Abstract
Metastatic Merkel cell carcinoma (MCC) and cutaneous squamous cell carcinoma (cSCC) are rare and both show impressive responses to immune checkpoint inhibitor treatment. However, at least 40% of patients do not respond to these expensive and potentially toxic drugs. Development of predictive biomarkers of response and rational, effective combination treatment strategies in these rare, often frail patient populations is challenging. This review discusses the pathophysiology and treatment of MCC and cSCC, with a particular focus on potential biomarkers of response to immunotherapy, and discusses how transfer learning using big data collected from patients with common tumours can be used in combination with deep phenotyping of rare tumours to develop predictive biomarkers and elucidate novel treatment targets.
Keywords: Immune checkpoint inhibitor, Machine learning, Merkel cell carcinoma, Squamous cell skin cancer
Highlights
-
•
Metastatic Merkel cell carcinoma and cutaneous squamous cell carcinoma are rare tumours.
-
•
Immunotherapy gives impressive responses but most patients do not survive long term.
-
•
Small patient numbers prevent extensive biomarker research in clinical trials.
-
•
Pooled data from common and rare tumours can be used to train neural networks.
-
•
In rare cancers, neural networks can help identify biomarkers and novel treatment targets.
Introduction
Merkel cell carcinoma (MCC) and cutaneous squamous cell carcinoma (cSCC) are rare in the metastatic setting. The treatment landscape for patients with inoperable or metastatic MCC and cSCC has changed rapidly since the introduction of immune checkpoint inhibitors (ICIs). International efforts have resulted in prospective, non-randomized, single-arm phase 2 studies evaluating ICIs in MCC and cSCC. Trials with the programmed death ligand 1 (PD-L1) antibody avelumab and the programmed death 1 (PD-1) antibodies pembrolizumab and cemiplimab have demonstrated objective response rates (ORRs, complete plus partial responses) of up to ~60% metastatic MCC and cSCC with subsets of patients achieving long-term disease control [[1], [2], [3], [4], [5]].
Despite impressive response rates, most patients with metastatic MCC and cSCC do not achieve long-term benefit from ICIs due to either primary or secondary resistance. For common ICI-sensitive tumours, efforts are focused on the identification of predictive response biomarkers and rational combinational strategies to enhance anti-cancer immune responses. Evidence is emerging that a complex set of tumour, patient and environmental factors govern the strength and timing of the anti-cancer immune response [6,7]. Understanding the role of these immune-modulating factors in metastatic MCC and cSCC is hampered by scarcity of data.
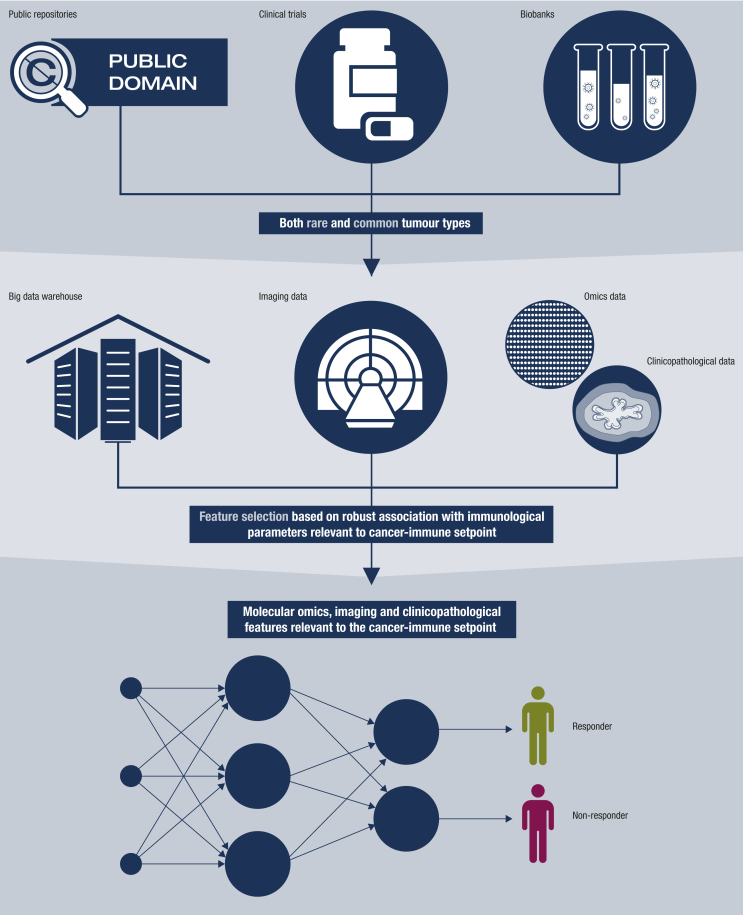
This review presents a brief overview of the pathophysiology and treatment of metastatic MCC and cSCC, focused on potential biomarkers of ICI response, and describes how big data from common tumours can be pooled with the limited available data from rare tumours to help move the treatment of patients with metastatic MCC and cSCC forward (Fig. 1). This is essential as, despite the successes of immunotherapy, the majority of these frail, elderly patients will still die from their disease, and patients who do not respond to therapy are potentially exposed to unnecessary toxicity.
Figure 1.
Schematic overview of how data from common tumours can be used in combination with machine learning to predict immune checkpoint inhibitor responses in rare tumours. A big-data warehouse is constructed by pooling data from public repositories, clinical trials and biobanks. Data consist of clinicopathological, multi-omics and imaging data from common and rare tumours. By applying appropriate statistical inference on this big-data warehouse, clinicopathological, omics and imaging features can be selected that are strongly associated with immunological parameters potentially relevant to the cancer-immune setpoint. These selected features have the highest likelihood of contributing to the accuracy of a predictive model for response to immunotherapy. By using only these selected features as input parameters, the relatively small-scale cohorts of patients treated with immunotherapy can be used to train an accurate and non-overfitted predictive model, which will ultimately improve patient selection for this treatment.
Merkel cell carcinoma: pathophysiology and treatment
MCC is an aggressive, rare skin tumour known for its rapid growth and its likelihood to metastasize. Individuals with a history of extensive sun exposure and immune suppression are at increased risk, and incidence increases with age [[8], [9], [10]].
MCC was initially thought to originate from skin mechanoreceptors called ‘Merkel cells’; however, the cell of origin of MCC is a matter of ongoing debate [11,12]. There are two main routes of pathogenesis for MCC, namely integration of the Merkel cell polyomavirus (MCPyV) into the genome and exposure to ultra-violet (UV) radiation. MCPyV is a common virus with 60–80% seroprevalence in healthy adults, and ~80% of MCCs are MCPyV-positive [13]. For malignant transformation, the virus must be clonally integrated into the host genome at a location resulting in durable expression of viral oncoproteins [14]. Non-virally-associated MCCs have a UV-radiation-associated mutational signature [15]. The mutational burden in these UV-associated MCCs is many times higher than in their virally-induced counterparts and at least as high as in cutaneous melanoma [15,16]. Just over 10% of MCC patients with localized disease develop visceral metastases, and the 5-year overall survival (OS) of these patients was <20% prior to the introduction of ICIs [17]. Data from retrospective series demonstrated that metastatic MCC is highly sensitive to platinum-based chemotherapy combinations with an ~75% overall response rate. However, resistance to chemotherapy occurs quickly with a median progression-free survival of only ~3 months [18]. Expression of viral antigens and high mutational load are both associated with anti-cancer immune responses in other tumour types, and this provided a clear rationale for performing studies with ICIs in metastatic MCC.
Avelumab was evaluated in an open-label, single-arm, multi-centre study in 88 patients with metastatic MCC after previous line(s) of chemotherapy (JAVELIN Merkel 200 Part A) [2]. The ORR was 33%. Among the responders (n = 29), 21 (74%) patients had an ongoing response at 1 year [3]. Of the patients responding at weeks 7 and 13, 90% were still alive 18 months after initiation of treatment, compared with 20–26% of the non-responders [19]. In parallel, avelumab was tested as first-line therapy in metastatic MCC (JAVELIN Merkel 200 Part B). At pre-planned interim analysis in 29 patients with at least 3 months of follow-up, the ORR was 62% with 14 of 18 patients with an ongoing response (78%) at the time of analysis [4]. Pembrolizumab was also evaluated as first-line therapy in patients (n = 50) with advanced MCC (KEYNOTE 017), and similar ORR (56%) and durable responses (79% at 2 years) were observed [20]. Avelumab and pembrolizumab both scored 3 (Table 1) on the ESMO-Magnitude of Clinical Benefit Scale Version 1.1 (ESMO-MCBS), which is a good result for a single-arm study lacking quality-of-life data [21].
Table 1.
Trials with immune checkpoint inhibitors in metastatic Merkel cell carcinoma (MCC) and cutaneous squamous cell carcinoma (cSCC)
| Merkel cell carcinoma |
ORR (95% CI) |
1 year OS |
ESMO-MCBS |
| Avelumab (anti-PD-L1) | |||
| First-line treatment (n = 29) | 62% (42–79) | Data not available | 3 |
| Second-line treatment for metastatic MCC (n = 88) | 33% (23–44) | 52% | 3 |
| Pembrolizumab (anti-PD-1) | |||
| First-line treatment for recurrent locally advanced or metastatic MCC (n = 50) |
56% (35–76) |
72% |
3 |
| Cutaneous squamous cell carcinoma |
ORR (95% CI) |
1 year OS |
ESMO-MCBS |
| Cemiplimab (anti-PD-1) | |||
| First- or second-line treatment for metastatic cSCC (n = 59) | 47% (34–61) | Data not available | 3 |
The ESMO-Magnitude of Clinical Benefit Scale Version 1.1 (ESMO-MCBS) is a tool for evaluation of the magnitude of benefit from clinical studies [21]. The maximum score for a single-arm study is 4 and can only be achieved when quality-of-life data are available.
ORR, objective response rate; CI, confidence interval; OS, overall survival; PD-L1, programmed death ligand 1; PD-1, programmed death 1.
Metastatic cutaneous squamous cell carcinoma: pathophysiology and treatment
cSCC is the second most common skin cancer in the primary setting, but is rare in the metastatic setting with only 2% of patients developing distant metastases [22]. Incidence increases with age, and other risk factors include immunosuppression and sun exposure [23]. cSCC is characterized by a high mutational burden of 50 mutations/Mb, which is approximately four to five times higher than in cutaneous melanoma [24,25]. Interestingly, beta genus human papilloma virus may be a co-factor in cSCC development; therefore, as in MCC, expression of viral antigens could play a role in sensitivity to ICIs [26]. Additionally, the NOTCH signalling pathway has been implicated in cSCC and may influence immune infiltration of these tumours [22,27,28]. In the case of inoperable recurrence or metastatic disease, median OS is < 5 months [29].
Metastatic cSCC can respond to various chemotherapeutic agents including cisplatin, docetaxel and cetuximab, a monoclonal antibody targeting the epidermal growth factor receptor. However, no standard exists due to lack of prospective studies [30].
The PD-1 antibody cemiplimab was studied in an open-label, single-arm, phase 1/2 trial in patients with locally advanced or metastatic cSCC [5]. In the phase 2 part of the trial in metastatic patients (n = 59), an ORR of 47% was achieved; of the 28 responding patients, 82% continued to have a response at the time of data cut-off and the duration of response was >6 months in 57%. Updated data after median follow-up of 17 months showed a 49% ORR [31]. Preliminary data on an additional group of metastatic cSCC patients (n = 56) treated with fixed-dose cemiplimab showed an ORR of 39% at median 8 months of follow-up with median duration of response not yet reached [31]. Cemiplimab scored 3 on the ESMO-MCBS with this single-arm study lacking quality-of-life data (Table 1).
Predicting response and resistance to immune checkpoint inhibitors
Tumour cells undergo molecular ‘rewiring’ during carcinogenesis to escape destruction by the immune system. The equilibrium between the complex set of patient, tumour and environmental factors that promote or suppress the anti-cancer immune response is known as the ‘cancer-immune setpoint’ [6]. The cancer-immune setpoint represents the threshold that must be surpassed for a person with cancer to respond to ICIs. Factors influencing the setpoint differ between patients and include degree of tumour foreignness, general immune status of the patient, immune cell infiltration of the tumour, expression of checkpoints and absence of inhibitory tumour metabolism [7]. This knowledge has not translated into reliable factors that clearly aid in selection of patients for immunotherapy across tumour types; however, a number of factors have some utility as single factors and may be of interest in MCC and cSCC [32].
PD-L1 expression
Immunohistochemical expression of PD-L1 in pre-treatment tumour biopsies is currently used to select patients with non-small-cell lung, breast and cervical cancer for treatment with ICIs [[33], [34], [35]]. However, its value in patient selection varies across tumour types, and patients without PD-L1 expression can respond to ICI treatment in all settings [36,37]. This is likely due to heterogeneity in the expression of PD-L1 between and within tumours and receptor turnover [38,39]. In a small study with whole-body, non-invasive PD-L1 imaging using zirconium-89-labelled atezolizumab, tumour uptake of the tracer predicted response in a group of patients including non-small-cell lung cancer, bladder cancer and triple-negative breast cancer, while immunohistochemical PD-L1 tumour expression failed to do so [40].
PD-L1 can be expressed by tumour cells and by infiltrating immune cells in the tumour micro-environment. In a retrospective study including 67 tumour specimens from 49 MCC patients, tumour PD-L1 expression was associated with improved survival [41]. Patients with chemotherapy-refractory MCC and positive immunohistochemical PD-L1 expression (1% staining threshold in tumour cells) in pre-treatment biopsies appeared to be more likely to respond to avelumab (ORR 35% in PD-L1-positive versus 19% in PD-L1-negative) [2,3]. In the first-line avelumab study in MCC, data on PD-L1 were not reported [4]. In the pembrolizumab MCC study, tumour PD-L1 expression was more frequent in MCPyV-positive tumours than in virus-negative tumours (71% versus 25%), but was not associated with response [1,20]. In both studies, durable responses were seen irrespective of tumour PD-L1 status. Although PD-L1 expression has been described as a potential prognostic marker in metastatic cSCC, it has not been reported whether response is related to PD-L1 expression status [5,[42], [43], [44]].
Tumour-infiltrating lymphocytes
The presence of tumour-infiltrating lymphocytes (TILs) is associated with better prognosis irrespective of treatment in many cancer types, including melanoma and MCC [45]. Increased presence of T cells at the invasive tumour margin and in the tumour prior to treatment, as well as an increase in tumour-infiltrating T-cells during treatment, are positively associated with response to pembrolizumab in patients with melanoma [46]. TILs specific to MCPyV oncoproteins are enriched in some MCPyV-positive MCCs and associated with enhanced expression of both PD-L1 and PD-1 [47,48]. The number of TILs is almost two times higher in MCPyV-positive MCC than in MCPyV-negative MCC, and high intratumoural T-cell counts were associated with improved survival in both MCPyV-positive and MCPyV-negative MCC [49]. In the preliminary analysis of the pembrolizumab trial, TILs were not significantly correlated with viral status or clinical response [1]. TILs have not been evaluated in cSCC.
Tumour mutational burden
The total number of mutations present in a tumour specimen is termed the ‘mutation load’ or ‘tumour mutational burden’ (TMB). Highly mutated tumours are more likely to harbour neo-antigens that can be recognized by the immune system. In several tumour types, a high TMB has been associated with improved response to both cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4) and PD-1 inhibition [50,51]. The measurement of TMB is complicated by the lack of a standard assessment method and intratumoural heterogeneity [52,53]. Moreover, patients with a low TMB can also respond to ICIs. MCPyV-negative MCC tumours have a 25-fold higher TMB than MCPyV-positive tumours, and are characterized by classic UV-signature mutations [15]. TMB was not measured in the clinical trials in MCC. The classic UV signature is also found in cSCC and TMB is generally high, but TMB was not associated with an ICI response in the cemiplimab study [5,42].
Viral status of the tumour
Viruses drive carcinogenesis in human cancers through diverse mechanisms including expression of viral oncoproteins. Oncogenic viral integration in tumour cells is thought to recruit immune effector cells into the tumour micro-environment and upregulate PD-1 and CTLA-4 immunosuppressive pathways, and therefore virally induced tumours may be more likely to respond to ICIs [54]. Expression of MCPyV large T-antigen by tumour cells can be measured by immunohistochemical analysis. In the second-line avelumab study, objective responses occurred in 12 of 46 (26%) patients who tested positive for MCPyV and in 11 of 31 (36%) patients who tested negative for MCPyV. This was determined using a monoclonal antibody specific for MCPyV large T-antigen [2]. The viral status of patients was not reported in the first-line avelumab study. In the MCC pembrolizumab study, expression of the MCPyV large T-antigen oncoprotein was determined by immunohistochemistry, and was complemented by assessment of the presence of serum antibodies or circulating T cells specific for MCPyV oncoproteins [1]. Responses to pembrolizumab were observed in both MCPyV-positive and MCPyV-negative MCC. As with TMB, viral status alone is unlikely to be a predictive marker. Virus-negative tumours have a UV mutational signature with an associated high mutational burden and therefore an alternative reason to respond to ICIs.
A strategy to improve immunotherapy treatment in patients with metastatic MCC and cSCC
Utilizing preclinical tumour models and small-scale patient cohorts, factors associated with the cancer-immune setpoint have been identified in common cancer types. However, only limited factors were studied in the clinical trials in MCC and cSCC, and none of the factors investigated have proven to be sufficient to select patients for immune checkpoint inhibition. Moreover, interesting potentially predictive factors, such as combinations of various markers, signatures from gene expression profiling and data on the diversity or contents of the patient's faecal microbiome, have not been investigated in these rare tumours. Accrual to new trials in MCC and cSCC will be challenging as patients now have access to immunotherapy as standard of care. The question arises how to decide which biomarkers or combination strategies should be prioritized in novel trials.
To improve understanding of the cancer-immune setpoint and to enable rational prioritization of potential biomarkers and treatment strategies in MCC and cSCC, the small-scale MCC and cSCC data must be combined with big data from common cancers. This requires a holistic approach that simultaneously incorporates a host of distinct but related clinicopathological, multi-omics and imaging features characterizing the patient, the tumour and the tumour microenvironment. This can be done most effectively by taking advantage of recent developments in machine learning algorithms capable of distilling complex multilayered information from multifactorial big data [55]. The first step would be to build a big-data warehouse containing clinicopathological, multi-omics and imaging data from patients with rare and common cancers [56] (Fig. 1).
Which big data should be entered into the data warehouse?
To construct the big-data warehouse, multiple types of data from both common and rare tumours must be combined. Firstly, clinicopathological data are required, including patient demographics, tumour type and stage, tumour characteristics including grade and routine immunohistochemical staining results, routine baseline laboratory tests, and anti-cancer treatments including response rates and survival data. Such clinical data have proven to have predictive and/or prognostic value in various settings in cancer patients [57,58]. Secondly, imaging data are required. These include raw data of standard-of-care imaging such as computed tomography and magnetic resonance imaging, as these data can be used to extract mineable high-dimension data, a process known as ‘radiomics’ [59]. 18F-labelled fluoro-2- deoxyglucose-positron emission tomography (PET) scans are used routinely for staging, but actually measure tumour glucose uptake and therefore contain information on the metabolic profile of the tumour [60]. Increasingly, studies using molecular PET imaging are being performed in small-scale clinical trials, and such scans provide whole-body data on drug distribution, expression of immune checkpoint proteins such as PD-L1 or PD-1, and immune-response-related factors such as interleukin-2 or presence of CD8 cells essential for an immune-related response [40,[61], [62], [63]]. Thirdly, multi-omics data (e.g. genomics, transcriptomics, proteomics, metabolomics and microbiomics) are desirable to characterize the patient's immune system, the tumour and the tumour microenvironment.
How can big data be collected?
Multiple sources can be used to fill the big-data warehouse, such as repositories in the public domain, past and ongoing large-scale and small-scale clinical trials, and data from biobanking initiatives. Public repositories such as the Cancer Genome Atlas, the Gene Expression Omnibus and ArrayExpress can provide a major contribution to this big-data warehouse [[64], [65], [66]]. Tens of thousands of genomic and transcriptomic profiles are available for a broad spectrum of rare and common cancer types in combination with a variable amount of metadata describing patient, tumour or treatment characteristics. Although the vast majority of these samples are from patients not treated with immunotherapy and therefore lack phenotypic data on anti-cancer immune responses, they are still an extremely valuable resource to model the cancer-immune set-point [67]. Currently, several gene-expression-based computational deconvolution methods (e.g. CIBERSORT) are available that can complement the available gene expression profiles with inferred immunological parameters describing the type, fraction or functionality of immune cells present within the tumour microenvironment [68]. This is relevant as, ultimately, immune cells present in the tumour microenvironment effectuate the anti-cancer immune response. Therefore, these inferred immunological parameters can be used as proxy phenotypes for the cancer-immune set-point. A major challenge in using data from public repositories is that metadata are described by unstructured text for most individual samples. To address this, an array of tools can be used to identify relevant samples and extract relevant metadata, such as machine learning algorithms used in natural language processing (e.g. context-unaware deep learning and latent semantic indexing), expression signature-based classifier tools and crowdsourcing tools (Table 2). The resulting annotations can be combined with a manual integration and curation step to produce high-quality consensus sample annotations, enabling the identification of relevant samples and relevant metadata.
Table 2.
Examples of tools for sample identification in public repositories
| Automatic text mining tools | |
| Zooma/OntoCat |
http://www.ebi.ac.uk/spot/zooma/ |
| Expression signature-based classifier tools | |
| GEMENI | http://genomics.wpi.edu/gemini/ |
| SPIED3 | http://www.spied.org.uk/cgi-bin/HGNC-SPIED3.1.cgi |
| ProfileChaser | http://profilechaser.ucsf.edu/ |
| ExpressionBlast | http://www.expression.cs.cmu.edu/ |
| SEEK |
http://seek.princeton.edu |
| Crowdsourcing tools | |
| Search Tag Analyze Resource | http://stargeo.org/ |
| CREEDS | http://amp.pharm.mssm.edu/creeds/ |
| ADEPTUS | http://acgt.cs.tau.ac.il/adeptus/ |
Selection of tools that can be used to identify relevant samples in public repositories.
Large randomized and small translational clinical trials on immunotherapeutic interventions in common and rare tumours are a rich data source which should be publicly accessible for research. In addition, a wealth of data is available in registries, biobanking initiatives and, occasionally, single expert-centre case series for rare tumours [69,70]. To enable ongoing collection of as much data as possible from patients with metastatic MCC and cSCC, it is essential that all of these patients are seen at a specialized centre at least once after diagnosis. In this way, suitable patients can be preferentially enrolled in clinical trials, either with novel (combination) treatment strategies, or biomarker or imaging studies. Patients not eligible for clinical trials can provide clinical data and biomaterials before and during treatment with standard-of-care therapies. Concerns about travelling distance to centres for older and frail patients with rare tumours are real [71]. Considering providing the actual treatment closer to home, and continuing contact with the specialized centre for follow-up can mitigate this. At time of progression, biomaterials can be collected again and second-line treatment can be considered.
Importing data into the warehouse should be in accordance with the findable, accessible, interoperable and reusable (FAIR) principles. In addition, ethical, legal and social issues regarding these data sources and their use are essential to consider [72,73].
Utilization of the big-data warehouse
The big-data warehouse can be used to develop a neural network (NN) capable of predicting response to immunotherapy in both common cancers and rare tumours such as MCC and cSCC [74] (Fig. 1). Such a predictive NN can use, for example, omics, and classical clinicopathological and imaging features as input parameters.
However, the number of samples treated with immunotherapy and available for training such a predictive NN is relatively small for rare cancers when compared with the huge number of potential input parameters (e.g. 20 000 gene expression profiles). This can result in overfitting, which happens when a predictive NN learns the detail and noise in the training data to the extent that it negatively impacts the performance of the predictive NN on new data [75]. Therefore, to avoid overfitting of such a predictive NN, a key step is to reduce the architectural complexity of the predictive NN. This can be done by reducing the number of input parameters by feature selection [76]. Data from rare and common cancers in the big-data warehouse can be used for feature selection. By applying appropriate statistical inference, clinicopathological, omics and imaging features associated with (inferred) immunological parameters potentially relevant to the cancer-immune setpoint can be identified reliably. This step enables selection of features with the most robust associations that have the highest likelihood of contributing to the accuracy of the predictive NN for predicting the response to immunotherapy. Another solution to unlock the relatively small data of common and rare cancers treated with ICIs for predictive NNs is the concept of transfer learning, which is a machine learning method where a model developed for a task is re-used as the starting point for a model on a second task. For example, generative adversarial nets or autoencoders can be used to extract a biologically relevant latent space from the tens of thousands of expression profiles generated for common and rare tumour types available within the public domain [77,78]. This will reduce the dimensionality of the data, for example, from 20 000 genes to 100 latent variables. These latent variables could be interpreted as the activity of biological signalling pathways or processes. Subsequently, using these 100 latent variables as input parameters for another model might enable the development of a less-complex predictive NN capable of predicting the response to immunotherapy. Such a predictive NN can be trained using training sets of limited size. These training sets can be formed by pooling and using all available data from series of patients with rare or common cancers treated with immunotherapy. In this way the relatively small-scale cohorts of patients treated with immunotherapy can be used to train an accurate and non-overfitted predictive model, which will ultimately improve patient selection for this treatment. The features identified during model training may contain targetable components that potentially modify the cancer-immune setpoint, and are of interest to enhance anti-cancer immune responses in patients with rare or common cancers.
Discussion and conclusions
The number of approved ICIs and their indications are expanding rapidly and there are >2000 active clinical trials underway in which immunotherapy strategies are being investigated [79]. Extreme responses to ICIs have been observed in several rare cancers (e.g. MCC, cSCC, subgroups of sarcoma and natural killer T-cell lymphoma) [80,81]. However, in general, with the increasing number of indications, the average percentage of patients per disease (subtype) type responding to treatment with ICIs is decreasing [82]. There is a large unmet need for biomarkers predictive of response and data to support rational combinational strategies in both rare and more common tumours.
According to the RARECAREnet database on the epidemiology of rare cancers in Europe, rare cancers are those with an incidence rate of <6 per 100 000 per year in the European population [83]. In total, rare tumours comprise almost 25% of all new cancer diagnoses and therefore form a substantial data source. The way forward for rare cancers, such as metastatic MCC and cSCC, may be to pool data from both common and rare tumours to construct an NN capable of predicting response to immunotherapy in individual rare and common cancers [84,85]. Clinicopathological data, as well as raw images from scans from large-scale and small-scale clinical trials and single and multicentre biobanking initiatives, should be made available. To enhance the quantity of data on rare cancers, centralization of patient care should be supported and patients with rare cancers should be seen at least once at a specialized centre. The pooled data as well as the predictive NN should be widely accessible. Provision of data could be coupled to use of the database and NN to encourage data sharing and ensure continuous input into the model. Once the NN is available, the next step is to use it in a prospective clinical trial with ICIs to determine how accurately it can predict response.
In conclusion, the way forward for rare cancers such as metastatic MCC and cSCC may be through pooling of data with data from more common immunotherapy-sensitive tumours in order to allow construction of NNs capable of predicting response to immunotherapeutic strategies.
Funding
This research was supported by the Netherlands Organization for Scientific Research (NWO-VENI grant 916-16025 to RSNF) and the Dutch Cancer Society (RUG 2013-5960 to RSNF, RUG 2016-/10034 to EGEdV, RUG 2017-1/10913 to MJ).
Disclosure
Dr Jalving has served on advisory boards for Merck, BMS, Novartis, Pierre Fabre, Tesaro and AstraZeneca, and has received speakers' fees from Sanofi and Abbvie. EGEdV reports Institutional Financial Support for her advisory role from Daiichi Sankyo, Merck, NSABP, Pfizer, Sanofi, Synthon and Institutional Financial Support for clinical trials or contracted research from Amgen, AstraZeneca, Bayer, Chugai Pharma, CytomX Therapeutics, G1 Therapeutics, Genentech, Nordic Nanovector, Radius Health, Regeneron, Roche, Synthon, all outside the submitted work. EEV is medical director of the executive board of the Netherlands Cancer Institute and as such legally responsible for all contracts with pharma.
Declaration of interests
The authors have declared no conflicts of interest.
References
- 1.Nghiem P.T., Bhatia S., Lipson E.J., et al. PD-1 blockade with pembrolizumab in advanced Merkel cell carcinoma. N Engl J Med. 2016;374:2542–2552. doi: 10.1056/NEJMoa1603702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaufman H.L., Russell J., Hamid O., et al. Avelumab in patients with chemotherapy-refractory metastatic Merkel cell carcinoma: a multicentre, single group, open-label, phase 2 trial. Lancet Oncol. 2016;17:1374–1385. doi: 10.1016/S1470-2045(16)30364-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaufman H.L., Russell J.S., Hamid O., et al. Updated efficacy of avelumab in patients with previously treated metastatic Merkel cell carcinoma after ≥1 year follow-up: JAVELIN Merkel 200, a phase 2 clinical trial. J Immunother Cancer. 2018;6:4–7. doi: 10.1186/s40425-017-0310-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D’Angelo S.P., Russell J., Lebbé C., et al. Efficacy and safety of first-line avelumab treatment in patients with stage IV metastatic Merkel cell carcinoma: a preplanned interim analysis of a clinical trial. JAMA Oncol. 2018;4 doi: 10.1001/jamaoncol.2018.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Migden M.R., Rischin D., Schmults C.D., et al. PD-1 blockade with cemiplimab in advanced cutaneous squamous-cell carcinoma. N Engl J Med. 2018;379:341–351. doi: 10.1056/NEJMoa1805131. [DOI] [PubMed] [Google Scholar]
- 6.Chen D.S., Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 7.Blank C.U., Haanen J.B., Ribas A., Schumacher T.N. The ‘cancer immunogram’. Science. 2016;352:658–660. doi: 10.1126/science.aaf2834. [DOI] [PubMed] [Google Scholar]
- 8.Harms K.L., Healy M.A., Nghiem P., et al. Analysis of prognostic factor from 9387 Merkel cell carcinoma cases forms the basis for the new 8th edition AJCC staging system. Ann Surg Oncol. 2016;23:3564–3571. doi: 10.1245/s10434-016-5266-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stang A., Becker J.C., Nghiem P., Ferlay J. The association between geographic location and incidence of Merkel cell carcinoma in comparison to melanoma: an international assessment. Eur J Cancer. 2018;94:47–60. doi: 10.1016/j.ejca.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kieny A., Cribier B., Meyer N., et al. Epidemiology of Merkel cell carcinoma. A population-based study from 1985 to 2013, in Northeastern France. Int J Cancer. 2019;144:741–745. doi: 10.1002/ijc.31860. [DOI] [PubMed] [Google Scholar]
- 11.Lemasson G., Coquart N., Lebonvallet N., et al. Presence of putative stem cells in Merkel cell carcinomas. J Eur Acad Dermatol Venereol. 2012;26:789–795. doi: 10.1111/j.1468-3083.2011.04132.x. [DOI] [PubMed] [Google Scholar]
- 12.Sauer C.M., Haug A.M., Chteinber E., et al. Reviewing the current evidence supporting early B-cells as the cellular origin of Merkel cell carcinoma. Crit Rev Oncol Hematol. 2017;116:99–105. doi: 10.1016/j.critrevonc.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 13.Spurgeon M.E., Lambert P.F. Merkel cell polyomavirus: a newly discovered human virus with oncogenic potential. Virology. 2013;435:118–130. doi: 10.1016/j.virol.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng H., Shuda M., Chang Y., Moore P.S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harms P.W., Vats P., Verhaegen M.E., et al. The distinctive mutational spectra of polyomavirus-negative Merkel cell carcinoma. Cancer Res. 2015;75:3720–3727. doi: 10.1158/0008-5472.CAN-15-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madore J., Wong S.Q., Waldeck K., et al. UV-associated mutations underlie the etiology of MCV-negative Merkel cell carcinomas. Cancer Res. 2015;75:5228–5235. doi: 10.1158/0008-5472.CAN-15-1877. [DOI] [PubMed] [Google Scholar]
- 17.van Veenendaal L.M., van Akkooi A.C.J., Verhoef C., et al. Merkel cell carcinoma: clinical outcome and prognostic factors in 351 patients. J Surg Oncol. 2018;117:1768–1775. doi: 10.1002/jso.25090. [DOI] [PubMed] [Google Scholar]
- 18.Iyer J.G., Blom A., Doumani R., et al. Response rates and durability of chemotherapy among 62 patients with metastatic Merkel cell carcinoma. Cancer Med. 2016;5:2294–2301. doi: 10.1002/cam4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D’angelo S., Hunger M., Brohl A., et al. Early objective respons to avelumab treatment is associated with improved overall survival in patients with metastatic Merkel cell carcinoma. Cancer Immunol Immunother. 2019;68:609–618. doi: 10.1007/s00262-018-02295-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nghiem P., Bhatia S., Lipson E.J., et al. Durable tumor regression and overall survival in patients with advanced Merkel cell carcinoma receiving pembrolizumab as first-line therapy. J Clin Oncol. 2019;37:693–702. doi: 10.1200/JCO.18.01896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cherny N.I., Dafni U., Bogaerts J., et al. ESMO-magnitude of clinical benefit scale version 1.1. Ann Oncol. 2017;28:2340–2366. doi: 10.1093/annonc/mdx310. [DOI] [PubMed] [Google Scholar]
- 22.Thompson A.K., Kelley B.F., Prokop L.J., et al. Risk factors for cutaneous squamous cell carcinoma recurrence, metastasis, and disease-specific death: a systematic review and meta-analysis. JAMA Dermatol. 2016;152:419–428. doi: 10.1001/jamadermatol.2015.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ratushny V., Ridky T.W., Seykora J.T., et al. From keratinocyte to cancer: the pathogenesis and modeling of cutaneous squamous cell carcinoma. J Clin Invest. 2012;122:464–472. doi: 10.1172/JCI57415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexandrov L.B., Nik-Zainal S., Wedge D.C., et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inman G.J., Wang J., Nagano A., et al. The genomic landscape of cutaneous SCC reveals drivers and a novel azathioprine associated mutational signature. Nat Commun. 2018;9:3667. doi: 10.1038/s41467-018-06027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chahoud J., Semaan A., Chen Y., et al. Association between β-genus human papillomavirus and cutaneous squamous cell carcinoma in immunocompetent individuals – a meta-analysis. JAMA Dermatol. 2016;152:1354–1364. doi: 10.1001/jamadermatol.2015.4530. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L., Qin H., Wu Z., et al. Pathogenic genes related to the progression of actinic keratoses to cutaneous squamous cell carcinoma. Int J Dermatol. 2018;57:1208–1217. doi: 10.1111/ijd.14131. [DOI] [PubMed] [Google Scholar]
- 28.Al Labban D.A., Jo S.H., Ostano P., et al. Notch-effector CSL promotes squamous cell carcinoma by repressing histone demethylase KDM6B. J Clin Invest. 2018;128:2581–2599. doi: 10.1172/JCI96915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun L., Chin R.I., Gastman B., et al. Association of disease recurrence with survival outcomes in patients with cutaneous squamous cell carcinoma of the head and neck treated with multimodality therapy. JAMA Dermatol. 2019;155:442–447. doi: 10.1001/jamadermatol.2018.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trodello C., Pepper J.P., Wong M., Wysong A. Cisplatin and cetuximab treatment for metastatic cutaneous squamous cell carcinoma: a systematic review. Dermatol Surg. 2017;43:40–49. doi: 10.1097/DSS.0000000000000799. [DOI] [PubMed] [Google Scholar]
- 31.Rischin D., Lim A.M., Schmults C.D., et al. Phase 2 study of 2 dosing regimens of cemiplimab, a human monoclonal anti-PD-1, in metastatic cutaneous squamous cell carcinoma (mCSCC) Ann Oncol. 2019;30:v533–v563. [Google Scholar]
- 32.Nishino M., Ramaiya N.H., Hatabu H., Hodi F.S. Monitoring immune-checkpoint blockade: response evaluation and biomarker development. Nat Rev Clin Oncol. 2017;14:655–668. doi: 10.1038/nrclinonc.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mok T.S.K., Wu Y.L., Kudaba I., et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet. 2019;393:1819–1830. doi: 10.1016/S0140-6736(18)32409-7. [DOI] [PubMed] [Google Scholar]
- 34.Schmid P., Adams S., Rugo H.S., et al. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med. 2018;379:2108–2121. doi: 10.1056/NEJMoa1809615. [DOI] [PubMed] [Google Scholar]
- 35.Chung H.C., Ros W., Delord J.P., et al. Efficacy and safety of pembrolizumab in previously treated advanced cervical cancer: results from the phase II KEYNOTE-158 study. J Clin Oncol. 2019;37:1470–1478. doi: 10.1200/JCO.18.01265. [DOI] [PubMed] [Google Scholar]
- 36.Balar A.V., Galsky M.D., Rosenberg J.E., et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet. 2017;389:67–76. doi: 10.1016/S0140-6736(16)32455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daud A.I., Wolchok J.D., Robert C., et al. Programmed death-ligand 1 expression and response to the anti-programmed death 1 antibody pembrolizumab in melanoma. J Clin Oncol. 2016;34:4102–4109. doi: 10.1200/JCO.2016.67.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mclaughlin J., Han G., Schalper K.A., et al. Quantitative assessment of the heterogeneity of PD-L1 expression in non-small-cell lung cancer. JAMA Oncol. 2016;2:46–54. doi: 10.1001/jamaoncol.2015.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Madore J., Vilain R.E., Menzies A.M., et al. PD-L1 expression in melanoma shows marked heterogeneity within and between patients: implications for anti-PD-1/PD-L1 clinical trials. Pigm Cell Melanoma Res. 2015;28:245–253. doi: 10.1111/pcmr.12340. [DOI] [PubMed] [Google Scholar]
- 40.Bensch F., van der Veen E.L., Lub-de Hooge M.N., et al. 89Zr-atezolizumab imaging as a non-invasive approach to assess clinical response to PD-L1 blockade in cancer. Nat Med. 2018;24:1852–1858. doi: 10.1038/s41591-018-0255-8. [DOI] [PubMed] [Google Scholar]
- 41.Lipson E.J., Vincent J.G., Loyo M., et al. PD-L1 expression in the Merkel cell carcinoma microenvironment: association with inflammation, Merkel cell polyomavirus, and overall survival. Cancer Immunol Res. 2013;1:54–63. doi: 10.1158/2326-6066.CIR-13-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Midgen M.R., Khushalani N.I., Chang A.N.S., et al. Primary analysis of phase 2 results of cemiplimab, a human monoclonal anti-PD-1, in patients (pts) with locally advanced cutaneous squamous cell carcinoma (laCSCC) J Clin Oncol. 2019;37 6015–6015. [Google Scholar]
- 43.Kamiya S., Kato J., Kamiya T., et al. Association between PD-L1 expression and lymph node metastasis in cutaneous squamous cell carcinoma. Asia-Pac J Clin Oncol. 2018 doi: 10.1111/ajco.13102. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 44.García-Díez I., Hernández-Ruiz E., Andrades E., et al. PD-L1 expression is increased in metastasizing squamous cell carcinomas and their metastases. Am J Dermatopathol. 2018;40:647–654. doi: 10.1097/DAD.0000000000001164. [DOI] [PubMed] [Google Scholar]
- 45.Fu Q., Chen N., Ge C., et al. Prognostic value of tumor-infiltrating lymphocytes in melanoma: a systematic review and meta-analysis. OncoImmunology. 2019;8 doi: 10.1080/2162402X.2019.1593806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tumeh P.C., Harview C.L., Yearley J.H., et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Afanasiev O.K., Yelistratova L., Miller N., et al. Merkel polyomavirus-specific T cells fluctuate with Merkel cell carcinoma burden and express therapeutically targetable PD-1 and Tim-3 exhaustion markers. Clin Cancer Res. 2013;19:5351–5360. doi: 10.1158/1078-0432.CCR-13-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iyer J.G., Afanasiev O.K., McClurkan C. Merkel cell polyomavirus-specific CD8⁺ and CD4⁺ T-cell responses identified in Merkel cell carcinomas and blood. Clin Cancer Res. 2011;17:6671–6680. doi: 10.1158/1078-0432.CCR-11-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sihto H., Böhling T., Kavola H. Tumor infiltrating immune cells and outcome of Merkel cell carcinoma: a population-based study. Clin Cancer Res. 2012;18:2872–2881. doi: 10.1158/1078-0432.CCR-11-3020. [DOI] [PubMed] [Google Scholar]
- 50.Rizvi N.A., Hellmann M.D., Snyder A., et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Snyder A., Makarov V., Merghoub T., Yuan J., Zaretsky J.M., Desrichard A., et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qiu P., Poehlein C.H., Marton M., et al. Measuring tumor mutational burden (TMB) in plasma from mCRPC patients using two commercial NGS Assays. Nat Sci Rep. 2019;9:114. doi: 10.1038/s41598-018-37128-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Endris V., Buchhalter I., Allgäuer M., et al. Measurement of tumor mutational burden (TMB) in routine molecular diagnostics: in silico and real-life analysis of three larger gene panels. Int J Cancer. 2019;144:2303–2312. doi: 10.1002/ijc.32002. [DOI] [PubMed] [Google Scholar]
- 54.Song C., Wylie K.M., Wyczalkowski M.A., et al. Dynamic host immune response in virus-associated cancers. Commun Biol. 2019;2:109. doi: 10.1038/s42003-019-0352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rajkomar A., Dean J., Kohane I. Machine learning in medicine. N Engl J Med. 2019;380:1347–1358. doi: 10.1056/NEJMra1814259. [DOI] [PubMed] [Google Scholar]
- 56.Foran D.J., Chen W., Chu H., et al. Roadmap to a comprehensive clinical data warehouse for precision medicine applications in oncology. Cancer Inform. 2017;16:1–10. doi: 10.1177/1176935117694349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Motzer R.J., Bacik J., Schwartz L.H., et al. Prognostic factors for survival in previously treated patients with metastatic renal cell carcinoma. J Clin Oncol. 2004;22:454–463. doi: 10.1200/JCO.2004.06.132. [DOI] [PubMed] [Google Scholar]
- 58.Weide B., Martens A., Hassel J.C., et al. Baseline biomarkers for outcome of melanoma patients treated with pembrolizumab. Clin Cancer Res. 2016;22:5487–5496. doi: 10.1158/1078-0432.CCR-16-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krupinski E.A., Kinahan P., Riviere P. Artificial intelligence in medical imaging. J Med Imaging. 2018;6 doi: 10.1117/1.JMI.6.1.011001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Heer E.C., Brouwers A.H., Boellaard R., et al. Mapping heterogeneity in glucose uptake in metastatic melanoma using quantitative F-FDG PET/CT analysis. EJNMMI Res. 2018;8:101. doi: 10.1186/s13550-018-0453-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bensch F., Brouwers A.H., Lub-de Hooge M.N., et al. 89Zr-trastuzumab PET supports clinical decision making in breast cancer patients, when HER2 status cannot be determined by standard work up. Eur J Nucl Med Mol Imaging. 2018;45:2300–2306. doi: 10.1007/s00259-018-4099-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niemeijer A.N., Leung D., Huisman M.C., et al. Whole body PD-1 and PD-L1 positron emission tomography in patients with non-small-cell lung cancer. Nat Commun. 2018;9:4664. doi: 10.1038/s41467-018-07131-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koleva-Kolarova R.G., Greuter M.J.W., van Kruchten M., et al. The value of PET/CT with FES or FDG tracers in metastatic breast cancer: a computer simulation study in ER-positive patients. Br J Cancer. 2015;112:1617–1625. doi: 10.1038/bjc.2015.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weinstein J.N., Collisson E.A., Mills G.B., et al. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edgar R., Domrachev M., Lash A.E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rocca-Serra P., Brazma A., Parkinson H., et al. ArrayExpress: a public database of gene expression data at EBI. C R Biol. 2003;326:1075–1078. doi: 10.1016/j.crvi.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 67.Jiang P., Gu S., Pan D., et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24:1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Newman A.M., Liu C.L., Green M.R., et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iyer J.G., Parvathaneni K., Bhatia S., et al. Paraneoplastic syndromes (PNS) associated with Merkel cell carcinoma (MCC): a case series of 8 patients highlighting different clinical manifestations. J Am Acad Dermatol. 2016;75:541–547. doi: 10.1016/j.jaad.2016.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ugurel S., Spassova I., Wohlfarth J., et al. MHC class-I downregulation in PD-1/PD-L1 inhibitor refractory Merkel cell carcinoma and its potential reversal by histone deacetylase inhibition: a case series. Cancer Immunol Immunother. 2019;68:983–990. doi: 10.1007/s00262-019-02341-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jain R., Menzin J., Lachance K., et al. Travel burden associated with rare cancers: the example of Merkel cell carcinoma. Cancer Med. 2019;8:2580–2586. doi: 10.1002/cam4.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wilkinson M.D., Dumontier M., Mons B., et al. The FAIR guiding principles for scientific data management and stewardship. Sci Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hartman A.L., Jonker A.H., Parisi M.A., et al. Ethical, legal, and social issues (ELSI) in rare diseases: a landscape analysis from funders. Eur J Hum Genet. 2019 doi: 10.1038/s41431-019-0513-3. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kang T., Ding W., Zhang L., et al. A biological network-based regularized artificial neural network model for robust phenotype prediction from gene expression data. BMC Bioinform. 2017;18:565. doi: 10.1186/s12859-017-1984-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abdelhafiz D., Yang C., Ammar R., Nabavi S. Deep convolutional neural networks for mammography: advances, challenges and applications. BMC Bioinform. 2019;20(Suppl. 11):281. doi: 10.1186/s12859-019-2823-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen Z., Pang M., Zhao Z., et al. Feature selection may improve deep neural networks for the bioinformatics problems. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz763. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 77.Esteva A., Krupel B., Novoa R., et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang X., Dizaji K.G., Huang H. Conditional generative adversarial network for gene expression inference. Bioinformatics. 2018;34:i603–i611. doi: 10.1093/bioinformatics/bty563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tang J., Yu J.X., Hubbard-Lucey V.M., et al. The clinical trial landscape for PD1/PDl1 immune checkpoint inhibitors. Nat Rev Drug Discov. 2018;17:854–855. doi: 10.1038/nrd.2018.210. [DOI] [PubMed] [Google Scholar]
- 80.Kwong Y.K., Chan T.S.Y., Tan D., et al. PD1 blockade with pembrolizumab is highly effective in relapsed or refractory NK/T-cell lymphoma failing l-asparaginase. Blood. 2017;129:2437–2442. doi: 10.1182/blood-2016-12-756841. [DOI] [PubMed] [Google Scholar]
- 81.Veenstra R., Kostine M., Cleton-Jansen A.M., et al. Immune checkpoint inhibitors in sarcomas: in quest of predictive biomarkers. Lab Invest. 2018;98:41–50. doi: 10.1038/labinvest.2017.128. [DOI] [PubMed] [Google Scholar]
- 82.Haslam A., Prasad V. Estimation of the percentage of US patients with cancer who are eligible for and respond to checkpoint inhibitor immunotherapy drugs. JAMA Netw Open. 2019;2 doi: 10.1001/jamanetworkopen.2019.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gatta G., Capocaccia R., Botta L., et al. Burden and centralised treatment in Europe of rare tumours: results of RARECAREnet – a population-based study. Lancet Oncol. 2017;18:1022–1039. doi: 10.1016/S1470-2045(17)30445-X. [DOI] [PubMed] [Google Scholar]
- 84.Topol E. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25:44–56. doi: 10.1038/s41591-018-0300-7. [DOI] [PubMed] [Google Scholar]
- 85.Ross C., Swetlitz I. Stat news. 2018. IBM’s Watson supercomputer recommended ‘unsafe and incorrect’ cancer treatments, internal documents show.https://www.statnews.com/2018/07/25/ibm-watson-recommended-unsafeincorrect-treatments/ Available at: [Google Scholar]