Abstract
Skeletal malocclusions are common phenotypes in humans and have a strong influence on genetic factors. Transforming growth factor beta (TGFβ) controls numerous functions of the human body, including cell proliferation, differentiation, and migration. Thus, this study is aimed at evaluating whether genetic polymorphisms in TGFB1 and its receptor TGFBR2 are associated with mandibular retrognathism in German children and adolescents. Children and teenagers older than 8 years in the mixed or permanent dentition were included in this study. Patients with syndromes and facial trauma and patients with congenital alterations were excluded. Digital cephalometric tracings were performed using the anatomical landmarks point A, point B, sella (S), and nasion (N). Patients that have a retrognathic mandible (SNB < 78°) were selected as case group, and the patients with an orthognathic mandible (SNB = 78°– 82°) were selected as the control group. Genomic deoxyribonucleic acid (DNA) from saliva was used to evaluate four genetic polymorphisms in TGFB1 (rs1800469 and rs4803455) and TGBR2 (rs3087465 and rs764522) using real-time PCR. Chi-square or Fisher exact tests were used to compare gender, genotype, and allele distribution among groups. Genotype distribution was calculated in an additive and recessive model. Haplotype analysis was also performed. The established alpha of this study was 5%. A total of 146 patients (age ranging from 8 to 18 years) were included in this epidemiological genetic study. The genetic polymorphism rs3087465 in TGFBR2 was associated with mandibular retrognathism. Carrying the AA genotype in the rs3087465 polymorphism decreased the chance of having mandibular retrognathism (odds ratio = 0.25, confidence interval 95% = 0.06 to 0.94, p = 0.045). None of the haplotypes was associated with mandibular retrognathism (p > 0.05). In conclusion, we found that the genetic polymorphism rs3087465 in the promoter region of the TGFBR2 was associated with mandibular retrognathism in Germans.
1. Introduction
There are a wide variety of skeletal malocclusions (dentofacial deformities) in humans [1], and the frequency of each dentofacial deformity ranges according to the studied population/ethnicity [2]. One of the most common dental facial deformities is mandibular retrognathism [3], which is a facial alteration of the skeletal jaw-cranial base relationship. Retrognathism is characterized by a retruded position of the mandible as a result of an anomaly of the skeletal jaw-cranial base relationship [4]. This condition has a strong genetic background and some genes have been associated with mandibular retrognathism in humans from different populations in past years [3–6]. Some previous studies associated mandibular retrognathism with genetic polymorphisms in genes encoding Myosin IH (MYO1H) [5], Matrilin 1 (MATN1) [4], bone morphogenetic protein 2 (BMP2) [7], ADAM metallopeptidase with thrombospondin type 1 motif 9 (ADAMTS9) [6], and parathyroid hormone (PTH) and the vitamin-D-related genes: vitamin D receptor (VDR), cytochrome P450 family 24 subfamily A member 1 (CYP24A1), and cytochrome P450 family 27 subfamily B member 1 (CYP27B1) [8].
The transforming growth factor beta (TGFβ) family constitutes a group of three isoforms, TGFβ1, TGFβ2, and TGFβ3. Their structure is formed by interrelated dimeric polypeptide chains. Pleiotropic and redundant functions of the TGFβ family control several functions, such as cell proliferation, differentiation, and migration in all human tissues. The TGFβ family has numerous key roles in the bone tissue controlling physiological phenomena regarding maintenance of metabolic homeostasis [9]. TGFβ isoforms and their receptors, type I receptor (TGFβRI or ALK5) and type II receptor (TGFβRII or TGFBR2) play innumerous essential roles in endochondral and intramembranous ossification [10].
Several functional genetic polymorphisms were identified in TGFB1 (gene encoding TGFβ1) and TGFBR2 (gene encoding TGFβRII) and they were associated with higher serum or plasma level of TGFβ1 and enhanced transcriptional activity of TGFβRII [11]. It is possible that some of these functional genetic polymorphisms play a role in the establishment of maxillary and mandibular morphology leading to skeletal malocclusion phenotypes. Therefore, the present study evaluated if well-known functional genetic polymorphisms in TGFB1 and its receptor TGFBR2 are associated with mandibular retrognathism in Germans children and teenagers.
2. Materials and Methods
This study was approved by the Ethics Committee from the University of Regensburg (# 19-1549-101). Informed consent was obtained from all patients and/or their legal guardians (in the case of minors during the sample collection), and age-appropriate assent documents were also used for patients younger than 14 years. This project was made following the Helsinki Declaration. The Strengthening the Reporting of Genetic Association study (STREGA) statement checklist was followed to design the study and report the results. The chi-square test for sample size (power) calculation was performed by PASS 15 Power Analysis and Sample Size Software (NCSS, LLC. Kaysville, Utah, USA). Küchler et al.'s (2021) study was used to obtain the W effect size (0.225), with an alpha of 5% and power of 80% the test predicts 155 patients for this study.
This is a cross-sectional nested case-control study design. For this cross-sectional study, patients undergoing orthodontic treatment at private orthodontic practices in Regensburg-Germany and the University of Regensburg were screened. Children and teenagers of both sexes were recruited and they were consecutively included in this study from 2020 to 2021.
Patients with underling syndromes, adults (older than 18 years), patients with mandibular prognathism and congenital alterations including tooth agenesis (except for third molar agenesis), oral cleft patients, and patients with facial trauma were excluded. Only one patient per family was recruited. To minimize genetic and phenotypic variance and to maximize data interpretability, only patients with a Middle-European ancestry (at maximum one grandparent not from Middle Europe) were included. All patients included were children older than 8 years in the mixed or permanent dentition.
2.1. Retrognathic and Orthognathic Characterization
Digital lateral cephalograms from each patient's orthodontic record with the mandible in centric relationship were evaluated in this study.
The measurements were performed by two trained and calibrated examiners. Intraclass correlation coefficients (ICC) were used to calculate intra- and interexaminer reliability. Interexaminer reliability showed significant good agreement for both examiners (ICC, 0.98 - 0.95). Intraexaminer reliability also showed significant good agreement (ICC, 0.97 - 0.91).
Digital cephalometric were tracings performed using the software Ivoris® (Computer konkret AG, Falkenstein, Germany, version 8.2.15.110). The anatomical landmarks point A, point B, sella (S), and nasion (N) were determined manually using the cephalometric analysis software, and, subsequently, the angular measurements SNB and ANB were calculated (Figure 1).
Figure 1.
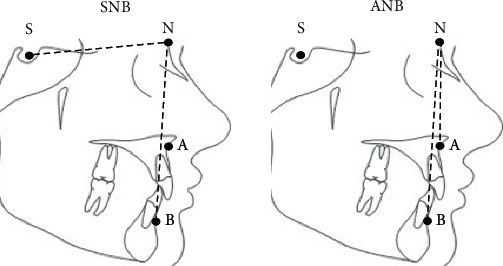
Lateral cephalometric landmarks and reference lines studied. Points: point A, point B, sella (S), and nasion (N).
Patients presenting a retrognathic mandible (SNB < 78°) were selected as a case group, while those presenting an orthognathic mandible (SNB = 78°– 82°) were selected as a control group. Patients having mandibular prognathism (SNB > 82°) were excluded.
2.2. Selection of Genetic Polymorphisms and Laboratorial Analysis
For the selection of the genetic polymorphisms, we searched the promoter, intronic, and coding genetic polymorphisms of the TGFB1 and TGFBR2 from the dbSNP database (http://www.ncbi.nlm.nih.gov/snp/). Only genetic polymorphisms with heterozygosity above 0.2 in the global population were considered. The classification of each genetic polymorphism as potentially functional (polymorphisms that can result in amino acid changes of the corresponding proteins or occurring in the promoter region of the gene and potentially affecting the expression of the gene or previously associated with other conditions and potentially clinically relevant) was also taken into consideration in the selection process. The characteristics and description of the genetic polymorphisms selected for this study are presented in Table 1.
Table 1.
Characteristics of the studied genetic polymorphisms.
| Gene | Polymorphism and base change | Comments |
|---|---|---|
|
TGFB1
Transforming growth factor beta 1 |
rs1800469 (T/C) | Polymorphism rs1800469 is located in the promoter region of the TGFB1 gene and has the function of regulating expression levels of protein TGFβ1 (affects gene transcriptional activity and serum levels of TGFβ1) [19, 20]. |
| rs4803455 (C/A) | The polymorphism rs4803455 is located in intron 2 of TGFB1 gene and was associated with a variety of different conditions (https://www.ncbi.nlm.nih.gov/snp/rs4803455). Moreover, it was suggested as a potential genetic marker for growth response to recombinant human growth hormone (r-hGH) treatment [38]. | |
|
| ||
|
TGFBR2
Transforming growth factor beta receptor 2 |
rs3087465(A/G) | Polymorphism rs3087465 is located in the promoter region of the gene and increases TGFβ type II receptor expression [26]. |
| rs764522 (G/C) | The polymorphism rs764522, which is located in 5 kb upstream in the promoter region of TGFBR2 increases TGFβ type II receptor expression [26]. | |
For the genotyping analysis, we used genomic DNA that was isolated from buccal epithelial cells collected using two cytobrushes placed in extraction solution (Tris-HCl 10 mmol/L, pH 7.8; EDTA 5 mmol/L; SDS 0.5%, 1 mL). Briefly, proteinase K (100 ng/mL) were added to each tube. Ammonium acetate was added to eliminate nondigested proteins, and the liquid was then centrifuged. DNA was precipitated with isopropanol, washed with ethanol. The DNA was quantified by spectrophotometry (Nanodrop 1000; Thermo Scientific, Wilmington, DE, USA) [12].
The selected genetic polymorphisms, which were described in Table 1, were blindly genotyped via real-time polymerase chain reaction (PCR) using the Mastercycler® ep realplex-S thermocycler (Eppendorf AG, Hamburg, Germany). The TaqMan technology was used. Initial denaturation at 95°C for 30 seconds, followed by 40 cycles of denaturation at 92°C for 5 seconds and annealing/extension at 60°C for 20 seconds. The 3.125 μL reaction volume contained 1.5 μL Master Mix, 0.125 μM TaqMan probe, and 4 ng DNA in 1.5 μM nuclease-free water. Assays and reagents were supplied by Applied Biosystems (Foster City, CA, USA). A negative control template was included in each reaction (each 96-well plate), in which the reaction mixture contains the reagents, but not the DNA. Additionally, 10% of the samples were randomly selected in order to repeat the analysis and showed 99% concordance.
Patients with not enough DNA or DNA samples that failed to be genotyped in the PCR analysis were excluded from the further analyses.
2.3. Statistical Analysis
Chi-squared test estimated the Hardy-Weinberg equilibrium (HWE) for each studied polymorphism (https://wpcalc.com/en/equilibrium-hardy-weinberg/).
Chi-squared or Fisher's exact tests compared gender, genotype, and allele distribution among groups and genotype distribution was calculated in an additive model and recessive model. Haplotype analysis was also performed. PLINK version 1.06 (https://zzz.bwh.harvard.edu/plink/ld.shtml) was used for the analysis with an established alpha of 5% (p < 0.05). The odds ratio and confidence interval 95% was calculated to investigate the chance of presenting mandibular retrognathism for the associated genetic polymorphism.
3. Results
A total of 168 patients were screened, two patients were excluded due to biological relations to included patients (siblings), one due to oral cleft, 13 due to age older than 18 years, and six patients due to mandibular prognathism. Finally, 146 patients (age ranging from 8 to 18 years) were included in this epidemiological genetic study. The characteristics of the included sample are presented in Table 2. Mean age in the mandibular retrognathism group was 11.56 years (standard deviation = 2.05), while the mean age in the mandibular retrognathism group was 12.66 years (standard deviation = 2.2). The SNB angle was significantly lower in the mandibular retrognathism group (ranging from 66.0° to 77.9°) than in the control group (ranging from 78.1° to 81.9°) (p = 0.0014).
Table 2.
Comparison of cephalometric variables between mandibular retrognathism and orthognathic mandible.
| Variables | Orthognathic mandible (n = 50) |
Mandibular retrognathism (n = 96) | p value |
|---|---|---|---|
| Gender,n(%) | |||
| Male | 29 (58.0%) | 45 (46.8%) | 0.202 |
| Female | 21 (42.0%) | 51 (53.2%) | |
| SNB (°) | |||
| Mean (SD) | 79.62 (SD 1.07) | 78.32 (SD 2.67) | 0.001∗ |
| ANB (°) | |||
| Mean (SD) | 3.48 (SD 2.75) | 4.26 (SD 2.27) | 0.068 |
Note: SD: standard deviation; ∗statistically significant difference (p < 0.05); n: number of individuals; %: percent; °: degrees.
All the genetic polymorphisms assessed were within the Hardy-Weinberg equilibrium (chi‐squareHWE = 0.452 for rs1800469, chi‐squareHWE = 0.309 for rs4803455, chi‐squareHWE = 3.17 for rs3087465, and chi‐squareHWE = 0.161 for rs764522). For the rs1800469 (A/G), rs4803455 (C/A), rs3087465 (A/G), and rs764522 (G/C) polymorphisms, success rates of genotyping were 95.9%, 90.4%, 95.9%, and 91.8%, respectively.
Table 3 shows the genotype and allele distributions and the association results in the allele distribution and genotype distribution in additive and recessive models. The only significantly associated polymorphism was rs3087465 in TGFBR2. Patients that carry the AA genotype in the polymorphism rs3087465 had significantly decreased the chance to have mandibular retrognathism (odds ratio = 0.25, confidence interval = 0.06 to 0.94, p = 0.045).
Table 3.
Genotype distribution among group and p values.
| Gene | rs | Genotype distribution and allele distributions, n (%) | p valueGenotype | p valueAllele | p valueRecessive | ||
|---|---|---|---|---|---|---|---|
| Genotype | Orthognathic mandible | Mandibular retrognathism | |||||
| TGFB1 | rs1800469 | TT | 5 (10.0) | 4 (4.4) | 0.310 | 0.525 | 0.199 |
| CT | 18 (36.0) | 41 (45.6) | |||||
| CC | 27 (54.0) | 45 (50.0) | |||||
| A | 28 (28.0) | 49 (27.2) | |||||
| G | 72 (72.0) | 131 (72.8) | |||||
| rs4803455 | CC | 10 (20.8) | 18 (21.4) | 0.740 | 0.692 | 0.478 | |
| CA | 27 (56.3) | 42 (50.0) | |||||
| AA | 11 (22.9) | 24 (28.6) | |||||
| C | 47 (48.0) | 78 (46.4) | |||||
| A | 49 (52.0) | 90 (57.3) | |||||
|
| |||||||
| TGFBR2 | rs3087465 | AA | 6 (12.0) | 3 (3.3) | 0.098 | 0.603 | 0.045∗ |
| AG | 21 (42.0) | 48 (53.3) | |||||
| GG | 23 (46.0) | 39 (43.3) | |||||
| A | 33 (33.0) | 54 (30.0) | |||||
| G | 67 (67.0) | 126 (70.0) | |||||
| rs764522 | GG | 0 (0.0) | 2 (2.3) | 0.575 | 0.525 | 0.302 | |
| GC | 11 (23.9) | 22 (25.0) | |||||
| CC | 35 (76.1) | 64 (72.7) | |||||
| G | 11 (12.0) | 26 (14.8) | |||||
| C | 81 (88.0) | 150 (85.2) | |||||
Note: ∗means statistically significant difference; n: number of individuals; %: percent; rs: the code of polymorphisms; TGFB1: transforming growth factor beta 1; TGFBR2: transforming growth factor beta receptor 2; C: cytosine; A: adenine; T: thymine; G: guanine.
The haplotype analysis for the polymorphisms in TGFB1 (rs4803455-rs1800469) and TGFBR2 (rs764522-rs3087465) is presented in Table 4. There was no statistically significant association (p > 0.05).
Table 4.
Haplotype analysis of the studied genetic polymorphisms.
| Gene | Haplotype | Frequency | p value | ||
|---|---|---|---|---|---|
| Orthognathic mandible | Mandibular retrognathism | ||||
| TGFB1 | rs4803455-rs1800469 | CT | 0.264 | 0.242 | 0.706 |
| AT | 0.027 | 0.025 | 0.930 | ||
| CC | 0.225 | 0.213 | 0.818 | ||
| AC | 0.482 | 0.517 | 0.586 | ||
|
| |||||
| TGFBR2 | rs764522-rs3087465 | GA | 0.084 | 0.103 | 0.610 |
| CA | 0.252 | 0.208 | 0.407 | ||
| GG | 0.035 | 0.049 | 0.602 | ||
| CG | 0.627 | 0.639 | 0.855 | ||
Note: TGFB1: transforming growth factor beta 1; TGFBR2: transforming growth factor beta receptor 2; C: cytosine; A: adenine; T: thymine; G: guanine.
4. Discussion
Some studies evaluating dentofacial deformities via cephalometric images in different populations/ethnicities have been performed in the past decades to investigate the genetic contribution of different skeletal malocclusions. Literature reviews showed that many genes involved in a range of functions are associated with skeletal malocclusions [13, 14], especially genes encoding growth factors and growth factor receptors [8, 15–17]. Growth factors are found in all tissues; they regulate local cell-to-cell metabolism and mediate cellular effects of different hormones. Bone matrix is a large reservoir for numerous growth factors that are regulators of bone remodeling processes [18]. Therefore, we hypothesized that functional genetic polymorphisms in TGFB1 and TGFBR2 could be involved in the etiology of mandibular retrognathism in Germans.
In our study, we explored two well-known genetic polymorphisms in TGFB1. Several polymorphisms have been described in the coding and regulatory sequences of the TGFB1 gene, including a promoter polymorphism involving a cytosine-to-thymine transition. The -509C/T functional promoter polymorphism (rs1800469) within the TGFB1 gene has been extensively assessed in different genetic epidemiological studies. Moreover, a number of studies have attempted to investigate whether the polymorphic variants in TGFB1 change TGFβ1 expression [19–21]. Another genetic polymorphism widely explored in different conditions is rs4803455 involving a C-to-A transition, which is located in intron 2 of TGFB1 and was therefore selected in our study. Although both variants (rs1800469 and rs4803455) with a known role were hypothesized as potential candidates for mandibular retrognathism, none of these genetic polymorphisms were associated with the phenotype in our study. However, it is possible that other polymorphisms in these genes could be involved in mandibular retrognathism.
TGFB initially binds its receptor, which is TGFBR2 and later transactivates TGFBR1, leading to the formation of a heterotetrameric receptor complex. TGFBR1 and TGFBR2 are members of the serine-threonine protein kinase family. TGFBR2 is a constitutive kinase, while TGFBR1 kinase is only activated after the formation of the TGFB/TGFBR2 complex [22]. Recently, there have been several studies investigating the association between genetic polymorphisms in TGFBR2 in various diseases, such as abdominal aortic aneurysm, papillary thyroid carcinoma, and end-stage renal disease, especially two promoter polymorphisms rs764522 (-1444C/G) and rs3087465 (-834A/G) [23–25]. Therefore, we decided to investigate these polymorphisms in our study. We observed that the AA genotype in polymorphism rs3087465 (-834A/G) acted as a protective factor for mandibular retrognathism. Interestingly, this genetic polymorphism located in a promoter region was previously associated with alterations in TGFβ type II receptor expression [26–28].
Although the sample size was a limitation that could lead to a type I error, our results raised a possibility of a novel candidate gene for mandibular retrognathism. It is interesting to mention that mutations in the TGFBR2 gene are associated with Marfan syndrome [29] and Loeys-Deitz syndrome [30]. The craniofacial phenotypes of these both syndromes included mandibular retrognathism as a common trait observed [31–33], which clearly suggests the role of genetic polymorphisms in TGFBR2 in nonsyndromic mandibular retrognathism. Also, studies with animal models have demonstrated that TGFBR2 plays a critical role in the formation of the intramembranous bone and endochondral bone and that TGFBR2 is crucial for skeletal development [34, 35] including craniofacial development [34, 36, 37]. Deletion of Tgfbr2 via Col2a1-Cre in mice caused several defects in the base of the skull [34]. Removal of TGFBR2 driven by Prx-Cre results in defects in the skull vault [37]. Mice with Tgfbr2 conditional gene ablation in the cranial neural crest have craniofacial anomalies including defects in mandibular development resulting in a smaller mandible [36].
To the best of our knowledge, this is the first study to investigate genes involved in mandibular retrognathism in Germans; however, it is important to emphasize that these findings must be validated in independent larger cohorts. Another important aspect to be discussed is that we investigated children and teenagers. This approach was also performed before. The study from Sasaki et al. [15] investigated the association between a genetic polymorphism in the gene encoding growth hormone receptor (GHR) and mandibular prognathism in children aged 3 to 13 years. The authors concluded that P561T in GHR may affect mandibular growth during early childhood.
Briefly, our results add novel information regarding the genetic contribution to mandibular retrognathism etiology suggesting rs3087465 (-834A/G) in TGFBR2 as candidate gene, additionally to the previously genes suggested in studies from different populations: MYO1H [5], MATN1 [4], BMP2 [7], ADAMTS9 [6], PTH, VDR, CYP24A1, and CYP27B1 [8]. Once our understanding of the nature of the genetic influences improves, we will be able to provide a clearer idea of how genes and environmental factors interact to influence mandibular retrognathism in humans.
5. Conclusion
The genetic polymorphism rs3087465 in the promoter region of the TGFBR2 was associated with mandibular retrognathism in Germans. Determining the factors affecting mandibular growth will contribute to early diagnosis and treatment of mandibular retrognathism.
Acknowledgments
The authors thank all participants of the study. This study was financed in part by the Alexander-von-Humboldt-Foundation (Küchler/Kirschneck accepted on July 4th, 2019) and by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior–Brasil (CAPES)–Finance Code 001.
Data Availability
The data generated during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.van den Braber W., van der Bilt A., van der Glas H., Rosenberg T., Koole R. The influence of mandibular advancement surgery on oral function in retrognathic patients: a 5-year follow-up study. Journal Oral and Maxillofacial Surgery . 2006;64(8):1237–1240. doi: 10.1016/j.joms.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 2.Kavitha L., Karthik K. Comparison of cephalometric norms of caucasians and non-caucasians: a forensic aid in ethnic determination. Journal of Forensic Dental Sciences . 2012;4(1):53–55. [PMC free article] [PubMed] [Google Scholar]
- 3.Küchler E. C., Reis C. L. B., Carelli J., et al. Potential interactions among single nucleotide polymorphisms in bone- and cartilage-related genes in skeletal malocclusions. Orthodontics & Craniofacial Research . 2021;24(2):277–287. doi: 10.1111/ocr.12433. [DOI] [PubMed] [Google Scholar]
- 4.Balkhande P. B., Lakkakula B. V. K. S., Chitharanjan A. B. Relationship between matrilin-1 gene polymorphisms and mandibular retrognathism. American Journal of Orthodontics and Dentofacial Orthopedics . 2018;153(2):255–261. doi: 10.1016/j.ajodo.2017.06.023. [DOI] [PubMed] [Google Scholar]
- 5.Arun R. M., Lakkakula B. V., Chitharanjan A. B. Role of myosin 1H gene polymorphisms in mandibular retrognathism. Journal of Orthodontics and Dentofacial Orthopedics . 2016;149(5):699–704. doi: 10.1016/j.ajodo.2015.10.028. [DOI] [PubMed] [Google Scholar]
- 6.Wang C., Ni Z., Cai Y., Zhou Y., Chen W. Association of polymorphism rs67920064 in ADAMTS9 gene with mandibular retrognathism in a Chinese population. Medical Science Monitor . 2020;26:p. e925965. doi: 10.12659/MSM.925965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Küchler E. C., Hannegraf N. D., Lara R. M., et al. Investigation of genetic polymorphisms in BMP2, BMP4, SMAD6, and RUNX2 and persistent apical periodontitis. Journal of Endodontics . 2021;47(2):278–285. doi: 10.1016/j.joen.2020.11.014. [DOI] [PubMed] [Google Scholar]
- 8.Küchler E. C., Reis C. L. B., Marañón-Vásquez G., et al. Parathyroid hormone gene and genes involved in the maintenance of vitamin d levels association with mandibular retrognathism. Journal of Personalized Medicine . 2021;11(5):p. 369. doi: 10.3390/jpm11050369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poniatowski L. A., Wojdasiewicz P., Gasik R., Szukiewicz R. Transforming growth factor beta family: insight into the role of growth factors in regulation of fracture healing biology and potential clinical applications. Mediators of Inflammation . 2015;2015:17. doi: 10.1155/2015/137823.137823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen G., Deng C., Li Y. P. TGF-beta and BMP signaling in osteoblast differentiation and bone formation. International Journal of Biological Sciences . 2012;8(2):272–288. doi: 10.7150/ijbs.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin G., Deng Y., Miao R., et al. TGFB1 and TGFBR2 functional polymorphisms and risk of esophageal squamous cell carcinoma: a case-control analysis in a Chinese population. Journal of Cancer Research and Clinical Oncology . 2008;134(3):345–351. doi: 10.1007/s00432-007-0290-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Küchler E. C., Tannure P. N., Falagan-Lotsch P., Lopes T. S., Granjeiro J. M., Amorim L. M. F. Buccal cells DNA extraction to obtain high quality human genomic DNA suitable for polymorphism genotyping by PCR-RFLP and real-time PCR. Journal of Applied Oral Science . 2012;20(4):467–471. doi: 10.1590/S1678-77572012000400013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uribe L. M. M., Miller S. F. Genetics of the dentofacial variation in human malocclusion. Orthodontics & Craniofacial Research . 2015;18(1):91–99. doi: 10.1111/ocr.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dehesa-Santos A., Iber-Diaz P., Iglesias-Linares A. Genetic factors contributing to skeletal class III malocclusion: a systematic review and meta-analysis. Clinical Oral Investigations . 2021;25(4):1587–1612. doi: 10.1007/s00784-020-03731-5. [DOI] [PubMed] [Google Scholar]
- 15.Sasaki Y., Satoh K., Hayasaki H., Fukumoto S., Fujiwara T., Nonaka K. The P561T polymorphism of the growth hormone receptor gene has an inhibitory effect on mandibular growth in young children. European Journal of Orthodontics . 2009;31(5):536–541. doi: 10.1093/ejo/cjp017. [DOI] [PubMed] [Google Scholar]
- 16.Jiang Q., Mei L., Zou Y., et al. Genetic polymorphisms in FGFR2 underlie skeletal malocclusion. Journal of Dental Research . 2019;98(12):1340–1347. doi: 10.1177/0022034519872951. [DOI] [PubMed] [Google Scholar]
- 17.Rodrigues A. S., Teixeira E. C., Antunes L. S., et al. Association between craniofacial morphological patterns and tooth agenesis-related genes. Progress in Orthodontics . 2020;21(1):p. 9. doi: 10.1186/s40510-020-00309-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lind M. Growth factors: possible new clinical tools: a review. Acta Orthopaedica Scandinavica . 1996;67(4):407–417. doi: 10.3109/17453679609002342. [DOI] [PubMed] [Google Scholar]
- 19.Grainger D. J., Heathcote K., Chiano M., et al. Genetic control of the circulating concentration of transforming growth factor type beta1. Human Molecular Genetics . 1999;8(1):93–97. doi: 10.1093/hmg/8.1.93. [DOI] [PubMed] [Google Scholar]
- 20.Grainger D. J. TGF-beta and atherosclerosis in man. Cardiovascular Research . 2007;74(2):213–222. doi: 10.1016/j.cardiores.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 21.Du L., Gong T., Yao M., Dai H., Ren H. G., Wang H. Contribution of the polymorphism rs1800469 of transforming growth factor β in the development of myocardial infarction: meta-analysis of 5460 cases and 8413 controls (MOOSE-compliant article) Medicine . 2019;98(26):p. e15946. doi: 10.1097/MD.0000000000015946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rechtman M. M., Nakaryakov A., Shapira K. E., Ehrlich M., Henis Y. I. Different domains regulate homomeric and heteromeric complex formation among type I and type II transforming growth factor-β receptors. Journal of Biological Chemistry . 2009;284(12):7843–7852. doi: 10.1074/jbc.M809215200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biros E., Norman P. E., Jones G. T., et al. Meta-analysis of the association between single nucleotide polymorphisms in TGF-β receptor genes and abdominal aortic aneurysm. Atherosclerosis . 2011;219(1):218–223. doi: 10.1016/j.atherosclerosis.2011.07.105. [DOI] [PubMed] [Google Scholar]
- 24.Choe B. K., Kim S. K., Park H. J., et al. Polymorphisms of TGFBR2 contribute to the progression of papillary thyroid carcinoma. Molecular & Cellular Toxicology . 2012;8(1):1–8. doi: 10.1007/s13273-012-0001-0. [DOI] [Google Scholar]
- 25.Ki H. J., Kim S. Y., Lee S. H., et al. Transforming growth factor-β receptor 2 gene polymorphisms are associated with end-stage renal disease. Kidney Research and Clinical Practice . 2015;34(2):93–97. doi: 10.1016/j.krcp.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vitiello G. A. F., Amarante M. K., Banin-Hirata B. K., et al. Transforming growth factor beta receptor II (TGFBR2) promoter region polymorphism in Brazilian breast cancer patients: association with susceptibility, clinicopathological features, and interaction with TGFB1 haplotypes. Breast Cancer Research and Treatment . 2019;178(1):207–219. doi: 10.1007/s10549-019-05370-1. [DOI] [PubMed] [Google Scholar]
- 27.Vitiello G. A. F., Guembarovski R. L., Hirata B. K. B., et al. Transforming growth factor beta 1 (TGFβ1) polymorphisms and haplotype structures have dual roles in breast cancer pathogenesis. Journal of Cancer Research and Clinical Oncology . 2018;144(4):645–655. doi: 10.1007/s00432-018-2585-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hadj-Ahmed M., Ghali R. M., Bouaziz H., et al. Transforming growth factor beta 1 polymorphisms and haplotypes associated with breast cancer susceptibility: a case-control study in Tunisian women. Tumor Biology . 2019;41(8):p. 1010428319869096. doi: 10.1177/1010428319869096. [DOI] [PubMed] [Google Scholar]
- 29.Mizuguchi T., Collod-Beroud G., Akiyama T., et al. Heterozygous TGFBR2 mutations in Marfan syndrome. Nature Genetics . 2004;36(8):855–860. doi: 10.1038/ng1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loeys B. L., Chen J., Neptune E. R., et al. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nature Genetics . 2005;37(3):275–281. doi: 10.1038/ng1511. [DOI] [PubMed] [Google Scholar]
- 31.Coster P., Pauw G., Martens L., Paepe A. Craniofacial structure in Marfan syndrome: a cephalometric study. American Journal of Medical Genetics Part A . 2004;131(3):240–248. doi: 10.1002/ajmg.a.30393. [DOI] [PubMed] [Google Scholar]
- 32.Johnson C. M., Spruiell B., Wiesen C., Pimenta L. A., Vann W., Frazier-Bowers S. A. Craniofacial characterization of Marfan syndrome. Orthodontics & Craniofacial Research . 2019;22(S1):56–61. doi: 10.1111/ocr.12295. [DOI] [PubMed] [Google Scholar]
- 33.Almpani K., Liberton D. K., Jani P., et al. Loeys-Dietz and Shprintzen-Goldberg syndromes: analysis of TGF-β-opathies with craniofacial manifestations using an innovative multimodality method. Journal of Medical Genetics . 2021;2021:107695. doi: 10.1136/jmedgenet-2021-107695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baffi M. M., Slattery E., Sohn P., Moses H. L., Chytil A., Serra R. Conditional deletion of the TGF-β type II receptor in Col2a expressing cells results in defects in the axial skeleton without alterations in chondrocyte differentiation or embryonic development of long bones. Developmental Biology . 2004;276(1):124–142. doi: 10.1016/j.ydbio.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 35.Seo H. S., Serra R. Deletion of Tgfbr2 in Prx1-cre expressing mesenchyme results in defects in development of the long bones and joints. Developmental Biology . 2007;310(2):304–316. doi: 10.1016/j.ydbio.2007.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ito Y., Yeo J. Y., Chytil A., et al. Conditional inactivation of TGFBR2 in cranial neural crest causes cleft palate and calvaria defects. Development . 2003;130(21):5269–5280. doi: 10.1242/dev.00708. [DOI] [PubMed] [Google Scholar]
- 37.Seo H. S., Serra R. Tgfbr2 is required for development of the skull vault. Developmental Biology . 2009;334(2):481–490. doi: 10.1016/j.ydbio.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clayton P., Chatelain P., Tatò L., et al. A pharmacogenomic approach to the treatment of children with GH deficiency or Turner syndrome. European Journal of Endocrinology . 2013;169(3):277–289. doi: 10.1530/EJE-13-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data generated during the current study are available from the corresponding author on reasonable request.


