Abstract
Fourier transform infrared (FTIR) spectroscopy in combination with the partial least squares (PLS) multivariative statistical technique was used for quantitative analysis of the poly(β-hydroxybutyrate) (PHB) contents of bacterial cells. A total of 237 replicate spectra from 34 samples were obtained together with gas chromatography-determined reference PHB contents. Using the PLS regression, we were able to relate the infrared spectra to the reference PHB contents, and the correlation coefficient between the measured and predicted values for the optimal model with a standard error of prediction of 1.49% PHB was 0.988. With this technique, there are no solvent requirements, sample preparation is minimal and simple, and analysis time is greatly reduced; our results demonstrate the potential of FTIR spectroscopy as an alternative to the conventional methods used for analysis of PHB in bacterial cells.
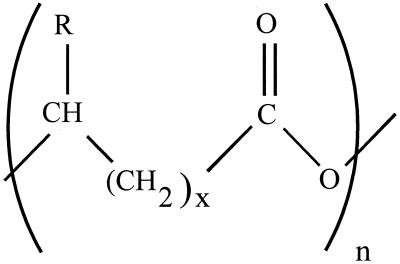
Many prokaryotic microorganisms synthesize polyhydroxyalkanoates (PHAs) as carbon and energy reserves (2, 5, 8, 25, 26), and poly(β-hydroxybutyrate) (PHB) is the most common PHA. The general chemical structure of these compounds is shown in Fig. 1.
FIG. 1.
General chemical structure of PHAs. For PHB, x is 1 and R is CH3.
The composition of the alkyl side chain (the R group) and the number of methylene units together determine the identity of the monomer. For PHB, the number of methylene units is 1 and the R group is CH3. A number of organisms have been developed in an effort to optimize the yield of PHB, and recently accumulations of PHB at levels up to 80% of dry cell weight have been obtained under optimum growth conditions (25). PHB is an attractive alternative to the environmentally unfriendly petrochemically derived plastics because its copolymer with polyhydroxyvalerate is a biodegradable molecule with properties similar to those of polyolefins, such as polypropylene and polyethylene. For this reason industrial fermentation of PHB has been the focus of a number of studies (1, 38, 39), while other studies have been directed towards the use of PHB-producing organisms in the breakdown of wastewater (32, 42).
The most common method available at present for analysis of PHAs in bacterial cells is gas chromatography (GC) (6, 22, 31). The GC method involves hydrolysis and subsequent methanolysis or propanolysis of the PHAs in whole cells in the presence of sulfuric acid and chloroform. This method is time-consuming and laborious and involves extensive use of solvents. Other methods of PHA analysis include gravimetry, infrared (IR) spectroscopy of chemically extracted PHB (23), fluorimetry (10), and cell carbon analysis (37). To aid in the development of more efficient fermentation processes and to monitor production, rapid feedback on the state of fermentation in terms of the PHA content of cells is required.
Fourier transform infrared (FTIR) spectroscopy is a routine chemical technique used to study molecular structure, but when it is applied to a large collection of intact microbial cells, the resulting spectra reflect the total biochemical composition of the cells (28). IR spectroscopy can thus provide a total, simultaneous chemical analysis. The observed bands in spectra of PHA-producing bacteria are due to the major cellular constituents, such as proteins, lipids, polysaccharides, nucleic acids, and PHAs (9, 15, 19, 20, 28). Because FTIR spectra can be considered chemical profiles of samples, the spectra can be used to predict the concentration of an analyte (e.g., the concentration of PHB). FTIR spectroscopy has been used previously to monitor water-soluble extracellular analytes in fermentation systems for ethanol (13, 30), lactic acid (12, 30), and glucose and acetic acid (11) and, very recently, for quantification of a recombinant protein (27). In a recent study Schuster et al. (35) used FTIR spectroscopy to monitor the physiological state of bacterial cells as an indirect method to determine the stage of fermentation. Quantification of PHB and major cell components by using dispersive IR spectroscopy and evaluation of data by using an absorption coefficient at selected wave number values have been developed by Zagreba et al. (43) and have been used for PHB analysis by Savenkova et al. (33). Grube et al. (16) have recently used this method for quantitative analysis of major cell components. In this study we monitored an intracellular analyte, PHB. For quantification, the spectra had to be calibrated with known reference values for the analyte, as determined by an established method, such as GC. This allowed the development of mathematical models in which the spectra were related to the analyte concentrations. The most common multivariative methods used for this purpose are the principal-component regression (PCR) and partial least-squares (PLS) methods.
In a recent study Hong et al. investigated identification of different PHAs in whole bacterial cells by using FTIR spectroscopy (21), but to our knowledge no work has been carried out previously on quantification of PHAs in intact cells. FTIR spectroscopy also has the following advantages: very small samples are required (∼0.4 mg of biomass); speed (analysis time, ∼30 min); no solvent is required; and minimal sample manipulation is required.
We describe here a study of intracellular PHA contents in which we used FTIR spectroscopy and multivariative statistics, and we show that this combination method is a promising alternative to the conventional methods used for PHB analysis.
MATERIALS AND METHODS
Bacterial strains and plasmid DNA.
PHB accumulation was studied in a recombinant Escherichia coli DH5α clone containing the Acinetobacter sp. pha locus on plasmid pJKD1425. E. coli(pJKD1425) carried all of the genes required for PHB biosynthesis under the control of the Acinetobacter promoters (34).
Culture methods.
E. coli was maintained for the long term on a 33% glycerol solution at −70°C and was subcultured weekly on Luria-Bertani (LB) agar plates. For maintenance of the plasmid, 100 μg of ampicilin per cm3 was added as required.
Seeder cultures were prepared in 10-cm3 portions of LB medium (0.5 g of yeast extract per liter, 1 g of tryptone per liter, 1 g of NaCl per liter) supplemented with 0.5% glucose and 100 μg of ampicilin per cm3 by inoculating the medium with single colonies from agar plates. The cultures were then grown aerobically at 37°C for 24 h.
E. coli shake flasks (120 rpm) containing 100 cm3 of LB medium supplemented with 0.5% glucose and 100 μg of ampicilin per cm3 were inoculated with 2-cm3 portions of a 24-h-old seeder culture, and the preparations were incubated aerobically at 37°C for 24 to 48 h.
Sample preparation.
Over the course of the culture period, 34 samples with a range of PHB contents were collected aseptically and analyzed by IR spectroscopy and GC. For the IR spectroscopy analysis, 1 cm3 of a culture was collected and centrifuged at 13,000 × g for 5 min. The growth medium was removed by washing, resuspension in isotonic saline, and further centrifugation. The final pellet was resuspended in 80 μl of isotonic saline, and 20 μl of this solution was deposited onto a type KRS-5 (thallium bromide-iodide crystal) IR-transparent substrate. Additional serial dilutions of the remaining solution were prepared, and 20-μl portions were deposited. The deposits were dried in a vacuum desiccator for 10 to 15 min before spectra were acquired immediately.
For the GC analysis, 10-20-cm3 portions of cultures were collected at the same time that samples were collected for the IR spectroscopy analysis. These samples were centrifuged at 13,000 × g for 5 min, the supernatants were removed, and the remaining pellets were freeze-dried and stored at −20°C until analysis.
Spectral acquisition.
Spectra were recorded with a Bruker model IFS-55 FTIR spectrometer coupled to a Bruker IR microscope fitted with a liquid N2-cooled mercury-cadmium-tellurium detector. The Bruker system was controlled with an IBM-compatible PC running OPUS, version 2.2, software. Absorbance spectra were collected at wavenumber values between 3,650 and 700 cm−1 with spectral resolution of 8 cm−1, and 10 scans were coadded and averaged. A Blackman-Harris four-term apodization function was used along with a zero-filling factor of 2.
To minimize differences between spectra due to baseline shifts, the spectra were baseline corrected by using the Rubber Band algorithm of the OPUS, version 2.2, software and 200 baseline points and excluding the CO2 bands. Spectra were normalized to the amide I band at 1,654 cm−1 to account for any differences in deposit thickness. Six to 12 spectra were recorded for each sample deposit to assess precision and to ensure that representative spectra of each sample deposit were collected.
PHB reference analysis.
The PHB reference measurements were obtained by using a modified acidic methanolysis method of Braunegg et al. (6). The modification was found to be necessary because individual weighing of approximately 3 to 4 mg of biomass resulted in relatively high errors even when an analytical balance was used. To reduce the errors associated with weighing of the samples, the following sample transfer method was used. An accurately weighed 20- to 30-mg portion of freeze-dried cell mass was placed in a polytetrafluoroethylene-lined screw-cap test tube and crushed to a fine powder. To this was added a volume of chloroform which resulted in a freeze-dried cell mass concentration of 3 to 4 mg/cm3 of chloroform, and the resulting solution was sonicated for ca. 30 min or until the solution was completely homogenized. One cubic centimeter of this solution was then placed in another polytetrafluroethylene-lined screw-cap test tube, to which were added 0.85 cm3 of methanol containing ca. 0.5 mg of benzoic acid per cm3 as an internal standard and 0.15 ml of concentrated sulfuric acid. The reaction mixture was heated at 100°C for 2 h and cooled rapidly; then 1 cm3 of distilled water was added, and the mixture was vortexed for 1 min. The organic and aqueous layers were allowed to separate, and 0.5 μl of the lower organic layer was injected into the GC. The GC used was a Varian model 3700 GC equipped with a DB-Wax capillary column (15 m by 0.53 mm; internal film thickness, 1.0 μm) coupled to a Hewlett-Packard model 3396A integrator. The injector and flame ionization detector temperatures were set at 200 and 250°C, respectively. The temperature was set at 80°C and then increased at a rate of 10°C/min until the maximum temperature, 200°C, was reached. Under these conditions, the retention times for the PHB derivative (methyl 3-hydroxybutyric acid) and the benzoic acid internal standard were 4.4 and 5.8 min, respectively.
Data analysis.
Two multivariative statistical methods, the PCR and PLS methods, were employed to create calibration models to relate the spectra obtained to the reference measurements obtained. The regression coefficients and the standard errors of prediction (SEP) for the PLS regression models were used as measurements of the “goodness of fit.” The SEP is the standard deviation of the spread of the errors (the difference between the measured and predicted values). The SEP is calculated from a leave-out-one cross-validation (7, 14, 17, 24). These statistical analyses were performed by using the Unscrambler 6.11 Camo ASA computer software package.
Derivative spectra were also used in the preprocessing of data prior to PLS regression. Spectral derivatives can remove remaining spectral contributions from a sample deposit that are not corrected for by the baseline correction and normalization functions. The spectral derivative technique can also be considered a pseudoresolution enhancement technique, because the derivatives are able to highlight slight variations in the slopes and contours of bands and hence increase the accessible spectral information. These two advantages of spectral derivatives, therefore, can combine to reduce the number of factors required to obtain a minimum SEP by removing the need for extra factors to model spectral variations that are not related to the analyte of interest.
RESULTS AND DISCUSSION
The IR spectrum of a sample represents its total chemical composition, because every chemical compound in the sample makes its own distinct contribution to the absorbance spectrum. The distinctness of an individual spectrum, which is determined by the chemical structure of each component and the degree to which each component contributes to the spectrum, is directly related to the concentrations of the components of the sample. It is on this basis that quantification of analytes is performed.
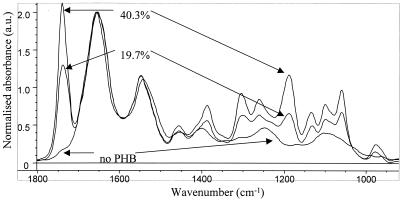
The representative spectra in Fig. 2 are results of the cumulative absorbances for all of the chemical species present, which resulted in the relatively broad spectral features that are typical of biological samples. These spectra are dominated by the absorbances for the major cellular constituents, namely, PHB (in the case of PHB-producing microorganisms) and cellular proteins. The major PHB bands are the intense ester carbonyl stretch at 1738 to 1728 cm−1 and a number of strong bands at wavenumber values between 1,450 and 1,000 cm−1 due to methyl (CH3) and methylene (CH2) deformations and C-O stretches. The major spectral features due to the organism itself are the protein absorbances apparent as strong features in the lower trace of Fig. 2. These are the amide I band at 1,654 cm−1, which is due primarily to the amide carbonyl stretching vibration, and the amide II band at 1,540 cm−1, which is due mostly to N-H bending vibrations. A more detailed description of band assignments is presented in Table 1.
FIG. 2.
Representative spectra of samples with different PHB contents and a sample containing no PHB. a.u., arbitrary units.
TABLE 1.
Band assignments
| Wavenumber value (cm−1) | Assignmenta | Comments |
|---|---|---|
| ∼1,735 | νC⩵O of ester functional groups primarily from lipids, fatty acids, and PHB (18, 40, 45)b | Exact position of PHB absorbance depends on the degree of crystallinity of the PHB |
| ∼1,650 | νC⩵O of amides associated with proteins (29, 40) | Usually called the amide I band; may also contain contributions from C⩵C stretches of olefinic and aromatic compounds |
| ∼1,540 | δ(N H) of amides associated with proteins (29, 40) | Usually called the amide II band; may also contain contributions from C⩵N stretches |
| ∼1,455 | δas (CH3) and δas (CH2) of proteins (44) | Positions of these assignments can vary; contributions also from PHB |
| ∼1,398 | δs (CH3) and δs (CH2) of proteins, νs(C O of COO−) groups (29, 44) | Positions of these assignments can vary; contributions also from PHB |
| ∼1,242 | νas (P⩵O) of the phosphodiester backbone of nucleic acids (DNA and RNA) (29, 41) | May also be due to the presence of phosphorylated proteins and polyphosphate storage products |
| ∼1,080 | νs (P⩵O) of the phosphodiester backbone of nucleic acids (DNA and RNA) (29, 41) | May also be due to the presence of phosphorylated proteins and polyphosphate storage products |
| 1,200–900 | ν(C O C) of polysaccharides (41, 44) | Contributions also from PHB |
νas, asymmetric stretch; νs, symmetric stretch; δas, asymmetric deformation (bend); δs, symmetric deformation (bend).
The numbers in parentheses are reference numbers.
Spectral reproducibility.
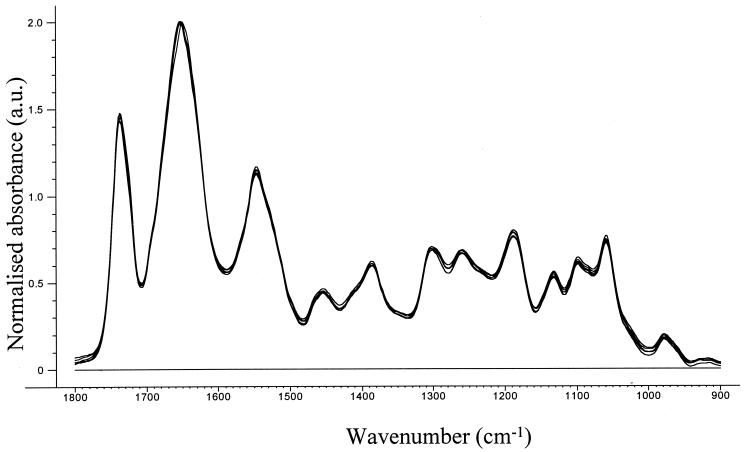
Six to 12 replicate spectra obtained from different areas within each sample deposit were recorded to ensure that representative spectra were collected from each deposit. These spectra were then baseline corrected and normalized to the amide I band. The replicate spectra exhibited little variation, as shown in Fig. 3, in which six spectra are overlaid together.
FIG. 3.
Six replicate spectra from the same deposit of a sample containing 27.3% PHB. All spectra have been baseline corrected and normalized to the amide I band. a.u., arbitrary units.
The baseline correction function attempts to correct for the spectral artifacts caused by variations in the scattering, diffraction, and refraction properties of the samples as the IR beam passes through a sample. The normalization function attempts to correct for differences in spectral absorbances due to different sample thicknesses within the same deposit. Normalization is performed to the amide I band because there is no spectral contribution from PHB in this spectral region. It has been shown that the protein content per cell does not vary substantially for a wide range of PHB contents, and so the protein content of the cells provides an ideal internal standard for normalization (26).
The manual sample preparation procedure results in unavoidable irregularities in deposition from sample to sample and even within a sample. The differences add to the total error of the model by contributing a certain amount of imprecision. The precision of values predicted from an IR multivariative model is calculated by using replicate spectral measurements. A PLS regression was performed with all 237 replicate spectra from 34 samples, which allowed us to calculate the average predicted value for each sample, the standard deviation, and the relative standard deviation (RSD) for the predicted values for the replicates of each sample. The resulting data could then be used to assess the reproducibility of the IR spectra. An analysis of the spread of the predictions for replicate spectra for each sample showed that only one replicate spectrum out of the entire data set was an outlier; i.e., the predicted value was more than 2 standard deviations from the mean. This was found to be the case for the regressions performed with the spectra themselves, as well as the regressions performed with first and second derivatives of the spectra.
The replicate spectra in Fig. 3 can thus be considered highly reproducible and show that baseline correction and normalization almost completely accounted for and corrected for sample deposit heterogeneity.
To detect outliers that were due to errors in the reference measurements, as opposed to errors in IR measurements, the replicate spectra for each sample were averaged, and a PLS regression (performed with spectra and first and second derivative spectra) was performed by using the averages instead of the replicate spectra. The results showed that there was one consistent outlier (more than 2 SEP from the reference value), which was removed from the data set; this left 33 samples together with their replicates. An examination of the raw spectra for the outlier sample revealed that they all had unusually high absorbance values. The mercury-cadmium-tellurium detector used in this study is nonlinear at high absorbances, and so only spectra with maximum absorbances of <0.7 should have been used in the analysis.
PLS regression results.
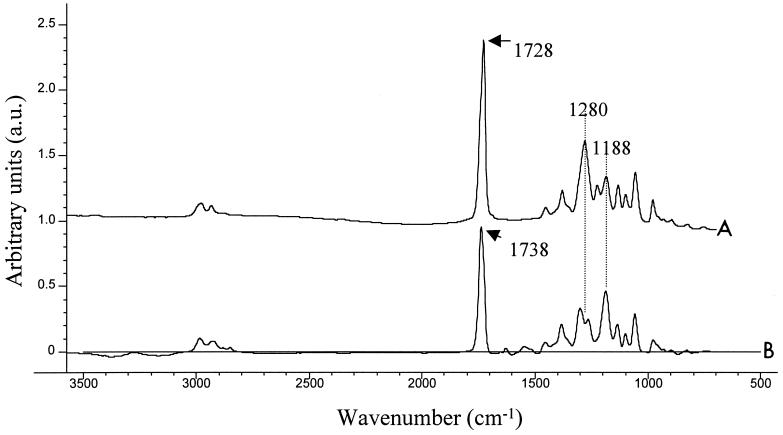
Due to the long computational times required, whole-spectrum PLS regressions were not performed for the replicate spectrum data sets. However, a PLS regression performed with the whole spectra of averaged replicates (33 spectra, one from each sample) resulted in data that were virtually identical to data from PLS regression performed with the 1,800- to 900-cm−1 spectral region. This was due to the fact that virtually all of the PHB spectral information is in the 1,800- to 900-cm−1 region. The results of this PLS regression are shown in Table 2. A regression coefficient plot, which shows the variables which correlated most closely with the PHB content, and a solution cast spectrum of pure PHB are shown in Fig. 4. The regression coefficient plot, which is essentially a mathematical extraction of the PHB spectrum from the spectra of the whole organism, shows that the spectral range from 1,800 to 900 cm−1 contains most of the chemical information related to PHB.
TABLE 2.
Summary of PLS results
| Data | Spectra
|
First derivative
spectra
|
Second derivative spectra
|
|||
|---|---|---|---|---|---|---|
| r | SEP (%) | r | SEP (%) | r | SEP (%) | |
| Replicate spectrum data set, 1,800-900 cm−1 | 0.983 | 1.77 (9)a | 0.983 | 1.76 (7) | 0.982 | 1.81 (6) |
| Replicate spectrum data set (outlier removed), 1,800-900 cm−1 | 0.988 | 1.49 (10) | 0.988 | 1.50 (7) | 0.987 | 1.58 (10) |
| Averaged replicate spectra, 3,650-700 cm−1 | 0.975 | 2.12 (2) | 0.980 | 1.89 (1) | 0.981 | 1.84 (2) |
| Averaged replicate spectra (outlier removed) 3,650-700 cm−1 | 0.981 | 1.89 (3) | 0.985 | 1.68 (1) | 0.985 | 1.68 (2) |
| Averaged replicate spectra, 1,800-900 cm−1 | 0.977 | 2.05 (1) | 0.980 | 1.88 (1) | 0.982 | 1.83 (2) |
| Averaged replicate spectra (outlier removed), 1,800-900 cm−1 | 0.982 | 1.84 (2) | 0.985 | 1.68 (1) | 0.985 | 1.68 (2) |
The values in parentheses are the numbers of PLS factors required to obtain a minimum SEP.
FIG. 4.
Spectrum of a solution-cast film of pure PHB (line A) and regression coefficient plot from a PLS regression performed with entire underivatized spectra (line B). The major spectral differences are indicated.
Figure 4 also shows the differences between the solution-cast spectrum of PHB and the plot of regression coefficients. The major difference is the shift of the position of the ester carbonyl stretch in the solution cast spectrum of PHB from 1,728 to 1,738 cm−1 in the regression coefficient plot. There are also a number of other differences in the intensities of bands, particularly those at 1,280 and 1,188 cm−1, as indicated in Fig. 4. These differences are due to differences in the physical state of the PHB. The solution cast PHB crystallizes almost instantly after solvent evaporation, while the PHB in the cells remains in an amorphous state even after complete drying. This observation confirms the results of recent studies on the physical state of PHB in vivo, which showed that PHB is in an amorphous state (3, 36). The PHB in cells, however, does slowly begin to crystallize 10 to 15 min after drying. This causes spectral changes with time, which means that spectra of samples must be obtained at the same time after complete drying. FTIR spectroscopy has also been used to investigate and even predict the degree of crystallinity of PHB samples (4).
An initial PLS regression performed with the entire replicate spectrum data set before outlier removal (237 spectra from 34 samples) resulted in a relatively poor correlation coefficient (between predicted and measured values) of 0.983 and an SEP of 1.77% PHB (nine factors). These results were improved by outlier removal, as described above. The analyses in which we used the raw and spectral first derivative provided the best results; the correlation coefficient and SEP were 0.988 and 1.49% PHB (10 factors), respectively, for the raw spectrum PLS regression. The SEP in this optimized model was only 17% greater than the standard error of calibration, which can be considered the theoretical “best” fit. The results obtained by using first and second derivatives showed no further improvement, although slightly fewer factors were required. This indicates that spectral normalization and baseline removal remove most of the spectral variation not related to the analytes of interest. This shows that the validation samples from the leave-out-one cross-validation analysis were predicted almost as well as the calibration samples were, which indicated that the model was valid and robust. These results and the results of other PLS regressions are summarized in Table 2. The optimal number of factors in a particular model was chosen by examining a plot of SEP versus number of factors.
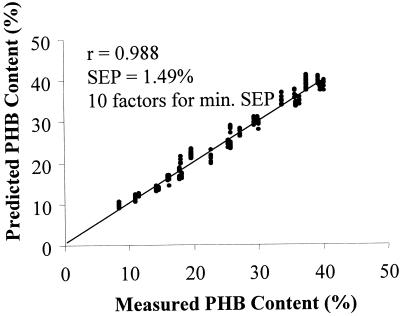
A plot of predicted PHB content versus measured PHB content is shown in Fig. 5, which graphically shows the excellent fit. PCR was also employed as an additional multivariative technique to relate the spectral information to the analyte concentrations. The results proved to be practically identical to the PLS results.
FIG. 5.
Plot of measured versus predicted PHB contents from the optimized PLS regression model.
It is not possible to measure the absolute accuracy of the IR method compared to the accuracy of the reference method without resorting to more fundamental reference analysis. Such fundamental analysis would be almost impossible as it would involve creating known standard samples of PHB in bacteria. It is, however, possible to estimate the role which imprecision in the IR and reference methods plays in determining the overall accuracy of the method. A comparison of the RSD of the replicate spectra per sample to the RSD of the triplicate reference measurements showed that in general the IR method was more precise, indicating that most of the error with the model is due to errors in reference measurements. Grube et al. (16) obtained similar results for quantification of major cell components.
In this study we demonstrated the ability of FTIR spectroscopy in combination with multivariative statistics to quantitatively determine the PHB contents of bacterial cells, particularly recombinant E. coli cells. To further improve the attractiveness of the technique, investigations into the possibility of using one data set for spectra for analysis of PHB from different organisms (i.e., samples with different biological backgrounds) are being conducted. This could mean that one model derived from spectra of different organisms could be used to predict the PHB contents of different bacterial species. This technique has several advantages, including no solvent requirement, minimal, simple sample preparation, and greatly reduced analysis time (ca. 30 min from sampling to quantification), and shows that FTIR spectroscopy can be used as an alternative to the conventional methods used to analyze PHB in bacterial cells. The technique, which is also useful as a rapid screening tool for new organisms, is not limited to analysis of PHB fermentations and could also be useful in analyses of other fermentation products.
ACKNOWLEDGMENTS
We thank Victor Guzman for assistance with the GC analysis and J. K. Davies, Department of Microbiology, Monash University, for supplying the recombinant E. coli.
REFERENCES
- 1.Ackermann J U, Babel W. Approaches to increase the economy of the PHB production. Polym Degrad Stab. 1998;59:183–186. [Google Scholar]
- 2.Anderson A J, Dawes E A. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev. 1990;54:450–472. doi: 10.1128/mr.54.4.450-472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnard G N, Sanders J K M. The poly-β-hydroxybutyrate granule in vivo. J Biol Chem. 1989;264:3286–3291. [PubMed] [Google Scholar]
- 4.Bloembergen S, Holden D A, Hamer G H, Bluhm T L, Marchessault R H. Studies of composition and crystallinity of bacterial poly(β-hydroxybutyrate-co-β-hydroxyvalerate) Macromolecules. 1986;19:2865–2871. [Google Scholar]
- 5.Braunegg G, Lefebvre G, Genser K F. Polyhydroxyalkanoates, biopolymers from renewable resources: physiological and engineering aspects. J Biotechnol. 1998;65:127–161. doi: 10.1016/s0168-1656(98)00126-6. [DOI] [PubMed] [Google Scholar]
- 6.Braunegg G, Sonnleitner B, Lafferty R M. A rapid gas chromatographic method for the determination of poly-β-hydroxybutyric acid in bacterial biomass. Eur J Appl Microbiol Biotechnol. 1978;6:29–37. [Google Scholar]
- 7.Brereton R G. Chemometrics. Applications of mathematics and statistics to laboratory systems. New York, N.Y: Ellis Horwood Limited; 1993. [Google Scholar]
- 8.Choi J, Lee S Y. Factors affecting the economics of polyhydroxyalkanoate production by bacterial fermentation. Appl Microbiol Biotechnol. 1999;51:13–21. [Google Scholar]
- 9.Curk M C, Peladan F, Hubert J C. Fourier transform infrared (FTIR) spectroscopy for identifying Lactobacillusspecies. FEMS Microbiol Lett. 1994;123:241–248. [Google Scholar]
- 10.Degelau A, Scheper T, Bailey J E, Guske C. Fluorometric measurement of poly-β hydroxybutyrate in Alcaligenes eutrophusby flow cytometry and spectrofluorometry. Appl Microbiol Biotechnol. 1995;42:653–657. [Google Scholar]
- 11.Doak D L, Phillips J A. In situ monitoring of an Escherichia colifermentation using a diamond composition ATR probe and mid-infrared spectroscopy. Biotechnol Prog. 1999;15:529–539. doi: 10.1021/bp990039b. [DOI] [PubMed] [Google Scholar]
- 12.Fayolle P, Daniel P, Corrieu G. Monitoring of fermentation processes producing lactic acid bacteria by mid-infrared spectroscopy. Vib Spectrosc. 1997;14:247–252. [Google Scholar]
- 13.Gallignani M, Garrigues S, de la Guardia M. Derivative Fourier transform infrared spectrometric determination of ethanol in alcoholic beverages. Anal Chim Acta. 1994;287:275–283. [Google Scholar]
- 14.Geladi P, Kowalski B R. Partial least-squares regression: a tutorial. Anal Chim Acta. 1986;185:1–17. [Google Scholar]
- 15.Goodacre R, Timmins E M, Rooney P J, Rowland J J, Kell D B. Rapid identification of Streptococcus and Enterococcusspecies using diffuse reflectance-absorbance Fourier transform infrared spectroscopy and artificial neural networks. FEMS Microbiol Lett. 1996;140:233–239. doi: 10.1016/0378-1097(96)00186-3. [DOI] [PubMed] [Google Scholar]
- 16.Grube M, Zagreba J, Gromozova E, Fomina M M. Comparative investigation of the macromolecular composition of mycelia forms Thielavia terrestrisby infrared spectroscopy. Vib Spectrosc. 1999;19:301–306. [Google Scholar]
- 17.Haaland D M, Thomas E V. Partial least squares methods for spectral analysis. 1. Relation to other quantitative calibration methods and the extraction of qualitative information. Anal Chem. 1988;60:1193–1202. [Google Scholar]
- 18.Hedrick D B, Nivens D E, Stafford C, White D C. Rapid differentiation of archaebacteria from eubacteria by diffuse reflectance Fourier-transform IR spectroscopic analysis of lipid preparation. J Microbiol Methods. 1991;13:67–73. [Google Scholar]
- 19.Helm D, Labischinski H, Naumann D. Elaboration of a procedure for identification of bacteria using Fourier transform IR spectral libraries: a stepwise correlation approach. J Microbiol Methods. 1991;14:127–142. [Google Scholar]
- 20.Helm D, Labischinski H, Schallen G, Naumann D. Classification and identification of bacteria by Fourier-transform infrared spectroscopy. J Gen Microbiol. 1991;137:69–79. doi: 10.1099/00221287-137-1-69. [DOI] [PubMed] [Google Scholar]
- 21.Hong K, Sun S, Tian W, Chen G Q, Huang W. A rapid method for detecting bacterial polyhydroxyalkonates in intact cells by Fourier transform infrared spectroscopy. Appl Microbiol Biotechnol. 1999;51:523–526. [Google Scholar]
- 22.Huijberts G N M, van der Wal H, Wilkinson C, Eggink G. Gas-chromatographic analysis of poly(3-hydroxyalkanoates) in bacterial. Biotechnol Lett. 1994;8:187–192. [Google Scholar]
- 23.Juttner R R, Lafferty R M, Knackmuss H J. A simple method for the determination of poly-β-hydroxybutric acid in microbial biomass. Eur J Appl Microbiol. 1975;1:233–237. [Google Scholar]
- 24.Kemsley E K. Discriminant analysis of high-dimensional data: a comparison of principal component analysis and partial least squares data reduction methods. Chemom Intell Lab Syst. 1996;33:47–61. [Google Scholar]
- 25.Lee S Y. Bacterial polyhdroxyalkanoates. Biotechnol Bioeng. 1996;49:1–14. doi: 10.1002/(SICI)1097-0290(19960105)49:1<1::AID-BIT1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 26.Lee S Y. Plastic bacteria? Progress and prospects for polyhydroxyalkanoate production in bacteria. TIBTECH. 1996;14:431–438. [Google Scholar]
- 27.McGovern A C, Kara E R, Kell B V, Goodacre D B. Rapid analysis of the expression of heterologous proteins in Escherichia coliusing pyrolysis mass spectrotrometry and Fourier transform infrared spectroscopy with chemometrics: application to alpha 2-interferon production. J Biotechnol. 1999;72:157–167. doi: 10.1016/s0168-1656(99)00128-5. [DOI] [PubMed] [Google Scholar]
- 28.Naumann D, Helm D, Labischinski H. Microbiological characterisation by FTIR spectroscopy. Nature. 1991;351:81–82. doi: 10.1038/351081a0. [DOI] [PubMed] [Google Scholar]
- 29.Nelson W H. Modern techniques for rapid microbiological analysis. New York, N.Y: VCH Publishers; 1991. [Google Scholar]
- 30.Picque D, Lefier D, Grappin R, Corrieu G. Monitoring of fermentation by infrared spectrometry. Anal Chim Acta. 1993;279:67–72. [Google Scholar]
- 31.Riis V, Mai W. Gas chromatographic determination of poly-β-hydroxybutyric acid in microbial biomass after hydrochloric acid propanolysis. J Chromatogr. 1988;445:285–289. [Google Scholar]
- 32.Satoh H, Iwamoto Y, Mino T, Matsuo T. Activated sludge as a possible source of biodegradable plastic. Water Sci Technol. 1998;38:103–109. [Google Scholar]
- 33.Savenkova L, Bonartseva G, Gertsberg Z, Beskina N, Zagreba J, Grube M. Nitrogen fixation and poly-3-hydroxybutyrate accumulation by some Azotobactergenus bacteria strains. Kcs Proc Latvian Acad Sci Sect B. 1994;562/563:89–92. [Google Scholar]
- 34.Schembri M A, Bayly R C, Davies J K. Phosphate concentration regulates transcription of the Acinetobacterpolyhydroxyalkanoic acid biosynthetic genes. J Bacteriol. 1995;177:4501–4507. doi: 10.1128/jb.177.15.4501-4507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuster K C, Mertens F, Gapes J R. FTIR spectroscopy applied to bacterial cells as a novel method for monitoring complex biotechnological processes. Vib Spectrosc. 1999;19:467–477. [Google Scholar]
- 36.Song J J, Yoon S C, Yu S M, Lenz R W. Differential scanning calorimetric study of poly(3-hydroxyoctanoate) inclusions in bacterial cells. Int J Biol Macromol. 1998;23:165–173. doi: 10.1016/s0141-8130(98)00046-4. [DOI] [PubMed] [Google Scholar]
- 37.Stenholm H, Song S, Eriksen H T, Iverson J J L. Indirect estimation of poly-β-hydroxybutyric acid by cell carbon analysis. Biotechnol Tech. 1998;12:451–454. [Google Scholar]
- 38.Wang F, Lee S Y. High cell density culture of metabolically engineered Escherichia colifor the production of poly(3-hydroxybutyrate) in a defined medium. Biotechnol Bioeng. 1998;58:325–328. [PubMed] [Google Scholar]
- 39.Wegen R J V, Ling Y, Middleberg A P J. Industrial production of polyhydroxyalkanoates using Escherichia coli: an economic analysis. Chem Eng Res Design. 1998;76:417–426. [Google Scholar]
- 40.Williams D H, Fleming I. Spectroscopic methods in organic chemistry. 5th ed. London, United Kingdom: McGraw-Hill International (UK) Ltd.; 1996. [Google Scholar]
- 41.Wong P T T, Wong R H, Caputo T A, Godwin T A, Rigas B. Infrared spectroscopy of exfoliated human cervical cells: evidence of extensive structural changes during carcinogenesis. Proc Natl Acad Sci USA. 1988;88:10988–10992. doi: 10.1073/pnas.88.24.10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu P H, Chua H, Huang A L, Lo W, Chen G Q. Conversion of food industrial wastes into bioplastics. Appl Biochem Biotechnol. 1998;70–72:603–614. doi: 10.1007/BF02920172. [DOI] [PubMed] [Google Scholar]
- 43.Zagreba J, Savenkov, Ginovska M. Microbial conversions—fundamentals and application aspects. Riga, Latvia: Zinatre; 1990. IR-spectrophotometric control of chemical composition of microbial biomass; pp. 139–147. [Google Scholar]
- 44.Zeroual W, Choisy C, Doglia S M, Bobichon H, Angiboust J F, Manfait M. Monitoring of bacterial growth and structural analysis as probed by FT-IR spectroscopy. Biochim Biophys Acta. 1994;1222:171–178. doi: 10.1016/0167-4889(94)90166-x. [DOI] [PubMed] [Google Scholar]
- 45.Zeroual W, Manfait M, Choisy C. FT-IR spectroscopy study of perturbations induced by antibiotic on bacteria (Escherichia coli) Pathol Biol. 1995;43:300–305. [PubMed] [Google Scholar]