Abstract
Background
To date, studies into the bone marrow (BM) immune microenvironment have been limited due to reliance on the analysis of BM aspirates in which the microenvironmental context is lost. GeoMX™ digital spatial profiling (DSP) is a new technique developed for the analysis of formalin-fixed paraffin-embedded tissue samples which allows high multiplex analysis of protein expression in multiple user-defined regions within a tissue section. We examined the applicability of this technique to the analysis of protein expression in diagnostic BM trephine samples.
Materials and methods
Archival BM trephines were obtained from patient groups (normal, myelodysplasia and aplastic anaemia). Regions of interest in each section were identified by dual CD3+/CD45+ immunohistochemistry staining to identify immune infiltrates, and DSP was applied.
Results
Due to variability in cell number within regions of interest and differing cellular composition of the BM trephines, raw protein expression counts were normalised by internal controls and nuclei count to determine the expression level of each protein within each region of interest. In heat map analysis using Spearman's rank correlation, aplastic anaemia samples clustered away from both normal and myelodysplasia samples, demonstrating significant differences in their BM immunology.
Conclusions
GeoMX™ DSP is an innovative new technique that, for the first time, allows the analysis of archival BM trephines at an unprecedented level of detail. It will allow investigations in large cohorts of patients with haematological malignancies to identify new biomarkers, new mechanisms of disease pathogenesis and new drug targets.
Key words: bone marrow, digital spatial profiling, GeoMX, immune microenvironment, immunohistochemistry, trephine
Highlights
-
•
Bone marrow trephines are a ready source of archival material.
-
•
Processing methods have limited their use in translational research.
-
•
GeoMX™ digital spatial profiling allows high multiplex analysis of bone marrow.
-
•
Large high-powered retrospective studies of bone marrow in blood cancers are now possible.
Introduction
Immunity, cancer biology and therapy are intimately linked.1 Identification of immune signatures within archival tumour biopsies has led to an appreciation of the prognostic impact of host immune responses, and ultimately to the identification of targets such as PD-1/PD-L1 and CTLA-4 for the development of immunotherapy and clinical application.2 Whilst host immunity has been implicated in the pathogenesis, prognosis and mechanisms of action of drug therapy in haematological malignancies,3, 4, 5, 6, 7, 8 analysis of the tumour microenvironment in haematological conditions in primary patient samples has been limited. To date, assessments of immune function in haematological conditions have been limited to the study of mouse models or primary patient peripheral blood samples or bone marrow (BM) aspirates, which do not fully represent the BM microenvironment due to physical disruption and/or variable degrees of peripheral blood contamination.9, 10, 11
BM trephines are collected routinely from the majority of patients with either malignant or non-malignant haematological disorders as part of their diagnostic assessment. While for some conditions, such as chronic myeloid leukaemia and acute lymphoblastic leukaemia, regular BM trephines have been replaced by molecular techniques for disease monitoring,12 BM trephines are often used for therapeutic monitoring for disorders without a readily identifiable and tractable molecular lesion, such as aplastic anaemia or myelofibrosis. Therefore, there is a large sample archive of BM trephines collected from patients at diagnosis and throughout their disease course which represents a largely untapped sample set for microenvironmental analysis. However, current diagnostic and research analyses are limited to anatomic morphology and semi-quantitative immunohistochemistry (IHC) analysis, thus limiting the detailed analysis of this sample set.
In solid malignancies, formalin-fixed paraffin-embedded (FFPE) pathology specimens have been a source of primary patient tissue for genomic studies including gene expression arrays.13 However, BM trephines are not processed as standard FFPE tissue. They require a decalcification step, usually using acid-based solutions, which fragments genomic material,14 limiting the research potential of these samples. Whilst DNA or RNA isolation has been achieved successfully in some studies where EDTA decalcification has been used,15,16 this is not a standard process in many diagnostic pathology laboratories due to the increased processing time,17 and is therefore not an option for many sample sets. In addition, genomic studies are of limited use in examining the spatial context of cells/tissues, and are an impractical means of undertaking large cohort analysis of the BM microenvironment due to the requirement for substantial amounts of input material.
Multiplex IHC on archival BM trephines may be a means to allow these samples to be used in translational studies. Indeed, in research applications, multiplex IHC has been applied successfully to solid organ biopsies with over 60 colour IHC described in colorectal cancer FFPE tissue,18 although most studies described to date are limited to ≤12 colours due to secondary antibody specificity requirements and spectral overlap of directly conjugated fluorescent primary antibodies. Other systems including Opal™ (PerkinElmer, Inc., Waltham, MA) overcome issues of secondary antibody species cross-reactivity but are currently limited to seven colour analyses. As these systems have developed, there has been a concurrent improvement in quantification and image analysis systems,19 such that these analyses are now quantitative across a large range, moving beyond the semi-quantitative methods widely used in diagnostic pathology. The use of metal-ion-tagged antibodies combined with mass spectroscopy in breast cancer FFPE tissue has overcome overlapping emission spectra and increased multiplex capacity to over 100 targets,20,21 but at the cost of very low throughput (24 h per sample) and destruction of the tissue sample.
The limitations of existing approaches have led to the recent development of a multiplex IHC technique using antibodies tagged with DNA barcodes via an ultraviolet (UV)-cleavable linker. Developed by NanoString Technologies, Inc. (Seattle, WA), GeoMX™ digital spatial profiling (DSP) allows the simultaneous digital quantitative analysis of over 80 proteins within user-defined regions of interest (ROIs), which are selected using standard fluorescent IHC on a single slide leaving the sample undamaged.22,23 The DSP technique was initially utilised on FFPE biopsy samples of solid tumours.24, 25, 26 Realising its potential to overcome the limitations of existing genomic or IHC approaches in the analysis of archival BM trephines, we examined, for the first time, the applicability of DSP to the analysis of the immune microenvironment in archival BM trephine samples.
Materials and methods
Patient samples
This study was approved under a waiver of consent by the Melbourne Health Human Research Ethics Committee. At the time of sample collection (2012–2015), BM trephines were processed using standard diagnostic laboratory practice of fixation in B5 for a minimum of 2 h, decalcification for 2 h in 10% formic acid/3% hydrochloric acid and paraffin embedding. Four-micrometre sections were obtained from identified archival BM trephine blocks from Melbourne Health Pathology. Cellularity and differential count data were obtained from the pathology reports on each sample.
GeoMX™ digital spatial profiling
BM trephine sections underwent DSP at NanoString Technologies laboratories under an early technology access programme, as described recently.22,26,27 ROI selection and subsequent statistical and correlative analyses were undertaken in the Australian Cancer Research Foundation translational research laboratory. Briefly, a multiplexed cocktail of primary antibodies, each with a unique, UV-photocleavable indexing oligo (Table 1), mouse anti-human CD3E antibody clone UMAB54 (Origene Technologies, Inc., Rockville, MD) conjugated to AF647 using AF647 antibody labelling kit (Thermo Fisher Scientific, Waltham, MA), rabbit anti-human CD45 antibody clone D9M81 (Cell Signalling Technology, Inc., Danvers, MA) conjugated to AF594 using AF594 antibody labelling kit (Thermo Fisher Scientific) and Syto83 nucleic acid stain (Thermo Fisher Scientific), was applied to a slide-mounted BM trephine tissue section. The tissue area of interest was located using fluorescence imaging, and ROIs 12 × 300 μm in diameter were selected based on dual CD3/CD45 staining to optimise assessment of immune infiltration of the BM microenvironment. ROIs were processed sequentially by focusing UV light through each ROI, and the released indexing oligos were aspirated. Once all ROIs were processed, indexing oligos were hybridised to NanoString optical barcodes for ex-situ digital counting and subsequently analysed with an nCounter® Analysis System (NanoString Technologies, Inc.). Mean CD3 and CD45 fluorescence in ROIs was determined using ImageJ 1.52q.
Table 1.
Markers used for digital spatial profiling analysis
| AKT | CD14 | CD45 | Granzyme B | Pan-CK | STAT3 |
| B7-H3 | CD19a | CD45RO | Histone H3 | PD-1 | STINGb |
| B7-H4b | CD163b | CD56 | HLA-DRb | PD-L1 | VISTA |
| Bcl-2 | CD20 | CD66bb | Ki67 | p-STAT3 | Mouse IgG |
| Beta-2-microglobulin | CD3 | CD68 | ICOSb | PTEN | Rabbit IgG |
| Beta-catenin | CD4 | CD8a | IDO-1b | OX40Lb | |
| CD11c2 | CD44 | FoxP3 | p-AKT | S6 |
AA, aplastic anaemia; MDS, myelodysplasia.
Analysed in normal, MDS1 and MDS2.
Analysed in AA1 and AA2 alone.
Housekeeping protein normalisation
Digital counts were analysed as follows: raw counts were first normalised to the average geometric mean of the internal spike in controls to account for system variation, and normalised to the geometric mean of histone H3 and S6 across all ROIs.
Nuclei count normalisation
For each ROI, total nuclei were counted using automated segmentation of fluorescent images. Briefly, each 16-bit nuclear image was converted to an 8-bit image. A binary mask was generated using the minimum cross entropy global thresholding method.28 In each 8-bit grayscale image, all of the local maxima were extracted and used as seeds in the marker-based watershed algorithm29 that separated cells within the same mask compartment. Small particles were removed after processing. All nuclei counts were compared with the original image, and only those that were confirmed were used for subsequent analysis. Digital counts were analysed as follows: raw counts were first normalised by the average geometric mean of the internal spike in controls to account for system variation, and values were normalised by nuclei count for each ROI.
Statistical analysis
Statistical analysis was performed using Prism Version 8.0.0 (GraphPad Software, Inc., La Jolla, CA). Volcano plots comparing samples were analysed using the Holm-Sidak method with alpha = 0.05. Differential expression of individual antigens was performed using Kruskal–Wallis analysis of variance with Dunn's multiple comparisons. Heat map analysis was performed using Heatmapper.30 ROIs were clustered according to average linkage and distance measured using Spearman's rank correlation.
Results
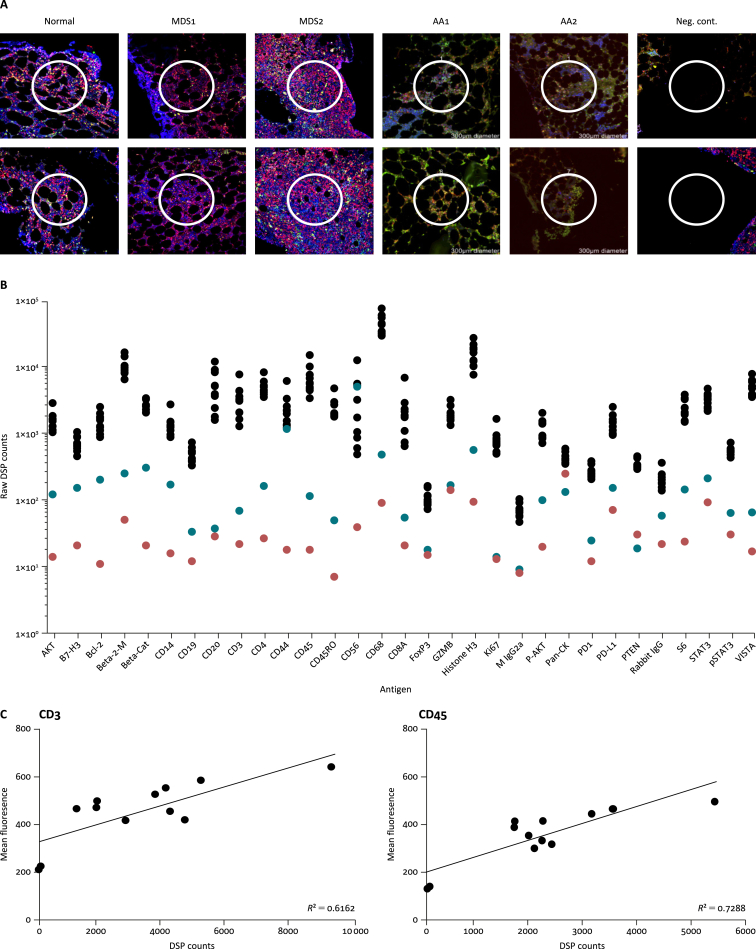
In a preliminary pilot study, archival diagnostic BM trephines were analysed from one patient with normal BM and two patients with myelodysplasia (MDS). The cellularity of these samples varied from 30% to 70% (Table 2). To focus the analysis on the immune microenvironment, 12 × 300-μm ROIs were selected in each sample based on dual CD45/CD3 staining (Figure 1A). The DSP panel used (Table 1) covers antigens of various immune subsets (e.g. CD14 – monocytes; CD3, CD4, CD8A – T cells; CD68 – M1 macrophages; FoxP3 – T regulatory cells). Raw counts from the DSP analysis ranged from 0 to 183 000, allowing determination of protein expression across a wide dynamic range (Figure 1B). To determine the level of non-specific staining, ROIs in the bony trabeculum and empty glass slide were included. For most antigens, the counts in the control ROIs were below the counts of the tissue ROIs. CD44 and CD56 counts in the decalcified trabeculum ROIs were similar to the tissue ROIs, suggesting that these antigens are not affected by the decalcification process. There were also detectable DSP counts in the glass slide ROI, suggesting a level of background staining that is variable between antibodies. As these samples are archival, confirmation of the level of marker expression using alternate samples (such as BM aspirates) was not possible in this study. However, correlation between DSP and alternative analysis methods has been described recently, indicating strong correlation with standard IHC techniques.23,27 Indeed, when the mean CD3 and CD45 counts of the ROIs in the healthy BM sample were compared with DSP counts, good correlation was seen for both antigens (Figure 1C).
Table 2.
Patient and sample characteristics
| Normal | MDS1 | MDS2 | AA1 | AA2 | |
|---|---|---|---|---|---|
| Age at biopsy (years) | 22 | 60 | 56 | 57 | 25 |
| Cellularity (%) | 50 | 30 | 70 | 5 | <10 |
| Differential count (%) | |||||
| Immature myeloid cells | 23.0 | 31.0 | 49.0 | 4.0 | 20.0 |
| Neutrophils | 38.5 | 15.0 | 25.5 | 1.5 | 3.5 |
| Erythroid cells | 13.0 | 39.0 | 10.0 | 88.0 | 58.0 |
| Lymphocytes | 18.0 | 12.5 | 4.5 | 5.0 | 13.0 |
| Monocytes | 2.5 | 1.5 | 10.0 | 0.0 | 1.0 |
| Other | 5.0 | 1.0 | 1.0 | 1.5 | 4.5 |
AA, aplastic anaemia; MDS, myelodysplasia.
Figure 1.
Analysis of bone marrow (BM) trephines using digital spatial profiling (DSP).
(A) Representative regions of interest (ROIs) from each sample in the study. Using fluorescent immunohistochemistry [CD3 (green), CD45 (red) and nuclear stain Syto83 (blue)], 300-μm ROIs were selected in each tissue, and the expression levels of 31 proteins within each were determined by staining with nCounter barcode-tagged antibodies. (B) Raw DSP counts in the normal BM trephine. ROIs were placed within the tissue (black), decalcified trabeculum (blue) and blank slide (red) to determine the background level of staining for each antigen. (C) Correlation between ROI mean fluorescence and normalised DSP counts in the normal BM trephine. Neg. cont., negative control; Beta-2-M, beta-2-microglobulin; Beta-Cat, beta-catenin; GZMB, granzyme B; M IgG2a, mouse IgG2a; Pan-CK, pan-cytokeratin.
To determine the applicability of DSP to diseases with severe hypocellularity or where immune populations are decreased, two samples from patients with aplastic anaemia (AA) were analysed. These BM trephines had cellularities of 5% and <10%, respectively (Table 2). DSP was performed using CD3/CD45/nuclear fluorescent IHC markers with a DSP panel. Twelve 300-μm ROIs were selected for DSP and, despite the very low cellularity, DSP was successful with raw counts ranging from 0 to 166 000, demonstrating the sensitivity of this technique.
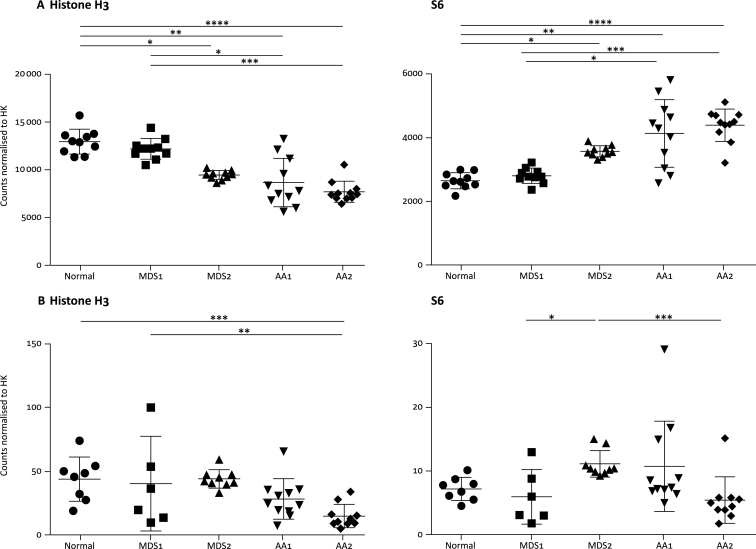
The high degree of variation in the cellularity of samples with ROIs of consistent sizes necessitated normalisation of the raw data. Normalisation by housekeeping proteins included in the DSP panel, histone H3 and S6 (Figure 2A), was not effective at normalising expression between samples, with a reciprocal relationship between the expression of these two proteins after normalisation. This may have been due to the heterogeneous nature of the tissue [especially differences in the proportions of neutrophil and erythroid populations (Table 2)], and therefore normalisation by nuclei count was performed. In instances where the nuclei count on image analysis did not agree with the visual count, these ROIs were excluded from downstream analysis. Nuclei normalisation in ROIs with confirmed counts resulted in more consistent expression levels of histone H3 and S6 (Figure 2B), suggesting that normalisation by nuclei count rather than by expression of housekeeping proteins will be useful in any tissues where there is a mixed cellular composition, or changes in cellular composition between comparator groups are predicted.
Figure 2.
Expression levels of housekeeping proteins histone H3 and S6 after normalisation.
(A) Digital spatial profiling (DSP) counts were normalised by internal controls and normalised by expression levels of housekeeping proteins. (B) DSP counts were normalised to housekeeping (HK) protein expression and normalised to nuclei count. Kruskal–Wallis analysis of variance with Dunn's multiple comparisons *≤0.05, **≤0.01, ***≤0.001, ****≤0.0001.
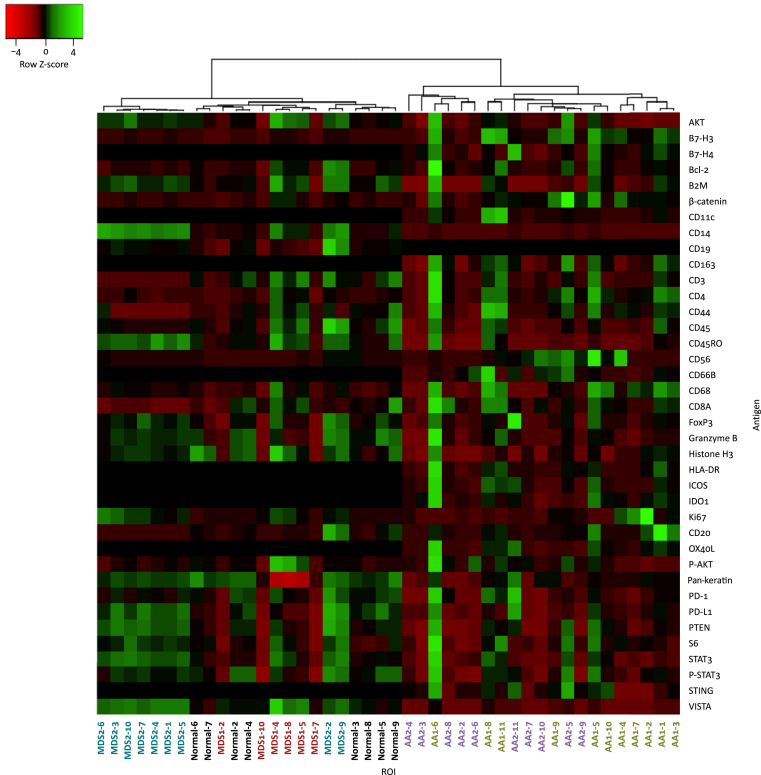
Heat map analysis using Spearman's rank correlation of the normalized data (Figure 3) showed that both AA samples had a distinct expression pattern and clustered together to the right, separate from both normal and MDS BM samples on the left. These results show that BM immunology in AA is significantly different from that present in either normal or MDS BM.
Figure 3.
Hierarchical clustering of nuclei normalised digital spatial profiling counts according to average linkage and Spearman's rank correlation.
For ease of visualisation on the x-axis, regions of interest (ROIs) from the same patient are the same colour. Black, normal; red, MDS1; blue, MDS2; green, AA1; purple, AA2.
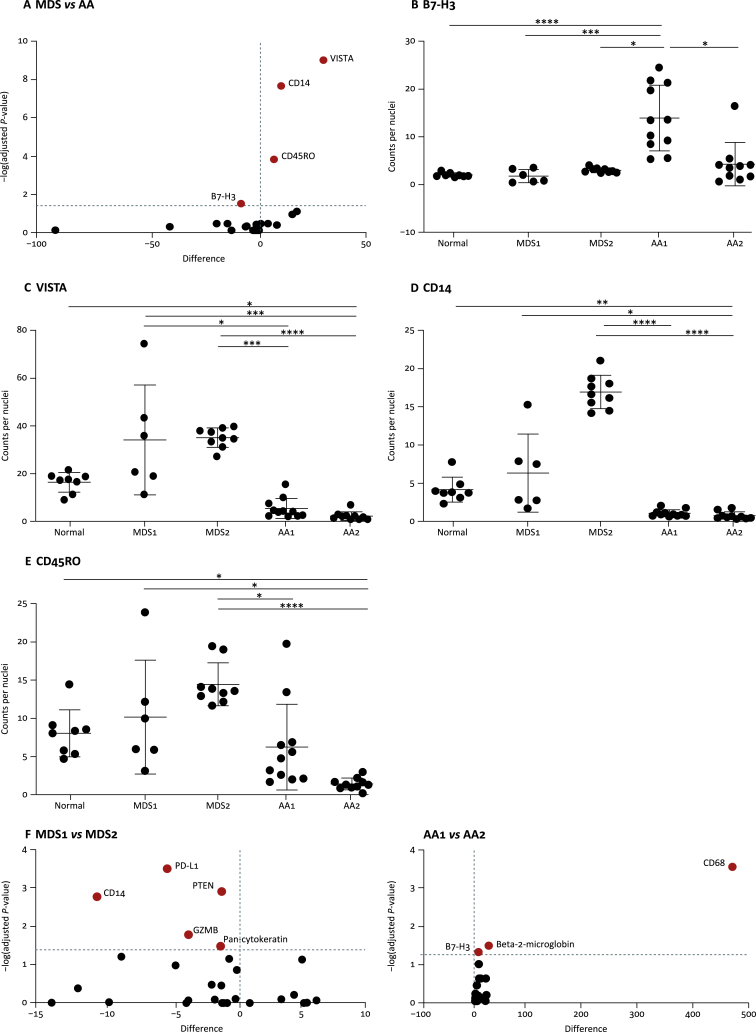
Volcano plot analysis was used to compare marker expression between AA and MDS samples (Figure 4A). AA samples had significantly increased expression of B7-H3 in one sample (Figure 4B), and significantly decreased expression of VISTA, CD14 and CD45RO in both samples compared with the MDS and normal samples (Figure 4C–E). Importantly, the variation in CD14 expression by DSP indicates differences in the proportion of monocytes, which is in agreement with the differential count performed when the sample was collected (Table 2). Differences could also be detected between the two MDS and AA samples using Volcano plot analysis (Figure 4F), with variation in the expression of PD-L1, PTEN, CD14 and granzyme B identified in MDS samples, and variation in the expression of CD68, beta-2-microglobulin and B7-H3 identified in AA samples, demonstrating important variations in disease immunology between patients with the same disease background.
Figure 4.
Nuclei normalised marker expression levels between samples.
(A) Volcano plot showing differential expression between patients with myelodysplasia (MDS) and aplastic anaemia (AA). The dotted horizontal line represents the adjusted P-value cut-off. Statistical significance determined using the Holm-Sidak method with alpha = 0.05. Points in red are those that achieved statistical significance. (B–E) Expression levels of B7-H3, VISTA CD14 and CD45RO across samples. Each point represents a region of interest within the sample. Kruskal–Wallis analysis of variance with Dunn's multiple comparisons *≤0.05, **≤0.01, ***≤0.001, ****≤0.0001. (F) Volcano plots showing differential expression within patients with MDS and AA. The dotted horizontal line represents the adjusted P-value cut-off. Statistical significance determined using the Holm-Sidak method with alpha = 0.05. Points in red are those that achieved statistical significance.
Discussion
The identification of microenvironmental immune markers within BM as either determinants of prognosis or as potential targets for therapy has been hampered by the lack of a readily applicable analysis platform for the multiplex assessment of BM trephines. While BM aspirates are available, they are logistically difficult to collect in large numbers, only allow the analysis of cell intrinsic factors, often have variable degrees of contamination with peripheral blood, and the microenvironmental context of the disease is lost. In contrast, archival BM trephine tissue exists in every single pathology department around the world with accompanying complete outcome data, making them an ideal resource for translational research. This study examined the feasibility of applying multiplex IHC using GeoMX™ DSP to analyse archival diagnostic BM trephine samples fixed in B5 and decalcified in an acid solution. Importantly, this study has shown that these archival samples, collected from patients between 2012 and 2015, can be used for analysis of the BM microenvironment, and demonstrated that specialised processing and storage of trephine samples at the time of collection is not required. The thousands of BM trephines stored worldwide with their accompanying patient outcome data provide the opportunity to perform retrospective analysis of the BM microenvironment in haematological diseases to provide a deeper understanding of disease biology, and identify new biomarkers and therapeutic targets which could be validated subsequently in prospective studies.
This study explored internal normalisation methods across samples. Despite the consistent size of selected ROIs, the variable cellularity of the samples (5–70%) meant that the number of cells analysed differed significantly within the individual ROIs of each sample and collectively between each sample. While the DSP panel includes the proteins histone H3 and S6 for normalisation, this study found that they were ineffective for normalising the data. This was possibly due to significant variation in the expression of ribosomal proteins across cells of different blood lineages,31 including between neutrophils and erythroid cells which showed a high level of variation between samples in this study. In addition, electron microscopy of AA BM has shown that erythroid cells have significant nuclear injury (pyknosis, karyolysis and karyorrhexis), with nuclear irregularities also seen in monocytes/macrophages and granulocytes,32 which may affect the levels of histone H3 in AA samples. Taken together, this demonstrates that both tissue heterogeneity and disease pathology may influence the effectiveness of normalisation by protein expression. In contrast, normalisation by nuclei count within the ROI is not biased by either of these issues, and effectively normalised expression between the different disease states. Therefore, we recommend that nuclei count should be used for normalisation for any comparisons within disease states (e.g. responders vs non responders), between disease stages (e.g. diagnosis vs multiply relapsed refractory) or between diagnostic groups as these states will likely affect the protein expression and cellular composition of the BM compartment.
Although analysis of these five samples was primarily aimed at testing the technical feasibility of the DSP analysis platform in BM trephine sample analysis, this study also demonstrated that this technique can provide insights into microenvironmental differences in patients with haematological disorders. For example, changes in CD45RO and CD14 expression were found in samples from patients with MDS, suggesting that alterations in T-cell memory and monocyte lineages may drive different clinical outcomes of MDS. Increased monocyte populations have been described previously as a biomarker of disease progression in MDS.33 In AA samples, upregulation of the costimulatory molecule B7-H3 and downregulation of the T-cell checkpoint molecule VISTA suggests an ongoing environment of excess costimulation and a deficiency of downregulation of T-cell immune checkpoints, respectively. VISTA knockout mice have been described to have an accumulation of spontaneously activated T cells with hyper-production of inflammatory cytokines and a lower threshold for self-antigens.34 The present study is the first to show loss of VISTA expression in AA samples, and may represent a mechanism of disease pathogenesis where T cells of AA patients become – and are allowed to remain – self-reactive, resulting in the destruction of hematopoietic stem cells. Given the heterogeneity between patients and diseases, these findings clearly require examination in larger, specifically designed, appropriately powered studies and validation using alternative methods. However, despite this, the study findings indicate that DSP has the utility as a potentially powerful means to undertake detailed analysis of archival BM trephines.
Compared with other multiplex IHC techniques in what is a rapidly evolving field, DSP does not require that the samples undergo multiple rounds of chemical stripping necessary in the use of conventional multicolour IHC which can lead to sample degradation. The high multiplicity of analysis is complemented by the ability to have any antibody labelled for DSP analysis, allowing customisation for a wide range of targets and diseases. The major limitation of the DSP technique is the inability to obtain single cell resolution without specifically placing an ROI on each cell. The recently described CODEX technology,20 which combines both single cell resolution and high multiplex IHC, may overcome this current limitation. Direct comparison between these techniques in any tissue is yet to be described and would be of great interest.
Selection of the tissue samples and the ROIs for DSP analysis requires careful consideration. In diseases that cause low cellularity of the BM (such as AA), finding sufficient regions for analysis can be challenging; therefore, care must be taken during initial sample selection to ensure the suitability of the tissue for analysis. Furthermore, the tissue should be assessed to determine the level of peripheral blood contamination, which may artificially increase the counts of some markers, and the level of tissue preservation, which may affect the ability to determine the nuclei count. For these reasons, examination of a tissue section stained with haematoxylin and eosin is advisable prior to DSP analysis.
The output digital count data from DSP can be manipulated using conventional bioinformatics techniques developed for the analysis of microarray and gene expression data. However, in order to fully utilise this technology, statistical methods that account for the multiple analysis regions per sample need to be developed. These regions are neither repeated measures nor independent samples, and methods that account for this need to be developed.
GeoMX™ DSP will allow, for the first time, analysis of BM trephines from retrospective cohorts of uniformly treated patients at every stage of their treatment with full outcome data for which cryopreserved BM samples are limited or unavailable. It adds to the toolbox of techniques available to haematological researchers for studies into new biomarkers, new drug targets and new mechanisms of disease response, leading to improved treatment choices and personalised treatments for all patients with haematological diseases.
Availability of data
Raw data for this study can be accessed at https://doi.org/10.17632/92835h4bz7.1.
Acknowledgements
The authors wish to thank Melbourne Health Pathology for their assistance in sourcing the samples used in this study. The authors would also like to thank Alison Van Schoiack and Yan Liang from NanoString Technologies, Inc. for technical assistance.
Funding
This work was supported, in part, by NanoString Technologies, Inc. under an Early Technology Access Program and by Fight Cancer Foundation, Australia (no grant number).
Disclosure
NanoString Technologies, Inc. provided RK with a travel stipend to present this work at an international conference which did not influence the content or conclusions of this paper.
References
- 1.Ogino S., Galon J., Fuchs C.S., Dranoff G. Cancer immunology – analysis of host and tumor factors for personalized medicine. Nat Rev Clin Oncol. 2011;8:711–719. doi: 10.1038/nrclinonc.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoos A. Development of immuno-oncology drugs – from CTLA4 to PD1 to the next generations. Nat Rev Drug Discov. 2016;15:235–247. doi: 10.1038/nrd.2015.35. [DOI] [PubMed] [Google Scholar]
- 3.Wang C., Yang Y., Gao S., et al. Immune dysregulation in myelodysplastic syndrome: clinical features, pathogenesis and therapeutic strategies. Crit Rev Oncol Hematol. 2018;22:123–132. doi: 10.1016/j.critrevonc.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 4.Kawano Y., Roccaro A.M., Ghobrial I.M., Azzi J. Multiple myeloma and the immune microenvironment. Curr Cancer Drug Targets. 2017;17:806–818. doi: 10.2174/1568009617666170214102301. [DOI] [PubMed] [Google Scholar]
- 5.Glenthoj A., Orskov A.D., Hansen J.W., Hadrup S.R., O'Connell C., Gronbaek K. Immune mechanisms in myelodysplastic syndrome. Int J Mol Sci. 2016;17(6):944. doi: 10.3390/ijms17060944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tamura H. Immunopathogenesis and immunotherapy of multiple myeloma. Int J Hematol. 2018;107:278–285. doi: 10.1007/s12185-018-2405-7. [DOI] [PubMed] [Google Scholar]
- 7.Sutton L.A., Rosenquist R. The complex interplay between cell-intrinsic and cell-extrinsic factors driving the evolution of chronic lymphocytic leukemia. Semin Cancer Biol. 2015;34:22–35. doi: 10.1016/j.semcancer.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 8.Forconi F., Moss P. Perturbation of the normal immune system in patients with CLL. Blood. 2015;126:573–581. doi: 10.1182/blood-2015-03-567388. [DOI] [PubMed] [Google Scholar]
- 9.Dean J., McCarthy D., Golden-Mason L., O'Farrelly C. Trephine biopsies are enriched for activated T/NK cells and cytotoxic T cells. Immunol Lett. 2005;99:94–102. doi: 10.1016/j.imlet.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 10.Batinic D., Marusic M., Pavletic Z., et al. Relationship between differing volumes of bone marrow aspirates and their cellular composition. Bone Marrow Transplant. 1990;6:103–107. [PubMed] [Google Scholar]
- 11.Loken M.R., Chu S.C., Fritschle W., Kalnoski M., Wells D.A. Normalization of bone marrow aspirates for hemodilution in flow cytometric analyses. Cytometry B Clin Cytom. 2009;76:27–36. doi: 10.1002/cyto.b.20429. [DOI] [PubMed] [Google Scholar]
- 12.Garces-Eisele J. Molecular biology strategies to detect residual disease. Hematology. 2012;17(Suppl. 1):S66–S68. doi: 10.1179/102453312X13336169155691. [DOI] [PubMed] [Google Scholar]
- 13.Scott D.W., Wright G.W., Williams P.M., et al. Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood. 2014;123:1214–1217. doi: 10.1182/blood-2013-11-536433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wickham C.L., Sarsfield P., Joyner M.V., Jones D.B., Ellard S., Wilkins B. Formic acid decalcification of bone marrow trephines degrades DNA: alternative use of EDTA allows the amplification and sequencing of relatively long PCR products. Mol Pathol. 2000;53:336. doi: 10.1136/mp.53.6.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wickham C.L., Boyce M., Joyner M.V., et al. Amplification of PCR products in excess of 600 base pairs using DNA extracted from decalcified, paraffin wax embedded bone marrow trephine biopsies. Mol Pathol. 2000;53:19–23. doi: 10.1136/mp.53.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagasaka T., Lai R., Chen Y.Y., et al. The use of archival bone marrow specimens in detecting B-cell non-Hodgkin's lymphomas using polymerase chain reaction methods. Leuk Lymphoma. 2000;36:347–352. doi: 10.3109/10428190009148856. [DOI] [PubMed] [Google Scholar]
- 17.Fend F., Tzankov A., Bink K., et al. Modern techniques for the diagnostic evaluation of the trephine bone marrow biopsy: methodological aspects and applications. Prog Histochem Cytochem. 2008;42:203–252. doi: 10.1016/j.proghi.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Gerdes M.J., Sevinsky C.J., Sood A., et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc Natl Acad Sci U S A. 2013;110:11982–11987. doi: 10.1073/pnas.1300136110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamilton N. Quantification and its applications in fluorescent microscopy imaging. Traffic. 2009;10:951–961. doi: 10.1111/j.1600-0854.2009.00938.x. [DOI] [PubMed] [Google Scholar]
- 20.Angelo M., Bendall S.C., Finck R., et al. Multiplexed ion beam imaging of human breast tumors. Nat Med. 2014;20:436–442. doi: 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giesen C., Wang H.A., Schapiro D., et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11:417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 22.Decalf J., Albert M.L., Ziai J. New tools for pathology: a user's review of a highly multiplexed method for in situ analysis of protein and RNA expression in tissue. J Pathol. 2018;247(5):650–661. doi: 10.1002/path.5223. [DOI] [PubMed] [Google Scholar]
- 23.Van T.M., Blank C.U. A user's perspective on GeoMx™ digital spatial profiling. Immuno-Oncol Technol. 2019;1:11–18. doi: 10.1016/j.iotech.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amaria R.N., Reddy S.M., Tawbi H.A., et al. Neoadjuvant immune checkpoint blockade in high-risk resectable melanoma. Nat Med. 2018;24(11):1649–1654. doi: 10.1038/s41591-018-0197-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blank C.U., Rozeman E.A., Fanchi L.F., et al. Neoadjuvant versus adjuvant ipilimumab plus nivolumab in macroscopic stage III melanoma. Nat Med. 2018;24(11):1655–1661. doi: 10.1038/s41591-018-0198-0. [DOI] [PubMed] [Google Scholar]
- 26.Toki M.I., Merritt C.R., Wong P.F., et al. High-plex predictive marker discovery for melanoma immunotherapy-treated patients using digital spatial profiling. Clin Cancer Res. 2019;25(18):5503–5512. doi: 10.1158/1078-0432.CCR-19-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merritt C.R., Ong G.T., Church S., et al. High multiplex, digital spatial profiling of proteins and RNA in fixed tissue using genomic detection methods. bioRxiv. 2019 doi: 10.1101/559021. [DOI] [PubMed] [Google Scholar]
- 28.Li C.H., Lee C.K. Minimum cross entropy thresholding. Pattern Recognit. 1993;26:617–625. [Google Scholar]
- 29.Comaniciu D., Meer P. Mean shift: a robust approach toward feature space analysis. IEEE Trans Pattern Anal Mach Intell. 2002;24:603–619. [Google Scholar]
- 30.Babicki S., Arndt D., Marcu A., et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44:W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guimaraes J.C., Zavolan M. Patterns of ribosomal protein expression specify normal and malignant human cells. Genome Biol. 2016;17:236. doi: 10.1186/s13059-016-1104-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ru Y.X., Zhu X.F., Gao J.T., et al. Ultrastructural characteristics of nucleated cells in bone marrow of patients with acquired aplastic anemia. Ultrastruct Pathol. 2008;32:81–88. doi: 10.1080/01913120802063099. [DOI] [PubMed] [Google Scholar]
- 33.Keerthivasan G., Mei Y., Zhao B., et al. Aberrant overexpression of CD14 on granulocytes sensitizes the innate immune response in mDia1 heterozygous del(5q) MDS. Blood. 2014;124:780–790. doi: 10.1182/blood-2014-01-552463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L., Le Mercier I., Putra J., et al. Disruption of the immune-checkpoint VISTA gene imparts a proinflammatory phenotype with predisposition to the development of autoimmunity. Proc Natl Acad Sci U S A. 2014;111:14846–14851. doi: 10.1073/pnas.1407447111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data for this study can be accessed at https://doi.org/10.17632/92835h4bz7.1.