Abstract
First-generation adoptive T-cell transfer (ACT) administering tumor-infiltrating lymphocytes (TILs), and second-generation ACT using autologous T cells genetically modified to express tumor-specific T-cell receptors (TCRs) or chimeric antigen receptors (CARs) have both shown promise for the treatment of several cancers, including melanoma, leukemia and lymphoma. However, these treatments require labor-intensive manufacturing of the cell product for each patient, frequently utilize lentiviral or retroviral vectors to genetically modify the T cells, and have limited antitumor efficacy in solid tumors. Gene editing is revolutionizing the field of gene therapy, and ACT is at the forefront of this revolution. Gene-editing technologies can be used to re-engineer the phenotype of T cells to increase their antitumor potency, to generate off-the-shelf ACT products, and to replace endogenous TCRs with tumor-specific TCRs or CARs using homology-directed repair (HDR) donor templates. Adeno-associated viral vectors or linear DNA have been used as HDR donor templates. Of note, non-viral delivery substantially reduces the time required to generate clinical-grade reagents for manufacture of T-cell products—a critical step for the translation of personalized T-cell therapies. These technological advances in the field using gene editing open the door to the third generation of ACT therapies.
Keywords: Adoptive T-cell therapy, Gene editing in T cells, Non-viral gene editing in T cells
Highlights
-
•
CRISPR-Cas9 allows the generation of tumor-specific T cells for adoptive T-cell transfer (ACT).
-
•
Gene editing allows generation of off-the-shelf ACT products.
-
•
Gene editing can tailor T-cell phenotype and increase antitumor potency.
-
•
Non-viral gene editing is a requirement for personalized ACT.
-
•
Personalized third-generation ACT: gene-edited neoantigen-specific T cells.
- Gene editing with CRISPR-Cas9 allows generating tumor specific T cells for adoptive T cell transfer (ACT).
- Gene editing allows generating off-the-shelf ACT products.
- Gene editing can tailor T cell phenotype and increase antitumor potency.
- Non-viral gene editing is a requirement for personalized ACT.
- Personalized 3rd generation ACT: Gene-edited neoantigen specific T cells.
Adoptive T-cell therapy: how did we get here?
Adoptive T-cell transfer (ACT) has shown promise as a treatment option for patients with several types of cancer. The potential of this approach was initially demonstrated in clinical trials using tumor-infiltrating lymphocytes (TILs) in studies carried out primarily by Rosenberg et al. at the National Cancer Institute Surgery Branch. TILs were obtained from patients with metastatic melanoma, expanded ex vivo, and re-infused back into the patients in combination with interleukin 2 following a lymphodepleting conditioning regimen. This treatment led to response rates of 40–56% [1], [2], [3], [4], [5]. Twenty-four percent of these patients had complete responses, with a remarkable survival rate of 93% at 5 years [6]. These early clinical trials provided the proof of concept that complete eradication of a metastatic solid cancer can be achieved with ex vivo expanded, tumor-reactive T cells.
In order to receive adoptive TIL therapy, the patient is required to have pre-existing tumor-reactive T cells that can be obtained from a tumor resection or a biopsy and expanded ex vivo in sufficient numbers. These requirements limit the number of patients and cancer histologies that can be treated [7], [8], [9] (Table 1). To circumvent the need to isolate and expand the patient's own tumor-reactive T cells, the first technological revolution in the field involved the use of peripheral T cells genetically modified to target the tumor, expanding the potential of ACT. Three key discoveries made this development possible.
Table 1.
Characteristics of the different adoptive T-cell transfer (ACT) therapies.
| First-generation ACT: TIL therapy | Second-generation ACT: engineered T-cell therapy | Third-generation ACT: gene-edited T-cell therapy and personalized therapy | |
|---|---|---|---|
| Pre-existing immunity | Yes | No | Depends on the antigens targeted |
| Antigen targeted | Unknown | Tumor associated antigens: germline antigens, differentiation antigens, antigens overexpressed in tumors, common cancer-specific mutations, etc. | Any tumor-associated antigens or neoantigens |
| Limited to available HLAs | No | Yes (except for CAR T-cell therapies) | No |
| Cancer-specific receptors | Endogenous TCRs | Exogenous TCRs or CARs | Exogenous TCRs or CARs replacing T-cell endogenous TCRs |
| Therapeutic T-cell source | Expanded autologous TILs | Modified autologous peripheral T cells | Modified autologous or heterologous peripheral T cells or heterologous stem cells |
| Gene-modification method | None (although TILs can be modified with retroviral or lentiviral vectors to express cytokines, etc.) | Retroviral or lentiviral vectors, transposons | CRISPR/Cas9 with linear DNA or adeno-associated viral vectors |
CAR, chimeric antigen receptor; CRISPR, clustered regularly interspaced short palindromic repeats; Cas9, CRISPR-associated system; HLA, human leukocyte antigen; TCR, T-cell receptor; TIL, tumor infiltrating lymphocyte.
The first discovery was the identification of tumor-associated antigens that are shared among patients with different cancer histologies and are able to induce a cellular immune response [10], [11]. These tumor antigens are either non-mutated antigens expressed by tumors and select normal tissues, or antigens derived from aberrant intronic sequences, alternative open reading frames, or common cancer-specific mutations such as BRAFV600E and KrasG12D [10], [11]. The non-mutated antigens can be classified into two categories: lineage-specific antigens and germline antigens. The lineage-specific antigens are expressed by cancer cells and normal cells; for example, the melanosomal differentiation antigens (e.g. MART-1, tyrosinase, or gp100) expressed in melanocytes, or the CD19, a marker of B-cell lineage. The germline antigens, such as NY-ESO-1 and MAGE-A3, are expressed during fetal development but are restricted to germline cells in adults. These antigens, however, are often re-expressed in many types of cancer.
The second discovery was the identification of receptors that give tumor reactivity to T cells, leading to the ability to engineer artificial tumor-reactive receptors. This was achieved by isolating T-cell clones reactive against tumor antigens presented by the major histocompatibility complex (MHC) molecules and sequencing their T-cell receptors (TCRs). In the mid-90s, Cole et al. were the first to isolate TIL clones reactive against the melanoma shared antigen MART-1, sequence the TCRα and TCRβ chains, and express them in Jurkat cells to show epitope specificity [12], [13]. Another approach that was being pursued concurrently was to link the variable domain [single-chain variable fragment (scFv)] of monoclonal antibodies specific for cell surface cancer antigens to T-cell signal transduction domains to generate tumor-reactive chimeric antigen receptors (CARs). Eshhar et al. generated the first CAR by fusing the scFv from a monoclonal antibody with a transmembrane domain and the CD3z signaling domain [14].
The third advancement was the ability to clone the sequence of tumor-reactive TCRs and CARs into viral vectors, and use them to genetically modify peripheral blood lymphocytes generating new tumor-targeted T cells [15], [16], [17]. The need for viral vectors was based on the notorious difficulty of expressing foreign transgenes efficiently and stably in T cells, as physical methods had thus far yielded very low transfection efficiency in this cell type. In the years that followed, several TCRs for shared antigens and CARs were identified and subsequently cloned into lentiviral or retroviral vectors [[18], [19], [20], [21], [22]]. These advances overcame the early limitations of TIL therapy and allowed for the treatment of a broader range of patients. Moreover, the use of CARs bypasses the human leukocyte antigen (HLA) restriction limitation of TCRs (Table 1), as they target cell surface proteins expressed by cancer cells.
The first clinical trial using autologous engineered T cells targeted MART-1-positive metastatic melanoma in the context of the high-prevalence HLA allele, HLA-A*02:01 (expressed in ∼40% of the general population). In this trial, the antitumor responses observed included two complete responses among 15 patients [23]. Significant clinical responses have also been observed in other clinical trials utilizing T cells genetically modified with TCRs targeting MART-1, as well as those targeting NY-ESO-1 and other antigens. For example, clinical trials targeting NY-ESO-1-positive tumors revealed response rates of 55–66%, 50–61% and 80% in patients with melanoma, synovial cell sarcoma and multiple myeloma, respectively [24], [25], [26], [27], [28]. After two decades of preclinical research and dozens of clinical trials that led to the successful treatment of hundreds of patients, the US Food and Drug Administration has approved two CAR T-cell therapy products, tisagenlecleucel and axicabtagene ciloleucel, targeting the cell surface marker CD19 expressed by benign and malignant B cells. These treatments were initially developed by the teams of Carl June and Steven A. Rosenberg, respectively [111], [112]. The ACT of T cells genetically modified to express CD19 CARs led to dramatic response rates in adult and pediatric patients with refractory B-cell leukemias and lymphomas [29]. Examples of other CAR targets being tested in patients with solid tumors include IL13Rα2, GD2, HER2, mesothelin and EGFRvIII. However, solid tumors are more challenging to treat with gene-modified cell therapies than leukemias and lymphomas, mainly due to the heterogeneous expression of tumor antigens, the suppressive micro-environment, the limited T-cell trafficking to the tumor, and the lack of persistence of T cells [21], [22], [30].
Genetically modified T cells: from retroviral and lentiviral vectors to gene editing
Historically, lentiviral and retroviral vectors have been the viral vectors of choice to genetically modify T cells for ACT due to their ability to insert transgenes permanently into the T-cell genome. Despite demonstrated successes, well-recognized limitations include random integrations into the genome, variable copy number, non-optimal transgene expression by non-native promoters, and the potential for transgene silencing [31]. In addition to these limitations, a longstanding concern is the potential for malignant transformation resulting from viral vector integrations, despite the fact that no cases of oncogenic transformation have been reported following the administration of T cells genetically modified with these vectors to hundreds of patients. Recently, a patient was reported to have a major expansion from a single T-cell clone with a missense mutation of the methylcytosine dioxygenase TET2 gene in one allele and an insertion of the lentiviral vector expressing a CD19 CAR disrupting the other allele [32]. Although in this case, the loss of function of TET2 translated into an improved therapeutic effect, this serves as a reminder of the potential dangers of random integrations into the genome. Operationally, other major hurdles include the time and expense required for the generation of clinical-grade viral vectors. Moreover, despite the progress achieved in optimization of vector-manufacturing protocols for gene therapy over the last three decades, high titer retroviral and lentiviral vector preparations are still challenging to obtain following good manufacturing practices (GMP), and only a small number of academic centers and companies are able to produce them (Table 2).
Table 2.
Comparison between genetic modification using lentiviral or retroviral vectors and gene editing.
| Retroviral/lentiviral vectors |
Gene editing |
||
|---|---|---|---|
| Advantages | Disadvantages | Advantages | Disadvantages |
| High transduction efficiency and easy transduction procedures | Random integration into the genome | Endogenous regulation of transgenic TCRs/CARs. In CARs-T cells, endogenous regulation avoids overexpression, tonic signaling and T-cell exhaustion | Lower modification efficiency |
| Easy manufacturing on small scale | Variable transgene expression | Homogeneous expression | Off-target double-stranded breaks |
| Artificial regulation from viral vector promoter can lead to transgene silencing or overexpression | Knock-in and knock-out can be multiplexed. Potential to increase T cell potency and generate off-the-shelf T-cell products | Electroporation, Cas9, and HDR donor-induced toxicity. Decreased T-cell expansion | |
| Costly and time-consuming large-scale manufacturing at GMP/clinical grade | Viral and non-viral methods. The non-viral methods allow for fast reagent manufacturing at GMP/clinical level | ||
| No TCR mispairing | |||
| Lower cost with non-viral methods | |||
CAR, chimeric antigen receptor; GMP, good manufacturing practices; HDR, homology-directed repair; TCR, T-cell receptor.
Alternative methods to modify T cells have been considered, such as the use of transposons [33], [34], [35] or mRNA. However, both methods present several limitations. Transposons eliminate the need for viral vectors but still integrate randomly into the genome. Synthetic mRNA eliminates the risk of oncogenic integration at the expense of having only transient expression of the tumor-specific CARs or TCRs. To be effective, T cells genetically modified with mRNA would require repeated administration [36], as the transgene expression would be diluted by half with each T-cell division due to the lack of integration into the T-cell genome.
New gene-editing techniques are revolutionizing the field of gene therapy, and ACT is at the forefront of this revolution. Four different gene-editing tools have been used to modify T cells: zinc-finger nucleases (ZFNs), transcription activator-like nucleases (TALENs), MegaTALs (meganucleases fused to TALs), and clustered regularly interspaced short palindromic repeats/CRISPR-associated system (CRISPR/Cas9) [37], [38], [39], [40], [41]. Early work using ZFNs and TALENs demonstrated that knock-out of the endogenous TCRα and TCRβ chains could be combined with genetic modification of edited T cells. TCR-deficient T cells could be genetically modified using viral vectors [37], [40] or transposons [41] to enforce the expression of a tumor- or virus-specific TCR or CAR. Disruption of endogenous TCRs led to improved expression and functionality of transgenic TCRs and a significant increase in the in vivo antitumor activity of modified cells in mice. These were the first steps towards the generation of off-the-shelf adoptive T-cell products. Studies using TALENs also showed that gene editing could be multiplexed to disrupt multiple genes. One example was genetic knock-down of endogenous TCR and the surface protein CD52, with the goal of making gene-edited cells resistant to the cytotoxic anti-CD52 antibody alemtuzumab. Alemtuzumab could then be used concurrently or before ACT to lymphodeplete the host without eliminating the CD52-negative T cells [39]. This strategy with simultaneous lentiviral transduction with a CD19 CAR vector has been tested clinically [42]. Gene editing fulfills the promise of simultaneously knocking-out and knocking-in multiple genes of interest in a permanent and targeted fashion to redirect T-cell specificity, tailor their phenotype and improve antitumor activity.
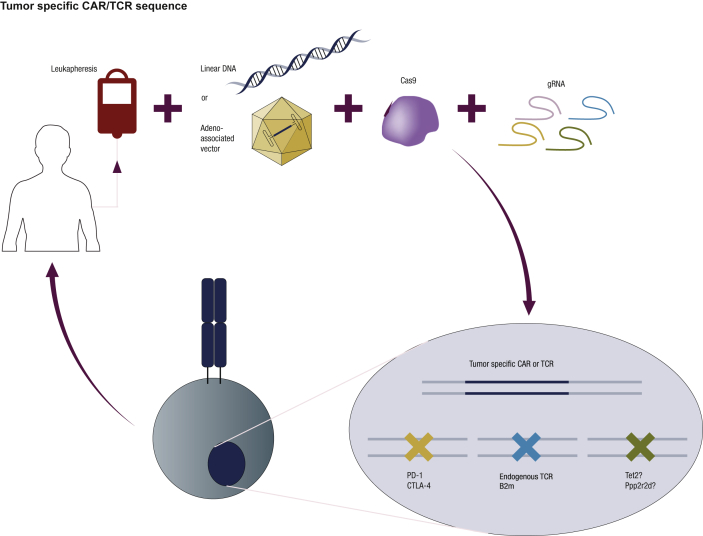
CRISPR-Cas9 is currently the favorite method within the gene-editing field given its technical simplicity, cost-efficiency and ability to be multiplexed [43], [44], [45]. In order to edit the T-cell genome, bulk peripheral blood mononuclear cells (PBMCs) or a sorted cell subset are activated and electroporated to deliver guide RNA (gRNA) and Cas9 as mRNA or a ribonucleoprotein (RNP) comprised of recombinant Cas9 protein complexed with gRNA (Figure 1). In both cases, the expression of Cas9 is transient, decreasing the toxicity of Cas9 mediated by off-target effects, and avoiding the described immunogenicity of Cas9 expression by host cells [46]. To achieve the end goal, gRNA targets Cas9 nuclease in the region of the genome to be modified, and Cas9 generates a double-stranded break (DSB). In order to repair the break, the cell uses either the non-homologous end joining (NHEJ) mechanism or homology-directed repair (HDR). NHEJ induces direct religation of the cleaved ends using an error-prone mechanism that introduces small nucleotide insertions and deletions leading, for the most part, to disruption of the targeted gene (knock-out). Alternatively, HDR requires a homologous strand donor template to induce DSB repair in a template-dependent manner, leading to gene correction or the addition of a gene of interest (knock-in). This technique leads to knock-out efficiencies of ≤98% depending on the targeted gene [47]. Single-stranded oligonucleotides, long linear single- or double-stranded DNA, and adeno-associated vectors (AAVs) can be used as donor DNA templates for HDR [48], [49], [50]. Large fragment integration efficiencies ≤57% can be obtained with this technique depending on the locus modified, the size of the insertion, and the type of donor DNA template used for HDR [48], [49].
Figure 1.
T-cell gene editing with multiplex knock-out and endogenous T-cell receptor (TCR) replacement. Peripheral blood mononuclear cells, or specific T-cell subsets, can be electroporated to deliver Cas9 protein and one or multiple gRNAs complexed as ribonucleoproteins (RNPs). To replace endogenous TCRs with tumor-specific TCRs or chimeric antigen receptors (CARs), double- or single-stranded DNA can be used as a homology-directed repair (HDR) template and delivered together with the RNPs. Alternatively, adeno-associated vectors can also be used as HDR templates by infecting the modified cells after electroporation. Gene editing combining knock-in and knock-out with multiple gRNAs will allow generation of off-the-shelf allogeneic tumor-targeted T cells (endogenous TCR-β2M-) or T cells with enhanced antitumor potency (PD-1-, CTLA-4-). Other candidate genes, such as TET2 and PPP2R2D, are being knocked-out to investigate whether their absence will increase the potency of T cells.
Eyquem et al. [48] reported a strategy to disrupt the TRAC locus and insert a CD19 CAR under its transcriptional control using CRISPR/Cas9 and an AAV vector as HDR donor template. They delivered gRNA and Cas9 mRNA by electroporation followed by AAV infection, resulting in a knock-in efficiency of ≤40%. Using this method of targeted integration, they demonstrated that CAR expression was homogenous and consistent among donors, and the edited T cells possessed increased antitumor potency in vivo in murine leukemia models compared with the cells genetically modified using retroviral vectors. This study revealed that the finely controlled transcriptional regulation of endogenous TCRs prevents tonic CAR signaling in the absence of antigen, and allows CAR internalization and recovery of expression upon antigen exposure. This physiological regulation of the CARs delays effector T-cell differentiation and exhaustion, leading to superior antitumor activity [48] (Table 2).
Shortly after these studies were reported, Roth et al. [49] went one step further in simplifying T-cell editing protocols, and showed that T cells can be genetically modified by co-electroporating RNP complexes and long linear single- or double-stranded DNA (ssDNA or dsDNA, respectively) as a template for HDR. It had been described previously that dsDNA is toxic for T cells at high concentrations because it activates DNA-sensing pathways [51]. However, the authors showed that co-delivery with RNP reduces the toxicity of dsDNA. Interestingly, the authors were able to replace the endogenous TCRs with tumor-specific TCRs. Using this approach, they created fully functional antitumor T cells with a knock-in efficiency of ∼12% and an almost complete knock-out of the endogenous TCRs. Upon co-culture with melanoma cells expressing matched HLA and antigen, the gene-edited T cells were able to secrete cytokines, degranulate, and kill tumor cells at the same level as T cells genetically modified using a retroviral vector. In an in vivo ACT model, the gene-edited T cells demonstrated antitumor activity against implanted tumors in mice. Roth et al. also showed that this technique can be multiplexed to insert genes in up to three loci [49]. This new method avoids the need for viral vectors that are labor intensive and expensive to generate on a large scale, potentially increasing the cost-efficiency of the ACT manufacturing process. More importantly, non-viral methods such as that described by Roth et al. [49] would allow rapid T-cell gene editing, as the gRNA and HDR DNA templates needed for gene modification can be generated within a matter of weeks. This is an absolute requirement for personalized ACT strategies tailored to the specific characteristics of each patient's cancer cells. Generation of viral vectors to express individual TCRs for each patient and the associated lot release testing would take several months with current technologies and regulations, which impedes their use for personalized approaches (Table 2).
Multiplex gene editing: towards best-in-class modified T cells
Multiplex CRISPR technology can be used to improve the features of T cells to become even more powerful therapies than those generated using the current standard methods. In contrast with current autologous therapies, several groups and companies are generating off-the-shelf allogeneic T cells (or universal gene-modified T cells). Off-the-shelf T cells would be particularly useful to treat those patients who, due to their disease or previous treatments, have difficulty obtaining PBMCs in sufficient number and quality to be modified ex vivo. Additionally, the use of off-the-shelf T cells has the potential to increase the cost-efficiency of the therapy by treating multiple patients with the same batch of T cells, recognizing shared antigens. Universal CAR T cells can be generated by knocking-out both endogenous TCRs and β2M (an essential component of the HLA complex) [52], [53]. Disruption of endogenous TCRs within universal T cells is required to avoid recognition of host antigens by transferred T cells, a response known as graft-versus-host disease. Concurrent disruption of HLA within universal T cells is required to avoid elimination of the transferred cells by the host immune response. Non-classical HLA molecules, such as HLA-E, could be overexpressed in universal T cells to avoid the rejection of these cells by natural killer cells [54]. Interestingly, the same multiplexing strategy could be used with hematopoietic stem cells or induced pluripotent stem cells to generate a continuous source for universal genetically modified T cells [55], [56].
Multiplex CRISPR methodology has also been used to increase T-cell potency. Knock-out of immune checkpoints such as CTLA-4, PD-1 or LAG-3, together with viral-driven expression of a CAR/TCR, has led to enhanced antitumor activity in murine models [53], [57], [58]. This strategy is currently being tested in clinical trials (Figure 1). The clinical success of immune checkpoint blockade therapy demonstrates the benefits of removing the breaks from T cells. However, the genetic and permanent disruption of these checkpoints in tumor-targeted T cells could lead to their uncontrolled expansion when administered in patients with cancer. The safety of this approach deserves further investigation.
Loss-of-function screens with pooled short hairpin RNAs (shRNAs) or CRISPRs are powerful techniques to identify genes required for certain functions, or genes that, when silenced, confer favorable features to the target cells. They are a very valuable source of information that allow for the discovery of new gene functions. A pooled shRNA screen in a murine ACT model in vivo showed that Ppp2r2d knock-down enhanced T-cell survival and proliferation upon TCR activation, cytokine production and antitumor activity in vivo [59]. Ppp2r2d is a regulatory subunit of the family of PP2A phosphatases that had not been studied previously in the context of T-cell regulation. Ppp2r2d inhibits cell entry into mitosis, induces the exit from mitosis, and plays a role in Bcl-2-associated agonist of cell death (BAD)-mediated apoptosis [59]. Recently, Shifrut et al. [60] developed SLICE, a new platform for genome-wide CRISPR loss-of-function screens in primary T cells. With this technology, four target genes were identified—CBLB, SOCS1, TCEB2 and RASA2—that enhanced the proliferation and in vitro cytotoxicity of tumor-specific T cells when knocked-out. CBLB mediates the ubiquitination of TCRs upon activation [61], and its inhibition enhances the antitumor activity of adoptively transferred CD8+ T cells [62]. SOCS1 is a negative regulator of the JAK/STAT pathway in T cells [63], and TCEB2 is a binding partner of SOCS1 [63]. RASA2 encodes for a GTPase-activating protein that stimulates the GTPase activity of wild-type Ras [64], but its role in regulating T-cell proliferation had not been previously described [60]. Remarkably, this powerful technique can be adapted to identify genetic perturbations that confer an advantage to T cells in specific contexts, such as in vivo persistence and antitumor activity, or suppressive micro-environments [60]. The use of these screens will unravel the T-cell regulation network and point to multiple genes that could be knocked-out to potentially improve the long-term antitumor activity of adoptively transferred T cells (Figure 1).
China is leading the race for clinical trials using CRISPR/Cas9-edited T cells. Several clinical trials are underway in China to test PD-1 knock-out open repertoire T cells for non-small cell lung cancer (NCT02793856) and esophageal cancer (NCT03081715), PD-1 knock-out Epstein–Barr virus (EBV)-specific cytotoxic T cells for EBV-associated malignancies (NCT03044743), universal CD19 CAR T cells (NCT03166878), universal bi-specific CD19 and CD20 or CD22 CAR T cells (NCT03398967), PD-1 knock-out mesothelin CAR T cells (NCT03747965), and PD-1 and TCR double knock-out mesothelin CAR T cells (NCT03545815). A more conservative approach in terms of safety has been undertaken in the USA, in which only one clinical trial is currently open and accruing patients to test T cells genetically modified with a viral vector expressing an NY-ESO-1 TCR (HLA-A2.1 restricted) and knocked-out for the endogenous TCR and PD-1 genes using CRISPR/Cas9 gene editing in patients with melanoma, multiple myeloma and sarcoma (NCT03399448, clinicaltrials.gov). All of these clinical trials are listed in clinicaltrials.gov.
Gene editing: challenges to overcome
Remarkably rapid progress has been achieved over the last few years towards clinical translation of the CRISPR/Cas9 gene-editing tools. However, there are still significant challenges that the field needs to address. The main concerns are the off-target effect of Cas9 due to the binding of gRNA to regions with imperfect complementarity, and the common single-nucleotide variants that can generate high-efficiency Cas9 off-target sites [65], [66], [67]. To avoid these issues, nucleases that require two gRNA to induce DSBs have been used, such as Cas9 nickases or the paired catalytically inactive Cas9 fused to the Fokl nucleases [68]. More recently, several high-fidelity Streptococcus pyogenes SpCas9 mutants were generated by either structure-guided protein engineering, in vivo screening, or directed evolution. These Cas9 mutants decrease the off-target DSBs substantially without decreasing the knock-out or knock-in efficacy [69], [70], [71], [72], [73]. An intense effort has also been placed on the discovery of additional CRISPR/Cas nucleases that can be used for gene editing and display improved or alternative properties. Among these, Cas12a allows targeting of T-rich protospacer adjacent motifs, produces staggered cuts, and has been optimized to increase on-target cuts and decrease off-target cuts [74]; certain Cas12b mutants display increased specificity compared with SpCas9 [75], [76]; and CasX generates staggered DSBs, is smaller than SpCas9, and is derived from a non-pathogenic organism [77]. Interestingly, modifications in certain positions of gRNA can improve cleavage specificity [78]. Finally, there is a need for improved tools to design CRISPR/Cas9 gRNAs and predict target efficiency and specificity. These tools should be based on empirical off-target profile data, and consider the individual's genomic variations and chromatin accessibility of the potential on-target and off-target sites [79], [80]. An additional concern for the therapeutic use of CRISPR/Cas9 tools is that large deletions and genomic re-arrangements have been observed at the targeted site in embryonic stem cells, hematopoietic progenitors, and cell lines [81], [82]. Despite the reported potential aberrant repairs and off-target toxicities, comforting results published by Roth et al. [49] showed a 0.01% off-target integration when using ssDNA as an HDR donor template. Although the techniques used cannot detect all the possible genomic alterations, the low random integration frequency is in stark contrast to the random and multiple integrations that occur with lentiviral and retroviral vectors. Regardless, thorough characterization of the genetically modified cells, not limited to the immediate vicinity of the targeted sites and predicted off-target sites, is needed to assess the safety and functionality of the new gene-editing approaches.
Another drawback of the use of CRISPR/Cas9 to edit T cells for ACT is the limited HDR efficiency. The HDR repair mechanism is restricted to the late S and G2 phases of the cell cycle [83], [84], and quiescent cells tend to use NHEJ to repair DSBs. Additionally, p53 can induce cell cycle arrest when DSBs are generated by the CRISPR/Cas9 system, precluding efficient HDR [85], [86]. Cell cycle synchronization [87], as well as mechanisms to inhibit NHEJ [84], [88], [89], [90], [91], apoptosis [92], p53-dependent cell cycle arrest [85], [86], or DNA sensing [93], have been reported to increase knock-in efficiencies under certain conditions. However, their role in T-cell editing has not been reported to date. Interestingly, a high-throughput drug screen identified multiple drugs that improve HDR efficiency in pluripotent stem cells [94]. Finally, Cas9 modifications, such as covalently linking the HDR template to Cas9 [95] or fusing the N-terminal fragment of the CtIP protein to Cas9 [96], have also been shown to increase HDR.
Third-generation adoptive T-cell therapies
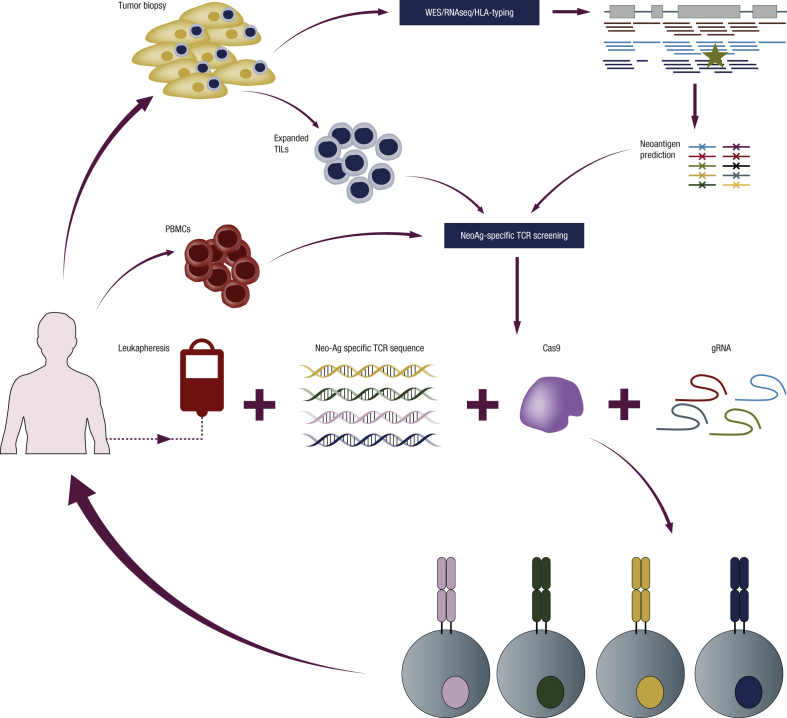
We are at the forefront of a new technological revolution that we believe will lead to third-generation adoptive cell therapies, changing, as happened with the second-generation ACT, the methods to modify T cells, the type of antigens targeted, and the methods to isolate TCRs. First, the new gene-editing technologies will allow replacement of endogenous TCRs with new tumor-specific receptors (TCRs or CARs) regulated under the transcriptional control of endogenous TCRs. The endogenous transcriptional control will lead to the generation of cells with functional features closer to a natural immune response and, likely, with longer lasting clinical responses that are not limited by the over-activation observed with some CARs [48], [97]. The use of endogenous regulation will avoid the silencing of TCR expression observed in some clinical trials [31]. Moreover, safety will likely increase by avoiding random viral vector integration into the genome, and by limiting possible TCR mispairing with endogenous TCR chains, as TCR mispairing could lead to new TCR reactivities and potential off-target toxicities [98]. Eliminating the need for viral vectors will simplify manufacturing processes and improve the cost-efficiency of these therapies. More importantly, it will significantly reduce the time to produce the GMP reagents needed, allowing the generation of truly personalized adoptive T-cell therapies specifically designed for each patient's tumor antigens and HLA types. Second, ACT protocols to date have mainly targeted shared antigens that are expressed by cancer cells but can also be expressed by cells in healthy tissues. Targeting non-synonymous cancer-specific mutations (or neoantigens) would avoid the toxicities related to the expression of the target antigen in normal tissues. In order to predict these neoantigens, tumors can be biopsied and sequenced, and the genomic and expression data can be used to predict which mutations could potentially give rise to new immunogenic antigens presented by one of the patient's HLAs [99]. Third, to identify TCRs targeting neoantigen mutations, TILs [100], [101], [102], [103] or PBMCs [104], [105] can be used. Several techniques to identify clones of T cells reactive against neoantigen mutations have been described based on the use of multimer peptide-MHC complexes (pMHC) [105], [106], [107] or dendritic cells expressing tandem minigenes encoding for the predicted mutations [103]. More recently, high-throughput techniques that allow identification of the antigen recognized by T-cell clones have been described based on the use of yeast pMHC libraries [108], trogocytosis [109] or new chimeric MHC molecules [110]. Once the T-cell clone is identified, the TCR can be sequenced and used to modify a large number of PBMCs. Accuracy, high-throughput power and applicability of these techniques still needs to be demonstrated. It is envisaged that the combination of these techniques with non-viral T-cell editing using CRISR-Cas9 will be at the center of the new ACT technological revolution (Figure 2).
Figure 2.
Personalized adoptive T-cell therapy using gene editing. To perform personalized adoptive T-cell therapy, a biopsy of the patient's tumor can be subjected to whole-exome sequencing and RNA sequencing and identify the most immunogenic neoantigens using bioinformatic prediction algorithms. T cells specific for the predicted neoantigens can be isolated from the patient's tumor infiltrating lymphocytes (TILs) or peripheral blood mononuclear cells (PBMCs) using multimer peptide-MHC complexes (pMHC) libraries or dendritic cells transduced with neoantigen tandem minigenes, among other techniques. The two chains of the T-cell receptors (TCRs) can then be sequenced from single cells. Linear DNA encoding for these TCRs can be synthesized and electroporated together with gRNA and Cas9 into PBMCs from the same patient to generate neoantigen-specific T cells. These gene-edited T cells can be expanded and re-infused back into the patient. Multiplex knock-out can be used to increase the potency of T cells.
Funding
This study was funded, in part, by the Parker Institute for Cancer Immunotherapy, National Institutes of Health Grants R35 CA197633 and P01 CA168585, the Ressler Family Fund and support from Ken and Donna Schultz (to AR).
Disclosure
Dr. Puig-Saus has no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Dr. Ribas reports personal fees from consulting with Amgen, Chugai, Genentech-Roche, Novartis, Merck, and being a member of the scientific advisory board of Arcus, Bioncotech, Compugen, Cytomx, Five Prime, FLX-Bio, Merus, Rgenix, PACT Pharma, Tango Therapeutics.
Acknowledgements
The authors acknowledge the contributions of Dr. Paula Kaplan-Lefko for critical review of the article, and Dr. Katie Campbell for assistance with figure design.
References
- 1.Dudley M.E., Wunderlich J.R., Yang J.C., Hwu P., Schwartzentruber D.J., Topalian S.L., et al. A phase I study of nonmyeloablative chemotherapy and adoptive transfer of autologous tumor antigen-specific T lymphocytes in patients with metastatic melanoma. J Immunother. 2002;25:243–251. doi: 10.1097/01.CJI.0000016820.36510.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dudley M.E., Wunderlich J.R., Yang J.C., Sherry R.M., Topalian S.L., Restifo N.P., et al. Adoptive cell transfer therapy following non-myeloablative but lymphodepleting chemotherapy for the treatment of patients with refractory metastatic melanoma. J Clin Oncol. 2005;23:2346–2357. doi: 10.1200/JCO.2005.00.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dudley M.E., Yang J.C., Sherry R., Hughes M.S., Royal R., Kammula U., et al. Adoptive cell therapy for patients with metastatic melanoma: evaluation of intensive myeloablative chemoradiation preparative regimens. J Clin Oncol. 2008;26:5233–5239. doi: 10.1200/JCO.2008.16.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goff S.L., Dudley M.E., Citrin D.E., Somerville R.P., Wunderlich J.R., Danforth D.N., et al. Randomized, prospective evaluation comparing intensity of lymphodepletion before adoptive transfer of tumor-infiltrating lymphocytes for patients with metastatic melanoma. J Clin Oncol. 2016;34:2389–2397. doi: 10.1200/JCO.2016.66.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenberg S.A., Packard B.S., Aebersold P.M., Solomon D., Topalian S.L., Toy S.T., et al. Use of tumor-infiltrating lymphocytes and interleukin-2 in the immunotherapy of patients with metastatic melanoma. A preliminary report. N Engl J Med. 1988;319:1676–1680. doi: 10.1056/NEJM198812223192527. [DOI] [PubMed] [Google Scholar]
- 6.Rosenberg S.A., Yang J.C., Sherry R.M., Kammula U.S., Hughes M.S., Phan G.Q., et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res. 2011;17:4550–4557. doi: 10.1158/1078-0432.CCR-11-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goff S.L., Smith F.O., Klapper J.A., Sherry R., Wunderlich J.R., Steinberg S.M., et al. Tumor infiltrating lymphocyte therapy for metastatic melanoma: analysis of tumors resected for TIL. J Immunother. 2010;33:840–847. doi: 10.1097/CJI.0b013e3181f05b91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall M.L., Liu H., Malafa M., Centeno B., Hodul P.J., Pimiento J., et al. Expansion of tumor-infiltrating lymphocytes (TIL) from human pancreatic tumors. Journal for ImmunoTherapy of Cancer. 2016;4:61. doi: 10.1186/s40425-016-0164-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee H.J., Kim Y.-A., Sim C.K., Heo S.-H., Song I.H., Park H.S., et al. Expansion of tumor-infiltrating lymphocytes and their potential for application as adoptive cell transfer therapy in human breast cancer. Oncotarget. 2017;8:113345–113359. doi: 10.18632/oncotarget.23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coulie P.G., Van Den Eynde B.J., Van Der Bruggen P., Boon T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Canc. 2014;14:135–146. doi: 10.1038/nrc3670. [DOI] [PubMed] [Google Scholar]
- 11.Ilyas S., Yang J.C. Landscape of tumor antigens in T cell immunotherapy. J Immunol. 2015;195:5117–5122. doi: 10.4049/jimmunol.1501657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole D.J., Weil D.P., Shamamian P., Rivoltini L., Kawakami Y., Topalian S., et al. Identification of MART-1-specific T-cell receptors: T cells utilizing distinct T-cell receptor variable and joining regions recognize the same tumor epitope. Cancer Res. 1994;54:5265–5268. [PubMed] [Google Scholar]
- 13.Cole D.J., Weil D.P., Shilyansky J., Custer M., Kawakami Y., Rosenberg S.A., et al. Characterization of the functional specificity of a cloned T-cell receptor heterodimer recognizing the MART-1 melanoma antigen. Cancer Res. 1995;55:748–752. [PubMed] [Google Scholar]
- 14.Eshhar Z., Waks T., Gross G., Schindler D.G. Specific activation and targeting of cytotoxic lymphocytes through chimeric single chains consisting of antibody-binding domains and the gamma or zeta subunits of the immunoglobulin and T-cell receptors. Proc Natl Acad Sci Unit States Am. 1993;90:720–724. doi: 10.1073/pnas.90.2.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clay T.M., Custer M.C., Sachs J., Rosenberg S. a., Nishimura M.I., Hwu P., et al. Efficient transfer of a tumor antigen-reactive TCR to human peripheral blood lymphocytes confers anti-tumor reactivity. J Immunol. 1999;196:507–513. [PubMed] [Google Scholar]
- 16.Hughes M.S., Yu Y.Y., Dudley M.E., Zheng Z., Robbins P.F., Li Y., et al. Transfer of a TCR gene derived from a patient with a marked antitumor response conveys highly active T-cell effector functions. Hum Gene Ther. 2005;16:457–472. doi: 10.1089/hum.2005.16.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pogulis R.J., Pease L.R. A retroviral vector that directs simultaneous expression of alpha and beta T cell receptor genes. Hum Gene Ther. 1998;9:2299–2304. doi: 10.1089/hum.1998.9.15-2299. [DOI] [PubMed] [Google Scholar]
- 18.Cohen C.J., Zheng Z., Bray R., Zhao Y., Sherman L.A., Rosenberg S.A., et al. Recognition of fresh human tumor by human peripheral blood lymphocytes transduced with a bicistronic retroviral vector encoding a murine anti-p53 TCR. J Immunol. 2005;175:5799–5808. doi: 10.4049/jimmunol.175.9.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgan R.A., Dudley M.E., Yu Y.Y.L., Zheng Z., Robbins P.F., Theoret M.R., et al. High efficiency TCR gene transfer into primary human lymphocytes affords avid recognition of melanoma tumor antigen glycoprotein 100 and does not alter the recognition of autologous melanoma antigens. J Immunol. 2003;171:3287–3295. doi: 10.4049/jimmunol.171.6.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y., Zheng Z., Robbins P.F., Khong H.T., Rosenberg S.A., Morgan R.A. Primary human lymphocytes transduced with NY-ESO-1 antigen-specific TCR genes recognize and kill diverse human tumor cell lines. J Immunol. 2005;174:4415–4423. doi: 10.4049/jimmunol.174.7.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fesnak A.D., June C.H., Levine B.L. Engineered T cells: the promise and challenges of cancer immunotherapy. Nat Rev Canc. 2016;16:566–581. doi: 10.1038/nrc.2016.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tran E., Robbins P.F., Rosenberg S.A. Final common pathway of human cancer immunotherapy: targeting random somatic mutations. Nat Immunol. 2017;18:255–262. doi: 10.1038/ni.3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan R.A., Dudley M.E., Wunderlich J.R., Hughes M.S., Yang J.C., Sherry R.M., et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D’angelo S.P., Melchiori L., Merchant M.S., Bernstein D., Glod J., Kaplan R., et al. Antitumor activity associated with prolonged persistence of adoptively transferred NY-ESO-1c259T cells in synovial sarcoma. Cancer Discov. 2018;8:944–957. doi: 10.1158/2159-8290.CD-17-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nowicki T.S., Berent-Maoz B., Cheung-Lau G.C., Huang R.R., Wang X., Tsoi J., et al. A pilot trial of the combination of transgenic NY-ESO-1-reactive adoptive cellular therapy with dendritic cell vaccination with or without ipilimumab. Clin Cancer Res. 2018;25:2096–2108. doi: 10.1158/1078-0432.CCR-18-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rapoport A.P., Stadtmauer E.A., Binder-Scholl G.K., Goloubeva O., Vogl D.T., Lacey S.F., et al. NY-ESO-1-specific TCR-engineered T cells mediate sustained antigen-specific antitumor effects in myeloma. Nat Med. 2015;21:914–921. doi: 10.1038/nm.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robbins P.F., Kassim S.H., Tran T.L.N., Crystal J.S., Morgan R.A., Feldman S.A., et al. A pilot trial using lymphocytes genetically engineered with an NY-ESO-1-reactive T-cell receptor: long-term follow-up and correlates with response. Clin Cancer Res. 2015;21:1019–1027. doi: 10.1158/1078-0432.CCR-14-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robbins P.F., Morgan R.A., Feldman S.A., Yang J.C., Sherry R.M., Dudley M.E., et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J Clin Oncol. 2011;29:917–924. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.June C.H., Sadelain M. Chimeric antigen receptor therapy. N Engl J Med. 2018;379:64–73. doi: 10.1056/NEJMra1706169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long K.B., Young R.M., Boesteanu A.C., Davis M.M., Melenhorst J.J., Lacey S.F., et al. CAR T cell therapy of non-hematopoietic malignancies: detours on the road to clinical success. Front Immunol. 2018;9:2740. doi: 10.3389/fimmu.2018.02740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burns W.R., Zheng Z., Rosenberg S.A., Morgan R.A. Lack of specific γ-retroviral vector long terminal repeat promoter silencing in patients receiving genetically engineered lymphocytes and activation upon lymphocyte restimulation. Blood. 2009;114:2888–2899. doi: 10.1182/blood-2009-01-199216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fraietta J.A., Nobles C.L., Sammons M.A., Lundh S., Carty S.A., Reich T.J., et al. Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature. 2018;558:307–312. doi: 10.1038/s41586-018-0178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang X., Guo H., Kang J., Choi S., Zhou T.C., Tammana S., et al. Sleeping Beauty transposon-mediated engineering of human primary T cells for therapy of CD19+lymphoid malignancies. Mol Ther. 2008;16:580–589. doi: 10.1038/sj.mt.6300404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kebriaei P., Singh H., Huls M.H., Figliola M.J., Bassett R., Olivares S., et al. Phase I trials using Sleeping Beauty to generate CD19-specific CAR T cells. J Clin Investig. 2016;126:3363–3376. doi: 10.1172/JCI86721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng P.D., Cohen C.J., Yang S., Hsu C., Jones S., Zhao Y., et al. Efficient nonviral Sleeping Beauty transposon-based TCR gene transfer to peripheral blood lymphocytes confers antigen-specific antitumor reactivity. Gene Ther. 2009;16:1042–1049. doi: 10.1038/gt.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y., Moon E., Carpenito C., Paulos C.M., Liu X., Brennan A.L., et al. Multiple injections of electroporated autologous T cells expressing a chimeric antigen receptor mediate regression of human disseminated tumor. Cancer Res. 2010;70:9053–9061. doi: 10.1158/0008-5472.CAN-10-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berdien B., Mock U., Atanackovic D., Fehse B. TALEN-mediated editing of endogenous T-cell receptors facilitates efficient reprogramming of T lymphocytes by lentiviral gene transfer. Gene Ther. 2014;21:539–548. doi: 10.1038/gt.2014.26. [DOI] [PubMed] [Google Scholar]
- 38.Osborn M.J., Webber B.R., Knipping F., Lonetree C.L., Tennis N., DeFeo A.P., et al. Evaluation of TCR gene editing achieved by TALENs, CRISPR/Cas9, and megaTAL nucleases. Mol Ther. 2016;24:570–581. doi: 10.1038/mt.2015.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Philip L.P.B., Schiffer-Mannioui C., Le Clerre D., Chion-Sotinel I., Derniame S., Potrel P., et al. Multiplex genome-edited T-cell manufacturing platform for "off-the-shelf" adoptive T-cell immunotherapies. Cancer Res. 2015;75:3853–3864. doi: 10.1158/0008-5472.CAN-14-3321. [DOI] [PubMed] [Google Scholar]
- 40.Provasi E., Genovese P., Lombardo A., Magnani Z., Liu P.Q., Reik A., et al. Editing T cell specificity towards leukemia by zinc finger nucleases and lentiviral gene transfer. Nat Med. 2012;18:807–815. doi: 10.1038/nm.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torikai H., Reik A., Liu P.Q., Zhou Y., Zhang L., Maiti S., et al. A foundation for universal T-cell based immunotherapy: T cells engineered to express a CD19-specific chimeric-antigen-receptor and eliminate expression of endogenous TCR. Blood. 2012;119:5697–5705. doi: 10.1182/blood-2012-01-405365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qasim W., Zhan H., Samarasinghe S., Adams S., Amrolia P., Stafford S., et al. Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci Transl Med. 2017 Jan 25;9(374) doi: 10.1126/scitranslmed.aaj2013. pii: eaaj2013. [DOI] [PubMed] [Google Scholar]
- 43.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Charlesworth C.T., Deshpande P.S., Dever D.P., Camarena J., Lemgart V.T., Cromer M.K., et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat Med. 2019;25:249–254. doi: 10.1038/s41591-018-0326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seki A., Rutz S. Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells. J Exp Med. 2018;215:985–997. doi: 10.1084/jem.20171626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eyquem J., Mansilla-Soto J., Giavridis T., Van Der Stegen S.J.C., Hamieh M., Cunanan K.M., et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543:113–117. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roth T.L., Puig-Saus C., Yu R., Shifrut E., Carnevale J., Li P.J., et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature. 2018;559:405–409. doi: 10.1038/s41586-018-0326-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schumann K., Lin S., Boyer E., Simeonov D.R., Subramaniam M., Gate R.E., et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc Natl Acad Sci Unit States Am. 2015;112:10437–10442. doi: 10.1073/pnas.1512503112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hornung V., Latz E. Intracellular DNA recognition. Nat Rev Immunol. 2010;10:123–130. doi: 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- 52.Liu X., Zhang Y., Cheng C., Cheng A.W., Zhang X., Li N., et al. CRISPR-Cas9-mediated multiplex gene editing in CAR-T cells. Cell Res. 2017;27:154–157. doi: 10.1038/cr.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ren J., Liu X., Fang C., Jiang S., June C.H., Zhao Y. Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition. Clin Cancer Res. 2017;23:2255–2266. doi: 10.1158/1078-0432.CCR-16-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Torikai H., Reik A., Soldner F., Warren E.H., Yuen C., Zhou Y., et al. Toward eliminating HLA class i expression to generate universal cells from allogeneic donors. Blood. 2013;122:1341–1349. doi: 10.1182/blood-2013-03-478255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seet C.S., He C., Bethune M.T., Li S., Chick B., Gschweng E.H., et al. Generation of mature T cells from human hematopoietic stem and progenitor cells in artificial thymic organoids. Nat Methods. 2017;14:521–530. doi: 10.1038/nmeth.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Themeli M., Kloss C.C., Ciriello G., Fedorov V.D., Perna F., Gonen M., et al. Generation of tumor-targeted human T lymphocytes from induced pluripotent stem cells for cancer therapy. Nat Biotechnol. 2013;31:928–933. doi: 10.1038/nbt.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ren J., Zhang X., Liu X., Fang C., Jiang S., June C.H., et al. A versatile system for rapid multiplex genome-edited CAR T cell generation. Oncotarget. 2017;8:17002–17011. doi: 10.18632/oncotarget.15218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y., Zhang X., Cheng C., Mu W., Liu X., Li N., et al. CRISPR-Cas9 mediated LAG-3 disruption in CAR-T cells. Front Med. 2017;11:554–562. doi: 10.1007/s11684-017-0543-6. [DOI] [PubMed] [Google Scholar]
- 59.Zhou P., Shaffer D.R., Alvarez Arias D.A., Nakazaki Y., Pos W., Torres A.J., et al. In vivo discovery of immunotherapy targets in the tumour microenvironment. Nature. 2014;506:52–57. doi: 10.1038/nature12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shifrut E., Carnevale J., Tobin V., Roth T.L., Woo J.M., Bui C.T., et al. Genome-wide CRISPR screens in primary human T cells reveal key regulators of immune function. Cell. 2018;175 doi: 10.1016/j.cell.2018.10.024. 1985-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Voisinne G., García-Blesa A., Chaoui K., Fiore F., Bergot E., Girard L., et al. Co-recruitment analysis of the CBL and CBLB signalosomes in primary T cells identifies CD5 as a key regulator of TCR-induced ubiquitylation. Mol Syst Biol. 2016;12:876. doi: 10.15252/msb.20166837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hinterleitner R., Gruber T., Pfeifhofer-Obermair C., Lutz-Nicoladoni C., Tzankov A., Schuster M., et al. Adoptive transfer of siRNA Cblb-silenced CD8 + T lymphocytes augments tumor vaccine efficacy in a B16 melanoma model. PLoS One. 2012;7 doi: 10.1371/journal.pone.0044295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liau N.P.D., Laktyushin A., Lucet I.S., Murphy J.M., Yao S., Whitlock E., et al. The molecular basis of JAK/STAT inhibition by SOCS1. Nat Commun. 2018;9:1558. doi: 10.1038/s41467-018-04013-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arafeh R., Qutob N., Emmanuel R., Keren-Paz A., Madore J., Elkahloun A., et al. Recurrent inactivating RASA2 mutations in melanoma. Nat Genet. 2015;47:1408–1410. doi: 10.1038/ng.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang L., Grishin D., Wang G., Aach J., Zhang C.Z., Chari R., et al. Targeted and genome-wide sequencing reveal single nucleotide variations impacting specificity of Cas9 in human stem cells. Nat Commun. 2014;5:5507. doi: 10.1038/ncomms6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guilinger J.P., Thompson D.B., Liu D.R. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Casini A., Olivieri M., Petris G., Montagna C., Reginato G., Maule G., et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat Biotechnol. 2018;36:265–271. doi: 10.1038/nbt.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen J.S., Dagdas Y.S., Kleinstiver B.P., Welch M.M., Sousa A.A., Harrington L.B., et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550:407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vakulskas C.A., Dever D.P., Rettig G.R., Turk R., Jacobi A.M., Collingwood M.A., et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat Med. 2018;24:1216–1224. doi: 10.1038/s41591-018-0137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kleinstiver B.P., Sousa A.A., Walton R.T., Tak Y.E., Hsu J.Y., Clement K., et al. Engineered CRISPR–Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat Biotechnol. 2019;37:276–282. doi: 10.1038/s41587-018-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Strecker J., Jones S., Koopal B., Schmid-Burgk J., Zetsche B., Gao L., et al. Engineering of CRISPR-Cas12b for human genome editing. Nat Commun. 2019;10:212. doi: 10.1038/s41467-018-08224-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Teng F., Cui T., Feng G., Guo L., Xu K., Gao Q., et al. Repurposing CRISPR-Cas12b for mammalian genome engineering. Cell Discovery. 2018;4:63. doi: 10.1038/s41421-018-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu J.-J., Orlova N., Oakes B.L., Ma E., Spinner H.B., Baney K.L.M., et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature. 2019;566:218–223. doi: 10.1038/s41586-019-0908-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ryan D.E., Taussig D., Steinfeld I., Phadnis S.M., Lunstad B.D., Singh M., et al. Improving CRISPR-Cas specificity with chemical modifications in single-guide RNAs. Nucleic Acids Res. 2018;46:792–803. doi: 10.1093/nar/gkx1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wilson L.O.W., O'Brien A.R., Bauer D.C. The current state and future of CRISPR-Cas9 gRNA design tools. Front Pharmacol. 2018;9:749. doi: 10.3389/fphar.2018.00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kosicki M., Tomberg K., Bradley A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rezza A., Jacquet C., Le Pillouer A., Lafarguette F., Ruptier C., Billandon M., et al. Unexpected genomic rearrangements at targeted loci associated with CRISPR/Cas9-mediated knock-in. Sci Rep. 2019;9:3486. doi: 10.1038/s41598-019-40181-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hustedt N., Durocher D. The control of DNA repair by the cell cycle. Nat Cell Biol. 2017;19:1–9. doi: 10.1038/ncb3452. [DOI] [PubMed] [Google Scholar]
- 84.Yang D., Scavuzzo M.A., Chmielowiec J., Sharp R., Bajic A., Borowiak M. Enrichment of G2/M cell cycle phase in human pluripotent stem cells enhances HDR-mediated gene repair with customizable endonucleases. Sci Rep. 2016;6:21264. doi: 10.1038/srep21264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haapaniemi E., Botla S., Persson J., Schmierer B., Taipale J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med. 2018;24:927–930. doi: 10.1038/s41591-018-0049-z. [DOI] [PubMed] [Google Scholar]
- 86.Ihry R.J., Worringer K.A., Salick M.R., Frias E., Ho D., Theriault K., et al. P53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med. 2018;24:939–946. doi: 10.1038/s41591-018-0050-6. [DOI] [PubMed] [Google Scholar]
- 87.Lin S., Staahl B.T., Alla R.K., Doudna J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014;3 doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 89.Gwiazda K.S., Grier A.E., Sahni J., Burleigh S.M., Martin U., Yang J.G., et al. High efficiency CRISPR/Cas9-mediated gene editing in primary human T-cells using mutant adenoviral E4orf6/E1b55k "helper" proteins. Mol Ther. 2016;24:1570–1580. doi: 10.1038/mt.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., Ploegh H.L. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pinder J., Salsman J., Dellaire G. Nuclear domain 'knock-in' screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 2015;43:9379–9392. doi: 10.1093/nar/gkv993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li X.L., Li G.H., Fu J., Fu Y.W., Zhang L., Chen W., et al. Highly efficient genome editing via CRISPR-Cas9 in human pluripotent stem cells is achieved by transient BCL-XL overexpression. Nucleic Acids Res. 2018 Nov 2;46(19):10195–10215. doi: 10.1093/nar/gky804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu H., Zhang H., Wu X., Ma D., Wu J., Wang L., et al. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature. 2018;563:131–136. doi: 10.1038/s41586-018-0629-6. [DOI] [PubMed] [Google Scholar]
- 94.Yu C., Liu Y., Ma T., Liu K., Xu S., Zhang Y., et al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16:142–147. doi: 10.1016/j.stem.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Aird E.J., Lovendahl K.N., St Martin A., Harris R.S., Gordon W.R. Increasing Cas9-mediated homology-directed repair efficiency through covalent tethering of DNA repair template. Communications Biology. 2018;1:54. doi: 10.1038/s42003-018-0054-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Charpentier M., Khedher A.H.Y., Menoret S., Brion A., Lamribet K., Dardillac E., et al. CtIP fusion to Cas9 enhances transgene integration by homology-dependent repair. Nat Commun. 2018;9:1133. doi: 10.1038/s41467-018-03475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Long A.H., Haso W.M., Shern J.F., Wanhainen K.M., Murgai M., Ingaramo M., et al. 4-1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat Med. 2015;21:581–590. doi: 10.1038/nm.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bendle G.M., Linnemann C., Hooijkaas A.I., Bies L., De Witte M.A., Jorritsma A., et al. Lethal graft-versus-host disease in mouse models of T cell receptor gene therapy. Nat Med. 2010;16:565–570. doi: 10.1038/nm.2128. [DOI] [PubMed] [Google Scholar]
- 99.Hundal J., Carreno B.M., Petti A.A., Linette G.P., Griffith O.L., Mardis E.R., et al. pVAC-Seq: a genome-guided in silico approach to identifying tumor neoantigens. Genome Med. 2016;8:11. doi: 10.1186/s13073-016-0264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Robbins P.F., Lu Y.C., El-Gamil M., Li Y.F., Gross C., Gartner J., et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19:747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tran E., Ahmadzadeh M., Lu Y.C., Gros A., Turcotte S., Robbins P.F., et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science. 2015;350:1387–1390. doi: 10.1126/science.aad1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tran E., Robbins P.F., Lu Y.-C., Prickett T.D., Gartner J.J., Jia L., et al. T-cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med. 2016;375:2255–2262. doi: 10.1056/NEJMoa1609279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tran E., Turcotte S., Gros A., Robbins P.F., Lu Y.C., Dudley M.E., et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gros A., Parkhurst M.R., Tran E., Pasetto A., Robbins P.F., Ilyas S., et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med. 2016;22:433–438. doi: 10.1038/nm.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Van Rooij N., Van Buuren M.M., Philips D., Velds A., Toebes M., Heemskerk B., et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;31:e439–442. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hadrup S.R., Bakker A.H., Shu C.J., Andersen R.S., van Veluw J., Hombrink P., et al. Parallel detection of antigen-specific T-cell responses by multidimensional encoding of MHC multimers. Nat Methods. 2009;6:520–526. doi: 10.1038/nmeth.1345. [DOI] [PubMed] [Google Scholar]
- 107.Linnemann C., Heemskerk B., Kvistborg P., Kluin R.J.C., Bolotin D.A., Chen X., et al. High-throughput identification of antigen-specific TCRs by TCR gene capture. Nat Med. 2013;19:1534–1541. doi: 10.1038/nm.3359. [DOI] [PubMed] [Google Scholar]
- 108.Gee M.H., Han A., Lofgren S.M., Beausang J.F., Mendoza J.L., Birnbaum M.E., et al. Antigen identification for orphan T cell receptors expressed on tumor-infiltrating lymphocytes. Cell. 2018;172:549–563. doi: 10.1016/j.cell.2017.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li G., Bethune M.T., Wong S., Joglekar A.V., Leonard M.T., Wang J.K., et al. T cell antigen discovery via trogocytosis. Nat Methods. 2019;16:183–190. doi: 10.1038/s41592-018-0305-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Joglekar A.V., Leonard M.T., Jeppson J.D., Swift M., Li G., Wong S., et al. T cell antigen discovery via signaling and antigen-presenting bifunctional receptors. Nat Methods. 2019;16:191–198. doi: 10.1038/s41592-018-0304-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kochenderfer J.N., Wilson W.H., Janik J.E., Dudley M.E., Stetler-Stevenson M., Feldman S.A., et al. Eradication of B-lineage cells and regression of lymphoma in a patient treated with autologous T cells genetically engineered to recognize CD19. Blood. 2010;116(20):4099–4102. doi: 10.1182/blood-2010-04-281931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Porter D.L., Levine B.L., Kalos M., Bagg A., June C.H. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. New England J Med. 2011;365(8):725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]