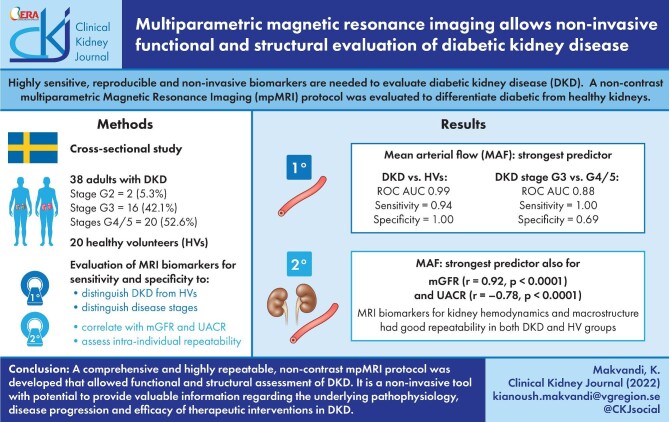
ABSTRACT
Background
We sought to develop a novel non-contrast multiparametric MRI (mpMRI) protocol employing several complementary techniques in a single scan session for a comprehensive functional and structural evaluation of diabetic kidney disease (DKD).
Methods
In the cross-sectional part of this prospective observational study, 38 subjects ages 18‒79 years with type 2 diabetes and DKD [estimated glomerular filtration rate (eGFR) 15‒60 mL/min/1.73 m2] and 20 age- and gender-matched healthy volunteers (HVs) underwent mpMRI. Repeat mpMRI was performed on 23 DKD subjects and 10 HVs. By measured GFR (mGFR), 2 DKD subjects had GFR stage G2, 16 stage G3 and 20 stage G4/G5. A wide range of MRI biomarkers associated with kidney haemodynamics, oxygenation and macro/microstructure were evaluated. Their optimal sensitivity, specificity and repeatability to differentiate diabetic versus healthy kidneys and categorize various stages of disease as well as their correlation with mGFR/albuminuria was assessed.
Results
Several MRI biomarkers differentiated diabetic from healthy kidneys and distinct GFR stages (G3 versus G4/G5); mean arterial flow (MAF) was the strongest predictor (sensitivity 0.94 and 1.0, specificity 1.00 and 0.69; P = .04 and .004, respectively). Parameters significantly correlating with mGFR were specific measures of kidney haemodynamics, oxygenation, microstructure and macrostructure, with MAF being the strongest univariate predictor (r = 0.92; P < .0001).
Conclusions
A comprehensive and repeatable non-contrast mpMRI protocol was developed that, as a single, non-invasive tool, allows functional and structural assessment of DKD, which has the potential to provide valuable insights into underlying pathophysiology, disease progression and analysis of efficacy/mode of action of therapeutic interventions in DKD.
Keywords: biomarkers, chronic kidney disease, diabetic kidney disease, magnetic resonance imaging (MRI), multiparametric
Graphical Abstract
Graphical Abstract.
INTRODUCTION
Chronic kidney disease (CKD) associated with diabetes mellitus (DM), i.e. diabetic kidney disease (DKD), occurs in ∼30–40% of patients with type 2 DM (T2DM) [1, 2]. DKD is associated with increased morbidity and mortality and is the leading cause of kidney failure [end-stage kidney disease (ESKD)] worldwide [1, 2]. With the increasing prevalence of T2DM, the global DKD burden is expected to increase [1, 2]. To prevent ESKD, early identification of patients at high risk of DKD progression, enabling timely treatment, is important. However, predicting the evolution of DKD remains difficult due to its highly variable progression, particularly in T2DM [1].
Established kidney biomarkers such as estimated glomerular filtration rate (eGFR) and proteinuria provide only a rough estimation of overall kidney damage [3]. Measured GFR (mGFR), using plasma clearance of filtration markers, more accurately measures kidney function than eGFR but is time-consuming and cumbersome for screening and ambulatory care [4]. Moreover, as the kidney can partially compensate for lost function, significant kidney fibrosis may occur without a measurable change in GFR [5]. As regards proteinuria, intra-individual variations are considerable [6]. Thus these routinely used biomarkers neither provide insights into underlying DKD pathophysiology and degree of anatomical damage nor allow risk stratification [7, 8]. Various DKD phenotypes are well recognized and other pathways to ESKD independent of albuminuria have been postulated, suggesting different pathophysiology not readily detected by conventional biomarkers [9]. While kidney biopsy is essential for assessing kidney pathophysiology in DKD [10], it is not suitable for long-term, serial monitoring of the dynamic process of disease progression or response to therapy, due to its invasive nature, an increased risk of bleeding in uremic patients [11] and susceptibility to sampling errors. Therefore, kidney biopsy is not routinely performed in clinical practice for DKD diagnosis or monitoring [12]. There is a clear need for new, sensitive, reproducible and non-invasive biomarkers to enhance our understanding of DKD pathophysiology, progression and efficacy/modes of action of therapeutic interventions in clinical practice/trials.
Novel, non-contrast magnetic resonance imaging (MRI) techniques are potential methods to non-invasively assess and quantify functional and pathophysiological changes in CKD [13–17]. Employing such complementary MRI techniques simultaneously in multiparametric MRI (mpMRI) may provide more comprehensive information across individual kidney compartments on microstructure (including kidney fibrosis and inflammation), macrostructure (kidney volume), oxygenation and haemodynamic measurements of renal artery blood flow and perfusion [13, 18].
In a cross-sectional study using an mpMRI in CKD patients (excluding DKD), several MRI biomarkers correlated with albuminuria, eGFR and histopathological measures of interstitial fibrosis [19]. Nevertheless, more studies identifying the most appropriate MRI biomarkers of kidney function and structure and their technical and clinical validation are required before mpMRI can be adopted in clinical practice [14, 19]. To our knowledge, no study has specifically investigated the use of mpMRI in DKD, although individual MRI techniques have been evaluated to detect early changes and renal blood flow in DKD [20–23].
In this kidney mpMRI study we evaluated a range of MRI biomarkers for their optimal sensitivity and specificity in distinguishing DKD subjects from healthy volunteers (HVs) and between different disease stages. Secondary aims were to analyse intra-individual repeatability of MRI biomarkers to capture natural biological variability, assess the correlation of each MRI biomarker to conventional biomarkers of kidney function and damage [i.e. mGFR, urine albumin:creatinine ratio (UACR)] and develop a bifactorial model to predict mGFR using MRI biomarkers.
MATERIALS AND METHODS
Study design and participants
This cross-sectional and longitudinal observational, investigator-driven, single-centre study conducted at Sahlgrenska University Hospital (SU), Gothenburg, Sweden was approved by the Regional Ethics Review Board and carried out according to the Helsinki Declaration of 1975 (revised 2013). Subjects with T2DM and DKD and HVs were enroled during the same period after obtaining written informed consent from all subjects.
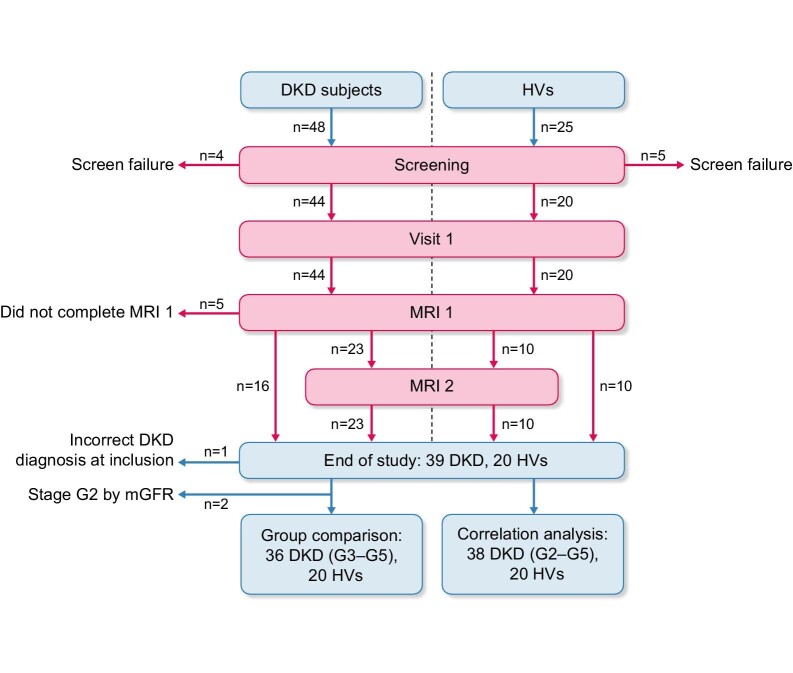
Between November 2016 and June 2018, 48 subjects with T2DM and DKD from the outpatient clinics of the Nephrology Department at SU/affiliated hospitals (n = 25) and primary care clinics in Gothenburg (n = 23) were screened by reviewing medical records (Fig. 1). Eligible subjects with T2DM and DKD were identified by medical history, UACR and eGFR using the Chronic Kidney Disease Epidemiology Collaboration creatinine equation [24] within the last 3 months of screening. The diagnosis of DKD was per treating physician's assessment. The inclusion/exclusion criteria are stated in Table 1. DKD subjects were stratified by mGFR into four GFR stages (G2‒G5) as per the Kidney Disease: Improving Global Outcomes (KDIGO) classification of CKD [25].
Figure 1:
Study flow chart. A total of 48 DKD subjects were screened: 4 did not fulfil the eligibility criteria and 5 did not complete the study, leaving a total of 39 DKD subjects at the end of the study. Of these, 36 subjects were included in the group comparison and 38 subjects in the correlation analysis. During the same enrolment period, 25 HVs were screened and 20 HVs were included and completed the study.
Table 1.
Inclusion/exclusion criteria
| Inclusion criteria | Exclusion criteria |
|---|---|
| DKD subjects | |
| Signed informed consent | Cause for the impaired kidney function diagnosed primarily to be other than DKD (histologic or clinical) |
| Age 18–79 years | History of renal transplant |
| T2DM with clinical diagnosis of DKD and an eGFR of 15–60 mL/min/1.73 m2 | Patients with congestive heart failure (NYHA class IV) |
| UACR ˃3 mg/mmol in the first morning urine sample | Pregnancy |
| An eGFR decline of <10 mL/min/1.73 m2 over the last 6 months | Any contraindication for MRI examination (e.g. pacemaker, severe claustrophobia) |
| Stable dose of antidiabetic treatment for >1 month | Allergy to iodine-based contrast agents |
| BMI 18–35 | Involved in the planning or execution of this study |
| Healthy volunteers | |
| Signed informed consent | HIV/hepatitis B or C positive |
| Age 18–79 years | Evidence of any active or chronic disease following a detailed medical history including but not limited to T2DM, congestive heart failure (NYHA 3 and 4) and hypertension |
| eGFR >70 mL/min/1.73 m2 | Pregnancy |
| UACR <3 mg/mmol | Any contraindication for MRI examination (e.g. pacemaker, severe claustrophobia) |
| BMI 18–35 | Allergy to iodine-based contrast agents |
| Not receiving any medical treatment | Involved in the planning or execution of this study |
HIV: human immunodeficiency virus; NYHA: New York Heart Association classification.
The control group was 25 age- (±5 years) and gender-matched HVs who were screened and recruited by the Clinical Trial Centre of the SU via local advertisement, as per the inclusion/exclusion criteria in Table 1.
Clinical/laboratory assessments
Demographic data, medical history, physical examination [including body mass index (BMI)], blood samples and the first-morning urine sample for determination of UACR were collected at the baseline visit. The mGFR was evaluated using the iohexol clearance test as described in the Supplementary Methods [26].
Kidney mpMRI examination
All subjects underwent a kidney mpMRI examination within 3 weeks following the mGFR assessment; 33 subjects (10 HVs and 23 DKD subjects) underwent a second MRI examination 2‒3 weeks (maximum 6 weeks) after the first MRI examination. Subjects drank 250 mL of water before the MRI scan. The MRI examination was performed on a Philips Achieva dStream 3T (Philips Healthcare, Best, The Netherlands) without any contrast agents, with a total scan time of 50 min. MRI biomarkers associated with kidney haemodynamics, oxygenation and macro- and microstructure were evaluated (Table 2) using the image acquisition/analysis protocols described in the Supplementary Methods.
Table 2.
Imaging variables measured in the mpMRI protocol
| Variable | Assessment |
|---|---|
| Kidney haemodynamics [13, 38, 39] | |
| Phase contrast (PC) MRI | |
| Peak systolic and diastolic velocity (cm/s) | Peak blood velocity in the renal artery in systole and diastole |
| RARI (no unit) | Measures the resistance of renal arterial flow to the kidney |
| MAF (mL/min) | Mean blood flow in the renal artery |
| Global perfusion (mL/min/100 g) | Mean renal flow per 100 g of kidney tissue |
| ASL (mL/100 g/min) [38] | Uses the magnetic labelling of water in arterial blood as an endogenous tracer to generate maps of regional kidney perfusion |
| Kidney macrostructure [40] | |
| Kidney volume (mL) | Volume of kidney parenchyma from T2-weighted structural images |
| Kidney BOLD MRI [22, 41] | |
| BOLD R2* (per s) | Allows indirect assessment of tissue oxygenation based on the paramagnetic properties of endogenous deoxyhaemoglobin |
| Kidney microstructure [19, 26, 42] | |
| ADC (mm2/s × 10−3) | Relates to interstitial fibrosis by measuring the restriction of water displacement seen on diffusion-weighted imaging |
| IVIM parameters (per s) [19] Fast component of diffusion (pseudo-perfusion) D* (mm2/s × 10−3) Slow component of diffusion D (mm2/s × 10−3) f (%) |
IVIM separates the intracellular water diffusion (D) and vascular perfusion (D*) components of the ADC measurement |
| R1 (per s) [26] | Longitudinal nuclear magnetic resonance relaxation rate (R1) of water reflects the molecular environment, e.g. viscosity, fibrosis and inflammation (interstitial oedema, cellular swelling) R1 = 1/T1 |
| MTC (%) [43] | Measurement of the energy interaction of large macromolecules and bulk water protons, which has been shown to correlate with fibrosis |
Statistical analysis
The ability of each MRI biomarker to discriminate between the DKD and HV groups was evaluated using a univariate logistic regression analysis with the DKD and HV groups as dependent variables and the MRI biomarker as an independent variable. The assessment of discrimination of the fitted logistic regression model was done via the receiver operating characteristics (ROC) curve and the area under the curve (AUC). The Youden cut-off and its sensitivity and specificity were calculated [27]. The same logistic regression methods and analyses were used to assess discrimination between stages G3 and G4/G5.
The intra-individual repeatability of the MRI biomarkers was assessed using the intra-individual coefficient of variance (CV) and the intraclass correlation coefficient (ICC) type (2, 1) based on the values from the repeated MRI measurements [28]. Each MRI biomarker's linear association with mGFR and UACR was evaluated using univariate linear regression with mGFR or UACR as a dependent variable and the MRI biomarker as an independent variable. The Pearson correlation with mGFR and the Spearman correlation with UACR were calculated to evaluate the association with each MRI biomarker.
Bivariate MRI predictors of mGFR were evaluated using a predefined approach; each pair of MRI predictors with a P-value <.10 in the univariate linear regression analysis was entered into a stepwise linear regression model. A significance level of .05 in model improvement was required to allow variables into the model. Because reduced GFR is known to correlate with increased UACR [29], bivariate predictors of UACR were examined using mGFR as one variable and adding one MRI endpoint using linear regression in the DKD subjects. The HVs were excluded from this analysis, as they had no significant UACR values. Continuous data are expressed using mean [standard deviation (SD)] or median [interquartile range (IQR)] and categorical data as numbers (percentages). Two-sided P-values <.05 were considered significant. Statistical analyses were done using SAS version 9.4 software (SAS Institute, Cary, NC, USA). Sample size calculation is described in the Supplementary Methods.
RESULTS
As in Fig. 1, 44/48 screened DKD subjects were found to be eligible. Of these, two classified as stages G3 and G4 by eGFR at screening were found by mGFR to have stages G2 (mGFR 62 and 64 mL/min/1.73 m2) and G5 (mGFR 11 and 12 mL/min/1.73 m2), resulting in 42 subjects in stages G3–G5. A total of 23 subjects underwent two MRI scans, 16 one scan and the remaining 5 could not complete MRI due to claustrophobia. A total of 39 subjects completed the study (n = 2 stage G2 and n = 37 stages G3–G5), of whom 1 stage G4 subject was excluded due to a revised diagnosis of hydronephrosis as a contributory cause of CKD, based on the MRI findings and past history. Thus the analysis set included 38 DKD subjects (stages G2–G5) for correlation analysis and 36 for group comparisons (16 in stages G3 versus 20 in G4/G5) (Fig. 1). Of the 25 screened HVs, 20 met the inclusion criteria and were included in the analysis set. Of these, 10 underwent two MRI scans (Fig. 1).
As regards the demographic and baseline clinical characteristics (Table 3), the main differences between DKD subjects and HVs were in the proportion of males (83% versus 75%), systolic blood pressure (150 versus 133 mmHg) and BMI (27.9 versus 25.5 kg/m2).
Table 3.
Baseline characteristics of the DKD and HV groups
| Variable | HVs (n = 20) | Stage G3 (n = 16) | Stages G4/G5 (n = 20) | Stages G3‒G5 (n = 36) |
|---|---|---|---|---|
| Age (years) | 66.7 (6.2) | 68.9 (5.6) | 68.3 (5.6) | 68.6 (5.5) |
| Ethnicity (Caucasian), n (%) | 20 (100) | 16 (100) | 18 (90) | 34 (94) |
| Gender (male), n (%) | 15 (75) | 12 (75) | 18 (90) | 30 (83) |
| BMI (kg/m2) | 25.5 (3.1) | 28 (3.3) | 27.8 (3.0) | 27.9 (3.1) |
| Systolic blood pressure (mmHg) | 133 (12) | 147 (21) | 153 (22) | 150 (21) |
| Diastolic blood pressure (mmHg) | 80.4 (7.3) | 80.1 (9.4) | 78 (7.9) | 78.9 (8.5) |
| Haemoglobin (g/L) | 145 (12.0) | 134 (11.6) | 133.2 (16.1) | 134 (14.1) |
| Creatinine (µmol/L) | 82 (12) | 135 (29) | 230 (54) | 188 (65) |
| UACR ratio (mg/mmol) | 1.2 (0.7) | 34.1 (44.3) | 85 (81.0) | 62.4 (71.1) |
| eGFR (mL/min/1.73 m2) | 79.1 (7.2) | 45.3 (10.7) | 25 (5.6) | 34 (13.1) |
| mGFR (mL/min/1.73 m2) | 81.5 (9.2) | 40.4 (6.7) | 21.8 (4.7) | 30.1 (10.9) |
| HbA1c (mmol/mol) | 35.6 (3.3) | 54.2 (11.6) | 62.2 (14.1) | 58.6 (13.5) |
| Use of RAAS blockers, n (%) | 0.0 (0) | 12 (75) | 14 (70) | 26 (72) |
| Hypertension, n (%) | 0.0 (0) | 14 (88) | 20 (100) | 34 (94) |
Data are presented as mean (SD) unless stated otherwise. RAAS: renin–angiotensin–aldosterone system. GFR stages were stratified by mGFR.
Differentiation between HVs and DKD subjects with MRI biomarkers
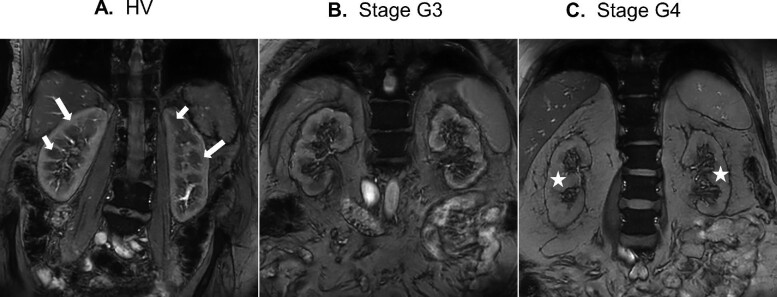
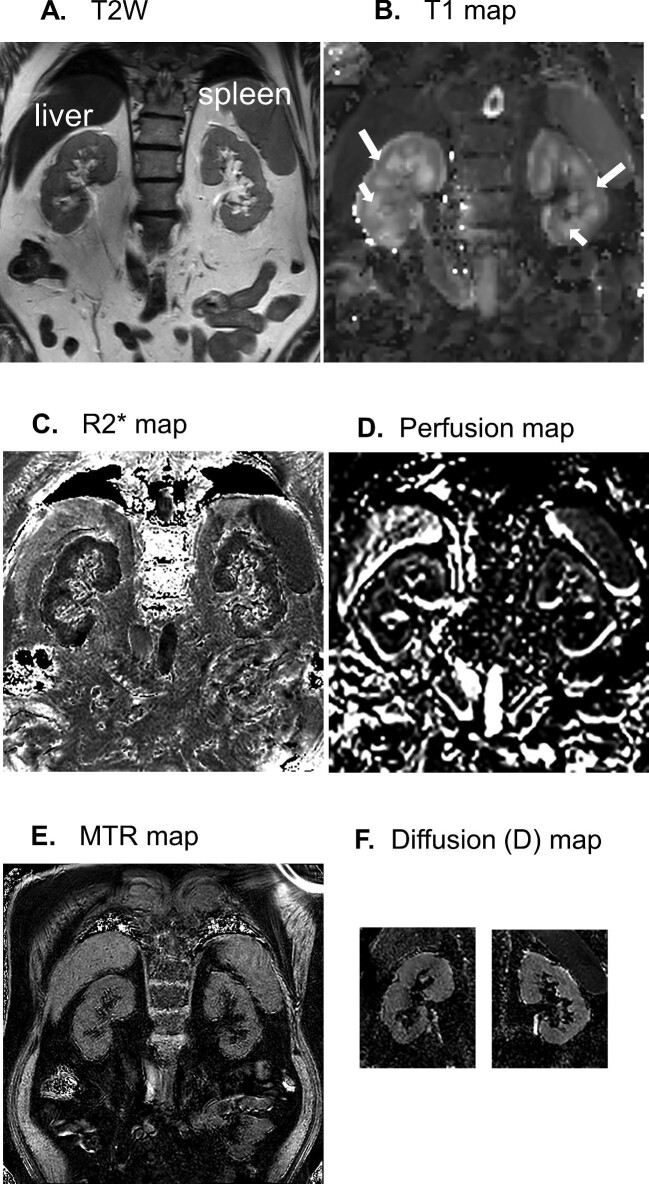
Anatomical T1-weighted MRI showed clear corticomedullary contrast in HVs, reduced contrast in stage G3 subjects and virtually no contrast in the stage G4/G5 subjects (example shown in Fig. 2). Representative mpMRIs are shown in Fig. 3. Examples of renal blood flow velocity profiles are shown in Fig. 4.
Figure 2:
Anatomical T1-weighted image of (A) a typical HV, (B) a DKD subject with GFR stage G3 and (C) a DKD subject with GFR stage G4. Long white arrows indicate the renal cortex, short white arrows indicate the renal medulla and stars indicate the regions with loss of corticomedullary contrast.
Figure 3:
Representative images from a G3 subject showing (A) T-weighted (T2W) image, (B) T1 map, (C) R2* map, (D) perfusion map (ASL), (E) MTR (magnetic transfer ratio) map and (F) D (tissue diffusion coefficient) map. Note that the D maps were created following realignment of a cropped version of the DWI data set and therefore a D map of the whole slice is not available. DWI:diffusion-weighted imaging. Long white arrows indicate the renal cortex and short white arrows indicate the renal medulla.
Figure 4:
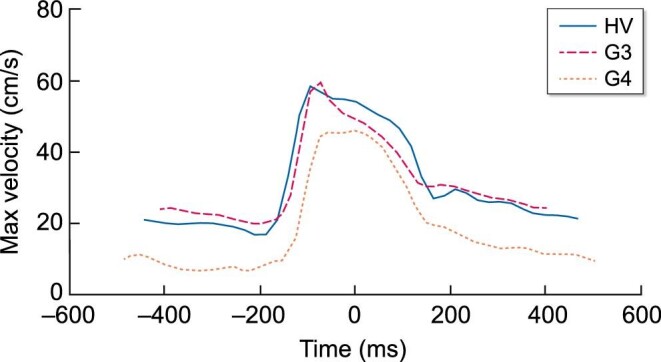
Representative phase contrast flow profiles through the cardiac cycle showing maximum velocity in the renal artery from a HV, stage G3 and stage G4 subject.
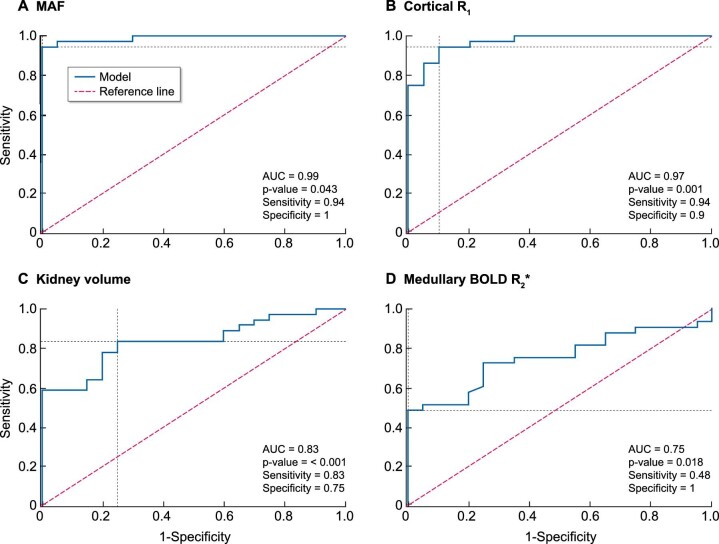
The strongest predictive properties differentiating HVs and DKD subjects were haemodynamic MRI biomarkers [MAF, end-diastolic velocity (EDV), renal artery resistive index (RARI), arterial spin labelling (ASL) perfusion cortex and global perfusion] with sensitivity and specificity for cut-off values of 0.81‒0.94 and 0.90‒1.00, respectively (Table 4). The strongest predictor was MAF, with a Youden cut-off value of 803 mL/min/1.73 m2, a sensitivity of 0.94 and a specificity of 1.00 (Table 3, Fig. 5).
Table 4.
Descriptive statistics and predictive properties (ROC AUC) of univariate imaging markers to distinguish between DKD subjects (stages G3‒G5) and HVs, including respective specificity and sensitivity for Youden cut-off values
| HVs | DKD (stages G3‒G5) | Prediction performance | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Variable | n | Mean (SD) | n | Mean (SD) | ROC AUC | Cut-off | Sensitivity | Specificity | *P-value |
| Kidney haemodynamics | |||||||||
| MAF (mL/min/1.73 m2) | 20 | 993 (126) | 36 | 538 (161) | 0.99 | <803 | 0.94 | 1.00 | .04 |
| PSV (cm/s) | 20 | 54.3 (8.28) | 36 | 50.5 (15.0) | 0.62 | <48.3 | 0.53 | 0.85 | .30 |
| EDV (cm/s) | 20 | 17.0 (3.9) | 36 | 8.75 (3.6) | 0.94 | <12.6 | 0.89 | 0.90 | .0005 |
| RARI | 20 | 0.68 (0.06) | 36 | 0.82 (0.06) | 0.94 | >0.73 | 0.94 | 0.85 | .0001 |
| ASL perfusion cortex (mL/min/100 g) | 20 | 164 (36.8) | 35 | 81.1 (40.7) | 0.93 | <125 | 0.86 | 0.90 | .0001 |
| Global perfusion (mL/min/100 g) | 20 | 458 (54) | 36 | 311 (83) | 0.94 | <380 | 0.81 | 0.95 | .001 |
| Kidney macrostructure | |||||||||
| Kidney volume (mL/1.73 m2) | 20 | 218 (26) | 36 | 176 (38) | 0.83 | <206 | 0.83 | 0.75 | .0008 |
| Kidney oxygenation | |||||||||
| BOLD R2* cortex (per s) | 20 | 17.3 (1.4) | 33 | 17.1 (1.4) | 0.52 | <16.6 | 0.36 | 0.80 | .53 |
| BOLD R2* medulla (per s) | 20 | 26.0 (2.3) | 33 | 23.5 (3.7) | 0.75 | <22.6 | 0.48 | 1.00 | .002 |
| Kidney microstructure | |||||||||
| ADC cortex (10–3 mm2/s) | 20 | 2.52 (0.19) | 34 | 2.31 (0.21) | 0.79 | <2.37 | 0.71 | 0.85 | .002 |
| ADC medulla (10–3 mm2/s) | 20 | 2.33 (0.18) | 34 | 2.19 (0.24) | 0.71 | <2.21 | 0.65 | 0.75 | .03 |
| R1 cortex (per s) | 20 | 0.72 (0.03) | 36 | 0.63 (0.04) | 0.97 | <0.69 | 0.94 | 0.90 | .001 |
| R1 medulla (per s) | 20 | 0.55 (0.02) | 36 | 0.55 (0.02) | 0.58 | <0.55 | 0.56 | 0.65 | .34 |
| IVIM D cortex (10−3 mm2/s) | 20 | 2.14 (0.24) | 34 | 1.91 (0.26) | 0.75 | <1.98 | 0.65 | 0.80 | .006 |
| IVIM D medulla (10−3 mm2/s) | 20 | 2.04 (0.27) | 34 | 1.85 (0.20) | 0.72 | <2.05 | 0.91 | 0.45 | .01 |
| IVIM D* cortex (10−3 mm2/s) | 19 | 56 (73.3) | 32 | 98 (351) | 0.48 | >19.4 | 0.72 | 0.37 | .62 |
| IVIM D* medulla (10−3 mm2/s) | 19 | 198 (459) | 31 | 190 (768) | 0.55 | <165 | 0.90 | 0.26 | .96 |
| IVIM f cortex (%) | 19 | 13.5 (5.7) | 32 | 15.2 (5.7) | 0.60 | >15 | 0.53 | 0.74 | .32 |
| IVIM f medulla (%) | 19 | 12.3 (5.1) | 31 | 14.5 (5.7) | 0.62 | >13.5 | 0.61 | 0.74 | .17 |
| MTR cortex (%) | 20 | 23 (1.9) | 36 | 21.8 (2.2) | 0.66 | <21.7 | 0.56 | 0.80 | .06 |
| MTR medulla (%) | 20 | 24.5 (2.13) | 36 | 23.7 (2.43) | 0.57 | <22.5 | 0.42 | 0.85 | .27 |
| Biochemistry | |||||||||
| eGFR (mL/min/1.73 m2) | 20 | 76.4 (7.76) | 36 | 34.9 (13.8) | 0.99 | <60 | 0.97 | 1.00 | .02 |
| UACR (mg/mmol) | 20 | 1.17 (0.54) | 36 | 67 (83.3) | 1.00 | >1.9 | 1.00 | 1.00 | .02 |
*P-value of OR = 1 for the variable (Wald test) in the model. Note that P-values are inappropriate in cases when there is complete (or quasi-complete) separation of data points, i.e. when sensitivity and specificity = 1 (or close to 1).
Figure 5:
ROC curves predicting separation of HVs versus DKD subjects using MRI measures: (A) MAF, (B) cortical R1, (C) kidney volume and (D) medullary BOLD R2*.
HVs and DKD subjects could also be distinguished using MRI biomarkers of kidney microstructure [R1 cortex, apparent diffusion coefficient (ADC) cortex and medulla and intravoxel incoherent motion (IVIM) slow diffusion cortex and medulla], macrostructure (kidney volume) and oxygenation in the medulla assessed with blood oxygen level-dependent (BOLD) R2* (Table 4). Moreover, BOLD R2* values were higher in the medulla versus cortex in both HVs (cortex 17.3 ± 1.4/s, medulla 26.0 ± 2.3/s) and DKD subjects (cortex 17.1 ± 1.4/s, medulla 23.5 ± 3.7/s), indicative of lower oxygenation in the medulla than the cortex (Table 4).
Differentiation between stages G3 and G4/G5 in DKD subjects with MRI biomarkers
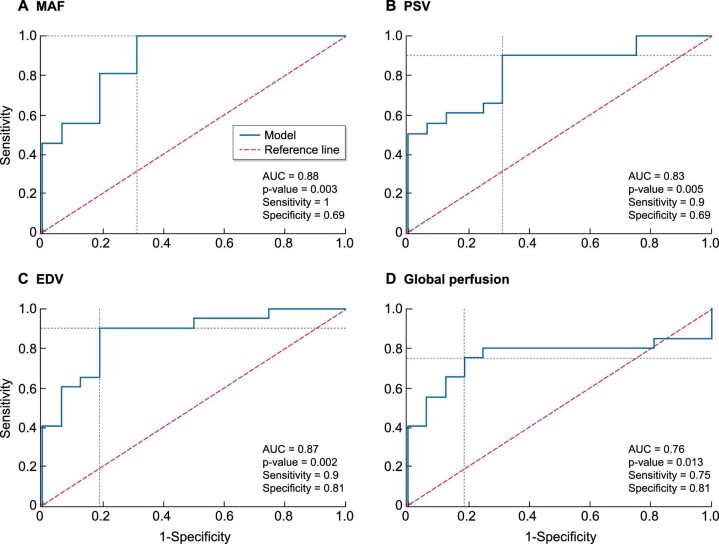
The strongest predictive properties to differentiate between stages G3 and G4/G5 were haemodynamic MRI biomarkers—MAF, peak systolic velocity (PSV), EDV and global perfusion measurements and all showed a ROC AUC >0.75 (Table 5), the best predictor again being MAF (Fig. 6). Youden cut-off values for separating the two groups were 605 mL/min/1.73 m2 for MAF, 53.2 cm/s for PSV and 8.35 cm/s for EDV, with sensitivities and specificities of 0.90‒1.00 and 0.69‒0.81, respectively (Table 5). MRI biomarkers for kidney microstructure also enabled stage G3 to be distinguished from G4/G5, where R1 in the cortex showed the highest predictive properties (ROC AUC = 0.76) (Table 5).
Table 5.
Descriptive statistics and predictive properties (ROC AUC) of univariate imaging markers to distinguish between stage G3 and stages G4/G5 in subjects with DKD, including respective specificity and sensitivity for Youden cut-off values
| Stage G3 | Stages G4/G5 | Prediction performance | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Variable | n | Mean (SD) | n | Mean (SD) | ROC AUC | Cut-off | Sensitivity | Specificity | *P-value |
| Kidney haemodynamics | |||||||||
| MAF (mL/min/1.73 m2) | 16 | 653 (133) | 20 | 447 (118) | 0.88 | <605 | 1.00 | 0.69 | .004 |
| PSV (cm/s) | 16 | 59.8 (14.2) | 20 | 43.1 (11.1) | 0.83 | <53.2 | 0.90 | 0.69 | .005 |
| EDV (cm/s) | 16 | 11.3 (3.2) | 20 | 6.72 (2.5) | 0.87 | <8.35 | 0.90 | 0.81 | .002 |
| RARI | 16 | 0.81 (0.06) | 20 | 0.84 (0.06) | 0.68 | >0.84 | 0.60 | 0.81 | .11 |
| ASL perfusion cortex (mL/min/100 g) | 16 | 96.4 (39.7) | 19 | 68.2 (37.8) | 0.72 | <79 | 0.79 | 0.69 | .05 |
| Global perfusion (mL/min/100 g) | 16 | 352 (43) | 20 | 278 (93) | 0.76 | <305 | 0.75 | 0.81 | .01 |
| Kidney macrostructure | |||||||||
| Kidney volume (mL/1.73 m2) | 16 | 185 (27.8) | 20 | 169 (43.4) | 0.63 | <155 | 0.40 | 0.94 | .20 |
| Kidney oxygenation | |||||||||
| BOLD R2* cortex (per s) | 15 | 17.2 (1.6) | 18 | 17 (1.25) | 0.51 | <16.1 | 0.28 | 0.87 | .56 |
| BOLD R2* medulla (per s) | 15 | 24.5 (3.7) | 18 | 22.8 (3.6) | 0.64 | <23.5 | 0.72 | 0.60 | .19 |
| Kidney microstructure | |||||||||
| ADC cortex (10–3 mm2/s) | 15 | 2.37 (0.17) | 19 | 2.27 (0.22) | 0.67 | <2.32 | 0.68 | 0.73 | .18 |
| ADC medulla (10–3 mm2/s) | 15 | 2.21 (0.24) | 19 | 2.17 (0.24) | 0.59 | <2.21 | 0.74 | 0.60 | .58 |
| R1 cortex (per s) | 16 | 0.65 (0.03) | 20 | 0.62 (0.04) | 0.76 | <0.62 | 0.50 | 0.94 | .02 |
| R1 medulla (per s) | 16 | 0.55 (0.02) | 20 | 0.54 (0.02) | 0.70 | <0.55 | 0.80 | 0.63 | .06 |
| IVIM D cortex (10−3 mm2/s) | 15 | 1.96 (0.17) | 19 | 1.87 (0.32) | 0.62 | <1.91 | 0.63 | 0.73 | .30 |
| IVIM D medulla (10−3 mm2/s) | 15 | 1.86 (0.17) | 19 | 1.85 (0.23) | 0.51 | <1.72 | 0.37 | 0.80 | .87 |
| IVIM D* cortex (10−3 mm2/s) | 15 | 48.2 (48.5) | 17 | 143 (482) | 0.42 | >2010 | 0.06 | 1.00 | .52 |
| IVIM D* medulla (10−3 mm2/s) | 14 | 48.7 (58.6) | 17 | 306 (1035) | 0.53 | >19.5 | 0.76 | 0.50 | .69 |
| IVIM f cortex (%) | 15 | 14.7 (5.13) | 17 | 15.6 (6.3) | 0.51 | >12.3 | 0.82 | 0.33 | .66 |
| IVIM f medulla (%) | 14 | 14.6 (5.59) | 17 | 14.3 (5.9) | 0.52 | <12 | 0.47 | 0.71 | .89 |
| MTR cortex (%) | 16 | 21.6 (2.41) | 20 | 22 (2.0) | 0.54 | >20.7 | 0.75 | 0.38 | .57 |
| MTR medulla (%) | 16 | 23.6 (2.35) | 20 | 23.9 (2.6) | 0.52 | >22.2 | 0.70 | 0.44 | .75 |
| Biochemistry | |||||||||
| eGFR (mL/min/1.73 m2) | 16 | 46.3 (12.1) | 20 | 25.9 (6.3) | 0.96 | <31 | 0.85 | 0.94 | .008 |
| UACR (mg/mmol) | 16 | 33 (47) | 20 | 94.2 (97) | 0.72 | >97 | 0.45 | 0.94 | .05 |
GFR stages were stratified by mGFR.
*P-value of OR = 1 for the variable (Wald test) in the model. Note that P-values are inappropriate in cases when there is complete (or quasi-complete) separation of data points, i.e. when sensitivity and specificity = 1 (or close to 1).
Figure 6:
ROC curves predicting separation of DKD subjects with GFR stage G3 versus stages G4/G5 using MRI biomarkers of kidney haemodynamics: (A) MAF, (B) PSV, (C) EDV and (D) global perfusion. GFR stages were stratified by mGFR.
The eGFR also showed strong predictive properties for distinguishing between stages G3 and G4/G5, with high sensitivity and specificity, whereas the UACR showed a lower ROC AUC (0.72), with sensitivity of 0.45 and specificity of 0.94 for the cut-off value of 97 mg/mmol (Table 5).
MRI biomarker repeatability in HVs and DKD subjects
Intra-individual CV values were lowest for RARI (2%), R1 in the cortex and medulla (2%) and for cortical and medullary BOLD R2* (≤5%), suggesting good repeatability. The CV values ranged between 2% and 12% for all MRI biomarkers except ASL perfusion cortex (CV 33%) and IVIM fast diffusion (D*) and perfusion fraction (f) measurements (CV >25%) (Table 6). The ICC values were ≥89% for MAF, PSV, EDV, RARI, global perfusion, cortical R1, medullary BOLD R2* and kidney volume (Table 6).
Table 6.
Intra-individual CV and ICC values for the evaluation of data repeatability of MRI biomarkers in HVs and DKD subjects
| Variable | CV | ICC |
|---|---|---|
| Kidney haemodynamics | ||
| MAF (mL/min/1.73 m2) | 0.07 | 0.97 |
| PSV (cm/s) | 0.09 | 0.90 |
| EDV (cm/s) | 0.12 | 0.96 |
| RARI | 0.02 | 0.96 |
| ASL perfusion cortex (mL/min/100 g) | 0.33 | 0.71 |
| Global perfusion (mL/min/100 g) | 0.09 | 0.92 |
| Kidney macrostructure | ||
| Kidney volume (mL/1.73 m2) | 0.07 | 0.89 |
| Kidney oxygenation | ||
| BOLD R2* cortex (per s) | 0.04 | 0.74 |
| BOLD R2* medulla (per s) | 0.05 | 0.90 |
| Kidney microstructure | ||
| ADC cortex (10−3 mm2/s) | 0.06 | 0.66 |
| ADC medulla (10−3 mm2/s) | 0.05 | 0.66 |
| R1 cortex (per s) | 0.02 | 0.94 |
| R1 medulla (per s) | 0.02 | 0.49 |
| IVIM D cortex (10−3 mm2/s) | 0.11 | 0.29 |
| IVIM D medulla (10−3 mm2/s) | 0.09 | 0.37 |
| IVIM D* cortex (10−3 mm2/s) | 4.97 | 0.00 |
| IVIM D* medulla (10−3 mm2/s) | 5.30 | 0.00 |
| IVIM f cortex (%) | 0.30 | 0.21 |
| IVIM f medulla (%) | 0.25 | 0.46 |
| MTR cortex (%) | 0.08 | 0.31 |
| MTR medulla (%) | 0.11 | 0.01 |
| Biochemistry | ||
| eGFR (mL/min/1.73 m2) | 0.06 | 0.99 |
| UACR (mg/mmol) | 0.28 | 0.96 |
Correlation of MRI biomarkers with mGFR and UACR
MRI biomarkers with strong predictive properties distinguishing both HVs and DKD subjects and GFR stages were also strongly correlated with mGFR, the reference measurement. There were statistically significant correlations (P < .05) between mGFR and MRI biomarkers: kidney haemodynamics (MAF, end-diastolic and PSV, RARI, cortical and global perfusion), kidney microstructure [cortical and medullary ADC, IVIM slow diffusion (D) in the cortex and medulla and cortical R1], oxygenation (medullary BOLD R2*) and macrostructure (kidney volume). The mGFR correlation to RARI was negative, indicating an increase in renal arterial resistance with kidney function decline (Table 7).
Table 7.
The correlation between mGFR and UACR and MRI biomarkers of kidney haemodynamics, micro- and macrostructure and oxygenation
| mGFR | UACR | UACR (bivariate analysis with mGFR + imaging variable) | ||||
|---|---|---|---|---|---|---|
| Variable | Pearson correlation (r) | P-value | Spearman correlation (r) | P-value | Pearson correlation (r2) | P-value |
| mGFR | –0.81 | <.0001 | 0.15 | .02 | ||
| Kidney haemodynamics | ||||||
| MAF (mL/min/1.73 m2) | 0.92 | <.0001 | –0.78 | <.0001 | 0.16 | .63 |
| PSV (cm/s) | 0.32 | .02 | –0.28 | .03 | 0.15 | .95 |
| EDV (cm/s) | 0.79 | <.0001 | –0.75 | <.0001 | 0.23 | .07 |
| RARI | –0.76 | <.0001 | 0.70 | <.0001 | 0.21 | .13 |
| ASL perfusion cortex (mL/min/100 g) | 0.70 | <.0001 | –0.65 | <.0001 | 0.15 | .51 |
| Global perfusion (mL/min/100 g) | 0.75 | <.0001 | –0.63 | <.0001 | 0.16 | .59 |
| Kidney macrostructure | ||||||
| Kidney volume (mL/1.73 m2) | 0.61 | <.0001 | –0.50 | <.0001 | 0.16 | .49 |
| Kidney oxygenation | ||||||
| BOLD R2* cortex (per s) | 0.07 | .59 | –0.11 | .41 | 0.14 | .82 |
| BOLD R2* medulla (per s) | 0.35 | .008 | –0.34 | .01 | 0.23 | .06 |
| Kidney microstucture | ||||||
| ADC cortex (10–3 mm2/s) | 0.48 | .0002 | –0.37 | .005 | 0.21 | .36 |
| ADC medulla (10–3 mm2/s) | 0.28 | .03 | –0.27 | .05 | 0.23 | .20 |
| R1 cortex (per s) | 0.78 | <.0001 | –0.82 | <.0001 | 0.36 | .003 |
| R1 medulla (per s) | 0.20 | .13 | –0.33 | .01 | 0.25 | .04 |
| IVIM D cortex (10−3 mm2/s) | 0.44 | .0007 | –0.27 | .04 | 0.30 | .03 |
| IVIM D medulla (10−3 mm2/s) | 0.35 | .007 | –0.23 | .08 | 0.24 | .15 |
| IVIM D* cortex (10−3 mm2/s) | –0.09 | .54 | –0.12 | .38 | 0.19 | .47 |
| IVIM D* medulla (10−3 mm2/s) | –0.00 | 1.00 | –0.18 | .19 | 0.15 | .44 |
| IVIM f cortex (%) | –0.17 | .23 | 0.09 | .54 | 0.37 | .005 |
| IVIM f medulla (%) | –0.21 | .13 | 0.17 | .22 | 0.19 | .17 |
| MTR cortex (%) | 0.23 | .09 | –0.26 | .05 | 0.15 | .74 |
| MTR medulla (%) | 0.13 | .34 | –0.14 | .30 | 0.15 | .82 |
A univariate linear regression was used to evaluate the linear association of each MRI biomarker with mGFR and UACR (mGFR or UACR as a dependent variable, MRI biomarker as an independent variable). Bivariate predictors of UACR were examined using mGFR as one variable and adding one MRI endpoint using linear regression only in the DKD subjects.
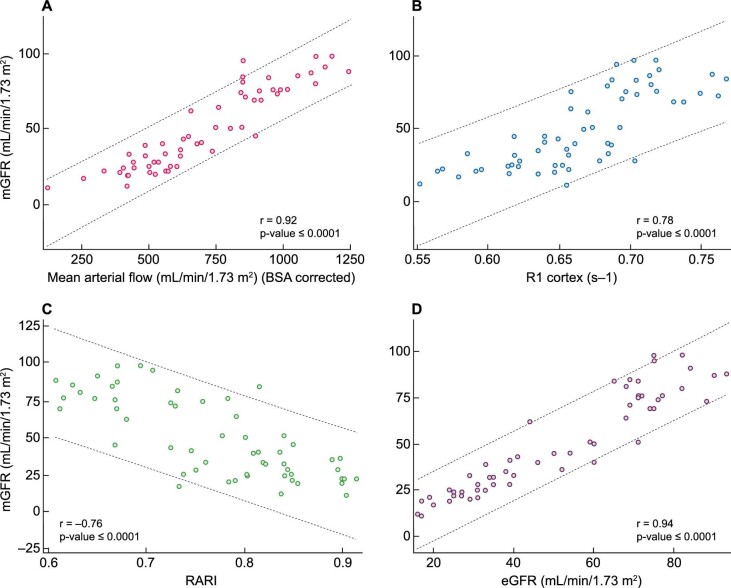
The strongest univariate MRI predictor of mGFR was MAF (r = 0.92; P < .0001) (Fig. 7). A slight improvement in the prediction occurred when combining MAF with ASL perfusion cortex, RARI, ADC cortex, R2* medulla, D or R1 cortex (r = 0.92–0.93), where all measurements with a P-value <.05 were included in the model.
Figure 7:
Scatter plots for mGFR versus MRI measures: (A) MAF, (B) cortical R1, (C) RARI and (D) eGFR.
The univariate analysis showed that several MRI biomarkers correlated significantly with UACR (Table 7). However, UACR correlated negatively with mGFR (Spearman correlation coefficient –0.81; P < .0001). Bivariate prediction of UACR using the linear regression with mGFR as one variable and adding one MRI endpoint showed that only R1 cortex, R1 medulla, D cortex and IVIM f cortex significantly improved prediction of UACR (Table 7). Many other parameters that initially correlated with UACR in the univariate analysis were no longer significant in the bivariate analysis (Table 7). In particular, haemodynamic parameters and kidney volume appeared to be linked to UACR via their effect on mGFR.
DISCUSSION
This is the first study, to our knowledge, evaluating the optimal sensitivity and specificity of an array of MRI biomarkers using mpMRI in DKD as opposed to a mixed CKD group [19, 30] and demonstrating intra-individual repeatability within DKD. Importantly, a robust, non-contrast mpMRI protocol was developed that allowed comprehensive evaluation of kidney haemodynamics, micro- and macrostructure and oxygenation and, as a single, non-invasive tool, distinguished HVs from DKD and various GFR stages (G3 versus G4/G5). Several MRI biomarkers correlated strongly with mGFR and/or UACR, indicating their biological relevance while providing additional information on underlying pathophysiology.
The MRI biomarkers with the strongest predictive properties to differentiate HVs from DKD subjects were specific for kidney haemodynamics, particularly MAF, RARI and EDV. In accordance with disease pathophysiology, MAF was lower in DKD subjects than HVs. Global perfusion and ASL measure kidney perfusion; both showed highly significant decreases in DKD subjects versus in HVs, as expected. Global perfusion, however, was greater than that reported by Buchanan et al. [19] in a non-diabetic CKD population, which may be explained by the fact that they calculated global perfusion using whole kidney volume rather than kidney parenchymal volume. Likewise, MRI biomarkers associated with kidney macrostructure (e.g. kidney volume) and microstructure (e.g. R1 and ADC in the cortex and R2* in the medulla) were able to differentiate HVs from DKD subjects. Not surprisingly, we found a significant decrease in parenchyma volume in DKD subjects versus HVs. Haemodynamic MRI biomarkers were also able to distinguish between stages G3 and G4/G5, particularly MAF, PSV, EDV and global perfusion, as was the microstructure MRI biomarker R1.
As far as we are aware, the haemodynamic MRI biomarkers EDV, PSV and RARI have only been evaluated in the renal arteries of healthy individuals [31]. In this study, EDV and RARI enabled HVs to be differentiated from DKD subjects, and both EDV and PSV could distinguish DKD subjects with stage G3 versus stages G4/G5. These are novel MRI biomarkers that may provide additional insights into DKD pathophysiology.
Some MRI biomarkers were not found to be directly related to kidney function, including IVIM parameters (D* and f) and the magnetization transfer ratio (MTR). The latter was reasonably repeatable, thereby showing potential for clinical trials; however, its correlation coefficient to mGFR was poor and it showed no significant correlations to other MRI, plasma or urine biomarkers when corrected for multiple comparisons. There are mixed positive [32] and negative [33] reports regarding the correlation of MTR with kidney fibrosis in animal models. The D* and f IVIM MRI measures were the least repeatable and may reflect kidney perfusion fluctuations during the cardiac cycle, and similar variability in these parameters has been demonstrated [34, 35]. Their inability to predict CKD may reflect a lack of precision per se as opposed to being unimportant for the underlying biology.
Most MRI biomarkers in our HV group were similar to those reported previously (Table 8) with some differences. In our HVs, MAF was slightly lower than that reported for a healthy population [36], although our subjects were 67 ± 6 years old and blood flow is expected to decrease with age. Others have even reported much lower values in healthy individuals [34]. Our kidney macrostructure evaluation in HVs shows a total parenchyma for both kidneys slightly smaller than the whole kidney volume reported in live kidney donors using computed tomography (CT) [37] and that of the total cortical volume in another CT study [38]. The slightly smaller volume may be due to the older age of HVs compared with kidney donors in the aforementioned study [37] and the well-known reduction in kidney size with age [39]. Moreover, we measured parenchyma volume, not total kidney volume. Overall, these data are consistent with previous studies in healthy populations and confirm our MRI protocol validity.
Table 8.
Comparison of MRI biomarkers between study values and literature values
| Study reference | Current study | Buchanan et al. [19] | de Boer et al. [34] | |||||
|---|---|---|---|---|---|---|---|---|
| Study subject summary | 20 HVs, median age 67.0 years (IQR 52.0–78.0) | 36 DKD, stages G3‒G5 | 20 HVs + 36 DKD | 22 CKD subjects, stages G3 and G4 | 19 HVs, median age 49.0 years (IQR 45.0–57.0) | |||
| Variable | Unit | Mean ± SD | Mean ± SD | CV (%) | Mean ± SD or median (IQR) | CV (%) | Mean ± SD | CV (%) |
| Kidney haemodynamics | ||||||||
| MAF | mL/min/1.73 m2 | 993 ± 126a | 538 ± 161a | 7 | 490 (170)b | 18 | 388 ± 147c | 13 |
| ASL perfusion cortex | mL/min/100 g | 164 ± 36.8 | 81.1 ± 40.8 | 33 | 71 (50) | 23 | 340 ± 51 | 10 |
| Global perfusion | mL/min/100 g | 458 ± 53.8 | 311 ± 82.9 | 9 | 130 (110) | 18 | Not measured | Not measured |
| Kidney macrostructure | ||||||||
| Kidney volume | mL/1.73 m2 | 218 ± 25.7 | 176 ± 37.7 | 7 | 170 (39)b | 3.8 | Not measured | Not measured |
| Kidney oxygenation | ||||||||
| BOLD R2* cortex | per s | 17.3 ± 1.35 | 17.1 ± 1.41 | 4 | 20.0 ± 3.2 | 4.6 | 19.3 ± 2.1 | 6.1 |
| BOLD R2* medulla | per s | 26.0 ± 2.31 | 23.5 ± 3.70 | 5 | 33.0 ± 8.0 | 6.8 | 26.2 ± 2.7 | 5.8 |
| Kidney microstructure | ||||||||
| ADC cortex | 10−3 mm2/s | 2.52 ± 0.19 | 2.31 ± 0.21 | 6 | 2.0 (0.2) | 5.3 | Not measured | Not measured |
| ADC medulla | 10−3 mm2/s | 2.33 ± 0.18 | 2.19 ± 0.24 | 5 | 2.0 ± 0.2 | 14 | Not measured | Not measured |
| R1 cortex | per s | 0.72 ± 0.03 | 0.63 ± 0.04 | 2 | 0.64 ± 0.06 | 2.9 | 0.66 ± 0.05 | 5.1 |
| R1 medulla | per s | 0.55 ± 0.02 | 0.55 ± 0.02 | 2 | 0.57 ± 0.03 | 3.9 | 0.53 ± 0.03 | 2.8 |
| IVIM D cortex | 10−3 mm2/s | 2.14 ± 0.24 | 1.91 ± 0.26 | 11 | 1.7 ± 0.2 | 7.7 | 2.1 ± 0.1 | 6.7 |
| IVIM D medulla | 10−3 mm2/s | 2.04 ± 0.27 | 1.85 ± 0.20 | 9 | 1.8 ± 0.2 | 22 | 1.9 ± 0.1 | 7.2 |
| IVIM f cortex | % | 13.5 ± 5.71 | 15.2 ± 5.70 | 30 | Not measured | Not measured | 10 ± 3 | 24 |
| IVIM f medulla | % | 12.3 ± 5.11 | 14.5 ± 5.66 | 25 | Not measured | Not measured | 13 ± 3 | 18 |
aTotal for both kidneys.
bNot corrected for body surface area.
cPer kidney.
We also tested the intra-individual repeatability of MRI biomarkers in both groups to evaluate them as monitoring biomarkers. Haemodynamic and kidney macrostructure measures (specifically R1 and ADC in the cortex and R2* in the medulla) had good repeatability, while ASL was less repeatable. This is the first study to demonstrate intra-individual repeatability of an mpMRI protocol in DKD, suggesting that these MRI biomarkers are well suited for clinical trials.
Some MRI biomarkers correlated with established biochemical surrogate biomarkers of kidney function and damage, namely GFR and UACR [40]. The GFR is the volume of fluid filtered from glomeruli, representing plasma flow from the glomerulus into Bowman's space. The tight coupling between MAF and mGFR in our study is therefore predictable [41], albeit greater than that found in a mixed CKD population [19], and reflects the precision of this imaging technique. Indeed, the strong correlation between MAF and mGFR made it difficult to meaningfully improve mGFR prediction by adding an independent imaging predictor in a bivariable model. Both global perfusion and ASL were also highly correlated with mGFR, although intra-individual repeatability for global perfusion was much better than for ASL. The purpose of this work is not to replace readily available GFR measurement routines with an expensive imaging technique, but to provide additional mechanistic data that may be useful to understand the pathophysiology of the underlying disease as well as the efficacy/mechanisms of action of novel drugs.
EDV, RARI and cortical R1 also correlated with mGFR here, such that EDV decreased and RARI (resistance) increased with declining kidney function. RARI measured with Doppler ultrasound increases with and is an independent risk factor for worsening kidney function in both CKD and DKD [42–44]. The sodium-glucose cotransporter 2 inhibitor (SGLT2i) dapaglifozin significantly reduces RARI measured by ultrasound in individuals with T2DM [45]. Given the good repeatability of EDV and RARI here, these MRI biomarkers may find application in clinical trials of novel therapies designed to reduce kidney fibrosis and subsequent renal arterial resistance.
Several MRI biomarkers, including MAF and cortical R1, correlated significantly with UACR. Buchanan et al. [19] also saw significant correlations between cortical T1 (T1 = 1/R1), MAF and several other MRI biomarkers with log urine protein:creatinine ratio (UPCR). Since UACR or UPCR correlated with mGFR in both our study and that of Buchannan et al. [19], we checked if a bivariate model of UACR combining an MRI measure with mGFR could statistically improve UACR prediction compared with mGFR alone. Only R1 (both cortical and medullary) and cortical IVIM D and f values remained significant UACR predictors. Thus they may be valuable tools to understand and monitor kidney damage beyond the decline in kidney function as DKD progresses.
Blood flow and oxygen levels in the kidney cortex and medulla are seriously impaired in moderate–severe kidney disease [46], and kidney tissue hypoxia is regarded as the common pathway for all CKD. Since renal blood flow influences both oxygen supply and demand, evaluating oxygen levels independently of perfusion is essential [47]. BOLD MRI is the only non-invasive method to measure kidney oxygen levels, and a high kidney cortical R2* (corresponding to low oxygenation) by BOLD MRI was shown to predict poor outcomes in some studies [15, 48, 49]. In our study, using mpMRI, cortical R2* did not correlate with GFR or UACR, in line with other studies [19, 50]. Some of the MRI biomarkers may be more suitable as prognostic biomarkers. However, due to the cross-sectional nature of this part of our study, we were unable to explore any prognostic correlation.
Our study has several strengths. A comprehensive, non-contrast mpMRI protocol allowed investigation of multiple aspects of kidney function and structure in a single 50-min sitting. A well-characterized population of individuals with T2DM and DKD and HVs were included. A standardized hydration protocol before MRI ensured uniform hydration in all. Additionally, mGFR enabled an accurate evaluation of kidney function. Finally, repeat MRI in half of the subjects allowed evaluation of the repeatability of MRI biomarkers. Limitations include a lack of a histologic ‘gold standard’ to validate MRI findings, a lack of generalizability to non-Swedish populations and the inclusion of DKD subjects based only on a clinical diagnosis. Thus some subjects may have had DKD together with hypertensive nephrosclerosis due to coexisting hypertension, as often occurs in a real-world setting. We also want to point out that in the absence of a non-diabetic CKD population, our findings may not be specific for DKD but may represent CKD in general.
In conclusion, for the first time a comprehensive and robust non-contrast mpMRI protocol was developed that allows non-invasive functional and structural evaluation of DKD. This novel approach has the potential to provide valuable insights into underlying pathophysiology, disease progression and analysis of efficacy/mode of action of therapeutic interventions in DKD and therewith improve management and prognosis of these patients. Studies assessing the influence of interventions such as SGLT2i and renin–angiotensin–aldosterone system blockers on MRI biomarkers are currently under way. The ongoing longitudinal part of our study will explore prognostic associations between MRI biomarkers and disease progression to provide further insights into DKD pathogenesis and risk stratification. Further studies with biopsy validation will be important to investigate whether our mpMRI protocol can non-invasively identify other/additional non-DKDs in patients presumed to have DKD, a question of high clinical relevance.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Erica De Coursey and Stig Eriksson at the MRI Unit, Department of Radiology, SU, Gothenburg, Sweden for performing mpMRI examinations; Lotta Sundström, Inger Olander and all physicians at Njurmedicin, SU, Gothenburg, Sweden and subjects who participated in this study. We are thankful to Eleanor Cox from the Sir Peter Mansfield Imaging Centre, Nottingham, UK, for providing software for analysis of T1 and ASL images and to Grażyna Söderbom for her technical editorial assistance.
Contributor Information
Kianoush Makvandi, Department of Molecular and Clinical Medicine/Nephrology, Institute of Medicine, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden.
Paul D Hockings, Antaros Medical, Molndal, Sweden; MedTech West, Chalmers University of Technology, Gothenburg, Sweden.
Gert Jensen, Department of Molecular and Clinical Medicine/Nephrology, Institute of Medicine, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden.
Tim Unnerstall, Department of Radiology, Sahlgrenska University Hospital, Gothenburg, Sweden.
Henrik Leonhardt, Department of Radiology, Sahlgrenska University Hospital, Gothenburg, Sweden.
Lisa V Jarl, Antaros Medical, Molndal, Sweden.
Camilla Englund, Antaros Medical, Molndal, Sweden.
Susan Francis, Sir Peter Mansfield Imaging Centre, University of Nottingham, Nottingham, UK.
Anna K Sundgren, Late-Stage Development, Cardiovascular, Renal and Metabolism, BioPharmaceuticals R&D, AstraZeneca, Gaithersburg, MD, USA.
Johannes Hulthe, Antaros Medical, Molndal, Sweden.
Seema Baid-Agrawal, Department of Molecular and Clinical Medicine/Nephrology, Institute of Medicine, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden; Transplant Center, Sahlgrenska University Hospital, Gothenburg, Sweden.
CONFLICT OF INTEREST STATEMENT
S.B.A. received research support for this study from Antaros Medical and AstraZeneca. P.D.H., L.V.J., C.E. and J.H. are employees of Antaros Medical. A.K.S. is an employee of AstraZeneca. All other authors declare that there are no relationships or activities that might bias, or be perceived to bias, their contributions to this article and the results presented herein have not been published previously in whole or part, except in abstract format.
FUNDING
The study was funded by Antaros Medical and AstraZeneca.
AUTHORS’ CONTRIBUTIONS
All named authors meet the International Committee of Medical Journal Editors’ criteria for authorship for this article. All authors assisted in the concept and design of the study, providing intellectual content of critical importance, revised and gave approval for the final version of the manuscript to be published. S.B.A. is the principal investigator. She participated in data acquisition, analysis and interpretation, supervision and conduct of the overall study, writing of the original draft of the article with input from others. K.M. participated in data acquisition, analysis and interpretation and writing of the original draft of the article with input from others. P.D.H. developed the MRI protocol together with S.F., participated in data analysis and interpretation and contributed to writing of the original draft of the article. G.E. participated in data acquisition, analysis and interpretation. T.U. and H.L. participated as radiologists in the MRI data analysis and interpretation and identified and reported incidental findings on MRI scans. S.F. developed the MRI protocol and participated in data analysis and interpretation.
DATA AVAILABILITY STATEMENT
The data underlying this article cannot be shared publicly to protect the privacy of individuals who participated in the study. The data will be shared upon reasonable request to the corresponding author.
REFERENCES
- 1. Fu H, Liu S, Bastacky SIet al. Diabetic kidney diseases revisited: a new perspective for a new era. Mol Metab 2019; 30: 250–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alicic RZ, Rooney MT, Tuttle KR. Diabetic kidney disease: challenges, progress, and possibilities. Clin J Am Soc Nephrol 2017; 12: 2032–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Luis-Lima S, Porrini E. An overview of errors and flaws of estimated GFR versus true GFR in patients with diabetes mellitus. Nephron 2017; 136: 287–91 [DOI] [PubMed] [Google Scholar]
- 4. Stevens LA, Levey AS. Measured GFR as a confirmatory test for estimated GFR. J Am Soc Nephrol 2009; 20: 2305–2313 [DOI] [PubMed] [Google Scholar]
- 5. Jufar AH, Lankadeva YR, May CNet al. Renal functional reserve: from physiological phenomenon to clinical biomarker and beyond. Am J Physiol Regul Int Comp Physiol 2020; 319: R690–R702 [DOI] [PubMed] [Google Scholar]
- 6. Waikar SS, Rebholz CM, Zheng Zet al. Biological variability of estimated GFR and albuminuria in CKD. Am J Kidney Dis 2018; 72: 538–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Inker LA, Levey AS, Pandya Ket al. Early change in proteinuria as a surrogate end point for kidney disease progression: an individual patient meta-analysis. Am J Kidney Dis 2014; 64: 74–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tuttle KR, Bakris GL, Bilous RWet al. Diabetic kidney disease: a report from an ADA Consensus Conference. Diabetes Care 2014; 37: 2864–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pugliese G, Penno G, Natali Aet al. Diabetic kidney disease: new clinical and therapeutic issues. Joint position statement of the Italian Diabetes Society and the Italian Society of Nephrology on “The natural history of diabetic kidney disease and treatment of hyperglycemia in patients with type 2 diabetes and impaired renal function”. Nutr Metab Cardiovasc Dis 2019; 29: 1127–1150 [DOI] [PubMed] [Google Scholar]
- 10. Tervaert TW, Mooyaart AL, Amann Ket al. Pathologic classification of diabetic nephropathy. J Am Soc Nephrol 2010; 21: 556–563 [DOI] [PubMed] [Google Scholar]
- 11. Luciano RL, Moeckel GW. Update on the Native Kidney Biopsy: Core Curriculum 2019. Am J Kidney Dis 2019; 73: 404–415 [DOI] [PubMed] [Google Scholar]
- 12. Stanton RC. Clinical challenges in diagnosis and management of diabetic kidney disease. Am J Kidney Dis 2014; 63: S3–S21 [DOI] [PubMed] [Google Scholar]
- 13. Selby NM, Blankestijn PJ, Boor Pet al. Magnetic resonance imaging biomarkers for chronic kidney disease: a position paper from the European Cooperation in Science and Technology Action PARENCHIMA. Nephro/Dial Transplant 2018; 33: ii4–ii14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mahmoud H, Buchanan C, Francis STet al. Imaging the kidney using magnetic resonance techniques: structure to function. Curr Opin Nephrol Hypertens 2016; 25: 487–493 [DOI] [PubMed] [Google Scholar]
- 15. Pruijm M, Milani B, Pivin Eet al. Reduced cortical oxygenation predicts a progressive decline of renal function in patients with chronic kidney disease. Kidney Int 2018; 93: 932–940 [DOI] [PubMed] [Google Scholar]
- 16. Villa G, Ringgaard S, Hermann Iet al. Phase-contrast magnetic resonance imaging to assess renal perfusion: a systematic review and statement paper. Magn Reson Mater Phys Biol Med 2020; 33: 3–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wolf M, de Boer A, Sharma Ket al. Magnetic resonance imaging T1- and T2-mapping to assess renal structure and function: a systematic review and statement paper. Nephrol Dial Transplant 2018; 33: ii41–ii50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Caroli A, Pruijm M, Burnier Met al. Functional magnetic resonance imaging of the kidneys: where do we stand? The perspective of the European COST Action PARENCHIMA. Nephrol Dial Transplant 2018; 33: ii1–ii3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Buchanan CE, Mahmoud H, Cox EFet al. Quantitative assessment of renal structural and functional changes in chronic kidney disease using multi-parametric magnetic resonance imaging. Nephrol Dial Transplant 2020; 35: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cakmak P, Yagci AB, Dursun Bet al. Renal diffusion-weighted imaging in diabetic nephropathy: correlation with clinical stages of disease. Diagn Interv Radiol 2014; 20: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Feng YZ, Ye YJ, Cheng ZYet al. Non-invasive assessment of early stage diabetic nephropathy by DTI and BOLD MRI. Br J Radiol 2020; 93: 20190562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mora-Gutierrez JM, Garcia-Fernandez N, Slon Roblero MFet al. Arterial spin labeling MRI is able to detect early hemodynamic changes in diabetic nephropathy. J Magn Reson Imaging 2017; 46: 1810–1817 [DOI] [PubMed] [Google Scholar]
- 23. Brown RS, Sun MRM, Stillman IEet al. The utility of magnetic resonance imaging for noninvasive evaluation of diabetic nephropathy. Nephrol Dial Transplant 2020; 35: 970–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Levey AS, Stevens LA, Schmid CHet al. A new equation to estimate glomerular filtration rate. Ann Intern Med 2009; 150: 604–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Levey AS, de Jong PE, Coresh Jet al. The definition, classification, and prognosis of chronic kidney disease: a KDIGO Controversies Conference Report. Kidney Int 2011; 80: 17–28 [DOI] [PubMed] [Google Scholar]
- 26. Krutzen E, Back SE, Nilsson-Ehle Iet al. Plasma clearance of a new contrast agent, iohexol: a method for the assessment of glomerular filtration rate. J Lab Clin Med 1984; 104: 955–961 [PubMed] [Google Scholar]
- 27. Fluss R, Faraggi D, Reiser B. Estimation of the Youden Index and its associated cutoff point. Biom J 2005; 47: 458–472 [DOI] [PubMed] [Google Scholar]
- 28. Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol Bull 1979; 86: 420–428 [DOI] [PubMed] [Google Scholar]
- 29. Gu S, Wang A, Ning Get al. Insulin resistance is associated with urinary albumin-creatinine ratio in normal weight individuals with hypertension and diabetes: the REACTION study. J Diabetes 2020; 12: 406–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cox EF, Buchanan CE, Bradley CRet al. Multiparametric renal magnetic resonance imaging: validation, interventions, and alterations in chronic kidney disease. Front Physiol 2017; 8: 696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Keegan J, Patel HC, Simpson RMet al. Inter-study reproducibility of interleaved spiral phase velocity mapping of renal artery haemodynamics. J Cardiovasc Magn Reson 2015; 17: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jiang K, Ponzo TA, Tang Het al. Multiparametric MRI detects longitudinal evolution of folic acid-induced nephropathy in mice. Am J Physiol Renal Physiol 2018; 315: F1252–F1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li A, Xu C, Liang Pet al. Role of chemical exchange saturation transfer and magnetization transfer MRI in detecting metabolic and structural changes of renal fibrosis in an animal model at 3T. Korean J Radiol 2020; 21: 588–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de Boer A, Harteveld AA, Stemkens Bet al. Multiparametric renal MRI: an intrasubject test-retest repeatability study. J Magn Reson Imaging 2021; 53: 859–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wittsack HJ, Lanzman RS, Quentin Met al. Temporally resolved electrocardiogram-triggered diffusion-weighted imaging of the human kidney: correlation between intravoxel incoherent motion parameters and renal blood flow at different time points of the cardiac cycle. Invest Radiol 2012; 47: 226–230 [DOI] [PubMed] [Google Scholar]
- 36. Field Mj, Pollock CA, Harris DC. Glomerular filtration in acute kidney injury. In: The Renal System: Basic Science and Clinical Conditions. New York: Churchill Livingstone, 2010; 57–68 [Google Scholar]
- 37. Fananapazir G, Benzl R, Corwin MTet al. Predonation volume of future remnant cortical kidney helps predict postdonation renal function in live kidney donors. Radiology 2018; 288: 153–157 [DOI] [PubMed] [Google Scholar]
- 38. Halleck F, Diederichs G, Koehlitz Tet al. Volume matters: cT-based renal cortex volume measurement in the evaluation of living kidney donors. Transpl Int 2013; 26: 1208–1216 [DOI] [PubMed] [Google Scholar]
- 39. Emamian SA, Nielsen MB, Pedersen JFet al. Kidney dimensions at sonography: correlation with age, sex, and habitus in 665 adult volunteers. Am J Roentgenol 1993; 160: 83–86 [DOI] [PubMed] [Google Scholar]
- 40. Levey AS, Gansevoort RT, Coresh Jet al. Change in albuminuria and GFR as end points for clinical trials in early stages of CKD: a scientific workshop sponsored by the National Kidney Foundation in collaboration with the US Food and Drug Administration and European Medicines Agency. Am J Kidney Dis 2020; 75: 84–104 [DOI] [PubMed] [Google Scholar]
- 41. Clodagh M, Martyn CS, Dhanda R. Renal physiology: blood flow, glomerular filtration and plasma clearance. Anaesth Intensive Care Med 2018; 19: 254–257 [Google Scholar]
- 42. Hanamura K, Tojo A, Kinugasa Set al. The resistive index is a marker of renal function, pathology, prognosis, and responsiveness to steroid therapy in chronic kidney disease patients. Int J Nephrol 2012; 2012: 139565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim JH, Lee SM, Son YKet al. Resistive index as a predictor of renal progression in patients with moderate renal dysfunction regardless of angiotensin converting enzyme inhibitor or angiotensin receptor antagonist medication. Kidney Res Clin Pract 2017; 36: 58–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sistani SS, Alidadi A, Moghadam AAet al. Comparison of renal arterial resistive index in type 2 diabetic nephropathy stage 0–4. Eur J Transl Myol 2019; 29: 8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Solini A, Giannini L, Seghieri Met al. Dapagliflozin acutely improves endothelial dysfunction, reduces aortic stiffness and renal resistive index in type 2 diabetic patients: a pilot study. Cardiovasc Diabetol 2017; 16: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pruijm M, Milani B, Burnier M. Blood oxygenation level-dependent MRI to assess renal oxygenation in renal diseases: progresses and challenges. Front Physiol 2016; 7: 667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bane O, Mendichovszky IA, Milani Bet al. Consensus-based technical recommendations for clinical translation of renal BOLD MRI. Magn Reson Mater Phys Biol Med 2020; 33: 199–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sugiyama K, Inoue T, Kozawa Eet al. Reduced oxygenation but not fibrosis defined by functional magnetic resonance imaging predicts the long-term progression of chronic kidney disease. Nephrol Dial Transplant 2020; 35: 964–970 [DOI] [PubMed] [Google Scholar]
- 49. Zhou H, Yang M, Jiang Zet al. Renal hypoxia: an important prognostic marker in patients with chronic kidney disease. Am J Nephrol 2018; 48: 46–55 [DOI] [PubMed] [Google Scholar]
- 50. Prasad PV, Li LP, Thacker JMet al. Cortical perfusion and tubular function as evaluated by magnetic resonance imaging correlates with annual loss in renal function in moderate chronic kidney disease. Am J Nephrol 2019; 49: 114–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article cannot be shared publicly to protect the privacy of individuals who participated in the study. The data will be shared upon reasonable request to the corresponding author.