Abstract
Maximal oxygen uptake (VO2max) is a direct measure of human cardiorespiratory fitness and is associated with health. However, the molecular determinants of interindividual differences in baseline (intrinsic) VO2max, and of increases of VO2max in response to exercise trainxìng (ΔVO2max), are largely unknown. Here, we measure ~5,000 plasma proteins using an affinity-based platform in over 650 sedentary adults before and after a 20-week endurance-exercise intervention and identify 147 proteins and 102 proteins whose plasma levels are associated with baseline VO2max and ΔVO2max, respectively. Addition of a protein biomarker score derived from these proteins to a score based on clinical traits improves the prediction of an individual’s ΔVO2max. We validate findings in a separate exercise cohort, further link 21 proteins to incident all-cause mortality in a community-based cohort and reproduce the specificity of ~75% of our key findings using antibody-based assays. Taken together, our data shed light on biological pathways relevant to cardiorespiratory fitness and highlight the potential additive value of protein biomarkers in identifying exercise responsiveness in humans.
Oxygen uptake (VO2) represents a measure of the body’s capacity to supply oxygen to skeletal muscle to perform physical work. VO2 reflects the integration of multiple organ systems and cellular processes, including pulmonary ventilation, oxygen carrying capacity and transport through the circulatory system, cardiac output, central nervous system recruitment of motor units, oxygen diffusion and extraction at the capillary–skeletal muscle level, as well as mitochondrial respiration. VO2max defines the limits of these processes and is thus widely considered the gold-standard measure of cardiorespiratory fitness (CRF)1,2.
It is thus not surprising that VO2max (as a direct measure of CRF) has been firmly established as a powerful prognostic marker of cardiovascular disease (CVD) and all-cause mortality3. VO2max’s inverse relationship with CVD and mortality risk applies to both its baseline measure (intrinsic VO2max4,5) and capacity to improve VO2max through regular physical activity (acquired or adaptive VO2max; ΔVO2max)6,7. Consequently, there has been significant interest in characterizing the relative contributions of different organ systems to VO2max. Several lines of evidence point to cardiac output and oxygen delivery as being the principal determinants of VO2max8,9; however, even the precise contributions of these processes, including oxygen diffusion, convection and mitochondrial oxidative capacity, are not fully resolved10,11.
Furthermore, both baseline measures of VO2max and ΔVO2max appear to vary greatly in the general population. In the HERITAGE Family Study, a subgroup of 429 apparently healthy but sedentary members of family units, who were of European descent, underwent direct measurements of baseline VO2max through cardiopulmonary exercise testing (CPET) on 2 separate days, and the s.d. (9 ml O2 kg−1 min−1) was ~29% of the mean (31 ml O2 kg−1 min−1) after adjustment for age, sex, body mass and body composition12. Similarly, among 720 HERITAGE participants who completed the supervised 20-week endurance-exercise training programme, the s.d. was 53% of the mean change in VO2max. Interestingly, there was no relationship between baseline and ΔVO2max in this group (r2 = 0.011). This suggests that these traits may have different biologic underpinnings and underscores our inability to predict VO2max ‘trainability’ using existing clinical factors13.
Given our incomplete understanding of the biologic basis of CRF and its close relationship to long-term health outcomes, uncovering the molecular determinants of VO2max may provide insights into the mechanistic links between physical fitness and well-being. Indeed, this has become an important goal of the medical community. Prior efforts to characterize both baseline and acquired CRF at the molecular level have included genetic analyses, transcriptomic profiling of skeletal muscle and plasma metabolomics14–16. Although biochemical profiling of plasma proteins has yielded insights into differences in substrate metabolism among different fitness states in animal models17 and has provided biologic ‘snapshots’ of human metabolism18, few data exist regarding plasma proteomics profiling of CRF in humans, particularly in the context of exercise training. These limitations are in part due to the technical challenges involved in capturing the highly dynamic range of circulating proteins. Advancements in aptamer-based profiling methods now allow for the high-throughput measurement of over 5,000 proteins19. This technology spans a dynamic range of at least 7 orders of magnitude (~100 fM–1 μM) with demonstrated high assay reproducibility across both hospital- and population-based cohorts20,21, and was recently applied in the HERITAGE study22.
Here, we sought to compare the circulating proteomic profiles of baseline VO2max as well as its adaptation to an exercise programme by applying a large-scale, affinity-based platform in more than 650 healthy but sedentary participants before and after a 20-week supervised endurance-exercise training intervention. We hypothesized that plasma protein signatures associated with VO2max would reflect its integrative biology and highlight proteins related to skeletal muscle, hematopoiesis and the vascular system, among other determinants of CRF. Further, given that clinical traits are weakly correlated with VO2max changes following exercise training, we anticipated that the addition of plasma proteins would improve the capacity to predict VO2max responsiveness. Finally, given that both baseline VO2max as well its capacity to change in response to exercise training are associated with future risk of death, we tested whether plasma proteins related to these measures would be associated with incident all-cause mortality in a separate population-based study.
Results
HERITAGE participant characteristics.
The HERITAGE cohort was composed of adult parents and their biologic offspring. The mean (s.d.) age of the full cohort (n = 745) used for baseline VO2max analyses was 34.3 (13.4) years; 288 were African American (39%), 409 were women (55%) and 503 were offspring (68%). Mean (s.d.) baseline VO2max was 2,345 (726) ml min−1. Among the participants with VO2max measurements before and after exercise training (n = 654), the mean ΔVO2max was 383 (203) ml O2 min−1 (Table 1).
Table 1 |.
HERiTAGE cohort clinical characteristics
| Clinical characteristics | Participants with baseline Vo2max (n = 745) | Participants with baseline and post-training Vo2max (n = 654) |
|---|---|---|
| Age, mean (s.d.), years | 34.3 (13.4) | 34.8 (13.6) |
| Female, n (%) | 409 (54.9) | 361 (55.2) |
| European descent, n (%) | 457 (61.3) | 424 (64.8) |
| BMI, median (interquartile range), kg/m2 | 25.5 (22.4–29.7) | 25.5 (22.5–29.7) |
| Maximal oxygen uptake, mean (s.d.), ml min−1 | ||
| Baseline | 2,345 (726) | 2,348 (732.5) |
| Change after exercise training | - | 383 (202.8) |
| SBP, mean (s.d.), mmHg | 119 (12.0) | 119 (11.8) |
| DBP, mean (s.d.), mmHg | 69 (8.9) | 68 (8.8) |
| Resting heart rate, mean (s.d.) | 65 (8.9) | 65 (8.9) |
Mean (s.d.) and median (25–75%) values are shown.
Plasma proteins associated with baseline levels of VO2max.
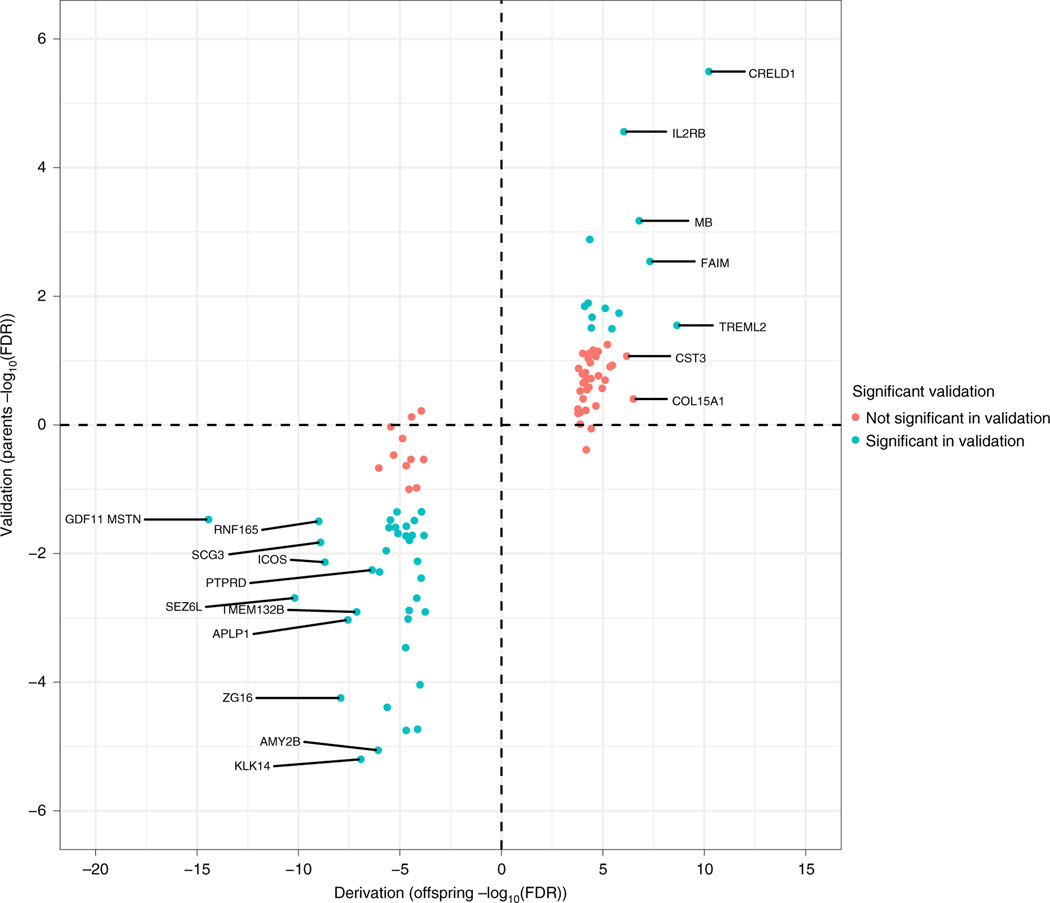
We measured ~5,000 proteins using a multiplexed, single-stranded DNA aptamer (SOMAmers) assay (Supplementary Table 1). We first tested for age- and sex-adjusted protein associations with baseline VO2max in the offspring generation (n = 503) and then sought to replicate our findings in the parent generation (n = 242). We identified 94 proteins that were associated with VO2max in the offspring by using a false-discovery rate (FDR) threshold of <1%. Fifty of 94 proteins were associated with VO2max in the parents at nominal significance (P < 0.05) and 90/94 were directionally consistent (Fig. 1). We subsequently collapsed these subgroups for all further analyses.
Fig. 1 |. Proteins associated with baseline Vo2max among offspring and parents.
Protein associations (FDR < 1%) using linear regression were determined first in the offspring cohort (n = 433). We subsequently validated 50/94 proteins in the cohort of parents (n = 221; P < 0.05). Ninety of 94 proteins were directionally consistent, indicated by quadrant (increase in parents and offspring, upper right; decrease in parents and offspring, lower left).
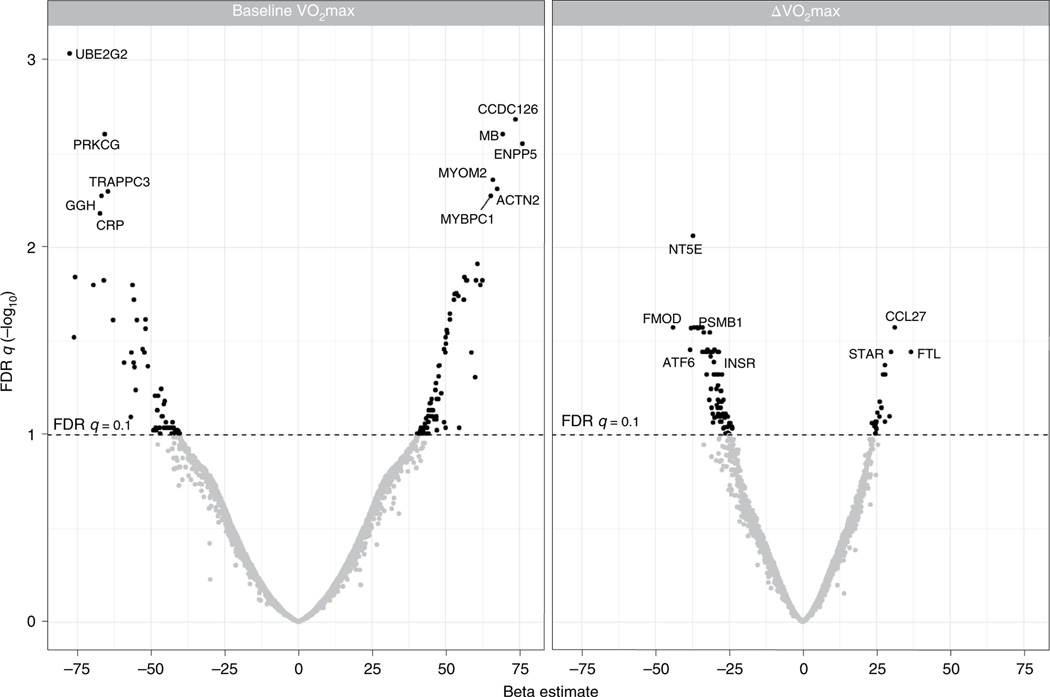
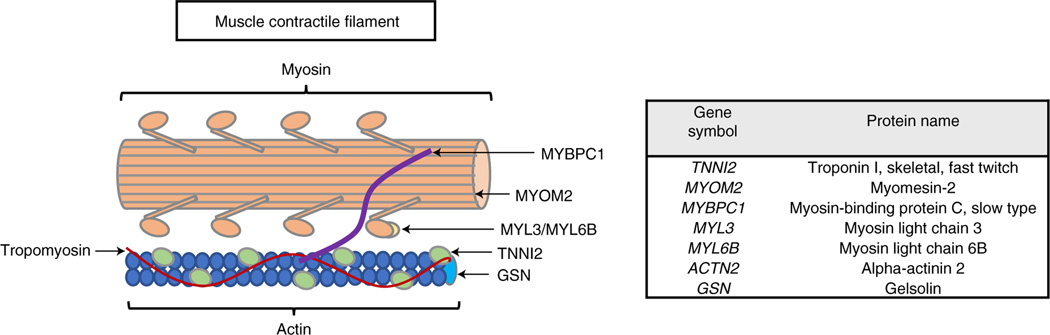
In the full cohort, we identified 147 circulating proteins that were associated with baseline VO2max (Fig. 2), including 85 proteins that were positively associated and 62 proteins negatively associated in analyses that were adjusted for age, sex, body mass index (BMI) and race (Supplementary Table 2). Proteins positively associated with baseline VO2max spanned organ systems and biologic processes relevant to CRF including angiogenesis (for example extracellular matrix protein 1 (ECM1) and anthrax toxin receptor 2 (ANTXR2)), coagulation and hematopoiesis (for example, complement decay-accelerating factor (DAF) and tetranectin (TN)) and lipid metabolism (for example apolipoprotein F (APOF) and lipase member K (LIPK)). Interestingly, we found a large number of circulating proteins related to striated muscle structure and function (Fig. 3 and Supplementary Table 3). These included actin and myosin stabilizing molecules (for example, alpha-actinin 2 (ACTN2) and myomesin-2 (MYOM2)); proteins involved in muscle contraction (for example, troponin-I (TNNI2) and myosin-binding protein C (MYBPC1)); and two essential myosin light-chain elements (MYL3 and MYL6B) that regulate force production during muscular cross-bridge cycles. We also identified several muscle-isoform-specific enzymes involved in glycolysis in plasma, including beta-enolase (ENOB), ALDOA, phosphoglycerate mutase 1 (PGAM1) and 2 (PGAM2) and lactate dehydrogenase alpha (LDHA) and beta (LDHB).
Fig. 2 |. Plasma proteins associated with baseline and ΔVo2max.
Protein relationships to baseline VO2max (ml O2 min−1) in a linear regression model adjusted for age, sex, BMI and race. The value of leptin’s relationship with baseline VO2max extends beyond the scale.
Fig. 3 |. Muscle proteins positively associated with baseline Vo2max.
Left, muscle filament depiction highlighting the proteins positively associated with baseline VO2max that participate in striated muscle structure and/or function. Myosin-binding protein slow-skeletal isoform (MYBPC1) regulates myosin–actin cross-bridge formation. Troponin I (TNNI2) inhibits actin-activated myosin ATPase activity. Gelsolin (GSN) is an actin severing protein. Myosin light-chain elements (MYL3 and MYL6B) regulate mechano-enzymatic function of myosin. Alpha-actinin 2 (ACTN2; not shown) and myomesin-2 (MYOM2) are actin and myosin stabilizing proteins. Right, table of names of proteins depicted at left and their associated gene symbols.
These baseline cross-sectional analyses also identified several well-known markers of metabolic dysregulation known to be positively associated with adiposity, including leptin, CRP and insulin, which were inversely associated with baseline VO2max. Thus, we adjusted for additional measures of body composition—body fat percentage and fat-free mass—to further examine the role of adiposity in our results. We found that the relationships between these proteins and VO2max were no longer significant after adjustment for body fat percentage but remained significant after adjustment for fat-free mass (Supplementary Table 4). In contrast to these markers of metabolic dysregulation, the striated muscle proteins described above (and in Supplementary Table 3) maintained their correlation with baseline VO2max after adjustment for body fat percentage but not fat-free mass, suggesting that their association with CRF may proceed through their relationship to lean body mass.
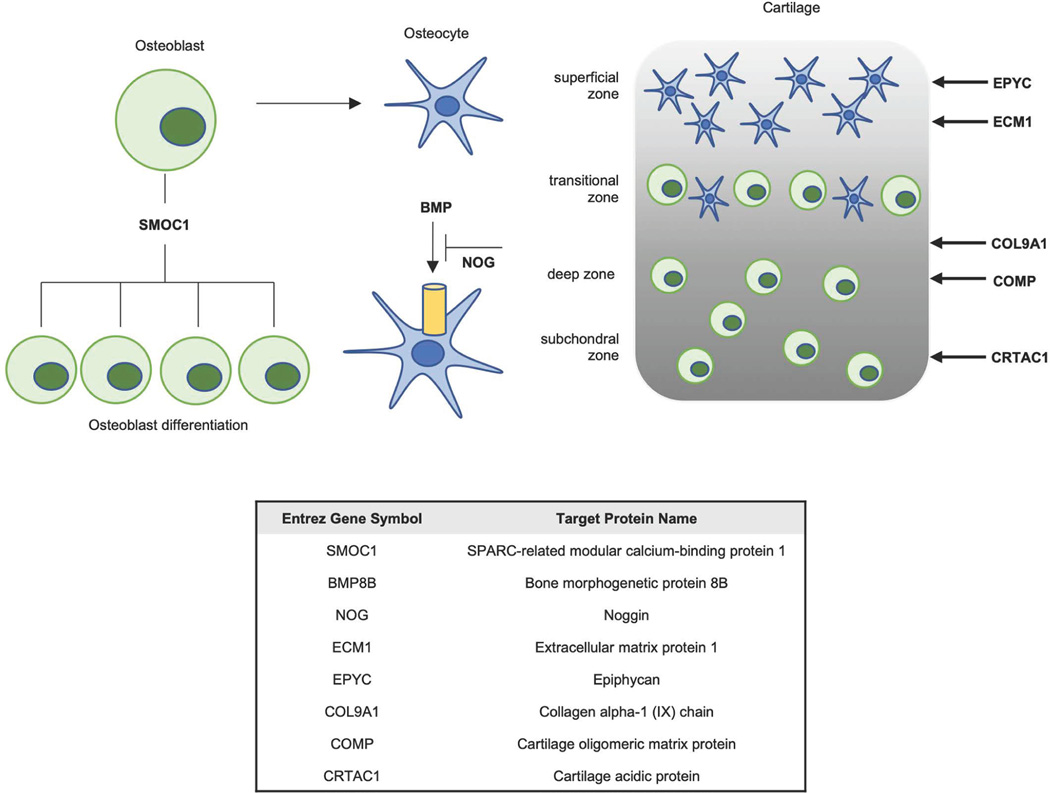
Among the 85 proteins positively associated with baseline VO2max following multivariate adjustment, 25 were known to be secreted based on UniProt Consortium data (Supplementary Table 2). The group of secreted proteins included multiple proteins related to bone homeostasis, including members of osteoblast differentiation (SPARC-related modular calcium binding protein 1 (SMOC1)), bone metabolism via TGF-ß signalling (NOG, bone morphogenic protein 8B (BMP8B)) and structural components of hyaline cartilage (COL9A1, COMP, EPYC; Extended Data Fig. 1).
Test results for the interaction of generation, sex and race on protein–VO2max relationships are shown in Supplementary Table 5. Although we identified 23 protein–generation interactions at nominal significance (P value < 0.05; highlighted in Supplementary Table 2), all were directionally consistent among parents and offspring. Similarly, all 20 protein X sex interactions were directionally consistent among males and females. Only Tartrate-resistant acid phosphatase type 5 (ACP5) and Neural cell adhesion molecule L1-like protein (NRCAM) were directionally different among the 29 protein-VO2max associations that were different between racial groups, with both ACP5 and NRCAM having a positive association with VO2max among African Americans and negative association among Caucasians (ACP5, β = 2.5 and −93.6, respectively; P for interaction = 0.003; NRCAM, β = 21.1 and −10.6, respectively; P for interaction = 0.02). All data have been made available and are available through the NIH Common Fund Molecular Transducers of Physical Activity Consortium (MoTrPAC; https://motrpac-data.org/related-studies/heritage-proteomics).
Validation of baseline VO2max findings in an external cohort.
To further assess the generalizability of our findings, we performed a similar proteomics screen in a separate cohort of abdominally obese individuals who were enroled in a dose–response trial of endurance exercise23. Participants in the validation study subgroup were older (mean age = 47) and had larger body mass (median BMI = 32.8) than HERITAGE participants. A higher percentage of the validation study subgroup was female (71%), and all participants were of European descent (Supplementary Table 6). Of the top 147 proteins associated with baseline VO2max in HERITAGE, 107 were available in the validation dataset. Seventy-nine proteins were directionally consistent, and 24 met statistical significance in the validation cohort in a linear regression model adjusted for age, sex and BMI (P < 0.05; Supplementary Table 7).
Proteins associated with VO2max changes to exercise training.
We found 102 baseline proteins that were associated with ΔVO2max in a linear regression model adjusted for age, sex, BMI, race and the baseline level of VO2max (Supplementary Table 8). The proteins with the strongest associations with ΔVO2max included: 5′ nucleotidase (NT5E), a cell-surface protein that hydrolyses extracellular nucleotides into membrane permeable nucleosides and in which cognate gene variants have been associated with premature arterial calcification24; IL-22 binding protein (IL22RA2), a soluble receptor whose ligand is involved in insulin and glucose homeostasis25; and fibromodulin (FMOD), a secreted protein that has been implicated in tissue repair and myogenic regulation through its interaction with myostatin26.
A generation–protein interaction on ΔVO2max was found for four proteins, with hepcidin (LEAP1) having directionally different associations among parents and offspring (Supplementary Table 9). Eleven proteins demonstrated a sex–protein interaction, with β−1,3-galactosyltransferase (B3GALT1) and triggering receptor expressed on myeloid cells 1 (TREM1) having directionally different associations among males and females. Among the 18 proteins that demonstrated a race–protein interaction on ΔVO2max, 6 demonstrated directionally different associations among African Americans and those of European descent: C–C motif chemokine 27 precursor (CCL27), retinal rod rhodopsin-sensitive cGMP 3′,5′-cyclic phosphodiesterase subunit delta (PDE6D), phosphatidylinositol polyphosphate 5-phosphatase type IV (INP5E), plexin-A1 (PLXA1), pleiotropin (PTN), and EGF-like repeat and discoidin I-like domain-containing protein 3 (EDIL3).
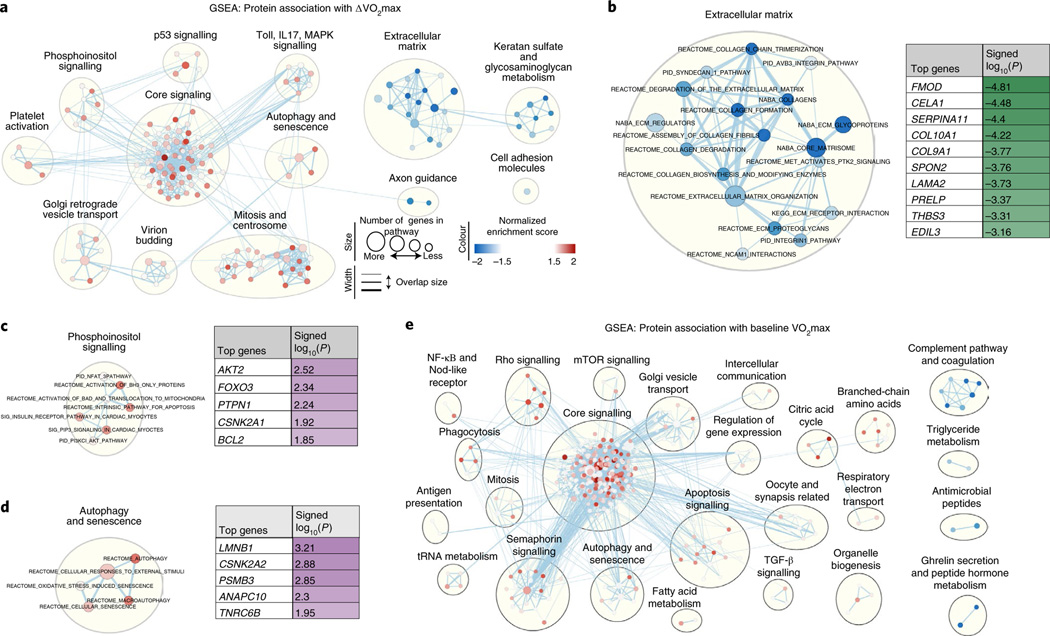
We next performed gene set enrichment analysis (GSEA) to further elucidate biochemical pathways among this set of proteins, as well as those previously identified in the baseline VO2max analyses (Supplementary Tables 10 and 11, respectively). Proteins negatively associated with ΔVO2max were most enriched for ECM-related proteins (the ‘matrisome’)27 (Fig. 4a,b). Positively associated proteins, however, were enriched for core signalling pathways that include platelet-derived growth factor receptor, neurotrophin and hepatocyte growth factor pathway signalling, among others (Fig. 4a,c,d). These biochemical pathways contrast with those enriched after GSEA was applied to proteins ranked by their association with baseline VO2max (Fig. 4e).
Fig. 4 |. GSEA for proteins associated with ΔVo2max or baseline Vo2max.
Overview of overrepresented biological pathways and their connectivity using Cytoscape v3.7.1. a, Network visualization of GSEA results using the complete dataset of protein–ΔVO2max associations. Red dots indicate pathways with over-represented positive protein–ΔVO2max associations, and blue dots indicate over-represented negative protein–ΔVO2max associations. A larger circle size denotes a larger number of genes in a pathway, and darker shades indicate a higher degree of enrichment. Clusters indicate biological pathways with shared proteins and biological function. b–d, Selected clusters of biological pathways with annotation, from a; the top contributing proteins to enrichment score are shown in a table. e, Network visualization of GSEA results using the complete dataset of protein–baseline VO2max associations.
We also compared the group of proteins associated with baseline VO2max with those associated with adaptive VO2max changes to exercise training and found minimal overlap between the two groups. Only five proteins, T132B, ATF6A, COL9A1, INS and PIANP, were associated with both baseline VO2max and ΔVO2max.
Plasma proteins improve prediction of ΔVO2max responses.
Given the vast heterogeneity in VO2max changes that occur with exercise training, as described above, and that clinical factors account for a limited amount of the variance in VO2max trainability15, we sought to determine whether baseline plasma proteins could improve our ability to predict VO2max changes in response to exercise training. Because baseline VO2max and VO2max changes with exercise training are minimally correlated, we tested to see whether proteins could help predict VO2max changes relative to one’s baseline VO2max level (ΔVO2max/baseline VO2max). We selected a relative VO2max change threshold of 15%, given that the median value among the cohort was ~16% (4.9 ml O2 kg−1 min−1) and a 15% change represented > 1 metabolic equivalent (1 MET), a clinically meaningful unit that has been related to >10% relative risk reduction in CVD and all-cause mortality in a series of longitudinal cohorts3.
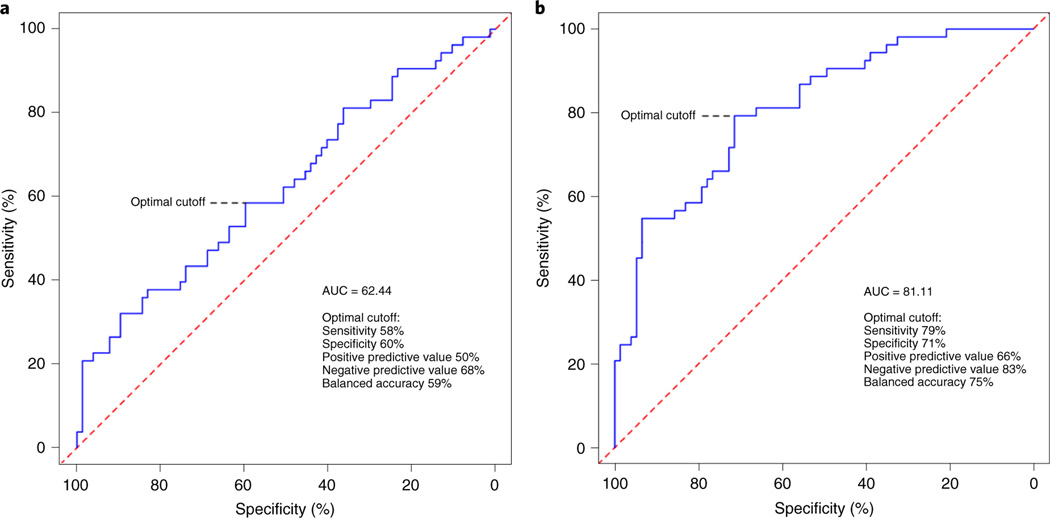
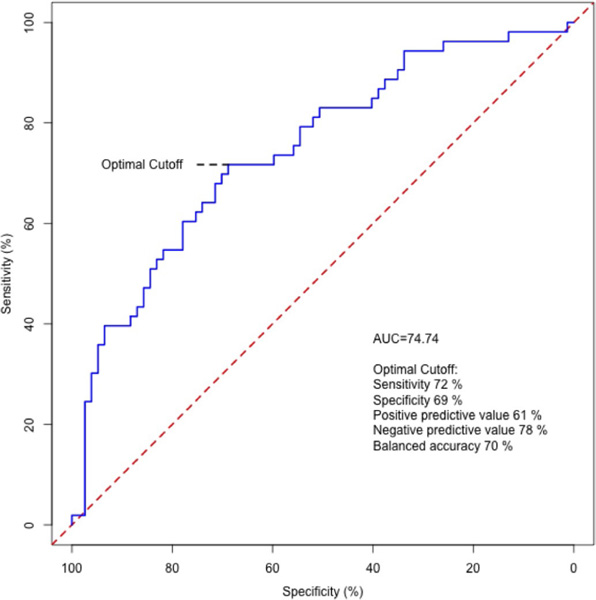
We first performed receiver–operating characteristic (ROC) analyses using a clinical trait model that included age, sex, race and BMI for relative VO2max changes > 15%. The area under of the curve (AUC) was 0.62 (P = 0.91) (Fig. 5). Feature selection and elastic net regression modelling of the 5,000 proteins yielded a final panel of 56 proteins (Supplementary Table 12). We next added our protein panel to the clinical trait model, and the AUC significantly increased to 0.81 (P = 0.00018). With regard to the operator characteristics, we found 79% sensitivity, 71% specificity, positive predictive value of 66% and negative predictive value of 83% for relative VO2max changes > 15%. In a subsequent model that included the same clinical traits but only the group of proteins that both overlapped with an antibody-based proteomics platform (see ‘Complementary data to support aptamer specificity’) and demonstrated moderate to strong correlation between both platforms (7/10 proteins; SELE, TCL1A, COMP, CREG1, STC1, IL1RL2, LILRA2; ρ = 0.41–0.91), the operator characteristics were similar but performed slightly worse (AUC = 0.75, Extended Data Fig. 2), suggesting that there is added information provided by the remaining protein targets in our main model.
Fig. 5 |. ROC curves for relative Vo2max changes with exercise training > 15%.
a, The clinical trait score (age, sex, BMI and race) had a modest AUC. b, Addition of the protein score significantly improved the AUC. The sensitivity, specificity, positive predictive value, negative predictive value and accuracy at the optimal cutoff are included.
Association of VO2max-related proteins and mortality.
We previously performed proteomics profiling in the Framingham Heart Study (FHS) Offspring Study using first a 1.1 k-plex (n = 821 participants) and then an updated 1.3 k-plex version (n = 1,092) of the aptamer-based proteomics platform used in HERITAGE28,29. The clinical characteristics of the FHS sample are presented in Supplementary Table 13. Among the 102 proteins that were associated with ΔVO2max in HERITAGE, 20 were available in both batches of FHS. Thirty-six out of the 147 proteins associated with baseline VO2max were available in the FHS.
Of 1,909 FHS participants, 551 died after a mean (s.d.) follow-up of 13.6 (5.6) years. In age- and sex-adjusted analyses, 12 out of 36 proteins associated with baseline VO2max and 9 out of 20 proteins associated with ΔVO2max were also associated with incident all-cause mortality (FDR q < 0.1; Table 2). We next performed stepwise regression using these protein sets (12 and 9 proteins, respectively) to estimate the percentage variation in all-cause mortality explained by each protein beyond age, sex and batch. Among the proteins associated with baseline VO2max, gelsolin (GSN) was the most significantly associated with all-cause mortality (hazard ratio (HR), 0.71; FDR q = 9.1 × 10−13) and explained 3.4% of the variation beyond age and sex. Among proteins associated with ΔVO2max, macrophage metalloelastase (MMP12) was the most significantly associated with all-cause mortality (HR, 1.34; FDR q = 1.2 × 10−7), explaining 1.8% of the variation in outcome.
Table 2 |.
Proteins associated with baseline or ΔVo2max in HERiTAGE and all-cause mortality in the FHS offspring Study
| Gene name | Protein name | Adjusted HR | 95% Cl | FDR q value | Variation explained by protein (%) | |
|---|---|---|---|---|---|---|
| Baseline VO2max | ||||||
| GSNa | Gelsolin | 0.71 | 0.65 | 0.78 | 9.1 × 10–13 | 3.00 |
| CRPa | C-reactive protein | 1.24 | 1.13 | 1.36 | 8.2 × 10–5 | |
| B2Ma | β2-microglobulin | 1.21 | 1.09 | 1.33 | 1.6 × 10–3 | 1.00 |
| ECM1a | Extracellular matrix protein 1 | 0.84 | 0.77 | 0.93 | 2.9 × 10–3 | |
| MBa–c | Myoglobin | 0.87 | 0.79 | 0.96 | 1.7 × 10–2 | 0.22 |
| FCGR3Ba–c | Low-affinity immunoglobulin gamma Fc region receptor III-B | 1.13 | 1.04 | 1.23 | 1.7 × 10–2 | |
| ACP5a–c | Tartrate-resistant acid phosphatase type 5 | 1.14 | 1.03 | 1.27 | 3.1 × 10–2 | 0.17 |
| PLGa | Plasminogen | 0.90 | 0.82 | 0.98 | 4.4 × 10–2 | 0.45 |
| NRCAMa,b | Neuronal cell adhesion molecule | 0.90 | 0.83 | 0.98 | 4.6 × 10–2 | - |
| CFBa | Complement factor B | 1.11 | 1.01 | 1.22 | 5.4 × 10–2 | - |
| ENPP7a–c | Ectonucleotide pyrophosphatase/phosphodiesterase family member 7 | 1.11 | 1.01 | 1.21 | 5.4 × 10–2 | - |
| NRXN3a | Neurexin-3-ß | 0.90 | 0.83 | 0.99 | 5.4 × 10–2 | - |
| ΔVO2max | ||||||
| MMP12a–c | Macrophage metalloelastase | 1.34 | 1.22 | 1.48 | 1.2 × 10–7 | 1.80 |
| FAPa–c | Prolyl endopeptidase FAP | 0.78 | 0.72 | 0.85 | 3.8 × 10–7 | |
| ANGPT2a–c | Angiopoietin-2 | 1.21 | 1.10 | 1.33 | 6.7 × 10–4 | 0.47 |
| STC1a–c | Stanniocalcin-1 | 1.19 | 1.09 | 1.30 | 1.8 × 10–3 | 0.74 |
| CCL27a,b | C-C motif chemokine 27 | 1.16 | 1.06 | 1.28 | 7.3 × 10–3 | - |
| IL11RAa | Interleukin-11 receptor subunit α | 0.86 | 0.79 | 0.94 | 7.3 × 10–3 | 0.54 |
| ERBB3a,b | Receptor tyrosine-protein kinase erbB-3 | 0.86 | 0.78 | 0.94 | 8.3 × 10–3 | 0.21 |
| ACANa,c | Aggrecan | 0.87 | 0.80 | 0.96 | 1.7 × 10–2 | - |
| IMDH2 | Inosine-5'-monophosphate dehydrogenase | 1.12 | 1.03 | 1.23 | 3.3 × 10–2 | - |
Cox proportional hazards analysis was performed for both the proteins associated with baseline VO2max and those associated with VO2max and all-cause mortality, adjusting for age, sex and batch. Proteins from each analysis that were statistically significant (FDR q < 0.1) were brought forwards in stepwise regression. The percent variation in all-cause mortality beyond age and sex is listed in the final column for those proteins retained in the final model.
Aptamer specificity supported by pQTLs and/or MS-based proteomics in population-based data (Supplementary Table 15).
Aptamer targets available for comparison on Olink Explore platform in HERITAGE subset (n = 88).
Proteins with Spearman correlation > 0.4 on aptamer and antibody-based platforms in HERITAGE subset.
Complementary data to support aptamer specificity.
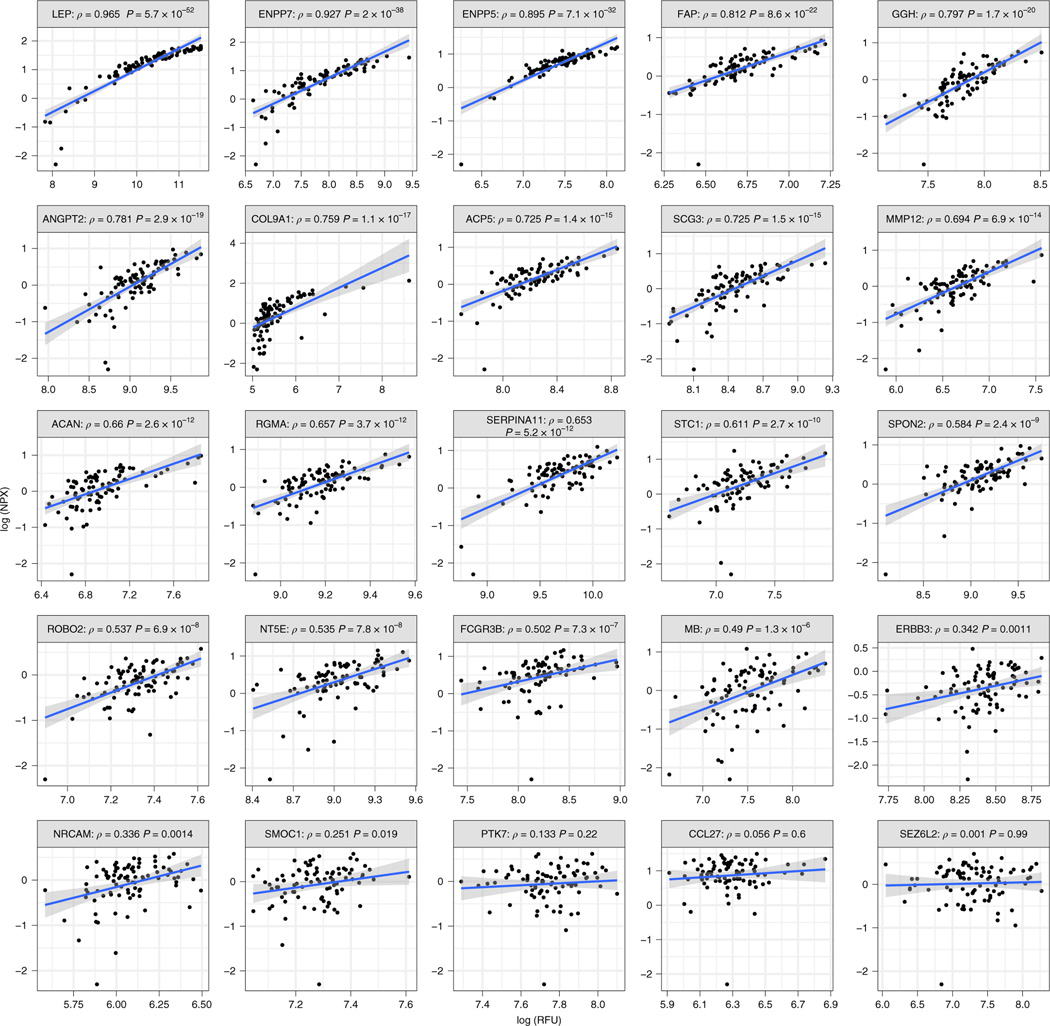
We tested the reproducibility of our top aptamer-based findings in HERITAGE specific samples using Olink’s antibody-based proteomics platform (Olink Explore). Clinical characteristics of the random sample from HERITAGE are shown in Supplementary Table 14. Among the 21 proteins significantly associated with incident all-cause mortality, 12 protein targets were available on both platforms. Nine out of 12 of the protein targets were highly correlated. In addition, among the top protein targets associated with either baseline or ΔVO2max that did not overlap with our all-cause mortality findings (Supplementary Table 15 and Tables 2 and 3 in Supplementary Data), an additional 13 proteins were available on both platforms. Ten of 13 assays demonstrated strong correlations. Taken together, 19 out of 25 of our top aptamer-based protein findings from HERITAGE were well correlated with an equivalent antibody-based assay (both sets of protein correlations shown in Fig. 6).
Fig. 6 |. Spearman’s correlations between aptamer-based and antibody-based assays among top findings.
Spearman’s correlations between protein levels measured by an aptamer-based method (log (RFU); x axis) and antibody-based method (log (NPX); y axis).
In addition, we leveraged mass spectrometry (MS)-based and genetic assays to support the specificity of the aptamer assays for our most significant findings. Among the 21 proteins significantly associated with incident all-cause mortality, genome-wide significant associations at cis loci (within 1 Mb of the transcription start site for the cognate gene of the protein) were identified for 17, consistent with the specificity of the aptamer–protein relationship. Aptamer specificity for two additional proteins (B2M and MB) was confirmed by MS30. Among the top 25 findings in both our baseline VO2max and ΔVO2max analyses, 23 and 24 were available for testing across genetic and MS-based analyses, respectively. The specificity of 11/23 proteins associated with baseline VO2max and 12/24 proteins associated with ΔVO2max was supported by these tests (Supplementary Table 15).
Discussion
VO2max—as a direct measure of CRF—reflects the body’s ability to transfer oxygen to skeletal muscle during sustained physical activity, and is thus a quantifiable measure of functional capacity. It has emerged as an important prognostic marker of future health risk that adds value beyond traditional risk factors3. While both baseline VO2max and the adaptive changes in VO2max in response to regular exercise provide valuable information about health status, these traits are largely unrelated to each other, a fact that underscores our limited understanding of their biologic basis and links to long-term health outcomes. Here, we performed large-scale plasma proteomic profiling in over 650 individuals with directly measured VO2max before and after an endurance-exercise intervention to illuminate the biochemical features of baseline CRF and its adaptation to regular exercise. These analyses produced four notable findings. First, there was a broad and diverse set of circulating proteins associated with both baseline VO2max levels and its changes in response to exercise training. Second, there was minimal overlap between the proteomic profiles of these distinct clinical traits. Third, the addition of a plasma protein score to baseline clinical traits improved the predictive accuracy of clinically significant improvements in VO2max to exercise training. Finally, key proteins that are correlated with baseline VO2max or ΔVO2max were also associated with incident all-cause mortality in a separate population-based cohort.
Proteins are important regulators of biologic processes and, like CRF, reflect an individual’s current health state as well as future risks22. The plasma proteome encompasses proteins from all tissues, making it an attractive medium to study the integrative biology of CRF. Indeed, we identified circulating proteins that spanned many of the organ systems involved in determining VO2max, including the nervous, musculoskeletal, pulmonary, haematologic and circulatory systems. These included tissue-specific, structural and functional proteins (for example, striated muscle, Fig. 3) and proteins with signal peptide sequences (for example, secreted proteins; Supplementary Table 2), as well as several proteins of uncertain function or not predicted to be secreted. Although these latter proteins may reflect tissue leakage or aberrantly secreted proteins, recent evidence suggests that traditional annotation methods may not fully account for proteins released into circulation via extracellular vesicles31. Indeed, our finding that a number of glycolytic enzymes, including fructose bisphosphate aldolase A (ALDOA), β-enolase 3 (ENO3) and lactate dehydrogenase (LDHB and LDHA), were present in the blood are consistent with those from Whitham et al.31, who demonstrated a rise in plasma levels during acute bouts of exercise. The mechanistic relevance of these findings remains unknown, and additional research is needed to understand whether these enzymes have unanticipated functional effects in circulation or are biomarkers of physiologic states.
Among a group of classically secreted proteins, we identified several relevant to bone homeostasis that were positively associated with baseline VO2max (Extended Data Figure 1). This group included BMP8B, an adipokine that regulates cartilage and bone development and has also been shown to induce brown-adipose-tissue thermogenesis32 and adipocyte neurovascular remodelling33, and SMOC1, a regulator of osteoblast differentiation relevant in physiologic cardiac hypertrophy34. We cannot localize the tissue origin of these circulating proteins, but our findings highlight the emerging paradigm of bone as an important endocrine organ involved in tissue crosstalk and exercise adaptation and motivate further interrogation of our data35.
Few data describing the plasma proteomic profiles of baseline VO2max exist22,36, and to our knowledge this is the first study to investigate large-scale proteomic relationships with longitudinal VO2max adaptations. Santos-Parker and colleagues36 performed aptamer-based proteomics using a smaller-scale (1.1 k-plex) platform among a group of 47 sedentary or exercise-trained young men and women, and older men. The authors performed gene network and gene ontology (GO)-based annotation to identify biological processes associated with those in the exercise-trained state. More recently, Williams et al.22 applied aptamer-based proteomic profiling in HERITAGE to generate a predictive model of cross-sectional VO2max based on 115 proteins, using a training set that included 50% of samples from participants at baseline and 50% after completing exercise training.
While there was overlap among some of the broad biologic processes identified by Santos-Parker et al. (for example, autophagy and vasculogenesis) or individual proteins found by Williams et al. (~23% of our findings overlapped), our baseline VO2max findings differed from these for several reasons. First, in contrast to these studies, our analyses were performed separately using only pretraining or post-training measures of VO2max. Our baseline analyses did not include values obtained after the HERITAGE exercise intervention, which may reflect adaptive changes in VO2max, a trait that is uncorrelated to its intrinsic value13. In addition, we used absolute values of VO2max (ml O2 min−1) and adjusted for clinical characteristics in contrast to the univariate analyses of weight-adjusted VO2max (ml O2 kg−1 min−1) performed by Williams et al.22. Adjustments for age, sex and race probably significantly contributed to the differences between our groups’ findings, owing to their relationships with CRF as previously documented and underscored in our interaction analyses37–39.
Interestingly, when we performed additional adjustments for body-composition measures, we found that proteins closely associated with adiposity (for example, C-reactive protein, leptin and insulin) were no longer significant after adjusting for body fat percentage, but remained highly associated with VO2max in a model adjusted for fat-free mass, similar to our main model using BMI. Although the main influence of body mass on VO2max is mediated by fat-free mass, these data support prior findings that adipose tissue may contribute to VO2max beyond differences in lean body weight40. Overall, there was modest overlap between the proteins related to baseline VO2max in the models adjusted for body fat percentage and fat-free mass compared with the BMI-adjusted model (61 proteins, 48% overlap and 15 proteins, 56% overlap, respectively), whereas there was only one common protein (insulin-like growth factor binding protein 1 (IGFBP1)) among the fat-free-mass-adjusted and body-fat-percentage-adjusted models (Supplementary Table 4). These findings, coupled with the attenuation of striated-muscle-specific protein associations with baseline VO2max after adjustment for lean body mass, highlight the importance of using standardized body size and composition adjustments for VO2max when comparing results across studies.
We believe that the limited number of derivation proteins that achieved statistical significance in the external validation cohort reflects the large differences in sample size between the two studies (n = 745 in HERITAGE versus n = 91 in the validation study) and the directional consistency of protein–VO2max relationships (79/107) better reflects the stability of our findings across these studies. Further, given the known age- and body-size-related effects on proteomic profiles, as demonstrated in HERITAGE, we believe that large differences in the clinical characteristics between the two studies—even after restricting the validation cohort to age- and BMI-specific limits—impact the interpretation of our findings. We encourage additional validation of our findings; however, we are unaware of any other longitudinal, large-scale proteomic studies that include directly measured VO2max at the moment.
The distinct proteomic profiles of baseline VO2max and its exercise-induced changes that we observed are consistent with prior clinical observations demonstrating a lack of correlation between these traits13,14. The molecular mechanisms that underlie these differences are not well understood, and prior efforts to characterize CRF using candidate gene analyses41, gene-expression data for skeletal muscle42 and genome-wide association (GWAS) studies43 have been limited by small sample sizes, lack of replication and the inherent challenges in applying reductionist strategies to describe a complex trait.
Using GSEA, we found nonrandom associations with baseline VO2max in pathways related to hematopoiesis and angiogenesis (pathway participants included: chitinase 1 (CHIT1), haeme oxygenase 2 (HMOX2), cAMP-dependent protein kinase A (PRKACA), extracellular matrix protein 1 (ECM1)), the complement and coagulation systems (CD55, complement factor B precursor (CFB), cofilin-1 (CFI), plasminogen precursor (PLG), heparin cofactor 2 (SERPIND1)) and metabolic processes, including glycolysis, as described above (Supplementary Table 11a,b). These findings are consistent with those recently published from HERITAGE using integrative genomic analyses from GWAS and skeletal-muscle expression data in participants of European descent44. There, Ghosh et al. identified several gene loci that highlighted key determinants of CRF that we found using GSEA and through manual annotation (for example, skeletal muscle function (SGCG, DMRT2), cardiovascular physiology (CASQ2, ATE1) and hematopoiesis (PICALM)).
In contrast to our baseline VO2max findings, we observed pathway enrichment reflecting proteins involved in extracellular matrix regulation (collagen alpha-1 (III) chain (COL3A1), COL9A1 COL10A1, aggrecan core protein (ACAN) and macrophage metalloelastase (MMP12)), key signalling pathways (for example, platelet-derived growth factor receptor B (PDGFRB) and hypoxia-induced factor 1 (HIF-1) signalling) and autophagy (for example, guanine nucleotide exchange factor (VAV3), cofilin-1 (CFL1)), among others, that were related to VO2max responses to the exercise programme (Supplementary Table 10). These pathways were also present in a group of 16 over-represented Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in GSEA previously performed using GWAS from HERITAGE45. While many of the proteins encoded by the relevant genes from HERITAGE genomic analyses were intracellular and were not captured on our plasma proteomics platform, our shared findings regarding relevant pathways point to possible biologic underpinnings that reflect or possibly mediate the differences between these two traits. Ongoing efforts to incorporate additional molecular profiling data in the study of fitness traits, including the NIH-sponsored initiative, Molecular Transducers of Physical Activity Consortium (MoTrPAC: NCT03960827), will further advance our understanding of these processes.
We also identified five circulating proteins that were associated with both VO2max traits. Although variants in TMEM132B have been associated with lean body mass46, and insulin may also be correlated with both traits through its relationship to body composition, the relationships of COL9A1, PIANP and ATF6A with VO2max are unclear and remain the subject of future exploration.
Our protein biomarker analyses highlight the current lack of predictive capacity for exercise-induced VO2max responses and the potential for large-scale plasma protein profiling for biomarker discovery. Although individual clinical traits such as age, sex, race and BMI have all been shown to influence VO2max, their collective ability to predict a clinically meaningful response in VO2max to exercise training was modest, and no other readily available biomarkers exist. The addition of our protein score helped identify at a high percentage (negative predictive value = 83%) those individuals unable to modestly improve their VO2max despite undergoing a standardized, supervised exercise training programme. If validated in an external cohort, these findings would help with the early identification of individuals who may benefit from alternative lifestyle interventions or additional therapeutics to improve their CRF.
Finally, our observation that plasma proteins related to both baseline VO2max and its trainability are also associated with future mortality risk highlights the potential value of biochemical profiling to better understand the mechanistic links between CRF and long-term health outcomes. The strongest relationship among both sets of proteins was gelsolin (Table 2), both a secreted and intracellular protein with multiple cellular functions. Gelsolin was positively associated with baseline VO2max (β = 56.3; FDR = 0.014) and inversely associated with incident all-cause mortality (HR = 0.71; 95% CI, 0.65–0.78), explaining ~3% of the variation in mortality after adjustment for age and sex in stepwise regression. Prior groups have linked lower plasma gelsolin levels to adverse outcomes in people with sepsis47 and end-stage renal disease48, and most recently higher gelsolin levels were associated with a decreased risk of congestive heart failure after adjusting for established risk factors49. Our data demonstrating its inverse association with all-cause mortality in a large population-based cohort extend these findings. Whether gelsolin is a biomarker or potential mediator of CRF and long-term health remains unclear. Gelsolin’s most well-studied role relates to intracellular actin filament severing and cytoskeletal remodelling50; however, its secreted form predominantly comes from striated muscle and has been shown to function as an extracellular scavenger of actin51 and inflammatory intermediates52, as well as a participant in signal transduction pathways relevant to CRF, including the PI3K pathway53. Additional research into gelsolin’s role in cardiometabolic health is warranted by these recent findings.
There are several limitations to our work. First, HERITAGE is a single-arm study and thus VO2max adaptations may reflect unmeasured factors beyond the exercise-training stimuli. Leisure-time physical activity was not measured; however, all participants were sedentary for 6 months prior to enrolment. The aptamer-based platform that we utilized targets ~5,000 proteins; however, this technology is biased towards circulating proteins and does not provide complete coverage of the plasma proteome. Further, affinity-based assays, such as aptamer technology, are subject to nonspecific binding and may have limitations in their performance in response to post-translational protein modifications54. To address these concerns, we measured protein levels of 25 of our top findings using an orthogonal, antibody-based platform in a random subset of 88 HERITAGE samples and found that 18 out of 25 protein targets were correlated with our aptamer-based results. Among the 7 proteins with a Spearman correlation < 0.5, 2 aptamer targets (SMOC1 and ERBB3) have variants in cis (located within 1 Mb of the transcription start site of the gene encoding the protein) that are highly associated with protein levels in internal HERITAGE genetic-protein analyses (SMOC1, P = 5.9 × 10–8; ERBB3, P = 2.16 × 10–6). In addition, five (CCL27, PTK7, SMOC1, NRCAM and ERBB3) aptamer measurements had cis genotype-protein quantitative trait loci (cis-pQTL) relationships from publicly available and existing population-based human genetics studies, and one protein (MB) was validated using a multiple-reaction-monitoring MS-based method (Supplementary Table 15). Although we cannot resolve the reason for the lack of a stronger correlation between these target proteins, these additional data support the specificity of our aptamer-based findings. Ultimately, we recognize the need for additional confirmation to validate the remaining analytes in the platform. Efforts to do so are ongoing22, and all of our primary data have been made available to the broader scientific community for subsequent efforts. The proteomics platform includes a broad group of proteins; however, we are unable to identify their tissue origin. We limited the number of adjustments in our analyses relating proteins to all-cause mortality because our central goal was to assess the presence of shared protein biology between CRF and long-term health outcomes, thus these findings cannot explain the specific mechanisms through which this occurs nor can they be used as biomarkers of risk prediction without additional work. We also limited our analyses of VO2max changes to linear methods, thus there may be additional insights yielded by using nonlinear methods. Our tests for interaction among race, sex and generation among protein– VO2max relationships may not have been sufficiently powered, and our use of nominal statistical significance may have yielded false positive results, particularly given that the great majority of interactions were directionally consistent between groups.
In summary, we identified a large number of circulating proteins that are associated with VO2max and highlight distinct profiles that exist for its baseline state as well as its adaptation to endurance exercise training. While our findings highlight specific proteins and biochemical pathways associated with these traits, further analyses of these data should yield additional biologic insights and motivate studies in model systems to both identify the sources of these proteins and evaluate their functional significance.
Methods
HERITAGE Family Study.
The HERITAGE Family Study design and its participants have been described55. Briefly, family units of African Americans and people of European descent, totalling 763 sedentary participants (62% of European descent) between the ages of 17 and 65 years, were enroled in a 20-week training study of graded endurance exercise training across 4 clinical centres in the United States and Canada. Participants were healthy but sedentary over the previous 3 months and were free from apparent cardiometabolic disease. A total of 745 participants who had baseline measures of VO2max and plasma samples were included in cross-sectional analyses, whereas 654 participants who completed exercise training and had complete data were used for longitudinal analyses. Written informed consent was obtained from all participants in the HERITAGE Family Study. HERITAGE study consent was reviewed and the research performed in these analyses was approved by Beth Israel Deaconess Medical Center’s institutional review board.
Cardiopulmonary exercise testing and VO2max.
Two maximal CPETs were performed on separate days, at least 48 hours apart, before and after the 20-week exercise training programme, using a cycle ergometer (model 800S, SensorMedics) connected to a metabolic cart (model 2900, SensorMedics). Standard gas-exchange measures were obtained as an average of 20-second intervals. The criteria used for the attainment of VO2max were defined as: a respiratory exchange ratio >1.1, plateau in VO2 uptake (change of <100 ml/min in the last 3 consecutive 20-second averages) and a HR within 10 beats/minute of the maximal level predicted by age. All participants met at least one of these criteria in one of the two tests12, but most met two or more56. The average of the two measurements before and after exercise training were used as VO2max unless the values differed by more than 5%, in which case the higher value was used. The correlation between VO2max measurements between the two tests (r = 0.97), coefficient of variations (CVs, 5%) and reproducibility among clinical centres were excellent57. We used absolute (ml O2 min−1) rather than weight-adjusted (ml O2 kg−1 min−1) measures of VO2max so that body mass changes that occurred after exercise testing were not incorporated into our assessment of ΔVO2max.
Exercise training protocol and plasma sampling.
Participants exercised 3 times per week for 20 weeks, beginning at 30 minutes/session and increasing to 50 minutes/session for the final 6 weeks of the programme. Exercise intensity increased from the heart rate associated with 55% VO2max obtained during baseline CPET to the heart rate associated with 75% VO2max over the final 8 weeks of training. Cycle ergometers were electronically programmed to maintain a training heart rate by adjusting the power output. Each exercise session for all participants was continuously monitored by trained staff. Fasting plasma samples were collected in EDTA tubes from peripheral intravenous catheters prior to the beginning of the exercise training programme and at 24 hours following completion of the final exercise session.
Proteomic profiling.
Aptamer-based method.
Detailed analytic methods of the SOMAscan assay have been described19–21. Briefly, archived plasma samples stored at −80 °C from HERITAGE were diluted in 3 different concentrations (40%, 1% and 0.05%) and incubated with a mixture of fluorescently labelled single-stranded DNA aptamers (~5,000 SOMAmer). Plasma samples had either 0 freeze–thaw cycles or 1 freeze–thaw cycle prior to proteomics profiling. Protein–aptamer complexes were isolated from unbound or nonspecifically-bound proteins using a two-step, streptavidin-bead-based immobilization process. Aptamers eluted from the target proteins were quantified using the degree of fluorescence on a DNA microarray chip. Samples were normalized to 12 hybridization control sequences within each microarray and across plates, using the median signal for each dilution. We have previously reported median intra- and interassay CVs for the SOMAscan assay of ~5% (ref. 58).
Antibody-based method.
We subsequently performed additional proteomics profiling using an antibody-based technology (Olink) on a random sample (n = 88) from the HERITAGE study to determine the reproducibility of our aptamer-based results. Briefly, the Olink plasma extension assay technology uses DNA oligonucleotide-labelled antibody pairs to bind target proteins; 384 assays are performed on 4 separate panels with different dilutions for different dynamic ranges of target proteins (total proteins assayed = 1,536). After incubation with plasma samples, the oligonucleotide pairs hybridize and are extended by DNA polymerase to create a unique DNA barcode that is subsequently read out using next-generation sequencing. The median intra-assay CV for the 1,536 proteins was 10.25%, as assessed by multiple replicates of a pooled sample included in the experiment.
Genome-wide association studies.
We also leveraged existing GWASs of proteins to help to determine aptamer specificity. Genotypes were available for 1,421 participants in the Malmo Diet and Cancer Study and 759 participants in the FHS with existing SOMAscan data59. A meta-analysis of genome-wide association analyses was performed to identify variants associated with circulating protein levels within 1 MB of the cognate gene, which were considered cis. Analyses were conducted on unrelated individuals. The methods used to generate publicly available genetics analyses for SOMAscan data have been described30,60,61.
Framingham Heart Study.
Participants in the FHS Offspring cohort who attended the fifth examination (1991–1995) and who had previously underwent plasma proteomic profiling with the SOMAscan single-stranded DNA aptamer-based platform (1.1 or 1.3 k-plex assays) were included in this study28,29. A total of 1,909 participants were included in analyses. Clinical characteristics were obtained from FHS investigators.
Validation cohort.
The clinical characteristics and methods to derive baseline VO2max from this randomized clinical exercise trial have been described23. Briefly, 300 sedentary adults with abdominal obesity were randomized into 3 exercise arms and a control group. Of the 217 participants who completed the 24-week exercise intervention, 216 had baseline VO2max data and were available as a validation cohort. Given substantial differences—by design—in clinical characteristics between the validation and HERITAGE study cohorts, we restricted our analysis to subjects in the validation study with BMI < 40 and age < 55 (n = 91), to more closely approximate HERITAGE participants.
Statistical analysis.
Baseline clinical characteristics of participants in the HERITAGE Family Study, validation study and FHS are reported as means ± s.d., proportions, or medians (interquartile range) according to visual inspection of normality. A two-sample Student’s t-test was used to compare cases and controls in FHS. All protein values were natural-logarithmically transformed for subsequent analyses. Correlations between aptamer-based and antibody-based proteomics assays were assessed using the Spearman correlation coefficient. Linear regression was performed to determine the relationship between baseline protein values and both baseline VO2max (ml O2 min−1) as well as the changes in VO2max (ΔVO2max, post-training VO2max – pretraining VO2max). Covariates in regression models included age, sex and baseline values of BMI, body fat percentage, fat-free mass (kg),and VO2max (Δ model only). Protein levels were standardized to mean = 0 and multiples of 1 s.d. We used the Benjamini–Hochberg procedure to correct for multiple comparisons and employed a FDR < 0.1 to determine statistical significance for these hypothesis-generating analyses.
We tested for the interactions of generation, sex and race with protein level on baseline- and ΔVO2max and adjusted for the other covariates, given previously reported differences in VO2max trainability among these groups13.
To evaluate the predictive utility of protein biomarkers for relative VO2max changes (ΔVO2max/baseline VO2max) after exercise training, we performed the following analyses. First, we implemented a clinical trait model that included age, sex, race and BMI for relative VO2max changes > 15%. We then added more than 5,000 proteins to train a more comprehensive model. The maximum number of missing values per protein within the entire dataset was ≤7, and the total number of missing values was <2%. The data were randomly split into a training set (80% of cohort) that uses crossvalidation and a test set (20%) that was not used for model development. All preprocessing steps were first applied to the training set. The same steps were then carried out for the test set. We used a k-nearest neighbour algorithm to impute missing values (k = 10)62. All continuous variables were zero-centred and scaled (s.d. = 1). Scaling in the test set was applied using the same scaling factors calculated from the training set. The initial set of more than 5,000 predictors (proteins, age, sex, race and BMI) was reduced using a constraint-based feature selection algorithm for identifying minimal feature subsets (MMPC algorithm63). We then fit elastic net logistic regression models on the basis of the remaining predictors. The hyperparameters of the elastic net were optimized for the AUC using a global optimization algorithm. Receiver–operating characteristics of the protein score were subsequently calculated, with sensitivity, specificity, positive predictive value and negative predictive value generated. The training performance in the results is the result of repeated tenfold cross validation within the 80% training datasets.
GSEA using the full proteomics dataset was performed using the Molecular Signatures Database canonical pathways collection (MSigDB, http://software.broadinstitute.org/gsea/msigdb/collections.jsp), which includes a total of 2,199 curated gene sets from domain experts64. Signed log-transformed P values were computed from the regression models using the coefficient estimates and P values for protein–VO2max associations. The full proteomic dataset was then ranked by their signed P values and used as input for GSEA (v4.0.3, with default parameters). GSEA results were exported to Cytoscape for visualization with the Enrichment Map tool using the following thresholds for gene set significance (P < 0.05, FDR q < 0.15, overlap index > 0.5)65.
For the FHS participants, we performed Cox proportional-hazard regression to model all-cause mortality using the proteins that were significantly associated with baseline or ΔVO2max and also available in FHS. In age-, sex- and batch-adjusted models, proteins that were associated with baseline or ΔVO2max using a FDR q < 0.1 were brought forward for stepwise regression to estimate the percentage variation in all-cause mortality explained by each protein. Cis variants were identified using a linear regression model to assess the associations of variants with proteins that had statistically significant relationships with baseline and ΔVO2max; statistical significance was set at P < 5 × 10−8. All statistical analyses were performed using R version 3.6.2 (R Core Team, R Foundation for Statistical Computing).
Extended Data
Extended Data Fig. 1 |. Secreted proteins positively related to bone homeostasis and baseline Vo2max.
Functional representation of proteins’ role in bone metabolism and homeostasis. Left and middle: SMOC1 regulates osteoblast differentiation. BMPs are related to bone formation via the TGF-β pathway and are mediated by extracellular signalling molecules such as NOG. Right: simplified schematic of proteins related to cartilage formation and their location within cartilage tissue.
Extended Data Fig. 2 |. Receiver-operating characteristic curve for relative Vo2max changes with exercise training > 15% using overlapping targets between aptamer- and antibody-based proteomic platforms.
7/10 overlapping proteins on both platforms demonstrated moderate-strong correlation (SELE, TCL1A, COMP, CREG1, STC1, IL1RL2, LILRA2; ρ = 0.41–0.91) and were used in modeling.
Supplementary Material
Acknowledgements
This study is supported by the National Institute of Health grants K23 HL150327–01A1 (J.M.R.); R01 HL132320; HL133870 (R.E.G.); U24 DK112340 (R.E.G., S.A.C.), R01 HL45670, HL47317, HL47321, HL47323 and HL47327 (all in support of the HERITAGE Family Study); NR019628 (M.A.S., R.E.G.); and HL146462 (M.A.S.). C.B. is partially funded by the John W. Barton Sr. Chair in Genetics and Nutrition. S.G. and C.B. are partially supported by the NIH-funded COBRE grant (NIH 8 P30GM118430–01). S.G. is supported in part by 2 U54 GM104940 from the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health, which funds the Louisiana Clinical and Translational Science Center. This research was also supported by the National Medical Research Council, Ministry of Health, Singapore (WBS R913200076263) to S.G. D.S. is supported with a doctoral scholarship from the German Academic Scholarship Foundation (Studienstiftung des deutschen Volkes).
Footnotes
Reporting Summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Reprints and permissions information is available at www.nature.com/reprints.
Competing interests
S.A.C. is a member of the scientific advisory boards of Kymera, PTM BioLabs and Seer and is a scientific advisor to Pfizer and Biogen. The other authors declare no competing interests.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s42255-021-00400-z.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s42255-021-00400-z.
Data availability
Deidentified, individual-level proteomics and phenotypic data that support the HERITAGE findings within this paper are available at https://motrpac-data.org/related-studies/heritage-proteomics. Overlapping aptamer-based and antibody-based proteomics data on the HERITAGE sample are included Supplementary Data Table 1. GWAS summary statistics for FHS and JHS are available through restricted access via the database of Genotypes and Phenotypes (dbGaP), a publicly available resource developed to archive data from human studies of genotype–phenotype relationships and can be accessed here (https://www.ncbi.nlm.nih.gov/gap/; FHS accession number: phs000363.v19.p13; JHS accession number: phs000964). FHS proteomics data have also been deposited in dbGaP and are available through the same accession number. JHS proteomics data have been deposited in the JHS Data Coordinating Center and are being deposited in dbGaP (accession number: phs002256.v1.p1); pending its receipt in dbGaP, all JHS data are available from the JHS Data Coordinating Center on request (JHSccdc@umc.edu). In addition, proteogenetics findings (precise SNP IDs) included in Supplementary Table 15 from FHS/MDCS meta-analysis and JHS have been provided in Tables 2 and 3 in the Supplementary Data, respectively. Additional data supporting the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Hawley JA, Hargreaves M, Joyner MJ & Zierath JR Integrative biology of exercise. Cell 159, 738–749 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Hawkins MN, Raven PB, Snell PG, Stray-Gundersen J & Levine BD Maximal oxygen uptake as a parametric measure of cardiorespiratory capacity. Med. Sci. Sports Exerc. 39, 103–107 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Ross R et al. Importance of assessing cardiorespiratory fitness in clinical practice: a case for fitness as a clinical vital sign: a scientific statement from the american heart association. Circulation 134, e653–e699 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Myers J et al. Exercise capacity and mortality among men referred for exercise testing. N. Engl. J. Med. 346, 793–801 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Mora S et al. Ability of exercise testing to predict cardiovascular and all-cause death in asymptomatic women: a 20-year follow-up of the lipid research clinics prevalence study. JAMA 290, 1600–1607 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Blair SN et al. Changes in physical fitness and all-cause mortality. A prospective study of healthy and unhealthy men. JAMA 273, 1093–1098 (1995). [PubMed] [Google Scholar]
- 7.Clausen JSR, Marott JL, Holtermann A, Gyntelberg F & Jensen MT Midlife cardiorespiratory fitness and the long-term risk of mortality: 46 years of follow-p. J. Am. Coll. Cardiol. 72, 987–995 (2018). [DOI] [PubMed] [Google Scholar]
- 8.di Prampero PE & Ferretti G Factors limiting maximal oxygen consumption in humans. Respir. Physiol. 80, 113–127 (1990). [DOI] [PubMed] [Google Scholar]
- 9.González-Alonso J & Calbet JA Reductions in systemic and skeletal muscle blood flow and oxygen delivery limit maximal aerobic capacity in humans. Circulation 107, 824–830 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Wagner PD CrossTalk proposal: diffusion limitation of O2 from microvessels into muscle does contribute to the limitation of V̇O2max. J. Physiol. 593, 3757–3758 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joyner MJ & Coyle EF Endurance exercise performance: the physiology of champions. J. Physiol. 586, 35–44 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouchard C et al. Familial resemblance for VO2max in the sedentary state: the HERITAGE family study. Med. Sci. Sports Exerc. 30, 252–258 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Skinner JS et al. Age, sex, race, initial fitness, and response to training: the HERITAGE Family Study. J. Appl. Physiol. 90, 1770–1776 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Williams CJ et al. Genes to predict VO. BMC Genomics 18, 831 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarzynski MA, Ghosh S & Bouchard C Genomic and transcriptomic predictors of response levels to endurance exercise training. J. Physiol. 595, 2931–2939 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis GD et al. Metabolic signatures of exercise in human plasma. Sci. Transl. Med. 2, 33ra37 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Overmyer KA et al. Maximal oxidative capacity during exercise is associated with skeletal muscle fuel selection and dynamic changes in mitochondrial protein acetylation. Cell Metab. 21, 468–478 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wewer Albrechtsen NJ et al. Plasma proteome profiling reveals dynamics of inflammatory and lipid homeostasis markers after Roux-En-Y gastric bypass surgery. Cell Syst. 12, 601–612(2018). [DOI] [PubMed] [Google Scholar]
- 19.Jacob J et al. Application of large scale aptamer-based proteomic profiling to ‘planned’ myocardial infarctions. Circulation 137, 1270–1277 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gold L et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 5, e15004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim CH et al. Stability and reproducibility of proteomic profiles measured with an aptamer-based platform. Sci. Rep. 8, 8382 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams SA et al. Plasma protein patterns as comprehensive indicators of health. Nat. Med. 25, 1851–1857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ross R, Hudson R, Stotz PJ & Lam M Effects of exercise amount and intensity on abdominal obesity and glucose tolerance in obese adults: a randomized trial. Ann. Intern. Med. 162, 325–334 (2015). [DOI] [PubMed] [Google Scholar]
- 24.St Hilaire C et al. NT5E mutations and arterial calcifications. N. Engl. J. Med. 364, 432–442 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasnain SZ et al. Glycemic control in diabetes is restored by therapeutic manipulation of cytokines that regulate beta cell stress. Nat. Med. 20, 1417–1426 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Lee EJ et al. Fibromodulin: a master regulator of myostatin controlling progression of satellite cells through a myogenic program. FASEB J. 30, 2708–2719 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Hynes RO & Naba A Overview of the matrisome — an inventory of extracellular matrix constituents and functions. Cold Spring Harb. Perspect. Biol. 4, a004903 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ngo D et al. Aptamer-based proteomic profiling reveals novel candidate biomarkers and pathways in cardiovascular disease. Circulation 134, 270–285 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ko D et al. Proteomics profiling and risk of new-onset atrial fibrillation: Framingham Heart Study. J. Am. Heart Assoc 8, e010976 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emilsson V et al. Co-regulatory networks of human serum proteins link genetics to disease. Science 361, 769–773 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitham M et al. Extracellular Vesicles Provide a Means for Tissue Crosstalk during Exercise. Cell Metab. 27, 237–251.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Whittle AJ et al. BMP8B increases brown adipose tissue thermogenesis through both central and peripheral actions. Cell 149, 871–885 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pellegrinelli V et al. Adipocyte-secreted BMP8b mediates adrenergic-induced remodeling of the neuro-vascular network in adipose tissue. Nat. Commun. 9, 4974 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seldin MM et al. A strategy for discovery of endocrine interactions with application to whole-body metabolism. Cell Metab. 27, 1138–1155 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karsenty G & Olson EN Bone and muscle endocrine functions: unexpected paradigms of inter-organ communication. Cell 164, 1248–1256 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos-Parker JR, Santos-Parker KS, McQueen MB, Martens CR & Seals DR Habitual aerobic exercise and circulating proteomic patterns in healthy adults: relation to indicators of healthspan. J. Appl. Physiol. 125, 1646–1659 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang CY et al. Cardiorespiratory fitness levels among US adults 20–49 years of age: findings from the 1999–2004 national health and nutrition examination survey. Am. J. Epidemiol. 171, 426–435 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Swift DL et al. Low cardiorespiratory fitness in African Americans: a health disparity risk factor? Sports Med. 43, 1301–1313 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fleg JL et al. Accelerated longitudinal decline of aerobic capacity in healthy older adults. Circulation 112, 674–682 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Abe T, Loenneke JP & Thiebaud RS Fat-free adipose tissue mass: impact on peak oxygen uptake (VO2peak) in adolescents with obesity. Sports Med. 49, 9–15 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Bray MS et al. The human gene map for performance and health-related fitness phenotypes: the 2006–2007 update. Med. Sci. Sports Exerc. 41, 35–73 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Timmons JA et al. Using molecular classification to predict gains in maximal aerobic capacity following endurance exercise training in humans. J. Appl. Physiol. 108, 1487–1496 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bouchard C et al. Genomic predictors of the maximal O2 uptake response to standardized exercise training programs. J. Appl. Physiol. 110, 1160–1170 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghosh S et al. Exploring the underlying biology of intrinsic cardiorespiratory fitness through integrative analysis of genomic variants and muscle gene expression profiling. J. Appl. Physiol. 126, 1292–1314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghosh S et al. Integrative pathway analysis of a genome-wide association study of (V)O2max response to exercise training. J. Appl. Physiol. 115, 1343–1359 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carvalho-Silva D et al. Open Targets Platform: new developments and updates two years on. Nucleic Acids Res. 47, D1056–D1065 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee PS et al. Plasma gelsolin depletion and circulating actin in sepsis: a pilot study. PLoS ONE 3, e3712 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee PS et al. Plasma gelsolin and circulating actin correlate with hemodialysis mortality. J. Am. Soc. Nephrol. 20, 1140–1148 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Egerstedt A et al. Profiling of the plasma proteome across different stages of human heart failure. Nat. Commun. 10, 5830 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Witke W et al. Hemostatic, inflammatory, and fibroblast responses are blunted in mice lacking gelsolin. Cell 81, 41–51 (1995). [DOI] [PubMed] [Google Scholar]
- 51.Lee WM & Galbraith RM The extracellular actin-scavenger system and actin toxicity. N. Engl. J. Med. 326, 1335–1341 (1992). [DOI] [PubMed] [Google Scholar]
- 52.Goetzl EJ et al. Gelsolin binding and cellular presentation of lysophosphatidic acid. J. Biol. Chem. 275, 14573–14578 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Li GH, Arora PD, Chen Y, McCulloch CA & Liu P Multifunctional roles of gelsolin in health and diseases. Med Res. Rev. 32, 999–1025 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Baird GS & Hoofnagle AN A novel discovery platform: aptamers for the quantification of human proteins. Clin. Chem. 63, 1061–1062 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Bouchard C et al. The HERITAGE family study. Aims, design, and measurement protocol. Med. Sci. Sports Exerc. 27, 721–729 (1995). [PubMed] [Google Scholar]
- 56.Skinner JS et al. Heart rate versus %VO2max: age, sex, race, initial fitness, and training response — HERITAGE. Med. Sci. Sports Exerc. 35, 1908–1913 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Skinner JS et al. Reproducibility of maximal exercise test data in the HERITAGE family study. Med. Sci. Sports Exerc. 31, 1623–1628 (1999). [DOI] [PubMed] [Google Scholar]
- 58.Candia J et al. Assessment of variability in the SOMAscan assay. Sci. Rep. 7, 14248 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benson MD et al. The genetic architecture of the cardiovascular risk proteome. Circulation 137, 1158–1172 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suhre K et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat. Commun. 8, 14357 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun BB et al. Genomic atlas of the human plasma proteome. Nature 558, 73–79 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Batista GEAPA & Monard MC An analysis of four missing data treatment methods for supervised learning. Appl. Artif. Intell. 17, 519–533 (2003). [Google Scholar]
- 63.Tsamardinos I, Brown LE & Aliferis CF The max-min hill-climbing Bayesian network structure learning algorithm. Mach. Learn. 65, 31–78 (2006). [Google Scholar]
- 64.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Merico D, Isserlin R, Stueker O, Emili A & Bader GD Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS ONE 5, e13984 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deidentified, individual-level proteomics and phenotypic data that support the HERITAGE findings within this paper are available at https://motrpac-data.org/related-studies/heritage-proteomics. Overlapping aptamer-based and antibody-based proteomics data on the HERITAGE sample are included Supplementary Data Table 1. GWAS summary statistics for FHS and JHS are available through restricted access via the database of Genotypes and Phenotypes (dbGaP), a publicly available resource developed to archive data from human studies of genotype–phenotype relationships and can be accessed here (https://www.ncbi.nlm.nih.gov/gap/; FHS accession number: phs000363.v19.p13; JHS accession number: phs000964). FHS proteomics data have also been deposited in dbGaP and are available through the same accession number. JHS proteomics data have been deposited in the JHS Data Coordinating Center and are being deposited in dbGaP (accession number: phs002256.v1.p1); pending its receipt in dbGaP, all JHS data are available from the JHS Data Coordinating Center on request (JHSccdc@umc.edu). In addition, proteogenetics findings (precise SNP IDs) included in Supplementary Table 15 from FHS/MDCS meta-analysis and JHS have been provided in Tables 2 and 3 in the Supplementary Data, respectively. Additional data supporting the findings of this study are available from the corresponding author upon reasonable request.