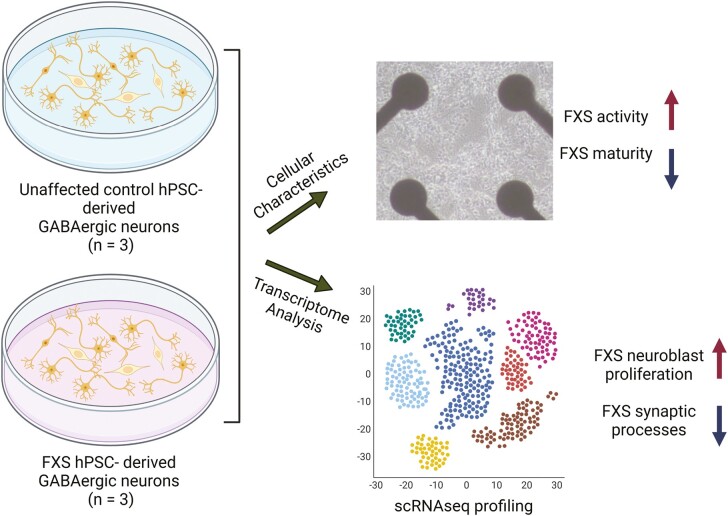
Abstract
Fragile X Syndrome (FXS), the leading monogenic cause of intellectual disability and autism spectrum disorder, is caused by expansion of a CGG trinucleotide repeat in the 5ʹ-UTR of the Fragile X Mental Retardation-1 (FMR1) gene. Epigenetic silencing of FMR1 results in loss of the Fragile X Mental Retardation Protein (FMRP). Although most studies to date have focused on excitatory neurons, recent evidence suggests that GABAergic inhibitory networks are also affected. To investigate human GABAergic neurogenesis, we established a method to reproducibly derive inhibitory neurons from multiple FXS and control human pluripotent stem cell (hPSC) lines. Electrophysiological analyses suggested that the developing FXS neurons had a delay in the GABA functional switch, a transition in fetal development that converts the GABAA channel’s function from depolarization to hyperpolarization, with profound effects on the developing brain. To investigate the cause of this delay, we analyzed 14 400 single-cell transcriptomes from FXS and control cells at 2 stages of GABAergic neurogenesis. While control and FXS cells were similar at the earlier time point, the later-stage FXS cells retained expression of neuroblast proliferation-associated genes and had lower levels of genes associated with action potential regulation, synapses, and mitochondria compared with controls. Our analysis suggests that loss of FMRP prolongs the proliferative stage of progenitors, which may result in more neurons remaining immature during the later stages of neurogenesis. This could have profound implications for homeostatic excitatory-inhibitory circuit development in FXS, and suggests a novel direction for understanding disease mechanisms that may help to guide therapeutic interventions.
Keywords: fragile X syndrome, autism, GABAergic neurons, neurogenesis, human iPSCs, hESCs
Graphical Abstract
Significance Statement.
The timing of cell-cell interactions is fundamental to embryonic development; this report is about a delay at a critical stage of nerve cell development in human pluripotent stem cells carrying a mutation that causes Fragile X syndrome (FXS), an inherited intellectual disability. Three FXS cell lines showed an intrinsic delay in a developmental milestone, a polarity switch in the cells’ response to GABA that has profound effects on the developing brain. Our report sets the stage for the investigation of therapeutic strategies to prevent or reverse the effects of the developmental delay.
Introduction
Fragile X syndrome (FXS) is the leading monogenic cause of intellectual disability and autism spectrum disorder (ASD), affecting as many as one in 3600 in the general population.1 FXS is closely linked to ASD; 60% of males and 20% of females who have been diagnosed with FXS meet the diagnostic criteria of ASD.2, 3 It is also an X-linked disease caused by the expansion of a trinucleotide repeat in the 5ʹ UTR region of the FMR1 (Fragile X Mental Retardation-1) gene. This results in hypermethylation of its locus, silencing the transcription of FMR1. Fragile X Mental Retardation Protein (FMRP, encoded by FMR1) has numerous roles in regulating RNA metabolism and translation; it directly binds to both mRNA and translational machinery.4 FMRP recognizes the quadruplex RNA structure,5 regulates RNA stability, binds to ribosomal proteins, and stalls ribosomal translocation.6–8 Loss of FMRP results in upregulated global protein synthesis in FXS cells.9
Previous mechanistic studies have mainly focused on the Fmr1 knockout mouse in which synaptogenesis and circuit formation are disrupted by loss of FMRP.10–14 These studies focused on excitatory neurons because the predominant theory for the cause of FXS (the mGluR5 theory15) centered on this subtype. As a result, much less is known about deficits in inhibitory neurons in FXS. Studies using the Fmr1 knockout mouse model have suggested fewer GABAergic synapses,6 fewer fast spiking interneurons,16 reduced expression of GABAA receptor subunits,17 and downregulation of transcription of Gad2 (glutamate decarboxylase 2, also known as Gad65) and Gad1 (glutamate decarboxylase 1, also known as Gad67).18 The failure of mGluR5-based therapeutics for FXS in clinical trials has prompted a re-examination of the importance of inhibitory neurons. Interestingly, recent studies have shown excitatory/inhibitory imbalance in Fmr1 knockout mice19 and report that the mice tend to have functionally impaired interneurons.20
Perturbation in GABA signaling also occurs early in development in Fmr1 knockout mice; increased excitability was observed during critical period development and was thought to be due to a delay in the GABA functional switch that occurs during cortical development.14,21,22 The GABA functional switch is characterized by a change in the effects of GABA in the developing brain, by which the neurotransmitter is initially excitatory, but then switches to being inhibitory due to a change in the type of K-Cl cotransporter expressed by developing neurons.
There are considerable differences between rodent and primate GABAergic neuron subtypes.23,24 To test whether FXS-specific phenotypes exist in developing human inhibitory neurons and to focus on the disease-specific aspects of GABAergic neurogenesis without confounding variables associated with individual hPSC cell lines, we developed a protocol that would reproducibly differentiate both control and FXS-affected human pluripotent stem cells into multiple GABAergic neuron subtypes. To analyze the cultures, we focused on characteristics of individual cells within the cultures, using gene expression and electrophysiological analysis methods. We studied a collection of 3 affected and 3 control hPSC lines (control and FXS hiPSCs, and an FMR1 gene-edited hESC line and its isogenic control line). In this report, the edited hESCs and the FXS-derived iPSCs are referred to as “FXS cells”. We found that FXS neuronal networks showed higher activity during maturation relative to controls, suggesting a delay in the developmental switch in GABA response from excitatory to inhibitory function. To characterize GABAergic neurogenesis at the gene expression level, we profiled 14,400 single-cell transcriptomes at early and later stages of neurogenesis in vitro. At the earlier time point, FXS and control cells were similar in cell type composition and gene expression profiles, but significant cell type-specific differences in gene expression emerged by the later time points. Cell stage-specific differentially expressed genes (DEGs) revealed enhanced neuroblast proliferation in FXS progenitors at the later stages, and a decrease in mature neuronal characteristics in FXS GABAergic neurons. Overall, our study reveals the specific physiological and transcriptional events underlying the delays in GABAergic neuron differentiation and maturation that are intrinsic to human FXS.
Materials and Methods
Cell Lines
This study included 6 human pluripotent stem cell (PSC) lines in all experiments (Supplementary Table S1): 2 FXS patient-derived iPSCs (SC105iPS6, SC215iPS1), 2 male control PSCs (a non-disease hESC line SAB1-13D and an iPSC line SC714iPS517 derived from a healthy subject), and an isogenic pair of male hESC lines (WA01 (H1)) in which one line has a targeted loss of FMRP expression.25 SC105iPS6, SC215iPS1, SAB1-13D, and SC714iPS517 were characterized in our previous study.26
Maintenance of Pluripotent Stem Cells
All hPSCs were maintained with mTeSR1 medium (STEMCELL Technologies) on Geltrex extracellular matrix (Life Technologies). Upon reaching 80% confluency, cells were passaged routinely at 1:3—1:5 ratio with dispase (Life Technologies). During passage, cell aggregates were gently triturated 3-5 times using Pasteur pipettes.
Differentiation of Inhibitory Neurons from Pluripotent Stem Cells
hPSCs were dissociated with Accutase (Life Technologies) and plated on Geltrex-coated 12-well plates at 2.1 × 105 cell/cm2 in E6 (STEMCELL Technologies) medium, supplemented with 10 µM rock inhibitor Y-27632 (REPROCELL). The next day, Y-27632 was withdrawn, and neural induction was initiated in E6 medium in the presence of 10 µM SB421542 (TGF-β receptor kinase inhibitor; Tocris), 100 nM LDN193189 (BMP signaling inhibitor; Tocris), and 2 µM XAV939 (WNT pathway inhibitor; REPROCELL), referred to as “neural induction medium”. On day 10 of differentiation, neural progenitors were passaged as aggregates, in a manner similar to passaging hPSCs, with a 1:3 ratio, and were replated on poly-d-lysine- and laminin-coated 12-well plates in a progenitor expansion medium supplemented with Y-27632. The progenitor expansion medium is a 1-to-1 ratio of DMEM/F12 and Neurobasal medium, with 1× N2 supplement and 1 × B27 supplement without vitamin A, 0.5 mg/mL BSA, 1× Pen-Strep, and 100 µM β-mercaptoethanol (referred to as “progenitor medium”, all from Life Technologies). Progenitor expansion was allowed from days 10 to 25, during which cells were replated as aggregates weekly. On day 25 of differentiation, cells were dissociated with Accutase, and seeded on 24-well plates at 1.3 × 105 cell/cm2 in neuronal maturation medium supplemented with 10 µM Y-27632. Maturation medium is composed of Neurobasal Plus, 1× B27 plus, 1× N2 (all from Life Technologies), 100 µM cAMP (Sigma), 20 ng/mL BDNF (Peprotech), 20 ng/mL GDNF (Peprotech), and 200 nM ascorbic acid (Sigma).
Quantitative PCR (qRT-PCR) and Principal Component Analysis on the qRT-PCR Dataset
RNA was extracted using the RNeasy Plus kit (Qiagen). cDNA was synthesized with 200 ng input RNA with QuantiTect Reverse Transcription Kit (Qiagen). Quantitative PCR was performed using the TaqMan Gene Expression assay. All assays were carried out based on manufacturers’ recommendations. TaqMan probes are listed in Supplementary Table S2. Delta CT was normalized using GAPDH. Visualization of qRT-PCR expression data was done in R (3.6) with the ggplot2 package. Principal component analysis was performed using the prcomp function in R.
Immunocytochemistry and Image Analysis
Cells were fixed in 4% paraformaldehyde for 15 minutes at room temperature and permeabilized with 0.1% Triton X-100, 1% BSA, and 10% donkey serum in PBS. Primary and secondary antibodies are listed in Supplementary Tables S3 and S4. All imaging was performed on a Keyence BZ-X710 All-in-one Fluorescence Microscope.
Quantification of KCC1-positive and KCC2-positive neurons used the following strategy: image preprocessing was performed using the OpenCV module, noise was removed using the medianBlur function, and images were binarized using the adaptiveThreshold function with the ADAPTIVE_THRESH_GAUSSIAN_C option. To quantify KCC1 or KCC2 intensity, mean intensity was calculated for the denoised images. To quantify the percentages of KCC-positive and KCC2-positive cells, the number of pixels that overlaid with MAP2-positive pixels were normalized by the total number of MAP2-positive pixels.
Microelectrode Array Recording and Analysis
On day 42 of neural differentiation, cells were dissociated by Accutase and replated on poly-d-lysine and laminin coated MEA plates at 5.5 × 105 cells/cm2 (Axion, CytoView 48 wells). Cells were cultured in BrainPhys medium (STEMCELL Technologies) supplemented with 1× B27 plus and 1× N2 (both from Life Technologies), 100 µM cAMP (Sigma), 20 ng/mL BDNF (Peprotech), 20 ng/mL GDNF (Peprotech), and 200 nM ascorbic acid (Sigma), and half of the medium was replaced every 2-3 days. Spontaneous activity recording was performed every 5 days from day 47 to day 77 using the Maestro system (Axion). Only wells with at least 2 active electrodes for at least 40% of the recording time were included for the subsequent analysis. Spike list and metadata were imported to the R package meaRtools27 for subsequent analysis. Mean firing rate, mean interspike intervals, spike train tiling coefficient, burst frequency, mean frequency in burst, and total spikes in burst were analyzed using default parameters. Three batches of differentiation (all 6-cell lines, 8 replicates per cell line) were recorded and included in the analysis.
Patch-Clamp Recording
Electrophysiological recordings were performed on all 6-cell lines, with 10-15 cells per cell line on day 52 or day 62 of differentiation in a small-volume recording chamber providing laminar solution flow (Warner Instruments, Hamden, CT). A coverslip with the plated cells was placed in a recording chamber perfused with 32 °C artificial cerebrospinal fluid (ACSF) with the following composition (mM): 124 NaCl, 2.5 KCl, 1.25 NaH2PO4, 26 NaHCO3, 2 CaCl2, 2 MgCl2, and 10 glucose (95% O2/5% CO2; all chemicals from Sigma-Aldrich, St. Louis, MO). Cells were visualized with infrared differential interference contrast (IR-DIC) using a water immersion objective (Nikon Instruments Inc., Melville, NY). Patch pipettes were pulled from borosilicate glass tubing (1.5 mm outer diameter, 0.3 mm wall thickness) on a horizontal puller (P-97; Sutter Instrument, Novato, CA). Recordings made with an Axopatch 700B amplifier (Molecular Devices, Sunnyvale, CA) were low-pass filtered at 3 kHz and digitized on-line at 20 kHz using National Instruments PCI-MIO-16-E4 multifunctional board controlled by the data acquisition software DASYLab 6.0 (Dasytec, Amherst, NH).
To record miniature inhibitory postsynaptic currents (mIPSCs), the recording chamber was perfused with ACSF supplemented with the Na+ channel blocker tetrodotoxin citrate (Tocris Bio-Techne, Minneapolis, MN) at a concentration 0.5 µM, to block neuronal firing. For mIPSC recordings, pipette electrodes were filled with a solution with the following composition (mM): 155 cesium methanesulfonate, 2 CsCl, 0.1 CaCl2, 0.5 EGTA, 5 HEPES buffer, 2 MgATP, 0.3 GTP sodium salt hydrate, 2 phosphocreatine; pH was adjusted to 7.2 with CsOH (all chemicals from Sigma-Aldrich, St. Louis, MO). Resistance of the electrodes submerged in ACSF varied from 4 to 7 MΩ. Access resistance before recordings was <25 MΩ and did not change more than 10% during the experiments. Series resistance was compensated by 70% (with 20 µs lag values). Recordings were performed at 0 mV holding voltage. To ensure that mIPSCs were specific for the GABAA receptors, occasionally recordings of mIPSCs were followed by perfusion with the GABAA receptor blocker (+)-bicuculline (Tocris Bio-Techne, Minneapolis, MN) at a concentration 20 µM. 0.5-3 minutes long recordings of mIPSCs were analyzed using cDasyLab for recordings and a custom script for analysis.
Single-cell Library Preparation
Cultures were dissociated at 2 stages of differentiation: early in neurogenesis on day 22, and late in neurogenesis on either day 42 or day 48. We used the Chromium single-cell RNA-seq library preparation instrument and protocol (10X Genomics). This platform enables the analysis of large numbers of cells at high capture efficiency (up to 65%)28 and acceptably small doublet rates (~0.9% for 1000 cell input).
Sample Multiplexing
To mitigate several known limitations of single-cell sequencing technology, including sample-specific batch effects, detection of cell doublets, and the cost of generating massive datasets, we used the “cell hashing” protocol.29 We used oligo-tagged antibodies against ubiquitously expressed surface proteins (BD Single-Cell Multiplexing Kit), to uniquely label cells from distinct samples, which were subsequently pooled. This allowed multiplexing of cells from all 6-cell lines pooled into a single-cell library. Similar to a multi-species experiment, this allows higher removal of doublet rates (by the multiplexing factor) and decreases reagent and labor costs. Using this cost-efficient library preparation method, we aimed for 1000 cells and 50 million reads (50 000 reads per cell) for each sample at each time point.
Pipeline and QC Overview
We performed primary and secondary analysis of scRNA-seq data using CellRanger, an analysis suite consisting of multiple pipelines for end-to-end analysis of single-cell data. In brief, it uses a custom-built wrapper around Illumina’s bcl2fastq to demultiplex raw base calls. This is followed by removal of duplicates using unique molecular identifier (UMI) counting. The preprocessed samples are aligned using STAR, which performs splicing-aware alignment of reads to the genome. Aligned reads are then bucketed into exonic and intronic regions using a reference gene transfer format (GTF) file. Finally, read counts per gene are calculated, and these values are used for downstream estimation of differentially expressed genes. Cells with fewer than 200 different gene transcripts were filtered out from downstream analysis. After initial filtering, 4132 cells from control cultures with a mean UMI of 1270 and 5492 cells from FXS cultures with a mean UMI of 1378 passed QC on day 22. For the days 42-48 dataset, 1765 cells from control cultures with a mean UMI of 1427, and 2434 cells from FXS cultures with a mean UMI of 1011 passed QC.
Integration and Clustering of scRNA-seq Datasets by iNMF (Integrative Non-negative Matrix Factorization)
The R LIGER (linked inference of genomic experimental relationships) package was used to implement iNMF. Normalization and scaling were performed with the default settings. Variable genes were chosen with a variance threshold of 0.1, and iNMF was performed on variable gene expression using the optimizeALS function with k = 24 and lambda = 5. In the shared factor space, a neighborhood graph was built based on shared factor loading patterns. The Louvain community detection clustering algorithm was then used to find clusters across the two stages of neurogenesis. This was computed using the quantileAlignSNF function in the LIGER package with resolution = 0.3. Cluster annotation was performed by assessing marker gene expression patterns and top genes in factors associated with each cluster. To compare the difference in factor loadings across FXS and control, the mean factor loadings of both FXS and control were calculated at each time point.
Correlation of genes and maturation score
The maturation score was calculated based on published methods.30 Briefly, non-linear dimensionality reduction was performed on the cell distance matrix using R package DiffusionMap to obtain the first 50 dimensions. The first 3 dimensions, which each accounted for at least 4% of the total variance, were selected and a principal curve (R package princurve, smoother = “lowess”, f = 1/3) was fitted. The beginning of the curve was assigned such that maturation score was negatively correlated with the expression of vimentin (VIM), a canonical marker of neural progenitors. The maturation score of a cell was measured by the arc length along the principal curve.
Differential Expression and GSEA (Gene Set Enrichment Analysis)
For each cluster at each time point, expression of each gene was fitted to a hurdle model using R package MAST: zlm(~ group + cngeneson) in which “group” is a variable specifying control or FXS and “cngeneson” is the gene detection rate. To identify genes differentially expressed in FXS, a likelihood-ratio test was performed by dropping the group variable. Differentially expressed genes were selected if the fold change was at least 1.1 and FDR (false discovery rate) was less than 0.05.
Gene ontology (GO) biological process terms were downloaded from the Molecular Signatures Database (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp). To test for enrichment, the coefficients of the group variable from the hurdle model belonging to various GO terms of interest were compared to the background using the gseaAfterBoot function in MAST. GO terms were considered to be enriched if the adjusted P-value was less than 0.05.
Overrepresentation Test
ASD-associated genes were obtained from the Gene Scoring Module of the SFARI database (https://gene.sfari.org/database/gene-scoring/). To determine whether DEGs from each cluster and time point were significantly enriched for ASD genes, the P-value of the hypergeometric test was calculated in R by calling phyper(s, S-s, N-K, K, lower.tail = FALSE), in which “s” is number of overlapped DEGs that overlaps with ASD genes, “S” is the total number of ASD genes, “N” is the number of expressed genes in the scRNAseq dataset and “K” is the total number of DEGs.
Other Comparisons Between FXS and Control Cells
For other comparisons between FXS and control samples, such as for immunocytochemistry (ICC), multielectrode array (MEA), and patch-clamp data analysis, the Mann-Whitney U test was performed using the wilcox.test function in R.
Results
Reproducible Differentiation of hPSCs into GABAergic Inhibitory Neurons
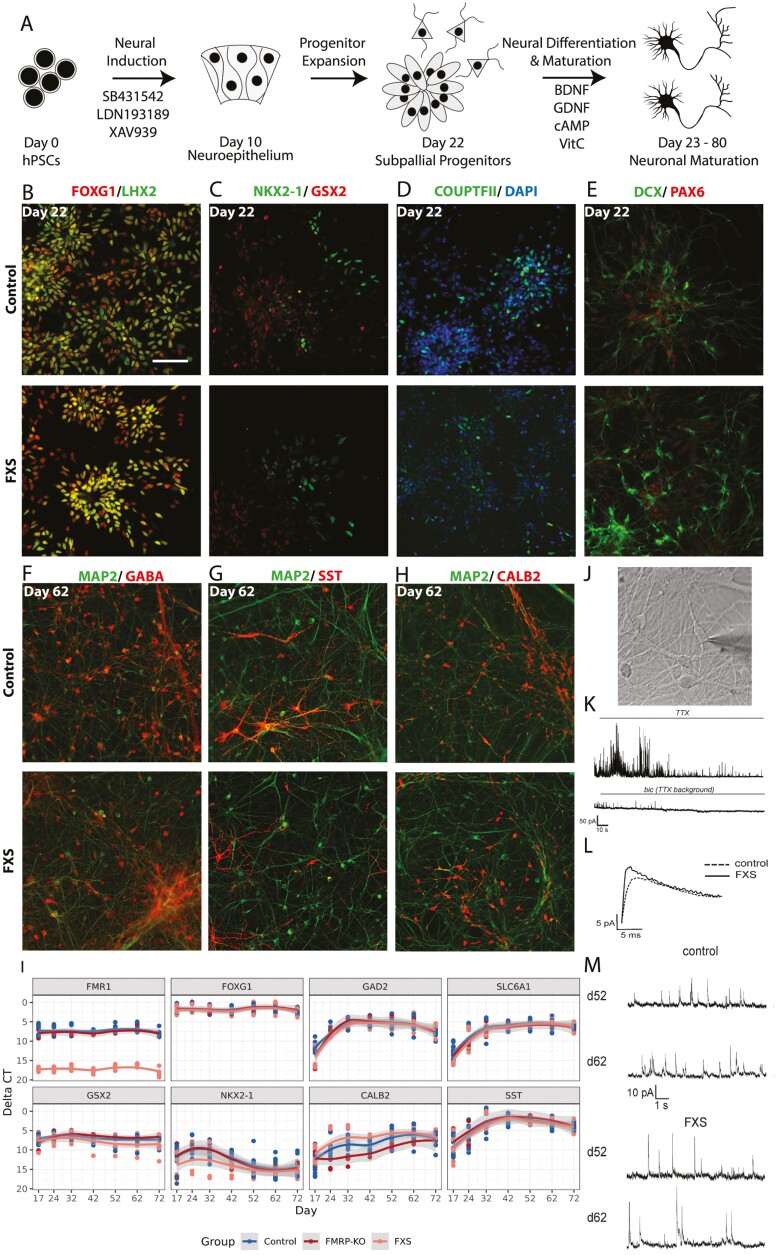
To study GABAergic neuronal development, we established a reproducible protocol to generate GABAergic neurons from hPSCs. GABAergic inhibitory neurons originate from the ganglionic eminence (GE), which is a transient structure in the ventral telencephalon that guides cell and axon migration. Although protocols for the generation of medial ganglionic eminence (MGE)-derived inhibitory neurons have been reported,31,32 there is less information about producing other types of telencephalic inhibitory neurons, such as those from the caudal ganglionic eminence (CGE) and the lateral ganglionic eminence (LGE). Thus, to include a wider variety of inhibitory subtypes, we set out to pattern neural progenitors to a mixed fate of MGE and CGE/LGE progenitors. Morphogens such as SHH, FGF, and WNT are important in specifying the dorsal-ventral axis of the telencephalon.33,34 We hypothesized that fine-tuning the dose and timing of these morphogens could lead to a reproducible protocol to generate inhibitory neurons derived from mixed GE fates. Because higher SHH concentration signals the cells to adopt a more ventral fate, manipulation of SHH would be expected to push cells toward a distinct GE fate rather than a mixture of MGE and CGE/LGE fates. Therefore, we did not optimize SHH concentration but tested several conditions of WNT and FGF2 treatment instead (Fig. 1A, Supplementary Fig. S1). We extracted RNA from each condition on differentiation day 30, a time when areal identity of neural progenitors is largely determined, and quantified the cell types with the expression of canonical markers for the dorsal telencephalon (cortex), MGE, and CGE/LGE by qRT-PCR.
Figure 1.
A directed differentiation protocol yields inhibitory neurons representing a mixed ganglionic eminence fate that form functional GABAergic synapses. (A) In addition to dual SMAD inhibition, additional WNT inhibition was also applied during neural induction. On day 10, neural progenitors were withdrawn from small molecule treatments, and progenitors expanded from day 10 to day 22. Cells were dissociated on day 22 and seeded as single cells for terminal differentiation and maturation in the presence of neurotrophic factors BDNF, GDNF, cAMP, and vitamin C. Representative immunocytochemistry images of differentiating cultures from one control cell line (SAB1-13D) and one FXS cell line (SC105iPS6) are shown in B-H. (B-E) At progenitor stage on day 22, this protocol yielded ventral forebrain progenitor cells. Bar: 100 µm. (F-H) At neuronal maturation stage on day 62 of differentiation, inhibitory neurons derived from ventral forebrain progenitors dominated the cultures, as marked by GABA, SST, and CALB2 antibodies. (I) Expression of key GE lineage markers were assayed by qRT-PCR at differentiation days 17, 24, 32, 42, 52, 62, and 72. Three separate batches of differentiations were included. (J) Phase-contrast image of a patch-clamped neuron. (K) Recorded traces illustrate the experimental paradigm. mIPSCs were recorded in the presence of tetrodotoxin (TTX), that blocks action potentials, and bicuculline, a GABAA receptor blocker, was used at the end of each recording to confirm inhibitory postsynaptic currents. (L) Superimposed mIPSC events obtained by averaging individual events of control and FXS neurons. (M) Representative mIPSC traces of control (n = 21) and FXS neurons (n = 17) on days 52 and 62. Scale bar (B-H) = 100 μm.
None of the conditions we tested altered telencephalon identity, as evidenced by FOXG1 expression. However, as expected, areal identity along the dorsal-ventral axis was sensitive to the duration and timing of application of WNT inhibition and FGF2 treatment (Supplementary Fig. S1D). When FGF2 was added during the progenitor proliferation phase, the excitatory lineage genes (EMX1, EOMES, SLC17A7, TBR1) were upregulated, and the inhibitory lineages genes (GAD2, GSX2, NKX2-1) were downregulated. The duration of WNT inhibition by XAV939 also drastically shifted the dorsal-ventral identity. While XAV939 treatment from day 0-4 showed only modest differences from untreated cells, day 0-9 treatment drastically shifted the cell culture to a more ventral identity, even without additional SHH. This was evidenced by the upregulation of NKX2-1 while maintaining the expression of CGE/LGE marker GSX2. Along the same trend, excitatory lineage markers were drastically reduced with this treatment. Therefore, with WNT inhibition throughout the neural induction phase during differentiation days 0-9 without additional SHH and FGF2 treatments, we were able to derive cell cultures that were dominated by a mixture of GE identity cells.
To test for reproducibility, 3 batches of differentiations were performed on all 6 cell lines included in this study. Total RNA was extracted at 8 time points: days 9, 17, 24, 32, 42, 52, 62, and 72, and the expression of key marker genes was measured by qRT-PCR (Supplementary Fig. S2A). Overall, each cell line showed similar expression patterns across batches. FMR1 was never expressed in FXS hiPSC-derived cells but was highly expressed in control cells throughout the time period examined. As previously reported,25 the genetically engineered WA01 hESC line used here (referred to as FMRP-KO) does transcribe FMR1, but because of a premature stop signal does not produce the protein product, FMRP (Supplementary Fig. S3). FOXG1 and LHX2 were also highly expressed throughout the time period, thereby validating telencephalon identity. Inhibitory neuron progenitor genes (GSX2, NKX2-1) peaked at day 24, consistent with their roles in progenitor specification and expansion. The expression of inhibitory neuronal subtype genes (CALB2, RELN, SST, NPY, SLC6A1, GAD2) peaked at day 42 and then plateaued, indicating that neurogenesis likely proceeded until day 42. PAX6 is expressed in pallial progenitors but is also detected in LGE and CGE progenitors and a subset of LGE-derived interneurons.23 Thus, it was not surprising that PAX6 expression remained high throughout the course of differentiation. The expression of excitatory lineage markers SC17A7, EOMES, EMX1, and TBR1 was low compared with inhibitory markers. The excitatory marker expression peaked at about day 17 but slowly decreased to near zero with time, suggesting that at the end of neurogenesis, the cell culture consisted mostly of inhibitory neurons (Fig. 1 and Supplementary Fig. S1).
To examine the effects of disease status, differentiation day, or batch on our data, we performed principal component analysis (PCA) on our marker gene expression panel (Supplementary Fig. S2B). The first principal component (PC) explained approximately 39.3% of the variance and correlated highly with differentiation day. However, we did not observe clustering by batch or disease status across our first 4 principal components (cumulative 77.31% variance explained). Overall, these results show that the inhibitory neuron derivation protocol generated a mixed population of GE-derived inhibitory neurons and was largely resistant to disease status and batch-to-batch differences.
No Difference in Distribution of Neuronal Subtypes Between FXS and Control
We further characterized cell identity via immunocytochemistry on 3 control and 3 FXS cell lines. On day 22, 70%-80% of cells expressed the telencephalon markers FOXG1 and LHX2 in both FXS and control lines (Fig. 1B). Cells positive for at least one of the GE markers (NKX2-1, GSX2, COUPTFII, and PAX6) (Fig. 1C, E) accounted for most of the cells in the cultures. On day 62, most of the neurons were GABA-positive (Fig. 1F). Inhibitory neuron subtypes such as SST-expressing and CALB2-expressing inhibitory neurons were abundant (Fig. 1G, H). No significant difference was observed by qRT-PCR between FXS and controls in marker genes FOXG1, GAD2, SLC6A1, GSX2, NKK2-1, CALB, or SST (Fig. 1I).
Both FXS and Control Neurons Formed Functional GABAergic Synapses
We tested whether FXS and control GABAergic neurons formed functional synapses by performing patch-clamp recording for all 6-cell lines, with 10-15 cells per cell line on day 52 or day 62 of differentiation. Under voltage clamp mode with intracellular recording, we measured miniature postsynaptic currents (Fig. 1J-M) on days 52 and 62 of differentiation. No excitatory events were detected, further confirming that our protocol yielded a predominantly inhibitory neuronal population. Upon bicuculline administration, which blocks GABAA receptors, all postsynaptic events disappeared, showing that the postsynaptic events detected were mediated by GABAA receptors. FXS- and control-derived inhibitory neurons did not show differences in miniature inhibitory postsynaptic current (mIPSC) frequency or mIPSC amplitudes in the small dataset we tested (Supplementary Fig. S4A, C). However, mIPSC rise time was significantly faster in FXS neurons on day 62 (Supplementary Fig. S4B). Together, these data indicate that FXS and control neurons both formed functional GABAergic synapses in culture, and suggest a difference in GABA receptor channel dynamics between FXS and control neurons.
Increased Activity in FXS PSC-Derived Inhibitory Neurons
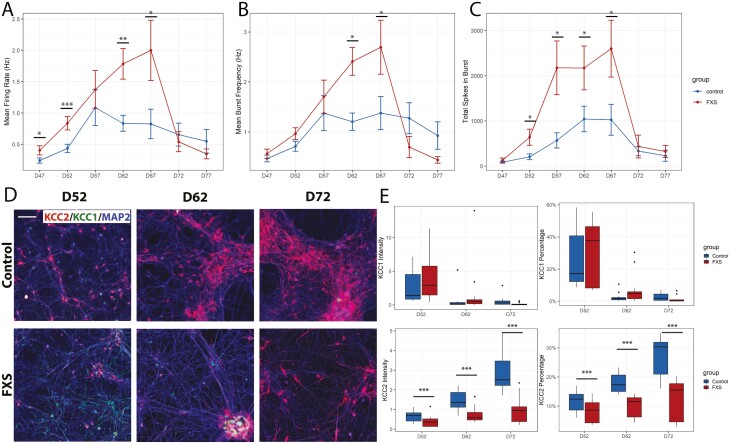
To test whether there were differences in neuronal activities in FXS hPSC-derived inhibitory neurons, we recorded spontaneous activity from 8 replicates per group of 3 FXS and 3 control cultures every 5 days from days 47 to 77 using microelectrode array (MEA) (Fig. 2A-C). To control for cell density on the MEA, we plated cells at 5.5 × 105 cells/cm2 on day 42. Early in neurogenesis GABA functions as an excitatory neurotransmitter, and as expected, both control and FXS were active on MEA (Supplementary Fig. S5). However, FXS cells were more active from days 47 to 67; they fired more frequently (Fig. 2A), burst more frequently (Fig. 2B) and had a greater number of spikes in bursts (Fig. 2C). The data shown in Fig. 2 are the results of 3 separate differentiations.
Figure 2.
Functional and immunocytochemical analyses suggest functional delay in FXS inhibitory neuronal culture. (A-C) Spontaneous firing activity was measured from days 47 to 77 in FXS and control cultures. FXS cultures showed reduced active spiking activity across most time points as measured by mean firing rate (A). Burst frequency and the number of total spikes per burst are significantly higher in FXS cultures at the indicated time points (B, C). Three control and 3 FXS cell lines were assessed, including an FMR1 knockout hESC line and isogenic control. Wilcoxon signed-rank tests were performed. Immunocytochemistry was performed with KCC1, KCC2, and MAP2 on days 52, 62, and 72 of neuronal culture with FXS and control cell cultures. Representative images are shown in (D). Two batches of differentiations were included, with 3 control and 3 FXS cell lines (including a pair of isogenic control and FMR1 knockout). Mean intensity, and percentage of positive pixels normalized to MAP2 were measured for KCC1 and KCC2 (E). While KCC1+neurons were not variable between control and FXS cultures, the number of KCC2+ neurons was greater than control at all time points. * P < .05, ** P < .01, *** P < .001. Scale bar (D) = 100 μm.
As the neurons matured, we expected to see an increase in spontaneous spiking at first, followed by a decrease in activity as GABA changed from functionally excitatory to inhibitory. This is consistent with what we observed; both the mean firing rate of active electrodes (Fig. 2A) and mean bursting (Fig. 2B, C) first increased over time, then decreased. Importantly, activity peaked in controls on day 57, whereas the peak of activity in FXS cells occurred 10 days later, consistent with the idea that the GABA functional switch was delayed in maturing FXS inhibitory neuron cultures.
Aberrant KCC1-KCC2 Transition in Maturing FXS Inhibitory Neurons
We further investigated the delay in the GABA functional switch in human FXS inhibitory neurons by examining the expression of two key potassium/chloride cotransporters that regulate intracellular Cl− concentration and the GABA switch. In immature neurons, the action of KCC1 (K+/Cl− cotransporter 1; SLC12A4, solute carrier family 12 member 4) maintains a high intracellular Cl− concentration and GABA is excitatory. As neurons mature, expression of KCC2 (K+/Cl− cotransporter 2; SLC12A5, solute carrier family 12 member 5), initiates the GABA functional switch. We performed immunocytochemistry to detect KCC1 and KCC2 antigens on days 52, 62, and 72 in control and FXS hPSC-derived inhibitory neurons. KCC1+ neurons and KCC2+ neurons were quantified for each group per time point (Fig. 2D, E).
Overall, there was little difference in KCC1 expression between control and FXS. On day 52, there were comparable percentages of KCC1+ neurons and mean KCC1 intensity in both FXS and control (Fig. 2E). On days 62 and 72, KCC1+ neurons decreased sharply to almost 0 in both control and FXS.
We also observed a steady increase of KCC2 expression from days 52 to 72 in both control and FXS cultures (Fig. 2E). However, the magnitude of increase was much larger in control cultures compared with in FXS and at every time point, there were significantly more KCC2+ neurons in control compared to FXS cultures.
Overall, our observations are consistent with an aberrant transition from KCC1 to KCC2 protein expression in the FXS maturing GABAergic neuronal culture, supporting the idea that the delay in the decrease of neuronal activity is the result of a delayed GABA functional switch in FXS GABAergic neurons during maturation.
Multiple GABAergic Neuron Subtypes were Generated in Both FXS and Control Cultures
The study of GABAergic neural development in FXS has been hindered by an inability to derive multiple GABAergic lineage subtypes. To better examine cellular subpopulations in our cultures, we performed single-cell RNA-seq on early (day 22) and late (days 42-48) stages of neurogenesis. To integrate data from the two stages of differentiation, we used LIGER to perform integrative non-negative matrix factorization (NMF) ,35 which allows for single-cell gene expression matrices to be decomposed into both shared and dataset-specific metagenes, which are referred to as “factors”. Each factor represents a set of genes that co-varies and is generally associated with a cell type-specific or a biological program-specific signature.
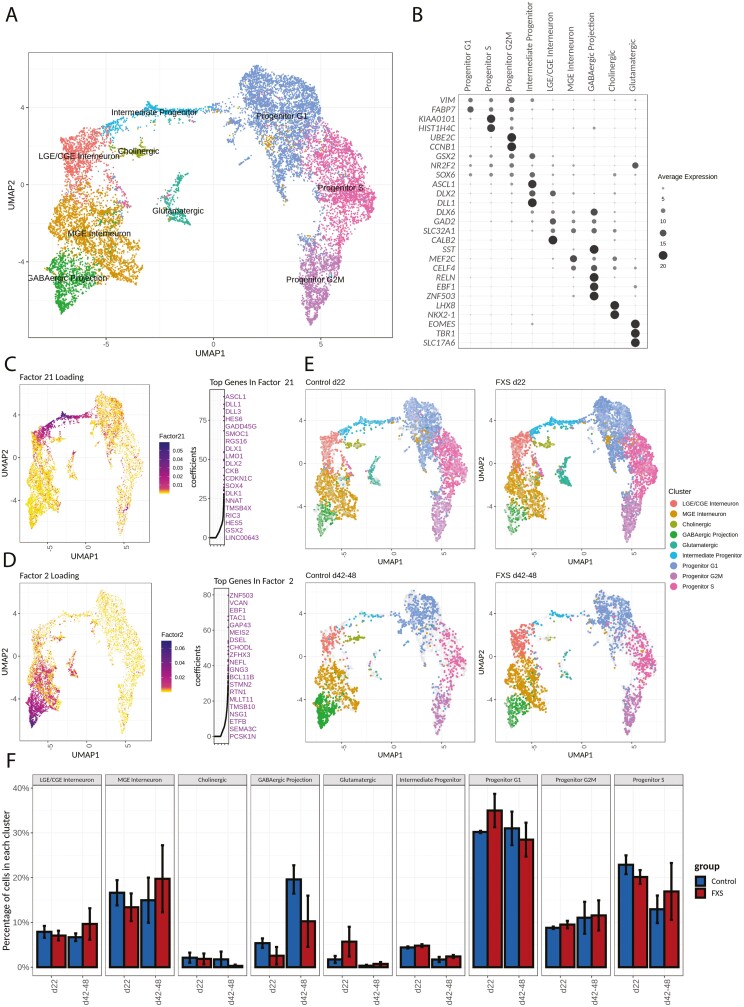
Joint clustering of the single-cell datasets was performed based on the similarity of shared factor patterns across the early and late culture time points. We annotated clusters (Fig. 3A) based on expression of canonical markers as well as the major contributing genes identified by NMF analysis (Supplementary Table S5). A clear separation was seen between neuronal and progenitor clusters. Forebrain marker FOXG1 (Supplementary Fig. S6B), together with the pan-GE markers MEIS2 and DLX2 (Supplementary Fig. S6C), were broadly expressed in all clusters, while expression of DLX6, a marker for developing and mature inhibitory neurons, was more restricted to the neuronal clusters. GABAergic neurons comprised the majority of the neuronal population based on GAD2 and SLC32A1 expression (Fig. 3B and Supplementary Fig. S6H), an observation that is in agreement with our ICC and patch-clamp results. Interestingly, progenitor clusters were not distinguished by GE regional identity; GE region-specific progenitor markers (GSX2, NR2F2, NKX2-1) did not show clear separation among progenitor groups (Supplementary Fig. S6D, E). Instead, progenitor clusters were segregated by cell cycle phase, demonstrated by the expression of cell cycle genes such as KIAA0101, HIST1H4C, UBE2C, and CCNB1 (Fig. 3B). Similarly, intermediate progenitors were marked by the expression of ASCL1, DLX2, DLL1, HES6 (Fig. 3B, C).
Figure 3.
Single-cell RNA sequencing reveals diverse cell types present in FXS and control hPSC-derived inhibitory neuron cell cultures. (A) Unbiased clustering of scRNA-seq data. Cell types were annotated according to both expression of known marker genes (B) and factor loadings relevant to each cell cluster (CD). Progenitor clusters are predominantly separated by cell cycle phase. The neuronal population is composed predominantly of GABAergic neurons and a small percentage of glutamatergic neurons. (B) Expression of genes specific to each cluster. (C, D) Non-Negative Matrix Factorization was used to delineate factors of weighted genes and their associations with each cluster. For each factor shown in C-D, factor loadings are color coded in the UMAP representation on the left, while the top 20 genes with highest coefficients are marked on the right. (C) Top genes in Factor 21, such as ASCL1, HES6, and DLX2, are markers for intermediate progenitors. (D) Top genes in Factor 2, such as EBF1, ZNF503, and BCL11B, are associated with GABAergic projection neurons. (E) UMAP representation of cells from either control or FXS samples at 2 different time points. The gray background represents the rest of cells in the complete dataset. (F) Quantification of cluster composition at each time point. Progenitor populations decreased as the neuronal population increased over time. Intermediate progenitor and glutamatergic neurons were a minor cell population.
Louvain clustering identified 3 clusters of GABAergic neurons (Fig. 3A, B): a cluster with high expression of CALB2 and NPY but not SST, suggestive of CGE and LGE interneurons (Supplementary Fig. S6D), a second cluster that showed expression of SST, SOX6 and MEF2C (Supplementary Fig. S6E) characteristic of MGE interneurons, and a third cluster that was associated with factor 2, whose highest coefficient genes included ZNF503 and EBF1 (Fig. 3D, Supplementary Fig. S6G), which are markers for GABAergic projection neurons. A small cluster showed highly specific expression of both NKX2-1 and LHX8 (Fig. 3B). Genetic fate mapping has shown that LHX8+/NKX2-1+ cells predominantly generate cholinergic neurons.30 We therefore annotated this cluster as “cholinergic cells”.
A small cluster of glutamatergic excitatory neurons could be identified by the presence of excitatory lineage markers EOMES, TBR1, and SLC17A6 (Fig. 3B, Supplementary Fig. S6I) that were only sparsely present on days 42-48 relative to day 22 (Fig. 3E, F). This is in agreement with our qRT-PCR measurements that indicated that expression of excitatory lineage markers peaked at about day 24 and gradually diminished with time (see Supplementary Fig. S2A). Quantification of the number of cells per cluster per time point showed an increase in the proportion of GABAergic neuronal clusters and a decrease in progenitor clusters on days 42-48 compared to day 22. However, at both time points, there was no significant difference in cluster distribution between FXS and control (Fig. 3E, F). Furthermore, pseudo-bulk RNA-seq data were generated for each sample, and the correlation between samples was computed. At each time point, correlation between samples was above 0.86 (Supplementary Fig. S7), indicating high consistency in differentiation among different cell lines. Overall, cell type analysis from single-cell profiling was consistent with the qRT-PCR and ICC data that indicated that both FXS and control hPSCs generated subtypes of GABAergic neurons that represented MGE-, LGE-, and CGE-derived cells.
Shared and Cluster-Specific Differentially Expressed Genes (DEGs) in FXS
To understand cell type-specific differences between FXS and control, we performed differential gene expression analysis at the two time points representing early (day 22) and late (days 42-48) stages of neurogenesis. For each time point, cells from each cluster were compared separately. Gene expression profiles were fitted with a hurdle model that accounts for the probability of gene dropout, cellular gene detection rate, and disease effect using MAST.36 A likelihood ratio test was performed by dropping the disease variable in the hurdle model to detect genes that were differentially expressed between clusters. Genes were considered differentially expressed relative to control if there was at least a 10% change in gene expression with a false discovery rate less than 0.05.
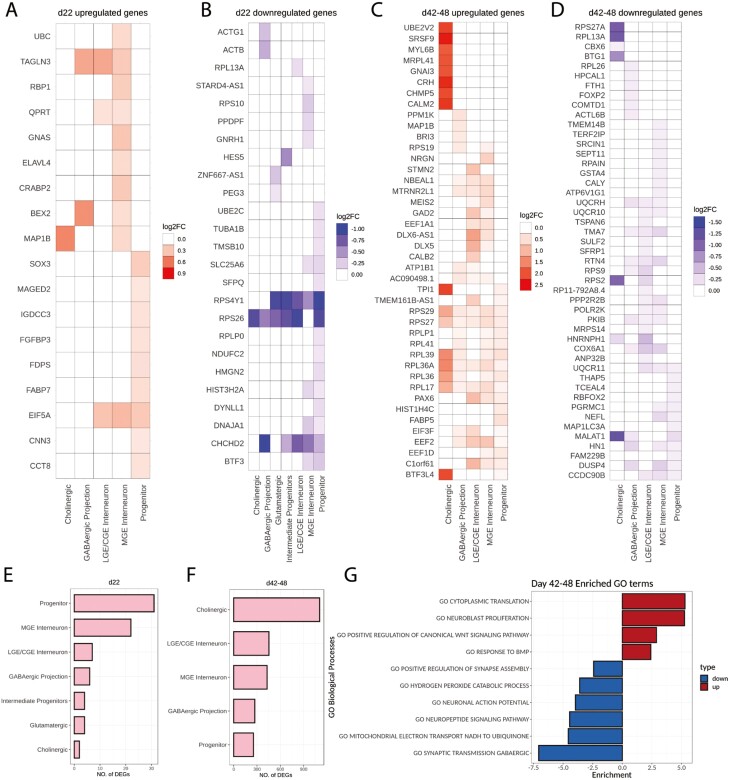
On day 22, only 4 differentially expressed genes (DEGs) were shared between progenitor and neuronal clusters: EIF5A, a translation initiation factor, was broadly upregulated in FXS; RPS4Y1, RPS26, and CHCHD2 were broadly downregulated in FXS (Fig. 4A, B). CHCHD2 is associated with neuronal migration and was identified as an FXS candidate gene in our previous study26 as well as a downregulated gene in multiple interneuron clusters in autism brain.37TAGLN3, upregulated in multiple neuronal clusters, is a neuron-specific protein that regulates neurite growth.38 Most DEGs in FXS cells were cell cluster specific on day 22, and many have functions in protein translation (EIF5A, RPL13A, RPS10, and RPLP0) and cytoskeleton remodeling (TAGLN3, MAP1B, ACTG1, ACTB, TUBA1B, TMSB10, and DYNLL1). However, no gene ontology (GO) terms were significantly enriched in these DEGs. Overall, the differences between FXS and control were minor at the earlier time point.
Figure 4.
Differentially expressed genes include both shared and cell type- and time point-specific genes. (A-D) Top 10 differentially expressed genes in each cluster are shown by log2 fold change value. Red indicates upregulation while blue indicates downregulation. Many genes, such as a group coding for ribosomal proteins, showed a shared pattern of upregulation across cell clusters. (E, F) The number of differentially expressed genes increased dramatically from the early time point (day 22) (E) to the later time point (days 42-48) (F). (G) GO biological process terms enriched in DEG genes at the later time point (days 42-48).
At the later stage of neurogenesis, the differences between FXS and control became more pronounced. At this stage, many DEGs in FXS were shared between progenitor and neuronal clusters. Notably, among the most significant differentially expressed genes (Fig. 4C) was a group of ribosome genes (RPS29, RPS27, RPLP1, RPL41, RPL39, RPL36A, RPL17, EIF3F, EEF2, and EEF1D) that were universally upregulated in FXS across clusters. This suggests that translation was broadly upregulated in FXS cells across cell types. Apparent differential expression of ribosomal genes sometimes can be caused by artificial sampling of outlier samples that have more ribosomal reads than expected. To test if this were the case in our dataset, ribosomal protein read percentages were plotted across samples at each time point (Supplementary Fig. S8). We found the percentage of reads belonging to ribosome genes was similar across different samples for each time point, which suggests that the differential expression of ribosomal genes on days 42-48 represents a biological difference between FXS and control cells and not a sequencing artifact.
Many of the FXS-associated downregulated genes were also shared across clusters (Fig. 4D). Among them, many (UQCRH, UQCR10, COX6A1, UQCR11, and CCDC90B) are nuclear-encoded mitochondrial genes that have important roles in electron transfer through the respiratory chain.
Burden analyses demonstrated that not only did the number of DEGs increase dramatically from day 22 (<30 genes) to days 42-48 (300-1000 genes), but that certain cell types were more affected than others (Fig. 4E, F). For example, progenitors had an enrichment of DEGs at day 22, whereas the most affected cell types at days 42-48 were the cholinergic cells, followed by the GABAergic interneurons. Gene set enrichment analysis (GSEA) on the days 42-48 DEGs (Fig. 4G) revealed that the most significant upregulated GO biological processes in FXS (days 42-48) were cytoplasmic translation, neuroblast proliferation, and WNT and BMP signaling, while downregulated processes included neuronal functions such as action potential and synaptic processes, as well as mitochondrial functions (hydrogen peroxide and electron transport) (Fig. 4G).
Overrepresentation of Autism Risk Genes Among FXS DEGs
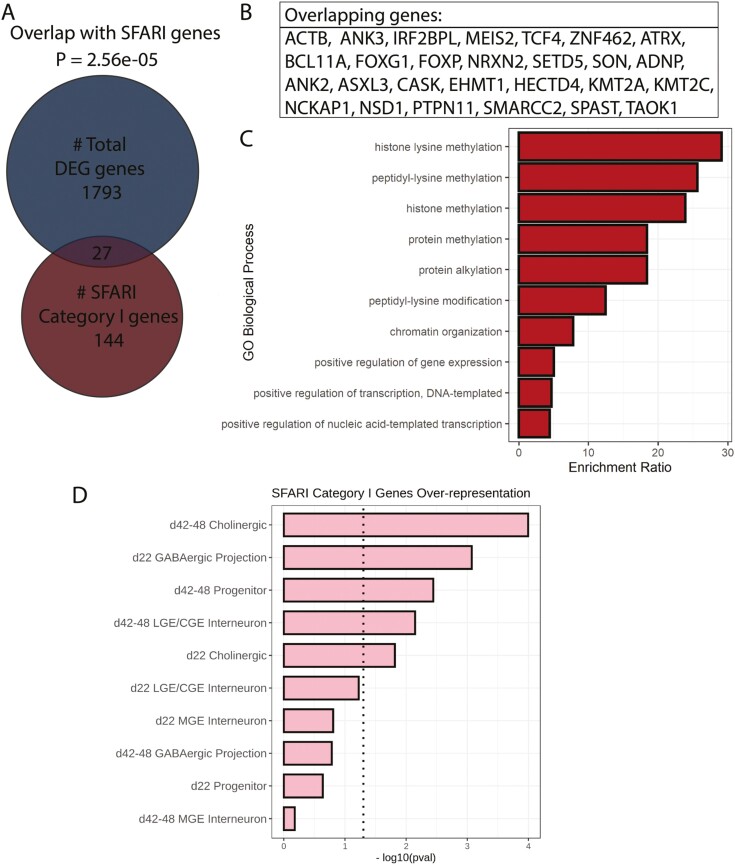
Because of the significant overlap between manifestations of FXS and ASD (autism spectrum disorder), we asked whether genes that are strongly implicated in autism (SFARI category I) were enriched in the FXS DEGs. We found a significant overlap between FXS DEGs and ASD risk genes (P = 2.56e–05, hypergeometric test): among the 1793 DEGs in FXS, 27 overlap with SFARI (category I) genes (Fig. 5A, B). Interestingly, many of these genes are involved in epigenetic modification (Fig. 5C). Enrichment here was also cluster specific. ASD risk genes were significantly enriched in the GABAergic projection neurons and the cholinergic clusters on day 22 as well as the progenitor and the GABAergic CGE/LGE interneuron and the cholinergic clusters on days 42-48 (Fig. 5D). A list of overlapping genes between ASD risk genes and FXS DEGs is provided in Supplementary Table S6.
Figure 5.
Sets of differentially expressed genes were enriched with autism-associated genes, especially at later time points. (A) Overall, 27 differentially expressed genes overlap with 144 Category I genes from the SFARI database. An over-representation test was carried out by hypergeometric test and P-value of overlap by chance is 2.56e-05. (B) List of overlapping genes between the DEGs in this study and the SFARI Category I genes. (C) GO analysis of the overlapping genes showed their involvement in the histone lysine methylation process. (D) Overlap between SFARI genes and DEGs by cell type and time points. Dotted line indicates statistical significance (P < .05).
Downregulated Genes in FXS Progenitors Were Negatively Correlated with Maturation at Days 42-48
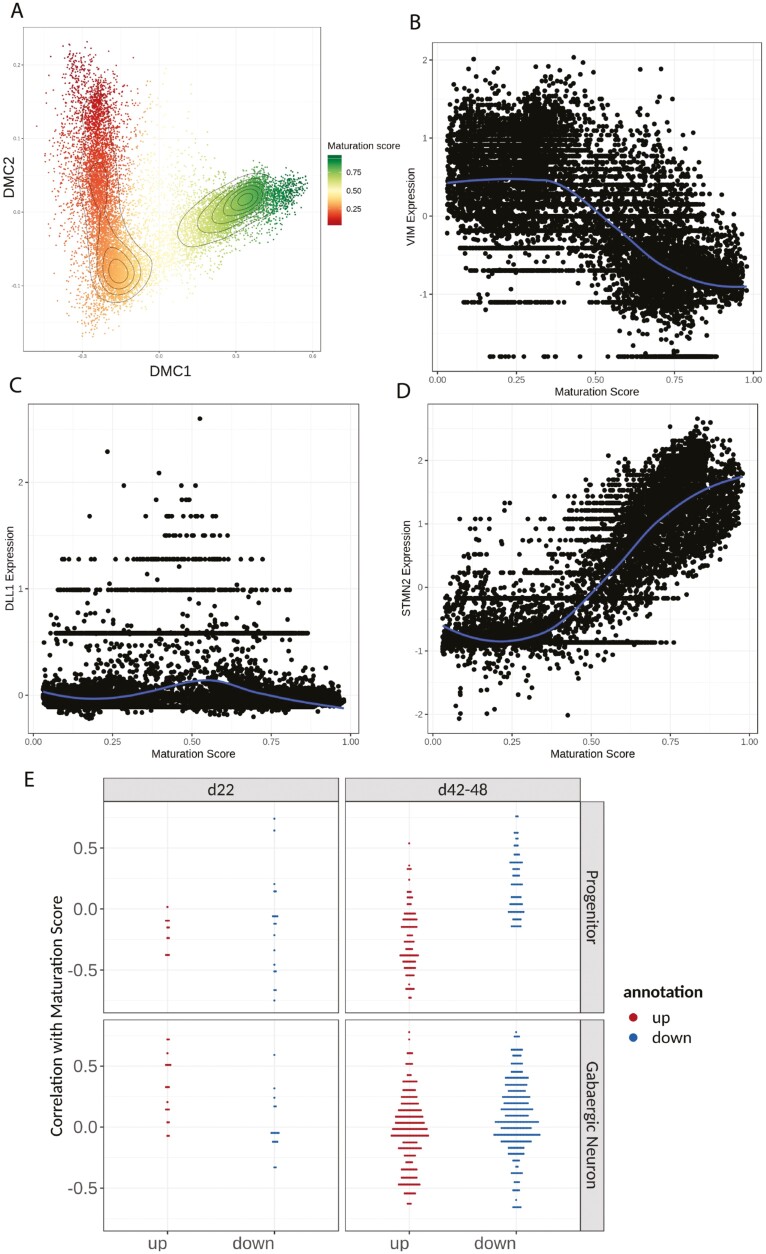
Our GSEA revealed an upregulation of progenitor-related processes and downregulation of neuronal processes in FXS at early and late stages of neurogenesis, respectively. Next, we determined whether DEGs from progenitor or neuronal clusters in FXS were correlated with maturation. The maturation score reflects the differentiation trajectory from progenitor to neuron. We first calculated the maturation score of each cell (Fig. 6A, Methods).30 As expected, progenitor marker VIM had the highest expression in cells with low maturation scores (Fig. 6B), while intermediate progenitor marker DLL1 showed highest expression in cells with mid-range maturation score (Fig. 6C), and neuronal marker STMN2 showed the highest expression in cells with a high maturation score (Fig. 6D).
Figure 6.
Maturation score based on differentiation trajectory showed FXS cells had lower maturation scores in GABAergic neuron clusters on day 48. (A) Diffusion map representation of the scRNA-seq dataset based on the first 2 components. Colors are coded by the maturation score. (B-D) Scatter plots of normalized expression (centered around 0) of key marker genes. The gene expression from each cell was plotted against the maturation score. (B) VIM marks neural progenitors and shows the highest expression in cells with low maturation scores. (C) DLL1 marks intermediate progenitors, and its expression peaked in cells that have mid-level maturation scores. (D) STMN2 marks neurons and has the highest expression in cells with high maturation scores. (E) Correlation of gene expression to maturation score was plotted against upor downregulated genes. On day 22, the correlation of DEG genes were randomly distributed. On days 42-48, especially in the progenitor cluster, upregulated genes in FXS were negatively correlated with the maturation score, while downregulated genes in FXS were positively correlated with maturation score.
We calculated Spearman correlation between maturation score and gene expression. The list of DEGs and their correlation with maturation is provided in Supplementary Table S7. On day 22, downregulated genes in the FXS progenitor cluster were uniformly distributed while upregulated genes were more negatively correlated with the maturation score (Fig. 6E). In the GABAergic neuronal group, both upregulated and downregulated genes were positively correlated with the maturation score. In contrast, at the later stage of neurogenesis, especially in the progenitor cluster, upregulated genes in FXS were preferentially negatively correlated with the maturation score, while the downregulated genes in FXS were preferentially positively correlated with the maturation score. These data suggest that at the later stage of neurogenesis, FXS cells were in a less mature state and the difference was most prominent in FXS neural progenitors.
Factor Analysis Revealed Differences in Biological Processes Associated with Maturation in FXS at the Late Stage of Neurogenesis
Using non-negative matrix factorization, we decomposed the scRNA-seq expression matrix such that each cell could be described as a linear combination of biologically interpretable factors (metagenes). Factor loadings are defined as the coefficients of these linear combinations. Factors with significant differences in factor loadings between FXS and control cells represent disease-specific changes in biological programs. Supplementary Fig. S9 shows the results of our factor loading analysis. On day 22 of differentiation, we found that 5 out of the 24 factors displayed a significant difference in mean factor loadings ≥25% between FXS and controls (adjusted P-value < 0.05, Bonferonni correction). However, the absolute loading values for these factors were small, indicating that there were few differences between FXS and control cells on day 22.
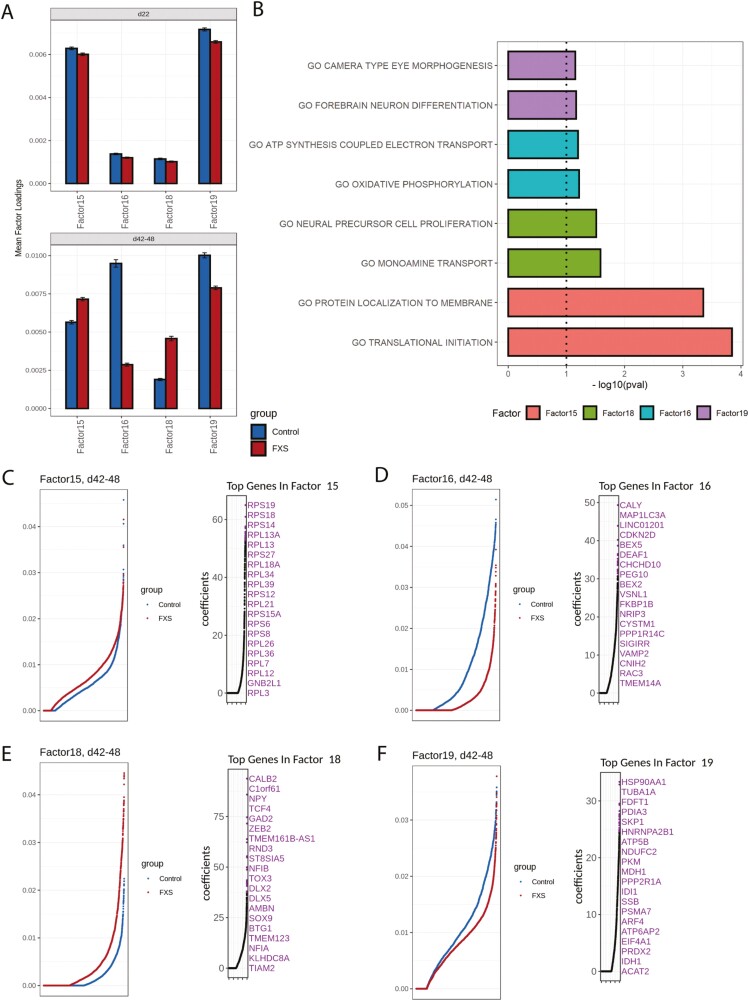
However, at the later stage of differentiation (days 42-48), 11 of the 24 factors showed a significant difference in mean factor loadings ≥25% (adjusted P-value < 0.05, Bonferonni correction). Factors 15, 16, 18, and 19 were particularly interesting because the mean loadings were high and the difference in mean loadings between FXS and control were large (Fig. 7A), indicating large differences between FXS and control in the biological processes associated with these factors. Moreover, these factors were not different from control at the early time point, suggesting that these gene sets became dysregulated later in neurogenesis. For factor 15, in which FXS had higher mean loadings than control, the top genes were ribosome subunits (Fig. 7C). As expected, GSEA revealed that this factor is enriched with GO terms such as “translational initiation” and “protein localization to membrane” (Fig. 7B), suggesting that protein translation may be upregulated in FXS compared with control at the later time point, which agrees with findings that translational activity is increased in cells lacking FMRP.9 FXS cells also showed higher mean loadings in factor 18. Among the top genes in factor 18 are key regulators of interneuron precursors and newly determined interneurons (DLX2, DLX5, TCF4, ZEB2) (Fig. 7E). The top GO term associated with factor 18 is “neural precursor cell proliferation” (Fig. 7B). Together, these analyses show that at the later time point, biological processes associated with precursor proliferation and protein translation were more prominent in FXS relative to controls.
Figure 7.
Factors associated with FXS cells revealed differences in cellular processes and maturation state. (A) Mean factor loadings from control (blue) and FXS (red) samples are shown at different time points for factors 15, 16, 18, and 19. Difference was minor at the early time point (day 22), but increased on the later time point (days 42-48). Factors 15 and 18 showed higher mean loading in FXS samples, while Factors 16 and 19 showed consistently lower mean loadings in FXS samples at the later time point. (B) Top GO biological process terms associated with factors that showed the most loading difference between FXS and control. (C-F) For each factor shown, loadings from either control or FXS cells were ranked and plotted on the left, and the top 20 genes with the highest coefficients from the corresponding factor are marked on the right. (C) Factor 15 showed a group of ribosomal genes, suggesting increased translation processes in FXS cells. (D) Top genes in Factor 16 showed a group of genes responsible for synaptic vesicular transport (CALY, VSNL1, VAMP2), indicating more mature neuronal processes. (E) Factor 18 showed genes associated with early-born GABAergic neurons (DLX2, DLX5), indicating younger neurons in FXS cells. (F) Top genes in Factor 19 belong to a group of genes responsible for energy metabolism including ATP5F1B, NDUFC2, PKM, ACAT2, MDH1, and IDH1.
FXS cells also showed significant decreases in mean loadings of factors 16 and 19. Top genes in factor 16, such as CALY, MAP1LC3A, CNIH2, VAMP2, RAC3, and VSNL1, are genes important for cytoskeleton remodeling and vesicular transport (Fig. 7D). Interestingly, the top GO terms associated with factor 16 is “ATP synthesis” and “oxidative phosphorylation”, which are required to provide energy for vesicular transport (Fig. 7B). Top genes in factor 19 are involved in energy metabolism processes such as glycolysis (PKM, MDH1, and IDH1) and lipid metabolism (ACAT2) (Fig. 7F). Therefore, on days 42-48, FXS cells showed lower levels of processes associated with more mature neurons, such as energy metabolism and synaptic vesicle transport.
Overall, at the later time point, biological processes associated with progenitors were higher in FXS compared to control, while metabolic processes associated with maturing neurons were lower. Factor loading analyses were consistent with our DEG and GO analyses, and indicated that pathways involved in neuronal maturation were perturbed in FXS cells. These data also highlight defects in energy production and mitochondrial function in FXS.
Discussion
Human iPSCs are a valuable complement to animal models for understanding the unique characteristics of human diseases.39 However, differences in the individual genetic backgrounds or epigenetic variations of iPSC lines can confound interpretation of results, leading to mistaken attribution of experimental differences to the disease state. Ideally, to have confidence that observed differences are due to the disease rather than the irrelevant variance of cell lines, results should be obtained from multiple independent disease-specific and control cell lines. Toward this end, we optimized the timing and extent of morphogen perturbation to develop a reliable method for generation of multiple subtypes of GABAergic neurons from hPSCs. Our single-cell transcriptome analysis demonstrates high reproducibility over the culture time course among the 3 FXS lines and among the 3 non-disease control lines.
Delayed Maturation of FXS GABAergic Neurons
The 3-cell lines that lacked FMRP, 2 from FXS patients and a FMRP-knockout hESC line, all showed similar characteristics in vitro, indicating that the absence of the protein was responsible for the observed differences from control. At the earlier time point studied by gene expression profiling, when progenitors dominated the cultures, the lack of the protein had no obvious effect on profiles of the cells and the FXS lines were not discernably different from the control cells. As the cells differentiated, both the control and the FXS cells became electrically active as detected by MEA analysis. Patch-clamp recording of the cells showed that GABA was the neurotransmitter underlying the synaptic activity. At later time points, the activity of the FXS cells persisted, while activity of the control cells diminished, a phenomenon that is consistent with the developmental switch in GABA polarity that normally occurs during inhibitory GABAergic neurogenesis in vivo. The GABA polarity switch is a dramatic event during cortical neurogenesis, when GABA changes from being an excitatory neurotransmitter to an inhibitory signal. The polarity switch is believed to have profound effects during cortical development, affecting neuronal migration and dendritic development and establishing the excitatory/inhibitory balance that is critical to postnatal cortical function.40
Our single-cell transcriptome analysis supports the idea that FXS cells (the FXS iPSCs and FMRP knockout hESC line) experienced an intrinsic developmental delay in their differentiation. We profiled cells at 2 stages of neurogenesis in vitro, and found that while there was little difference between control and disease-specific cells at the earlier stage of neurogenesis, there were striking differences at the later stage. The most prominent difference, indicated by both factor loading analysis and differentially expressed genes, was the upregulation of genes related to neuroblast proliferation and newly born neurons in the FXS cultures. In agreement with this observation, we also found a downregulation of genes related to more mature neuronal processes such as action potential regulation and synaptic processes in FXS cells. These results suggest that based on transcriptome signature at the late stage of neurogenesis, the FXS GABAergic neurodevelopment was at a less mature stage. This could mean that FXS GABAergic progenitors may be stalled at a younger state and continue to generate immature inhibitory neurons for a longer period of time, leading to dysfunctional neural networks.
Immunocytochemical analysis provided further evidence for delayed maturity of the FXS cells and the involvement of the GABA polarity switch. We determined that as the neurons matured, differences in the expression of a key modulator that underlies the GABA functional polarity switch emerged. KCC2 (encoded by SLC12A5), a developmentally regulated neuron-specific K+/Cl− cotransporter, has been shown to mediate the inhibitory action of GABA. Loss of KCC2 in disease and injury is thought to contribute to excitotoxicity of neurons.41 We observed that while KCC2 increased with time in culture of both FXS and control neurons, there were fewer KCC2-positive cells in the FXS cultures.
Taken together, our results implicate a delay in the developmental switch from GABA’s excitatory to inhibitory function as a consistent intrinsic characteristic of cells lacking FMRP.
Mitochondrial Dysfunction
In addition to the developmental delay in the FXS cells, our study also found a downregulation of genes related to energy metabolism and mitochondria function in FXS GABAergic neurons. The cause and effect of the mitochondria dysfunction is not clear; it could be an underlying cause of the developmental delay or it could be the result of the more immature state of FXS neurons. There is evidence of a link between loss of FMRP and mitochondrial function. Obesity and type II diabetes are common comorbidities of FXS.42 Furthermore, a reduced NAD+/NADH ratio and deficient oxidative phosphorylation were found in a Drosophila Fmr1 knockout model.43 A recent study demonstrated aberrant cellular metabolism resulted from ATP synthase c-subunit leakage in both human and mouse FXS cells.44 Much like the delay in the GABA functional switch, our data suggest that there is a delay in the metabolic switch from glycolysis to oxidative phosphorylation that occurs during normal neurogenesis.45
Our study points to an aberrant neurogenic origin of the delayed GABA hyperpolarizing action in FXS hPSC-derived GABAergic neurons. Increased translation processes and decreased mitochondria function are associated with this change. Therefore, our study provides evidence for an alternative hypothesis of neurodevelopmental delay in FXS. In humans, neurogenesis continues after birth,46,47 so this opens the door for novel therapeutic strategies that could mitigate the symptoms of the disease if applied early enough in brain development.
Supplementary Material
Acknowledgments
We thank the members of the Loring lab at Scripps Research, especially Ha Tran, Candace Lynch, Aditi Shankar, Suzanne Peterson, Lucas James, and Andres Bratt-Leal, for technical support and intellectual feedback.
Contributor Information
Ai Zhang, Department of Molecular Medicine, Scripps Research, La Jolla, CA, USA; Skaggs Graduate School of Chemical and Biological Sciences, Scripps Research, La Jolla, CA, USA; Aspen Neuroscience, Inc. San Diego, CA, USA.
Irina Sokolova, Department of Immunology and Microbiology, Scripps Research, La Jolla, CA, USA.
Alain Domissy, Center for Computational Biology, Scripps Research, La Jolla, CA, USA.
Joshua Davis, Department of Molecular Medicine, Scripps Research, La Jolla, CA, USA.
Lee Rao, Department of Molecular Medicine, Scripps Research, La Jolla, CA, USA; Biological and Medical Informatics Graduate Program, University of California, San Francisco, CA, USA.
Kagistia Hana Utami, Department of Physiology, National University of Singapore, Singapore; Translational Laboratory in Genetic Medicine, Agency for Science, Technology and Research (A*STAR), Singapore.
Yanling Wang, Department of Molecular Medicine, Scripps Research, La Jolla, CA, USA; Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, IL, USA.
Randi J Hagerman, MIND Institute, University of California Davis, Sacramento, CA, USA.
Mahmoud A Pouladi, Department of Physiology, National University of Singapore, Singapore; Translational Laboratory in Genetic Medicine, Agency for Science, Technology and Research (A*STAR), Singapore; British Columbia Children’s Hospital Research Institute, Department of Medical Genetics, University of British Columbia, Vancouver, Canada.
Pietro Sanna, Department of Immunology and Microbiology, Scripps Research, La Jolla, CA, USA.
Michael J Boland, Institute for Genomic Medicine, Columbia University Irving Medical Center, New York, NY, USA; Department of Neurology, Columbia University, New York, NY, USA.
Jeanne F Loring, Department of Molecular Medicine, Scripps Research, La Jolla, CA, USA.
Funding
This work was supported by the National Institutes of Health (R33MH087925) and California Institute for Regenerative Medicine (TR3-05603, RT3-07655 DISC2-09073, DISC2P-11595, TR1-01250, RT1-01108, GC1R-06673-A). M.A.P. was supported by a BC Children’s Hospital Research Institute Investigator Grant IGAP Award, and a Scholar Award from the Michael Smith Foundation for Health Research. We thank the Scripps Research Skaggs Graduate School of Chemical and Biological Sciences for support of A.Z.
Conflict of Interest
A.Z. is currently an employee and stockholder in Aspen Neurosciences, Inc. and J.F.L. is founder and a stockholder in the company. R.H. received funding from Zynerba Pharmaceuticals to carry out a treatment study in fragile X syndrome.
The accession number for the single-cell RNA sequencing data reported in this paper is GEO: GSE198138.
References
- 1. Cornish K, Turk J, Hagerman R.. The fragile X continuum: new advances and perspectives. J Intellect Disabil Res. 2008;52(Pt 6):469-482. [DOI] [PubMed] [Google Scholar]
- 2. Lewis P, Abbeduto L, Murphy M, et al. Cognitive, language and social-cognitive skills of individuals with fragile X syndrome with and without autism. J Intellect Disabil Res. 2006;50(Pt 7):532-545. [DOI] [PubMed] [Google Scholar]
- 3. McDuffie A, Abbeduto L, Lewis P, et al. Autism spectrum disorder in children and adolescents with fragile X syndrome: within-syndrome differences and age-related changes. Am J Intellect Dev Disabil 2010;115(4):307-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pasciuto E, Bagni C.. SnapShot: FMRP interacting proteins. Cell 2014;159(1):218-2e1. [DOI] [PubMed] [Google Scholar]
- 5. Suhl JA, Chopra P, Anderson BR, Bassell GJ, Warren ST.. Analysis of FMRP mRNA target datasets reveals highly associated mRNAs mediated by G-quadruplex structures formed via clustered WGGA sequences. Hum Mol Genet. 2014;23(20):5479-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. La Fata G, Gartner A, Dominguez-Iturza N, et al. FMRP regulates multipolar to bipolar transition affecting neuronal migration and cortical circuitry. Nat Neurosci. 2014;17(12):1693-1700. [DOI] [PubMed] [Google Scholar]
- 7. Li Y, Stockton ME, Bhuiyan I, et al. MDM2 inhibition rescues neurogenic and cognitive deficits in a mouse model of fragile X syndrome. Sci Transl Med. 2016;8(336):336ra-33661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Richter JD, Zhao X.. The molecular biology of FMRP: new insights into fragile X syndrome. Nat Rev Neurosci. 2021;22(4):209-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jacquemont S, Pacini L, Jonch AE, et al. Protein synthesis levels are increased in a subset of individuals with fragile X syndrome. Hum Mol Genet. 2018;27(21):3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hou L, Antion MD, Hu D, et al. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron 2006;51(4):441-454. [DOI] [PubMed] [Google Scholar]
- 11. Napoli I, Mercaldo V, Boyl PP, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell 2008;134(6):1042-1054. [DOI] [PubMed] [Google Scholar]
- 12. Edbauer D, Neilson JR, Foster KA, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 2010;65(3):373-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Darnell JC, Van Driesche SJ, Zhang C, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 2011;146(2):247-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He Q, Nomura T, Xu J, Contractor A.. The developmental switch in GABA polarity is delayed in fragile X mice. J Neurosci. 2014;34(2):446-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bear MF, Huber KM, Warren ST.. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27(7):370-377. 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 16. Selby L, Zhang C, Sun QQ.. Major defects in neocortical GABAergic inhibitory circuits in mice lacking the fragile X mental retardation protein. Neurosci Lett. 2007;412(3):227-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gantois I, Vandesompele J, Speleman F, et al. Expression profiling suggests underexpression of the GABA(A) receptor subunit delta in the fragile X knockout mouse model. Neurobiol Dis. 2006;21(2):346-357. [DOI] [PubMed] [Google Scholar]
- 18. D’Hulst C, Heulens I, Brouwer JR, et al. Expression of the GABAergic system in animal models for fragile X syndrome and fragile X associated tremor/ataxia syndrome (FXTAS). Brain Res. 2009;1253:176-183. [DOI] [PubMed] [Google Scholar]
- 19. Moskalyuk A, Van De Vijver S, Verstraelen P, et al. Single-Cell and Neuronal Network Alterations in an In Vitro Model of Fragile X Syndrome. Cereb Cortex. 2020;30(1):31-46. [DOI] [PubMed] [Google Scholar]
- 20. Castagnola S, Caz.areth J, Lebrigand K, et al. Agonist-induced functional analysis and cell sorting associated with single-cell transcriptomics characterizes cell subtypes in normal and pathological brain. Genome Res. 2020;30(11):1633-1642. 10.1101/gr.262717.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harlow EG, Till SM, Russell TA, et al. Critical period plasticity is disrupted in the barrel cortex of FMR1 knockout mice. Neuron 2010;65(3):385-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. He Q, Arroyo ED, Smukowski SN, et al. Critical period inhibition of NKCC1 rectifies synapse plasticity in the somatosensory cortex and restores adult tactile response maps in fragile X mice. Mol Psychiatry. 2019;24(11):1732-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hansen DV, Lui JH, Flandin P, et al. Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat Neurosci. 2013;16(11):1576-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ma T, Wang C, Wang L, et al. Subcortical origins of human and monkey neocortical interneurons. Nat Neurosci. 2013;16(11):1588-1597. [DOI] [PubMed] [Google Scholar]
- 25. Utami KH, Skotte NH, Colaco AR, et al. Integrative Analysis Identifies Key Molecular Signatures Underlying Neurodevelopmental Deficits in Fragile X Syndrome. Biol Psychiatry. 2020;88(6):500-511. [DOI] [PubMed] [Google Scholar]
- 26. Boland MJ, Nazor KL, Tran HT, et al. Molecular analyses of neurogenic defects in a human pluripotent stem cell model of fragile X syndrome. Brain: a journal of neurology 2017;140(3):582-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gelfman S, Wang Q, Lu YF, et al. meaRtools: An R package for the analysis of neuronal networks recorded on microelectrode arrays. PLoS Comput Biol. 2018;14(10):e1006506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zheng GX, Terry JM, Belgrader P, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stoeckius M, Zheng S, Houck-Loomis B, et al. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol 2018; 19(1): 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mayer C, Hafemeister C, Bandler RC, et al. Developmental diversification of cortical inhibitory interneurons. Nature 2018;555(7697):457-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maroof AM, Keros S, Tyson JA, et al. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell 2013;12(5):559-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nicholas CR, Chen J, Tang Y, et al. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell 2013;12(5):573-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gunhaga L, Marklund M, Sjodal M, et al. Specification of dorsal telencephalic character by sequential Wnt and FGF signaling. Nat Neurosci. 2003;6(7):701-707. [DOI] [PubMed] [Google Scholar]
- 34. Paek H, Gutin G, Hebert JM.. FGF signaling is strictly required to maintain early telencephalic precursor cell survival. Development. 2009;136(14):2457-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Welch JD, Kozareva V, Ferreira A, et al. Single-Cell Multi-omic Integration Compares and Contrasts Features of Brain Cell Identity. Cell 2019;177(7):1873-87 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Finak G, McDavid A, Yajima M, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Velmeshev D, Schirmer L, Jung D, et al. Single-cell genomics identifies cell type-specific molecular changes in autism. Science 2019;364(6441):685-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pape M, Doxakis E, Reiff T, et al. A function for the calponin family member NP25 in neurite outgrowth. Dev Biol. 2008;321(2):434-443. [DOI] [PubMed] [Google Scholar]
- 39. Sharma A, Sances S, Workman MJ, Svendsen CN.. Multi-lineage Human iPSC-Derived Platforms for Disease Modeling and Drug Discovery. Cell Stem Cell 2020;26(3):309-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ganguly K, Schinder AF, Wong ST, Poo M.. GABA itself promotes the developmental switch of neuronal GABAergic responses from excitation to inhibition. Cell 2001;105(4):521-532. [DOI] [PubMed] [Google Scholar]
- 41. Tang X, Kim J, Zhou L, et al. KCC2 rescues functional deficits in human neurons derived from patients with Rett syndrome. Proc Natl Acad Sci USA. 2016;113(3):751-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dy ABC, Tassone F, Eldeeb M, et al. Metformin as targeted treatment in fragile X syndrome. Clin Genet. 2018;93(2):216-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weisz ED, Towheed A, Monyak RE, et al. Loss of Drosophila FMRP leads to alterations in energy metabolism and mitochondrial function. Hum Mol Genet. 2018;27(1):95-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Licznerski P, Park HA, Rolyan H, et al. ATP Synthase c-Subunit Leak Causes Aberrant Cellular Metabolism in Fragile X Syndrome. Cell 2020;182(5):1170-85 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shin J, Berg DA, Zhu Y, et al. Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell 2015;17(3):360-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Malik S, Vinukonda G, Vose LR, et al. Neurogenesis continues in the third trimester of pregnancy and is suppressed by premature birth. J Neurosci. 2013;33(2):411-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schork AJ, Won H, Appadurai V, et al. A genome-wide association study of shared risk across psychiatric disorders implicates gene regulation during fetal neurodevelopment. Nat Neurosci. 2019;22(3):353-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.