Abstract
Pancreatic ductal adenocarcinoma (PDAC) is characterized by extensive desmoplasia, which challenges the molecular analyses of bulk tumor samples. Here we FACS-purified epithelial cells from human PDAC and normal pancreas and derived their genome-wide transcriptome and DNA methylome landscapes. Clustering based on DNA methylation revealed two distinct PDAC groups displaying different methylation patterns at regions encoding repeat elements. Methylationlow tumors are characterized by higher expression of endogenous retroviral (ERV) transcripts and dsRNA sensors which leads to a cell intrinsic activation of an interferon signature (IFNsign). This results in a pro-tumorigenic microenvironment and poor patient outcome. Methylationlow/IFNsignhigh and Methylationhigh/IFNsignlow PDAC cells preserve lineage traits, respective of normal ductal or acinar pancreatic cells. Moreover, ductal-derived KrasG12D/Trp53−/− mouse PDACs show higher expression of IFNsign compared to acinar-derived counterparts. Collectively, our data point to two different origins and etiologies of human PDACs, with the aggressive Methylationlow/IFNsignhigh subtype potentially targetable by agents blocking intrinsic IFN-signaling.
Keywords: Pancreatic cancer, Methylation, Interferon, Stroma
Introduction
With an overall 5-year survival rate of about 9%, pancreatic ductal adenocarcinoma (PDAC) still belongs to the cancer entities with the poorest prognosis (Cancer Research UK and American Cancer Society) (1). A hallmark of PDAC is the immense contribution of desmoplastic stroma to the total tumor mass. Typically, epithelial PDAC cells represent only a minority of cells (as low as 10%) within the tumor mass. The extensive stromal presence represents a major technical challenge when aiming to interrogate the specific molecular signals contributed by the neoplastic epithelial cell compartment from data generated using bulk tumor samples. Some studies have addressed this question in the field of gene expression using sophisticated bioinformatics, labour-intensive laser microdissection (2,3) and, more recently, single cell sequencing approaches (4–7). PDAC methylation studies rely, so far, mostly on bulk tumor analyses (8) or comparisons with cell lines (9) and xenografts (10–12), and have focused on predefined regions of the genome. Here, we report whole genome bisulfite sequencing combined with transcriptome and functional analyses of FACS-purified human PDAC and normal pancreas epithelial cells. We provide insights on how the altered DNA methylome landscape of PDAC cells impact tumor aggressiveness, and suggest the existence of different cells-of-origin for two distinct PDAC subsets.
Results
Transcriptome and methylome of purified human PDAC and normal pancreas epithelial cells
We aimed to determine the genome-wide DNA methylation and transcriptomic landscapes of purified epithelial cancer cells present within human PDAC tumor masses. To avoid confounding effects of stromal cells in PDAC tumors, we established a protocol to FACS-isolate epithelial cells (as EpCAM+/CD45−) from primary untreated PDAC tumors (PDAC-Ep, n = 7). For comparison, we FACS-isolated epithelial cells (as EpCAM+/CD45−, mainly comprised of ductal and acinar cells) from tumor-free adjacent (normal) pancreatic tissues (NP-Ep, n = 6) (Fig. 1A, Supplementary Table S1, and Extended Data Fig. 1). Using low-input technologies, we derived the transcriptomes (RNA-seq (13)) and the DNA methylomes (tagmentation-based whole genome bisulfite sequencing – TBWGBS (14)) of these epithelial cells (Fig. 1A; Supplementary Table S2; Supplementary Table S3).
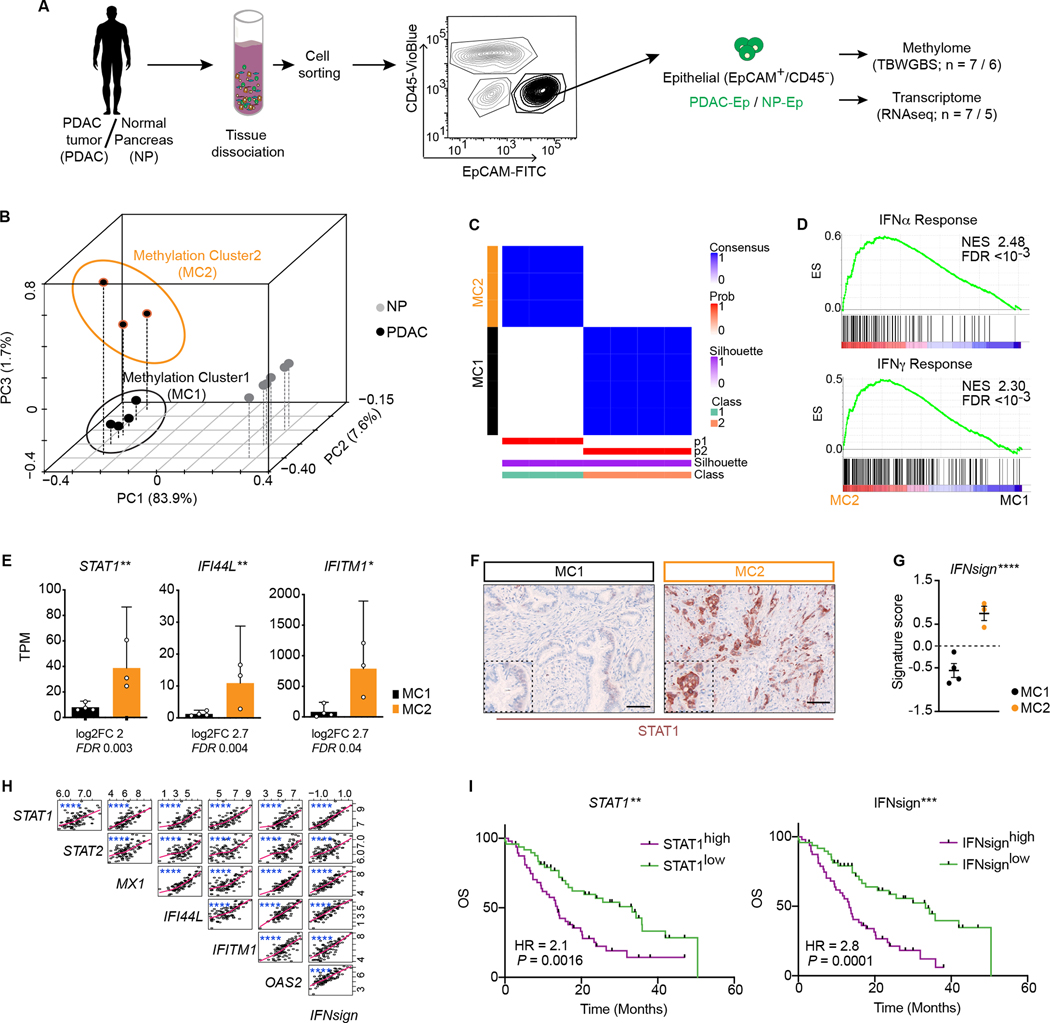
Figure 1. Methylation patterns of epithelial cells identify two groups of tumors associated to IFN signaling and survival.
A, Scheme depicting strategy for tissue processing and isolation of epithelial cells. B, Principal component analysis using PDAC-Ep vs NP-Ep DMRs. Percentage indicates proportion of variance explained by each component. C, Consensus clustering of PDAC-Ep samples. D, Gene set enrichment analyses (GSEA) of IFN response signatures in MC2 vs MC1. ES, enrichment score; NES, normalized enrichment score; FDR, false discovery rate. E, mRNA levels (transcript per million, TPM) of indicated genes. Data are mean + 95% CI; two-tailed Wald-test. F, Representative staining of STAT1 in one MC1 and one MC2 sample. G, Interferon signature score. Lines are mean ± 95% s.e.m.; paired (of mean expression of each gene in the signature) two-tailed t-test. H, Cross-correlations between indicated genes in patient cohort. Each dot is one patient. r, Spearmańs rank correlation coefficient. I, Kaplan-Meier curves display overall survival (OS) of patients with low or high expression of STAT1 or IFNsign according to median. HR, Hazard ratio; logrank-test. H-I: Data source: (25). * FDR/P < 0.05; ** FDR/P < 0.01; *** P < 0.001; **** P < 0.0001.
As expected, isolated epithelial cells showed high expression of EPCAM and very low or undetectable expression of stromal markers (CD45, PDGFRB and VWF) (Supplementary Fig. S1A). Compared to NP-Ep, PDAC-Ep cells showed robust expression of KRT19 and decreased expression of SMAD4, as expected for PDAC cancer cells (15,16) (Supplementary Fig. S1B). 48.7% of the RNA sequenced reads from PDAC-Ep cells harboured KRAS mutations whereas no KRAS mutations were found in NP-Ep RNA reads (Supplementary Fig. S1C). Altogether, these data demonstrate the purity of the isolated epithelial cell populations. Additionally, while we cannot exclude that in some PDACs EpCAM− epithelial tumor cells co-exist, only 0% to 8% mutant KRAS reads were detected in RNAseq data from paired EpCAM−/CD45− samples (which comprise endothelial cells, vascular endothelial, nerve tissue as well as potentially epithelial cells that have lost EpCAM expression) indicating that our sorting strategy captured the majority of the PDAC epithelial cells (Supplementary Fig. S1D).
The methylome analysis identified 56,177 regions showing differential DNA methylation (differentially methylated regions - DMRs) between PDAC-Ep and NP-Ep (cut-off 40% differences in methylation; Supplementary Fig. S1D; Supplementary Table S4). These DMRs covered 1.78 × 107 bp of the genome. Of these, a larger proportion was hypomethylated in PDAC-Ep compared to NP-Ep cells than hypermethylated (66% vs 34%, respectively) (Supplementary Fig. S1E). This reflects the different global distribution of DNA methylation levels of single CpGs (CGs) along the genomes of these samples (Supplementary Fig. S1F). The majority of DMRs (79%) were localised outside CpG islands (CGIs) (10%) or CGI-shores (11%) (Supplementary Fig. S1G). The vast majority of CGIs showed a gain of methylation in PDAC-Ep, while the regions outside of CGIs showed a marked loss of methylation. As these non-CGI sites make up the majority in the genome, a global hypomethylation pattern is observed in PDAC-Ep (Supplementary Fig. S1H).
Next, we correlated the methylomes and transcriptomes by closest gene analysis (13,17–19). Only 13% of the DMRs (7,397) correlated with up- or down- regulation of gene expression (FDR < 0.05; Supplementary Fig. S2A–C). Enrichment analysis of the DMRs correlating to gene expression changes revealed deregulated pathways including axon guidance, WNT, TGF-beta, NOTCH signaling and apoptosis, confirming previous reports (8–10) (Supplementary Table S5). DMRs correlating to differential gene expression were also mainly hypomethylated in PDAC-Ep compared to NP-Ep (69%; Supplementary Fig. S2B). The 7,397 DMRs correlating with changes in gene expression could be classified into four groups according to the direction of the changes (Supplementary Fig. S2B). Inverse correlation between DNA methylation and gene expression (hyper/loss and hypo/gain) comprised the majority of DMRs (63.5%). These DMRs were located closer to transcription start sites (TSS) and had a larger overlap with promoter regions (Supplementary Fig. S2B,C upper plots). Hypermethylated regions showed a bigger overlap to CGIs and enhancers compared to hypomethylated regions (Supplementary Fig. S2B,C lower plots). The hypermethylation/loss of expression group included genes of the pancreatic progenitor lineage (NR5A2, PDX1, PROX1; Supplementary Fig. S2D,E and data not shown). Pancreatic progenitor genes were also included in the hypomethylation/loss of expression group (e.g. ONECUT1), which also comprised genes involved in tight junctions (e.g. CDH5). Gain of expression correlating to loss of methylation (hypo/gain) occurred in genes previously reported to be upregulated in PDAC, such as SERPINB5 (20), genes involved in cytoskeletal organization (ITGB1, ACTB) as well as genes related to inflammatory response (CCL20, CCL22, IL1A) (Supplementary Fig. S2F–H and data not shown). Lastly, hypermethylation/gain of expression occurred in gene body regions distant to TSS enriched in CGI (Supplementary Fig. S2B–C) and it was found in genes related to embryonic development (HOXA2, HOXA3, HOXB9) and matrix metalloproteases (MMPs) including MMP9 (Supplementary Fig. S2I,J). In summary, we provide a comprehensive data set comprising whole genome methylome and transcriptome data of freshly isolated pure human PDAC and normal pancreatic epithelial cells. Our data confirm, but also massively expand the number of known DMRs and associated transcriptional changes in purified PDAC epithelial cells to the genome-wide level (Supplementary Table S4, Supplementary Fig. S2K) (8–10,21).
Methylation profiling separates two PDAC clusters associated to IFN signaling and survival
The large amount of DMRs identified between PDAC-Ep and NP-Ep cells illustrate the profound changes that occur at the level of DNA methylation in epithelial cells during PDAC carcinogenesis. Interestingly, by performing principal component analysis using all PDAC-Ep vs NP-Ep DMRs, the tumor samples separated into two clearly distinguishable groups: Methylation Cluster 1 (MC1) and Methylation Cluster 2 (MC2) (Fig. 1B). A separation of the PDAC-Ep samples was also revealed by unsupervised principal component analysis (Supplementary Fig. S3A). The existence of two PDAC methylation groups was further validated by consensus clustering (Fig. 1C and Extended Data Fig.2). Comparison of the transcriptomes of MC1 and MC2 PDAC cells identified 320 differentially expressed genes (FDR<0.01; Supplementary Fig. S3B; Supplementary Table S6). As expected, no changes in expression of EPCAM or KRT19 were observed, indicating that the presence of the two groups was not due to contamination with other cell types (Supplementary Fig. S3C). Gene set enrichment analysis (GSEA (22,23)) using the hallmark gene sets revealed a marked enrichment for IFN-alpha and IFN-gamma response signatures in MC2 samples (Fig. 1D; Supplementary Fig. S3D). Among other genes, the transcription factor signal transducer and activator of transcription 1 (STAT1), a master regulator of IFN signaling, as well as IFI44L and IFITM1 were highly upregulated in MC2 compared to MC1 tumor cells (Fig. 1E,F and Supplementary Fig. S3E). Interestingly, several immune checkpoint modulators were also higher expressed in MC2 as compared to MC1 (Supplementary Fig. S3F). To further analyse the status of IFN signaling in public PDAC datasets, we built a 47-gene signature (IFNsign) by combining genes enriched in MC2 from both IFN-alpha and IFN-gamma response gene sets (see methods for details) (Supplementary Fig. S3G). As expected, the IFNsign was robustly overexpressed in MC2 compared to MC1 (Fig. 1G and Supplementary Fig. S3H). Analysis of expression datasets from human PDAC tumors revealed a strong correlation between the expression of different individual IFN-related genes and our IFNsign (Fig. 1H and Supplementary Fig. S4A). These data demonstrate that an entire IFN program is highly expressed in a subset of PDAC patients. Importantly, patients with an IFNsignhigh or STAT1high status showed a significantly overall worse survival compared to patients with low expression of this IFN program or STAT1 (Fig. 1I and Supplementary Fig. S4B). Of note, there were no significant differences on tumor epithelial content between IFNsign high and low patients (Supplementary Fig. S4C). Increased immune infiltration was associated with IFNsignhigh tumors, but this was not linked to changes in survival (Supplementary Fig. S4D–E). Altogether, these data suggest that the differences observed in survival are linked to the differential activation of an IFN program in PDAC epithelial cells.
Previous studies, mostly performed by bulk analysis, have investigated the possibility to stratify PDACs by differential gene expression signatures (2,12,24–26). While some tumors might have a blended correlation to subtypes (27,28), associations between the expression of the progenitor-like/classical subtype signature with more favorable, and the squamous-/basal-/quasi-mesenchymal-like subtype signature with poor patient outcome, have been intensely investigated (2,12,24–27,29,30). MC1 samples showed enrichment for progenitor-like/classical signatures, while MC2 samples were enriched for squamous-/basal-like gene signatures (Supplementary Fig. S4F). Accordingly, STAT1 and IFNsign expression were higher in tumors classified as squamous-/basal-like compared to those classified as progenitor-like/classical (Supplementary Fig. S4G,H). Together, these results demonstrate an association of an activated IFN program specifically within the PDAC epithelial cell compartment and the previously described squamous-/basal-like subtype. Nevertheless, STAT1 or IFNsign expression levels were able to further stratify patients with tumors classified as progenitor-like or classical subtype in a more favorable or poor outcome group (Supplementary Fig. S4I,J) suggesting both overlapping and complementary stratification power of the IFN signature.
In summary, genome-wide methylome and transcriptome analysis of purified tumor cells revealed two groups of PDACs with distinct methylation landscapes connected to the activation of a differential and cell autonomous IFN program and poor overall survival.
IFNsignhigh status correlates with low DNA methylation at non-CGI sites and upregulation of repeat-derived transcripts
Next, we explored if the observed differences in the expression of IFN-related genes in PDAC epithelial cells could be explained by differential methylation of the loci encoding IFN target genes themselves. Towards this aim, we first determined DMRs between MC2/IFNsignhigh and MC1/IFNsignlow PDAC-Ep (inter-tumor DMRs, Supplementary Table S7). We observed no association between changes in methylation and changes in expression of IFN-related genes. Moreover, from the 3,070 DMRs identified between MC1 and MC2, only 114 (3.7%) were located close to differentially expressed genes and the correlation did not reach significance. These data suggest that the major differences in DNA methylation between tumor groups (>3,000 DMRs between MC1 and MC2) do not directly influence gene expression. Accordingly, DMRs between MC1 and MC2 were located at larger distances to TSS compared to DMRs between normal and PDAC epithelial cells (Supplementary Fig. S5A).
We next performed a genome-wide distribution analysis of DNA methylation, which revealed an overall lower methylation level of MC2 PDAC cells compared to MC1 ones (Fig. 2A and Supplementary Fig. S5B,C). Global hypomethylation is a known epigenetic alteration occurring in some cancers (31,32) and it is typically associated to large genomic regions that coincide with lamina-associated, late-replicating regions termed partially methylated domains (PMDs) (33). Indeed, PMD estimation in our cohort revealed that around 47% of the genome of MC2 PDAC cells was occupied by PMDs, in contrast to only 15% in MC1 samples (Supplementary Fig. S5C,D). While MC1-PMDs seemed to have a rather random genomic distribution, as shown by the poor overlap of the PMDs of each individual MC1 sample, MC2-PMDs were highly conserved as shown by a strong overlap between the individual MC2-PMDs (Supplementary Fig. S5E,F). These data highlight that hypomethylation in PDAC epithelial cells does not equally occur in all patients.
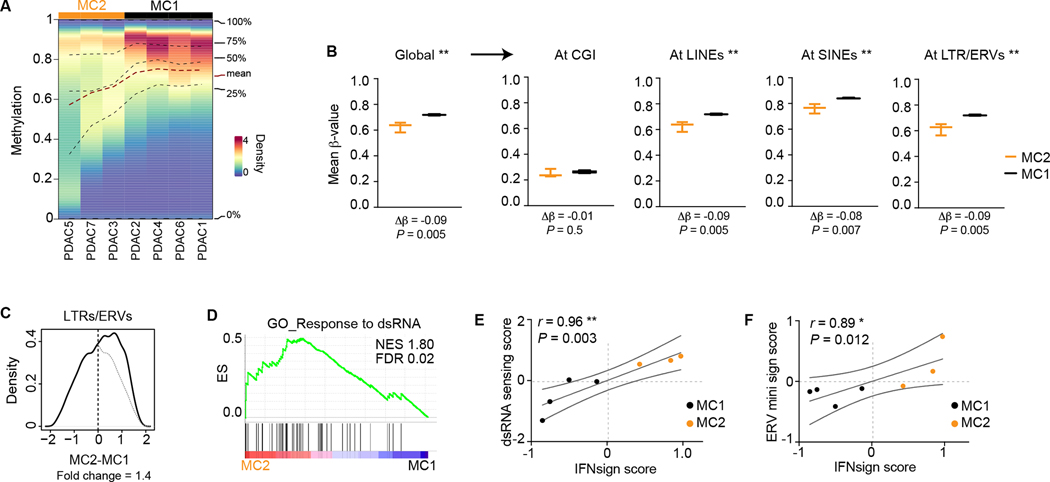
Figure 2. Hypomethylation at non-CGI sites and dsRNA sensing accompanies IFNsignhigh tumors.
A, Methylation density plot. Lines indicate quartiles and mean values. B, Global methylation and mean methylation of CGs at different genomic features calculated per sample and plotted per group. n = 3 (MC2), 4 (MC1). Boxes extend from 25th to 75th percentiles with line representing median. Whiskers indicate minimum and maximum values. Two-tailed t-test. C, Distribution of expression difference of indicated repetitive elements between MC2 and MC1 (see methods for details). Fold change of area under the curve is shown. Gray line mirror projection of the line at negative values. D, Gene set enrichment analyses in MC2 vs MC1 samples. ES, enrichment score; NES, normalized enrichment score; FDR, false discovery rate. E-F, Correlations between IFN signature score and dsRNA sensing score (E) or ERV expression score (F). r, Spearmańs rank correlation coefficient; ** P < 0.01, * P < 0.05.
Since we noticed that hypomethylation in MC2 did also occur outside PMDs (Supplementary Fig. S5G), we next explored methylation levels by genomic location. Methylation at CGI sites showed no major difference between MC2 and MC1 (Fig. 2B). In fact, although clustering based on CGI methylation did separate normal from PDAC samples, this was not sufficient to discriminate tumor groups (Supplementary Fig. S5H and Extended Data Fig. 2). In striking contrast, clustering using regions outside CGIs effectively discriminated MC1 and MC2 tumor groups (Supplementary Fig. S5I–K and Extended Data Fig. 2). More specifically, MC2 and MC1 remarkably differed in their methylation levels at sites harboring repetitive elements including LINEs (long interspersed nuclear elements), SINEs (short interspersed nuclear elements) and LTR/ERVs (long terminal repeats/endogenous retroviruses) all of which were consistently lower methylated in MC2/IFNsignhigh PDAC cells (Fig. 2B). These results highlight the value of using whole genome analyses to include areas beyond regions with high CGI content.
Global demethylation imposed by pharmacological inhibition of DNA methyltransferases or histone deacetylases has been reported to induce transcriptional derepression of repeats. Especially LTR/ERV transcripts can form dsRNA structures that activate intracellular pattern recognition receptors (PRR) leading to increased IFN signaling (34–37). Analysis of the transcriptome of MC1 and MC2 samples revealed that transcripts derived from LTR/ERV and LINE but not SINE sequences, showed a higher enrichment in MC2/IFNsignhigh compared to MC1/IFNsignlow PDAC cells (Fig. 2C and Supplementary Fig. S6A). Moreover, MC2 samples were enriched in gene signatures related to dsRNA response (Fig. 2D and Supplementary Fig. S6B). The level of the IFNsign correlated with the expression of dsRNA recognition receptors and with the expression of a subset of ERVs (Fig. 2E,F and Supplementary Fig. S6C,D). In summary, based on methylation of purified epithelial cells, we have identified a subset of PDAC tumors showing lower methylation at repeat sites, increased expression of LTR/ERV and LINE-derived transcripts, an engaged dsRNA response and high IFN signaling.
Validation of MC1 and MC2 subgroups in an independent cohort
To validate our data in an independent cohort, we analysed samples from additional 11 PDAC patients (Supplementary Table S1). From these, we determined the transcriptomes of their purified EpCAM+ epithelial cells as for the initial seven samples described above. In addition, PDAC samples were orthotopically transplanted into NSG mice and the PDAC methylomes of the 10 PDXs were determined using Infinium MethylationEPIC arrays (validation set; Supplementary Fig. S7A) which, compared to previous array platforms, additionally covers mainly non-CGI sites (38). 10 PDX tumors derived from the initial MC1/IFNsignlow and MC2/ IFNsignhigh samples were also profiled as controls (discovery set; Supplementary Fig. S7A). As previously observed using WGBS data, consensus clustering of the discovery PDX samples confirmed the separation between MC1 and MC2 tumors based on methylation at non-CGI but not at CGIs (Supplementary Fig. S7B,C). Strikingly, three of the new tumors clustered with the three MC2/IFNsignhigh PDX samples while eight clustered with the four MC1/IFNsignlow samples (Supplementary Fig. S7B,C). As observed with TBWGBS, also the PDX MC2 PDACs showed: (a) lower methylation at LINE, SINE and LTR/ERV sites (Supplementary Fig. S7D), (b) enrichment in expression of IFN signatures and increased STAT1 and ERV expression compared to MC1 tumors (Supplementary Fig. S7E–I). Association of the MC1 and MC2 methylation clusters to the previously described classical/progenitor-like and basal-like/squamous subtypes existed although it was less notable than the association to the STAT1 status (Supplementary Fig. S7B).
Collectively, these data validate the MC1 and MC2 PDAC groups and provide further evidence for the link between hypomethylation-mediated derepression of repeat elements and the activation of the IFN pathway in a total of 18 human PDACs (12 MC1/IFNsignlow and 6 MC2/IFNsignhigh).
Cell autonomous activation of IFN-signaling in primary PDAC cells
To expand our findings, we investigated a panel of PDAC patient derived primary cell lines (PDCs) (39) by expression and MethylationEPIC array analyses. We could identify IFNsignhigh and IFNsignlow PDCs according to RNA and protein expression (Fig. 3A and Supplementary Fig. S8A). IFNsignhigh cells were enriched in IFN-alpha and gamma response gene sets (Fig. 3B) as well as in previously reported squamous-like/basal/quasimesenchymal-like subtype signatures (Supplementary Fig. S8B). In agreement with the poor overall survival associated to IFNsignhigh patients (Fig. 1I), mice injected orthotopically with IFNsignhigh PDCs succumbed earlier (i.e. reached endpoint criteria) (Fig. 3C), formed higher grade tumors (Supplementary Fig. S8C) and displayed a higher incidence of lung metastasis formation compared to mice injected with IFNsignlow PDCs (Fig. 3D, Supplementary Fig. S8D,E). These results provide functional in vivo evidence that IFNsignhigh PDAC cells are more aggressive compared to IFNsignlow tumor cells.
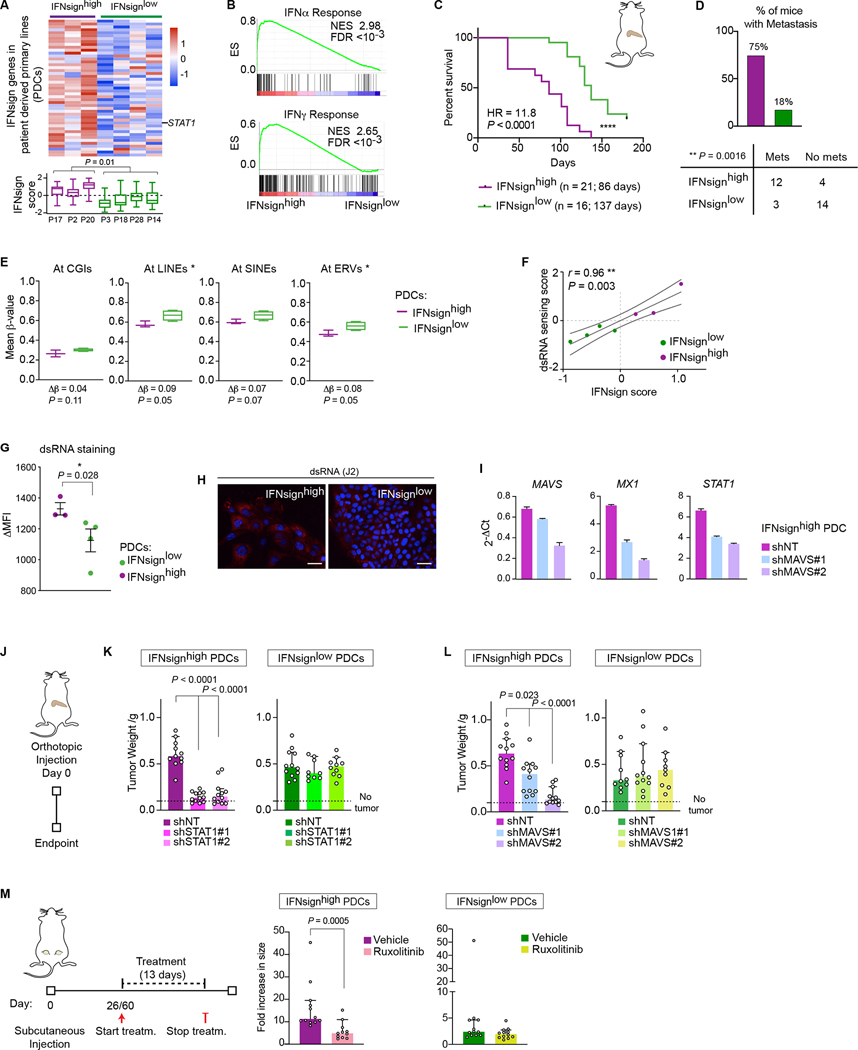
Figure 3. Inhibition of dsRNA sensing or JAK/STAT pathway results in reduced tumor growth in an IFN-status dependent manner.
A, Expression of IFNsign genes in patient-derived PDAC cells (PDCs) (above). IFNsign score in the same cells (below). Two-tailed nested t-test. B, Gene set enrichment analyses (GSEA) of IFN response signatures in PDCs according to their IFNsign status. ES, enrichment score; NES, normalized enrichment score; FDR, false discovery rate. C, Kaplan-Meier curve of mice injected with IFNsignhigh (n = 3 cell lines with 5 or 6 mice per line. Total n = 16) or IFNsignlow (n = 4 cell lines with 5 or 6 mice per line. Total n = 21) PDCs. Mice were sacrificed when humane end point was reached. Median survival per group is indicated. HR, Hazard ratio; logrank-test. D, Mice from each group in (A) with macroscopic lung metastases. E, PDCs mean methylation of CpGs at indicated regions. Values are calculated per sample and plotted per group. Two-tailed Fisher-test. F, Correlation between IFNsign and dsRNA sensing scores. r, Spearmańs rank correlation coefficient. G, Delta of median fluorescence intensity of dsRNA staining (J2 antibody) vs isotype in PDCs. Each dot represents the mean of two independent experiments per PDC. Data are mean ± s.e.m. One-tailed Mann-Whitney test. H, Representative immunofluorescence picture of dsRNA staining in one IFNsignhigh (PACO20) and one IFNsignlow (PACO3) PDC. Scale bar = 50μm. I, mRNA expression of indicated genes in MAVS knock-down (KD) PDCs (PACO2) measured by RT-qPCR and relative to HPRT1. Data are mean + s.e.m. J, KD PDCs were injected orthotopicaly and tumors were analysed at endpoint. K-L, Size of orthotopic tumors grown from PDCs upon knock-down of STAT1 (K) or MAVS (L) or non targeting (NT) control. STAT1KD: IFNsignhigh (PACO2): n = 11 (NT), 12 (#1), 12 (#2); IFNsignlow (PACO3): n = 11 (NT), 11 (#1), 10 (#2). MAVSKD: IFNsignhigh (PACO2): n = 12 (NT), 12 (#1), 12 (#2); IFNsignlow (PACO3): n = 11 (NT), 11 (#1), 10 (#2). M, Fold increase growth of established subcutaneous tumors treated with vehicle or Ruxolitinib. Data show tumor growth normalized to start of treatment. IFNsignhigh (PACO2): n = 12 (Vehicle), 11 (Ruxolitinib). IFNsignlow (PACO3): n = 12 (Vehicle), 12 (Ruxolitinib). (K-M) Data are median + 95% CI. Two-tailed Mann-Whitney test. (A,E) Boxes extend from 25th to 75th percentiles with line representing median. Whiskers indicate minimum and maximum values. * P < 0.05; ** P < 0.01; **** P < 0.0001.
DNA methylation analyses showed that IFNsignhigh PDCs had lower genome-wide methylation compared to IFNsignlow PDCs with strongest differences at sites containing LTR/ERV and LINE repeats (Fig. 3E and Supplementary Fig. S8F). Treatment of IFNsignlow PDCs with the DNA demethylating agent decitabine (5-Aza-2’-Deoxycytidine; 5-Aza-CdR) was sufficient to induce the expression of IFN-related genes (Supplementary Fig. S8G,H). Importantly, IFNsign expression in PDCs also correlated with the expression of dsRNA sensors and signatures of response to dsRNA (Fig. 3F and Supplementary Fig. S8I) and indeed higher levels of cytoplasmic dsRNA were detected in IFNsignhigh PDCs (Fig. 3G,H). Finally, knock-down of MAVS, a master regulator of dsRNA response, using two differentially efficient shRNAs, caused a proportional downregulation of IFN response genes (Fig. 3I). Together with the data obtained from the 10 PDXs and primary tumors, these data show that IFNsignhigh status in PDAC epithelial cells correlates with lower DNA methylation and higher expression of repetitive elements/dsRNA and indicate that the IFNhigh status in MC2 type PDACs is sustained in a tumor cell autonomous manner. Interestingly, type I and II IFNs were below detection limit in concentrated conditioned medium (CM) of PDCs in both IFNsignhigh and IFNsignlow PDCs. Blocking of type III IFNs receptor (IFNLR1) did not reduce the expression of IFN-related genes in PDCs (Supplementary Fig. S9A,B) and neither affected the growth of PDCs in co-cultures with stellate cells (StC) (see below, Supplementary Fig. S9C). These data exclude that secreted IFNs themselves are responsible for the robust activation of the IFN pathway in PDCs and are consistent with a cell intrinsic and ERVs/dsRNA mediated mechanism of IFN activation.
Inhibition of IFN signaling impairs PDAC tumor growth
We next interrogated if the disruption of IFN signaling would impact tumor growth in vivo. Knock-down of STAT1 reduced the expression of IFN-related genes only in IFNsignhigh, but not IFNsignlow PDCs (Supplementary Fig. S10A). Strikingly, STAT1 downregulation impaired orthotopic tumor growth of IFNsignhigh, but not of IFNsignlow cells in mice (Figure 3J,K). Similarly, downregulation of MAVS negatively affected the in vivo growth of IFNsignhigh (Fig. 3I,L) but not IFNsignlow tumors (Fig. 3L and Supplementary Fig. S10B). These results suggest that PDAC patients bearing IFNsignhigh tumors could benefit from inhibition of the STAT1 pathway. To test this hypothesis, we established subcutaneous tumor models using both PDC types. When tumors were palpable, mice were treated with the FDA approved JAK/STAT inhibitor ruxolitinib. Ruxolitinib treatment reduced the expression of IFN-related genes in vivo (Supplementary Fig. S10C) and significantly impaired tumor growth of IFNsignhigh cells while it did not interfere with the growth of IFNsignlow PDCs (Fig. 3M). These data establish a preclinical proof-of-concept that tailored inhibition of IFN signaling might be beneficial for patients with MC2-type PDAC.
IFNsignhigh PDAC epithelial cells reprogram the stroma towards tumor-promotion
We next sought for a possible explanation contributing to the observed higher aggressiveness of MC2 tumors and the poorer survival of these patients. Cancer-associated fibroblasts (CAFs) are typically the most abundant stromal cell population in PDAC (40). The existence of different types of CAFs with diverse or even opposing roles has been recognized in different types of cancer (41) including, more recently, PDAC (42–45). The existence of two main subset of PDAC-CAFs, the inflammatory (iCAFs) (46) and the myofibroblastic (myCAFs) has been shown in mouse models and human samples (5,47,48). We aimed to investigate if IFNsignhigh/low PDAC epithelial cells could differentially educate or re-program the fibroblastic stroma, which in turn may impact tumor aggressiveness. Towards this end, we treated normal human pancreatic stellate cells (StCs) with tumor cell conditioned medium (CM) derived from IFNsignhigh or IFNsignlow PDCs, or CM from StCs as control, and analysed changes in gene expression (Fig. 4A). Indeed, transcriptomes of StCs exposed to IFNsignhigh CM clustered apart from the ones treated with IFNsignlow CM or control medium (Fig. 4A). The top enriched signatures in IFNsignhigh CM treated StCs were IFN-alpha and IFN-gamma response programs that included classical IFN-related genes (such as MX1 or IFI44L) (Fig. 4B and Supplementary Fig. S11A). Along with classical IFN-related genes, CM of IFNsignhigh tumor cells also induced the expression of inflammatory programs (Supplementary Fig. S11B) and pro-inflammatory cytokines (e.g. CCL2, IL8, IL1A, IL1B), some of them previously shown to be expressed by iCAFs (Supplementary Fig. S11A,C). In line with these data, MC2 PDAC patients showed a trend towards higher systemic inflammation as measured by C-reactive protein levels in serum (Supplementary Fig. S11D).
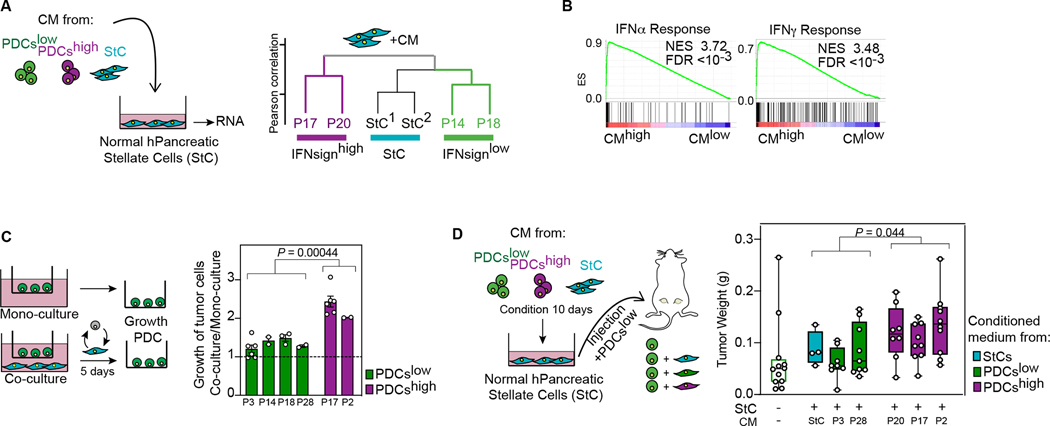
Figure 4. IFNsignhigh tumor cells activate stromal cells in a tumor promoting manner.
A, Dendogram of expression profile of stellate cells (StC) treated with different condition media. B, GSEA of indicated signatures of StCs treated with conditioned media from IFNsignhigh vs IFNsignlow cells. ES, enrichment score; NES, normalized enrichment score; FDR, false discovery rate. C, Growth of PDCs co-cultured with StCs (10,000 cells) relative to mono-culture growth. Data are mean ± s.e.m.; each dot is an independent experiment. Two-tailed nested t-test. D, Weight of tumors arising from injection of tumor cells with or without preconditioned StCs. Every dot is one tumor. Boxes extend from 25th to 75th percentiles with line representing median. Whiskers go from smallest to largest value. Two-tailed nested t-test.
To functionally test the impact of this re-programming of StCs mediated by IFNsignhigh PDAC cell, we first assessed the in vitro growth of tumor cells alone (mono-culture) versus in indirect co-culture with StCs. IFNsignhigh PDCs increased the expression of both IFN and pro-inflammatory cytokines in co-cultured StCs as previously shown in the CM experiments (Supplementary Fig. S11E). IFNsignhigh cells grown in presence of StC expanded more than IFNsignlow cells (Fig. 4C and Supplementary Fig. S11F) suggesting a stronger positive feed-forward loop provided by IFNsignhigh-activated StCs. Treatment with ruxolitinib partially reduced this growth promoting effect in the co-culture setting while it had no effect on PDCs or StC alone (Supplementary Fig. S11G,H). Finally, to test the functional consequences of this reprogramming in vivo, we co-injected IFNsignlow PDC cells together with StCs that had been pre-treated either with IFNsignhigh, IFNsignlow or StC control CM, into recipient mice and monitored PDAC tumor growth (Fig. 4D and Supplementary Fig. S11I,J). In line with our in vitro results, StCs reprogrammed by IFNsignhigh CM supported PDC-mediated tumor growth to a greater extent compared to StCs reprogrammed by IFNsignlow CM or control CM (Fig. 4D and Supplementary Fig. S11J).
In summary, our data demonstrate that IFNsignhigh cells elicit a distinct activation program in normal StCs towards a pro-inflammatory and tumor-promoting phenotype, which includes the upregulation of several molecules previously assigned to pro-tumorigenic microenvironments (Supplementary Fig. S11K) (48,49). These data provide an explanation for the higher aggressiveness and overall poorer survival of patients with MC2 PDAC tumors.
Methylation traits and transcriptomic data suggest two different types of cells-of-PDAC origin
Despite a markedly different DNA methylome between tumor and normal cells, specific methylation signatures retained from normal cells may exist in tumors, a characteristic previously used to trace back the potential cell-of-cancer origin (50–55). The healthy adult human exocrine pancreas is comprised of two main epithelial cell types, with acinar cells comprising the majority of the pancreatic tissue (70–60%), and ductal cells representing 20–30% (56). Both cell types arise from a common progenitor during pancreas development (57,58). During transformation, acinar cells can dedifferentiate and/or transdifferentiate to progressively acquire ductal-cell characteristics (so called acinar to ductal metaplasia - ADM) (57,59,60). Although genetic mouse models have shown that both acinar (through ADM) and ductal cells can give rise to PDAC lesions (61–66), the cell-of-origin for human PDAC remains highly controversial (63,67,68).
We investigated if the DNA methylome of MC1/IFNsignlow and MC2/IFNsignhigh tumors contained remnant footprints of healthy ductal or acinar cells. For this, and since our initial FACS sorted EpCAM+ NP-Ep cells (Fig. 1) are comprised of an unknown representation of ductal and acinar cells, we compiled a total of new 20 samples derived from 10 healthy human pancreas donors. This collection comprised (1) primary acinar and ductal cells isolated by FACS (69) (n = 4 samples), (2) FACS-purified ductal (n = 3 samples) and dedifferentiated acinar cells (n = 3 samples) after in vitro culture of an exocrine mixed population, (3) the original mixed exocrine population of these samples (59,70) (n = 3 samples), and (4) normal bulk pancreas tissue (which is comprised of a mix of ductal and acinar cells with possibly few endocrine cells; n = 3 samples) (Supplementary Fig. 12A). We extracted the DNA from these samples, performed Infinium MethylationEPIC arrays, and supplemented these data with a previously published methylation dataset of human acinar and ductal cells (n = 4) (71). Integration of the data revealed that acinar and ductal cells have very distinct methylation patterns and that these epigenetic traits are maintained during the early stages of acinar dedifferentiation (Supplementary Fig. S12B–C). Normal bulk pancreas showed an intermediate profile with slightly higher similarity to acinar/dedifferentiated acinar cells consistent with the observation that normal pancreas consists of a majority of acinar cells (Supplementary Fig. S12B–C) (56).
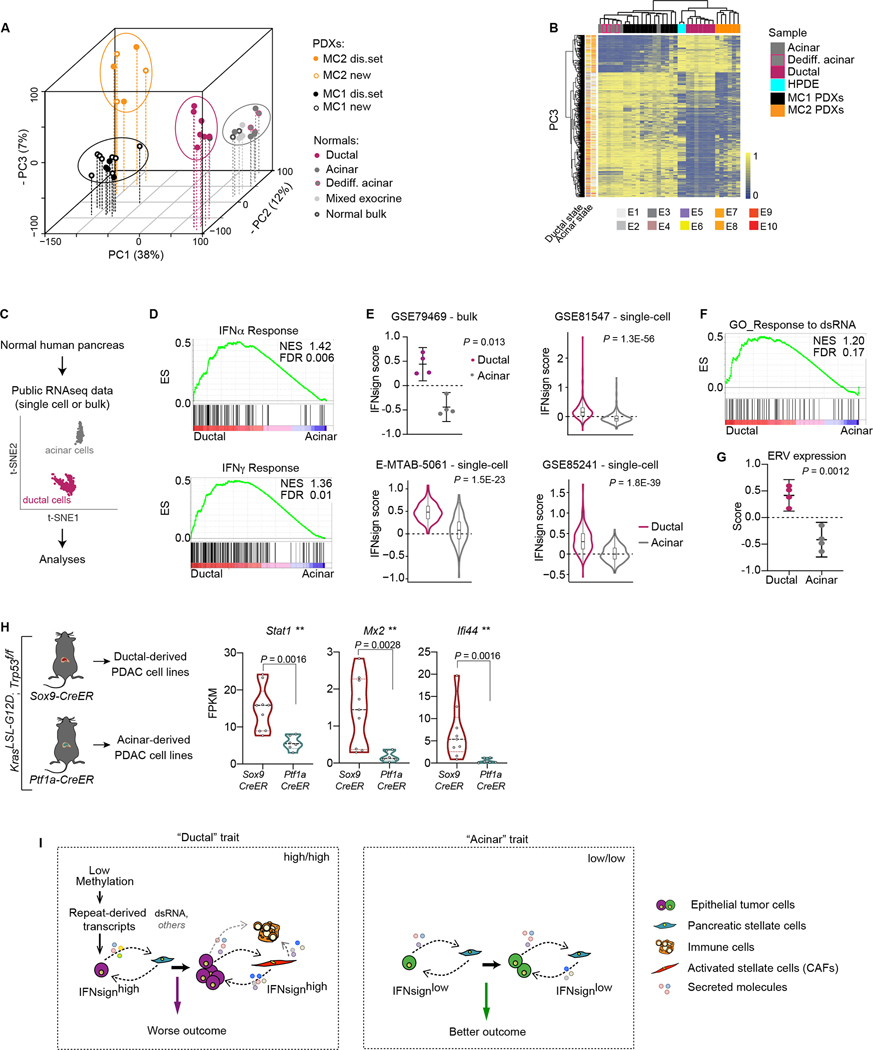
Next, we joined this new methylome dataset of normal pancreas cell types with our collection of 18 PDAC tumors (Supplementary Fig. S7A). Unsupervised clustering of the complete EPIC array dataset separated MC2/IFNsignhigh from MC1/IFNsignlow tumors in PC3, as previously observed with the WGBS data (Fig. 5A, Fig. 1B). At the global level, the methylomes of PDAC-Ep cells largely diverged from that of normal epithelial cells regardless of the tumor group (MC1 or MC2) or of the normal epithelial type (acinar or ductal). This was seen in PC1, which explained the largest proportion of the variance between samples (38%) (Fig. 5A). This large tumor to normal epithelial cell methylome divergence was further corroborated when using the top differentially methylated sites (DMSs) between acinar and ductal or MC1 and MC2 samples (Supplementary Fig. S13A and B, respectively). While PC1 could clearly be associated to the differences between normal and PDAC-Ep cells, both PC2 and PC3 separated MC1 from MC2 as well as the normal acinar from ductal cells suggesting that, although globally different, both type of PDAC-Ep cells share some similarities with both acinar and ductal cells at the level of DNA methylation (Fig. 5A, Supplementary Fig. S13C,D). Indeed, when checking the methylation status of mature acinar and ductal markers in the tumor samples, all tumors showed a profile similar to that of acinar cells as they showed increased methylation of ductal genes, and a profile similar to ductal cells as they showed increased methylation of acinar genes (Supplementary Fig. S13E). These data indicate that, during transformation, tumor cells massively rearrange their DNA methylome resulting in a pattern that strongly diverges from the methylation profiles of mature normal acinar or ductal cells. Conserved lineage related traits associated to cell-of-origin might therefore be obscured in the tumors by the profound changes happening at the global level.
Figure 5. Methylation, expression and mouse models data suggest a ductal relation of MC2 tumors.
A, Principal component analysis of normal and PDX samples. Percentages indicate proportion of variance explained by each component. MC1-PDXs: n = 12 (2 technical replicates); MC2: n = 6; Normal bulk: n = 3; Acinar: n = 4; Dedifferentiated acinar: n = 3; Ductal: n = 7. B, Clustering of samples using the top 1,000 CGs driving PC3 and including the ductal line HPDE (blue). C, Illustration of data analyzed in panels D-G. Adapted from (76). D, GSEA analyses in ductal vs acinar samples (GSE79469). ES, enrichment score; NES, normalized enrichment score; FDR, false discovery rate. E, Interferon signature score. GSE79469: n = 4 ductal, 4 acinar; Lines are mean ± 95% CI; paired two-tailed t-test. GSE81547: n = 389 ductal, 411 acinar; Mann-Whitney test. E-MTAB-5061: n = 135 ductal, 112 acinar; Mann-Whitney test. GSE85241: n = 258 ductal, 282 acinar; Mann-Whitney test. F, GSEA analysis in ductal vs acinar samples (GSE79469). ES, enrichment score; NES, normalized enrichment score; FDR, false discovery rate. G, ERV expression signature score. GSE79469: n = 4 ductal, 4 acinar; Lines are mean ± 95% CI. Paired two-tailed t-test. H, Normalized expression of indicated genes (FPKM - fragments per kilobase of transcript per million mapped reads) in mouse cell lines from ductal [n = 9 cell lines from 9 tumors (8 mice)] or acinar [n = 6 cell lines from 6 tumors (4 mice)] derived PDAC mouse tumors. Black dotted line indicates median. Colored dotted lines indicate upper and lower quartiles. Mann-Whitney test. I, Model.
To further decipher this, we reasoned that methylation marks linked to a ductal cell-of-origin and maintained during transformation should also be present in HPDE cells, an immortal cell line derived from normal pancreatic ductal cells (72,73). The methylome of HPDE cells was indeed similar to that of normal ductal cells at CGs associated to classical acinar and ductal markers (Supplementary Fig. S13F). Strikingly, HPDE cells cluster with normal ductal cells when using the top 1,000 CGs driving PC3 but not with those of PC2, suggesting that the information on ductal identity is contained in PC3 (Fig. 5B and Supplementary Fig. S13G). Methylation at these CGs also associated MC2/IFNsignhigh tumors to ductal and HPDE cells while MC1/IFNsignlow tumors intermingled with normal acinar and dedifferentiated acinar cells (Fig. 5B and Supplementary Fig. S13B). Additionally, annotation of these CGs to chromatin states identified in purified normal human acinar and ductal cells (69), revealed that PC3-CGs were significantly more associated to acinar, and specially, ductal regulatory regions (i.e. enhancers) than CGs driving other components of the PCA (Supplementary Fig. S13H). Collectively, these data strengthen the hypothesis that PC3 can distinguish between acinar and ductal identity independent of transformation and uncover commonalities at the level of DNA methylation between MC2/IFNsignhigh PDAC epithelial cells and healthy as well as immortalized cells of ductal origin. Conversely, MC1/IFNsignlow PDAC cells show higher similarities to healthy acinar cells in PC3.
To further explore a possible relationship between ductal cells and MC2/IFNsignhigh tumors, we interrogated the status of IFN signaling in healthy human acinar and ductal cells at the transcriptomic level by analysing publicly available gene expression data of acinar and ductal cells at the bulk and single-cell level (Fig. 5C) (74–77). GSEA revealed an enrichment of IFN-related signatures in ductal compared to acinar cells in all datasets (Fig. 5D and Supplementary Fig. S14A–C). Accordingly, expression of the IFNsign and individual IFN-related genes was consistently higher in healthy ductal compared to acinar cells (Fig. 5E and Supplementary Fig. S14D). Ductal cells also showed enrichment for dsRNA response signatures (Fig. 5F and Supplementary Fig. S14B,C) and indeed showed higher expression of repeats, including LTR/ERVs (Fig. 5G and Supplementary Fig. 14E). Interestingly, normal ductal cells also expressed higher levels of the gene signature associated to basal-like PDAC subtype (Supplementary Fig. S14F), which is also enriched in MC2/IFNsignhigh tumor epithelial cells (Supplementary Fig. S4F and Supplementary Fig. S7B). Genes upregulated in MC2/IFNsignhigh tumors but not part of the IFNsign were also enriched in normal ductal cell (Supplementary Fig. S14G). Collectively, these data indicate a broader similarity of MC2/IFNsignhigh tumors to ductal cells at the transcriptomic level.
Taken together, transcriptomic and DNA methylome data link MC2/IFNsignhigh tumors to ductal cells while MC1/IFNsignlow tumors are more similar to acinar cells. These data support the possibility that MC1-like tumors may preferentially arise from acinar, while MC2-like PDACs originate from ductal cells in the exocrine pancreas.
To explore this possibility further, we analyzed the expression of IFN-response genes in a panel of mouse PDAC cell lines derived from two genetically engineered mouse models both driven by KrasG12D and Trp53 loss, but targeted either to ductal or acinar cells (63) (Fig. 5H and Supplementary Fig. S15A). Strikingly, PDAC cells from ductal-derived tumors (Sox9CreER; KrasLSL-G12D; Trp53f/f mice) expressed significantly higher levels of IFN-related genes and dsRNA sensors compared to the ones derived from tumors of acinar origin (Ptf1aCreER; KrasLSL-G12D; Trp53f/f mice) (Fig. 5H and Supplementary Fig. S15A).
Collectively, our data derived from human normal and cancerous pancreatic epithelial cells, as well as from mouse transgenic models point to a ductal origin for MC2 Methylationlow/IFNsignhigh PDACs, while MC1 Methylationhigh/IFNsignlow tumors appear rather derived from acinar cells (Fig. 5I).
Discussion
Based on whole genome DNA methylome analysis of purified epithelial tumor cells isolated from fresh human PDAC tissue, and analyses of PDAC-derived primary PDXs and PDCs models, our data uncovered an unexpected link between hypomethylation at repeat-elements, the activation of an IFN program, the generation of a pro-inflammatory tumor promoting microenvironment, poor prognosis and a ductal cell of origin in a subset of human PDAC tumors (Fig. 5I). Expression of IFN-related genes (78,79) or repeats (80) have previously been noted in PDAC supporting our observations here. However, these descriptive observations had not yet been linked to differential DNA methylation, cell-of-origin, pro-inflammatory microenvironment or PDAC aggressiveness. Activation of IFN signaling through intracellular recognition of derepressed endogenous repeats has been described upon treatment with epigenetic drugs such as decitabine or HDAC inhibitors, a phenomenon termed viral mimicry (34,35,81,82). This viral mimicry is typically linked to growth arrest mainly due to the cytotoxic effects exerted by IFN signaling (83). The extent and make-up of the repeat expression and IFN activation acutely induced by epigenetic drugs may, however, differ from the chronic situation we identified here in untreated naïve MC2-type PDACs. Interestingly, TP53 function has been shown to suppress mobilization and expression of retrotransposons, while loss of TP53 can overcome the IFN-mediated apoptosis induced by deregulated repeat expression in mouse embryonic fibroblasts (84,85). Indeed, TP53 mutations have a higher incidence in squamous/basal-like PDAC tumors (26,29,30), which associate with the MC2-type. This phenomenon seems to be also related to a ductal origin of PDAC as only PDACs derived from KrasG12D/Trp53−/− ductal, but not acinar cells, show high expression of the IFN network (Fig. 5g and Supplementary Fig. 15). Additional mechanisms such as histone modifications or the joint action of certain transcription factors might control repeat expression and mobilization beyond DNA methylation. Additionally, this control might differ between normal and tumor cells as we observed that ductal cells have higher methylation levels than acinar cells despite bearing higher expression of repeats. Further studies in the field of genome regulation will shed light on the molecular details of these complex mechanisms.
IFNsignhigh/MC2 tumor cells activate and re-program stellate cells much more robustly compared to IFNsignlow/MC1 tumor cells, generating a pro-inflammatory microenvironment which ultimately promotes tumor growth. This process initiated by hypomethylated repeat expression links to the known important roles of the PDAC stroma for tumor cell survival and growth (46,48,49,86). The IFN-mediated stromal activation in MC2 PDACs may be conceptually related to the situation in colorectal cancers, where epithelial cells escape TGFβ − mediated cytostatic effects by mutating SMAD4 or TGFβ receptor, while benefiting from the tumor promoting effect of TGFβ on the stromal microenvironment (87).
Activation of IFN signaling has been recently proposed as a mechanism to increase immune checkpoint blockade (ICB) response (88–90). Indeed, IFNsignhigh tumor cells expressed higher level of several immune checkpoint molecules (Supplementary Fig. S3F). Although we could not evaluate the functional contribution of the adaptive immune system to the IFNsignhigh/low scenario in the immunodeficient PDX mouse models, this observation suggests potentially targetable differences between the tumor subgroups. However, a sustained chronic IFN activation status, similar to the one we observe in IFNsignhigh tumors, has also been reported to cause ICB resistance (91). Thus, approaches to use ICB to potentially treat IFNsignhigh/low PDACs deserve further functional investigation. Possibly, efficient inhibition of intrinsic interferon signaling may be an approach to target these tumors with little side effects on normal cells (92).
The cell of origin in human PDAC has been debated for decades. PDAC mouse models originated from acinar cells (61,93) and data from tracing experiments (60,94) show that mutant acinar cells transdifferentiate to a ductal-like phenotype (with mixed characteristics of embryonic progenitor cells) (67) to form precancerous lesions called Pancreatic Intraepithelial Neoplasia (PanINs), which ultimately progress into PDAC. Mouse models where PDAC originates from ductal cells have been challenging as these cells seem more refractory to transformation needing additional insults, such as TP53 mutations to progress towards PDAC (65,66). However, as we and others showed, ductal-derived PDAC models associate to worse outcome and show significantly fewer and, if at all, only late stage PanINs (63,64). Starting from ductal cells, an acinar-ductal metaplasia would not be required and classical PanIN formation may not occur in such PDAC subtypes. Indeed, little or no occurrence of PanIN lesions has also been observed in some PDAC patients and was associated to poor survival (95–97). Future studies need to extent such observations to serial samples and association to specific molecular features of MC1 and MC2 type tumors. In addition to our analysis on resectable PDAC tumors, it will be interesting to investigate these patterns also in the spectrum of unresectable tumors as well as metastatic lesions. This would be of special interest in light of the poor effects achieved by the combination of Ruxolitinib with Capecitabine to treat metastatic PDAC patients (98,99).
Our understanding in pancreatic cancer biology has exponentially increased in the past years. Numerous studies have contributed to our knowledge of PDAC genetics and transcriptomics and new studies start to shed light into the epigenomics of PDAC (11,12,100). Our data show that differential DNA methylation of repeat-derived genetic elements results in the execution of an epithelial cell-autonomous IFN expression program, which promotes a pro-inflammatory and tumor-promoting stromal feed-forward loop leading to increased aggressiveness of pancreatic cancer. Moreover, we suggest that the basis for Methylationlow/IFNsignhigh (MC2) and Methylationhigh/IFNsignlow (MC1) PDAC phenotypes is already laid down in the lineage identity of the ductal and acinar cells of the normal pancreas. Stable epigenetic traits imprinted in the cells-of-origin could be used to stratify different types of PDAC and may improve risk stratification and provide novel targeting opportunities.
Methods
PDAC and Normal Pancreas samples for epithelial isolation
Tissue samples were obtained from patients who received partial pancreatoduodenectomy at the Department of General, Visceral and Transplantation Surgery, University of Heidelberg. Patients were part of the HIPO-project. The study was approved by the ethical committee of the University of Heidelberg (case number S-206/2011 and EPZ-Biobank Ethic Vote #301/2001) and conducted in accordance with the Helsinki Declaration; written informed consent was obtained from all patients. Fixed sections were evaluated by two independent pathologists (W.W. and A.M) and only samples which did not contain normal tissue contamination were included in the study.
Normal samples were resected from sites distal to tumor lesions. A slide section from the processed sample was fixed for pathological evaluation. Hematoxilin & Eosin staining was performed in fixed sections and slides were evaluated by two independent pathologists (W.W. and A.M). Only histologically normal samples were included in the study. Patient and tumor characteristics are summarized in Supplementary Table S1.
Donor pancreata were isolated and processed by the Beta Cell Bank of the JDRF Centre for Beta Cell Therapy in Diabetes (Brussels, Belgium) affiliated to the Eurotransplant Foundation, and written informed consent for use of donor material for research was obtained according to Belgian laws; or were obtained from organ procurement centers at USA (including IIDP, NDPRI, IIAM, nPOD) from de-identified post-mortem samples and were excluded from the IRB Review per 45CFR56 and NIH policy.
Human tissue dissociation and flow cytometry sorting
Tumor tissue:
Tumor tissues were minced using a razor blade and dissociated using the human Tumor Dissociating Kit (Milteny Biotec) following manufactureŕs instructions. Dissociated samples were sequentially filtered through cell strainers of 100 μm → 70 μm → 40 μm (BD Falcon). To lyse erythrocytes, the cell pellet was resuspended in ACK lysing buffer (Lonza) and incubated 2 min at room temperature. After one wash with HBSS (Thermo Fisher) cells were pelleted and resuspended in Tumor Staining Buffer (HBSS + 1% BSA + 2mM EDTA) and blocked for 10 min at room temperature with FcR blocking reagent (Miltenly Biotec). Next, the conjugated antibodies (see below) were added and the sample was stained for 15 min at 4°C.
Normal tissue:
Normal samples were first dislodged by injecting fetal bovine serum (FBS) in the tissue. Next, tissues were minced using a razor blade. Pieces were placed in a 50 ml falcon with 7 ml of G Solution(101) (1 l HBSS + 0.9 g glucose + 47.6 μM CaCl2) and incubated on ice for 1 min. G solution was removed and the process was repeated 2–3 times until the G Solution was clear. Pieces were added to the Digestion Mix Solution (Advanced DMEM/F12 + 25mM HEPES + 1% BSA + Trypsin Inhibitor (1:10; Sigma Aldrich) + enzymes from the Tumor Dissociating Kit (Milteny Biotec)) and incubated at 37°C for 15 min. Digested tissue was homogenized by pipetting and sequentially filtered using cell strainers of 100 μm → 70 μm → 40 μm (BD Falcon). Digestion was stopped by adding one volume of FBS. If some tissues fragments were still left, the digestion was repeated and the filtered fractions were pooled. Cells were pelleted, resupended in ACK lysing buffer (Lonza) and incubated 2 min at room temperature. Cells were washed using Wash Buffer (HBSS + 1% BSA + 1% FCS + 1mM EDTA + Trypsin inhibitor + 2mM MgCl2) and pelleted. Pellet was resuspended in Normal Staining Buffer (Wash Buffer + 2mM CaCl2) + 2.5 μg/ml of DNase (Sigma Aldrich) and FcR blocking reagent (Miltenly Biotec) and incubated for 15 min at room temperature. Next, the conjugated antibodies (see below) were added and the sample was stained for 15 min at 4°C.
Staining antibodies: EPCAM-FITC (1:11 dilution; clone HEA-125; Miltenyi Biotec); CD45-VioBlue (1:11 dilution; clone 5B1; Milteny Biotec). Propidium Iodide was used to exclude dead cells. Epithelial cells were defined as EPCAM-FITC+/CD45-VioBlue−. CAFs were defined as EPCAM-FITC−/CD45−VioBlue-. An example of the gating strategy can be found in Supplementary information.
For DNA extraction sorted cells were collected into ice-cold CO2 independent medium (Thermo Fisher), pelleted and stored at −80°C. For RNA extraction cells were sorted into RNA lysis buffer (Arcturus PicoPure RNA isolation Kit, Life Technologies) and stored at −80°C. Single cell suspensions were sorted using a FACSFusion systems (BD Biosciences).
DNA isolation and TBWGBS
Genomic DNA from sorted cells was isolated using the QIAamp DNA Micro Kit (Qiagen) according to the manufacturer’s instructions. Tagmentation based WGBS was performed as described previously (102) using 30 ng genomic DNA as input. Four sequencing libraries with different barcodes were generated per sample and pooled in equimolar amounts to a 10 nM pool. Each pool was sequenced paired-end, 125 bp, on two lanes of a HiSeq2000 sequencer (Illumina).
Mapping of whole genome bisulfite sequencing data and methylation calling
The TWGBS data were processed as described (14): The human reference genome (hg19) was in silico transformed for both the top strand (C to T) and bottom strand (G to A) using MethylCtools (55). Before alignment, trimming of adaptor sequences was performed using SeqPrep (https://github.com/jstjohn/SeqPrep). The first read in each read pair was then C to T converted and the second read in the pair was G to A converted. The converted reads were aligned to a combined reference of the transformed top strands (C to T) and bottom strands (G to A) using BWA (bwa-0.6.2-tpx) (103) with default parameters but with disabling the quality threshold parameter for read trimming (-q) of 20 and the Smith-Waterman for the unmapped mate (-s). After alignment, reads were converted back to the original states, and reads mapping to the antisense strand of the respective reference were removed. Duplicate reads were marked, and the complexity was determined using Picard MarkDuplicates (http://broadinstitute.github.io/picard/). Reads with alignment scores of less than 1 were filtered before subsequent analysis. Total genome coverage was calculated using the total number of bases aligned from uniquely mapped reads over the total number of mappable bases in the genome. At each cytosine position, reads that maintain the cytosine status were considered methylated, and reads in which cytosine was converted to thymine were considered unmethylated. Only bases with Phred-scaled quality score of ≥ 20 were considered. In addition, the five base pairs at the two ends of the reads were excluded from methylation calling according to M-bias plot quality control. For the TWGBS libraries, the first nine base pairs of the second read and the final nine base pairs before the adaptor of the first read were excluded from methylation calling.
Calling of DMRs
The raw counts of methylated and unmethylated reads for each CpG site from different libraries were merged for each sample. R (version 3.4.2) and BSmooth (104) (version 1.14.0) were used with default parameters for smoothing and visualization purposes. Calling of DMRs was performed using DSS (105) (version 2.26.0) with default parameters for comparison between normal and tumor samples as well as comparing MC1 and MC2.
Annotation
Gene-related annotations were obtained from Gencode v19. Genomic annotations for DNase, CGIs, and repeats (SINEs, LINEs, ERVs) were obtained from UCSC table browser. CGI-shores were defined as 2kb upstream and downstream flanking CGI. Chromatin states (15 states model) from normal pancreas from Roadmap dataset (106) were used to annotate DMRs to enhancers. EnhG, EnhBiv, Enh were merged and defined as “Enhancer”. The value for the overlap between one DMR and one genomic feature is the fraction of base pairs of the DMR that overlap to the genomic feature.
Overlap differentially methylated genes from different studies
Genes identified as differentially methylated between PDAC and normal pancreas in previous studies were obtained from Nones et al. (8). The overlap of genes was illustrated using Venn Diagram (107).
Calculation of global beta values and beta values at CpG islands and repeats
Based on CpG island, LINE, SINE and ERV annotations, mean beta values were calculated for each single feature in each sample. Beta values for each annotation category per sample were derived by calculating a mean beta value of all features of the same annotation category.
For the global beta value distribution plots the distribution of genome-wide smoothed beta values per sample was plotted using frequency polygons. ggplot(…) + geom_freqpoly(binwidth = 0.01). In case of group wise distribution, the mean of the group was calculated.
3D Methylation-based Principal Component Analysis (PCA)
Based on all DMRs between normal vs tumor, a beta value matrix was created and a PCA using R (version 3.4.2) and prcomp(...) was performed (Fig. 1B). For an unbiased approach (Supplementary Fig. S3A) the genomes were organized into windows of 5kb and filtered for a mean CpG coverage of 10 reads in each sample and window. 10,000 most variable windows were used to perform PCA.
Methylation Density Plots and 2D Principal Component Analysis (PCA)
Density plots were generated by the densityHeatmap() function in the ComplexHeatmap package(108). For each column that represents one sample in the plot, colours were mapped to the density values in the corresponding distribution. The black dashed lines correspond to the five quantiles of the distributions and the red dashed lines correspond to the mean value of the distributions. For 2D PCA plotting, CpG sites were randomly sampled by a probability of 0.001. For this subset of CpG sites, the top 2000 CpG sites with the highest variance were selected to perform the PCA analysis.
Estimation of Partially Methylated Domains (PMDs)
Genome-wide methylation profiles are represented as long vectors of numeric values, where the order of data points corresponds to chromosomal CpG positions. The Bayesian framework and dynamic programming approach of fastseg (version 1.20.0) (109) were used to segment the genome based on the distribution of methylations. As fastseg does not take into account the distance between CpG sites but rather only considers the order of CpG sites, each chromosome was split into blocks if the gap between two adjacent CpG sites was larger than 100kb. Blocks containing less than 50 CpG sites were removed.
While fastseg can predict segment borders, each segment must still be classified based on its methylation. To do so, the distribution of both the mean and the standard deviation of methylation in the segments were examined (Supplementary Methods Figure 1A). On each plot, there are at least two clear clusters, which correspond to unmethylated and fully methylated segments. We marked segments with mean methylation ≤ 0.1 and sd ≤ 0.15 as unmethylated and segments with mean methylation ≥ 0.7 and sd ≥ 0.2 as fully methylated (indicated by red rectangles in the above plots). The remaining segments were defined as intermediately methylated segments and PMDs were detected among them.
Since the intermediately methylated segments are more heterogeneous (mean methylation ranging from 0.1 to 0.7), it is expected that there are clusters of segments, which are close to each other. The gap ratio was defined to measure the relative distance to neighbouring segments in relation to the width of the segment itself. For a certain segment i, the gap ratio denoted as ri is defined as ri = di/wi where di is the minimal distance to the two neighbouring segments and wi is the width of the segment i. Segments with a smaller gap ratio tend to be close to the neighbouring segments and need to be merged into a larger segment.
To select a proper cutoff of the gap ratio to merge neighbouring segments, the distribution of gap ratios for all samples were computed (Supplementary Methods Figure 1B).
On the left side of the distributions the high peaks correspond to small gap ratios, where segments are highly close t each other. Thus, we set a cutoff of gap ratio ≤ 0.4, and if two segments were close enough, they were merged into a long segment keeping the gaps in between. Rainfall plotting (Supplementary Methods Figure 1C) demonstrates that the process successfully merges the cluster.
Finally, the width distribution of intermediately methylated merged segments was computed and a threshold for the width of a PMD was set to ≥10kb.
Hilbert plots
Hilbert curve visualization was performed by the HilbertCurve R package (110). The Hilbert curves for both PMDs and methylation profiles were constructed with level 9. The averaging mode for the methylation was “absolute”.
UpSet plot
UpSet plots were generated by the UpSet() function of the ComplexHeatmap package. The intersection size of two sets of genomic regions was defined as the total number of base pairs of the intersected regions and the union size of two sets of genomic regions was defined as the total number of base pairs of the merged regions. Mode “distinct” was used to calculate the size of each combination of sets.
Isolation of human normal acinar, ductal, de-differentiated acinar and mixed exocrine samples
Ductal and acinar cells from fresh tissue were isolated as previously described using HPx1 (acinar) and CD133 (ductal) markers (74).
To obtain ductal and de-differentiated acinar cells from in vitro cultures exocrine preparations were cultured as previously described (59,70) in Advanced RPMI 1640 medium containing 5% heat-inactivated fetal bovine serum and 1% penicillin-streptomycin solution (Life Technologies) under 5% CO2 atmosphere at 37 °C. FITC-conjugated UEA-1 (Ulex Europeaus Agglutinin-1, Sigma, Overijse, Belgium) lectin labeling of acinar cells was performed according to Houbracken et al. and Baldan et al. (59,70). Exocrine cell fraction labeled with UEA-1 was kept in suspension culture. At day 4 of culture, cell clusters were dissociated following the protocol of Baldan et al. (59). Cells were stained with carbohydrate antigen 19.9 (mouse monoclonal anti-human CA19.9, Dako, Heverlee, Belgium). Alexa Fluor 647 anti-mouse (Jackson Laboratory, Westgrove, PA, USA) was used as secondary antibody. Analysis and cell sorting were performed on a BD FACSAria (BD Biosciences). Viable, single cells were gated based on forward and side scatter.
Infinium MethylationEPIC DNA methylation arrays
Genomic DNA was extracted from two independent replicates of HPDE cells using the DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer’s instructions. PDX genomic DNA was extracted using Allprep DNA/RNA/Protein Mini Kit (Qiagen). Genomic DNA from acinar, ductal, de-differentiated acinar or mixed exocrine samples was extracted from frozen pellets using QIAamp DNA Micro Kit (Qiagen) according to manufacturer’s instructions. Additionally, three commercial genomic DNA preps were used (#CD564011 (Origene); #CD564404 (Origene); #A712202 (BioChain)).
DNA methylation data were generated using Infinium MethylationEPIC BeadChip arrays at the Genomics and Proteomics Core Facility of the DKFZ or were previously available (GSE122126) (71). Processing of the data was done with the RnBeads Package (version 2.4.0) for R (111) using default settings. Beta values were normalized using the BMIQ (Beta MIxture Quantile dilation) (112) method. Hierarchical clustering was performed by the pheatmap() function using default parameters. Annotation to acinar and ductal enhancers was done using previously published ChromHMM data from isolated human acinar and ductal cells (69). Calculations were performed with R version 3.6.0 in R-Studio.
RNA isolation and RNA sequencing
Total RNA isolation and RNA sequencing were performed as previously described (113): RNA was extracted using the ARCTURUS PicoPure RNA Isolation Kit (Life Technologies) including a DNase treatment step (Qiagen) following manufacturer’s instructions. cDNA was generated with 1 ng of total RNA and thirteen cycles of amplification using the SMARTer Ultra Low RNA Kit for Illumina sequencing (Clontech) according to manufacturer’s instructions. The sequencing libraries were generated using the NEB Next ChIP-Seq kit (New England Biolabs) according the manufacture’s instructions. The quality of total RNA, cDNA and library were monitored throughout the process using a 2100 Bioanalyzer (Agilent). Sequencing was done in a HiSeq2000 sequencer (Illumina) with three samples per lane, paired-end 100bp.
RNA-seq data processing
RNA-seq reads were aligned to the hg19 reference genome using the STAR alignment software (version 2.3.0e) (114), with the merged transcriptome of Gencode v19 and lincRNA catalog annotations (115). Counts of reads mapped to exons were estimated by htseq-count (116). Expression values normalized using DEseq2 (117) were used for heatmap visualization. On the heatmap, genes were scaled by z-score scaling. Heatmaps were generated by the ComplexHeatmap R package(108). TPM values were calculated by normalizing to the sum of the length of non-overlapping exons for each gene and to the library size for each sample.
For expression of repeats featureCount (version 1.5.3) (118) was applied on the merged annotation of Gencode v19 and RepeatMasker (UCSC) considering three classes or repetitive elements: SINEs, LINEs, and LTRs. The --fracOverlap parameter was set to 0.5 to enforce that each given single read is counted as in a repeat only if more than 50% base pairs of the read overlap to it. TPM normalization was applied to repeats expression using a library size estimated by the total reads mapped to genes. The expression difference between group 1 and group 2 was defined as (μ1-μ2)/σ, where μ1 and μ2 are the mean in group 1 and group 2, respectively, and σ is the standard deviation by pooling values in the two groups.
The association of samples to previously described PDAC subtypes was based on published signatures (Supplementary Table S8) (2,24,119) using the following method: Assume a signature matrix has n signature genes and K subgroups. The signature scores in subgroup k is presented as a vector denoted as sk. For sample i in another expression matrix where the expression vector is denoted as xi with the same set of genes as the signatures (also in the same order), we calculate the Euclidean distance denoted as dik between xi and sk. Over all signature vectors, dik forms a vector denoted as di. We order the vector di decreasingly and take the first two highest values di(1) and di(2) and define the difference score as ai = (di(1)−di(2))/μ where μ is the mean of vector di. To get a significant large ai, data points in each sk are randomly permutated nr times (e.g nr = 1000), following the process above. The random difference score in permutation j is calculated denoted as aijrandom. The p-value for the difference score to be significantly large is: pi = 1/1000 j∑1000I(rijrandom > ri) where I(expr) = 1 if expr is evaluated as true, or else I(expr) = 0. If pi < 0.05, the subgroup with the smallest distance to sample i is assigned, or else sample i is labeled as no subgroup is assigned.
Correlation methylation - expression
For each DMR, Spearman correlation test was applied to the mean methylation of the DMR and the expression value of the gene with the nearest TSS to that DMR. The significant correlations were filtered by FDR ≤ 0.05.
Analyses of public datasets
PDAC data
Normalized expression data, subtype annotation, tumor cellularity (QPURE) estimation and survival data were obtained from the supplementary information accompanying the Bailey et al. publication (25). Normalized expression and survival data were downloaded from NCBI GEO with accession number GSE21051. TCGA pancan normalized values from PDAC samples were downloaded from The Cancer Genome Atlas database and non-PDAC samples were excluded prior to the analyses. TCGA subtyping was used as defined in Connor et al. 2017 (27). Kaplan-Meier curves were calculated with GraphPad Prism 8 software (Bailey et al., GSE21051, (25)) or in KM Plotter (TCGA, (120)). For correlation between expression of genes and of genes with IFNsign, the average IFN signature expression for each patient was calculated and correlations were computed via Spearman correlation (R).
Normal acinar and ductal RNA expression data
Normalized RPKM values for bulk RNA-seq data of acinar and ductal cells were obtained from accession number GSE57973. Single cell RNA-seq preprocessed data as raw count values for the studies with the accession numbers GSE81547, E-MTAB-5061 and GSE85241 were downloaded and further processed per study using R (version 3.4.2) and Seurat (121) (version 2.3.0). The analysis used healthy control samples only. Log normalization was applied using NormalizeData() and highly variable genes (HVGs) defined using FindVariableGenes() with default parameters. Data was further normalized using linear regression via the function ScaleData(), regressing for the number of unique molecular identifiers (UMIs) in all datasets as well as patient origin for GSE81547, patient origin for E-MTAB-5061, patient and library origin for GSE85241. For downstream analysis, the first ten principal components were used. Cells were defined as ductal or acinar based on authors annotation, for study GSE81547 and E-MTAB-5061. In case of unavailable cell annotation (GSE85241) cells were clustered into acinar or ductal using FindClusters(dims.use = 1:10, resolution = 0.8, ) and the expression of genes defined in the original study as markers of acinar or ductal cell. A mean expression value for each cell was calculated based on an IFN signature and individual genes were visualized as violin plots, additionally applying a Mann-Whitney test.
Acinar and ductal methylation data
EPIC methylation array data from isolated acinar and ducal cells were obtained from accession number GSE122126, and normalized and processed together with the data generated in house.
Human cells
Tumor patient-derived cell lines (PDCs) have been previously reported (PACO2, PACO17, PACO14, PACO18) (39) or have been similarly derived (PACO3, PACO20, PACO28). PDCs were maintained in CSC medium which contains advanced Dulbecco’s modified Eagle’s medium–nutrient mixture F-12 (DMEM/F12; Life Technologies) with N2 supplement (Life Technologies), 50 ng/ml basic fibroblast growth factor (bFGF; Peprotech), 20 ng/ml epidermal growth factor (EGF; Peprotech), 10 ng/ml LONG R3 insulin-like growth factor–I (IGF-I) (Sigma), 100 μM β-mercaptoethanol (Life Technologies), 2 μg/ml heparin (Sigma) and grown in Primaria plates (BD) and used at maximum passage 15. PDCs were categorized according to the expression of IFN signature genes as: IFNsignhigh: PACO2, PACO17, PACO20. IFNsignlow: PACO3, PACO14, PACO18, PACO28.
Human pancreatic Stellate cells (StC) were purchased from ScienCell Research Laboratories and cultured using poly-L-Lysine coated (Sigma-Aldrich) tissue culture flasks and SteCM medium (ScienCell) with 2% serum. StC were used for a maximum of 6 passages.
Human pancreatic ductal epithelial (HPDE) cells (73) were obtained from American Type Culture Collection (ATCC), cultured according to the supplier’s information and used at passage 14.
All cell lines used were authenticated monthly by single-nucleotide polymorphism profiling and tested for mycoplasma contamination (both by Multiplexion).
Conditioned medium (CM)
PDCs were seeded in CSC complete medium. After 24 hours cells were washed once with PBS (Sigma-Aldrich) and incubated in CSC-reduced medium (CSC minus bFGF, EGF and IGF) for 1h. Subsequently, fresh CSC reduced medium was added for 48 h. CM was harvested and spinned at 300 × g for 10 min to remove cellular debris. PDCs were harvested and counted to ensure equal cell numbers. StC were preincubated for 1h with CSC-reduced medium and treated with PDC CM or StC CM for 12 h prior to RNA extraction. For long term conditioning, StC were treated with PDC CM or StC CM every 3 days during 10 days.
Co-cultures
PDCs were seeded in 0.4 μm pore trans-wells (Becton Dickinson). Cell numbers were adapted experimentally to ensure equal values during the experiment. The same number of cells was seeded in a transparent 96-well plate to monitor cell confluence during the experiment. 24h later, pancreatic StC were seeded onto companion plates (Becton Dickinson) coated with poly-L-lysine (Sigma-Aldrich). The next day, StC and PDCs were preincubated in CSC reduced medium for 1h prior to setting the co-culture. After two days of co-culture, 50% of the medium was replaced with fresh CSC reduced medium. After four days of co-culture, transwells were placed on a new plate and cell amount was estimated using CellTiter-Blue stock solution (Promega) following manufaturer’s instructions.
Ruxolitinib (Focus Biomolecules) was added daily at a final concentration of 100nM.
Human IFN lambda Receptor 1 (IFNLR1) extracellular domain (clone MMHLR-1; PBL assay science) or Mouse IgG1 Iso Control (clone P3.6.2.8.1; eBioscience) were used at a final concentration of 0.5μg/ml. IFNLR1 or IgG control were added on day 1 and 3 of co-culture. Recombinant IL28A or IL29 were used at 10ng/ml.
5-Aza-CdR treatment
PDCs were treated daily with 500 nM of 5-Aza-2′-deoxycytidine (5-Aza-CdR; Santa Cruz) or DMSO as control for five days. On day sixth proteins and RNA were extracted.
Cytokine treatment
PDCs were seeded in Primaria 96 wells (Corning) and treated every two days for a total of five days with recombinant human IL8, IL1A or IL1B (Peprotech) or 0.1% BSA PBS. Cell amount was estimated using CellTiter-Blue stock solution (Promega) following manufacturer’s instructions.
IFN ELISAs
CM from 5×106 cells was collected after 48h. Cellular debris was excluded by centrifugation at 1.500rpm. 12ml of tumor media or only media as control were concentrated to 1.2mL using Amicon columns (3,000NMWL, Merck Millipore) following manufactureŕs instructions. The following ELISÁs kits were used as indicated by the manufacturer: Human IFN alpha Platinum ELISA (BMS216, Affymetrix Bioscience), VeriKine Human IFN Beta ELISA kit (41410, pbl assay science), Human INFγ ELISA kit (EHIFNG, Invitrogen), Human IL-29/IFN-λ1 (DY7246, R&D Systems), and Human IL-28A/IFN-λ2 (DY1587, R&D Systems).
Generation of knock-down cells
STAT1 and MAVS knock-downs were generated with a miR-E (122) StagBFPEP lentiviral vector, a modified version of the original SGEP vector kindly provided by Johannes Zuber (IMP – Research Institute of Molecular Pathology GmbH, Vienna) in which the constitutively-expressed GFP protein is replaced by the tagBFP protein. miR-E sh oligonucleotides were designed using the shERWOOD algorithm tool (123): Non-silencing control: 5’-TGCTGTTGACAGTGAGCGCTCTCGCTTGGGCGAGAGTAAGTAGTGAAGCCACAGATGTACTTACTCTCGCCCAAGCGAGATTGCCTACTGCCTCGGA-3’; shSTAT1 #1: 5’-TGCTGTTGACAGTGAGCGCCAGAAAGAGCTTGACAGTAAATAGTGAAGCCACAGATGTATTTACTGTCAAGCTCTTTCTGTTGCCTACTGCCTCGGA-3’; shSTAT1 #2: 5’-TGCTGTTGACAGTGAGCGCTCAGAGCACAGTGATGTTAGATAGTGAAGCCACAGATGTATCTAACATCACTGTGCTCTGAATGCCTACTGCCTCGGA-3’; shMAVS #1: 5’-TGCTGTTGACAGTGAGCGCCCAAGTTGCCAACTAGCTCAATAGTGAAGCCACAGATGTATTGAGCTAGTTGGCAACTTGGATGCCTACTGCCTCGGA-3’; shMAVS #2: 5’- TGCTGTTGACAGTGAGCGAACCAATCCAGCACCATCCAAATAGTGAAGCCACAGATGTATTTGGATGGTGCTGGATTGGTGTGCCTACTGCCTCGGA-3’. Oligonucleotides were amplified by PCR using the Q5 High-Fidelity DNA Polymerase (New England Biolabs). The following primers were used for PCR amplification: miRE-Xho-fw: 5’-TGAACTCGAGAAGGTATATTGCTGTTGACAGTGAGCG-3’; miRE-EcoOligo-rev: 5’-TCTCGAATTCTAGCCCCTTGAAGTCCGAGGCAGTAGGC-3’. PCR products containing shGene and non-silencing miR-Es were subcloned into the StagBFPEP recipient vector via EcoRI-HF and XhoI restriction sites. Lentiviral particles were produced by transfecting HEK293T cells with StagBFPEP-miR-E vectors together with plasmids encoding for the packaging proteins pMD2G and psPAX2 using Polyethylenimine (PEI; Polyscience). Cells were transduced at MOI5.
Gene expression microarrays
Total RNA from PDCs or treated StCs was isolated using the RNeasy kit (Qiagen) according to the manufacturer’s instructions. Gene expression analyses were performed using Illumina HumanHT12v4 BeadChips at the Genomics and Proteomics Core Facility of the DKFZ.
Gene set enrichment analysis (GSEA)
GSEA (23) was conducted using the GSEA desktop application and the gene sets downloaded from the Broad Institute (Hallmarks) or indicated in supplementary information using 2000 permutations. For microarray data, quantile-normalized data was used as input. For RNA-seq data, ranked log2 fold change values calculated by DESeq2 (117) were used. Genes with NA FDR values were excluded from the analyses. IFNsign comprises genes from Interferon-alpha response and Interferon-gamma response Hallmark gene sets with a rank metric score > 1 in MC2 vs MC1 samples.
Genesets used in the study are provided in Supplementary Table S8.
Immunofluorescence
PDCs cells were seeded on 8-chamber culture slides (Falcon) coated with poly-L-Lysine (Sigma-Aldrich) and grown to 50–70% confluency. Cells were fixed in 4% freshly depolymerized formaldehyde for 10 min, permeabilized with 0.25% (vol/vol) Triton X-100 (Sigma) for 20 min and blocked with 1% BSA for 1 h. J2 antibody for dsRNA(124) (Scicons J2 from English and Scientific Consulting Kft, Hungary, 10010200) or mouse IgG2a k isotype control (eBM2a; eBioscience) were incubated overnight at 4 °C (20ug/ml), and detected by fluorescence using a goat anti-mouse IgG, F(ab’)2 dylight 649 (1:300 dilution; Jackson ImmunoResearch Laboratories). Slides were mounted using ProLong Antifade GOLD with DAPI (Life Technologies), as described by the manufacturer. Pictures were taken using a Nikon Eclipse Ti microscope.
FACS dsRNA
100,000 PDCs were fixed with Cytofix/Cytoperm solution (BD) for 10 min on ice. After washing with Permwash (BD), cells were stained with J2 antibody (Scicons J2 from English and Scientific Consulting Kft, Hungary, 10010200) or mouse IgG2a k isotype control (eBM2a; eBioscience) overnight at 4°C (2 ug/ml). The day after, cells were washed with Permwash (BD) and stained using a goat anti-mouse conjugated to APC (1:200 dilution; R&D, F0101B). Quantification of the difference between the median fluorescent intensity of J2 staining and isotype staining was calculated on singlets for each cell line (ΔMFI). An example of the gating strategy is shown in Supplementary information. Two independent experiments were performed per cell line except for PACO20 for which only one replicate was available. FACS analyses were performed on a LSR2 (BD Biosciences) and data analysis was done with FlowJo software version 10.0.3 (FlowJo, LCC).
Western Blot
Whole-cell lysates of PACO cells were prepared using RIPA lysis buffer (Cell Signaling Technology) with 1 mM PMSF (Sigma), 1 mM EDTA and 1mM Halt Protease-Phosphatase Inhibitor Cocktail (Pierce). Protein concentration was determined by the Pierce BCA assay (Thermo Scientific). Proteins were resolved on 4–12% TGX gels (Criterion, Bio-Rad) with TGS (Tris-Glycine-SDS) running buffer (Bio-Rad) and blotted on PVDF membranes (Trans-Blot TURBO, Bio-Rad). Membranes were blocked for 1 h in TBS containing 0.1% (v/v) Tween-20 and 5% (w/v) BSA. Primary antibodies against MDA5 (D74E4) (1/1,000; Cell Signaling, #5321), STAT1 (42H3) (1/1,000; Cell Signaling, #9175), phospho-STAT1 (Tyr701) (58D6) (1/1,000; Cell signaling, #9167), Phospho-STAT1 (Ser727) (1/1,000; Cell signaling, #9177), and MX1 (E-5) (1/1,000; Santa Cruz, sc-271024) were incubated overnight at 4°C in blocking solution. Secondary antibodies (anti-rabbit-IgG horseradish peroxidase-linked antibody [1/10,000 in TBS containing 0.3% Tween-20, Cell Signaling, #7074] or mouse IgG binding protein HRP [1/10,000 in TBS containing 0.1% Tween-20, Santa Cruz, sc-516102]) were incubated for 1 h at room temperature. Membranes were washed in TBS-Tween 0.1% and immunocomplexes were detected using the ECL kit (Amersham International) and acquired with a ChemiDoc XRS+ System (Bio-Rad). Uncropped images en in Extended Data Fig. 3.
Real-time quantitative PCR
Total RNA was extracted using the miRNeasy mini kit (Qiagen) and reverse transcribed using the high-capacity cDNA reverse transcription kit (Applied Biosystems) combined with RNase-Free DNase (Qiagen) treatment according to the manufacturer’s protocol. RT–qPCR was performed and analysed on ViiA7 (Applied Biosystems) using Power SYBR Green PCR Master Mix (Applied Biosystems; Supplementary Fig. S10A, Fig. 3I and Supplementary Fig. 10B, S8H) or TaqMan gene expression assay (Applied Biosystems; Supplementary Fig. S11A,E, I and S10C). The following primers and TaqMan probes (Applied Biosystems) were used to acquire expression data with the Viia 7 Real-Time PCR System (Applied Biosystems): Primers: HPRT1 (Fw: 5’ - CCTGGCGTCGTGATTAGTGAT - 3’; Rev: 5’ - AGACGTTCAGTCCTGTCCATA - 3’); OAS2 (Fw: 5’ - GGAGCTTCCTGATTGGCAGA - 3’; Rev: 5’ - ATGTAGGGT GGCAAGCACTG - 3’); MX1 (Fw: 5’ - CTGCACAGGTTGTTCTCAGC - 3’; Rev: 5’ - GTTTCCGAAGTGGACATCGCA - 3’); STAT1 (Fw: 5’ - CAGCTTGACTCAAAATTCCTGGA - 3’; Rev: 5’ - TGAAGATTACGCTTGCTTTTCCT - 3’); MAVS (Fw: 5’ - AGGAGACAGATGGAGACACA - 3’; Rev: 5’ - CAGAACTGGGCAGTACCC - 3’); Taqman Probes: PPIA (Hs04194521_s1); GAPDH (Hs99999905_m1); HPRT1 (Hs02800695_m1); MX1 (Hs00895608_m1); IFI44L (Hs00915292_m1); CCL2 (Hs00234140 m1); OAS2 (Hs00942643_m1); STAT1 (Hs01013996_m1); IL8 (Hs00174103_m1); IL1A (Hs00174092_m1); IL1B (Hs01555410_m1).
Mice
NOD.Prkdcscid.Il2rgnull (NSG) mice were bred and housed under specific pathogen-free conditions at the central animal facility of the German Cancer Research Center (DKFZ). Female mice were used for the studies. All animal experiments were approved by the governmental committee for animal experimentation (Regierungspräsidium Karlsruhe; G39/13, G115/19, G213/18).
Orthotopic experiment
For the orthotopic tumor growth of PDCs experiment (Fig. 3C,J,L) 200,000 cells were mixed with Matrigel (2mg/ml; BD) and injected into the mice’s pancreas. Engraftment of tumors and subsequent growth were monitored by regular palpation of the implantation site. For experiment shown in Fig. 3C mice were killed as soon as they reached abortion criteria defined in the procedure (human end point).
StC/PDC co-injection
50,000 IFNsignlow cells (PACO3) were mixed with 100,000 StC, which had been previously conditioned with StC or PDC medium and injected subcutaneously in HBSS. Mice were killed 90 days after injection.
Ruxolitinib treatment
100,000 IFNsignhigh (PACO2) or IFNsignlow (PACO3) PDCs were subcutaneously injected in mice. When tumors reached a size of approximately 50 mm3 (26 or 60 days respectively) mice were randomly assigned to each treatment group (n = 6 per group. One tumor from the PACO2 Ruxolitinib group was excluded from the analysis because no data on initial size of the tumor could be taken). Vehicle (1% DMSO in 0.1% NaCl) or Ruxolitinib (8.7 mg/Kg in 0.1% NaCl; TargetMol) were administered daily by peritoneal injection during 13 days. Mice were killed 3 days after last administration. Tumors were measured with a calliper and volume was calculated according to the formula (length x height x width) x (π/6). Fold increase was normalized to size at starting of treatment.
1° PDX
Primary xenografts were established by implanting 1–2mm3 pieces of patient tumors onto the pancreas of NSG mice.
Mouse PDAC cells
To generate mouse PDAC cell lines, tumors identified at necropsy in Sox9-CreER;KrasLSL-G12D;Trp53f/f;R26RYFP and Ptf1aCreER;KrasLSL-G12D;Trp53f/f;R26RYFP mice were dissociated and plated using previously described culture conditions (101). RNA was isolated from low passage cells grown on a collagen gel and RNA-sequencing libraries were generated using the TruSeq kit (Illumina) on a Neoprep system (Illumina) and then sequenced using a NextSeq device (UBC Biomedical Research Centre Next Generation Sequencing Service) at 30–50x coverage. Paired end reads were aligned to the mouse genome (UCSC mm9) using STAR alignment (114) and Fragments Per Kilobase of transcript, per Million mapped reads (FPKM) data was generated with Cufflinks on the Basespace (Illumina) platform.
Immunohistochemistry
Tumor specimens were fixed in 10% formalin overnight and embedded in paraffin. For immunohistochemistry, slides were deparaffinized and rehydrated. Antigen retrieval was enhanced by boiling in a steam pot at pH 6 in Dako Target Retrieval Solution (Dako) for 15 min, followed by cooling for 30 min and washing in distilled water. Nonspecific binding was blocked by using the Linaris Avidin/Biotin blocking Kit (Vector Labs) according to the manufacturer’s instructions. Slides were incubated with primary antibodies for 30 min, rinsed in PBS-T (PBS with 0.5% Tween-20), incubated for 20 min with the appropriate secondary antibody using the Dako REAL Detection System (Dako) and rinsed in PBS-T. After blocking of endogenous peroxidase and incubation with streptavidin–horseradish peroxidase (HRP) (20 min at room temperature), slides were developed with AEC (Dako) and counterstained with hematoxylin. All antibodies were diluted in Dako antibody diluent. Anti-STAT1 (dilution 1:100; HPA000982, Sigma), anti-SERPINB5 (dilution 1:75; HPA020136, Sigma).
Statistical analysis.
For the in vitro experiments, indicated biological replicates were used or grouped analyses were carried out. For the in vivo treatment experiments, six mice per group were used. Hence, for the reported differences, the sample size used gave sufficient power to call them reliable. For the in vivo co-injection the number of injected mice was chosen according to availability of cells and, therefore, grouped analysis was carried out. Quantitative results were analyzed by one-way analysis of variance (one-way ANOVA), two-sided unpaired Mann-Whitney U test or two-sided unpaired Student’s or nested t-test as indicated in the figure legends, using GraphPad Prism 8 software. For RNA-seq expression data of sorted epithelia cells two-tailed Wald test was calculated by DEseq2 package (117). Survival analysis was performed by using the Mantel-Cox log-rank test and the hazard-ratio (logrank) test using the GraphPad Prism 8 software. Estimation of variation within each group was determined by s.e.m. or 95% CI as indicated in the figure legends. Levels of significance were determined as follows: * p<0.05; ** p<0.01; *** p<0.001; **** p<0.0001.
Data availability
Gene expression microarray data of PDCs, StCs treated with CM and normalized Infinium MethylationEPIC data have been deposited at Gene Expression Omnibus database under accession numbers GSE134092, GSE134093, and GSE134217, respectively. Processed high-throughput data (i.e. full DMR calling and differential gene expression from RNA-seq data) are provided as supplementary tables. Sequence data and methylation raw files have been deposited at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession number EGAS00001004660. Other raw data are available upon reasonable request.
Extended Data
Extended Data Figure 1 -.
FACS plots
Extended Data Figure 2 -.
Consensus Clustering: Collect partition resluts from 20 tested methods
Extended Data Figure 3 -.
Scans of uncropped Western blot membranes with size marker
Supplementary Material
Significance:
The mutational landscapes of PDAC alone cannot explain the observed interpatient heterogeneity. We identified two PDAC subtypes characterized by differential DNA methylation, preserving traits from normal ductal/acinar cells associated to IFN-signaling. Our work suggests that epigenetic traits and the cell of origin contribute to PDAC heterogeneity.
Acknowledgements
We thank the EPZ-Biobank and surgery teams, especially M. Schenk and M. Fischer, for excellent technical assistance regarding tissue specimens, and M. Bähr for excellent technical assistance on TBWGBS library preparation. We thank the next-generation sequencing (NGS) unit of the Genomics Core Facility at DKFZ for providing NGS and related services, the microarray unit and all members of the Flow cytometry Core Facility at DKFZ for excellent support, the members of the Central Animal Laboratory at DKFZ for animal husbandry. We thank DKFZ-HIPO for technical support and funding. We thank ODCF, especially J. Kerssemakers for support on data deposition. We thank A. Descot, M. Saini, F. M. Zickgraf and C. Eisen for discussions and U. Rochlitzer with help translating the mouse experimental protocols.
Grant support:
This work was supported by: HIPO-015A (A.T., M.R.S., E.E., O.S., T.H., R.E., W.W); the Dietmar Hopp Foundation (A.T.); the German Ministry of Science and Education (BMBF) e:Med program for systems biology (PANC-STRAT consortium, grant no. 01ZX1305C and 01ZX1605C; A.T., M.R.S., R.E., O.S., and W.W.); the German Ministry of Science and Education (BMBF) (grant no. 031L0076A; B.B.); the DKFZ-NCT program NCT3.0 (A.T., M.R.S., M.S., R.E., N.A.G., and O.S. - NCT3.0_2015.17 PrecO-Panc); the DKTK Joint Funding Program (A.T., M.R.S., W.W.); the Pancreatic Cancer Canada Foundation Innovation Grant, Pancreas Centre BC, CIHR Open Operating and New Investigator grants to (J.L.K), and a University of British Columbia Faculty of Medicine Graduate Award (A.YL.L.); the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation, Project ID 329628492 – SFB 1321, S02P; K.S. and W.W.); the German Research Foundation (M.M.G.; GA 1818/ 2–1). The collection and processing of the specimens via PancoBank was supported by Heidelberger Stiftung Chirurgie. I.R. is a recipient of an Odysseus I fellowship of the Fund for Scientific Research Flanders (FWO). E.E. was a recipient of an EMBO long-term fellowship (ALTF 344–2013).
Abbreviations
- PDAC
Pancreatic ductal adenocarcinoma
- GSEA
Gene set enrichment analysis
- TBWGBS
Tagmentation-based whole genome bisulfite sequencing
- CG
CpG
- CGI
CpG island
- DMR
Differentially methylated region
- TSS
Transcription start site
- MC1
Methylation cluster 1
- MC2
Methylation cluster 2
- PDCs
Patient derived cells
- StCs
Stellate cells
Footnotes
Conflict of interest: The authors declare no conflict of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin Wiley; 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SGH, Hoadley KA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maurer C, Holmstrom SR, He J, Laise P, Su T, Ahmed A, et al. Experimental microdissection enables functional harmonisation of pancreatic cancer subtypes. Gut. 2019;68:1034–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elyada E, Bolisetty M, Laise P, Flynn WF, Courtois ET, Burkhart RA, et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov. 2019;9:1102–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernard V, Semaan A, Huang J, Anthony San Lucas F, Mulu FC, Stephens BM, et al. Single-cell transcriptomics of pancreatic cancer precursors demonstrates epithelial and microenvironmental heterogeneity as an early event in neoplastic progression. Clin Cancer Res. American Association for Cancer Research Inc.; 2019;25:2194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moncada R, Barkley D, Wagner F, Chiodin M, Devlin JC, Baron M, et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat Biotechnol Nature Research; 2020;38:333–42. [DOI] [PubMed] [Google Scholar]
- 7.Peng J, Sun BF, Chen CY, Zhou JY, Chen YS, Chen H, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res Springer US; 2019;29:725–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nones K, Waddell N, Song S, Patch AM, Miller D, Johns A, et al. Genome-wide DNA methylation patterns in pancreatic ductal adenocarcinoma reveal epigenetic deregulation of SLIT-ROBO, ITGA2 and MET signaling. Int J Cancer. 2014;135:1110–8. [DOI] [PubMed] [Google Scholar]
- 9.Vincent A, Omura N, Hong S-M, Jaffe A, Eshleman J, Goggins M. Genome-wide analysis of promoter methylation associated with gene expression profile in pancreatic adenocarcinoma. Clin Cancer Res. 2011;17:4341–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan AC, Jimeno A, Lin SH, Wheelhouse J, Chan F, Solomon A, et al. Characterizing DNA methylation patterns in pancreatic cancer genome. Mol Oncol Elsevier B.V; 2009;3:425–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lomberk G, Blum Y, Nicolle R, Nair A, Gaonkar KS, Marisa L, et al. Distinct epigenetic landscapes underlie the pathobiology of pancreatic cancer subtypes. Nat Commun Springer US; 2018;9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicolle R, Blum Y, Marisa L, Loncle C, Gayet O, Moutardier V, et al. Pancreatic Adenocarcinoma Therapeutic Targets Revealed by Tumor-Stroma Cross-Talk Analyses in Patient-Derived Xenografts. Cell Rep. 2017;21:2458–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cabezas-Wallscheid N, Klimmeck D, Hansson J, Lipka DB, Reyes A, Wang Q, et al. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell. 2014;15:507–22. [DOI] [PubMed] [Google Scholar]
- 14.Wang Q, Gu L, Adey A, Radlwimmer B, Wang W, Hovestadt V, et al. Tagmentation-based whole-genome bisulfite sequencing. Nat Protoc. 2013;8:2022–32. [DOI] [PubMed] [Google Scholar]
- 15.Schussler MH, Skoudy A, Ramaekers F, Real FX. Intermediate Filaments as Differentiation Markers of Normal Pancreas and Pancreas Cancer. Am J Pathol. 1992;140:559–68. [PMC free article] [PubMed] [Google Scholar]
- 16.Chromosome H, Hahn SA, Schutte M, Hoque ATMS, Moskaluk CA, Costa LT, et al. DPC4, A Candidate Tumor Suppressor Gene at. Science (80- ). 1994;107247:21–4. [DOI] [PubMed] [Google Scholar]
- 17.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol Nature Publishing Group; 2010;28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delacher M, Imbusch CD, Weichenhan D, Breiling A, Hotz-Wagenblatt A, Träger U, et al. Genome-wide DNA-methylation landscape defines specialization of regulatory T cells in tissues. Nat Immunol. 2017;18:1160–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlereth K, Weichenhan D, Bauer T, Heumann T, Giannakouri E, Lipka D, et al. The transcriptomic and epigenetic map of vascular quiescence in the continuous lung endothelium. Elife. eLife Sciences Publications Ltd; 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhasin MK, Ndebele K, Bucur O, Yee EU, Otu HH, Plati J, et al. Meta-analysis of transcriptome data identifies a novel 5-gene pancreatic adenocarcinoma classifier. Oncotarget. 2016;7:23263–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato N, Fukushima N, Maitra A, Matsubayashi H, Yeo CJ, Cameron JL, et al. Discovery of novel targets for aberrant methylation in pancreatic carcinoma using high-throughput microarrays. Cancer Res. 2003;63:3735–42. [PubMed] [Google Scholar]
- 22.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet Nature Publishing Group; 2003;34:267–73. [DOI] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. National Academy of Sciences; 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey P, Chang DK, Nones K, Johns AL, Patch A-M, Gingras M-C, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. Nature Publishing Group; 2016;531:47–52. [DOI] [PubMed] [Google Scholar]
- 26.Raphael BJ, Hruban RH, Aguirre AJ, Moffitt RA, Yeh JJ, Stewart C, et al. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2017;32:185–203.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Connor AA, Denroche RE, Jang GH, Lemire M, Zhang A, Chan-Seng-Yue M, et al. Integration of Genomic and Transcriptional Features in Pancreatic Cancer Reveals Increased Cell Cycle Progression in Metastases. Cancer Cell. 2019;35:267–282.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayashi A, Fan J, Chen R, Ho Y, Makohon-Moore AP, Lecomte N, et al. A unifying paradigm for transcriptional heterogeneity and squamous features in pancreatic ductal adenocarcinoma. Nat Cancer. Springer US; 2020;1:59–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan-Seng-Yue M, Kim JC, Wilson GW, Ng K, Figueroa EF, O’Kane GM, et al. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nat Genet. 2020;52:231–40. [DOI] [PubMed] [Google Scholar]
- 30.Puleo F, Nicolle R, Blum Y, Cros J, Marisa L, Demetter P, et al. Stratification of Pancreatic Ductal Adenocarcinomas Based on Tumor and Microenvironment Features. Gastroenterology. 2018;155:1999–2013.e3. [DOI] [PubMed] [Google Scholar]
- 31.Ehrlich M, Wang RY. Eukaryotic DNA. 1981;212. [DOI] [PubMed] [Google Scholar]
- 32.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. [DOI] [PubMed] [Google Scholar]
- 33.Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, et al. letters Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina – associated domains. Nat Genet Nature Publishing Group; 2011;44:40–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roulois D, Yau HL, Pugh TJ, Brien O, Carvalho DD De, Roulois D, et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts Article DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell. Elsevier Inc.; 2015;162:961–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell. 2015;162:974–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brocks D, Schmidt CR, Daskalakis M, Jang HS, Shah NM, Li D, et al. DNMT and HDAC inhibitors induce cryptic transcription start sites encoded in long terminal repeats. Nat Genet. 2017;49:1052–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leonova KI, Brodsky L, Lipchick B, Pal M, Novototskaya L, Chenchik A a, et al. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc Natl Acad Sci U S A. 2013;110:E89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moran S, Arribas C, Esteller M. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics. 2016;8:389–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noll EM, Eisen C, Stenzinger A, Espinet E, Muckenhuber A, Klein C, et al. CYP3A5 mediates basal and acquired therapy resistance in different subtypes of pancreatic ductal adenocarcinoma. Nat Med. 2016;22:278–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feig C, Gopinathan A, Neesse A, Chan DS, Cook N, Tuveson DA. The Pancreas Cancer Microenvironment. 2012;18:4266–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Augsten M Cancer-associated fibroblasts as another polarized cell type of the tumor microenvironment. Front Oncol. 2014;4:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Özdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25:719–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherman MH, Yu RT, Engle DD, Ding N, Atkins AR, Tiriac H, et al. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell. Elsevier Inc; 2014;159:80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, et al. Stromal Elements Act to Restrain, Rather Than Support, Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2014;25:735–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Djurec M, Graña O, Lee A, Troulé K, Espinet E, Cabras L, et al. Saa3 is a key mediator of the protumorigenic properties of cancer-associated fibroblasts in pancreatic tumors. Proc Natl Acad Sci U S A. 2018;115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Erez N, Truitt M, Olson P, Hanahan D. Article Cancer-Associated Fibroblasts Are Activated in Incipient Neoplasia to Orchestrate Tumor-Promoting Inflammation in an NF- k B-Dependent Manner. Cancer Cell. Elsevier Ltd; 2010;17:135–47. [DOI] [PubMed] [Google Scholar]
- 47.Öhlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS, Ponz-Sarvise M, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. The Rockefeller University Press; 2017;214:579–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biffi G, Oni TE, Spielman B, Hao Y, Elyada E, Park Y, et al. IL1-Induced JAK/STAT Signaling Is Antagonized by TGFβ to Shape CAF Heterogeneity in Pancreatic Ductal Adenocarcinoma. Cancer Discov. 2019;9:282–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi Y, Gao W, Lytle NK, Huang P, Yuan X, Dann AM, et al. Targeting LIF-mediated paracrine interaction for pancreatic cancer therapy and monitoring. Nature. Springer US; 2019;569:131–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kulis M, Queirós AC, Beekman R, Martín-Subero JI. Intragenic DNA methylation in transcriptional regulation, normal differentiation and cancer. Biochim. Biophys. Acta - Gene Regul. Mech 2013. page 1161–74. [DOI] [PubMed] [Google Scholar]
- 51.Kulis M, Heath S, Bibikova M, Queirós AC, Navarro A, Clot G, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44:1236–42. [DOI] [PubMed] [Google Scholar]
- 52.Oakes CC, Seifert M, Assenov Y, Gu L, Przekopowitz M, Ruppert AS, et al. DNA methylation dynamics during B cell maturation underlie a continuum of disease phenotypes in chronic lymphocytic leukemia. Nat Genet Nature Publishing Group; 2016;48:253–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bormann F, Rodríguez-Paredes M, Lasitschka F, Edelmann D, Musch T, Benner A, et al. Cell-of-Origin DNA Methylation Signatures Are Maintained during Colorectal Carcinogenesis. Cell Rep. 2018;23:3407–18. [DOI] [PubMed] [Google Scholar]
- 54.Rodríguez-Paredes M, Bormann F, Raddatz G, Gutekunst J, Lucena-Porcel C, Köhler F, et al. Methylation profiling identifies two subclasses of squamous cell carcinoma related to distinct cells of origin. Nat Commun Springer US; 2018;9:577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hovestadt V, Jones DTW, Picelli S, Wang W, Kool M, Northcott PA, et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature. 2014;510:537–41. [DOI] [PubMed] [Google Scholar]
- 56.Bouwens L, Pipeleers DG. Extra-insular beta cells associated with ductules are frequent in adult human pancreas. Diabetologia. 1998;41:629–33. [DOI] [PubMed] [Google Scholar]
- 57.Puri S, Folias AE, Hebrok M. Plasticity and dedifferentiation within the pancreas: Development, homeostasis, and disease. Cell Stem Cell. Elsevier Inc.; 2015;16:18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jennings RE, Berry AA, Strutt JP, Gerrard DT, Hanley NA. Human pancreas development. Dev Company of Biologists Ltd; 2015;142:3126–37. [DOI] [PubMed] [Google Scholar]
- 59.Baldan J, Houbracken I, Rooman I, Bouwens L. Adult human pancreatic acinar cells dedifferentiate into an embryonic progenitor-like state in 3D suspension culture. Sci Rep. 2019;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strobel O, Dor Y, Alsina J, Stirman A, Lauwers G, Trainor A, et al. In Vivo Lineage Tracing Defines the Role of Acinar-to-Ductal Transdifferentiation in Inflammatory Ductal Metaplasia. Gastroenterology. W.B. Saunders; 2007;133:1999–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guerra C, Schuhmacher AJ, Cañamero M, Grippo PJ, Verdaguer L, Pérez-Gallego L, et al. Chronic Pancreatitis Is Essential for Induction of Pancreatic Ductal Adenocarcinoma by K-Ras Oncogenes in Adult Mice. Cancer Cell. Cell Press; 2007;11:291–302. [DOI] [PubMed] [Google Scholar]
- 62.Navas C, Hernández-Porras I, Schuhmacher AJ, Sibilia M, Guerra C, Barbacid M. EGF Receptor Signaling Is Essential for K-Ras Oncogene-Driven Pancreatic Ductal Adenocarcinoma. Cancer Cell. Cell Press; 2012;22:318–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee AYL, Dubois CL, Sarai K, Zarei S, Schaeffer DF, Sander M, et al. Cell of origin affects tumour development and phenotype in pancreatic ductal adenocarcinoma. Gut. 2019;68:487–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ferreira RMM, Sancho R, Messal HA, Nye E, Spencer-Dene B, Stone RK, et al. Duct- and Acinar-Derived Pancreatic Ductal Adenocarcinomas Show Distinct Tumor Progression and Marker Expression. Cell Rep ElsevierCompany.; 2017;21:966–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bailey JM, Hendley AM, Lafaro KJ, Pruski MA, Jones NC, Alsina J, et al. P53 mutations cooperate with oncogenic Kras to promote adenocarcinoma from pancreatic ductal cells. Oncogene. Nature Publishing Group; 2016;35:4282–8. [DOI] [PubMed] [Google Scholar]
- 66.Von Figura G, Fukuda A, Roy N, Liku ME, Morris Iv JP, Kim GE, et al. The chromatin regulator Brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nat Cell Biol. Nature Publishing Group; 2014;16:255–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rooman I, Real FX. Pancreatic ductal adenocarcinoma and acinar cells: a matter of differentiation and development? Gut. 2012;61:449–58. [DOI] [PubMed] [Google Scholar]
- 68.Storz P. Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nat. Rev. Gastroenterol. Hepatol Nature Publishing Group; 2017. page 296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arda HE, Tsai J, Rosli YR, Giresi P, Bottino R, Greenleaf WJ, et al. A Chromatin Basis for Cell Lineage and Disease Risk in the Human Pancreas. Cell Syst Elsevier Inc.; 2018;7:310–322.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Houbracken I, De Waele E, Lardon J, Ling Z, Heimberg H, Rooman I, et al. Lineage tracing evidence for transdifferentiation of acinar to duct cells and plasticity of human pancreas. Gastroenterology. W.B. Saunders; 2011;141:731–741.e4. [DOI] [PubMed] [Google Scholar]
- 71.Moss J, Magenheim J, Neiman D, Zemmour H, Loyfer N, Korach A, et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun Springer US; 2018;9:5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ouyang H, Mou LJ, Luk C, Liu N, Karaskova J, Squire J, et al. Immortal human pancreatic duct epithelial cell lines with near normal genotype and phenotype. Am J Pathol. 2000;157:1623–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Furukawa T, Duguid W, Rosenberg L, Viallet J, Galloway D, Tsao M. Long-term culture and immortalization of epithelial cells from normal adult human pancreatic ducts transfected by the E6E7 gene of human papilloma virus 16. Am J Pathol. 1996;148:1763–70. [PMC free article] [PubMed] [Google Scholar]
- 74.Arda HE, Li L, Tsai J, Torre EA, Rosli Y, Peiris H, et al. Age-Dependent Pancreatic Gene Regulation Reveals Mechanisms Governing Human β Cell Function. Cell Metab Cell Press; 2016;23:909–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Enge M, Arda HE, Mignardi M, Beausang J, Bottino R, Kim SK, et al. Single-Cell Analysis of Human Pancreas Reveals Transcriptional Signatures of Aging and Somatic Mutation Patterns. Cell. Elsevier Inc.; 2017;171:321–330.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Palasantza A, Eliasson P, Kasper M, Sandberg R, Palasantza A, Eliasson P, et al. Single-Cell Transcriptome Profiling of Human Pancreatic Islets in Health and Type 2 Diabetes. Cell Metab. 2016;24:593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Muraro MJ, Dharmadhikari G, Gr??n D, Groen N, Dielen T, Jansen E, et al. A Single-Cell Transcriptome Atlas of the Human Pancreas. Cell Syst. 2016;3:385–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Monsurrò V, Beghelli S, Wang R, Barbi S, Coin S, Di Pasquale G, et al. Anti-viral state segregates two molecular phenotypes of pancreatic adenocarcinoma: potential relevance for adenoviral gene therapy. J Transl Med. 2010;8:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moerdyk-Schauwecker M, Shah NR, Murphy AM, Hastie E, Mukherjee P, Grdzelishvili VZ. Resistance of pancreatic cancer cells to oncolytic vesicular stomatitis virus: Role of type I interferon signaling. Virology. Elsevier; 2013;436:221–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, et al. Aberrant Overexpression of Satellite Repeats in Pancreatic and Other Epithelial Cancers. Science (80- ). 2011;331:593–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sheng W, LaFleur MW, Nguyen TH, Chen S, Chakravarthy A, Conway JR, et al. LSD1 Ablation Stimulates Anti-tumor Immunity and Enables Checkpoint Blockade. Cell. Elsevier Inc.; 2018;174:549–563.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Izquierdo-Bouldstridge A, Bustillos A, Bonet-Costa C, Aribau-Miralbés P, García-Gomis D, Dabad M, et al. Histone H1 depletion triggers an interferon response in cancer cells via activation of heterochromatic repeats. Nucleic Acids Res. 2017;45:11622–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Parker BS, Rautela J, Hertzog PJ. Antitumour actions of interferons: Implications for cancer therapy. Nat Rev Cancer. Nature Publishing Group; 2016;16:131–44. [DOI] [PubMed] [Google Scholar]
- 84.Kastenhuber ER, Lowe SW. Putting p53 in Context. Cell. Elsevier Inc.; 2017;170:1062–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Leonova KI, Brodsky L, Lipchick B, Pal M, Novototskaya L, Chenchik AA, et al. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc Natl Acad Sci U S A. 2013;110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Somerville TD, Biffi G, Daßler-Plenker J, Hur SK, He X-Y, Vance KE, et al. Squamous trans-differentiation of pancreatic cancer cells promotes stromal inflammation. Elife. 2020;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Calon A, Espinet E, Palomo-Ponce S, Tauriello DVF, Iglesias M, Céspedes MV, et al. Dependency of Colorectal Cancer on a TGF-β-Driven Program in Stromal Cells for Metastasis Initiation. Cancer Cell. 2012;22:571–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ishizuka JJ, Manguso RT, Cheruiyot CK, Bi K, Panda A, Iracheta-Vellve A, et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature. Nature Publishing Group; 2019;565:43–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cañadas I, Thummalapalli R, Kim JW, Kitajima S, Jenkins RW, Christensen CL, et al. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat Med. 2018;24:1143–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Goel S, Decristo MJ, Watt AC, Brinjones H, Sceneay J, Li BB, et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature. Nature Publishing Group; 2017;548:471–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Benci JL, Xu B, Qiu Y, Wu TJ, Dada H, Twyman-Saint Victor C, et al. Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell. Elsevier; 2016;167:1540–1554.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Minn AJ. Interferons and the Immunogenic Effects of Cancer Therapy. Trends Immunol Elsevier Ltd; 2015;36:725–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Habbe N, Shi G, Meguid RA, Fendrich V, Esni F, Chen H, et al. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad Sci U S A. National Academy of Sciences; 2008;105:18913–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Means AL, Meszoely IM, Suzuki K, Miyamoto Y, Rustgi AK, Coffey RJ, et al. Pancreatic epithelial plasticity mediated by acinar cell transdifferentiation and generation of nestin-positive intermediates. Development. The Company of Biologists Ltd; 2005;132:3767–76. [DOI] [PubMed] [Google Scholar]
- 95.Hassid B, Lucas AL, Salomao M, Weng C, Liu F, Khanna L, et al. Absence of pancreatic intraepithelial neoplasia predicts poor survival after resection of pancreatic cancer. Pancreas. Lippincott Williams and Wilkins; 2014;43:1073–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Oda Y, Aishima S, Morimatsu K, Shindo K, Fujino M, Mizuuchi Y, et al. Pancreatic intraepithelial neoplasia in the background of invasive ductal carcinoma of the pancreas as a prognostic factor. Histopathology. Blackwell Publishing Ltd; 2014;65:389–97. [DOI] [PubMed] [Google Scholar]
- 97.Miyazaki T, Ohishi Y, Miyasaka Y, Oda Y, Aishima S, Ozono K, et al. Molecular Characteristics of Pancreatic Ductal Adenocarcinomas with High-Grade Pancreatic Intraepithelial Neoplasia (PanIN) Are Different from Those without High-Grade PanIN. Pathobiology. S. Karger AG; 2017;84:192–201. [DOI] [PubMed] [Google Scholar]
- 98.Hurwitz HI, Uppal N, Wagner SA, Bendell JC, Beck JT, Wade SM, et al. Randomized, double-blind, phase II study of ruxolitinib or placebo in combination with capecitabine in patients with metastatic pancreatic cancer for whom therapy with gemcitabine has failed. J Clin Oncol. 2015;33:4039–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hurwitz H, Van Cutsem E, Bendell J, Hidalgo M, Li CP, Salvo MG, et al. Ruxolitinib + capecitabine in advanced/metastatic pancreatic cancer after disease progression/intolerance to first-line therapy: JANUS 1 and 2 randomized phase III studies. Invest New Drugs. Springer New York LLC; 2018;36:683–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Diaferia GR, Balestrieri C, Prosperini E, Nicoli P, Spaggiari P, Zerbi A, et al. Dissection of transcriptional and cis -regulatory control of differentiation in human pancreatic cancer. EMBO J. 2016;35:595–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Reichert M, Takano S, Heeg S, Bakir B, Botta GP, Rustgi AK. Isolation, culture and genetic manipulation of mouse pancreatic ductal cells. Nat Protoc. 2013;8:1354–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Weichenhan D, Wang Q, Adey A, Wolf S, Shendure J, Eils R, et al. Tagmentation-Based Library Preparation for Low DNA Input Whole Genome Bisulfite Sequencing. In: Tost J, editor. DNA Methylation Protoc New York, NY: Springer New York; 2018. page 105–22. [DOI] [PubMed] [Google Scholar]
- 103.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hansen KD, Langmead B, Irizarry RA. BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol. 2012;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Park Y, Wu H. Differential methylation analysis for BS-seq data under general experimental design. Bioinformatics. 2016;32:1446–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Heberle H, Meirelles VG, da Silva FR, Telles GP, Minghim R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. BioMed Central Ltd.; 2015;16:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–9. [DOI] [PubMed] [Google Scholar]
- 109.Klambauer G, Schwarzbauer K, Mayr A, Clevert DA, Mitterecker A, Bodenhofer U, et al. Cn.MOPS: Mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic Acids Res. 2012;40:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gu Z, Eils R, Schlesner M. HilbertCurve: An R/Bioconductor package for high-resolution visualization of genomic data. Bioinformatics. Oxford University Press; 2016;32:2372–4. [DOI] [PubMed] [Google Scholar]
- 111.Assenov Y, Müller F, Lutsik P, Walter J, Lengauer T, Bock C. Comprehensive analysis of DNA methylation data with RnBeads. Nat Methods. 2014;11:1138–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cabezas-Wallscheid N, Klimmeck D, Hansson J, Lipka DB, Reyes A, Wang Q, et al. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell. 2014;15:507–22. [DOI] [PubMed] [Google Scholar]
- 114.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Anders S, Pyl PT, Huber W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liao Y, Smyth GK, Shi W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30. [DOI] [PubMed] [Google Scholar]
- 119.Somerville TDD, Xu Y, Miyabayashi K, Tiriac H, Cleary CR, Maia-Silva D, et al. TP63-Mediated Enhancer Reprogramming Drives the Squamous Subtype of Pancreatic Ductal Adenocarcinoma. Cell Rep. 2018;25:1741–1755.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nagy Á, Lánczky A, Menyhárt O, Gyorffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5:1704–13. [DOI] [PubMed] [Google Scholar]
- 123.Knott SR V, Maceli A, Erard N, Chang K, Marran K, Zhou X, et al. A computational algorithm to predict shRNA potency. Mol Cell. 2015;56:796–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schonborn J, Oberstraß J, Breyel E, Tittgen J, Schumacher J, Lukacs N. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res. 1991;19:2993–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene expression microarray data of PDCs, StCs treated with CM and normalized Infinium MethylationEPIC data have been deposited at Gene Expression Omnibus database under accession numbers GSE134092, GSE134093, and GSE134217, respectively. Processed high-throughput data (i.e. full DMR calling and differential gene expression from RNA-seq data) are provided as supplementary tables. Sequence data and methylation raw files have been deposited at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession number EGAS00001004660. Other raw data are available upon reasonable request.