr(VI) exposure-transformed cells [BEAS-2B-Cr(VI)]. B and C, Relative total RNA m6A levels measured by using the EpiQuik m6A RNA Methylation Quantification Kit. The total RNA m6A levels in Cr(VI)-transformed cells (B) or chromate-exposed mouse lung tissues (C) are expressed relative to the passage-matched control cells (means ± SD, n = 3) (B) or vehicle control-exposed mouse lungs (means ± SD, n = 6) (C), respectively. *p < .05.
We next sought to determine whether chronic Cr(VI) exposure also increases total RNA m6A modification levels in mouse lung tissues. Male A/J mice were exposed to a vehicle control (PBS) or CaCrO4 (100 μg per mouse) via oropharyngeal aspiration once per week for 26 weeks and euthanized 44 weeks after the last chromate exposure (Zeidler-Erdely et al., 2020). By using the EpiQuik m6A RNA Methylation Quantification Assay, it was determined that the total RNA m6A levels in chronical chromate-exposed mouse lung tissues are significantly higher than that in vehicle PBS-exposed mouse lung tissues (Figure 1C). Collectively, these results indicate that chronic Cr(VI) exposure alters functional RNA modifications as evidenced by the significantly higher levels of total RNA m6A modification in chronic Cr(VI) exposure-transformed human bronchial epithelial cells and in chronic chromate-exposed mouse lung tissues.
Chronic Cr(VI) Exposure Upregulates the RNA Methyltransferase METTL3 Expression
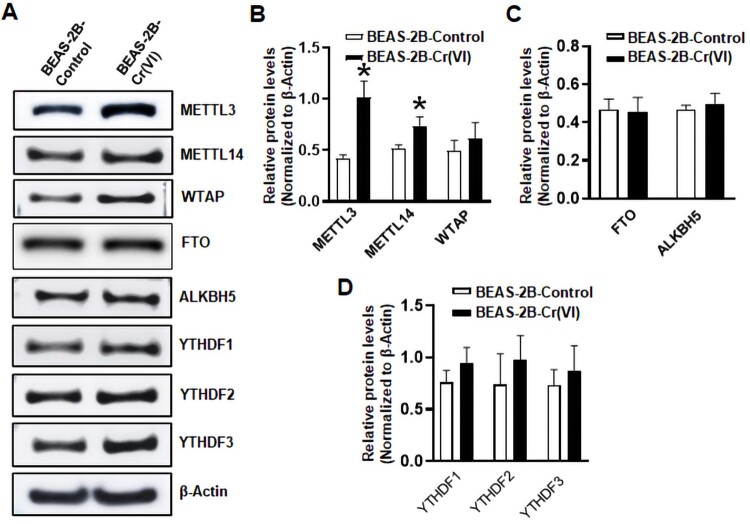
We next explored the mechanism of how chronic Cr(VI) exposure increases total RNA m6A levels. The RNA m6A modification level is dynamically regulated by the m6A writers, erasers, and its interaction with reader proteins (Uddin et al., 2021). Our Western blot analysis revealed that the protein levels of methyltransferase METTL3 and METTL14 in the m6A writer complex are significantly higher in Cr(VI)-transformed BEAS-2B cells than in the passage-matched control BEAS-2B cells (Figs. 2A and B). Moreover, a significant increase of METTL3 protein levels is also detected in Cr(VI)-transformed 16HBE cells (Supplementary Figure 2A). However, the protein levels of WTAP, another important component in the m6A writer complex, the m6A erasers FTO and ALKBH5, and the m6A readers YTHDF1-3 are not dramatically changed in Cr(VI)-transformed BEAS-2B cells (Figs. 2A–D). Similar results are also observed in Cr(VI)-transformed 16HBE cells (Supplementary Figs. 2A–C).
Figure 2.
The RNA methyltransferase methyltransferase like-3 expression levels are significantly upregulated in chronic hexavalent chromium [Cr(VI)] exposure-transformed human bronchial epithelial cells. A, Representative Western blot analysis images for the selected N6-methyladenosine (m6A) writers, erasers, and reader proteins in passage-matched control cells and Cr(VI)-transformed cells. B–D, The quantitative results of Western blot analysis of m6A writers, erasers, and reader proteins in passage-matched control cells and Cr(VI)-transformed cells. The corresponding Western blot protein band intensities were quantified using the ImageJ software and normalized by the intensity of the β-actin protein band (means ± SD, n = 3). *p < .05.
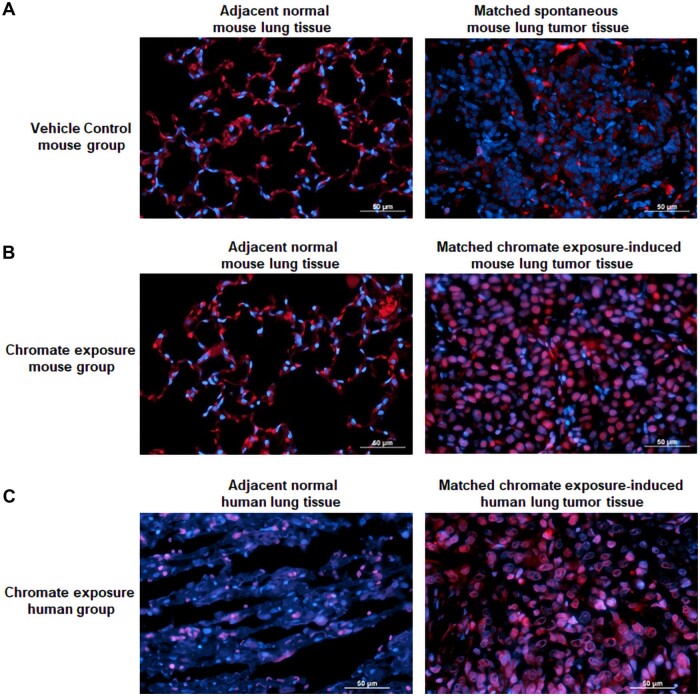
We further determined whether chronic Cr(VI) exposure upregulates METTL3 expression levels in mice and humans. As mentioned above, male A/J mice were exposed to a vehicle control (PBS) or CaCrO4 (100 μg per mouse) via oropharyngeal aspiration once per week for 26 weeks and euthanized 44 weeks after the last chromate exposure (Zeidler-Erdely et al., 2020). Due to the long period (77 weeks) of this experiment, the mice in the vehicle control treatment group also developed spontaneous lung tumors. However, no increase of METLL3 protein levels is detected in mouse spontaneous lung tumors, compared with the adjacent normal lung tissues (Figure 3A). In sharp contrast, the METTL3 protein levels are drastically increased in chronic chromate exposure-caused mouse lung tumors (Figure 3B). Moreover, a great increase in METTL3 protein levels is also detected in a 40-year old nonsmoker male chromate worker exposed to chromate for 22 years (Figure 3C). These results along with the results presented in Figure 1C suggest that chronic Cr(VI) exposure may also upregulate the methyltransferase METTL3 expression levels in mouse and human lungs and increases total RNA m6A modification levels in lung tissues.
Figure 3.
The RNA methyltransferase methyltransferase like-3 (METTL3) expression levels are significantly upregulated in chronic hexavalent chromium [Cr(VI)] exposure-caused mouse and human lung tumor tissues. A–C, The representative overlaid immunofluorescence images of METTL3-positive nuclear staining in red fluorescence with nuclear DNA 4′6-diamidino-2-phenylindole staining in blue fluorescence. Six-week-old male A/J mice were exposed to a vehicle control (50 µl, PBS) (A) or 100 µg of calcium chromate (50 µl, suspended in PBS) (B) via oropharyngeal aspiration once a week for 26 weeks and sacrificed 44 weeks after last chromate exposure. Human lung tissues (C) were from a 40-year-old nonsmoker male chromate worker exposed to chromate for 22 years. Similar staining results were obtained from another 2 control group mice, 2 chromate exposure group mice, and 2 chromate workers. Scale bar: 50 µm.
Inhibiting Methyltransferase Activities Reduces the Transformed Phenotypes of Cr(VI)-Transformed Cells
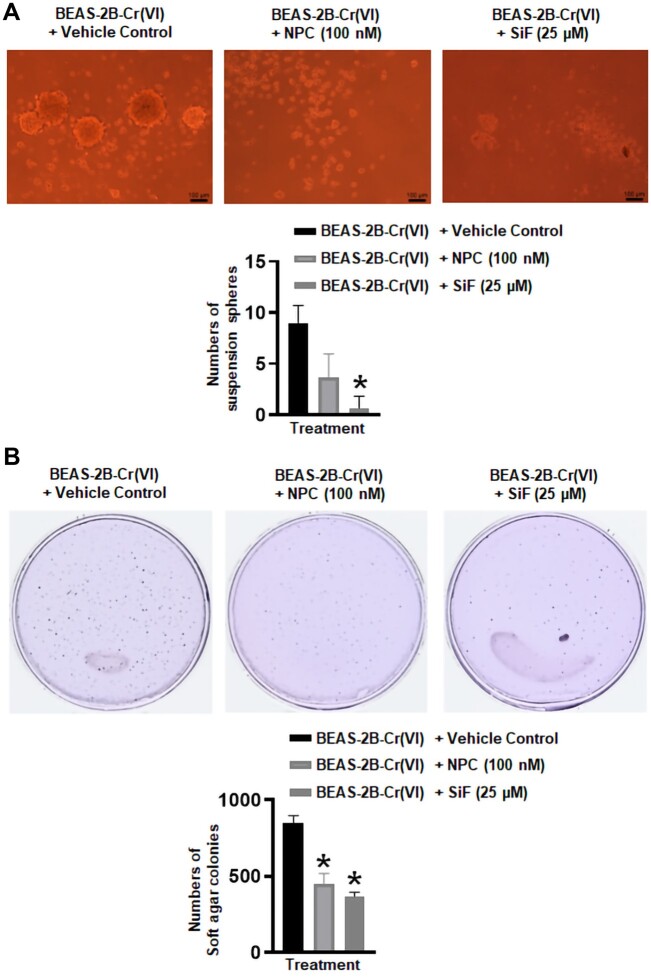
We next explored the potential role of METTL3 upregulation in Cr(VI)-induced cell transformation and tumorigenesis. We first used 2 broad methyltransferase inhibitors (neplanocin A [NPC] and sinefungin [SiF]) to inhibit METTL3 and other methyltransferase activities and determined their effects on the transformed phenotypes of Cr(VI)-transformed cells. It was found that treatment with 100 nM of NPC that have no significant effects on Cr(VI)-transformed cell monolayer growth greatly reduce the number of suspension spheres formed by Cr(VI)-transformed BEAS-2B cells (Figure 4A). Similar results were achieved with another methyltransferase inhibitor SiF (25 µM) treatment. In addition, NPC or SiF treatment also significantly decreased the number of suspension spheres formed by Cr(VI)-transformed 16HBE cells (Supplementary Figure 3). These results suggest that the activities of METTL3 and/or other methyltransferases are important for maintaining chronic Cr(VI) exposure-induced CSC-like property. In addition, treatment with 100 nM of NPC or 25 µM of SiF also significantly reduced the number of soft agar colonies formed by Cr(VI)-transformed BEAS-2B cells (Figure 4B). Together, these results suggest an important role of METTL3 and/or other methyltransferases in maintaining the transformed phenotypes of Cr(VI)-transformed cells.
Figure 4.
Pharmacological inhibitions of the methyltransferase activities significantly reduce the transformed phenotypes of hexavalent chromium [Cr(VI)]-transformed cells. A, Representative suspension culture sphere images and the quantitative results (means ± SD, n = 3) of the methyltransferase inhibitor treatment in Cr(VI)-transformed cells. Cells were treated once with a vehicle control, NPC (neplanocin A, 100 nM), or SiF (sinefungin, 25 µM) when cells were seeded into the 24 well-plate. The suspension spheres were photographed and counted (if >100 µm) 10 days after the cell seeding. B, Representative soft agar colony plate images and the quantitative results (means ± SD, n = 3) of the methyltransferase inhibitor treatment in Cr(VI)-transformed cells. Cells were treated once with a vehicle control, NPC (100 nM), or SiF (25 µM) when cells were seeded into the 6-cm dish. The soft agar colony plates were photographed and the colonies were counted (if > 100 µm) 4 weeks after the cell seeding. *p < .05, compared with the vehicle control-treated cells.
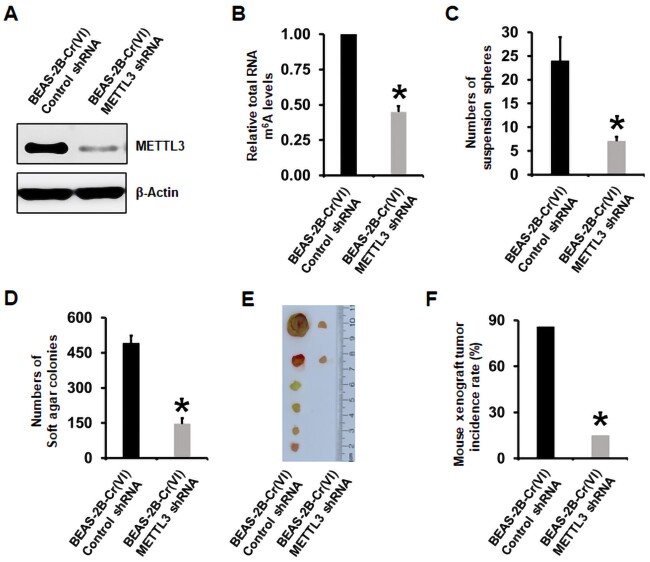
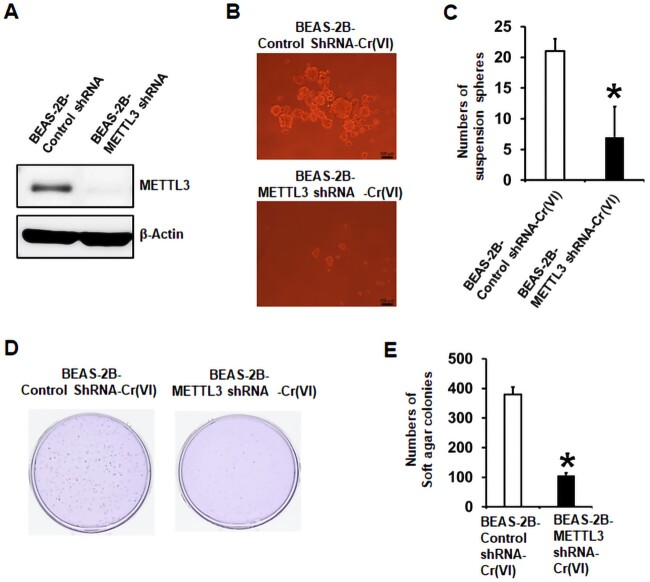
METTL3 Knockdown in Cr(VI)-Transformed Cells Reduces their m6A Levels, Transformed Phenotypes, and Tumorigenicity
To more specifically determine the role of METTL3 upregulation in chronic Cr(VI) exposure-induced cell transformation, CSC-like property, and tumorigenesis, we next took a genetic approach to specifically knockdown METTL3 expression by using METTL3-specific targeting shRNAs. We first knocked down METTL3 expression in Cr(VI)-transformed BEAS-2B and 16HBE cells. The METTL3 knockdown efficiencies were confirmed by Western blot analysis (Figure 5A and Supplementary Figure 4A). It was found that METTL3 knockdown significantly decreased total RNA m6A levels in Cr(VI)-transformed cells (Figure 5B and Supplementary Figure 4B), confirming that METTL3 upregulation plays a critical role in increasing total RNA m6A modification levels by chronic Cr(VI) exposure. Knockdown of METTL3 also significantly reduced the number of suspension spheres and soft agar colonies formed by Cr(VI)-transformed cells (Figs. 5C and D; Supplementary Figure 5). Moreover, METTL3 knockdown significantly decreased the tumorigenicity of Cr(VI)-transformed BEAS-2B cells (Figs. 5E and F): the tumor incidence rate was reduced from 85.7% in mice injected with BEAS-2B-Cr(VI)-Control shRNA cells to 25% (p < .05) in mice injected with BEAS-2B-Cr(VI)-METTL3 shRNA cells.
Figure 5.
Stable knockdown of methyltransferase like-3 (METTL3) in hexavalent chromium [Cr(VI)]-transformed cells significantly reduces their total RNA N6-methyladenosine (m6A) levels, transformed phenotypes, and tumorigenicity. A, Representative Western blot image showing the METTL3 knockdown efficiency in Cr(VI)-transformed cells. B, Effect of METTL3 knockdown in Cr(VI)-transformed cells on their total RNA m6A levels measured by using the EpiQuik m6A RNA Methylation Quantification Kit. The results are expressed relative to the control shRNA cells (means ± SD, n = 3). (C and D) Effect of METTL3 knockdown in Cr(VI)-transformed cells on their suspension culture sphere formation (C) or soft agar colony formation (D) (means ± SD, n = 3). E and F, Effect of METTL3 knockdown in Cr(VI)-transformed cells on their tumor forming capability in nude mice. *p < .05.
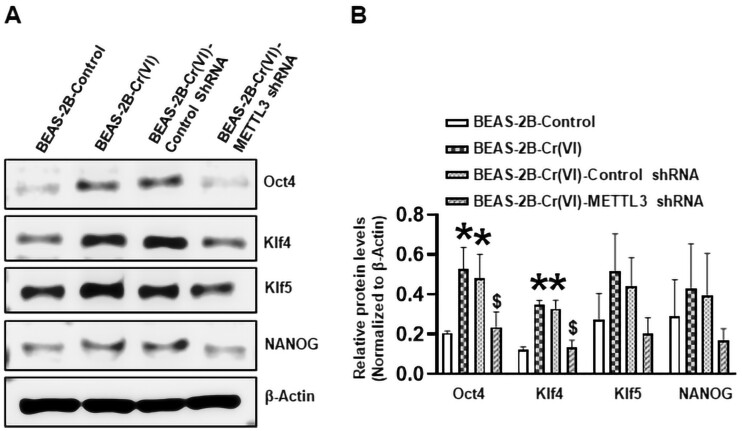
Our recent studies showed that the expression levels of several CSC markers are increased in Cr(VI) and other carcinogen-transformed cells (Lin et al., 2021; Wang et al., 2019a) Western blot was performed to determine the effect of METTL3 knockdown on the expression levels of several CSC markers. As shown in Figure 6, the expression levels of CSC markers Oct4 and Klf4 are significantly increased in Cr(VI)-transformed cells. Stably expressing a control shRNA in Cr(VI)-transformed cells did not significantly affect the levels of these markers. In contrast, stably expressing a METTL3 targeting shRNA significantly reduced the levels of Oct4 and Klf4 (Figure 6). Collectively, these results indicate that chronic Cr(VI) exposure upregulates METTL3 expression to increase total RNA m6A modification levels, which plays an important role in maintaining chronic Cr(VI) exposure-induced CSC-like property, and tumorigenesis.
Figure 6.
Stable knockdown of methyltransferase like-3 (METTL3) in hexavalent chromium [Cr(VI)]-transformed cells reduces their protein levels of several cancer stem cell marks. A, Representative images of Western blot analysis of selected cancer stem cell marks in passage-matched control cells, Cr(VI)-transformed cells and Cr(VI)-transformed cells expressing control nontargeting shRNA or METTL3 targeting shRNA. B, The quantitative results of Western blot analysis of selected cancer stem cell marks in passage-matched control cells, Cr(VI)-transformed cells, and Cr(VI)-transformed cells expressing control nontargeting shRNA or METTL3 targeting shRNA. The corresponding Western blot protein band intensities were quantified using the ImageJ software and normalized by the intensity of the β-actin protein band (means ± SD, n = 3). * p < .05, compared with BEAS-2B-Control. $p < .05, compared with BEAS-2B-Cr(VI) or BEAS-2B-Cr(VI)-Control shRNA cells.
METTL3 Knockdown in Parental BEAS-2B Cells Reduces the Capability of Cr(VI) to Induce Cell Transformation and CSC-Like Property
After demonstrating that METTL3 upregulation plays important roles in maintaining the m6A modification levels and the malignant phenotypes of cells already transformed by Cr(VI). We next sought to determine whether METTL3 plays a role in the process of cell transformation by chronic Cr(VI) exposure. We stably knocked down METTL3 expression in parental BEAS-2B cells and a drastic METTL3 knockdown was confirmed by Western blot analysis (Figure 7A). BEAS-2B-Control shRNA cells and BEAS-2B-METTL3 shRNA knockdown cells were continuously exposed to 0.25 µM of Cr(VI) (K2Cr2O7) for 20 weeks for cell transformation. It was found that a 20-week of Cr(VI) exposure causes transformation of BEAS-2B-Control shRNA cells (Figs. 7B–E) to an extent similar to the transformation of BEAS-2B cells by Cr(VI) reported in our recent publications (Wang et al., 2018, 2019a). However, METTL3 knockdown significantly reduced the capability of chronic Cr(VI) exposure to induce cell transformation and CSC-like property as evidenced by the much fewer suspension spheres and soft agar colonies formed by Cr(VI)-exposed METLL3 knockdown cells (Figs. 7B–E). Western blot analysis revealed that METTL3 expression levels are greatly lower in BEAS-2B-METTL3 shRNA-Cr(VI) cells than in BEAS-2B-Control shRNA-Cr(VI) cells (Supplementary Figure 6). These results suggest that METTL3 upregulation contributes causally to chronic Cr(VI) exposure-induced cell malignant transformation and CSC-like property.
Figure 7.
Stable knockdown of methyltransferase like-3 (METTL3) in parental BEAS-2B cells significantly reduces the capability of chronic hexavalent chromium [Cr(VI)] exposure to induce cell transformation and cancer stem cell-like property. A, Representative Western blot image showing the METTL3 knockdown efficiency in parental BEAS-2B cells. (B and C) Representative images of (B) and quantitative results (means ± SD, n = 3) of (C) suspension spheres formed by chronic Cr(VI)-exposed (20 weeks) BEAS-2B cells with nontargeting control shRNA or METTL3 targeting shRNA. D and E, Representative images of (B) and quantitative results (means ± SD, n = 3) of (C) soft agar colonies formed by chronic Cr(VI)-exposed (20 weeks) BEAS-2B cells with nontargeting control shRNA or METTL3 targeting shRNA. *p < .05. BEAS-2B cells with nontargeting control shRNA or METTL3 targeting shRNA were continuously exposed to 0. 25 µM of Cr(VI) (K2Cr2O7) for 20 weeks for cell transformation as described in Materials and Methods section. At the end of cell transformation, cells were collected for suspension sphere formation (B and C) and soft agar colony formation (D and E) assays.
DISCUSSION
The importance of epitranscriptomic dysregulations in cancer has been evidenced by recent studies, showing that dysregulations in RNA modifications, particularly the m6A modification, are critically involved in cancer progression and resistance to cancer therapies (Barbieri and Kouzarides, 2020; Huang et al., 2020; Liu et al., 2019; Uddin et al., 2021). However, very few studies have been done to determine the effect of environmental exposure on the epitranscriptome or the role of RNA modification dysregulations in environmental carcinogenesis especially in metal carcinogenesis (Yang, 2020). Environmental carcinogens such as metal carcinogens are common and important cancer etiology factors. A better understanding on the mechanism of environmental carcinogenesis will significantly improve our capabilities to more efficiently prevent and treat cancer. The findings from this study strongly suggest that environmental exposure could change cellular epitranscriptome, and that dysregulations of RNA modifications especially the m6A modification could play important roles in environmental carcinogenesis.
Cr(VI) is a common environmental carcinogen that humans are constantly exposed to; however, the mechanism of Cr(VI) carcinogenesis has not been well understood. Previous studies on the mechanism of Cr(VI) carcinogenesis mostly focus on its genotoxic effects. This is due to the fact that Cr(VI) undergoes a series of metabolic reductions inside cells to generate reactive Cr metabolites and reactive oxygen species, which produce various genotoxic effects that may play important roles in Cr(VI) carcinogenesis (Holmes et al., 2008; Nickens et al., 2010; Salnikow and Zhitkovich, 2008; Zhitkovich, 2011). In addition, previous studies showed that Cr(VI) exposure also causes nongenotoxic effects such as epigenetic changes, as evidenced by the increased DNA methylation and dysregulated histone protein posttranslational modifications in Cr(VI)-exposed cells and Cr(VI) exposure-caused human lung cancer tissues (Brocato and Costa, 2013; Chervona et al., 2012). Together, previous studies indicate that Cr(VI) exposure is capable of altering DNA and histone protein modifications, which may play important roles in Cr(VI) carcinogenesis. However, it is currently unknown whether Cr(VI) exposure changes RNA modifications and whether RNA modification dysregulations play critical roles in Cr(VI) carcinogenesis. This study provides novel evidence demonstrating that, in addition to its previously reported genotoxic and epigenetic effects, chronic Cr(VI) exposure also causes significant changes in the epitranscriptome, opening new avenues for studying the mechanism of Cr(VI) carcinogenesis.
The evidence presented in this study showing the epitranscriptomic effect of chronic Cr(VI) exposure is multifaceted: (1) It was first found that the levels of total RNA m6A modification, the most prevalent modification in eukaryotic mRNAs, are significantly upregulated in 2 Cr(VI)-transformed human bronchial epithelial cell lines (BEAS-2B and 16HBE). The increase of m6A modification levels in Cr(VI)-transformed cells was initially revealed by the m6A microarray analysis, which was further confirmed by using a m6A RNA Methylation Quantification Assay to directly measure total RNA m6A levels. (2) It was next determined that chronic Cr(VI) exposure also greatly increases the m6A levels in mouse lung tissues. This was achieved by directly measuring the m6A levels from total RNAs extracted from vehicle control- and Cr(VI)-exposed mouse lung tissues. (3) The strong positive IF staining of the m6A writer METTL3 protein in chronic Cr(VI) exposure-caused mouse and human lung tumor tissues provided additional evidence supporting that chronic Cr(VI) exposure also changes the epitranscriptome in lung tissues. Interestingly, the positive IF staining of METTL3 in vehicle control group mouse spontaneous lung tumors is not increased compared with the adjacent normal lung tissues, emphasizing the importance of METTL3 upregulation in Cr(VI)-induced lung tumorigenesis. Given the recent exciting findings showing that increased m6A modification plays curial roles in cancer (Barbieri and Kouzarides, 2020; Huang et al., 2020; Liu et al., 2019; Uddin et al., 2021), our findings suggest that increased m6A modification may represent a novel and important mechanism by which chronic Cr(VI) exposure causes lung cancer.
How does chronic Cr(VI) exposure increase m6A modification levels? Like DNA and protein modifications, the levels of m6A modification are also dynamically regulated by the corresponding writers (the m6A writer complex), erasers (m6A demethylases), and readers (the m6A interacting proteins) (Uddin et al., 2021; Yang et al., 2018; Yue et al., 2015). We found that the protein levels of METTL3, the major catalytic methyltransferase in the m6A writer complex, are significantly increased in chronic Cr(VI) exposure-transformed human bronchial epithelial cells, Cr(VI) exposure-induced mouse and human lung tumor tissues. These are nicely correlated with the increased levels of m6A in Cr(VI)-transformed cells and Cr(VI)-exposed mouse lungs. However, the protein levels of the m6A erasers (FTO and ALKBH5) and major reader proteins (YTHDF1-3) are not significantly changed in Cr(VI)-transformed cells, suggesting that chronic Cr(VI) exposure increases m6A levels likely mainly by upregulating the METTL3 expression. Indeed, this is supported by the findings showing that stably knocking down METTL3 expression in Cr(VI)-transformed cells significantly reduces their m6A levels.
Do upregulation of METTL3 expression and increases of m6A modification play a critical role in chronic Cr(VI) exposure-induced cell transformation, CSC-like property and tumorigenesis? We addressed this important question by using 3 approaches. First, we used pharmacological inhibitors of methyltransferases and showed that inhibition of methyltransferase activities significantly reduces the transformed phenotypes of Cr(VI)-transformed cells, which suggest that methyltransferase activities are critically involved in maintaining the transformed phenotypes of Cr(VI)-transformed cells. Second, we used a genetic approach to specifically knockdown METTL3 expression in Cr(VI)-transformed cells. Knockdown of METTL3 significantly reduced Cr(VI)-induced CSC-like property and tumorigenesis. The findings from both the pharmacological inhibitor treatment and METTL3 shRNA knockdown approaches demonstrated an important role of METLL3 upregulation in maintaining the malignant phenotypes of Cr(VI)-transformed cells. Third, we further determined the role of METTL3 in the process of Cr(VI) inducing cell transformation and CSC-like property by stably knocking down METTL3 expression in parental nontransformed BEAS-2B cells. Excitingly, it was found that METTL3 knockdown significantly reduces the capability of chronic Cr(VI) exposure to induce cell transformation and generate CSC-like cells. Collectively, these findings indicate that METTL3 upregulation and increases of m6A modification are not only critically involved in maintaining the malignant phenotypes of Cr(VI)-transformed cells, but are also needed for Cr(VI) exposure to promote cell transformation and produce CSC-like property.
In summary, we found that chronic Cr(VI) exposure causes cellular epitranscriptomic dysregulation as evidenced by significantly increasing total RNA m6A modification levels in Cr(VI)-exposed human bronchial epithelial cells and mouse lung tissues. It was further determined that Cr(VI) exposure increases RNA m6A modification levels by upregulating the expression of a key RNA methyltransferase METTL3, which plays important role in chronic Cr(VI) exposure-induced cell transformation, CSC-like property and tumorigenesis. Given the increasingly recognized crucial roles of RNA m6A modification and METTL3 in cancer, these findings suggest a critical role for epitranscriptomic dysregulation in Cr(VI) carcinogenesis.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
FUNDING
This work was supported by the National Institutes of Environmental Health Sciences (R01ES026151, R01ES029496, R01ES029942, and 1R01ES032787).
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Supplementary Material
Contributor Information
Zhishan Wang, Division of Cancer Biology, Department of Medicine, MetroHealth Medical Center, Case Western Reserve University School of Medicine, Cleveland, Ohio 44109, USA.
Mohammad Burhan Uddin, Center for Environmental and Systems Biochemistry, University of Kentucky College of Medicine, Lexington, Kentucky 40536, USA.
Jie Xie, Department of Toxicology and Cancer Biology, University of Kentucky College of Medicine, Lexington, Kentucky 40536, USA.
Hua Tao, Division of Cancer Biology, Department of Medicine, MetroHealth Medical Center, Case Western Reserve University School of Medicine, Cleveland, Ohio 44109, USA.
Patti C Zeidler-Erdely, Health Effects Laboratory Division, National Institute for Occupational Safety and Health, Morgantown, West Virginia 26508, USA.
Kazuya Kondo, Department of Oncological Medical Services, Graduate School of Biomedical Sciences, Tokushima University Graduate School, Tokushima City 770-8509, Japan.
Chengfeng Yang, Division of Cancer Biology, Department of Medicine, MetroHealth Medical Center, Case Western Reserve University School of Medicine, Cleveland, Ohio 44109, USA.
Disclaimer: Mention of brand name does not constitute product endorsement. The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the National Institute for Occupational Safety and Health, Centers for Disease Control and Prevention.
REFERENCES
- Ali A. H., Kondo K., Namura T., Senba Y., Takizawa H., Nakagawa Y., Toba H., Kenzaki K., Sakiyama S., Tangoku A. (2011). Aberrant DNA methylation of some tumor suppressor genes in lung cancers from workers with chromate exposure. Mol. Carcinog. 50, 89–99. [DOI] [PubMed] [Google Scholar]
- Agency for Toxic Substances and Disease Registry (ATSDR). (2012). Toxicological Profile for Chromium. U.S. Department of Health and Human Services, Public Health Service, Agency for Toxic Substances and Disease Registry, Atlanta, GA. Available at: https://www.atsdr.cdc.gov/toxprofiles/tp7.pdf. Accessed January 18, 2022.
- Agency for Toxic Substances and Disease Research (ATSDR). (2019). Top 20 Hazardous Substances: ATSDR/EPA Priority List for 2017. U.S. Department of Health and Human Services Public Health Service/U.S. Environmental Protection Agency. Available at: https://www.atsdr.cdc.gov/SPL/index.html#2019spl. Accessed January 18, 2022.
- Barbieri I., Kouzarides T. (2020). Role of RNA modifications in cancer. Nat. Rev. Cancer 20, 303–322. [DOI] [PubMed] [Google Scholar]
- Boccaletto P., Bagiński B. (2021). MODOMICS: An operational guide to the use of the RNA modification pathways database. Methods Mol. Biol. 2284, 481–505. [DOI] [PubMed] [Google Scholar]
- Brocato J., Costa M. (2013). Basic mechanics of DNA methylation and the unique landscape of the DNA methylome in metal-induced carcinogenesis. Crit. Rev. Toxicol. 43, 493–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P., Machnicka M. A., Purta E., Piatkowski P., Baginski B., Wirecki T. K., de Crécy-Lagard V., Ross R., Limbach P. A., Kotter A., et al. (2018). MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q. Y., Murphy A., Sun H., Costa M. (2019). Molecular and epigenetic mechanisms of Cr(VI)-induced carcinogenesis. Toxicol. Appl. Pharmacol. 377, 114636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervona Y., Arita A., Costa M. (2012). Carcinogenic metals and the epigenome: Understanding the effect of nickel, arsenic, and chromium. Metallomics 4, 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G., Abdallah W. M., Foley J. M., Jackson K. W., Clarke M. F., Kawamura M. J., Wicha M. S. (2003). In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 17, 1253–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M., Harada B. T., Behm M., He C. (2018). RNA modifications modulate gene expression during development. Science 361, 1346–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A. L., Wise S. S., Wise J. P. Sr. (2008). Carcinogenicity of hexavalent chromium. Indian J. Med. Res. 128, 353–372. [PubMed] [Google Scholar]
- Huang H., Weng H., Chen J. (2020). M6A Modification in coding and non-coding RNAs: Roles and therapeutic implications in cancer. Cancer Cell 37, 270–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries B., Wang Z., Oom A. L., Fisher T., Tan D., Cui Y., Jiang Y., Yang C. (2014). MicroRNA-200b targets protein kinase Cα and suppresses triple-negative breast cancer metastasis. Carcinogenesis 35, 2254–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries B., Wang Z., Yang C. (2016). The role of microRNAs in metal carcinogen-induced cell malignant transformation and tumorigenesis. Food Chem. Toxicol. 98, 58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Agency for Research on Cancer (IARC). (1990). Chromium, nickel and welding. IARC monographs on the Evaluation of Carcinogenic Risks to Humans. World Health Organization, Lyon, France, Vol. 49, pp. 49–256. [Google Scholar]
- Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y. G., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo K., Takahashi Y., Hirose Y., Nagao T., Tsuyuguchi M., Hashimoto M., Ochiai A., Monden Y., Tangoku A. (2006). The reduced expression and aberrant methylation of p16(INK4a) in chromate workers with lung cancer. Lung Cancer 53, 295–302. [DOI] [PubMed] [Google Scholar]
- Li Y., Xiao Y., Lin H. P., Reichel D., Bae Y., Lee E. Y., Jiang Y., Huang X., Yang C., Wang Z. (2019). In vivo β-catenin attenuation by the integrin α5-targeting nano-delivery strategy suppresses triple negative breast cancer stemness and metastasis. Biomaterials 188, 160–172. [DOI] [PubMed] [Google Scholar]
- Lin H. P., Wang Z., Yang C. (2021). LncRNA DUXAP10 upregulation and the hedgehog pathway activation are critically involved in chronic cadmium exposure-induced cancer stem cell-like property. Toxicol. Sci. 184, 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Harada B. T., He C. (2019). Regulation of gene expression by N6-methyladenosine in cancer. Trends Cell Biol. 29, 487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. D., Jaffrey S. R. (2014). The dynamic epitranscriptome: N 6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 15, 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickens K. P., Patierno S. R., Ceryak S. (2010). Chromium genotoxicity: A double-edged sword. Chem. Biol. Interact. 188, 276–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree I. A., Evans M. E., Pan T., He C. (2017). Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saletore Y., Meyer K., Korlach J., Vilfan I. D., Jaffrey S., Mason C. E. (2012). The birth of the epitranscriptome: Deciphering the function of RNA modifications. Genome Biol. 13, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salnikow K., Zhitkovich A. (2008). Genetic and epigenetic mechanisms in metal carcinogenesis and cocarcinogenesis: Nickel, arsenic, and chromium. Chem. Res. Toxicol. 21, 28–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Wei J., He C. (2019). Where, when, and how: Context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74, 640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi M., Kondo K., Soejima S., Kajiura K., Kawakita N., Toba H., Kawakami Y., Yoshida M., Takizawa H., Tangoku A. (2020). Chromate exposure induces DNA hypermethylation of the mismatch repair gene MLH1 in lung cancer. Mol. Carcinog. 59, 24–31. [DOI] [PubMed] [Google Scholar]
- Uddin M. B., Wang Z., Yang C. (2020). Dysregulations of functional RNA modifications in cancer, cancer stemness and cancer therapeutics. Theranostics 10, 3164–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M. B., Wang Z., Yang C. (2021). The m6A RNA methylation regulates oncogenic signaling pathways driving cell malignant transformation and carcinogenesis. Mol. Cancer 20, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. S., Wang Z., Yang C. (2021). Dysregulations of long non-coding RNAs - The emerging “lnc” in environmental carcinogenesis. Semin. Cancer Biol. 76, 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Humphries B., Xiao H., Jiang Y., Yang C. (2013). Epithelial to mesenchymal transition in arsenic-transformed cells promotes angiogenesis through activating β-catenin-vascular endothelial growth factor pathway. Toxicol. Appl. Pharmacol. 271, 20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Humphries B., Xiao H., Jiang Y., Yang C. (2014). MicroRNA-200b suppresses arsenic-transformed cell migration by targeting protein kinase Cα and Wnt5b-protein kinase Cα positive feedback loop and inhibiting Rac1 activation. J. Biol. Chem. 289, 18373–18386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Li Y., Xiao Y., Lin H. P., Yang P., Humphries B., Gao T., Yang C. (2019b). Integrin a9 depletion promotes β-catenin degradation to suppress triple-negative breast cancer tumor growth and metastasis. Int. J. Cancer 145, 2767–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Lin H. P., Li Y., Tao H., Yang P., Xie J., Maddy D., Kondo K., Yang C. (2019a). Chronic hexavalent chromium exposure induces cancer stem cell-like property and tumorigenesis by increasing c-Myc expression. Toxicol. Sci. 172, 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wu J., Humphries B., Kondo K., Jiang Y., Shi X., Yang C. (2018). Upregulation of histone-lysine methyltransferases plays a causal role in hexavalent chromium-induced cancer stem cell-like property and cell transformation. Toxicol. Appl. Pharmacol. 342, 22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Yang C. (2019). Metal carcinogen exposure induces cancer stem cell-like property through epigenetic reprograming: A novel mechanism of metal carcinogenesis. Semin. Cancer Biol. 57, 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Yang J., Fisher T., Xiao H., Jiang Y., Yang C. (2012). Akt activation is responsible for enhanced migratory and invasive behavior of arsenic-transformed human bronchial epithelial cells. Environ. Health Perspect. 120, 92–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Yang P., Xie J., Lin H. P., Kumagai K., Harkema J., Yang C. (2020). Arsenic and benzo[a]pyrene co-exposure acts synergistically in inducing cancer stem cell-like property and tumorigenesis by epigenetically down-regulating SOCS3 expression. Environ. Int. 137, 105560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Li Y., Tao H., Humphries B., Li A., Jiang Y., Yang C., Luo R., Wang Z. (2018). Integrin α5 down-regulation by miR-205 suppresses triple negative breast cancer sternness and metastasis by inhibiting the Src/Vav2/Rac1 pathway. Cancer Lett. 433, 199–209. [DOI] [PubMed] [Google Scholar]
- Yang C. (2020). ToxPoint: Dissecting functional RNA modifications in responses to environmental exposure-mechanistic toxicology research enters a new era. Toxicol. Sci. 174, 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Wu J., Zhang R., Zhang P., Eckard J., Yusuf R., Huang X., Rossman T. G., Frenkel K. (2005). Caffeic acid phenethyl ester (CAPE) prevents transformation of human cells by arsenite (As) and suppresses growth of As-transformed cells. Toxicology 213, 81–96. [DOI] [PubMed] [Google Scholar]
- Yang Y., Hsu P. J., Chen Y. S., Yang Y. G. (2018). Dynamic transcriptomic m6A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 28, 616–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y., Liu J., He C. (2015). RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 29, 1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccara S., Ries R. J., Jaffrey S. R. (2019). Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 20, 608–624. [DOI] [PubMed] [Google Scholar]
- Zeidler-Erdely P. C., Falcone L. M., Antonini J. M., Fraser K., Kashon M. L., Battelli L. A., Salmen R., Trainor T., Grose L., Friend S., et al. (2020). Tumorigenic response in lung tumor susceptible A/J mice after sub-chronic exposure to calcium chromate or iron (III) oxide. Toxicol. Lett. 334, 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Tan Y. S., Haslam S. Z., Yang C. (2010). Perfluorooctanoic acid effects on steroid hormone and growth factor levels mediate stimulation of peripubertal mammary gland development in C57BL/6 mice. Toxicol. Sci. 115, 214–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Wang Z., Jiang Y., Yang C. (2011). Inactivation of Rac1 reduces Trastuzumab resistance in PTEN deficient and insulin-like growth factor I receptor overexpressing human breast cancer SKBR3 cells. Cancer Lett. 313, 54–63. [DOI] [PubMed] [Google Scholar]
- Zheng G., Dahl J. A., Niu Y., Fedorcsak P., Huang C. M., Li C. J., Vågbø C. B., Shi Y., Wang W. L., Song S. H., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhitkovich A. (2011). Chromium in drinking water: Sources, metabolism, and cancer risks. Chem Res. Toxicol. 24, 1617–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Costa M. (2020). Metals and molecular carcinogenesis. Carcinogenesis 41, 1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.