Abstract
Mechanistic study and precision treatment of primary liver cancer (PLC) are hindered by marked heterogeneity, which is challenging to recapitulate in any given liver cancer mouse model. Here, we report the generation of 25 mouse models of PLC by in situ genome editing of hepatocytes recapitulating 25 single or combinations of human cancer driver genes. These mouse tumors represent major histopathological types of human PLCs and could be divided into three human-matched molecular subtypes based on transcriptomic and proteomic profiles. Phenotypical characterization identified subtype- or genotype-specific alterations in immune microenvironment, metabolic reprogramming, cell proliferation, and expression of drug targets. Furthermore, single-cell analysis and expression tracing revealed spatial and temporal dynamics in expression of pyruvate kinase M2 (Pkm2). Tumor-specific knockdown of Pkm2 by multiplexed genome editing reversed the Warburg effect and suppressed tumorigenesis in a genotype-specific manner. Our study provides mouse PLC models with defined genetic drivers and characterized phenotypical heterogeneity suitable for mechanistic investigation and preclinical testing.
Mouse liver tumor model panel reveals diverse human-matched and therapy-related phenotypical and molecular characters.
INTRODUCTION
Primary liver cancer (PLC) is the third leading cause of cancer-related mortality worldwide, and treatments are very limited (1). Hepatocellular carcinoma (HCC) accounts for 75 to 85% of PLC, and intrahepatic cholangiocarcinoma (ICC) makes up 10 to 15%, besides other rare types. Thirty percent to 40% of HCC patients may be eligible for surgical resection or transplantation. However, for advanced HCCs, only a few multikinase inhibitors providing very limited survival benefits are available (2, 3). However, immunotherapy using checkpoint inhibitors, such as anti–PD-1 and anti–PD-L1, has shown promising results for advanced HCC (4). Noteworthy, a combination of anti–PD-L1 and anti-VEGF (vascular endothelial growth factor) antibodies results in better efficacy than sorafenib (5). ICC is more difficult to diagnose and to treat compared with HCC and has worse prognosis (6). While much hope has been placed in the identification of novel targets through molecular profiling, it was obscured by the heterogeneity in the cause, genetics, and phenotypes of PLC. Mutations of cancer driver genes are major triggers of liver tumorigenesis. Recent investigations of the HCC and ICC genomes had revealed vastly different mutational landscape among patients and identified recurrently affected gene loci, such as TERT promoter, TP53, CTNNB1, MYC, and RAS (7–11). These findings not only highlighted the need for functional characterization of these aberrations but also suggested a daunting challenge in developing preclinical models.
Hydrodynamic force generated by pressurized injection of solution into the blood vessel could breach the endothelium and the closely associated hepatocyte plasma membrane. This approach was successfully used in mice to deliver transposons and CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats–CRISPR-associated protein 9) systems into hepatocytes. Hepatocytic genome could thus be edited by transposon integration or CRISPR-Cas9–mediated nonhomologous end joining (12). Such an approach has revealed susceptibility of mature hepatocytes to malignant transformation (13, 14). In this study, we have generated a plasmid toolbox for the expression of transgenes or short hairpin RNAs (shRNAs) in Piggybac (PB) transposons, as well as single-guide RNAs (sgRNAs) and Cas9 for in situ genome editing of mouse hepatocytes. Selection of genetic combinations for genome editing was guided by the mutational landscape of human PLC. We thus produced primary mouse liver tumors in 25 genotypes representing major histopathological types of PLC. Further molecular profiling identified three human-matched molecular subtypes of mouse tumors reflecting heterogeneity and dynamics in treatment-related phenotypes. By multiplexed genome editing, we demonstrated a genotype-specific essential role of pyruvate kinase M2 (Pkm2) in metabolic reprogramming and liver tumorigenesis. We thus provide an integrated platform including an expandable plasmid toolbox for in situ genome editing, a biobank of mouse liver tumors, and corresponding multiomic datasets, which allows further investigation and targeting of heterogeneous phenotypes of liver cancer.
RESULTS
In situ genome editing generates a panel of genetically defined mouse liver tumors
To induce liver tumors representing heterogeneous phenotypes as a result of diverse genetic drivers, we first consulted the mutational landscape of HCC. Twenty-two potential driver genes were selected by mutational frequency from seven human HCC cohorts with diverse etiological factors and ethnic origins (table S1). FGF19 and CCND1 in the same amplicon were included as a single-event F/C. Mutations in the TERT promoter region were omitted because of long telomeres in laboratory mouse strains (15). NRAS was mutated in about 0.8% of HCC (11, 16), while KRAS was mutated in as high as 30% of ICC (10). We used NRAS (labeled as RAS below) since it was successfully used to induce liver tumorigenesis in a similar setting (17). Genetic combinations of two events were chosen on the basis of the pattern of co-occurrence (fig. S1A). Since expression of myristoylated AKT together with the intracellular domain of the NOTCH1 receptor (NICD) has been reported to induce ICC from mouse hepatocytes (18), we included this combination to increase phenotypical diversity. Activation of oncogenes was mimicked by transgenes carried by PB transposons, which encoded epitope-tagged wild-type or constitutively active proteins (Fig. 1A and fig. S1B). Expression of transgenes was confirmed by immunoblotting and immunohistochemistry (IHC) (fig. S1, C and D). Inactivation of tumor suppressors was mimicked by CRISPR-Cas9–mediated knockout (Fig. 1A). Cas9 and sgRNAs were coexpressed from a plasmid (fig. S1E), and for genotypes involving two tumor suppressors, liver-specific heterozygous Cas9 transgenic mice were used to increase the efficiency of gene editing. sgRNAs were evaluated by the T7 endonuclease 1 (T7E1) mismatch detection assay in tissue culture (fig. S1F). For Tp53, a germline knockout strain that is free of liver tumor at 6 months of age was also used (19).
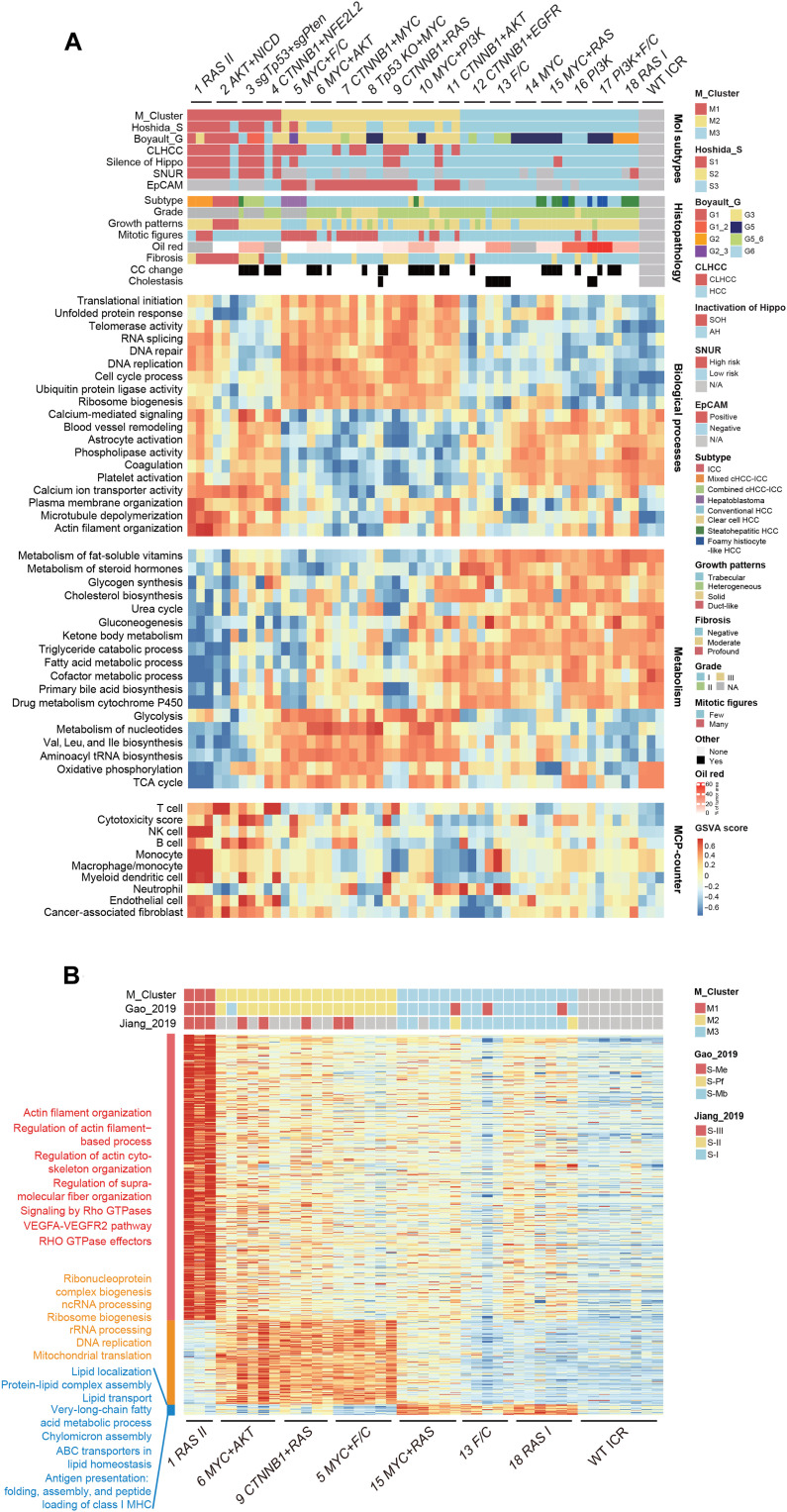
Fig. 1. Generation of genetically defined mouse liver tumors by in situ genome editing.
(A) Overview of model generation and characterization. For model characterization, the number of samples analyzed for each genotype was labeled. (B) Genetic combinations used for making mouse liver tumor models. Frequencies were calculated from pooled Memorial Sloan Kettering (MSK), Asan Medical Center (AMC), and TCGA cohorts. Genes marked in red are oncogenic, and genes in blue are tumor suppressive. Successful models are underlined and darker shaded in the pie graph. (C) Cumulative ratio of tumorigenesis at the indicated time after injection. (D) Size and number of nodules induced by different genetic combinations. Violin plots showing the median (black solid line) and distribution of sizes. Box plots showing the median and interquartile range of nodule number. Typical nodule patterns in anti-HA IHC are shown on the right. Scale bar, 5 mm.
Each plasmid combination was hydrodynamically injected to an average of 12 mice, and 2 to 3 mice were sacrificed per month, or when symptoms of tumorigenesis such as abdominal enlargement were evident. While 11 single or combinations of plasmids did not elicit tumorigenesis, 25 successfully induced liver tumors, including 5 single genes, 5 Tp53+X combinations, 6 CTNNB1+X combinations, 6 MYC+X combinations, 2 PI3K+X combinations, and AKT+NICD (table S2). These genetic alterations were found in 29.8% of human HCC (Fig. 1B). Fourteen combinations induced tumors in >40% of mice (high efficiency; table S2). As expected, single driver genes were less efficient; for instance, neither CTNNB1 nor MYC induced tumorigenesis by 6 months. However, the CTNNB1+MYC combination drastically induced tumorigenesis in all mice within 2 months (Fig. 1C). In general, MYC combinations drove fast tumorigenesis, followed by those containing CTNNB1. Among Tp53 combinations, Tp53 KO+MYC reached 80% efficiency at 2 months, while sgTp53+sgPten reached 40% efficiency at 8 months, but other sgTp53 combinations showed much lower efficiency or failed (Fig. 1C).
We thereafter referred to tumors by their driver genes as genotypes. Representative livers are shown in fig. S2A. Expression of driver genes was confirmed by IHC staining of the epitope tag (fig. S2B). Five IHC-negative genotypes were confirmed by mRNA expression (fig. S2C). Noteworthy, the expression of these genes could be detected by IHC soon after injection (fig. S1D), suggesting subdued protein expression during tumorigenesis. Successful knockout of tumor suppressors was determined by T7E1 assay using tumor samples (fig. S2D). Editing of the Tp53 locus in sgTp53+sgRb1 tumors was confirmed by genomic DNA sequencing, although it evaded T7E1 detection possibly because of the nature of indels (insertion or deletion mutations; fig. S2D). The size and number of nodules on liver sections were quantified from anti-HA (hemagglutinin) tag stainings or, when not available, from hematoxylin and eosin (H&E) stainings (Fig. 1D). Nodules were defined by a size of 0.04 mm2, which was about 50 cells, or larger on paraffin sections (fig. S2E). By k-means clustering analysis, three tumorigenesis patterns were found (fig. S2F). Group 1 was featured by one to three large tumors per section in most cases, and group 3 was featured by many small nodules. Group 2 was intermediate in number and size. Notably, two distinct patterns were induced by RAS. RAS I was the major type with many nodules in sizes of normal distribution, and RAS II was found in 10% of mice as an overwhelmingly large and stiff tumor (Fig. 1D and fig. S2A), which were characterized separately below. Together, a panel of genetically defined mouse liver tumors in 25 genotypes was generated by in situ genome editing.
Molecular profiling revealed three subtypes of mouse liver tumors
Fourteen high-efficiency genotypes and three low-efficiency genotypes with sufficient samples (fig. S3A) were subjected to transcriptomic and proteomic profiling, whole-exome sequencing (WES), and histopathological analysis. Eight low-efficiency genotypes, including sgTp53+sgRb1, sgTp53+JAK1, sgTp53+MET, CTNNB1+sgArid2, MYC+sgCdkn2a, MYC+EGFR, PI3K+sgRb1, and EGFR, were not further analyzed because of insufficient number of samples. Genotype-centered characteristics are summarized in Table 1. For transcriptomic analysis by RNA sequencing (RNA-seq), three tumors for each genotype, but two for CTNNB1+NFE2L2, were used. Unsupervised clustering indicated similar transcriptomic profiles within genotypes (fig. S3B and table S3). Because of the similarity of sham-operated Institute of Cancer Research (ICR) strain and C57/BL6 strain controls, only ICR datasets were used for further analysis. We compared the transcriptome of each genotype to The Cancer Genome Atlas (TCGA) cohort of human HCC using the transcriptome overlap measure (TROM) method for cross-species analysis (20). The results indicated that each genotype was similar to 0.9 to 27.8% of human HCC, with an average of 13.0% (Table 1 and fig. S3C).
Table 1. Summary of mouse liver tumor models.
NA, not applicable.
| # | Genotype |
Tumori genesis rate |
Tumor
latency (months) |
Tumori genesis pattern* |
Histopathological
subtype |
HCC
grade |
M_
Cluster |
Compare to human HCC | Features | |||||
|
Transcriptome
Hoshida_S |
Transcriptome
Boyault_G |
Proteome
Gao_2019 |
Proteome
Jiang_2019 |
% of TCGA cohort with
similar transcriptome by TROM |
||||||||||
| 18 | RAS | I | 73.7% (I:II = 9:1) |
5 | 3 | Steatohepatitic HCC | II | M3 | S3 | G2 | S-Mb | S-I | 1.0% | Small tumor, liver metabolic function, lipid storage, glycogen storage |
| 1 | II | 1 or 2 | Mixed cHCC-ICC | NA | M1 | S1 | G1/G3 | S-Me | S-III | 20.9% | Large tumor, hepatocyte dedifferentiation, profound fibrosis, immune cell infiltration, proliferation, focal AFP, silence of Hippo, SNUR high risk |
|||
| 2 | AKT+NICD | 100.0% | 2 | Mainly 3 | ICC | NA | M1 | S1/S3 | G1 | NA | NA | 14.4% | Hepatocyte transdifferentiation, profound fibrosis, immune cell infiltration, silence of Hippo, SNUR high risk |
|
| 3 | sgTp53+sgPten | 57.1% | 8 | Mainly 1 | Combined cHCC-ICC | NA | M1 | S1 | G1_2/G6 | NA | NA | 14.4% | Hepatocyte dedifferentiation, profound fibrosis, Treg infiltration, lipid storage, silence of Hippo, SNUR high risk |
|
| 4 | CTNNB1+NFE2L2 | 23.1% | 12 | 1 | Conventional HCC | II | M1 | S1/S3 | G3/G6 | NA | NA | 15.0% | Immune cell infiltration | |
| 5 | MYC+F/C | 57.7% | 1 | 1 | Hepatoblastoma | NA | M2 | S1/S2 | G2/G3 | S-Pf | Partial S-III | 27.8% | Extramedullary hematopoiesis, proliferation, global AFP, EpCAM signature |
|
| 6 | MYC+AKT | 73.1% | 1 | Mainly 3 | Conventional HCC | II | M2 | S3 | G3 | S-Pf | Partial S-III | 15.0% | AFP, EpCAM signature | |
| 7 | CTNNB1+MYC | 100.0% | 2 | 2 or 3 | Conventional HCC | II-III | M2 | S2 | G3/G5_6 | NA | NA | 12.6% | Proliferation, focal AFP, EpCAM signature |
|
| 8 | Tp53 KO+MYC | 64.3% | 2 | 1 or 2 | Conventional HCC | III | M2 | S2/S3 | G5/G3 | NA | NA | 22.2% | Proliferation, global AFP, EpCAM signature, CNV |
|
| 9 | CTNNB1+RAS | 91.7% | 2 | 2 or 3 | Conventional HCC | II | M2 | S2 | G3 | S-Pf | Partial S-III | 19.7% | Bile-filled pseudoglands, AFP, EpCAM signature, moderate fibrosis, silence of Hippo |
|
| 10 | MYC+PI3K | 71.4% | 2 | Mainly 1 | Conventional, clear cell, or steatohepatic HCC | II-III | M2 | S3 | G3/G5 | NA | NA | 8.6% | Lipid storage, proliferation, global AFP |
|
| 11 | CTNNB1+AKT | 40.0% | 5 | Mainly 2 | Conventional HCC | II-III | M2 | S2/S3 | G3 | NA | NA | 24.3% | Moderate fibrosis, AFP, EpCAM signature |
|
| 12 | CTNNB1+EGFR | 23.7% | 3 | Mainly 2 | Conventional HCC | II | M3 | S3 | G5/G6 | NA | NA | 5.7% | Global AFP | |
| 13 | F/C | 50.0% | 9 | Mainly 1 | Conventional HCC | I-II | M3 | S3 | G5/G6 | S-Mb | S-I | 8.4% | Lipid storage, cholestasis, SNV, Ctnnb1 mutation |
|
| 14 | MYC | 21.4% | 9 | 1 or 2 | Conventional HCC | II | M3 | S3 | G5 | NA | NA | 2.4% | ||
| 15 | MYC+RAS | 72.0% | 2 | Mainly 3 | Steatohepatitic HCC | II | M3 | S3 | G5 | S-Mb | S-I | 0.9% | AFP | |
| 16 | PI3K | 43.8% | 6 | Mainly 2 | Conventional, foamy histiocyte-like, or steatohepatic HCC |
II-III | M3 | S3 | G6 | NA | NA | 12.4% | Lipid storage | |
| 17 | PI3K+F/C | 72.7% | 5 | Mainly 2 | Foamy histiocyte-like HCC |
II | M3 | S3 | G5 | NA | NA | 8.4% | Lipid storage, cholestasis, SNV, Ctnnb1 mutation |
|
*Tumorigenesis pattern: 1, one to three large tumors per section; 3, many small nodules; 2, intermediate in number and size.
Hierarchical consensus clustering revealed the three transcriptomic subtypes M1-M3 among 53 mouse liver tumors (Fig. 2A and fig. S3D). Multidimensional scaling (MDS) plot confirmed distinctions of the three subtypes (fig. S3E). mRNA signatures of subtypes were identified by a supervised analysis (table S3). We next applied M signatures to a previously reported dataset of 56 mouse liver tumors generated by N-nitrosodiethylamine (DEN), TAK1ΔHEP (TAK1), MUP-uPA + high-fat diet (MUP), and Stelic Animal Model (STAM) methods (21). By the nearest template prediction (NTP) analysis, STAM tumors were heterogeneous with M1, M2, or M1/2 features, likely due to extremely high mutation frequency in this model (fig. S3F). TAK1 tumors were M1 or M2 or could not be classified. Tumors induced by DEN were all M3, and MUP tumors were mostly M1. These results suggested M signatures as widely applicable tools in classifying mouse models of liver cancer.
Fig. 2. Molecular profiling revealed three subtypes of mouse liver tumors.
(A) Transcriptomic subtypes and their histopathological features. Heatmaps were organized by M cluster grouping for molecular subtypes, histopathology, and mRNA signatures of biological processes, mRNA signatures of metabolism, and cell types by MCP-counter. Tumors are in columns, grouped by the M cluster membership. (B) Proteomic subtypes of mouse liver tumors. Heatmap depicts relative abundance (Z score) of signature proteins (row) in tumors (column). Classification of tumors using the M proteomic signatures and human HCC proteomic signatures is indicated on top. Biological functions of signature proteins were analyzed by enrichment of MsigDB ontologies.
Proteomic profiles classified human HCC into subgroups associated with clinical and molecular attributes (22, 23). We selected seven genotypes representing M1-M3 subtypes for proteomic analysis using a label-free technique on a mass spectrometry (MS) platform that has characterized a large cohort of human HCC under similar quality control (22). Detection of the proteome was stable across samples (fig. S4A). On average, 3724 proteins per tumor and 3165 proteins per normal liver tissue were identified (fig. S4B and table S4). Hierarchical consensus clustering identified three proteomic subgroups (Fig. 2B and fig. S4C). Proteomic or transcriptomic profiles resulted in identical allocation of tumors (Fig. 2, A and B); thus, the proteomic subtypes were still referred to as M1-M3. Pairing of transcriptomic and proteomic profiles revealed 6091 mRNA-protein pairs with an overall positive correlation (median r = 0.45; fig. S4D and table S4), and 3119 pairs (51.2%) had significant positive correlation [r > 0, false discovery rate (FDR) < 0.05]. Among 1138 genes with no correlation in their mRNA and protein expression levels (−0.2 < r < 0.2, FDR > 0.05), 252 genes were differential to normal liver on the protein level, highlighting the extra strength of proteomic analysis (fig. S4E and table S4). Proteins involved in mitochondrial respiratory electron transport were repressed on the protein level in all subtypes. A similar observation has been made in human HCC (23). Nontranscriptional inhibition of mitochondrial respiration genes found in Gao’s HCC cohort and our mouse tumors was confirmed in an independent cohort of HCC (fig. S4F), which thus might be a common feature of liver tumorigenesis. Together, three molecular subtypes of mouse liver tumors were revealed by transcriptomic and proteomic profiles.
Mouse liver tumors exhibit human-matched subtype-specific phenotypes
We next determined the disease relevance of mouse M subtypes. Human HCC could be subgrouped according to transcriptomic signatures, for example, using the Hoshida S1-S3 and Boyault G1-G6 standards (24, 25), and by proteomic signatures into the Gao’s S-Me, S-Pf, and S-Mb or Jiang’s S-I, S-II, and S-III subgroups (22, 23). Using the NTP method (26), all mouse tumors could be classified by the S1-S3 or G1-G6 transcriptomic signatures (Fig. 2A and table S3). Similarly, profiled mouse tumors were enriched for at least one of the human HCC proteomic signatures (Fig. 2B and table S4). Furthermore, unsupervised clustering of a mixture of mouse and human tumors revealed grouping by matching subtypes rather than species (fig. S5). These findings indicate the relevance of mouse M1-M3 subtypes to human HCC. Relationships of specific M subtypes to human molecular subtypes were further discussed below.
M1 includes four genotypes and was transcriptomically similar to human S1 or G1-G2 (Fig. 2A). The human S1 subgroup was featured by activation of a cholangiocarcinoma-like HCC (CLHCC) signature, which predicts poor prognosis (27). Most M1 tumors were also molecularly classified as CLHCC (Fig. 2A). This observation recapitulated pathological features revealed by histological analysis, for which mice were assigned into specific PLC subtypes by evaluating the histopathological features of the five largest tumors on H&E-stained liver sections (Figs. 2A and 3A and table S5). Three of the four genotypes in M1 exhibited pathological features of ICC. While AKT+NICD tumors were well-differentiated ICC as previously reported (18), both RAS II and sgTp53+sgPten tumors were combined HCC and ICC (cHCC-ICC), a rare type of PLC showing both hepatocellular and biliary epithelial differentiation (28). RAS II was mixed-type cHCC-ICC, in which HCC and ICC components were mixed without clear boundaries. sgTp53+sgPten tumors were mostly combined-type cHCC-ICC with clearly defined areas of HCC and ICC in the same tumor. Compound mutation of TP53 and PTEN was also found in combined-type cHCC-ICC (29). M1 tumors were also enriched for a signature of Hippo pathway inactivation (Fig. 2A), which was reported to induce dedifferentiation of mature hepatocytes (30, 31). Note that the hydrodynamic injection method mostly transfects hepatocytes according to previous studies (32), suggesting dedifferentiation or transdifferentiation as the underlying mechanism for the formation of ICC traits. Consistently, both HCC compartment, as marked by the expression of hepatocyte nuclear factor 4 α (Hnf4a), and ICC compartment, as marked by the expression of keratin 19 (Krt19), were positive for the expression of the driver gene or its downstream marker gene (Fig. 3B). Furthermore, HA+Hnf4a+Krt19+ cells were identified in Ras II and sgTp53+sgPten tumors (Fig. 3B), suggesting a transition state during transdifferentiation.
Fig. 3. M1 tumors were featured by dedifferentiation, fibrosis, and immune infiltration.
(A) Representative images of histopathological subtypes of mouse and human liver tumors. Scale bars, 80 μm. H, human; M, mouse. (B) Traits of hepatocyte dedifferentiation. Multiplex IHC staining was done. pAKT marked inactivation of Pten. Scale bars, 50 μm. (C) Degree of fibrosis in mouse liver tumors. Sirius red staining was performed on liver sections, and representative tumors in three genotypes are shown for each degree. Scale bars, 80 μm. (D) Immune infiltration in mouse liver tumors. Liver sections were stained with anti-CD45, anti-CD3, anti-FOXP3, and anti-F4/80 antibodies by IHC. Representative strong and weak stainings are shown on the left, and quantifications are on the right. Scale bars, 100 μm. Results are representative of at least three independent experiments.
Proteomic signatures allocated RAS II of the M1 subtype into the human S-III or S-Me subgroups (Fig. 2B). S-III is distinguished by aggressive characteristics including activation of integrin and Rho guanosine triphosphatase (GTPase) pathways (22). Consistently, the M1 proteomic signature was most notably featured by actin filament organization and Rho signaling (Fig. 2B). Furthermore, the M1 subtype was also enriched for focal adhesion and cytoskeletal genes on the transcriptional level (Fig. 2A). Remodeling of the cytoskeleton is often accompanied by altered extracellular matrix. Consistently, fibrosis was most profound in the ICC-related M1 subtype as revealed by Sirius red staining (Figs. 2A and 3C). Mild fibrosis was also found in three CTNNB1-containing genotypes.
In human HCC, the immune-high subtype was associated with poor differentiation, positive Krt19, and S1 or G2 transcriptomic subclasses (33). Analysis of transcriptomic profiles using the Microenvironment Cell Populations-counter (MCP-counter) method for estimating the abundance of tissue-infiltrating immune cells (34) indicated that the M1 subtype was highly immune cell infiltrated (Fig. 2A). IHC staining confirmed marked infiltration of CD45+ cells in 98.7% of M1 subtype tumors (Fig. 3D). Furthermore, macrophages, T cells, and immunosuppressive regulatory T cells (Tregs) were also enriched in M1 tumors (Fig. 3D), which was consistent with proteomic findings of S-III tumors (22). These findings suggested a direct role of driver genes in the establishment of distinct tumor immune microenvironment. Together, M1 mouse liver tumors were similar to those of respective human subtypes with features of hepatocyte dedifferentiation and ICC-like properties, remodeling of the cytoskeleton and related fibrosis, and heavy infiltration of immune cells.
The M2 subtype was featured by highly proliferative hepatic stem cell–like HCC
M2 includes seven genotypes driven by CTNNB1 or MYC combinations, which were mostly conventional HCC of the World Health Organization (WHO) standard (35), except a few clear cell or steatohepatitic HCC of the MYC+PI3K genotype, and the MYC+F/C genotype as hepatoblastoma (HB) (Fig. 2A). HCCs were further graded according to the Edmondson-Steiner system (36), which indicated most tumors in grade II, except Tp53 KO+MYC tumors mostly in grade III (Fig. 2A and fig. S6A). Notably, genotypes involving activation of the phosphatidylinositol 3-kinase (PI3K) pathway were prone to steatosis regardless of molecular subtype, which was further confirmed by oil red staining (Fig. 2A and table S5). MYC+F/C tumors exhibited features of embryonal epithelial type HB (Fig. 3A). Notably, MYC+F/C tumors were characterized by extramedullary hematopoiesis, which was also found in many HBs (37). IHC staining confirmed abundant Ly76+ nucleated erythroblasts and anucleate mature erythrocytes, B220+ B cells, MPO+ granulocytes, immature Ly6G+ neutrophils lacking nuclear fragmentation, and, less abundantly, CD34+ hematopoietic stem cells (fig. S6B). Transcriptomic analysis indicated that MYC+F/C tumors resembled the robust Cluster 2 (rC2) and C2A subtype HB (fig. S6C), which were highly proliferative (38).
M2 tumors were transcriptomically similar to human S2-S3 or G3 and fell into proteomic subgroups S-Pf (Fig. 2, A and B). M2 tumors strongly enrich for a signature of epithelial cell adhesion molecule (EpCAM)–positive HCC (Fig. 2A). In addition, IHC staining indicated high expression of α-fetoprotein (AFP) in global or focal patterns in M2 tumors (fig. S6D). Double positivity of EpCAM and AFP expression defines a hepatic stem cell–like HCC subtype with poor prognosis (39), and these are features of the human S2 subtype. Thus, M2 tumors feature a state of stemness different from dedifferentiation found in M1. An exception is the CTNNB1+RAS genotype, which was pathologically featured by intratumor pseudoglands (sometimes lined with HA+Krt19+ epithelium) filled with bile (fig. S6, E and F) and was transcriptionally also enriched for CLHCC signatures (Fig. 2A). While a similar phenotype has been reported in tumors induced by CTNNB1+MET using a similar strategy (40), we did not observe a similar phenotype in other CTNNB1 combination–induced tumors. These findings suggest that cooperation between CTNNB1 and a common downstream signaling of RAS and MET, for example, the mitogen-activated protein kinase pathway, might be responsible for the phenotype. The stemness of M2 was accompanied by strong proliferation, as observed by high mitotic figure in histopathological analysis (Fig. 2A). Furthermore, transcriptomic and proteomic profiles consistently identified strong activation of protein synthesis, DNA replication, and cell cycle progression in M2 tumors (Fig. 2, A and B, and tables S3 and S4). Thus, the M2 subtype was featured by highly proliferative hepatic stem cell–like HCC.
The M3 subtype was featured by retention of liver function and liver metabolism
The M3 subtype contained tumors driven by single genes and three other combinations, which were transcriptomically similar to human S3 or G5-G6, and fell into proteomic subgroups S-Mb and S-I (Fig. 2, A and B). These human subtypes were highly overlapping and were characterized by retention of liver functions, especially liver-specific metabolic pathways, and thus have the best prognosis compared to their sister subtypes (22, 23, 41). Consistently, proteins of lipid metabolism were most abundant in M3 signature proteins (Fig. 2B). Notably, steatohepatitic HCCs were mostly found in the M3 subgroup, and foamy histiocyte-like HCCs were found in two PI3K-containing genotypes (Figs. 2A and 3A). Furthermore, analysis of the transcriptome also revealed enrichment of liver-specific functions such as metabolism of ketone body, vitamin, steroid hormone, drug, and bile acid (Fig. 2A). Consistently, MDS plot confirmed a closer distance of M3 to normal liver tissue (fig. S3E). As expected, using prognosis-related human HCC subtyping signatures, such as the Seoul National University Recurrence (SNUR) signature of early recurrence (42), CLHCC, Silence of Hippo, and EpCAM signatures, M3 tumors were classified as the better prognostic group (Fig. 2A). Markedly different pathological characters were found in the two types of RAS nodules. In contrast to RAS II, which was mixed cHCC-ICC with profound cell proliferation, the M3 genotype RAS I was like conventional HCC with accumulation of fat and glycogen (fig. S6G). Higher protein level of RAS was found in type I nodules (fig. S6, G and H). Thus, divergent phenotypes may be caused by RAS-induced premalignant senescence program and differential cell death–related microenvironment (43, 44). Together, M3 mouse liver tumors were characterized by liver function retention and liver metabolism.
Hotspot mutation of Ctnnb1 in M3 tumors
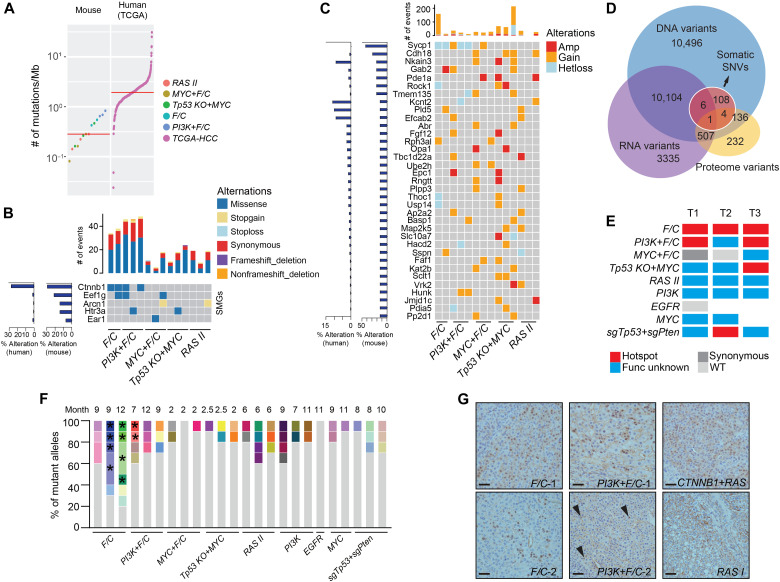
By pathological analysis, we observed cholestasis in two M3 genotypes F/C and PI3K+F/C (Fig. 2A and fig. S7A). In human HCC, cholestasis is a feature of CTNNB1 mutant tumors possibly due to dysregulation of bile salt transporters (45). However, such a phenotype was not observed in CTNNB1-containing genotypes. Nevertheless, we found activation of liver-specific and canonical Wnt/β-catenin target genes in F/C and, to a less degree, in PI3K+F/C tumors (fig. S7B). Since mutation of CTNNB1 is also a genomic feature of the human S3 and the overlapping G5-G6 subtypes of human HCC, we suspected that endogenous Ctnnb1 may be mutated in F/C and PI3K+F/C tumors. To explore the genomic landscape, WES was performed on a total of 14 mouse tumors in five genotypes (200× coverage) and their matched para-tumor tissues (100×) (fig. S8A and table S6). Somatic mutations were identified by pairwise comparison. Mutation rates were moderately higher in F/C and PI3K+F/C tumors but in a similar range between genotypes with a median of 0.3 mutations per megabase (Fig. 4A). This rate was lower than that observed in human HCC (1.9/Mb) (11) or other mouse models (2 to 122/Mb) (21). In somatic mutation profiles, excessive T>G transversions were found in RAS II and Tp53 KO+MYC tumors (fig. S8B). Clustering of mouse tumors and human cohorts based on weights of the human cancer mutational signatures revealed a general dissimilarity (fig. S8C). However, consistent with the function of Tp53, DNA damage repair–related signatures 15 and 30 were enriched in Tp53 KO+MYC tumors. Eleven significantly mutated genes (SMGs; FDR < 0.2) were identified from 24,306 genes evaluated using MuSiC, and 5 SMGs were conserved in human (Fig. 4B). CTNNB1 was found recurrently mutated in F/C and PI3K+F/C tumors, and the variant allele fraction (VAF) was between 0.132 and 0.425. VAF for Eef1g, Arcn1, Htr3a, and Ear1 was lower, varying between 0.048 and 0.176. EEF1G, ARCN1, and HTR3A were also altered in 0.5 to 1.6% of the TCGA cohort (11). Copy number variations (CNVs) were evaluated using Control-FREEC (46). CNVs were overall infrequent, and chromosome losses or gains were only observed in Tp53 KO+MYC tumors (fig. S8D). In contrast, F/C and PI3K+F/C tumors were low in CNVs but instead had a higher ratio of single-nucleotide variants (SNVs) and indels (Fig. 4, B and C), demonstrating differential roles of driver genes in shaping the mutational landscape of liver tumors. Thirty-five genes were recurrently affected by CNVs, among which 21 were also identified in human HCC at a frequency higher than 1% (Fig. 4C).
Fig. 4. Mutational profiles of mouse liver tumors.
(A) Mutational burden of mouse liver tumors and human HCC (the TCGA cohort, 358 cases) by WES. Red line indicates the median. (B and C) SMGs (B) and genes with CNV (C) in mouse tumors. Alteration frequencies in mouse tumors are indicated on the left in bar graphs. Numbers of events found in individual tumors are indicated on top. (D) Overlap of nonsynonymous SNVs and single–amino acid variants detected in WES, RNA-seq, and LC-MS/MS. (E) Hotspot mutations of Ctnnb1 exon 3 in specific genotypes. Exon 3 of Ctnnb1 was cloned and sequenced for one to three tumors in each of the nine genotypes, and the presence of hotspot mutations is indicated in red. (F) Diversity of Ctnnb1 alleles in mouse tumors. Different alleles are indicated by color. Alleles from a single tumor sharing a common mutation are indicated by asterisks. (G) Nuclear localization of Ctnnb1 in mouse tumors. Two tumors for each of F/C and PI3K+F/C genotypes are presented. RAS I was a negative control, and CTNNB1+RAS was a positive control. Arrowheads point to nuclear Ctnnb1 in PI3K+F/C tumor 2. Scale bars, 40 μm.
Six hundred and forty-eight proteomic variants corresponding to 2.6% of DNA and RNA variants were identified in mouse tumors (Fig. 4D), in a range similar to that in human HCC (1.8%) (23). Mutation of Ctnnb1 exon 3, which encodes the degradation motif, was the only variant confirmed on all levels. Further cloning and sequencing of exon 3 from tumors of nine genotypes revealed human cancer hotspot mutations in additional genotypes, but still mostly in F/C and PI3K+F/C tumors (Fig. 4E and fig. S8E). While muscular transgenic expression of FGF19 was reported to cause liver tumors with Ctnnb1 mutation (47), the absence of Ctnnb1 hotspot mutations in MYC+F/C tumors suggested an insufficient role of FGF19. Various mutant alleles were commonly found in a single tumor (Fig. 4F). Furthermore, various SNVs in addition to a common SNV or indel were found, suggesting further mutation on a founder mutant allele. Staining confirmed subclonal nuclear accumulation of Ctnnb1 protein in F/C and PI3K+F/C tumors (Fig. 4G), which was similar to that found in human HCC (48). These results suggest that both signals induced by FGF19 and long tumor latency were contributing to mutation of Ctnnb1 exon 3, a shared feature of human S3 HCC and mouse M3 liver tumors. Together, mouse liver tumors have genotype-specific features of genomic alterations.
Spatial and temporal dysregulation of drug targets in mouse liver tumors
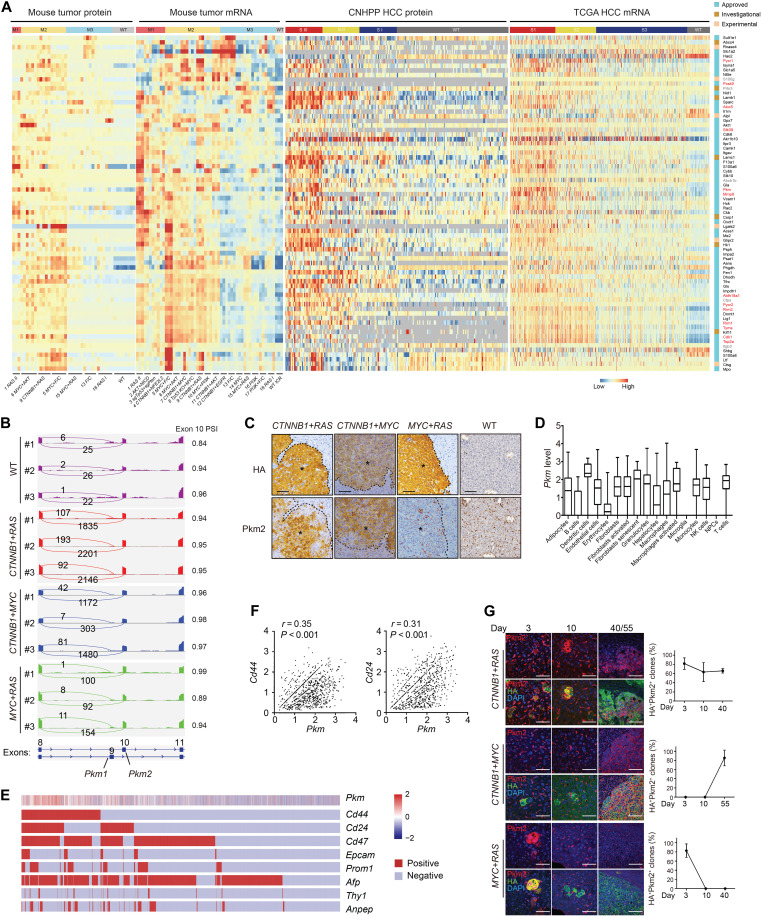
Molecular targeted therapies are highly demanded for PLC. We analyzed the expression of 2892 drug targets documented by DrugBank (49). Seventy-three drug targets were found significantly dysregulated on mRNA or protein level in at least one of the genotypes (FDR < 0.01; Fig. 5A and table S7). Sixty eight of these drug targets were also aberrantly expressed in human HCC on the protein or mRNA level (FDR < 0.01), while 13 of them have been identified by previous proteomic profiling of human HCC (fig. S9A) (22, 23). In general, abnormal expressions of drug targets were detected on both mRNA and protein levels in human and mouse. Notably, misexpression was more prevalent in mouse M1/M2 and the matching human S1/S2 subtypes, which were clinically more aggressive. Thus, our mouse models not only provided new candidate drug targets for PLC but also may serve as a platform for preclinical testing.
Fig. 5. Spatial and temporal dysregulation of drug targets in mouse liver tumors.
(A) Significantly deregulated drug targets (log2FC > 5, FDR < 0.01 for protein or log2FC > 2, FDR < 0.01 for mRNA) (row) in mouse tumors. Expression of homologs in human HCC is also shown. Genes in red were previously identified in human HCC by proteomic profiling. (B) Expression of Pkm isoforms in tumors. RNA-seq read coverage across Pkm exons 8 to 11 of samples in triplicates. PSI for exon 10 (Pkm2) is shown on the right. (C) Aberrant expression of Pkm2 protein in mouse tumors. Tumors indicated by asterisks. Scale bars, 80 μm. (D) Quantification of Pkm expression in different cell populations of CTNNB1+RAS tumors. Box plots showing medians and interquartile ranges of log-transformed normalized UMI counts of Pkm. (E) Expression of Pkm in hepatocytes. Single hepatocytes were grouped by expression of eight CSC markers, and relative abundance (Z score) of Pkm in each cell (column) is indicated on top. (F) Pkm correlated with Cd44 and Cd24 on the mRNA level. Pearson correlation r values and P values are indicated. (G) Representative multiplexed IHC of Pkm2 and HA expression in lesions 3 to 55 days after injection. Scale bars, 50 μm in days 3 and 10 and 100 μm in day 40/55. Portions of Pkm2+ lesions were quantified from three mice at each time point for each genotype. Results are representative of three independent experiments.
We then focused on Pyruvate kinase muscle (Pkm) for the following reasons: First, it was 1 of the 13 drug targets deregulated in both human and mouse tumors on mRNA and protein levels (Fig. 5A). Second, among deregulated metabolic processes, glycolysis was one of a few enhanced ones (Fig. 2A), with Pkm as a rate-limiting enzyme, which is important to consider for therapeutic inhibition. Third, deregulated expression of PKM in cancer was widely documented, but the functional roles remain debated (50), which could be due to heterogeneity. Thus, our models may be useful in resolving the issue. Alternative splicing of PKM mRNA generates two distinct isoforms, PKM1 containing exon 9 and PKM2 including exon 10. Aberrant expression of PKM2 in association with poor prognosis has been reported in many cancers, including HCC, which we have confirmed (fig. S9, B and C). Analysis of RNA-seq data for the inclusion of isoform-specific exons confirmed that Pkm2 was the dominating isoform of elevated Pkm in mouse tumors, and CTNNB1+RAS, CTNNB1+MYC, and MYC+RAS represented high, intermediate, or low expression of Pkm2, respectively (Fig. 5B). This observation was further confirmed by reverse transcription polymerase chain reaction (RT-PCR)–based absolute quantification of mRNA levels using exon-specific primers (fig. S9D). Genotype-dependent up-regulation of Pkm2 was also verified by IHC (Fig. 5C). We noticed intratumor heterogeneity of Pkm2 expression on the protein level. To determine tumor-specific expression of Pkm on the single-cell level, we performed single-cell RNA-seq (scRNA-seq) using three CTNNB1+RAS tumors. An average of 22,468 mapped reads per cell from a total of 18,213 single cells were obtained. In addition, an expression matrix of 18,770 genes in 14,958 cells per tumor was retained with 4.378% mitochondrion DNA, 0.056% hemoglobin, 7526 mean unique molecular identifier (UMI) counts, 4246 median UMI counts, and 1744 median genes per cell. Two-dimensional maps generated by uniform manifold approximation and projection (UMAP) revealed 17 distinct cell populations. Identities of cell clusters were assigned by the SingleR algorithm using a collection of mouse bulk RNA-seq datasets as the reference compendium (51, 52), resulting in identification of hepatic tumor cells, immune cells, and other stromal cells (fig. S9E). Mapping of Pkm expression to these cell populations indicated a wide range of Pkm expression in different cells (fig. S9E). High expression of Pkm was found in most immune cells, fibroblasts, and endothelial cells (Fig. 5D). Unexpectedly, Pkm expression was heterogeneous and relatively low in tumor cells (Fig. 5D). It was reported that Pkm2 plays a functional role in cancer stem cells (CSCs) (53). To determine whether Pkm was selectively expressed in certain CSC populations, we clustered 4191 tumor cells according to gene expression enrichment of multiple CSC markers, including Cd24, Cd44, EpCAM, Prom1, Thy1, Anpep, Cd47, and Afp (Fig. 5E). The highest expression of Pkm was found in a Cd44+Cd24+Cd47+Afp+ population (Fig. 5E). Among all CSC markers examined, the mRNA level of Cd44 and Cd24 was significantly correlated with that of Pkm (r > 0.3, P < 0.001) (Fig. 5F). Thus, Pkm was preferentially expressed in CSC subpopulations besides stroma cells.
To determine the temporal dynamics of Pkm2 expression during tumor development, we further traced its expression from single tumor-initiating cells to established lesions. This experiment revealed markedly different dynamics between genotypes (Fig. 5G). While Pkm2 was constantly expressed during the establishment of CTNNB1+RAS lesions, it was only expressed from a later time point in CTNNB1+MYC lesions and was expressed at the beginning but quickly dimmed out in MYC+RAS lesions. This dynamic pattern was concealed in previous endpoint analyses. While CTNNB1 was reported to promote PKM2 transcription (54), our results suggest a more profound role of this function in established lesions. In contrast, RAS may induce Pkm2 expression at the tumor initiation stage. RAS alone also induced transient expression of Pkm2 during tumor initiation (fig. S9F). Together, this panel of mouse liver tumors revealed genotype- and subtype-specific abnormalities in expression of drug targets and demonstrated the ability in resolving spatial and temporal dynamics of the dysregulation.
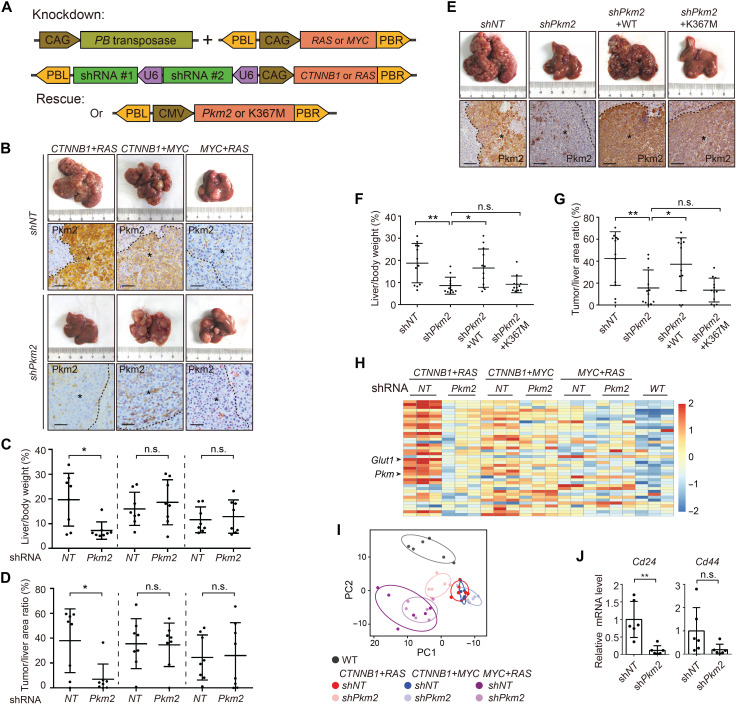
Multiplexed genome editing revealed genotype-dependent addiction to Pkm2
To determine the functional role of Pkm2 in tumor cells, we designed a multiplexed genome editing strategy, in which two Pkm2 shRNAs were expressed simultaneously with CTNNB1 or RAS (Fig. 6A). The shRNAs were designed to target a region specific to Pkm2, and nonspecific effect on Pkm1 was excluded (fig. S10A). Knockdown of Pkm2 potently suppressed CTNNB1+RAS tumors as indicated by liver/body weight ratio and quantification of tumor burden from liver sections (Fig. 6, B to D). However, a similar effect was absent in MYC+RAS or CTNNB1+MYC tumors. Successful knockdown of Pkm2 was confirmed by IHC stainings (Fig. 6B), and in CTNNB1+RAS tumors, simultaneous down-regulation of Pkm1 was observed (fig. S10B), possibly caused by indirect effect due to altered microenvironment. No compensatory expression of Pkm1 was observed (fig. S10B). Two experiments further confirmed that inhibition of CTNNB1+RAS tumorigenesis was due to specific ablation of Pkm2 activity. First, individual Pkm2 shRNAs similarly repressed CTNNB1+RAS tumors (fig. S10C). Second, tumorigenesis could be rescued by ectopic expression of wild-type but not the kinase-inactive K367M mutant Pkm2 (Fig. 6, E to G). These results demonstrated a genotype-dependent essential role of Pkm2 in liver tumorigenesis.
Fig. 6. Knockdown of Pkm2 suppressed tumorigenesis induced by CTNNB1+RAS.
(A) Illustration of plasmids used for Pkm2 knockdown and rescue in vivo. (B to D) Knockdown of Pkm2 suppressed CTNNB1+RAS tumors. Representative livers at 3 months after injection are shown. Knockdown of Pkm2 was confirmed by IHC staining (B). Scale bars, 80 μm. Asterisks indicate tumors. Tumorigenesis was quantified by liver/body weight ratio (C) and tumor/liver area ratio (D). (E to G) Reexpression of Pkm2 rescued liver tumorigenesis. Pkm2 wild type (WT) or a kinase-inactive K367M mutant was expressed for rescue. Experiments were similar to (B) to (D). Scale bars, 160 μm. (H) Heatmap shows glycolytic genes (row) in indicated samples (column). (I) Knockdown of Pkm2 changed the composition of cellular metabolites in CTNNB1+RAS tumors. Untargeted metabolomic profiling was performed by GC-TOF/MS with six biological repeats for each genotype. Groups were separated by principal components analysis. (J) Knockdown of Pkm2 reduced mRNA levels of CSC markers as determined by RT-PCR. Data are presented as means ± SD of six biological repeats. Results are representative of three independent experiments. P values were calculated by Student’s t test. *P < 0.05; **P < 0.01.
Insensitivity of MYC+RAS tumors to Pkm2 knockdown was likely due to low expression of Pkm2. However, no response of CTNNB1+MYC tumors was unexpected. We found that CTNNB1+RAS tumors were unique among M2 tumors in that they not only exhibited activation of glycolysis but also demonstrated strong down-regulation of genes related to the tricarboxylic acid (TCA) cycle and oxidative phosphorylation (Fig. 2A). However, sustained expression of TCA cycle and oxidative phosphorylation genes was found in CTNNB1+MYC tumors. Thus, energy metabolism was reprogrammed more completely in CTNNB1+RAS tumors toward a typical Warburg effect, which may have led to addiction to Pkm2 and glycolysis. Knockdown of Pkm2 normalized the expression of glycolytic genes and glucose transporter Glut1 and shifted cellular metabolite composition in CTNNB1+RAS tumors (Fig. 6, H and I, and fig. S10D). However, in CTNNB1+MYC tumors, these changes were not observed. In addition, knockdown of Pkm2 caused a marked reduction of Cd24 and Cd44 in CTNNB1+RAS tumors, suggesting impairment of the CSC compartment (Fig. 6J). Together, Pkm2 plays a genotype-specific role in metabolic reprogramming and liver tumorigenesis.
DISCUSSION
The heterogeneity of PLC requires therapies guided by improved patient classification. Despite substantial efforts in molecular subtyping of PLCs, clinical use was limited by the lack of druggable targets. Preclinical models that recapitulate the genomic and phenotypical heterogeneity of PLC have been crucial for target identification and preclinical testing. In contrast to previous mouse models exhibiting unpredictable heterogeneous molecular profiles, such as those observed in the STAM and TAK1 models (21), a panel of PLC mouse models with genotype-dependent phenotypes was made by in situ genome editing of hepatocytes. Genotype-centered molecular and histopathological characteristics, as well as relevance to human cancer, are summarized in Table 1. The stability of phenotypes relies on the use of human-guided top-ranked genetic combinations as tumorigenic drivers. An exception is the two phenotypes induced by RAS, which is likely due to interplay between differential RAS expression levels with the microenvironment. Although we could not rule out rare possibilities, the hydrodynamic injection method mainly transfects hepatocytes according to previous reports (32). However, versatile histopathological types beyond HCC were consistently observed in a genotype-dependent manner, including ICC, cHCC-ICC, and HB, demonstrating the plasticity of hepatocytes and the determinant role of driver genes. Molecular profiles separated mouse tumors into three human-matched molecular subtypes regardless of their histopathological characters. Shared molecular profiles had also been observed between human HCC and ICC (27, 55). These observations suggested an exciting possibility of common molecular targeted therapy beyond histopathological subtypes. Analysis of these mouse models also revealed distinct tumor immune microenvironments in association with molecular subtypes. Although further characterization would be important, it is reasonable to speculate that these models would be valuable in testing immunotherapies because of its orthotopic and mosaic nature in immunocompetent mice. However, care should also be taken in study design considering the impact of infrequent acquired mutations and human-mouse differences in the immune system.
How genetic alterations drive PLC phenotypes remains largely unknown. We demonstrate that, to a large extent, a one-gene one-phenotype relationship does not exist. Instead, subtype-shared features could be induced by completely different driver genes. For instance, characteristic activation of DNA replication and cell cycle in M2 tumors was not restricted to tumors driven by MYC combinations. On the contrary, MYC alone or when combined with RAS could not induce a similar change. These findings highlight the limitation of isolated genomic alteration events as biomarkers and further support classification of molecular phenotypes as a promising alternative. Functional interaction between driver genes may play an important role here. While the two-hit hypothesis of tumorigenesis was demonstrated by extensive experimental evidence, the precise mechanism is elusive for most cases. By mimicking clinically observed genetic combinations, our models provide a platform for the investigation of functional interaction of driver genes. Note that some phenotypes observed in human tumors may not be recapitulated by the expression of a respective mutant driver gene due to either interplay between multiple driver genes or differential expression levels. An example is the observation of cholestasis in genotypes of frequently acquired Ctnnb1 mutation but not in genotypes using CTNNB1 transgene. Use of new methods, for instance, single-nucleotide editing of endogenous oncogenes, may further improve the models. In addition, while both germline Tp53 KO mice (Tp53 KO+MYC) and sgTp53 (sgTp53+sgPten) were used in the current model panel, extra caution should be taken in interpreting the results from Tp53 KO mice since mutation in nontumor cells may affect the phenotype, and sgTp53 would be preferred in future investigations.
Another important finding is that driver gene expression and molecular phenotypes may be subjected to dynamic change during tumorigenesis, such as that observed for Pkm2. While stage-specific gene expression plays a central role during embryonic development, proof of this concept in tumorigenesis was limited by technical difficulty in pinpointing early lesions with a determined fate toward tumorigenesis. Nevertheless, in situ genome editing based on hydrodynamic injection creates a genetically marked minor population of tumor-initiating cells. This design unveiled genotype-dependent temporal dynamics of Pkm2 expression along the development of tumors. Such phenotypical dynamics is unlikely an exception but rather a norm of tumorigenesis that could be studied using a similar approach.
The mosaic nature of genome editing through hydrodynamic injection provides an additional advantage in investigating vital genes. For instance, PKM2 was shown to provide a growth advantage of tumor cells (56). However, due to a broad role in metabolism, germline knockout of Pkm2 leads to liver damage followed by tumorigenesis (57), which prevents determination of the tumor-specific role of Pkm2. However, multiplexed genome editing allows knockdown of Pkm2 only in tumor cells, leaving the para-tumor tissue intact. Therefore, we demonstrated that Pkm2, a long-debated molecule in cancer, is required for primary solid tumors in a context-dependent manner by maintaining the CSC compartment. Noteworthy, Pkm2 was reported dispensable in liver tumors induced by MYC (58). Tumors induced by MYC+RAS or CTNNB1+MYC were also independent of Pkm2, likely due to distinct metabolic reprogramming. These observations not only highlight the importance to stratify patients for precision treatment but also demonstrate the value of a model panel with diverse phenotypes, as well as tumor-restricted multiplexed genome editing in preclinical studies. To this end, subtype- or genotype-specific alterations of drug targets revealed by this study could be further investigated. Noteworthy, cabozantinib, a multikinase inhibitor approved for HCC treatment, was found to lead to decreased Pkm2 expression and stable disease in mouse liver tumors induced by c-MET+CTNNB1 or AKT+c-MET (59). The therapeutic value of PKM2 as a target is worthy of further investigation.
Recently, Molina-Sánchez and colleagues (60) reported a study of mouse liver tumors in nine MYC- or CYNNB1-containing genotypes generated using a similar approach. They carefully investigated the influence of MYC expression levels on β-catenin activity as a mechanism of intertumor heterogeneity. However, we are more focused on comprehensive characterization of a representative panel as a widely applicable resource for basic and translational research of liver cancer. To that end, we established and characterized double amounts of genotypes by histopathological and transcriptomic profiling and additionally performed proteomic profiling and WES, which led to in-depth depiction of subtype features and M transcriptomic signatures generally applicable for classification of mouse liver tumors. Notably, there were only two overlapping genotypes between the two studies. Nevertheless, both studies support the use of mouse liver tumor panels generated by in vivo genome editing to tackle the heterogeneity problem of liver cancer. Furthermore, while the Molina-Sánchez study demonstrated the feasibility of primary cell lines from mouse tumors in evaluating drug response in vitro, we demonstrated a multiplexed genome editing strategy in functional characterization of drug targets in vivo. Thus, the studies are complementary in demonstrating the translational application of this kind of mouse liver tumor panels.
In summary, we provide an integrated resource comprising an expandable plasmid toolbox for in situ genome editing of mouse hepatocytes, a biobank of mouse liver tumors in different genotypes, and multiomic datasets revealing human-matched subtypes of mouse liver tumors. Our data can be a valuable resource for mechanistic study of heterogeneous abnormalities in liver cancer and for the evaluation of novel therapies in a preclinical setting, which may thus fill the gap in translating molecular subtyping to clinics.
MATERIALS AND METHODS
Mice and hydrodynamic injection
All animal experiments were conducted in accordance with protocols approved by the Zhejiang University Animal Care and Use Committee. Four-week-old male ICR mice were purchased from Shanghai SLAC Laboratory Animal Company. Tp53 KO mice were a gift from Y. Cang. Cre-dependent Rosa26 Cas9 knockin mice from Z. Meng were crossed with Albumin-Cre mice from Y. Cang to obtain hepatocyte-specific Cas9 transgenic mice. Hydrodynamic injection was performed as previously described (61). Briefly, mice were anesthetized by isoflurane, and then plasmid DNA suspended in sterile Ringer’s solution in a volume equal to 10% of the body weight was injected in 5 to 7 s via the tail vein of mice. The amount of injected DNA was 50 μg of transposon plasmids together with 10 μg of PB transposase plasmids (40 μg + 15 μg in addition to 20 μg of Pkm2 plasmids in rescue experiments) or 50 μg of pX330 plasmids encoding Cas9 and sgRNAs for model construction. Animals were monitored for up to a year. Animals were euthanized at specific time points after injection or when symptoms of tumorigenesis such as abdominal enlargement were evident. Livers were pictured and weighted, and tissues were then fixed or frozen for further processing.
H&E staining and IHC
Mouse livers were fixed in 10% neutral-buffered formalin for 24 hours at room temperature and processed for paraffin embedding according to standard protocols. For frozen sections, livers were directly embedded in optimum cutting temperature compound and stored at −80°C. The tissue sections were deparaffinized and rehydrated through dimethylbenzene and graded ethanol. Tissue sections were stained with H&E using standard protocols. For IHC analysis, sections were boiled for 40 min in retrieval solution or treated with proteinase K (20 μg/ml) for antigen retrieval. Subsequently, sections were stained with specific antibodies using the avidin-biotin complex system (Vector Laboratories). Table S8 details the antibodies used for IHC. Signal was detected using the ABC kit, and 3,3′-diaminobenzidine (DAB) (Vector Laboratories) was used as the substrate for color development. Sections were counterstained with hematoxylin solution. Sirius red and oil red stainings were performed using respective kits (purchased from Servicebio) following the manufacturers’ instructions.
Fluorescent multiplexed IHC
Fluorescent multiplexed IHC was performed with the Opal 7-color Manual IHC Kit (Akoya, NEL861001KT) according to the manufacturer’s protocol. In brief, slides were deparaffinized, microwave-treated in epitope retrieval buffer for 45 s at 100% power and an additional 15 min at 20% power, blocked in Opal Antibody Diluent/Block at room temperature for 10 min, and incubated with the specific primary antibody overnight at 4°C, 10 min with the secondary horseradish peroxidase (HRP)–conjugated antibody Polymer HRP Ms + Rb at room temperature, and 10 min with Opal fluorophore working solution. Slides were rinsed between staining steps with 1× tris-buffered saline with Tween 20 buffer and stripped between each round of staining via microwave treatment in antigen retrieval buffer. After the final microwave treatment, slides were stained with 4′,6-diamidino-2-phenylindole (DAPI) for 10 min followed by mounting. Images were acquired with a confocal microscope (LSM 880, Zeiss). Table S8 lists the antibodies used.
Histopathological review
Two to five mice of each genotype were subjected to histopathological review. H&E-stained slides were fully scanned into digital images for review. Images without genotype information were reviewed independently by pathologists S.W. and X.Z. Before slide review, pathological characteristics to be evaluated were discussed and criteria were set as described below mainly according to the WHO standard or specific reports. The five largest nodules (all nodules if less than five) on each section were evaluated. For heterogeneity in the degree of a phenotype, the highest degree was noted. For heterogeneity in phenotypical categories, if any category could not reach a minimum 70% of nodules, “heterogeneous” was recorded.
According to pathological characters, each mouse tumor was classified into one specific type including HB, ICC, HCC, or cHCC-ICC. For HCC, subtypes were determined on the basis of WHO classification (35). Mixed- and combined-type cHCC-ICC were determined as described (29, 62).
Tumor grade was scored on the basis of the features of cytoplasm, cytological atypia, Nuclear/Cytoplasmic (N/C) ratio, mitotic figures, and growth patterns according to the Edmondson-Steiner system (36). More specifically, grade I tumors showed eosinophilic cytoplasm with no cytological atypia, with near-normal N/C ratio and few mitotic figures, with bile droplets seen occasionally, and with trabecular growth patterns. Grade II tumors showed mild cytological atypia and high N/C ratio, with distinct nucleolus and many mitotic figures. Grade III tumors showed moderate cytological atypia with basophilic cytoplasm, with high N/C ratio and large and irregular nuclei, and with solid growth patterns. Grade IV tumors showed marked cytological atypia with scanty cytoplasm, and tumor cells were in spindle, irregular, and disordered arrangements. Special pathological features including hyaline bodies, Mallory bodies, cholestasis, macrovascular steatosis, and clear cell change were observed and scored as none or yes.
Pictures of H&E-stained human liver tumor sections with specific histopathological features were found from pathological databases of the Second Affiliated Hospital, Zhejiang University School of Medicine (Hangzhou, China) and the Children’s Hospital, Zhejiang University School of Medicine (Hangzhou, China). Samples were collected from years 2012 to 2018 with informed consent according to the Second Affiliated Hospital Ethics Committee and the Children’s Hospital Ethics Committee. The diagnosis of liver cancer was confirmed by pathological examinations.
Quantification and statistical analysis
H&E- or IHC-stained sections were scanned with a digital section scanner (KF-PRO-005) or Olympus VS120 at ×10 to ×40 magnification. Quantification was performed with ImageJ, and GraphPad was used for statistical analysis. For quantification of nodule size and number from HA or H&E stainings, nodules of >0.04 mm2 were quantified by ImageJ. K-means clustering method was used to distinguish three groups according to size and number by R package factoextra. Oil red stainings were quantified as the percentage of signal-positive area per field of view for three to five random views. Sirius red staining was quantified as the percentage of signal-positive area in total nodule area per section. IHC staining of CD45 and F4/80 was quantified as the percentage of signal-positive area in the 10 largest nodules (if available) per section. For quantification of CD3 and FOXP3, CD3+ and FOXP3+ cells were counted per square millimeter. For quantification of AFP, the IHC profile plug-in of ImageJ was used to determine the percentage contribution of four intensity categories, which were denoted as 0 (negative), 1 (low positive), 2 (positive), and 3 (high positive). Then, H score was calculated as (1 × % low positive) + (2 × % positive) + (3 × % high positive). The 10 largest nodules (if available) in each section were quantified for five mice.
Statistical analysis for comparing two experimental groups was performed using Student’s t tests. P value of <0.05 was considered statistically significant. Differences are labeled n.s. for not significant, * for P < 0.05, ** for P < 0.01, and *** for P < 0.001. Analyses were performed with Prism 8 (GraphPad Software). Kaplan-Meier curves were used to analyze survival.
WES and analysis
Genomic DNA was isolated with the TIANamp Genomic DNA Kit (Tiangen). DNA quality was verified by the following two methods: (i) DNA degradation and contamination were monitored on 1% agarose gels; (ii) DNA concentration was measured using the Qubit DNA Assay Kit and the Qubit 2.0 Fluorometer (Life Technologies). WES was performed by Novogene (Beijing, China). Exome capture was performed using the Agilent SureSelect XT Mouse All Exon Kit (Agilent Technologies) according to the manufacturer’s protocols. After capture and enrichment with index tags, products were purified using AMPure XP system (Beckman Coulter, Beverly, USA) and quantified using the Agilent high sensitivity DNA assay on the Agilent Bioanalyzer 2100 System. DNA libraries were subjected to 150–base pair (bp) paired-end sequencing on the Illumina NovaSeq platform. Following raw data quality control processing, high-quality clean data were aligned to the reference genome mm10 by BWA software (63) and Samblaster (64). Sambamba was used to mark PCR duplicates. SAMtools (1.0) was used to call single-nucleotide polymorphisms and indels. Somatic SNV, indel, and CNV were detected by muTect (1.1.4), Strelka (v1.0.13), and Control-FREEC (v6.7), respectively. SMGs were determined by MuSiC (Q < 0.05) (65). All genes with somatic mutations in mouse tumors were further searched for known driver genes of human cancer consolidated from five sources (66–70). Only variants with “pass” status were considered for further analysis. Mutation spectrum and mutation signatures in tumor samples were analyzed by non-negative matrix factorization (0.22) (71) based on the frequency of mutations with 96 categories. Documented 30 mutational signatures were obtained from COSMIC v3.1. The sources of somatic mutations were as follows: exome sequencing data from mouse liver tumor models (total N = 14, from RAS II, PI3K+F/C, Tp53 KO+MYC, F/C, and MYC+F/C, N = 3, 3, 3, 2, and 3, respectively) and human HCC (total N = 1222, from LIHC-INSERM, LIHC-AMC, LIHC-TCGA, and LINC-JP, N = 241, 231, 358, and 392, respectively) (7, 11, 72, 73). The proportion of mutations for each of the signatures within each genotype of mouse tumors and human HCC cohorts was calculated on the basis of their total mutation burden. Tumor mutation burden (TMB) was defined as the total number of nonsynonymous somatic coding mutations within exonic regions. To calculate TMB, the total number of nonsynonymous somatic coding mutations counted was divided by the size of the coding sequence region captured by the HGSC VCRome 2.1 (LIHC-TCGA) or SureSelect XT Mouse All Exon Kit (mouse liver tumors). TMB was calculated with the software maftools (R package), which was log10-transformed, and a scatterplot was subsequently generated by maftools (R package). VAF was calculated using the PyClone method as described (74). Oncoprint was used to visualize multiple genomic alteration events by ComplexHeatmap (R package). Percentage of alteration events for each gene and the total number of mutations for each sample are shown. Genetic alterations of human HCC were analyzed and visualized by cBioPortal and OncoPrint, respectively.
RNA-seq and analysis
RNA was isolated with TRIzol reagent (Takara). RNA-seq was performed by Novogene (Beijing, China). RNA quality was verified by the following four methods: (i) RNA degradation and contamination were monitored on 1% agarose gels; (ii) RNA purity was checked using the NanoPhotometer spectrophotometer (IMPLEN); (iii) RNA concentration was measured using the Qubit RNA Assay Kit and the Qubit 2.0 Fluorometer (Life Technologies); (iv) RNA integrity was assessed using the RNA Nano 6000 Assay Kit and the Bioanalyzer 2100 System (Agilent Technologies). A total of 1 μg of RNA per sample was used as input material. Sequencing libraries were generated using the NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, USA) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. In brief, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in the NEBNext First Strand Synthesis Reaction Buffer (5×). First-strand complementary DNA (cDNA) was synthesized using random hexamer primers and M-MuLV Reverse Transcriptase [ribonuclease (RNase) H minus]. Second-strand cDNA synthesis was subsequently performed using DNA polymerase I and RNase H. In the reaction buffer, deoxynucleotide triphosphates (dNTPs) with deoxythymidine triphosphate (dTTP) were replaced by deoxyuridine triphosphate (dUTP). Remaining overhangs were converted into blunt ends via exonuclease/polymerase treatment. After 3′ adenylation, NEBNext Adaptors with a hairpin loop structure were ligated to the DNA fragments to prepare for hybridization. To preferentially select cDNA fragments of 250 to 300 bp, the library fragments were purified using an AMPure XP system (Beckman Coulter). Size-selected, adaptor-ligated cDNA was treated with 3 μl of USER Enzyme (NEB) at 37°C for 15 min followed by 5 min at 95°C before PCR. PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers, and Index (X) Primer. At last, products were purified (AMPure XP system), and library quality was assessed on the Agilent Bioanalyzer 2100 system. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using the TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina NovaSeq platform, and 150-bp paired-end reads were generated. A reference genome (mm10) index was built using Hisat2 v2.0.5, and paired-end clean reads were aligned to the reference genome using Hisat2 v2.0.5. Raw count data per gene were calculated using featureCounts v1.5.0-p3. Then, FPKM (fragments per kilobase of transcript per million mapped reads) of each gene was calculated on the basis of the length of the gene and read count mapped to this gene. Transcription levels quantified by FPKM were filtered to remove genes whose expression was quantified as zero in more than 75% of the tumor samples. A pseudo FPKM value of 1 was added to each element before the statistical test, and gene quantifications were subsequently log2-transformed. The DEseq2 (as implemented in R software) was used to assess genes that were differentially expressed between tumors and normal liver, followed by multiple testing using the Benjamini-Hochberg procedure. Genes with Benjamini-Hochberg–adjusted P < 0.05 and fold change [expressed as log2 (ratio of average FPKM in tumor versus normal groups)] ≥ 1 were considered differentially expressed. The batch effect of RNA-seq data was evaluated by principal components analysis and corrected with ComBat in SVA (R package, www.bioconductor.org/packages/release/bioc/html/sva.html).
For shNT and shPkm2 samples, RNA isolation and quality control procedures were the same as above, and RNA-seq was performed by Vazyme Biotech (Nanjing, China). A total of 1 μg of RNA per sample was used as input material. Sequencing libraries were generated using the VAHTS Stranded mRNA-seq Library Prep Kit for Illumina (Vazyme, NR602) following the instructions. Total RNA was purified using poly-T oligo-attached magnetic beads. The products were fragmented using divalent cations under elevated temperature in Vazyme Frag/Prime Buffer. The fragments were produced into first-strand cDNA by reverse transcriptase and random primers. Strand specificity is achieved by replacing dTTP with dUTP, and second-strand cDNA synthesis was subsequently performed using DNA polymerase I and RNase H. Then, cDNA fragments were end repaired with the addition of a single “A” base at the 3′ end of each strand, ligated with the special sequencing adapters (Vazyme, N803). The products were purified with VAHTSTM DNA Clean Beads (Vazyme, N411) to get the appropriate size for sequencing. PCR was performed, and aimed products were purified finally. Library concentration was measured and preliminarily quantified by the Qubit RNA Assay Kit in Qubit 3.0. Insert size was assessed using the Agilent Bioanalyzer 2100 system and qualified by Step One Plus Real-Time PCR system (ABI, USA). The clustering of the index-coded samples was performed on the cBot Cluster Generation System (Illumina, USA), and then library preparations were sequenced on an Illumina HiSeq X Ten platform and 150-bp paired-end module. Raw reads were filtered by removing reads containing adapter, poly-N, and low-quality reads for subsequent analysis. The reference genome index was built using Bowtie2 (v2.2.9), and paired-end clean reads were aligned to the reference genome (mm10) using TopHat (v2.1.1). The mapped reads of each sample were assembled using Cufflinks (v2.2.1) with a reference-based approach. FPKMs for coding genes were calculated by Cuffdiff (v1.3.0) for each sample.
Alternative splicing analysis and percentage of inclusion calculation
For alternative splicing analysis, RNA-seq bam files were imported into Integrative Genome Browser (IGV). Reads were mapped to mouse reference genome (mm10) and form a Sashimi plot to reflect the read coverage of target exon.
The percentage of inclusion (PSI) statistics was used to quantify the inclusion evidence of exon skipping and for the alternative use of 5′ or 3′ splicing sites. For exon-skipping events, PSI considering splice-junction reads (S1 and S2) of all isoforms in the region between two constitutive exons (C1 and C2) reflected values of individual alternative exons. PSI = (S1 + S2)/sum (C1 ~ C2).
Quantitative proteomic analysis
Minced liver tissues were subjected to protein extraction in lysis buffer [1% sodium deoxycholate, 10 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), 40 mM 2-chloroacetamide, and 100 mM tris-HCl (pH 8.5)] supplemented with protease inhibitors (Thermo Fisher Scientific). Lysates were centrifuged at 16,000g for 10 min at 4°C. Supernatants were digested with sequencing-grade trypsin. Tryptic peptides were dried in a vacuum concentrator (Thermo Fisher Scientific) and then analyzed by liquid chromatography–MS/MS (LC-MS/MS).
Peptide samples were loaded onto a trap column (100 μm × 2 cm, homemade; particle size, 3 μm; pore size, 120 Å; SunChrom, USA), separated by a heated homemade silica microcolumn (150 μm × 30 cm; particle size, 1.9 μm; pore size, 120 Å; SunChrom, USA) with a gradient of 6 to 40% mobile phase B (acetonitrile and 0.1% formic acid) at a flow rate of 600 nl/min for 150 min. LC-MS/MS was performed on an Orbitrap Fusion mass spectrometer using an Orbitrap mass analyzer at a mass resolution of 120,000 (Thermo Fisher Scientific, Rockford, IL, USA) coupled with an Easy-nLC 1000 nanoflow LC system using an ion trap analyzer with the AGC target at 5 × 103 and maximum injection time at 35 ms (Thermo Fisher Scientific). The MS/MS analysis was performed under a data-dependent mode. One full scan was followed by up to 20 data-dependent MS/MS scans with higher-energy collision dissociation (normalized collision energy of 35%). Dynamic exclusion time was set with 25 s.
All raw data from MS were submitted to Firmiana (75), which were then searched against the 2013 version of the National Center for Biotechnology Information (NCBI) human Refseq protein database with Mascot 2.3 search engine (Maxtrix Science Inc.). The mass tolerances for precursor and product ions were 20 parts per million and 0.5 Da, respectively. Allowed missed cleavages were up to two. Fixed modification of the search was set to cysteine carbamidomethylation. Variable modifications included N-acetylation and oxidation of methionine. Charges of precursor ions were limited to +2, +3, and +4. The protein-level FDR was limited to 1%. Protein quantification was done by calculating iBAQ values, which were then converted into iFOT values by dividing the iBAQ value of each protein by the sum of all iBAQ values in the same sample (76).
Single-cell RNA sequencing and analysis
Fresh liver tumors from three mice were mechanically dissociated and digested in 5 ml of RPMI 1640 medium with collagenase/dispase (2 mg/ml) and 0.001% deoxyribonuclease I, conducted by TDK1 standard procedure in a GentleMACS Octo instrument (Miltenyi Biotec). Digestion was stopped by adding 5 ml of RPMI 1640 with 2% fetal bovine serum (FBS), and tissue pieces were removed by using a 70-μm strainer. Cell suspension was centrifuged at 450g for 5 min, and cell pellet was resuspended in ACK lysis buffer and incubated for 5 min to lyse red blood cells. Dead cells were cleaned by adding 3 ml of 30% Percoll and centrifuged at 450g for 5 min, and cell pellet was resuspended in 2% FBS/phosphate-buffered saline. To reduce the fraction of immune cells in the sample, single-cell suspensions were stained with anti-CD45 for fluorescence-activated cell sorting, and CD45-low cells were harvested up to 106 cells. Cell viability of ≥80% was confirmed by trypan blue staining.
For scRNA-seq, chromium microfluidic chips were loaded with cell suspension with 3′ chemistry and barcoded with a 10× Chromium Controller (10X Genomics). RNA was reverse-transcribed from the barcoded cells, and sequencing libraries were constructed with reagents from a Chromium Single Cell 3′ v3 reagent kit (10X Genomics) according to the manufacturer’s instructions. Sequencing was performed with NovaSeq according to the manufacturer’s instructions (Illumina). CellRanger 4.0.0 pipeline (10X Genomics) was used to demultiplex and map raw reads to mouse reference genome mm10 using default parameters. Filtering, barcode counting, and UMI counting were conducted by CellRanger 3.1.0. It used the Chromium cellular barcodes to generate feature barcode matrices, determine clusters, and perform gene expression analysis and achieved 3993 median UMI counts and 1628 median genes per cell. Genes were filtered when expressing a cell number of <3. Cells with a detected gene number of <200 or >6500, mitochondrion gene percentage of <30, hemoglobin gene percentage of <5, and doublet nucleus were removed by package Seurat (v3.1.0) and DoubletFinder. Data were subsequently log-normalized (divided by the total expression and amplified scaling factor 10,000) before further analyses. For clustering, highly variable genes were (top 2000) selected and the principal components based on those genes were used to build a graph, which was segmented with a resolution of 0.6. UMAP was demonstrated by Loupe browser software (10X Genomics). Annotation of cell types was performed using SingleR 1.0.0 with default parameters using a collection of mouse bulk RNA-seq datasets as the reference compendium (51, 52).
mRNA-protein correlation
Spearman correlation coefficient was applied to measure the correlation between mRNA expression and protein abundance for each mRNA-protein pair across 22 samples. In addition, P value corresponding to the correlation coefficient was computed and adjusted by the FDR correction. Significance of the correlation pair was determined, based on an FDR cutoff of 0.05. Interaction networks were generated by Cytoscape v.3.7.21 (77) from differentially expressed mRNA or proteins.
Transcriptomic and proteomic subtype identification in mouse tumors
Hierarchical consensus clustering was performed on transcriptomic or proteomic expression matrix of tumors to generate subgroups. Before the consensus clustering analysis, we first performed a centered log ratio transformation to facilitate the interpretation of the expression data. The top 5000 variable (by median absolute deviation) genes or proteins were filtered to run consensus clustering. Consensus clustering was implemented on the ConsensusClusterPlus R package (78) with the following settings: number of repetitions = 1000 bootstraps; pItem = 0.8 (resampling 80% of any sample); pFeature = 1 (resampling 100% of any gene/protein); and hierarchical clustering with up to six clusters. The number of clustering was determined by the average pairwise consensus matrix within consensus clusters and the delta plot of the relative change in the area under the cumulative distribution function (CDF) curve. We selected three clusters as the best solution for the consensus matrix with k = 3 or k = 4 deemed to be a cleanest separation among clusters in both transcriptome and proteome, but the consensus CDF and delta plot exhibited that there was little increase in area for k = 3 compared to k = 4. On the basis of the evidence above, transcriptomic and proteomic data were both clustered into three groups.
Identification of subtype signature genes or proteins
To identify molecular signatures of M1-M3 subtypes, we compared genes and proteins expressed in each subtype against those in other subtypes and normal tissue. The statistical significance was calculated by R/Bioconductor package DEseq2 v.1.26.0. For each comparative analysis, the genes/proteins were required to be expressed in at least 25% of the samples in one of the subtypes. To define subtype signature genes/proteins, the following cutoff criteria were used: (i) All Benjamini-Hochberg–adjusted P values should be less than 0.001 for mRNA and 0.05 for protein compared to the other subtypes and normal tissue; (ii) fold change [expressed as log2(ratio of average mRNA expression/protein abundance between subtypes)] ≥ 1. Signature proteins were subjected to pathway analyses using R/Bioconductor package clusterProfiler46 v.3.14.1 (79).
Expression of human HCC signatures and pathway activities in mouse tumors
The NTP algorithm was applied to stratify mouse tumors by using previously defined signature genes/proteins and its derived mRNA/protein expression matrix. All statistical analyses were performed in R (v.3.6.2), and a significance level of FDR < 0.05 was used. GSVA (R package) (80) and MCP-counter (R package) (34) were used to calculate GSVA scores and immune infiltration scores for different molecular functions and immune signatures.
Analysis of drug targets
Targets of drugs approved or under development were downloaded from DrugBank. Deregulation was defined as meeting the following cutoff criteria in at least one of the genotypes. For drug targets: (i) mRNA fold change [expressed as log2 (ratio of average mRNA expression in one genotype to that in normal tissue)] > 2, and Benjamini-Hochberg–adjusted P values less than 0.01, or (ii) protein fold change [expressed as log2 (ratio of average protein abundance in one genotype to that in normal tissue)] > 5, and Benjamini-Hochberg–adjusted P values less than 0.01.
Human-mouse transcriptome similarity
We used TROM algorithm (TROM R package) (20) to assess the similarity between HCC mouse models and TCGA-LIHC patients. We use the following settings to select associated orthologs of molecular characteristic genes: Z scores > 1.5 and subsequently comparing the biological samples by testing the overlap of their associated genes. TROM scores were calculated by the following formula
A larger TROM score represents greater similarity. A significant similarity was defined as TROM score > 1.3 (Bonferroni-corrected P value of <0.05).
Analysis of metabolites by MS
Untargeted metabolomic profiling was performed on the XploreMET platform using gas chromatography time-of-flight MS (GC-TOF/MS) system by Metabo-Profile Biotechnology Co. Ltd. (Shanghai, China) as previously described (81). Briefly, 50 mg of each sample was homogenized in 50 μl of 50% prechilled methanol and 10 μl of internal standard with 25 mg of prechilled zirconium oxide beads and centrifuged at 14,000g for 20 min at 4°C. Supernatant was transferred to an autosampler vial. Chloroform/methanol (175 μl; 3:1) was added and centrifuged at 14,000g for 20 min. Supernatant was evaporated briefly by a CertrilVap vacuum concentrator and lyophilized by a FreeZone freeze-dryer. The derivatization and injection of samples were carried on a robotic multipurpose sample MPS2 (Gerstel, Muehlheim, Germany) with dual heads. Samples were derivatized with 50 μl of methoxyamine (20 mg/ml in pyridine) at 30°C for 2 hours, and 50 μl of N-methyl-N-(trimethylsilyl)trifluoroacetamide (1% Trimethylchlorosilane) was added for another hour by sample preparation head. Meanwhile, samples were injected with head. The flow rate of gas was 1.0 ml/min by the carrier helium. The transfer interface and injection temperature were 270°C, while the source was 220°C. Electron impact ionization (70 eV) in the full-scan model (mass/charge ratio, 50 to 500) was used for measurement. A metabolite database ADAP was used to compare the retention indices and MS data with known structure reference standards, which were commercially purchased. ADAP was also used for raw MS data processing.
For targeted metabolome, samples were homogenized and centrifuged, and the supernatants were combined and subjected to automated sample derivatization and separation using a robotic multipurpose sample MPS2 (Gerstel, Muehlheim, Germany) with a double head. Metabolites were quantified using GC-TOF/MS system operating in electron ionization mode (Pegasus HT, Leco Corp., St. Joseph, MO, USA). The 73 individual bile acid standards were prepared in methanol, ultrapure water, or sodium hydroxide solution at a concentration of 5 mM. The concentrations of individual bile acids were 2500, 500, 250, 50, 10, 2.5, or 1 nM. Each 180-μl acetonitrile/methanol standard was added to 5 μl of samples in a tube and shaken at 1500g for 20 min. The mixture was centrifuged, and 20 μl of supernatant was taken and mixed well with 50 μl of mobile phase. After brief shaking for 5 min and centrifugation at 13,500g for 20 min, supernatant was transferred to a 96-well plate for LC-MS analysis. Data were processed with proprietary software XploreMET (v2.0, Metabo-Profile, Shanghai, China).
Tissue culture and transfection
H2.35 cells [American Type Culture Collection (ATCC), CRL-1995] were cultured in Dulbecco’s minimum essential medium (DMEM; Gibco) containing 10% FBS (YEASEN), penicillin/streptomycin (50 μg/ml; Gibco), and 200 nM dexamethasone. Human embryonic kidney 293T (ATCC, CRL-11268) and Neuro-2a cells (ATCC, CCL-131) were grown in DMEM containing 10% FBS and penicillin/streptomycin (50 μg/ml) at 37°C and 5% CO2 atmosphere. The above cells were routinely examined with Myco-Blue Mycoplasma Detector (Vazyme), and no mycoplasma pollution was found. Transfection was performed using YEASEN transfection reagent according to the manufacturer’s protocols.
Detection of indels by T7E1 assay
For in vitro efficiency test, cells transfected with pX330 plasmids were treated with puromycin (3 μg/ml; InvivoGen) for 48 hours, and genomic DNA was extracted using a TIANamp Genomic DNA kit following the manufacturer’s instructions. For detection of indels in tumors, fresh tumor samples were used to extract genomic DNA. The specific targeted regions were PCR-amplified using the PrimeSTAR HS DNA Polymerase (Takara). Purified PCR products were heat-denatured and reannealed using a thermocycler to produce DNA heteroduplexes. DNA heteroduplexes were incubated with T7 endonuclease I (NEB) for 30 min at 37°C. Electrophoresis in 2% agarose gel was carried out, and bands were quantified by ImageJ. For each lane, the fraction of the PCR product cleaved (fcut) was calculated as (fcut) = (b + c)/(a + b + c), where a is the integrated intensity of the undigested PCR product, and b and c are the integrated intensities of each cleavage product. Then, indel occurrence can be estimated with the following formula, based on the binomial probability distribution of duplex formation: indel (%) = 100 × (1 − ).
Quantitative PCR
RNA of liver samples was extracted using an RNAiso Plus kit (Takara), and cDNA was synthesized using PrimeScript RT Master Mix (Takara). qPCR was performed with the Hifair III One Step RT-qPCR SYBR Green Kit (YEASEN) on a Bio-Rad CFX96 system. Succinate dehydrogenase complex flavoprotein subunit A (Sdha) transcript levels were used as an internal control. For the absolute quantification of Pkm1 and Pkm2 mRNA levels, standard curves of the gradient dilution of Pkm1 and Pkm2 DNA templates and threshold values (Ct) were determined by RT-PCR. The primers for qPCR are listed in table S8.
Immunoblotting
Immunoblotting was performed using standard protocol. Briefly, cells were lysed with 1% SDS lysis buffer, and protein concentration was determined using the BCA Assay Kit (Thermo Fisher Scientific). Samples were resolved by SDS–polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes (Millipore), and blotted with the desired antibodies. Protein expression was detected by ECL Detection Reagent.
Construction of plasmids for in situ genome editing
The CAG-PB transposase and PB (CAG-RFP) DS plasmids were previously described (82). PB-CAG-HA-CTNNB1-S33/37A, PB-CAG-Myri-AKT-HA, PB-CAG-EGFR-L858R-HA, PB-CAG-HA-MYC, PB-CAG-HA-NRAS-G12V, and PB-CAG-Myri-HA-PIK3CA-H1047R were constructed by cloning the coding regions into the Spe I and Bam HI sites of the PB (CAG-RFP) DS vector. PB-CAG-JAK1-S703I-Ollas and PB-CAG-Ollas-NFE2L2-T80K plasmids were constructed by amplifying respective cDNAs from pLVX vectors and ligated into the Spe I–digested PB (CAG-) DS empty vector. The PB-EF1α-MET-HA, PB-EF1α-HA-NICD, PB-CMV-Ollas-CCND1-FGF19-HA, PB-CMV-Ollas-Pkm2, and PB-CMV-Ollas-Pkm2-K367M plasmids were constructed by excising the corresponding fragments from pLVX vectors and ligated into PB (CAG-RFP) DS plasmid cut by Eco RI. gRNAs targeting Rb1, Arid1a, Axin1, Rps6ka3, Ncor1, Arid2, and Cdkn2a were designed by the Zhang laboratory CRISPR Design Tool, and the gRNAs targeting Tp53 and Pten were reported (14). Annealed oligonucleotides were ligated into the pEP-KO vector (a gift from X. Zongping) digested with Sap I or pX330-U6-Chimeric_BB-CBh-hSpCas9 vector (Addgene, #42230) digested with Bbs I. To make the PB-CAG-HA-CTNNB1-S33/37A-U6-shNT-U6-shNT, PB-CAG-HA-CTNNB1-S33/37A-U6-shPkm2#1-U6-shPkm2#1, PB-CAG-HA-CTNNB1-S33/37A-U6-shPkm2#2-U6-shPkm2#2, PB-CAG-HA-CTNNB1-S33/37A-U6-shPkm2#1-U6-shPkm2#2, PB-CAG-HA-NRAS-G12V-U6-shNT-U6-shNT, and PB-CAG-HA-NRAS-G12V-U6-shPkm2#1-U6-shPkm2#2 plasmids, shRNAs targeting mouse Pkm2 were first designed by the Broad Institute Genetic Perturbation Platform Web portal. Annealed oligonucleotides were ligated into the pLKO.1 vector (Addgene, #10807) digested with Age I and Eco RI. Two individual U6-shRNA fragments, including two targeting Pkm2 or two tandem repeats of U6-shNT, were then PCR-amplified from the pLKO.1 vector and were then ligated with Swa I–digested PB-CAG-HA-CTNNB1-S33/37A or PB-CAG-HA-NRAS-G12V plasmids using Gibson assembly. The sequences of sgRNA and shRNA are available in table S8.
Acknowledgments
We thank Y. Cang for Tp53 KO and Albumin-Cre mice, Z. Meng for Cre-dependent Rosa26 Cas9 knockin mice, T. Xu and X. Wu for the piggyBac system, L. Shen and Q. Zhou for advice on bioinformatic analysis, L. Chen for assistance on T7E1 assay, and the core facility of the Life Sciences Institute for technical assistance.
Funding: This work was supported by grants to B.Z. from the National Natural Science Foundation of China Key Project (81730069), the National Key R&D Program of China (2017YFA0504502), the National Natural Science Foundation of China General Project (31970726), Natural Science Foundation of Zhejiang key project (LZ21C070002), and the Fundamental Research Funds for the Central Universities.
Author contributions: B.Z. conceived the project, and B.Z. and T.L. designed the study. M.T., Y. Zhao, J.Z., D.G., S.H., Y. Zhang, G.Z., S.L., C.W., H.Y., X.C., and L.L. performed experiments and analyzed data. S.W. and X.Z. performed pathological evaluation. J.Q., M.L., and N.Z. designed and performed proteomic experiments. X.B., J.J., and X.-H.F. provided reagents and conceptual advice. B.Z., M.T., Y. Zhao, and J.Z. wrote the manuscript with comments from all authors.
Competing interests: A patent regarding the generation and application of an HB mouse model was filed by Zhejiang University on 30 September 2021 with serial number 202111162460.0, and B.Z., M.T., and Y. Zhang are listed as coinventors of this patent application (pending). The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The raw data of WES and RNA-seq have been deposited in NCBI Sequence Read Archive (SRA) with accession code PRJNA674008. Data for proteomic profiling by MS/MS can be obtained from iProX database (https://iprox.org/) under accession number IPX0002646000. Reagents related to this paper including plasmids can be provided by the corresponding authors pending scientific review and a completed material transfer agreement. Requests should be submitted to the corresponding authors.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S8
REFERENCES AND NOTES
- 1.Forner A., Reig M., Bruix J., Hepatocellular carcinoma. Lancet 391, 1301–1314 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Llovet J. M., Ricci S., Mazzaferro V., Hilgard P., Gane E., Blanc J. F., de Oliveira A. C., Santoro A., Raoul J. L., Forner A., Schwartz M., Porta C., Zeuzem S., Bolondi L., Greten T. F., Galle P. R., Seitz J. F., Borbath I., Häussinger D., Giannaris T., Shan M., Moscovici M., Voliotis D., Bruix J., Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 359, 378–390 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Ray E. M., Sanoff H. K., Optimal therapy for patients with hepatocellular carcinoma and resistance or intolerance to sorafenib: Challenges and solutions. J. Hepatocell. Carcinoma 4, 131–138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahipal A., Tella S. H., Kommalapati A., Lim A., Kim R., Immunotherapy in hepatocellular carcinoma: Is there a light at the end of the tunnel? Cancers (Basel) 11, 1078 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finn R. S., Qin S., Ikeda M., Galle P. R., Ducreux M., Kim T. Y., Kudo M., Breder V., Merle P., Kaseb A. O., Li D., Verret W., Xu D. Z., Hernandez S., Liu J., Huang C., Mulla S., Wang Y., Lim H. Y., Zhu A. X., Cheng A. L., Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N. Engl. J. Med. 382, 1894–1905 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Blechacz B., Gores G. J., Cholangiocarcinoma: Advances in pathogenesis, diagnosis, and treatment. Hepatology 48, 308–321 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulze K., Imbeaud S., Letouzé E., Alexandrov L. B., Calderaro J., Rebouissou S., Couchy G., Meiller C., Shinde J., Soysouvanh F., Calatayud A. L., Pinyol R., Pelletier L., Balabaud C., Laurent A., Blanc J. F., Mazzaferro V., Calvo F., Villanueva A., Nault J. C., Bioulac-Sage P., Stratton M. R., Llovet J. M., Zucman-Rossi J., Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 47, 505–511 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujimoto A., Furuta M., Totoki Y., Tsunoda T., Kato M., Shiraishi Y., Tanaka H., Taniguchi H., Kawakami Y., Ueno M., Gotoh K., Ariizumi S. I., Wardell C. P., Hayami S., Nakamura T., Aikata H., Arihiro K., Boroevich K. A., Abe T., Nakano K., Maejima K., Sasaki-Oku A., Ohsawa A., Shibuya T., Nakamura H., Hama N., Hosoda F., Arai Y., Ohashi S., Urushidate T., Nagae G., Yamamoto S., Ueda H., Tatsuno K., Ojima H., Hiraoka N., Okusaka T., Kubo M., Marubashi S., Yamada T., Hirano S., Yamamoto M., Ohdan H., Shimada K., Ishikawa O., Yamaue H., Chayama K., Miyano S., Aburatani H., Shibata T., Nakagawa H., Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat. Genet. 48, 500–509 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Totoki Y., Tatsuno K., Covington K. R., Ueda H., Creighton C. J., Kato M., Tsuji S., Donehower L. A., Slagle B. L., Nakamura H., Yamamoto S., Shinbrot E., Hama N., Lehmkuhl M., Hosoda F., Arai Y., Walker K., Dahdouli M., Gotoh K., Nagae G., Gingras M. C., Muzny D. M., Ojima H., Shimada K., Midorikawa Y., Goss J. A., Cotton R., Hayashi A., Shibahara J., Ishikawa S., Guiteau J., Tanaka M., Urushidate T., Ohashi S., Okada N., Doddapaneni H., Wang M., Zhu Y., Dinh H., Okusaka T., Kokudo N., Kosuge T., Takayama T., Fukayama M., Gibbs R. A., Wheeler D. A., Aburatani H., Shibata T., Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat. Genet. 46, 1267–1273 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Nakamura H., Arai Y., Totoki Y., Shirota T., Elzawahry A., Kato M., Hama N., Hosoda F., Urushidate T., Ohashi S., Hiraoka N., Ojima H., Shimada K., Okusaka T., Kosuge T., Miyagawa S., Shibata T., Genomic spectra of biliary tract cancer. Nat. Genet. 47, 1003–1010 (2015). [DOI] [PubMed] [Google Scholar]
- 11. Cancer Genome Atlas Research Network, comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 169, 1327–1341.e23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., Zhang F., Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X., Calvisi D. F., Hydrodynamic transfection for generation of novel mouse models for liver cancer research. Am. J. Pathol. 184, 912–923 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xue W., Chen S., Yin H., Tammela T., Papagiannakopoulos T., Joshi N. S., Cai W., Yang G., Bronson R., Crowley D. G., Zhang F., Anderson D. G., Sharp P. A., Jacks T., CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 514, 380–384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prowse K. R., Greider C. W., Developmental and tissue-specific regulation of mouse telomerase and telomere length. Proc. Natl. Acad. Sci. U.S.A. 92, 4818–4822 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harding J. J., Nandakumar S., Armenia J., Khalil D. N., Albano M., Ly M., Shia J., Hechtman J. F., Kundra R., el Dika I., Do R. K., Sun Y., Kingham T. P., D’Angelica M. I., Berger M. F., Hyman D. M., Jarnagin W., Klimstra D. S., Janjigian Y. Y., Solit D. B., Schultz N., Abou-Alfa G. K., Prospective genotyping of hepatocellular carcinoma: Clinical implications of next-generation sequencing for matching patients to targeted and immune therapies. Clin. Cancer Res. 25, 2116–2126 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlson C. M., Frandsen J. L., Kirchhof N., McIvor R. S., Largaespada D. A., Somatic integration of an oncogene-harboring Sleeping Beauty transposon models liver tumor development in the mouse. Proc. Natl. Acad. Sci. U.S.A. 102, 17059–17064 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan B., Malato Y., Calvisi D. F., Naqvi S., Razumilava N., Ribback S., Gores G. J., Dombrowski F., Evert M., Chen X., Willenbring H., Cholangiocarcinomas can originate from hepatocytes in mice. J. Clin. Invest. 122, 2911–2915 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacks T., Remington L., Williams B. O., Schmitt E. M., Halachmi S., Bronson R. T., Weinberg R. A., Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4, 1–7 (1994). [DOI] [PubMed] [Google Scholar]
- 20.Li W. V., Chen Y., Li J. J., TROM: A testing-based method for finding transcriptomic similarity of biological samples. Stat. Biosci. 9, 105–136 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dow M., Pyke R. M., Tsui B. Y., Alexandrov L. B., Nakagawa H., Taniguchi K., Seki E., Harismendy O., Shalapour S., Karin M., Carter H., Font-Burgada J., Integrative genomic analysis of mouse and human hepatocellular carcinoma. Proc. Natl. Acad. Sci. U.S.A. 115, E9879–E9888 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Y., Sun A., Zhao Y., Ying W., Sun H., Yang X., Xing B., Sun W., Ren L., Hu B., Li C., Zhang L., Qin G., Zhang M., Chen N., Zhang M., Huang Y., Zhou J., Zhao Y., Liu M., Zhu X., Qiu Y., Sun Y., Huang C., Yan M., Wang M., Liu W., Tian F., Xu H., Zhou J., Wu Z., Shi T., Zhu W., Qin J., Xie L., Fan J., Qian X., He F.; Chinese Human Proteome Project (CNHPP) Consortium , Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature 567, 257–261 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Gao Q., Zhu H., Dong L., Shi W., Chen R., Song Z., Huang C., Li J., Dong X., Zhou Y., Liu Q., Ma L., Wang X., Zhou J., Liu Y., Boja E., Robles A. I., Ma W., Wang P., Li Y., Ding L., Wen B., Zhang B., Rodriguez H., Gao D., Zhou H., Fan J., Integrated proteogenomic characterization of hbv-related hepatocellular carcinoma. Cell 179, 561–577.e22 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Hoshida Y., Nijman S. M. B., Kobayashi M., Chan J. A., Brunet J. P., Chiang D. Y., Villanueva A., Newell P., Ikeda K., Hashimoto M., Watanabe G., Gabriel S., Friedman S. L., Kumada H., Llovet J. M., Golub T. R., Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 69, 7385–7392 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyault S., Rickman D. S., de Reyniès A., Balabaud C., Rebouissou S., Jeannot E., Hérault A., Saric J., Belghiti J., Franco D., Bioulac-Sage P., Laurent-Puig P., Zucman-Rossi J., Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology 45, 42–52 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Hoshida Y., Nearest template prediction: A single-sample-based flexible class prediction with confidence assessment. PLOS ONE 5, e15543 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woo H. G., Lee J. H., Yoon J. H., Kim C. Y., Lee H. S., Jang J. J., Yi N. J., Suh K. S., Lee K. U., Park E. S., Thorgeirsson S. S., Kim Y. J., Identification of a cholangiocarcinoma-like gene expression trait in hepatocellular carcinoma. Cancer Res. 70, 3034–3041 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Connell L. C., Harding J. J., Shia J., Abou-Alfa G. K., Combined intrahepatic cholangiocarcinoma and hepatocellular carcinoma. Chin. Clin. Oncol. 5, 66 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Xue R., Chen L., Zhang C., Fujita M., Li R., Yan S.-M., Ong C. K., Liao X., Gao Q., Sasagawa S., Li Y., Wang J., Guo H., Huang Q.-T., Zhong Q., Tan J., Qi L., Gong W., Hong Z., Li M., Zhao J., Peng T., Lu Y., Lim K. H. T., Boot A., Ono A., Chayama K., Zhang Z., Rozen S. G., Teh B. T., Wang X. W., Nakagawa H., Zeng M.-S., Bai F., Zhang N., Genomic and transcriptomic profiling of combined hepatocellular and intrahepatic cholangiocarcinoma reveals distinct molecular subtypes. Cancer Cell 35, 932–947.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sohn B. H., Shim J. J., Kim S. B., Jang K. Y., Kim S. M., Kim J. H., Hwang J. E., Jang H. J., Lee H. S., Kim S. C., Jeong W., Kim S. S., Park E. S., Heo J., Kim Y. J., Kim D. G., Leem S. H., Kaseb A., Hassan M. M., Cha M., Chu I. S., Johnson R. L., Park Y. Y., Lee J. S., Inactivation of hippo pathway is significantly associated with poor prognosis in hepatocellular carcinoma. Clin. Cancer Res. 22, 1256–1264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yimlamai D., Christodoulou C., Galli G. G., Yanger K., Pepe-Mooney B., Gurung B., Shrestha K., Cahan P., Stanger B. Z., Camargo F. D., Hippo pathway activity influences liver cell fate. Cell 157, 1324–1338 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu F., Song Y., Liu D., Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 6, 1258–1266 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Kurebayashi Y., Ojima H., Tsujikawa H., Kubota N., Maehara J., Abe Y., Kitago M., Shinoda M., Kitagawa Y., Sakamoto M., Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology 68, 1025–1041 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Becht E., Giraldo N. A., Lacroix L., Buttard B., Elarouci N., Petitprez F., Selves J., Laurent-Puig P., Sautès-Fridman C., Fridman W. H., de Reyniès A., Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 17, 218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagtegaal I. D., Odze R. D., Klimstra D., Paradis V., Rugge M., Schirmacher P., Washington K. M., Carneiro F., Cree I. A.; The WHO Classification of Tumours Editorial Board , The 2019 WHO classification of tumours of the digestive system. Histopathology 76, 182–188 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edmondson H. A., Steiner P. E., Primary carcinoma of the liver: A study of 100 cases among 48,900 necropsies. Cancer 7, 462–503 (1954). [DOI] [PubMed] [Google Scholar]
- 37.von Schweinitz D., Schmidt D., Fuchs J., Welte K., Pietsch T., Extramedullary hematopoiesis and intratumoral production of cytokines in childhood hepatoblastoma. Pediatr. Res. 38, 555–563 (1995). [DOI] [PubMed] [Google Scholar]
- 38.Cairo S., Armengol C., de Reyniès A., Wei Y., Thomas E., Renard C. A., Goga A., Balakrishnan A., Semeraro M., Gresh L., Pontoglio M., Strick-Marchand H., Levillayer F., Nouet Y., Rickman D., Gauthier F., Branchereau S., Brugières L., Laithier V., Bouvier R., Boman F., Basso G., Michiels J. F., Hofman P., Arbez-Gindre F., Jouan H., Rousselet-Chapeau M. C., Berrebi D., Marcellin L., Plenat F., Zachar D., Joubert M., Selves J., Pasquier D., Bioulac-Sage P., Grotzer M., Childs M., Fabre M., Buendia M. A., Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell 14, 471–484 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Yamashita T., Forgues M., Wang W., Kim J. W., Ye Q., Jia H., Budhu A., Zanetti K. A., Chen Y., Qin L. X., Tang Z. Y., Wang X. W., EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 68, 1451–1461 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Tao J., Xu E., Zhao Y., Singh S., Li X., Couchy G., Chen X., Zucman-Rossi J., Chikina M., Monga S. P. S., Modeling a human hepatocellular carcinoma subset in mice through coexpression of met and point-mutant β-catenin. Hepatology 64, 1587–1605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.S. H. Lee, S. Y. Yim, J. J. Shim, J. S. Lee, in Hepatocellular Carcinoma: Translational Precision Medicine Approaches, Y. Hoshida, Ed. (Humana Press, 2019), pp. 109–123. [PubMed] [Google Scholar]
- 42.Woo H. G., Park E. S., Cheon J. H., Kim J. H., Lee J. S., Park B. J., Kim W., Park S. C., Chung Y. J., Kim B. G., Yoon J. H., Lee H. S., Kim C. Y., Yi N. J., Suh K. S., Lee K. U., Chu I. S., Roskams T., Thorgeirsson S. S., Kim Y. J., Gene expression-based recurrence prediction of hepatitis B virus-related human hepatocellular carcinoma. Clin. Cancer Res. 14, 2056–2064 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Seehawer M., Heinzmann F., D’Artista L., Harbig J., Roux P. F., Hoenicke L., Dang H., Klotz S., Robinson L., Doré G., Rozenblum N., Kang T. W., Chawla R., Buch T., Vucur M., Roth M., Zuber J., Luedde T., Sipos B., Longerich T., Heikenwälder M., Wang X. W., Bischof O., Zender L., Necroptosis microenvironment directs lineage commitment in liver cancer. Nature 562, 69–75 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang T. W., Yevsa T., Woller N., Hoenicke L., Wuestefeld T., Dauch D., Hohmeyer A., Gereke M., Rudalska R., Potapova A., Iken M., Vucur M., Weiss S., Heikenwalder M., Khan S., Gil J., Bruder D., Manns M., Schirmacher P., Tacke F., Ott M., Luedde T., Longerich T., Kubicka S., Zender L., Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 479, 547–551 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Calderaro J., Couchy G., Imbeaud S., Amaddeo G., Letouzé E., Blanc J. F., Laurent C., Hajji Y., Azoulay D., Bioulac-Sage P., Nault J. C., Zucman-Rossi J., Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J. Hepatol. 67, 727–738 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Boeva V., Popova T., Bleakley K., Chiche P., Cappo J., Schleiermacher G., Janoueix-Lerosey I., Delattre O., Barillot E., Control-FREEC: A tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 28, 423–425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicholes K., Guillet S., Tomlinson E., Hillan K., Wright B., Frantz G. D., Pham T. A., Dillard-Telm L., Tsai S. P., Stephan J. P., Stinson J., Stewart T., French D. M., A mouse model of hepatocellular carcinoma: Ectopic expression of fibroblast growth factor 19 in skeletal muscle of transgenic mice. Am. J. Pathol. 160, 2295–2307 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zucman-Rossi J., Benhamouche S., Godard C., Boyault S., Grimber G., Balabaud C., Cunha A. S., Bioulac-Sage P., Perret C., Differential effects of inactivated Axin1 and activated beta-catenin mutations in human hepatocellular carcinomas. Oncogene 26, 774–780 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Wishart D. S., Feunang Y. D., Guo A. C., Lo E. J., Marcu A., Grant J. R., Sajed T., Johnson D., Li C., Sayeeda Z., Assempour N., Iynkkaran I., Liu Y., Maciejewski A., Gale N., Wilson A., Chin L., Cummings R., Le D., Pon A., Knox C., Wilson M., DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 46, D1074–D1082 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dayton T. L., Jacks T., Vander Heiden M. G., PKM2, cancer metabolism, and the road ahead. EMBO Rep. 17, 1721–1730 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aran D., Looney A. P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R. P., Wolters P. J., Abate A. R., Butte A. J., Bhattacharya M., Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benayoun B. A., Pollina E. A., Singh P. P., Mahmoudi S., Harel I., Casey K. M., Dulken B. W., Kundaje A., Brunet A., Remodeling of epigenome and transcriptome landscapes with aging in mice reveals widespread induction of inflammatory responses. Genome Res. 29, 697–709 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Z., Li M., Zhang L., Zhao H., Şahin Ö., Chen J., Zhao J. J., Songyang Z., Yu D., Oncogenic kinase-induced PKM2 tyrosine 105 phosphorylation converts nononcogenic PKM2 to a tumor promoter and induces cancer stem-like cells. Cancer Res. 78, 2248–2261 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang J., Cao R., Zhang Y., Xia Y., Zheng Y., Li X., Wang L., Yang W., Lu Z., PKM2 dephosphorylation by Cdc25A promotes the Warburg effect and tumorigenesis. Nat. Commun. 7, 12431 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chaisaingmongkol J., Budhu A., Dang H., Rabibhadana S., Pupacdi B., Kwon S. M., Forgues M., Pomyen Y., Bhudhisawasdi V., Lertprasertsuke N., Chotirosniramit A., Pairojkul C., Auewarakul C. U., Sricharunrat T., Phornphutkul K., Sangrajrang S., Cam M., He P., Hewitt S. M., Ylaya K., Wu X., Andersen J. B., Thorgeirsson S. S., Waterfall J. J., Zhu Y. J., Walling J., Stevenson H. S., Edelman D., Meltzer P. S., Loffredo C. A., Hama N., Shibata T., Wiltrout R. H., Harris C. C., Mahidol C., Ruchirawat M., Wang X. W.; TIGER-LC Consortium , Common molecular subtypes among asian hepatocellular carcinoma and cholangiocarcinoma. Cancer Cell 32, 57–70.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Christofk H. R., Vander Heiden M. G., Harris M. H., Ramanathan A., Gerszten R. E., Wei R., Fleming M. D., Schreiber S. L., Cantley L. C., The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230–233 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Dayton T. L., Gocheva V., Miller K. M., Israelsen W. J., Bhutkar A., Clish C. B., Davidson S. M., Luengo A., Bronson R. T., Jacks T., Vander Heiden M. G., Germline loss of PKM2 promotes metabolic distress and hepatocellular carcinoma. Genes Dev. 30, 1020–1033 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Méndez-Lucas A., Li X., Hu J., Che L., Song X., Jia J., Wang J., Xie C., Driscoll P. C., Tschaharganeh D. F., Calvisi D. F., Yuneva M., Chen X., Glucose catabolism in liver tumors induced by c-MYC can be sustained by various PKM1/PKM2 ratios and pyruvate kinase activities. Cancer Res. 77, 4355–4364 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shang R., Song X., Wang P., Zhou Y., Lu X., Wang J., Xu M., Chen X., Utpatel K., Che L., Liang B., Cigliano A., Evert M., Calvisi D. F., Chen X., Cabozantinib-based combination therapy for the treatment of hepatocellular carcinoma. Gut 70, 1746–1757 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Molina-Sánchez P., de Galarreta M. R., Yao M. A., Lindblad K. E., Bresnahan E., Bitterman E., Martin T. C., Rubenstein T., Nie K., Golas J., Choudhary S., Bárcena-Varela M., Elmas A., Miguela V., Ding Y., Kan Z., Grinspan L. T., Huang K.-L., Parsons R. E., Shields D. J., Rollins R. A., Lujambio A., Cooperation between distinct cancer driver genes underlies intertumor heterogeneity in hepatocellular carcinoma. Gastroenterology 159, 2203–2220.e14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo X., Zhao Y., Yan H., Yang Y., Shen S., Dai X., Ji X., Ji F., Gong X. G., Li L., Bai X., Feng X. H., Liang T., Ji J., Chen L., Wang H., Zhao B., Single tumor-initiating cells evade immune clearance by recruiting type II macrophages. Genes Dev. 31, 247–259 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Allen R. A., Lisa J. R., Combined liver cell and bile duct carcinoma. Am. J. Pathol. 25, 647–655 (1949). [PMC free article] [PubMed] [Google Scholar]
- 63.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Faust G. G., Hall I. M., SAMBLASTER: Fast duplicate marking and structural variant read extraction. Bioinformatics 30, 2503–2505 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dees N. D., Zhang Q., Kandoth C., Wendl M. C., Schierding W., Koboldt D. C., Mooney T. B., Callaway M. B., Dooling D., Mardis E. R., Wilson R. K., Ding L., MuSiC: Identifying mutational significance in cancer genomes. Genome Res. 22, 1589–1598 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kandoth C., McLellan M. D., Vandin F., Ye K., Niu B., Lu C., Xie M., Zhang Q., McMichael J. F., Wyczalkowski M. A., Leiserson M. D. M., Miller C. A., Welch J. S., Walter M. J., Wendl M. C., Ley T. J., Wilson R. K., Raphael B. J., Ding L., Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tamborero D., Gonzalez-Perez A., Perez-Llamas C., Deu-Pons J., Kandoth C., Reimand J., Lawrence M. S., Getz G., Bader G. D., Ding L., Lopez-Bigas N., Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci. Rep. 3, 2650 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vogelstein B., Papadopoulos N., Velculescu V. E., Zhou S., Diaz L. A., Kinzler K. W., Cancer genome landscapes. Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sondka Z., Bamford S., Cole C. G., Ward S. A., Dunham I., Forbes S. A., The COSMIC cancer gene census: Describing genetic dysfunction across all human cancers. Nat. Rev. Cancer 18, 696–705 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martínez-Jiménez F., Muiños F., Sentís I., Deu-Pons J., Reyes-Salazar I., Arnedo-Pac C., Mularoni L., Pich O., Bonet J., Kranas H., Gonzalez-Perez A., Lopez-Bigas N., A compendium of mutational cancer driver genes. Nat. Rev. Cancer 20, 555–572 (2020). [DOI] [PubMed] [Google Scholar]
- 71.Alexandrov L. B., Nik-Zainal S., Wedge D. C., Campbell P. J., Stratton M. R., Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 3, 246–259 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ahn S. M., Jang S. J., Shim J. H., Kim D., Hong S. M., Sung C. O., Baek D., Haq F., Ansari A. A., Lee S. Y., Chun S. M., Choi S., Choi H. J., Kim J., Kim S., Hwang S., Lee Y. J., Lee J. E., Jung W. R., Jang H. Y., Yang E., Sung W. K., Lee N. P., Mao M., Lee C., Zucman-Rossi J., Yu E., Lee H. C., Kong G., Genomic portrait of resectable hepatocellular carcinomas: Implications of RB1 and FGF19 aberrations for patient stratification. Hepatology 60, 1972–1982 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Zhang J., Baran J., Cros A., Guberman J. M., Haider S., Hsu J., Liang Y., Rivkin E., Wang J., Whitty B., Wong-Erasmus M., Yao L., Kasprzyk A., International Cancer Genome Consortium Data Portal—A one-stop shop for cancer genomics data. Database (Oxford) 2011, bar026 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roth A., Khattra J., Yap D., Wan A., Laks E., Biele J., Ha G., Aparicio S., Bouchard-Côté A., Shah S. P., PyClone: Statistical inference of clonal population structure in cancer. Nat. Methods 11, 396–398 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feng J., Ding C., Qiu N., Ni X., Zhan D., Liu W., Xia X., Li P., Lu B., Zhao Q., Nie P., Song L., Zhou Q., Lai M., Guo G., Zhu W., Ren J., Shi T., Qin J., Firmiana: Towards a one-stop proteomic cloud platform for data processing and analysis. Nat. Biotechnol. 35, 409–412 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Ni X., Tan Z., Ding C., Zhang C., Song L., Yang S., Liu M., Jia R., Zhao C., Song L., Liu W., Zhou Q., Gong T., Li X., Tai Y., Zhu W., Shi T., Wang Y., Xu J., Zhen B., Qin J., A region-resolved mucosa proteome of the human stomach. Nat. Commun. 10, 39 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T., Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wilkerson M. D., Hayes D. N., ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics 26, 1572–1573 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu G., Wang L. G., Han Y., He Q. Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hanzelmann S., Castelo R., Guinney J., GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pan L., Qiu Y., Chen T., Lin J., Chi Y., Su M., Zhao A., Jia W., An optimized procedure for metabonomic analysis of rat liver tissue using gas chromatography/time-of-flight mass spectrometry. J. Pharm. Biomed. Anal. 52, 589–596 (2010). [DOI] [PubMed] [Google Scholar]
- 82.Ding S., Wu X., Li G., Han M., Zhuang Y., Xu T., Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122, 473–483 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Tables S1 to S8