Abstract
Better biomarkers to predict death early in acute liver failure (ALF) are needed. To that end, we obtained early (study day 1) and later (day 3) serum samples from transplant-free survivors (n = 28) and nonsurvivors (n = 30) of acetaminophen-induced ALF from the NIH-sponsored Acute Liver Failure Study Group and from control volunteers (n = 10). To identify proteins that increase early in serum during ALF, we selected individuals from this cohort for whom alanine aminotransferase was lower on day 1 than day 3, indicating a time point before peak injury (n = 10/group). We then performed untargeted proteomics on their day 1 samples. Out of 1682 quantifiable proteins, 361 were ≥ 4-fold elevated or decreased in ALF patients versus controls and 16 of those were further elevated or decreased ≥ 4-fold in nonsurvivors versus survivors, indicating potential to predict death. Interestingly, 1 of the biomarkers was lactate dehydrogenase (LDH), which is already measured in most clinical laboratories. To validate our proteomics results and to confirm the prognostic potential of LDH, we measured LDH activity in all day 1 and 3 samples from all 58 ALF patients. LDH was elevated in the nonsurvivors versus survivors on both days. In addition, it had prognostic value similar to the model for end-stage liver disease and outperformed the King’s College Criteria, while a combination of model for end-stage liver disease and LDH together outperformed either alone. Finally, bioinformatics analysis of our proteomics data revealed alteration of numerous signaling pathways that may be important in liver regeneration. Overall, we conclude LDH can predict death in APAP-induced ALF.
Keywords: drug-induced liver injury, acute liver injury, liver regeneration, biomarkers
Acute liver failure (ALF) is a syndrome of encephalopathy, coagulopathy, and multi-organ dysfunction caused by loss of liver function secondary to acute liver injury. It is a relatively rare condition. Indeed, the true incidence of ALF is unknown, and estimates vary widely around the world (Bower et al., 2007; Ho et al., 2014; Hoofnagle et al., 1995; Thanapirom et al., 2019; Weiler et al., 2020). In the United States, crude estimates range from 5.5 to 31.2 cases per million population per year (or around 1600–10 000 cases annually; Bower et al., 2007; Hoofnagle et al., 1995). However, an often-cited figure is 2000 total cases per year. Although it is rare, ALF can be extremely devastating: Despite recent improvements in patient outcomes, overall mortality remains high at around 25–30% (Reuben et al., 2016). Currently, the major life-extending treatment for ALF is a liver transplant. However, clinicians must decide quickly when a transplant is needed because the time from hospital admission or onset of ALF to death is on the order of days (Ostapowicz et al., 2002; Shakil et al., 2000).
Unfortunately, current liver biomarkers help little with the decision to perform a transplant. The primary biomarker of liver injury is serum alanine aminotransferase (ALT). Although ALT is sensitive for detection of liver injury, it has limited prognostic value (Karvellas et al., 2017; Kuroda et al., 2021; McGill and Jaeschke, 2014). Although liver function tests like bilirubin, prothrombin time (PT), and the international normalized ratio (INR) correlate better with poor outcomes, they often peak late in ALF progression when a liver transplant is no longer feasible. Finally, prognostic scores like the model for end-stage liver disease (MELD), which includes bilirubin and INR, are helpful but lack sufficient sensitivity and specificity to principally guide patient care (De Clercq et al., 2021). Thus, the identification and validation of a new prognostic biomarker or panel of biomarkers that can predict death early in ALF, before the peak of injury and dysfunction, could dramatically improve the care of these critically ill patients by allowing early activation of resources necessary for liver transplantation.
Here, we used untargeted proteomics analysis to identify candidate biomarkers to predict poor outcomes in patients with acetaminophen (APAP)-induced ALF. We then selected one of those biomarkers, lactate dehydrogenase (LDH), for further testing and confirmation. Importantly, LDH is routinely measured in many clinical laboratories and could be immediately implemented as a simple prognostic biomarker in ALF in routine practice if our results can be further verified. Finally, we used a reverse translational approach with Ingenuity Pathway Analysis (IPA) and subsequent Upstream Analysis to explore the mechanistic significance of our untargeted proteomics results.
MATERIALS AND METHODS
Acute Liver Failure Study Group samples
Serum samples from 28 random transplant-free survivors and 30 random nonsurvivors of APAP-induced ALF were obtained from the Acute Liver Failure Study Group (ALFSG) biorepository. ALF was diagnosed by ALFSG investigators and defined as INR ≥ 1.5, hepatic encephalopathy, duration of illness < 26 weeks, and absence of chronic liver disease. APAP toxicity was determined to be the etiology based on a combination of patient-reported history of APAP overdose, a detectable APAP level documenting ingestion, and aminotransferase level of ≥ 1000 IU/l. Due to hepatic encephalopathy, consent was obtained from next of kin. Samples were centrifuged at each ALFSG study site to obtain serum and stored at −80°C for later distribution and analysis. Demographic and laboratory data provided with the samples included daily values for serum ALT, aspartate aminotransferase (AST), total bilirubin (Tbili), PT, and creatinine (Cre) during hospitalization; age; sex; race; and ethnicity. Internal review board (IRB) approval was obtained at each ALFSG study site and the study was conducted in accordance with the 1975 Declaration of Helsinki.
Volunteer subjects
Ten volunteers without liver disease and with reported recent therapeutic APAP exposure were recruited at the University of Arkansas for Medical Sciences (UAMS) in Little Rock, Arkansas, USA. Recent therapeutic exposure was considered useful to control for the effects of APAP itself, though not essential for the study. Each subject was informed of the potential risks and benefits of the study and signed a consent form. After enrollment, a blood sample was collected from each subject and serum was separated by centrifugation. The study protocol was reviewed and approved by the UAMS IRB and the study was conducted in accordance with the 1975 Declaration of Helsinki.
Untargeted proteomics
Abundant serum proteins were depleted with HighSelect Top14 resin (Thermo) according to the manufacturer’s instructions. Proteins were reduced and alkylated prior to digestion with sequencing grade modified porcine trypsin (Promega) using S-Trap columns (Protifi). Tryptic peptides were then separated by reverse phase XSelect CSH C18 2.5 um resin (Waters) on an in-line 150 × 0.075 mm column using an UltiMate 3000 RSLCnano system (Thermo). Peptides were eluted using a 60 min gradient from 98:2 to 65:35 buffer A:B ratio (buffer A = 0.1% formic acid, 0.5% acetonitrile; buffer B = 0.1% formic acid, and 99.9% acetonitrile). Eluted peptides were ionized by electrospray (2.2 kV) followed by mass spectrometric analysis on an Orbitrap Exploris 480 mass spectrometer (Thermo). To assemble a chromatogram library, 6 gas-phase fractions were acquired on the Orbitrap Exploris with 4 m/z DIA spectra (4 m/z precursor isolation windows at 30 000 resolution, normalized automatic gain control (AGC) target 100%, maximum inject time 66 ms) using a staggered window pattern from narrow mass ranges using optimized window placements. Precursor spectra were acquired after each DIA duty cycle, spanning the m/z range of the gas-phase fraction (ie, 496–602 m/z, 60 000 resolution, normalized AGC target 100%, maximum injection time 50 ms). For wide-window acquisitions, the Orbitrap Exploris was configured to acquire a precursor scan (385–1015 m/z, 60 000 resolution, normalized AGC target 100%, maximum injection time 50 ms) followed by 50× 12 m/z DIA spectra (12 m/z precursor isolation windows at 15 000 resolution, normalized AGC target 100%, maximum injection time 33 ms) using a staggered window pattern with optimized window placements. Precursor spectra were acquired after each DIA duty cycle. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD029576.
Proteomics data analysis
Following data acquisition, data were searched using an empirically corrected library and a quantitative analysis was performed to obtain a comprehensive proteomic profile. Proteins were identified and quantified using EncyclopeDIA (Searle et al., 2018) and visualized with Scaffold DIA using 1% false discovery thresholds at both the protein and peptide level. Protein exclusive intensity values were assessed for quality using an in-house ProteiNorm app, a tool for systematic evaluation of normalization methods, imputation of missing values and comparisons of multiple differential abundance methods (Graw et al., 2020). Normalization methods evaluated included log2 normalization (Log2), median normalization (Median), mean normalization (Mean), variance stabilizing normalization (Huber et al., 2002), quantile normalization (Bolstad, 2021), cyclic loess normalization (Ritchie et al., 2015), global robust linear regression normalization (Chawade et al., 2014), and global intensity normalization (Global Intensity; Chawade et al., 2014). The individual performance of each method was evaluated and verified by comparing of the following metrics: total intensity, pooled intragroup coefficient of variation, pooled intragroup median absolute deviation, pooled intragroup estimate of variance, intragroup correlation, sample correlation heatmap (Pearson), and log2-ratio distributions. The normalized data were used to perform statistical analysis using linear models for microarray data (limma) with empirical Bayes (eBayes) smoothing to the standard errors (Ritchie et al., 2015). Proteins with a false discovery rate (FDR)-adjusted p value < .05 and a fold change > 2 were generally considered significant. For biomarker identification, we focused on biomarkers elevated ≥ 4-fold in ALF survivors versus controls and further elevated ≥ 4-fold in nonsurvivors versus survivors of ALF to ensure more robust results. Analyzed data are available to academic researchers upon request.
LDH measurement
LDH activity was measured using a standard kinetic assay based on the loss of NADH absorbance in the reaction mixture. Briefly, each serum sample was diluted in 1× phosphate-buffered saline and mixed with 60 mM potassium phosphate buffer (pH 7.5) containing 0.7 mM pyruvate and 216 mM NADH in at least 1 well of a 96-well plate. The plate was then placed into a UV/Vis spectrophotometer and absorbance at 340 nm was monitored over 3 min in 11 s intervals at 37°C. Activity was calculated using the Beer-Lambert equation and expressed in international units per liter (U/l).
Pathway analysis
Pathway analysis and subsequent Upstream Analysis of the untargeted proteomics data were performed using IPA software (Qiagen, Germantown, Maryland). LogFC cutoffs of −1 to 1 and a p value cutoff of .05 were used in the initial core analysis.
Statistical analyses
Sensitivity, specificity, likelihood ratios, and posttest probabilities were calculated using standard equations (Deacon, 2009). Data normality was tested using the Shapiro-Wilk test. For normally distributed data, groups were compared using Student’s t test. For nonnormally distributed data, groups were compared using a t test on ranks. Logistic regression was used to screen for associations between biomarkers and outcome, and receiver operating characteristic (ROC) curves were used to visualize the associations. The equation to combine MELD score and LDH values was MELD-LDH = −5.844 + (0.682*log(LDH)) + (2.702*log(MELD)), derived using multiple logistic regression and modified to optimize results. All statistical analyses were performed in SigmaPlot 12.5 (Systat, San Jose, California) or R 3.6.1 (R Foundation for Statistical Computing, Vienna, Austria).
RESULTS
Participant Demographics
Table 1 provides a summary of demographics and clinical laboratory data from the days 1 and 3 samples for all ALF survivors and nonsurvivors, as well as data from control subjects.
Table 1.
All Participant Demographics and Laboratory Data
| Parameter | Control Volunteers | S | NS |
|---|---|---|---|
| N | 10 | 28 | 30 |
| Female (%) | 5 (50) | 19 (68) | 20 (67) |
| Median age (range) | 45 (23–66) | 31 (19–70) | 36 (18–67) |
| Race and ethnicity | |||
| White, non-Hispanic (%) | 8 (80) | 26 (93) | 27 (90) |
| Black, non-Hispanic (%) | 2 (20) | 1 (3.5) | 2 (6.7) |
| White, Hispanic (%) | 0 (0) | 1 (3.5) | 1 (3.3) |
| Other (%) | 0 (0) | 0 (0) | 0 (0) |
| Peak ALT (U/l), mean ± SE | 17 ± 2 | 4513 ± 920 | 3406 ± 531 |
| Peak Tbili (mg/dl), mean ± SE | NA | 5.5 ± 0.6 | 8.6 ± 1.2 |
| Peak Cre (mg/dl), mean ± SE | NA | 3.4 ± 0.6 | 3.4 ± 0.3 |
| Peak MELD, mean ± SE | NA | 28 ± 2 | 39 ± 1 |
| Days from enrollment to Peak ALT, mean ± SE | NA | −0.52 ± 0.2 | −0.27 ± 0.2 |
Abbreviations: S, survivors; NS, nonsurvivors; NA, not available.
Discovery Proteomics
To identify candidate biomarkers with potential to predict poor outcomes in ALF, we performed untargeted proteomics analysis of the day 1 samples from a subset of 10 participants from each group. Individuals in the subsets were chosen because their day 1 samples were collected before ALT peaked during hospitalization, indicating that they represent an early time point in the progression of ALF, to increase the likelihood that our candidate biomarkers will have utility early in the course of acute liver injury. Table 2 provides demographic and laboratory data for these subgroups.
Table 2.
Untargeted Proteomics Subgroup Demographics and Laboratory Data
| Parameter | Control Volunteers | S | NS |
|---|---|---|---|
| n | 10 | 10 | 10 |
| Sex (% female) | 5 (50) | 5 (50) | 5 (50) |
| Age (median, range) | 45 (23–66) | 32 (19–46) | 33 (18–67) |
| Race and ethnicity | |||
| White, non-Hispanic (%) | 8 (80) | 8 (80) | 7 (70) |
| Black, non-Hispanic (%) | 2 (20) | 1 (10) | 2 (20) |
| White, Hispanic (%) | 0 (0) | 1 (10) | 1 (10) |
| Other (%) | 0 (0) | 0 (0) | 0 (0) |
| Peak ALT (U/l) (mean ± SE) | 17 ± 2 | 8275 ± 1494 | 7493 ± 992 |
Abbreviations: S, survivors; NS, nonsurvivors.
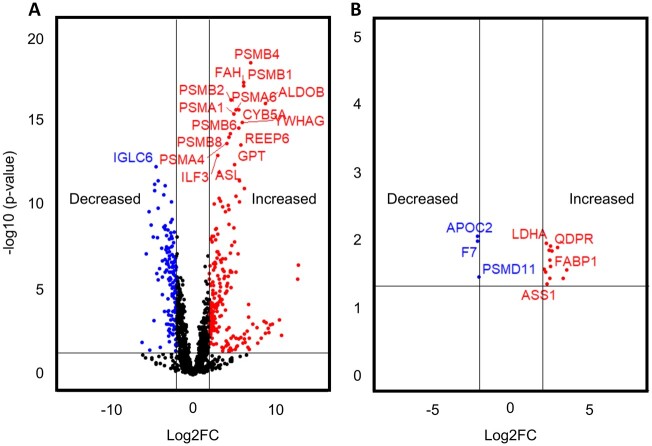
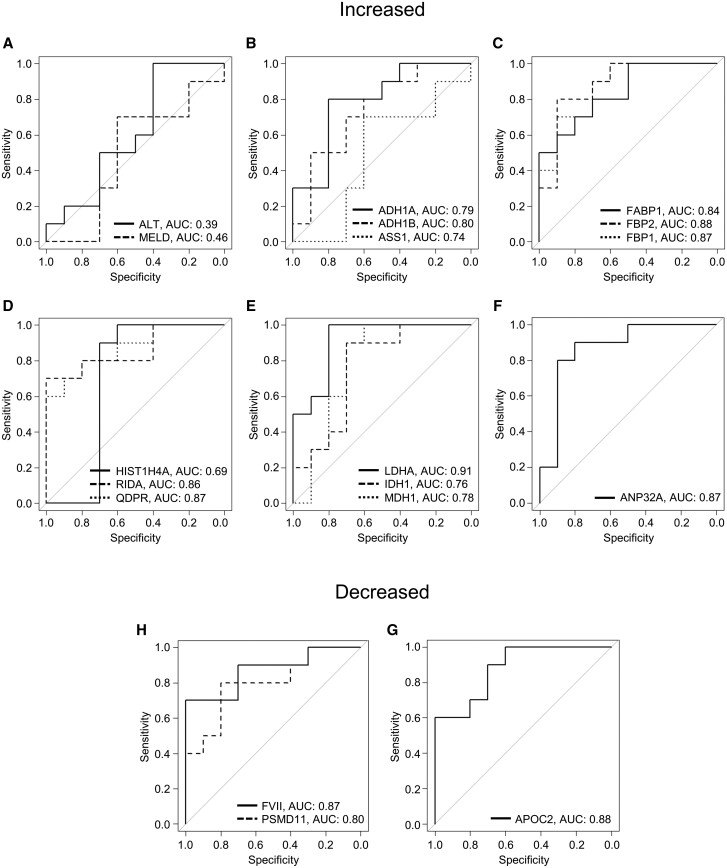
Using untargeted proteomics, we were able to quantify 1682 proteins across all serum samples from the ALF patients and volunteers. To ensure identification of the most robust and promising biomarker candidates, we excluded proteins that were < 4-fold elevated or < 4-fold decreased in serum from ALF survivors compared with healthy volunteers, leaving 221 elevated and 140 decreased (Figure 1A). Tables 3 and 4 list the top 20 proteins in each category (increased and decreased). From the remaining proteins, we further excluded those that were < 4-fold elevated or <4-fold decreased in samples from ALF nonsurvivors compared with survivors. This left us with 13 biomarker candidates that were at least 4-fold elevated in ALF versus controls and again 4-fold higher in ALF nonsurvivors versus survivors, and another 3 biomarker candidates that were at least 4-fold decreased in the ALF survivors versus controls and again 4-fold decreased in nonsurvivors versus survivors (Figure 1B). Table 5 lists all 16 of these biomarker candidates. ROC curve analysis further revealed that all 16 had better overall sensitivity and specificity for death (as indicated by greater areas under the curve [AUC]) than either the commonly used MELD score or presentation ALT in the ALF patients (Figure 2).
Figure 1.
Untargeted proteomics revealed 16 biomarker candidates that differentiate nonsurvivors and survivors. Day 1 serum samples from survivors (n = 10) and nonsurvivors (n = 10) of acetaminophen-induced acute liver failure (ALF) and healthy controls (n = 10) were subjected to untargeted proteomics. A, Volcano plot displaying results for ALF survivors versus volunteer controls. B, Volcano plot displaying the 13 proteins that were ≥ 4-fold elevated in ALF survivors versus control and ≥ 4-fold further elevated in nonsurvivors versus survivors and the 3 proteins that were ≥ 4-fold decreased in ALF survivors versus control and ≥ 4-fold further decreased in nonsurvivors versus survivors.
Table 3.
Top 20 Proteins Elevated in Serum in ALF Patients Versus Control Subjects
| Protein | Log2FC | Adjusted p Value |
|---|---|---|
| MAVS | 15.2 | 3.2 × 10-9 |
| CTPS2 | 12.8 | 3.2 × 10−7 |
| EIF4E | 12.7 | 2.2 × 10−6 |
| IGHV3-72 | 10.8 | 4.7 × 10−3 |
| SRSF1 | 10.5 | 5.9 × 10−4 |
| ITGB2 | 9.65 | 1.0 × 10−3 |
| LACTB2 | 9.38 | 1.1 × 10−3 |
| TTC36 | 9.27 | 3.3 × 10−3 |
| AP2B1 | 9.15 | 1.8 × 10−3 |
| RAB8B | 8.90 | 8.7 × 10−4 |
| RAB8A | 8.90 | 8.7 × 10−4 |
| APOL2 | 8.87 | 3.4 × 10−3 |
| ALDOB | 8.83 | 9.3 × 10−17 |
| CES1P1 | 8.69 | 6.9 × 10−4 |
| SEPT9 | 8.30 | 6.7 × 10−3 |
| LILRB2 | 7.91 | 1.3 × 10−2 |
| RPL6 | 7.84 | 1.6 × 10−3 |
| EHD1 | 7.30 | 1.0 × 10−3 |
| PSMB4 | 7.04 | 3.5 × 10−19 |
| C19orf12 | 6.90 | 7.3 × 10−3 |
Abbreviations: Log2FC, log base 2 fold-change; ALF, acute liver failure.
Table 4.
Top 20 Proteins Decreased in Serum in ALF Patients Versus Control Subjects
| Protein | Log2FC | Adjusted p-value |
|---|---|---|
| NARS | −6.15 | 1.3 × 10−2 |
| CYP3A4 | −5.64 | 7.2 × 10−8 |
| OSGEP | −5.33 | 2.3 × 10−10 |
| LRRC59 | −5.27 | 3.4 × 10−2 |
| F7 | −5.09 | 1.5 × 10−9 |
| SPP2 | −4.84 | 7.4 × 10−9 |
| CUL4A | −4.71 | 6.4 × 10−4 |
| INHBC | −4.64 | 5.7 × 10−12 |
| RARRES2 | −4.62 | 1.3 × 10−11 |
| IGLC6 | −4.46 | 4.8 × 10−13 |
| VTN | −4.40 | 1.0 × 10−6 |
| APOC4 | −4.28 | 8.0 × 10−9 |
| HPSE | −4.23 | 2.5 × 10−6 |
| CFHR5 | −4.23 | 1.4 × 10−10 |
| PROC | −4.21 | 3.5 × 10−12 |
| PGP | −4.09 | 5.5 × 10−7 |
| EIF2S2 | −4.01 | 2.4 × 10−2 |
| CFHR4 | −3.79 | 3.4 × 10−8 |
| FARP1 | −3.71 | 1.9 × 10−5 |
| APOF | −3.70 | 1.7 × 10−6 |
Abbreviations: Log2FC, log base 2 fold-change; ALF, acute liver failure
Table 5.
Candidate Biomarker Ranking
| Protein | AUC | Specificity (%) at 90% Sensitivity | Sensitivity (%) at 90% Specificity | PTP+ (%) at 90% Sensitivity |
|---|---|---|---|---|
| Increaseda | ||||
| LDHA/LDH-M | 0.91 | 80 | 60 | 66 |
| FBP2 | 0.88 | 70 | 80 | 56 |
| ANP32A | 0.87 | 80 | 80 | 66 |
| FBP1 | 0.87 | 70 | 70 | 56 |
| QDPR | 0.87 | 60 | 70 | 49 |
| RIDA | 0.86 | 40 | 70 | 39 |
| FABP1 | 0.84 | 50 | 60 | 44 |
| ADH1B | 0.8 | 50 | 40 | 44 |
| ADH1A | 0.79 | 30 | 30 | 44 |
| MDH1 | 0.78 | 70 | 30 | 56 |
| IDH1 | 0.76 | 70 | 30 | 56 |
| ASS1 | 0.74 | 50 | 50 | 44 |
| HIST1H4A | 0.69 | 70 | 0 | 56 |
| Decreasedb | ||||
| APOC2 | 0.88 | 70 | 60 | 56 |
| FVII | 0.87 | 70 | 70 | 56 |
| PSMD11 | 0.8 | 40 | 50 | 39 |
| Reference markers | ||||
| MELD | 0.47 | 20 | 13 | 31 |
| ALT | 0.39 | 0 | 10 | 0 |
PTP calculation is based on 30% pretest probability of death.
Abbreviations: PTP+, positive posttest probability.
Increased ≥4-fold in ALF survivors versus controls and ≥4-fold further in nonsurvivors versus survivors.
Decreased ≥4-fold in ALF survivors versus controls and ≥4-fold further in nonsurvivors versus survivors.
Figure 2.
Receiver operating characteristic (ROC) curves for the 16 biomarker candidates. Day 1 serum samples from survivors (n = 10) and nonsurvivors (n = 10) of acetaminophen-induced acute liver failure and healthy controls (n = 10) were subjected to untargeted proteomics. A–F, ROC curves for the elevated biomarkers. G–H, ROC curves for the decreased biomarkers.
To determine which of these 16 biomarker candidates are most promising for clinical use, we ranked them based on AUC and calculated specificity for death at 90% sensitivity, sensitivity at 90% specificity, and positive posttest probability at 90% sensitivity as well (Table 5). We were intrigued to discover that LDH-M (encoded by the LDHA gene) was among the biomarker candidates with greatest potential for clinical use as a prognostic marker to predict death in ALF based on these metrics. Importantly, clinical laboratories routinely measure total LDH activity in circulation. Furthermore, total LDH released into serum during liver injury should consist almost entirely of LDH-M. Most LDH in cells and serum exists as 1 of 5 tetrameric isoforms (LDH1 through LDH5), each composed of various combinations of 2 major subunits (LDH-M and LDH-H). The dominant tetramer in the liver is LDH-5, which is made up of 4 LDH-M subunits. It follows, therefore, that most or all LDH activity in serum during liver injury is from LDH-M.
Confirmation of LDH
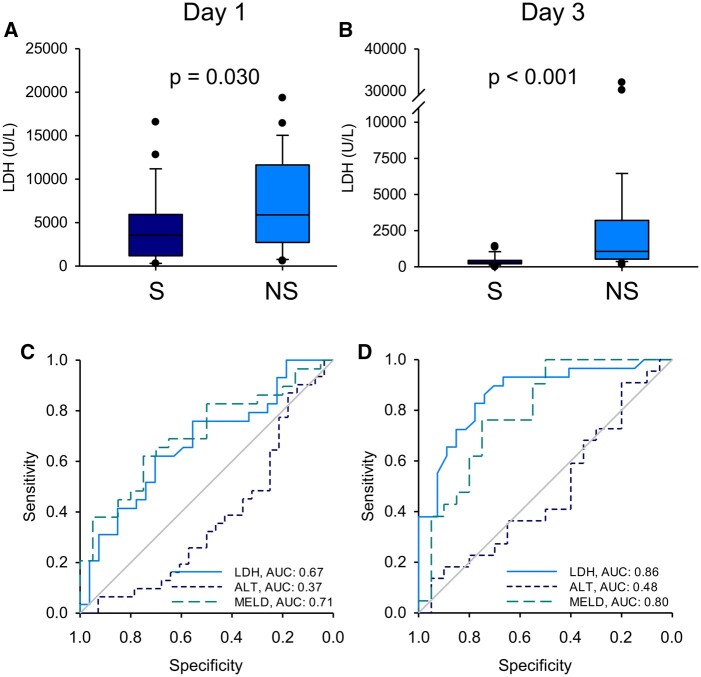
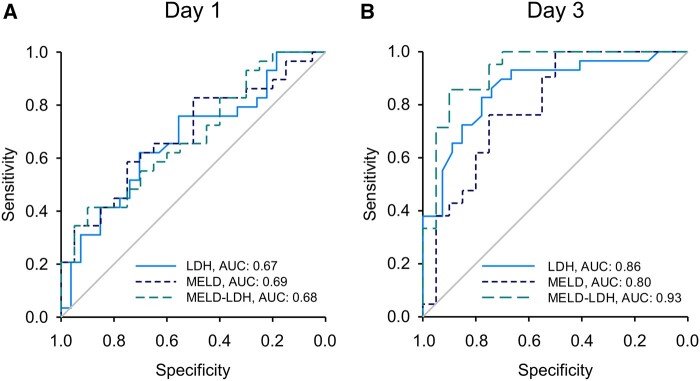
To further explore the prognostic utility of LDH, we measured total LDH activity in both days 1 and 3 serum samples from all 58 of the ALF patients. Consistent with our untargeted proteomics results, total LDH was significantly higher in serum samples from the nonsurvivors compared with the survivors on both days 1 and 3 (Figs. 3A and 3B). In addition, ROC analysis revealed that LDH performed similarly overall to the MELD score in our ALF cohort (Figs. 3C and 3D, and Table 6). To determine if the combination of LDH and MELD score can predict outcome better than MELD alone, we performed multiple logistic regression and tested a modified form of the resulting curve fit as a novel prognostic score. Indeed, the novel MELD-LDH score tended to outperform both LDH alone and MELD alone in the day 3 samples (Figure 4 and Table 6), though not on day 1. Finally, the sensitivity of LDH and MELD-LDH at 90% specificity compared favorably to the King’s College Criteria (KCC) as well, which had similar specificity of 90% (day 1) and 100% (day 3) but sensitivity of just 20% (day 1) and 19% (day 3). These data indicate that LDH may be a more convenient biomarker for prognosis in ALF than MELD or the KCC due to similar or even modestly better performance, without the need for multiple tests or calculation. They also demonstrate that the combination of MELD and LDH further improves prediction.
Figure 3.
Serum lactate dehydrogenase (LDH) activity was greater in the nonsurvivors of acetaminophen-induced acute liver failure than the survivors. Total LDH activity and alanine aminotransferase (ALT) were measured in serum from all nonsurvivors (n = 30) and survivors (n = 28) on study days 1 and 3. Model for end-stage liver disease (MELD) scores were also obtained when available (n = 20 for survivors and 29 for nonsurvivors). A, LDH activity on day 1. B, LDH activity on day 3. C, Receiver operating characteristic (ROC) curves for LDH, ALT, and MELD score on day 1. Area under the curve (AUC) values are displayed in the bottom right corner. D, ROC curves for LDH, ALT, and MELD score on day 3. AUC values are displayed in the bottom right corner. Differences in AUC values for MELD-LDH versus MELD and MELD-LDH versus LDH were statistically significant (p < .05) on day 3. Boxes show the 25th to 75th percentiles. Whiskers show the 10th and 90th percentile values. Lines show median values. Dots represent outliers.
Table 6.
Comparison of MELD-LDH With Other Metrics
| Metric | AUC (95% CI) | Specificity at 90% Sensitivity | Sensitivity at 90% Specificity |
|---|---|---|---|
| Day 1 | |||
| MELD | 0.69 | 15 | 31 |
| LDH | 0.67 | 22 | 34 |
| MELD-LDH | 0.68 | 30 | 41 |
| Day 3 | |||
| MELD | 0.80 | 55 | 43 |
| LDH | 0.86 | 67 | 62 |
| MELD-LDH | 0.93* | 75 | 86 |
Abbreviations: MELD, model for end-stage liver disease; LDH, lactate dehydrogenase; AUC, area under the curve.
p < .05 versus MELD or LDH alone.
Figure 4.
Serum lactate dehydrogenase (LDH) activity improved prediction of death in acetaminophen-induced acute liver failure. Total LDH activity and alanine aminotransferase were measured in serum from all nonsurvivors and survivors on study days 1 and 3. Model for end-stage liver disease (MELD) scores were also obtained when available (n = 20 for survivors and 29 for nonsurvivors). A, Receiver operating characteristic (ROC) curves for LDH, MELD, and MELD-LDH on day 1. B, ROC curves showing sensitivity and specificity for LDH, MELD, and MELD-LDH on day 3.
Upstream Analysis
To explore the mechanistic significance of our proteomics data, we next used IPA software to perform Upstream Analysis of our untargeted proteomics data. Upstream Analysis is a bioinformatics approach that looks at differences in gene expression or, in this case, protein levels between groups and identifies the likely signaling pathways that were activated or suppressed to cause those differences based on the prior literature. This approach revealed likely activation of 16 signaling pathways and likely deactivation or suppression of 8 others (Table 7). Interestingly, several of the pathways that appeared to be activated are known to be involved in cell proliferation and/or liver regeneration, warranting exploration in future studies.
Table 7.
Altered Signaling Pathways in Nonsurvivors Compared With Survivors
| Regulator | z-score | p Value |
|---|---|---|
| LKB1 (aka STK11) | 3.201 | 3.32 × 10−8 |
| AHR | 3.133 | 3.44 × 10−5 |
| FOXO3 | 2.939 | 1.52 × 10−3 |
| CD38 | 2.433 | 8.41 × 10−3 |
| IL-5 | 2.345 | 4.59 × 10−3 |
| CD28 | 2.333 | 1.62 × 10−2 |
| HIF1α | 2.332 | 4.06 × 10−3 |
| MLXIPL | 2.253 | 2.20 × 10−4 |
| INSR | 2.216 | 2.78 × 10−4 |
| α-catenin | 2.215 | 1.04 × 10−2 |
| SRF | 2.190 | 1.44 × 10−2 |
| MYC | 2.182 | 5.70 × 10−11 |
| CSF1 | 2.160 | 8.27 × 10−3 |
| THRB | 2.138 | 6.69 × 10−5 |
| ACOX1 | 2.111 | 5.56 × 10−7 |
| CBX5 | −2.000 | 3.89 × 10−2 |
| CLPP | −2.000 | 5.47 × 10−3 |
| TNFSF11 | −2.182 | 9.60 × 10−3 |
| CEBPB | −2.198 | 1.22 × 10−5 |
| MAP4K4 | −2.236 | 9.39 × 10−3 |
| KDM5A | −2.236 | 1.52 × 10−2 |
| OGA | −2.496 | 7.43 × 10−4 |
| GDF2 | −2.588 | 7.31 × 10−6 |
DISCUSSION
There is a critical need for better tools to predict death and therefore transplant-need early in ALF. This is particularly true for APAP-related injury because it evolves rapidly. One study found that most patients with APAP toxicity have either died, received a transplant, or recovered by day 5 after hospital admission (Reddy et al., 2016). Prognostic indexes such as the MELD score and the KCC are helpful, but insufficient. The MELD score, for example, has sensitivity and specificity of just 60–70% each for prediction of death in ALF at commonly used cutoff values (De Clercq et al., 2021). The KCC have better specificity, but similarly poor sensitivity (De Clercq et al., 2021). Newer scores have been developed with better performance than MELD or KCC, but include analytes that are not yet common. In addition, these indexes can be onerous for the clinician because they require consideration of multiple test results and sometimes manual calculation, as most clinical laboratories do not validate such scores nor provide them in-house. Identification of a single, widely available test that can predict death at least as well as current prognostic scores could facilitate faster life-saving decisions. In this study, we found that LDH, which is routinely measured in clinical laboratories, predicted death approximately as well as the MELD and KCC in our APAP-induced ALF cohort. Furthermore, our data indicate that the combined MELD-LDH score proposed herein improves prediction over either MELD or LDH alone. It is not clear why LDH would predict poor outcome better than similar enzyme markers like ALT, but one possibility is that some LDH is released by extrahepatic tissues and could therefore serve as an indicator of impending multi-organ dysfunction. In any case, these results should now be verified in larger patient cohorts that include other ALF etiologies. In addition, optimal cutoff values for LDH should be determined.
A point of technical innovation in our work is the use of untargeted proteomics directly in human samples. Surprisingly, although there has been considerable interest in identifying and developing novel prognostic biomarkers in ALF over the last 20 years (De Clercq et al., 2021), untargeted proteomics has never before been applied to patient samples for this purpose. One prior study did use an untargeted proteomics approach in a porcine model of ALF, but only one of the resulting markers was tested in humans (Wang et al., 2017). Interestingly, the latter biomarker was fructose-1,6-bisphosphatase 1 (FBP1), which was also elevated in nonsurvivors in our cohort. Other potential prognostic biomarkers of interest from our proteomics data include malate dehydrogenase 1 (MDH1), argininosuccinate synthetase 1 (ASS1), and fatty acid-binding protein 1 (FABP1). All 3 have been proposed as biomarkers to detect drug-induced liver injury (McGill et al., 2014; Schomaker et al., 2013; Vazquez et al., 2020), and there is evidence that FABP1 can also predict poor outcomes in ALF (Karvellas et al., 2017, 2021). Altogether, our proteomics data confirm these earlier results, and indicate that further exploration of FBP1 and FABP1 as prognostic biomarkers is also warranted.
Our data may also provide insight into mechanisms of APAP-induced liver injury and regeneration. We used Upstream Analysis to explore changes in signaling pathways between survivors and nonsurvivors. The validity of comparing cell signaling between these 2 groups using serum is based on 2 assumptions. First, we assume that hepatocellular damage results in release of most cell proteins into the extracellular milieu such that the serum proteins provide a glimpse of internal cell processes. Second, we assume that the extent of cell damage and protein release is similar between the 2 groups based on the fact that there was no significant difference in serum ALT between the survivors and nonsurvivors. Several interesting signaling mediators appeared to be activated in the nonsurvivors. For example, liver kinase b1 (LKB1) is thought of as a tumor suppressor that limits cell proliferation. Consistent with that idea, LKB1 KO mice have accelerated regeneration in the partial hepatectomy model (Maillet et al., 2018). So, it is possible that activation of LKB1 reduced regeneration in the nonsurvivors in this study, contributing to their decline. On the other hand, it was also reported that hepatocyte growth factor (HGF) treatment induces proliferation and phosphorylation of both LKB1 and AMPK in primary mouse hepatocytes and that knockdown of LKB1 reduces the HGF-induced AMPK phosphorylation and proliferation (Vázquez-Chantada et al., 2009). Those data indicate that LKB1-AMPK signaling can actually mediate hepatocyte proliferation in mice. Consistent with the latter, Merlen et al. (2014) also reported that AMPKα1 KO mice have delayed liver regeneration after partial hepatectomy. Similar to LKB1, there is evidence that the aryl hydrocarbon receptor (AhR) and Forkhead Box O3 (FOXO3) can suppress hepatocyte proliferation. Chronic activation or overactivation of AhR also reduces liver regeneration in vivo (Mitchell et al., 2006), while overexpression of FOXO3 reduces hepatocyte proliferation in vitro (Shizu et al., 2016). The role that each of the signaling mediators plays in APAP-induced liver injury and subsequent regeneration should be resolved in future studies.
Although our results are interesting, this study suffers from several major weaknesses. First, our ALF groups were relatively small. Additional studies with larger cohorts are needed. Second, our ALF groups included only those with APAP-induced ALF. It is not yet clear how the results would translate to non-APAP ALF. Third, we did not have information with regard to the use, timing, or regimen of NAC treatment for most of the patients. It is possible that some of the differences between groups can be explained in part by differences in treatment. That will need to be considered in future studies. Finally, serum is not a well-validated specimen type for exploration of cell signaling during liver injury, which could limit the accuracy of our Upstream Analysis results. Future work should address these deficiencies.
CONCLUSIONS
Overall, we conclude that LDH is a promising, widely available biomarker to predict poor outcome in APAP-induced ALF with performance roughly equivalent to the MELD score. Furthermore, in our cohort of ALF patients, the combination of MELD and LDH together predicted outcome modestly better than either MELD or LDH alone. In future human studies, we plan to verify our results using larger patient cohorts with a variety of ALF etiologies and to establish optimal LDH and MELD-LDH cutoff values for prognosis. Finally, we will explore the significance of our Upstream Analysis results using preclinical models of APAP-induced liver injury.
FUNDING
The ALFSG is funded by the National Institutes of Health (NIH) grant (U01DK058369 to W.M.L.). The IDeA National Resource for Quantitative Proteomics at UAMS is supported in part by NIH grant (R24GM137786). Additional funding was provided by a 2018 Pinnacle Research Award from the AASLD Foundation (to M.R.M.) and NIH grants (R01DK102142 to H.J.) and (T32GM106999 to J.H.V.; PI: Dr Paul Prather).
DECLARATION OF CONFLICTING INTERESTS
M.R.M. has consulted for Acetaminophen Toxicity Diagnostics, LLC, and GlaxoSmithKline.
Contributor Information
Joel H Vazquez, Department of Pharmacology and Toxicology, College of Medicine, University of Arkansas for Medical Sciences, Little Rock, Arkansas 72205, USA.
Stefanie Kennon-McGill, Department of Environmental and Occupational Health, Fay W. Boozman College of Public Health, University of Arkansas for Medical Sciences, Little Rock, Arkansas 72205, USA.
Stephanie D Byrum, Department of Biochemistry and Molecular Biology, College of Medicine, University of Arkansas for Medical Sciences, Little Rock, Arkansas 72205, USA.
Samuel G Mackintosh, Department of Biochemistry and Molecular Biology, College of Medicine, University of Arkansas for Medical Sciences, Little Rock, Arkansas 72205, USA.
Hartmut Jaeschke, Department of Pharmacology, Toxicology, and Therapeutics, University of Kansas Medical Center, Kansas City, Kansas 66160, USA.
D Keith Williams, Department of Biostatistics, College of Medicine, University of Arkansas for Medical Sciences, Little Rock, Arkansas 72205, USA.
William M Lee, Division of Digestive and Liver Diseases, Department of Internal Medicine, University of Texas Southwestern Med School, Dallas, Texas 75390, USA.
Jonathan A Dranoff, Division of Gastroenterology and Hepatology, Department of Internal Medicine, College of Medicine, University of Arkansas for Medical Sciences, Little Rock, Arkansas 72205, USA.
Mitchell R McGill, Department of Pharmacology and Toxicology, College of Medicine, University of Arkansas for Medical Sciences, Little Rock, Arkansas 72205, USA; Department of Environmental and Occupational Health, Fay W. Boozman College of Public Health, University of Arkansas for Medical Sciences, Little Rock, Arkansas 72205, USA.
The authors certify that all research involving human subjects was done under full compliance with all government policies and the Helsinki Declaration.
REFERENCES
- Bolstad B. (2021). preprocessCore: A Collection of Pre-Processing Functions. R package version 1.57.0. Available at: https://github.com/bmbolstad/preprocessCore. Accessed February 22, 2022.
- Bower W. A., Johns M., Margolis H. S., Williams I. T., Bell B. P. (2007). Population-based surveillance for acute liver failure. Am. J. Gastroenterol. 102, 2459–2463. [DOI] [PubMed] [Google Scholar]
- Chawade A., Alexandersson E., Levander F. (2014). Normalyzer: A tool for rapid evaluation of normalization methods for omics data sets. J. Proteome Res. 13, 3114–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon A. (2009). Chapter thirteen. In Calculations in Clinical Laboratory Sciences. (Sherwood R. A., Hooper J., Eds.), ACB Venture Publications, London, UK. [Google Scholar]
- De Clercq P., Geerts A., Vlierberghe H. V., Verhelst X. (2021). The utility of biomarkers in prognosis assessment of patients with acute liver failure. Hepatol. Res. 51, 750–757. [DOI] [PubMed] [Google Scholar]
- Graw S., Tang J., Zafar M. K., Byrd A. K., Bolden C., Peterson E. C., Byrum S. (2020). proteiNorm - A user-friendly tool for normalization and analysis of TMT and label-free protein quantification. ACS Omega 5, 25625–25633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C. M., Lee C. H., Wang J. Y., Lee P. H., Lai H. S., Hu R. H. (2014). Nationwide longitudinal analysis of acute liver failure in Taiwan. Medicine (Baltimore) 93, e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoofnagle J. H., Carithers R. L. Jr., Shapiro C., Ascher N. (1995). Fulminant hepatic failure: Summary of a workshop. Hepatology 21, 240–252. [PubMed] [Google Scholar]
- Huber W., von Heydebreck A., Sültmann H., Poustka A., Vingron M. (2002). Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18, S96–S104. [DOI] [PubMed] [Google Scholar]
- Karvellas C. J., Speiser J. L., Tremblay M., Lee W. M., Rose C. F.; Acute Liver Failure Study Group. (2017). Elevated FABP1 serum levels are associated with poorer survival in acetaminophen-induced acute liver failure. Hepatology 65, 938–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karvellas C. J., Speiser J. L., Tremblay M., Lee W. M., Rose C. F.; Acute Liver Failure Study Group. (2021). Elevated serum liver-type fatty acid binding protein levels in non-acetaminophen acute liver failure patients with organ dysfunction. Dig. Dis. Sci. 66, 273–283. [DOI] [PubMed] [Google Scholar]
- Kuroda H., Abe T., Fujiwara Y., Nagasawa T., Suzuki Y., Kakisaka K., Takikawa Y. (2021). Contrast‐enhanced ultrasonography‐based hepatic perfusion for early prediction of prognosis in acute liver failure. Hepatology 73, 2455–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet V., Boussetta N., Leclerc J., Fauveau V., Foretz M., Viollet B., Couty J. P., Celton-Morizur S., Perret C., Desdouets C. (2018). LKB1 as a gatekeeper of hepatocyte proliferation and genomic integrity during liver regeneration. Cell Rep 22, 1994–2005. [DOI] [PubMed] [Google Scholar]
- McGill M. R., Cao M., Svetlov A., Sharpe M. R., Williams C. D., Curry S. C., Farhood A., Jaeschke H., Svetlov S. I. (2014). Argininosuccinate synthetase as a plasma biomarker of liver injury after acetaminophen overdose in rodents and humans. Biomarkers 19, 222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill M. R., Jaeschke H. (2014). Mechanistic biomarkers in acetaminophen-induced hepatotoxicity and acute liver failure: From preclinical models to patients. Expert Opin. Drug Metab. Toxicol. 10, 1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlen G., Gentric G., Celton-Morizur S., Foretz M., Guidotti J. E., Fauveau V., Leclerc J., Viollet B., Desdouets C. (2014). AMPKα1 controls hepatocyte proliferation independently of energy balance by regulating Cyclin A2 expression. J. Hepatol. 60, 152–159. [DOI] [PubMed] [Google Scholar]
- Mitchell K. A., Lockhart C. A., Huang G., Elferink C. J. (2006). Sustained aryl hydrocarbon receptor activity attenuates liver regeneration. Mol. Pharmacol. 70, 163–170. [DOI] [PubMed] [Google Scholar]
- Ostapowicz G., Fontana R. J., Schiødt F. V., Larson A., Davern T. J., Han S. H. B., McCashland T. M., Shakil A. O., Hay E., Hynan L., et al. ; U.S. Acute Liver Failure Study Group. (2002). Results of a prospective study of acute liver failure at 17 tertiary care centers in the United States. Ann. Intern. Med. 137, 947–954. [DOI] [PubMed] [Google Scholar]
- Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., et al. (2019). The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy K. R., Ellerbe C., Schilsky M., Stravitz R. T., Fontana R. J., Durkalski V., Lee W. M.; Acute Liver Failure Study Group. (2016). Determinants of outcome among patients with acute liver failure listed for liver transplantation in the United States. Liver Transplant. 22, 505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuben A., Tillman H., Fontana R. J., Davern T., McGuire B., Stravitz R. T., Durkalski V., Larson A. M., Liou I., Fix O., et al. (2016). Outcomes in adults with acute liver failure between 1998 and 2013: An observational cohort study. Ann. Intern. Med. 164, 724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W., Smyth G. K. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomaker S., Warner R., Bock J., Johnson K., Potter D., Van Winkle J., Aubrecht J. (2013). Assessment of emerging biomarkers of liver injury in human subjects. Toxicol. Sci. 132, 276–283. [DOI] [PubMed] [Google Scholar]
- Searle B. C., Pino L. K., Egertson J. D., Ting Y. S., Lawrence R. T., MacLean B. X., Villén J., MacCoss M. J. (2018). Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry. Nat. Commun. 9, 5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakil A. O., Kramer D., Mazariegos G. V., Fung J. J., Rakela J. (2000). Acute liver failure: Clinical features, outcome analysis, and applicability of prognostic criteria. Liver Transplant. 6, 163–169. [DOI] [PubMed] [Google Scholar]
- Shizu R., Abe T., Benoki S., Takahashi M., Kodama S., Miayata M., Matsuzawa A., Yoshinari K. (2016). PXR stimulates growth factor-mediated hepatocyte proliferation by cross-talk with the FOXO transcription factor. Biochem. J. 473, 257–266. [DOI] [PubMed] [Google Scholar]
- Thanapirom K., Treeprasertsuk S., Soonthornworasiri N., Poovorawan K., Chaiteerakij R., Komolmit P., Phaosawasdi K., Pinzani M. (2019). The incidence, etiologies, outcomes, and predictors of mortality of acute liver failure in Thailand: A population-base study. BMC Gastroenterol. 19, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Chantada M., Ariz U., Varela-Rey M., Embade N., Martínez-Lopez N., Fernández-Ramos D., Gómez-Santos L., Lamas S., Lu S. C., Martínez-Chantar M. L., et al. (2009). Evidence for LKB1/AMP-activated protein kinase/endothelial nitric oxide synthase cascade regulated by hepatocyte growth factor, S-adenosylmethionine, and nitric oxide in hepatocyte proliferation. Hepatology 49, 608–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez J. H., Clemens M. M., Allard F. D., Yee E. U., Kennon-McGill S., Mackintosh S. G., Jaeschke H., Hambuchen M. D., McGill M. R. (2020). Identification of serum biomarkers to distinguish hazardous and benign aminotransferase elevations. Toxicol. Sci. 173, 244–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Sun Z., Jiang J., Wu D., Liu X., Xie Z., Chen E., Zhu D., Ye C., Zhang X., et al. (2017). Proteomic signature of acute liver failure: From discovery and verification in a pig model to confirmation in humans. Mol. Cell. Proteomics 16, 1188–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler N., Schlotmann A., Schnitzbauer A. A., Zeuzem S., Welker M. W. (2020). The epidemiology of acute liver failure: Results of a population-based study including 25 million state-insured individuals. Dtsch. Arztebl. Int. 117, 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]