Abstract
This study was undertaken in an effort to understand how the population structure of bacteria within terrestrial deep-subsurface environments correlates with the physical and chemical structure of their environment. Phylogenetic analysis was performed on strains of Arthrobacter that were collected from various depths, which included a number of different sedimentary units from the Yakima Barricade borehole at the U.S. Department of Energy's Hanford site, Washington, in August 1992. At the same time that bacteria were isolated, detailed information on the physical, chemical, and microbiological characteristics of the sediments was collected. Phylogenetic trees were prepared from the 39 deep-subsurface Arthrobacter isolates (as well as 17 related type strains) based on 16S rRNA and recA gene sequences. Analyses based on each gene independently were in general agreement. These analyses showed that, for all but one of the strata (sedimentary layers characterized by their own unifying lithologic composition), the deep-subsurface isolates from the same stratum are largely monophyletic. Notably, the layers for which this is true were composed of impermeable sediments. This suggests that the populations within each of these strata have remained isolated under constant, uniform conditions, which have selected for a particular dominant genotype in each stratum. Conversely, the few strains isolated from a gravel-rich layer appeared along several lineages. This suggests that the higher-permeability gravel decreases the degree of isolation of this population (through greater groundwater flow), creating fluctuations in environmental conditions or allowing migration, such that a dominant population has not been established. No correlation was seen between the relationship of the strains and any particular chemical or physical characteristics of the sediments. Thus, this work suggests that within sedimentary deep-subsurface environments, permeability of the deposits plays a major role in determining the genetic structure of resident bacterial populations.
One of the goals of microbial ecology is to determine how environmental factors have shaped the genetic structure of natural microbial populations, creating patterns of genetic variability and affecting evolutionary change. The physical and chemical conditions of the environment act directly on microbes and therefore play an important role in determining the genetic structure of their populations (although horizontal gene transfer also plays a role [20, 31, 47]). Noting the importance of environmental selection on bacteria, Maynard Smith (30) included in his model of bacterial population genetics the concept of ecotypic structure (different clones adapting to their surrounding environment).
A handful of studies examining free-living bacterial populations have explored the extent to which the environment has shaped the population structure. McArthur et al. (32) have shown that the amount of genetic diversity found in Burkholderia (formerly Pseudomonas) cepacia populations across a landscape gradient, as measured by multilocus enzyme electrophoresis, was directly proportional to levels of local environmental diversity. Conversely, a later study examined (by multilocus enzyme electrophoresis) the diversity within B. cepacia populations collected along a stream continuum and found that the population diversity could not be correlated to habitat diversity, nor was there a correlation between genetic and geographic patterns (50). The authors suggested that this was the case because fast mixing may predominate within the stream environment, in contrast to the more isolated soil environment of the previous study.
Comparable analyses have been performed on various marine bacterial populations. A study of marine Vibrio species, isolated from across the water column at locations throughout the world, found that most of the large taxonomic groups identified inhabited particular depth ranges, and the substrates available at these depths could be correlated with the substrates best utilized by the resident group (41). Similarly, studies on marine cyanobacterial (Prochlorococcus) populations found a correlation between depths that the populations inhabited and the light adaptation characteristics of the organisms, dividing the population into high-light- and low-light-adapted clades (11, 49). However, a study on populations of marine cyanobacteria of the genus Synechococcus found no such correlation (48). Also, Field et al. (12) found that analysis of 16S rRNA genes from members of the SAR11 cluster (an uncultured group of the bacterioplankton), cloned from across the water column in locations in both the Atlantic and Pacific Oceans, showed evidence of niche partitioning, with certain lineages showing highly depth-specific distributions.
Studies performed over the past decade have firmly established that microorganisms exist within deep-subsurface environments (100- to 1,000-m depth) (1–3, 5, 14, 15, 17, 23, 24, 33, 39, 42). The types of microbes present and their relative abundances can be quite variable (from below detection to 107 cells per g [dry weight] of sediment) (18). Because the deep-subsurface environments themselves represent a wide variety of geologic, hydrologic, and geochemical conditions, it is difficult to generalize the factors that will be important to the microbial ecology of all subsurface environments. However, it is recognized that factors such as sediment porosity and texture, nutrient availability, oxygen conditions, and degree of water saturation and rate of water flow will be important in determining the types of microbes present and their distribution, persistence, and activities at depth (18).
We wished to examine the genetic diversity within populations of terrestrial deep-subsurface bacteria, both to determine the population structure and to see if this structure could be correlated with the physical and chemical structure of the environment. We chose to study the bacterial population diversity in the deep subsurface of the U.S. Department of Energy (DOE) Hanford site, Washington State, because the distinct, varying lithofacies present over a narrow depth interval represent a variety of geochemically and geophysically distinct habitats. This site had been targeted by the DOE Deep-Subsurface Science Program for detailed geomicrobiological investigation. Sediment core samples were collected during drilling of the Yakima Barricade borehole (YBB) at the Hanford site (well no. 699-48-96) in August 1992. We chose, as our focus group for this study, isolates from the YBB identified as belonging to the genus Arthrobacter. Arthrobacter species are common in soils and are aerobic, high-G+C, chemoorganotrophic, gram-positive bacteria characterized by a rod-to-coccus morphology change as they enter stationary phase. Arthrobacter isolates were found to be present in fairly large numbers in this, as well as other (4, 39, 46), deep-subsurface environments.
We used phylogenetic sequence analysis of two genes, the 16S rRNA and recA genes, to help determine the relationships among the Arthrobacter species. The utility of 16S rRNA as a phylogenetic molecular tool is well recognized (13, 36, 37, 51). RecA is a relatively conserved protein from bacteria and is involved in DNA repair and homologous recombination. RecA protein and gene sequences have been used in a number of studies to determine bacterial phylogenies (9, 21, 28, 34, 52). These analyses show phylogenies that are largely congruent with those based on 16S rRNA gene sequences (9, 21, 28). Phylogenies based on the two gene sequences were used to confirm one another in our study. Also, since recA, as a protein-encoding sequence, is likely to show more variability at the nucleotide level than 16S rRNA gene sequences (since more fluctuation is allowed within protein-coding genes because of codon degeneracy), higher resolution of the more closely related species in trees based on recA might be expected. Thus, it was hoped that the use of recA might help to further distinguish differences within the group under study. Therefore, using phylogenies based on these two molecules, we sought to explore the genetic diversity of Arthrobacter species collected from various depths at the Hanford site in order to determine how the varying environmental conditions created by the different sedimentary facies may have played a role in shaping the population structure.
MATERIALS AND METHODS
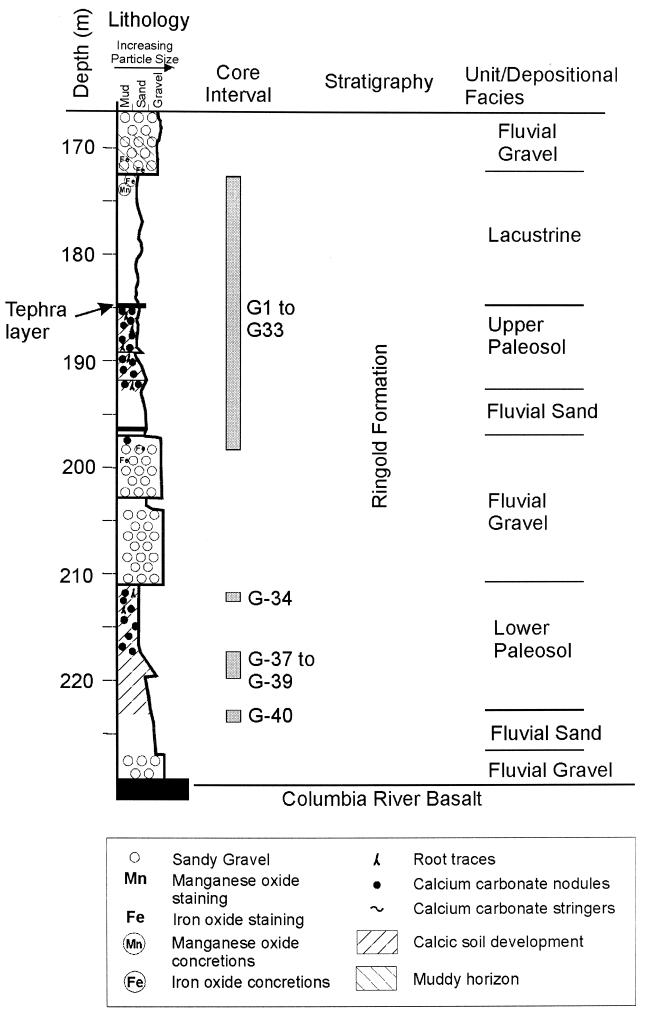
Description of the deep-subsurface site.
The Hanford site lies in the semiarid south-central portion of Washington State. The YBB is located in the western portion of the Hanford site in an area of low meteoric water recharge. The depth to the aquifer in this area is 100 m; groundwater recharge occurs laterally from uplands to the west, making this site hydrologically upstream of any contamination associated with Hanford operations. The subsurface samples used in this study were collected at the YBB in August 1992 as part of a project to explore in detail the microbiology, geohydrology, and geochemistry of the subsurface (6, 15, 16, 24, 33, 35). The sediment samples, taken as a set of cores at between 172.9 and 223.0 m, were well within the saturated zone. This interval is composed of a series of mostly fine-grained fluvial (river)-lacustrine (lake)-paleosol (ancient soil) deposits, approximately 6 million to 8 million years old, stratigraphically within the Ringold Formation, which overlies the Columbia River Basalt Group (7). A stratigraphic column summarizing the sedimentary lithofacies is shown in Fig. 1, which also indicates the areas from which core samples were taken and the lithologic descriptions of the sediments. The top of the interval sampled is composed of fine-grained lacustrine sediments (173- to 185-m depth) that are underlain by an approximately 5-cm layer of volcanic tuff (tephra). Between the tephra and the top of the basalt are represented two complete fluvial sequences, beginning with gravel at the base and grading up to sand and finally a paleosol. Thus, there is a paleosol layer (upper paleosol, 185 to 193 m), followed by an interval of fluvial sands (193 to 197 m), a coarse-grained fluvial sandy gravel (197 to 210.8 m), and another paleosol sequence (lower paleosol, 210.8 to 222.5 m) that is underlain by another fluvial sand and sandy gravel sequence. The lowermost fluvial sand and gravel units were not evaluated in this study.
FIG. 1.
Lithographic and stratigraphic column of the YBB. The areas from which core samples were taken are indicated, along with their respective sediment sample identifiers.
Drilling, tracer introduction, and soil sample collection.
Coring was performed using cable-tool drilling (a drilling method that relies on percussion to advance the borehole without the use of drilling fluids, which can contaminate core samples). Samples were collected using a 2.5-ft-long, 5-in.-outside-diameter split spoon lined with clear, sterilized lexan liners fitted inside the split spoon. Between core runs, 8-in. steel casing was advanced to the next core interval and cleaned out with a core barrel.
Fluorescent microspheres and potassium bromide were added during coring as particle and solute tracers, respectively, to help detect any contamination that may have infiltrated the inner core that was used for analysis (24). Examination for the presence of the tracers in the inner core material showed that no significant contamination existed in the material used for study (24). Once cores were recovered, they were immediately capped and processed on site in an argon-filled glove bag using flame-sterilized instruments. Processing included cutting away the core liner and paring away approximately 1 cm of the outer surface of the core to remove potentially contaminated material. The topmost approximately 12 to 50 cm of inner core material was then homogenized to provide a common sample with which microbiological and geochemical assays were performed, while the bottom approximately 10 to 12 cm was used for permeability and other physical-property measurements. Physical, chemical, and microbiological tests were done by various laboratories on portions of core samples following overnight shipment on ice of samples packed in argon-filled jars.
Physical and geochemical analyses of sediments.
Detailed physical and chemical analyses were performed on sediment samples (16, 24, 33). Lithologic descriptions and estimation of grain size distribution were made on site. Hydraulic conductivity measurements were performed by Core Petrophysics, Inc. (Houston, Tex.). The pH, Eh, and dissolved ions (measured using an ion chromatograph) were analyzed on expressed porewaters (extracted by centrifugation). Total organic carbon was measured by Huffman Laboratories (Golden, Colo.) as the difference between total carbon (measured by combustion) and carbonate carbon (measured by acidification).
Bacterial strains.
For isolation of subsurface strains, serial dilutions were made of homogenized sediment samples (stored at 4°C) in phosphate buffer and plated on a variety of media (2, 4). All colonies arising on plates were collected and preserved in the DOE Subsurface Microbial Culture Collection (SMCC) (D. Balkwill, Florida State University). The isolates, as part of the SMCC, were subjected to a battery of tests, including morphological and physiological characterizations as well as 16S rRNA similarity rank analysis (29) and sequence analysis (4). Isolates that were chosen for these analyses (Table 1) were any that were identified as being most closely related to Arthrobacter globiformis from sediment samples that were processed immediately or stored no longer than 3 to 8 weeks. Because the samples were not all equally screened for isolates, the relative distribution of Arthrobacter species used in this study should not be interpreted as the relative abundance of Arthrobacter species along the sampled interval. Deep-subsurface Arthrobacter isolates were grown at 25 to 30°C in nutrient broth (Difco Laboratories, Detroit, Mich.).
TABLE 1.
Physical and chemical characteristics of, and Arthrobacter isolates from, Hanford YBB sediment samples
| Lithofacies and depth interval (m) | Sediment sample | pHa | Ehb (mV) | NO3 (mg liter−1) | PO4 (mg liter−1) | SO4 (mg liter−1) | TOC (mg kg−1) | % Clay/ % silt/ % sand | Hydraulic conductivity (cm s−1) | Isolate(s) |
|---|---|---|---|---|---|---|---|---|---|---|
| Lacustrine unit | ||||||||||
| 172.9–173.8 (oxidized layer) | G1 | 7.8 | NDc | 2.53 | <0.10 | 12.43 | 300 | 18/76/6 | <1.0 × 10−9 | G960, G961, G964, G965, ZAT001, ZAT004, ZAT005, ZAT012, ZAT013 |
| 173.8–174.3 | G2 | 8.0 | ND | 3.71 | 0.55 | 14.08 | 310 | 19/79/2 | <1.0 × 10−9 | |
| 174.5–175.2 | G3 | 8.7 | 200 | 2.37 | <0.10 | 8.61 | 9,610 | 29/68/3 | ND | ZAT014 |
| 176.2–176.9 | G5 | 8.7 | 260 | 1.64 | <0.10 | 5.5 | 10,480 | 38/53/9 | ND | G962, G969, ZAT255 |
| 177.1–177.5 | G6 | 8.1 | ND | 1.45 | <0.10 | 4.68 | 11,690 | 33/56/11 | 2.9 × 10−7 | G915 |
| 177.5–178.4 | G7 | 8.4 | 200 | 1.46 | <0.10 | 3.94 | 10,860 | 41/57/2 | 5.7 × 10−8 | G963, G970 |
| 178.6–179.3 | G8 | 8.5 | 180 | 1.35 | <0.10 | 4.03 | 8,440 | 34/64/2 | ND | |
| 179.4–180.2 | G9 | 8.6 | 150 | 2.07 | <0.10 | 7.56 | 9,700 | 45/55/0 | ND | G966, G980 |
| 180.3–181.0 | G10 | 8.8 | 150 | 2.31 | <0.10 | 7.50 | 7,760 | 28/71/1 | 1.6 × 10−8 | G968, G979, ZAT200 |
| 181.1–181.6 | G11 | 8.5 | 160 | <0.03 | <0.10 | 23.07 | 8,250 | 28/72/0 | ND | |
| 181.8–182.3 | G12 | 8.7 | 130 | 2.11 | <0.10 | 12.15 | 6,500 | 30/70/0 | ND | |
| 182.3–183.0 | G13 | 8.9 | 190 | <0.03 | <0.10 | 17.41 | 6,190 | 29/71/0 | ND | ZAT262, ZAT263 |
| 183.1–183.9 | G14 | 8.8 | 180 | 4.94 | <0.10 | 15.55 | 5,370 | 36/64/0 | ND | ZAT351, ZAT352 |
| 184.6–184.8 | G16A | 9.0 | 160 | 0.14 | <0.10 | 73.38 | 12,180 | 35/63/2 | 1.1 × 10−7 | |
| Upper paleosol unit | ||||||||||
| 184.8–184.8 (tefra) | G16T | 9.5 | ND | ND | ND | ND | ND | 36/20/44 | 1.1 × 10−7 | ZAT277 |
| 184.8–185.0 | G16B | 8.6 | 160 | 2.91 | <0.10 | 3.64 | ND | 15/52/33 | 1.1 × 10−7 | |
| 185.2–186.0 | G17 | 7.0 | 50 | 1.88 | <0.30 | 41.33 | 870 | 35/58/7 | ND | |
| 186.1–186.9 | G18 | 7.6 | 50 | 2.45 | <0.10 | 72.62 | 990 | 45/52/3 | ND | |
| 187.5–188.4 | G20 | 7.3 | 80 | 0.48 | <0.10 | 140.1 | 2,090 | 51/34/15 | ND | |
| 188.7–189.6 | G21 | 6.3 | 80 | <0.03 | <0.10 | 202.2 | 7,410 | 10/75/15 | 4.5 × 10−7 | ZAT031 |
| 191.7–192.1 | G25 | 8.1 | 300 | <0.03 | <0.10 | 11.47 | 1,300 | 10/13/77 | ND | |
| 192.2–192.7 | G26 | 7.0 | 320 | 17.17 | <0.01 | 39.81 | 660 | 22/61/17 | <1.0 × 10−9 | |
| Fluvial sands unit | ||||||||||
| 192.9–193.6 | G27 | 6.6 | 300 | 3.01 | <0.10 | 12.58 | 250 | 36/29/35 | <1.0 × 10−9 | |
| 193.9–194.6 | G28 | 7.3 | 290 | 2.81 | <0.10 | 17.89 | 250 | 10/52/38 | ND | |
| 195.0–195.6 | G29 | 7.5 | 290 | 1.99 | <0.10 | 16.43 | 230 | 8/33/59 | ND | |
| 195.8–196.3 | G30 | 7.3 | 290 | <0.03 | <0.10 | 15.77 | 180 | 10/16/74 | 8.8 × 10−7 | |
| 196.3–197.0 | G31 | 8.8 | 290 | 2.06 | <0.10 | 15.00 | 460 | 10/40/50 | ND | G954, G986, G991 |
| Fluvial gravel unit | ||||||||||
| 197.6–197.7 | G33 | 7.0 | 280 | <0.03 | <0.10 | 10.97 | 160 | ND | ND (10−4)d | G919, G982, G984, G993 |
| 211.3–211.7 | G34 | ND | ND | ND | ND | ND | ND | ND | ND | |
| Lower paleosol unit | ||||||||||
| 217.5–217.7 | G37 | ND | ND | ND | ND | ND | ND | ND | ND | G950, G959, ZAT054, ZAT055, ZAT056 |
| 222.2–223.0 | G40 | ND | ND | 3.56 | <0.07 | 19.52 | ND | ND | ND |
Of expressed porewater.
Of slurry.
ND, not determined.
No data are available for the hydraulic conductivity of this fluvial gravel sequence, but a measurement within the fluvial gravel sequence overlying the lacustrine is on the order of 10−4 cm s−1, and this stratum is likely to have a similar permeability.
The following American Type Culture Collection (ATCC) and Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSM) type strains of Arthrobacter and related genera were also included in these analyses for comparison: Arthrobacter agilis (ATCC 966, DSM 20550), A. aurescens (ATCC 13344, DSM 20116), A. citreus (ATCC 11624, DSM 20133), A. globiformis (ATCC 8010, DSM 20124), A. histidinolovorans (ATCC 11442, DSM 20115), A. nicotianae (ATCC 15236, DSM 20123), A. nicotinovorans (ATCC 49919, DSM 420), A. oxydans (ATCC 14358, DSM 20119), A. pascens (ATCC 13346, DSM 20545), A. polychromogenes (ATCC 15216, DSM 20136), A. protophormiae (ATCC 19271, DSM 20168), A. sulfureus (ATCC 19098, DSM 20167), A. ureafaciens (ATCC 7562, DSM 20126), A. uratoxydans (ATCC 21749, DSM 20647), Micrococcus luteus (ATCC 381), and M. lylae (ATCC 27566, DSM 20315). The strains were obtained from the ATCC and were grown at 25 to 30°C in nutrient broth.
Primer design and PCR amplification, cloning, and sequencing of 16S rRNA and recA genes.
DNA was isolated from the Arthrobacter and Micrococcus strains by lysozyme digestion followed by extraction in phenol and chloroform in the presence of hexadecyltrimethylammonium bromide (38) or as described previously (4). PCR amplification and sequencing of 16S rRNA genes from subsurface Arthrobacter species were done as described by Balkwill et al. (4). 16S rRNA sequences for the type strains were obtained from GenBank. The GenBank accession numbers for these sequences are as follows: A. agilis, X80748; A. aurescens, X83405; A. citreus, X80737; A. globiformis, X80736; A. histidinolovorans, X83406; A. nicotianae, X80739; A. nicotinovorans, X80743; A. oxydans, X83408; A. pascens, X80740; A. polychromogenes, X80741; A. protophormiae, X80745; A. sulfureus, X83409; A. ureafaciens, X80744; A. uratoxydans, X83410; M. luteus, M38242; and M. lylae, X80750. These 16S rRNA sequences were determined for DSM type strains, except M. luteus 16S rRNA, for which the ATCC strain was used; we chose ATCC strains for recA sequence determination that matched these DSM strains (listed above). Also used in these analyses were sequences for the Streptomyces ambofaciens ATCC 23877 16S rRNA gene (GenBank accession number M27245) and the S. ambofaciens DSM 40697 recA gene (accession number Z30324).
Primers used in the PCR amplification and sequencing of the Arthrobacter recA genes are shown in Table 2. Since, prior to this study, no recA sequences had been determined for any Arthrobacter species, for initial analysis of recA, degenerate primers GPRA-FB and GPRA-R2, designed against highly conserved areas of the RecA protein, were used in the PCR to amplify an interior ∼350-bp fragment from total DNA of A. globiformis. This fragment was gel purified (Qiagen QIAquick Gel Purification Kit), cloned into the PCR product-cloning vector pCR2 (Invitrogen TA Cloning Kit), and sequenced. In order to amplify a larger region of recA for phylogenetic studies, GPRA-UF2 and GPRA-UR2 were designed against other conserved areas based on an alignment of the amino acid sequences of seven previously characterized recA genes (three from other high-G+C gram-positive genera) that were most related to the translation of the A. globiformis 350-bp fragment. Using these primers, successful amplification of an ∼830-bp band was obtained from a number of Arthrobacter species. These products were cloned as described above from A. globiformis and one of the deep-subsurface Arthrobacter species. Sequencing of these clones facilitated design of more specific primers, A19-F2 and A1-R, that were used to amplify and sequence many of the recA genes. The growing collection of sequences was used to design alternate primers (AU-F1 and AU-R1) that were used to amplify and sequence other Arthrobacter recA genes. AU-FM1 and AU-RM1 were used to complete the sequence in both directions. PCR products were gel purified and sequenced (200 fmol per reaction) on an Applied Biosystems model 373A automated DNA sequencer.
TABLE 2.
Primers developed for PCR and sequencing of recA genes
| Primer | Nucleotide (amino acid) positiona | Primer sequence |
|---|---|---|
| GPRA-FBb | 289–305 (96–101) | Glu His Ala Leu Asp Pro |
| 5′-GAR CAY GCN CTN GAY CC-3′ | ||
| Pro | ||
| GPRA-R2 | 638–619 (212–206) | Gly Gly Thr Thr Thr Glu Pro |
| 5′-CC SCC SGK SGT SGT YTC NGG-3′ | ||
| Ser | ||
| GPRA-UF2 | 67–86 (22–28) | Gly Lys Gly Ala Val Met Arg |
| 5′-GGS AAG GGS KCN GTN ATG CG-3′ | ||
| Ala Phe Lys | ||
| GPRA-UR2 | 910–893 (304–298) | Glu Lys Gly Gln Gly Leu Gln |
| 5′- C CTT SCC CTG SCC NAR YT-3′ | ||
| A19-F2 | 79–98 | 5′-GTCATGCGCCTGGGCGACGA-3′ |
| A1-R1 | 909–890 | 5′-CTTGCCCTGGCCGAGTTGGT-3′ |
| AU-F1 | 120–137 | 5′-CATYCCCACCGGHTCCAT-3′ |
| AU-R1 | 834–817 | 5′-CATGTCGATGATGCCGCC-3′ |
| AU-FM1 | 463–482 | 5′-GAAATCGAAGGCGACATGGG-3′ |
| AU-RM1 | 531–514 | 5′-ACGCAGGGCCTGGCTCAT-3′ |
Position relative to that in E. coli recA (A of initial ATG = position 1; amino acid numbering started after initial Met).
Primers identical to GPRA-FB and similar to GPRA-R2 (adapted here for the high G+C content of Arthrobacter) were used previously (8) to amplify recA genes from various low-G+C gram-positive bacteria.
Phylogenetic data analysis.
The 16S rRNA gene sequences were aligned, and after the ends of the sequences were trimmed to equal lengths, 1,160 bases (including gaps) of the sequences were included in the phylogenetic analyses (corresponding to nucleotide positions 197 to 1325 of the A. globiformis 16S rRNA gene). The recA sequences were aligned and trimmed to equal lengths, and 360 bases were included in the phylogenetic analyses (corresponding to nucleotide positions 312 to 669 of the Escherichia coli recA gene). Alignment of 16S rRNA sequences was performed by eye, using the secondary structure of A. globiformis 16S rRNA (Ribosomal Database Project) (29) as a guide. The recA nucleotide sequences were aligned using the program Clustal V (19), aided by the high conservation within the translations of the sequence. Analysis of sequence composition was done using the MEGA program (27). Phylogenetic analyses were done using the PHYLIP 3.572 package of programs (10), using both neighbor-joining/distance matrix (DNADIST and NEIGHBOR) and parsimony (DNAPARS) methods.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the 16S rRNA sequences generated for this report are as follows (SMCC strain, accession number): G915, AF197020; G919, AF197021; G950, AF197022; G954, AF197023; G959, AF197024; G960, AF197025; G961, AF197026; G962, AF197027; G963, AF197028; G964, AF197029; G965, AF197030; G966, AF197031; G968, AF197032; G969, AF197033; G970, AF197034; G979, AF197035; G980, AF197036; G982, AF197037; G984, AF197038; G986, AF197039; G991, AF197040; G993, AF197041; ZAT001, AF197042; ZAT004, AF197044; ZAT005, AF197045; ZAT012, AF197046; ZAT013, AF197047; ZAT014, AF197048; ZAT031, AF197049; ZAT054, AF197050; ZAT055, AF197051; ZAT056, AF197052; ZAT200, AF197053; ZAT255, AF197054; ZAT262, AF197055; ZAT263, AF197056; ZAT277, AF197057; ZAT351, AF196342; and ZAT352, AF196343. The GenBank accession numbers for the recA sequences generated for this report are as follows (SMCC or type strain, accession number): G915, AF214757; G919, AF214758; G950, AF214759; G954, AF214760; G959, AF214761; G960, AF214744; G961, AF214762; G962, AF214763; G963, AF214764; G964, AF214776; G965, AF214777; G966, AF214765; G968, AF214766; G969, AF214739; G970, AF214740; G979, AF214767; G980, AF214742; G982, AF214768; G984, AF214741; G986, AF214769; G991, AF214743; G993, AF214746; ZAT001, AF214745; ZAT004, AF214748; ZAT005, AF214747; ZAT012, AF214755; ZAT013, AF214770; ZAT014, AF214773; ZAT031, AF214756; ZAT054, AF214771; ZAT055, AF214772; ZAT056, AF214774; ZAT200, AF214775; ZAT255, AF214750; ZAT262, AF214754; ZAT263, AF214751; ZAT277, AF214753; ZAT351, AF214749; ZAT352, AF214752; A. agilis, AF214779; A. aurescens, AF214793; A. citreus, AF214781; A. globiformis, AF214780; A. histidinolovorans, AF214788; A. nicotianae, AF214792; A. nicovorans, AF214784; A. oxydans, AF214789; A. pascens, AF214786; A. polychromogenes, AF214785; A. protophormiae, AF214790; A. sulfureus, AF214787; A. ureafaciens, AF214782; A. uratoxydans, AF214791; M. luteus, AF214783; and M. lylae, AF214778.
RESULTS AND DISCUSSION
Sediment chemical, physical, and microbiological characteristics.
In this study, we explored the phylogenetic diversity of deep-subsurface Arthrobacter populations within several sedimentary lithofacies of the YBB at the DOE Hanford site. The strains were isolated from sediment samples taken at depths of 172.9 to 217.7 m (Fig. 1) during the August 1992 coring of the YBB. With the same sediment samples, detailed microbiological, physical, and chemical properties were analyzed. Table 1 details physical and chemical information for the sediment samples and indicates from which samples Arthrobacter species used in these analyses were isolated.
The sampled interval consists of varying lithologies, each with distinct physical and chemical characteristics. In general, total organic carbon (TOC) was highest in the lacustrine layer, except for the uppermost portion, which includes sample G1, at 172.9–173.8 m. The top 2 m of the lacustrine unit is an oxidized layer, characterized by iron and manganese oxides and low organic carbon, associated with highly oxidizing groundwater in the gravels immediately above the lacustrine sediments. TOC was also relatively low in the upper paleosol and in the fluvial sands and gravels. In general, the pH was slightly alkaline throughout the lacustrine interval and was more neutral to acidic throughout the upper paleosol and lower layers (except for fluvial sand sample G31 [196.3 to 197.0 m], at pH 8.8). The Eh indicated, in general, moderately oxidizing conditions throughout the interval, being somewhat less oxidizing in the upper paleosol. Overall, nitrate concentrations varied little, and phosphate concentrations were extremely low. Sulfate concentrations were relatively low throughout the lacustrine unit but began increasing in the upper paleosol, reaching a maximum within sample G21 (188.7 to 189.6 m), after which the concentration dropped off dramatically and remained low throughout the rest of the interval. Ammonium concentrations were very low in all samples, mostly below detection (data not shown). The salinity (major dissolved cations) (data not shown) was relatively constant throughout the sampled interval, with the exception of three samples (G2 [173.8 to 174.3 m], G17 [185.2 to 186.0 m], and G27 [192.9 to 193.6 m]) which exhibited lower cation concentrations than other samples (none of these were samples in which Arthrobacter species used in these studies originated). The lacustrine, fluvial sand (which was well cemented), and paleosol units have particularly low porosity and hydraulic conductivities, which inhibit movement of groundwater, nutrients, and bacteria through these layers. Thus, the lacustrine, upper paleosol, and fluvial sand units, combined, form a low-permeability hydraulic barrier between two highly permeable gravel layers. Extremely little chemical, physical, or microbial data are available for the lower paleosol unit.
Several groups have investigated the microbial characteristics of the sampled interval (16, 24, 33). In general, investigators have found microbial activities and numbers (both aerobic and anaerobic) to be very low across the interval. The numbers of cells and direct measurements of their metabolism were highest in the lacustrine layers, where TOC was highest, and were lowest in the fluvial sands, where TOC was also low (24). The persistence of organic matter in the lacustrine layer may be because of the lack of its availability to microbes, probably due to the extremely low permeability of this unit (24). The results of these investigations suggest that patchy aerobic and anaerobic microenvironments exist throughout these sediments, allowing low levels of various anaerobes as well as strict aerobes, including Arthrobacter species, to persist within the sediments.
Sequencing of 16S rRNA and recA genes from Arthrobacter strains.
The recA and 16S rRNA gene sequences were determined for 39 YBB deep-subsurface Arthrobacter species (Table 1). Seventeen Arthrobacter and related-genus type strains were also included in the analyses for comparison. The overall pairwise nucleotide sequence identity for deep-subsurface Arthrobacter strains for 16S rRNA genes was 95.3 to 100%, and that for recA was 78.1 to 100%. Occasionally, multiple deep-subsurface strains were found to have identical 16S rRNA gene sequences and identical recA gene sequences. However, these strains were not always isolates from the same sediment sample. For example, ZAT004 and ZAT013 (from the upper lacustrine facies, sediment sample G1) were found to have identical sequences for both genes, but their sequence identity is shared by ZAT054 (from the lower paleosol unit, sediment sample G37). Similarly, ZAT055 (from the lower paleosol unit, sediment sample G37) was found to have sequences identical to those from G984 and G993 (both from the fluvial gravel unit, sediment sample G33). Therefore, the fact that isolates exhibiting identical sequences may originate from different sediment samples indicates that the identities cannot simply be the result of isolating essentially identical organisms (originating from the breakup of microcolonies during homogenization of the sediment sample or from siblings that may have multiplied during the short-term storage of some sediment samples). The majority of nucleotide differences in recA sequences between deep-subsurface strains were synonymous substitutions: while there were 124 variable sites in the 360 nucleotide positions analyzed, there were only 17 variable sites in the 120 amino acid residues. Preliminary trees generated using the RecA protein sequences showed that there was not enough sequence variation in the amino acid sequence to be phylogenetically useful, and therefore the nucleic acid sequences of recA were used in the phylogenetic analyses.
Phylogenetic analyses.
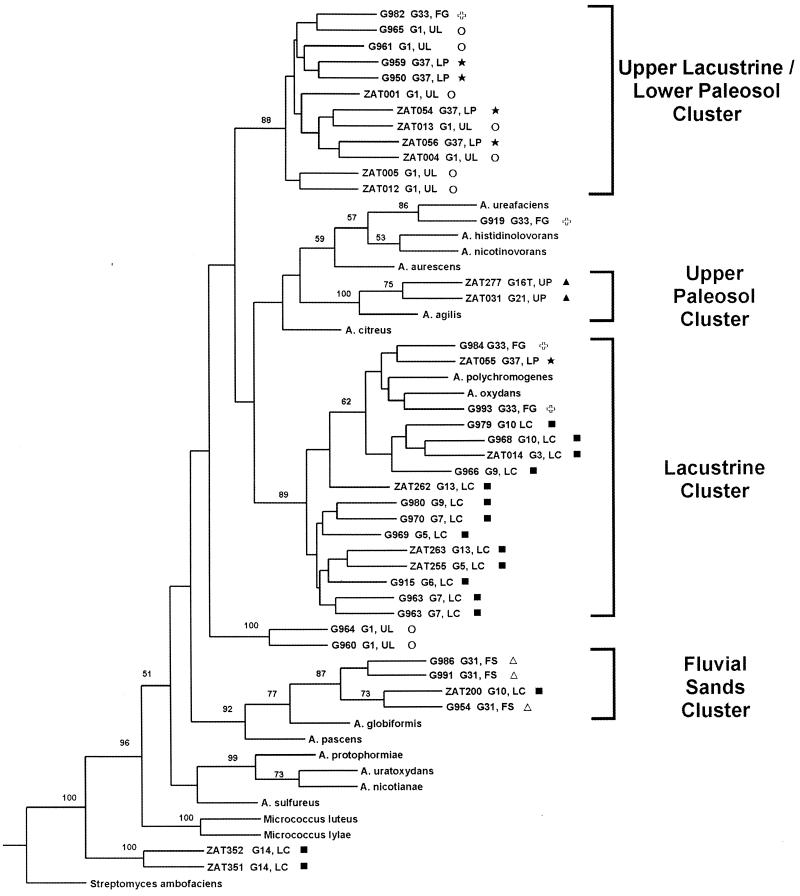
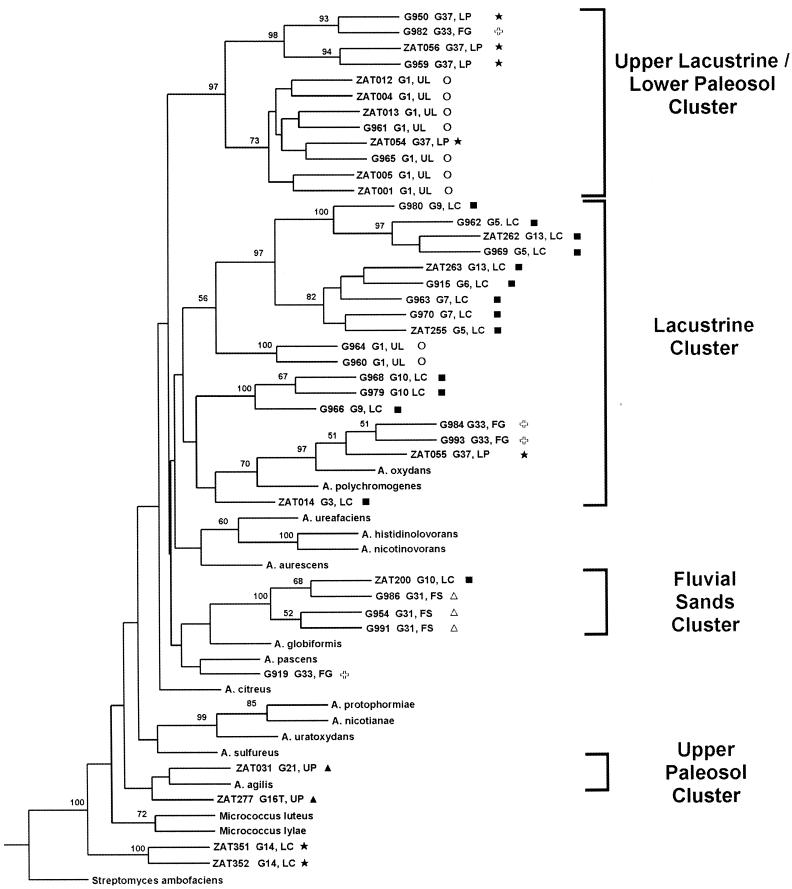
Phylogenetic trees based on 16S rRNA and recA gene sequences were generated by maximum-parsimony analyses using S. ambofaciens as an outgroup to root the trees (Fig. 2 and 3). Bootstrap analyses were performed, and bootstrap values of 50% or greater are shown at the appropriate nodes. Distance matrix analyses gave trees with topologies very similar to those obtained by the parsimony method for the two genes. The recA gene trees give relationships in general agreement with those generated by 16S rRNA analysis. This suggests that true phylogenetic relationships are being observed and that the relationships have not been obscured by any horizontal gene transfer that may have taken place. (Moreover, recent horizontal gene transfer within these deep-subsurface sediments is unlikely given the low cell numbers present and low permeability of sediments.) The differences that do exist between the 16S rRNA and recA gene trees are in the deeper (more ancestral) nodes of the trees. These deeper nodes in the recA gene trees have bootstrap values of less than 50%, which indicates that they are not statistically significant. There is more variability within the recA nucleotide sequences of the group than there is within the 16S rRNA gene sequences. (For instance, for our data sets the average percent sequence identities within clusters and between clusters based on 16S rRNA sequence analysis are 99.2 to 99.9% and 96.9 to 98.1%, respectively; the values based on recA sequence analysis are 89.4 to 100% and 85.6 to 89.1%, respectively.) This leads to the lower bootstrap values at the deeper nodes but higher resolution at the shallower nodes, as seen by the high bootstrap values on these nodes. Thus, within recA sequences there has been so much change between distantly related groups (between clusters) that these deeper relationships cannot be properly distinguished using recA, but they can be distinguished using the more conserved 16S rRNA sequences. However, the less distant relationships (within clusters) can be better resolved with recA than with 16S rRNA.
FIG. 2.
Phylogenetic tree based on parsimony analysis of 16S rRNA gene sequences for deep-subsurface and type strains of Arthrobacter species. For the deep-subsurface strains, the soil sample identifier and stratum from which they originate are indicated. FG and , fluvial gravel; FS and ▵, fluvial sand; LC and ■, lacustrine; LP and ★, lower paleosol; UL and ○, upper lacustrine; UP and ▴, upper paleosol. The tree is rooted using S. ambofaciens as an outgroup. Numbers at the nodes indicate the percentages of occurrence in 100 bootstrapped trees; only values that are 50% or greater are shown.
FIG. 3.
Phylogenetic tree based on parsimony analysis of recA gene sequences for deep-subsurface and type strains of Arthrobacter species. For the deep-subsurface strains, the soil sample identifier and stratum from which they originate are indicated. Abbreviations and symbols are as in Fig. 2. The tree is rooted using S. ambofaciens as an outgroup. Numbers at the nodes indicate the percentages of occurrence in 100 bootstrapped trees; only values that are 50% or greater are shown.
Previous phylogenetic analyses of Arthrobacter type strain species and related groups based on 16S rRNA have begun to clarify the relationships within this group and the relationships between this group and the closely related Micrococcus genus (25, 26, 43). Our analyses based on the 16S rRNA and recA gene sequences for the type strains are largely in accordance with these studies. Koch et al. (26), basing their phylogenetic analysis on 16S rRNA sequences, found that Arthrobacter species cluster within two groups that roughly match groups identified as having similar cell wall characteristics (as described in reference 22). Group II (A. sulfureus, A. nicotianae, A. protophormiae, and A. uratoxydans) and a subgroup of group I (A. histidinolovorans, A. nicotinovorans, A. ureafaciens, and A. aurescens) are preserved in both our 16S rRNA and recA trees. Also, as is seen in the trees of Koch et al. (26), our trees indicate that Arthrobacter (formerly Micrococcus) algis is included within the Arthrobacter radiation.
The relationships among the deep-subsurface strains were examined for trends relative to the structure of their environment. The strains fall into several monophyletic groups. Based on the distribution relative to type strains, these clusters may represent different species or slightly higher-order groups. For many of the strata, the majority of isolates from the same lithofacies were found to group within the same monophyletic cluster. This is true for the uppermost portion of the lacustrine unit (which we designate the upper lacustrine), as well as the lacustrine proper, the upper paleosol, the fluvial sands, and the lower paleosol. This trend is seen best in the 16S rRNA-based tree, and we have named the clades (in both the 16S rRNA and recA trees) based on the stratum of origin of the majority of isolates in each cluster of the 16S rRNA tree (Fig. 2 and 3).
The groups seen by 16S rRNA analysis are largely supported by analysis based on recA (compare Fig. 2 and 3). It should be noted, however, that in the recA tree the deepest node linking the lacustrine cluster is not supported by a high bootstrap value (Fig. 3). Moreover, in the tree based on distance matrix analysis of recA, the lacustrine cluster is divided in two, with some of the isolates being monophyletic with the upper lacustrine-lower paleosol clade and the others clustering with the fluvial sand clade (data not shown). However, since the node linking these two groups in the recA tree (Fig. 3) is not supported by a bootstrap value of greater than 50%, this branch point is not statistically significant. As discussed above, the recA trees can resolve relationships within clusters better than 16S rRNA trees can; thus, the upper lacustrine-lower paleosol clade, which appears as a single cluster on the 16S rRNA tree (Fig. 2), can be resolved into separate upper lacustrine and lower paleosol groups by the recA tree (Fig. 3).
The monophyletic clusters indicate that many of the individual stratigraphic units have a similar population throughout, and these populations are distinct from those found in adjacent units. All of the units whose isolates form clusters (i.e., upper lacustrine, lacustrine, paleosol, and fluvial sand) exhibit low permeability, indicating that they may be somewhat isolated strata, with the low permeability inhibiting migration of bacteria and nutrients into and out of these layers. The same low permeability likely inhibits the migration within the units as well. Thus, the most likely way to have the same genotype present throughout the entire interval (e.g., members of the lacustrine cluster were isolated from samples that span a 9-m interval) is to have persistence of a single genotype that was present throughout the layer when the sediment was more porous (i.e., during or soon after burial). This genotype would be selected for later due to the environmental conditions that became common throughout the interval following burial and compaction of the sediments. Thus, that genotype would become the major fraction of the present population. If the conditions are significantly different in a certain part of that layer, as they are in the upper lacustrine versus the rest of the lacustrine, then those conditions might select for a different dominant type. It is likely that the stratigraphic units became isolated shortly after burial and that the populations sampled have been resident in the sediments since the time of their deposition (6 million to 8 million years ago). However, it is not possible at present to tell precisely how long these populations have been dominant within their respective layers or to tell from where they originated or where they may have migrated.
The handful of isolates available from the fluvial gravel unit did not cluster together. The fluvial gravels tend to be more permeable than other layers within the Hanford site (see hydraulic conductivities in Table 1), suggesting that the increased permeability allows for changes in environmental conditions, which preclude selection for a particular genotype. Alternatively, this permeability may allow transport of species into and out of the cross section of the stratum that was sampled by the YBB.
As mentioned above, the majority of isolates from the upper lacustrine unit form a group separate from the rest of the lacustrine isolates. This sedimentary interval has a distinct oxidized, low-organic-matter character that makes it very different from the rest of the lacustrine interval. Thus, although the upper lacustrine unit has the same physical properties as the rest of the lacustrine interval, it has unique chemical properties that were caused by contact with the highly permeable gravels above (the groundwater from which oxidized the upper lacustrine sediments). The low permeability of the entire lacustrine interval likely limits the mobility of nutrients and bacteria within the layer. The fact that the two units have distinct populations may be caused by the difference in the chemical makeup of the upper lacustrine, which, if it has existed for a significant period of time, may have selected for a different member of the original lacustrine Arthrobacter population than the rest of the lacustrine. Alternatively, the difference in populations may be the result of migration of a different genotype into the upper layer that has been able to succeed in the different environment of that layer. That migration could have been aided by the same conditions (e.g., water flow) that caused this environment to become oxidized.
The two upper paleosol isolates are similar even though one was isolated from the seemingly very different environment of the thin tephra layer at the top of the upper paleosol unit. Perhaps this thin tephra interval has been influenced, chemically and/or microbiologically, by the layer below. This hypothesis is supported by the high sand content of this layer, which indicates that at some point it was much more permeable than the units above and below.
We found no particular physical or chemical characteristic common to the environments of isolates within any of the phylogenetic clusters. In addition, isolates from the two paleosols did not cluster together, indicating that facies type is not necessarily selecting for a particular genotype but rather that in different layers, even though they may be similar depositional environments, different genotypes were able to succeed. There is a monophylogenetic relationship for isolates from the upper lacustrine and lower paleosol intervals. These represent very different depositional environments. These strata may both contain low levels of organic matter: TOC is very low in the upper lacustrine (Table 1), and although no data are available for the lower paleosol, paleosols can be expected to have a low organic matter content (40). However, it may be that the phylogenetic relationship is not due to any single common physiochemical characteristic but instead is due to a complex interaction of a variety of selective forces that independently led to the success of a similar genotype in the different facies.
Although this study was limited to the use of culturable strains, we do not believe that the structure of the populations observed is an artifact of the use of isolates, since there was no difference in the way the isolates across the stratigraphic units were isolated. This study paves the way for future studies in which culturability is not a limitation. Because there have been recent advances in the ability to PCR amplify DNA directly from sediments, we suggest designing primers specific for the Arthrobacter group to PCR amplify and either sequence or perform restriction fragment length polymorphism analyses on DNA directly from the sediment samples. Analyses of the 16S rRNA gene alone could be used for this purpose, since it appears from this study to be a good molecule to sample the relationships across the entire group, and recA showed similar trends. However, use of the recA gene would provide a finer picture of the relationships among closely related strains.
Many studies of the population structure of pathogenic or symbiotic bacteria have been conducted, but relatively few such studies exist for free-living bacteria (examples of such studies include those reported in references 11, 12, 32, 41, and 48 to 50). For this subsurface site we have found that, similar to the case for the marine SAR11 cluster (12), the Arthrobacter populations of the YBB predominantly showed depth-specific (in this case, stratum-specific) distribution. The structure of the Arthrobacter populations from the majority of strata in this study is also similar to that observed for the B. cepacia soil populations analyzed by McArthur et al. (32). As in that study, we observed low genetic diversity (the formation of phylogenetic clusters) within areas of low environmental variability (within uniform, isolated strata). However, the structure of the fluvial gravel population appears to be more similar to that observed for the well-mixed stream environment in the study of B. cepacia stream populations (50), where greater environmental variability precludes selection for any particular genotype. Studies such as this on how local population structure is shaped by environmental factors may help us to understand, on a more global scale, the forces which direct microbial diversity of free-living bacteria and determine whether different bacteria are cosmopolitan in their distribution or are endemic to a particular geographic site (44, 45).
Conclusions.
We have sampled the diversity within Arthrobacter populations in a diverse assemblage of deep-subsurface strata. The genetic structure of the populations at the YBB appears to be predominantly created and controlled by the degree of physical isolation and physical-chemical homogeneity of individual sedimentary lithofacies. For the majority of facies, a single population, most likely one that was present at the time of sediment deposition or one that migrated into the layer after deposition and was most suited to survival in that particular sediment type, has persisted and become the dominant culturable Arthrobacter type. It will be interesting to see if similar relationships between the structures of the environment and resident microbial populations hold true for other genera within the YBB or for other deep-subsurface sites and how this population structure may be altered by human activities in the subsurface. An understanding of the structure of deep-subsurface bacterial populations and how this is related to the structure of the environment would help direct deep-subsurface bioremediation efforts, as well as help direct the search within the deep subsurface for microbes that possess unique, not previously documented, characteristics for biotechnology applications.
ACKNOWLEDGMENTS
We thank Brendan Bohannan for helpful discussions in the preparation of the manuscript.
This research was supported by the Deep Microbiology Subprogram of the Subsurface Science Program, Office of Energy Research, U.S. Department of Energy, under grants DE-FG02-93ER61680 (to R.V.M.) and DE-FG02-96ER62210 and DE-FG05-91ER61159 (to D.L.B.).
REFERENCES
- 1.Amy P S, Haldeman D L, Ringelberg D, Hall D H, Russell C. Comparison of identification systems for classification of bacteria isolated from water and endolithic habitats within the deep subsurface. Appl Environ Microbiol. 1992;58:3367–3373. doi: 10.1128/aem.58.10.3367-3373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balkwill D L. Numbers, diversity, and morphological characteristics of aerobic, chemoheterotrophic bacteria in deep subsurface sediments from a site in South Carolina. Geomicrobiol J. 1989;7:33–52. [Google Scholar]
- 3.Balkwill D L, Fredrickson J K, Thomas J M. Vertical and horizontal variations in the physiological diversity of the aerobic chemoheterotrophic bacterial microflora in deep southeast coastal plain subsurface sediments. Appl Environ Microbiol. 1989;55:1058–1065. doi: 10.1128/aem.55.5.1058-1065.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balkwill D L, Reeves R H, Drake G R, Reeves J Y, Crocker F H, King M B, Boone D R. Phylogenetic characterization of bacteria in the subsurface microbial culture collection. FEMS Microbiol Rev. 1997;20:201–216. doi: 10.1111/j.1574-6976.1997.tb00309.x. [DOI] [PubMed] [Google Scholar]
- 5.Boivin-Jahns V, Ruimy R, Bianchi A, Daumas S, Christen R. Bacterial diversity in a deep-subsurface clay environment. Appl Environ Microbiol. 1996;62:3405–3412. doi: 10.1128/aem.62.9.3405-3412.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandler D P, Brockman F J, Bailey T J, Fredrickson J K. Phylogenetic diversity of archaea and bacteria in a deep subsurface paleosol. Microb Ecol. 1998;36:37–50. doi: 10.1007/s002489900091. [DOI] [PubMed] [Google Scholar]
- 7.Department of Energy. Consultation draft, site characterization plan, reference repository location, Hanford Site, Washington. DOE/RW-0164. Vol. 1. Washington, D.C.: U.S. Department of Energy; 1988. [Google Scholar]
- 8.Duwat P, Ehrlich S D, Gruss A. A general method for cloning recAgenes of gram-positive bacteria by polymerase chain reaction. J Bacteriol. 1992;174:5171–5175. doi: 10.1128/jb.174.15.5171-5175.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisen J A. The RecA protein as a model molecule for molecular systematic studies of bacteria: comparison of trees of RecAs and 16S rRNAs from the same species. J Mol Evol. 1995;41:1105–1123. doi: 10.1007/BF00173192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felsenstein J. PHYLIP-phylogeny interference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 11.Ferris M J, Palenik B. Niche adaptation in ocean cyanobacteria. Nature. 1998;396:226–240. [Google Scholar]
- 12.Field K G, Gordon D, Wright T, Rappé M, Urbach E, Vergin K, Giovannoni S J. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl Environ Microbiol. 1997;63:63–70. doi: 10.1128/aem.63.1.63-70.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox G E, Stackebrandt E, Hespell R B, Gibson J, Maniloff J, Dyer T A, Wolfe R S, Balch W E, Tanner R S, Mgrum L J, Zablen L B, Blakemore R, Gupta R, Bonen L, Lewis B J, Sahl D A, Leuhrsen K H, Chen K N, Woese C R. The phylogeny of the prokaryotes. Science. 1980;209:457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- 14.Fredrickson J K, Garland T R, Hicks R J, Thomas J M, Li S W, McFadden K M. Lithotrophic and heterotrophic bacteria in deep subsurface sediments and their relation to sediment properties. Geomicrobiol J. 1989;7:53–66. [Google Scholar]
- 15.Fredrickson J K, Li S W, Brockman F J, Haldeman D L, Amy P S, Balkwill D L. Time-dependent changes in viable numbers and activities of aerobic heterotrophic bacteria in subsurface samples. J Microbiol Methods. 1995;21:253–265. [Google Scholar]
- 16.Fredrickson J K, McKinley J P, Nierzwicki-Bauer S A, White D C, Ringelberg D B, Rawson S A, Li S-M, Brockman F J, Bjornstad B N. Microbial community structure and biogeochemistry of Miocene subsurface sediments: implications for long-term microbial survival. Mol Ecol. 1995;4:619–626. [Google Scholar]
- 17.Ghiorse W C. Special issue on deep subsurface microbiology. Geomicrobiol J. 1989;7:1–136. [Google Scholar]
- 18.Ghiorse W C, Wilson J T. Microbial ecology of the terrestrial subsurface. Adv Appl Microbiol. 1988;33:107–172. doi: 10.1016/s0065-2164(08)70206-5. [DOI] [PubMed] [Google Scholar]
- 19.Higgins D G, Bleasby A J, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. CABIOS. 1991;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 20.Jain R, Rivera M C, Lake J A. Horizontal gene transfer among genomes: the complexity hypothesis. Proc Natl Acad Sci USA. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karlin S, Weinstock G M, Brendel V. Bacterial classifications derived from RecA protein sequence comparisons. J Bacteriol. 1995;177:6881–6893. doi: 10.1128/jb.177.23.6881-6893.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keddie R M, Collins M D, Jones D. Genus Arthrobacter Conn and Dimmick 1947, 300AL. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 2. Baltimore, Md: Williams and Wilkins; 1986. pp. 1228–1301. [Google Scholar]
- 23.Kieft T L, Amy P S, Brockman F J, Fredrickson J K, Bjornstad B N, Rosacker L L. Microbial abundance and activities in relation to water potential in the vadose zones of arid and semiarid sites. Microb Ecol. 1993;26:59–78. doi: 10.1007/BF00166030. [DOI] [PubMed] [Google Scholar]
- 24.Kieft T L, Fredrickson J K, McKinley J P, Bjornstad B N, Rawson S A, Phelps T J, Brockman F J, Pfiffner S M. Microbiological comparisons within and across contiguous lacustrine, paleosol, and fluvial subsurface sediments. Appl Environ Microbiol. 1995;61:749–757. doi: 10.1128/aem.61.2.749-757.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koch C, Rainey F A, Stackebrandt E. 16S rDNA studies on members of Arthrobacter and Micrococcus: an aid for their future taxonomic restructuring. FEMS Microbiol Lett. 1994;123:167–172. [Google Scholar]
- 26.Koch C, Schumann P, Stackebrandt E. Reclassification of Micrococcus agilis (Ali-Chohen 1889) to genus Arthrobacter agilis comb. nov. and emendation of the genus Arthrobacter. Int J Syst Bacteriol. 1995;45:837–839. doi: 10.1099/00207713-45-4-837. [DOI] [PubMed] [Google Scholar]
- 27.Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetics analysis, version 1.01. University Park: The Pennsylvania State University; 1993. [Google Scholar]
- 28.Lloyd A T, Sharp P M. Evolution of the recAgene and the molecular phylogeny of bacteria. J Mol Evol. 1993;37:399–407. doi: 10.1007/BF00178869. [DOI] [PubMed] [Google Scholar]
- 29.Maidak B L, Cole J R, Parker J C T, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maynard Smith J. The population genetics of bacteria. Proc R Soc Lond B Biol Sci. 1991;245:37–41. [Google Scholar]
- 31.Maynard Smith J, Smith N H, O'Rourke M, Spratt B G. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McArthur J V, Kovacic D A, Smith M H. Genetic diversity in natural populations of a soil bacterium across a landscape gradient. Proc Natl Acad Sci USA. 1988;85:9621–9624. doi: 10.1073/pnas.85.24.9621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKinley J P, Stevens T O, Fredrickson J K, Zachara J M, Colwell F S, Wagnon K B, Smith S C, Rawson S A, Bjornstad B N. Biogeochemistry of anaerobic lacustrine and paleosol sediments within an aerobic unconfined aquifer. Geomicrobiol J. 1997;14:23–39. [Google Scholar]
- 34.Nowak A, Kur J. Genomic species typing of acinetobacters by polymerase chain reaction amplification of the recAgene. FEMS Microbiol Lett. 1995;130:327–332. doi: 10.1111/j.1574-6968.1995.tb07739.x. [DOI] [PubMed] [Google Scholar]
- 35.Ogram A, Sun W, Brockman F J, Fredrickson J K. Isolation and characterization of RNA from low-biomass deep-subsurface sediments. Appl Environ Microbiol. 1995;61:763–768. doi: 10.1128/aem.61.2.763-768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olsen G J. Phylogenetic analysis using ribosomal RNA. Methods Enzymol. 1988;164:793–812. doi: 10.1016/s0076-6879(88)64084-5. [DOI] [PubMed] [Google Scholar]
- 37.Olsen G J, Woese C R, Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peloquin L, Greer C W. Cloning and expression of the polychlorinated biphenyl-degradation gene cluster from Arthrobacter M5 and comparison to analogous genes from gram negative bacteria. Gene. 1993;125:35–40. doi: 10.1016/0378-1119(93)90742-l. [DOI] [PubMed] [Google Scholar]
- 39.Phelps T. Special issue: deep subsurface microbiology. J Microbiol Methods. 1995;21:225–328. [Google Scholar]
- 40.Retallack G J. Soils of the past. Boston, Mass: Unwin Hyman; 1990. [Google Scholar]
- 41.Simidu U, Tsukamoto K. Habitat segregation and biochemical activities of marine members of the family Vibrionaceae. Appl Environ Microbiol. 1985;50:781–790. doi: 10.1128/aem.50.4.781-790.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinclair J L, Ghiorse W C. Distribution of aerobic bacteria, protozoa, algae, and fungi in deep subsurface sediments. Geomicrobiol J. 1989;7:15–31. [Google Scholar]
- 43.Stackebrandt E, Koch C, Gvozdiak O, Schumann P. Taxonomic dissection of the genus Micrococcus: Kocuria gen. nov., Nesterenkonia gen. nov., Kytococcus gen. nov., Dermacoccus gen. nov., and MicrococcusCohn 1872 gen. emend. Int J Syst Bacteriol. 1995;45:682–692. doi: 10.1099/00207713-45-4-682. [DOI] [PubMed] [Google Scholar]
- 44.Stanley J T. Bacterial biodiversity: a time for place. ASM News. 1999;65:681–687. [Google Scholar]
- 45.Stanley J T, Gosink J J. Poles apart: biodiversity and biogeography of sea ice bacteria. Annu Rev Microbiol. 1999;53:189–215. doi: 10.1146/annurev.micro.53.1.189. [DOI] [PubMed] [Google Scholar]
- 46.Stim K P. A phylogenetic analysis of micro-organisms isolated from subsurface environments. Mol Ecol. 1995;4:1–10. doi: 10.1111/j.1365-294x.1995.tb00186.x. [DOI] [PubMed] [Google Scholar]
- 47.Syvanen M. Horizontal gene transfer: evidence and possible consequences. Annu Rev Genet. 1994;28:237–261. doi: 10.1146/annurev.ge.28.120194.001321. [DOI] [PubMed] [Google Scholar]
- 48.Toledo G, Palenik B. Synechococcus diversity in the California Current as seen by RNA polymerase (rpoC1) gene sequences of isolated strains. Appl Environ Microbiol. 1997;63:4298–4303. doi: 10.1128/aem.63.11.4298-4303.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.West N J, Scanlan D J. Niche-partitioning of Prochlorococcuspopulations in a stratified water column in the eastern North Atlantic Ocean. Appl Environ Microbiol. 1999;65:2585–2591. doi: 10.1128/aem.65.6.2585-2591.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wise M G, Shimkets L J, McArthur J V. Genetic structure of a lotic population of Burkholderia (Pseudomonas) cepacia. Appl Environ Microbiol. 1995;61:1791–1798. doi: 10.1128/aem.61.5.1791-1798.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou J, Spratt B G. Sequence diversity within the argF, fbp and recA genes of natural isolates of Neisseria meningitidis: interspecies recombination within the argFgene. Mol Microbiol. 1992;6:2135–2146. doi: 10.1111/j.1365-2958.1992.tb01387.x. [DOI] [PubMed] [Google Scholar]