ABSTRACT
Monkeypox virus (MPXV), a zoonotic virus endemic to the African continent, has been reported in 33 non-endemic countries since May 2022. We report an almost complete genome of the first confirmed case of MPXV in Brazil. Shotgun metagenomic sequencing was completed in 18 hours, from DNA extraction to consensus sequence generation.
Keywords: Viral metagenomics, Monkeypox virus, West African clade, Genomic surveillance, Brazil
INTRODUCTION
Monkeypox virus (MPXV) is a double-stranded DNA zoonotic virus with a 197-kb genome, member of the Orthopoxvirus (OPV) genus and Poxviridae family, which also includes smallpox virus that causes smallpox 1 . MPXV was first identified in a 9-month-old boy in 1970 in the Democratic Republic of Congo. Since then, several outbreaks of monkeypox have been reported in the African continent, where 1,408 suspected cases and 66 deaths were reported in 2022 alone 1,2 . MPXV is currently classified into two lineages, the West and Central African clades, although a novel classification into clades 1, 2 and 3 has recently been proposed 3 . The MPXV Clade 3 includes most human outbreaks from 2017, 2018 and 2022, and can be further divided into lineages A, A.1, A.1.1, and B.1 3 . In early May 2022, cases of MPXV were detected in the UK and Portugal, most of them with no known travel history to endemic countries. As of June 9, 2022, 1,240 cases have been confirmed in 33 countries on all continents 4 , most of them in European countries and the United States 5 . Available epidemiological and contact tracing data revealed that most cases are associated with men who have sex with men (MSM) 3 . The first two MPXV cases from South America were reported from Argentina on May 27, 2022 6 . Both reported cases traveled to Spain 2 and 9 days prior to case notification, and the two MPXV partial genomes (942 bp) were made available as the first monkeypox sequences from Latin America. As of June, 8, 2022, Brazil had 8 suspected monkeypox infections in the States of Santa Catarina, Ceara, Mato Grosso do Sul, Rio Grande do Sul, Rondonia and Sao Paulo 7 .
Here we report an almost complete genome of the first confirmed case of monkeypox detected in Brazil. A skin swab of the lesions (vesicle and crust) was collected on June 7, 2022, at the Emilio Ribas Institute of Infectious Diseases from a 41-year-old male patient with a recent travel history to Portugal and Spain, and clinical manifestation suggestive of monkeypox signs and symptoms.
Ethical aspects
The authors applied the free and informed consent form that was signed by the patient, authorizing the collection of the biological sample and its use in the sequencing of the complete genome of the virus. In addition, the patient also consented to the sequencing results being released to the scientific community.
MATERIALS AND METHODS
Viral DNA was isolated from 200 μL of the recovered material using the QIAamp Viral DNA Mini Kit (Cat Nº 51304, Qiagen, Germany) according to the manufacturer’s instructions and eluting in 60 μL of elution buffer. DNA was quantified using fluorimetry with the Qubit dsDNA High Sensitivity Assay (Cat Nº Q32854, Life Technologies, Waltham, USA) on the Qubit 3.0 instrument (Life Technologies, USA). Shotgun metagenomics was performed using 10 ng of the extracted DNA and the Rapid PCR Barcoding kit (SQK-RPB004) - Oxford Nanopore Technologies (ONT, UK), adapted from the Rapid SMART-9N protocol 8 . PCR products were then purified using a 1:1 ratio of AMPure XP beads (Cat Nº A63881, Beckman Coulter, UK) and quantified.
MinION libraries were prepared using an input of 50 ng per sample, pooled in an equimolar way followed by the rapid adapter ligation. The final libraries were loaded onto FLO-MIN106 flow cells on the MinION device (ONT, UK) and sequenced using MinKNOW 1.15.1 with the standard 48-hour run script. FASTQ files were demultiplexed and trimmed using Guppy V5.0.16 1 (ONT, UK), and the barcoded FASTQ files were aligned and mapped to the reference genome (GenBank accession Nº MN648051) using minimap2 version 2.28.0 9 and converted to a sorted BAM file using SAMtools 10 . NanoStat version 1.1.2 1 10,11 was used to compute the number of raw reads and minimum contig length to cover 50 percent of the genome (N50) of the aligned reads. Tablet 1.19.05.2827 was used for genome visualization and to compute the number of mapped reads, percentage of genome coverage, and coverage depth 12 . Variants were detected with medaka_variants and the consensus sequence was built using medaka_consensus (ONT, UK). Genome regions with <20x coverage were masked.
The dataset was aligned to the reference genome using MAFFT version 7·453 13 . Maximum likelihood analyses of almost complete and complete genome sequences were conducted using IqTree2 14 under a Jukes Cantor nucleotide substitution model.
RESULTS
Our sequencing run generated a total of 954,284 reads, with 784,000 reads with quality score > 8. The average depth was 277.7x, covering 100% of the viral genome with at least 1 read and an N50 of 4,493. Complete MPXV genome sequence has been submitted to NCBI GenBank accession number ON751962, and raw data can also be found in our project’s dedicated GitHub repository 15 . To contextualize the novel monkeypox genome, we downloaded 102 whole-genome sequences from NCBI GenBank (Supplementary Table S1). Of these, 81 were genomes from the ongoing MPXV B.1 multi-country outbreak.
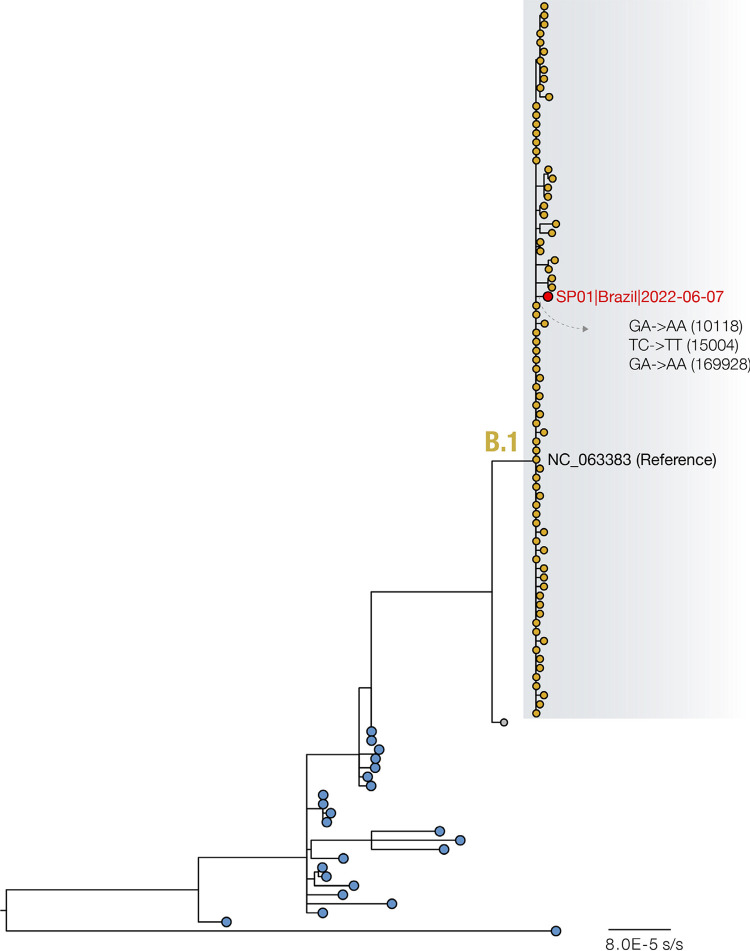
The complete genome sequences revealed that this first case of MPXV in Brazil clustered within the newly proposed B.1 lineage, and was closely related to sequences from Portugal, Germany, USA and Spain (Figure 1). We identified 3 unique SNPs compared to the updated US-CDC genome (Accession Nº ON563414.3): GA->AA (10118; non-synonymous), TC->TT (15004; synonymous) and GA->AA (169928; non-synonymous). The G->A and C->T mutations such as those observed in the new Brazilian MPXV genome, may be an effect of APOBEC3 deaminase editing, as previously suggested 16 .
Figure 1. Maximum likelihood phylogeny with 103 whole genome MPXV sequences, including 102 available in NCBI GenBank up to June 9, 2022 (see Supplementary Table S1 for the Accession Numbers used in this study). Highlighted genomes (yellow tips), including the SP01 described in this study (red tip), belong to the newly proposed B.1 lineage3. An interactive visualization of the phylogeny presented here can be found at Argimón et al.17 .
We then evaluated whether the recommended diagnostic RT-qPCR primers were suitable for the identification of the newly sequenced virus genome 2 . Investigating putative mismatches in the binding sites of primers and probes for both the generic MPXV and the specific B.1 MPXV detection sets 2,16 , we found two mutations in the binding regions for forward (T2676C) and reverse (G2608A) primers of the generic set. These mutations are located six and eight nucleotides away from the 3’OH end of the forward and reverse primers, respectively, and are unlikely to affect the sensitivity of this primer set, indicating that existing primers are capable of detecting infections from the current outbreak.
DISCUSSION
Here we describe the genome of the first monkeypox cases in Brazil. Our study contributes to the rapid genomic surveillance of MPXV, which is imperative to contextualize and better understand the epidemiology, transmission patterns, evolution of the virus, its adaptation to human transmission, and continuous evaluation of laboratory and clinical diagnostic methods.
CONCLUSION
The almost complete genome sequencing of this first Brazilian case of MPXV was crucial to confirm the diagnosis by means of a fast and reliable sequencing technique, to ensure the molecular diagnosis of future cases by PCR and characterize the newly proposed B.1 lineage 3 .
ACKNOWLEDGMENTS
This study was supported by the Medical Research Council-São Paulo Research Foundation (FAPESP) CADDE partnership award (MR/S0195/1 and FAPESP18/143890). We acknowledge support from the Wellcome Trust and Royal Society (Sir Henry Dale Fellowship 204311/Z/16/Z), Bill & Melinda Gates Foundation (INV-034540 and INV-034652), the Wellcome ARTIC network (206298), the Rede Corona-ômica BR MCTI/FINEP affiliated to RedeVírus/MCTI (FINEP 01.20.0029.000462/20, CNPq 404096/2020-4) and CNPq/MCTI funding 402794/2020-6.
Footnotes
Supplementary Material available from: https://doi.org/10.48331/scielodata.KOUBE9
REFERENCES
- 1.Bunge EM, Hoet B, Chen L, Lienert F, Weidenthaler H, Baer LR, et al. The changing epidemiology of human monkeypox: a potential threat? A systematic review. PLoS Negl Trop Dis. 2022;16:e0010141. doi: 10.1371/journal.pntd.0010141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Organização Pan-Americana da Saúde [cited 2022 Jun 20];Diretrizes laboratoriais para detecção e diagnóstico da infecção pelo vírus da varíola do macaco. https://www.paho.org/pt/documentos/diretrizes-laboratoriais-para-deteccao-e-diagnostico-da-infeccao-pelo-virus-da-variola
- 3.Happi C, Adetifa I, Mbala P, Njouom R, Nakoune E, Happi A, et al. Urgent need for a non-discriminatory and non-stigmatizing nomenclature for monkeypox virus. [cited 2022 Jun 20];Virological.org. doi: 10.1371/journal.pbio.3001769. https://virological.org/t/urgent-need-for-a-non-discriminatory-and-non-stigmatizing-nomenclature-for-monkeypox-virus/853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kraemer MU, Tegally H, Pigott DM, Dasgupta A, Sheldon J, Wilkinson E, et al. Tracking the 2022 monkeypox outbreak with epidemiological data in real-time. Lancet Infect Dis. 2022 doi: 10.1016/S1473-3099(22)00359-0. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention monkeypox and orthopoxvirus outbreak global map. 2022. [cited 2022 Jun 20]. https://www.cdc.gov/poxvirus/monkeypox/response/2022/world-map.html
- 6.Argentina. Ministerio de Salud Salud informa que el resultado de la muestra PCR tomada por ANLIS Malbrán al primer caso sospechoso de viruela símica dio positivo. [cited 2022 Jun 20]. https://www.argentina.gob.ar/noticias/salud-informa-que-el-resultado-de-la-muestra-pcr-tomada-por-anlis-malbran-al-primer-caso
- 7.Brasil. Ministério da Saúde Informe diário sala de situação nacional de monkeypox: no 16, 07/06/22. [cited 2022 Jun 20]. https://www.gov.br/saude/pt-br/composicao/svs/resposta-a-emergencias/sala-de-situacao-de-saude/sala-de-situacao-de-monkeypox/atualizacao-dos-casos-no-brasil/card-diario-no-16-7-06-22/view
- 8.Claro IM, Ramundo MS, Coletti TM, Silva CA, Valenca IN, Candido DS, et al. Rapid viral metagenomics using SMART-9N amplification and nanopore sequencing. Wellcome Open Res. 2021;6:241. doi: 10.12688/wellcomeopenres.17170.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.GitHub Create statistic summary of an Oxford Nanopore read dataset. [cited 2022 Jun 20]. https://github.com/wdecoster/nanostat
- 12.Milne I, Stephen G, Bayer M, Cock PJ, Pritchard L, Cardle L, et al. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 2013;14:193–202. doi: 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
- 13.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogeneticiInference in the genomic era. Mol Biol Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.GitHub CADDE-CENTRE/Monkeypox. [cited 2022 Jun 20]. https://github.com/CADDE-CENTRE/Monkeypox
- 16.Virological.org Discussion of on-going MPXV genome sequencing. [cited 2022 Jun 20]. https://virological.org/t/discussion-of-on-going-mpxv-genome-sequencing/802
- 17.Argimón S, Abudahab K, Goater RJ, Fedosejev A, Bhai J, Glasner C, et al. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom. 2016;2:e000093. doi: 10.1099/mgen.0.000093. [DOI] [PMC free article] [PubMed] [Google Scholar]