Abstract
Immune checkpoint blockade (ICB) unleashes immune cells to attack tumors, thereby inducing durable clinical responses in many cancer types. The number of patients responding to ICB is modest, however, and combination treatments are likely needed to overcome the multifaceted suppressive pathways active in the tumor microenvironment (TME). The development of precision immuno-oncology (IO) strategies allowing to identify the optimal treatment of each patient upfront is therefore a pivotal question in the field of cancer immunotherapy. Although single-parameter biomarkers can enrich for response to ICB, their predictive capacity is far from perfect and their clinical utility is complicated by their continuous nature and the difficulty to determine cut-offs that reliably distinguish responding patients from those without clinical benefit. The antitumor immune response that is induced or reinvigorated by immunotherapy is a complex cascade of events requiring the interplay of multiple cell types. To move towards precision IO, it is therefore essential to understand for each individual patient at which level(s) the antitumor immune response failed and how it can be therapeutically restored. Holistic approaches to profile human tumor microenvironments and treatment-induced responses may help to identify critical rate-limiting factors of antitumor immunity. These factors need to be translated into clinically applicable multimodal predictors that allow for the selection of the best IO treatment. This review discusses strategies to (i) create such holistic views of antitumor immunity, (ii) identify measurable parameters capturing the complexity of a patient’s immune status, and (iii) facilitate the incorporation of precision IO research in the clinic.
Key words: cancer immunotherapy, biomarkers, personalized cancer therapy, immune checkpoint inhibition, ex vivo tumor models, precision oncology
Highlights
-
•
Holistic assessment of patient-specific immune profiles to identify new biomarkers.
-
•
Multimodal predictors better reflect the complexity of antitumor immune responses.
-
•
Combination of technical approaches to find key parameters influencing ICB response.
-
•
Exploitation of in vivo and ex vivo cancer models to profile response dynamics.
Introduction
Cancer immunotherapy, particularly immune checkpoint blockade (ICB), has revolutionized the treatment landscape for many cancer types.1 By blocking the interaction between inhibitory receptors such as programmed cell death protein 1 (PD-1), cytotoxic T lymphocyte-associated protein 4 (CTLA-4), and their ligands, tumor-specific T cells can be reinvigorated to mediate tumor regression.2,3 Although some clinical responses are long-lasting, response rates overall remain modest. To elicit the full potential of immunotherapy, alternative and/or combination treatments are required to overcome the multitude of primary and secondary resistance mechanisms. Hence, strategies to develop precision immuno-oncology (IO) treatments as well as to preselect patients for current therapies, but also for future personalized treatments, are urgently needed.
Although many biomarkers have been selected based on a solid biological rationale, they often show limited or no predictive potential in informing treatment decisions.4 Historically, biomarkers were developed by assessing tumor cell characteristics such as molecular features or pathways relevant for the treatment. Translation of this approach to ICB has proven challenging, as this treatment does not directly target cancer cells, but rather is aimed at restoring the defective antitumor immune response. As this response can fail at manifold levels, it is likely that one single biomarker is not sufficient to capture the reason of failure across patients. By the same token, it is conceivable that to successfully restore antitumor immunity in each patient, individual immunotherapy strategies are necessary. This review will focus on the development of precision IO approaches discussing (i) strategies to identify multimodal biomarkers providing a holistic reflection of the patient’s antitumor immune response, (ii) (pre)clinical approaches to identify precision IO biomarkers and treatment strategies, and (iii) opportunities and challenges for precision IO research.
Single parameter IO biomarkers
The first parameter that has been assessed for its value to predict response to PD-1/programmed death-ligand 1 (PD-L1) blockade is PD-L1 due to its critical role in this inhibitory pathway. In addition, PD-L1 expression was suggested to mirror the activity of tumor-specific T cells due to its adaptive expression following interferon-γ (IFN-γ) in most cell types.1,5 Both cancer cells and immune cells express PD-L1, and while expression on both cell types was associated with objective response,6,7 the clinical relevance of these expression patterns remains to be determined. Numerous clinical studies assessing PD-L1 as a biomarker have shown that clinical responses are enriched in patients with PD-L1-high tumors,8,9 but also demonstrated that some patients with PD-L1-low tumors benefit from ICB and vice versa.2,10,11 In addition, genomic aberrations at the PD-L1 locus on chromosome 9p24.1 have been found to result in increased PD-L1 expression in several cancer types, such as Hodgkin’s lymphoma, mediastinal large B-cell lymphoma, and in both non-small-cell lung cancer (NSCLC) and small-cell lung cancer.12 Importantly, Hodgkin’s lymphomas, which harbor PD-L1 locus alterations in up to 97%, show particularly high response rates to PD-1 blockade.13, 14, 15, 16 In some cancer types, as for instance pancreatic and hepatocellular cancer, epigenetic regulation of PD-L1 expression has also been reported, though its impact on clinical response has not yet been explored.17,18
A second parameter that was associated with clinical benefit upon ICB is the number of mutations in the tumor exome, i.e. the tumor mutational burden (TMB).19, 20, 21 This observation was supported by the finding that a subgroup of TMB-high tumors, characterized by mismatch repair deficiency or microsatellite instability, shows high response rates to ICB.22,23 Somatic mutations in the tumor can give rise to so-called neoantigens, which are specific to the tumor and can be recognized by tumor-killing T cells. Hence, a high TMB should increase the likelihood of the cancer cells being eradicated by T cells.24 As observed for PD-L1, however, some tumor types with high TMB are not responsive to ICB or, vice versa, tumor types with low TMB have better response rates than would be predicted based on the number of mutations.19,21,25,26
A third parameter showing predictive value for ICB response is the presence of tumor infiltrating lymphocytes (TILs). The prognostic value of tumor immune infiltration has been demonstrated in many cancer types.27, 28, 29 The first standardized approach to measure the immune infiltrate in colorectal cancers (CRCs) was reported by the ‘immunoscore’, which quantified the density of CD8+ and memory T cells.29 Whereas the value of the immunoscore for predicting response upon ICB is still under evaluation,30 the importance of TIL infiltration for ICB response has been demonstrated in many studies.31, 32, 33, 34 Nevertheless, immune infiltration is no guarantee for response to ICB and may further depend on the type, state, and activity of the infiltrating immune cells.35,36
One critical challenge to fully exploit the above discussed biomarkers for ICB is presented by the continuous nature of these parameters. Thus, consistent and harmonized approaches for assessment and the definition of cut-offs for each biomarker are needed, such as for example proposed for the assessment of TILs37, 38, 39 or for the standardized quantification of PD-L1 by digital pathology approaches.40 Furthermore, to improve the predictive value of these biomarkers, it might be beneficial to apply them in combination, as for example demonstrated by a number of studies that showed better identification of ICB responders by various combinations of TILs, PD-L1, and/or TMB as compared with each marker alone.20,31,41,42 Next to efforts that aim at a better detection of ICB responders, strategies to upfront identify patients without benefit to ICB and in need for alternative or combination therapies should also be developed. Such combination therapies may include both IO-IO combinations (e.g. anti-PD-1 and/or anti-CTLA-4 combined with other immunotherapeutics) or IO drugs in combination with antiangiogenics or chemotherapies. Importantly, biomarkers for response to ICB monotherapy may not necessarily be predictive for combination treatments and need therefore to be assessed separately. As an ultimate goal, the field should aspire to personalize cancer immunotherapy, meaning that a set of specific biomarkers can be tested for each individual patient that identifies the best treatment (combination) from a selection of potential therapeutic options. To achieve this, multimodal predictors are required that capture the complexity of the (therapeutically) induced or reinvigorated antitumor immune response.
Holistic assessment of antitumor immune profiles
An antitumor immune response that ultimately leads to the effective elimination of cancer cells consists of a series of stepwise events that need to be initiated, sustained, and expanded in the tumor. This process has been described as the cancer immunity cycle.43 As this process is absent or dysfunctional in cancer patients, immunotherapy aims at initiating or restoring the cancer immunity cycle. Tumors harbor a plethora of cellular components, however, which influence the distinct steps in the cancer immunity cycle via stimulatory and suppressive signals and differ between cancer types and patients. Therefore, it is crucial to gain a broad understanding of the complex set of cancer cell-intrinsic, immune, and stromal characteristics that modulate the antitumor immune response in the context of immunotherapy. In addition to factors derived from the tumor microenvironment (TME), it is important to be aware that factors related to host immunity such as germline genetics, microbiome, environmental exposure, pharmacological agents, or systemic inflammation may also impact a patient’s immune profile, and thereby antitumor immunity.35
Over the past years, the idea to create a holistic view of the antitumor immune status in each individual cancer patient has been proposed within the frameworks of the cancer-immune set point35 and the cancer immunogram.44 Both concepts suggest appraising an individual cancer patient’s immune profile by assessing multiple biological parameters that reflect rate-limiting elements of antitumor immunity. These elements have been deployed by the cancer-immune set point to evaluate the balance between factors that stimulate or inhibit antitumor immunity.35 The set point thereby reflects the threshold that must be exceeded for a cancer patient to respond to immunotherapy. Key factors that were proposed to be integrated within the cancer-immune set point relate to the cancer immunity cycle and reflect the impact of cancer-intrinsic properties, therapeutic agents, environmental factors, microbiome, and host genetics. By the same token, to profile antitumor immunity, the cancer immunogram proposed seven groups of parameters reflecting the immunogenicity of the tumor and the capability of the immune system to reject the tumor.44 More specifically, these groups relate to tumor foreignness, tumor sensitivity to immune effectors, absence of inhibitory tumor metabolism, general immune status, absence of soluble mediators, absence of checkpoints, and immune cell infiltration. Analysis of multiple parameters for each group may help to estimate the overall state of the antitumor immune response and the most likely reason for its failure in each individual patient.
The frameworks of the cancer-immunity set point and cancer immunogram have been recently employed to create a ‘tumor personality test’ to guide treatment decisions.45,46 Bagaev et al.46 used 29 functional gene signatures representing key properties such as oncogenic signaling, angiogenesis, immunosuppression, and proliferation for the TME classification of >10 000 cancer patients. The obtained information was coalesced into a more simplistic discrimination of four TME subtypes with conserved relationships between immune and stroma activity across 20 cancer types. Importantly, these TME subtypes were prognostic and predictive of immunotherapy response. These observations were translated into a visualization tool integrating genomic and microenvironmental profiling that, though further prospective validation is required, may reflect a first step towards the development of more nuanced biomarkers.
Collectively, the above-described concepts all share the idea of harnessing multiple parameters that allow deconvolution and organization of the torrent of stimulatory and inhibitory signals present in the tumor and/or host to predict immunotherapy response. The challenge remains, however, to find measurable parameters that can provide a comprehensive overview of the immune status for each patient and be translated into a set of clinically applicable biomarkers. To achieve this goal, it is essential to understand for each individual patient which element(s) of the antitumor response is/are defective, and how these elements can specifically be ‘repaired’. To this end, technical approaches are required that allow in-depth profiling of human TMEs as well as obtaining information on the mechanisms underpinning response or resistance in (pre)clinical model systems (Figure 1).
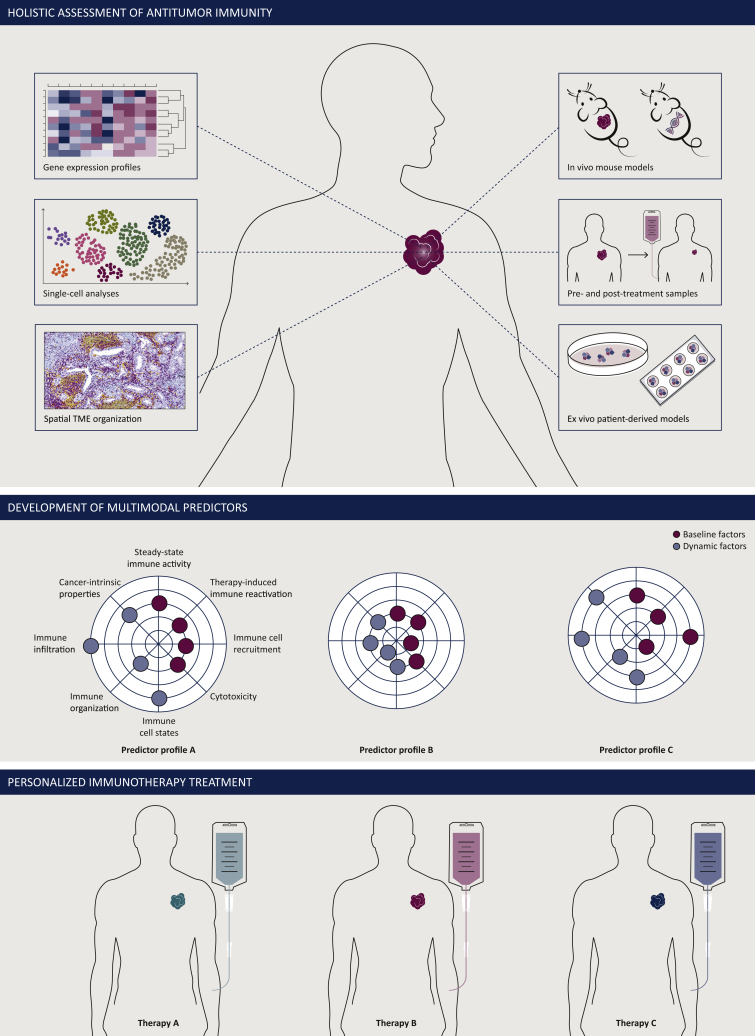
Figure 1.
Development of multimodal predictors and personalized immunotherapies by holistic assessment of antitumor immunity.
To deconvolute and organize the wide spectrum of elements impacting on antitumor immunity, a holistic approach may be employed. Combination of different multi-omic technologies and preclinical models should allow to obtain a broad overview of rate-limiting elements of the antitumor immune response (top). These data are then translated into a clinically applicable multimodal predictor reflecting each patient’s individual cancer immune profile (middle). Ultimately, this predictor will allow to identify personalized immunotherapy treatment of each patient (bottom).
Technologies to identify key elements of antitumor immunity
The human TME is a complex ecosystem harboring cancer, immune, and stromal cell populations that acquire various cell states. Thus, approaches are required that can provide broad and ideally unbiased information on the presence, state, and function of these cells in the TME both during steady-state and upon treatment with immunotherapy. Such approaches may include gene expression analyses, single-cell technologies, multiplex imaging approaches, and preclinical in vivo and ex vivo models.
To capture the broader state of the TME, tumor transcriptomes have been assessed for predicting ICB responsiveness using a set of selected genes. Thereby, independent computational predictors based on gene expression profiles (GEPs) relating to tumor inflammation,47 immune checkpoint expression,48 cytolytic T cell activity,49,50 or T-cell dysfunction and exclusion (TIDE51) have been evaluated in specific clinical cohorts treated with ICB. In addition, GEPs associated with ICB resistance including transcriptional signatures related to innate anti-PD-1 resistance,52 and T-cell exclusion and immune evasion53 have been established. While most of these studies have been limited to a few small patient cohorts, efforts are being undertaken to harmonize data from multiple trials to find new features that predict response or resistance across cancer types. For instance, the TIDE signature, which combines factors underlying the two mechanisms of T-cell dysfunction and exclusion, has integrated and modeled data from 189 human cancer studies with 33 197 samples.51 More recently, Litchfield and colleagues54 carried out a pan-cancer meta-analysis on 1008 ICB-treated patients from 12 individual cohorts. Importantly, the developed 11-parameter predictor was derived from both transcriptomic and whole-exome data and carried out significantly better than TMB alone to identify ICB responders across cancer types.
While the predictive value of tumor immune infiltrates has clearly been demonstrated, not only the presence, but also the state of tumor-infiltrating immune cells may be exploited to predict response to ICB. Single-cell technologies such as single-cell RNA sequencing (scRNAseq) and cytometry by time-of-flight have been employed to assess cell states of tumor-infiltrating immune cells. As the crucial role of T cells in immunotherapy response is well established, most efforts have so far focused on dissecting the intratumoral T-cell pool.55, 56, 57, 58, 59, 60 These studies have revealed that tumor-infiltrating T cells can acquire a wide range of cell states.61 Of note, the dysfunctional CD8+ T-cell pool has been found to contain a spectrum of cell states ranging from a predysfunctional, stem-like state to late-dysfunctional cells. A number of studies have demonstrated that the predysfunctional CD8+ T-cell subset, characterized by expression of the transcription factor Tcf1, is essential for durable response to ICB in tumor-bearing mice.57, 58, 59 TCF1+ T cells have also been identified in the blood and tumors of human melanoma patients,57 and have been associated with response to anti-PD-1 in two independent human melanoma cohorts.60 In contrast to the predysfunctional TCF1+ T cells, late-dysfunctional T cells lose their proliferative capacity, display high expression of inhibitory receptors such as PD-1, CTLA-4, and TIM-3, and secrete CXCL13. Moreover, they show increased capacity for tumor recognition.10,56,62,63 CD8+ T cells expressing high levels of PD-1 or of both PD-1 and CTLA-4 were found to be predictive for anti-PD-1 response in NSCLC and melanoma.2,62 Recently, the potential of such late-dysfunctional T cells, termed PD-1T TILs, to predict immunological response to PD-1 blockade has been demonstrated for a range of cancer types.36 Collectively, these data highlight the potential of certain T cell states as biomarkers for anti-PD-1 response. In addition, technical approaches that directly ‘measure’ the tumor recognition potential of T cells, both in the TME and the blood, may be of great interest, particularly as it has been shown that only a modest percentage of intratumoral T cells recognize tumor antigens.64,65
More comprehensive profiling of tumor immune infiltrates has revealed that in addition to T cells, also the presence of B cells has predictive value in a number of tumor types including melanoma, renal cell carcinoma, and sarcoma.66, 67, 68 Notably, intratumoral B cells frequently reside within tertiary lymphoid structures (TLS), which have also been associated with ICB response (see below). In addition, recent data from murine models suggest that PD-1 blockade may exert a major effect via myeloid cells, as based on experiments where PD-1 expression is selectively absent on these cells.69 To date, however, it is not clear whether this observation can be translated to patients and whether the presence of certain myeloid cell states has predictive potential for ICB.
Next to the state of immune cells, also the localization and organization of the immune infiltrate within a tumor seems to be of importance for its capability to respond to immunotherapy. For instance, by subdividing T cells based on their location in the tumor, CD8+ T-cell infiltration in the tumor core and at the invasive margin was found to be predictive for response to PD-1 blockade in melanoma.32 In addition, the proximity of CD8+ T cells and tumor cells, and of PD-1+ and PD-L1+ cells, respectively, has been associated with response.32,70 Newer technologies such as high-dimensional imaging of tumor tissue by imaging mass cytometry (IMC),71 multiplexed ion beam imaging by time of flight72 or co-detection by indexing (CODEX)73 are being used to spatially map TMEs and identify more complex patterns of immune infiltrates in tumors. Such approaches have identified two types of spatial immune infiltration patterns, so-called mixed versus compartmentalized infiltrates, that relate to distinct clinical outcomes in breast cancer and head and neck squamous cell carcinoma.72,74 Hoch et al.75 employed IMC with combined RNA and protein co-staining to identify heterogeneous chemokine patches in human melanoma that sculpted the local immune infiltration landscape. A recent study in CRC pursued the same idea, namely to profile specialized immune niches instead of quantifying cell types or cell states within a tumor.76 Using CODEX, nine distinct cellular neighborhoods were identified that related to different specialized tumor areas. Importantly, the study could demonstrate that the prognostic value of specific cell types depended on the neighborhood and was not observed when the overall frequency of the same cell type was analyzed. Whereas it is conceivable that the organization and context of the cellular neighborhood may also influence the capacity of immune cells to be reactivated by immunotherapy, this question needs to be addressed in future studies.
One specific type of immune niche that has recently gained attention is formed by TLS.77,78 TLS arise in the context of chronic tissue inflammation and are organized immune cell clusters harboring specialized cellular components. Characteristically, they consist of an inner zone with B cells surrounded by T cells, similar to the lymph follicles in secondary lymphoid organs. Importantly, the formation of TLS in tumors has been associated with better prognosis and response to ICB in a number of cancer types.66, 67, 68 In addition to the predictive value of the baseline presence of TLS in a tumor, it has been shown that ICB increases the number and size of TLS in humans and mice.67,79 It is still unclear, however, whether TLS are induced as a bystander phenomenon of an ongoing antitumor response or whether they actively contribute to tumor control and immunotherapy response.
Taken together, the above observations suggest that the integration of multi-omic analyses of human TMEs may leverage the identification of multimodal predictors of immunotherapy response. In addition, to assess potential biomarkers at the level of cell types or cell states, specialized immune niches may also be exploited both for response prediction and for the development of personalized treatments. It is conceivable that the context of such local infiltrates may alter the relevance and particularly the function of a cell state in a tumor. Thus, to fully understand how this coordinated behavior of immune infiltrates mediates tumor control and how it can be initiated or boosted by immunotherapies, it is vital to develop preclinical tumor models that can recapitulate the nuanced interactions between TME components and capture the dynamics of the treatment response at a patient-specific level.
Preclinical modeling of immunotherapy responses
Preclinical models, such as two-dimensional (2D) and 3D in vitro cultures and animal models, are extensively used to study drug responses and to identify the mechanisms underlying those responses. The large majority of these models has been established from a cancer cell-centric view, however, and are therefore often not optimally suited to study the perturbation of cancer-immune-stroma interactions by immunotherapies. Particularly, the maintenance of immune and stromal compartments poses a substantial challenge in many models, which the field has started to address.80
Mouse tumor models have provided translational utility over many years and have greatly improved our understanding of antitumor immunity. Different types of mouse models, including syngeneic models, genetically engineered mouse models (GEMMs) and patient-derived xenograft (PDX) models, have been exploited for this purpose. Immunocompetent syngeneic models are most frequently used to study responses upon ICB as they are low cost and relatively straightforward to use. Importantly, whereas some immunological mechanisms are conserved between mice and humans, as for instance the expression and role of immune checkpoints, critical differences in other components of the immune system, such as myeloid cells exist which causes species-specific—and sometimes model-specific—interactions between cancer and immune cells.81 To reflect a more physiological tumor growth, mice have been genetically engineered with oncogenic drivers for different cancer types. Whereas these GEMMs have been essential to improve our understanding of the genetic alterations that underlie cancer development and progression, the response rates of these models to ICB have so far been modest. The lack of substantial antitumor immune responses may be accounted for by the low mutation rate and neoantigen burden that many GEMMs exhibit.80 To overcome this limitation, tumors in GEMMs have been programmed to express de novo neoepitopes, as for instance in the NINJA mouse model.82 To better capture the intertumor and intratumor heterogeneity of human cancers, PDX models, in which pieces of patient tumors are grown in immunodeficient mice, have been created. While these models accurately predict response to targeted therapies,83, 84, 85 one important limitation of PDX models for immunotherapy is the replacement of the human tumor stroma by the one of the mouse, which leads to the loss of tumor-stroma interactions due to mismatched mouse and human ligands and receptors.86 Using a PDX model that was humanized by transfer of CD34+ human umbilical blood stem cells, distinct responses to PD-1 blockade in microsatellite instable and stable CRC PDXs could, however, be observed.87 Other models, such as MITRG and MISTRG mice express several human cytokines, which lead to the development of human natural killer cells, macrophages, and monocytes from engrafted CD34+ hematopoietic stem cells, but still lack functional mature human lymphocytes.88 While the development of mouse models with a humanized adaptive immune system is challenging,89,90 such models may greatly improve the use of PDXs for immune-oncology and specifically immunotherapy biomarker research.
To complement murine studies, analyses of sequential tumor biopsies have been used to monitor treatment-induced changes in human tumors. Comparison of pre- and on-treatment biopsies of melanoma patients undergoing anti-PD-1 treatment revealed that responding tumors showed more pronounced increases in different immune cell populations and induction of gene programs related to immune activation.91 Similar observations have been made in other studies using different technical approaches to compare longitudinal tumor samples from patients receiving either anti-PD-1 monotherapy or combined PD-1 and CTLA-4 blockade. Accordingly, the induction of a downstream IFN-γ -cell clones have been observed in responding patients.67,92, 93, 94 As in most of these studies the on-treatment samples were collected a few weeks to months after treatment start, however, they have limited utility to study early dynamics of the induced response. A recent study in advanced melanoma showed that already after one cycle of neoadjuvant anti-PD-1 treatment, 30% of patients had a complete or major pathological response, underscoring the importance of early changes in intratumoral immune reactivation.95 Thus, patient-derived ex vivo tumor systems could bridge the gap between mouse models and human samples to shed light on the early dynamics of immunotherapy-induced immune responses.
The development of technologies to culture or grow individual human tumors in 3D cultures such as patient-derived organoids (PDOs) or patient-derived explants (PDEs) has created new tools to boost precision medicine. Particularly, PDOs are now widely being used for drug screening purposes.96 PDOs are multicellular organotypic structures that recapitulate the features as well as the behavior of the original tumor tissue.97, 98, 99 PDOs can be established both from stem cells and from cancer biopsies, and can be expanded, passaged, and cryopreserved for further use.100, 101, 102, 103 The value of PDOs to test the sensitivity of individual tumors to chemotherapy and targeted drugs has been reported in a number of cancer types.104, 105, 106, 107 PDOs typically contain only cancer cells, however, and are therefore not suited to test immunotherapeutic drugs. To overcome this limitation, two different approaches have been exploited: systems that reconstitute PDOs with immune and/or stromal cells, and ‘en bloc’ PDO and PDE models that preserve all intratumoral cellular compartments.108
In reconstituted PDO systems, tumor organoids are co-cultured with exogenously added immune components, often autologous peripheral blood lymphocytes. Such reconstituted cultures treated with PD-1 blockade have, for instance, been successfully used to generate tumor-reactive CD8+ and CD4+ T cells in CRC.109 The co-culture of PDOs with distinct immune and stromal cell subsets could help to better understand the role of specific TME components and their interaction with cancer cells for antitumor immunity. The addition of single or a few selected immune cell populations may not, however, be sufficient to capture the complexity of the TME and its response to immunotherapy. As an alternative approach, several methods to culture the TME ‘en bloc’ and preserve immune, stromal, and cancer cell populations ex vivo have been established. Using a microfluidic setup, patient-derived organotypic tumor spheroids showed induction of chemokine secretion upon ex vivo ICB.110 Air-liquid-interface PDOs obtained from different cancer types responded to ex vivo PD-1 blockade by induction of IFN-γ.111 Similar observations have been made following anti-PD-L1 treatment in an ex vivo organ culture system that used fragmented tumor clusters derived from NSCLC specimens.112 Using a PDE model of patient-derived tumor fragments, it was demonstrated that immunological responses captured by T-cell activation as well as the secretion of multiple cytokines and chemokines by the TME can predict clinical responses to PD-1 blockade.36 Notably, combination of ex vivo cultures with the analysis of baseline tumor properties allowed to distinguish distinct TME subgroups across cancer types that were associated with either response or resistance to PD-1 blockade and to identify predictive markers such as TLS.36,112
Whereas the conceptual idea behind all these ex vivo systems is similar, they differ in a number of factors including tissue size, addition of growth factors and supplements, culture time, and preservation of immune architecture, which need to be considered when choosing a model (Table 1). Importantly, it has been demonstrated that, similar to mouse models, ex vivo models can be used to perturb immunotherapy responses, for instance by inhibiting T-cell receptor or IFN-γ signaling, thereby allowing to identify critical cellular components or signaling pathways that underlie those treatment-induced responses.36 Whereas so far, mostly changes in cytokine and chemokine secretion, expression of cytotoxic genes, or flow cytometry-based detection of T-cell activation markers have been assessed to measure ICB responses ex vivo, PDOs and PDEs may be combined with multiple endpoint analyses including scRNAseq or high-dimensional imaging, thereby providing insights into local immunotherapy responses at single-cell resolution. In case sufficient material can be obtained, multiple treatments can be tested in the same tumor, offering the possibility to directly compare the therapeutic efficacy of different mono- or combination therapies. There are a number of challenges for ex vivo models that need to be addressed in future research. For instance, the lack of self-renewal of ‘en bloc’ ex vivo cultures makes them dependent on the amount of available patient material, which is often difficult to obtain. Therefore, large biobanks with viable tumor material need to be established (see below). Another problem is caused by the lack of systemic immunity in current models which limits studies to the intratumoral immune response. Thus, efforts aiming at modeling immune cell recruitment to PDOs/PDEs will be of great value. While genome editing has successfully been employed in tumor-only organoids,113,114 the application of functional genetic screens particularly in ‘en bloc’ models has not yet been reported, but would be of value to examine the effect of specific genes on the antitumor immune response. Taken together, patient-derived ex vivo models which recapitulate treatment responses observed in patients can complement preclinical in vivo models and analyses of longitudinal patient samples, and are a powerful tool to support precision IO approaches.
Table 1.
Comparison of human ‘en bloc’ patient-derived organoid/patient-derived explant models
| Model | Patient-derived organotypic tumor spheroids | Air-liquid interface patient-derived organoids | Ex vivo organ culture | Patient-derived tumor fragments |
|---|---|---|---|---|
| Culture type | Submerged in collagen matrix with microfluidics | Air-liquid interface | Submerged in medium | Submerged in collagen-Matrigel matrix |
| Tissue fragmentation | Mincing, filtration (100 μM and 40 μM filters) | Mincing | Manual dissection, followed by mechanical and enzymatical dissociation, filtration (450 μm filter) | Manual dissection |
| Size | 40-100 μm | 40-100 μm | 30-450 μm | 1-2 mm |
| Culture time | 6 days | >1 month | 5 days | 2 days |
| Medium supplements | NA | 50% Wnt3a, RSPO1, Noggin, nicotinamide, N-acetylcysteine, B-27 without vitamin A, A83-01, gastrin, SB-202190, EGF | Transferrin-insulin-selenium mix, amphotericin, gentamycin, non-essential amino acids | Non-essential amino acids, sodium pyruvate |
| Maintenance of cancer morphology | Yes | Yes | Yes | NA |
| Maintenance of immune infiltrate | Yes | Yes | Yes | Yes |
| Maintenance of immune organization | NA | NA | NA | Yes |
| Type of ICB tested | Anti-PD-1, anti-CTLA-4, anti-PD-1 + anti-CTLA-4 | Anti-PD-1 | Anti-PD-L1, anti-PD-L1 + anti-CTLA-4 | Anti-PD-1 |
| Characterization of ICB response | Broad cytokine and chemokine profiling RNAseq |
RT-PCR (IFN-γ, granzyme B, perforin) | RT-PCR (IFN-γ), IHC (granzyme B) | Broad cytokine and chemokine profiling Flow cytometry |
| Perturbation | NA | NA | NA | Inhibition of TCR signaling and IFN-γ signaling |
CTLA-4, cytotoxic T lymphocyte-associated protein 4; ICB, immune checkpoint blockade; IFN, interferon; NA, no data available; PD-1, programmed cell death protein 1; PD-L1, programmed death-ligand 1; RNAseq, RNA sequencing; TCR, T-cell receptor.
Opportunities and challenges for precision IO research
To support the development of holistic predictors that allow to fully personalize immunotherapy treatment, a number of challenges need to be addressed by the field (Figure 2). For instance, the assessment of clinical specimens is a pivotal requirement to understand interpatient heterogeneity and to tailor the best treatment to each individual patient. To this end, large sample numbers are needed that allow to discriminate specific signals from the noise caused by heterogeneity. The collection of patient samples is particularly challenging for viable tumor tissue and requires sufficient infrastructure as well as a multidisciplinary team of oncologists, surgeons, pathologists, and researchers to establish a robust pipeline that ensures good material quality.
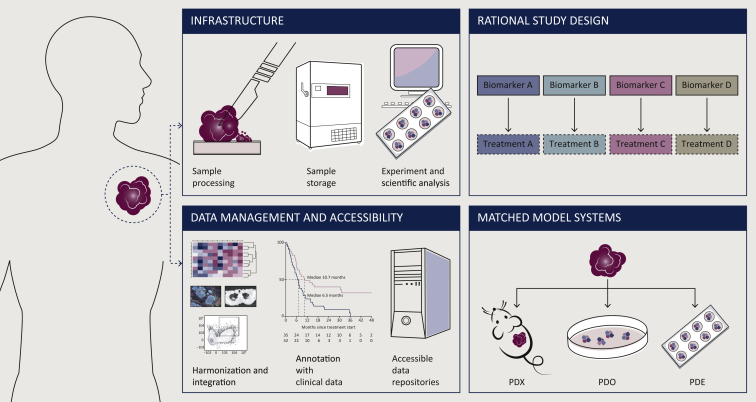
Figure 2.
Challenges for precision immuno-oncology research.
The identification of precision biomarkers and personalized treatments comes with a number of opportunities and challenges. These include (i) dedicated infrastructure and personnel to establish a robust and efficient pipeline for patient material collection and processing, (ii) the creation of data repositories for the establishment of large, well-structured, harmonized, and clinically annotated datasets, (iii) exploitation of synergies between distinct preclinical model systems matched for individual patients, and (iv) integrating personalized treatment approaches into the design of clinical studies.
PDE, patient-derived explant; PDO, patient-derived organoid; PDX, patient-derived xenograft.
A second challenge is presented by the massive increase in data resulting from the numerous clinical and preclinical biomarker studies. Data-driven approaches such as artificial intelligence, machine learning, and predictive modeling are powerful tools for precision biomarker research. Such approaches are currently widely exploited for identification of image-based biomarkers using radiomics-115,116 or digital pathology-based approaches.117,118 Likewise, the use of machine learning applications for investigating fundamental biological processes has been described119 and may therefore also provide a powerful tool for deconvoluting high-dimensional genomic or proteomic data aimed at biomarker discovery. Datasets that are large enough and sufficiently clinically annotated for this purpose are, however, currently limited. Therefore, the creation of data repositories providing access to large, well-structured, and fully annotated datasets is of critical importance. Computational approaches to standardize, integrate, and harmonize data from different immunotherapy trials may help to create such large datasets.51,54 As an example of such an effort, the Cancer Immune Monitoring and Analysis Centers and Cancer Immunologic Data Commons (CIMAC-CIDC) Network was established in 2017. This network consisting of four academic centers is aimed at facilitating biomarker identification, integration, and comparability by performing comprehensive tumor immunoprofiling with validated and harmonized assays across different centers.120 All data are integrated and collected in a large database for biomarker analysis, and assay protocols are published for future use by the larger community. Similarly, data obtained from multi-omic and spatial technologies, using distinct file formats and metadata, need to be collected in databases and standardized. Novel computational approaches to exploit these large datasets are required and may be inspired by other disciplines, as for instance recently demonstrated by the implementation of streamlined multispectral imaging protocols developed in astroscience for multiplex immunofluorescence TME analyses.121
While preclinical models are essential to understand the dynamics of immunotherapy responses and to link them to features of the TME, no single model can fully capture the complexity of the human tumor landscape and the heterogeneity between cancer types and individual patients. Thus, it is important to understand the specific strengths and limitations of each model system and to combine them if possible. Particularly, the setup of matched in vivo and ex vivo platforms, for instance by creating PDX and PDO/PDE from the same tumor, may be synergistic and strengthen the specificity of findings when consistently observed across different models.11 In addition, as mouse models usually lack the heterogeneity that is observed in human tumors, efforts that leverage a set of distinct mouse models reflecting different subtypes of human disease could improve the translation of observations from mice to patients. Such an effort has recently been reported in a study where a set of GEM melanoma models representative for a variety of human melanoma subtypes showed diverse responsiveness to ICB in line with clinical observations.122 Thus, the development of such murine tumor model cohorts that capture various molecular and phenotypic features of human cancer subtypes will help to gain new insights into mechanisms of response or resistance to immunotherapy and to foster personalization of treatment.
Finally, novel designs of immunotherapy trials may be considered to support and rationalize the development of personalized treatments. In current studies, patients are often stratified into groups based on the presence or absence of a biomarker, but receive all the same study treatment. Alternative approaches have recently been exploited, where distinct treatments are tested in patient groups selected on specific molecular alterations or immune signatures, as for instance in the BISCAY, DRUP, and DONIMI trials.123, 124, 125, 126 While varying success of this approach has been observed in these studies, this may be improved by a better (preclinical) identification of predictors to stratify patients for different treatments. Alternatively, a back-and-forth approach between small investigator-driven trials and preclinical analysis of tumor samples, as for instance recently suggested by the Lombard Street Approach for neoadjuvant melanoma, may help to tailor immunotherapy combinations and identify new treatment combinations for nonresponding patients in a relatively short time.127 A first example of an approach that directly bases treatment recommendations including, but not restricted to, immunotherapy on extensive multilevel tumor profiling is the Tumor Profiler Study.128 In contrast to other studies that are solely based on genomic tumor profiling, this study combines fast diagnostic and exploratory analyses using bulk and single-cell genomic, transcriptomic, and spatial profiling, as well as ex vivo drug response assays. This approach allows both to generate an individual tumor profile for each patient within 4 weeks that is used for clinical treatment decisions, and to collect a large set of data that can be further explored for hypothesis-generating research. Importantly, such studies that directly link tumor properties to the outcome of a specific treatment may ultimately allow to develop companion diagnostic tests (being either single or composite biomarkers) with regulatory value.
Conclusions
Reactivation of antitumor immunity by immunotherapies can induce deep and durable responses, however, its benefit is still limited to a subset of cancer patients. Based on the multifaceted signals that modulate antitumor immune responses, personalized strategies are required to find the optimal treatment of each patient. To this end, the development of holistic multimodal predictors of response that capture critical rate-limiting steps of antitumor immunity is crucial. Preclinical animal and human model systems need to be established that fulfill the specific requirements to model immunotherapy treatments outside of the patients. Finally, considerations with regard to optimization of sample acquisition, data collection, and computational analysis, as well as preclinical and clinical trial design, should facilitate precision IO research, and ultimately, the development of easy applicable and affordable biomarker tests to fully personalize immunotherapy.
Acknowledgments
Funding
This work was supported by a KWF Young Investigator grant [grant number 12046] to DST.
Disclosure
DST received research funding from Bristol Myers Squibb and Asher Biotherapeutics, outside of the current work. All other authors have declared no conflicts of interest.
Given their role as Associate Editor, D.S. Thommen had no involvement in the peer review of this article and has no access to information regarding its peer review. Full responsibility for the editorial process for this article was delegated to John Haanen, Editor in Chief of the Journal.
References
- 1.Pardoll D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daud A.I., Wolchok J.D., Robert C., et al. Programmed death-ligand 1 expression and response to the anti-programmed death 1 antibody pembrolizumab in melanoma. J Clin Oncol. 2016;34(34):4102–4109. doi: 10.1200/JCO.2016.67.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barber D.L., Wherry E.J., Masopust D., et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439(7077):682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 4.Lu S., Stein J.E., Rimm D.L., et al. Comparison of biomarker modalities for predicting response to PD-1/PD-L1 checkpoint blockade: a systematic review and meta-analysis. JAMA Oncol. 2019;5(8):1195–1204. doi: 10.1001/jamaoncol.2019.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mellman I., Coukos G., Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480(7378):480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Topalian S.L., Hodi F.S., Brahmer J.R., et al. Safety, activity, and immune correlates of anti–PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herbst R.S., Soria J.C., Kowanetz M., et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doroshow D.B., Bhalla S., Beasley M.B., et al. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat Rev Clin Oncol. 2021;18(6):345–362. doi: 10.1038/s41571-021-00473-5. [DOI] [PubMed] [Google Scholar]
- 9.Niu M., Yi M., Li N., Luo S., Wu K. Predictive biomarkers of anti-PD-1/PD-L1 therapy in NSCLC. Exp Hematol Oncol. 2021;10(1):1–13. doi: 10.1186/s40164-021-00211-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliveira G., Stromhaug K., Klaeger S., et al. Phenotype, specificity and avidity of antitumour CD8+ T cells in melanoma. Nature. 2021;596:119–125. doi: 10.1038/s41586-021-03704-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robert C., Schachter J., Long G.V., et al. Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med. 2015;372(26):2521–2532. doi: 10.1056/NEJMoa1503093. [DOI] [PubMed] [Google Scholar]
- 12.Sun C., Mezzadra R., Schumacher T.N. Regulation and function of the PD-L1 checkpoint. Immunity. 2018;48(3):434–452. doi: 10.1016/j.immuni.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Younes A., Santoro A., Shipp M., et al. Nivolumab for classical Hodgkin’s lymphoma after failure of both autologous stem-cell transplantation and brentuximab vedotin: a multicentre, multicohort, single-arm phase 2 trial. Lancet Oncol. 2016;17(9):1283–1294. doi: 10.1016/S1470-2045(16)30167-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roemer M.G.M., Advani R.H., Ligon A.H., et al. PD-L1 and PD-L2 genetic alterations define classical Hodgkin lymphoma and predict outcome. J Clin Oncol. 2016;34(23):2690–2697. doi: 10.1200/JCO.2016.66.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Armand P., Shipp M.A., Ribrag V., et al. Programmed death-1 blockade with pembrolizumab in patients with classical Hodgkin lymphoma after brentuximab vedotin failure. J Clin Oncol. 2016;34(31):3733–3739. doi: 10.1200/JCO.2016.67.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ansell S.M., Lesokhin A.M., Borrello I., et al. PD-1 Blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med. 2015;372(4):311–319. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao G., Jin L.L., Liu C.Q., et al. EZH2 negatively regulates PD-L1 expression in hepatocellular carcinoma. J Immunother Cancer. 2019;7(1):1–15. doi: 10.1186/s40425-019-0784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu C., Paschall A.V., Shi H., et al. The MLL1-H3K4me3 axis-mediated PD-L1 expression and pancreatic cancer immune evasion. J Natl Cancer Inst. 2017;109(6):1–12. doi: 10.1093/jnci/djw283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snyder A., Makarov V., Merghoub T., et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rizvi N.A., Hellmann M.D., Snyder A., et al. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yarchoan M., Hopkins A., Jaffee E.M. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377(25):2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le D.T., Durham J.N., Smith K.N., et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le D.T., Uram J.N., Wang H., et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fumet J.-D., Truntzer C., Yarchoan M., Ghiringhelli F. Tumour mutational burden as a biomarker for immunotherapy: current data and emerging concepts. Eur J Cancer. 2020;131:40–50. doi: 10.1016/j.ejca.2020.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paz-Ares L., Dvorkin M., Chen Y., et al. Durvalumab plus platinum–etoposide versus platinum–etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomised, controlled, open-label, phase 3 trial. Lancet. 2019;394(10212):1929–1939. doi: 10.1016/S0140-6736(19)32222-6. [DOI] [PubMed] [Google Scholar]
- 26.Goodman A.M., Kato S., Bazhenova L., et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16(11):2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pagès F., Kirilovsky A., Mlecnik B., et al. In situ cytotoxic and memory T cells predict outcome in patients with early-stage colorectal cancer. J Clin Oncol. 2009;27(35):5944–5951. doi: 10.1200/JCO.2008.19.6147. [DOI] [PubMed] [Google Scholar]
- 28.Mlecnik B., Tosolini M., Kirilovsky A., et al. Histopathologic-based prognostic factors of colorectal cancers are associated with the state of the local immune reaction. J Clin Oncol. 2011;29(6):610–618. doi: 10.1200/JCO.2010.30.5425. [DOI] [PubMed] [Google Scholar]
- 29.Galon J., Angell H.K., Bedognetti D., Marincola F.M. The continuum of cancer immunosurveillance: prognostic, predictive, and mechanistic signatures. Immunity. 2013;39(1):11–26. doi: 10.1016/j.immuni.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 30.Galon J., Fox B.A., Bifulco C.B., et al. Immunoscore and immunoprofiling in cancer: an update from the melanoma and immunotherapy bridge 2015. J Transl Med. 2016;14(1):1–6. doi: 10.1186/s12967-016-1029-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang A.C., Postow M.A., Orlowski R.J., et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 2017;545(7652):60–65. doi: 10.1038/nature22079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tumeh P.C., Harview C.L., Yearley J.H., et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pagès F., Mlecnik B., Marliot F., et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018;391(10135):2128–2139. doi: 10.1016/S0140-6736(18)30789-X. [DOI] [PubMed] [Google Scholar]
- 34.Kaseb A.O., Vence L., Blando J., et al. Immunologic correlates of pathologic complete response to preoperative immunotherapy in hepatocellular carcinoma. Cancer Immunol Res. 2019;7(9):1390–1395. doi: 10.1158/2326-6066.CIR-18-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen D.S., Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541(7637):321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 36.Voabil P., de Bruijn M., Roelofsen L.M., et al. An ex vivo tumor fragment platform to dissect response to PD-1 blockade in cancer. Nat Med. 2021;27:1250–1261. doi: 10.1038/s41591-021-01398-3. [DOI] [PubMed] [Google Scholar]
- 37.Hendry S., Salgado R., Gevaert T., et al. Assessing tumor-infiltrating lymphocytes in solid tumors: a practical review for pathologists and proposal for a standardized method from the International Immunooncology Biomarkers Working Group: part 1: assessing the host immune response, TILs in invasive breast carcinoma and ductal carcinoma in situ, metastatic tumor deposits and areas for further research. Adv Anat Pathol. 2017;24(5):235–251. doi: 10.1097/PAP.0000000000000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merino D.M., McShane L.M., Fabrizio D., et al. Establishing guidelines to harmonize tumor mutational burden (TMB): in silico assessment of variation in TMB quantification across diagnostic platforms: phase I of the Friends of Cancer Research TMB Harmonization Project. J Immunother Cancer. 2020;8(1):1–14. doi: 10.1136/jitc-2019-000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stenzinger A., Allen J.D., Maas J., et al. Tumor mutational burden standardization initiatives: recommendations for consistent tumor mutational burden assessment in clinical samples to guide immunotherapy treatment decisions. Genes Chromosom Cancer. 2019;58(8):578–588. doi: 10.1002/gcc.22733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koelzer V.H., Gisler A., Hanhart J.C., et al. Digital image analysis improves precision of PD-L1 scoring in cutaneous melanoma. Histopathology. 2018;73(3):397–406. doi: 10.1111/his.13528. [DOI] [PubMed] [Google Scholar]
- 41.Yu J., Huang C., Sun Y., et al. Effect of laparoscopic vs open distal gastrectomy on 3-year disease-free survival in patients with locally advanced gastric cancer: the CLASS-01 randomized clinical trial. JAMA. 2019;321(20):1983–1992. doi: 10.1001/jama.2019.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Althammer S., Tan T.H., Spitzmüller A., et al. Automated image analysis of NSCLC biopsies to predict response to anti-PD-L1 therapy. J Immunother Cancer. 2019;7(1):1–12. doi: 10.1186/s40425-019-0589-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen D.S., Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39(1):1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 44.Blank C.U., Haanen J.B., Ribas A., Schumacher T.N. The “cancer immunogram” visualizing the state of cancer–immune system interactions may spur personalized therapy. Science. 2016;352(6286):658–660. doi: 10.1126/science.aaf2834. [DOI] [PubMed] [Google Scholar]
- 45.Houlahan K.E., Curtis C. A tumor “personality” test to guide therapeutic decision making. Cancer Cell. 2021;39(6):747–749. doi: 10.1016/j.ccell.2021.04.018. [DOI] [PubMed] [Google Scholar]
- 46.Bagaev A., Kotlov N., Nomie K., et al. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell. 2021;39(6):845–865.e7. doi: 10.1016/j.ccell.2021.04.014. [DOI] [PubMed] [Google Scholar]
- 47.Ayers M., Lunceford J., Nebozhyn M., et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Auslander N., Zhang G., Lee J.S., et al. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat Med. 2018;24(10):1545–1549. doi: 10.1038/s41591-018-0157-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oh D.Y., Kwek S.S., Raju S.S., et al. Intratumoral CD4+ T cells mediate anti-tumor cytotoxicity in human bladder cancer. Cell. 2020;181(7):1612–1625.e13. doi: 10.1016/j.cell.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Allen E.M., Miao D., Schilling B., et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350(6257):207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang P., Gu S., Pan D., et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24(10):1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hugo W., Zaretsky J.M., Sun L., et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165(1):35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jerby-Arnon L., Shah P., Cuoco M.S., et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell. 2018;175(4):984–997.e24. doi: 10.1016/j.cell.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Litchfield K., Reading J.L., Puttick C., et al. Meta-analysis of tumor- and T cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell. 2021;184(3):596–614.e14. doi: 10.1016/j.cell.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brummelman J., Mazza E.M.C., Alvisi G., et al. High-dimensional single cell analysis identifies stemlike cytotoxic CD8+T cells infiltrating human tumors. J Exp Med. 2018;215(10):2520–2535. doi: 10.1084/jem.20180684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H., van der Leun A.M., Yofe I., et al. Dysfunctional CD8 T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell. 2019;176(4):775–789.e18. doi: 10.1016/j.cell.2018.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siddiqui I., Schaeuble K., Chennupati V., et al. Intratumoral Tcf1 + PD-1 + CD8 + T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity. 2019;50(1):195–211.e10. doi: 10.1016/j.immuni.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 58.Miller B.C., Sen D.R., Al Abosy R., et al. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol. 2019;20(3):326–336. doi: 10.1038/s41590-019-0312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kurtulus S., Madi A., Escobar G., et al. Checkpoint blockade immunotherapy induces dynamic changes in PD-1 − CD8 + tumor-infiltrating T cells. Immunity. 2019;50(1):181–194.e6. doi: 10.1016/j.immuni.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sade-Feldman M., Yizhak K., Bjorgaard S.L., et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 2018;175(4):998–1013.e20. doi: 10.1016/j.cell.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van der Leun A.M., Thommen D.S., Schumacher T.N. CD8+ T cell states in human cancer: insights from single-cell analysis. Nat Rev Cancer. 2020;20(4):218–232. doi: 10.1038/s41568-019-0235-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thommen D.S., Koelzer V.H., Herzig P., et al. A transcriptionally and functionally distinct PD-1 + CD8 + T cell pool with predictive potential in non-small-cell lung cancer treated with pd-1 blockade. Nat Med. 2018;24(7):994–1004. doi: 10.1038/s41591-018-0057-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Caushi J.X., Zhang J., Ji Z., et al. Transcriptional programs of neoantigen-specific TIL in anti-PD-1-treated lung cancers. Nature. 2021;596(7870):126–132. doi: 10.1038/s41586-021-03752-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scheper W., Kelderman S., Fanchi L.F., et al. Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat Med. 2019;25(1):89–94. doi: 10.1038/s41591-018-0266-5. [DOI] [PubMed] [Google Scholar]
- 65.Simoni Y., Becht E., Fehlings M., et al. Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature. 2018;557(7706):575–579. doi: 10.1038/s41586-018-0130-2. [DOI] [PubMed] [Google Scholar]
- 66.Cabrita R., Lauss M., Sanna A., et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature. 2020;577(7791):561–565. doi: 10.1038/s41586-019-1914-8. [DOI] [PubMed] [Google Scholar]
- 67.Helmink B.A., Reddy S.M., Gao J., et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 2020;577(7791):549–555. doi: 10.1038/s41586-019-1922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petitprez F., de Reynies A., Keung E.Z., et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature. 2020;577:556–560. doi: 10.1038/s41586-019-1906-8. [DOI] [PubMed] [Google Scholar]
- 69.Strauss L., Mahmoud M.A.A., Weaver J.D., et al. Targeted deletion of PD-1 in myeloid cells induces antitumor immunity. Sci Immunol. 2020;5(43) doi: 10.1126/sciimmunol.aay1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gide T.N., Silva I.P., Quek C., et al. Close proximity of immune and tumor cells underlies response to anti-PD-1 based therapies in metastatic melanoma patients. Oncoimmunology. 2019;9(1):1659093. doi: 10.1080/2162402X.2019.1659093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Giesen C., Wang H.A.O., Schapiro D., et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11(4):417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 72.Keren L., Bosse M., Marquez D., et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell. 2018;174(6):1373–1387.e19. doi: 10.1016/j.cell.2018.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goltsev Y., Samusik N., Kennedy-Darling J., et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell. 2018;174(4):968–981.e15. doi: 10.1016/j.cell.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blise KE, Sivagnanam S, Banik GL, Coussens LM, Goecks J. Single-cell spatial proteomics analyses of head and neck squamous cell carcinoma reveal tumor heterogeneity and immune architectures associated with clinical outcome. bioRxiv. Published online 2021. https://doi.org/10.1101/2021.03.10.434649. [DOI] [PMC free article] [PubMed]
- 75.Hoch T, Schulz D, Eling N, Gómez JM, Levesque MP, Bodenmiller B. Multiplexed imaging mass cytometry of chemokine milieus in metastatic melanoma characterizes features of response to immunotherapy. bioRxiv. Published online 2021:2021.07.29.454093. [DOI] [PubMed]
- 76.Schürch C.M., Bhate S.S., Barlow G.L., et al. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell. 2020;182(5):1341–1359.e19. doi: 10.1016/j.cell.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sautès-Fridman C., Lawand M., Giraldo N.A., et al. Tertiary lymphoid structures in cancers: prognostic value, regulation, and manipulation for therapeutic intervention. Front Immunol. 2016;7:1–11. doi: 10.3389/fimmu.2016.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schumacher T.N., Thommen D.S. Tertiary lymphoid structures in cancer. Science. 2022;375(6576) doi: 10.1126/science.abf9419. [DOI] [PubMed] [Google Scholar]
- 79.Rodriguez A.B., Peske J.D., Woods A.N., et al. Immune mechanisms orchestrate tertiary lymphoid structures in tumors via cancer-associated fibroblasts. Cell Rep. 2021;36(3):109422. doi: 10.1016/j.celrep.2021.109422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McCarthy C.E., Zahir N., Eljanne M., Sharon E., Voest E.E., Palucka K. Developing and validating model systems for immuno-oncology. Cancer Cell. 2021;39(8):1018–1022. doi: 10.1016/j.ccell.2021.05.017. [DOI] [PubMed] [Google Scholar]
- 81.Kuo C.J., Voest E., Parrini M.C. Models for immuno-oncology research. Cancer Cell. 2020;38(2):145–147. doi: 10.1016/j.ccell.2020.07.010. [DOI] [PubMed] [Google Scholar]
- 82.Damo M., Fitzgerald B., Lu Y., et al. Inducible de novo expression of neoantigens in tumor cells and mice. Nat Biotechnol. 2021;39(1):64–73. doi: 10.1038/s41587-020-0613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fiebig H.H., Schuchhardt C., Henss H., Fiedler L., Löhr G.W. Comparison of tumor response in nude mice and in the patients. Behring Inst Mitt. 1984;74:343–352. [PubMed] [Google Scholar]
- 84.Sanmamed M.F., Chester C., Melero I., Kohrt H. Defining the optimal murine models to investigate immune checkpoint blockers and their combination with other immunotherapies. Ann Oncol. 2016;27(7):1190–1198. doi: 10.1093/annonc/mdw041. [DOI] [PubMed] [Google Scholar]
- 85.Einarsdottir BO, Bagge RO, Bhadury J, et al. Melanoma patient-derived xenografts accurately model the disease and develop fast enough to guide treatment decisions. 2014;5(20):B38-B38. [DOI] [PMC free article] [PubMed]
- 86.Patton E.E., Mueller K.L., Adams D.J., et al. Melanoma models for the next generation of therapies. Cancer Cell. 2021;39(5):610–631. doi: 10.1016/j.ccell.2021.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Capasso A., Lang J., Pitts T.M., et al. Characterization of immune responses to anti-PD-1 mono and combination immunotherapy in hematopoietic humanized mice implanted with tumor xenografts. J Immunother Cancer. 2019;7(1):1–16. doi: 10.1186/s40425-019-0518-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rongvaux A., Willinger T., Martinek J., et al. Development and function of human innate immune cells in a humanized mouse model. Nat Biotechnol. 2014;32(4):364–372. doi: 10.1038/nbt.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Forsberg E.M.V., Lindberg M.F., Jespersen H., et al. HER2 CAR-T cells eradicate uveal melanoma and T-cell therapy–resistant human melanoma in IL2 transgenic NOD/SCID IL2 receptor knockout mice. Cancer Res. 2019;79(5):899–904. doi: 10.1158/0008-5472.CAN-18-3158. [DOI] [PubMed] [Google Scholar]
- 90.Jespersen H., Lindberg M.F., Donia M., et al. Clinical responses to adoptive T-cell transfer can be modeled in an autologous immune-humanized mouse model. Nat Commun. 2017;8(1):707. doi: 10.1038/s41467-017-00786-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Riaz N., Havel J.J., Makarov V., et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171(4):934–949.e15. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yost K.E., Satpathy A.T., Wells D.K., et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat Med. 2019;25(8):1251–1259. doi: 10.1038/s41591-019-0522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Blank C.U., Rozeman E.A., Fanchi L.F., et al. Neoadjuvant versus adjuvant ipilimumab plus nivolumab in macroscopic stage III melanoma. Nat Med. 2018;24(11):1655–1661. doi: 10.1038/s41591-018-0198-0. [DOI] [PubMed] [Google Scholar]
- 94.Grasso C.S., Tsoi J., Onyshchenko M., et al. Conserved interferon-γ signaling drives clinical response to immune checkpoint blockade therapy in melanoma. Cancer Cell. 2020;38(4):500–515.e3. doi: 10.1016/j.ccell.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang A.C., Orlowski R.J., Xu X., et al. A single dose of neoadjuvant PD-1 blockade predicts clinical outcomes in resectable melanoma. Nat Med. 2019;25(3):454–461. doi: 10.1038/s41591-019-0357-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Veninga V., Voest E.E. Tumor organoids: opportunities and challenges to guide precision medicine. Cancer Cell. 2021;39:1190–1201. doi: 10.1016/j.ccell.2021.07.020. [DOI] [PubMed] [Google Scholar]
- 97.Blokzijl F., De Ligt J., Jager M., et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 2016;538(7624):260–264. doi: 10.1038/nature19768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sachs N., Clevers H. Organoid cultures for the analysis of cancer phenotypes. Curr Opin Genet Dev. 2014;24:68–73. doi: 10.1016/j.gde.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 99.Weeber F., Van De Wetering M., Hoogstraat M., et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc Natl Acad Sci U S A. 2015;112(43):13308–13311. doi: 10.1073/pnas.1516689112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sachs N., de Ligt J., Kopper O., et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172(1-2):373–386.e10. doi: 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 101.Sato T., Stange D.E., Ferrante M., et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141(5):1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 102.Van De Wetering M., Francies H.E., Francis J.M., et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161(4):933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yan H.H.N., Siu H.C., Law S., et al. A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell Stem Cell. 2018;23(6):882–897.e11. doi: 10.1016/j.stem.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 104.Ooft S.N., Weeber F., Dijkstra K.K., et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci Transl Med. 2019;11(513) doi: 10.1126/scitranslmed.aay2574. [DOI] [PubMed] [Google Scholar]
- 105.Kopper O., de Witte C.J., Lõhmussaar K., et al. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat Med. 2019;25(5):838–849. doi: 10.1038/s41591-019-0422-6. [DOI] [PubMed] [Google Scholar]
- 106.de Witte C.J., Espejo Valle-Inclan J., Hami N., et al. Patient-derived ovarian cancer organoids mimic clinical response and exhibit heterogeneous inter- and intrapatient drug responses. Cell Rep. 2020;31(11):107762. doi: 10.1016/j.celrep.2020.107762. [DOI] [PubMed] [Google Scholar]
- 107.Vlachogiannis G., Hedayat S., Vatsiou A., et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359(6378):920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yuki K., Cheng N., Nakano M., Kuo C.J. Organoid models of tumor immunology. Trends Immunol. 2020;41(8):652–664. doi: 10.1016/j.it.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dijkstra K.K., Cattaneo C.M., Weeber F., et al. Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell. 2018;174(6):1586–1598.e12. doi: 10.1016/j.cell.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jenkins R.W., Aref A.R., Lizotte P.H., et al. Ex vivo profiling of PD-1 blockade using organotypic tumor spheroids. Cancer Discov. 2018;8(2):196–215. doi: 10.1158/2159-8290.CD-17-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Neal J.T., Li X., Zhu J., et al. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175(7):1972–1988.e16. doi: 10.1016/j.cell.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kamer I., Bab-Dinitz E., Zadok O., et al. Immunotherapy response modeling by ex-vivo organ culture for lung cancer. Cancer Immunol Immunother. 2021;70(8):2223–2234. doi: 10.1007/s00262-020-02828-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Artegiani B., Hendriks D., Beumer J., et al. Fast and efficient generation of knock-in human organoids using homology-independent CRISPR–Cas9 precision genome editing. Nat Cell Biol. 2020;22(3):321–331. doi: 10.1038/s41556-020-0472-5. [DOI] [PubMed] [Google Scholar]
- 114.Teriyapirom I., Batista-Rocha A.S., Koo B.K. Genetic engineering in organoids. J Mol Med. 2021;99(4):555–568. doi: 10.1007/s00109-020-02029-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Guerrisi A., Russillo M., Loi E., et al. Exploring CT texture parameters as predictive and response imaging biomarkers of survival in patients with metastatic melanoma treated with PD-1 inhibitor nivolumab: a pilot study using a delta-radiomics approach. Front Oncol. 2021;11:1–12. doi: 10.3389/fonc.2021.704607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Faron A., Opheys N.S., Nowak S., et al. Deep learning-based body composition analysis predicts outcome in melanoma patients treated with immune checkpoint inhibitors. Diagnostics (Basel) 2021;1-9:2314. doi: 10.3390/diagnostics11122314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Barrera C., Velu P., Bera K., et al. Computer-extracted features relating to spatial arrangement of tumor infiltrating lymphocytes to predict response to nivolumab in non-small cell lung cancer (NSCLC) J Clin Oncol. 2018;36(suppl 15) 12115-12115. [Google Scholar]
- 118.Wu J., Lin D. A review of artificial intelligence in precise assessment of programmed cell death-ligand 1 and tumor-infiltrating lymphocytes in non−small cell lung cancer. Adv Anat Pathol. 2021;28:439–445. doi: 10.1097/PAP.0000000000000322. [DOI] [PubMed] [Google Scholar]
- 119.Pezoulas V.C., Hazapis O., Lagopati N., et al. Machine learning approaches on high throughput NGS data to unveil mechanisms of function in biology and disease. Cancer Genomics Proteomics. 2021;18(5):605–626. doi: 10.21873/cgp.20284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chen X., Dong Z., Hubbell E., et al. Prognostic significance of blood-based multi-cancer detection in plasma cell-free DNA. Clin Cancer Res. 2021;27(15):4221–4229. doi: 10.1158/1078-0432.CCR-21-0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Berry S., Giraldo N.A., Green B.F., et al. Analysis of multispectral imaging with the AstroPath platform informs efficacy of PD-1 blockade. Science. 2021;372(6547) doi: 10.1126/science.aba2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pérez-Guijarro E., Yang H.H., Araya R.E., et al. Multimodel preclinical platform predicts clinical response of melanoma to immunotherapy. Nat Med. 2020;26(5):781–791. doi: 10.1038/s41591-020-0818-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Powles T., Carroll D., Chowdhury S., et al. An adaptive, biomarker-directed platform study of durvalumab in combination with targeted therapies in advanced urothelial cancer. Nat Med. 2021;27(5):793–801. doi: 10.1038/s41591-021-01317-6. [DOI] [PubMed] [Google Scholar]
- 124.van der Velden D.L., Hoes L.R., van der Wijngaart H., et al. The drug rediscovery protocol facilitates the expanded use of existing anticancer drugs. Nature. 2019;574(7776):127–131. doi: 10.1038/s41586-019-1600-x. [DOI] [PubMed] [Google Scholar]
- 125.Hoes L.R., van der Wijngaart H., van Berge Henegouwen J.M., et al. 594P The drug rediscovery protocol (DRUP): results of the first 500 treated patients. Ann Oncol. 2020;31:S498. [Google Scholar]
- 126.Reijers I.L.M., Dimitriadis P., Rozeman E.A., et al. Personalized combination of neoadjuvant domatinostat, nivolumab and ipilimumab in macroscopic stage III melanoma patients stratified according to the interferon-gamma signature: the DONIMI study. J Clin Oncol. 2020;38(suppl 15) :TPS10087-TPS10087. [Google Scholar]
- 127.Versluis J.M., Thommen D.S., Blank C.U. Rationalizing the pathway to personalized neoadjuvant immunotherapy: the Lombard Street Approach. J Immunother Cancer. 2020;8(2):1–8. doi: 10.1136/jitc-2020-001352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Irmisch A., Bonilla X., Chevrier S., et al. The tumor profiler study: integrated, multi-omic, functional tumor profiling for clinical decision support. Cancer Cell. 2021;39(3):288–293. doi: 10.1016/j.ccell.2021.01.004. [DOI] [PubMed] [Google Scholar]