Abstract
Isolation of Vibrio hollisae strains, particularly from the environment, is rare. This may be due, in part, to the difficulty encountered when using conventional biochemical tests to identify the microorganism. In this study, we evaluated whether two particular genes may be useful for the identification of V. hollisae. The two genes are presumed to be conserved among the bacterial species (gyrB) or among the species of the genus Vibrio (toxR). A portion of the gyrB sequence of V. hollisae was cloned by PCR using a set of degenerate primers. The sequence showed 80% identity with the corresponding Vibrio parahaemolyticus gyrB sequence. The toxR gene of V. hollisae was cloned utilizing a htpG gene probe derived from the V. parahaemolyticus htpG gene, which is known to be linked to the toxR gene in V. hollisae. The coding sequence of the cloned V. hollisae toxR gene had 59% identity with the V. parahaemolyticus toxR coding sequence. The results of DNA colony hybridization tests using the DNA probes derived from the two genes of V. hollisae indicated that these gene sequences could be utilized for differentiation of V. hollisae from other Vibrio species and from microorganisms found in marine fish. PCR methods targeting the two gene sequences were established. Both PCR methods were shown to specifically detect the respective target sequences of V. hollisae but not other organisms. A strain of V. hollisae added at a concentration of 1 to 102 CFU/ml to alkaline peptone water containing a seafood sample could be detected by a 4-h enrichment incubation in alkaline peptone water at 37°C followed by quick DNA extraction with an extraction kit and 35-cycle PCR specific for the V. hollisae toxR gene. We conclude that screening of seafood samples by this 35-cycle, V. hollisae toxR-specific PCR, followed by isolation on a differential medium and identification by the above htpG- and toxR-targeted PCR methods, can be useful for isolation from the environment and identification of V. hollisae.
Vibrio hollisae is a halophilic bacterium that was first described in 1982 by Hickman and collaborators (6). In the majority of cases reported thus far, V. hollisae has been isolated from clinical sources (1, 2, 5, 6, 13, 15, 18). Individuals who had no underlying illness and consumed seafood such as raw shellfish were infected in many cases. To our knowledge, there are only two reports on the isolation of V. hollisae from environmental samples, i.e., from a coastal fish and an oyster (9, 19).
The exact pathogenic mechanism of V. hollisae is not known. However, this organism has been shown to produce a hemolysin similar to that of the thermostable direct hemolysin of Vibrio parahaemolyticus (19, 32), which is considered a major enterotoxic factor of the organism (20). The gene encoding thermostable direct hemolysin (tdh) is distributed in a minor population of environmental strains of V. parahaemolyticus and rare strains of Vibrio mimicus and Vibrio cholerae (non-O1 strains) isolated from diarrheal patients (23). Unlike the findings with these three Vibrio species, all clinical and environmental strains of V. hollisae that others and a member of our group have examined so far carried the tdh gene (19, 21, 22). A hypothesis on past horizontal transfer of the tdh gene among Vibrio species was proposed (23). V. hollisae may have served as a source of the tdh gene in interspecies horizontal transfer between Vibrio species in the marine environment (22, 23). However, few environmental strains of V. hollisae were available for examination. This is a critical point of the hypothesis. To examine the hypothesis and to assess the public health significance of V. hollisae in the environment, it is imperative to isolate more strains of V. hollisae from the environment and examine them for the presence or absence of the tdh gene.
The paucity of the isolation of V. hollisae from clinical specimens may be due in part to poor growth of this bacterium on selective isolation media for enteric pathogens such as thiosulfate-citrate-bile salts-sucrose agar and MacConkey agar (1, 2, 5, 7). It is also biochemically inert, so definitive identification by conventional biochemical tests requires NaCl added to a final concentration of 1% (2, 6, 7) and some clinical strains fail to be identified by the API 20E system (1, 2). It is more difficult to isolate V. hollisae from the marine environment and identify it because the environmental samples contain many nonpathogenic Vibrio species and related but unestablished organisms. In addition, optimum culture conditions for isolation and identification may not be the same for clinical and environmental strains. The public health significance of V. hollisae may be underestimated due to the paucity of reports of isolation from both clinical and environmental sources. If there were a better method for isolation and identification of V. hollisae, it would be useful for isolation and subsequent study of this organism. In this study, we examined whether identification of V. hollisae by PCR is possible. For this purpose, the ideal situation was to target the nucleotide sequence that is conserved and that reflects phylogenetic relationship. The sequences of rRNA genes are usually utilized for this purpose, but the interspecies homology values of rRNA genes of Vibrio species are so high that these gene sequences are not suitable (11, 27). Recently, the gyrB gene encoding the B subunit of DNA gyrase was utilized for identification of V. parahaemolyticus by PCR (28). Members of our group and others demonstrated that the toxR gene sequence is also useful for the identification of V. parahaemolyticus by PCR (10). We presumed that the toxR gene is a global regulatory gene possessed by the members of the genus Vibrio because the toxR gene was detected in V. cholerae, V. parahaemolyticus, Vibrio fisheri, and Vibrio vulnificus and the gene sequences were analyzed (12, 14, 16, 26). We therefore cloned and sequenced the gyrB and toxR genes of V. hollisae and demonstrated that these gene sequences can be utilized for identification of V. hollisae by PCR. We then evaluated whether the established PCR method could be applied directly to seafood samples to facilitate screening of V. hollisae-bearing samples.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used in this study are listed in Table 1. The bacterial strains belonging to various species of the genus Vibrio were from our laboratory stock strains or strains supplied by other workers for this study. The gyrB gene and the toxR gene of V. hollisae were cloned from clinical strains 89A 1962 and 525-82 (1, 22), respectively. A spontaneous streptomycin-resistant mutant of V. hollisae 525-82, a clinical strain, was obtained by plating a concentrated overnight culture of the wild-type strain onto Luria-Bertani (LB) agar containing 50 μg of streptomycin per ml. The mutant strain was designated 525-82SmR.
TABLE 1.
Results of the tests for the gyrB and toxR genes
| Species | Sourcea or strain no. | Result of detectionb by:
|
|||
|---|---|---|---|---|---|
| DNA probec
|
PCR
|
||||
| gyrB gene | toxR gene | gyrB gened | toxR genee | ||
| Vibrio hollisae | C | + (11/11) | + (11/11) | + (11/11) | + (11/11) |
| RIMD 2221011f | + | + | + | + | |
| Vibrio cholerae O1 | C | +W(2/2) | − (0/2) | − (0/2) | − (0/2) |
| Vibrio cholerae O1 | NIH 35A3 | +W | − | − | − |
| Vibrio cholerae O139 | MO45 | +W | − | − | − |
| Vibrio cholerae non-O1, non-O139 | AM2 | +W | − | − | − |
| Vibrio parahaemolyticus | C | +W(4/4) | − (0/4) | − (0/4) | − (0/4) |
| Vibrio fluvialis | C | +W(2/2) | − (0/2) | − (0/2) | − (0/2) |
| Vibrio furnisii | RIMD 2223001 | +W | − | − | − |
| Vibrio damsela | C | +W(2/2) | − (0/2) | − (0/2) | − (0/2) |
| Vibrio metschnikovii | C | − (0/2) | − (0/2) | − (0/2) | − (0/2) |
| Vibrio mimicus | RIMD 2218001 | +W | − | − | − |
| Vibrio vulnificus | RIMD 2219009 | +W | − | − | − |
| Vibrio alginolyticus | E | +W(3/3) | − (0/3) | − (0/3) | − (0/3) |
| Vibrio cincinnatiensis | ATCC 35912 | +W | − | − | − |
| Vibrio carchariae | ATCC 35084 | +W | − | − | − |
| Vibrio anguillarum | PT-87050 | − | − | − | − |
| Vibrio ordalii | ATCC 33509 | +W | − | − | − |
| Vibrio campbellii | ATCC 25920 | +W | − | − | − |
| Vibrio harveyi | ATCC 14126 | +W | − | − | − |
| Vibrio nereis | ATCC 25917 | +W | − | − | − |
| Vibrio pelagius | ATCC 25916 | +W | − | − | − |
| Vibrio splendidus | ATCC 33125 | +W | − | − | − |
| Vibrio tubiashii | ATCC 19109 | − | − | − | − |
| Vibrio aestuarianus | ATCC 35048 | − | − | − | − |
| Vibrio mediterranei | ATCC 43341 | +W | − | − | − |
| Vibrio orientalis | ATCC 33934 | +W | − | − | − |
| Vibrio diazotrophicus | ATCC 33466 | +W | − | − | − |
| Vibrio gazogenes | ATCC 29988 | +W | − | − | − |
| Vibrio proteolyticus | NCMB 1326 | +W | − | − | − |
| Vibrio logei | ATCC 15382 | +W | − | − | − |
| Vibrio nigripulchritudo | ATCC 27043 | − | − | − | − |
| Vibrio navarrensis | ATCC 51183 | +W | − | − | − |
| Vibrio mytili | ATCC 51288 | − | − | − | − |
| Vibrio iliopiscarius | HUFP 9601 | +W | − | − | − |
| Vibrio ichthyoenteri | IFO 15847 | +W | − | − | − |
| Vibrio penaeicida | IFO 15640 | − | − | − | − |
| Unknown (non-Vibrio hollisae)g | E | NTh | − (0/74) | − (0/74) | − (0/74) |
| Escherichia coli | MC1061 | +W | − | − | − |
C, clinical source; E, environmental source.
+, detected; +W, weak reaction detected; −, not detected. For species of which multiple strains were tested, the number of positive strains/the number of tested strains is given in parentheses.
DNA probes specific to the V. hollisae gyrB and V. hollisae toxR genes (described in the text) were used.
Results obtained when primer pairs HG-F1 and HG-R2, HG-F2 and HG-R1, and HG-F2 and HG-R2 were employed.
Results obtained when primer pair HT-F3 and HT-R2 was employed.
Same as KUMA871, an isolate from a coastal fish (19).
Seventy-four strains isolated from marine fish (explained in the text).
NT, not tested.
The strains of unknown identification were isolated from marine fishes caught in a coastal area of Japan in the summer months of 1995 to 1997. The intestinal contents, skin surface, or gill surface of 10 species of coastal fish was smeared onto the agar medium with which V. hollisae was previously isolated from a coastal fish (19). The inoculated agar medium was incubated at 25°C, and representative purple colonies, i.e., mannitol and maltose nonfermentors, were selected. Some green but no yellow colonies were included for comparison. These strains were examined for selected biochemical characteristics by standard methods except that the NaCl concentration of the test medium was adjusted to 1% (7, 8). Escherichia coli MC1061 was described previously (3).
gyrB sequence determination.
The gyrB sequence was amplified by PCR using the PCR primers UP-1 and UP-2r designed by Yamamoto and Harayama (31). The 20-μl reaction mixture contained 6 μl of purified cellular DNA of V. hollisae 89A 1962 (0.175 μg/ml), 2 μl of each of the PCR primers (10 μM) 2 μl of 10× buffer containing 20 mM MgCl2 (ExTaq buffer; Takara, Shiga, Japan), 1 μl (0.5 U) of Taq polymerase (ExTaq; Takara), 2 μl of each deoxynucleotide triphosphate (2.5 mM), and 5 μl of distilled water. The amplification conditions were 30 cycles of amplification consisting of denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 2 min, followed by one cycle of 72°C for 7 min. Amplification was performed in a Genius thermal cycler (model FGENO5TY; Techne Ltd., Oxford, Cambridge, England). The amplicon of 1.3 kb was cloned into a vector plasmid, pGEM-T Easy (Promega Corp., Madison, Wis.), using E. coli MC1061 as the host. The nucleotide sequences of both strands of the cloned fragment were determined with the ABI-PRISM BigDye Terminator Cycle Sequencing Ready Reaction kit and an ABI-PRISM 310 genetic analyzer (Applied Biosystems Division, Perkin-Elmer, Foster City, Calif.)
toxR sequence determination.
The approach used in cloning the V. hollisae toxR gene was a design based on the possibility that the toxR gene and the htpG gene encoding a heat shock protein were located contiguously in the chromosome (discussed below). First, a htpG-bearing restriction fragment of cellular DNA of V. hollisae 525-82 was identified and cloned using the V. parahaemolyticus htpG-specific gene probe as follows. The KpnI digest of cellular DNA of V. hollisae 525-82 was size fractionated using agarose gel electrophoresis, and 5.9-kb DNA fragments were cloned into KpnI-cleaved pUC118 (29) in E. coli MC1061. The transformants containing the recombinant plasmid, pKON20, were detected by the DNA colony blot hybridization method under stringent conditions (21) with the V. parahaemolyticus htpG-specific probe.
Next, a toxR-bearing subfragment within the insert was sought. A restriction endonuclease map of the insert of pKON20 was constructed. The pKON20 was digested with appropriate restriction enzymes and was then examined by Southern blot hybridization with the DNA probe specific to the V. parahaemolyticus toxR gene under reduced-stringency conditions (with 31% formamide) as described previously (24). A 2.7-kb PstI-ScaI DNA fragment, one of the subfragments that gave a weak hybridization signal, was subcloned into the PstI-HincII sites of pUC119 vector plasmid (29), resulting in pKON22. The nucleotide sequence of the insert was determined as described above.
DNA probes.
One of the recombinant plasmids that were confirmed to carry the cloned V. hollisae gyrB sequence (explained below) was named pVAR1 and used for preparation of the V. hollisae gyrB-specific DNA probe. NotI recognition sites flanked the insertion sites in pVAR1. Therefore, pVAR1 was digested with NotI enzyme and the 1.3-kb insert was isolated and used as V. hollisae gyrB-specific probe DNA. To prepare a V. parahaemolyticus htpG-specific DNA probe, a 1.5-kb HindIII fragment internal to the V. parahaemolyticus htpG gene was isolated from a recombinant plasmid, pKTN170 (M. Nishibuchi, J. Okuda, and K. Kumagai, unpublished data.), and used as the probe DNA. The V. parahaemolyticus toxR-specific probe DNA which is internal to the V. parahaemolyticus toxR coding region was prepared by isolating a 687-bp DNA fragment from pKTN118 as described previously (14). The V. hollisae toxR-specific probe DNA was 349-bp amplicons prepared by PCR using pKON22 as the template and primers HT-F1 and HT-R2 (described below). The composition of the PCR reaction mixture and the amplification conditions were the same as those described below for the V. hollisae toxR-specific PCR. The probe DNAs were labeled by the random priming method with 32P-labeled dCTP (4).
DNA colony hybridization test.
The strains belonging to V. hollisae, V. cholerae, V. parahaemolyticus, Vibrio fluvialis, Vibrio furnisii, Vibrio damsela, Vibrio metchnikovii, V. mimicus, V. vulnificus, Vibrio alginolyticus, Vibrio anguillarum, and E. coli and those of unknown identification were grown overnight at 25°C using LB agar containing 1% NaCl. The other strains belonging to various species of the genus Vibrio were grown to reasonable colony sizes on marine agar 2216 (Difco Laboratories, Detroit, Mich.) at 25°C. DNA colony blots were prepared as described previously (21). The DNA colony hybridization test was performed under high-stringency conditions (in a solution containing 50% formamide) as described previously (21).
V. hollisae gyrB-specific PCR.
The strains belonging to V. hollisae, V. cholerae, V. parahaemolyticus, V. fluvialis, V. furnisii, V. damsela, V. metschnikovii, V. mimicus, V. vulnificus, V. alginolyticus, and E. coli were grown in LB broth containing 1% NaCl at 37°C with shaking (160 rpm) overnight. V. anguillarum PT-87050 and the strains of unknown identification were grown in LB broth containing 1% NaCl at 25°C with shaking (160 rpm) overnight. The other strains belonging to various species of the genus Vibrio were grown to good turbidity in marine broth 2216 (Difco Laboratories) at 25°C with shaking (160 rpm). One milliliter of the broth culture was boiled for 5 min, and the supernatant was obtained by centrifugation (13,000 rpm) on a tabletop centrifuge (Centrifuge 5415C; Eppendorf, Hamburg, Germany) at room temperature. The supernatant was diluted 10-fold in distilled water. The diluted supernatant was used as the template for PCR amplification.
The nucleotide sequence of a part of the V. hollisae gyrB gene determined in this study (described below) was compared with the corresponding V. parahaemolyticus gyrB sequence. The regions that were not well conserved between the two sequences were selected. The V. hollisae gyrB sequences of these regions were examined by a computer program for designing PCR primers (Oligo 4.05; National Bioscience, Inc., Plymouth, Minn.) Pairs of two forward and two reverse primers thus designed were evaluated. The forward primers were HG-F1 (5′-GCTCTGTCGGAAAAACTTGA-3′ [positions 100 to 119]) and HG-F2 (5′-CAGATTTATCGCATGGGTGT-3′ [positions 154 to 173]), and the reverse primers were HG-R1 (5′-TTGCATCGATACTTCTACTG-3′ [positions 501 to 482]) and HG-R2 (5′-ATGCTCAAAATGGAACACAG-3′ [positions 462 to 443]). The positions indicated above correspond to those of the gyrB sequence deposited in the DDJB, EMBL, and GenBank databases (see below).
The PCR reaction mixture consisted of 2 μl of DNA template (supernatant of the boiled culture diluted 1:10), 1 μl of 2.5 mM deoxynucleotide triphosphate, 5 μl of each of the primers (2 μM) 0.1 μl of Taq polymerase (Taq DNA polymerase in storage buffer A [5 U/μl] Promega Corp.) 2 μl of 10× buffer (thermophilic DNA polymerase 10× buffer, magnesium free; Promega Corp.), 1.2 μl of 25 mM MgCl2, and 3.7 μl of distilled water. The amplification conditions were 30 cycles of amplification consisting of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min, followed by one cycle of 72°C for 7 min. Amplification was performed in a Genius thermal cycler (model FGENO5TY). Ten microliters of the reaction mixture was resolved by electrophoresis in 2% agarose gel to detect the amplicon of the expected size.
V. hollisae toxR-specific PCR.
The supernatant of the boiled culture of the test strain was obtained and diluted as described for the V. hollisae gyrB-specific PCR above, and the diluted supernatant was used as the DNA template. The broth culture obtained in the spike experiment (described below) was boiled, diluted (1:10), and used as the DNA template in the same manner unless otherwise specified.
The nucleotide sequence of the V. hollisae toxR gene determined in this study (described below) was compared with those of the V. parahaemolyticus toxR, V. cholerae toxR, and V. fischeri toxR genes (14, 16, 26). Three forward primers and two reverse primers specific to the V. hollisae toxR sequence were selected from the regions not conserved among the toxR sequences of four species. The forward primers were HT-F1 (5′-GAGACCGCAGCGAAAACGGAG-3′ [positions 391 to 411]), HT-F2 (5′-AACGGAGCCTGAGCTTCTTTCC-3′ [positions 405 to 426]), and HT-F3 (5′-CTGCCCAGACACTCCCTCTTC-3′ [positions 434 to 454]). The reverse primers were HT-R1 (5′-GCTGCCATTGAGAAATGCGTGC-3′ [positions 685 to 664]) and HT-R2 (5′-CTCTTTCCTTACCATAGAAACCG-3′ [positions 739 to 717]). The positions indicated above correspond to those of the toxR sequence deposited in the DDJB, EMBL, and GenBank databases (see below).
The PCR reaction mixture consisted of 10 μl of DNA template (diluted supernatant of the boiled culture [1:10]), 5 μl of 10× buffer containing 20 mM MgCl2 (ExTaq buffer; Takara), 4 μl of 2.5 mM deoxynucleotide triphosphate, 1 μl of each primer (20 μM) 0.25 μl of Taq polymerase (ExTaq, Takara), and 28.75 μl of distilled water. The amplification conditions were, unless otherwise specified, 24 cycles of amplification consisting of denaturation at 95°C for 1 min, annealing at 62°C for 1.5 min, and extension at 72°C for 1.5 min, followed by one cycle of 72°C for 5 min. Amplification was performed in a DNA Thermal Cycler 480 (Perkin-Elmer Co.). Ten microliters of the reaction mixture was resolved by electrophoresis in 2% agarose gel to detect the amplicon of the expected size.
Spike experiments.
A 40-g seafood sample was put into 400 ml of alkaline-peptone water (APW; 1% peptone, pH 8.6), the mixture was incubated at 37°C for 5 to 10 min and homogenized by vigorous shaking for 2 min, and 30-ml aliquots of the homogenate were placed in sterile containers. V. hollisae 525-82SmR grown in LB broth to the mid-log phase was added to the aliquoted homogenate at the concentrations of 0, 10, 102, 103, 104, 105, and 106 CFU/ml, and the homogenate was mixed well. Samples for PCR assay (1 ml) and for plate count (1 ml) were taken immediately as zero-hour samples, and the rest were incubated at 37°C without shaking. Samples for the PCR assay and plate count were taken again after 4 h of incubation, and then the rest were incubated at 15°C. The samples for plate count were also obtained 4.5 h later from the samples incubated at 15°C. Unless otherwise specified, the sample for PCR assay was subjected to DNA extraction by the boiling method described above for the V. hollisae toxR-specific PCR.
Mixtures of selected homogenate and 525-82SmR after 4 h of enrichment incubation at 37°C were also used to compare the DNA extraction methods for PCR temperate preparation. Triplicate 1-ml samples were prepared for each test. One of the samples was treated by the boiling method described above. The remaining two samples were centrifuged (14,000 rpm) on the tabletop centrifuge (model 5415C, Eppendorf) for 5 min, the pellet was obtained and suspended in 0.1 ml of saline, and DNA was extracted and dissolved in 0.1 ml of distilled water. DNA was extracted by a method based on chaotropic extraction with NaI followed by ethanol precipitation using a DNA extraction kit (code no. 295-50201; Wako Pure Chemical Industries, Ltd., Osaka, Japan) or by chaotropic extraction followed by DNA absorption onto silica-coated magnetic beads (MagExtractor-Genome-; TOYOBO Co. Ltd., Osaka, Japan). These DNA extractions were performed according to the manufacturers' specifications.
PCR amplification with primer pair HT-F3 and HT-R2 was carried out as described for the V. hollisae toxR-specific PCR except that the total reaction volume was reduced to 20 μl (the volume of each component was reduced accordingly), Taq polymerase (ExTaq; Takara) was replaced by Taq DNA polymerase in storage buffer A (Promega Corp.), and not only 24 but also 35 thermal cycles were employed.
The sample for plate count was diluted appropriately in sterile saline and 0.1 ml of the sample was inoculated in triplicate onto the isolation medium (19) and onto the same medium with 100 μg of streptomycin per ml. Purple colonies on the agar medium were counted after overnight incubation at 37°C. The average colony count per sample was determined. When needed, representative purple colonies (up to 20 colonies per plate) were examined by V. hollisae toxR-specific PCR as described above.
Nucleotide sequence accession numbers.
The nucleotide sequence data of the gyrB and toxR genes of V. hollisae reported in this paper will appear in the DDJB, EMBL, and GenBank nucleotide sequence databases with the accession numbers AB027462 and AB027503, respectively.
RESULTS
gyrB gene sequence analysis.
We were able to amplify a selected region of the V. hollisae gyrB gene by applying a set of degenerate, universal PCR primers to the cellular DNA of V. hollisae 89A 1962. The amplified sequence was 1259 bp long and fairly homologous with the corresponding sequences of the reported gyrB genes of gram-negative bacteria (62 to 80% identity values). The V. parahaemolyticus gyrB sequence exhibited the highest overall identity value. The results indicated that the cloned sequence is the expected region of the V. hollisae gyrB gene.
toxR gene sequence analysis.
A 2.7-kb PstI-ScaI fragment was cloned from the cellular DNA of V. hollisae 525-82 as described in Materials and Methods, and the nucleotide sequence was determined. The sequence contained an open reading frame of 292 amino acids, and this open reading frame was identical in length with that of the V. parahaemolyticus toxR coding sequence (14). The two nucleotide sequences shared 59% nucleotide sequence identity. The identity value was almost comparable to that between the V. cholerae toxR gene and the V. parahaemolyticus toxR gene. Deduced amino acid sequences of V. parahaemolyticus ToxR and V. cholerae ToxR shared a region of high homology (80% identity) at the N terminus, which was presumed to be a DNA binding domain, although the identity over the entire sequence was 55% (14). The identities for the putative DNA binding domain region (91 residues) and the entire coding region between the amino acid sequences deduced from the V. hollisae open reading frame and from the V. parahaemolyticus toxR gene were 68 and 47%, respectively (data not shown). We therefore concluded that the open reading frame of V. hollisae 525-82 is the coding region of the V. hollisae toxR gene.
Examination of Vibrio strains by V. hollisae gyrB and V. hollisae toxR probes.
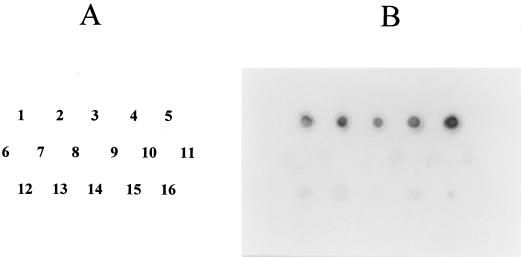
DNA fragments internal to the V. hollisae gyrB and toxR genes were employed as the probe DNAs in DNA colony hybridization tests to examine the distribution of homologous sequences in the strains belonging to 33 species of genus Vibrio, and 74 strains of unknown identification (Table 1). The strains of unknown identification were isolated from coastal fishes using the medium designed for isolation of V. hollisae. However, the results of the tests for selected biochemical characteristics, which appear to be important for identification of V. hollisae strains (6, 8), indicated that these strains are likely to be distinct from V. hollisae (Table 2). V. hollisae strains exhibited strong hybridization signals with the V. hollisae gyrB-specific probe, whereas most strains belonging to other Vibrio spp. and E. coli MC1061 showed distinctly weaker hybridization signals and the other strains of Vibrio spp. exhibited no reaction. The representative hybridization signals are shown in Fig. 1. On the other hand, only V. hollisae strains demonstrated hybridization signals with the V. hollisae toxR-specific probe. No other test strains, including the presumed non-V. hollisae strains of fish origin, showed any sign of hybridization with this probe. The representative hybridization patterns are presented in Fig. 2.
TABLE 2.
Comparison of the biochemical characteristics of the strains isolated from marine fish with those of V. hollisae strainsa
| Species | No. of strains | Color of the colony on the isolation mediumb | TSI | H2S (on TSI) | Oxidase | Motility | Lysine decarboxylase | Arginine dihydrolase | Ornithine decarboxylase | Indole |
|---|---|---|---|---|---|---|---|---|---|---|
| V. hollisae | 2c | Purple | K/Ad | − | + | − | − | − | − | + |
| Unknowne | 5 | Purple | K/A | − | + | − | − | − | − | − |
| 3 | Purple | K/A | − | + | − | − | + | − | − | |
| 25 | Purple | K/A | − | + | − | − | + | − | − | |
| 1 | Purple | K/A | − | − | + | − | + | − | − | |
| 15 | Purple | K/A | − | − | − | − | + | − | − | |
| 1 | Purple | K/A | − | − | − | − | − | − | − | |
| 2 | Purple | K/A | − | − | − | + | + | − | − | |
| 13 | Green | K/A | − | + | − | − | + | − | − | |
| 6 | Green | K/A | − | + | − | + | + | + | − | |
| 3 | Green | K/A | + | + | − | + | + | + | − |
TSI, triple sugar iron.
The agar medium designed for isolation of V. hollisae (19).
A clinical strain, 525-82, and an environmental strain, RIMD 2221011. The characteristics of these strains were consistent with those of V. hollisae described in references 7, 9, and 19.
K/A, alkaline top (red) and acid bottom (yellow).
Seventy-four strains isolated from marine fish (explained in the text).
FIG. 1.
DNA colony hybridization test with the V. hollisae gyrB gene probe for strains of V. hollisae, other Vibrio species, and E. coli. (A) Location of the inoculated strains (numbers correspond to dot positions in panel B): 1 to 5, V. hollisae strains 89A 1961, 89A 1960, 89A 4206, 91A 2120, and RIMD 2221011, respectively; 6, V. cholerae O139 MO45; 7, V. cholerae O1 NIH 35A3; 8, V. anguillarum PT-87050; 9, V. alginolyticus AM2; 10, V. mimicus RIMD 2218001; 11, V. vulnificus RIMD 2219009; 12, V. fluvialis RIMD 2220002; 13, V. furnisii RIMD 2223001; 14, V. damsela RIMD 2222001; 15, V. parahaemolyticus WP1; 16, E. coli MC1061. (B) Hybridization signals of the test strains detected on the X-ray film.
FIG. 2.
DNA colony hybridization test with the V. hollisae toxR gene probe for strains of V. hollisae and other Vibrio species. (A) Location of the inoculated strains (numbers correspond to dot positions in panel B): 1, V. cholerae O1 NIH41; 2, V. cholerae O1 NIH41; 3, V. cholerae O139 MO45; 4, V. cholerae non-O1, non-O139 AM21; 5, V. hollisae 525-82; 6, V. anguillarum PT-87050; 7, V. alginolyticus 219; 8, V. alginolyticus 220; 9, V. mimicus RIMD 2218001; 10, V. vulnificus RIMD 2219009; 11, V. fluvialis RIMD 2220002; 12, V. furnisii RIMD 2223001; 13, V. damsela RIMD 2222001; 14, V. metchnikovii RIMD 2208006; 15 to 24, V. hollisae strains 85A 7503, 89A 1961, 89A 1960, 89A 4206, 91A 2120, 93A 5688, SJ 90, FO 93, CDC 9039-81, and 4047, respectively. (B) Hybridization signals of the test strains detected on the X-ray film.
Examination of Vibrio strains by V. hollisae gyrB- and V. hollisae toxR-targeted PCR.
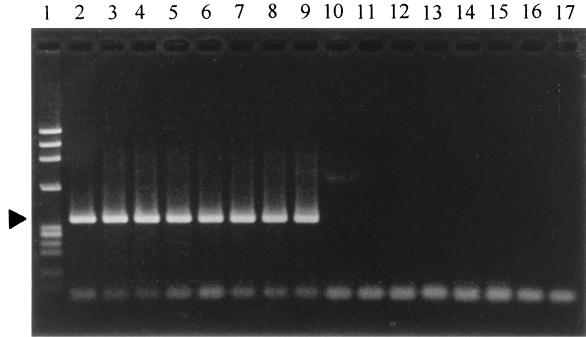
The strains listed in Table 1 were examined by the PCR methods that were designed for specific detection of V. hollisae. Two forward primers (HG-F1 and HG-F2) and two reverse primers (HG-R1 and HG-R2) were designed for detection of the V. hollisae gyrB gene as described in Materials and Methods. Four pairs of the primers were tested using the amplification conditions described in Materials and Methods. Three primer pairs (HG-F1 and HG-R2, HG-F2 and HG-R1, and HG-F2 and HG-R2) yielded the results shown in Table 1, and thus the PCR using any of these primer pairs was judged to be specific to the V. hollisae gyrB gene. Amplicon patterns of selected strains are shown in Fig. 3. The V. hollisae gyrB gene in 3 of 11 clinical strains of V. hollisae could not be detected when the primer pair HG-F1 and HG-R1 was employed under the conditions described in Materials and Methods, although a false-positive result was not obtained with any of the test strains listed in Table 1 (data not shown in Table 1). We did not pursue further the optimal amplification conditions for the primer pair HG-F1 and HG-R1.
FIG. 3.
Detection of the V. hollisae gyrB gene by PCR. Primer pair HG-F1 and HG-R2 and the amplification conditions and gel electrophoresis method described in Materials and Methods were employed. Lanes: 1, molecular weight markers (φX 174 phage DNA digested with HaeIII); 2 to 9, V. hollisae strains 89A 1961, 89A 1960, 89A 4206, 91A 2120, 93A 5688, SJ 90, FO 93, and RIMD 2221011, respectively; 10, V. anguillarum PT-87050; 11, V. cholerae O1 NIH41; 12, V. fluvialis RIMD 2220002; 13, V. furnisii RIMD 2223001; 14, V. parahaemolyticus WP1; 15, V. vulnificus RIMD 2219009; 16, V. alginolyticus AM2; 17, V. damsela RIMD 2222001. The position of the specific amplicons (363 bp) is indicated by the solid triangle.
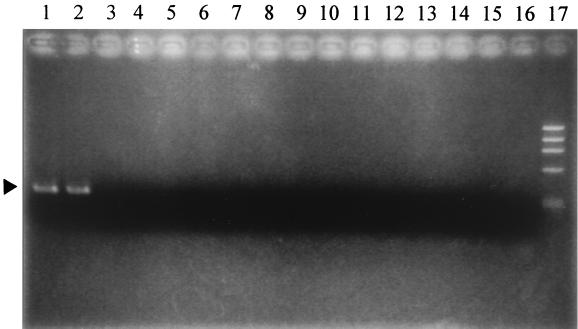
Three forward primers (HT-F1, HT-F2, and HT-F3) and two reverse primers (HT-R1 and HT-R2) were designed for detection of the V. hollisae toxR gene as described in Materials and Methods. Initially, six pairs of these primers were tested using a positive control, V. hollisae 525-82, and a negative control, V. parahaemolyticus AQ3815, with varying amplification conditions. As a result, the primer pair HT-F3 and HT-R2 and the amplification conditions described in Materials and Methods were found to be the best of all tested combinations in clearly distinguishing the V. hollisae toxR gene from the V. parahaemolyticus toxR gene (data not shown). This PCR protocol was applied to examine all test strains and was shown to be specific to the V. hollisae toxR gene (Table 1). A photograph illustrating the amplicon patterns of representative strains is shown in Fig. 4.
FIG. 4.
Detection of the V. hollisae toxR gene by PCR. Primer pair HT-F3 and HT-R2 and the amplification conditions and gel electrophoresis method described in Materials and Methods were employed. Lanes 1 and 2, V. hollisae strains 89A 1961 and 89A 1960, respectively; 3, Vibrio mediterranei ATCC 43341; 4, Vibrio orientalis ATCC 33934; 5, Vibrio diazotrophicus ATCC 33466; 6, V. fluvialis NCTC 11327; 7, Vibrio gazogenes ATCC 29988; 8, Vibrio proteolyticus NCMB 1326; 9, Vibrio nigripulchritudo ATCC 27043; 10, Vibrio cincinnatiensis ATCC 35912; 11, Vibrio navarrensis ATCC 51183; 12, Vibrio mytili ATCC 51288; 13, Vibrio ordalii ATCC 33509; 14, Vibrio ichthyoenteri IFO 15847; 15, Vibrio penaeicida IFO 15640; 16, Vibrio splendidus ATCC 33125; 17, molecular weight markers (φX 174 phage DNA digested with HaeIII). The position of the specific amplicons (306 bp) is indicated by the solid triangle.
Detection by V. hollisae toxR-specific PCR and isolation of V. hollisae added to seafood samples.
Each of various seafood samples was homogenized in APW, and the homogenate was not spiked or was artificially contaminated with V. hollisae 525-82SmR at 10, 102, 103, 104, 105, and 106 CFU/ml. The sensitivity of the V. hollisae toxR-specific PCR method for detecting 525-82SmR by direct examination of the inoculated homogenate before and after 4 h of enrichment incubation at 37°C was determined. The PCR samples were examined by PCR employing not only 24 but also 35 thermal cycles. All seafood samples without 525-82SmR yielded negative results. The lowest initial levels of 525-82SmR that gave a positive PCR result are shown in Table 3. The detection limits of the 24-cycle PCR were 105 CFU/ml or above and 103 CFU/ml or above, respectively, before and after the enrichment incubation. The detection limits of 35-cycle PCR ranged between 103 and 106 CFU/ml and between 10 and 104 CFU/ml, respectively, before and after the enrichment incubation.
TABLE 3.
Detection of V. hollisae 525-82SmR spiked into APW containing seafood by V. hollisae toxR-specific PCR and by incubation with isolation medium
| Seafood sample | Time relative to 4-h incubation | Lowest initial level of 525-82SmR (CFU/ml) detectable by PCRa
|
Concn (CFU/ml) of purple colonies in APW-seafood homogenateb
|
||||
|---|---|---|---|---|---|---|---|
| Not spiked
|
Spiked
|
||||||
| 24 cycles | 35 cycles | Vh-M | Vh-M + SM | Vh-M | Vh-M + SM | ||
| Oysterc | Before | 106 | 105 | 0 | 0 | 2.72 × 103 | 2.67 × 103 |
| After | 103 | 102 (5.30) | 0 | 0 | 7.17 × 104 | 3.80 × 105 | |
| Coastal fish 1d | Before | >106 | 104 | 1.80 × 102 | 0 | 1.43 × 103 | 1.85 × 103 |
| After | 104 | 102 (0.556) | 3.00 × 103 | 0 | 3.20 × 105 | 1.84 × 105 | |
| Coastal fish 2e | Before | 106 | 103 | 1.07 × 102 | 0 | 5.93 × 102 | 6.07 × 102 |
| After | 104 | 10 (2.54) | 0 | 0 | 9.33 × 103 | 2.37 × 104 | |
| Squidf | Before | >106 | 105 | 3.50 × 102 | 0 | 1.01 × 104 | 3.79 × 103 |
| After | 104 | 102 (1.27) | 3.00 × 104 | 0 | 1.46 × 105 | 1.86 × 105 | |
| Ivory shellg | Before | 105 | 103 | 9.33 × 102 | 0 | 3.00 × 103 | 3.73 × 102 |
| After | 104 | 102 (0.250) | 1.03 × 104 | 0 | 7.33 × 103 | 1.87 × 104 | |
| Octopusf | Before | >106 | 105 | 4.67 × 10 | 0 | 4.60 × 102 | 9.20 × 102 |
| After | 105 | 103 (1.00) | 3.33 × 102 | 0 | 8.67 × 103 | 8.67 × 103 | |
| Shrimph | Before | >106 | 106 | 5.47 × 102 | 2.20 × 102 | 7.87 × 102 | 2.23 × 102 |
| After | 105 | 104 (1.40) | 7.07 × 104 | 5.67 × 102 | 6.5 × 104 | 9.33 × 103 | |
Values in parentheses are ratios of the number of purple colonies on Vh-M supplemented with streptomycin to the number of those on Vh-M recovered from the indicated enrichment culture.
Purple colonies detected on isolation medium. Vh-M, the agar medium designed for isolation of V. hollisae (19). Vh-M + SM, Vh-M supplemented with SM (100 μg/ml). Homogenate was not spiked (not inoculated) or was spiked with 525-82SmR (103 CFU/ml).
Crassostrea gigas.
Trichiurus lepturus.
Sillago japonica.
Not identified to the species level.
Babylonia japonica.
Penaeus monodon.
Sensitivities of the 35-cycle PCR for the octopus and shrimp samples even after a 4-h enrichment were relatively low (detectable initial levels, 103 and 104 CFU/ml, respectively [Table 3]). Washing the bacterial cells and other particles in these samples by repeated centrifugation did not increase the sensitivity of the PCR (data not shown). Therefore, two commercially available DNA extraction kits were compared to assess whether PCR templates prepared by the kits can achieve higher PCR sensitivity than that achieved by the boiling method. Oyster, octopus, and shrimp samples were examined (Table 4). The two DNA extraction kits, based on chaotropic extraction followed by ethanol precipitation (method 2) or by absorption onto silica-coated magnetic beads (method 3), gave higher sensitivities (by 2 log units) than did the boiling of the enrichment broth (original method, i.e., method 1) for the oyster sample. Method 3 also increased the PCR sensitivity by 2 log units for the octopus and shrimp samples whereas method 2 did not. An initial level of 10 or 102 CFU/ml of 525-82SmR could be detected for coastal fish, squid, and ivory shell samples when DNA extraction method 3 was employed (data not shown in Table 4).
TABLE 4.
Comparison of DNA extraction methods for the 35-cycle PCR to detect V. hollisae 525-82SmR after a 4-h enrichment in APW containing a seafood sample
| Seafood samplea | DNA extraction methodb | Lowest initial level (CFU/ml) of 525-82SmR detectable by PCR |
|---|---|---|
| Oyster | 1 | 102 |
| 2 | 1 | |
| 3 | 1 | |
| Octopus | 1 | 103 |
| 2 | 103 | |
| 3 | 10 | |
| Shrimp | 1 | 104 |
| 2 | 104 | |
| 3 | 102 |
See the footnotes c, f, and h of Table 3 for details.
1, boiling; 2, chaotropic extraction followed by ethanol precipitation; 3, chaotropic extraction followed by absorption onto silica-coated magnetic beads.
In the spike experiment whose results are presented in Table 3, the number of V. hollisae cells recoverable as purple colonies on the V. hollisae isolation medium from the enrichment culture at each sampling time was also examined. The streptomycin-resistant mutant strain, 525-82SmR, was used, and selection on isolation medium with or without streptomycin added was designed to distinguish the colonies of 525-82SmR from false-positive colonies (purple colonies formed by non-V. hollisae organisms that ferment neither mannitol nor maltose). The seafood samples not inoculated with 525-82SmR contained purple-colony-forming organisms at concentrations of 0 to 933 CFU/ml of homogenate (Table 3). The seafoods other than the shrimp did not yield purple colonies on the medium with streptomycin (Table 3), verifying that the number of purple colonies on V. hollisae isolation medium (Vh-M) supplemented with streptomycin (SM) can be used to estimate the concentration of 525-82SmR in the samples other than the shrimp. As the level of the 525-82SmR inoculum increased to 10 and 102 CFU/ml, the number of purple colonies on Vh-M supplemented with SM increased similarly (data not shown). The numbers of purple colonies on Vh-M and Vh-M supplemented with SM became similar for most samples if the initial inoculum level of 525-82SmR was 103 CFU/ml (Table 3).
Examination by the V. hollisae toxR-specific PCR method of representative purple colonies confirmed that nearly all purple colonies on both media recovered before and after enrichment incubation were V. hollisae 525-82SmR if the initial inoculum level of V. hollisae 525-82SmR was 103 CFU/ml or more (data not shown). The ratio of the number of purple colonies on Vh-M supplemented with SM to that on Vh-M for the samples that contained the lowest initial 525-82SmR level detectable by the V. hollisae toxR-specific PCR was calculated (Table 3) for discussion. They ranged from 0.250 to 5.30, indicating that, for these samples, 25% or more of the purple colonies on Vh-M apparently contained 525-82SmR.
DISCUSSION
We analyzed the gyrB homologue of V. hollisae in this study. Although the entire V. hollisae gyrB gene sequence was not analyzed, high homologies between the determined sequence and reported gyrB sequences of gram-negative bacteria allowed us to conclude that the cloned amplicon was the expected region of the V. hollisae gyrB gene. The 73 to 80% identity that was detected between the V. hollisae gyrB sequence and the corresponding regions of V. parahaemolyticus, V. alginolyticus, and E. coli (data not shown) can explain the weak reactions observed in the DNA colony hybridization test. These and the hybridization results listed in Table 1 suggest that the divergence in the gyrB sequence between V. hollisae strains and other test strains would be in the range of approximately 20 to 27% for weakly positive strains and more for negative strains. The divergence was sufficient for differentiation of V. hollisae from other organisms by the PCR method (Table 1 and Fig. 3).
As was reported for V. cholerae (25), the htpG gene was located immediately upstream of the toxR gene in V. parahaemolyticus (M. Nishibuchi, J. Okuda, and K. Kumagai, unpublished data). We therefore presumed that the toxR-htpG gene arrangement is universal in vibrios. The homology between the V. cholerae htpG gene and the V. parahaemolyticus htpG gene was considerably higher than the homology between the toxR genes of the two species (14; M. Nishibuchi, J. Okuda, and K. Kumagai, unpublished data). Our attempt to utilize the V. parahaemolyticus htpG gene probe for cloning the V. hollisae toxR gene was successful. The result supports our hypothesis on the general toxR-htpG gene arrangement. This approach would be useful for cloning the toxR genes of other vibrios.
The overall nucleotide sequence identity between V. hollisae and V. parahaemolyticus was much lower for the toxR gene (59%) than for the gyrB gene (80%). In addition, the conserved region encoding the putative DNA binding domain of ToxR was not included in the V. hollisae toxR gene probe. The sequences of the PCR primers for V. hollisae toxR detection were selected from within the probe sequence. This explains why both the DNA probe method and the PCR method were shown to be successful for specific detection of the V. hollisae toxR gene (Table 1; Fig. 2 and 4).
The primary purpose of this study was to develop easy-to-perform and reliable genetic methods for identification of V. hollisae among the strains isolated from the marine environment. The test strains included not only the standard strains of Vibrio spp. but also organisms presumed to be non-V. hollisae isolated from marine fishes. The PCR results served to confirm the presumptive classification of the fish isolates by brief biochemical characterization. No discrepancy was seen between the results obtained by two PCR methods targeting the two different genes (Table 1). We conclude therefore that both PCR methods are equally useful for specific detection of V. hollisae. PCR methods targeted to the gyrB and toxR genes were shown to be useful for identification of V. parahaemolyticus in previous studies (10, 28) and of V. hollisae in this study. The same approach would be applicable to establishment of the PCR-based identification method for other species of the genus Vibrio.
Our ultimate goal is to isolate V. hollisae strains from the environment. Our previous approach was inoculation of the seafood samples onto the isolation medium (Vh-M) followed by conventional biochemical tests of the isolated strains (19). The PCR methods established in this study can replace the standard biochemical tests and facilitate the identification process. We also evaluated whether the screening process can be further facilitated by direct application of the V. hollisae toxR-specific PCR method to the seafood samples. Our idea was to screen V. hollisae-bearing seafood samples before attempting isolation of the candidate strains on Vh-M. It would obviate plating of the V. hollisae-negative enrichment culture and subsequent screening and identification and greatly reduce the workload. It is essential, then, that this direct PCR method have a high sensitivity. In addition, the V. hollisae strain in the sample has to be maintained in the enrichment broth so that the strain can be recovered after examination by PCR. The sensitivities of the V. hollisae toxR-specific PCR with 24 and 35 thermal cycles for LB broth cultures of six selected V. hollisae strains were 105 CFU/ml (40 CFU/PCR reaction) and 103 to 104 CFU/ml (0.4 to 4 CFU/PCR reaction), respectively, (data not shown). Equal sensitivities were observed in the direct PCR detection of V. hollisae 525-82SmR in some seafood homogenates before enrichment culture, but sensitivities were lower for some seafood samples (Table 3), suggesting that some seafood homogenates inhibit the PCR reaction to some degrees. Enrichment culture of the seafood sample in APW for 4 h and examination by the 35-cycle PCR allowed detection of 10 or 102 CFU/ml 525-82SmR (initial level) in seafood homogenates other than octopus and shrimp homogenates. Since none of the seafood homogenates inhibited the growth of 525-82SmR in APW (Table 3), the low sensitivities of PCR observed with the octopus and shrimp homogenates were attributable to these tissues inhibiting the PCR reaction. The effect of potential PCR inhibitors in these samples could be removed by employing the DNA extraction kit based on chaotropic extraction followed by absorption onto silica-coated magnetic beads (Table 4, method 3). Thus, inclusion of this DNA extraction method allowed detection of 1 to 102 CFU/ml 525-82SmR (initial level) regardless of the kind of seafood sample.
We chose a short-term enrichment because preliminary experiments suggested that non-V. hollisae organisms in some seafood samples can considerably overgrow if enrichment is carried out for longer than 4 h. Twenty-five percent or more of the purple colonies on Vh-M, which resulted from inoculation of 10 to 105 CFU/ml of 525-82SmR in seafood homogenates, apparently contained 525-82SmR after the 4-h enrichment incubation (Table 3). The reason for the result that the number of purple colonies on Vh-M supplemented with SM exceeded that on Vh-M (the ratios in parentheses in Table 3 became more than 1) is not known but it may be partly due to growth inhibition among the organisms, including non-purple colonies, on Vh-M. Approximately 4.5 h, including the chaotropic and silica-magnetic bead DNA extraction step, was needed to obtain the PCR result. After the 4.5-h lag, the number of V. hollisae 525-82SmR CFU recoverable from the enrichment broth was reduced only by about one log unit and were still ca. 103 CFU/ml or more if the enrichment culture was maintained at 15°C during PCR examination (data not shown). The results suggest that screening of the purple colonies recovered from the 35-cycle PCR-positive enrichment culture for V. hollisae may not require intensive search of the purple colonies. From these results, we conclude that a 4-h enrichment incubation of seafood samples in APW, followed by the chaotropic and silica-magnetic bead DNA extraction and examination with the 35-cycle V. hollisae toxR-specific PCR is a sensitive screening method. V. hollisae cells can be recovered from the enrichment culture onto the screening medium after detection of V. hollisae-positive sample by PCR. The colonies can then be examined by the PCR methods for the V. hollisae toxR and V. hollisae gyrB genes, as described above. We propose that this approach can be an effective method to isolate V. hollisae from the environment.
V. hollisae may be present as viable but nonculturable cells in the environment. Such cells may be observed by the fluorescent antibody staining method if a V. hollisae-specific antibody becomes available. Alternatively, the methods shown to be useful to resuscitate viable but nonculturable cells of vibrios (17, 30) can be included in the above enrichment step. This approach may be helpful in recovering V. hollisae in the viable but nonculturable form and will be included in our future project.
ACKNOWLEDGMENTS
This research was supported in part by the COE program on “Making Regions: Proto-Areas, Transformations and New Formations in Asia and Africa,” by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture, Japan, and by the “Research for the Future” program of The Japan Society for the Promotion of Science (JSPS-RFTF97L00706).
V. Vuddhakul and J. Okuda contributed equally to this study.
We thank the Research Center for Emerging Infectious Diseases, Osaka University, and Satoru Matsuoka of Ehime Prefectural Fish Disease Control Center for providing bacterial strains. We are grateful to Yohko Takeda for technical assistance.
REFERENCES
- 1.Abbott S L, Janda J M. Severe gastroenteritis associated with Vibrio hollisae infection: report of two cases and review. Clin Infect Dis. 1994;18:310–312. doi: 10.1093/clinids/18.3.310. [DOI] [PubMed] [Google Scholar]
- 2.Carnahan A M, Harding J, Watsky D, Hansman S. Identification of Vibrio hollisae associated with severe gastroenteritis after consumption of raw oysters. J Clin Microbiol. 1994;32:1805–1806. doi: 10.1128/jcm.32.7.1805-1806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 4.Feinberg A P, Vogelstein B. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 5.Gras-Rouzet S, Donnio P Y, Juguet F, Plessis P, Minet J, Avril J L. First European case of gastroenteritis and bacteremia due to Vibrio hollisae. Eur J Clin Microbiol Infect Dis. 1996;15:864–866. doi: 10.1007/BF01691217. [DOI] [PubMed] [Google Scholar]
- 6.Hickman F W, Farmer III J J, Hollis D G, Fanning G R, Steigerwalt A G, Weaver R E, Brenner D J. Identification of Vibrio hollisae sp. nov. from patients with diarrhea. J Clin Microbiol. 1982;15:395–401. doi: 10.1128/jcm.15.3.395-401.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janda J M, Powers C, Bryant R G, Abbott S L. Current perspectives on the epidemiology and pathogenesis of clinically significant Vibrio spp. Clin Microbiol Rev. 1988;1:245–267. doi: 10.1128/cmr.1.3.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelly M T, Hickman-Brenner F W, Farmer J J., III . Vibrio. In: Balows A, Hausler W J Jr, Herrmann K L, Isenberg H D, Shadomy H J, editors. Manual of clinical microbiology. 5th ed. Washington, D.C.: American Society for Microbiology; 1991. pp. 384–395. [Google Scholar]
- 9.Kelly M T, Stroh E M D. Occurrence of Vibrionaceae in natural and cultivated oyster populations in the Pacific Northwest. Diagn Microbiol Infect Dis. 1988;9:1–5. doi: 10.1016/0732-8893(88)90054-5. [DOI] [PubMed] [Google Scholar]
- 10.Kim Y B, Okuda J, Matsumoto C, Takahashi N, Hashimoto S, Nishibuchi M. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J Clin Microbiol. 1999;37:1173–1177. doi: 10.1128/jcm.37.4.1173-1177.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kita-Tsukamoto K, Oyaizu H, Nanba K, Shimidu U. Phylogenetic relationships of marine bacteria, mainly members of the family Vibrionaceae, determined on the basis of 16S rRNA sequences. Int J Syst Bacteriol. 1993;43:8–19. doi: 10.1099/00207713-43-1-8. [DOI] [PubMed] [Google Scholar]
- 12.Lee S E, Shin S H, Kim S Y, Kim Y R, Shin D H, Chung S S, Lee Z H, Lee J Y, Jeong K C, Choi S H, Rhee J H. Vibrio vulnificus has the transmembrane transcription activator ToxRS stimulating the expression of the hemolysin gene vvhA. J Bacteriol. 2000;182:3405–3415. doi: 10.1128/jb.182.12.3405-3415.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levine W C, Griffin P M the Gulf Coast Vibrio Working Group. Vibrio infections on the Gulf Coast: results of first year of regional surveillance. J Infect Dis. 1993;167:479–483. doi: 10.1093/infdis/167.2.479. [DOI] [PubMed] [Google Scholar]
- 14.Lin Z, Kumagai K, Baba K, Mekalanos J J, Nishibuchi M. Vibrio parahaemolyticus has a homolog of the Vibrio cholerae toxRS operon that mediates environmentally induced regulation of the thermostable direct hemolysin gene. J Bacteriol. 1993;175:3844–3855. doi: 10.1128/jb.175.12.3844-3855.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowry P W, McFarland L M, Threefoot H K. Vibrio hollisae septicemia after consumption of catfish. J Infect Dis. 1986;154:730–731. doi: 10.1093/infdis/154.4.730. [DOI] [PubMed] [Google Scholar]
- 16.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 17.Mizunoe, Y., S. N. Wai, T. Ishikawa, A. Takade, and S. Yoshida. Resuscitation of viable but nonculturable cells of Vibrio parahaemolyticus induced at low temperature under starvation. FEMS Microbiol. Lett. 186:115–120. [DOI] [PubMed]
- 18.Morris J G, Jr, Miller H G, Wilson R, Tacket C O, Hollis D G, Hickman F W, Weaver R E, Blake P A. Illness caused by Vibrio damsela and Vibrio hollisae. Lancet. 1982;i:1294–1297. doi: 10.1016/s0140-6736(82)92853-7. [DOI] [PubMed] [Google Scholar]
- 19.Nishibuchi M, Doke S, Toizumi S, Umeda T, Yoh M, Miwatani T. Isolation from a coastal fish of Vibrio hollisae capable of producing a hemolysin similar to the thermostable direct hemolysin of Vibrio parahaemolyticus. Appl Environ Microbiol. 1988;54:2144–2146. doi: 10.1128/aem.54.8.2144-2146.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishibuchi M, Fasano A, Russel R G, Kaper J B. Enterotoxigenicity of Vibrio parahaemolyticus with and without genes encoding thermostable direct hemolysin. Infect Immun. 1992;60:3539–3545. doi: 10.1128/iai.60.9.3539-3545.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishibuchi M, Ishibashi M, Takeda Y, Kaper J B. Detection of the thermostable direct hemolysin gene and related DNA sequences in Vibrio parahaemolyticus and other Vibrio species by the DNA colony hybridization test. Infect Immun. 1985;49:481–486. doi: 10.1128/iai.49.3.481-486.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishibuchi M, Janda J M, Ezaki T. The thermostable direct hemolysin gene (tdh) of Vibrio hollisae is dissimilar in prevalence to and phylogenetically distant from the tdh genes of other vibrios: implications in the horizontal transfer of the tdh gene. Microbiol Immunol. 1996;40:59–65. doi: 10.1111/j.1348-0421.1996.tb03304.x. [DOI] [PubMed] [Google Scholar]
- 23.Nishibuchi M, Kaper J B. Thermostable direct hemolysin gene of Vibrio parahaemolyticus: a virulence gene acquired by a marine bacterium. Infect Immun. 1995;63:2093–2099. doi: 10.1128/iai.63.6.2093-2099.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishibuchi M, Taniguchi T, Misawa T, Khaeomanee-Iam V, Honda T, Miwatani T. Cloning and nucleotide sequence of the gene (trh) encoding the hemolysin related to the thermostable direct hemolysin of Vibrio parahaemolyticus. Infect Immun. 1989;57:2691–2697. doi: 10.1128/iai.57.9.2691-2697.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parsot C, Mekalanos J J. Expression of ToxR, the transcriptional activator of the virulence factors in Vibrio cholerae, is modulated by the heat shock response. Proc Natl Acad Sci USA. 1990;87:9898–9902. doi: 10.1073/pnas.87.24.9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reich K A, Schoolnik G K. The light organ symbiont Vibrio fisheri possesses a homolog of the Vibrio cholerae transmembrane transcriptional activator ToxR. J Bacteriol. 1994;176:3085–3088. doi: 10.1128/jb.176.10.3085-3088.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruimy R, Breittmayer V, Elbaze P, Lafay B, Boussemart O, Gauthier M, Christine R. Phylogenetic analysis and assessment of the genera Vibrio, Photobacterium, Aeromonas, and Plesiomonas deduced from small-subunit rRNA sequences. Int J Syst Bacteriol. 1994;44:416–426. doi: 10.1099/00207713-44-3-416. [DOI] [PubMed] [Google Scholar]
- 28.Venkateswaran K, Dohmoto N, Harayama S. Cloning and nucleotide sequence of the gyrB gene of Vibrio parahaemolyticus and its application in detection of this pathogen in shrimp. Appl Environ Microbiol. 1998;64:681–687. doi: 10.1128/aem.64.2.681-687.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 30.Wai S N, Moriya T, Kondo K, Misumi H, Amako K. Resuscitation of Vibrio cholerae O1 strain TSI-4 from a viable but nonculturable state by heat shock. FEMS Microbiol Lett. 1996;136:187–191. doi: 10.1111/j.1574-6968.1996.tb08047.x. [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto S, Harayama S. PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl Environ Microbiol. 1995;61:1104–1109. doi: 10.1128/aem.61.3.1104-1109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoh M, Honda T, Miwatani T. Purification and partial characterization of a Vibrio hollisae hemolysin that relates to the thermostable direct hemolysin of Vibrio parahaemolyticus. Can J Microbiol. 1986;32:632–636. doi: 10.1139/m86-118. [DOI] [PubMed] [Google Scholar]