Abstract
Functional genetic screens by CRISPR-Cas9 allow for the unbiased discovery of proteins causally involved in complex biological processes. In recent years, this approach has been used by multiple laboratories to uncover a range of tumor cell regulators determining immune sensitivity. In this review, we provide an overview of genetic screens carried out both in vitro and in vivo. By comparative analysis we highlight commonly identified proteins and pathways that are key in establishing tumor-intrinsic immune susceptibility. Together, these screens demonstrated the importance of the antigen presentation, interferon-γ, tumor necrosis factor and autophagy pathways in governing sensitivity of tumor cells to immune attack. Moreover, they underline the complex interplay between tumor cells and their microenvironment, providing both fundamental and clinically relevant insights into the mechanisms of tumor immune resistance.
Key words: immunotherapy, immune checkpoint blockade, CRISPR-Cas9, genetic screen, therapy resistance
Highlights
-
•
CRISPR-Cas9 screens shed light on tumor-intrinsic mechanisms of immune sensitivity.
-
•
Different screen settings highlight tumor-intrinsic and environmental influences.
-
•
Effects of IFN-γ and antigen presentation pathways depend on environmental contexts.
-
•
Cellular context impacts how TNF and autophagy pathways affect immune sensitivity.
-
•
Potential therapeutic targets identified in the TNF, autophagy and IFN-γ pathways.
Introduction
The advent of CRISPR-Cas9 technology has revolutionized daily laboratory practice by allowing for the targeted inactivation of genes of interest.1 This technology is also of use in pooled genetic screens. There, instead of knocking out single genes, cells are transduced with a single guide RNA (sgRNA) library, targeting many different genes. This transduction is carried out at a low multiplicity of infection to generate a large pool of cells, each of which harbors a single and distinct genetic perturbation. Empowering subsequent analyses, screens are commonly designed such that at least hundreds of cells carry the same sgRNA, also called library coverage. The next step in the screen is to apply a specific biological or pharmacological pressure and determine the relative fitness (or any other trait of interest) of each of the perturbed cells in response to this treatment. For quantification of the phenotype, an inventory is made of the frequency of each sgRNA in the pool of cells before and after selection. Since every sgRNA represents a specific DNA sequence, it can serve as a cellular barcode. By deep sequencing the sgRNA sequences present in each cell, the effect of a particular genetic inactivation on the phenotype of interest can be assessed.2
In recent years, we and others have used this powerful functional genetic screening approach to understand the process of tumor cell-intrinsic immune resistance.3, 4, 5, 6, 7, 8, 9 This tool has proven its merit for the current challenges of immunotherapy, in particular that of immune checkpoint blockade (ICB). By antibody blockade of the inhibitory T-cell checkpoints programmed cell death protein 1 (PD-1) and cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4), ICB can cause durable clinical responses for cancer patients with various tumor indications.10, 11, 12, 13, 14, 15, 16, 17 Despite this success, however, the majority of patients still fail to respond durably to ICB treatment, commonly owing to intrinsic or acquired resistance.14,15,17,18 What has become clear is the contribution of CD8+ T cells to the clinical efficacy of ICB therapies (Table 1). Resistance to ICB correlates with a lack of CD8+ T-cell infiltration in the tumor, both in preclinical models and in clinical samples.19, 20, 21, 22, 23 When present within the tumor, CD8+ T cells can recognize and attack only cells that present cognate, and sufficiently foreign, antigenic peptides within the context of major histocompatibility complex (MHC) class I. In line with this, both the absence of (clonal) (neo)antigens and the loss of the cellular machinery required for proper antigen presentation (AP) are associated with reduced T-cell reactivity and lack of response to ICB therapy.17,24, 25, 26, 27, 28, 29, 30, 31, 32 But even tumors with actionable mutations and intact AP can resist attack by infiltrated CD8+ T cells and ICB therapy, by avoiding T-cell effector molecules such as interferon-γ (IFN-γ), tumor necrosis factor (TNF) and granzymes.4,9,22,32, 33, 34, 35, 36, 37, 38, 39
Table 1.
Common T-cell resistance mechanisms
| Type of resistance | Example |
|---|---|
| Lack of T-cell infiltration | Genetically driven, active T-cell exclusion19,23 |
| Lack of actionable antigens | Low expression of antigenic transcripts26,27,29 |
| Defects in antigen presentation | Loss of B2M heterozygosity30 B2M mutations29,32,66 |
| Insensitivity to T cell effector molecules | IFN-γ pathway mutations32,36,39 Caspase 8 mutations22 |
IFN-γ, interferon-γ.
By using genetic screens in the context of immunotherapy resistance, investigators can forward our understanding of immune-oncology in two distinct ways. Firstly, these screens can, in an unbiased and comparative way, confirm and rank the importance of already established, immunologically relevant pathways. Secondly, they can reveal novel immunotherapeutic targets within established or novel pathways. Here, we will first provide a summarized overview of the genetic screens that aimed to identify factors determining tumor-intrinsic resistance and sensitivity to CD8+ T-cell attack. We will subsequently discuss in detail their reproducibility by overlapping the hits between the different genetic screens, highlighting common tumor-intrinsic pathways of resistance and vulnerability. We then continue by placing these findings in a broader context by discussing the biological roles of some of the key tumor cell regulators of immune sensitivity that have been identified in these screens, the complexity of the communication between tumor cells and their microenvironment, as well as limitations and potential opportunities for clinical applications.
Functional screens to identify regulators of immune sensitivity
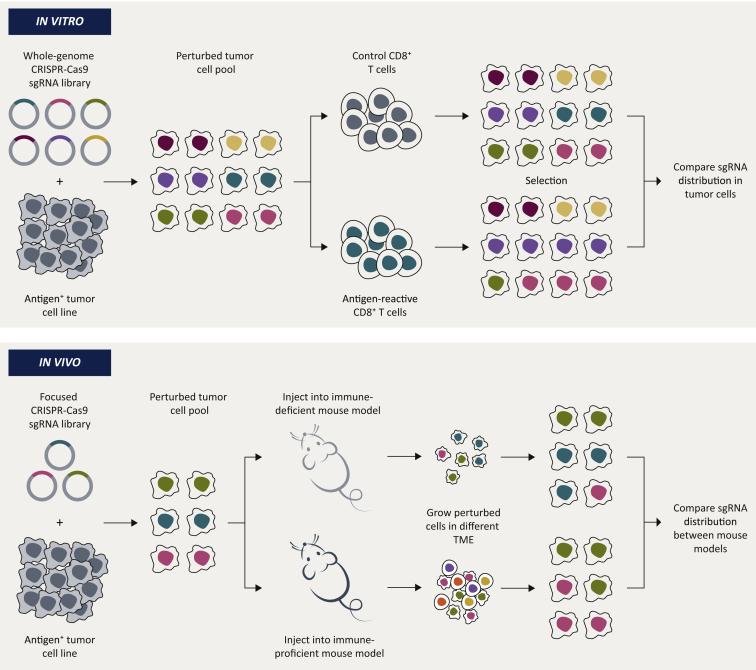
To identify tumor cell regulators of immune sensitivity, some research groups chose to carry out in vitro screens, whereas others aimed to identify immune modulators by carrying out screens in vivo. Each approach has its own merit, as we will discuss (Figure 1 and Table 2).
Figure 1.
General setup of CRISPR/Cas9 screens to uncover regulators of immune sensitivity in tumor cells.
sgRNA, single guide RNA; TME, tumor microenvironment.
Table 2.
Strengths and weaknesses of in vitro and in vivo screening systems
| In vitro screens | In vivo screens |
|---|---|
| Whole-genome scale sgRNA libraries | Focused sgRNA libraries |
| Limited number of cell types involved in immune selection | Complex and rich mixture of cell types that drive the immune selection |
| Easily scalable, allows for high-quality, high-coverage screens | Poorly scalable, generally poor(er) sensitivity |
| Allows only for the addition of locally acting (immuno)therapies | Allows for the addition of locally and systemically acting (immuno)therapies |
| Reductionistic | Holistic, allows for the discovery of emergent traits |
| Flexible; enables highly defined (genetic) screening setting | More difficult to establish highly defined (genetic) screening setting |
sgRNA, single guide RNA.
In vitro screens
One of the biggest advantages of carrying out screens in vitro is that they enable the investigator to engineer a highly defined experimental setting. This reductionist approach allows for homogeneous genetic and cellular conditions. For example, many cell types are known to contribute to the response to ICB, including CD8+ T cells, regulatory T cells, B cells, dendritic cells and natural killer (NK) cells.40,41 It is nearly impossible to recapitulate the effects of this complex mixture of cells in vitro, and therefore most laboratories carried out their screens using only (homogeneous) CD8+ T cells as a way of treating their mutagenized pool of tumor cells. At first sight this may seem like a limiting approach, but CD8+ T cells are key determinants of ICB efficacy.42,43 This approach may therefore be key in uncovering new, important mechanisms of immune sensitivity. Furthermore, an in vitro setting makes it relatively straightforward to scale up, thereby allowing the use of whole-genome sgRNA libraries (called library complexity). Lastly, an in vitro screening setup usually allows for a more exhaustive identification of hits, as one can carefully define and optimize the experimental conditions. This permits a deep, unbiased cataloguing of key tumor-intrinsic determinants in a relatively simple and agnostic manner. Below, we will discuss some of these in vitro screens, labeled by the first author of the corresponding publications (Table 3).
Table 3.
Screens used for in vitro overlap analyses.
| Cell line | Tumor type | Immune Attack | Library | Sensitivity/resistance | Publication | Organism |
|---|---|---|---|---|---|---|
| B16F10-OVA | Melanoma | OT-I CD8+ T cells | Brie | Resistance | Kearney et al.4 | Mus musculus |
| Renca-HA | Renal adenocarcinoma | CL4 CD8+ T cells | Mouse Toronto Knockout | Both | Lawson et al.5 | Mus musculus |
| EMT6-HA | Mammary carcinoma | CL4 CD8+ T cells | Mouse Toronto Knockout | Both | Lawson et al.5 | Mus musculus |
| CT26-HA | Colon carcinoma | CL4 CD8+ T cells | Mouse Toronto Knockout | Both | Lawson et al.5 | Mus musculus |
| 4T1-HA | Mammary carcinoma | CL4 CD8+ T cells | Mouse Toronto Knockout | Both | Lawson et al.5 | Mus musculus |
| MC38-OVA | Melanoma | OT-I CD8+ T cells | Mouse Toronto Knockout | Both | Lawson et al.5 | Mus musculus |
| B16F10-OVA | Melanoma | OT-I CD8+ T cells | Mouse Toronto Knockout | Both | Lawson et al.5 | Mus musculus |
| B16F10-OVA | Melanoma | OT-I CD8+ T cells | Brie | Both | Pan et al.7 | Mus musculus |
| Mel624-NY-ESO+ | Melanoma | ESO CD8+ T cells | GeCKOv2 | Resistance | Patel et al.8 | Homo sapiens |
| D10-MART-1+IFNGR1KO | Melanoma | MART-1 CD8+ T cells | GeCKOv2 | Both | Vredevoogd et al.9 | Homo sapiens |
OVA, ovalbumin.
Patel
Patel and colleagues8 exposed Mel624 human melanoma cells transduced with a whole-genome CRISPR-Cas9 library to NY-ESO-1-specific CD8+ T cells. With this screen, they found that, primarily, the loss of APLNR caused resistance. Illustrating the robustness of the screen, they also identified sgRNAs targeting B2M, TAP1, TAPBP (all AP proteins), STAT1 and JAK1 (both IFN-γ-signaling proteins) to have the same (expected) effect. Through immunoprecipitation experiments, the authors found that APLNR binds to JAK1. Using recombinant IFN-γ, they showed that normally, APLNR promotes the IFN-γ-dependent signal transduction of JAK1. The decrease in tumor-intrinsic IFN-γ signaling upon APLNR inactivation in turn resulted in the reduced expression of the above-mentioned proteins involved in AP to CD8+ T cells, rendering tumor cells functionally resistant to immune pressure, both in vitro and in vivo. The authors also identified a number of mutations in APLNR in ICB-treated patient tumors which, when reconstituted in their in vitro tumor model, resulted in resistance to T-cell-mediated killing.
Pan
Pan and colleagues7 put CRISPR-Cas9-mutagenized B16F10 murine melanoma cells under selection of Pmel-reactive CD8+ T cells. In doing so, they found not only that, expectedly, the ablation of Jak1, Stat1, Ifngr1 (all IFN-γ-signaling proteins), B2m, Tap1 and Tap2 (all AP proteins) caused resistance. They also discovered that, conversely, the loss of Arid2, Pbrm1 and Brd7 sensitized tumors to T-cell attack. These findings were validated in a second genome-wide screen in which B16F10 cells expressing the model (high affinity) antigen ovalbumin (OVA) were challenged with OT-I CD8+ T cells. This demonstrated that these genes operate independently of the relative affinity of the tumor antigen targeted by the CD8+ T cells.44,45 ARID2, PB1 (encoded by Pbrm1) and BRD7 are all part of PBAF, a SWI/SNF family chromatin remodeling complex.46 Pan and colleagues7 showed that PBAF complex component-deficient tumor cells have reduced expression of MTORC1 target genes and an altered metabolic state. Furthermore, these cells displayed a stronger response to IFN-γ through the enhanced chromatin accessibility of IFN-γ-stimulated genes. As a result, PBAF-deficient tumor cells were more sensitive to T-cell attack both in vitro and in vivo. Importantly, it was also shown that patients whose tumors express low levels of ARID2 had a survival benefit, though only in the context of an inflammatory tumor microenvironment (TME), again implying an immune response-modulating function of the PBAF complex.
Kearney
Kearney and colleagues4 carried out a large number of screens using two murine tumor cell lines and distinct CD8+ T-cell populations to identify factors that, when knocked out, cause resistance to CD8+ T-cell attack. Library-transduced MC38-OVA colon carcinoma cells were initially treated with OT-I CD8+ T cells and, through this approach, it was found that they became resistant to CD8+ T-cell killing by avoiding IFN-γ signaling (ablation of Jak1 and Ifngr1), TNF signaling (ablation of Casp8 and Tnfrsf1a) or by preventing AP (B2m or Tap1 inactivation). The authors continued by carrying out a similar genetic screen but treated the MC38-OVA cells with perforin 1 (Prf1) knockout OT-I CD8+ T cells, allowing for the assessment of the relative dependency of these evasion pathways on perforin-mediated killing. In the absence of perforin, tumor cells could still avoid CD8+ T-cell killing by limiting intrinsic TNF and IFN-γ signaling, but the protective effect of perturbed AP was lost when the tumor cells were treated with Prf1-/- OT-I CD8+ T cells. Similar findings were made when MC38-OVA cells were replaced by B16F10-OVA melanoma cells for the genetic screens. From these data, the authors gathered, and later confirmed both in vitro and in vivo, that TNF and IFN-γ from T cells can not only affect tumor cells directly under attack, but also those that are not directly attacked by CD8+ T cells (also known as bystander killing47, 48, 49, 50). In a later publication, the authors also showed that their screen identified depletion hits in the TNF pathway; Rnf31 and Rbck.51
Vredevoogd
Having observed that many IFN-γ signaling components were identified in these in vitro genetic screens, we chose to specifically interrogate the IFN-γ-independent tumor signaling networks for modulators of immune sensitivity. We challenged whole-genome library-perturbed, IFNGR1-/- D10 human melanoma cells with MART-1-specific CD8+ T cells.9 The loss of a number of TNF pathway proteins, particularly those residing in the pro-survival arm acting downstream of the TNF receptor, sensitized tumors to MART-1 CD8+ T-cell killing, including TRAF2, BIRC2, MAP3K7, CFLAR, IKBKG and TBK1. We showed that TRAF2 inactivation lowered the tumor TNF cytotoxicity threshold by promoting the onset of RIPK1-dependent apoptosis, thereby sensitizing tumor cells both in vitro and in vivo to immune attack. Supporting these preclinical findings, in patient tumors we found that there is an immune selection against loss of functional TRAF2. This finding was recently validated and expanded in a meta-analysis of several patient cohorts26 and suggests that tumors carrying TRAF2 mutations undergo immune editing to avoid T-cell killing. Extending our initial findings regarding TRAF2, we also demonstrated that the combined inhibition of two hits from the screen, TRAF2 and BIRC2 (which form a complex), cooperated with ICB in eliminating melanoma upon adoptive T-cell transfer in mice.
Lawson
Lawson and colleagues5 carried out parallel whole-genome CRISPR-Cas9 screens in a number of murine tumor cell lines, to systematically catalogue immune sensitivity modifiers. Aside from identifying known resistance mechanisms including loss of the AP machinery or IFN-γ signaling, they found that perturbations in autophagy, in particular the loss of Atg12, sensitized tumor cells to CD8+ T-cell challenge. They continued by showing that this sensitization was dependent on TNF, since neutralizing TNF antibodies reverted the Atg12 knockout (KO) phenotype. In a reverse genetic screen in Atg12 KO cells, the authors observed that the knockout of Tnfrsf1a mediates resistance in Atg12 KO cells, but not in parental cells, again providing evidence that the lack of Atg12 sensitizes to TNF. This finding could also be validated pharmacologically, as an inhibitor of autophagy, autophinib, sensitized 41 different tumor cell lines to the cytotoxic activities of IFN-γ and TNF.
In vivo screens
The advantages of in vivo screens are almost entirely opposite to those of in vitro screens: they allow for the assessment of immune resistance mechanisms in model systems that better resemble the patient situation, since the TME, with its complex components and dynamics, is an integral part of the screening system. In contrast to in vitro systems, the use of animal models in genetic screening also enables the administration of immunotherapies, whether or not acting systemically, including GVAX and anti-CTLA-4.52, 53, 54 Contrary to in vitro screens, however, maintaining sufficient complexity of the pool of perturbed cells for reliably calling hits is more challenging in vivo. Therefore, more focused libraries, based on prior information on the pathways or gene sets likely involved in the phenotype of interest, are commonly used instead of whole-genome libraries (Tables 2 and 4).
Table 4.
Screens used for in vivo overlap analyses
| Cell line | Tumor type | Immune attack | Library | Sensitivity/resistance | Publication | Organism |
|---|---|---|---|---|---|---|
| B16F10 | Melanoma | GVAX + anti-PD-1 + anti-CTLA-4 + spontaneous immunity | Custom (based on in vitro pathways) | Both | Manguso et al.6 | Mus musculus |
| EMT6 | Mammary carcinoma | Spontaneous immunity | Custom (targeting immune-relevant genes) | Both | Lawson et al.5 | Mus musculus |
| Renca | Renal adenocarcinoma | anti-PD-1 + anti-CTLA-4 + spontaneous immunity | Custom (Manguso library6) | Both | Dubrot et al.3 | Mus musculus |
CTLA-4, cytotoxic T-lymphocyte-associated antigen 4; PD-1, programmed cell death protein 1.
Manguso
Manguso and colleagues6 carried out an in vivo immune modulator screen, using a focused library comprising sgRNAs targeting 2368 genes including kinases, cell surface proteins and immune factors.12 They compared the relative sgRNA distribution of this CRISPR library in B16F10-Cas9 cells injected into Tcra-/- mice (animals lacking the TCR alpha chain and therefore unable to apply CD4 and CD8 T-cell-directed pressure) to those injected into normal, C57BL/6 mice treated with GVAX (a granulocyte–macrophage colony-stimulating factor-enriched tumor vaccine), anti-CTLA-4 therapy and anti-PD-1 therapy. They obtained multiple hits, some of which expectedly caused resistance, such as the loss of IFN-γ signaling (Stat1, Jak1, Ifngr1, Ifngr2 and Jak2). In addition, they identified a number of TNF pathway hits that, instead, sensitized tumors to immune attack (including Birc2 and Ripk1). The authors chose to focus on Ptpn2, loss of which boosted IFN-γ, and to some extent IFN-α/β, signaling in response to immune challenge. Another hit originating from this screen, Adar1, was found to limit the IFN-dependent activity of two separate antitumor pathways in response to double-stranded RNA sensing: PKR-induced growth arrest and MDA5-induced immune infiltration.55
Lawson
Having carried out their systematic in vitro screens in multiple cell types, Lawson and colleagues5 established a focused library based on the hits from those screens to perturb EMT6 mammary carcinoma cells. They injected this cell pool into either immunodeficient NCG mice (lacking T, B and NK cells) or immunocompetent BALB/c mice and compared the relative frequency of perturbed cells. The authors observed, similar to their in vitro screens, that defects in autophagy sensitized tumor cells to immune challenge, including the loss of Atg12. Contrary to the expectation and their own in vitro screens, they also found the loss of IFN signal transducer Jak1 to render tumors in BALB/c mice highly sensitive to immune pressure, although this finding was not investigated further.
Dubrot
Dubrot and colleagues3 carried out an in vivo genetic screen using the Manguso library6 in renal carcinoma Renca cells. They compared the relative sensitization of perturbed cells injected into immunodeficient NSG mice with that in wildtype BALB/c mice treated with a combination of anti-CTLA-4 and anti-PD-1 therapy. The authors found the loss of Atg5 to sensitize tumors to immune pressure, in agreement with the work by Lawson and colleagues.5 Dubrot observed that ablation of several genes involved in AP (Tap1, Tap2 and B2m) sensitized tumors in immunocompetent mice. These AP components, responsible for loading and presenting antigens by MHC-I, are required for CD8+ T cells to recognize and kill tumor cells.56,57 The authors, expectedly, found that NK cells are required for the disposal of AP-deficient tumors. NK cells do not rely on MHC-I to recognize tumors, but are regulated by several inhibitory and activating cell surface receptors, including NKG2D.58, 59, 60 The investigators found that Renca tumors, in contrast to the B16F10 cells used in the original Manguso screen,6 express high levels of activating NK cell ligands which, in an NKG2D-dependent fashion, sensitized AP-deficient tumors to NK cell attack.
Common hits between the different screens
After highlighting some of the key genes and mechanisms determining immune response uncovered in these screens, we next assessed the findings at a more global level. Specifically, we catalogued the results of the majority of the in vitro and in vivo screens (Tables 3 and 4, respectively), in order to find commonalities and potential discrepancies between them.
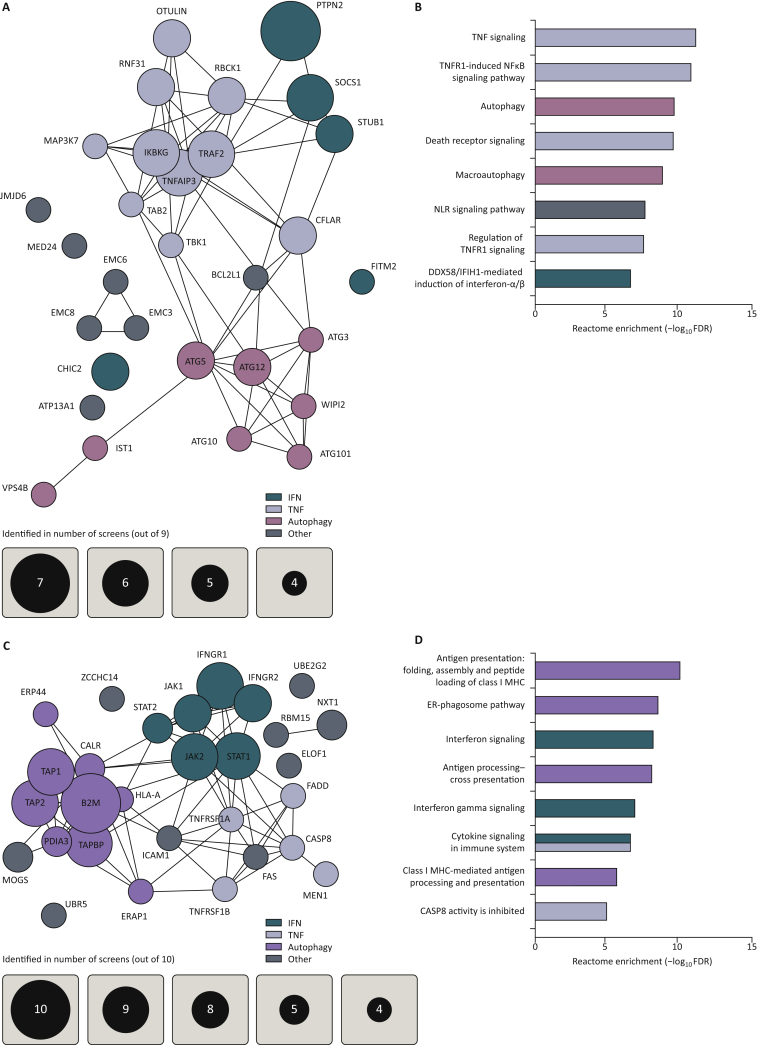
The different in vitro screens show many overlapping hits illustrating their robustness and reproducibility among different laboratories, sgRNA libraries and biological systems. The screens together identified a number of TNF, IFN-γ and autophagy pathway genes that, upon genetic ablation, sensitize tumor cells to CD8+ T-cell killing. A number of these hits were already discussed in detail in some of the screening papers, for example PTPN2 and TRAF26,9 (Figure 2A and B). Other genes, such as STUB1 and CHIC2, have only more recently been characterized in detail, also by us.61, 62, 63 Intriguingly, the loss of a cluster of previously undescribed genes, EMC3, EMC6 and EMC8, also seems to sensitize to CD8+ T-cell challenge. When looking at genes whose knockout drives immune resistance, again many genes show up as common hits. This brings a high confidence level to the observations that the loss of AP components, IFN-γ signaling, TNF receptors and TNF-dependent caspase pathways cause resistance to CD8+ T-cell killing (Figure 2C and D).
Figure 2.
In vitro screens identify common sensitivity and resistance hits.
(A) STRING148 clustering of significant hits determining tumor sensitivity to immune pressure in vitro, from at least four screens. Significance was determined by the original authors as P < 0.05 or FDR < 0.05. Murine gene symbols were translated to their human paralogs by SynGO.149 (B) Gene ontology enrichment scores of the eight top Reactome gene sets by Panther,150 based on in vitro sensitivity hits identified in at least three screens. Gene sets larger than 500 genes were excluded. (C) STRING148 clustering of significant hits determining tumor resistance to immune pressure in vitro, from at least four screens. Significance was determined by the original authors as P < 0.05 or FDR < 0.05. Murine gene symbols were translated to their human paralogs by SynGO.149 (D) Gene ontology enrichment scores of the eight top Reactome gene sets by Panther,150 based on in vitro sensitivity hits identified in at least three screens. Gene sets larger than 500 genes were excluded.
CASP8, caspase 8; ER, endoplasmic reticulum; FDR, false discovery rate; MHC, major histocompatibility complex; NFκB, nuclear factor-kappa B; NLR, NOD-like receptor; TNF, tumor necrosis factor; TNFR1, tumor necrosis factor receptor 1.
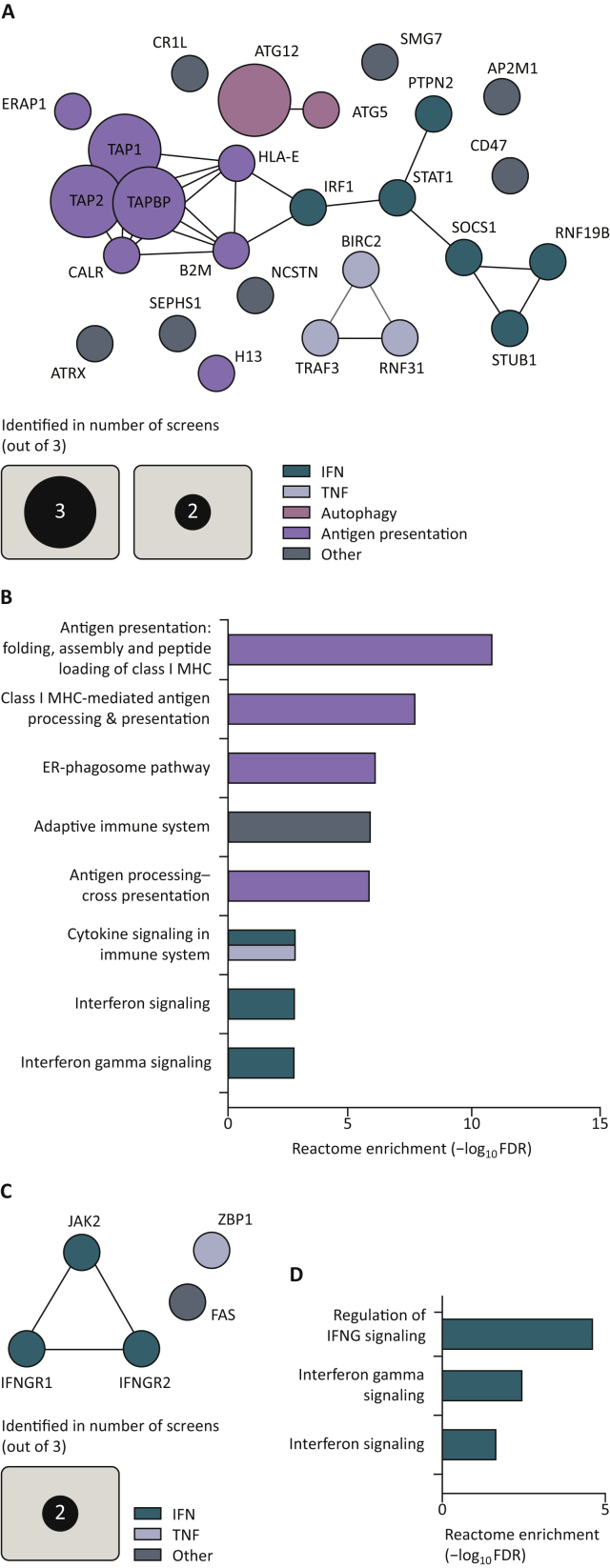
Analysis of the in vivo screens revealed, at times unexpectedly, an incomplete match with the findings made in the in vitro screens. Autophagy-related and TNF signaling hits were found to sensitize tumors to immune challenge, similar to what was observed in vitro. The loss of antigen-presentation machinery (e.g. TAP1, TAP2 and B2M), however, seemed to render tumors more sensitive to immune-mediated clearance in vivo, which is in contrast to what was seen in vitro with CD8+ T-cell screens. Additionally, inactivation of genes positively regulating IFN signaling sensitized in vivo tumors to immune attack, again in contrast to the in vitro findings. This included STAT1, which is required to successfully relay IFN-γ and IFN-α/β signals64 (Figure 3A and B). Puzzlingly, the loss of receptors for IFN-γ, IFNGR1 and IFNGR2 were found to reduce immune sensitivity of the queried tumors in vivo in the Manguso and Dubrot screens, whereas they enhanced sensitivity in the screen of Lawson and colleagues3,5,6 (Figure 3C and D). Below, we will discuss for each of the major pathways identified, translational implications of the findings made in the genetic screens, while also offering a biological context (Figure 4).
Figure 3.
In vivo screens identify common sensitivity hits and common resistance hits.
(A) Clustering analysis of significant in vivo sensitivity hits identified in at least two screens. Significance was determined by the original authors as P < 0.05 or FDR < 0.05. Murine gene symbols were translated to their human paralog by SynGO.149 Clustering was carried out by STRING.148 (B) Gene ontology enrichment scores of the eight top Reactome gene sets by Panther,150 based on in vivo sensitivity hits identified in at least two screens. Gene sets larger than 500 genes were excluded. (C) Clustering analysis of significant in vivo resistance hits identified in at least two screens. Significance was determined by the original authors as P < 0.05 or FDR < 0.05. Murine gene symbols were translated to their human paralog by SynGO.149 Clustering was carried out by STRING.148 (D) Gene ontology enrichment scores of the only significant Reactome gene sets by Panther,150 based on in vivo sensitivity hits identified in at least two screens. Gene sets larger than 500 genes were excluded.
ER, endoplasmic reticulum; FDR, false discovery rate; IFN, interferon; IFNG, interferon-γ; MHC, major histocompatibility complex; TNF, tumor necrosis factor.
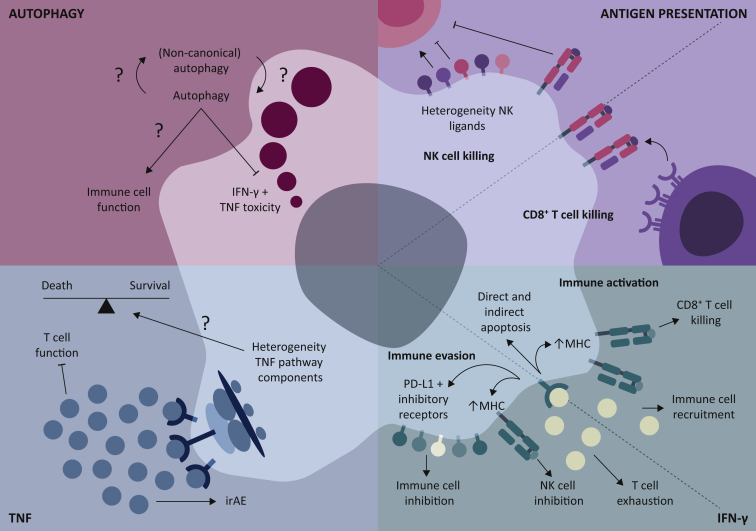
Figure 4.
Biological and translational implications of the genes and pathways identified through CRISPR/Cas9 genetic screening for immune regulators in tumor cells.
For the antigen presentation and IFN-γ pathways, the environmental (specific effects on specific cell types) and cellular (heterogeneity in expression of key signaling proteins) contexts are important. For the autophagy and the TNF pathways, this is mostly the cellular context.
IFN- γ, interferon-γ; irAE, immune-related adverse events; MHC, major histocompatibility complex; NK, natural killer cells; PD-L1, programmed death-ligand 1; TNF, tumor necrosis factor.
Biological and translational implications
Antigen presentation
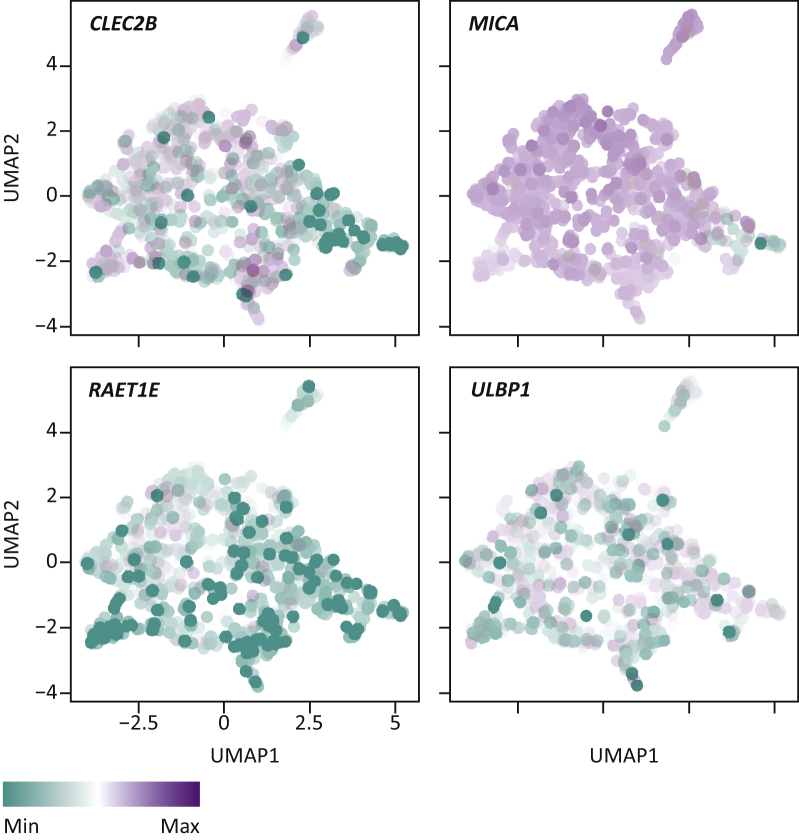
The finding that the loss of AP causes resistance in in vitro screens is expected, because it constitutes a strict requirement for CD8+ T cells to recognize foreign, in this case malignant, cells.56,57 The finding that loss of the same proteins can also cause sensitivity in vivo can be readily explained: in vivo, cells lacking productive AP can also be attacked by NK cells, as the loss of AP relieves their inhibition by MHC-I molecules.30,65 Tumors lacking AP machinery, however, can become resistant to ICB,30,66,67 which raises the question why, in human tumors, NK cells apparently fail to eradicate AP-deficient tumors. One possibility is that despite the loss of AP, NK cells receive too few activating signals, such as MICA (an MHC homolog not presenting antigens), or are still inhibited by other proteins, such as HLA-E (a non-classical MHC-I molecule). Intriguingly, the expression of these ligands differs per tumor (cell line), and may therefore serve as tumor-intrinsic NK cell sensitivity modulators3,68, 69, 70, 71 (Figure 5). Another mechanism by which AP-deficient tumors may survive is NK cell exhaustion. Much like CD8+ T cells72, NK cells, too, lose functionality upon chronic stimulation, which may in part be PD-1-dependent.73, 74, 75, 76 Lastly, CD8+ T cells and NK cells provide reciprocal stimulation, and the lack of CD8+ T-cell activity could negatively affect NK cell function and vice versa.77, 78, 79 Future investigations will have to shed light on the relative contributions of these mechanisms to ICB resistance of AP-deficient human tumors.
Figure 5.
Natural killer cell ligand heterogeneity in the CCLE database.
UMAP analysis of RNA expression of natural killer (NK) cell ligands in the Cancer Cell Line Encyclopedia (CCLE) database.151 Example expression values are given for CLEC2B, MICA, RAET1E and ULBP1. Each datapoint corresponds to a single cell line analyzed.
IFN-γ signaling
In vitro, inactivation of essential genes for IFN-γ signal transduction, such as IFNGR1, IFNGR2, JAK1, JAK2 and STAT1, were recurrently identified as screen hits causing resistance to CD8+ T-cell pressure.4,5,7,8 In fact, IFNGR1 was identified as a resistance hit in all screens but our own, which was carried out in IFNGR1-/- cells.9 This finding highlights the common and homogeneous engagement of, and reliance on, IFN-γ to establishing immune sensitivity in vitro. Consistent with this, the loss of negative regulators of IFN-γ signal transduction, such as SOCS1 and PTPN2, were often found as sensitizing hits in vitro.5,6 When moving into an in vivo context, however, it becomes less predictable how perturbations of the IFN-γ pathway affect tumor immune sensitivity. Probably the most striking example of this is the loss of IFNGR1/2, which causes sensitivity in some in vivo screens,5 but resistance in others.3,6
To understand these seemingly paradoxical findings, it is important to distinguish antitumor effects elicited by IFN-γ from those which, in certain conditions, may benefit the tumor. IFN-γ signaling has several direct antitumor effects, such as the induction of both cell cycle arrest64,80, 81, 82, 83, 84 and apoptosis.83,85,86 Additionally, IFN-γ signaling increases tumor susceptibility to other T cell effector molecules like FasL and TNF-related apoptosis-inducing ligand (TRAIL).87,88 At the same time, IFN-γ negatively affects the tumor indirectly. These activities include enhanced expression of multiple components of the AP pathway, including the increased expression of MHC-I,89,90 while simultaneously inducing the expression of lymphocyte-attracting chemokines like CXCL9, CXCL10 and CXCL11.91, 92, 93 Further underpinning the antitumor role of IFN-γ are clinical studies showing that a high transcriptional signature for IFN-γ signaling is associated with response to ICB,94, 95, 96 while loss of core components of the signaling pathway, like JAK1, can cause resistance to ICB both in preclinical models and patient tumors.6,32,35,39,97
In contrast, another important facet of IFN-γ signaling is exploited by tumor cells to evade immune surveillance. Firstly, IFN-γ-induced programmed death-ligand 1 (PD-L1), arguably the most prominent IFN-γ-mediated immune evasion mechanism and well-established therapeutic target,15,98, 99, 100 binds to its receptor PD-1 on multiple immune cells and inhibits their activities.74,98,99,101 As discussed above, IFN-γ also elevates the levels of MHC-I expression, which, while essential for CD8+ T-cell-mediated tumor control, simultaneously acts as an inhibitory signal for NK cells.65 Lastly, its secreted nature allows IFN-γ to reach far beyond the immediate cell-cell interaction interface to mediate more global immune-suppressive effects.48,49 One example is the induced expression of the enzyme indoleamine 2,3-dioxygenase 1 (IDO1), which metabolizes and degrades the essential amino acid tryptophan, thus limiting T-cell function.86,102 The loss of tryptophan from the TME can also result in altered peptide presentation, potentially allowing tumors to escape immune surveillance.103 Together, these protumor effects of IFN-γ fuel the potential of tumors to escape immune control. In support of this, there are documented examples of preclinical models and patient tumors with deleterious mutations in the IFN-γ pathway that have a better response to ICB than matched controls.5,104, 105, 106, 107
To reconcile these opposing effects of IFN-γ signaling and ultimately aid clinical practice, it is important to better understand the context in which IFN-γ signaling shows a beneficial or detrimental effect on immune responses, whether or not in the context of ICB. In a highly CD8+ T cell-dependent model (B16F10 melanoma expressing the model antigen SIY), Williams and colleagues107 found that the loss of IFN-γ pathway components JAK1 and IFN-γ-R2 sensitized tumors to immune attack in vivo. The authors found that the absence of IFN-γ-induced PD-L1 was the cause for the sensitivity; restoration of PD-L1 expression by overexpression reverted the sensitive phenotype.107
These findings would predict that in a similarly CD8+ T cell-dependent model, but in the context of immunotherapy, the loss of tumor-intrinsic IFN-γ signaling would shift the balance towards resistance, since that should cancel the protective effect of IFN-γ-induced PD-L1. This prediction is supported by independent preclinical studies.6,105 It raises the question why this type of resistance is not universal.104, 105, 106 One potential explanation is the relative activity of NK cells within different tumors (or tumor models). In models in which NK cells are more active than they are in B16F10-SIY, such as the Renca tumor,3,5 loss of IFN-γ pathway activity would sensitize to NK cell killing by preventing IFN-γ-induced MHC class I induction.5,55,107 This is in line with findings from genetic screens for NK cell-specific immune dependencies, where loss of tumor-intrinsic IFN-γ pathway components sensitized to NK cell killing.108,109 ICB in these NK cell-dependent models should, if anything, only increase the activity of NK cells towards the tumor and thus further sensitize IFN-γ-insensitive tumors to NK cell attack.3 From these relatively reductionist models, one could argue that IFN-γ-insensitive tumors are generally more sensitive to immune attack, except in CD8+ T cell-dependent models treated with ICB (Figure 6).
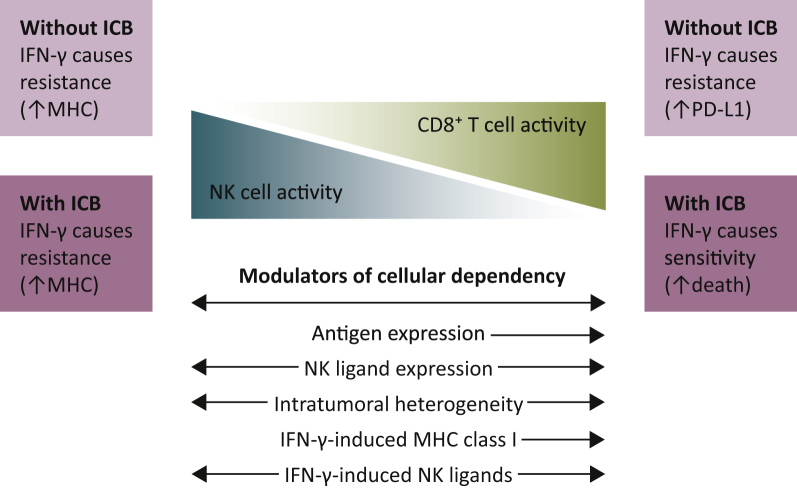
Figure 6.
IFN-γ activity and modulators in tumors with different immune cell reliance.
In NK cell-dependent tumor models, IFN-γ was shown to cause resistance through the up-regulation of MHC class I, irrespective of ICB treatment. Conversely, in highly CD8+ T cell-dependent models, IFN-γ has more disparate effects depending on ICB treatment. In particular, in the absence of ICB, IFN-γ causes PD-L1-mediated immune resistance. PD-L1 blocking antibodies break this resistance, leaving largely antitumor effects of IFN-γ.
ICB, immune checkpoint blockade; IFN-γ, interferon-γ; MHC, major histocompatibility complex; NK, natural killer; PD-L1, programmed death-ligand 1.
This is, however, an incomplete picture in clinical practice, as patient tumors are unlikely to be exclusively CD8+ T cell- or NK cell-dependent. Additionally, what is currently poorly understood is how these situations change as a function of the dynamics of an immune attack on the tumor. While IFN-γ-insensitive tumors are perhaps more sensitive to NK cells due to their lower MHC class I levels, IFN-γ is known to induce many NK ligands, both inhibitory and activating, and may alter NK activity after an initial challenge.110 In addition, the tumor may also undergo immune editing which may again alter cellular dependencies in a dynamic process.31 For example, tumor cells expressing productive CD8+ T cell antigens come under selective pressure to lose either their antigen(s) or MHC-I expression (or other factors sensitizing to immune attack), changing the relative engagement of the tumor from mainly CD8+ T cells to mainly NK cells.26,29,30
Another important aspect to understand the consequences of IFN-γ signaling is that of tumor heterogeneity. Aside from the intertumor heterogeneity in NK ligand expression that may determine whether IFN-γ pathway mutant tumors are able to recruit and activate NK cells,3,68, 69, 70, 71 mutational analyses of patient tumors show a diverse range of mutations in downstream components of the IFN-γ pathway that may (subtly) affect signaling in heterogeneous ways.32,36,39 Intratumoral heterogeneity is important as well. In an elegant study, Williams and colleagues107 demonstrated that while IFN-γ pathway mutant tumors are in principle more sensitive to immune pressure in vivo, they can be protected by PD-L1 expressed on the surface of neighboring wildtype cells when intermixed. Further complicating this matter is the fact that IFN-γ not only targets the tumor, but also immune cells. For example, IFN-γ promotes both CD8+ T-cell expansion, by skewing T-cell memory development, and contraction, by directly killing targeted T cells.111, 112, 113 At the same time, this cytokine also contributes to CD8+ T-cell exhaustion.104,105 Furthermore, IFN-γ can both limit and promote the attraction of CD8+ T cells to the tumor.93,107
Taken together, these findings paint a nuanced and complex picture of IFN-γ signaling: it can simultaneously provide stimulatory and inhibitory signals to different effector cell types driving antitumor immunity. At the same time, IFN-γ creates a selective environment for tumor-intrinsic resistance mechanisms to emerge.104 Thus, deepening our understanding of the context-dependent and cell type-specific effects of IFN-γ signaling will be required to provide a more granular view on the question why a strong IFN-γ response on the one hand is associated with ICB response,32,36,94,96 and with a lack of response in other cases.104,106 Ultimately, this may enable us to more successfully exploit modulation of IFN-γ signaling to aid therapeutic outcome.
TNF signaling
The TNF signaling pathway is highly structured and compartmentalized; it bifurcates early after receptor engagement into pro-survival and pro-death arms.114 In the screens we analyzed, the loss of one of the receptors of TNF, TNFRSF1A, and the inactivation of the initiator of TNF-dependent apoptosis, caspase 8, were commonly shown to cause resistance to immune attack,4,5,9 in line with our knowledge regarding the pro-death arm of the TNF pathway.115, 116, 117 Conversely, loss of key components of the pro-survival arm of the TNF signaling pathway, including TRAF2, TNFAIP3 and IKBKG, resulted in enhanced sensitivity to T-cell attack, again in line with our understanding of TNF signal relay.4,5,9,114,118
This bifurcation and compartmentalization of the TNF signaling pathway may constitute an interesting translational opportunity, in that it allows for the precise targeting of its pro-survival arm. Additionally, perturbations in the TNF pathway seem to behave consistently with this bifurcated model, and our general understanding (whether causing resistance or sensitivity), in both in vitro and in vivo settings,4,5,9 unlike the IFN-γ pathway. This implies that the major effects of tumor-intrinsic TNF perturbation are limited to the cell that is actually perturbed, again, unlike IFN-γ.
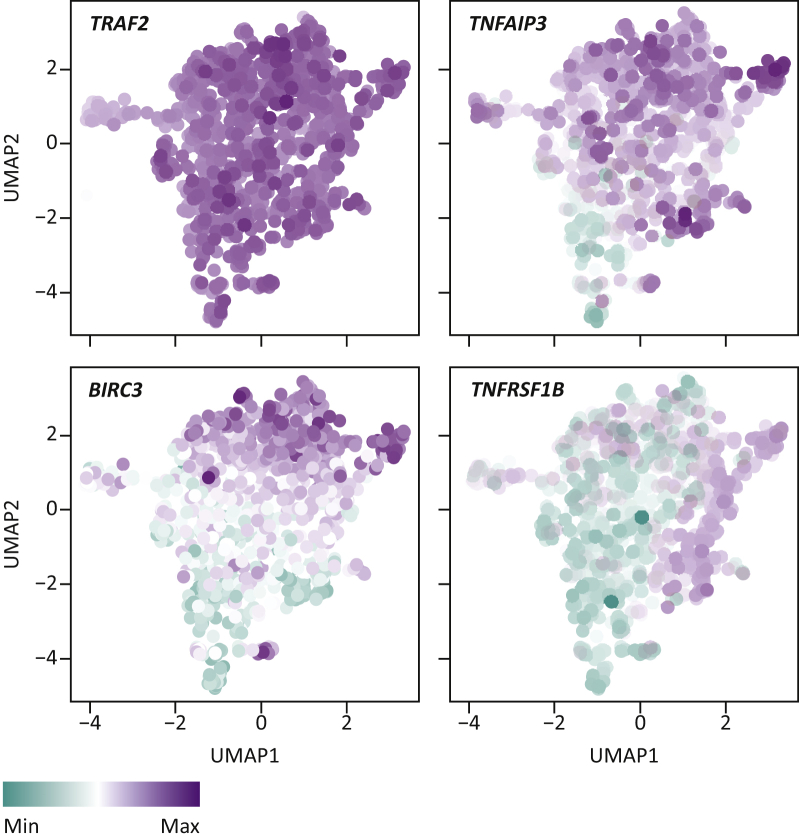
Despite consistently matching our understanding of the protumor and antitumor effects of TNF signaling, what is currently unclear is whether the TNF pathway has one general signaling node that can be broadly used as a therapeutic target. To illustrate this point, in the parallel screens carried out by Lawson and colleagues,5 TRAF2 KO was found to sensitize all six cell lines tested, except B16F10, while the loss of CFLAR sensitized all but MC38 cells and, differently still, BIRC2 inactivation sensitized none of the cell lines tested, except for EMT6.5 We and others have also observed such cell line-dependent sensitivity mechanisms: some cell lines depend heavily on TRAF2 to become resistant to TNF, while others rely more on, or even require, the activity of cIAP1/2.9,33,119 Although these proteins are thought to signal linearly, and in fact interact, these findings suggest that TRAF2 and cIAP1/2 incorporate and transmit TNF input signals not always through canonical signaling.114 Another example is the role of caspase 8: while this protein is thought to be required for canonical TNF-mediated apoptosis, its loss has also been shown to predispose leukemia cells to TNF-mediated necroptosis, further illustrating differential TNF signaling in different cell types.120,121 This heterogeneity in TNF signal relay, that is the differential dependence on different proteins for eventual TNF signal output, may at least in part be due to the differential expression of TNF pathway components, as can be observed from the Cancer Cell Line Encyclopedia (CCLE) (Figure 7), and/or by other (epi)genetic mechanisms. Nonetheless, these findings suggest that the TNF pathway, specifically its pro-survival arm, may be an attractive target for therapeutic translation. Further studies should aid in determining at which level in the pathway pharmacologic intervention provides the most effective and common clinical benefit.
Figure 7.
TNF signaling pathway heterogeneity in the CCLE database.
UMAP analysis of RNA expression of TNF pathway components in the Cancer Cell Line Encyclopedia (CCLE) database.151 Example expression values are given for TRAF2, TNFAIP3, BIRC3 and TNFRSF1B. Each datapoint corresponds to a single cell line analyzed.
TNF, tumor necrosis factor.
The use of TNF-neutralizing antibodies for patients experiencing ICB-induced immune-related adverse events (irAE) provides us with real-world clinical data and interesting insights into the clinical relevance of TNF in patient tumors. Multiple groups have reported that TNF neutralization administered after irAE onset did not affect or only marginally affected immunotherapy response.122, 123, 124, 125, 126 Supporting these clinical data, in murine models, prophylactic blockade of TNF ameliorated irAE, while preserving immunotherapy response.127, 128, 129 In contrast, Verheijden et al.130 found an association between anti-TNF treatment and decreased survival of melanoma patients receiving ICB. Most clinical data would be consistent with the idea that TNF alone is insufficient to mount a meaningful antitumor effect in tumors. We showed that the relatively low levels of TNF in untreated tumors and tumors not responding to ICB may contribute to this.9 Additionally, the relatively low antitumor activity of TNF may be due to genetic alterations: we found that in ICB-treated tumors where TNF levels are high, an accumulation of non-synonymous TNF pathway mutations correlated with a lack of response to treatment.9
Another relevant facet of TNF signaling in patient tumors is how it affects other cells in the TME and throughout the body. As we discussed above, from antibody neutralization studies it appeared that TNF can mediate severe irAEs.122, 123, 124, 125, 126,130 A phase I clinical trial with the second mitochondrial-derived activator of caspase (SMAC)-mimetic birinapant (targeting cIAP1 in the pro-survival TNF pathway) suggested that it is well tolerable,131 although it remains to be determined how this will develop in conjunction with ICB. Additionally, Bertrand and colleagues127,128 demonstrated in murine models that TNF can limit both CD4+ T cell and CD8+ T cell accumulation in the tumor, in part by directly driving activation-induced cell death in the affected cells, thereby limiting ICB effectivity. This notwithstanding, we conclude that the screening data align generally well with observations from the clinic, indicating the requirement, and opportunity, for specific tumor-intrinsic perturbations of the pro-survival arm of the TNF signaling pathway to unleash its tumoricidal potential.
Autophagy
Perturbation of autophagy was commonly observed to increase sensitivity to immune attack, both in vitro and in vivo.3,5,7,9,132 While the mechanism by which this occurs was not entirely elucidated, it was shown that this was at least in part due to enhanced sensitivity to TNF, underscoring its role in determining tumor susceptibility to immune killing. This finding supports an earlier report showing that inhibition of autophagy enhanced TNF-dependent liver injury, by promoting the activity of caspase 8.133 This finding may also be therapeutically exploited, given the fact that inhibitors of the autophagic machinery are available and seem to have some in vivo activity in murine models.134, 135, 136 One complicating matter of using inhibitors of autophagy, however, is the dependency of other cell types on this process. For example, CD8+ T cells deficient in autophagy seem to have enhanced effector function, but are less capable of forming long-term memory, thus potentially limiting this pharmaceutical approach.137, 138, 139 Additionally, Lawson and colleagues5 demonstrated that the loss of a single autophagy gene results in enhanced sensitivity to T-cell attack, but that the loss of multiple genes in fact reduces sensitivity to T-cell attack.5 Why and how this suppression and masking, as termed by Lawson and colleagues,5 of some autophagy components by others occurs, is unknown. Pharmaceutical intervention in the autophagic activity, through this suppression, could therefore also result in resistance to immune attack, as was seen in some cell lines.5 This differential effect of autophinib in different cell lines may also be indicative of cellular heterogeneity, and the reliance of cell lines on different components of the autophagic machinery, as was observed in the genetic screens (Figure 2). It is clear then, that more research is necessary to fully comprehend the translational value of these findings.
Conclusions and future outlook
The genetic screens above highlight the power and relevance of genetic screens both for increasing our understanding of tumor-intrinsic immune resistance mechanisms and for identifying new immunotherapeutic targets. What has also become apparent, however, is that both the cellular (the heterogeneity of TNF signal relay, NK cell ligand expression and different effects of autophagy perturbation in tumor and T cells) and environmental contexts (the differential effects of IFN-γ and AP in vitro and in vivo) in which these screens were carried out influence the screen outcomes. A major step forward would be the development of a large-scale, standardized screening approach and database for immune dependencies, much like what has been done previously for genetic fitness dependencies in tumor cells.140,141 Extending beyond that, the CRISPR-Cas9 technology has fueled the generation of new in vivo models that enable carrying out screens in immune cells to understand their limitations and unravel mechanisms for potential clinical exploitation, like multiple groups have started doing.142, 143, 144, 145, 146, 147 These efforts complement the tumor-centric approaches discussed here. Together, these screening strategies will conceivably contribute to more effective and rational ICB (combination) treatments, allowing more patients to durably respond.
Acknowledgments
Funding
This work is supported by the Dutch Cancer Society, Netherlands [grant number NKI 2014-7241] and by base funding from Oncode Institute.
Disclosure
DSP is co-founder, shareholder, and advisor of Immagene. DSP and DWV filed for patents covering the use of TRAF2 and cIAP1/2 inhibitors in cancer. GA has declared no conflicts of interest.
References
- 1.Cong L., Ran F.A., Cox D., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shalem O., Sanjana N.E., Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16(5):299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubrot J., Lane-Reticker S.K., Kessler E.A., et al. In vivo screens using a selective CRISPR antigen removal lentiviral vector system reveal immune dependencies in renal cell carcinoma. Immunity. 2021;54(3):571–585.e6. doi: 10.1016/j.immuni.2021.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Kearney C.J., Vervoort S.J., Hogg S.J., et al. Tumor immune evasion arises through loss of TNF sensitivity. Sci Immunol. 2018;3(23):eaar3451. doi: 10.1126/sciimmunol.aar3451. [DOI] [PubMed] [Google Scholar]
- 5.Lawson K.A., Sousa C.M., Zhang X., et al. Functional genomic landscape of cancer-intrinsic evasion of killing by T cells. Nature. 2020;586(7827):120–126. doi: 10.1038/s41586-020-2746-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manguso R.T., Pope H.W., Zimmer M.D., et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. 2017;547(7664):413–418. doi: 10.1038/nature23270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan D., Kobayashi A., Jiang P., et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science. 2018;359(6377):770–775. doi: 10.1126/science.aao1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel S.J., Sanjana N.E., Kishton R.J., et al. Identification of essential genes for cancer immunotherapy. Nature. 2017;548(7669):537–542. doi: 10.1038/nature23477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vredevoogd D.W., Kuilman T., Ligtenberg M.A., et al. Augmenting immunotherapy impact by lowering tumor TNF cytotoxicity threshold. Cell. 2019;178(3):585–599.e15. doi: 10.1016/j.cell.2019.06.014. [DOI] [PubMed] [Google Scholar]
- 10.Eggermont A.M.M., Blank C.U., Mandala M., et al. Adjuvant pembrolizumab versus placebo in resected stage III melanoma. N Engl J Med. 2018;378(19):1789–1801. doi: 10.1056/NEJMoa1802357. [DOI] [PubMed] [Google Scholar]
- 11.Forde P.M., Chaft J.E., Smith K.N., et al. Neoadjuvant PD-1 blockade in resectable lung cancer. N Engl J Med. 2018;378(21):1976–1986. doi: 10.1056/NEJMoa1716078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodi F.S., O'Day S.J., McDermott D.F., et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larkin J., Chiarion-Sileni V., Gonzalez R., et al. Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med. 2019;381(16):1535–1546. doi: 10.1056/NEJMoa1910836. [DOI] [PubMed] [Google Scholar]
- 14.Seidel J.A., Otsuka A., Kabashima K. Anti-PD-1 and anti-CTLA-4 therapies in cancer: mechanisms of action, efficacy, and limitations. Front Oncol. 2018;8(MAR):86. doi: 10.3389/fonc.2018.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Topalian S.L., Hodi F.S., Brahmer J.R., et al. Safety, activity, and immune correlates of anti–PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber J., Mandala M., Del Vecchio M., et al. Adjuvant nivolumab versus ipilimumab in resected stage III or IV melanoma. N Engl J Med. 2017;377(19):1824–1835. doi: 10.1056/NEJMoa1709030. [DOI] [PubMed] [Google Scholar]
- 17.Yarchoan M., Hopkins A., Jaffee E.M. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377(25):2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma P., Hu-Lieskovan S., Wargo J.A., Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168(4):707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jerby-Arnon L., Shah P., Cuoco M.S., et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell. 2018;175(4):984–997.e24. doi: 10.1016/j.cell.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang P., Gu S., Pan D., et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24(10):1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joyce J.A., Fearon D.T. T cell exclusion, immune privilege, and the tumor microenvironment. Science. 2015;348(6230):74–80. doi: 10.1126/science.aaa6204. [DOI] [PubMed] [Google Scholar]
- 22.Rooney M.S., Shukla S.A., Wu C.J., Getz G., Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1-2):48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spranger S., Bao R., Gajewski T.F. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature. 2015;523(7559):231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 24.Alexandrov L.B., Nik-Zainal S., Wedge D.C., et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodman A.M., Kato S., Bazhenova L., et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16(11):2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Litchfield K., Reading J.L., Puttick C., et al. Meta-analysis of tumor- and T cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell. 2021;184(3):596–614.e14. doi: 10.1016/j.cell.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGranahan N., Furness A.J.S., Rosenthal R., et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paulson K.G., Voillet V., McAfee M.S., et al. Acquired cancer resistance to combination immunotherapy from transcriptional loss of class I HLA. Nat Commun. 2018;9(1):3868. doi: 10.1038/s41467-018-06300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenthal R., Cadieux E.L., Salgado R., et al. Neoantigen-directed immune escape in lung cancer evolution. Nature. 2019;567(7749):479–485. doi: 10.1038/s41586-019-1032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sade-Feldman M., Jiao Y.J., Chen J.H., et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat Commun. 2017;8(1):1–11. doi: 10.1038/s41467-017-01062-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schumacher T.N., Schreiber R.D. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 32.Zaretsky J.M., Garcia-Diaz A., Shin D.S., et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375(9):819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kearney C.J., Lalaoui N., Freeman A.J., Ramsbottom K.M., Silke J., Oliaro J. PD-L1 and IAPs co-operate to protect tumors from cytotoxic lymphocyte-derived TNF. Cell Death Differ. 2017;24(10):1705–1716. doi: 10.1038/cdd.2017.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen P.L., Roh W., Reuben A., et al. Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 2016;6(8):827–837. doi: 10.1158/2159-8290.CD-15-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dighe A.S., Richards E., Old L.J., Schreiber R.D. Enhanced in vivo growth and resistance to rejection of tumor cells expressing dominant negative IFNγ receptors. Immunity. 1994;1(6):447–456. doi: 10.1016/1074-7613(94)90087-6. [DOI] [PubMed] [Google Scholar]
- 36.Gao J., Shi L.Z., Zhao H., et al. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell. 2016;167(2):397–404.e9. doi: 10.1016/j.cell.2016.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khazen R., Müller S., Gaudenzio N., Espinosa E., Puissegur M.P., Valitutti S. Melanoma cell lysosome secretory burst neutralizes the CTL-mediated cytotoxicity at the lytic synapse. Nat Commun. 2016;7(1):1–15. doi: 10.1038/ncomms10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shankaran V., Ikeda H., Bruce A.T., et al. IFNγ, and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410(6832):1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 39.Shin D.S., Zaretsky J.M., Escuin-Ordinas H., et al. Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov. 2017;7(2):188–201. doi: 10.1158/2159-8290.CD-16-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sade-Feldman M., Yizhak K., Bjorgaard S.L., et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 2018;175(4):998–1013.e20. doi: 10.1016/j.cell.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y., Zhang Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol. 2020;17(8):807–821. doi: 10.1038/s41423-020-0488-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahmadzadeh M., Johnson L.A., Heemskerk B., et al. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood. 2009;114(8):1537–1544. doi: 10.1182/blood-2008-12-195792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tumeh P.C., Harview C.L., Yearley J.H., et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hogquist K.A., Jameson S.C., Heath W.R., Howard J.L., Bevan M.J., Carbone F.R. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76(1):17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 45.Overwijk W.W., Theoret M.R., Finkelstein S.E., et al. Tumor regression and autoimmunity after reversal of a functionally tolerant state of self-reactive CD8+ T cells. J Exp Med. 2003;198(4):569–580. doi: 10.1084/jem.20030590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan Z., Cui K., Murray D.M., et al. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005;19(14):1662–1667. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ando K., Hiroishi K., Kaneko T., et al. Perforin, Fas/Fas ligand, and TNF-alpha pathways as specific and bystander killing mechanisms of hepatitis C virus-specific human CTL. J Immunol. 1997;158(11):5283–5291. [PubMed] [Google Scholar]
- 48.Hoekstra M.E., Bornes L., Dijkgraaf F.E., et al. Long-distance modulation of bystander tumor cells by CD8+ T-cell-secreted IFN-γ. Nat Cancer. 2020;1(3):291–301. doi: 10.1038/s43018-020-0036-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thibaut R., Bost P., Milo I., et al. Bystander IFN-γ activity promotes widespread and sustained cytokine signaling altering the tumor microenvironment. Nat Cancer. 2020;1(3):302–314. doi: 10.1038/s43018-020-0038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang B., Karrison T., Rowley D.A., Schreiber H. IFN-γ- and TNF-dependent bystander eradication of antigen-loss variants in established mouse cancers. J Clin Invest. 2008;118(4):1398–1404. doi: 10.1172/JCI33522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Freeman A.J., Vervoort S.J., Michie J., et al. HOIP limits anti-tumor immunity by protecting against combined TNF and IFN-gamma-induced apoptosis. EMBO Rep. 2021:e53391. doi: 10.15252/embr.202153391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dranoff G., Jaffee E., Lazenby A., et al. Vaccination with irradiated tumor cells engineered to secrete murine granulocyte-macrophage colony-stimulating factor stimulates potent, specific, and long-lasting anti-tumor immunity. Proc Natl Acad Sci U S A. 1993;90(8):3539–3543. doi: 10.1073/pnas.90.8.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kvistborg P., Philips D., Kelderman S., et al. Anti-CTLA-4 therapy broadens the melanoma-reactive CD8+ T cell response. Sci Transl Med. 2014;6(254):254ra128. doi: 10.1126/scitranslmed.3008918. [DOI] [PubMed] [Google Scholar]
- 54.Sotomayor E.M., Borrello I., Tubb E., Allison J.P., Levitsky H.I. In vivo blockade of CTLA-4 enhances the priming of responsive T cells but fails to prevent the induction of tumor antigen-specific tolerance. Proc Natl Acad Sci U S A. 1999;96(20):11476–11481. doi: 10.1073/pnas.96.20.11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ishizuka J.J., Manguso R.T., Cheruiyot C.K., et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature. 2019;565(7737):43–48. doi: 10.1038/s41586-018-0768-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dupage M., Mazumdar C., Schmidt L.M., Cheung A.F., Jacks T. Expression of tumour-specific antigens underlies cancer immunoediting. Nature. 2012;482(7385):405–409. doi: 10.1038/nature10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neefjes J., Jongsma M.L.M., Paul P., Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011;11(12):823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 58.Dokun A.O., Kim S., Smith H.R.C., Kang H.S.P., Chu D.T., Yokoyama W.M. Specific and nonspecific NK cell activation during virus infection. Nat Immunol. 2001;2(10):951–956. doi: 10.1038/ni714. [DOI] [PubMed] [Google Scholar]
- 59.Gilfillan S., Ho E.L., Cella M., Yokohama W.M., Colonna M. NKG2D recriuts two distinct adapters to trigger NK cell activation and costimulation. Nat Immunol. 2002;3(12):1150–1155. doi: 10.1038/ni857. [DOI] [PubMed] [Google Scholar]
- 60.Smyth M.J., Cretney E., Kelly J.M., et al. Activation of NK cell cytotoxicity. Mol Immunol. 2005;42(4 SPEC. ISS.):501–510. doi: 10.1016/j.molimm.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 61.Apriamashvili G., Vredevoogd D.W., Krijgsman O., et al. Loss of ubiquitin ligase STUB1 amplifies IFNγ-R1/JAK1 signaling and sensitizes tumors to IFNγ. bioRxiv. 2020 doi: 10.1101/2020.07.07.191650. [DOI] [Google Scholar]
- 62.Ng S., Lim S., Nyi Sim A.C., et al. STUB1 is an intracellular checkpoint for interferon gamma sensing. bioRxiv. 2020 doi: 10.1101/2020.12.14.420539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rebeyev N. Cambridge Institute for Medical Research; Cabridge: 2020. CHIC2 and STUB1 regulate interferon-γ receptor cell surface expression [doctoral thesis] [DOI] [Google Scholar]
- 64.Bromberg J.F., Horvath C.M., Wen Z., Schreiber R.D., Darnell J.E. Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon α and interferon γ. Proc Natl Acad Sci U S A. 1996;93(15):7673–7678. doi: 10.1073/pnas.93.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kärre K., Ljunggren H.G., Piontek G., Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319(6055):675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 66.Gettinger S., Choi J., Hastings K., et al. Impaired HLA class I antigen processing and presentation as a mechanism of acquired resistance to immune checkpoint inhibitors in lung cancer. Cancer Discov. 2017;7(12):1420–1435. doi: 10.1158/2159-8290.CD-17-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khong H.T., Restifo N.P. Natural selection of tumor variants in the generation of “tumor escape” phenotypes. Nat Immunol. 2002;3(11):999–1005. doi: 10.1038/ni1102-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bauer S., Groh V., Wu J., et al. Activation of NK cells and T cells by NKG2D, a receptor for stress- inducible MICA. Science. 1999;285(5428):727–729. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 69.Herberman R.B., Holden H.T. Natural cell-mediated immunity. Adv Cancer Res. 1978;27:305–377. doi: 10.1016/s0065-230x(08)60936-7. [DOI] [PubMed] [Google Scholar]
- 70.lo Monaco E., Tremante E., Cerboni C., et al. Human leukocyte antigen E contributes to protect tumor cells from lysis by natural killer cells. Neoplasia. 2011;13(9):822–830. doi: 10.1593/neo.101684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moretta L., Bottino C., Pende D., Vitale M., Mingari M.C., Moretta A. Different checkpoints in human NK-cell activation. Trends Immunol. 2004;25(12):670–676. doi: 10.1016/j.it.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 72.Thommen D.S., Schumacher T.N. T cell dysfunction in cancer. Cancer Cell. 2018;33(4):547–562. doi: 10.1016/j.ccell.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Coudert J.D., Scarpellino L., Gros F., Vivier E., Held W. Sustained NKG2D engagement induces cross-tolerance of multiple distinct NK cell activation pathways. Blood. 2008;111(7):3571–3578. doi: 10.1182/blood-2007-07-100057. [DOI] [PubMed] [Google Scholar]
- 74.Hsu J., Hodgins J.J., Marathe M., et al. Contribution of NK cells to immunotherapy mediated by PD-1/PD-L1 blockade. J Clin Invest. 2018;128(10):4654–4668. doi: 10.1172/JCI99317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oppenheim D.E., Roberts S.J., Clarke S.L., et al. Sustained localized expression of ligand for the activating NKG2D receptor impairs natural cytotoxicity in vivo and reduces tumor immunosurveillance. Nat Immunol. 2005;6(9):928–937. doi: 10.1038/ni1239. [DOI] [PubMed] [Google Scholar]
- 76.Tripathy S.K., Keyel P.A., Yang L., et al. Continuous engagement of a self-specific activation receptor induces NK cell tolerance. J Exp Med. 2008;205(8):1829–1841. doi: 10.1084/jem.20072446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Crouse J., Xu H.C., Lang P.A., Oxenius A. NK cells regulating T cell responses: mechanisms and outcome. Trends Immunol. 2015;36(1):49–58. doi: 10.1016/j.it.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 78.Fehniger T.A., Cooper M.A., Nuovo G.J., et al. CD56bright natural killer cells are present in human lymph nodes and are activated by T cell-derived IL-2: a potential new link between adaptive and innate immunity. Blood. 2003;101(8):3052–3057. doi: 10.1182/blood-2002-09-2876. [DOI] [PubMed] [Google Scholar]
- 79.Waggoner S.N., Cornberg M., Selin L.K., Welsh R.M. Natural killer cells act as rheostats modulating antiviral T cells. Nature. 2012;481(7381):394–398. doi: 10.1038/nature10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Buard A., Vivo C., Monnet I., Boutin C., Pilatte Y., Jaurand M.C. Human malignant mesothelioma cell growth: activation of janus kinase 2 and signal transducer and activator of transcription 1α for inhibition by interferon-γ. Cancer Res. 1998;58(4):840–847. [PubMed] [Google Scholar]
- 81.Chen B., He L., Savell V.H., Jenkins J.J., Parham D.M. Inhibition of the interferon-γ/signal transducers and activators of transcription (STAT) pathway by hypermethylation at a STAT-binding site in the p21(WAF1) promoter region. Cancer Res. 2000;60(12):3290–3298. [PubMed] [Google Scholar]
- 82.Chin Y.E., Kitagawa M., Su W.C.S., You Z.H., Iwamoto Y., Fu X.Y. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21WAF1/CIP1 Mediated by STAT1. Science. 1996;272(5262):719–722. doi: 10.1126/science.272.5262.719. [DOI] [PubMed] [Google Scholar]
- 83.Chin Y.E., Kitagawa M., Kuida K., Flavell R.A., Fu X.Y. Activation of the STAT signaling pathway can cause expression of caspase 1 and apoptosis. Mol Cell Biol. 1997;17(9):5328–5337. doi: 10.1128/mcb.17.9.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mandal M., Bandyopadhyay D., Goepfert T.M., Kumar R. Interferon-induces expression of cyclin-dependent kinase-inhibitors p21(WAF1) and p27(Kip1) that prevent activation of cyclin-dependent kinase by CDK-activating kinase (CAK) Oncogene. 1998;16(2):217–225. doi: 10.1038/sj.onc.1201529. [DOI] [PubMed] [Google Scholar]
- 85.Detjen K.M., Farwig K., Welzel M., Wiedenmann B., Rosewicz S. Interferon γ inhibits growth of human pancreatic carcinoma cells via caspase-1 dependent induction of apoptosis. Gut. 2001;49(2):251–262. doi: 10.1136/gut.49.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Malik S.T.A., Knowles R.G., East N., Lando D., Stamp G., Balkwill F.R. Antitumor activity of gamma-interferon in ascitic and solid tumor models of human ovarian cancer. Cancer Res. 1991;51(24):6643–6649. [PubMed] [Google Scholar]
- 87.Siegmund D., Wicovsky A., Schmitz I., et al. Death receptor-induced signaling pathways are differentially regulated by gamma interferon upstream of caspase 8 processing. Mol Cell Biol. 2005;25:6363–6379. doi: 10.1128/MCB.25.15.6363-6379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu X., Fu X.Y., Plate J., Chong A.S.-F. IFN-γ induces cell growth inhibition by fas-mediated apoptosis: requirement of STAT1 protein for up-regulation of Fas and FasL expression. Cancer Res. 1998;58(13):2832–2837. [PubMed] [Google Scholar]
- 89.Früh K., Yang Y. Antigen presentation by MHC class I and its regulation by interferon γ. Curr Opin Immunol. 1999;11(1):76–81. doi: 10.1016/s0952-7915(99)80014-4. [DOI] [PubMed] [Google Scholar]
- 90.Ikeda H., Old L.J., Schreiber R.D. The roles of IFNγ in protection against tumor development and cancer immunoediting. Cytokine Growth Factor Rev. 2002;13(2):95–109. doi: 10.1016/s1359-6101(01)00038-7. [DOI] [PubMed] [Google Scholar]
- 91.Cole K.E., Strick C.A., Paradis T.J., et al. Interferon-inducible T cell alpha chemoattractant (I-TAC): a novel non- ELR CXC chemokine with potent activity on activated T cells through selective high affinity binding to CXCR3. J Exp Med. 1998;187(12):2009–2021. doi: 10.1084/jem.187.12.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liao F., Rabin R.L., Yannelli J.R., Koniaris L.G., Vanguri P., Farber J.M. Human mig chemokine: biochemical and functional characterization. J Exp Med. 1995;182(5):1301–1314. doi: 10.1084/jem.182.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Luster A.D., Leder P. IP-10, a -C-X-C- chemokine, elicits a potent thymus-dependent antitumor response in vivo. J Exp Med. 1993;178(3):1057–1065. doi: 10.1084/jem.178.3.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ayers M., Lunceford J., Nebozhyn M., et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu D.D., Schilling B., Liu D.D., et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat Med. 2019;25(12):1916–1927. doi: 10.1038/s41591-019-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rozeman E.A., Hoefsmit E.P., Reijers I.L.M., et al. Survival and biomarker analyses from the OpACIN-neo and OpACIN neoadjuvant immunotherapy trials in stage III melanoma. Nat Med. 2021;27(2):256–263. doi: 10.1038/s41591-020-01211-7. [DOI] [PubMed] [Google Scholar]
- 97.Kaplan D.H., Shankaran V., Dighe A.S., et al. Demonstration of an interferon γ-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci U S A. 1998;95(13):7556–7561. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dong H., Strome S.E., Salomao D.R., et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8(8):793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 99.Freeman G.J., Long A.J., Iwai Y., et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192(7):1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brahmer J.R., Drake C.G., Wollner I., et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J Clin Oncol. 2010;28(19):3167–3175. doi: 10.1200/JCO.2009.26.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Benson D.M., Bakan C.E., Mishra A., et al. The PD-1/PD-L1 axis modulates the natural killer cell versus multiple myeloma effect: a therapeutic target for CT-011, a novel monoclonal anti-PD-1 antibody. Blood. 2010;116(13):2286–2294. doi: 10.1182/blood-2010-02-271874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fallarino F., Grohmann U., Vacca C., et al. T cell apoptosis by tryptophan catabolism. Cell Death Differ. 2002;9(10):1069–1077. doi: 10.1038/sj.cdd.4401073. [DOI] [PubMed] [Google Scholar]
- 103.Bartok O., Pataskar A., Nagel R., et al. Anti-tumour immunity induces aberrant peptide presentation in melanoma. Nature. 2021;590(7845):332–337. doi: 10.1038/s41586-020-03054-1. [DOI] [PubMed] [Google Scholar]
- 104.Benci J.L., Xu B., Qiu Y., et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell. 2016;167(6):1540–1554.e12. doi: 10.1016/j.cell.2016.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Benci J.L., Johnson L.R., Choa R., et al. Opposing functions of interferon coordinate adaptive and innate immune responses to cancer immune checkpoint blockade. Cell. 2019;178(4):933–948.e14. doi: 10.1016/j.cell.2019.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hellmann M.D., Nathanson T., Rizvi H., et al. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell. 2018;33(5):843–852.e4. doi: 10.1016/j.ccell.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Williams J.B., Li S., Higgs E.F., et al. Tumor heterogeneity and clonal cooperation influence the immune selection of IFN-γ-signaling mutant cancer cells. Nat Commun. 2020;11(1):1–14. doi: 10.1038/s41467-020-14290-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Freeman A.J., Vervoort S.J., Ramsbottom K.M., et al. Natural killer cells suppress T cell-associated tumor immune evasion. Cell Rep. 2019;28(11):2784–2794.e5. doi: 10.1016/j.celrep.2019.08.017. [DOI] [PubMed] [Google Scholar]
- 109.Zhuang X., Veltri D.P., Long E.O. Genome-wide CRISPR screen reveals cancer cell resistance to NK cells induced by NK-derived IFN-γ. Front Immunol. 2019;10:2879. doi: 10.3389/fimmu.2019.02879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Aquino-López A., Senyukov V.V., Vlasic Z., Kleinerman E.S., Lee D.A. Interferon gamma induces changes in natural killer (NK) cell ligand expression and alters NK cell-mediated lysis of pediatric cancer cell lines. Front Immunol. 2017;8:391. doi: 10.3389/fimmu.2017.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Badovinac V.P., Tvinnereim A.R., Harty J.T. Regulation of antigen-specific CD8+ T cell homeostasis by perforin and interferon-γ. Science. 2000;290(5495):1354–1357. doi: 10.1126/science.290.5495.1354. [DOI] [PubMed] [Google Scholar]
- 112.Sercan Ö., Stoycheva D., Hämmerling G.J., Arnold B., Schüler T. IFN-γ receptor signaling regulates memory CD8 + T cell differentiation. J Immunol. 2010;184(6):2855–2862. doi: 10.4049/jimmunol.0902708. [DOI] [PubMed] [Google Scholar]
- 113.Sercan Ö., Hämmerling G.J., Arnold B., Schüler T. Cutting edge: innate immune cells contribute to the IFN-γ-dependent regulation of antigen-specific CD8 + T cell homeostasis. J Immunol. 2006;176(2):735–739. doi: 10.4049/jimmunol.176.2.735. [DOI] [PubMed] [Google Scholar]
- 114.Chen G., Goeddel D.V. TNF-R1 signaling: a beautiful pathway. Science. 2002;296(5573):1634–1635. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- 115.Lin Y., Devin A., Rodriguez Y., Liu Z.G. Cleavage of the death domain kinase RIP by Caspase-8 prompts TNF-induced apoptosis. Genes Dev. 1999;13(19):2514–2526. doi: 10.1101/gad.13.19.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Varfolomeev E.E., Schuchmann M., Luria V., et al. Targeted disruption of the mouse Caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity. 1998;9(2):267–276. doi: 10.1016/s1074-7613(00)80609-3. [DOI] [PubMed] [Google Scholar]
- 117.Wang L., Du F., Wang X. TNF-α induces two distinct caspase-8 activation pathways. Cell. 2008;133(4):693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 118.Yeh W.-C., Shahinian A., Speiser D., et al. Early lethality, functional NF-κB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity. 1997;7(5):715–725. doi: 10.1016/s1074-7613(00)80391-x. [DOI] [PubMed] [Google Scholar]
- 119.Dufva O., Koski J., Maliniemi P., et al. Integrated drug profiling and CRISPR screening identify essential pathways for CAR T-cell cytotoxicity. Blood. 2020;135(9):597–609. doi: 10.1182/blood.2019002121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Brumatti G., Ma C., Lalaoui N., et al. The caspase-8 inhibitor emricasan combines with the SMAC mimetic birinapant to induce necroptosis and treat acute myeloid leukemia. Sci Transl Med. 2016;8(339):339ra69. doi: 10.1126/scitranslmed.aad3099. [DOI] [PubMed] [Google Scholar]
- 121.McComb S., Aguadé-Gorgorió J., Harder L., et al. Activation of concurrent apoptosis and necroptosis by SMAC mimetics for the treatment of refractory and relapsed ALL. Sci Transl Med. 2016;8(339):339ra70. doi: 10.1126/scitranslmed.aad2986. [DOI] [PubMed] [Google Scholar]
- 122.Arriola E., Wheater M., Karydis I., Thomas G., Ottensmeier C. Infliximab for IPILIMUMAB-related colitis-letter. Clin Cancer Res. 2015;21(24):5642–5643. doi: 10.1158/1078-0432.CCR-15-2471. [DOI] [PubMed] [Google Scholar]
- 123.Badran Y.R., Cohen J.V., Brastianos P.K., Parikh A.R., Hong T.S., Dougan M. Concurrent therapy with immune checkpoint inhibitors and TNFα blockade in patients with gastrointestinal immune-related adverse events. J Immunother Cancer. 2019;7(1):226. doi: 10.1186/s40425-019-0711-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Johnson D.H., Zobniw C.M., Trinh V.A., et al. Infliximab associated with faster symptom resolution compared with corticosteroids alone for the management of immune-related enterocolitis. J Immunother Cancer. 2018;6(1):103. doi: 10.1186/s40425-018-0412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lesage C., Longvert C., Prey S., et al. Incidence and clinical impact of anti-TNFα treatment of severe immune checkpoint inhibitor-induced colitis in advanced melanoma: the Mecolit Survey. J Immunother. 2019;42(5):175–179. doi: 10.1097/CJI.0000000000000268. [DOI] [PubMed] [Google Scholar]
- 126.Weber J.S., Larkin J.M.G., Schadendorf D., et al. Management of gastrointestinal (GI) toxicity associated with nivolumab (NIVO) plus ipilimumab (IPI) or IPI alone in phase II and III trials in advanced melanoma (MEL) J Clin Oncol. 2017;35(suppl 15):9523. [Google Scholar]
- 127.Bertrand F., Rochotte J., Colacios C., et al. Blocking tumor necrosis factor α enhances CD8 T-cell-dependent immunity in experimental melanoma. Cancer Res. 2015;75(13):2619–2628. doi: 10.1158/0008-5472.CAN-14-2524. [DOI] [PubMed] [Google Scholar]
- 128.Bertrand F., Montfort A., Marcheteau E., et al. TNFα blockade overcomes resistance to anti-PD-1 in experimental melanoma. Nat Commun. 2017;8(1):2256. doi: 10.1038/s41467-017-02358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Perez-Ruiz E., Minute L., Otano I., et al. Prophylactic TNF blockade uncouples efficacy and toxicity in dual CTLA-4 and PD-1 immunotherapy. Nature. 2019;569(7756):428–432. doi: 10.1038/s41586-019-1162-y. [DOI] [PubMed] [Google Scholar]
- 130.Verheijden R.J., May A.M., Blank C.U., et al. Association of anti-TNF with decreased survival in steroid refractory ipilimumab and Anti-PD1-treated patients in the Dutch melanoma treatment registry. Clin Cancer Res. 2020;26(9):2268–2274. doi: 10.1158/1078-0432.CCR-19-3322. [DOI] [PubMed] [Google Scholar]
- 131.Amaravadi R.K., Schilder R.J., Martin L.P., et al. A phase I study of the SMAC-mimetic birinapant in adults with refractory solid tumors or lymphoma. Mol Cancer Ther. 2015;14(11):2569–2575. doi: 10.1158/1535-7163.MCT-15-0475. [DOI] [PubMed] [Google Scholar]
- 132.Orvedahl A., McAllaster M.R., Sansone A., et al. Autophagy genes in myeloid cells counteract IFNγ-induced TNF-mediated cell death and fatal TNF-induced shock. Proc Natl Acad Sci U S A. 2019;116(33):16497–16506. doi: 10.1073/pnas.1822157116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Amir M., Zhao E., Fontana L., et al. Inhibition of hepatocyte autophagy increases tumor necrosis factor-dependent liver injury by promoting caspase-8 activation. Cell Death Differ. 2013;20(7):878–887. doi: 10.1038/cdd.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fu Y., Hong L., Xu J., et al. Discovery of a small molecule targeting autophagy via ATG4B inhibition and cell death of colorectal cancer cells in vitro and in vivo. Autophagy. 2019;15(2):295–311. doi: 10.1080/15548627.2018.1517073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.McAfee Q., Zhang Z., Samanta A., et al. Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc Natl Acad Sci U S A. 2012;109(21):8253–8258. doi: 10.1073/pnas.1118193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Robke L., Laraia L., Carnero Corrales M.A., et al. Phenotypic identification of a novel autophagy inhibitor chemotype targeting lipid kinase VPS34. Angew Chem Int Ed Engl. 2017;56(28):8153–8157. doi: 10.1002/anie.201703738. [DOI] [PubMed] [Google Scholar]
- 137.DeVorkin L., Pavey N., Carleton G., et al. Autophagy regulation of metabolism is required for CD8 + T cell anti-tumor immunity. Cell Rep. 2019;27(2):502–513.e5. doi: 10.1016/j.celrep.2019.03.037. [DOI] [PubMed] [Google Scholar]
- 138.Puleston D.J., Zhang H., Powell T.J., et al. Autophagy is a critical regulator of memory CD8+ T cell formation. Elife. 2014;3:e03706. doi: 10.7554/eLife.03706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Xu X., Araki K., Li S., et al. Autophagy is essential for effector CD8 + T cell survival and memory formation. Nat Immunol. 2014;15(12):1152–1161. doi: 10.1038/ni.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Behan F.M., Iorio F., Picco G., et al. Prioritization of cancer therapeutic targets using CRISPR–Cas9 screens. Nature. 2019;568(7753):511–516. doi: 10.1038/s41586-019-1103-9. [DOI] [PubMed] [Google Scholar]
- 141.Tsherniak A., Vazquez F., Montgomery P.G., et al. Defining a cancer dependency map. Cell. 2017;170(3):564–576.e16. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chen Z., Arai E., Khan O., et al. In vivo CD8+ T cell CRISPR screening reveals control by Fli1 in infection and cancer. Cell. 2021;184(5):1262–1280.e22. doi: 10.1016/j.cell.2021.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dong M.B., Wang G., Chow R.D., et al. Systematic immunotherapy target discovery using genome-scale in vivo CRISPR screens in CD8 T cells. Cell. 2019;178(5):1189–1204.e23. doi: 10.1016/j.cell.2019.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gurusamy D., Henning A.N., Yamamoto T.N., et al. Multi-phenotype CRISPR-Cas9 Screen Identifies p38 Kinase as a Target for Adoptive Immunotherapies. Cancer Cell. 2020;37(6):818–833.e9. doi: 10.1016/j.ccell.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Henriksson J., Chen X., Gomes T., et al. Genome-wide CRISPR screens in T helper cells reveal pervasive crosstalk between activation and differentiation. Cell. 2019;176(4):882–896.e18. doi: 10.1016/j.cell.2018.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shifrut E., Carnevale J., Tobin V., et al. Genome-wide CRISPR screens in primary human T cells reveal key regulators of immune function. Cell. 2018;175(7):1958–1971.e15. doi: 10.1016/j.cell.2018.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wei J., Long L., Zheng W., et al. Targeting REGNASE-1 programs long-lived effector T cells for cancer therapy. Nature. 2019;576(7787):471–476. doi: 10.1038/s41586-019-1821-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Szklarczyk D., Gable A.L., Lyon D., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Koopmans F., van Nierop P., Andres-Alonso M., et al. SynGO: an evidence-based, expert-curated knowledge base for the synapse. Neuron. 2019;103(2):217–234.e4. doi: 10.1016/j.neuron.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mi H., Muruganujan A., Huang X., et al. Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0) Nat Protoc. 2019;14(3):703–721. doi: 10.1038/s41596-019-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ghandi M., Huang F.W., Jané-Valbuena J., et al. Next-generation characterization of the cancer cell line encyclopedia. Nature. 2019;569(7757):503–508. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]