Abstract
Induced pluripotent stem cell (iPSC)-derived kidney organoids are a potential tool for the regeneration of kidney tissue. They represent an early stage of nephrogenesis and have been shown to successfsully vascularize and mature further in vivo. However, there are concerns regarding the long-term safety and stability of iPSC derivatives. Specifically, the potential for tumorigenesis may impede the road to clinical application. To study safety and stability of kidney organoids, we analyzed their potential for malignant transformation in a teratoma assay and following long-term subcutaneous implantation in an immune-deficient mouse model. We did not detect fully functional residual iPSCs in the kidney organoids as analyzed by gene expression analysis, single-cell sequencing and immunohistochemistry. Accordingly, kidney organoids failed to form teratoma. Upon long-term subcutaneous implantation of whole organoids in immunodeficient IL2Ry−/−RAG2−/− mice, we observed tumor formation in 5 out of 103 implanted kidney organoids. These tumors were composed of WT1+CD56+ immature blastemal cells and showed histological resemblance with Wilms tumor. No genetic changes were identified that contributed to the occurrence of tumorigenic cells within the kidney organoids. However, assessment of epigenetic changes revealed a unique cluster of differentially methylated genes that were also present in undifferentiated iPSCs. We discovered that kidney organoids have the capacity to form tumors upon long-term implantation. The presence of epigenetic modifications combined with the lack of environmental cues may have caused an arrest in terminal differentiation. Our results indicate that the safe implementation of kidney organoids should exclude the presence of pro-tumorigenic methylation in kidney organoids.
Keywords: induced pluripotent stem cell, kidney organoids, cancer, kidney development, transplantation
Graphical Abstract
Upon long-term subcutaneous implantation of human kidney organoids, we observed tumor formation in 5 out of 103 implanted organoids. These tumors showed histological resemblance with Wilms tumor. Epigenetic analysis revealed a unique cluster of differentially methylated genes that was also present in undifferentiated iPSCs. Our results indicate that kidney organoids have the capacity to form tumors upon long-term implantation.
Significance Statement.
Human induced pluripotent stem cell (iPSC)-derived kidney organoids have been the focus of attention due to their expansive potential for disease modeling, drug screening, and regenerative medicine. Their clinical applicability is accompanied by the question of whether safety issues may arise in the long term. The authors demonstrate that kidney organoids failed to develop teratomas, in contrast to iPSCs. However, they discovered that kidney organoids had the ability to form tumors. These tumors contained immature blastemal cells, with similarities with the pediatric cancer Wilms tumor. Epigenetic analyses revealed a unique cluster of differentially methylated genes in the tumors that were also present in iPSCs. These findings support a stringent evaluation of pro-tumorigenic epigenetic changes in kidney organoids prior to clinical implementation.
Introduction
There is an urgent need for novel therapies for patients with kidney failure.1 Current treatment options for kidney failure are dialysis or transplantation. However, dialysis is associated with high mortality and high morbidity, while transplantation is limited by the availability of donor organs.1 Regeneration of the kidney in patients with kidney failure could potentially replace the need for dialysis and transplantation. Human kidney organoids derived from induced pluripotent stem cells (iPSCs) form a potential source for the regeneration of injured kidney tissue.2 Over the past years, various methods to generate complex kidney structures from human iPSCs have been reported.3,4,5,6,7 These methods attempt to recreate embryonic kidney development in a sequential manner by stimulating the Wnt-, FGF-, and/or BMP pathways to induce kidney differentiation.8 Kidney organoids from established protocols have been shown to resemble embryonic first- or second-trimester kidneys.9,10 Upon implantation in animal models, they have been shown to vascularize and mature successfully, thereby establishing their potential as a regenerative therapy.11,12,13,14,15 However, it remains unclear whether a fully adult phenotype can be achieved in this setting.15,16
The use of progenitor cells for regenerative medicine is accompanied by safety concerns with respect to tumorigenesis.17 Like embryonic stem cells, iPSCs possess intrinsic qualities of self-renewal and pluripotency which is achieved by the induction of important pluripotency factors, such as POU5F1, MYC, SOX2, and ZFP42.18 Upon implantation in an undifferentiated state, iPSCs form teratomas.19 It is therefore imperative that contamination of kidney organoids with residual pluripotent cells is avoided. In addition, iPSCs and iPSC-derivatives are vulnerable to the accumulation of (epi)genetic changes, which may also account for tumorigenic risk.20
Provided that residual iPSCs are successfully eliminated and the risk of teratoma formation is therefore minimized, the possibility remains that partly differentiated progenitor cells undergo tumorigenic transformation. Kidney organoids represent an early stage of nephrogenesis and contain renal progenitors that are expected to complete mesenchymal-epithelial transition (MET) upon vascularization. However, the absence of appropriate environmental cues or the presence of the aforementioned genomic imbalances can lead to an arrest in differentiation. Subsequently, kidney tumors may arise due to the prolonged maintenance and uncontrolled proliferation of renal progenitors.
Several studies concerning the safety of other pluripotent stem cell derivatives, such as neural cells, do report the occurrence of teratoma or tumor formation.21,22,23,24,25,26,27,28 Even though the characteristics of tumors varied between studies due to differences in protocols, cell lines and follow-up periods, tumors were generally described as an expansion of undifferentiated neural precursors and thought to be capable of damaging and compressing host tissue.24,26 In some cases, tumors were observed even after a pre-selection for a purified precursor population.27,28 These results underscore the importance of stringent assessment into the safety of iPSC-derivatives. Therefore, the aim of this study is to examine the tumorigenic risk of human iPSC-derived kidney organoids. To do so, we examined in detail residual iPSCs within the organoid structures, studied the occurrence of genetic and epigenetic deviations during iPSC propagation and organoid formation, analyzed potential teratoma formation and studied safety and stability of kidney organoids following long-term subcutaneous implantation in an immune-deficient mouse model. This is the first study to show the occurrence of tumor formation in kidney organoids as a result of epigenetic modifications.
Results
Kidney Organoids Do Not Contain Residual iPSCs
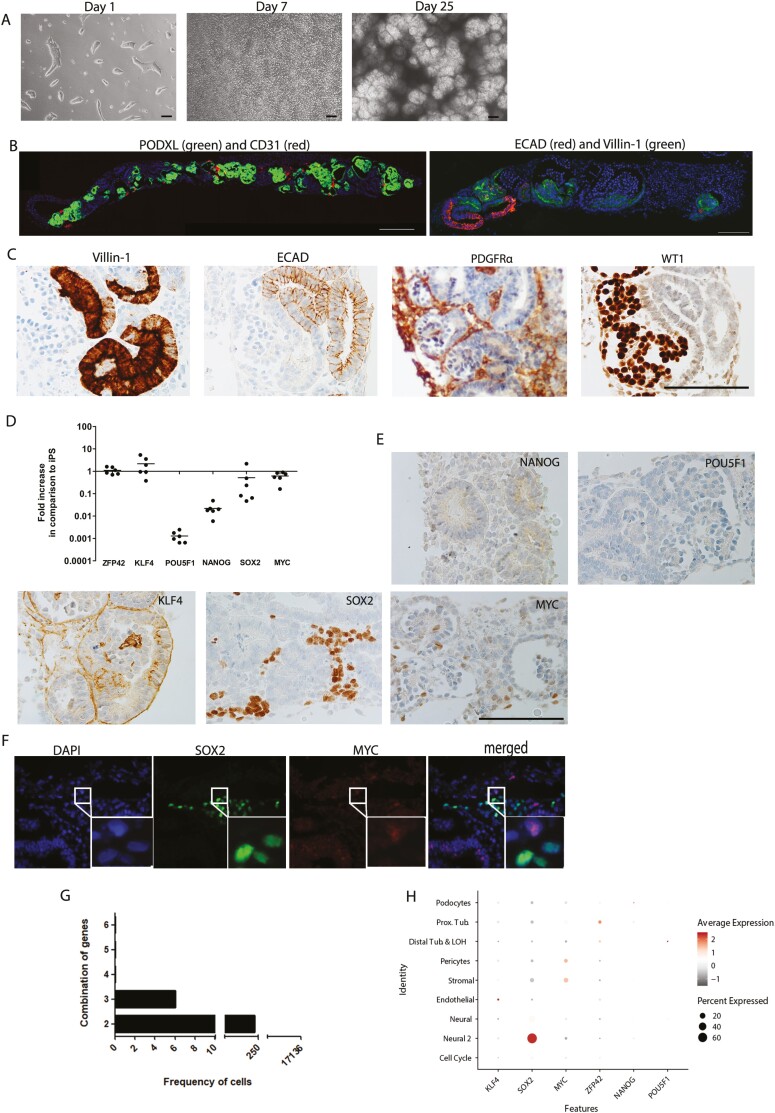
On day 25 of culture of iPSCs under kidney inducing conditions, complex structures had developed containing glomerular cells, tubular and endothelial cells (Fig. 1A and B). Immunohistochemistry demonstrated the presence of Villin-1+ and ECAD+ proximal and distal tubular cells, respectively (Fig. 1C). A PDGFRα+ stromal population and WT1+ glomerular cells could also be observed (Fig. 1B). To determine the presence of residual iPSCs in day 25 organoids, we first analyzed the dynamics of gene expression of pluripotency markers during differentiation. Bulk mRNA analysis demonstrated that KLF4 and ZFP42 remained stable up to the end of the culture period in comparison to undifferentiated iPSCs while POU5F1, NANOG, SOX2 and MYC mRNA expression was downregulated (Fig. 1D). Immunohistochemistry indicated that protein expression of NANOG and POU5F1 was absent in the organoids (Fig. 1E). KLF4 was present in a membranous pattern without nuclear positivity, while SOX2 and MYC displayed nuclear distribution (Fig. 1E) without co-localization (Fig. 1F), suggesting MYC- and SOX2-expressing cells represent distinct populations of non-pluripotent cells. To further explore the presence of residual iPSCs in kidney organoids, co-expression of multiple pluripotency markers in individual cells was examined by scRNAseq data of a day 25 organoid. No cells could be detected that co-expressed all the pluripotent markers simultaneously (Fig. 1G). Combinations of any 3 pluripotent genes could be detected in 6 out of 17 136 cells, while combined expression of 2 pluripotent genes could be found in 236 cells or 1.38% of the total amount of cells. SOX2 was the most frequently expressed pluripotency gene (Supplementary Table S2) and could specifically be found in an off-target neural cluster (Fig. 1H). In conclusion, we did not detect residual iPSCs in the organoids. However, the expression of some pluripotent markers suggests the presence of less differentiated or off-target cell types within the kidney organoids. To compare our findings with other published datasets, we used the DevKidCC tool and examined the expression of 6 pluripotency markers (KLF4, SOX2, MYC, REX1, NANOG, and OCT4) in kidney organoid cell clusters of 7 publications using similar kidney organoid differentiation protocols. Similar to our findings, expression of some pluripotency markers was detected in particular cell populations in all studies, but no clusters of cells expressing all pluripotency markers were found, confirming our finding that kidney organoids do not contain residual iPSC (Supplementary Fig. S1).
Figure 1.
Kidney organoids contain no residual iPSC. (A) Representative bright-field images of kidney organoid culture at different time points of differentiation. Scale bars, 100 µm. (B) Double immunofluorescence staining of whole organoid sections at the endpoint of differentiation (day 25). Left: PODXL (green) and CD31 (red). Scale bar, 100 µm. Right: ECAD (red) and Villin-1 (green). Scale bar, 200 µm. (C) Immunohistochemical analysis for markers of proximal tubular cells (Villin-1), distal tubular cells (ECAD), renal interstitium (PDGFR-α), and glomerular cells (WT1). Scale bar, 100 µm. (D) Gene expression analysis of pluripotency markers. Plots depict gene expression changes relative to undifferentiated iPSC. Data are represented as mean ± SEM (n = 6 from 2 experiments). (E) Immunohistochemical analysis for pluripotency markers NANOG, POU5F1, KLF4, SOX2, and MYC. Scale bar, 100 µm. (F) Double immunofluorescence staining for SOX2 (green) and MYC (red) showing absence of co-localization. (G) Graph depicting the frequency of cells in kidney organoid containing multiple expression parameters. (H) Dot plot visualization to depict the expression of the pluripotency marker per cluster.
Kidney Organoids Contain Cells With Tumorigenic Potential
We next evaluated the tumorigenic risk of iPSCs and iPSC-derived kidney organoids by performing a teratoma assay in NOD-SCID mice (Fig. 2A). Either iPSCs- or organoid aggregates were injected in 4 locations in the flanks of the mice. After 5 weeks, iPSC aggregates exhibited significant growth in 7 out of 8 locations, with tissue weights varying from 0.3 to 1.08 g (Fig. 2B). The tissues were irregular in shape and contained a necrotic center filled with clear fluid. We characterized the tissues by H&E and observed the presence of 3 germ layers: endoderm, ectoderm, and mesoderm, evidencing the teratoma nature of the tissues (Fig. 2C). A cluster of SOX2+ NANOG+ POU5F1+ cells could be visualized indicating the presence of pluripotent cells (Fig. 2D). Moreover, there were cells positive for either SOX2 or KLF4 immunohistochemistry (Fig. 2E). MYC and CD30, a marker for malignant embryonic stem cell-derived tumors, were negative (Supplementary Figure S2). At the same time point, we also analyzed tissues harvested from mice injected with organoid aggregates. No tumor or teratoma formation could be detected. At 10 weeks we did observe a tumor of 0.6 cm in 1 out of 8 locations in the mice injected with organoid aggregates. Visual inspection of other organs showed no signs of metastatic growth and the harvested tissue was irregular in shape yet did not contain a necrotic center. H&E showed only the presence of derivatives from the mesodermal lineage, namely cartilage, mesenchymal, and kidney-specific structures thereby suggesting a lack of teratoma formation (Fig. 3A). The structures were surrounded by a large population of small round cells that seemed to contribute to the growth of the tumor (Fig.3B). The tissue contained cytoplasmic and nuclear protein expression of POU5F1 and KLF4, respectively. Moreover, there was no co-localization of POU5F1 and KLF4. Importantly, there were no cells that expressed NANOG, SOX2, and MYC (Fig. 3C), indicating that there were no cells that expressed multiple pluripotency markers simultaneously. This is in contrast with the cells in iPSC-derived teratomas, that co-expressed multiple pluripotency markers (SOX2+ NANOG+ POU5F1). The lack of co-localization of pluripotency markers suggests that there are no remaining pluripotent cells in the kidney organoid-derived tumors.
Figure 2.
Teratoma assay confirms the ability of iPSC to form teratomas. (A) A schematic overview of the teratoma assay. (B) Weights of iPSC- and organoid-derived teratoma. (C) Histochemical staining (H&E) of iPSC-derived teratoma containing 3 germ layers; endoderm (cavity lining epithelium), ectoderm (pigment epithelium containing melanin granules) and mesoderm (adipose/ stromal tissue). Scale bar, 100 µm. (D) Immunohistochemical analysis for markers of pluripotency, respectively NANOG, SOX2, and POU5F1 (black arrows indicate positive cells). Scale bar, 100 µm. (E) Immunohistochemical analysis for SOX2 and KLF4, respectively in iPSC-derived teratoma. Scale bar, 100 µm.
Figure 3.
Teratoma assay demonstrates the absence of teratoma formation in kidney organoids, in contrast to iPSCs. (A) Histochemical staining (H&E) of organoid-derived tissue containing mesodermal tissues such as cartilage (black arrow), mesenchymal cells (asterisk), and renal cells (asterisk indicates lumen of tubuli), respectively. Scale bars, 100 µm. (B) Histochemical staining (H&E) of the population of small round cells in organoid-derived tumor contributing to the growth of the tumor. Scale bars, 100 µm. (C) Immunohistochemical analysis for markers of pluripotency, respectively KLF4, POU5F1, NANOG, SOX2, and MYC. Scale bars, 100 µm.
A Proportion of In Vivo Implanted Kidney Organoids Displays Tumor Formation
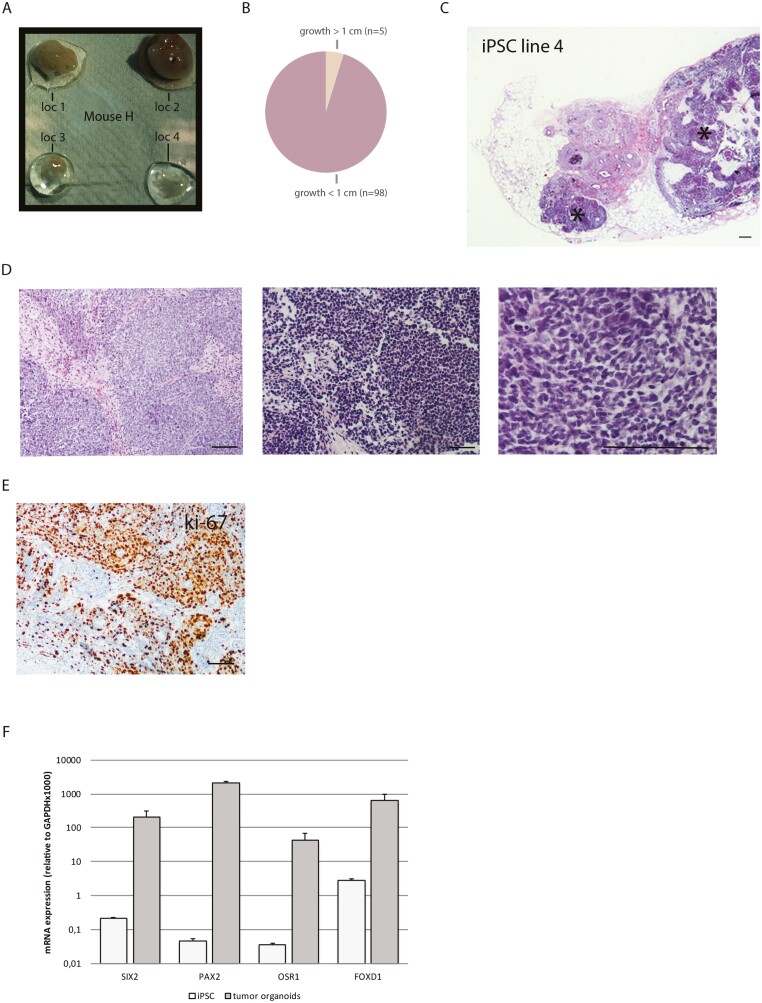
Clinical application of kidney organoids most likely would involve implantation of whole organoids instead of aggregates or single cells. Therefore, we subcutaneously implanted whole organoids into an immune-deficient BALB/c IL2Ry−/−RAG2−/− mouse model to study the tumorigenic potential of kidney organoids after vascularization and further maturation (Fig. 4A). Preimplantation organoids displayed a normal phenotype with multiple nephron structures (Fig. 4B). After a follow-up of 2 months, the xenografts were collected and analyzed by H&E and immunohistochemistry. We confirmed the presence of nephron structures by the presence of Villin-1+ proximal tubular cells and vascularized glomeruli as observed by the presence of erythrocytes in the glomerular tufts (Fig. 4C). Also, the brush border localization of NHE3 and NKCC2 in the tubular structures supported sustained kidney differentiation (Fig. 4D). Several nephron structures could be visualized in transition to a fibrotic state and this observation is in agreement with recent data reporting stromal overgrowth (Fig. 4E).16,29 Moreover, the implanted organoids contained cartilage nodules of various sizes (Fig. 4F). One hundred and three organoids derived from 4 iPSC lines were implanted in 6 independent experiments with consistent observations of nephron structures and other tissues of the mesodermal lineage such as cartilage and mesenchymal tissue. On 5 occasions we witnessed the presence of tissue growths reaching a size of 1 cm and unique histological observations in organoid implantation sites belonging to 1 of the 4 iPSC lines (Supplementary Table S3, Fig. 5A and B).
Figure 4.
A subcutaneous implantation model shows survival and engraftment of kidney organoids after 2 months. (A) A schematic overview of subcutaneous implantations of whole kidney organoids. (B) H&E staining of pre-implantation organoid (iPSC line 4) at 2 different magnifications. Scale bars indicate 100 µm. (C) Immunohistochemical staining for proximal tubular structures (Villin-1) and erythrocytes present in glomerular structures of organoids 2 months after implantation. Scale bar, 100 µm. (D) Immunohistochemistry for markers of the proximal tubuli (NHE3) and the Loop of Henle (NKCC2) in the implanted kidney organoid (as indicated by black arrows). Scale bar, 100 µm. (E) Histochemical staining (H&E) of consecutive slides depicting the fibrotic transition of glomerular structures. Scale bar, 100 µm. (F) Histochemical staining (H&E) of cartilage nodules within implanted kidney organoid. Scale bar, 100 µm.
Figure 5.
Kidney organoids have the capacity to form tumors. (A) Representative photo of implanted kidney organoids found in mouse H. (B) Pie chart demonstrating the frequency of >1 cm growths of the total number of implanted organoids. (C) Histochemical staining (H&E) of tumor from iPSC line 4 containing small round cells (black asterisk). Scale bar represents 100 µm. (D) Images of areas with small round cells in 2 tumors (left and middle, H&E staining). Zoomed in image of the first tumor. Scale bars represent 100 µm. (E) Immunohistochemistry for Ki-67 to demonstrate high proliferation of cells within the tumor. Scale bar represents 100 µm. (F) mRNA expression of the nephron progenitor markers SIX2, PAX2, OSR1, and FOXD1 in tumor organoids compared to undifferentiated iPSC. Data are expressed as a ratio with GAPDH × 1000 with SE. n = 2.
Macroscopically, these tissues were devoid of any necrosis or any fluid matter. Organoids from iPSC line 4 contained large clusters of small round cells adjacent to the nephron structures as observed by H&E (Fig. 5C and D). These clusters were dispersed throughout the tissue and displayed mitotic activity as observed by the high proportion of ki-67+ cells within the clusters (Fig. 5E). Despite the tissue malformation, the organoids demonstrated highly elevated mRNA expression of the nephron progenitor markers SIX2, PAX2, OSR1 and FOXD1 compared to iPSC (Fig. 5F).
Kidney Organoid-Derived Tumors Resemble Wilms Tumor
Characterization of the malformed tissues derived from kidney organoids in the teratoma assay and the organoid implantation studies was performed using immunohistochemistry. The tumor cells were surrounded by reactive stroma, as indicated by the presence of αSMA (Fig. 6A). Staining for the Wilms tumor markers WT1, CD56, and CD5730 was also performed due to the histological resemblance of the organoid-derived tumors to Wilms tumor (Fig. 6B-D). While CD57 protein expression could not be detected, WT1 and CD56 showed specific nuclear and cytoplasmic reactivity in all the cells within the clusters, respectively (Fig. 6B and C). Similar observations were made in the organoid-derived tumor observed in the teratoma assay (Supplementary Fig. S3). CD56 was found to be absent in the tubule structures (Supplementary Fig. S4). Chromogranin A and synaptophysin were negative in the tissues excluding a neuro-endocrine origin despite CD56 positivity (data not shown). There was no evidence of NANOG+ or MYC+ pluripotent cells in the tumor tissue (Supplementary Fig. S5).
Figure 6.
Kidney organoid-derived tumors resemble Wilms tumors. (A-D) Immunohistochemical staining in 2 organoid-derived tumors for αSMA, WT1, CD56, and CD57, respectively. Left panel shows staining of one tumor, right panel of another tumor. Scale bars, 100 µm.
Tumorigenic Kidney Organoids Do Not Acquire Chromosomal Abnormalities
Subsequently, we analyzed whether nephron progenitors had acquired chromosomal abnormalities that initiated oncogenic transformation and subsequent expansion. Therefore, we performed a Global Screening Array on 2 explanted tumor organoids and their parent iPSCs (line 4) to check for copy number variants and allelic changes in the cells that may have occurred during the differentiation process or during the implantation period. The explanted derivatives were identical to their parent iPSC line as shown by a comparison of the allelic calls (Supplementary Table S4). Furthermore, no major copy number variations or allelic changes could be found (Fig. 7A), reducing the likelihood that genetic changes were at the basis of tumorigenic tissue formation.
Figure 7.
Genetic changes are excluded as the cause of kidney organoid-derived tumor formation. (A). Global screening array (GSA) shows no major copy number variations or allelic changes in iPSCs and implanted organoids of line 4 (tumor organoid 1 and tumor organoid 2). (B) Heatmap representation of unsupervised hierarchical clustering based on differentially methylated regions (DMRs) between control organoids of line 4, tumor organoids of line 4, 3 unrelated iPSC (iPSC 1-3), tumorigenic iPSC line 4 (iPSC 4), and non-tumorigenic iPSC line 3 (iPSC 5) (fold change ≥ 2). A region that is hypomethylated in tumor organoids compared to control organoids that resemble the methylation status of iPSC is marked by an arrow. (C) GO enrichment analysis based on the selection of overlapping genes associated with the DMRs between control organoids and tumor organoids. Dot plot visualization shows the top 10 most significant processes. Dot size indicates the number of significant genes in the given pathway and color represents significance. (D) Table view of top 10 most significant processes identified including enrichment, P value, and affected genes.
Tumorigenic Kidney Organoids Demonstrate Epigenetic Changes
Next, we examined whether epigenetic alterations could have induced a tumorigenic phenotype in the organoids. We studied genome-wide DNA methylation using MeD-seq.31 MeD-seq is a novel and efficient technique that utilizes the restriction enzyme LPnPI, which specifically cuts 16 bp downstream from methylated and/or hydroxymethylated CpGs. By doing so the technique focuses on the sequencing resources to methylated regions only allowing identification of differential methylated regions (DMRs) at single-nucleotide resolution.31 The samples were divided into 3 groups and consisted of 3 implanted control organoids (iPSC line 4), 3 tumor organoids (iPSC line 4), 3 unrelated control iPSC lines, tumor-forming iPSC line 4, and non-tumor forming iPSC line 3. Hierarchical clustering of the tissues was performed based on the differentially methylated regions found genome-wide both in intra- and intergenic regions (Fig. 7B). The control and tumorigenic organoids clearly clustered away from the iPSC, as expected. Interestingly, the heat map revealed a distinct cluster of significantly hypomethylated DMRs that was highly similar in both tumor organoids and iPSCs. In order to determine which biological pathways this cluster represented; we performed a gene ontology (GO) analysis on the overlapping genes associated with the DMRs (Fig. 7C and D). Methylation patterns of DMRs and their chromosomal locations were depicted using the Integrative Genomics Viewer (IGV) (Supplementary Fig. S6). The top pathway involved the GO term nephrin family interactions and overlap between the genes in this pathway (ACTN4, WASL, CASK) and the DMRs were present in the gene bodies.
Experimental Procedures
iPSC Culture
Four human iPSC lines from 3 donors (2 different clones were derived from one donor) from healthy primary skin fibroblasts (Supplementary Table S1) were reprogrammed as described earlier, using a single, multicistronic lentiviral vector encoding POU5F1, SOX2, KLF4, and MYC.32 Written informed consent was obtained in accordance with the Medical Ethics Committee of the Erasmus University Medical Center (MEC-2017-248).33 Frozen cells were thawed and cultured in E8 medium (Thermo Fisher Scientific, Bleiswijk, The Netherlands) on Geltrex (Gibco, Waltham, USA). TrypLE Select (Thermo Fisher Scientific) was used to make iPSC single suspensions before initiation of differentiation.
Kidney Organoid Differentiation
The differentiation protocol was adapted from Takasato et al34 and described earlier.35 In brief, human iPSCs at a density of 20 000-25 000 per cm2 were cultured in a monolayer in STEMdiff APEL2 medium (APEL2) (STEMCELL Technologies, Vancouver, Canada) supplemented with 8 µM CHIR99021 (R&D Systems, Minneapolis, USA), 5% protein-free hybridoma medium II (PFHMII, Thermo Fisher Scientific) and 1% antibiotic-antimycotic (Thermo Fisher Scientific) for 3-4 days. Recombinant human FGF9 (200 ng/mL; R&D Systems) and heparin (1 μg/mL; Sigma-Aldrich) were added after 3-4 days. After 7 days of monolayer culture, the cells were pulsed for 1 h with 5 μM CHIR99021, pelleted, and transferred onto a Transwell membrane (0.4 μm pore polyester membrane, Corning, New York, USA). For 5 days, the organoids were stimulated with 200 ng/mL recombinant human FGF9 and 1 μg/mL heparin after which growth factors were removed.
Quantitative PCR
The High Pure RNA Isolation Kit (Roche Life Sciences, Indianapolis, USA) was used to extract mRNA from iPSCs and organoids. cDNA was produced from 500 ng mRNA with random primers (Promega Benelux B.V.). Gene expression was quantified using TaqMan Gene Expression Assays-on demand (all Applied Biosystems). Expression levels were normalized to GAPDH as a reference gene.
Genomic DNA Isolation
Explanted control and tumor organoids from iPSC line 4 were minced into small pieces prior to isolation. DNA was isolated using the QIAamp DNA Micro kit (Qiagen, Venlo, the Netherlands), according to the manufacturer’s protocol. Concentration and degradation of the isolated DNA were assessed with the Agilent 2200 TapeStation (Agilent Technologies, Amstelveen, the Netherlands).
Immunohistochemistry and Immunofluorescence
Formalin-fixed paraffin-embedded organoids were cut into four-micron sections. They were stained with H&E (HE600, Ventana Medical Systems, Tucson, AZ, USA). An automated staining system (Ventana Benchmark ULTRA) was used for immunohistochemistry using an ultraView or optiView universal DAB detection Kit. The tissue samples were incubated with WT1 (Clone 6F-H2) (Cell Marque Rocklin, CA, USA), ECAD (Clone 36) (Ventana Medical Systems), Villin-1 (Clone EPR3491) (Abcam, Cambridge, UK), PODXL (Clone EPR9518) (Abcam), CD31 (Clone JC70) (Cell Marque), NKCC2 (L224) (Stressmarq, Victoria, BC, Canada), NHE3 (Stressmarq), CD56 (Clone MRQ-42) (Ventana Medical Systems), CD57 (Clone NK-1) (Ventana Medical Systems), ki-67 (Clone 30-9) (Ventana Medical Systems), NANOG (R&D Systems), KLF4 (R&D Systems), SOX2 (Clone SP76) (Ventana Medical Systems), MYC (Clone Y69) (Ventana Medical Systems), POU5F1 (Clone MRQ-10) (Ventana Medical Systems), CD30 (Clone Ber-H2) (Ventana Medical Systems, nephrin (R&D Systems), PDGFR-α (R&D Systems). Double staining of SOX2 and MYC was performed by incubating the samples with MYC followed by detection with Red610 (#760-245, Ventana) and SOX2 for 30 minutes at 37 °C followed by detection with FAM (#760-243, Ventana). Adult kidney tissue and tonsil were used as positive controls. Negative controls were obtained by the omitting primary antibody. Double staining of PODXL and CD31 was performed by incubating sections with CD31 followed by detection with Red610 (#760-245, Ventana) and PODXL for 30 minutes at 37 °C followed by detection with FAM (#760-243, Ventana).
Single-cell RNA Sequencing (scRNA-seq)
Single-cell suspensions of an in vitro cultured organoid from iPSC line 4 was generated as previously published.35 Single-cell libraries were prepared using the Chromium Single Cell 3’ Reagent Kit v3 (10× Genomics, Pleasanton, CA, USA). Next-generation sequencing (28-8-0-91 cycles) was carried out on an Illumina NovaSeq6000 platform (Illumina, San Diego, USA). Raw data were processed into FASTQ files. Raw sequences were inspected for their quality using FastQC (version v0.11.5). Reads containing sequence information were mapped to the GRCh38 human reference genome. Generation of BAM files and filtered gene-barcode matrices was accomplished using the Cell Ranger Software (version 3.0.2) (10× Genomics). The single cell transcriptomics datasets have been deposited to the Sequence Read Archive (SRA) data repository under accession number GSE165266.
Analysis
Analysis was done using Seurat (v3.2.0)36 and as previously published.35 Marker gene lists were generated to find differentially expressed genes between clusters, with a log fold change above 0.25. Annotation of clusters to cell types was done by manual inspection of the features defining each cluster and comparison to previous studies.5,9,10,37 Visualizations using the DotPlot function graphically represent the percentage of expression per cluster by dot diameter and average nonzero expression in log2 scale by dot color; data was not shown if the percentage of expressing cells (pct. exp.) <0.01. To identify cells matching multiple expression parameters, queries were done using the WhichCells function. Expression level cutoffs were set to >0.1.
Teratoma Assay
Kidney organoids were incubated with Collagenase Type IV (Thermofisher) at 37 °C for 10 minutes to obtain aggregates of cells. After washing, the cells were re-suspended in cold advanced DMEM/F12 medium and then mixed directly with Matrigel (Corning) in the ratio of 1:1 with a total volume of 100-200 μL iPSCs were dissociated using 0.5 mM EDTA dissociation buffer (Gibco) into aggregates. Female nude mice (8-12 weeks old) were subcutaneously injected in the flanks in 4 pockets with either undifferentiated iPSC (2 lines of 2 different donors; 4 injection sites, so 4 replicates per donor) or organoid aggregates at a concentration of 0.5 × 106 to 0.75 × 106 cells. Teratoma formation was monitored weekly and mice were sacrificed at week 5 or 10 or in case the tumor size reached 1 cm. All animal experiments were approved by the Dutch Central Committee on the Ethics of Animal Experiments (AVD101002017867).
Global Screening Array
gDNA from 2 tumor organoids and their parent iPSC (line 4) was used for the Global screening array (GSAv3) (Illumina, San Diego, USA). The global screening array was used according to standard protocols, followed by an analysis in GenomeStudio software (Illumina). The GenomeStudio Final reports were used to analyze and visualize the results in Nexus Discovery (BioDiscovery, California, USA). A report resolution of 50 kb was used to analyze the data for chromosomal aberrations. To compare the different samples at the SNP level, the array final reports were selected as input for an R script. Using statistics, we determined whether the allelic calls of the iPSCs and implanted tissues matched, mismatched, or failed.
Methylated DNA Sequencing (MeD-seq)
Sample Preparations
MeD-seq analyses were essentially carried out as previously described.31 In brief, 11 DNA samples, including 3 control organoids, 3 tumor organoids, and 5 iPSC lines (including the tumor-forming iPSC line), were digested by LpnPI (New England Biolabs, Ipswich, MA, USA), resulting in snippets of 32bp around a fully methylated recognition site that contains a CpG. These short DNA fragments were further processed using the ThruPlex DNA-seq 96D kit (Rubicon Genomics Ann Arbor, MI, USA). Stem-loop adapters were blunt-end ligated to repaired input DNA and amplified to include dual indexed barcodes using a high-fidelity polymerase to generate an indexed Illumina NGS library. The amplified end product was purified on a Pippin HT system with 3% agarose gel cassettes (Sage Science, Beverly, MA, USA). Multiplexed samples were sequenced on Illumina HiSeq2500 systems for single read of 50bp according to manufacturer’s instructions. Dual indexed samples were de-multiplexed using bcl2fastq software (Illumina, San Diego, CA, USA).
MeD-seq Data Analysis
Data processing was carried out using specifically created scripts in Python. Raw fastq files were subjected to Illumina adaptor trimming and reads were filtered based on LpnPI restriction site occurrence between 13 and 17 bp from either 5ʹ or 3ʹ end of the read. Reads that passed the filter were mapped to hg38 using bowtie2. Genome-wide individual LpnPI site scores were used to generate read count scores for the following annotated regions: transcription start sites (TSS, 1 kb before and 1 kb after), CpG-islands and gene bodies (1kb after TSS till TES). Gene and CpG-island annotations were downloaded from ENSEMBL (www.ensembl.org). Detection of differentially methylated regions (DMRs) was performed between 3 datasets containing the regions of interest (TSS, gene body or CpG-islands) using the χ2 test on read counts. Significance at a P-value of <.05 was called by either Bonferroni or FDR using the Benjamini-Hochberg procedure.
In addition, a genome-wide sliding window was used to detect sequentially differentially methylated LpnPI sites. Statistical significance was called between LpnPI sites in predetermined groups using the χ2 test. Neighboring significantly called LpnPI sites were binned and reported. Annotation of the overlap of genome-wide detected DMRs was reported for TSS, CpG-islands, and gene body regions. DMR thresholds were based on LpnPI site count, DMR sizes (in bp), and fold changes of read counts as mentioned in the figure legends before performing hierarchical clustering. The differentially methylated datasets generated and analyzed during the current study have been deposited to the Sequence Read Archive (SRA) data repository under accession number GSE165266.
Gene Ontology Analysis
Bioinformatics analyses were performed in R (version 4.0.2). Gene classification was done by mapping gene IDs to the human terms using the webtool Gene Ontology. Unannotated genes and genes with non-root annotations were excluded from the analysis. Pathway identification and enrichment analysis were performed with pathfindR (version 1.5.1)38 using the Reactome gene set (version 70) and the Biogrid protein-protein interaction database (version 1.1.1). Briefly, genes with their associated P-values and log fold-change values derived from the methylation sequencing analysis were mapped onto the Reactome gene set. A search of active pathways was performed, and the resulting active pathways were sorted based on their pathway score (quartile threshold = 0.80). Enrichment analyses were then performed using the gene IDs in each of the active pathways. To determine whether certain genes are enriched, the hypergeometric distribution was assumed, and P-values were calculated. Adjustment of P-values was done using the Bonferroni method. Enrichment results were then filtered by an adjusted P-value threshold of .05.
Subcutaneous Implantations
Whole 25-day-old kidney organoids that demonstrated robust nephron structure formation were subcutaneously implanted into 4 dorsal pockets (2 per pocket) in immune-deficient mice (BALB/c IL2Ry−/− RAG2−/−DKO).39 Geltrex (50 µL) was added along with each organoid. All implanted organoids were of approximately the same size. Eight weeks after surgery, the animals were sacrificed after which the implanted organoids were removed from the murine tissue. The harvested organoids were fixed in 4% paraformaldehyde for immunohistochemistry or frozen at −80 °C for gene expression analysis. The experiments were approved by the national Animal Ethical Committee of the Netherlands (licence number AVD101002016635).
Discussion
A thorough understanding of the tumorigenic potential of human iPSC-derived kidney organoids is crucial for the advancement of this promising tool in regenerative medicine into the clinical arena. Here, we confirm the absence of fully functional residual iPSCs in kidney organoids and accordingly show the lack of teratoma formation from kidney organoids. Yet, kidney organoids demonstrated the capacity to form tumors upon long-term subcutaneous implantation. These tumors contained a proportion of immature blastemal cells and showed protein expression of WT1 and CD56 and no immunostaining of epithelial markers ECAD and Villin-1. The absence of pluripotent cells in the tumor tissue refuted the possibility of a malignant transformation of residual iPSCs. This conclusion was supported by the absence of typical histological characteristics of a teratoma. The high expression of WT1 and CD56, and the histological phenotype of the tumors demonstrate some degree of similarity between the organoid-derived tumors and Wilms tumor. Wilms tumor is the most common pediatric kidney tumor and specifically illustrates the consequences of dysregulated nephrogenesis.40 In Wilms tumor metanephric mesenchymal cells fail to terminally differentiate but continue to proliferate, resulting in a disorganized tumor containing blastemal, epithelial, and stromal elements.24 Mutations are often the underlying cause of Wilms tumor formation, and although tumor-forming kidney organoids and their parent iPSC did not contain chromosomal abnormalities, we did not analyze the presence of point mutations that may explain tumor-formation. Nevertheless, our results demonstrate that kidney organoids could potentially serve as a model for studying the formation of Wilms tumor. Such studies should include the genetic component of Wilms tumors and may make use of cell fate tracking tools that allow for tracing the origin of tumorigenic cells in kidney organoids.
Analysis of epigenetic profiles of the tumorigenic organoids revealed the likely cause for tumor formation. Indeed, we discovered a unique cluster of hypomethylated DMRs in the DNA methylation profile of tumorigenic organoids that largely overlapped with iPSCs and grouped away from control organoids. The epigenetic resemblance of tumorigenic organoids with iPSCs corresponds with the observation that proliferating cells within the tumors possess a less differentiated or a more progenitor-like phenotype than the unaffected cells within control organoids. The epigenetic changes may have conferred a growth advantage to the cells. For example, there is evidence that high mobility group protein B1 (HMGB1), one of the differentially methylated genes between tumor organoids and control organoids, is known to inhibit apoptosis and induce proliferation. Therefore, increased activity due to promotor hypomethylation of HMGB1 may play a role in the maintenance and expansion of the progenitor population.41 Interestingly, the top differentially methylated genes detected by GO analysis are implicated in kidney function, which is in agreement with the kidney-specific phenotype of the progenitors in the tumorigenic organoids. Although the functional consequences of gene body demethylation are unclear,42 it is possible that disrupted expression levels of these genes impaired further kidney maturation. Besides the contribution of altered methylation patterns, a lack of environmental cues from supportive cell populations may also have prevented progression to terminal differentiation.
Interestingly, in addition to blastemal tissue, the tumorigenic organoids also contained, maturing nephrons. In normal kidney development, a balance between nephron progenitor renewal and differentiation dictates the final number of nephrons necessary for proper kidney function.43 It may be advantageous to have highly proliferative nephron progenitors to achieve a sufficient yield of nephrons. However, the persistent abundance of ki-67+ cells could indicate that proliferation will continue indefinitely and suggests that the cells will maintain a progenitor state instead of committing to a nephron fate. As shown in recent studies, a future solution to reduce tumorigenic risk may be to pre-emptively eliminate cells that show a less mature and more proliferative phenotype (without compromising the architecture of kidney organoids).44,45,46
We observed a rare occurrence of tumor formation in a large number of implanted organoids (104 in total), and the observed tumors were derived from one of the 4 studied iPSC lines. Therefore, we cannot exclude the possibility that this specific iPSC line may have an intrinsic predisposition toward tumor formation. However, the iPSC line did not show any major chromosomal abnormalities and behaved phenotypically similar to the other lines. It is therefore possible that a risk for tumor formation is applicable across iPSC lines and their derivatives. Nevertheless, tumorigenic risks may vary across organoids derived from different iPSC lines. For potential future therapeutic implantation of organoids, it would be valuable to screen iPSC-derived organoids for their tumorigenic potential.
Although this study clearly demonstrates the tumorigenic risk of kidney organoids, some potential limitations should be mentioned. First, subcutaneous implantation may provide a different environment for the maturation of kidney organoids than renal subcapsular implantation. However, studies using subcapsular implantations of kidney organoids used shorter time points and may therefore be insufficient to evaluate the long-term safety of kidney organoids.11,13 The subcapsular implantation model is also restricted to the implantation of one or half an organoid per mouse and there is thus a smaller amount of tissue that can potentially form tumors. Subcutaneous implantation, on the other hand, forms a relatively efficient approach to implant a large number of organoids and also allows for easy identification and removal of tumors. Secondly, a major challenge in the field is the significant variation in kidney organoids from different iPSC lines, batches, and experiments.37 The variability in kidney organoid phenotype may potentially also affect the tumorigenic potential of organoids. In our study we found tumor formation in some organoids from one iPSC line in one experiment and we cannot rule out that variability in organoid status was at the basis of the tumorigenic potential of organoids. Nevertheless, our study demonstrates that the possibility that organoids form tumors exists. More research will be required to find out the exact cause.
In conclusion, even though we usually observed successful engraftment, vascularization and maturation of kidney organoids, the rare observation of tumor formation highlights the current challenges in the safe implementation of kidney organoids as a therapy. Importantly, we show that epigenetic changes may be at the basis of tumorigenic transformation of kidney organoids. As the field of regenerative medicine is slowly moving toward clinical application of iPSC-derivatives, it is crucial to identify possible safety issues associated with kidney organoids.
Supplementary Material
Acknowledgements
We would like to thank the Department of Pathology for the help in processing samples. We would also like to thank Dr. Mehrnaz Ghazvini for providing the iPSC data for the MeD-seq analysis.
Author Contributions
A.S.S.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing; Z.D., W.C., T.B., S.K., J.B., W.V.I., E.B. B.E.: collection and assembly of data; H.T.M., R.B.: collection of data, data analysis and interpretation; A.D.K, Q.P., L.O., M.C.G: data analysis and interpretation; E.H., C.B., J.G., M.H.: conception and design, data analysis and interpretation, manuscript writing. All authors approved the final version of the manuscript.
Conflict of Interest
R.B., J.B., W.V.I. and J.G. are shareholders in Methylomics B.V., a company that applies MeD-seq to develop methylation markers for cancer staging.
Data Availability
The datasets generated and analyzed during the current study are available in the NCBI Gene Expression Omnibus under accession number GSE165266 and GSM5029240.
References
- 1. Jha V, Garcia-Garcia G, Iseki K, et al. Chronic kidney disease: global dimension and perspectives. Lancet. 2013;382:260-272. [DOI] [PubMed] [Google Scholar]
- 2. Shankar AS, Hoorn EJ, Gribnau J, et al. Current state of renal regenerative therapies. Transplantation. 2019;103(2):250-61. [DOI] [PubMed] [Google Scholar]
- 3. Morizane A, Kikuchi T, Hayashi T, et al. MHC matching improves engraftment of iPSC-derived neurons in non-human primates. Nat Commun. 2017;8(1):385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Takasato M, Er PX, Chiu HS, et al. Generating of kidney organoids from human pluripotent stem cells. Nat Protoc. 2016;11(9):1681-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garreta E, Prado P, Tarantino C, et al. Fine tuning the extracellular environment accelerates the derivation of kidney organoids from human pluripotent stem cells. Nat Mater. 2019;18(4):397-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Freedman BS, Brooks CR, Lam AQ, et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat Commun. 2015;6:8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Taguchi A, Nishinakamura R.. Higher-order kidney organogenesis from pluripotent stem cells. Cell Stem Cell. 2017;21(6):730-746.e6. [DOI] [PubMed] [Google Scholar]
- 8. Takasato M, Little MH.. The origin of the mammalian kidney: implications for recreating the kidney in vitro. Development. 2015;142:1937-1947. [DOI] [PubMed] [Google Scholar]
- 9. Wu H, Uchimura K, Donnelly EL, Kirita Y, Morris SA, Humphreys BD.. Comparative analysis and refinement of human PSC-derived kidney organoid differentiation with single-cell transcriptomics. Cell Stem Cell. 2018;23(6):869-881.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Combes AN, Zappia L, Er PX, Oshlack A, Little MH.. Single-cell analysis reveals congruence between kidney organoids and human fetal kidney. Genome Med. 2019;11(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van den Berg CW, Ritsma L, Avramut MC, et al. Renal subcapsular transplantation of PSC-derived kidney organoids induces neo-vasculogenesis and significant glomerular and tubular maturation in vivo. Stem Cell Reports. 2018;10(3):751-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bantounas I, Ranjzad P, Tengku F, et al. Generation of functioning nephrons by implanting human pluripotent stem cell-derived kidney progenitors. Stem Cell Reports. 2018;10(3):766-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sharmin S, Taguchi A, Kaku Y, et al. Human induced pluripotent stem cell-derived podocytes mature into vascularized glomeruli upon experimental transplantation. J Am Soc Nephrol. 2016;27(6):1778-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tanigawa S, Islam M, Sharmin S, et al. Organoids from nephrotic disease-derived iPSCs identify impaired NEPHRIN localization and slit diaphragm formation in kidney podocytes. Stem Cell Reports. 2018;11(3):727-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Subramanian A, Sidhom EH, Emani M, et al. Single cell census of human kidney organoids shows reproducibility and diminished off-target cells after transplantation. Nat Commun. 2019;10(1):5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nam SA, Seo E, Kim JW, et al. Graft immaturity and safety concerns in transplanted human kidney organoids. Exp Mol Med. 2019;51(11):1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee AS, Tang C, Rao MS, Weissman IL, Wu JC.. Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat Med. 2013;19(8):998-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861-872. [DOI] [PubMed] [Google Scholar]
- 19. Hentze H, Soong PL, Wang ST, Phillips BW, Putti TC, Dunn NR.. Teratoma formation by human embryonic stem cells: evaluation of essential parameters for future safety studies. Stem Cell Res. 2009;2(3):198-210. [DOI] [PubMed] [Google Scholar]
- 20. Ben-David U, Benvenisty N.. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat Rev Cancer. 2011;11(4):268-277. [DOI] [PubMed] [Google Scholar]
- 21. Brederlau A, Correia AS, Anisimov SV, et al. Transplantation of human embryonic stem cell-derived cells to a rat model of Parkinson’s disease: effect of in vitro differentiation on graft survival and teratoma formation. Stem Cells. 2006;24(6):1433-1440. [DOI] [PubMed] [Google Scholar]
- 22. Cai J, Yang M, Poremsky E, Kidd S, Schneider JS, Iacovitti L.. Dopaminergic neurons derived from human induced pluripotent stem cells survive and integrate into 6-OHDA-lesioned rats. Stem Cells Dev. 2010;19(7):1017-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nori S, Okada Y, Yasuda A, et al. Grafted human-induced pluripotent stem-cell-derived neurospheres promote motor functional recovery after spinal cord injury in mice. Proc Natl Acad Sci USA. 2011;108(40):16825-16830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aubry L, Bugi A, Lefort N, Rousseau F, Peschanski M, Perrier AL.. Striatal progenitors derived from human ES cells mature into DARPP32 neurons in vitro and in quinolinic acid-lesioned rats. Proc Natl Acad Sci USA. 2008;105(43):16707-16712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roy NS, Cleren C, Singh SK, Yang L, Beal MF, Goldman SA.. Functional engraftment of human ES cell-derived dopaminergic neurons enriched by coculture with telomerase-immortalized midbrain astrocytes. Nat Med. 2006;12(11):1259-1268. [DOI] [PubMed] [Google Scholar]
- 26. Seminatore C, Polentes J, Ellman D, et al. The postischemic environment differentially impacts teratoma or tumor formation after transplantation of human embryonic stem cell-derived neural progenitors. Stroke. 2010;41(1):153-159. [DOI] [PubMed] [Google Scholar]
- 27. Arnhold S, Klein H, Semkova I, Addicks K, Schraermeyer U.. Neurally selected embryonic stem cells induce tumor formation after long-term survival following engraftment into the subretinal space. Invest Ophthalmol Vis Sci. 2004;45(12):4251-4255. [DOI] [PubMed] [Google Scholar]
- 28. Doi D, Morizane A, Kikuchi T, et al. Prolonged maturation culture favors a reduction in the tumorigenicity and the dopaminergic function of human ESC-derived neural cells in a primate model of Parkinson’s disease. Stem Cells. 2012;30(5):935-945. [DOI] [PubMed] [Google Scholar]
- 29. Kumar Gupta A, Sarkar P, Wertheim JA, Pan X, Carroll TJ, Oxburgh L.. Asynchronous mixing of kidney progenitor cells potentiates nephrogenesis in organoids. Commun Biol. 2020;3(1):231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vasei M, Moch H, Mousavi A, Kajbafzadeh AM, Sauter G.. Immunohistochemical profiling of Wilms tumor: a tissue microarray study. Appl Immunohistochem Mol Morphol. 2008;16(2): 128-134. [DOI] [PubMed] [Google Scholar]
- 31. Boers R, Boers J, de Hoon B, et al. Genome-wide DNA methylation profiling using the methylation-dependent restriction enzyme LpnPI. Genome Res. 2018;28(1):88-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Warlich E, Kuehle J, Cantz T, et al. Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. Mol Ther. 2011;19(4):782-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. de Esch CE, Ghazvini M, Loos F, et al. Epigenetic characterization of the FMR1 promoter in induced pluripotent stem cells from human fibroblasts carrying an unmethylated full mutation. Stem Cell Reports. 2014;3(4):548-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Takasato M, Er PX, Chiu HS, Little MH.. Generation of kidney organoids from human pluripotent stem cells. Nat Protoc. 2016;11(9):1681-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shankar AS, Du Z, Mora HT, et al. Human kidney organoids produce functional renin. Kidney Int. 2021;99(1):134-147. [DOI] [PubMed] [Google Scholar]
- 36. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R.. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Phipson B, Er PX, Combes AN, et al. Evaluation of variability in human kidney organoids. Nat Methods. 2019;16(1):79-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ulgen E, Ozisik O, Sezerman OU.. pathfindR: an R package for comprehensive identification of enriched pathways in Omics data through active subnetworks. Front Genet. 2019;10(858): 1-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Farrell E, Both SK, Odörfer KI, et al. In-vivo generation of bone via endochondral ossification by in-vitro chondrogenic priming of adult human and rat mesenchymal stem cells. BMC Musculoskelet Disord. 2011;12(31):1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hohenstein P, Pritchard-Jones K, Charlton J.. The yin and yang of kidney development and Wilms’ tumors. Genes Dev 2015;29(5):467-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang S, Zhang Y.. HMGB1 in inflammation and cancer. J Hematol Oncol. 2020;13(116):1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G.. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26(4):577-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kobayashi A, Valerius MT, Mugford JW, et al. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell. 2008;3(2):169-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kojima K, Miyoshi H, Nagoshi N, et al. Selective ablation of tumorigenic cells following human induced pluripotent stem cell-derived neural stem/progenitor cell transplantation in spinal cord injury. Stem Cells Transl Med. 2019;8(3):260-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee MO, Moon SH, Jeong HC, et al. Inhibition of pluripotent stem cell-derived teratoma formation by small molecules. Proc Natl Acad Sci USA. 2013;110(35):E3281-E3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ben-David U, Gan QF, Golan-Lev T, et al. Selective elimination of human pluripotent stem cells by an oleate synthesis inhibitor discovered in a high-throughput screen. Cell Stem Cell. 2013;12(2):167-179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are available in the NCBI Gene Expression Omnibus under accession number GSE165266 and GSM5029240.