Abstract
Denitrifying bacteria capable of degrading halobenzoates were isolated from various geographical and ecological sites. The strains were isolated after initial enrichment on one of the monofluoro-, monochloro-, or monobromo-benzoate isomers with nitrate as an electron acceptor, yielding a total of 33 strains isolated from the different halobenzoate-utilizing enrichment cultures. Each isolate could grow on the selected halobenzoate with nitrate as the terminal electron acceptor. The isolates obtained on 2-fluorobenzoate could use 2-fluorobenzoate under both aerobic and denitrifying conditions, but did not degrade other halobenzoates. In contrast, the 4-fluorobenzoate isolates degraded 4-fluorobenzoate under denitrifying conditions only, but utilized 2-fluorobenzoate under both aerobic and denitrifying conditions. The strains isolated on either 3-chlorobenzoate or 3-bromobenzoate could use 3-chlorobenzoate, 3-bromobenzoate, and 2- and 4-fluorobenzoates under denitrifying conditions. The isolates were identified and classified on the basis of 16S rRNA gene sequence analysis and their cellular fatty acid profiles. They were placed in nine genera belonging to either the α-, β-, or γ-branch of the Proteobacteria, namely, Acidovorax, Azoarcus, Bradyrhizobium, Ochrobactrum, Paracoccus, Pseudomonas, Mesorhizobium, Ensifer, and Thauera. These results indicate that the ability to utilize different halobenzoates under denitrifying conditions is ubiquitously distributed in the Proteobacteria and that these bacteria are widely distributed in soils and sediments.
Halogenated aromatic compounds are widely distributed in the environment as a result of their widespread use as herbicides, insecticides, fungicides, solvents, fire retardants, pharmaceuticals, and lubricants. Chlorinated compounds are used more frequently than fluorinated or brominated compounds (23). Several of these chemicals cause considerable environmental pollution and human health problems due to their persistence and toxicity. In addition, a large number of halogenated organic compounds are naturally produced by plants, marine organisms, insects, bacteria, fungi, and mammals and released into the environment (11).
Biodegradation of numerous halogenated aromatic compounds has been reported under both aerobic and anaerobic conditions (for reviews, see references 6, 13, 14, 22, and 26). Halobenzoates have been studied as model chemicals for biodegradation of halogenated aromatic compounds, and their aerobic degradation pathways in different bacteria are well characterized (for reviews, see references 6, 7, 13, 21, 26, and 35). The capability to degrade halobenzoates under aerobic conditions has been found in diverse bacteria isolated from various geographical and ecological sources.
The observed functional and taxonomic diversity of halobenzoate-degrading bacteria under aerobic conditions raises questions about the diversity of these catabolic properties under anaerobic conditions. Investigations with enrichment cultures under different anaerobic electron acceptor conditions have demonstrated halobenzoate degradation coupled to denitrification, sulfidogenesis, iron reduction, or methanogenesis (10, 15, 19, 34). Under methanogenic conditions, halobenzoate degradation is initiated by reductive dehalogenation (30), and this process has been extensively studied (6, 14). Under denitrifying conditions, different halobenzoate isomers can be degraded by enrichment cultures established with soils and sediments from various geographical locations (15, 16, 34). Anaerobic utilization of halobenzoate was dependent on and coupled to denitrification, and denitrifying consortia could be maintained with halobenzoate as the sole carbon source. Only one denitrifying chlorobenzoate- and bromobenzoate-degrading bacterium has previously been isolated (17), and the strain was identified as a genomovar of the species Thauera aromatica (28). In addition, a number of fluorobenzoate-degrading denitrifying bacteria have been isolated (25, 31, 34). Our previous work with enrichment cultures suggested that denitrifying bacteria capable of degrading halobenzoates are widespread in different environments; however, there is little information on their taxonomic or biochemical diversity. It is thus of interest to examine this functional group of denitrifying bacteria in detail. In this paper, isolation of pure cultures from the enrichment cultures established with sediments and soils from several different geographical and ecological sites was pursued. The objective was to systematically examine growth on halobenzoate as a sole carbon source, nitrate dependency of halobenzoate degradation, substrate specificity, and taxonomic diversity of halobenzoate-degrading denitrifying bacteria.
MATERIALS AND METHODS
Enrichment cultures.
Sediment and soil samples were collected from various environmental and geographical locations, including estuarine sediments (Arthur Kill, N.J.; Lubeck, Maine; Hopewell Rock, Canada; Alma, Canada; and Shihwa Lake, Korea), freshwater sediments (Passion Puddle, New Brunswick, N.J.; Blue Mountain Lake, N.Y.; Lake Vesijärvi, Finland; Kyungan River, Korea; and Anyang River, Korea), and soils (Rutgers Golf Course, Piscataway, N.J., field soil, Wyoming; El Yunque Forest, Puerto Rico; Guanica Forest, Puerto Rico; Viikki, Finland; and Salpausselkä, Finland). Samples were transferred in sealed glass jars and stored at 4°C until used. Strict anaerobic techniques for medium preparation, culture handling, and sampling were followed throughout the enrichment process. Initial enrichment cultures were established by using 10% (vol/vol) sediment or soil slurry in a minimal salts medium with 30 mM nitrate (32) under argon. One-hundred-milliliter aliquots of the slurry were dispensed in 160-ml serum bottles, sealed with rubber stoppers, and crimped with aluminum seals under a headspace of argon. Halobenzoates (2-fluorobenzoate, 3-fluorobenzoate, 4-fluorobenzoate, 2-chlorobenzoate, 3-chlorobenzoate, 4-chlorobenzoate, 2-bromobenzoate, 3-bromobenzoate, or 4-bromobenzoate; >97% purity; Aldrich Chemical Co., Milwaukee, Wis.) were fed from deoxygenated stock solutions to a final concentration of 100 μM. All enrichment cultures, including autoclaved sterile controls, were incubated in the dark at 30°C. The utilization of halobenzoates was monitored over time, and halobenzoates were refed when depleted. A total of halobenzoates of 1 mmol/liter−1 was fed before cultures were subcultured.
Isolation of halobenzoate-degrading denitrifying bacteria.
The primary enrichment cultures on 2-fluorobenzoate, 4-fluorobenzoate, 3-chlorobenzoate, 4-chlorobenzoate, 3-bromobenzoate, and 4-bromobenzoate were serially diluted up to 10−3 after 1 mM halobenzoate had been degraded. After substrate loss was observed, the 10−3 dilution samples were streaked onto minimal salts (32) plates solidified with Noble agar (Difco, Detroit, Mich.) containing 10 mM nitrate as the electron acceptor and 1 mM benzoate in combination with either 1 mM 2-fluorobenzoate, 4-fluorobenzoate, 3-chlorobenzoate, 4-chlorobenzoate, 3-bromobenzoate, or 4-bromobenzoate as the carbon sources. The plates were incubated in an anaerobic jar under argon at 30°C. After 2 weeks of incubation, single colonies were picked with a sterile capillary tube and restreaked onto fresh minimal salts plates. This isolation procedure was repeated until the colony morphology appeared homogeneous under a dissecting microscope. The purity of isolates was checked on minimal salts plates containing 1 mM succinate and 10 mM nitrate.
To confirm the utilization of halobenzoates, isolates were pregrown on minimal salts plates containing 10 mM nitrate, 1 mM benzoate, and 1 mM succinate under an atmosphere of argon. Cells were then transferred to 10 ml of liquid minimal salts medium (32) and tested for the degradation of halobenzoate (100 μM) under denitrifying conditions.
Growth of isolates on halobenzoate.
To test for growth on halobenzoates under denitrifying conditions, cells were pregrown in liquid medium with 30 mM nitrate as electron acceptor, 5 mM succinate, and 200 μM halobenzoate as carbon and energy sources. The cells were then collected by centrifugation, washed twice with fresh medium, and inoculated in Hungate test tubes (Bellco, Vineland, N.J.) containing 10 ml of fresh liquid medium with 10 mM nitrate and different concentrations of halobenzoates (0, 0.2, 0.5, and 1 mM). Duplicate tubes were prepared for each concentration. Cultures without substrates were used as controls. Growth was monitored by measuring the optical density at 620 nm directly from the Hungate test tubes with a Spectronic 20 spectrophotometer (Bausch and Lomb, Rochester, N.Y.).
To determine the nitrate dependency of halobenzoate degradation, cells were inoculated in Hungate test tubes with or without 0.5 mM halobenzoate or 10 mM nitrate. Each condition was tested in duplicate tubes and growth of cells was monitored as described above.
Substrate utilization.
The utilization of benzoate, hydroxybenzoates, and halobenzoates (>97% purity; Aldrich Chemical Co.) was tested with a dense cell suspension of each isolate after growth in liquid medium with 5 mM succinate and 200 μM halobenzoate under denitrifying conditions. The cultures were then fed 200 μM test substrate and incubated under either aerobic or denitrifying conditions. After 14 days of incubation, the loss of substrates was determined by high-performance liquid chromatography (HPLC).
Determination of cellular fatty acids.
All of the isolates were grown with 10 mM succinate and 30 mM nitrate under anaerobic conditions for 24 h as described previously (27). Cells were harvested by centrifugation, and the cellular fatty acids were saponified, methylated, and extracted by the procedure of the SHERLOCK Microbial Identification System (MIDI, Inc., Newark, Del.). The cellular fatty acid composition was then determined by gas chromatography with an HP 5890 series II gas chromatograph (Hewlett Packard, Palo Alto, Calif.) and the SHERLOCK Microbial Identification System.
16S rRNA gene amplification, sequencing, and phylogenetic analysis.
The genomic DNAs of isolates were extracted and purified as described previously (29). PCR amplification of the 16S rRNA gene of each isolate was performed with a GeneAmp PCR system 2400 as described previously (29). The partial sequence (600 nucleotides) of the 16S rRNA gene was obtained by using internal 16S rRNA oligonucleotide sequencing primers 27F and 685R (18) and an ABI 377A automated sequencer (Perkin-Elmer).
The reference 16S rRNA gene sequences of the species Acidovorax avenaeT (AF078759), Paracoccus denitrificansT (Y16927), Ensifer adhaerensT (AF191739), Ochrobactrum anthropiT (D12794), Bradyrhizobium elkaniiT (U35000), Pseudomonas stutzeriT (U25432), Thauera aromaticaT (X77118), and Azoarcus tolulyticusT (L33694) were obtained from the GenBank database (National Center for Biotechnology Information, National Library of Medicine, Bethesda, Md.) and were aligned by using the PILEUP program of the Genetics Computer Group (GCG) software package (2).
The phylogenetic analysis was carried out according to Kimura's two-parameter method (20) and neighbor-joining topology (24). The SEQBOOT program was used to obtain the confidence level for neighbor-joining analysis with a 100 bootstrap data set (5).
Analytical methods.
All enrichment cultures were monitored periodically for the loss of halobenzoate and nitrate. Approximately 1 ml of well-mixed slurry was sampled with sterile syringes preflushed with argon, centrifuged in a microcentrifuge, and filtered through a 0.45-μm-pore-size filter (Millipore). Samples from pure culture studies were centrifuged if necessary, filtered, and frozen (−20°C) until analyzed. Halobenzoates and other aromatic compounds were analyzed by high-performance liquid chromatography (model LC-10AS; Shimadzu Corp., Kyoto, Japan) with a reverse-phase C18 column (Sperisorb 4.6 by 250 nm, 5-μm particle size; Phenomenex, Torrance, Calif.) and UV detection at 280 nm as previously described (34). The concentrations of nitrate and nitrite were monitored with an ion chromatography system (Dionex DX-100, Sunnyvale, Calif.) with conductivity detection as previously described (34).
Nucleotide sequence accession number.
The 16S rRNA sequences for the halobenzoate-degrading isolates have been deposited in GenBank under accession numbers AF229856 to AF229888.
RESULTS
Halobenzoate degradation by enrichment cultures.
The utilization of halobenzoates by denitrifying enrichment cultures established with sediment and soil from different geographical and ecological sites was observed (data not shown). In general, similar activities were observed with inocula from diverse sources, including estuarine and freshwater sediments and agricultural and forest soils. Complete loss of 2-fluorobenzoate, 4-fluorobenzoate, 3-chlorobenzoate, 4-chlorobenzoate, 3-bromobenzoate, and 4-bromobenzoate was observed within 30 to 70 days in most cultures. Stable cultures were maintained by repeated feeding of these halobenzoates. 3-Fluorobenzoate was not degraded in any of the enrichment cultures. Slow utilization of 2-chlorobenzoate was only observed in the enrichment cultures from Passion Puddle, N.J., and Kyungan River, Korea, but the activity was not maintained during subsequent refeeding. In contrast to 2-chlorobenzoate, utilization of 2-bromobenzoate was observed within 60 to 90 days in about half of the enrichment cultures, and activity was maintained after refeeding.
Isolation of halobenzoate-degrading denitrifying bacteria.
The halobenzoate-degrading enrichments were subcultured, and isolation of denitrifying bacteria was pursued. Using a combination of benzoate and halobenzoate as the carbon sources, a large number of isolates were obtained from the active enrichment cultures of each halobenzoate. They were all capable of degrading benzoate under denitrifying conditions, and 33 isolates out of a total of 122 isolates were able to degrade halobenzoates under denitrifying conditions. These isolates were obtained mainly from enrichment cultures of 2-fluorobenzoate, 4-fluorobenzoate, 3-chlorobenzoate, or 3-bromobenzoate. The strain designations and sources of isolates are listed in Table 1. No 2-bromobenzoate-degrading bacteria could be isolated from the 2-bromobenzoate-utilizing enrichment cultures, although a number of benzoate-degrading denitrifying bacteria were isolated (data not shown). Neither 4-chlorobenzoate- nor 4-bromobenzoate-degrading isolates were obtained from any of the enrichment cultures after repeated attempts. Further studies were focused on the 33 isolates capable of degrading 2-fluorobenzoate, 4-fluorobenzoate, 3-chlorobenzoate, and 3-bromobenzoate under denitrifying conditions.
TABLE 1.
Geographical and ecological origins and doubling time of isolates and closely matched species by 16S rRNA sequence
| Substrate | Strain | Ecological and geographical source | Doubling time (h)a | Closely matched species | % Similarityb |
|---|---|---|---|---|---|
| 2-Fluorobenzoate | 2FB1 | Agricultural soil, Wyo. | 20 | Pseudomonas oleovorans | 99.7 |
| 2FB2 | Estuarine sediment, Arthur Kill, N.J. | 21 | Azoarcus tolulyticus | 99.8 | |
| 2FB3 | Pond sediment, Passion Puddle, N.J. | 26 | Bradyrhizobium elkanii | 98.2 | |
| 2FB4 | River sediment, Kyungan, Korea | NDc | Acidovorax avenae | 96.7 | |
| 2FB5 | River sediment, Kyungan, Korea | 11 | Acidovorax avenae | 95.0 | |
| 2FB6 | Agricultural soil, Wyo. | 15 | Azoarcus tolulyticus | 99.6 | |
| 2FB7 | River sediment, Kyungan, Korea | 21 | Acidovorax avenae | 95.4 | |
| 2FB8 | River sediment, Kyungan, Korea | ND | Ensifer adhaerens | 97.4 | |
| 2FB9 | Agricultural soil, Viikki, Finland | ND | Pseudomonas stutzeri | 97.0 | |
| 2FB10 | Agricultural soil, Viikki, Finland | ND | Ochrobactrum anthropi | 99.2 | |
| 2FB11 | Estuarine sediment, Shihwa, Korea | ND | Pseudomonas stutzeri | 97.9 | |
| 4-Fluorobenzoate | 4FB1 | Estuarine sediment, Arthur Kill, N.J. | 58 | Thauera aromatica | 99.3 |
| 4FB2 | Estuarine sediment, Arthur Kill, N.J. | 35 | Thauera aromatica | 99.3 | |
| 4FB3 | Estuarine sediment, Arthur Kill, N.J. | 151 | Pseudomonas stutzeri | 99.0 | |
| 4FB4 | Agricultural soil, Wyo. | 95 | Pseudomonas stutzeri | 99.7 | |
| 4FB5 | Agricultural soil, Wyo. | 18 | Pseudomonas stutzeri | 99.7 | |
| 4FB6 | Agricultural soil, Wyo. | 58 | Ensifer adhaerens | 99.3 | |
| 4FB7 | Estuarine sediment, Arthur Kill, N.J. | 51 | Pseudomonas stutzeri | 98.4 | |
| 4FB8 | Pond sediment, Passion Puddle, N.J. | ND | Paracoccus denitrificans | 100.0 | |
| 4FB9 | Pond sediment, Passion Puddle, N.J. | ND | Ochrobactrum anthropi | 99.2 | |
| 4FB10 | Estuarine sediment, Shihwa, Korea | 36 | Azoarcus tolulyticus | 99.9 | |
| 4FB11 | Marine sediment, Hopewell Rock, Canada | 114 | Mesorhizobium loti | 96.9 | |
| 4FB12 | River sediment, Anyang, Korea | ND | Ochrobactrum anthropi | 98.9 | |
| 4FB13 | Agricultural soil, Viikki, Finland | ND | Ochrobactrum anthropi | 96.8 | |
| 4FB14 | River sediment, Kyungan, Korea | ND | Ochrobactrum anthropi | 98.9 | |
| 3-Chlorobenzoate | 3CB2 | Estuarine sediment, Arthur Kill, N.J. | 34 | Thauera aromatica | 99.3 |
| 3CB3 | Agricultural soil, Wyo. | 37 | Thauera aromatica | 98.9 | |
| 3CB4 | Estuarine sediment, Arthur Kill, N.J. | 133 | Ochrobactrum anthropi | 99.2 | |
| 3CB5 | Agricultural soil, Wyo. | 131 | Ochrobactrum anthropi | 99.2 | |
| 3CB6 | Estuarine sediment, Shihwa, Korea | 117 | Pseudomonas stutzeri | 97.5 | |
| 3CB7 | River sediment, Kyungan, Korea | 147 | Pseudomonas stutzeri | 97.9 | |
| 3-Bromobenzoate | 3BB1 | Agricultural soil, Wyo. | 20 | Thauera aromatica | 98.9 |
| 3BB2 | Estuarine sediment, Arthur Kill, N.J. | 38 | Pseudomonas stutzeri | 98.9 |
Doubling time was measured for growth on halobenzoates under denitrifying conditions.
The sequence similarities were measured with 600 bp of 16S rRNA gene sequences and compared with the type strains of each species by using the olddistance program in the GCG package.
ND, not determined.
Growth and requirement of nitrate for halobenzoate degradation.
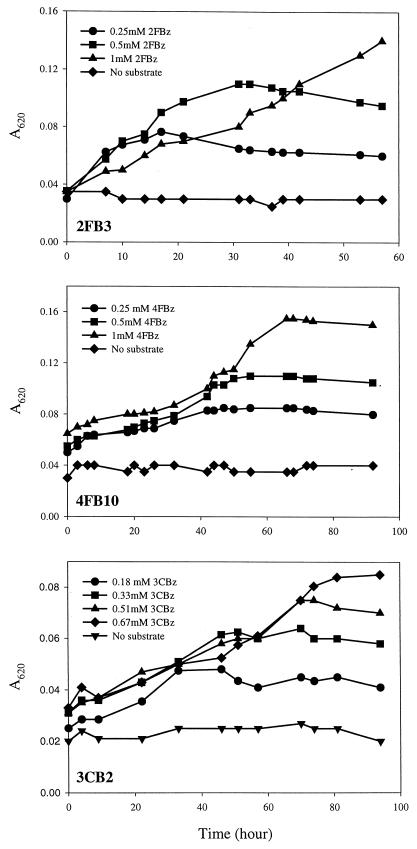
Growth on the respective halobenzoates under denitrifying conditions was verified for each strain. For example, the growth of strains 2FB3, 4FB10, and 3CB2 on different concentrations of halobenzoates is shown in Fig. 1. The doubling time of each strain was calculated from the growth curve on the respective halobenzoate (Table 1). The isolates from 2-fluorobenzoate enrichment cultures had doubling times from 11 to 26 h. These growth rates were higher than those of the other isolates growing on 4-fluorobenzoate, 3-chlorobenzoate, or 3-bromobenzoate. The isolates from 4-fluorobenzoate enrichment cultures had doubling times ranging from 18 to 58 h, except for strains 4FB3, 4FB4, and 4FB11, which were over 95 h. Strains 3CB2 and 3CB3 grew faster on 3-chlorobenzoate under denitrifying conditions than the other 3-chlorobenzoate-degrading strains. Strains 3BB1 and 3BB2 growing on 3-bromobenzoate under denitrifying conditions had doubling times of 20 and 38 h, respectively.
FIG. 1.
Growth of strains 2FB3, 4FB10, and 3CB2 on 2-fluorobenzoate (2FBz), 4-fluorobenzoate (4FBz), and 3-chlorobenzoate (3CBz), respectively, under denitrifying conditions. Different concentrations of halobenzoate were spiked in denitrifying medium containing 10 mM nitrate.
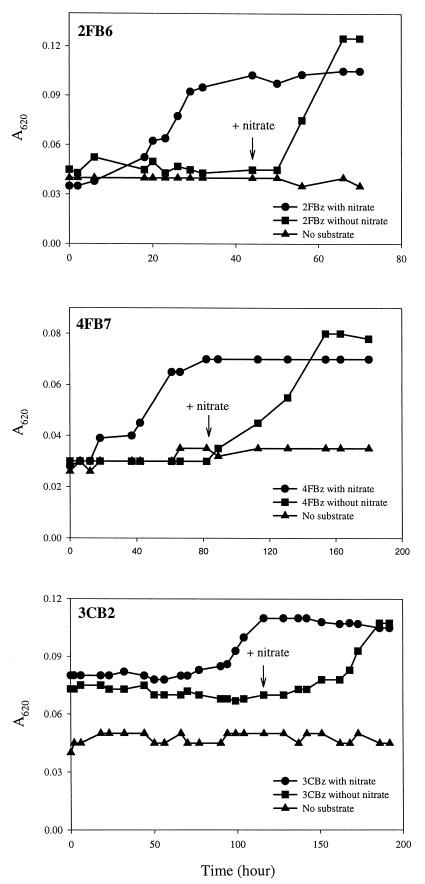
The anaerobic degradation and growth on halobenzoate were dependent on the presence of nitrate, as shown for strains 2FB6, 4FB10, and 3CB2 (Fig. 2). Without nitrate, the isolates did not grow on halobenzoate, but they did grow after the medium was spiked with nitrate. Similar results were obtained for the other halobenzoate-degrading isolates (data not shown). In addition, tests of alternative electron acceptors (10 mM NO3−, NO2−, SO42−, SO32−, S2O32−, SeO42−, or SeO32−, or in the presence of oxygen) with succinate as a carbon source showed that the isolates could grow with O2, NO3−, or NO2− as an electron acceptor (data not shown).
FIG. 2.
Nitrate-dependent growth of strains 2FB6, 4FB7, and 3CB2 on 2-fluorobenzoate (2FBz), 4-fluorobenzoate (4FBz), and 3-chlorobenzoate (3CBz) (0.5 mM), respectively. Nitrate-free cultures were spiked with 10 mM nitrate as indicated by the arrow.
Substrate specificity.
The test of substrate specificity with benzoate, hydroxybenzoates, and halobenzoates showed that all of the isolates degraded benzoate, 3-hydroxybenzoate, and 4-hydroxybenzoate under both denitrifying and aerobic conditions (Table 2). Most of the strains could use 2-hydroxybenzoate under aerobic conditions, but none degraded it under denitrifying conditions. The isolates from the 2-fluorobenzoate enrichment cultures appeared to be limited in their substrate ranges, and with the exception of two strains, did not utilize other halobenzoates under either denitrifying or aerobic conditions. In contrast, most strains from the 4-fluorobenzoate enrichment cultures could use 2-fluorobenzoate and 4-fluorobenzoate under both denitrifying and aerobic conditions. Interestingly, 3-chlorobenzoate and 3-bromobenzoate degradation under denitrifying conditions was observed with two strains isolated on fluorobenzoate. The isolates from the 3-chlorobenzoate and 3-bromobenzoate enrichment cultures used 2-fluorobenzoate under both denitrifying and aerobic conditions and utilized 4-fluorobenzoate, 3-chlorobenzoate, and 3-bromobenzoate under denitrifying conditions.
TABLE 2.
Substrate utilization by halobenzoate-degrading isolates under aerobic and denitrifying conditions
| Strain | Substrate utilization (aerobic/denitrifying conditions)a
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bz | 2OHBz | 3OHBz | 4OHBz | 2FBz | 3FBz | 4FBz | 2CBz | 3CBz | 4CBz | 2BBz | 3BBz | 4BBz | |
| 2FB1 | +/+ | +/− | +/+ | +/+ | +/+ | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− |
| 2FB2 | +/+ | +/− | +/+ | +/+ | +/+ | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− |
| 2FB3 | +/+ | +/− | +/+ | +/+ | +/+ | +/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− |
| 2FB4 | +/+ | +/− | +/+ | +/+ | +/+ | +/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− |
| 2FB5 | +/+ | +/− | +/+ | +/+ | −/+ | −/− | +/− | −/− | −/− | −/− | −/− | −/− | −/− |
| 2FB6 | +/+ | +/− | +/+ | +/+ | +/+ | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− |
| 2FB7 | +/+ | +/− | +/+ | +/+ | +/+ | −/− | −/+ | −/− | −/− | −/− | −/− | −/− | −/− |
| 2FB8 | +/+ | +/− | +/+ | +/+ | +/+ | +/− | +/− | −/− | −/− | −/− | −/− | −/− | −/− |
| 2FB9 | +/+ | +/− | +/+ | +/+ | −/+ | +/− | +/+ | −/− | −/− | −/− | −/− | −/− | −/− |
| 2FB11 | +/+ | +/− | +/+ | +/+ | +/+ | +/− | +/− | −/− | −/− | −/− | −/− | −/− | −/− |
| 4FB1 | +/+ | +/− | +/+ | +/+ | −/+ | +/− | +/+ | −/− | −/− | −/− | −/− | −/− | −/− |
| 4FB2 | +/+ | −/− | +/+ | +/+ | −/+ | −/− | +/+ | −/− | −/− | −/− | −/− | −/− | −/− |
| 4FB3 | +/+ | +/− | +/+ | +/+ | −/− | −/− | +/+ | −/− | −/− | −/− | −/− | −/− | −/− |
| 4FB4 | +/+ | +/− | +/+ | +/+ | +/+ | +/− | +/+ | −/− | −/− | −/− | −/− | −/− | −/− |
| 4FB5 | +/+ | +/− | +/+ | +/+ | +/− | +/− | +/+ | −/− | −/+ | −/− | −/− | −/+ | −/− |
| 4FB6 | +/+ | +/− | +/+ | +/+ | +/+ | +/− | +/+ | −/− | −/+ | −/− | −/− | −/+ | −/− |
| 4FB7 | +/+ | +/− | +/+ | +/+ | −/− | +/− | −/+ | −/− | −/− | −/− | −/− | −/− | −/− |
| 4FB10 | +/+ | −/− | +/+ | +/+ | −/+ | −/− | −/+ | −/− | −/− | −/− | −/− | −/− | −/− |
| 4FB11 | +/+ | +/− | +/+ | +/+ | +/+ | −/− | +/+ | −/− | −/− | −/− | −/− | −/− | −/− |
| 3CB2 | +/+ | −/− | +/+ | +/+ | +/+ | −/− | −/+ | −/− | −/+ | −/− | −/− | −/+ | −/− |
| 3CB3 | +/+ | +/− | +/+ | +/+ | +/+ | +/− | −/+ | −/− | −/+ | −/− | −/− | −/+ | −/− |
| 3CB4 | +/+ | +/− | +/+ | +/+ | +/+ | +/− | −/+ | −/− | −/+ | −/− | −/− | −/+ | −/− |
| 3CB5 | +/+ | +/− | +/+ | +/+ | +/+ | −/− | −/+ | −/− | −/+ | −/− | −/− | −/+ | −/− |
| 3CB6 | +/+ | +/− | +/+ | +/+ | +/+ | −/− | +/+ | −/− | −/+ | −/− | −/− | −/+ | −/− |
| 3CB7 | +/+ | +/− | +/+ | +/+ | +/+ | −/− | +/+ | −/− | −/+ | −/− | −/− | −/+ | −/− |
| 3BB1 | +/+ | +/− | +/+ | +/+ | +/+ | −/− | −/+ | −/− | −/+ | −/− | −/− | −/+ | −/− |
| 3BB2 | +/+ | +/− | +/+ | +/+ | +/+ | −/− | −/+ | −/− | −/+ | −/− | −/− | −/+ | −/− |
+, complete loss of substrate; −, less than 10% loss of substrate. Substrate loss was determined by HPLC after incubation for 14 days. Bz, benzoate; OHBz, hydroxybenzoate; FBz, fluorobenzoate; CBz, chlorobenzoate; BBz, bromobenzoate.
Cellular fatty acid analysis.
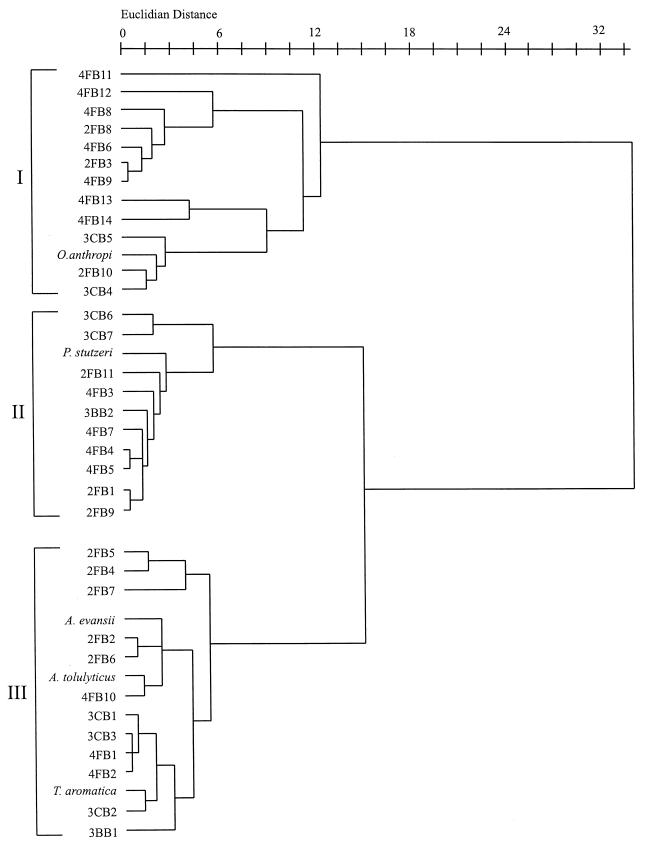
The halobenzoate-degrading strains were grown on succinate under denitrifying conditions, and cellular fatty acids were extracted and analyzed by gas chromatography. Cluster analysis grouped the 33 strains into three clusters on the basis of their fatty acid profiles (Fig. 3). Cluster I was separated from clusters II and III at a Euclidian distance of over 30, while clusters II and III diverged at a Euclidian distance of 14. Ochrobactrum anthropi NCTC 12168T and Pseudomonas stutzeri ATCC 17588T, used as reference strains, were included in clusters I and II, respectively, while strains from the genera Azoarcus and Thauera were located in cluster III. There was no specific grouping of the strains based on either isolation substrate or source of inoculum. Each of these main clusters contained 2-fluorobenzoate-, 4-fluorobenzoate-, and 3-chlorobenzoate-degrading isolates.
FIG. 3.
Dendrogram of halobenzoate-degrading isolates on the basis of their cellular fatty acid profiles. Isolates were grown with 10 mM succinate and 30 mM nitrate for 24 h in the atmosphere of argon.
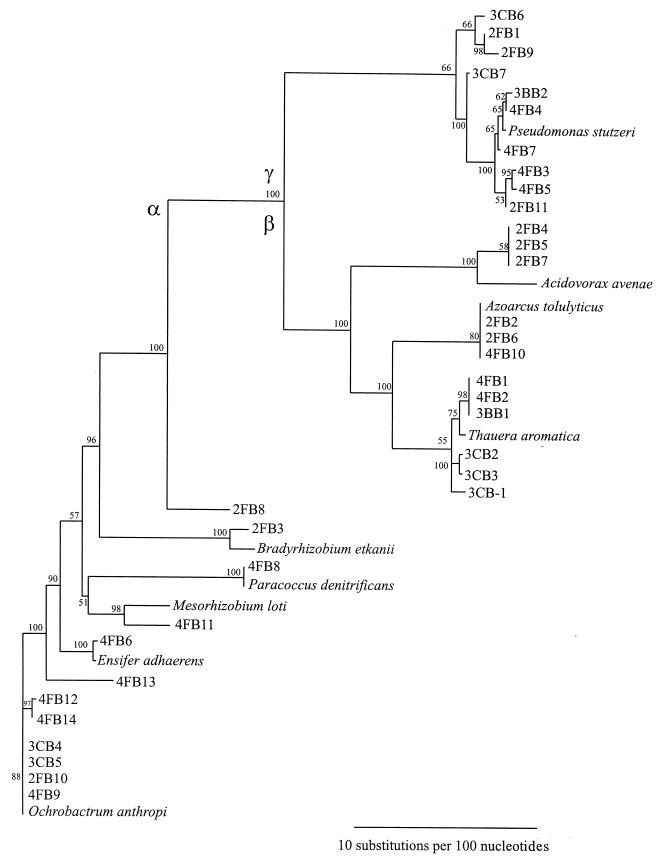
Phylogenetic analysis.
Phylogenetic analysis was done on the basis of the partial 16S rRNA sequences (600 nucleotides) of all isolates. The 33 strains were grouped into three clusters (Fig. 4 and Table 1) in agreement with fatty acid analysis. The closely matched species of the isolates were searched in the GenBank database, and the percent similarities with type strains of each species were calculated by using the olddistance program in the GCG package (Table 1). The strains in cluster I belonged to the genera Paracoccus, Mesorhizobium, Ensifer, Ochrobactrum, and Bradyrhizobium in the α-subunit of the Proteobacteria. The strains in cluster II were confirmed to belong to the genus Pseudomonas in the γ-subunit of Proteobacteria, which was also supported by fatty acid analysis. Strains in cluster III were identified as belonging to the genera Azoarcus, Acidovorax, and Thauera in the β-subunit of the Proteobacteria.
FIG. 4.
Phylogenetic relationship of halobenzoate-degrading isolates. The tree was constructed with 600 bases of the 16S rRNA gene by using a neighbor-joining tree and a 100 bootstrap for the confidence level.
The data show that the halobenzoate-degrading strains are widely distributed in the Proteobacteria. 2-Fluorobenzoate-degrading isolates were widely assigned to the α-, β-, and γ-branches in the genera Ochrobactrum, Bradyrhizobium, Acidovorax, Azoarcus, and Pseudomonas, while the isolates from the 4-fluorobenzoate enrichment cultures were members of the genera Paracoccus, Ensifer, Mesorhizobium, Ochrobactrum, Thauera, Azoarcus, and Pseudomonas. The isolates obtained from 3-chlorobenzoate or 3-bromobenzoate enrichment cultures were placed in the genera Ochrobactrum, Thauera, and Pseudomonas.
DISCUSSION
A total of 33 isolates capable of degrading halobenzoates under denitrifying conditions were obtained from enrichment cultures utilizing 2-fluorobenzoate, 4-fluorobenzoate, 3-chlorobenzoate, and 3-bromobenzoate. The results of our experiments show that organisms able to degrade halobenzoates under denitrifying conditions can be isolated from enrichment cultures, and, regardless of the halogenated substrate used in each enrichment, they belong to different branches of the Proteobacteria. With regard to the geographic and ecological characteristics of the sample sources, the results also indicate the widespread occurrence of these organisms. For example, the strains of the genera Ochrobactrum and Pseudomonas had diverse origins as well as halobenzoate-degrading capacities. In addition, strains of the genus Thauera were obtained from the estuarine sediment of Arthur Kill and field soil from Wyoming by using 4-fluorobenzoate, 3-chlorobenzoate, or 3-bromobenzoate, and strains of the genus Azoarcus capable of degradation of fluorobenzoates under denitrifying conditions were obtained from the estuarine sediments of Arthur Kill and Shihwa Lake (Table 1). Therefore, the diversity of species and various halobenzoate degradation patterns were independent of the geographical and ecological sources of the isolates.
Although 4-chlorobenzoate and 4-bromobenzoate were readily degraded in enrichment cultures with almost all inocula tested, repeated attempts did not result in isolation of pure cultures capable of degrading either 4-chlorobenzoate or 4-bromobenzoate. Previously, anaerobic transformation of 4-chlorobenzoate was demonstrated with pure cultures in the presence of nitrate. The coryneform bacterium strain NTB-1 was capable of converting 4-chlorobenzoate to 4-hydroxybenzoate under strict anaerobic conditions in the presence of nitrate, although hydroxybenzoate was not degraded further in the absence of oxygen (12, 36). Thauera aromatica strain T1, which is capable of degrading toluene and 4-hydroxybenzoate under denitrifying conditions, could use 4-chlorobenzoate as a carbon source under denitrifying conditions after insertion of a cosmid carrying the 4-chlorobenzoate dehalogenase genes from Pseudomonas sp. strain CBS3 (1). Although these pure culture studies indicate the possibility of obtaining isolates from the 4-chlorobenzoate-utilizing enrichment cultures, the degradation of 4-chlorobenzoate may be carried out by the combined activity of the consortia, or the halobenzoate-degrading bacteria may require specific nutrients provided by other members of the consortium. However, it is not clear why the nutritional requirements of the 4-chlorobenzoate- and 4-bromobenzoate-degrading bacteria would be substantially different from those of the isolates obtained on the other halobenzoates.
Halobenzoate degradation under denitrifying conditions was affected by both the halogen substituents and the substitution position. 2-Chlorobenzoate and 3-fluorobenzoate were not degraded by either enrichment cultures or pure cultures, although 3-chlorobenzoate, 3-bromobenzoate, 2-fluorobenzoate, and 4-fluorobenzoate were readily degraded. The negative results could be due to the regioselectivities of enzymes capable of degrading halobenzoates or to the toxicities of these compounds. In fact, inhibition of 2-fluorobenzoate degradation was demonstrated in the presence of 3-fluorobenzoate under denitrifying conditions, although benzoate utilization was not affected (34). The recalcitrance of 2-chlorobenzoate and 2-bromobenzoate under denitrifying conditions is in agreement with previous enrichment culture studies (15, 16), although initial loss of 2-bromobenzoate was observed in some of the enrichment cultures. However, the activity of these 2-bromobenzoate-utilizing cultures was difficult to maintain during subculturing (unpublished results).
Selective enrichment followed by isolation yielded diverse halobenzoate-degrading denitrifying bacteria; nonetheless, enrichment may limit and bias the diversity of organisms obtained. Previous studies of the isolation of aerobic benzoate or 2,4-dichlorophenoxyacetate-degrading bacteria after selective enrichment showed that the numbers of isolates were limited and the isolates formed restricted groups (3, 4). Similar results were also seen with toluene-degrading denitrifiers isolated from various geographical locations (8, 33). The isolates were tightly grouped together, regardless of their geographical and ecological origins, and assigned to the genus Azoarcus (37). Later, these strains were differentiated into three closely related species: Azoarcus tolulyticus, Azoarcus toluvorans, and Azoarcus toluclasticus (27). A study of phenol-degrading denitrifiers isolated from different geographical sources also showed a tight grouping of isolates in the genus Azoarcus (33). However, we isolated diverse groups of bacteria after enrichment in spite of the selective conditions represented by halobenzoate as a substrate and nitrate as an electron acceptor. For instance, four 4-fluorobenzoate-degrading isolates obtained from the same enrichment of Arthur Kill sediment (Table 1) were identified as either Thauera aromatica or Pseudomonas stutzeri, and these strains also had different growth rates. 3-Chlorobenzoate-degrading isolates obtained from a Wyoming soil sample were assigned to either Thauera aromatica or Ochrobactrum anthropi as well.
The isolation of bacteria from enrichment cultures could impose limits on the study of microbial diversity, because enrichment cultures create artificial conditions, which may not be favorable to some members of the microbiota. However, many strains of bacteria have been isolated after enrichments that increased the population and stimulated the slow-growing organisms. This is particularly true of microorganisms capable of degrading recalcitrant compounds, and enrichment cultivation is still important for the study of microbial populations with respect to taxonomic and functional diversity.
Halobenzoates are used as substrates by various bacteria under aerobic conditions (13). A considerable taxonomic diversity of aerobic bacteria capable of degrading halobenzoates was observed from the isolates from contaminated sites as well as from pristine environments (9). We have shown that under denitrifying conditions, halobenzoates were utilized by strains belonging to various branches of the Proteobacteria, and the microbial populations with this biochemical capability appear to be ubiquitously distributed in the environment. Thus, the taxonomic diversity of halobenzoate-degrading denitrifying bacteria has been addressed by traditional enrichment and isolation techniques in this study, although the limitation of enrichment is still present. Further studies of these isolates will unveil the functional diversity of halobenzoate degradation under denitrifying conditions.
ACKNOWLEDGMENTS
This work was supported by grants from the United States Environmental Protection Agency (R822457) and the Office of Naval Research (N00014-94-0434 and N0014-99-1-0761).
REFERENCES
- 1.Coschigano P W, Häggblom M M, Young L Y. Metabolism of both 4-chlorobenzoate and toluene under denitrifying conditions by a constructed bacterial strain. Appl Environ Microbiol. 1994;60:989–995. doi: 10.1128/aem.60.3.989-995.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunbar J, White S, Forney L. Genetic diversity through the looking glass: effect of enrichment bias. Appl Environ Microbiol. 1997;63:1326–1331. doi: 10.1128/aem.63.4.1326-1331.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunbar J, Wong D C L, Yarus M J, Forney L J. Autoradiographic method for isolation of diverse microbial species with unique catabolic traits. Appl Environ Microbiol. 1996;62:4180–4185. doi: 10.1128/aem.62.11.4180-4185.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 6.Fetzner S. Bacterial dehalogenation. Appl Microbiol Biotechnol. 1998;50:633–657. doi: 10.1007/s002530051346. [DOI] [PubMed] [Google Scholar]
- 7.Fetzner S, Lingens F. Bacterial dehalogenases: biochemistry, genetics, and biotechnological applications. Microbiol Rev. 1994;58:641–685. doi: 10.1128/mr.58.4.641-685.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fries M R, Zhou J, Chee-Sanford J, Tiedje J M. Isolation, characterization, and distribution of denitrifying toluene degraders from a variety of habitats. Appl Environ Microbiol. 1994;60:2802–2810. doi: 10.1128/aem.60.8.2802-2810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fulthorpe R R, Rhodes A N, Tiedje J M. High levels of endemicity of 3-chlorobenzoate-degrading soil bacteria. Appl Environ Microbiol. 1998;64:1620–1627. doi: 10.1128/aem.64.5.1620-1627.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genthner B R S, Price II W A, Pritchard P H. Anaerobic degradation of chloroaromatic compounds in aquatic sediments under a variety of enrichment conditions. Appl Environ Microbiol. 1989;55:1466–1471. doi: 10.1128/aem.55.6.1466-1471.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gribble G W. The natural production of chlorinated compounds. Environ Sci Technol. 1994;28:310A–319A. doi: 10.1021/es00056a712. [DOI] [PubMed] [Google Scholar]
- 12.Groenewegen P, van den Tweel W J, de Bont J A M. Anaerobic bioformation of 4-hydroxybenzoate from 4-chlorobenzoate by the coryneform bacterium NTB-1. Appl Microbiol Biotechnol. 1992;36:541–547. [Google Scholar]
- 13.Häggblom M M. Microbial breakdown of halogenated aromatic pesticides and related compounds. FEMS Microbiol Rev. 1992;103:29–72. doi: 10.1111/j.1574-6968.1992.tb05823.x. [DOI] [PubMed] [Google Scholar]
- 14.Häggblom M M, Milligan P W. Anaerobic biodegradation of halogenated pesticides: influence of alternate electron acceptors. In: Bollag J M, Stotzky G, editors. Soil biochemistry. New York, N.Y: Marcel Dekker; 2000. pp. 1–34. [Google Scholar]
- 15.Häggblom M M, Rivera M D, Young L Y. Influence of alternative electron acceptors on the anaerobic biodegradability of chlorinated phenols and benzoic acids. Appl Environ Microbiol. 1993;59:1162–1167. doi: 10.1128/aem.59.4.1162-1167.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Häggblom M M, Rivera M D, Young L Y. Anaerobic degradation of halogenated benzoic acids coupled to denitrification observed in a variety of sediment and soil samples. FEMS Microbiol Lett. 1996;144:213–219. doi: 10.1111/j.1574-6968.1996.tb08533.x. [DOI] [PubMed] [Google Scholar]
- 17.Häggblom M M, Young L Y. Anaerobic degradation of 3-halobenzoates by a denitrifying bacterium. Arch Microbiol. 1999;171:230–236. doi: 10.1007/s002030050704. [DOI] [PubMed] [Google Scholar]
- 18.Johnson J L. Similarity analysis of rRNAs. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C.: American Society for Microbiology; 1994. pp. 683–700. [Google Scholar]
- 19.Kazumi J, Häggblom M M, Young L Y. Diversity of anaerobic microbial processes in chlorobenzoate degradation: nitrate, iron, sulfate and carbonate as electron acceptors. Appl Microbiol Biotechnol. 1995;43:929–936. doi: 10.1007/BF02431930. [DOI] [PubMed] [Google Scholar]
- 20.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 21.Leisinger T, Bader R. Microbial dehalogenation of synthetic organohalogen compounds: hydrolytic dehalogenases. Chimia. 1993;47:116–121. [Google Scholar]
- 22.Neilson A. A review: the biodegradation of halogenated organic compounds. J Appl Bacteriol. 1990;69:445–470. doi: 10.1111/j.1365-2672.1990.tb01536.x. [DOI] [PubMed] [Google Scholar]
- 23.Rossberg M, Lendle W, Togel A, Dreher E-L L E, Rassaeerts H, Kleinschmidt P, Strack H, Beck U, Lipper K-A, Trokelson T R, Loser E, Beutel K K. Chlorinated hydrocarbon. In: Gerhartz W, editor. Ullmann's encyclopedia of industrial chemistry. Weinheim, Germany: VCH; 1986. pp. 233–398. [Google Scholar]
- 24.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. J Mol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 25.Schennen U, Braun K, Knackmuss H-J. Anaerobic degradation of 2-fluorobenzoate by benzoate-degrading, denitrifying bacteria. J Bacteriol. 1985;161:321–325. doi: 10.1128/jb.161.1.321-325.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slater J H, Bull A T, Hardman D J. Microbial dehalogenation. Biodegradation. 1995;6:181–189. doi: 10.1007/BF00700463. [DOI] [PubMed] [Google Scholar]
- 27.Song B, Häggblom M M, Zhou J, Tiedje J M, Palleroni N J. Taxonomic characterization of denitrifying bacteria that degrade aromatic compounds and description of Azoarcus toluvorans sp. nov. and Azoarcus toluclasticus sp. nov. Int J Syst Bacteriol. 1999;49:1129–1140. doi: 10.1099/00207713-49-3-1129. [DOI] [PubMed] [Google Scholar]
- 28.Song B, Palleroni N J, Häggblom M M. Description of strain 3CB-1, a genomovar of Thauera aromatica, capable of degrading 3-chlorobenzoate coupled to nitrate reduction. Int J Syst Evol Microbiol. 2000;50:551–558. doi: 10.1099/00207713-50-2-551. [DOI] [PubMed] [Google Scholar]
- 29.Song B, Young L Y, Palleroni N J. Identification of denitrifier strain T1 as Thauera aromatica and proposed for emendation of the genus Thauera definition. Int J Syst Bacteriol. 1998;48:889–894. doi: 10.1099/00207713-48-3-889. [DOI] [PubMed] [Google Scholar]
- 30.Suflita J M, Horowitz A, Shelton D R, Tiedje J M. Dehalogenation: a novel pathway for the anaerobic biodegradation of haloaromatic compounds. Science. 1982;218:1115–1117. doi: 10.1126/science.218.4577.1115. [DOI] [PubMed] [Google Scholar]
- 31.Taylor B F, Hearn W L, Pincus S. Metabolism of monofluoro- and monochlorobenzoates by a denitrifying bacterium. Arch Microbiol. 1979;122:301–306. doi: 10.1007/BF00411295. [DOI] [PubMed] [Google Scholar]
- 32.Tschech A, Fuchs G. Anaerobic degradation of phenol by pure cultures of newly isolated denitrifying pseudomonads. Arch Microbiol. 1987;148:213–217. doi: 10.1007/BF00414814. [DOI] [PubMed] [Google Scholar]
- 33.van Schie P M, Young L Y. Isolation and characterization of phenol-degrading denitrifying bacteria. Appl Environ Microbiol. 1998;64:2432–2438. doi: 10.1128/aem.64.7.2432-2438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vargas C, Song B, Camps M, Häggblom M M. Anaerobic degradation of fluorinated aromatic compounds. Appl Microbiol Biotechnol. 2000;53:342–347. doi: 10.1007/s002530050032. [DOI] [PubMed] [Google Scholar]
- 35.Winter B, Zimmermann W. Degradation of halogenated aromatics by actinomycetes. Met Ions Biol Syst. 1992;28:157–203. [Google Scholar]
- 36.Zaitsev G M, Karasevich Y N. Preparatory metabolism of 4-chlorobenzoic and 2,4-dichlorobenzoic acids in Corynebacterium sepedonicum. Mikrobiologiya. 1985;54:282–285. [Google Scholar]
- 37.Zhou J, Fries M R, Chee-Sanford J C, Tiedje J M. Phylogenetic analyses of a new group of denitrifiers capable of anaerobic growth on toluene and description of Azoarcus tolulyticus sp. nov. Int J Syst Bacteriol. 1995;45:500–506. doi: 10.1099/00207713-45-3-500. [DOI] [PubMed] [Google Scholar]