Abstract
Over the last few decades, immunotherapy has shown significant therapeutic efficacy in a broad range of cancer types. Antitumor immune responses are contingent on the recognition of tumor-specific antigens, which are termed neoantigens. Tumor neoantigens are ideal targets for immunotherapy since they can be recognized as non-self antigens by the host immune system and thus are able to elicit an antitumor T-cell response. There are an increasing number of studies that highlight the importance of tumor neoantigens in immunoediting and in the sensitivity to immune checkpoint blockade. Therefore, one of the most fundamental tasks in the field of immuno-oncology research is the identification of patient-specific neoantigens. To this end, a plethora of computational approaches have been developed in order to predict tumor-specific aberrant peptides and quantify their likelihood of binding to patients' human leukocyte antigen molecules in order to be recognized by T cells. In this review, we systematically summarize and present the most recent advances in computational neoantigen prediction, and discuss the challenges and novel methods that are being developed to resolve them.
Key words: neoantigens, immunotherapy, personalized medicine
Highlights
-
•
Tumors have the ability to acquire immune escape mechanisms.
-
•
Tumor-specific aberrant peptides (neoantigens) can elicit an immune response by the host immune system.
-
•
The identification of neoantigens is one of the most fundamental tasks in the field of immuno-oncology research.
-
•
A plethora of computational approaches have been developed in order to predict patient-specificneoantigens.
Introduction
Conventional treatment of malignant tumors is based upon surgery, chemotherapy, and radiation therapy, each of which has its advantages and drawbacks. Surgical procedures cannot always ensure the complete removal of tumor cells, and recent studies show that the inflammatory response to a post-operative infection can increase the risk of tumor recurrence in cancer through the release of proinflammatory mediators.1,2 Radiation therapy and chemotherapy can elicit acquired resistance by different mechanisms, including multidrug resistance, suppression of apoptosis, altered drug metabolism, and enhanced DNA repair and gene amplification.3,4
Immunotherapy that harnesses the power of the immune system to target malignant cells has emerged in recent years and is showing remarkable results in clinical trials. One of the major drawbacks of immunotherapy is that tumor cells can evolve immunoevasive and immunosuppressive phenotypes, thus achieving immune escape. Immunosuppressive tumor cells can express membrane proteins like the programmed death-ligand 1 protein which binds to its receptor [programmed cell death protein 1 (PD-1)] on activated T cells and delivers a signal that inhibits T-cell receptor (TCR)-mediated activation of interleukin-2 production and T-cell proliferation.5,6 The 2018 Nobel Prize in physiology and medicine award winners, James P. Allison and Tasuku Honjo, have shown that the PD-1 blockade is effective against many types of tumors because it enhances the antitumor activity of cytotoxic T-lymphocytes, which recognize various tumor-specific antigens (TSAs),7,8 and these findings formed the basis of the immune checkpoint inhibition (ICI) therapy.
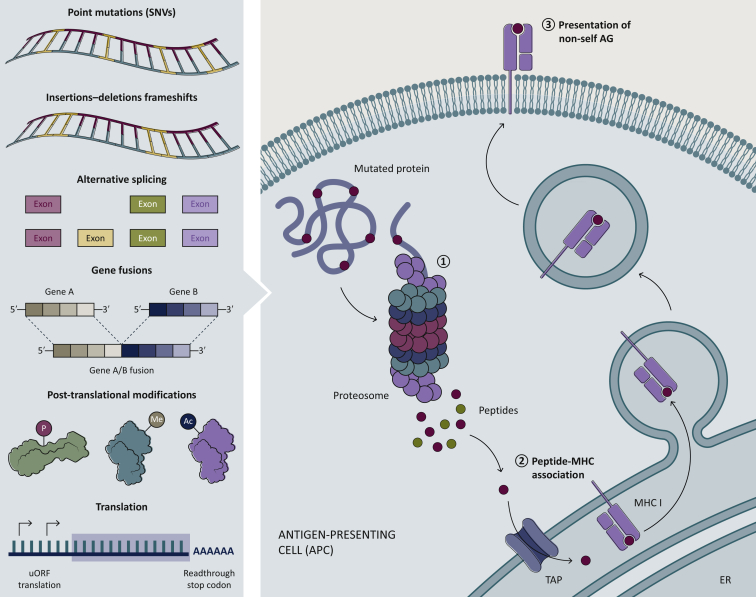
The inherent genetic instability of tumor cells leads to the occurrence of a large number of non-synonymous somatic mutations that are not present in healthy tissue. Expression of these tumor-specific mutations will produce aberrant proteins which will subsequently be proteolytically cleaved by the proteasome (Figure 1). The resulting mutated peptides are then transferred to the endoplasmic reticulum (ER) lumen through the transporter associated with antigen processing (TAP) complex where they will be made available for binding to major histocompatibility complex class I (MHC-I; in vertebrates) molecules or the human leukocyte antigen class I (HLA-I; in humans) within the peptide loading complex.9 These peptides, named neoantigens, are defined as the tumor-specific mutated peptides that are presented on the membrane of malignant cells via the HLA-I protein complex and are not subjected to central or peripheral tolerance, thus being capable of inducing CD8+ T-cell mediated antitumor responses.10,11 Over the years, mouse cancer models and strong correlative clinical data provided definitive experimental evidence of how targeting neoantigens can result in a positive response to immune-mediated therapies.12, 13, 14 Whereas HLA-I molecules are expressed by most nucleated cells and primarily present endogenously-derived peptide antigens to CD8+ T cells, HLA class II (HLA-II) molecules are predominantly expressed by professional antigen presenting cells (pAPCs) and present antigenic peptides—mainly generated from exogenous proteins—to CD4+ T cells. Despite the fact that HLA-II is constitutively expressed only on pAPCs, there is evidence that HLA-II expression can be induced by interferon-γ in many other cell types, including tumor cells15 (HLA-II-positive malignant cells). Briefly, once HLA-II chains are complexed with the invariant chain (Ii) protein and the co-chaperone HLA-DM in the ER, the complex buds off in a vesicle that fuses with endosomes. Subsequently, HLA-II is loaded with endocytosed exogenous peptides or endogenously-derived peptides originating from autophagy.16 Stabilized peptide-HLA-II complexes are presented to CD4+ T cells on the cell surface. Tumor-specific HLA-II expression has been associated with improved prognosis and response to immunotherapy in humans,17, 18, 19, 20 and increased tumor rejection in murine models.21,22
Figure 1.
Sources of non-self neoantigens. Neoantigens originate from mutated proteins expressed only in cancer cells.
These non-self antigens can derive from a number of different events at the gene, transcript, or protein level, such as point mutations (SNV), small insertions or deletions (indels), alternative splicing and fusion of genes. But also translation errors and post-transcriptional modifications can lead to aberrant proteins. These aberrant proteins are then processed by the proteasome and cleaved into shorter peptides. The transporter associated with antigen processing (TAP) brings these peptides to the endoplasmic reticulum, where they are loaded on to the major histocompatibility complex (MHC) molecule. The peptide–MHC complex is then transported to the cell surface and presented to T cells.
AG, antigen; ER, endoplasmic reticulum; SNVs, single nucleotide variations; uORF, upstream open reading frame.
Despite promising results and the increasing interest in immunotherapy, however, there are many technical challenges and questions that arise from the very nature of tumors and their ability to acquire immune escape mechanisms. In addition to immunosuppression, the immunoevasive attributes of tumor cells reside in the weak immunogenicity (defined as the ability of a peptide bound to an MHC molecule to induce adaptive immune responses) of most neoantigens. Therefore, the identification of immunogenic neoantigens is a pivotal step in the field of immuno-oncology research and plays an instrumental role in the development of novel immunotherapeutic approaches.
Advances in next generation sequencing (NGS) techniques has permitted improved identification of tumor-specific neoantigens and a better understanding of tumor–immune system interactions. Sequencing depth, quality of tumor tissue, the source of the sequencing material, and other factors, however, still pose a major challenge in the process of in silico neoantigen prediction. Moreover, the analysis of high-throughput sequencing data is a daunting task and requires a high level of bioinformatics expertise. A typical neoantigen prediction computational workflow can be summarized into three steps: (i) variant calling and inference of tumor-specific mutated peptides, (ii) HLA typing, and (iii) HLA binding affinity prediction and filtering/prioritization of neoantigens. The main focus of this review is to (i) highlight important information regarding the neoantigen landscape and the approaches available for mining this information for each class of neoantigens, (ii) present the latest developments and advances regarding the algorithms and computational frameworks available for the identification and prioritization of neoantigens that have emerged since our last survey, 23 and (iii) briefly discuss the technical caveats of the available methods, and also address some important biological questions that need to be addressed in order to develop methods that can predict the immunogenicity of neoantigens.
The tumor antigen landscape
Tumor cells express a broad spectrum of antigens including TSAs (or neoantigens), tumor-associated antigens (TAAs), and cancer germline antigens (CGAs). TAAs and CGAs are not expressed exclusively by tumor cells, but can also be found on the surface of cells residing within normal tissue.24,25 Due to their expression in healthy tissue, targeting such antigens would pose two issues: (i) poor results due to central immunological tolerance mechanisms, and more importantly (ii) increased risk for cross-reactivity with structurally related self-peptides, and off-target toxicities.26,27 Unlike TAAs and CGAs, neoantigens (TSAs) are expressed only by tumor cells and can thus be considered akin to truly foreign peptides, as they are completely absent from normal tissue, and therefore represent an ideal immunotherapy target since they can be recognized as non-self by the host immune system.28 Despite these theoretical advantages, however, neoantigen-specific approaches cannot completely eliminate the risk of autoimmunity. This is exemplified by the fact that neoantigens derived from single nucleotide variations (SNVs) can exhibit high resemblance to their normal counterparts and thus neoantigen-specific T cells can be cross-reactive with the non-mutated peptides.29,30
Sources of neoantigens
Neoantigens are cancer-specific aberrant peptides that can be recognized as non-self and which can elicit an immune response by the host immune system. These aberrations can result from several types of genomic or transcriptome-based alterations and post-translational modifications (PTMs) in tumors. The most well characterized neoantigens are the result of non-synonymous somatic mutations such as SNVs, small insertions, deletions (indels), frameshift mutations, or other genomic rearrangements, such as gene fusions.31, 32, 33 Neoantigens can also arise from post-transcriptional aberrations, including cancer-specific alternative exon splicing,34 intron retention,35 and premature transcription ending. Finally, another less explored source of neoantigens are cancer-specific post-translational protein modifications, such as methylation, phosphorylation, acetylation, and glycosylation36,37 (Figure 1). Due to the considerable cost and difficulties presented by experimental methods used for the identification of PTMs, recently many computational methods like GPS-Lipid,38 and MusiteDeep39 have been developed for predicting PTMs. To date, PTM-derived neoantigens have not been a significant focus of recent immuno-oncology research, and therefore there is an urgent need to expand the characterization of post-translational modified HLA-bound peptides as well as the repertoire of TCR that recognize these modified peptides. Technological advances in deep RNAseq gene expression analysis, whole-cell, and MHC-elute mass spectrometry (MS) peptide detection will be essential for the discovery of neoantigens of this class.40
Depending on the neoantigen class in question, different approaches and sequencing techniques must be utilized in order to elucidate the diverse repertoire of tumor-specific mutations.
SNVs and small indels
Peptides derived from SNVs and small indels belong to the most commonly studied category of neoantigens, mainly due to the fact that the methods involved in the identification of these mutations are well established, maintained, and easily accessible, but also because SNVs and small indels were considered to be the major source of neoantigens until recently. The problem with SNV-derived neoantigens is that they can exhibit significant similarity to their normal counterparts and thus only a small percentage of these putative neoantigens appear to be immunogenic.
In order to assess TSAs derived from SNVs and small indels, it is important that whole exome sequencing (WES) or whole genome sequencing (WGS) reads of tumor and matched normal DNA samples are used. Typically, the reads will first go through quality control and if required they can be processed to remove low-quality base calls and residual sequencing adapters. The reads are then aligned to a reference genome using a short read aligner. According to the GATK Best Practices workflow,41 it is recommended that the BAM files should undergo additional processing before variant calling: i.e. identify redundant reads and base quality score recalibration (BQSR) which adjusts the base quality scores of the reads using an empirical error model which can be carried out using tools such as GATK4. In addition, if it is not an integral element of the variant caller, read realignment around known indels using GATK3 can be carried out in order to reduce alignment errors. With the support and evidence of the aligned reads (BAM files) from tumor and normal tissue, numerous variant callers can detect somatic variants that are present in tumor samples. Most commonly, these callers use either Bayesian inference or traditional statistical models combined with specific filters. Some examples of variant calling tools include MuTect/MuTect2,42 VarScan2,43 Manta,44 SomaticSniper,45 FreeBayes,46 and Strelka.47 Since there is no universal ‘gold-standard’ tool and due to discrepancies among variant callers, finding a single best caller for various datasets is considered impractical. One solution to this issue is to combine the results from individual callers either by majority voting48 or with consensus approaches.49 Once the variants are identified, the final step would be to annotate them and produce the resulting mutated protein sequences; the most commonly used tools for this process are the Ensembl variant effect prediction (VEP)50 and SnpEff.51
Alternative splicing variants
Mutations in splice sites or splicing factors, exon skipping, intron retention, and a variety of post-transcriptional modifications can produce splice variants which have been suggested to be particularly relevant for cancer types with low tumor mutational burden but harboring splice factor mutations.52 These events are detectable only at the transcriptome level and can be quantified using RNA sequencing (RNA-seq) data. This class of neoantigens, however, includes non-mutated peptides and given that potential off-target effects of cell-based immunotherapies may have drastic consequences, there are several questions regarding the specificity and cross-reactivity of the predicted alternative splicing (AS)-derived neoantigens. In a recent study, the authors developed a proteogenomic strategy to identify cancer-restricted non-mutated antigens using medullary thymic epithelial cells (mTECs) as ‘normal control’.53 This was based on the unique characteristic of mTECs to express peripheral antigens, which contributes to the establishment of T-cell self-tolerance. mTECs display a high level of AS and RNA editing, further expanding the broad repertoire of self antigens in the thymus.54, 55, 56 We anticipate that the method can be complemented by the incorporation of RNA-seq data of normal tissues from the Genotype-Tissue Expression (GTEx) project.
There are two major approaches applied for AS event analysis: (i) isoform-based and (ii) count-based strategies.57 The first step for isoform-based methods is the reconstruction of full-length transcripts and then, based on the sequencing reads supporting these transcripts, the estimation of their relative abundances. Once the abundances are estimated, statistical testing is applied in order to identify the differential expression of the reconstructed transcripts between conditions (e.g. between tumor and normal samples). Tools using isoform-based methods include Trinity,58 Scripture,59 Cufflinks,60 Cuffdiff2,61 EBSeq,62 StringTie,63 and DiffSplice.64 These approaches rely heavily on accurate transcript quantification and may be affected by the sequencing depth and the read length.
Count-based methods can be further divided into exon-based and event-based approaches.
Exon-based methods seek to assign read counts to different features (such as exons or junctions) instead of reconstructing full-length transcripts. These approaches tend to be more robust in terms of differentially expressed exons/junctions between conditions, but their limitation lies with the fact that such approaches are incapable of identifying the type of splicing event occurring in a gene. Tools using exon-based methods include DEXseq,65 SplicingCompass,66 edgeR,67 and limma.68 Finally, event-based approaches seek to quantify directly the splicing events by measuring the fraction of mRNAs expressed from a gene containing a specific form of an AS event.69 The problem with this approach is that it is not designed to accommodate the varying uncertainty of isoform expression across isoform groups. Consequently, its application for isoform inference results in reduced power for some classes of isoforms and increased false discovery rate for others. Several tools use event-based approaches, including MAJIQ,70 SplAdder,71 rMATS,72 SUPPA2,73 MISO,74 and dSpliceType.75
Gene fusions
Recurrent balanced rearrangements, most commonly translocations, have been shown to represent important early steps in the initiation of carcinogenesis.76, 77, 78 These rearrangements usually exert their action either by deregulation of gene expression in one of the breakpoints or with the creation of a hybrid gene through the fusion of parts of two genes.79 The expression of fusion genes results in chimeric proteins, which have the potential to be highly immunogenic due to their difference from their normal counterparts, especially in terms of peptides that include the fusion junction point and both the neighboring breakpoint regions. Fusion events can be identified using WGS or RNA-seq data. In principle, WES data can also be used for fusion events detection, albeit WGS provides the most comprehensive and unbiased characterization of genomic alterations in genomes.80 Gene fusion predictors using targeted captured DNA data include BreakID81 and GRIDSS2.82 Expressed gene fusions, however, are only detectable with RNA-seq data, which require less storage space, and analysis time.
Fusion transcript prediction algorithms follow two strategies, which are broadly defined as (i) assembly-first and (ii) mapping-first approaches. Assembly-first methods perform de novo assembly of reads into longer transcripts and proceed then to identify chimeric transcripts that are consistent with recurrent balanced rearrangements. Such methods allow for the exploration of fusion transcripts that are not well represented by the reference genome sequence, or novel fusion transcripts that are entirely absent from the reference genome. The major drawback of the assembly-first methods is that they exhibit low sensitivity when compared with read mapping methods in most cases.83 Assembly-first tools include TrinityFusion58,84 and JAFFA-assembly.85
Mapping-first approaches align the sequencing reads directly to the reference genome and proceed then to identify those reads composed of segments which map in a non-linear way to two different locations of the reference genome (split reads), and read pairs from the same fragment whose alignments to the reference genome have distance and/or orientation that differ from the expected if the fragment was contiguous to the reference genome (discordant reads). Such methods require the number of supporting reads to correlate with the expression of the genes involved in the fusion. True predictions usually have a balanced number of split and discordant reads. Events with only discordant reads or without discordant reads and only split reads having anchors in just one gene are frequently artifacts. These methods exhibit increased sensitivity, but their limitation is their inability to identify novel fusion transcripts and intragenic deletions (deletions within a gene are difficult to distinguish from ordinary splicing in RNA-Seq data). The two top performing and most widely used mapping-first tools are Arriba86 and STAR-Fusion.87
HLA typing
The HLA complex is one of the most gene-dense and polymorphic regions in the human genome which encodes key components of the human adaptive immune system.88 The HLA region consists of six classical HLA genes, more specifically HLA-A, -B, -C for HLA class I and HLA-DR, -DQ, -DP for HLA class II. HLA polymorphisms occur in domains responsible for epitope binding, and thus the overall immune repertoire is exponentially broadened. Until recently, only CD8+ T cells and therefore only HLA class I molecules were considered to have an important role in neoantigen recognition, but there is increasing evidence that the majority of the immunogenic tumor mutanome is recognized by CD4+ T cells,89 and thus HLA class II neoantigens can also elicit immune responses to cancer. Starting from the year 1998, the European Molecular Biology Laboratory-European Bioinformatics Institute (EMBL-EBI) created a publicly available, and curated database containing serologically defined HLA antigens and their genes/alleles defined by nucleotide sequences (IMGT/HLA database).90
The first step for almost every HLA typing tool is the mapping of reads to the exonic and intronic regions of the HLA genes, as defined in the IMGT/HLA database. Then different approaches can be taken for the identification of the HLA types. Briefly, some tools create a list of HLA alleles by selecting those with no missed exons and no more than one mismatch and then proceed to form pairs of HLA alleles (e.g. A∗01:01:01 and A∗01:01:02) from this list. For each pair, a score is calculated (applying a scoring scheme based on multiple sequence alignment91) and the pair with the best score is reported as the final result. Other tools construct a binary hit matrix for all the reads mapping to at least one HLA allele and assuming that the correct HLA genotype explains the highest number of mapped reads, they formulate an integer linear programming optimization in order to find an optimal solution. Finally, some tools use a graph-based alignment approach to ensure increased read mapping sensitivity and then seek to find maximum likelihood estimates of abundance through an expectation-maximization algorithm. Most tools accept RNA-seq, WES, and WGS data as input and can carry out HLA class I typing and/or class II typing. One of the best performing tools for HLA class I typing is OptiType92 followed by Polysolver,93 while for class I and II typing the two best performing tools are HLA-HD94 and HISAT genotype.95
HLA class I typing tools have been researched extensively and can achieve high sensitivity. The use of long reads has been proposed in order to increase HLA typing accuracy, and while the results look promising, there are still several limitations, mainly attributed to the high background signal.96,97 Using long but noisy sequencing reads for HLA typing requires the development of novel bioinformatics solutions distinct from those designed for shorter but more accurate reads, and moreover, current HLA typing tools can reach 99% sensitivity for common HLA class I alleles using short paired-end reads.98 This percentage, however, does not represent the whole biological truth. This is because HLA allele sequences may only be partially available in the IMGT/HLA repository; for example, so far 3644 alleles have been classified for HLA-A and although all alleles of HLA-A have known sequences for exons 2 and 3, only 383 alleles have full-length sequences available.99 This problem is not restricted to HLA class I only, as HLA class II typing is less investigated, being evident by the number of named alleles for each HLA gene in the IMGT/HLA database: as of October 2021, the IMGT/HLA database contains 23 002 entries with known sequences for HLA class I whereas it contains only 8673 entries with known sequences for class II. Other facts that add to this problem are the dimeric nature of the functional HLA class II complex and the copy number variation of one of the loci (HLA-DRB) that makes this region exceptionally convoluted. Furthermore, the IMGT/HLA database includes the most frequent HLA alleles in the human population, which proves problematic in the case of rare or novel HLA alleles and subsequently leads to an increased false negative discovery rate.
Therefore, specialized HLA typing [polymerase chain reaction (PCR)- or NGS-based] using either long/mid-range PCR-based isolation, or hybridization-based capture methods, will be superior in a clinical setting.100 Nonetheless, while PCR-based HLA typing using sequence-specific primers, sequence-specific oligonucleotide probes, and Sanger sequencing-based typing methods have significantly improved HLA typing resolution, there are several caveats, including time-consuming protocols, low throughput, unphased data, and ambiguity.101
HLA binding affinity prediction
Among the various processes described so far in this review, the major determinant of neoantigen presentation is the binding of the tumor-specific epitopes to the HLA molecules. Therefore, computational predictors that discriminate HLA binding from non-binding peptides are critical. Typically, these predictors utilize MS identified HLA eluted ligands (EL) or binding affinity data deposited in the Immune Epitope Database (IEDB),102 the SysteMHC Atlas,103 the proteomics identifications database (PRIDE),104 or other publicly available MS-based immunopeptidomic datasets, to train machine learning (ML) classifiers. Early developed tools use linear regression-based methods to predict HLA peptide binding affinity. The problem with this approach is that it operates under the assumption that the contribution of individual residues to the overall binding affinity is linear in nature, which is rarely the case since the correlation between neighboring peptide residues can also affect the HLA binding. In order to account for this non-linear relationship, current ML classifiers utilize artificial neural networks (ANNs). For example, a feedforward neural network can simulate the contribution of each peptide residue type by adapting the weights of locally connected, one-dimensional convolutional layers in order to capture the complex interactions between HLA binding residues.
Allele-specific methods train a model for each HLA allele and learn the binding patterns of each allele separately. Enough experimentally validated ligands are available for only a few hundreds of HLA alleles, however, which represents only a small fraction of the HLA alleles observed in the human population. To address this issue, pan-allele predictors have been introduced that allow for interpolation between ligands, but also between receptors. The input of these algorithms consists of both the sequence of the ligand and the sequence of the HLA allele’s binding site, and thus they are powerful at capturing correlations between amino acids in the HLA binding site and in the ligand. It is noted that in principle, pan-specific methods can predict binders of any HLA allele with known protein sequence,105 which implies that truly novel HLA alleles might pose an issue when it comes to binding affinity predictions. The most widely used allele-specific binding affinity predictors are NetMHC106 and MHCflurry107 for HLA-I, and NetMHCII,108 and mixMHC2pred109 for HLA-II. Among the top performing pan-specific HLA binding affinity predictors are MHCflurry 2.099 and NetMHCpan110 for HLA class I and NetMHCIIpan110 for HLA class II. Due to the different training approaches, allele-specific methods outperform the pan-specific methods for HLA molecules where sufficient data are available to accurately characterize the binding motif, and pan-specific methods outperform the allele-specific methods when data are scarcer. It has been shown, however, that consensus approaches combining both methods can improve the binding affinity prediction accuracy.108,111
Although ANNs have addressed the non-linear nature of the peptide-HLA binding process, there are known limitations to the methods depending on the datasets each method uses to train its internal ML algorithm. Typically, these ML algorithms fall into two broad categories, with the first being ML classifiers trained on binding affinity data. This can limit substantially the prediction power, since only the binding event is modeled and no other biological feature involved in the process is accounted for. In order to resolve this issue, the second category of ML classifiers are trained on combined binding affinity and MS-based EL data. Despite the major improvements in the quality of immunopeptidomics data, there are still several technical restrictions to overcome. The MS obtained spectra are compared with in silico generated spectra of peptides from protein sequence databases with MS search tools (spectra searches). One limitation is that the spectra search is limited to the available databases, which are usually restricted to the annotated human proteome. To address this limitation, dedicated proteogenomics computational pipelines for customized reference databases have been developed to expand the search space beyond the canonical human proteome.112,113 Second, peptides that have features that make them incompatible with ionization might not be detected with standard methods.114 Finally, the antibodies employed during the immunopurification process of peptide-HLA complexes in EL assays are mostly pan-specific, which may eventually result in multiallelic data. More recent ML algorithms seek to annotate the EL datasets and deconvolute the multiallelic to single allelic data before they employ them to train the predictors.110,115,116 Another promising approach in order to solve this issue is the monoallelic strategy for profiling the HLA peptidome which leverages cell lines expressing a single HLA allele and optimized immunopurifications.117,118
Filtering and prioritization of neoantigens
Early day neoantigen prediction methods targeted binding affinity, measured in half-maximal inhibitory concentration (IC50), for the filtering and prioritization of neoantigens. As a rule of thumb, every peptide exhibiting an IC50 <500 nM was considered a ‘candidate’ and the remaining peptides (with IC50 >500 nM) would be filtered out, then the putative neoantigens would be prioritized according to the IC50 values from the lowest (strong binders) to highest (weak binders). As the methods evolved, the concept of neoantigen ranking scores was introduced for the classification of peptides into strong and weak binders. In brief, the binding affinity predictions are scored and ranked compared with a set of random natural HLA binding peptides in order to address the inherent bias of certain molecules towards higher/lower mean predicted affinities.110
Due to the great diversity and the stochastic nature of the T-cell immune response, however, a single value associated with a part of the whole process can hardly provide sufficient information to accurately model the complex tumor–immune interactions. In this context, systematic integration of multiple features into a unified neoantigen prioritization algorithm would yield increased classification accuracy. These features extend beyond the characteristics of HLA binding and presentation, including clonality of the neoantigen,119 amino acid characteristics like ‘hydrophobicity’,120 ‘polarity and charged value’,121 ‘molecular size'',122 ‘entropy of peptides’,123 and promiscuity of HLA molecules which was shown to be correlated with bad prognosis after ICI therapy.124 Most recently developed algorithms125, 126, 127, 128 measure the information gain from such features by utilizing feature selection processes,129 and then proceed to train ML classifiers on the basis of the selected immunogenicity features. Nevertheless, a major downside of ML approaches is overfitting, and their performance can be significantly affected by the quantity and quality of the training datasets. Ideally, in order to avoid this issue, ML classifiers should be trained on large positive (e.g. experimentally validated immunogenic epitopes) and negative (non- immunogenic) datasets. Unfortunately, there is still a lack of such comprehensive positive/negative datasets. Peptide-MHC multimers130 or yeast display assays131 leverage the isolation of antigen-specific T cells. Together with single-cell TCR sequencing, immunogenic peptides can be identified and characterized at a large scale,130,131 although at high costs and technical challenges which may limit their application. The combination of these technologies and the integration of structural modeling information can further improve the classification accuracy, as has been shown by recently developed tools like Net-TCR 2.0132 or PRIME.133
In a recent study, the Tumor Neoantigen Selection Alliance of the Parker Institute for Cancer Immunotherapy identified key components of tumor epitope immunogenicity.134 According to the study, these components can be classified into ‘presentation’ and ‘recognition’ features of the immunogenic peptides. The first category encompasses features that are associated with effective antigen presentation, namely HLA binding affinity, expression of the originating gene (‘tumor abundance’), expected duration of peptide-HLA interaction (‘binding stability’), and peptide hydrophobicity. The second category involves peptide features considered to be associated with immunogenicity among peptides that have the highest likelihood of being presented. Two features were identified: (i) ‘agretopicity’135, 136, 137 which is the ratio of mutant binding affinity to wild-type binding affinity and (ii) ‘foreignness’138, 139, 140 which is the probability of TCR recognition as inferred by the homology of the tumor peptide to known pathogenic peptides in the IEDB.
Future perspectives
In-depth understanding of tumor–immune interactions
Although peptide-HLA binding has been researched extensively and is in fact one of the best characterized processes in neoantigen presentation, there are several caveats impeding the accurate and unbiased elucidation of the complex relationships between the tumor and the immune system. This may be attributed to the aforementioned inherent technical biases in MS data141 and also due to our poor understanding of the nature of the tumor-immune system interactions, which limits the modeling capabilities especially in terms of T-cell recognition. In order to increase the robustness of the models, some methods integrate information that spans beyond the process of peptide-HLA binding, like proteasomal cleavage sites, TAP transportation, and ER loading. Although this seemed promising at first, in practice the gain in accuracy is marginal,23,142 and most computational pipelines consider this step optional. We expect that integrating immunogenicity features such as binding stability, peptide hydrophobicity, agretopicity, and foreignness into computational pipelines will enable increased accuracy in predicting immunogenic neoantigens.
Exploration of the immunological ‘dark matter’
So far, cancer research has mostly focused on mutations that alter protein coding sequences. There is increasing evidence suggesting, however, that non-canonical and cryptic peptides also contribute to the HLA peptidome. The emergence of proteogenomics has radically revolutionized our perspective of the cancer proteome by identifying peptides encoded by all reading frames of any genomic region.53,143, 144, 145 Moreover, experiments involving ribosome profiling provided strong evidence for pervasive translation outside of annotated protein coding genes.146 Using approaches derived from statistical physics, it has become possible to quantify transcriptome-wide motif usage in human and murine non-coding RNAs, determining that most have motif usage consistent with the coding genome.147 In a recent study, Liepe et al.148 report evidence that a large fraction of HLA class I ligands are spliced together by the proteasome from two different fragments of the same protein, due to the proteasome-catalyzed peptide splicing process. Although proteasomal splicing is a controversial subject and there are numerous published concerns regarding the findings of the aforementioned study,149, 150, 151 in a way, it highlighted our poor understanding of the biological processes involved and the challenges that remain regarding the computational identification of peptides that are not encoded in the proteome. Exploring the uncharted waters of the immunological ‘dark matter’ may uncover the contribution of proteins derived from non-canonical sources to the cancer immune repertoire, and expand the range of putative neoantigens.
Ease of access and deployment of computational pipelines
Dependency issues, version control, lack of scalability, and inconsistencies between development and production environments are only a few of the problems a researcher has to resolve in order to perform computational analysis in general and to predict neoantigens specifically. Software container technology, such as Docker (https://www.docker.com) and Singularity (https://sylabs.io), along with package and environment management systems like Conda (https://conda.io) have revolutionized the practice of software development and deployment. These technologies enable the containerization of software along with its required dependencies in a sanitized environment, thus avoiding any software conflicts, and ensure portability and reproducibility across different information technology platforms in healthcare systems and the cloud.
Finally, the deployment of the multiple components required by a bioinformatics pipeline for neoantigen prediction in an orchestrated manner can prove to be a daunting task and usually requires a high level of bioinformatics expertise. Workflow management systems such as Nextflow,152 Snakemake,153 Airflow (https://airflow.apache.org/), and CWL154 allow the development of scalable and reproducible data analysis workflows. Most of the current bioinformatics pipelines (Table 1) are deposited as open source projects in repositories like GitHub (https://github.com/) and can be ported locally and deployed by researchers with minimal effort.
Table 1.
Computational tools and pipelines used in/for neoantigen prediction
| Name | Neoantigen types | Input data | Neoantigen class | Repository (if available) | |
|---|---|---|---|---|---|
| Neoantigen prediction pipelines | NextNEOpi98 | SNVs, indels, gene fusions | WES/WGS and RNA-seq or WES/WGS only, as raw FASTQ or BAM files | Class I and II | https://github.com/icbi-lab/nextNEOpi |
| NeoFuse175 | Gene fusions | RNA-seq FASTQ files | Class I and II | https://github.com/icbi-lab/NeoFuse | |
| Antigen.garnish140 | SNVs, indels, gene fusions | VCF of mutations, gene fusions, or transcripts or peptide sequences | Class I and II | https://github.com/andrewrech/antigen.garnish | |
| CloudNeo176 | SNVs | VCF of somatic mutations and BAM (DNA- or RNA-seq) | Class I | https://github.com/TheJacksonLaboratory/CloudNeo | |
| DeepHLApan177 | SNVs | CSV files | Class I | https://github.com/jiujiezz/deephlapan | |
| Epidisco178 | SNVs, indels, splice variants, gene fusions | WES and RNA-seq FASTQ files | Class I | https://github.com/hammerlab/epidisco | |
| INTEGRATE-neo179 | Gene fusions | RNA-seq or WGS FASTQ files | Class I | https://github.com/ChrisMaherLab/INTEGRATE-Neo | |
| MuPeXI180 | SNVs, indels | VCF of somatic mutations and precomputed expression data | Class I | https://github.com/ambj/MuPeXI | |
| Neoantimon181 | SNVs, indels, structural variants | VCF of somatic mutations or file of mutant RNA sequences, and precomputed HLA types | Class I and II | https://github.com/hase62/Neoantimon | |
| neoANT-HILL182 | SNVs, indels | VCF of somatic mutations, RNA-seq data (BAM or FASTQ files) | Class I | https://github.com/neoanthill/neoANT-HILL | |
| NeoFlow183 | SNVs, indels | VCF of somatic mutations, DNA- or RNA-seq FASTQ files, MS data in MGF format | Class I | https://github.com/bzhanglab/neoflow | |
| Neopepsee126 | SNVs | VCF of somatic mutations, RNA-seq FASTQ files, and HLA types | Class I | https://sourceforge.net/p/neopepsee/wiki/Home/ | |
| NeoPredPipe184 | SNVs, indels | VCF of somatic mutations and HLA types | Class I and II | https://github.com/MathOnco/NeoPredPipe | |
| NeoepitopePred | SNVs, gene fusions | WGS FASTQ files or WGS, WES or RNA-seq BAM files | Class I | https://stjudecloud.github.io/docs/guides/genomics-platform/analyzing-data/neoepitope/ | |
| Neoepiscope185 | SNVs, indels | VCF of somatic mutations, mapped DNA-seq reads (BAM), and HLA alleles | Class I and II | https://github.com/pdxgx/neoepiscope | |
| nf-core/epitopeprediction186 | SNVs, indels | VCF of somatic mutations | Class I and II | https://github.com/nf-core/epitopeprediction | |
| OpenVax187 | SNVs | FASTQ from WES and RNA-seq | Class I | ||
| ProGeo-neo188 | SNVs | VCF of somatic mutations, RNA-seq FASTQ files | Class I | https://github.com/kbvstmd/ProGeo-neo | |
| ProTECT189 | SNVs | DNA- and RNA-seq FASTQ files. Alternatively, precomputed BAM and/or VCF files | Class I and II | https://github.com/BD2KGenomics/protect | |
| pTuneos190 | SNVs, indels | FASTQ from WES and RNA-seq. Alternatively, VCF of somatic mutations, expression data, copy number, and tumor cellularity information | Class I | https://github.com/bm2-lab/pTuneos | |
| pVACtools191 | SNVs, indels, gene fusions | VCF of somatic mutations, expression/coverage information from DNA- and RNA-seq (pVACseq), gene fusions (pVACfuse), and HLA types. | Class I and II | https://github.com/griffithlab/pVACtools | |
| ScanNeo192 | Indels | Mapped RNA-seq reads (BAM) | Class I | https://github.com/ylab-hi/ScanNeo | |
| TIminer193 | SNVs | VCF of somatic mutations, RNA-seq FASTQ files | Class I | https://icbi.i-med.ac.at/software/timiner/timiner.shtml | |
| TSNAD194 | SNVs, indels | WES FASTQ files | Class I | https://github.com/jiujiezz/tsnad | |
| Vaxrank195 | SNVs, indels | VCF of somatic mutations, mapped RNA-seq reads (BAM), and HLA types | Class I | https://github.com/openvax/vaxrank | |
| TruNeo196 | SNVs, indels | WES and RNA-seq FASTQ files | Class I | https://github.com/yucebio/TruNeo |
ANN, artificial neural network; CSV, comma separated values; DCNN, deep convoluted neural network; HLA, human leukocyte antigen; GBDT, gradient boosted decision trees; GLM, generalized linear model; MGF, mascot generic format; MHC, major histocompatibility complex; MS, mass spectrometry; RNA-seq, RNA sequencing; SMM, stabilized matrix method; SNVs, single nucleotide variations; VCF, variant call format; WES, whole exome sequencing; WGS, whole genome sequencing.
Conclusion
The advancements in immuno-oncology have been impressive over the past 30 years; however, there are several bottlenecks and open questions that need to be resolved before immunotherapy can become one of the major pylons of cancer treatment. Computational approaches, like neoantigen prediction, will likely play a key role in unlocking the potential of immunotherapies like adoptive T-cell therapy or neoantigen vaccination, which showed significant tumor regression and even durable responses in patients with melanoma, cholangiocarcinoma, or glioblastoma.155, 156, 157, 158 Integration of computational methods in clinical settings will thus pave the way for personalized medicine.
Acknowledgments
Funding
This work was supported by the European Research Council (ERC) [grant number 786295] to ZT. ZT is a member of the German Research Foundation (DFG) project TRR 241(INF).
Disclosure
The authors have declared no conflicts of interest.
References
- 1.Beecher S.M., O'Leary D.P., McLaughlin R., Kerin M.J. The impact of surgical complications on cancer recurrence rates: a literature review. Oncol Res Treat. 2018;41:478–482. doi: 10.1159/000487510. [DOI] [PubMed] [Google Scholar]
- 2.Alieva M., van Rheenen J., Broekman M.L.D. Potential impact of invasive surgical procedures on primary tumor growth and metastasis. Clin Exp Metastasis. 2018;35:319–331. doi: 10.1007/s10585-018-9896-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mansoori B., Mohammadi A., Davudian S., Shirjang S., Baradaran B. The different mechanisms of cancer drug resistance: a brief review. Adv Pharm Bull. 2017;7:339–348. doi: 10.15171/apb.2017.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willers H., Azzoli C.G., Santivasi W.L., Xia F. Basic mechanisms of therapeutic resistance to radiation and chemotherapy in lung cancer. Cancer J. 2013;19:200–207. doi: 10.1097/PPO.0b013e318292e4e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freeman G.J., Long A.J., Iwai Y., et al. Engagement of the Pd-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192:1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharpe A.H., Pauken K.E. The diverse functions of the PD1 inhibitory pathway. Nat Rev Immunol. 2018;18:153–167. doi: 10.1038/nri.2017.108. [DOI] [PubMed] [Google Scholar]
- 7.Curran M.A., Montalvo W., Yagita H., Allison J.P. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc Natl Acad Sci. 2010;107:4275–4280. doi: 10.1073/pnas.0915174107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwai Y., Hamanishi J., Chamoto K., Honjo T. Cancer immunotherapies targeting the PD-1 signaling pathway. J Biomed Sci. 2017;24:26. doi: 10.1186/s12929-017-0329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joyce S. Immunoproteasomes edit tumours to escape immune recognition. Eur J Immunol. 2015;45:3241–3245. doi: 10.1002/eji.201546100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee C.H., Yelensky R., Jooss K., Chan T.A. Update on tumor neoantigens and their utility: why it is good to be different. Trends Immunol. 2018;39:536–548. doi: 10.1016/j.it.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schumacher T.N., Scheper W., Kvistborg P. Cancer neoantigens. Annu Rev Immunol. 2019;37:173–200. doi: 10.1146/annurev-immunol-042617-053402. [DOI] [PubMed] [Google Scholar]
- 12.Efremova M., Rieder D., Klepsch V., et al. Targeting immune checkpoints potentiates immunoediting and changes the dynamics of tumor evolution. Nat Commun. 2018;9:32. doi: 10.1038/s41467-017-02424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gubin M.M., Zhang X., Schuster H., et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schreiber R.D., Old L.J., Smyth M.J. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 15.Kambayashi T., Laufer T.M. Atypical MHC class II-expressing antigen-presenting cells: can anything replace a dendritic cell? Nat Rev Immunol. 2014;14:719–730. doi: 10.1038/nri3754. [DOI] [PubMed] [Google Scholar]
- 16.Valečka J., Almeida C.R., Su B., Pierre P., Gatti E. Autophagy and MHC-restricted antigen presentation. Mol Immunol. 2018;99:163–170. doi: 10.1016/j.molimm.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Johnson D.B., Estrada M.V., Salgado R., et al. Melanoma-specific MHC-II expression represents a tumour-autonomous phenotype and predicts response to anti-PD-1/PD-L1 therapy. Nat Commun. 2016;7:10582. doi: 10.1038/ncomms10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forero A., Li Y., Chen D., et al. Expression of the MHC class II pathway in triple-negative breast cancer tumor cells is associated with a good prognosis and infiltrating lymphocytes. Cancer Immunol Res. 2016;4:390–399. doi: 10.1158/2326-6066.CIR-15-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schumacher T., Bunse L., Pusch S., et al. A vaccine targeting mutant IDH1 induces antitumour immunity. Nature. 2014;512:324–327. doi: 10.1038/nature13387. [DOI] [PubMed] [Google Scholar]
- 20.Aarntzen E.H.J.G., De Vries I.J.M., Lesterhuis W.J., et al. Targeting CD4+ T-helper cells improves the induction of antitumor responses in dendritic cell-based vaccination. Cancer Res. 2013;73:19–29. doi: 10.1158/0008-5472.CAN-12-1127. [DOI] [PubMed] [Google Scholar]
- 21.Mortara L., Castellani P., Meazza R., et al. CIITA-induced MHC class II expression in mammary adenocarcinoma leads to a Th1 polarization of the tumor microenvironment, tumor rejection, and specific antitumor memory. Clin Cancer Res. 2006;12:3435–3443. doi: 10.1158/1078-0432.CCR-06-0165. [DOI] [PubMed] [Google Scholar]
- 22.Baskar S., Clements V.K., Glimcher L.H., Nabavi N., Ostrand-Rosenberg S. Rejection of MHC class II-transfected tumor cells requires induction of tumor-encoded B7-1 and/or B7-2 costimulatory molecules. J Immunol. 1996;156:3821–3827. [PubMed] [Google Scholar]
- 23.Hackl H., Charoentong P., Finotello F., Trajanoski Z. Computational genomics tools for dissecting tumour–immune cell interactions. Nat Rev Genet. 2016;17:441–458. doi: 10.1038/nrg.2016.67. [DOI] [PubMed] [Google Scholar]
- 24.Kalejs M., Erenpreisa J. Cancer/testis antigens and gametogenesis: a review and ‘brain-storming’ session. Cancer Cell Int. 2005;5:4. doi: 10.1186/1475-2867-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L., Goedegebuure S.P., Gillanders W.E. Preclinical and clinical development of neoantigen vaccines. Ann Oncol. 2017;28:xii11–xii17. doi: 10.1093/annonc/mdx681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stone J.D., Harris D.T., Kranz D.M. TCR affinity for p/MHC formed by tumor antigens that are self-proteins: impact on efficacy and toxicity. Curr Opin Immunol. 2015;33:16–22. doi: 10.1016/j.coi.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan R.Y., Chung W.H., Chu M.T., et al. Recent development and clinical application of cancer vaccine: targeting neoantigens. J Immunol Res. 2018;2018 doi: 10.1155/2018/4325874. e4325874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen D.S., Mellman I. Elements of cancer immunity and the cancer–immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 29.Joseph C.G., Darrah E., Shah A., et al. Association of the autoimmune disease scleroderma with an immunologic response to cancer. Science. 2014;343:152–157. doi: 10.1126/science.1246886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van den Berg J.H., Gomez-Eerland R., van de Wiel B., et al. Case report of a fatal serious adverse event upon administration of T cells transduced with a MART-1-specific T-cell receptor. Mol Ther. 2015;23:1541–1550. doi: 10.1038/mt.2015.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turajlic S., Litchfield K., Xu H., et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol. 2017;18:1009–1021. doi: 10.1016/S1470-2045(17)30516-8. [DOI] [PubMed] [Google Scholar]
- 32.Smith C.C., Selitsky S.R., Chai S., Armistead P.M., Vincent B.G., Serody J.S. Alternative tumour-specific antigens. Nat Rev Cancer. 2019;19:465–478. doi: 10.1038/s41568-019-0162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang W., Lee K.W., Srivastava R.M., et al. Immunogenic neoantigens derived from gene fusions stimulate T cell responses. Nat Med. 2019;25:767–775. doi: 10.1038/s41591-019-0434-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoyos L.E., Abdel-Wahab O. Cancer-specific splicing changes and the potential for splicing-derived neoantigens. Cancer Cell. 2018;34:181–183. doi: 10.1016/j.ccell.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smart A., Margolis C., Pimentel H., et al. Intron retention as a novel source of cancer neoantigens in cancer. Nat Biotechnol. 2018;36:1056–1058. doi: 10.1038/nbt.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engelhard V.H., Altrich-Vanlith M., Ostankovitch M., Zarling A.L. Post-translational modifications of naturally processed MHC-binding epitopes. Curr Opin Immunol. 2006;18:92–97. doi: 10.1016/j.coi.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 37.Malaker S.A., Penny S.A., Steadman L.G., et al. Identification of glycopeptides as posttranslationally modified neoantigens in leukemia. Cancer Immunol Res. 2017;5:376–384. doi: 10.1158/2326-6066.CIR-16-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie Y., Zheng Y., Li H., et al. GPS-Lipid: a robust tool for the prediction of multiple lipid modification sites. Sci Rep. 2016;6:28249. doi: 10.1038/srep28249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang D., Liu D., Yuchi J., et al. MusiteDeep: a deep-learning based webserver for protein post-translational modification site prediction and visualization. Nucleic Acids Res. 2020;48:W140–W146. doi: 10.1093/nar/gkaa275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purcell A.W., Ramarathinam S.H., Ternette N. Mass spectrometry-based identification of MHC-bound peptides for immunopeptidomics. Nat Protoc. 2019;14:1687–1707. doi: 10.1038/s41596-019-0133-y. [DOI] [PubMed] [Google Scholar]
- 41.van der Auwera G.A., Carneiro M.O., Hartl C., et al. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43 doi: 10.1002/0471250953.bi1110s43. 11.10.1-11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cibulskis K., Lawrence M.S., Carter S.L., et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koboldt D.C., Zhang Q., Larson D.E., et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen X., Schulz-Trieglaff O., Shaw R., et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 45.Larson D.E., Harris C.C., Chen K., et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28:311–317. doi: 10.1093/bioinformatics/btr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garrison E., Marth G. Haplotype-based variant detection from short-read sequencing. arxiv 12073907 Q-Bio. 2012 [Google Scholar]
- 47.Saunders C.T., Wong W.S.W., Swamy S., Becq J., Murray L.J., Cheetham R.K. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 48.Ewing A., Houlahan K., Hu Y., et al. Combining tumor genome simulation with crowdsourcing to benchmark somatic single-nucleotide-variant detection. Nat Methods. 2015;12:623–630. doi: 10.1038/nmeth.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goode D.L., Hunter M.A., Doyle M.A., et al. A simple consensus approach improves somatic mutation prediction accuracy. Genome Med. 2013;5:90. doi: 10.1186/gm494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McLaren W., Gil L., Hunt S.E., et al. The ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cingolani P., Platts A., Wang L.L., et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Biernacki M.A., Bleakley M. Neoantigens in hematologic malignancies. Front Immunol. 2020;11:121. doi: 10.3389/fimmu.2020.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laumont C.M., Vincent K., Hesnard L., et al. Noncoding regions are the main source of targetable tumor-specific antigens. Sci Transl Med. 2018;10 doi: 10.1126/scitranslmed.aau5516. eaau5516. [DOI] [PubMed] [Google Scholar]
- 54.Derbinski J., Schulte A., Kyewski B., Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol. 2001;2:1032–1039. doi: 10.1038/ni723. [DOI] [PubMed] [Google Scholar]
- 55.Danan-Gotthold M., Guyon C., Giraud M., Levanon E.Y., Abramson J. Extensive RNA editing and splicing increase immune self-representation diversity in medullary thymic epithelial cells. Genome Biol. 2016;17:219. doi: 10.1186/s13059-016-1079-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takahama Y., Ohigashi I., Baik S., Anderson G. Generation of diversity in thymic epithelial cells. Nat Rev Immunol. 2017;17:295–305. doi: 10.1038/nri.2017.12. [DOI] [PubMed] [Google Scholar]
- 57.Mehmood A., Laiho A., Venäläinen M.S., McGlinchey A.J., Wang N., Elo L.L. Systematic evaluation of differential splicing tools for RNA-seq studies. Brief Bioinform. 2020;21:2052–2065. doi: 10.1093/bib/bbz126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grabherr M.G., Haas B.J., Yassour M., et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guttman M., Garber M., Levin J.Z., et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trapnell C., Williams B.A., Pertea G., et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leng N., Dawson J.A., Thomson J.A., et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29:1035–1043. doi: 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu Y., Huang Y., Du Y., et al. DiffSplice: the genome-wide detection of differential splicing events with RNA-seq. Nucleic Acids Res. 2013;41:e39. doi: 10.1093/nar/gks1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anders S., Reyes A., Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22:2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aschoff M., Hotz-Wagenblatt A., Glatting K.H., Fischer M., Eils R., König R. SplicingCompass: differential splicing detection using RNA-Seq data. Bioinformatics. 2013;29:1141–1148. doi: 10.1093/bioinformatics/btt101. [DOI] [PubMed] [Google Scholar]
- 67.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ritchie M.E., Phipson B., Wu D., et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alamancos G.P., Pagès A., Trincado J.L., Bellora N., Eyras E. Leveraging transcript quantification for fast computation of alternative splicing profiles. RNA. 2015;21:1521–1531. doi: 10.1261/rna.051557.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Green C.J., Gazzara M.R., Barash Y. MAJIQ-SPEL: web-tool to interrogate classical and complex splicing variations from RNA-Seq data. Bioinformatics. 2018;34:300–302. doi: 10.1093/bioinformatics/btx565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kahles A., Ong C.S., Zhong Y., Rätsch G. SplAdder: identification, quantification and testing of alternative splicing events from RNA-Seq data. Bioinformatics. 2016;32:1840–1847. doi: 10.1093/bioinformatics/btw076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shen S., Park J.W., Lu Z.X., et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci U S A. 2014;111:E5593–E5601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trincado J.L., Entizne J.C., Hysenaj G., et al. SUPPA2: fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol. 2018;19:40. doi: 10.1186/s13059-018-1417-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Katz Y., Wang E.T., Airoldi E.M., Burge C.B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Deng N., Zhu D. In: Bioinformatics Research and Applications. ISBRA 2014. Lecture Notes in Computer Science. Basu M., Pan Y., Wang J., editors. Springer; Cham: 2014. dSpliceType: a multivariate model for detecting various types of differential splicing events using RNA-seq. 8492:322-333. [Google Scholar]
- 76.Rabbitts T.H. Chromosomal translocations in human cancer. Nature. 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 77.Rowley J.D. Chromosome translocations: dangerous liaisons revisited. Nat Rev Cancer. 2001;1:245–250. doi: 10.1038/35106108. [DOI] [PubMed] [Google Scholar]
- 78.Aplan P.D. Causes of oncogenic chromosomal translocation. Trends Genet. 2006;22:46–55. doi: 10.1016/j.tig.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mitelman F., Johansson B., Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 80.Schröder J., Kumar A., Wong S.Q. In: Murray S.S., editor. Vol. 1908. Springer; New York: 2019. Overview of fusion detection strategies using next-generation sequencing. (Tumor Profiling). 125-138. [Google Scholar]
- 81.Jin L., Yin Y., Chen W., et al. BreakID: genomics breakpoints identification to detect gene fusion events using discordant pairs and split reads. Bioinformatics. 2019;35:2859–2861. doi: 10.1093/bioinformatics/bty1070. [DOI] [PubMed] [Google Scholar]
- 82.Cameron D.L., Baber J., Shale C., et al. GRIDSS2: comprehensive characterisation of somatic structural variation using single breakend variants and structural variant phasing. Genome Biol. 2021;22:202. doi: 10.1186/s13059-021-02423-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Haas B.J., Dobin A., Li B., Stransky N., Pochet N., Regev A. Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Genome Biol. 2019;20:213. doi: 10.1186/s13059-019-1842-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Haas B.J., Papanicolaou A., Yassour M., et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Davidson N.M., Majewski I.J., Oshlack A. JAFFA: high sensitivity transcriptome-focused fusion gene detection. Genome Med. 2015;7:43. doi: 10.1186/s13073-015-0167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Uhrig S., Ellermann J., Walther T., et al. Accurate and efficient detection of gene fusions from RNA sequencing data. Genome Res. 2021;31:448–460. doi: 10.1101/gr.257246.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Haas B.J., Dobin A., Stransky N. STAR-Fusion: fast and accurate fusion transcript detection from RNA-seq. bioRxiv. 2017:120295. doi: 10.1101/120295. [DOI] [Google Scholar]
- 88.Trowsdale J., Knight J.C. Major histocompatibility complex genomics and human disease. Annu Rev Genomics Hum Genet. 2013;14:301–323. doi: 10.1146/annurev-genom-091212-153455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kreiter S., Vormehr M., van de Roemer N., et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520:692–696. doi: 10.1038/nature14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Robinson J., Halliwell J.A., McWilliam H., Lopez R., Parham P., Marsh S.G. The IMGT/HLA database. Nucleic Acids Res. 2013;41:D1222–D1227. doi: 10.1093/nar/gks949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chatzou M., Magis C., Chang J.M., et al. Multiple sequence alignment modeling: methods and applications. Brief Bioinform. 2016;17:1009–1023. doi: 10.1093/bib/bbv099. [DOI] [PubMed] [Google Scholar]
- 92.Szolek A., Schubert B., Mohr C., Sturm M., Feldhahn M., Kohlbacher O. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics. 2014;30:3310–3316. doi: 10.1093/bioinformatics/btu548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shukla S.A., Rooney M.S., Rajasagi M., et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat Biotechnol. 2015;33:1152–1158. doi: 10.1038/nbt.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kawaguchi S., Higasa K., Shimizu M., Yamada R., Matsuda F. HLA-HD: an accurate HLA typing algorithm for next-generation sequencing data. Hum Mutat. 2017;38:788–797. doi: 10.1002/humu.23230. [DOI] [PubMed] [Google Scholar]
- 95.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu C. A long road/read to rapid high-resolution HLA typing: the nanopore perspective. Hum Immunol. 2021;82:488–495. doi: 10.1016/j.humimm.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu C., Yang X., Duffy B.F., et al. High-resolution HLA typing by long reads from the R10.3 Oxford nanopore flow cells. Hum Immunol. 2021;82:288–295. doi: 10.1016/j.humimm.2021.02.005. [DOI] [PubMed] [Google Scholar]
- 98.Rieder D., Fotakis G., Ausserhofer M., et al. nextNEOpi: a comprehensive pipeline for computational neoantigen prediction. Bioinformatics. 2021:btab759. doi: 10.1093/bioinformatics/btab759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.O'Donnell T.J., Rubinsteyn A., Laserson U. MHCflurry 2.0: improved pan-allele prediction of MHC class I-presented peptides by incorporating antigen processing. Cell Syst. 2020;11:42–48.e7. doi: 10.1016/j.cels.2020.06.010. [DOI] [PubMed] [Google Scholar]
- 100.Kishore A., Petrek M. Next-generation sequencing based HLA typing: deciphering immunogenetic aspects of sarcoidosis. Front Genet. 2018;9:503. doi: 10.3389/fgene.2018.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wittig M., Anmarkrud J.A., Kässens J.C., et al. Development of a high-resolution NGS-based HLA-typing and analysis pipeline. Nucleic Acids Res. 2015;43:e70. doi: 10.1093/nar/gkv184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vita R., Overton J.A., Greenbaum J.A., et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015;43:D405–D412. doi: 10.1093/nar/gku938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shao W., Pedrioli P.G.A., Wolski W., et al. The SysteMHC Atlas project. Nucleic Acids Res. 2018;46:D1237–D1247. doi: 10.1093/nar/gkx664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vizcaíno J.A., Csordas A., del-Toro N., et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang L., Udaka K., Mamitsuka H., Zhu S. Toward more accurate pan-specific MHC-peptide binding prediction: a review of current methods and tools. Brief Bioinform. 2012;13:350–364. doi: 10.1093/bib/bbr060. [DOI] [PubMed] [Google Scholar]
- 106.Lundegaard C., Lamberth K., Harndahl M., Buus S., Lund O., Nielsen M. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Res. 2008;36:W509–W512. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.O'Donnell T.J., Rubinsteyn A., Bonsack M., Riemer A.B., Laserson U., Hammerbacher J. MHCflurry: open-source class I MHC binding affinity prediction. Cell Syst. 2018;7:129–132.e4. doi: 10.1016/j.cels.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 108.Jensen K.K., Andreatta M., Marcatili P., et al. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology. 2018;154:394–406. doi: 10.1111/imm.12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Racle J., Michaux J., Rockinger G.A., et al. Robust prediction of HLA class II epitopes by deep motif deconvolution of immunopeptidomes. Nat Biotechnol. 2019;37:1283–1286. doi: 10.1038/s41587-019-0289-6. [DOI] [PubMed] [Google Scholar]
- 110.Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48:W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Karosiene E., Lundegaard C., Lund O., Nielsen M. NetMHCcons: a consensus method for the major histocompatibility complex class I predictions. Immunogenetics. 2012;64:177–186. doi: 10.1007/s00251-011-0579-8. [DOI] [PubMed] [Google Scholar]
- 112.Yadav M., Jhunjhunwala S., Phung Q.T., et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515:572–576. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 113.Kalaora S., Barnea E., Merhavi-Shoham E., et al. Use of HLA peptidomics and whole exome sequencing to identify human immunogenic neo-antigens. Oncotarget. 2016;7:5110–5117. doi: 10.18632/oncotarget.6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ma B. Challenges in Computational analysis of mass spectrometry data for proteomics. J Comput Sci Technol. 2010;25:107–123. [Google Scholar]
- 115.Alvarez B., Reynisson B., Barra C., et al. NNAlign_MA; MHC peptidome deconvolution for accurate MHC binding motif characterization and improved T-cell epitope predictions. Mol Cell Proteomics. 2019;18:2459–2477. doi: 10.1074/mcp.TIR119.001658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Reynisson B., Barra C., Kaabinejadian S., Hildebrand W.H., Peters B., Nielsen M. Improved prediction of MHC II antigen presentation through integration and motif deconvolution of mass spectrometry MHC eluted ligand data. J Proteome Res. 2020;19:2304–2315. doi: 10.1021/acs.jproteome.9b00874. [DOI] [PubMed] [Google Scholar]
- 117.Pyke R.M., Mellacheruvu D., Dea S., et al. Precision neoantigen discovery using large-scale immunopeptidomes and composite modeling of MHC peptide presentation. Mol Cell Proteomics. 2021 doi: 10.1016/j.mcpro.2021.100111. In press. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 118.Abelin J.G., Keskin D.B., Sarkizova S., et al. Mass spectrometry profiling of HLA-associated peptidomes in mono-allelic cells enables more accurate epitope prediction. Immunity. 2017;46:315–326. doi: 10.1016/j.immuni.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McGranahan N., Furness A.J., Rosenthal R., et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chowell D., Krishna S., Becker P.D., et al. TCR contact residue hydrophobicity is a hallmark of immunogenic CD8+ T cell epitopes. Proc Natl Acad Sci U S A. 2015;112:E1754–E1762. doi: 10.1073/pnas.1500973112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Patronov A., Doytchinova I. T-cell epitope vaccine design by immunoinformatics. Open Biol. 2013;3:120139. doi: 10.1098/rsob.120139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dintzis H.M., Dintzis R.Z., Vogelstein B. Molecular determinants of immunogenicity: the immunon model of immune response. Proc Natl Acad Sci U S A. 1976;73:3671–3675. doi: 10.1073/pnas.73.10.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu M.K.P., Hawkins N., Ritchie A.J., et al. Vertical T cell immunodominance and epitope entropy determine HIV-1 escape. J Clin Invest. 2013;123:380–393. doi: 10.1172/JCI65330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Manczinger M., Koncz B., Balogh G.M., et al. Negative trade-off between neoantigen repertoire breadth and the specificity of HLA-I molecules shapes antitumor immunity. Nat Cancer. 2021;2:950–961. doi: 10.1038/s43018-021-00226-4. [DOI] [PubMed] [Google Scholar]
- 125.Li G., Iyer B., Prasath V.B.S., Ni Y., Salomonis N. DeepImmuno: deep learning-empowered prediction and generation of immunogenic peptides for T-cell immunity. Brief Bioinform. 2021;22:bbab160. doi: 10.1093/bib/bbab160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kim S., Kim H.S., Kim E., et al. Neopepsee: accurate genome-level prediction of neoantigens by harnessing sequence and amino acid immunogenicity information. Ann Oncol. 2018;29:1030–1036. doi: 10.1093/annonc/mdy022. [DOI] [PubMed] [Google Scholar]
- 127.Wang G., Wan H., Jian X., et al. INeo-Epp: a novel T-Cell HLA class-I immunogenicity or neoantigenic epitope prediction method based on sequence-related amino acid features. BioMed Res Int. 2020;2020:1–12. doi: 10.1155/2020/5798356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Smith C.C., Chai S., Washington A.R., et al. Machine-learning prediction of tumor antigen immunogenicity in the selection of therapeutic epitopes. Cancer Immunol Res. 2019;7:1591–1604. doi: 10.1158/2326-6066.CIR-19-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Azhagusundari B., Thanamani A.S. Feature selection based on information gain. IJITEE. 2013;2:4. [Google Scholar]
- 130.Bentzen A.K., Marquard A.M., Lyngaa R., et al. Large-scale detection of antigen-specific T cells using peptide-MHC-I multimers labeled with DNA barcodes. Nat Biotechnol. 2016;34:1037–1045. doi: 10.1038/nbt.3662. [DOI] [PubMed] [Google Scholar]
- 131.Gee M.H., Han A., Lofgren S.M., et al. Antigen identification for orphan T cell receptors expressed on tumor-infiltrating lymphocytes. Cell. 2018;172:549–563.e16. doi: 10.1016/j.cell.2017.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Montemurro A., Schuster V., Povlsen H.R., et al. NetTCR-2.0 enables accurate prediction of TCR-peptide binding by using paired TCRα and β sequence data. Commun Biol. 2021;4:1–13. doi: 10.1038/s42003-021-02610-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schmidt J., Smith A.R., Magnin M., et al. Prediction of neo-epitope immunogenicity reveals TCR recognition determinants and provides insight into immunoediting. Cell Rep Med. 2021;2:100194. doi: 10.1016/j.xcrm.2021.100194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wells D.K., van Buuren M.M., Dang K.K., et al. Key parameters of tumor epitope immunogenicity revealed through a consortium approach improve neoantigen prediction. Cell. 2020;183:818–834.e13. doi: 10.1016/j.cell.2020.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Capietto A.-H., Jhurjhunwala S., Pollock S.B., et al. Mutation position is an important determinant for predicting cancer neoantigens. J Exp Med. 2020;217 doi: 10.1084/jem.20190179. e20190179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ghorani E., Rosenthal R., McGranahan N., et al. Differential binding affinity of mutated peptides for MHC class I is a predictor of survival in advanced lung cancer and melanoma. Ann Oncol. 2018;29:271–279. doi: 10.1093/annonc/mdx687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Duan F., Duitama J., Al Seesi S., et al. Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J Exp Med. 2014;211:2231–2248. doi: 10.1084/jem.20141308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Balachandran V.P., Luksza M., Zhao J.N., et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551:512–516. doi: 10.1038/nature24462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Łuksza M., Riaz N., Makarov V., et al. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature. 2017;551:517–520. doi: 10.1038/nature24473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Richman L.P., Vonderheide R.H., Rech A.J. Neoantigen dissimilarity to the self-proteome predicts immunogenicity and response to immune checkpoint blockade. Cell Syst. 2019;9:375–382.e4. doi: 10.1016/j.cels.2019.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gfeller D., Bassani-Sternberg M. Predicting antigen presentation—what could we learn from a million peptides? Front Immunol. 2018;9:1716. doi: 10.3389/fimmu.2018.01716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Paul S., Karosiene E., Dhanda S.K., et al. Determination of a predictive cleavage motif for eluted major histocompatibility complex class II ligands. Front Immunol. 2018;9:1795. doi: 10.3389/fimmu.2018.01795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Laumont C.M., Daouda T., Laverdure J.-P., et al. Global proteogenomic analysis of human MHC class I-associated peptides derived from non-canonical reading frames. Nat Commun. 2016;7:10238. doi: 10.1038/ncomms10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chong C., Muller M., Pak H.S., et al. Integrated proteogenomic deep sequencing and analytics accurately identify non-canonical peptides in tumor immunopeptidomes. Nat Commun. 2020;11:1293. doi: 10.1038/s41467-020-14968-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Attig J., Young G.R., Hosie L., et al. LTR retroelement expansion of the human cancer transcriptome and immunopeptidome revealed by de novo transcript assembly. Genome Res. 2019;29:1578–1590. doi: 10.1101/gr.248922.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ingolia N.T., Brar G.A., Stern-Ginossar N., et al. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014;8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tanne A., Muniz L.R., Puzio-Kuter A., et al. Distinguishing the immunostimulatory properties of noncoding RNAs expressed in cancer cells. Proc Natl Acad Sci U S A. 2015;112:15154–15159. doi: 10.1073/pnas.1517584112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Liepe J., Marino F., Sidney J., et al. A large fraction of HLA class I ligands are proteasome-generated spliced peptides. Science. 2016;354:354–358. doi: 10.1126/science.aaf4384. [DOI] [PubMed] [Google Scholar]
- 149.Rolfs Z., Solntsev S.K., Shortreed M.R., Frey B.L., Smith L.M. Global identification of post-translationally spliced peptides with neo-fusion. J Proteome Res. 2019;18:349–358. doi: 10.1021/acs.jproteome.8b00651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mylonas R., Beer I., Iseli C., et al. Estimating the contribution of proteasomal spliced peptides to the HLA-I Ligandome. Mol Cell Proteomics. 2018;17:2347–2357. doi: 10.1074/mcp.RA118.000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Erhard F., Dölken L., Schilling B., Schlosser A. Identification of the cryptic HLA-I immunopeptidome. Cancer Immunol Res. 2020;8:1018–1026. doi: 10.1158/2326-6066.CIR-19-0886. [DOI] [PubMed] [Google Scholar]
- 152.Di Tommaso P., Chatzou M., Floden E.W., Barja P.P., Palumbo E., Notredame C. Nextflow enables reproducible computational workflows. Nat Biotechnol. 2017;35:316–319. doi: 10.1038/nbt.3820. [DOI] [PubMed] [Google Scholar]
- 153.Köster J., Rahmann S. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics. 2012;28:2520–2522. doi: 10.1093/bioinformatics/bts480. [DOI] [PubMed] [Google Scholar]
- 154.Crusoe M.R., Abeln S., Iosup A., et al. Methods included: standardizing computational reuse and portability with the common workflow language. arXiv 2105.07028 [cs.DC] 2021 https://arxiv.org/abs/2105.07028. [Google Scholar]
- 155.Ott P.A., Hu Z., Keskin D.B., et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547:217–221. doi: 10.1038/nature22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Keskin D.B., Anaandappa A.J., Sun J., et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature. 2019;565:234–239. doi: 10.1038/s41586-018-0792-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tran E., Turcotte S., Gros A., et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lu Y.-C., Yao X., Crystal J.S., et al. Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin Cancer Res. 2014;20:3401–3410. doi: 10.1158/1078-0432.CCR-14-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Orenbuch R., Filip I., Comito D., Shaman J., Peer I., Rabadan R. arcasHLA: high-resolution HLA typing from RNAseq. Bioinformatics. 2020;36:33–40. doi: 10.1093/bioinformatics/btz474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ka S., Lee S., Hong J., et al. HLAscan: genotyping of the HLA region using next-generation sequencing data. BMC Bioinformatics. 2017;18:258. doi: 10.1186/s12859-017-1671-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Li H., Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Boegel S., Lower M., Schafer M., et al. HLA typing from RNA-Seq sequence reads. Genome Med. 2012;4:102. doi: 10.1186/gm403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bai Y., Wang D., Fury W. In: HLA Typing. Methods in Molecular Biology. Boegel S., editor. Humana Press; New York, NY: 2018. PHLAT: inference of high-resolution HLA types from RNA and whole exome sequencing. 1802:193-201. [DOI] [PubMed] [Google Scholar]
- 164.Liu C., Yang X., Duffy B., et al. ATHLATES: accurate typing of human leukocyte antigen through exome sequencing. Nucleic Acids Res. 2013;41:e142. doi: 10.1093/nar/gkt481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang Y.-Y., Mimori T., Khor S.-S., et al. HLA-VBSeq v2: improved HLA calling accuracy with full-length Japanese class-I panel. Hum Genome Var. 2019;6:1–5. doi: 10.1038/s41439-019-0061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Warren R.L., Choe G., Freeman D.J., et al. Derivation of HLA types from shotgun sequence datasets. Genome Med. 2012;4:95. doi: 10.1186/gm396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dilthey A.T., Mentzer A.J., Carapito R., et al. HLA∗LA—HLA typing from linearly projected graph alignments. Bioinformatics. 2019;35:4394–4396. doi: 10.1093/bioinformatics/btz235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Phloyphisut P., Pornputtapong N., Sriswasdi S., Chuangsuwanich E. MHCSeqNet: a deep neural network model for universal MHC binding prediction. BMC Bioinformatics. 2019;20:270. doi: 10.1186/s12859-019-2892-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang H., Lund O., Nielsen M. The PickPocket method for predicting binding specificities for receptors based on receptor pocket similarities: application to MHC-peptide binding. Bioinformatics. 2009;25:1293–1299. doi: 10.1093/bioinformatics/btp137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Fleri W., Paul S., Dhanda S.K., et al. The immune epitope database and analysis resource in epitope discovery and synthetic vaccine design. Front Immunol. 2017;8:278. doi: 10.3389/fimmu.2017.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Liu Z., Cui Y., Xiong Z., Nasiri A., Zhang A., Hu J. DeepSeqPan, a novel deep convolutional neural network model for pan-specific class I HLA-peptide binding affinity prediction. Sci Rep. 2019;9:794. doi: 10.1038/s41598-018-37214-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sarkizova S., Klaeger S., Le P.M., et al. A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nat Biotechnol. 2020;38:199–209. doi: 10.1038/s41587-019-0322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chen B., Khodadoust M.S., Olsson N., et al. Predicting HLA class II antigen presentation through integrated deep learning. Nat Biotechnol. 2019;37:1332–1343. doi: 10.1038/s41587-019-0280-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Shao X.M., Bhattacharya R., Huang J., et al. High-throughput prediction of MHC Class I and II Neoantigens with MHCnuggets. Cancer Immunol Res. 2020;8:396–408. doi: 10.1158/2326-6066.CIR-19-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Fotakis G., Rieder D., Haider M., Trajanoski Z., Finotello F. NeoFuse: predicting fusion neoantigens from RNA sequencing data. Bioinformatics. 2020;36:2260–2261. doi: 10.1093/bioinformatics/btz879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bais P., Namburi S., Gatti D.M., Zhang X., Chuang J.H. CloudNeo: a cloud pipeline for identifying patient-specific tumor neoantigens. Bioinformatics. 2017;33:3110–3112. doi: 10.1093/bioinformatics/btx375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wu J., Wang W., Zhang J., et al. DeepHLApan: a deep learning approach for neoantigen prediction considering both HLA-peptide binding and immunogenicity. Front Immunol. 2019;10:2559. doi: 10.3389/fimmu.2019.02559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Rubinsteyn A., Kodysh J., Hodes I., et al. Computational pipeline for the PGV-001 neoantigen vaccine trial. Front Immunol. 2018;8:1807. doi: 10.3389/fimmu.2017.01807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zhang J., Mardis E.R., Maher C.A. INTEGRATE-neo: a pipeline for personalized gene fusion neoantigen discovery. Bioinformatics. 2017;33:555–557. doi: 10.1093/bioinformatics/btw674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bjerregaard A.-M., Nielsen M., Hadrup S.R., Szallasi Z., Eklund A.C. MuPeXI: prediction of neo-epitopes from tumor sequencing data. Cancer Immunol Immunother. 2017;66:1123–1130. doi: 10.1007/s00262-017-2001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hasegawa T., Hayashi S., Shimizu E., et al. Neoantimon: a multifunctional R package for identification of tumor-specific neoantigens. Bioinformatics. 2020;36:4813–4816. doi: 10.1093/bioinformatics/btaa616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Coelho A.C.M.F., Fonseca A.L., Martins D.L., Lins P.B.R., da Cunha L.M., de Souza S.J. neoANT-HILL: an integrated tool for identification of potential neoantigens. BMC Med Genomics. 2020;13:30. doi: 10.1186/s12920-020-0694-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wen B., Li K., Zhang Y., Zhang B. Cancer neoantigen prioritization through sensitive and reliable proteogenomics analysis. Nat Commun. 2020;11:1759. doi: 10.1038/s41467-020-15456-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Schenck R.O., Lakatos E., Gatenbee C., Graham T.A., Anderson A.R.A. NeoPredPipe: high-throughput neoantigen prediction and recognition potential pipeline. BMC Bioinformatics. 2019;20:264. doi: 10.1186/s12859-019-2876-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wood M.A., Nguyen A., Struck A.J., et al. Neoepiscope improves neoepitope prediction with multivariant phasing. Bioinformatics. 2020;36:713–720. doi: 10.1093/bioinformatics/btz653. [DOI] [PubMed] [Google Scholar]
- 186.Ewels P., Peltzer A., Fillinger S., et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol. 2020 doi: 10.1038/s41587-020-0439-x. [DOI] [PubMed] [Google Scholar]
- 187.Kodysh J., Rubinsteyn A. In: Bioinformatics for Cancer Immunotherapy. Methods in Molecular Biology. Boegel S., editor. Vol. 2120. Humana; New York, NY: 2020. OpenVax: an open-source computational pipeline for cancer neoantigen prediction. 147-160. [DOI] [PubMed] [Google Scholar]
- 188.Li Y., Wang G., Tan X., et al. ProGeo-neo: a customized proteogenomic workflow for neoantigen prediction and selection. BMC Med Genomics. 2020;13:52. doi: 10.1186/s12920-020-0683-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Rao A.A., Madejska A.A., Pfeil J., Paten B., Salama S.R., Haussler D. ProTECT—prediction of T-Cell epitopes for cancer therapy. Front Immunol. 2020;11:483296. doi: 10.3389/fimmu.2020.483296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zhou C., Wei Z., Zhang Z., et al. pTuneos: prioritizing tumor neoantigens from next-generation sequencing data. Genome Med. 2019;11:67. doi: 10.1186/s13073-019-0679-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Hundal J., Kiwala S., McMichael J., et al. pVACtools: a computational toolkit to identify and visualize cancer neoantigens. Cancer Immunol Res. 2020;8:409–420. doi: 10.1158/2326-6066.CIR-19-0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wang T.Y., Wang L., Alam S.K., Hoeppner L.H., Yang R. ScanNeo: identifying indel-derived neoantigens using RNA-Seq data. Bioinformatics. 2019;35:4159–4161. doi: 10.1093/bioinformatics/btz193. [DOI] [PubMed] [Google Scholar]
- 193.Tappeiner E., Finotello F., Charoentong P., Mayer C., Rieder D., Trajanoski Z. TIminer: NGS data mining pipeline for cancer immunology and immunotherapy. Bioinformatics. 2017;33:3140–3141. doi: 10.1093/bioinformatics/btx377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhou Z., Lyu X., Wu J., et al. TSNAD: an integrated software for cancer somatic mutation and tumour-specific neoantigen detection. R Soc Open Sci. 2017;4:170050. doi: 10.1098/rsos.170050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Rubinsteyn A., Hodes I., Kodysh J., Hammerbacher J. Vaxrank: a computational tool for designing personalized cancer vaccines. bioRxiv. 2018:142919. doi: 10.1101/142919. [DOI] [Google Scholar]
- 196.Tang Y., Wang Y., Wang J., et al. TruNeo: an integrated pipeline improves personalized true tumor neoantigen identification. BMC Bioinform. 2020;21:532. doi: 10.1186/s12859-020-03869-9. [DOI] [PMC free article] [PubMed] [Google Scholar]