Abstract
Two genes are synthetic lethal if mutations in both genes result in impaired cell viability, while mutation of either gene does not affect the cell survival. The potential usage of synthetic lethality (SL) in anticancer therapeutics has attracted many researchers to identify synthetic lethal gene pairs. To include newly identified SLs and more related knowledge, we present a new version of the SynLethDB database to facilitate the discovery of clinically relevant SLs. We extended the first version of SynLethDB database significantly by including new SLs identified through Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) screening, a knowledge graph about human SLs, a new web interface, etc. Over 16 000 new SLs and 26 types of other relationships have been added, encompassing relationships among 14 100 genes, 53 cancers, 1898 drugs, etc. Moreover, a brand-new web interface has been developed to include modules such as SL query by disease or compound, SL partner gene set enrichment analysis and knowledge graph browsing through a dynamic graph viewer. The data can be downloaded directly from the website or through the RESTful Application Programming Interfaces (APIs). Database URL: https://synlethdb.sist.shanghaitech.edu.cn/v2.
Introduction
Synthetic lethality (SL), initially described in Drosophila as recessive lethality (1), is a type of gene–gene interaction such that the perturbation of both genes causes the loss of cell viability, while the perturbation of either gene alone will not affect the cell viability (2). SL offers a strategy for cancer medicine by identifying new antibiotic or therapeutic targets (3, 4, 5). By inhibiting the SL partner of a gene with cancer-specific alteration, we can kill cancer cells and spare normal cells, thereby reducing the side effect of the treatment (6, 7). To discover SL gene pairs as a gold mine of cancer drug targets, researchers have applied various techniques, including chemical screening (8), RNA interference (RNAi) screening (9, 10, 11, 12, 13), Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) screening (14, 15) and bioinformatics methods (16, 17, 18, 19).
The first version of SynLethDB released in 2016 contains 34 089 SL gene pairs and is the first comprehensive database of SLs (20). It collects SL pairs for human and four model species, i.e. mouse, fruit fly, worm and yeast, from biochemical assays, public databases (21, 22), computational predictions (23) and text mining. In addition, it provides a statistical analysis module to evaluate the druggability and efficacy of SL pairs upon drug treatments by analyzing the large-scale drug sensitivity data. Recently, SynLethDB has been used as ground-truth SL data in various studies. For example, Liany et al. (24), Cai et al. (25) and Das et al. (26) used SynLethDB to train and test their computational SL prediction methods. Hu et al. (27) used SynLethDB to evaluate their method for de novo identification of synergistic optimal control nodes as candidate targets for combination therapy. Wang et al. (28) used the SLs in SynLethDB to investigate the link between SL interactions and drug sensitivity of cancer cells. Cui et al. used the SL data from SynLethDB in their web-based tool called siGCD (29) for analysis and visualization of the interactions among genes, cells and drugs associated with survival in human cancers.
Many CRISPR-based screening experiments have been conducted after 2015 and generated a large amount of data. Combinatorial CRISPR-based screening has been used to study genetic interactions, including the identification of SL interactions (14, 15, 30, 31, 32, 33). Computational methods such as GEMINI (34) were proposed to identify SL gene pairs from these screening data. GEMINI is a variational Bayesian approach proposed to identify SLs from combinatorial CRISPR screens. Data-driven method ISLE (17) searches in the lab-identified candidate SLs by tumor molecular profiles, patient clinical data and gene phylogeny relations to find out the clinical SLs. These wet-lab experiments and computational methods provided further evidence for some existing SLs or discovered new SLs that had not been included in the first version of SynLethDB.
To discover SL-based anticancer drug targets and clinical SLs, it is highly desirable to consider the relationships among SLs, cancers and drugs. Several studies combined SLs with the information about cancers and cancer–drug interactions to discover cancer-specific SLs for new cancer therapies. SL-BioDP (35) provides an online tool based on a data-driven method to predict SL interactions by mining cancer’s genomic and chemical interactions. However, it only supports the prediction of SL partners of the 623 genes belonging to 10 hallmark cancer pathways and 18 types of cancers. Synthetic Lethality Knowledge Graph (SLKG) (36) is a knowledge graph that contains seven kinds of relationships among genes, cancers and drugs. Unlike SL-BioDP, SLKG collects SL pairs from literature and existing databases instead of by prediction. Moreover, SLKG is also used to identify the best repurposable drug candidates and drug combinations. In addition to the relationships among SLs, drugs and cancers, their various features are also useful for discovering SLs and anticancer therapy. Taking the features of genes as an example, the co-expression, Gene Ontology (GO) semantic similarity and shared pathways between genes are commonly used features for predicting SLs (16, 26, 37, 38, 39). In addition, several tools have been developed to curate these features, such as GO terms and pathways associated with specific genes, anatomies and symptoms of cancers and the side effects and pharmacologic classes of drugs. For example, the Hetio package from Hetionet (40) provides a way to integrate different resources into a single data structure. We are motivated to use these tools to construct an integrative knowledge graph to better describe SL pairs.
In this paper, we present SynLethDB 2.0 to include newly discovered SLs and provide more related knowledge to help identify clinically relevant SLs (Figure 1). It is a significant expansion of the first version by adding 16 781 new SL gene pairs and integrating a biomedical knowledge graph, including 10 kinds of biomedical entities other than gene and 26 kinds of relationships for drug discovery other than SL. The 37 341 entities and 1 405 652 relationships were used to create a knowledge graph and stored in a graph database. A user-friendly website interface with new functionalities for data browsing, visualization and analysis has also been developed for users to browse the data and knowledge graph in SynLethDB. For example, users can search SLs by a disease or a compound, perform pathway or GO term enrichment for SL partners of a gene and inspect the connections between two genes in an interactive viewer.
Figure 1.
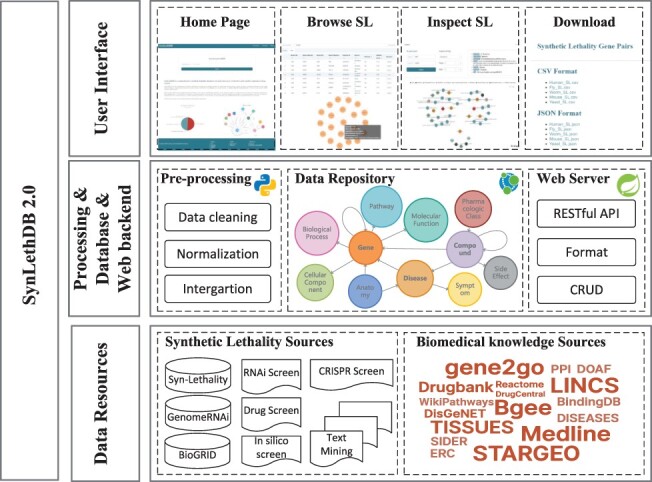
Architecture of SynLethDB 2.0. The bottom layer shows the data sources of SLs and other biomedical knowledge. The middle layer shows the data preprocessing steps, database storage and web server. The top layer shows the main functional modules of the user interface.
Materials and methods
Data sources
The new version of SynLethDB contains 50 868 SL pairs which include 35 943 of Homo sapiens, 381 of Mus musculus, 439 of Drosophila melanogaster, 105 of Caenorhabditis elegans and 14 000 of Saccharomyces cerevisiae. The first source of the new SL pairs is the research papers on identifying SLs via wet-lab experiments. Using the ‘synthetic lethal’ as a keyword for searching in PubMed, 293 related papers published during years from 2015 to 2019 were extracted for further manual collection of new SL pairs. The second source is public databases containing SL data such as GenomeRNAi (21) and BioGRID (22). The third source is the SL pairs predicted from wet-lab screen data by computational methods such as GEMINI (34). For each SL pair, we annotated its species, references to PubMed as supporting evidence, data source type, cell lines and confidence score. Synthetic rescue (SR) means mutation in one gene rescues the cell from lethality or growth defect caused by a mutation in another gene (41). It is related to drug resistance (42) and can be seen as the opposite relationship to SL. We collected 16 207 SR gene pairs and 5798 non-synthetic lethal (non-SL) gene pairs from the above three sources, which can be used as negative samples to train SL prediction models. Non-synthetic lethal pairs could be SR or other relationships. Some gene pairs show up in both SL and non-SL datasets, depending on the different cell lines or cancer types.
In addition to the above three kinds of gene pairs, we added 24 types of relationships between genes and other entities (e.g. drugs and cancers). These relationships include gene–compound associations, gene–cancer associations and other features about genes, cancers and drugs. We manually obtained a list of 53 cancers and curated these relationships from public databases with Python scripts from the open-source project of Hetionet (40). First, we used the Python script from Hetionet to collect the relationships from data sources. Hetionet collects the relationships between genes, drugs and diseases. We added the relationships among GO terms, pathways and SL genes into the dataset. Every type of relationship was processed into an independent CSV file at first and then integrated into the Neo4j database for persistent storage with the package Py2Neo. Finally, we constructed a knowledge graph to describe human SL gene pairs and the other 26 types of relationships, named SynLethKG (Synthetic Lethality Knowledge Graph).
Data quality improvement
In addition to collecting the data about SLs, we have also improved the annotation quality of SL gene pairs. First, we collected the SL entries from different sources into one TSV format file to facilitate subsequent unified processing. Second, we completed the missing identifiers of the genes. With annotation packages from Bioconductor, which provide genome annotations for different species, we completed the missing Entrez ID of a gene by its gene symbol or completed the missing gene symbol by its Entrez ID. Third, we deleted entries that still lacked gene IDs or gene symbols. These entries lacked gene IDs or gene symbols because they contained incorrect gene symbols or IDs, which may be due to recording errors from the original sources. After that, we downloaded the latest version of the gene annotations from the Gene Entrez database (43) on the National Center for Biotechnology Information (NCBI) FTP site and then deleted the SL entries that contain genes deprecated by the current NCBI Gene Entrez database. Lastly, we removed duplicate SL entries that have the same genes and PubMed IDs. The SL entries that contain the gene SL pair but were from different sources are merged into one entry.
Furthermore, unlike in the first version of the database where SLs were stored in the form of records in a table, in the new graph database SLs are stored as undirected edges between two gene nodes. Hence, only one SL entry can be stored between a pair of genes. The species, references to PubMed, supporting evidence, cell lines and other relevant information about an SL entry are stored as properties of the edge, and the gene annotation information is stored as the node properties.
Construction of graph database
In the previous version of SynLethDB, we used the relational database management system, MySQL, to store the data. In this version, we chose to use a graph database system, Neo4j, to store SL pairs and related biomedical knowledge. Graph database is more suitable for many-to-many relationships. The relational database computes the relationships at query time through expensive operations such as JOIN. By contrast, the graph database stores the relationships as edges which processes and queries the relationships more efficiently. We used the Java framework of Spring Data Neo4j, as middleware for object-graph mapping and data persistence. All the queries are accessible to users through the front-end interface in the form of Representational State Transfer (REST) API using Hypertext Application Language as the media type.
The front end of SynLethDB is a single-page application built using VueJS and Element UI. When changing the tabs, only the required content is updated instead of the whole page, enabling faster responses. It allows us to cache searching queries from users and create a better user experience until the web session is updated. Interactive and expandable graph viewers are developed with the ECharts JavaScript library to visualize the query results as connections in the graph database.
We used Nginx as a reverse proxy to hide the real host of SynLethDB for web security. In the deployment of SynLethDB, we followed the microservices architecture to get a higher scalability and reduce downtime through fault isolation. The database, web interface and web server are all hosted in independent docker containers and arranged by Docker Compose. These services can be easily migrated, automatically deployed and quickly restored, which ensures high accessibility of SynLethDB.
Confidence scores of SL pairs
The SLs in our database were collected from different sources, including manually checked publications, existing databases, computational predictions and text mining. According to the types of sources, we took two steps, i.e. quantification and integration, to calculate the final confidence scores, following the strategy of SynLethDB 1.0 (20). The main differences from the previous version are in the individual scores in the quantification step and the weight factors in the integration step.
In the quantification step, since an SL gene pair could be identified by using different experimental or computational methods, an individual quantitative score is assigned to each type of evidence. In this new version, to incorporate the new source of CRISPR screening, we reset the individual scores as shown in Table 1. If there are multiple pieces of evidence of the same type supporting an SL record, we adopted the probability disjunction formula to combine the individual scores as follows:
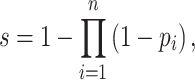 |
(1) |
where pi is the individual score for the ith evidence and s is the combined quantitative score of a specific type of source as listed in Table 1.
Table 1.
Quantitative Scores Assigned to SLs According to Experimental Methods.
| Method | Score |
|---|---|
| CRISPR interference | 0.85 |
| Drug inhibition | 0.75 |
| RNAi | 0.75 |
| Low throughput | 0.80 |
| High throughput | 0.50 |
In the integration step, we integrated the scores of different types of sources into a normalized confidence score for every SL pair. Different weights were assigned according to the source types. The integration formula for the final confidence score of an SL pair is
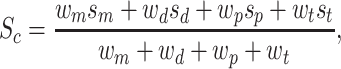 |
(2) |
where the default values of wm, wd, wp and wt were set to 0.8, 0.5, 0.3 and 0.2, as the weight factors of biochemical experiment, existing databases, computational prediction and text mining, respectively. Note that users can customize these weight values according to their own experience or preference when querying and ranking the SLs on the web interface.
Gene set enrichment analysis
Given a gene g, let G denote the set of all SL partner genes of g. The enrichment analysis is to find out the pathways and GO terms from each of the three ontologies (i.e. biological process, molecular function and cellular component) that occur significantly more frequently than randomly in the gene set G. We implemented two enrichment analysis methods based on the degree information and P-value, respectively.
Degree-based gene set enrichment analysis
An SLPR (Synthetic Lethality PageRank) score inspired by PageRank (44) was computed for each pathway or GO term associated with the gene set G. The pathways and GO terms can be ranked based on their SLPR scores. A larger SLPR score means that a pathway or GO term is more closely associated with the gene set. The SLPR score is defined as:
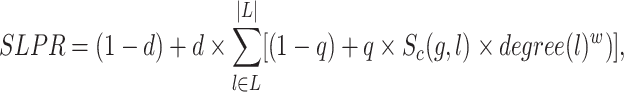 |
(3) |
where d is a damping factor set to 0.85, q is another damping factor set to 0.8 and w is set to −1 to reflect a negative correlation. For a specific pathway or GO term, L represents the subset of genes in set G that are directly connected with it. Given a gene 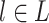 ,
, 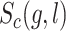 is the confidence score of the SL pair (g, l), degree(l) is the number of pathways or GO terms associated with l.
is the confidence score of the SL pair (g, l), degree(l) is the number of pathways or GO terms associated with l.
P-value-based gene set enrichment analysis
Assume that M is the number of genes in G and N is the number of genes having SL partners in the whole database. Given a specific pathway or GO term, n is the total number of genes associated with it and m is the number of genes in G associated with it. To show the enrichment of the gene set G with the pathway or GO term, we calculate a P-value as follows (45):
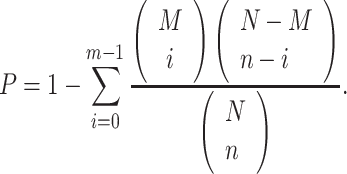 |
(4) |
Thus, we attain a list of pathways or GO terms sorted in order of the P-values. A smaller P-value means that G is more enriched with the given pathway or GO term.
SynLethDB 2.0 portal
A user-friendly web interface has been developed for SynLethDB to facilitate data visualization, analysis and interpretation. Compared with SynLethDB 1.0, SynLethDB 2.0 provides more ways for searching and browsing SLs. For example, users may wish to start with a type of cancer to find SLs associated with the cancer. Thus, in SynLethDB 2.0, in addition to searching by genes, users can also search SLs by a disease name. SynLethDB 2.0 also allows users to browse the part of knowledge graph related to an SL gene pair with a graph viewer to help understand the mechanism underlying the SL. Besides, it allows users to customize the integration weights of confidence scores so that those SLs more reliable or interesting to users would be prioritized at the top of the list of query results. In addition to the enrichment analysis tool based on P-values, SynLethDB 2.0 also provides a gene set enrichment analysis tool based on the graph degree, which ranks pathways and GO terms by their relevance to a gene set inferred based on the network topology of the knowledge graph.
On the home page of the website of SynLethDB, we provide a general introduction to the database, as well as the search bar for looking up SLs by gene symbols or gene IDs. Other functionalities of SynLethDB can be accessed by menu tabs on the website as follows.
Searching and browsing the SLs
In the first version of SynLethDB, users could only search for SLs by genes. In this new version, we collected 14 116 gene–cancer relationships and 56 921 gene–compound relationships for those genes involved in SLs from DisGeNET (46), DrugBank (47) and BindingDB (48). Based on these new data, we offer two new options for searching, namely, ‘search SL by disease’ and ‘search SL by compound’, and provide the autocomplete function to the list of all available cancers or compounds in SynLethDB. The searching results are shown in a table viewer.
Customizable confidence scores for SLs
A confidence score reflects an SL’s credibility based on its sources, which can be used to rank SLs. As mentioned earlier, we use a two-step scoring procedure (i.e. quantification and integration) to assign a confidence score based on the sources of the SL. In the quantification step, we assigned the quantitative scores to SL pairs according to their experimental methods as shown in Table 1. In the integration step, we provide default values for the weight factors but allow users to customize these weights to facilitate them to extract the SLs of a certain type of source that they are most interested in. When searching and browsing SLs by genes, users can adjust the weight factors of source types and rank results by the confidence scores.
Searching and browsing the knowledge graph SynLethKG
SynLethKG contains relationships that describe various features for genes, cancers and drugs. With the ‘Inspect SL’ functionality, all these relationships are categorized by their node types and can be browsed through an interactive graph viewer. Starting with SL genes to be inspected, users only need to click on the node they are about to inspect, and the graph viewer can fetch and visualize the results. The type of relationships and the number of edges to be displayed can be specified by the users. Properties of the nodes and edges, such as data sources and entity IDs, can also be viewed through an infobox in the upper right corner.
Gene set enrichment analysis of SL partners
We developed two methods for gene set enrichment analysis based on P-values and node degrees, respectively. Both methods take a gene symbol as input and conduct gene set enrichment analysis for the SL partners of this gene. The output includes the rank of the pathways and GO terms separately. A higher ranking of a pathway or a GO term indicates that the SL partners of this gene are more enriched with this pathway or GO term. The P-value-based enrichment analysis tool ranks the results by P-value calculated in Equation (4), and a lower P-value corresponds to higher ranking. Meanwhile, the degree-based enrichment analysis tool ranks the pathways and GO terms based on the SLPR score as calculated in Equation (3), and a higher SLPR score corresponds to a higher ranking.
Data access and download
We provide a download page to make it easy for users to retrieve a large amount of data. All the SL gene pairs are classified by species and can be downloaded in either CSV or JSON format. We provide the files of SynLethKG in the formats of CSV, JSON and GraphML for users to download. In particular, the datasets in GraphML format can be imported to other software tools such as Gephi and Cytoscape for analysis and visualization. For users who prefer the triplet format, we also provide a CSV file that contains all the relationships in the format (source, relationship, target). All the data can be freely accessed and downloaded without a login requirement. RESTful Application Programming Interfaces (APIs) are also provided for users to access and analyze the data by running the scripts in programming languages such as Python and R.
User manual
To lower the learning curve for new users of SynLethDB, we offer a web page containing a user manual, which gives an introduction to every functionality of SynLethDB, as well as examples of using the web interface, RESTful APIs and SynLethKG.
Results
Comparison with other databases
In this section, we compare SynLethDB 2.0 with existing databases of SL. SLKG (36) is a knowledge graph about SL, and it focuses on drug repositioning for tumor-specific treatments based on the concepts of SL and synthetic dosage lethality (SDL). It contains the relationships among genes, drugs and cancers. There are 19 987 SLs and 3039 SDLs in SLKG. Compared with SLKG, SynLethKG is focused on collecting existing SLs and related knowledge, and it includes more types of relationships and a more comprehensive list of SLs. SynLethDB 2.0 contains 35 943 human SLs and, in addition to relationships among genes, drugs and cancer types, it contains the relationships between genes and pathways, drugs and pharmacologic classes and so on. SynLethDB 1.0 is the first comprehensive database of SL. Based on that, SynLethDB 2.0 is even more comprehensive and user-friendly, as we have made extensive and important updates to the database in the following aspects.
Firstly, SynLethDB 2.0 is arguably the most up-to-date and most comprehensive database for SL. It contains 50 868 SL pairs in total, almost doubling the number of SL pairs in SynLethDB 1.0. In particular, SynLethDB 2.0 contains 35 943 human SLs, 381 mouse SLs, 439 fly SLs, 14 000 yeast SLs and 105 worm SLs as shown in Table 2. The number of human SLs in SynLethDB 2.0 is almost 1.8 times that in SynLethDB 1.0. Consistent with SynLethDB 1.0, we also provide the HUGO Gene Nomenclature Committee gene symbols, Entrez gene IDs, PubMed IDs of its original publications, types of sources and the confidence score calculated according to the sources for each SL pair in SynLethDB 2.0. Note that we updated the confidence scores by considering new sources of SLs such as CRISPR screening and allowing user-defined weight factors.
Table 2.
Comparison of Statistics Between Two Versions of SynLethDB.
| SynLethDB 1.0 | SynLethDB 2.0 | |
|---|---|---|
| # Human SLs | 19 952 | 35 943 |
| # Mouse SLs | 366 | 381 |
| # Fly SLs | 423 | 439 |
| # Worm SLs | 105 | 105 |
| # Yeast SLs | 13 241 | 14 000 |
| KG | No | Yes |
| Annotation | SLs only | Yes |
| Offline dataset | Yes | Yes |
| RESTful APIs | No | Yes |
Secondly, SynLethDB 2.0 provides more types of biomedical knowledge. SynLethDB 1.0 comprises mainly the SL relationships between genes. By adding the knowledge graph SynLethKG, SynLethDB 2.0 contains much more types of entities and relationships, including biological processes, pathways, molecule functions and cellular components for genes, pharmacologic classes and side effects for drugs, symptoms and anatomies for cancers. Overall, there are 37 341 entities (nodes) and 1 405 652 relationships (edges) in SynLethKG as shown in Table 3. The types of relationships and their numbers are listed in Table 4. In addition, SynLethDB 2.0 retains the annotations of SLs from SynLethDB 1.0 and corrects them. It also adds annotations to the nodes and edges in SynLethKG, such as the name of entity, the data source and the link to entity in the original data source, and other annotations such as the organisms of genes and the thresholds used when extracting the relationships. Therefore, SynLethDB 2.0 can provide users with more comprehensive annotations for the entries and relationships. Table 5 shows the number of each type of entities in SynLethKG and the average numbers of annotations and relationships of each kind of entities. The average number of relationships for each type of nodes was counted by adding the numbers of incident edges among all the nodes and dividing the sum by the total number of nodes.
Table 3.
Statistics About the Knowledge Graph SynLethKG
| # genes | 9856 | |
|---|---|---|
| Human SLs | # interactions | 35 943 |
| Density | 0.07% | |
| # entity types | 11 | |
| SynLethKG | # relationship types | 27 |
| # nodes | 37 341 | |
| # edges | 1 405 652 |
Table 4.
Numbers of the Relationships in SynLethKG.
| Type | # Edges |
|---|---|
| (Anatomy, downregulates, Gene) | 31 |
| (Anatomy, expresses, Gene) | 358 005 |
| (Anatomy, upregulates, Gene) | 26 |
| (Compound, binds, Gene) | 11 453 |
| (Compound, causes, Side Effect) | 135 063 |
| (Compound, downregulates, Gene) | 17 506 |
| (Compound, palliates, Cancer) | 42 |
| (Compound, resembles, Compound) | 5500 |
| (Compound, treats, Cancer) | 282 |
| (Compound, upregulates, Gene) | 13 573 |
| (Cancer, associates, Gene) | 7708 |
| (Cancer, downregulates, Gene) | 988 |
| (Cancer, localizes, Anatomy) | 1444 |
| (Cancer, presents, Symptom) | 1048 |
| (Cancer, resembles, Cancer) | 106 |
| (Cancer, upregulates, Gene) | 1263 |
| (Gene, covaries, Gene) | 16 985 |
| (Gene, interacts, Gene) | 87 103 |
| (Gene, non-synthetic lethal, Gene) | 2831 |
| (Gene, participates, Biological Process) | 393 049 |
| (Gene, participates, Cellular Component) | 59 054 |
| (Gene, participates, Molecular Function) | 65 207 |
| (Gene, participates, Pathway) | 41 790 |
| (Gene, regulates, Gene) | 147 639 |
| (Gene, synthetic lethal, Gene) | 35 943 |
| (Gene, synthetic rescue, Gene) | 895 |
| (Pharmacologic Class, includes, Compound) | 1118 |
Table 5.
Statistics About the Entities in SynLethKG.
| Labels (n) | Size | Avg_Anna | Avg_Relb |
|---|---|---|---|
| SideEffect | 5664 | 5.00 | 23.85 |
| Gene | 14 100 | 8.00 | 112.99 |
| BiologicalProcess | 12 141 | 5.00 | 32.37 |
| Compound | 1898 | 7.00 | 100.12 |
| MolecularFunction | 3012 | 5.00 | 21.65 |
| Anatomy | 390 | 6.64 | 921.81 |
| CellularComponent | 1619 | 5.00 | 36.48 |
| Pathway | 2069 | 5.00 | 20.63 |
| Symptom | 325 | 5.00 | 3.224 |
| PharmacologicClass | 357 | 6.00 | 3.13 |
| Cancer | 53 | 5.00 | 245.04 |
The average number of annotations of each type of nodes.
The average number of relationships of each type of nodes.
Thirdly, SynLethDB 2.0 provides additional ways to access the data. In SynLethDB 1.0, users can only access the SLs by searching a gene name as the query. In SynLethDB 2.0, users can search by names of drugs or cancers. In addition to searching from the web interface, users can also download the raw dataset from the website. Besides, SynLethDB 2.0 provides RESTful APIs which allow users to access the data through different programming languages like Python and R as well as command line.
SynLethKG for cancer driver genes
In SynLethKG, we collected various relationships for the genes involved in the SL pairs, including gene–gene relationships (gene expression covariation, gene interaction and gene regulation), GO annotations and pathways as shown in Table 4. In particular, SynLethKG has 14 100 genes, 12 141 biological processes, 3012 molecular functions, 1619 cellular components, 2026 pathways, etc. as nodes and their relationships as edges in Table 5.
Moreover, SynLethKG also contains 9959 relationships between the genes and 53 cancers from DisGeNET database (46) and 42 532 relationships between the genes and 1898 compounds from DrugBank database (47). For cancers, 325 symptoms and 390 anatomies are included as entities to describe the cancer features. For drugs, 357 pharmacologic classes and 5664 side effects are included as entities to describe drug features. Based on the same strategy as in SL-BioDP (35), we counted the numbers of cancer driver genes and genes from hallmark cancer pathways in 32 cancer types from The Cancer Genome Atlas (TCGA) contained in SynLethKG, as well as the numbers of their SL partners and related drugs.
Figure 2 shows the numbers of cancer driver genes, their SL partners and related drugs. We can observe that several cancers have quite a few SL pairs and drugs related to their driver genes, including BLCA (Bladder urothelial carcinoma), BRCA (Breast invasive carcinoma), CESC (Cervical squamous cell carcinoma and endocervical adenocarcinoma), COADREAD (Colorectal adenocarcinoma), HNSC (Head and neck squamous cell carcinoma), LGG (Brain lower grade glioma), LIHC (Liver hepatocellular carcinoma), SKCM (Skin cutaneous melanoma) and UCEC (Uterine corpus endometrial carcinoma). Figure 2 demonstrates that our database contains useful information about many genes, SLs and drugs related to cancers, making it a powerful tool for data-driven discovery and analysis of anticancer drug targets.
Figure 2.
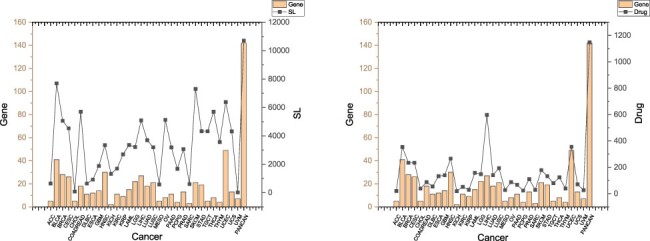
SLs and drugs in SynLethKG for driver genes of 32 cancers and pan-caner. The bar chart shows the numbers of cancer driver genes in SynLethKG. In the left figure, the line chart represents the numbers of SLs containing the cancer driver genes in SynLethKG, and in the right figure, the line chart represents the numbers of drugs associated with the driver genes in SynLethKG.
Case study
To demonstrate how to use SynLethDB 2.0 to discover drug targets, let us do a case study of searching SL partner genes of BRCA1 in breast cancer through the web interface as shown in Figure 3. First, with the ‘Search SL by disease’ module, we choose ‘breast cancer’ as the disease and select the relationship ‘Disease Associates Gene’. Then, the first line of the results shows that breast cancer is associated with breast cancer associated gene 1 (BRCA1). By clicking the ‘Search SL’ button in the ‘Function’ column, we searched for the SL partner genes of BRCA1. The result shows that poly (ADP-ribose) polymerase 2 (PARP2) is an SL partner of BRCA1 with a high confidence score (0.87). Hence we choose this SL pair for further inspection. By clicking the ‘Inspect’ button in the ‘Function’ column, we can browse more knowledge about this SL pair. Different types of biomedical relationships can be browsed by clicking the nodes in the graph. For example, we can see that both BRCA1 and PARP2 are associated with the ‘ovarian cancer’ and ‘breast cancer’ diseases, and BRCA1 participates in the ‘DNA Damage Response’ pathway, consistent with the literature. The annotations of any node or edge can be viewed in the infobox at the upper right corner by hovering the mouse over the node or edge. We hovered the mouse over the edge between ‘BRCA1’ and ‘PARP2’, and the infobox displayed the annotations of the SL relationship between ‘BRCA1’ and ‘PARP2’. The ‘pubmed_id’ attribute shows the PubMed IDs of papers which reported this SL; ‘cell_line’ shows the cell lines or cancer types in which this SL has been verified; ‘statistic_score’ is the confidence score of the SL; ‘unbiased’ indicates whether a relationship is bidirectional (when its value is true) or unidirectional (when its value is false); ‘source’ shows that this SL is collected from the Decipher project and Syn-Lethality database. As BRCA1 is known to be downregulated in breast cancer and PARP2 is an SL partner gene of BRCA1, we searched for compounds that downregulate PARP2 as candidate drugs for breast cancer. As shown in the knowledge graph at the lower left corner of Figure 3, Rucaparib, Talazoparib, Niraparib and Olaparib all bind to PARP2. On the other hand, we can also search for SLs related to Rucaparib, Talazoparib, Niraparib and Olaparib using the ‘Search SL by compound’ option. In this way, we can find that PARP2 is indeed a drug target, as shown in the right half of Figure 3. Through this case study, we can see how to use the basic functionalities of SynLethDB 2.0 through the web interface, which can be used to explore potential anticancer drug targets based on SL or analyze biological mechanisms behind SLs.
Figure 3.

A case study on BRCA1 in breast cancer. Using ‘Search SL by disease’, the SL genes associated with the disease will be shown. BRCA1 is a gene that has SL partners and is downregulated in breast cancer. Click the ‘Search SL’ button, and it shows that PARP2 is an SL partner of BRCA1 with a high confidence score (0.87). Inspecting this pair of SL genes, we notice that BRCA1 and PARP2 are both associated with the ‘ovarian cancer’ and ‘breast cancer’ diseases, and BRCA1 participates in the ‘DNA Damage Response’ pathway. The infobox at the upper right corner shows the annotations of the SL. The synthetic lethal relationship between BRCA1 and PARP2 has been reported in the literature on human cancer and cell lines including A549, PC3 and MDA468. The result that breast cancer downregulates BRCA1 and PARP2 is an SL partner of BRCA1 indicates that PARP2 is a drug target for breast cancer. Rucaparib, Talazoparib, Niraparib and Olaparib all bind with PARP2. Using ‘Search SL by compound’, we also identify PARP2 as a potential drug target of Rucaparib, Talazoparib, Niraparib and Olaparib.
Discussion and conclusion
With the development of RNAi and CRISPR screening technologies, data about SL have increased rapidly in the past few years. We have been continuously collecting SL data, integrating them into SynLethDB and improving the annotation quality. In this version, we have integrated more biomedical knowledge about human SLs into a knowledge graph called SynLethKG. The additional knowledge can provide more features for SL prediction and improve the performance of the predictive model. A similar procedure can be applied to predicting drugs based on SLs. We also provided a new web interface with online services for data browsing, visualization and analysis. For instance, ‘Search SL by disease’ can facilitate SL-based cancer drug discovery. The ‘SL inspect’ functionality displays relationships between a pair of SL genes from multiple sources in one intuitive graph. Enrichment analysis tools help analyze the most relevant pathways and GO of a gene’s SL partners. SynLethDB has been used as a source of training or testing datasets by many computational methods for SL prediction, and this new version of SynLethDB provides a larger and more comprehensive dataset for these methods. In addition, we realize that the data in SynLethDB are enriched with SLs of some hub genes, such as Kirsten ras proto-oncogene, because they are more experimentally studied. This kind of data skewness may introduce some bias, which makes a model learn superficial patterns and achieve inflated performance.
The overall goal of SynLethDB is to increase the understanding of SL mechanisms and to facilitate drug discovery. In the future, we will continue to collect new SLs and enhance the functionalities of the database. For instance, we will add genomics data and cell line annotations to make SLs more context-specific. In addition, we can create more efficient path queries based on the graph database to find the pathways shared between SL pairs and interactions between SLs and drugs.
Contributor Information
Jie Wang, School of Information Science and Technology, ShanghaiTech University, 393 Middle Huaxia Road, Pudong, Shanghai 201210, China.
Min Wu, Institute for Infocomm Research, Agency for Science, Technology and Research (A*STAR), 1 Fusionopolis Way, Singapore 138632, Singapore.
Xuhui Huang, School of Computing, National University of Singapore, Computing 1, 13 Computing Drive, Singapore 117417, Singapore.
Li Wang, School of Information Science and Technology, ShanghaiTech University, 393 Middle Huaxia Road, Pudong, Shanghai 201210, China.
Sophia Zhang, College of Agriculture and Life Sciences, Cornell University, 260 Roberts Hall, Ithaca, NY 14853, USA.
Hui Liu, School of Computer Science and Technology, Nanjing Tech University, 30 Puzhu Road, Nanjing 211816, China.
Jie Zheng, School of Information Science and Technology, ShanghaiTech University, 393 Middle Huaxia Road, Pudong, Shanghai 201210, China; Shanghai Engineering Research Center of Intelligent Vision and Imaging, 393 Middle Huaxia Road, Pudong, Shanghai 201210, China.
Acknowledgements
We would like to thank William R. Sellers for kindly answering our questions about synthetic lethal gene pairs identified by GEMINI from CRISPR screens and sharing the data.
Funding
ShanghaiTech university startup grant.
Conflict of interest
The authors declare that they have no competing interests.
Author contributions statement
J.Z., M.W. and H.L. conceived the study. J.W. and S.Z. collected the data and performed the analysis. J.W., X.H. and L.W. developed the SynLethKG knowledge graph and the SynLethDB database. J.W. drafted the manuscript with critical input from J.Z. and M.W. All authors reviewed the manuscript.
References
- 1. Dobzhansky T. (1946) Genetics of natural populations. XIII. Recombination and variability in populations of Drosophila pseudoobscura. Genetics, 31, 269–290. doi: 10.1093/genetics/31.3.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. O’Neil N.J., Bailey M.L. and Hieter P. (2017) Synthetic lethality and cancer. Nat. Rev. Genet., 18, 613–623. doi: 10.1038/nrg.2017.47 [DOI] [PubMed] [Google Scholar]
- 3. Hartwell L.H., Szankasi P., Roberts C.J. et al. (1997) Integrating genetic approaches into the discovery of anticancer drugs. Science, 278, 1064–1068. doi: 10.1126/science.278.5340.1064 [DOI] [PubMed] [Google Scholar]
- 4. Bryant H.E., Schultz N., Thomas H.D. et al. (2005) Specific killing of BRCA2-deficient tumours with inhibitors of poly (ADP-ribose) polymerase. Nature, 434, 913–917. doi: 10.1038/nature03443 [DOI] [PubMed] [Google Scholar]
- 5. Roemer T. and Boone C. (2013) Systems-level antimicrobial drug and drug synergy discovery. Nat. Chem. Biol., 222, 222–231. doi: 10.1038/nchembio.1205 [DOI] [PubMed] [Google Scholar]
- 6. Kaelin W.G. (1999) Choosing anticancer drug targets in the postgenomic era. J. Clin. Invest., 104, 1503–1506. doi: 10.1172/JCI8888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kaelin W.G. (2005) The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer, 5, 689–698. doi: 10.1038/nrc1691 [DOI] [PubMed] [Google Scholar]
- 8. Heinzel A., Marhold M., Mayer P. et al. (2019) Synthetic lethality guiding selection of drug combinations in ovarian cancer. PLoS ONE, 14, e0210859. doi: 10.1371/journal.pone.0210859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. O’Hare T., Zaberezhnyy V., Williams R.T. et al. (2010) Wnt/Ca2+/NFAT signaling maintains survival of Ph+ leukemia cells upon inhibition of Bcr-Abl. Cancer Cell, 18, 74–87. doi: 10.1016/j.ccr.2010.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bartz S.R., Zhang Z., Burchard J. et al. (2006) Small interfering RNA screens reveal enhanced cisplatin cytotoxicity in tumor cells having both BRCA network and TP53 disruptions. Mol. Cell. Biol., 26, 9377–9386. doi: 10.1128/MCB.01229-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chang J.-G., Chen C.-C., Wu Y.-Y. et al. (2016) Uncovering synthetic lethal interactions for therapeutic targets and predictive markers in lung adenocarcinoma. Oncotarget, 7, 73664–73680. doi: 10.18632/oncotarget.12046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Luo J., Emanuele M.J., Li D. et al. (2009) A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell, 137, 835–848. doi: 10.1016/j.cell.2009.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blank J.L., Liu X.J., Cosmopoulos K. et al. (2013) Novel DNA damage checkpoints mediating cell death induced by the NEDD8-activating enzyme inhibitor MLN4924. Cancer Res., 73, 225–234. doi: 10.1158/0008-5472.CAN-12-1729 [DOI] [PubMed] [Google Scholar]
- 14. Han K., Jeng E.E., Hess G.T. et al. (2017) Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat. Biotechnol., 35, 463–474. doi: 10.1038/nbt.3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shen J.P., Zhao D., Sasik R. et al. (2017) Combinatorial CRISPR–Cas9 screens for de novo mapping of genetic interactions. Nat. Methods., 14, 573–576. doi: 10.1038/nmeth.4225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jerby-Arnon L., Pfetzer N., Waldman Y.Y. et al. (2014) Predicting cancer-specific vulnerability via data-driven detection of synthetic lethality. Cell, 158, 1199–1209. doi: 10.1016/j.cell.2014.07.027 [DOI] [PubMed] [Google Scholar]
- 17. Lee J.S., Das A., Jerby-Arnon L. et al. (2018) Harnessing synthetic lethality to predict the response to cancer treatment. Nat. Commun., 9, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Srihari S., Singla J., Wong L. et al. (2015) Inferring synthetic lethal interactions from mutual exclusivity of genetic events in cancer. Biol. Direct, 10, 1–18. doi: 10.1186/s13062-015-0086-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hao Y., Zhang X., Chen Y. et al. (2016) Ranking novel cancer driving synthetic lethal gene pairs using TCGA data. Oncotarget, 7, 55352–55367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guo J., Liu H. and Zheng J. (2016) SynLethDB: synthetic lethality database toward discovery of selective and sensitive anticancer drug targets. Nucleic Acids Res., 44, D1011–D1017. doi: 10.1093/nar/gkv1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schmidt E.E., Pelz O., Buhlmann S. et al. (2013) GenomeRNAi: a database for cell-based and in vivo RNAi phenotypes, 2013 update. Nucleic Acids Res., 41, D1021–D1026. doi: 10.1093/nar/gks1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leung G. and McAdam R. et al. (2019) The BioGRID interaction database: update. Nucleic Acids Res., 47, D529–D541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ryan C.J., Lord C.J. and Ashworth A. (2014) DAISY: picking synthetic lethals from cancer genomes. Cancer Cell, 26, 306–308. doi: 10.1016/j.ccr.2014.08.008 [DOI] [PubMed] [Google Scholar]
- 24. Liany H., Jeyasekharan A. and Rajan V. (2020) Predicting synthetic lethal interactions using heterogeneous data sources. Bioinformatics, 36, 2209–2216. doi: 10.1093/bioinformatics/btz893 [DOI] [PubMed] [Google Scholar]
- 25. Cai R., Chen X., Fang Y. et al. (2020) Dual-dropout graph convolutional network for predicting synthetic lethality in human cancers. Bioinformatics, 36, 4458–4465. doi: 10.1093/bioinformatics/btaa211 [DOI] [PubMed] [Google Scholar]
- 26. Das S., Deng X., Camphausen K. et al. (2019) DiscoverSL: an R package for multi-omic data driven prediction of synthetic lethality in cancers. Bioinform., 35, 701–702. doi: 10.1093/bioinformatics/bty673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yuxuan H., Chen C., Ding Y. et al. (2019) Optimal control nodes in disease-perturbed networks as targets for combination therapy. Nat. Commun., 10, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang R., Han Y. and Zhao Z. et al. (2019) Link synthetic lethality to drug sensitivity of cancer cells. Brief. Bioinform., 20, 1295–1307. doi: 10.1093/bib/bbx172 [DOI] [PubMed] [Google Scholar]
- 29. Cui X.L., Han L., Liu Y. et al. (2021) siGCD: a web server to explore survival interaction of genes, cells and drugs in human cancers. Brief. Bioinform., 22. doi: 10.1093/bib/bbab058 [DOI] [PubMed] [Google Scholar]
- 30. Wong A.S.L., Choi G.C.G., Cui C.H. et al. (2016) Multiplexed barcoded CRISPR-Cas9 screening enabled by CombiGEM. Proc. Natl. Acad. Sci. U.S.A., 113, 2544–2549. doi: 10.1073/pnas.1517883113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao D., Badur M.G., Luebeck J. et al. (2018) Combinatorial CRISPR-Cas9 metabolic screens reveal critical redox control points dependent on the KEAP1-NRF2 regulatory axis. Mol. Cell, 69, 699–708. doi: 10.1016/j.molcel.2018.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang T., Yu H., Hughes N.W. et al. (2017) Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell, 168, 890–903. doi: 10.1016/j.cell.2017.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Steinhart Z., Pavlovic Z., Chandrashekhar M. et al. (2017) Genome-wide CRISPR screens reveal a Wnt–FZD5 signaling circuit as a druggable vulnerability of RNF43-mutant pancreatic tumors. Nat. Med., 23, 60–68. doi: 10.1038/nm.4219 [DOI] [PubMed] [Google Scholar]
- 34. Zamanighomi M., Jain S.S., Ito T. et al. (2019) GEMINI: a variational Bayesian approach to identify genetic interactions from combinatorial CRISPR screens. Genome Biol., 20, 1–10. doi: 10.1186/s13059-019-1745-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deng X., Das S., Valdez K. et al. (2019) Sl-biodp: multi-cancer interactive tool for prediction of synthetic lethality and response to cancer treatment. Cancers, 11, 1682. doi: 10.3390/cancers11111682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang B., Tang C., Yao Y. et al. (2021) The tumor therapy landscape of synthetic lethality. Nat. Commun., 12, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sinha S., Thomas D., Chan S. et al. (2017) Systematic discovery of mutation-specific synthetic lethals by mining pan-cancer human primary tumor data. Nat. Commun., 8, 1–13. doi: 10.1038/ncomms15580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu Y., Wu M., Liu C. et al. (2019) SL 2 MF: predicting synthetic lethality in human cancers via logistic matrix factorization. IEEE/ACM Trans. Comput. Biol. Bioinf., 17, 748–757. doi: 10.1109/TCBB.2019.2909908 [DOI] [PubMed] [Google Scholar]
- 39. Huang J., Wu M., Lu F. et al. (2019) Predicting synthetic lethal interactions in human cancers using graph regularized self-representative matrix factorization. BMC Bioinform., 20, 1–8. doi: 10.1186/s12859-019-3197-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Himmelstein D.S., Lizee A., Hessler C. et al. (2017) Systematic integration of biomedical knowledge prioritizes drugs for repurposing. eLife, 6, e26726. doi: 10.7554/eLife.26726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Höfken T. and Schiebel E. (2004) Novel regulation of mitotic exit by the Cdc42 effectors Gic1 and Gic2. Int. J. Cell Biol., 164, 219–231. doi: 10.1083/jcb.200309080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yunyan G., Wang R., Han Y. et al. (2018) A landscape of synthetic viable interactions in cancer. Brief. Bioinform., 19, 644–655. [DOI] [PubMed] [Google Scholar]
- 43. Maglott D., Ostell J., Pruitt K.D. et al. (2010) Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res., 39, D52–D57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Page L., Brin S., Motwani R. and Winograd T. (1999) The PageRank citation ranking: bringing order to the web. Technical report. Stanford InfoLab.
- 45. James Hung H.M., Robert T.O.N., Peter B. et al. (1997) The behavior of the p-value when the alternative hypothesis is true. Biometrics, 53, 11–22. [PubMed] [Google Scholar]
- 46. Piñero J., Ramírez-Anguita J.M., Saüch-Pitarch J. et al. (2020) The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res., 48, D845–D855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wishart D.S., Feunang Y.D., Guo A.C. et al. (2018) DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res., 46, D1074–D1082. doi: 10.1093/nar/gkx1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gilson M.K., Liu T., Baitaluk M. et al. (2016) BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res., 44, D1045–D1053. doi: 10.1093/nar/gkv1072 [DOI] [PMC free article] [PubMed] [Google Scholar]


